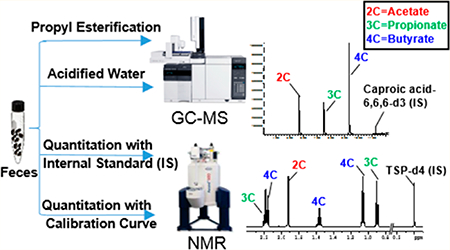
Abstract
Short chain fatty acids (SCFAs) are important regulators of host physiology and metabolism and may contribute to obesity and associated metabolic diseases. Interest in SCFAs has increased in part due to the recognized importance of how production of SCFAs by the microbiota may signal to the host. Therefore, reliable, reproducible, and affordable methods for SCFA profiling are required for accurate identification and quantitation. In the current study, four different methods for SCFA (acetic acid, propionic acid, and butyric acid) extraction and quantitation were compared using two independent platforms including gas chromatography coupled with mass spectrometry (GC−MS) and 1H nuclear magnetic resonance (NMR) spectroscopy. Sensitivity, recovery, repeatability, matrix effect, and validation using mouse fecal samples were determined across all methods. The GC−MS propyl esterification method exhibited superior sensitivity for acetic acid and butyric acid measurement (LOD < 0.01 μg mL−1, LOQ < 0.1 μg mL−1) and recovery accuracy (99.4%−108.3% recovery rate for 100 μg mL−1 SCFA mixed standard spike in and 97.8%−101.8% recovery rate for 250 μg mL−1 SCFAs mixed standard spike in). NMR methods by either quantitation relative to an internal standard or quantitation using a calibration curve yielded better repeatability and minimal matrix effects compared to GC−MS methods. All methods generated good calibration curve linearity (R2 > 0.99) and comparable measurement of fecal SCFA concentration. Lastly, these methods were used to quantitate fecal SCFAs obtained from conventionally raised (CONV-R) and germ free (GF) mice. Results from global metabolomic analysis of feces generated by 1H NMR and bomb calorimetry were used to further validate these approaches.
Graphical Abstract:
It is generally appreciated that compositional and functional changes in the gut microbiota promote or modify metabolic diseases.1,2 The gut microbiota impact host metabolism by producing or catabolizing metabolites (e.g., short chain fatty acids, SCFA), generating a barrier against pathogens3,4 and altering the physiological activity of the host.5,6 An important activity of the gut microbiota is fermentation of nondigestible dietary fibers to produce SCFAs. SCFAs are fatty acids with an aliphatic tail less than six carbons, and the primary fermentation-derived SCFAs are acetic acid (C2), propionic acid (C3), and butyric acid (C4). Ninety percent of SCFAs derived by bacterial fermentation are reabsorbed rapidly in the colon,7 utilized by the host as an energy source,8 and serve as anabolic substrates or precursors for biogenic synthesis.9 Acetate is the most abundant SCFA in the colon where it is transported to the liver and transformed into acetyl-CoA, a precursor for lipogenesis9 and gluconeogenesis.10 Further, acetic acid was reported to be involved in central appetite regulation for energy intake control.11,12 Propionic acid provides beneficial effects including antigluconeogenic,13,14 antilipogenic,14,15 anticholesterogenic,15,16anti-inflammatory,17,18 and anticarcinogenic17,19 activity. Butyric acid serves as a preferred nutrient for colonocytes20,21 and is implicated in colonic mucosa proliferation, intestinal lining integrity maintenance,21,22 colonic inflammation attenuation,23,24 and colonic cancer prevention23,25 and treatment.26,27 The discovery of the metabolic, immunological, and physiological implications of SCFAs has strengthened the demand for effective and precise quantitation approaches.
Multiple techniques have been employed for SCFA quantitation, including capillary electrophoresis,28,29 high pressure liquid chromatography coupled with ultraviolet detection (HPLC−UV),30,31 liquid chromatography coupled with mass spectrometry (LC−MS),32 gas chromatography coupled with both flame ionization (GC−FID) and mass spectrometry (GC−MS),33,34 and nuclear magnetic resonance (NMR) spectroscopy.35,36 Among these techniques, gas chromatography is most widely employed for its separation power, choice of detectors, and the relatively inexpensive cost of the instrumentation. Multiple extraction methods have been developed based on GC−MS techniques including acidified water extraction,37,38 organic reagent extraction,39 ultrafiltration,40,41 steam-distillation,42,43 vacuum evaporation stripping,44 purge and trap,45,46 headspace single-drop microextraction (HS-SDME),47 and derivatization to decrease polarity and improve volatility.48,49 NMR spectroscopy is commonly used in metabolomic studies due to its high reproducibility and simple preparation process.50 NMR spectroscopy has been used for SCFA profiling in extracts from mouse cecal content and feces.51,52 A basic analytical question is therefore raised regarding the complementarity and reproducibility of these methods. Or to what extent can current techniques (GC−MS or NMR) be combined to increase the analytical confidence and minimize irreproducible measurements. To compare the quantitative performance of GC−MS-based and NMR-based techniques for SCFAs measurement in biological samples, four common SCFA quantitation methods were assessed including GC−MS-based propyl esterification method,49 GC−MS-based acidified water extraction method,37,53 1H NMR-based quantitation relative to the reference compound sodium 3-trimethylsilyl [2,2,3,3-d4] propionate (TSP-d4), and 1H NMR based quantitation with calibration curve. Validation indices including sensitivity, recovery accuracy, repeatability, matrix effect, and biological concentrations were compared and evaluated across all methods. The methods were also applied to a comparison of conventional and germ-free mouse feces and further validated by NMR metabolic fingerprinting and bomb calorimetry.
METHODS
Animal Samples.
the method assessment experiment, feces obtained from conventionally raised C57BL/6J wild-type male mice (Jackson Laboratory, Bar Harbor, Maine) were pooled and measured. For method validation experiments, fresh feces were obtained from conventionally raised mice and germ free mice (The Pennsylvania State University, Gnotobiotic Facility).
GC−MS Analysis Procedure.
GC−MS Sample Preparation.
Fresh feces collected randomly from wild-type mice was pooled and spiked with 1-13C SCFAs at different concentration levels (250, 100, and 10 μg mL−1). For the propyl esterification method,49 50 mg of 1-13C SCFAs spiked feces were mixed with 1 mL of 0.005 M NaOH (containing 10 μg mL−1 internal standard caproic acid-6,6,6-d3), homogenized (Precellys, Bertin Technologies, Rockville, MD) at 6500 rpm, 1 cycle, 60 s, with 1.0 mm diameter zirconia/silica beads (BioSpec, Bartlesville, OK) added and then centrifuged (Eppendorf, Hamburg, Germany) at 13 200g, 4 °C, and 20 min. The supernatant was collected and mixed with an aliquot of 500 μL of 1-propanol/pyridine (v/v = 3:2) mixture. 100 μL of propyl chloroformate subsequently was added following a brief vortex for 1 min. Samples were derivatized in an incubator (Thermo Scientific, Marietta OH) at 60 °C for 1 h. The derivatized samples were extracted with a two-step hexane extraction (300 μL + 200 μL). A total 500 μL volume of extracts was obtained and stored at −20 °C for GC−MS quantitation. For the acidified water-extraction method, 50 mg of 1-13C SCFAs spiked fecal samples was mixed with 1 mL of HPLC water containing 10 μg mL−1 internal standard caproic acid-6,6,6-d3. The mixture then was homogenized, and the pH of the suspension was adjusted to 2−3 by adding 12 M HCl.37 The suspension was kept at room temperature for 10 min with occasional shaking then centrifuged at 13 200g, 4 °C, and 20 min. The supernatant was transferred to autosampler and stored at 4 °C for GC−MS analysis.
Experimental Condition for GC−MS Anlysis.
SCFAs were quantified with an Agilent 7890A gas chromatograph coupled with an Agilent 5975 mass spectrometer (Agilent Technologies Santa Clara, CA). The experimental conditions of propyl esterification method and acidified water extraction method are as previously described.49,53 See the Supporting Information for detailed experimental conditions.
GC−MS Spectra Data Processing.
All data were processed with Enhanced Chemstation (Agilent MSD chemstation) for mass spectra visualization, identification, and quantitation. More detailed data processing procedures can be found in the Supporting Information.
GC Method Validation.
Sensitivity.
Sensitivity was expressed by three indexes: limit of detection (LOD), limit of quantitation (LOQ), and linear range (LR). LOD represents the lowest detectable concentration of an individual SCFA in a sample with a signal-to-noise ratio greater than three. LOQ is the lowest concentration of SCFA in a sample which can be quantitatively determined with a signal-to-noise ratio above ten.54 LR is defined as the concentration range where the calibration curve displays linearity with correlation coefficient R2 > 0.99.
Precision.
Precision was expressed as recovery rate determined at three concentrations (250, 100, and 10 μg mL−1). Six fecal samples spiked with 1-13C SCFAs were analyzed. The fecal suspension without adding the SCFA standards was also analyzed to determine the initial amount of targeted compound present in the sample. Recovery rate was calculated by comparing calculated 1-13C standards concentration with the nominal concentration. Specifically, percent recovery (%R) was calculated using the following equation: %R = (quantified 1-13C SCFAs standards spiked in fecal sample − initial amount of 1-13C SCFAs present in the fecal samples)/ nominal standards conc. × 100.
Repeatability.
Repeatability was expressed by intraday and interday relative standard deviation (%RSD). Intraday repeatability was determined by running the same standard samples five times a day within a 24 h interval. Interday precision measurement was performed on the same samples on five different days. Mean and standard deviation of peak area of extracted ions for each injection were obtained. %RSD = standard deviation/mean ×100.
Matrix Effect.
The matrix effect was evaluated by comparing the response of the pure SCFA standards dissolved in extract solvent with the response of the SCFAs in the fecal matrix. Different concentrations of pure 1-13C SCFAs standards were spiked in either fecal suspensions or extract solvent. Unspiked samples were also analyzed to substrate the initial amount of SCFAs present in the samples out. The percent matrix effect (%ME) = (normalized area of SCFA in spiked fecal samples − normalized area of SCFAs of the unspiked fecal samples)/normalized area of SCFA in extract solvent.
Nuclear Magnetic Resonance (NMR) Spectroscopy.
SCFA Standard and Sample Preparation.
Fecal samples (50− 60 mg) were extracted with 1 mL of phosphate buffer (K2HPO4/NaH2PO4, 0.1 M, pH 7.4, 50% v/v D2O) containing 145.1 μM TSP-d4 as a chemical shift reference (δ 0.00). The samples were freeze−thawed three times with liquid nitrogen then homogenized (6500 rpm, 1 cycle, 60s) and centrifuged (11 180g, 4 °C, and 10 min). The supernatants were transferred to a new tube, and another 600 μL of PBS was added to the pellets followed by the same procedure described above. Fecal supernatants were combined and centrifuged (11 180g, 4 °C, and 10 min), then spiked with the SCFA standards at three final concentrations (250, 100, and 10 μg mL−1), and 600 μL of spiked fecal extract was transferred to a 5 mm NMR tube (Norell, Morganton, NC). Unspiked fecal supernatants were also analyzed to determine the initial amount of each SCFA presented in the sample.
1H NMR Spectroscopy, Spectral Data Processing, Analysis, and Validation.
1H NMR spectra of fecal extracts were acquired as previously described55 with a quantitative pulse sequence (repetition time ≥5T1). Quantitation analysis was performed based on either TSP-d4 reference with known concentration56 or calibration curve. More detailed 1H NMR acquisition, quantitation, and validation methodology can be found in the Supporting Information.
Method Validation and Application.
1H NMR-based SCFAs quantitation methods based on TSP-d4 as a reference or on the calibration curve were validated by measurement of accuracy, repeatability, sensitivity, and matrix effect. Further, the methods described were applied to investigate the SCFAs level and metabolic status differences between conventionally raised mice (CONV-R) and germ free mice (GF) combined with bomb calorimetry technique.
Data Analysis.
Graphical illustrations and statistical analyses were performed using Prism version 6. All data values were expressed as mean ± SEM. Statistical significance was defined as p < 0.05.
RESULTS AND DISCUSSION
Linearity and Sensitivity of GC−MS-Based and NMR-Based Methods.
The linearity (i.e., the correlation between numerical points), calibration range, LOD, LOQ, and calibration curve are extremely critical to assess the sensitivity and application of an analyticalmethod. As summarized in Table S-1, all calibration curves generated from 4 orders of magnitude of SCFA standard mixture demonstrated satisfactory linearity (R2 > 0.99) indicating that these methods could be employed extensively for quantifying SCFAs in biological samples with a wide range of concentrations. The GC−MS propyl esterification method yielded the lowest LOD and LOQ for acetic acid (LOD = 0.002 μg mL−1, LOQ = 0.02 μg mL−1) and butyric acid (LOD = 0.01 μg mL−1, LOQ = 0.09 μg mL−1). Propionate LOD/LOQ was not calculated as it could be detected in the background due to impurity in the derivatization solvent,49 whereas the GC−MS acidified water method (nonderivativation) showed a relatively high LOD and LOQ for acetic acid (LOD = 0.5 μg mL−1, LOQ = 5 μg mL−1), propionic acid (LOD = 0.8 μg mL−1, LOQ = 3 μg mL−1), and butyric acid (LOD = 0.2 μg mL−1, LOQ = 1 μg mL−1). LOD and LOQ detected were comparable with previous studies using the acidified water extraction method.37,53 Comparison of LOD and LOQ between derivatization and nonderivatization methods suggested that the improved volatility with derivatization increased the sensitivity compared to the nonderivatization method. Due to the high sensitivity of the derivatization approach, concentrations above 250 μg mL−1 saturated the detector thus compromising linearity. However, the acidified water method could quantify higher concentrations due to the relatively low sensitivity. Detectable SCFAs concentrations for the NMR method are approximately 2 μg mL−1, 100 fold less sensitive than GC−MS methods, which ranged from 0.002 to 0.01 μg mL−1. Quantifiable SCFAs levels by the NMR method started from 4 μg mL−1, significantly higher than GC−MS propyl esterification method which started from 0.02 μg mL−1.
Recovery and Matrix Effect of Measurement Based on Spiked Sample Matrix.
Recovery assessment was done by measuring a known amount of SCFA spiked into the biological matrix. Recovery is an informative indicator of whether the method presents satisfactory precision with interference from the biological matrix and other potential artifacts created during sample preparation and analysis. In the current study, 250, 100, and 10 μg mL−1 of 1-13C SCFAs mixture was spiked in pooled fecal extracts, and recovery was investigated by comparing the SCFA levels quantified by different methods and the expected concentration. GC−MS propyl esterification method was superior as almost 100% of 100 and 250 μg mL−1 spiked concentrations were recovered for all three SCFAs (Table 1). The GC−MS acidified water method showed better recovery at higher concentrations, ranging from 87.6% to 118.9%; however, it was inferior to the GC−MS propyl esterification method at lower concentrations. The two NMR-based methods showed very similar recovery, demonstrating the stability of the NMR-based technique. However, the recovery rate is inferior to the GC−MS propyl esterification method. The matrix effect is defined as the difference between the response of standards in solvents and those in a biological matrix which is known to interfere with the chromatographic efficiency and introduce error in the mass spectroscopy-based technique.57,58 Specific to the GC−MS-based technique, matrix components may cause enhancement59 or suppression60 of the analyte signal thus severely affecting quantitative analysis. Numerous factors might contribute to the matrix effect including matrix components, extraction reagents, the procedure itself, the purity of the reference standards, analyte physicochemical properties (stability, mass, charge, and concentration), column used, and myriad other factors. To minimize the matrix effect, both GC−MS based methods used reagents and standards of the highest grade available and the use of an internal standard to eliminate possible instrumentation errors. As shown in Table 1, the signal is suppressed by the matrix with GC−MS-based methods with a signal loss up to 24%, suggesting the accumulation of nonvolatile matrix components from fecal extracts like phospholipids and protein in the injection liner and column. The matrix effect introduced in NMR is usually due to pH, temperature, ionic strength, and protein content, resulting in peak position shifts and line width variations.61 The NMR quantitation method has reduced matrix effect compared to the GC−MS quantitation method, most likely due to the fact that it is less sensitive to matrix constituents and solvents and due to the simple extraction procedure (typically only requires dilution using phosphate buffer solution). In the present study, the NMR-based method demonstrated signal enhancement up to 18%, and the majority of matrix effects are around 10% signal enhancement, except 10 μg mL−1 spiked in butyric acid, which exhibited significant matrix effect (36% signal suppression), likely due to interference signals from the branched chain amino acids (valine, leucine, and isoleucine) with a resonance located δ 0.9, and compromised sensitivity in the low concentration range for the NMR-based quantitation method.
Table 1.
Recovery and Matrix Effect of SCFAs from Fecal Extracts Spiked with Different Concentration of SCFAs Standards by Different Quantitation Methodsa
| GC-MS propyl esterification method | |||||
|---|---|---|---|---|---|
| compounds | spiked amount (μg/mL) | amount recovered (μg/mL) | % recovery | % RSD | % matrix effect |
| acetic acid | 10 | 6.7 ± 1.0 | 66.9 | 14.9 | 76.3 |
| 100 | 108.3 ± 15.0 | 108.3 | 13.9 | 94.4 | |
| 250 | 254.5 ± 16.3 | 101.8 | 5.9 | 92.7 | |
| propionic acid | 10 | 7.2 ± 1.2 | 72.3 | 16.9 | 74.2 |
| 100 | 100.2 ± 17.6 | 100.2 | 17.5 | 84.2 | |
| 250 | 244.5 ± 36.6 | 97.8 | 15.0 | 92.7 | |
| butytic acid | 10 | 6.7 ± 1.1 | 67.0 | 11.2 | 77.7 |
| 100 | 99.4 ± 20.4 | 99.4 | 20.6 | 88.2 | |
| 250 | 249.7 ± 38.1 | 99.8 | 15.3 | 97.4 | |
| GC-MS acidified water method | |||||
|---|---|---|---|---|---|
| compounds | spiked amount (μg/mL) | amount recovered (μg/mL) | % recovery | % RSD | % matrix effect |
| acetic acid | 10 | 7.5 ± 1.9 | 74.8 | 25.1 | 116.8 |
| 100 | 108.7 ± 14.9 | 108.7 | 13.7 | 95.7 | |
| 250 | 226.1 ± 19.2 | 90.4 | 8.5 | 76.2 | |
| 10 | 7.5 ± 1.2 | 75.4 | 16.0 | 95.1 | |
| propionic acid | 100 | 106.3 ± 10.7 | 106.3 | 10.1 | 80.3 |
| 250 | 297.2 ± 23.1 | 118.9 | 7.8 | 76.2 | |
| 10 | 5.2 ± 0.4 | 52.2 | 6.8 | 89.6 | |
| butyric acid | 100 | 87.6 ± 17.0 | 87.6 | 20.6 | 76.2 |
| 250 | 291.7 ± 24.3 | 116.7 | 8.3 | 79.7 | |
| 1H NMR quantitation relative to TSP method | |||||
|---|---|---|---|---|---|
| compounds | spiked amount (μg/mL) | amount recovered (μg/mL) | % recovery | % RSD | % matrix effect |
| acetic acid | 10 | 8.4 ± 0.4 | 83.7 | 4.2 | 90.6 |
| 100 | 97.7 ± 4.3 | 97.7 | 4.4 | 118.5 | |
| 250 | 271.7 ± 16.0 | 108.7 | 5.9 | 111.7 | |
| 10 | 10.1 ± 0.6 | 101.4 | 6.0 | 107.2 | |
| propionic acid | 100 | 116.6 ± 5.5 | 116.6 | 4.7 | 116.1 |
| 250 | 332.1 ± 15.3 | 132.8 | 4.6 | 104.8 | |
| 10 | 5.4 ± 0.8 | 54.2 | 14.7 | 64.1 | |
| butyric acid | 100 | 92.9 ± 4.6 | 92.9 | 5.0 | 118.1 |
| 250 | 263.2 ± 14.7 | 105.3 | 5.6 | 106.8 | |
| 1H NMR quantitation with calibration curve method | ||||
|---|---|---|---|---|
| compounds | spiked amount (μg/mL) | amount recovered (μg/mL) | % recovery | % RSD |
| acetic acid | 10 | 8.6 ± 0.4 | 85.6 | 4.2 |
| 100 | 98.7 ± 5.1 | 98.7 | 5.2 | |
| 250 | 277.9 ± 16.3 | 111.2 | 5.9 | |
| 10 | 8.0 ± 0.48 | 80.2 | 6.0 | |
| 10 | 5.4 ± 0.8 | 54.1 | 14.7 | |
| butyric acid | 100 | 92.0 ± 4.7 | 92.0 | 5.1 |
| 250 | 263.0 ± 14.7 | 105.2 | 5.6 | |
Values are expressed as mean ± SD. n = 6 per group.
Intraday and Interday Repeatability Revealed Reliability of Different Methods.
Repeatability assessment is a critical aspect of investigating alternative analytical methods for reliable and robust quantitations. High repeatability also increases quantitative accuracy and efficiency by minimizing experimental replicates. Repeatability is assessed for each measurement method separately from replicated measurement on the same set of samples and described by within-subject relative standard deviation (RSD). In the current study, replicated measurements of different concentrations of SCFAs standard mixture were performed five times either within the same day (intraday) or on five different days (interday) by the four different measurement methods (Table S-2). Generally, the NMR-based techniques generated the smallest intraday and interday RSD ranging from 0.3% to 6.7%, with 80% of the RSD below 3%. The GC−MS propyl esterification method presented better intraday and interday repeatability compared to GC−MS acidified water method, except for propionic acid. Due to introduction of propionate to the background with the GC−MS propyl esterification solvent,49 propionic acid measurement was inconsistent at the lowest concentration 10 μg mL−1. The RSD for intraday and interday measurement reached 12.2% and 21.3%, respectively. Aside from the reduced impact from the matrix, the most attractive feature for NMR-based analytical techniques is the high repeatability. It is partially due to the unbiased detection nature of the NMR technique where the peak integrals relate directly to the chemical structure of the compound (number of protons giving rise to the peak in 1H NMR); thus, less interference from instrumentation and other external environmental factors would be introduced during intraday and interday repeated measurement. Further, the simple extraction process, nonvolatile extraction solution, and nondestructive detection platform maintain the maximum level of integrity and stability of the samples, contributing to the high repeatability especially during interday measurement. Conversely, MS-based methods are highly dependent on instrumentation (injector, column, inlet, and liner) and chromatographic conditions,62 sample preparation and storage (especially for highly volatile samples after derivatization), which raise issues with data repeatability and reproducibility between different research groups and research facilities.63 In the current study, GC−MS repeatability was improved (intraday and interday RSD of propyl esterification method ranging from 1.2% to 21.3% with 71% of the RSD below 7%, intraday and interday RSD of acidified water extraction method ranging from 2.4% to 12.6% with 67% of the RSD below 7%) through optimization of the instrument parameters, chromatographic conditions, normalization with internal standards as well as the addition of multiple washes pre and post injection and blank samples run between injections to eliminate carryover. However, extra attention for low level of propionic acid measurement using GC−MS propyl esterification methods is warranted (12.2% and 21.3% for intraday and interday RSD at 10 ug mL−1, respectively), and additional replicates or blank samples are requisite to minimize and/or understand background interference.
Application and Validation of SCFAs Quantitative Methods in Conventionally Raised (CONV-R) and Germ Free (GF) Mice Feces Study.
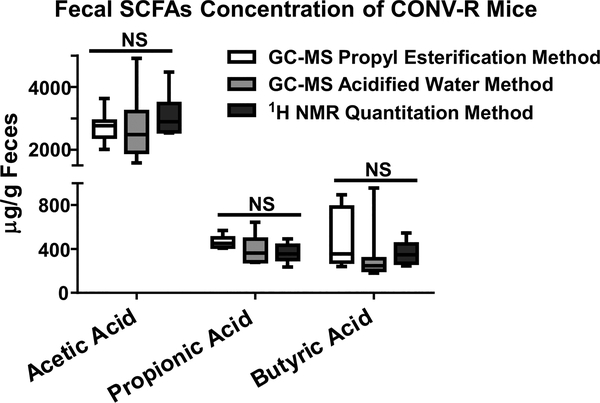
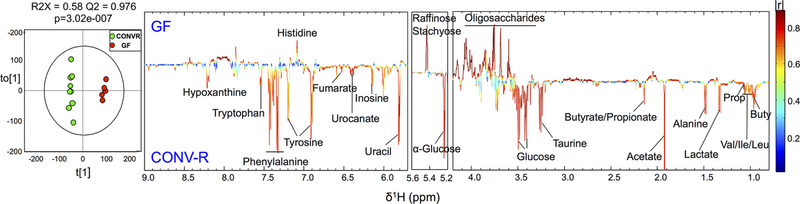
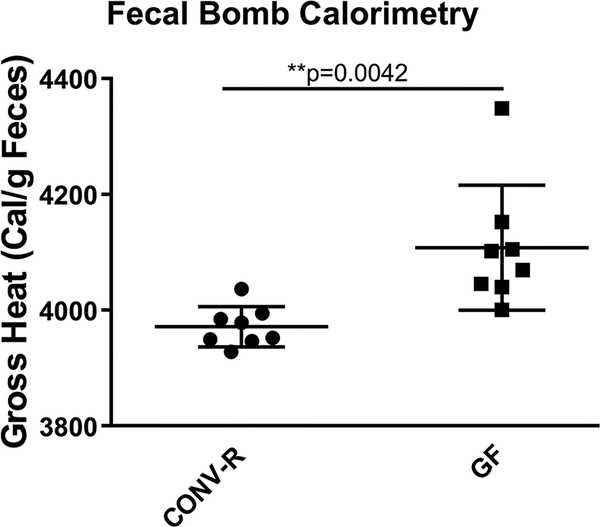
The SCFAs methods were used to quantify SCFAs levels in feces obtained from conventionally raised (CONV-R) and germ free (GF) mice. SCFAs were measurable but significantly lower in GF mice feces compared to CONV-R mice feces (p < 0.001, Figure S-5), consistent with the level and proportion of SCFAs in GF mice previously reported. For example, Hoverstad et al.64 reported the following concentrations using the GC−MS acidified water method with additional steps including vacuum distillation, alkalization, evaporation, and dissolution: acetic acid = 990 ± 380 μmol/kg (equivalent to 59.4 ± 22.8 μg/g), propionic acid = 17 ± 5.8 μmol/kg (equivalent to 1.3 ± 0.4 μg/g), and butyric acid = 7.1 ± 3.6 μmol/kg (equivalent to 0.6 ± 0.3 μg/g). For CONV-R mice, as shown in Figure 1, biological concentrations of SCFAs determined with GC−MS and NMR techniques were not significantly different, providing further validation for the applicability of the methods compared in the current study. The GC−MS acidified water method captured the broadest range of the biological concentration for all three SCFAs measured, indicating the greatest variation and least stability. The SCFAs concentrations quantified with different methods in this study (Figure 1 and Table S-3) were in good agreement with reported proportion and quantity of three primary SCFAs in fecal samples measured with other methods. For example, Lu et al.65 reported mouse fecal SCFAs concentrations using modified GC−MS acidified water direct injection method as acetic acid = 1468 ± 299 μg/g, propionic acid = 285 ± 94 μg/g, and butyric acid = 192 ± 55 μg/g; Garcia-Villalba et al. 39 reported the following SCFAs concentrations in rat fecal samples extracted with ethyl acetate: acetic acid = 2818.3 ± 720.7 μg/g, propionic acid= 268.5 ± 73.6 μg/g, and butyric acid ranging from 1387.8 ± 613.3 μg/g. In addition, the global metabolic profile between CONV-R and GF mice feces generated by 1H NMR (Figure 2) also revealed significant differences based on discriminant analysis (O-PLS-DA, described by R2X = 0.58, Q2 = 0.976) and CV-ANOVA test (p < 0.0001), supporting a dramatically different metabolic status imparted by the gut microbiota. Moreover, PLS-DA using only the SCFAs regions (the chemical shift [ppm] at 1.92 [acetate], 1.06 [propionate], and 0.9 [butyrate]) obtained via the 1H NMR approach were performed (Figure S-6). The groups are clearly and significantly separated by only SCFAs (R2X = 0.859, Q2 = 0.972, CV-ANOVA p < 0.0001), suggesting SCFAs themselves are sufficient to distinguish CONV-R and GF mice. GF mice showed a significantly lower level of fermentation end products SCFAs (acetate, propionate, and butyrate), branched chain amino acids (valine, leucine, and isoleucine), and bacterial-associated metabolites like taurine.66 Moreover, reduced glucose, phenylalanine, tyrosine, tryptophan, urocanate, hypoxanthine, inosine, uracil, and increased histidine were seen in feces of GF mice comparing to CONV-R mice, indicating altered glucose, amino acid, and nucleotide metabolism due to lack of microbial activity. Further, fermentation substrates like raffinose, stachyose, and many oligosaccharides were excreted in significantly higher levels into feces for GF mice, demonstrating the reduced metabolism of those substrates due to the absence of gut microbiota. Fecal energy quantitation was conducted with bomb calorimetry to further investigate the metabolic status in GF and CONV-R mice feces. The gross heat of feces from GF mice was 4110 ± 38.2 calorie per gram feces, approximately 140 calorie per gram higher than the gross heat of feces from CONV-R mice, which was 3970 ± 12.2 calorie per gram (Figure 3). Excessive fecal energy excretion in GF mice is consistent with high level of fecal dietary fibers determined by 1H NMR global metabolites profile. Because of the absence of bacterial fermentation in GF mice, host energy harvest capability and metabolic efficiency is reduced. Dietary fibers as potential energy sources are no longer accessible in the germ free gut and are excreted intact. The results are in good agreement with the previous studies from lean and obesity-resistant GF mice,67,68 suggesting the feasibility of the described methods for metabolic studies.
Figure 1.
Biological concentrations of fecal SCFAs in conventionally raised (CONV-R) mice (n = 10 per group) measured by GC−MS propyl esterification method, GC−MS acidified water method, and 1H NMR quantitation method. Values are expressed as the mean ±95% CI. Data was analyzed using ANOVA with Bonferroni’s correction. Biological concentrations measured by different methods were not significantly different.
Figure 2.
Global metabolic profiling of feces from germ free (GF) and conventionally raised (CONV-R) mice determined by 1H NMR. O-PLS-DA scores represent indicative power of models, p-value (CV-ANOVA) demonstrates the significance testing of OPLS model. Correlation coefficientcoded loadings plots for the models (right) from NMR spectra fecal extracts displaying changes and significance. The upward-pointed peak indicates the metabolite that peak represents (metabolite labeled) presented in higher level in GF mice. Peak pointing downward represents the metabolite presented more in CONV-R mice. The color-coded correlation coefficient indicates the significance of that change, the significance increases, as color gets warmer. A cutoff value of |r| > 0.754 (r > 0.754 and r < −0.754) is chosen for correlation coefficient as significant based on the discrimination significance (p ≤ 0.05).
Figure 3.
Quantitation of gross heat of feces from conventionally raised (CONV-R) and germ free (GF) mice by bomb calorimetry (n = 8 mice per group). Data was analyzed using a two-tailed Student t test. P < 0.05 was considered significant.
CONCLUSION
High-throughput metabolite profiling approaches including GC−MS and 1H NMR provide excellent platforms for quantitative detection of SCFAs in complex biological matrices. While MS-based methods, especially after derivatization, have incomparable sensitivity and precision, they can be influenced by matrix interference thus impacting repeatability. In practical applications, the GC−MS propyl esterification method is highly recommended for trace/ultratrace detection of SCFAs in biological fluid69 (plasma and urine) or intracellular SCFAs in cell culture and intestinal tissue. Because of the easier sample preparation procedure and short run time, the GC−MS acidified water method is most suitable for studies with large quantity, including samples from biodigesters or large-scale human studies. Alternatively, NMR-based methods, while exhibiting relatively low sensitivity, exhibit high repeatability and low matrix effect due to the nondestructive and noninvasive sample preparation and measurement technique. Additionally, NMR spectroscopy provides informative metabolic profiles of the overall metabolic characteristics; thus, it is best suited to measure cecal or fecal samples (higher SCFA concentration). In the CONV-R and GF mice feces study, the combination of GC−MS and NMR provided comprehensive and complementary views of SCFAs status and overall metabolic profile. Bomb calorimetry further confirmed the compromised energy harvesting capability of GF mice. Three mutually independent methods, GC−MS, NMR, and bomb calorimetry, led to consistent results, demonstrating the feasibility of the techniques used in metabolomics studies and the critical function that gut microbiota play in host energy balance and metabolic status.
Supplementary Material
ACKNOWLEDGMENTS
The authors wish to thank Anitha Vijay and Zak Kalikow for help with mice handling and sample collection.
Footnotes
The authors declare no competing financial interest.
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.analchem.7b00848.
Chemical and reagents used, detailed descriptions for each method, data acquisition, processing, and analysis; linearity and sensitivity of different methods (Table S-1); repeatability of different methods (Table S-2); biological concentrations of fecal SCFAs measured by different quantification methods (Table S-3); total ion chromatogram (Figure S-1) and fragmentation pattern and extract ion chromatogram (Figure S-2) of SCFA standard mixture by propyl esterification method and acidified water; 2D 1H−1H TOCSY (Figure S-3) and 2D 1H−13C HSQC (Figure S-4) NMR spectroscopy of mice feces; biological concentrations of SCFAs in GF and CONV-R fecal samples measured by GC−MS propyl esterification method (Figure S-4); PLS-DA of fecal SCFAs profiling from GF and CONV-R mice determined by 1H NMR. (PDF)
REFERENCES
- (1).Ley RE; Backhed F; Turnbaugh P; Lozupone CA; Knight RD; Gordon JI Proc. Natl. Acad. Sci. U. S. A. 2005, 102, 11070–11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2).Serino M; Luche E; Gres S; Baylac A; Berge M; Cenac C; Waget A; Klopp P; Iacovoni J; Klopp C; Mariette J; Bouchez O; Lluch J; Ouarne F; Monsan P; Valet P; Roques C; Amar J; Bouloumie A; Theodorou V; et al. Gut 2012, 61, 543–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3).Natividad JM; Verdu EF Pharmacol. Res. 2013, 69, 42–51. [DOI] [PubMed] [Google Scholar]
- 4).Sharma R; Young C; Neu J J. Biomed. Biotechnol. 2010, 2010, 305879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5).Blumberg R; Powrie F Sci. Transl. Med. 2012, 4, 137–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6).Hooper LV; Wong MH; Thelin A; Hansson L; Falk PG; Gordon JI Science 2001, 291, 881–884. [DOI] [PubMed] [Google Scholar]
- 7).Ruppin H; Bar-Meir S; Soergel KH; Wood CM; Schmitt MG Jr. Gastroenterology 1980, 78, 1500–1507. [PubMed] [Google Scholar]
- 8).Mcneil NI Am. J. Clin. Nutr. 1984, 39, 338–342. [DOI] [PubMed] [Google Scholar]
- 9).Zambell KL; Fitch MD; Fleming SE J. Nutr. 2003, 133, 3509–3515. [DOI] [PubMed] [Google Scholar]
- 10).Wolever TMS Falk Symp. 1994, 73, 251–259. [Google Scholar]
- 11).Byrne CS; Chambers ES; Morrison DJ; Frost G Int. J. Obes. 2015, 39, 1331–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12).Frost G; Sleeth ML; Sahuri-Arisoylu M; Lizarbe B; Cerdan S; Brody L; Anastasovska J; Ghourab S; Hankir M; Zhang S; Carling D; Swann JR; Gibson G; Viardot A; Morrison D; Thomas EL; Bell JD Nat. Commun. 2014, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13).Ximenes HM; Hirata AE; Rocha MS; Curi R; Carpinelli AR Cell Biochem. Funct. 2007, 25, 173–178. [DOI] [PubMed] [Google Scholar]
- 14).Chambers ES; Viardot A; Psichas A; Morrison DJ; Murphy KG; Zac-Varghese SE; MacDougall K; Preston T; Tedford C; Finlayson GS; Blundell JE; Bell JD; Thomas EL; Mt-Isa S; Ashby D; Gibson GR; Kolida S; Dhillo WS; Bloom SR; Morley W; et al. Gut 2015, 64, 1744–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15).Weitkunat K; Schumann S; Nickel D; Kappo KA; Petzke KJ; Kipp AP; Blaut M; Klaus S Mol. Nutr. Food Res. 2016, 60, 2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16).Anderson JW; Siesel AE Adv. Exp. Med. Biol. 1990, 270, 17–36. [DOI] [PubMed] [Google Scholar]
- 17).Hosseini E; Grootaert C; Verstraete W; Van de Wiele T Nutr. Rev. 2011, 69, 245–258. [DOI] [PubMed] [Google Scholar]
- 18).Tong LC; Wang Y; Wang ZB; Liu WY; Sun S; Li L; Su DF; Zhang LC Front. Pharmacol. 2016, 7, 253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19).Bindels LB; Porporato P; Dewulf EM; Verrax J; Neyrinck AM; Martin JC; Scott KP; Buc Calderon P; Feron O; Muccioli GG; Sonveaux P; Cani PD; Delzenne NM Br. J. Cancer 2012, 107, 1337–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20).Roediger WE Gastroenterology 1982, 83, 424–429. [PubMed] [Google Scholar]
- 21).Velazquez OC; Lederer HM; Rombeau JL Adv. Exp. Med. Biol. 1997, 427, 123–134. [PubMed] [Google Scholar]
- 22).Frankel W; Lew J; Su B; Bain A; Klurfeld D; Einhorn E; MacDermott RP; Rombeau J J. Surg. Res. 1994, 57, 210–214. [DOI] [PubMed] [Google Scholar]
- 23).Qiao Y; Qian J; Lu Q; Tian Y; Chen Q; Zhang YJ Surg. Res. 2015, 197, 324–330. [DOI] [PubMed] [Google Scholar]
- 24).Cushing K; Alvarado DM; Ciorba MA Clin. Transl. Gastroenterol. 2015, 6, e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25).Scharlau D; Borowicki A; Habermann N; Hofmann T; Klenow S; Miene C; Munjal U; Stein K; Glei M Mutat. Res., Rev. Mutat. Res 2009, 682, 39–53. [DOI] [PubMed] [Google Scholar]
- 26).Blouin JM; Penot G; Collinet M; Nacfer M; Forest C; Laurent-Puig P; Coumoul X; Barouki R; Benelli C; Bortoli S Int. J. Cancer 2011, 128, 2591–2601. [DOI] [PubMed] [Google Scholar]
- 27).Pouillart PR Life Sci. 1998, 63, 1739–1760. [DOI] [PubMed] [Google Scholar]
- 28).Arellano M; Jomard P; El Kaddouri S; Roques C; Nepveu F; Couderc F J. Chromatogr., Biomed. Appl. 2000, 741, 89–100. [DOI] [PubMed] [Google Scholar]
- 29).Arellano M; ElKaddouri S; Roques C; Couderc F; Puig P J. Chromatogr A 1997, 781, 497–501. [Google Scholar]
- 30).Stein J; Kulemeier J; Lembcke B; Caspary WF J. Chromatogr., Biomed. Appl 1992, 576, 53–61. [DOI] [PubMed] [Google Scholar]
- 31).de Sa LRV; de Oliveira MAL; Cammarota MC; Matos A; Ferreira-Leitao VS Int. J. Hydrogen Energy 2011, 36, 15177–15186. [Google Scholar]
- 32).Han J; Lin K; Sequeira C; Borchers CH Anal. Chim. Acta 2015, 854, 86–94. [DOI] [PubMed] [Google Scholar]
- 33).Bachmann C; Colombo JP; Beruter J Clin. Chim. Acta 1979, 92, 153–159. [DOI] [PubMed] [Google Scholar]
- 34).Murase M; Kimura Y; Nagata Y J. Chromatogr., Biomed. Appl 1995, 664, 415–420. [DOI] [PubMed] [Google Scholar]
- 35).Lewandowski ED; Chari MV; Roberts R; Johnston DL Am. J. Physiol. 1991, 261, H354–363. [DOI] [PubMed] [Google Scholar]
- 36).Jensen PR; Peitersen T; Karlsson M; In ‘t Zandt R; Gisselsson A; Hansson G; Meier S; Lerche MH J. Biol. Chem. 2009, 284, 36077–36082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37).Zhao G; Nyman M; Jonsson JA Biomed. Chromatogr. 2006, 20, 674–682. [DOI] [PubMed] [Google Scholar]
- 38).Hillman RE Clin Chem. 1978, 24, 800–803. [PubMed] [Google Scholar]
- 39).Garcia-Villalba R; Gimenez-Bastida JA; Garcia-Conesa MT; Tomas-Barberan FA; Carlos Espin J; Larrosa M J. Sep Sci 2012, 35, 1906–1913. [DOI] [PubMed] [Google Scholar]
- 40).Bjork JT; Soergel KH; Wood CM Gastroenterology 1976, 70, 864–864. [PubMed] [Google Scholar]
- 41).Vernia P; Breuer RI; Gnaedinger A; Latella G; Santoro ML Gastroenterology 1984, 86, 1557–1561. [PubMed] [Google Scholar]
- 42).Innocente N; Moret S; Corradini C; Conte LS J. Agric. Food Chem. 2000, 48, 3321–3323. [DOI] [PubMed] [Google Scholar]
- 43).Dejong C; Badings HT J. High Resolut. Chromatogr. 1990, 13, 94–98. [Google Scholar]
- 44).Ren YP; Wang JJ; Li XF; Wang XH J. Chem. Eng. Data 2012, 57, 46–51. [Google Scholar]
- 45).Boswell CE LC GC N. Am 2000, 18, 726. [Google Scholar]
- 46).Heikes DL; Jensen SR; Flemingjones ME J. Agric. Food Chem. 1995, 43, 2869–2875. [Google Scholar]
- 47).Chen Y; Li Y; Xiong Y; Fang C; Wang X J. Chromatogr A 2014, 1325, 49–55. [DOI] [PubMed] [Google Scholar]
- 48).Henningsson AM; Nyman EMGL; Bjorck IME J. Sci. Food Agric. 2002, 82, 385–393. [Google Scholar]
- 49).Zheng X; Qiu Y; Zhong W; Baxter S; Su M; Li Q; Xie G; Ore BM; Qiao S; Spencer MD; Zeisel SH; Zhou Z; Zhao A; Jia W Metabolomics 2013, 9, 818–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50).Savorani F; Rasmussen MA; Mikkelsen MS; Engelsen SB Food Res. Int. 2013, 54, 1131–1145. [Google Scholar]
- 51).Jacobs DM; Deltimple N; van Velzen E; van Dorsten FA; Bingham M; Vaughan EE; van Duynhoven J NMR Biomed. 2008, 21, 615–626. [DOI] [PubMed] [Google Scholar]
- 52).Cai J; Zhang L; Jones RA; Correll JB; Hatzakis E; Smith PB; Gonzalez FJ; Patterson AD J. Proteome Res. 2016, 15, 563–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53).Mulat DG; Feilberg A Talanta 2015, 143, 56–63. [DOI] [PubMed] [Google Scholar]
- 54).Currie LA Pure Appl. Chem. 1995, 67, 1699–1723. [Google Scholar]
- 55).Zhang L; Nichols RG; Correll J; Murray IA; Tanaka N; Smith PB; Hubbard TD; Sebastian A; Albert I; Hatzakis E; Gonzalez FJ; Perdew GH; Patterson AD Environ. Health Perspect 2015, 123, 679–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56).Dai H; Xiao C; Liu H; Tang H J. Proteome Res. 2010, 9, 1460–1475. [DOI] [PubMed] [Google Scholar]
- 57).Rogatsky E; Stein D J. Am. Soc. Mass Spectrom. 2005, 16, 1757–1759. [DOI] [PubMed] [Google Scholar]
- 58).Luigi Silvestro I. T. a. S. R. S. InTech 2013. [Google Scholar]
- 59).Hajslova J; Holadova K; Kocourek V; Poustka J; Godula M; Cuhra P; Kempny M J.Chromatogr A 1998, 800, 283–295. [DOI] [PubMed] [Google Scholar]
- 60).Hajslova J; Zrostlikova J J. Chromatogr A 2003, 1000, 181–197. [DOI] [PubMed] [Google Scholar]
- 61).Weljie AM; Newton J; Mercier P; Carlson E; Slupsky CM Anal. Chem. 2006, 78, 4430–4442. [DOI] [PubMed] [Google Scholar]
- 62).Barwick VJ J. Chromatogr A 1999, 849, 13–33. [Google Scholar]
- 63).Biber A; Scherer G; Hoepfner I; Adlkofer F; Heller WD; Haddow JE; Knight GJ Toxicol. Lett. 1987, 35, 45–52. [DOI] [PubMed] [Google Scholar]
- 64).Hoverstad T; Midtvedt T J. Nutr 1986, 116, 1772–1776. [DOI] [PubMed] [Google Scholar]
- 65).Lu Y; Fan C; Li P; Lu Y; Chang X; Qi K Sci. Rep. 2016, 6, 37589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66).Cook AM; Denger K Adv. Exp. Med. Biol. 2006, 583, 3–13. [DOI] [PubMed] [Google Scholar]
- 67).Gordon HA; Pesti L Bacteriol Rev. 1971, 35, 390–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68).Backhed F; Manchester JK; Semenkovich CF; Gordon JI Proc. Natl. Acad. Sci. U. S. A. 2007, 104, 979–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69).Singh V; Chassaing B; Zhang L; San Yeoh B; Xiao X; Kumar M; Baker MT; Cai J; Walker R; Borkowski K; Harvatine KJ; Singh N; Shearer GC; Ntambi JM; Joe B; Patterson AD; Gewirtz AT; Vijay-Kumar M Cell Metab. 2015, 22, 983–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.