Abstract
Background
Physical exercise can improve brain function by altering brain gene expression. The expression mechanisms underlying the brain’s response to exercise still remain unknown. miRNAs as vital regulators of gene expression may be involved in regulation of brain genes in response to exercise. However, as yet, very little is known about exercise-responsive miRNAs in brain.
Results
We constructed two comparative small RNA libraries of rat brain from a high-intensity intermittent swimming training (HIST) group and a normal control (NC) group. Using deep sequencing and bioinformatics analysis, we identified 2109 (1700 from HIST, 1691 from NC) known and 55 (50 from HIST, 28 from NC) novel candidate miRNAs. Among them, 34 miRNAs were identified as significantly differentially expressed in response to HIST, 16 were up-regulated and 18 were down-regulated. The results showed that all members of mir-200 family were strongly up-regulated, implying mir-200 family may play very important roles in HIST response mechanisms of rat brain. A total of 955 potential target genes of these 34 exercise-responsive miRNAs were identified from rat genes. Most of them are directly involved in the development and regulatory function of brain or nerve. Many acknowledged exercise-responsive brain genes such as Bdnf, Igf-1, Vgf, Ngf c-Fos, and Ntf3 etc. could be targeted by exercise-responsive miRNAs. Moreover, qRT-PCR and SABC immunohistochemical analysis further confirm the reliability of the expression of miRNAs and their targets.
Conclusions
This study demonstrated that physical exercise could induce differential expression of rat brain miRNAs and 34 exercise-responsive miRNAs were identified in rat brain. Our results suggested that exercise-responsive miRNAs could play important roles in regulating gene expression of rat brain in response to exercise.
Electronic supplementary material
The online version of this article (10.1186/s12867-019-0120-4) contains supplementary material, which is available to authorized users.
Keywords: Rat, miRNA, High-intensity intermittent swimming training, Exercise, Deep sequencing, Brain
Background
Accumulating evidence indicates that physical exercise helps maintain brain health and may profoundly benefit brain function [1–4], including the promotion of plasticity [2, 5], improvement of learning and memory performance [6, 7], mitigation of brain injury [8, 9] and reduction of brain disease risks (e.g., Alzheimer’s and Parkinson’s disease) [10, 11].
Previous studies have revealed that physical exercise elicits a differential gene expression pattern with significant changes in genes of relevance for brain function [12, 13]. For example, the expression level of brain-derived neurotrophic factor (BDNF) gene in various brain regions can be significantly increased after exercise [1, 14, 15], which is one of the major factors affecting on neuronal survival and differentiation as well as synaptic plasticity. Exercise can also rapidly increase gene expression of nerve growth factor (NGF) in specific areas of the brain [1]. Insulin-like growth factor 1 (IGF-1), a key regulator of reproductive neuroendocrine function, can be found at particularly high expression levels in brain after exercise [16, 17]. Moreover, many studies using animal models have demonstrated that physical exercise leads to differential expression of the brain genes encoding c-FOS [18, 19], NMDAR1 [20], VEGF [21, 22], Flk-1 [20], NT-3 [23], and VGF [24] etc. Such expression changes may contribute significantly to the beneficial effects of exercise [13]. However, it is still not clear how physical exercise alters gene expression profiling in brain.
The discovery of microRNA (miRNA) has greatly expanded our understanding of gene regulation mechanisms [25]. Many miRNAs have been identified from vertebrate brain and nervous system [26–28], and recent studies have demonstrated that miRNAs play critical roles in regulating gene expression during developmental processes like neurogenesis and neural differentiation and contribute to synaptic plasticity [29–33]. miRNAs also help protect against neurologic and psychiatric diseases [34, 35]. Interestingly, some miRNAs are also involved in regulating exercise-responsive brain genes, such as Bdnf [36], Igf-1 [37], and c-fos [38]. These evidences provide new clues to insight into the mechanisms how physical exercise changes gene expression pattern in brain. This prompts us to hypothesize that physical exercise may induce differential expression of miRNAs in brain, so that exercise-responsive miRNAs further regulate gene expression in brain. However, as yet, very little is known about exercise-responsive miRNAs in brain.
The aim of this study was to determine genome-wide expression profiles of rat brain miRNAs in response to physical exercise by deep sequencing and verify whether physical exercise can induce differential expression of miRNAs in rat brain, and to further identify and analyze exercise-responsive miRNAs in rat brain. Our findings in this study may pave the way for further understanding the molecular mechanisms how exercise affects brain function from the perspective of miRNA regulation.
Methods
Experimental animals and exercise program
Male Wister rats (R. norvegicus) used in this study were provided by the Lab Animal Center of Shandong LVYE Pharmaceutical Co., Ltd (Yantai, China). All rats were housed in an environment control room (temperature 20–24 °C; relative humidity 40–60%; 12:12 h light/dark cycle) in groups of four or five rats per cage with food and water available ad libitum. Prior to beginning swimming training experiments, all rats were acclimated to swimming exercise once a day for 5 days (10 min, 15 min, 30 min, 45 min, and 60 min/day, respectively). Two-month-old healthy male rats (200–220 g) were then randomly assigned to a high-intensity intermittent swimming training (HIST) group (n = 62) and a normal control (NC) group (n = 62). All rats from the HIST group were conducted to high-intensity intermittent swimming training bearing a weight equivalent to 5% of their body weight for 6 weeks. Swimming exercise was performed in a swimming bath with 60-cm water depth and 35 °C water. In a 6-week training program, ten sessions of swimming training were performed each day for 6 min each with a 4-min rest period between each session. During the rest period, weights were removed and the rats were blow-dried and placed back to their cages. The rats of the NC group were kept sedentary in cages without swimming training.
After the 6-week experimental period, the rats were euthanized immediately by cervical dislocation under sodium pentobarbital anesthesia. The brain tissues (cerebrum and cerebellum together as a whole) of each rat were immediately dissected and collected. The samples were frozen in liquid nitrogen and stored at − 80 °C for later use.
Small RNA library construction and deep sequencing
The brain tissues (comprised of cerebrum and cerebellum) of each rat were ground into power in liquid nitrogen. Total RNA was extracted using the TRIzol reagent (Invitrogen) following the manufacturer’s protocol. Fifteen RNA samples randomly selected from each group were mixed equally into HIST RNA pool and NC RNA pool, respectively. Small RNAs (sRNAs) of 16–30 nt were isolated and purified from the RNA pools by 15% denaturing PAGE. Subsequently, a 5′ RNA adaptor and a 3′ RNA adaptor were ligated to each sRNA using T4 RNA ligase. sRNAs were reversed transcribe into cDNA with SuperScript II Reverse Transcriptase (Invitrogen) and subjected to RT-PCR amplification. The RT-PCR products were further purified using 10% PAGE for constructing sRNA library. Lastly, HIST and NC sRNA libraries were sequenced in parallel by BGI (Shenzhen, China) using an Illumina/Solexa 1G Genome Analyzer (Illumina Inc., CA, USA).
Analysis of the deep-sequencing dataset
Raw sequencing reads from deep sequencing were first processed to obtain high-quality clean reads with the length of 18–30 nt through the elimination of the following: low-quality reads, reads with 5′ primer contaminants, reads without 3′ primer, reads without the insertion tag, reads with polyA, and reads shorter than 18 nt. The clean reads were mapped to the rat genome using SOAP (developed by BGI) [39] to analyze their distribution on the genome developed by BGI [39]. To eliminate all other non-miRNA, the clean sequences were aligned against the Rfam and Genbank databases to and annotate into several classes of non-miRNA sequences (including mRNA, rRNA, tRNA, snRNA, snoRNA, srpRNA, scRNA, and repeat-associated RNA) based on priority by using tag2annotation program (developed by BGI). After these non-miRNA sequences were removed, the remaining clean reads were used for further miRNA identification.
Identification of known miRNAs
The remaining sRNA sequences were searched against miRBase database (v18.0) to identify known miRNAs. The sRNA sequences matched to the known miRNAs of animal species deposited in miRBase database were considered to be known miRNAs in rat brain.
Prediction of novel candidate miRNAs
To identify potential novel candidate miRNAs, the unannotated sRNAs tags were mapped back to the rat genome using SOAP program. Only perfect alignments were retained for predicting novel candidate miRNAs. The mapped sequences and their flanking sequences together were first extracted from the rat genome as candidate precursor sequences of novel miRNAs. The MIREAP program (developed by BGI) was employed to identify potential novel miRNAs by exploring the hairpin-like structure, the Dicer cleavage sites and the minimum free energy of the extracted candidate precursor sequences. Secondary structures of the candidate precursor sequences were also checked using Vienna RNA Package. Sequences that met the following criteria were then considered to be miRNA precursors: (1) the sequence can fold into an appropriate and stable stem-loop hairpin secondary structure with a lower minimal free energy (MFE ≤ − 25.0 kcal/mol) and a higher minimal free energy index (MFEI ≥ 0.85); (2) the mature miRNA is present on one arm of the hairpin precursor; (3) the mature miRNA strand and its complementary strand (miRNA*) form a duplex with 2-nt 3′ overhangs; (4) base-pairing between the miRNA and the other arm of the hairpin has no more than 4 mismatches; (5) there is no more than one internal loop or bulge within the miRNA/miRNA* duplex, and the bulge or loop in size is less than 2 bases; (6) the number of mature miRNAs with predicted hairpin must be no fewer than 5 in the alignment results.
Differential expression analysis of miRNAs
To identify exercise-responsive miRNAs and to determine their genome-wide expression changes in response to HIST, we first compared the expression patterns of known and novel candidate miRNAs between HIST and NC libraries. miRNA counts in each libraries were first normalized to the number of transcripts per million (TPM). Then differential expression analysis of miRNAs between HIST and NC was performed using EXPR_SIG 3.0 tool (developed by BGI). Differential expression was considered to be significant when p-value < 0.05 and |log2(HIST/NC)| > 1. If the fold-change (log2(HIST/NC)) for a miRNA exceeded 1.0, it was considered to be up-regulated. If the fold-change was less than − 1.0, it was considered to be down-regulated miRNA.
Prediction of target genes of the differentially expressed miRNAs
Bioinformatic approaches have been used in many studies as an effective strategy to predict miRNA targets [40]. Potential target genes of the differentially expressed miRNAs were predicted against a set of rat cDNA sequences using RNAhybrid [41], miRanda [42], and TargetScan [43]. When one target for a given miRNA was similarly predicted by at least two of the three prediction tools, it was considered as a putative target gene of the given miRNA. To annotate the potential target genes, we performed a BLAST comparison their sequences against the NCBI databases or directly extracted gene information from the Rattus_norvegicus.gene_info file downloaded from NCBI FTP site (ftp://ftp.ncbi.nih.gov/gene/DATA/GENE_INFO/Mammalia/Rattus_norvegicus.gene_info).
qRT-PCR analysis
To further confirm the reliability of differential expressed analysis based on the sequencing results, twelve miRNAs (miR-4510, miR-182, miR-1839, miR-34c, miR-429, miR-122, miR-93, miR-212, miR-185, miR-7b, miR-483, and rno-miR-n048_5p) were selected to perform stem-loop qRT-PCR analysis.
Five rats were randomly sampled from each group for stem-loop qRT-PCR analysis. Each rat was an independent biological replicate. Small RNA (< 200 nt) was isolated from the brain region (consisted of cerebrum and cerebellum) of each rat using miRVana miRNA Isolation Kit (Ambion Inc., USA). First-strand cDNAs were synthesized with specific stem-loop RT primers (See Additional file 1: Table S1) using RevertAid First Stand cDNA Synthesis Kit (Thermo, Inc. USA) following the manufacturer’s protocol. Stem-loop qRT-PCR reactions were performed with miRNA-specific PCR primers (See Additional file 1: Table S1) and SYBR Green PCR mix (Qiagen) on a BioRad iCycler (BioRad, USA). U6 snRNA was used as an internal control and no-template qRT-PCR was used as negative control. Three technical replicates were done for each sample. The relative expression changes of miRNAs between HIST and NC were calculated using the 2−ΔΔCT method [44]. Statistical comparison analysis of miRNAs relative expression levels was performed using SPSS. The difference of miRNA expression level between HIST and NC was considered as significant when p < 0.05.
In addition, we also selected five target genes (c-Fos, Ncdn, Atrn, Ngf, and Ptn) of exercise-responsive miRNAs to validate their expression patterns via qRT-PCR. Among them, Ncdn, Atrn, Ngf, and Ptn were targeted, respectively, by miR-141↑, miR-183↑, miR-3897-3p↓, and miR-2881↑. c-Fos was targeted by miR-483↓ and miR-7b↓. All primers are listed in Additional file 1: Table S1. Statistical comparison analysis was the same as the above method.
SABC immunohistochemical analysis of c-Fos protein expression
Streptavidin-biotin complex (SABC) immunohistochemistry were applied to compare the expression of c-fos protein in rat brain tissues between HIST and NC. Rat brain tissues were cut into 5 μm sections using the freezing microtome. The sections were rinsed with 0.01 M PBS and incubated in turn with rabbit-anti-rat c-Fos polyclonal antibody (1:300; ZSGB-BIO company, China), biotinylated goat-anti-rabbit IgG (Boster Company, China), and streptavidin–biotin complex (SABC) (Boster Company, China) for 5 h, 30 min, and 30 min, respectively. The staining was visualized with diaminobenzidine (DAB) for 7–20 min at room temperature. The air-dried sections were dehydrated with graded ethanol and transparentized with xylene, and then sealed with neutral gum. Under the microscope (Olympus), the sections were observed and photographed for analyzing the density of c-Fos positive cells.
Results
Deep sequencing read analysis
Deep sequencing yielded a total of 28,667,031 HIST and 21,372,487 NC raw reads from HIST and NC libraries, respectively (Table 1). After filtering out low-quality reads, 3′ adaptor sequences were trimmed, contaminated reads were cleaned up, and reads shorter than 18 nt reads were removed, 27,877,781 HIST and 20,658,317 NC high-quality clean reads (≥ 18nt) were remained, corresponding to 1,066,655 HIST and 1,056,485 NC unique tags. The length distribution of sRNA sequences was similar in both libraries (Fig. 1). The majority (> 90%) of sRNA sequences were in the range of 18–25 nt. The 22-nt sequences were significantly more than those with other lengths, accounting for more than 30% of total clean reads. This length-distribution trend was consistent with the typical length-distribution trend of mature miRNAs in animals. Common sequences between HIST and NC libraries accounted for up to 95.7% of the total clean reads, representing 10.03% of all unique tags (Fig. 2). This indicated that common sRNAs between HIST and NC had higher expression levels than specific sRNAs. More than 70% of total clean sRNA reads from each library were mapped to the rat genome (Table 1), distributed across 21 chromosomes of rat.
Table 1.
Summary statistics of small RNA deep sequencing
| Library | Raw reads | High quality reads | Clean reads | Unique sRNAs | Total sRNAs mapped to rat genome | Unique sRNAs mapped to rat genome |
|---|---|---|---|---|---|---|
| HIST | 28,667,031 | 28,122,819 | 27,877,781 | 1,066,655 | 20,847,175 (74.78%) | 584,229 (54.77%) |
| NC | 21,372,487 | 20,988,298 | 20,658,317 | 1,056,485 | 14,488,689 (70.13%) | 594,634 (56.28%) |
Fig. 1.
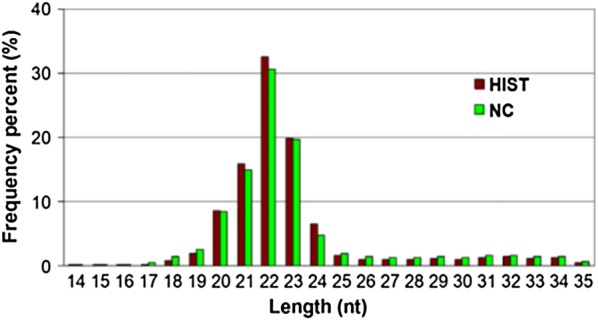
Length distribution of sRNA reads in HIST and NC libraries
Fig. 2.
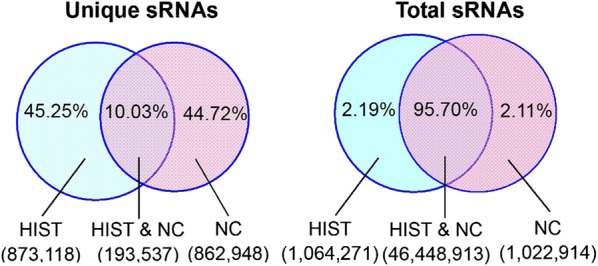
Summary of common and specific sRNA sequences between HIST and NC groups
Among the clean sequences, a total of 4,789,460 HIST and 4,883,792 NC reads were annotated into several classes of non-miRNA sequences (i.e., mRNA, rRNA, tRNA, snRNA, snoRNA, srpRNA, scRNA, repeat-associated elements, etc.) (listed in Table 2). After these non-miRNA sequences were removed, the remaining 23,088,321 HIST and 15,774,525 NC clean reads were used for further miRNA analysis.
Table 2.
Classification/annotation of small RNAs in HIST and NC libraries
| Category | HIST sRNAs | NC sRNAs | ||||||
|---|---|---|---|---|---|---|---|---|
| Unique | (%) | Redundant | (%) | Unique | (%) | Redundant | (%) | |
| rRNA | 142,588 | 13.37 | 2,856,748 | 10.25 | 151,074 | 14.30 | 3,330,834 | 16.12 |
| tRNA | 42,591 | 3.99 | 1,143,993 | 4.10 | 34,900 | 3.30 | 789,023 | 3.82 |
| scRNA | 796 | 0.07 | 8178 | 0.03 | 777 | 0.07 | 6108 | 0.03 |
| snRNA | 6533 | 0.61 | 60,646 | 0.22 | 6648 | 0.63 | 59,169 | 0.29 |
| snoRNA | 7540 | 0.71 | 265,995 | 0.95 | 7049 | 0.67 | 236,933 | 1.15 |
| srpRNA | 1619 | 0.15 | 20,165 | 0.07 | 1633 | 0.15 | 22,995 | 0.11 |
| exon_antisense | 3517 | 0.33 | 4665 | 0.02 | 2999 | 0.28 | 3922 | 0.02 |
| exon_sense | 180,213 | 16.90 | 226,294 | 0.81 | 179,548 | 16.99 | 219,397 | 1.06 |
| intron_antisense | 11,691 | 1.10 | 14,701 | 0.05 | 10,414 | 0.99 | 12,811 | 0.06 |
| intron_sense | 81,952 | 7.68 | 121,328 | 0.44 | 93,129 | 8.81 | 128,385 | 0.62 |
| Repeat | 50,441 | 4.73 | 66,747 | 0.24 | 57,451 | 5.44 | 74,215 | 0.36 |
| Unannotated | 537,174 | 50.00 | 23,088,321 | 83.00 | 510,863 | 48.00 | 15,774,525 | 76.00 |
| Total clean reads | 1,0,66,655 | 100 | 27,877,781 | 100 | 1,056,485 | 100 | 20,658,317 | 100 |
Identification of Known miRNAs in Rat Brain
To identify known miRNAs in rat brain, the sRNA sequences from each library were aligned against all known miRNAs of R. norvegicus as well as other 90 animal species in miRBase database. We identified 2109 (1700 in HIST and 1691 in NC) known miRNAs, belonging to 434 miRNA families (Additional file 2: Table S2). 22,240,775 (79.8%) HIST and 14,954,364 (72.4%) NC clean reads were, respectively identified as known miRNAs, suggesting that majority of the isolated small RNAs were known miRNAs. The two libraries shared 1282miRNAs, which included 214 miRNAs matched to known miRNAs of R. norvegicus (Fig. 3). In our study, the identification of so many brain miRNAs indicated that miRNAs were likely to participate widely in gene regulation in rat brain. Among the 2109 known miRNAs, 260 miRNAs were matched to the known miRNAs of R. norvegicus in miRBase 18.0, which corresponded to 144 miRNA families and accounted for 38.2% of all known miRNAs (680 miRNAs) of R. norvegicus. The other miRNAs were matched to known miRNAs of 64 other animal species and about half of these miRNAs were matched to known miRNAs of Mus musculus or Homo sapiens indicating that the majority of miRNAs identified in this study were conserved across different animal species.
Fig. 3.
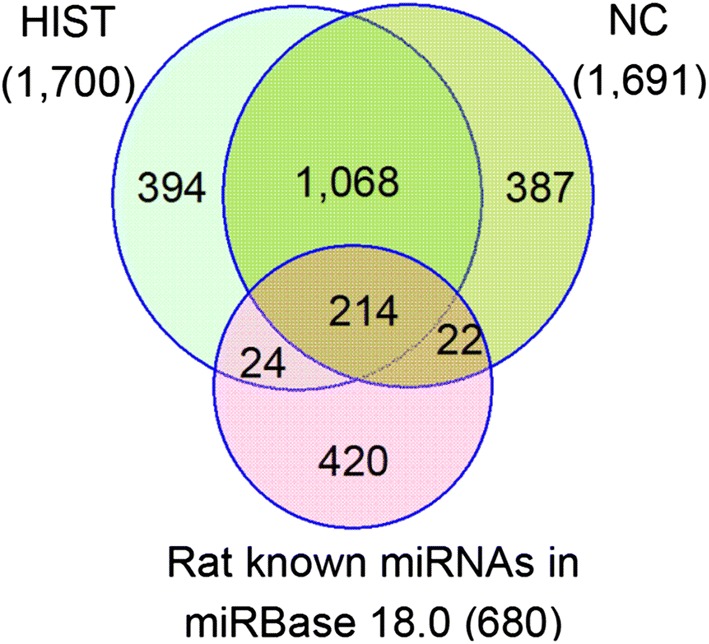
Co-expressed and specific-expressed known miRNAs between HIST and NC libraries and their matches to known rat miRNAs in miRBase 18.0
The known miRNAs in this study displayed significantly different expression levels as measured by the read counts, which ranged from one to more than one million copies (Additional file 2: Table S2). Twenty known miRNAs with the highest expression in the two libraries are listed in Table 3. Each of them is no less than 120,000 reads. However, 85 and 105 of 2109 miRNAs had only one copy in HIST and NC libraries, respectively. This indicated that Solexa sequencing technology is a powerful tool for identifying not only highly expressed miRNAs but also weakly expressed miRNAs. Among the identified 434 miRNA families, let-7 and mir-9 were two miRNA families with the highest expression in both libraries. In addition, thirty-five members of mir-154 family were identified in this study, more than those of other families.
Table 3.
The most abundant known miRNAs obtained from HIST and NC libraries
| miRNA name | Sequence | Length | Read count | miRNA family | miRNA orthologs | |
|---|---|---|---|---|---|---|
| HIST | NC | |||||
| let-7c | TGAGGTAGTAGGTTGTATGGTT | 22 | 2,314,478 | 1,205,674 | let-7 | hsa odi pma csa cin |
| miR-9a-5p | TCTTTGGTTATCTAGCTGTATGA | 23 | 1,889,962 | 1,416,714 | mir-9 | dme |
| let-7f | TGAGGTAGTAGATTGTATAGTT | 22 | 1,754,130 | 1,414,639 | let-7 | hsa |
| miR-378 | ACTGGACTTGGAGTCAGAAGGC | 22 | 1,544,707 | 1,452,656 | mir-378 | hsa ppy cfa |
| miR-143 | TGAGATGAAGCACTGTAGCT | 20 | 1,526,043 | 1,139,450 | mir-143 | rno mmu xtr pma |
| miR-182 | TTTGGCAATGGTAGAACTCACACCG | 25 | 1,638,730 | 326,627 | mir-182 | sko lgi cte mmu eca dre hsa spu |
| miR-9 | TCTTTGGTTATCTAGCTGTATG | 22 | 1,103,879 | 751,193 | mir-9 | egr sko bma xtr |
| let-7 | TGAGGTAGTAGGTTGTATAGTT | 22 | 804,332 | 490,827 | let-7 | cel aga sja spu |
| miR-30a | TGTAAACATCCTCGACTGGAAGC | 23 | 607,712 | 488,589 | mir-30 | hsa dre oan |
| miR-30d | TGTAAACATCCCCGACTGGAAGC | 23 | 594,387 | 413,656 | mir-30 | hsa pma oan bta |
| let-7b | TGAGGTAGTAGGTTGTGTGGTT | 22 | 638,961 | 307,685 | let-7 | hsa odi pma xtr cin |
| miR-127 | TCGGATCCGTCTGAGCTTGGC | 21 | 491,449 | 399,720 | mir-127 | mmu oar |
| miR-183 | TATGGCACTGGTAGAATTCACT | 22 | 623,293 | 125,748 | mir-183 | lgi mmu gga sko csa |
| let-7-5p | TGAGGTAGTAGGTTGTATAGT | 21 | 339,133 | 220,610 | let-7 | tca dme |
| miR-103a | AGCAGCATTGTACAGGGCTATGA | 23 | 280,569 | 274,894 | mir-103 | hsa pma |
| miR-24 | TGGCTCAGTTCAGCAGGAACAGT | 23 | 308,093 | 212,293 | mir-24 | hsa cfa |
| miR-26a | TTCAAGTAATCCAGGATAGG | 20 | 285,016 | 218,341 | mir-26 | hsa gga |
| miR-379 | TGGTAGACTATGGAACGTAGG | 21 | 272,494 | 226,457 | mir-379 | hsa |
| miR-99b | CACCCGTAGAACCGACCTTGC | 21 | 279,118 | 167,000 | mir-99 | mmu |
| let-7g | TGAGGTAGTAGTTTGTACAGTT | 22 | 227,155 | 185,745 | let-7 | mmu gga xtr dre |
Discovery of novel candidate miRNAs in the rat brain
According to the described criteria and methods above, 55 putative novel miRNAs were identified from both libraries: 50 from HIST and 28 from NC (Table 4 and Additional file 3: Table S3). Among them, 23 were co-expressed in both libraries, 27 were identified only in HIST and 5 were found only in NC. We also found that the 55 novel miRNAs had a size range of 20–24 nt, which was similar to other known animal miRNAs. They were derived from 66 putative precursor sequences with hairpin structures, varying from 69 to 96 nt in length. The minimum free energy (MFE) of these hairpins varied from − 63.7 to − 25.8 kcal/mol with an average of − 40.9 kcal/mol. Their minimum free energy index (MFEI) ranged from 0.85 to 2.05 with an average of 1.14, which were significantly higher than that for mRNAs (0.62–0.66), rRNAs (0.59), and tRNAs (0.64).
Table 4.
The most abundant novel candidate miRNAs identified from HIST and NC libraries
| miRNA name | Sequence | Length | Count | Precursor location | MFEa | MFEIb | |
|---|---|---|---|---|---|---|---|
| HIST | NC | ||||||
| rno-miR-n048_5p | CAGTGGTTTTACCCTATGGTAG | 22 | 2605 | 2040 | 19:37422715:37422794:+ | − 48.2 | 1.24 |
| rno-miR-n026_5p | AATGTGACTCAGCTATCTGAAC | 22 | 710 | 457 | 8:55466028:55466104:− | − 44.3 | 1.23 |
| rno-miR-n031_3p | TCAGTAGGCCAGACAGCAAGC | 21 | 588 | 453 | 11:36339323:36339403:− | − 39.6 | 0.86 |
| rno-miR-n044_5p | TGTGTTTTGTGTGTGTACATGT | 22 | 231 | 170 | 17:79266887:79266980:− | − 49.0 | 1.26 |
| 17:79268511:79268591:− | − 33.8 | 1.06 | |||||
| 17:79270548:79270628:− | − 38.0 | 1.19 | |||||
| 17:79273909:79273989:− | − 38.0 | 1.15 | |||||
| 17:79275714:79275805:− | − 40.3 | 1.12 | |||||
| 17:79276506:79276586:+ | − 38.0 | 1.15 | |||||
| 17:79281787:79281880:− | − 49.0 | 1.26 | |||||
| 17:79283453:79283546:− | -47.1 | 1.24 | |||||
| 17:79285357:79285437:− | − 33.8 | 1.06 | |||||
| 17:79277748:79277828:− | − 33.7 | 1.05 | |||||
| 17:79264421:79264501:− | − 38.0 | 1.19 | |||||
| rno-miR-n025_3p | ATGGTAATGGTGGTGGTGATGG | 22 | 213 | 108 | 8:64686682:64686763:+ | − 46.0 | 1.44 |
| rno-miR-n002_3p | CATAAGTGTAGAGAGTCTGTAGT | 23 | 122 | 59 | 1:131079839:131079925:+ | − 31.3 | 0.95 |
| rno-miR-n021_5p | TGGTTTACCGTCCCACATACA | 21 | 77 | 57 | 6:134389818:134389888:+ | − 43.5 | 1.45 |
| rno-miR-n022_3p | AAGGGCAAGCTCTCTTCGAGG | 21 | 46 | 36 | 6:134405230:134405322:+ | − 42.7 | 1.07 |
| rno-miR-n014_5p | TACAGTTAGACGTAGAGACCAT | 22 | 49 | 31 | 3:153140628:153140705:− | − 35.0 | 1.21 |
| rno-miR-n001_3p | CTCTAGCCAGGGCTTGACTGC | 21 | 48 | 24 | 1:116347377:116347469:− | − 53.3 | 0.95 |
| 1:116214989:116215081:+ | − 54.6 | 0.99 | |||||
aMFE denotes minimum free energy, and its unit is kcal/mol
bMFEI denotes minimum free energy index
These precursor sequences were distributed on all chromosomes except Chr13 (Additional file 3: Table S3). Chr1 and Chr17 contained more putative precursors than other chromosomes. The X chromosome had 4 of the 66 putative precursors. Among 55 novel candidate miRNAs, 53 had one unique genomic locus. However, rno-miR-n001-3p and rno-miR-n044_5p had multiple genomic loci. This indicated that different genomic loci might yield the same mature miRNA. Interestingly, rno-miR-n044_5p had 11 genomic loci, which distributed in cluster on Chr17. Further analysis revealed that all these 11 precursor sequences of rno-miR-n044_5p were almost identical. The existence of multi-copy miRNA genes suggests that the loss of function in one copy could be compensated by other copy or copies [45]. Because of the compensation mechanism, gene duplication can strengthen genetic robustness against null mutations to adapt the environment [45]. Meanwhile, this also implied that rno-miR-n044_5p might play pivotal regulatory roles in rat brain.
Compared with the known miRNAs, the predicted novel miRNAs exhibited much lower expression levels, ranging from 5 to 2605 copies (Additional file 3: Table S3). The majority of novel miRNAs (> 85%) were less than 100 reads. We found that 10 novel miRNAs with the highest expression were co-expressed in both libraries, together contributing more than 90% of the total expression of novel candidate miRNAs in each library (see Table 4). The reason that these novel miRNAs were not detected in previous studies could be that they express at the levels below the experimental detection threshold. Alternatively, they express only in some specific tissues at a particular developmental stage or under a specific induction.
Differential expression of brain miRNAs in response to HIST
Deep sequencing technology enables genome-wide expression patterns of miRNAs at unprecedented quantitative and qualitative accuracy [46]. In this study, we constructed genome-wide expression patterns of miRNAs from HIST and NC based on high-throughput Solexa sequencing datasets (Additional file 2: Table S2 and Additional file 3: Table S3).
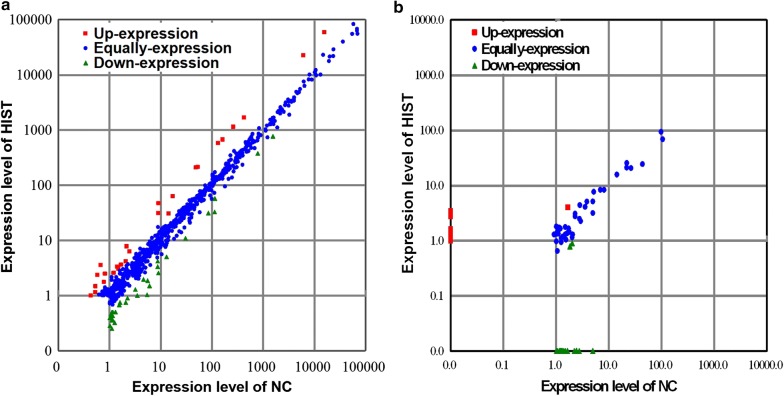
To investigate genome-wide expression changes of rat brain miRNAs in response to HIST and to screen exercise-responsive miRNAs in rat brain, differential expression analyses were performed to compare the expression patterns of known and novel miRNAs in rat brain between HIST and NC libraries based on the normalized reads. The majority of miRNAs showed similar expression patterns in both libraries. However, 31 known miRNAs and 3 novel miRNAs were identified as significantly differentially expressed between both libraries, with a more than two-fold change (|log2(HIST/NC)| > 1) and a p-value < 0.05. Among thirty-one differentially expressed known miRNAs, fifteen were up-regulated and sixteen were down-regulated (see Table 5, Fig. 4, and Additional file 4: Table S4). Of three differentially expressed novel miRNAs, rno-miR-n012_5p and rno-miR-n027_5p were down-regulated, and rno-miR-n006_3p was specifically expressed only in HIST library (see Table 5, Fig. 4, and Additional file 4: Table S4).
Table 5.
Significantly differentially expressed miRNAs of the rat brain in response to HIST
| miRNA name | HIST_Stda | NC_Std | log2(HIST/NC) | p-value | Sig.b | Markc |
|---|---|---|---|---|---|---|
| miR-122 | 1.87 | 5.57 | − 1.58 | 7.45E−03 | ** | Down |
| miR-1298 | 32.53 | 112.40 | − 1.79 | 6.38E−18 | ** | Down |
| miR-1343 | 0.75 | 2.08 | − 1.47 | 4.67E−02 | * | Down |
| miR-141 | 214.33 | 52.96 | 2.02 | 1.01E−15 | ** | Up |
| miR-141* | 3.55 | 0.68 | 2.39 | 3.24E−02 | * | Up |
| miR-182 | 58,782.66 | 15,810.92 | 1.89 | 0 | ** | Up |
| miR-183 | 22,358.06 | 6087.04 | 1.88 | 0 | ** | Up |
| miR-200a | 672.58 | 163.23 | 2.04 | 5.83E−46 | ** | Up |
| miR-200a* | 31.14 | 9.00 | 1.79 | 1.83E−03 | ** | Up |
| miR-200b | 1140.91 | 260.82 | 2.13 | 1.32E−81 | ** | Up |
| miR-200b* | 62.85 | 17.28 | 1.86 | 1.44E−05 | ** | Up |
| miR-200c | 1671.08 | 426.85 | 1.97 | 3.45E−103 | ** | Up |
| miR-263 | 7.75 | 2.18 | 1.83 | 3.27E−02 | * | Up |
| miR-263a-5p | 6.31 | 2.47 | 1.35 | 4.28E−02 | * | Up |
| miR-2881 | 30.88 | 14.38 | 1.10 | 1.56E−02 | * | Up |
| miR-34c | 770.29 | 1543.64 | − 1.00 | 8.67E−122 | ** | Down |
| miR-3897-3p | 1.04 | 5.42 | − 2.38 | 1.46E−02 | * | Down |
| miR-4154-3p | 3.34 | 8.76 | − 1.39 | 6.13E−03 | ** | Down |
| miR-429 | 582.83 | 132.63 | 2.14 | 5.56E−43 | ** | Up |
| miR-4466 | 1.97 | 4.60 | − 1.22 | 4.14E−02 | * | Down |
| miR-448 | 31.42 | 84.91 | − 1.43 | 6.27E−12 | ** | Down |
| miR-4492 | 1.00 | 3.49 | − 1.79 | 4.92E−02 | * | Down |
| miR-4497 | 10.98 | 30.54 | − 1.48 | 1.29E−05 | ** | Down |
| miR-4508 | 2.58 | 9.05 | − 1.81 | 2.33E−03 | ** | Down |
| miR-4510 | 46.78 | 9.10 | 2.36 | 6.53E−06 | ** | Up |
| miR-4651 | 1.51 | 6.05 | − 2.01 | 7.08E−03 | ** | Down |
| miR-483 | 4.23 | 8.71 | − 1.04 | 1.22E−02 | * | Down |
| miR-5128 | 0.90 | 2.23 | − 1.31 | 3.95E−02 | * | Down |
| miR-7b | 378.58 | 788.40 | − 1.06 | 8.66E−67 | ** | Down |
| miR-84a | 5.06 | 13.02 | − 1.36 | 3.24E−03 | ** | Down |
| miR-96 | 208.98 | 48.99 | 2.09 | 2.47E−16 | ** | Up |
| rno-miR-n006_3p | 1.51 | 0 | 7.24 | 8.58E−09 | ** | Up |
| rno-miR-n012_5p | 0.79 | 1.84 | − 1.22 | 1.89E−03 | ** | Down |
| rno-miR-n027_5p | 0.90 | 2.03 | − 1.18 | 3.94E−08 | ** | Down |
a‘HIST_Std’ and ‘NC_Std’ denote the normalized expression of miRNAs in the HIST and NC libraries, respectively
b‘*’ and ‘**’ indicate significance at 0.05 and 0.01 probability level, respectively
c‘Down’ and ‘Up’ mean down-regulated and up-regulated miRNAs, respectively
Fig. 4.
Scatter-plot graphs reveal the differential expression patterns of miRNAs between HIST and NC. a Scatter plot for known miRNAs. b Scatter plot for novel miRNAs
Our results of differential expression analysis strongly confirmed that HIST can significantly change the expression patterns of miRNAs in rat brain. This suggested that these exercise-responsive miRNAs may play key roles in regulating gene expression of rat brain in response to HIST.
miRNAs validation by stem-loop qRT-PCR
Twelve miRNAs with different expression levels were selected to perform stem-loop qRT-PCR analysis for validating their expression changes between HIST and NC. With the exception of miR-122, the other 11 miRNAs were successfully detected using stem-loop qRT-PCR. The lower expression level of miR-122 was the possible reason that it failed to be detected by stem-loop qRT-PCR.
The results of stem-loop qRT-PCR analysis showed that six miRNAs were significantly differentially expressed between HIST and NC, with a more than two-fold change (|log2(HIST/NC)| > 1) and a p-value < 0.05 (Fig. 5). Among them, three miRNAs (miR-4510, miR-429, and miR-182) were up-regulated and three miRNAs (miR-34c, miR-483, and miR-7b) were down-regulated. Five miRNAs (miR-212, miR-93, miR-1839, miR-185, and rno-miR-n048_5p) were not identified as significantly differentially expressed between both groups. The stem-loop qRT-PCR results were consistent with those of the differential expression analysis based on the deep sequencing data. Therefore, qRT-PCR analysis confirmed the reliability of the deep sequencing data and differential expression analysis.
Fig. 5.
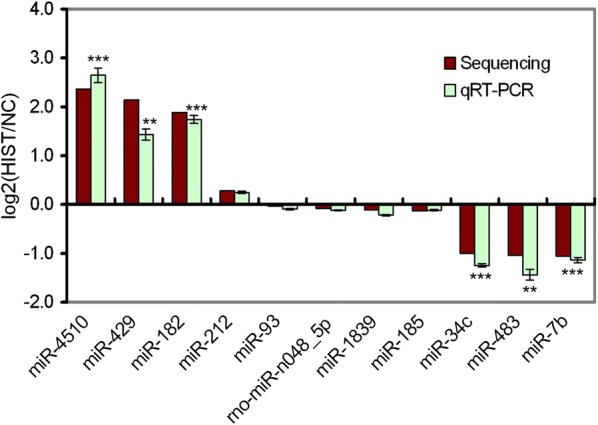
Validation of differentially expressed miRNAs by stem-loop qRT-PCR. The horizontal axis denotes the selected miRNAs for stem-loop qRT-PCR, and the vertical axis is the values of the Fold-change (log2(HIST/NC)) of miRNAs in response to HIST
Target prediction of differentially expressed miRNAs
To further understand the potential regulatory functions of these significantly differentially expressed miRNAs in rat brain between HIST and NC, their target genes were computationally predicted by RNAhybrid, miRanda and TargetScan. A total of 946 potential target genes of 31 differentially expressed known miRNAs were identified from a set of transcript sequences corresponding to rat genes (see Table 6 and Additional file 5: Table S5). The number of targets per known miRNA varied from 1 to 149 with an average of approximately 38. Moreover, we also predicted targets of 3 differentially expressed novel candidate miRNAs and obtained 9 potential targets (Additional file 5: Table S5). The number of targets per novel miRNA varied from 1 to 6 with an average of approximately 3.
Table 6.
The list of some target genes for exercise-responsive miRNAs in brain
| Target genes | Gene ID | Description | miRNAs |
|---|---|---|---|
| Igf1 | 24482 | Insulin-like growth factor 1 | miR-483, miR-96 |
| Nefh | 24587 | Neurofilament, heavy polypeptide | miR-4508 |
| Bdnf | 24225 | Brain-derived neurotrophic factor | miR-183, miR-2881, miR-34c, miR-3897-3p, miR-483 |
| Ngfr | 24596 | Nerve growth factor receptor | miR-141, miR-4492 |
| Rnf112 | 24916 | Brain ring finger protein 112 | miR-182 |
| Ptn | 24924 | Pleiotrophin | miR-2881 |
| Htr2c | 25187 | 5-Hydroxytryptamine (serotonin) receptor 2C | miR-4492 |
| Bcan | 25393 | Brevican (brain specific proteoglycan) | miR-2881 |
| Htr5a | 25689 | 5-Hydroxytryptamine (serotonin) receptor 5A | miR-483 |
| Ntf4 | 25730 | Neurotrophin 4 | miR-2881, miR-4492 |
| Neurod1 | 29458 | Neuronal differentiation 1 | miR-4492 |
| Vgf | 29461 | VGF nerve growth factor inducible | miR-2881, miR-3897-3p, miR-4492 |
| Htr2b | 29581 | 5-Hydroxytryptamine (serotonin) receptor 2B | miR-2881 |
| Cacnb1 | 50688 | Brain calcium channel, beta 1 subunit | miR-483 |
| Neurod2 | 54276 | Neuronal differentiation 2 | miR-3897-3p, miR-4492 |
| Ntrk1 | 59109 | Neurotrophic tyrosine kinase, receptor, type 1 | miR-3897-3p |
| Basp1 | 64160 | Brain abundant, membrane attached signal protein 1 | miR-2881, miR-3897-3p, miR-4492 |
| Smn1 | 64301 | Survival of motor neuron 1, telomeric | miR-4492 |
| Htr7 | 65032 | 5-Hydroxytryptamine (serotonin) receptor 7 | miR-2881 |
| Begain | 79146 | Brain-enriched guanylate kinase-associated protein | miR-3897-3p |
| Ntf3 | 81737 | Neurotrophin 3 | miR-200b, miR-429 |
| Atrn | 83526 | Membrane attracting | miR-183 |
| Ncdn | 89791 | Neurochondrin(neurite outgrowth protein) | miR-141 |
| Slc17a7 | 116638 | Brain solute carrier family 17, member 7 | miR-4154-3p |
| Nfasc | 116690 | Neurofascin | miR-182 |
| Baalc | 140720 | Brain and acute leukemia, cytoplasmic | miR-34c |
| Nlgn3 | 171297 | Neuroligin 3 | miR-84a |
| Nptx1 | 266777 | Neuronal pentraxin I | miR-182, miR-200b, miR-429 |
| Smndc1 | 287768 | Survival motor neuron domain containing 1 | miR-2881 |
| Nptx2 | 288475 | Neuronal pentraxin II | miR-4492 |
| Npdc1 | 296562 | Neural proliferation, differentiation and control, 1 | miR-4492 |
| Net1 | 307098 | Neuroepithelial cell transforming 1 | miR-4492 |
| Ngf | 310738 | Nerve growth factor (beta polypeptide) | miR-3897-3p |
| c-Fos | 314322 | c-fos oncogene | miR-483, miR-7b |
| Ppp1r1b | 360616 | Protein phosphatase 1, regulatory (inhibitor) subunit 1B | miR-1343, miR-2881, miR-4497, miR-4651 |
| Zc3h15 | 362154 | Zinc finger CCCH-type containing 15 in brain | miR-200c, miR-429 |
The results of target prediction indicated that these exercise-responsive miRNAs may participate in regulating many genes in rat brain. All of the predicted target genes encode proteins, and approximately one-third of them are transcription factors. Most of them are directly involved in the development and regulation of brain or nerve such as neurotrophin/neuropeptide, neural structure, neural signaling, synaptic protein, immune response, protein processing, metabolic enzyme, aging, and so on.
Neurotrophins are a class of essential proteins that induce the survival, neurogenesis, differentiation, development, and function of neurons. The neurotrophin family includes NGF, BDNF, NT-3, and NT-4 etc. NGF is critical for neuronal survival, maintenance and differentiation. Without it, neurons undergo apoptosis. NGF is a protein complex formed by three subunits (α, β, and γ). β is the biologically active subunit encoded by Ngf gene. Ngf gene was predicted to be targeted by miR-3897-3p↑. Moreover, Ngfr, a NGF receptor gene, was also targeted by miR-141↑ and miR-4492↓. Bdnf gene, encoding BDNF, was targeted by five exercise-responsive miRNAs (miR-183↑, miR-2881↑, miR-34c↓, miR-3897-3p↓, and miR-483↓). Ntf3 and Ntf4 encode the growth factors NT-3 and NT-4, respectively. They are expressed in certain neurons of the peripheral and central nervous system, and they help to support the survival and differentiation of existing neurons and promote the growth and differentiation of new neurons and synapses. Ntf3 was a candidate target gene of miR-200b↑ and miR-429↑, and Ntf4 was a candidate target of miR-2881↑ and miR-4492↓. This indicated that these exercise-responsive miRNAs participate in regulating the expression of neurotrophin genes in rat brain.
In addition, Vgf, a neuropeptide gene, was a shared candidate target gene of miR-2881↑, miR-4492↓, and miR-3897-3p↓. It may play a role in regulating energy homeostasis, metabolism [47] and synaptic plasticity [48] in rat brain. Ncdn (neurochondrin), a candidate gene targeted by miR-141↑, regulates neuronal synaptic plasticity. As an indirect marker of neuronal activity, c-Fos gene is the candidate target for miR-483↓ and miR-7b↓. Dopamine (DP) and 5-hydroxytryptamine (5-HT) are two important neurotransmitters. Ppp1r1b(DARPP32), a dopamine- and cyclic adenosine monophosphate-regulated phosphoprotein gene in rat brain, was a candidate target for four exercise-responsive miRNAs. 5-hydroxytryptamine receptor genes (Htr2b, Htr2c, Htr5a, and Htr7) were also candidate targets of exercise-responsive miRNAs. Among these predicted targets, Nptx1 and Nptx2 are involved in forming neural structure. This showed that these exercise-responsive miRNAs may play wide and important roles in the regulatory network of rat brain gene expression induced by HIST.
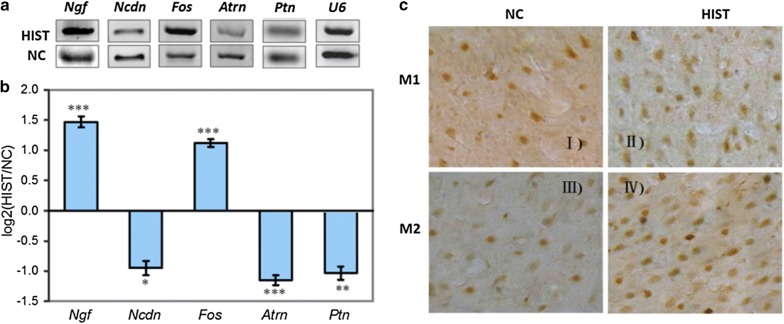
The qRT-PCR results of five candidate target genes showed that three genes (Ncdn, Atrn, and Ptn) were down-regulated and two genes (c-Fos and Ngf) were up-regulated in response to HIST (Fig. 6a, b). Expression analysis revealed that the expression patterns of all selected candidate target genes were negatively correlated with their corresponding miRNAs, verifying the accuracy of target prediction.
Fig. 6.
Validation of expression changes of miRNA targets in rat brain between HIST and NC by qRT-PCR and SABC immunohistochemistry. a Gel electrophoresis results of qRT-PCR for five target genes (Ngf, Ncdn, Fos, Atrn, and Ptn). Here, U6 snRNA was used as an internal control. b The expression fold changes of five target genes between HIST and NC detected by qRT-PCR. The vertical axis is the values of the Fold-change (log2(HIST/NC)) of target gene expression in response to HIST. c The immunohistochemistry results of brain c-fos protein expression changes between HIST and NC. Under the light microscope, c-Fos protein expression was detected in primary motor cortex (M1) and secondary motor cortex (M2) by the SABC immunohistochemistry techniques. II and IV are the c-Fos expression results in M1 and M2 regions of the HIST group, respectively. I and III are their corresponding controls. The number of Fos positive cells in M1 and M2 is significantly increased in response to HIST
SABC immunohistochemical analysis in rat brain confirmed that positive expression of c-Fos protein in HIST group was significantly higher than that in NC group (p < 0.01) (Fig. 6c). The number of c-Fos positive cells in HIST increased by a 2.1-fold in comparison to NC. By this token, the down-regulated expression of miR-483 and miR-7b, led to the expression increase of their target gene c-Fos at both mRNA and protein levels.
Discussion
As a behavioral stimulus, physical exercise can affect brain health and plasticity by altering gene expression in brain [2, 13]. Insight into the regulatory mechanisms of brain genes in response to exercise stimulation can help us understand how exercise acts on the regulatory networks controlling brain function. In the past few years, miRNAs have turned out to be new players in regulating gene expression of animals or plants response to stimuli or stresses [49]. Previous reports have revealed that exercise stimulation has effects on miRNA expression in skeletal muscle [50, 51] and ventricular function [52] etc. Here, we found that exercise can change miRNA expression profile in rat brain.
In this study, genome-wide identification of rat brain miRNAs (including 2109 known miRNAs and 55 novel miRNAs) greatly enriched the population of brain miRNAs in rat. This also implied that the number of miRNAs in rat brain is likely to be larger than previously thought. Many brain miRNAs reported in previous studies were also detected in our study. We found that these previously reported brain miRNAs were generally highly expressed in our study. For instance, miR-9, let-7, and miR-127 had expression levels greater than 10,000 TPM in both libraries. Most of the 2109 miRNAs identified in our study were not brain-specific miRNAs. In previous reports, they were also identified from other tissues across different animal species. For example, miR-378 identified in this study was also strongly expressed in the mammalian heart [53].
Differential expression analysis demonstrated that HIST significantly changed the expression patterns of miRNAs in rat brain. Thirty-four differentially expressed miRNAs were identified as exercise-responsive miRNAs, involving in regulating the expression of brain genes in response to exercise. We found that most of the previously reported brain miRNAs with high expression level (> 1000 TPM) were not generally identified as significantly differentially expressed in response to HIST. A reason for this difference may be that we used the strict threshold (|log2(HIST/NC)| > 1 and p-value < 0.05). Many miRNAs with 1.5- to 2.0-fold change and a p-value < 0.05 were not considered to be significantly differentially expressed in our study. Of course, we also found that a few of brain-enriched miRNAs (miR-182, miR-183, miR-200c, and miR-7b etc.), previously reported to be associated with brain function, were identified as differential expressed miRNAs in response to exercise. In addition, a few of specific-expressed miRNAs in HIST or NC were not identified as exercise-responsive miRNAs. This was because their expression levels were too low (TPM < 1) to meet the needs for differential expression analysis.
It was worth noting that all members of mir-200 family (miR-200a, miR-200b, miR-200c, miR-141, and miR-429), together with their asterisk miRNAs (miR-200a*, miR-200b*, and miR-141*) were identified as significantly differentially expressed with more than 3.5-fold change, accounting for 23.5% (8/34) of all differentially expressed miRNAs. Their expression levels were strongly up-regulated in response to HIST. This implied that mir-200 family plays regulatory roles in rat brain in response to HIST. In previous studies, they have also been identified in the different tissues of some animal species [54–57]. Choi et al. found that mir-200 family could regulate olfactory neurogenesis [58]. Lee et al. reported that miR-200b and miR-200c had the strong neuroprotective effect [59]. Here, we also found mir-200 family members were involved in the development and differentiation of the brain and nerve (see Additional file 5: Table S5). Therefore, we believed that further exploration of the roles of mir-200 family in brain may be very important to understand the complex regulatory mechanism of gene expression responsive to exercise in rat brain.
Most of the predicted target genes for the exercise-responsive miRNAs were directly related to the brain or nerve function in rat, implying our target prediction was relatively reliable. Target prediction revealed that a miRNA often had multiple target genes while a target was often targeted by multiple miRNAs. For example, Bdnf gene was targeted by two up-regulated and three down-regulated miRNAs. Hence, in most cases the relationship between a miRNA and its target gene(s) may not be simple one-to-one, implying that the rat brain is subject to a complex and flexible network of miRNA-mediated gene regulation. However, this also made it difficult to experimentally validate miRNA-target interactions. In this study, we only selected some simple miRNA-target pairs to verify the accuracy of target prediction by qRT-PCR.
Interestingly, many exercise-responsive genes, including Bdnf,Igf-1, Ngf, c-Fos, Ntf3, Ntf4, and Vgf etc., could also be targeted by exercise-responsive miRNAs (See Fig. 7). Igf-1 was targeted by miR-96 and miR-483, and c-Fos by miR-7b and miR-483. A previous study confirmed that up-regulated miR-7b inhibited the translation of c-Fos mRNA [38]. We also found that miR-483, a down-regulated miRNA, could simultaneously target three exercise-responsive genes: Bdnf, Igf-1, and c-Fos. These evidences revealed that the exercise-responsive miRNAs can participate in regulating the expression of the exercise-responsive genes in rat brain. Hereto, our results provided enough evidences to confirm that physical exercise can trigger differential expression of miRNAs in brain, so that exercise-responsive miRNAs can further stimulate changes in gene expression profile in brain.
Fig. 7.
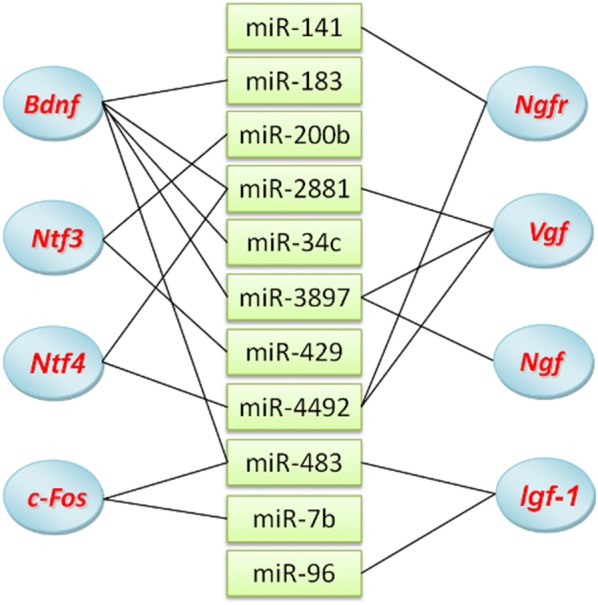
The corresponding relationship of the acknowledged exercise-responsive genes and the differentially expressed miRNAs
Exercise programs with different intensities can induce different gene expression profiles in brain [20]. Hence, we postulated that exercise programs with differing intensities might also lead to intensity-specific changes in miRNA expression patterns. In addition, many previous reports have shown that the gene expression induced by physical exercise varies in different brain regions. Therefore, we postulated that miRNA expression patterns in response to exercise might also be different in different brain regions. This study only explored the effect of HIST on miRNA expression profiles in specific brain region (cerebrum and cerebellum) of rats. In our laboratory, studies on miRNA expression in different brain regions responsive to exercise with different intensities are currently under the way. The regulatory mechanisms of brain gene expression in response to exercise may be complex, but miRNA-mediated gene regulation can provide the important clues to understand its complex mechanisms. In this study, we identified 34 exercise-responsive miRNAs from rat brain and analyzed their functions. Although these results are still preliminary, this may lay a foundation for further elucidation of the complex regulatory mechanisms in brain responsive to exercise from the perspective of miRNA regulation. Further analysis of the function of these exercise-responsive miRNAs and their regulatory network will be our next work.
Conclusion
A total of 2109 known and 55 novel candidate miRNAs were identified from the HIST and NC libraries of rat brain. Differential expression analysis showed that 34 HIST-responsive miRNAs were discovered, including 16 up-regulated and 18 down-regulated miRNAs. This study demonstrated that physical exercise could change the expression profiles of miRNAs in rat brain and that exercise-responsive miRNAs could regulate the expression of brain genes in response to exercise. Our findings lay a foundation for the further elucidation of the complex regulatory mechanisms responsive to exercise in the brain from the perspective of miRNA regulation.
Additional files
Additional file 1: Table S1. The primers used in qRT-PCR.
Additional file 2: Table S2. Known miRNAs identified in HIST and NC libraries.
Additional file 3: Table S3. Novel candidate miRNAs identified from HIST and NC libraries.
Additional file 4: Table S4. Significantly differentially expressed miRNAs between HIST and NC.
Additional file 5: Table S5. Target genes predicted for significantly differentially expressed miRNAs.
Authors’ contributions
YZ designed the study, performed the experiment and data analyses and wrote the manuscript. AZ designed the study, performed animal experiment and critically revised the manuscript. YW participated in the experiment and data analyses. PH, RZ, and SQ participated in animal experiment with swimming training. All authors read and approved the final manuscript.
Acknowledgements
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The datasets of miRNAs in the study are available from this article and its Additional files.
Consent for publication
Not applicable.
Ethics approval and consent to participate
All experimental procedures were performed in strict accordance with the guidelines for the care and use of laboratory animals of Yantai University and were approved by the Ethics Committee of Laboratory Animals of Yantai University (Permit Number: YTU-2010-0602).
Funding
This work was supported by the National Natural Science Foundation of China (Grant Nos. 31050009 and 31540030), the Natural Science Foundation of Shanxi Province of China (Grant No. 2011011034-1), and Yantai Science and Technology Development Project of China (Grant No. 2015YT06000238).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- HIST
high-intensity intermittent swimming training
- NC
normal control
- miRNA
microRNA
- sRNA
small RNA
- BDNF
brain-derived neurotrophic factor
- IGF-1
insulin-like growth factor 1
- NGF
nerve growth factor
- NT-3
neurotrophin-3
- NT-4
neurotrophin-4
- 5-HT
5-hydroxytryptamine
- DP
dopamine
- VEGF
vascular endothelial growth factor
- VGF
vascular growth factor
- NMDAR1
N-methyl-d-aspartate receptor 1
- BLAST
basic local alignment search tool
- TPM
transcripts per million
- MFE
minimum free energy
- MFEI
minimum free energy index
- RT-PCR
reverse-transcription PCR
- qRT-PCR
quantitative real-time reverse-transcription PCR
- SABC
streptavidin–biotin complex
- DAB
diaminobenzidine
Contributor Information
Yanhong Zhao, Email: zyhbob@163.com.
Anmin Zhang, Email: zam088@163.com.
Yanfang Wang, Email: wyf863@126.com.
Shuping Hu, Email: hushuping621022@126.com.
Ruiping Zhang, Email: zrp076@163.com.
Shuaiwei Qian, Email: qianshuaiwei999@yahoo.com.cn.
References
- 1.Neeper SA, Gomez-Pinilla F, Choi J, Cotman CW. Physical activity increases mRNA for brain-derived neurotrophic factor and nerve growth factor in rat brain. Brain Res. 1996;726(1–2):49–56. doi: 10.1016/0006-8993(96)00273-9. [DOI] [PubMed] [Google Scholar]
- 2.Cotman CW, Berchtold NC. Exercise: a behavioral intervention to enhance brain health and plasticity. Trends Neurosci. 2002;25(6):295–301. doi: 10.1016/S0166-2236(02)02143-4. [DOI] [PubMed] [Google Scholar]
- 3.Cotman CW, Berchtold NC, Christie LA. Exercise builds brain health: key roles of growth factor cascades and inflammation. Trends Neurosci. 2007;30(9):464–472. doi: 10.1016/j.tins.2007.06.011. [DOI] [PubMed] [Google Scholar]
- 4.van Praag H. Exercise and the brain: something to chew on. Trends Neurosci. 2009;32(5):283–290. doi: 10.1016/j.tins.2008.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pareja-Galeano H, Brioche T, Sanchis-Gomar F, Montal A, Jovani C, Martinez-Costa C, Gomez-Cabrera MC, Vina J. Impact of exercise training on neuroplasticity-related growth factors in adolescents. J Musculoskelet Neuronal Interact. 2013;13(3):368–371. [PubMed] [Google Scholar]
- 6.Albeck DS, Sano K, Prewitt GE, Dalton L. Mild forced treadmill exercise enhances spatial learning in the aged rat. Behav Brain Res. 2006;168(2):345–348. doi: 10.1016/j.bbr.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 7.van Praag H, Shubert T, Zhao C, Gage FH. Exercise enhances learning and hippocampal neurogenesis in aged mice. J Neurosci. 2005;25(38):8680–8685. doi: 10.1523/JNEUROSCI.1731-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Griesbach GS, Hovda DA, Gomez-Pinilla F, Sutton RL. Voluntary exercise or amphetamine treatment, but not the combination, increases hippocampal brain-derived neurotrophic factor and synapsin I following cortical contusion injury in rats. Neuroscience. 2008;154(2):530–540. doi: 10.1016/j.neuroscience.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stummer W, Weber K, Tranmer B, Baethmann A, Kempski O. Reduced mortality and brain damage after locomotor activity in gerbil forebrain ischemia. Stroke. 1994;25(9):1862–1869. doi: 10.1161/01.STR.25.9.1862. [DOI] [PubMed] [Google Scholar]
- 10.Tillerson JL, Caudle WM, Reveron ME, Miller GW. Exercise induces behavioral recovery and attenuates neurochemical deficits in rodent models of Parkinson’s disease. Neuroscience. 2003;119(3):899–911. doi: 10.1016/S0306-4522(03)00096-4. [DOI] [PubMed] [Google Scholar]
- 11.Friedland RP, Fritsch T, Smyth KA, Koss E, Lerner AJ, Chen CH, Petot GJ, Debanne SM. Patients with Alzheimer’s disease have reduced activities in midlife compared with healthy control-group members. Proc Natl Acad Sci U S A. 2001;98(6):3440–3445. doi: 10.1073/pnas.061002998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Molteni R, Ying Z, Gomez-Pinilla F. Differential effects of acute and chronic exercise on plasticity-related genes in the rat hippocampus revealed by microarray. Eur J Neurosci. 2002;16(6):1107–1116. doi: 10.1046/j.1460-9568.2002.02158.x. [DOI] [PubMed] [Google Scholar]
- 13.Tong L, Shen H, Perreau VM, Balazs R, Cotman CW. Effects of exercise on gene-expression profile in the rat hippocampus. Neurobiol Dis. 2001;8(6):1046–1056. doi: 10.1006/nbdi.2001.0427. [DOI] [PubMed] [Google Scholar]
- 14.Ferris LT, Williams JS, Shen CL. The effect of acute exercise on serum brain-derived neurotrophic factor levels and cognitive function. Med Sci Sports Exerc. 2007;39(4):728–734. doi: 10.1249/mss.0b013e31802f04c7. [DOI] [PubMed] [Google Scholar]
- 15.Saucedo Marquez CM, Vanaudenaerde B, Troosters T, Wenderoth N. High-intensity interval training evokes larger serum BDNF levels compared with intense continuous exercise. J Appl Physiol (1985). 2015;119(12):1363–1673. doi: 10.1152/japplphysiol.00126.2015. [DOI] [PubMed] [Google Scholar]
- 16.Trejo JL, Carro E, Torres-Aleman I. Circulating insulin-like growth factor I mediates exercise-induced increases in the number of new neurons in the adult hippocampus. J Neurosci. 2001;21(5):1628–1634. doi: 10.1523/JNEUROSCI.21-05-01628.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeon YK, Ha CH. Expression of brain-derived neurotrophic factor, IGF-1 and cortisol elicited by regular aerobic exercise in adolescents. J Phys Ther Sci. 2015;27(3):737–741. doi: 10.1589/jpts.27.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jee YS, Ko IG, Sung YH, Lee JW, Kim YS, Kim SE, Kim BK, Seo JH, Shin MS, Lee HH, et al. Effects of treadmill exercise on memory and c-Fos expression in the hippocampus of the rats with intracerebroventricular injection of streptozotocin. Neurosci Lett. 2008;443(3):188–192. doi: 10.1016/j.neulet.2008.07.078. [DOI] [PubMed] [Google Scholar]
- 19.Lee TH, Jang MH, Shin MC, Lim BV, Kim YP, Kim H, Choi HH, Lee KS, Kim EH, Kim CJ. Dependence of rat hippocampal c-Fos expression on intensity and duration of exercise. Life Sci. 2003;72(12):1421–1436. doi: 10.1016/S0024-3205(02)02406-2. [DOI] [PubMed] [Google Scholar]
- 20.Lou SJ, Liu JY, Chang H, Chen PJ. Hippocampal neurogenesis and gene expression depend on exercise intensity in juvenile rats. Brain Res. 2008;1210:48–55. doi: 10.1016/j.brainres.2008.02.080. [DOI] [PubMed] [Google Scholar]
- 21.Fabel K, Fabel K, Tam B, Kaufer D, Baiker A, Simmons N, Kuo CJ, Palmer TD. VEGF is necessary for exercise-induced adult hippocampal neurogenesis. Eur J Neurosci. 2003;18(10):2803–2812. doi: 10.1111/j.1460-9568.2003.03041.x. [DOI] [PubMed] [Google Scholar]
- 22.Uysal N, Kiray M, Sisman AR, Camsari UM, Gencoglu C, Baykara B, Cetinkaya C, Aksu I. Effects of voluntary and involuntary exercise on cognitive functions, and VEGF and BDNF levels in adolescent rats. Biotech Histochem. 2015;90(1):55–68. doi: 10.3109/10520295.2014.946968. [DOI] [PubMed] [Google Scholar]
- 23.Johnson RA, Mitchell GS. Exercise-induced changes in hippocampal brain-derived neurotrophic factor and neurotrophin-3: effects of rat strain. Brain Res. 2003;983(1–2):108–114. doi: 10.1016/S0006-8993(03)03039-7. [DOI] [PubMed] [Google Scholar]
- 24.Hunsberger JG, Newton SS, Bennett AH, Duman CH, Russell DS, Salton SR, Duman RS. Antidepressant actions of the exercise-regulated gene VGF. Nat Med. 2007;13(12):1476–1482. doi: 10.1038/nm1669. [DOI] [PubMed] [Google Scholar]
- 25.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 26.Hua D, Mo F, Ding D, Li L, Han X, Zhao N, Foltz G, Lin B, Lan Q, Huang Q. A catalogue of glioblastoma and brain MicroRNAs identified by deep sequencing. OMICS. 2012;16(12):690–699. doi: 10.1089/omi.2012.0069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim J, Krichevsky A, Grad Y, Hayes GD, Kosik KS, Church GM, Ruvkun G. Identification of many microRNAs that copurify with polyribosomes in mammalian neurons. Proc Natl Acad Sci USA. 2004;101(1):360–365. doi: 10.1073/pnas.2333854100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Olsen L, Klausen M, Helboe L, Nielsen FC, Werge T. MicroRNAs show mutually exclusive expression patterns in the brain of adult male rats. PLoS ONE. 2009;4(10):e7225. doi: 10.1371/journal.pone.0007225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krichevsky AM, King KS, Donahue CP, Khrapko K, Kosik KS. A microRNA array reveals extensive regulation of microRNAs during brain development. RNA. 2003;9(10):1274–1281. doi: 10.1261/rna.5980303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004;5(3):R13. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, Greenberg ME. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439(7074):283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- 32.Dogini DB, Ribeiro PA, Rocha C, Pereira TC, Lopes-Cendes I. MicroRNA expression profile in murine central nervous system development. J Mol Neurosci. 2008;35(3):331–337. doi: 10.1007/s12031-008-9068-4. [DOI] [PubMed] [Google Scholar]
- 33.Schratt G. Fine-tuning neural gene expression with microRNAs. Curr Opin Neurobiol. 2009;19(2):213–219. doi: 10.1016/j.conb.2009.05.015. [DOI] [PubMed] [Google Scholar]
- 34.Barbato C, Giorgi C, Catalanotto C, Cogoni C. Thinking about RNA? MicroRNAs in the brain. Mamm Genome. 2008;19(7–8):541–551. doi: 10.1007/s00335-008-9129-6. [DOI] [PubMed] [Google Scholar]
- 35.Satterlee JS, Barbee S, Jin P, Krichevsky A, Salama S, Schratt G, Wu DY. Noncoding RNAs in the brain. J Neurosci. 2007;27(44):11856–11859. doi: 10.1523/JNEUROSCI.3624-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mellios N, Huang HS, Grigorenko A, Rogaev E, Akbarian S. A set of differentially expressed miRNAs, including miR-30a-5p, act as post-transcriptional inhibitors of BDNF in prefrontal cortex. Hum Mol Genet. 2008;17(19):3030–3042. doi: 10.1093/hmg/ddn201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fenn AM, Smith KM, Lovett-Racke AE, Guerau-de-Arellano M, Whitacre CC, Godbout JP. Increased micro-RNA 29b in the aged brain correlates with the reduction of insulin-like growth factor-1 and fractalkine ligand. Neurobiol Aging. 2013;34(12):2748–2758. doi: 10.1016/j.neurobiolaging.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee HJ, Palkovits M, Young WS., 3rd miR-7b, a microRNA up-regulated in the hypothalamus after chronic hyperosmolar stimulation, inhibits Fos translation. Proc Natl Acad Sci USA. 2006;103(42):15669–15674. doi: 10.1073/pnas.0605781103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li R, Li Y, Kristiansen K, Wang J. SOAP: short oligonucleotide alignment program. Bioinformatics. 2008;24(5):713–714. doi: 10.1093/bioinformatics/btn025. [DOI] [PubMed] [Google Scholar]
- 40.Xu Z, Chen J, Li X, Ge J, Pan J, Xu X. Identification and characterization of microRNAs in channel catfish (Ictalurus punctatus) by using Solexa sequencing technology. PLoS ONE. 2013;8(1):e54174. doi: 10.1371/journal.pone.0054174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kruger J, Rehmsmeier M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006;34(Web Server issue):W451–W454. doi: 10.1093/nar/gkl243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. MicroRNA targets in Drosophila. Genome Biol. 2003;5(1):R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19(1):92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 45.Gu Z, Steinmetz LM, Gu X, Scharfe C, Davis RW, Li WH. Role of duplicate genes in genetic robustness against null mutations. Nature. 2003;421(6918):63–66. doi: 10.1038/nature01198. [DOI] [PubMed] [Google Scholar]
- 46.Osanto S, Qin Y, Buermans HP, Berkers J, Lerut E, Goeman JJ, van Poppel H. Genome-wide microRNA expression analysis of clear cell renal cell carcinoma by next generation deep sequencing. PLoS ONE. 2012;7(6):e38298. doi: 10.1371/journal.pone.0038298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hahm S, Mizuno TM, Wu TJ, Wisor JP, Priest CA, Kozak CA, Boozer CN, Peng B, McEvoy RC, Good P, et al. Targeted deletion of the Vgf gene indicates that the encoded secretory peptide precursor plays a novel role in the regulation of energy balance. Neuron. 1999;23(3):537–548. doi: 10.1016/S0896-6273(00)80806-5. [DOI] [PubMed] [Google Scholar]
- 48.Alder J, Thakker-Varia S, Bangasser DA, Kuroiwa M, Plummer MR, Shors TJ, Black IB. Brain-derived neurotrophic factor-induced gene expression reveals novel actions of VGF in hippocampal synaptic plasticity. J Neurosci. 2003;23(34):10800–10808. doi: 10.1523/JNEUROSCI.23-34-10800.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leung AK, Sharp PA. MicroRNA functions in stress responses. Mol Cell. 2010;40(2):205–215. doi: 10.1016/j.molcel.2010.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Davidsen PK, Gallagher IJ, Hartman JW, Tarnopolsky MA, Dela F, Helge JW, Timmons JA, Phillips SM. High responders to resistance exercise training demonstrate differential regulation of skeletal muscle microRNA expression. J Appl Physiol. 2011;110(2):309–317. doi: 10.1152/japplphysiol.00901.2010. [DOI] [PubMed] [Google Scholar]
- 51.Drummond MJ, McCarthy JJ, Fry CS, Esser KA, Rasmussen BB. Aging differentially affects human skeletal muscle microRNA expression at rest and after an anabolic stimulus of resistance exercise and essential amino acids. Am J Physiol Endocrinol Metab. 2008;295(6):E1333–E1340. doi: 10.1152/ajpendo.90562.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Soci UP, Fernandes T, Hashimoto NY, Mota GF, Amadeu MA, Rosa KT, Irigoyen MC, Phillips MI, Oliveira EM. MicroRNAs 29 are involved in the improvement of ventricular compliance promoted by aerobic exercise training in rats. Physiol Genomics. 2011;43(11):665–673. doi: 10.1152/physiolgenomics.00145.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ganesan J, Ramanujam D, Sassi Y, Ahles A, Jentzsch C, Werfel S, Leierseder S, Loyer X, Giacca M, Zentilin L, et al. MiR-378 controls cardiac hypertrophy by combined repression of mitogen-activated protein kinase pathway factors. Circulation. 2013;127(21):2097–2106. doi: 10.1161/CIRCULATIONAHA.112.000882. [DOI] [PubMed] [Google Scholar]
- 54.Chen PY, Manninga H, Slanchev K, Chien M, Russo JJ, Ju J, Sheridan R, John B, Marks DS, Gaidatzis D, et al. The developmental miRNA profiles of zebrafish as determined by small RNA cloning. Genes Dev. 2005;19(11):1288–1293. doi: 10.1101/gad.1310605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Coutinho LL, Matukumalli LK, Sonstegard TS, Van Tassell CP, Gasbarre LC, Capuco AV, Smith TP. Discovery and profiling of bovine microRNAs from immune-related and embryonic tissues. Physiol Genomics. 2007;29(1):35–43. doi: 10.1152/physiolgenomics.00081.2006. [DOI] [PubMed] [Google Scholar]
- 56.Lagos-Quintana M, Rauhut R, Meyer J, Borkhardt A, Tuschl T. New microRNAs from mouse and human. RNA. 2003;9(2):175–179. doi: 10.1261/rna.2146903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Peter ME. Let-7 and miR-200 microRNAs: guardians against pluripotency and cancer progression. Cell Cycle. 2009;8(6):843–852. doi: 10.4161/cc.8.6.7907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Choi PS, Zakhary L, Choi WY, Caron S, Alvarez-Saavedra E, Miska EA, McManus M, Harfe B, Giraldez AJ, Horvitz HR, et al. Members of the miRNA-200 family regulate olfactory neurogenesis. Neuron. 2008;57(1):41–55. doi: 10.1016/j.neuron.2007.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee ST, Chu K, Jung KH, Yoon HJ, Jeon D, Kang KM, Park KH, Bae EK, Kim M, Lee SK, et al. MicroRNAs induced during ischemic preconditioning. Stroke. 2010;41(8):1646–1651. doi: 10.1161/STROKEAHA.110.579649. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. The primers used in qRT-PCR.
Additional file 2: Table S2. Known miRNAs identified in HIST and NC libraries.
Additional file 3: Table S3. Novel candidate miRNAs identified from HIST and NC libraries.
Additional file 4: Table S4. Significantly differentially expressed miRNAs between HIST and NC.
Additional file 5: Table S5. Target genes predicted for significantly differentially expressed miRNAs.
Data Availability Statement
The datasets of miRNAs in the study are available from this article and its Additional files.