Abstract
Several genes and pathways associated with oral squamous cell carcinoma (OSCC) are significant in terms of early detection and prognosis. The objective of this literature review is to evaluate the current research on molecular pathways and genes involved in oral cancer. Articles on the genes involved in oral cancer pathways were evaluated to identify potential biomarkers that can predict survival. In total, 36 articles were retrieved from internet databases, including EBSCO Host, Google Scholar, PubMed, and Science Direct, using the keywords “biomarker of oral cancer,” “pathways of oral cancer,” “genes involved in oral cancer,” and “oral cancer pathways.” A total of 36 studies related to OSCC were chosen. Most of the studies used cell lines, while others used archival tissues, few studies followed up the cases. Three major interlinked pathways found were the nuclear factor kappa B (NF-kB), PI3K-AKT, and Wnt pathways. The commonly mutated genes were cyclin D1 (CCND1), Rb, p53, FLJ10540, and TC21. The NF-kB, PI3K-AKT, and Wnt pathways are most frequently involved in the molecular pathogenesis of oral cancer. However, the CCND1, Rb, p53, FLJ10540, and TC21 genes were found to be more accurate in determining patients’ overall survival. Polymerase chain reaction, immunohistochemistry, and immunoblotting were the commonly used detection methods.
Keywords: Biomarkers, molecular pathways, prognosis, survival, targeted therapy
Introduction
Cancer is a multistep process in which cell mutations accumulate and lead to initiation, progression, sustenance of proliferation, signaling, evasion of growth suppressors, avoidance of cell death, continual replication, angiogenesis, invasion, and metastasis. Cancer involves the mutation of several genes resulting in various pathway alterations.[1,2,3,4,5] Among the various head and neck cancer subtypes, approximately 10% comprise oral squamous cell carcinoma (OSCC).[6,7] It is necessary to elicit the genes and the pathways involved in oral cancer progression. Recent studies have examined the oncogenic signaling pathways, such as glycogen synthase kinase 3 (GSK-3β), c-Myc, AKT, β-catenin, p53, and nuclear factor kappa B (NF-kB), and suggested their roles in oral cancer progression. This systematic review aims to identify the most commonly involved genes and pathways in OSCC.
Methodology
A systematic review was conducted as per the STrengthening the Reporting of OBservational studies in Epidemiology (STROBE) [Table 1]. The key question was “which are the most commonly involved genes and pathways in OSCC?”
Table 1.
Methodology employed

Study design
A systematic review was conducted by considering all the published original research articles that emphasized the genes and associated pathways involved in head and neck cancer. We also included studies that evaluated the genes that can predict oral cancer prognosis.
Inclusion criteria
Articles that evaluated genes and pathways in OSCC were selected.
Exclusion criteria
Review articles, theses, personal judgments, book chapters, powerpoint presentations, and studies that did not validate their results were excluded. Studies on saliva and serum were also excluded as we concentrated on articles pertaining to patient tissues.
Information sources and search strategy
An automated online search of databases, such as EBSCO host, Google Scholar, PubMed, and NCBI, was performed using a combination of phrases and keywords such as “biomarker of oral cancer,” “pathways of oral cancer,” “genes involved in oral cancer,” and “oral cancer pathways.” The relevant articles were also cross-referenced to retrieve additional articles.
Study selection
We set the search strategy per the guidelines given by the STROBE. The initial selection was done by reviewing the titles followed by the abstracts. Articles that were believed to be suitable were re-evaluated by two authors (RSR and DA). Disagreements on the articles were resolved by consulting a third author (SVS).
Data collection process
The overall data collection involved the following information: authors, journals in which the article was published, year of publication, research focus, methodologies employed, results obtained, conclusions drawn, and future scope of research in the given field.
Synthesis of results
We summarized the results of the individual studies and listed the pathways involved as well as the survival rate of the patients. Genes involved in these pathways were also analyzed.
Results
Study selection
The articles were critically evaluated based on STROBE [Figure 1], and 36 of these were considered for the review.[8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43]
Figure 1.
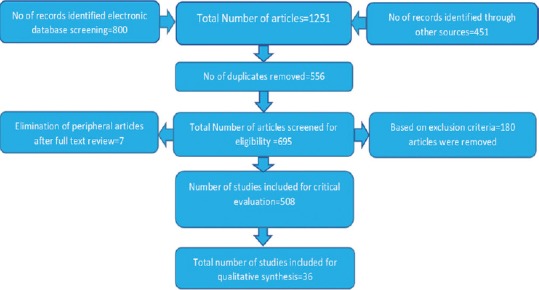
Study design
Study characteristics
Of the 36 selected studies, seven studies were published in 2000–2010 [Figure 2].[8,9,10,12,14,15,17,20,21,22,23,24,25,26,27,29,30,31,32,33,34,35,36,37,39,40,41,42,43]
Figure 2.
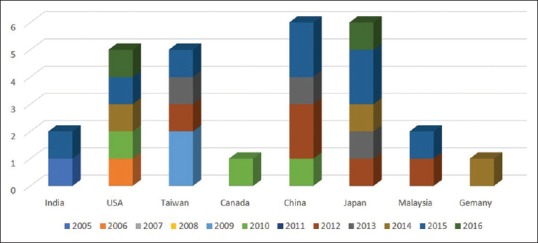
Articles publication frequency by different countries year wise (36 selected articles)
Synthesis of results
Of the 36 studies [Table 3], 34 studies emphasized the genes that regulated pathways in oral cancer progression:[8,9,10,11,12,14,15,16,17,18,19,20,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43]
Table 3.
Studies scrutinized for biomarker and gene expression
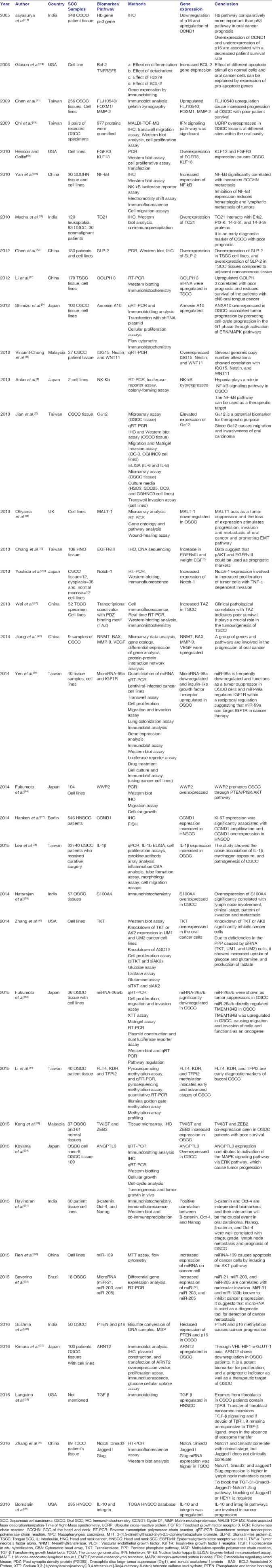
Twenty-one studies focused on genes and the associated pathways that explained the initiation, proliferation, and progression of OSCC.[10,11,14,16,18,19,20,23,24,25,26,27,28,29,30,31,34,35,37,39,43]
The pathways that were found to be commonly associated with OSCC included the PI3K-AKT pathway (seven studies)[10,11,13,14,18,29,39] and the NF-kB pathway (four studies).[8,20,26,39]
Three studies focused on genes associated with patient survival rate, including cyclin D1 (CCND1), Rb and p53, FLJ10540, and TC21.[17,19,28] Three studies showed the involvement of transforming growth factor-beta pathways in OSCC.[26,31,44]
The most commonly used techniques were polymerase chain reaction (PCR), immunoblotting assay, and immunohistochemistry (IHC). Other techniques employed were microarray assays, migration and Matrigel invasion assays, ELISA, transwell migration assays, flow cytometry, and luciferase assays.
Discussion
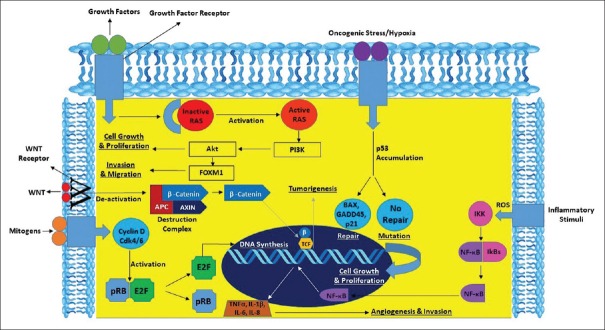
The potential pathways involved in the pathogenesis of oral cancer [Figure 3] and the potential biomarkers that predict patient survival must be explored to improve patient's quality of life.
Figure 3.
Interplay of genes and pathways in oral cancer
Genes and pathways involved in molecular pathogenesis influencing patient prognosis
There are 16 studies listed in Table 2 that show the involvement of various pathways and genes that predict patient's survival. The NF-kB, PI3K-AKT, and Wnt pathways and the CCND1, Rb, p53, FLJ10540, and TC21 genes are commonly mutated and can predict the patient's survival rate. The related studies and their summarized findings are described below:
Table 2.
Genes and pathways indicating the patient survival rate
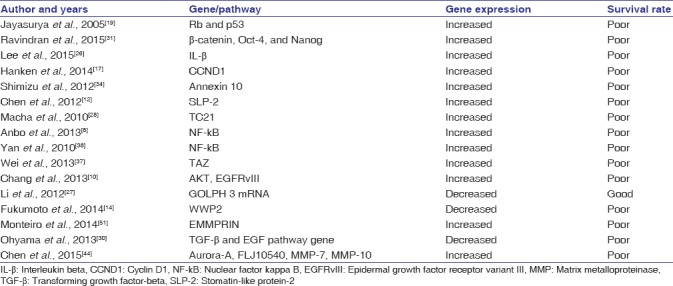
Rb and p53 pathways indicate poor survival in OSCC.[19] CCND1 IHC expression dictates the overall survival rate of patients in OSCC.[17] FLJ10540 expression contributes to a reduced patient survival rate.[11,44] TC21 expression serves as a poor prognostic indicator in OSCC.[28] An increased NF-kB pathway showed a low survival rate in head and neck squamous cell carcinoma (HNSCC).[38] Phosphorylated AKT, epidermal growth factor receptor variant III (EGFRvIII), and disease stage determine the survival of the patients.[10] In the present review, the most common pathways associated with OSCC patient's survival were the PI3K-protein kinase B (PKB)/AKT and NF-kB pathways.[8,10,38] It is important to identify these pathways, since their therapeutic interruption could possibly increase patient survival rates.
Cyclin D 1
CCND1 is a protein that regulates the cell cycle and is encoded by the CCND1 gene on chromosome 11q13. In a study conducted by Hanken et al., 2014,[17] CCND1 amplification and overexpression were associated with Ki-67 expression in HNSCC. There was overexpression of CCND1 that was associated with an increased relapse rate and lymph node metastasis, which dictated poor patient survival (79%).[17] In the phosphorylation of pRB, a major regulator of the G1/S transition of cell cycle CDK4/6-CCND complex is involved. The pRB phosphorylation results in the release of bound E2F transcription factors, premature transition into S-phase, and a resultant nondiploidy of DNA. In numerous epithelial malignancies, amplification of CCND1 locus, 11q13, has been reported. Approximately 30%–50% of OSCC tumors show amplification or increased protein expression.[45,46,47,48] CCND1 can be targeted through the inhibition of cyclin-dependent kinase 4.
Rb and p53 pathway
The Rb protein plays a key role in cell cycle regulation as its phosphorylation will release the brakes on the cell cycle with uncontrolled proliferation. Jayasurya et al., 2005[19] conducted a study on Rb and p53 pathways. CCND1, Rb, p16, CDK4, P21, Bcl-2, p53, and the proliferating cell nuclear antigen were analyzed in OSCC by IHC. Their results showed that the Rb and p53 pathways are interconnected. The Rb protein increased CCND1 and reduced p16 expression and showed a worse prognosis. The relative risk was found to be 2.92 for the Rb gene and 1.57 for p53. The Rb proteins, such as CCND1 were associated with the overall survival and remission of the patients compared to the p53 proteins. Overexpression of CCND1 and underexpression of p16 indicate poor survival.[19] Rb and p53 are the two key pathways that are frequently involved in the progression of OSCC. Some studies found that nearly 90% of cases involved one or both pathways, as well as abnormalities in either the Rb gene or the p53 gene. Some of the data suggest that these pathways are universally targeted in carcinogenesis and oral cancer.[19]
FLJ10450
Matrix metalloproteinase-2 (MMP-2) plays an essential role in tumor development and its expression correlates with metastasis in various cancers including OSCC. A study conducted by Chen et al., 2009[11] stated that FLJ10540 and FOXM1 upregulate the expression of MMP-2 and FLJ10540 plays a role in OSCC progression and metastasis. Increased expression of FLJ10540 in OSCC correlates with poor survival of the patients.[11] Reduction in the expression of FLJ10540 through siRNA inhibits the invasion and migration of the OSCC. Analysis showed a positive correlation of FLJ10540, FOXM1, and MMP-2 with aggressive behavior of OSCC. The findings suggest that FLJ10540 is an important prognostic factor besides a new therapeutic target in the FLJ10540/FOXM1/MMP-2 pathway for OSCC management.[11]
TC21
TC21 belongs to the Ras family, which is involved in various cell functions, including cellular proliferation, apoptosis, differentiation, vesicle trafficking, cytoskeletal organization, and nuclear transport. TC21 protein interacts with both Erk2 and PI3-K indicating the involvement of these pathways in TC21-mediated signaling in oral cancer cells. In the study conducted by Macha et al., 2010,[28] there was gradual increase of TC21 expression from normal tissue to leukoplakia and malignancy that was clinically significant (97.1%). They observed an increased TC21 expression in the early stages of OSCC (T1 + T2) than in the advanced stages (T3 + T4). Moreover, interaction of TC21 with Erk2, 14-3-3 σ, and PI3-K was also observed. Its interaction with Erk2, 14-3-3 σ, and PI3-K suggest that TC21 is a signaling pathway for oral cancer.[28] TC21 bypasses the inhibitory function of domain-negative Ras. TC21 is involved in both growth and morphologic transformation, suggesting that it plays major role in cellular growth through signaling pathways. Interestingly, it initiates the AKT, serine/threonine kinase pathways, which cause cell proliferation, survival, and transformation through various effectors, such as Bad, mTOR, and GSK-3β.[28] It is suggested that TC21 expression is an early event in carcinogenesis, and it coincides with poor prognosis of OSCC.
Nuclear factor kappa B pathway
NF-kB is involved in cell survival, cancer progression, inflammation, and immune response. Increase in NF-kB raises the Interleukin-6, which in turn increases the expression of signal transducer and activator of transcription. Enzyme cycloxygenase-2 (COX-2) is the target molecule of NF-kB, and it synthesizes prostaglandins from arachidonic acid. Overexpression of COX-2 induces cancer progression by increasing vascularity and cellular proliferation and the evasion of apoptosis processes.[38] Further metastasis releases vascular endothelial growth factors and MMP, which are inhibited by pyrrolidine dithiocarbonate. NF-kB contributes to tumor hematologic and lymphatic metastasis.[38] A similar study conducted by Anbo et al., 2013[8] observed that under hypoxic conditions, NF-kB is activated and tumor metastasis and growth is increased.[8] Similarly, the knockdown of NF-kB expression inhibited tumor cell proliferation. This indicates that an increase in NF-kB may result in reduced patient survival.
PI3K-protein kinase B/AKT pathway
The activated PKB/AKT pathway initiates various cellular functions, including growth, angiogenesis, metabolism, survival, proliferation, protein synthesis, apoptosis, and transcription.[49,50] The study conducted by Sushma et al., 2016[35] showed that methylation of PTEN and p16 is involved in the progression of OSCC, but promoter hypermethylation leads to inactivation of the PTEN and p16 genes, which activates AKT phosphorylation. Similarly, the study conducted by Fukumoto et al., 2014[14] showed an increase in WWP2 expression, which in turn reduced the expression of PTEN. Further, expression or upregulation of PI3K-PKB/AKT pathway induces tumor growth and results in poor prognosis.
Wnt signaling pathway
The study conducted by Ravindran et al., 2015[31] reported involvement of the Wnt/β-catenin pathway in the transformation of potentially malignant disorders to the cancerous stage in the oral cavity.[31] The Wnt/β-catenin pathway plays a major role in the carcinogenesis, and there is a significant correlation between metastatic grading and lymph node metastasis (P ≤ 0.001 for overall survival and <0.039 for disease-free survival).[31] The intracellular expression of β-catenin showed poor prognosis in overall survival of the patient. Moreover, β-catenin along with Nanog and Oct-4 expression showed a worse prognosis than β-catenin alone. The Wnt signaling pathway is often mutated, and Wnt/β-catenin pathway can be a potential target for anticancer therapy.
This review shows that among the various biomarkers and pathways, the most commonly involved in OSCC are Rb, p53, TC21, FLJ10540, CCND1, PI3K-AKT, NF-kB, and Wnt pathways. Only one study by Monteiro et al., 2014 showed increased EMMPRIN expression to correlate with poor prognosis and survival.[51] The Rb, p53, and PI3K/AKT pathways were associated with overall patient survival rate.
The present review considered the evaluation of biomarkers and pathways in tissues as it closely relates to their activity in oral cancer and eliminates false positives/negatives compared to serum/saliva. Evaluation of biomarker expression in the saliva/serum results in wide variability in the levels of potential OSCC salivary biomarkers in both noncancerous individuals and OSCC patients.[52] There is a need for further validation of OSCC serum/salivary biomarkers with individuals who have either a chronic oral inflammatory disease or other types of cancers but do not have OSCC. Following meticulous standardization, a comparative study of the expression of oral cancer biomarkers and pathways with correlation of their expression in tissues versus saliva/serum would be an interesting prospect to investigate. The present systematic review highlights the importance of therapeutic targeting of biomarkers and pathways involved in OSCC that could help improve prognosis.
Commonly employed techniques
PCR, Western blot analysis, IHC, microarray, gel electrophoresis, cell proliferation assays, immunofluorescence, proliferative assays, ELISA, and cell cycle assays are the most commonly used techniques. Among these, the most preferred technique was PCR as it is highly sensitive and specific. However, techniques such as IHC and Western blot analysis were frequently used to validate the results. Microarray assays were employed to validate large numbers of gene expression. Based on the analysis, PCR seems to be a reliable technique in determining the expression of proteins and upregulation of genes. A real-time PCR is advantageous over a semi-quantitative PCR as we can monitor the progress of a PCR reaction in real time. At the same time, a relatively small amount of PCR product (DNA, cDNA or RNA) can be quantified.
Conclusion
Although extensive studies have been conducted on these biomarkers and pathways using advanced technology, early diagnosis and treatment remain to be a pitfall. The findings of the present review emphasize the need for research in the domain of targeted cancer therapy. Therefore, it is essential to develop novel therapeutic mechanisms that can alter or inhibit these biomarkers and pathways.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
References
- 1.Rivlin N, Brosh R, Oren M, Rotter V. Mutations in the p53 tumor suppressor gene: Important milestones at the various steps of tumorigenesis. Genes Cancer. 2011;2:466–74. doi: 10.1177/1947601911408889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ribeiro IP, Barroso L, Marques F, Melo JB, Carreira IM. Early detection and personalized treatment in oral cancer: The impact of omics approaches. Mol Cytogenet. 2016;9:85. doi: 10.1186/s13039-016-0293-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Upadhyay P, Dwivedi R, Dutt A. Applications of next-generation sequencing in cancer. Curr Sci. 2014;107:795. [Google Scholar]
- 4.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 5.Coleman WB, Tsongalis GJ. Molecular mechanisms of human carcinogenesis. EXS. 2006;(96):321–49. doi: 10.1007/3-7643-7378-4_14. [DOI] [PubMed] [Google Scholar]
- 6.Curry JM, Sprandio J, Cognetti D, Luginbuhl A, Bar-ad V, Pribitkin E, et al. Tumor microenvironment in head and neck squamous cell carcinoma. Semin Oncol. 2014;41:217–34. doi: 10.1053/j.seminoncol.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 7.Moore SR, Johnson NW, Pierce AM, Wilson DF. The epidemiology of tongue cancer: A review of global incidence. Oral Dis. 2000;6:75–84. doi: 10.1111/j.1601-0825.2000.tb00105.x. [DOI] [PubMed] [Google Scholar]
- 8.Anbo N, Ogi K, Sogabe Y, Shimanishi M, Kaneko T, Dehari H, et al. Suppression of NF-κB/p65 Inhibits the proliferation in oral squamous cancer cells. J Cancer Ther. 2013;4:891–7. [Google Scholar]
- 9.Bornstein S, Schmidt M, Choonoo G, Levin T, Gray J, Thomas CR., Jr IL-10 and integrin signaling pathways are associated with head and neck cancer progression. BMC Genomics. 2016;17:38. doi: 10.1186/s12864-015-2359-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang KY, Tsai SY, Chen SH, Tsou HH, Yen CJ, Liu KJ, et al. Dissecting the EGFR-PI3K-AKT pathway in oral cancer highlights the role of the EGFR variant III and its clinical relevance. J Biomed Sci. 2013;20:43. doi: 10.1186/1423-0127-20-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen CH, Chien CY, Huang CC, Hwang CF, Chuang HC, Fang FM, et al. Expression of FLJ10540 is correlated with aggressiveness of oral cavity squamous cell carcinoma by stimulating cell migration and invasion through increased FOXM1 and MMP-2 activity. Oncogene. 2009;28:2723–37. doi: 10.1038/onc.2009.128. [DOI] [PubMed] [Google Scholar]
- 12.Chen S, Wang L, Li H, Lai J, Zhuang S, Zhang X, et al. Stomatin-like protein-2 is over-expressed during progression of clinically N0 tongue squamous cell cancer. Head Neck Oncol. 2012;4:71. [Google Scholar]
- 13.Chi LM, Lee CW, Chang KP, Hao SP, Lee HM, Liang Y, et al. Enhanced interferon signaling pathway in oral cancer revealed by quantitative proteome analysis of microdissected specimens using 16O/18O labeling and integrated two-dimensional LC-ESI-MALDI tandem MS. Mol Cell Proteomics. 2009;8:1453–74. doi: 10.1074/mcp.M800460-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fukumoto C, Nakashima D, Kasamatsu A, Unozawa M, Shida-Sakazume T, Higo M, et al. WWP2 is overexpressed in human oral cancer, determining tumor size and poor prognosis in patients: Downregulation of WWP2 inhibits the AKT signaling and tumor growth in mice. Oncoscience. 2014;1:807–20. doi: 10.18632/oncoscience.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fukumoto I, Hanazawa T, Kinoshita T, Kikkawa N, Koshizuka K, Goto Y, et al. MicroRNA expression signature of oral squamous cell carcinoma: Functional role of microRNA-26a/b in the modulation of novel cancer pathways. Br J Cancer. 2015;112:891–900. doi: 10.1038/bjc.2015.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gibson SA, Pellenz C, Hutchison RE, Davey FR, Shillitoe EJ. Induction of apoptosis in oral cancer cells by an anti-bcl-2 ribozyme delivered by an adenovirus vector. Clin Cancer Res. 2000;6:213–22. [PubMed] [Google Scholar]
- 17.Hanken H, Gröbe A, Cachovan G, Smeets R, Simon R, Sauter G, et al. CCND1 amplification and cyclin D1 immunohistochemical expression in head and neck squamous cell carcinomas. Clin Oral Investig. 2014;18:269–76. doi: 10.1007/s00784-013-0967-6. [DOI] [PubMed] [Google Scholar]
- 18.Henson BJ, Gollin SM. Overexpression of KLF13 and FGFR3 in oral cancer cells. Cytogenet Genome Res. 2010;128:192–8. doi: 10.1159/000308303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jayasurya R, Sathyan KM, Lakshminarayanan K, Abraham T, Nalinakumari KR, Abraham EK, et al. Phenotypic alterations in Rb pathway have more prognostic influence than p53 pathway proteins in oral carcinoma. Mod Pathol. 2005;18:1056–66. doi: 10.1038/modpathol.3800387. [DOI] [PubMed] [Google Scholar]
- 20.Jian SL, Hsieh HY, Liao CT, Yen TC, Nien SW, Cheng AJ, et al. Gα12 drives invasion of oral squamous cell carcinoma through up-regulation of proinflammatory cytokines. PLoS One. 2013;8:e66133. doi: 10.1371/journal.pone.0066133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang Q, Yu YC, Ding XJ, Luo Y, Ruan H. Bioinformatics analysis reveals significant genes and pathways to target for oral squamous cell carcinoma. Asian Pac J Cancer Prev. 2014;15:2273–8. doi: 10.7314/apjcp.2014.15.5.2273. [DOI] [PubMed] [Google Scholar]
- 22.Kimura Y, Kasamatsu A, Nakashima D, Yamatoji M, Minakawa Y, Koike K, et al. ARNT2 regulates tumoral growth in oral squamous cell carcinoma. J Cancer. 2016;7:702–10. doi: 10.7150/jca.14208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kong YH, Syed Zanaruddin SN, Lau SH, Ramanathan A, Kallarakkal TG, Vincent-Chong VK, et al. Co-expression of TWIST1 and ZEB2 in oral squamous cell carcinoma is associated with poor survival. PLoS One. 2015;10:e0134045. doi: 10.1371/journal.pone.0134045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koyama T, Ogawara K, Kasamatsu A, Okamoto A, Kasama H, Minakawa Y, et al. ANGPTL3 is a novel biomarker as it activates ERK/MAPK pathway in oral cancer. Cancer Med. 2015;4:759–69. doi: 10.1002/cam4.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Languino LR, Singh A, Prisco M, Inman GJ, Luginbuhl A, Curry JM, et al. Exosome-mediated transfer from the tumor microenvironment increases TGFβ signaling in squamous cell carcinoma. Am J Transl Res. 2016;8:2432–7. [PMC free article] [PubMed] [Google Scholar]
- 26.Lee CH, Chang JS, Syu SH, Wong TS, Chan JY, Tang YC, et al. IL-1β promotes malignant transformation and tumor aggressiveness in oral cancer. J Cell Physiol. 2015;230:875–84. doi: 10.1002/jcp.24816. [DOI] [PubMed] [Google Scholar]
- 27.Li H, Guo L, Chen SW, Zhao XH, Zhuang SM, Wang LP, et al. GOLPH3 overexpression correlates with tumor progression and poor prognosis in patients with clinically N0 oral tongue cancer. J Transl Med. 2012;10:168. doi: 10.1186/1479-5876-10-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Macha MA, Matta A, Sriram U, Thakkar A, Shukla NK, Datta Gupta S, et al. Clinical significance of TC21 overexpression in oral cancer. J Oral Pathol Med. 2010;39:477–85. doi: 10.1111/j.1600-0714.2009.00854.x. [DOI] [PubMed] [Google Scholar]
- 29.Natarajan J, Hunter K, Mutalik VS, Radhakrishnan R. Overexpression of S100A4 as a biomarker of metastasis and recurrence in oral squamous cell carcinoma. J Appl Oral Sci. 2014;22:426–33. doi: 10.1590/1678-775720140133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohyama Y, Kawamoto Y, Chiba T, Maeda G, Sakashita H, Imai K, et al. Inhibition of TGF-β and EGF pathway gene expression and migration of oral carcinoma cells by mucosa-associated lymphoid tissue 1. Br J Cancer. 2013;109:207–14. doi: 10.1038/bjc.2013.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ravindran G, Sawant SS, Hague A, Kingsley K, Devaraj H. Association of differential β-catenin expression with oct-4 and Nanog in oral squamous cell carcinoma and their correlation with clinicopathological factors and prognosis. Head Neck. 2015;37:982–93. doi: 10.1002/hed.23699. [DOI] [PubMed] [Google Scholar]
- 32.Ren Y, Zhu H, Chi C, Yang F, Xu X. MiRNA-139 regulates oral cancer tca8113 cells apoptosis through Akt signaling pathway. Int J Clin Exp Pathol. 2015;8:4588–94. [PMC free article] [PubMed] [Google Scholar]
- 33.Severino P, Oliveira LS, Andreghetto FM, Torres N, Curioni O, Cury PM, et al. Small RNAs in metastatic and non-metastatic oral squamous cell carcinoma. BMC Med Genomics. 2015;8:31. doi: 10.1186/s12920-015-0102-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shimizu T, Kasamatsu A, Yamamoto A, Koike K, Ishige S, Takatori H, et al. Annexin A10 in human oral cancer: Biomarker for tumoral growth via G1/S transition by targeting MAPK signaling pathways. PLoS One. 2012;7:e45510. doi: 10.1371/journal.pone.0045510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sushma PS, Jamil K, Kumar PU, Satyanarayana U, Ramakrishna M, Triveni B, et al. PTEN and p16 genes as epigenetic biomarkers in oral squamous cell carcinoma (OSCC): A study on South Indian population. Tumour Biol. 2016;37:7625–32. doi: 10.1007/s13277-015-4648-8. [DOI] [PubMed] [Google Scholar]
- 36.Vincent-Chong VK, Ismail SM, Rahman ZA, Sharifah NA, Anwar A, Pradeep PJ, et al. Genome-wide analysis of oral squamous cell carcinomas revealed over expression of ISG15, nestin and WNT11. Oral Dis. 2012;18:469–76. doi: 10.1111/j.1601-0825.2011.01894.x. [DOI] [PubMed] [Google Scholar]
- 37.Wei Z, Wang Y, Li Z, Yuan C, Zhang W, Wang D, et al. Overexpression of Hippo pathway effector TAZ in tongue squamous cell carcinoma: Correlation with clinicopathological features and patients’ prognosis. J Oral Pathol Med. 2013;42:747–54. doi: 10.1111/jop.12062. [DOI] [PubMed] [Google Scholar]
- 38.Yan M, Xu Q, Zhang P, Zhou XJ, Zhang ZY, Chen WT, et al. Correlation of NF-kappaB signal pathway with tumor metastasis of human head and neck squamous cell carcinoma. BMC Cancer. 2010;10:437. doi: 10.1186/1471-2407-10-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yen YC, Shiah SG, Chu HC, Hsu YM, Hsiao JR, Chang JY, et al. Reciprocal regulation of microRNA-99a and insulin-like growth factor I receptor signaling in oral squamous cell carcinoma cells. Mol Cancer. 2014;13:6. doi: 10.1186/1476-4598-13-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoshida R, Nagata M, Nakayama H, Niimori-Kita K, Hassan W, Tanaka T, et al. The pathological significance of notch1 in oral squamous cell carcinoma. Lab Invest. 2013;93:1068–81. doi: 10.1038/labinvest.2013.95. [DOI] [PubMed] [Google Scholar]
- 41.Li YF, Hsiao YH, Lai YH, Chen YC, Chen YJ, Chou JL, et al. DNA methylation profiles and biomarkers of oral squamous cell carcinoma. Epigenetics. 2015;10:229–36. doi: 10.1080/15592294.2015.1006506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang M, Chai YD, Brumbaugh J, Liu X, Rabii R, Feng S, et al. Oral cancer cells may rewire alternative metabolic pathways to survive from siRNA silencing of metabolic enzymes. BMC Cancer. 2014;14:223. doi: 10.1186/1471-2407-14-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang T, Liang L, Liu X, Wu JN, Chen J, Su K, et al. TGFβ1-smad3-jagged1-notch1-slug signaling pathway takes part in tumorigenesis and progress of tongue squamous cell carcinoma. J Oral Pathol Med. 2016;45:486–93. doi: 10.1111/jop.12406. [DOI] [PubMed] [Google Scholar]
- 44.Chen CH, Chang AY, Li SH, Tsai HT, Shiu LY, Su LJ, et al. Suppression of Aurora-A-FLJ10540 signaling axis prohibits the malignant state of head and neck cancer. Mol Cancer. 2015;14:83. doi: 10.1186/s12943-015-0348-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moreno-Bueno G, Rodríguez-Perales S, Sánchez-Estévez C, Hardisson D, Sarrió D, Prat J, et al. Cyclin D1 gene (CCND1) mutations in endometrial cancer. Oncogene. 2003;22:6115–8. doi: 10.1038/sj.onc.1206868. [DOI] [PubMed] [Google Scholar]
- 46.Niméus E, Baldetorp B, Bendahl PO, Rennstam K, Wennerberg J, Akervall J, et al. Amplification of the cyclin D1 gene is associated with tumour subsite, DNA non-diploidy and high S-phase fraction in squamous cell carcinoma of the head and neck. Oral Oncol. 2004;40:624–9. doi: 10.1016/j.oraloncology.2003.12.014. [DOI] [PubMed] [Google Scholar]
- 47.Rousseau A, Lim MS, Lin Z, Jordan RC. Frequent cyclin D1 gene amplification and protein overexpression in oral epithelial dysplasias. Oral Oncol. 2001;37:268–75. doi: 10.1016/s1368-8375(00)00114-7. [DOI] [PubMed] [Google Scholar]
- 48.Yuan B, Oechsli MN, Hendler FJ. A region within murine chromosome 7F4, syntenic to the human 11q13 amplicon, is frequently amplified in 4NQO-induced oral cavity tumors. Oncogene. 1997;15:1161–70. doi: 10.1038/sj.onc.1201269. [DOI] [PubMed] [Google Scholar]
- 49.Inoda S, Hirohashi Y, Torigoe T, Nakatsugawa M, Kiriyama K, Nakazawa E, et al. Cep55/c10orf3, a tumor antigen derived from a centrosome residing protein in breast carcinoma. J Immunother. 2009;32:474–85. doi: 10.1097/CJI.0b013e3181a1d109. [DOI] [PubMed] [Google Scholar]
- 50.Hemmings BA, Restuccia DF. PI3K-PKB/Akt pathway. Cold Spring Harb Perspect Biol. 2012;4:a011189. doi: 10.1101/cshperspect.a011189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Monteiro LS, Delgado ML, Ricardo S, Garcez F, do Amaral B, Pacheco JJ, et al. EMMPRIN expression in oral squamous cell carcinomas: Correlation with tumor proliferation and patient survival. Biomed Res Int 2014. 2014:905680. doi: 10.1155/2014/905680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cheng YS, Rees T, Wright J. A review of research on salivary biomarkers for oral cancer detection. Clin Transl Med. 2014;3:3. doi: 10.1186/2001-1326-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]