SUMMARY
Potential RNA drug targets for small molecules are found throughout the human transcriptome, yet small molecules known to elicit a pharmacological response by directly targeting RNA are limited to antibacterials. Herein, we describe AbsorbArray, a small molecule microarray-based approach that allows for unmodified small molecules, including FDA-approved drugs, to be probed for binding to RNA motif libraries in a massively parallel format. Several drug classes bind RNA including kinase and topoisomerase inhibitors. The latter avidly bound the motif found in the Dicer site of oncogenic microRNA (miR)-21 and inhibited its processing both in vitro and in cells. The most potent compound de-repressed a downstream protein target and inhibited a miR-21-mediated invasive phenotype. The compound’s activity was ablated upon overexpression of pre-miR-21. Target validation via Chemical Cross-Linking and Isolation by pull-down showed direct engagement of pre-miR-21 by the small molecule in cells, demonstrating RNAs should indeed be considered druggable.
In Brief
RNA is an emerging target for small molecules, but has it been an established one all along? Velagapudi et al. profiled the binding of medicines to thousands of RNA motifs, showing that broad drug classes bind RNA. Indeed, approved anticancer drugs target an oncogenic non-coding RNA, affecting its phenotype.
Graphical Abstract
INTRODUCTION
RNA plays important roles in nearly every cellular process and its dysregulation can contribute to disease (Calin and Croce, 2006; Cooper, et al., 2009). The Encyclopedia of DNA Elements (ENCODE) project and subsequent analyses showed that only 1–2% of our genome encodes for protein yet ca. 80% of it is transcribed into RNA (2012). Although the amount of functional non-coding RNA is controversial, it is clear that they play a myriad of important cellular roles.
The development of therapeutics that target RNA has mostly centered on using oligonucleotides. They are invaluable tools that have been advanced to multiple clinical trials. For example, in late 2016 an antisense agent was approved to treat spinal muscular atrophy, a devastating and heretofore incurable disease (Morrow, 2017). When antisense oligonucleotides are not directly delivered to the central nervous system (CNS), however, side effects have been observed, ranging from stimulating an immune response to thrombocytopenia (Frazier, 2015).
Small molecules could be a preferred modality for targeting RNA, however these compounds have been generally limited to antibiotics that target the three-dimensional structure of the ribosome (Blount and Breaker, 2006; Poehlsgaard and Douthwaite, 2005; Tenson and Mankin, 2006) or riboswitches (Blount and Breaker, 2006). Indeed, compounds targeting nucleic acids, including antisense, are known to bind to proteins as well (Ivanov, et al., 1998; Liang, et al., 2015). Compounds have been identified to target RNA by using various methods including screening, structure-based design, and sequence-based design, cementing RNA as a potentially broad small molecule drug target. One class of non-coding RNAs that has been extensively targeted by small molecules are viral RNAs. The first viral RNA regulatory elements shown to be bound and interfered with by small molecules were the HIV transactivation response (TAR) (Mei, et al., 1995) and Rev response element (RRE) (Zapp, et al., 1993). Other viral RNAs such as the hepatitis C virus internal ribosomal entry site (HCV IRES) and Influenza A have been focus of several targeting studies (Dibrov, et al., 2014; Lee, et al., 2014). Recently, several small molecules have been designed to target nucleotide repeat expansions including fragile X-associated tremor ataxia syndrome (FXTAS; CGG) (Yang, et al., 2016), myotonic dystrophy type 1 (DM1; CUG) (Gareiss, et al., 2008; Jahromi, et al., 2013; Pushechnikov, et al., 2009), myotonic dystrophy type 2 (DM2; CCUG) (Childs-Disney, et al., 2014), Huntington’s disease (HD; CAG) (Kumar, et al., 2012), and frontotemporal dementia and amyotrophic lateral sclerosis (c9FTD/ALD; G4C2) (Su, et al., 2014). A salient question is if small molecules that target non-coding RNAs can be identified and developed into medicines or chemical probes.
A way to evaluate the druggability of RNA is to study if known drugs bind RNA and affect physiologically important pathways. Perhaps non-coding human RNAs are targeted with approved drugs in their clinical settings, but these interactions have gone unidentified. The focus of this work is to study the potential of RNA as a small molecule drug target using known drug classes. Unmodified small molecule drugs were probed for binding RNA motifs using a non-covalent small molecule microarray (AbsorbArray) in a massively parallel library-versus-library screening approach dubbed two-dimensional combinatorial screening (2DCS) (Childs-Disney, et al., 2007; Disney, et al., 2008). Indeed, various drug classes including kinase inhibitors, pre-mRNA splicing modulators, and topoisomerase inhibitors bound RNA avidly. By mining Inforna, a sequence-based lead identification strategy for RNA (Velagapudi, et al., 2014), it was found that topoisomerase inhibitors bound the Dicer site of the microRNA (miR)-21 precursor (pre-miR-21). In cells, these compounds reduced mature miR-21 levels and modulated a miR-21-mediated invasive phenotype, as miR-21 is oncogenic. Importantly, Chemical Cross-Linking and Isolation by Pull-down [Chem-CLIP] (Guan and Disney, 2013; Rzuczek, et al., 2017; Yang, et al., 2015) studies revealed physical interactions between pre-miR-21 and the small molecule.
RESULTS
Development of AbsorbArray.
To develop an approach that allowed for unmodified compound libraries to be probed for binding to a library of RNA motifs, we looked to advances in small molecule microarrays. Small molecule microarrays have been invaluable to identify compounds that bind biomolecules, however compounds are immobilized by adding functional groups that allow for covalent attachment (Hergenrother, et al., 2000; MacBeath, et al., 1999; MacBeath and Schreiber, 2000). Such modifications could affect molecular recognition of the parent compound’s natural targets. Herein, we describe an approach named AbsorbArray (Figure 1) that allows small molecules to be non-covalently adhered onto a surface and probed for binding to a library of RNA motifs found in non-coding RNAs. By testing various conditions, we found that small molecules can be absorbed onto agarose-coated microarray surfaces when spotted onto hydrated surfaces that are subsequently dried (Figure S1).
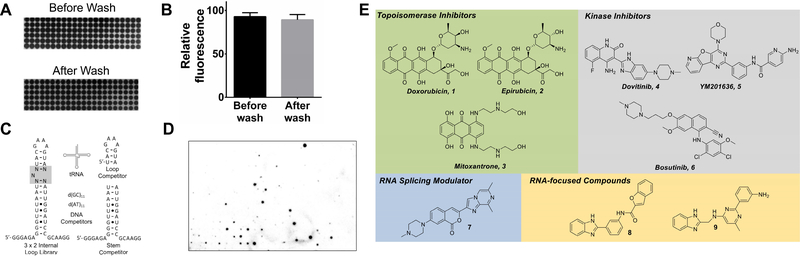
Figure 1:
The development of AbsorbArray and the RNA motif libraries used to study small molecule binding. A, representative image of an AbsorbArray chip after spotting mitoxantrone before and after washing. B, plot of the signal from mitoxantrone before and after washing. C, secondary structures of the nucleic acids used in this study. D, representative image of an AbsorbArray-enabled 2DCS screen (85.5 × 127.8 × 1.1 mm (width x height x thickness)). E, structures of various small molecules that are found to bind RNA in this AbsorbArray-enabled 2DCS experiment. See also Figure S1 and Table S1.
Identification of small molecules that bind RNA motifs via AbsorbArray and 2DCS.
AbsorbArray allows for unmodified small molecules to be retained on a surface after washing (Figure 1A & 1B) and then probed for binding to radiolabeled RNA motif libraries (Figure 1C & 1D). To validate the platform, we probed the binding of the following compounds to RNA motifs via AbsorbArray and 2DCS (Disney, et al., 2008): the NIH Clinical Collection (NIH-CC), a library of 727 pharmacologically active compounds used in human clinical trials; kinase inhibitors known to modulate RNA biology but not known to directly engage RNA (SYNlibrary95, Synkinase; n = 95); an RNA-focused library (n = 201) (Rzuczek, et al., 2015); and three compounds that modulate the alternative splicing of survival motor neuron 2 (SMN2) (Naryshkin, et al., 2014; Palacino, et al., 2015). As many of the compounds in these studies have been used in clinical trials, they provide a potentially rich source of drugs that can be repurposed to target RNA.
Briefly, the array was hybridized with a 32P-labeled RNA library displaying randomized nucleotides in the pattern of a 3×2 nucleotide internal loop (3×2 ILL; Figure 1C). The 1,024 member 3×2 ILL was chosen as it contains asymmetric internal loops and bulges present in various cellular RNAs that are of high importance, as described previously via a transcriptome-wide RNA structural analysis (Liu, et al., 2016). Incubation was completed in the presence of excess oligonucleotides that include tRNA, DNA, and mimics of the regions common to all library members (Figure 1C), ensuring interaction of the small molecules with the randomized region (Figure 1D). These studies defined a series of compounds and compound classes as RNA binders (Figure 1D), including topoisomerase inhibitors, kinase inhibitors, and splicing modulators (Figure 1E). Importantly, several unique small molecules (hits derived from the RNA-focused library) were discovered using this platform (Figure 1E).
The RNA motifs from 3×2 ILL that bound each compound were harvested from the array surface and sequenced. To score the relative affinities of the selected RNA motifs, an approach called High Throughput Structure-Activity Relationships Through Sequencing (HiT-StARTS) was applied (Velagapudi, et al., 2017). In HiT-StARTS, the frequency of each RNA bound to a small molecule is compared to its frequency in the starting library, as determined by RNA-seq analysis. This pooled population comparison quantifies the statistical significance of enrichment, or the parameter Zobs. HiT-StARTS quickly identifies binding and non-binding RNA motifs for a specific small molecule and minimizes false negatives and positives. Binding assays were completed between selected RNA motif-small molecule pairs, revealing high affinity and selective binding when Zobs >8 (Table 1), as observed previously (Velagapudi, et al., 2017) and highlighting the predictive value of HiT-StARTS. Fitness Scores are assigned for RNA binders by normalizing Zobs values to the highest Zobs for a given selection, with the best score assigned a value of 100 (Velagapudi, et al., 2014).
Table 1:
Global analysis of selected RNA-motif small molecule interactions and binding affinities of topoisomerase inhibitors to the A bulge in the Dicer site of pre-miR-21 and the corresponding fully paired RNA. Kd’s are reported in nM. See also Figure S2 and Table S1.
| Compound | Range of Zobs for Binders | Number of Binders | Range of Kd’s for Binders (nM) | A bulge | Base paired control |
|---|---|---|---|---|---|
| 1 | 28–8 | 371 | 20–550 | 58 ± 7.6 | >1000 |
| 2 | 37–8 | 281 | 6–35 | 24 ± 8.1 | 1080 ± 231 |
| 3 | 39–8 | 213 | 10–26 | 33 ± 3.3 | >1000 |
Inforna identifies oncogenic miRNA targets from AbsorbArray.
The RNA motifs identified by 2DCS of the NIH-CC were mined against the RNA motifs in all human miRNA precursors (Velagapudi, et al., 2014) to identify potential oncogenic miRNA targets. Several compounds were predicted to bind Drosha or Dicer processing sites of pre-miRNAs (Table S1). To further refine the RNA targets, we compared the relative expression profiles of the potential miRNA hits (Table S1) from publicly available databases using miRmine (Panwar, et al., 2017). miR-21 was found to be expressed 30–4,000-fold higher expression compared to other potential miRNA targets. As the oncomiR is upregulated in most cancers (Volinia, et al., 2006), contributes to various cancer phenotypes (Chan, et al., 2005; du Rieu, et al., 2010; Meng, et al., 2007; Seike, et al., 2009; Yan, et al., 2008), and has been a focus of various small molecule targeting efforts, miR-21 was further studied.
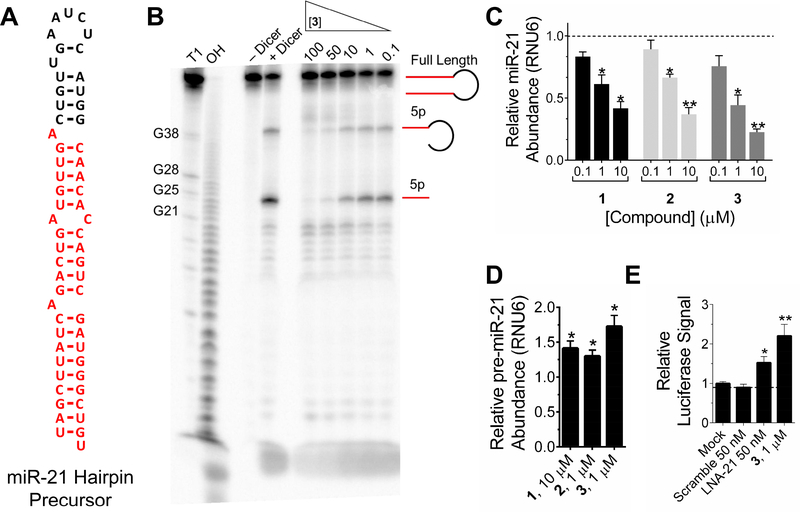
Inforna identified that topoisomerase inhibitors 1, 2 and 3 bind the A bulge in the Dicer site of the miR-21 hairpin precursor (Figure 2A). Compounds 1 and 3 were previously observed to stabilize single-stranded regions of RNA by NMR (Marcheschi, et al., 2009; Zheng, et al., 2009), supporting the hypothesis that these compounds can bind to the A bulge. We therefore measured the affinity of each compound to the A bulge of miR-21 (Figure S2), affording Kd’s of 58 ± 7.6 nM, 24 ± 8.1 nM, and 33 ± 3.3 nM for 1, 2, and 3, respectively (Table 1 & Figure S2B). Notably, binding was not saturable for compounds 1 and 3 to a fully base paired RNA up to 5000 nM; binding of 2 to the fully paired RNA was 28-fold weaker than to the A bulge, with a Kd of 1080 ± 231 nM (Table 1 & Figure S2C). Interestingly, 3 also binds the U bulge adjacent to the Dicer site with high affinity, as predicted by Inforna, with a dissociation constant of 46 ± 7.1 nM. Increased avidity of 3 is observed to an RNA that contains both the A and U bulges (Kd = 22 ± 8.3 nM) (Figure S2). Despite the dynamics of RNA structure, both the A and U bulge regions in the miR-21 hairpin precursor have been observed by NMR (Chirayil, et al., 2014). Additional binding analyses using microscale thermophoresis (MST) (Moon, et al., 2018; Seidel, et al., 2013), indicated the binding affinity of 3 to a miR-21 hairpin containing both the A and U bulge (miR-21 Hairpin; Figure S2D) and a miR-21 hairpin containing only the A bulge (miR-21 A Bulge; Figure S2E) as 500 and 300 nM, respectively. Notably, no binding was observed to a miR-21 base pair control without the A and U bulges (miR-21 Base Pair; Figure S2F). As 3 showed high affinity for both the A and U bulges found in miR-21, compound 3 was tested for inhibition of pre-miR-21 Dicer processing in vitro. A dose response was observed with an IC50 of ~3 μM (Figure 2B). The wild type pre-miR-21 (Pre-miR-21 WT) and a construct of pre-miR-21 without the U bulge (Pre-miR-21 U27 mutant) both had significant inhibition of in vitro Dicer processing with the addition of 3 μM of 3 (Figure S3). No significant in vitro Dicer processing of pre-miR-21 without an A bulge (Pre-miR-21 A22 mutant) was observed at 3 μM, indicating that compound 3 can more effectively inhibit biogenesis of the wild type pre-miR-21 (Pre-miR-21 WT) and of the pre-miR-21 construct without the A bulge (Pre-miR-21 U27 Mutant), and does not globally inhibit Dicer enzyme activity (Figure S3). Thus, this class of compounds, known to affect cellular processes by targeting DNA (Binaschi, et al., 1995), might also elicit anti-cancer effects by affecting the biogenesis of miR-21.
Figure 2:
Inhibition of Dicer processing of pre-miR-21 in vitro and in cells. A, secondary structure of the miR-21 hairpin precursor. B, representative gel image of the inhibition of Dicer processing of pre-miR-21 by 3. C, effect of 1, 2, and 3 on mature levels in triple negative breast cancer cell line MDA-MB-231. D, effect of 1, 2, and 3 on pre-miR-21 levels in triple negative breast cancer cell line MDA-MB-231. E, effect of 3, an LNA oligonucleotide targeting miR-21 (LNA-21; Exiqon, 4100689–101) and a control LNA (Scramble; Exiqon, 199006–102) on PTEN using a luciferase reporter assay. * indicates p<0.5; ** indicates p<0.01, as determined by a two-tailed Student t test. Data represents mean ± S.E.M. of biological triplicates. See also Figures S2 and S3.
Activity of lead compounds.
Compounds 1, 2, and 3 are readily taken up into cells and localize in both the cytoplasm and nucleus (Feofanov, et al., 1997; Shaul, et al., 2013). Therefore, the efficacy of the topoisomerase inhibitors for inhibiting biogenesis of miR-21 was tested in the triple negative breast cancer cell line MDA-MB-231 by measuring mature and pre-miR-21 levels by RT-qPCR. Indeed, all three compounds reduced levels of the mature miR-21 (Figure 2C) and increased levels of pre-miR-21 (Figure 2D). The IC50’s for 1, 2, and 3 for reducing mature miR-21 levels are approximately 5, 10, and 1 μM, respectively. As 3 most significantly increased pre-miR-21 levels (Figure 2D), it was further characterized for de-repressing a downstream protein target. Indeed, 3 de-repressed the downstream effect of miR-21 on the tumor suppressor phosphatase and tensin homologue deleted on chromosome 10 (PTEN) (Li, et al., 1997), using a previously validated luciferase reporter (O’Donnell, et al., 2005) (Figure 2E).
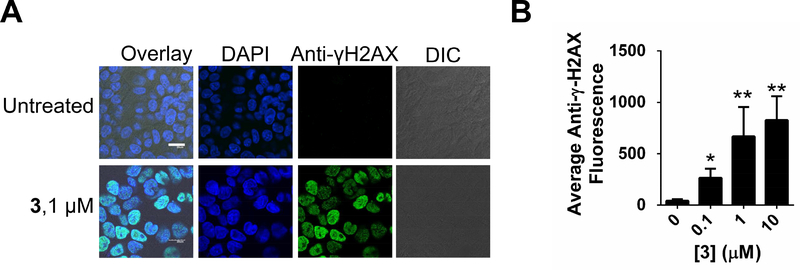
The most widely accepted mode of action for 3 is DNA intercalation, leading to cross-links and strand breaks (Lown, et al., 1984). We therefore studied if the concentration required to reduce mature miR-21 levels also causes DNA damage. DNA damage was assessed by imaging γ-H2AX foci that form in response to DNA double strand breaks (Pilch, et al., 2003; Rogakou, et al., 1999). These studies showed that both DNA damage and a reduction of miR-21 levels were observed at the active concentration of 1 μM (Figures 3 & S4A). We also tested the effect of 3 on topoisomerase II activity in vitro and found an inhibitory effect of 3 at 1 μM (Figure S4B-D).
Figure 3:
Effect of 3 on DNA damage. A, representative confocal microscopy images of the effect of 3 on DNA damage in MDA-MB-231 cells by staining for γ-H2AX foci. White scale bar indicates 20 μm. B, quantification of DNA damage upon treatment of MDA-MB-231 cells with 3. * indicates p<0.5; ** indicates p<0.01, as determined by a two-tailed Student t test. See also Figure S4.
Compound 3 inhibits a miR-21-mediated invasive phenotype.
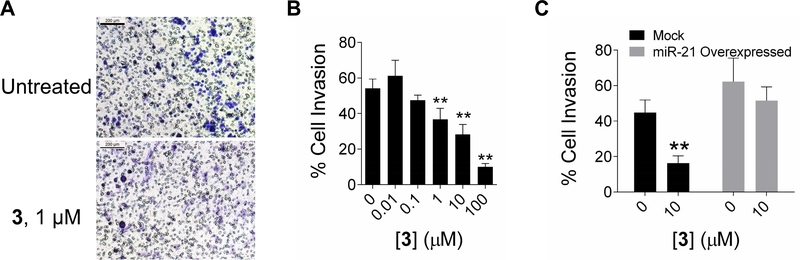
As previous studies have shown that miR-21 contributes to a migratory and invasive phenotype (Huang, et al., 2009; Zhu, et al., 2008), we next studied if inhibition of miR-21 by 3 was sufficient to induce a reversal of phenotype in MDA-MB-231 cells by using a Boyden chamber assay. Indeed, dose dependent inhibition of invasive phenotype was observed (Figures 4A & 4B). Importantly, the activity of 3 against an invasive phenotype is ablated by overexpression of pre-miR-21 (Figure 4C), indicating the effect is via a miR-21-mediated circuit.
Figure 4:
Evaluation of phenotypic effects of 3. A, representative images of reduced invasion observed in MDA-MB-231 cells after miR-21 inhibition through 3 treatment (1 μM). Cells were incubated for 16 h at 37 °C through Matrigel baseme nt membrane in a Boyden chamber assay. Invaded cells were fixed with 1% paraformaldehyde and stained with 0.1% crystal violet for visualization. Black scale bar indicates 200 μm. B, quantification of cell migration shown in panel A (2 biological replicates, 4 fields of view). C, effect on migration upon treatment with 3 (10 μM) when pre-miR-21 is overexpressed via transfection of a miR-21 plasmid in MDA-MB-231 cells. * indicates p<0.5; ** indicates p<0.01, as determined by a two-tailed Student t test.
Compound 3 binds pre-miR-21 in cells as determined by Chem-CLIP.
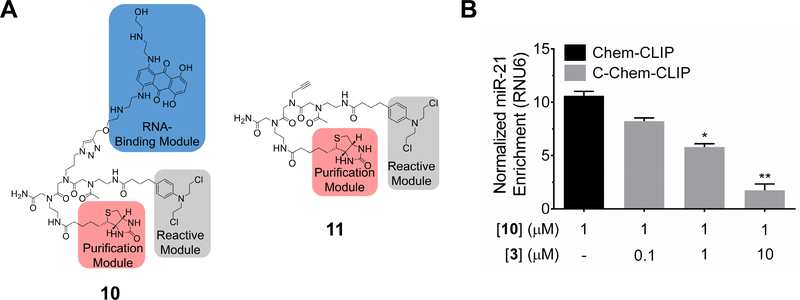
To further evaluate whether 3 directly engages pre-miR-21 in cells, we used Chem-CLIP, a small molecule-RNA profiling approach (Guan and Disney, 2013; Su, et al., 2014). A Chem-CLIP probe was synthesized by appending a biotin and a chlorambucil cross-linking module onto 3 to afford compound 10 (Figure 5A). Compound 10 cross-links with its cellular RNA targets, and the resulting small molecule-RNA conjugates are harvested by biotin capture. Both in vitro and in cells, 10 reacted with pre-miR-21 (Figures 5B & S5A-B). Importantly, co-addition of parent 3 and 10 in cells (Competitive Chem-CLIP [C-Chem-CLIP]) decreased the extent of pull-down of pre-miR-21 in a dose-dependent manner (Figure 5B), thus proving that 3 binds its RNA target in cells. Chem-CLIP was also performed on 11 (Figure 5A), which lacks the RNA-binding module; no enrichment of miR-21 observed in the pulled down fractions, whether in vitro or in cells, as expected (Figure S5A & S5C). Enrichment levels of miR-21 were normalized to levels of mature miR-21 after treatment with 10 or 11 (Figure S5D).
Figure 5:
Chem-CLIP target validation studies of 3 in MDA-MB-231 cells. A, structures of 10, 3-CA-Biotin, which is comprised of the RNA-binding module 3, a biotin purification module and a cross-linking chlorambucil (CA) module, and control compound 11 (Control CA-Biotin), which lacks the RNA-binding module. B, Chem-CLIP of pre-miR-21 using 1 μM of 10 in MDA-MB-231 cells and Competitive-Chem-CLIP (C-Chem-CLIP) of pre-miR-21 in MDA-MB-231 cells by using increasing concentrations of 3 to compete with 10 for binding (1 μM). * indicates p<0.5; ** indicates p<0.01, as determined by a two-tailed Student t test. Data represents mean ± S.E.M. of biological triplicates. See also Figure S5.
Selectivity of 3.
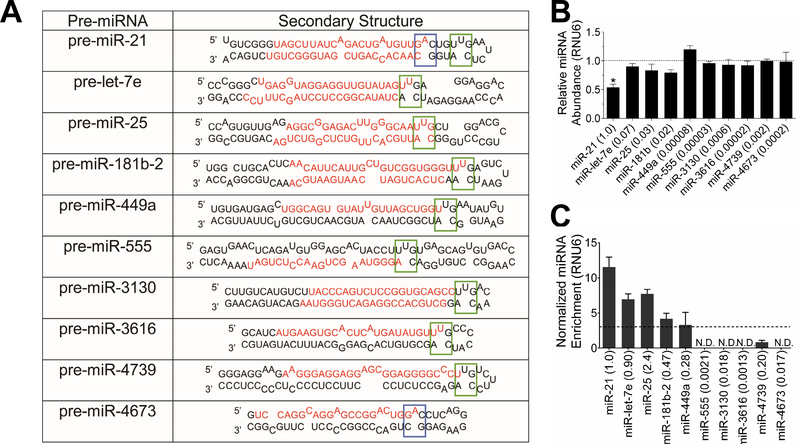
The selectivity of 3 for inhibiting miR-21 was measured by using RT-qPCR, in particular by studying its effect on miRNAs that contain the same motifs in pre-miR-21’s Dicer site, or RNA isoforms. Previous studies have identified that the expression levels of miRNAs will most likely be affected if the small molecule binds in a biologically important site, such as Dicer or Drosha processing sites (Velagapudi, et al., 2014). Therefore, we queried a database of miRNA secondary structures to identify precursor miRNAs that contain the same A and/or U bulges found in pre-miR-21’s Drosha or Dicer processing sites (Figure 6A). Despite the presence of the A and/or U bulges in Dicer processing sites of other pre-miRNAs, only levels of miR-21 were affected (Figure 6B).
Figure 6:
Selectivity studies of 3. A, pre-miRNAs that contain the same A (blue box) and/or U bulge (green box) found in the Dicer site of pre-miR-21. B, selectivity of 3 for miR-21 over other miRNAs with the same target A and/or U bulge, as determined by measuring mature miRNA levels by RT-qPCR. The numbers in parentheses after the miRNA name indicate relative expression levels as compared to miR-21. C, Pull-down of other miRNAs that contain A and/or U bulges by 10. The numbers in parentheses after the miRNA name indicates relative expression level as compared to miR-21. * indicates p <0.05; ** indicates p<0.01, as determined by a two-tailed Student t test; N.D. indicates mature miRNA levels were not detectable (Ct>32). Data represents mean ± S.E.M. of biological triplicates.
To determine if 3 does not inhibit levels of the other miRNAs studied because it does not bind the target in cells, we completed Chem-CLIP studies for the RNA isoforms (Figure 6C). Binding is observed to only two miRNAs, let-7e and miR-25; however, binding is not sufficient to reduce their mature levels. The binding site for 3 is located in the Dicer site of both pre-miRNAs (Liu, et al., 2016), however the lack of activity of 3 against let-7e and miR-25 could be traced to the fact that both let-7e and miR-25 are expressed at much lower levels compared to miR-21 (7% and 3%, respectively compared to miR-21). Collectively, these results suggest that the enhanced anti-miR-21 activity of 3 is due to the presence of two binding sites in pre-miR-21 (only a singular binding site is present in pre-let-7e and pre-miR-25) and its comparatively much higher expression level.
DISCUSSION
This study has shown that various drug classes have the capacity to bind RNA, including topoisomerase inhibitors that affect a key non-coding, oncogenic miRNA. It has been suggested previously that topoisomerase inhibitors including doxorubicin have non-topoisomerase II inhibitory activity that results in apoptosis in cancer cells (Swift, et al., 2006). Thus, our studies may be another example of compounds whose activity can be traced to affecting multiple pathways. Other classes of small molecules have been shown to target RNA in addition to their previously known target. For example, the target of the antibacterial roseoflavin, the flavin mononucleotide (FMN) riboswitch, was not uncovered until the discovery of riboswitches (Lee, et al., 2009). Aggressive activity in repurposing known drugs has revealed many additional targets across all types of biomolecules. It is likely that many drugs affect multiple pathways, some known and others unknown, to provide a therapeutic effect.
Extensive effort has been made to identify compounds that target miR-21 (Shortridge and Varani, 2015). Although several compounds have been shown to bind miR-21 in vitro (Connelly, et al., 2017), only a few inhibit miR-21 levels in cells (Gumireddy, et al., 2008; Naro, et al., 2015; Shortridge, et al., 2017). Most of the compounds identified were either transcriptional inhibitors or did not affect phenotype. Other studies showed that the aminoglycoside antibiotic streptomycin inhibited Dicer processing of pre-miR-21 and had a modest effect on apoptosis (about 25%) in MCF7 cells (Bose, et al., 2012). As the RNA-targeting small molecule field emerges, careful consideration must be paid to the effect of compound on phenotype and its correlation with target modulation. In this study, it was found that 3 inhibited levels of mature miR-21, concomitantly increased levels of pre-miR-21, and reversed the invasive phenotype caused by elevated expression of miR-21 in triple negative breast cancer cells. These effects are ablated upon overexpression of pre-miR-21, giving further support to 3’s mechanism of action.
The utility of these compounds to study miR-21 biology as chemical probes, however, is likely limited due to the multiple effects that the molecules have in cells. Other small molecules that have perhaps better selectivity are more useful chemical probes and include compounds targeting the ribosome and riboswitches (Carter, et al., 2000; Poehlsgaard and Douthwaite, 2005).
Of further interest is the observation that kinase inhibitors are a key compound class that target RNA. These compounds have garnered much attention as clinical candidates and many others are in pre-clinical development. It is also perhaps not surprising that these molecules target RNA given their penchant for having hydrogen bond donors and acceptors and the ability to stack with RNA bases. Thus, these molecules could be medicinally optimized to drug RNA and reach a clinical end point.
STAR Methods
Experimental Model and Subject Details:
All cells were grown at 37 oC and 5% CO2. MDA-MB-231 (ATCC: HTB-26) cells were cultured in Roswell Park Memorial Institute (RPMI) 1640 medium supplemented with 1% penicillin/streptomycin, 1% glutagro (Corning) and 10% fetal bovine serum (FBS) (complete growth medium). Cells were directly purchased from ATCC but were not authenticated.
Method Details:
General Nucleic Acids Methods:
All DNA oligonucleotides were purchased from Integrated DNA Technologies, Inc. (IDT) and used without further purification. The RNA competitor oligonucleotides and pre-miR-21 constructs for ITC were purchased from Dharmacon and de-protected according to the manufacturer’s standard procedure. Competitor oligonucleotides were used to ensure that RNA-small molecule interactions were confined to the randomized region of 3×2 nucleotide internal loop pattern). All aqueous solutions were made with nanopure water. The RNA library was transcribed by in vitro transcription from the corresponding DNA template.
Compound Libraries:
The 727-member NIH Clinical Collection (NIH-CC) was obtained from Scripps Molecular Screening Center. The NIH-CC is a small molecule library comprised of drugs that have been in Phase I or II clinical trials. Thus, compounds were selected for drug-likeness, bioavailability, and stability. The 95-member Kinase library (SYNLibrary-95, catalog # SYN-2103) was purchased from Synkinase. SYNLibrary-95 contains a kinase inhibitor library that recognizes 57 targets. Three pre-mRNA splicing regulators were obtained from the California Institute for Biomedical Research (Calibr). The 201-member RNA-focused compound library was obtained from Scripps Molecular Screening Center. These compounds were previously identified by performing chemical similarity search based on molecules that possess bis-benzimidazole or similar cores (Rzuczek, et al., 2015). Hit compounds were re-purchased in larger quantities for testing. Compound 1 was purchased from Cayman Chemicals, while 2 and 3 were purchased from Biotang, Inc.
Construction of AbsorbArray-assisted Small Molecule Microarrays:
Microarrays were constructed by pouring 15 mL of 1% molten agarose solution onto a glass slide [85.5 × 127.8 × 1.1 mm width x height x thickness)]. The agarose was then air dried for 1 h at room temperature to form a thick gel surface. A 200 nL aliquot of compounds (10 mM in DMSO) were then pinned into the agarose gel from a 384-well plate (Greiner; catalog #781201–906) using a Biomek NXP Laboratory Automation Workstation equipped with a 100 nL 384-pin floating head with diameter 0.787 mm and inter-pin distance of 4.5 mm. The compound-spotted slides were allowed to air dry completely to form a thin, invisible agarose layer. After drying, slides were washed three times with 1× PBST (1× PBS+0.1% Tween) followed by washing three times for 5 min with nanopure water and air dried completely. 2DCS of the final hit compounds were completed on microscopic slides [Fisher Scientific, 12550016; 75 × 25 × 1 mm (width x height x thickness); see Figure S1]. Quantification of compound spotting before and after washing was analyzed using Quantity One (BioRad).
RNA Selection:
2DCS selections were completed as previously described (Disney, et al., 2008). All oligonucleotides, including 5’-end 32P-labeled RNA library [prepared as described previously (Disney, et al., 2008)], competitor chase oligonucleotides (Figure 2D) and tRNA, were folded separately in 1× Hybridization Buffer (HB, 8 mM Na2HPO4, pH 7.0, 185 mM NaCl, and 1 mM EDTA) by heating at 90 °C for 2 min followed by cooling to room temperature on the bench top. The 5’-end 32P-labeled folded RNAs (~50,000 counts by Geiger counter) were mixed together and MgCl2 and bovine serum albumin (BSA) were added at 1 mM and 40 μg/mL final concentrations, respectively, in a total volume of 3000 μL. Prior to hybridization, microarrays were pre-equilibrated with 3000 μL of 1× HB supplemented with 1 mM MgCl2 and 40 μg/mL BSA (1× HB2) for 10 min at room temperature to prevent non-specific binding. After the slides were pre-equilibrated, HB2 was removed and the mixture of folded RNAs was applied to the microarray surface and distributed evenly across the array surface with a custom-cut piece of Parafilm (solution height ~2–3 mm). The slide was hybridized for approximately 45 min at room temperature. After incubation, the Parafilm was removed, and the slide was washed by submersion in 30 mL of HB for 5 s with gentle agitation three times. Excess buffer was removed completely from the slide, and the slide was dried at room temperature for 30 min. While compounds may diffuse from their initial spot, significant interspot diffusion of the RNA-ligand complexes were not observed. Although diffusion rates differ depending on the physical/chemical properties of each compound, washing of the plates before and after hybridization will remove complexes that have diffused out of the spotted area. The microarray was exposed to a phosphorimager screen and imaged using a Molecular Devices Typhoon phosphorimager. Then, the image was used as a template to harvest bound RNAs from the microarray surface. To harvest bound RNAs, 1 μL of nanopure water was added to each spot. After 30 s, the buffer was absorbed and the agarose gel at that spot was excised using a toothpick.
Reverse Transcription and PCR Amplification to Install Barcodes for RNA-seq:
The excised agarose was placed into a thin-walled PCR tube with 18 μL of water, 2 μL of 10x RQ1 DNase I buffer and 2 units of RQ1 RNase-free DNase (Promega). The solution was incubated at 37 °C for 2 h and then quenched by addition of 2 μL of 10x DNase stop solution (Promega). Samples were incubated at 65 °C for 10 min to completely in activate the DNase and then subjected to RT-PCR amplification to install a unique barcode. Reverse transcription reactions were completed in 1× RT buffer, 1 mM dNTPs, 5 μM RT primer (5’CCTCTCTATGGGCAGTCGGTGATCCTTGCGG ATCCAAT), 200 μg/mL BSA, 4 units of AMV reverse transcriptase, and 20 μL of DNase-treated selected RNAs. Samples were incubated at 60 °C for 1 h. A 20 μL aliquot of the RT reaction was added to 6 μL of 10× PCR Buffer, 4 μL of 100 μM forward primer including barcode (5’- CCATCTC ATCCCTGCGTGTCTCCGACTCAGxrefXXXXGATGGGAGAGGGTTTAAT where X represents unique barcode, GAT is the barcode adapter), 2 μL of 100 μM reverse primer, 0.6 μL of 250 mM MgCl2, and 2 μL of Taq DNA polymerase. Two-step PCR was performed at 95 °C for 1 min and 72 °C for 1 min. Aliquots of the RT-PCR product were checked every two cycles starting at cycle 10 on a non-denaturing 8% polyacrylamide gel stained with SYBR Green to ensure that background spots (excised from the array where compound was not delivered) were not amplified. RT-PCR products encoding selected RNAs were purified on a non-denaturing 8% polyacrylamide gel. Purity was assessed using a Bioanalyzer and samples were mixed in equal amounts and sequenced using an Ion Proton deep sequencer using PI chips (60–80 million reads).
In Vitro Dicer Processing Assay:
The miR-21 precursor (pre-miR-21) was 5’-end labeled with [γ−32P] ATP and T4 polynucleotide kinase as previously described (Velagapudi, et al., 2014). The RNA was then folded in 1× Reaction Buffer (Genlantis) by heating at 60 °C fo r 5 min and slowly cooling to room temperature, where it was then supplemented with 1 mM ATP and 2.5 mM MgCl2. Compound was added to the reaction mixture and the samples were allowed to incubate at room temperature for 15 min. Recombinant human Dicer enzyme (Genlantis) was added to a final concentration of 0.01 U/μL and the samples were incubated for an additional 30 min at 37 °C. Reactions were stopped by adding in 2× Gel Loading Buffer (8 M urea, 50 mM EDTA, 0.05% (w/v) bromophenol blue, 0.05% (w/v) xylene cyanol). To generate sequencing markers, pre-miR-21 was digested with RNase T1 (0.125 U/μL) in T1 Buffer (25 mM sodium citrate, pH 5, 7 M urea, and 1 mM EDTA) for 20 min at room temperature. An RNA hydrolysis ladder was prepared by incubating RNA in 1× RNA Hydrolysis Buffer (50 mM NaHCO3, 1 mM EDTA, pH 9.4) at 95 °C for 5 min. Cleavage product s were resolved on a denaturing 15% polyacrylamide gel, which was imaged using a Molecular Dynamics Typhoon phosphorimager and quantified with Bio-Rad’s QuantityOne software.
RNA Isolation and RT-qPCR:
MDA-MB-231 cells were cultured as described above and grown to 80% confluency in 24-well plates. Cells were treated with compound of interest for 24–48 h. Total RNA was then extracted with a Quick-RNA MiniPrep Kit (Zymo Research) per the manufacturer’s protocol. Approximately 200 ng of total RNA was used for reverse transcription reactions (RT), which were completed by using a miScript II RT Kit (Qiagen) per the manufacturer’s protocol. RT-qPCR was completed using a 7900HT Fast Real Time PCR System (Applied Biosystems), using Power SYBR Green Master Mix (Applied Biosystems). Primers for RT-qPCR were purchased from IDT or Eurofins (see Table S2) and used without further purification. Expression levels of RNAs were normalized to U6 small nuclear RNA or 18S rRNA.
PTEN Luciferase Assay:
MDA-MB-231 cells were grown in 48-well plates to ~60% confluency in complete growth medium. The cells were transiently co-transfected with 200 ng of a plasmid encoding the 3’ untranslated region (UTR) of PTEN fused to luciferase (O’Donnell, et al., 2005) and 40 ng of a plasmid encoding Renilla luciferase using Lipofectamine 2000 per the manufacturer’s protocol. At 5 h post-transfection, compounds were added in complete growth medium, and the cells were incubated for 48 h. Luciferase assays were completed based on a previously described protocol (Hampf and Gossen, 2006). Cells were lysed in 40 μL 1× Lysis Buffer (100 mM potassium phosphate buffer (pH 7.8), 0.2% Tween 20) for 10 min at room temperature. Next, 150 μL of 1× Firefly Luciferase Buffer (200 mM Tris–HCl (pH 8), 15 mM magnesium sulfate, 0.1 mM EDTA, 25 mM dithiothreitol, 1mM ATP, 200 μM coenzyme A, and 200 μM luciferin) and incubated for 2 min at room temperature. Luminescence signal was measured on a Biotek Flx800 plate reader. To measure Renilla luciferase activity, 150 μL of 1× Renilla Luciferase Buffer (25 mM sodium pyrophosphate, 10 mM sodium acetate, 15 mM EDTA, 500 mM sodium sulfate, 500 mM sodium chloride, 50 μM of 4-(6-Methyl-2-benzothiazolyl)benzeneamine (CAS# 92–36-4), and 4 μM benzyl-coelenterazine, pH 5.0) was added and incubated for 2 min at room temperature. Luminescence signal was measured on a Biotek Flx800 plate reader.
Imaging for DNA Damage:
The γ-H2AX immunofluorescence assay was used to assess DNA double strand breaks in cells.(Pilch, et al., 2003; Rogakou, et al., 1999) MDA-MB-231 cells were grown to ~80% confluence in a Mat-Tek 96-well glass bottom plate in growth medium. Cells were treated with 3 in complete growth medium for 24 hours. Cells were washed with 1x DPBS then fixed with 4% paraformaldehyde in 1× DPBS at 37 °C and 5% CO 2 for 10 minutes. Cells were washed 5 times with 1× DPBS and three times with 0.1% Triton X-100 in 1× DPBS for 5 min at 37 °C and 5% CO 2. Cells were then washed with 30% formamide in 2× SSC buffer for 10 min at room temperature and 2× SSC for 30 min at 37 °C and 5% CO 2. Cells were incubated with a 1:500 dilution of anti-phospho-Histone-H2A.X (Ser139) (clone JBW301, EMD Millipore) in 2× SSC for 1 h at 37 °C and 5% CO 2 then washed three times with 0.1% Triton X-100 in 1x DPBS for 5 min at 37 °C and 5% CO 2. Cells were the incubated with a 1:200 dilution of goat anti-mouse IgG-DyLight 488 conjugate (Thermo Fisher) in 2× SSC for 1 h at 37 °C and 5% CO2 then washed three times with 0.1% Triton X-100 in 1× DPBS for 5 min at 37 °C and 5% CO2. Cells were washed three times with 1× DPBS and then stained using a 1 μg/μL solution of DAPI in 1× DPBS for 5 min at 37 °C and 5% CO 2. Cells were washed five times in 1x DPBS and then imaged in 1× DPBS using an Olympus FluoView 1000 confocal microscope at 40× magnification. Anti-γ-H2AX fluorescence intensity was quantified using Olympus Fluoview software version 3.0. Average Anti-γ-H2AX fluorescence intensity measurements represent the average fluorescence intensity across eight images.
Boyden Chamber Invasion Assay:
Matrigel (Corning) was thawed overnight at 4 °C. M atrigel was diluted to 3 mg/mL of basement protein and then 100 μL was plated out into 24 well tissue culture inserts with 8 μm pore sizes (Greiner). Matrigel was incubated at 37 °C for 15 min and then placed back into 4 °C overnight. The followin g day, cells were cultured as described above and allowed to migrate towards complete growth media in the bottom well for 16 h. The media was vacuum aspirated and both hanging cell culture insert and bottom wells were washed twice with 1× PBS, gently shaking to mix. Excess liquid and cells inside the insert were removed with cotton swabs, after which 400 μL of 4% paraformaldehyde in 1× PBS was placed into the bottom well and incubated for 20 min at room temperature. The wells and inserts were washed twice with 1× PBS and then treated with 400 μL of 0.1% crystal violet solution in 1× PBS for 20 min at room temperature. Then wells and inserts were washed twice with water. Then wells and inserts were washed once with 1× PBS and dried with cotton swabs to remove extra stain and cells inside the insert. The inserts were air-dried and then analyzed by microscopy using a Leica DMI3000 B upright fluorescent microscope. Four different fields of view from each captured image were counted for crystal violet stained or unstained cells. Alternatively, MDA-MB-231 cells were transfected with a plasmid containing pre-miR-21 (Addgene) using Lipofectamine 2000 according to the manufacturer’s protocol and the experiment was performed as described above.
In vitro Chem-CLIP and C-Chem-CLIP:
Approximately 50000 counts of 32P 5’-end labeled precursor miR-21 was added to inactivated DMEM growth medium and folded at 95 °C for 45 s, then cooled on ice for 5 min. Compound 10 or 11 were added to folded RNA and incubated at 37 °C overnight. For C-Chem-CLIP, nucleic acids we re pre-treated with dilutions of 3 for 15 min at room temperature before Chem-CLIP probes were added. Streptavidin-agarose beads (Sigma-Aldrich, S1638; ≥15 μg/mL binding capacity) were washed three times with 1× PBS and resuspended in 1× PBS. Beads were added to the samples and incubated for 1 h at room temperature. Samples were centrifuged and the supernatant was transferred to a new tube. Beads were washed three times with 1× PBS with 0.1% (v/v) Tween-20 and centrifuged, with each wash being added to the supernatant tube. Radioactivity in the beads and the supernatant/wash tubes were quantified using a Beckman Coulter LS6500 Liquid Scintillation Counter.
Chem-CLIP and C-Chem-CLIP:
The MDA-MB-231 cells were grown to ~70% confluency as monolayers in 100 mm dishes. The cells were treated with 1 μM of 10 or 11 for 8 h. Total RNA was extracted using a Quick-RNA MiniPrep Kit (Zymo Research) per the manufacturer’s protocol. Approximately 30 μg of total RNA was then incubated with 100 μL of streptavidin-agarose beads (Sigma-Aldrich) and shaken for 1 h at room temperature. The solution was removed and beads washed six times with 300 μL of 1× PBS. The RNA bound to beads was released by heating at 65 °C for 20 min in 1× Elution Buffer (95% formamide, 10 mM EDTA, pH 8.2). Eluted RNA was then purified with a Quick-RNA MiniPrep Kit (Zymo Research) and used for subsequent RNA isolation and RT-qPCR as described above (RNA isolation and RT-qPCR). Normalized miRNA Enrichment of the measured RNA before and after pulldown was measured using equation 1:
| (1) |
where “Δ Ct before pulldown” is the difference between the Ct values for the RNA of interest and a housekeeping gene (U6 small nuclear RNA) in total RNA isolated from before pulldown cell lysate RNA and “Δ Ct after pulldown” is the difference between the Ct values for the RNA of interest and the same housekeeping gene after pulldown. Data was normalized to the levels of mature miR-21 measured after treatment with Chem-CLIP probe at the appropriate concentration.
In Vitro Topoisomerase Inhibition Assay:
Topoisomerase II inhibitory activity was measured using a Topoisomerase II Drug Screening Kit (TopoGEN, Inc.) per the manufacturer’s protocol. Dilutions of 1, 2, and 3 in water were added to 300 ng of DNA in 1× Complete Buffer (50 mM Tris-HCl pH 8, 150 mM NaCl, 10 mM MgCl2, 0.5 mM dithiothreitol, 30 μg/mL BSA, and 2 mM ATP), followed by addition of 7.5 U of Topoisomerase II enzyme. Samples were incubated at 37 °C for 30 min and stopped with 2 μL of 10% sodiu m dodecyl sulfate (SDS). Proteinase K (50 μg/mL) was added and incubated at 37 °C for 15 min. Topoisomers were separated on 1% agarose gels with or without 0.5 μg/mL ethidium bromide. Gels prepared with ethidium bromide were run in 1× TAE running buffer supplemented with 0.5 μg/mL ethidium bromide. Gels run without ethidium bromide were post stained with 0.5 μg/mL ethidium bromide. Both gels were destained in 1× TAE for 15 min and DNA products were visualized using a Bio-Rad Gel Doc XR+ imaging system (Figure S4).
Fluorescent Binding Affinity Measurements:
Dissociation constants were determined using an in-solution fluorescence-based assay. The RNA of interest was folded in 1× Assay Buffer (8 mM Na2HPO4, pH 7.0, 190 mM NaCl, 1 mM EDTA and 40 μg/mL BSA) by heating at 60 °C for 5 min and slowly cooling to room temperature. Small molecules were added to a final concentration of 250 nM for 3, or 2000 nM for 1 and 2. Serial dilutions (1:2) were then completed in 1× Assay Buffer supplemented with 250 nM of 3, or 2000 nM of 1 and 2. The solutions were incubated for 30 min at room temperature and then transferred to a 96-well plate and fluorescence intensity was measured. The change in fluorescence intensity as a function of RNA concentration was fit to equation 2:
| (2) |
where Bmax, is maximum specific binding; [RNA], is RNA concentration; h, is hill slope.
Microscale Thermophoresis (MST) Binding Measurements:
MST measurements were performed on a Monolith NT.115 system (NanoTemper Technologies) using the intrinsic fluorescent signal from compound 3. The samples were prepared in MST buffer containing 50 mM Tris-HCl, pH 7.4, 150mM NaCl,5 mM MgCl2 and 0.05 (v/v) % Tween-20. The concentration of compound was kept constant at 150 nM. The following RNA constructs were ordered from GE Healthcare Dharmacon for use in MST studies: Pre-miR-21 Full: GUUGACUGUUGAAUCUCAUGGCAAC; Pre-miR-21 A bulge: GUUGACUGUUGAAUCUCAAUGGCAAC; Pre-miR-21 Base Paired: GUUGACUGUUGAAUCUCAAUGGUCAAC. RNA was titrated in 1:1 dilutions beginning at 20 μM and then samples were filled into in premium-coated capillaries. The measurement was performed at 40 % LED and 20 to 80 % MST power, with a Laser-On time of 30 sec and Laser-Off time of 5 sec, detecting fluorescence at Ex: 605–645 nm, Em: 680–685 nm. The data were analyzed by thermophoresis analysis, and fitted by the quadratic binding equation in MST analysis software (NanoTemper Technologies). The dissociation constant was then determined using a single-site model to fit the curve.
Chemical Synthesis:
Abbreviations: Boc, tert-butyloxycarbonyl; oxyma, Ethyl (hydroxyimino)cyanoacetate; DCM, dichloromethane; DMAP, 4-Dimethylaminopyridine; DMF, N,N-dimethylformamide; DIC, N,N’-Diisopropylcarbodiimide; DIEA, N,N-Diisopropylethylamine; MS(ESI), mass spectrometry (electrospray ionization); HATU, 1-[Bis(dimethylamino)methylene-]1H-1,2,3-trizaolo[4,5-b]pyridinium 3-oxide hexafluorophosphate; HOAt, 1-hydroxy-7-azabenzotriazole; HPLC, high performance liquid chromatography; MeOH, methanol; NMR, nuclear magnetic resonance; TEA, Triethylamine; TFA, trifluoroacetic acid; Ts, p-toluenesulfonyl.
Synthesis of N1-(2-(prop-2-yn-1-yloxy)ethyl)ethane-1,2-diamine (3a)
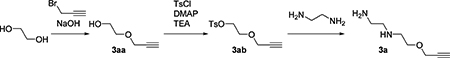
Both 2-(prop-2-yn-1-yloxy)ethan-1-ol (3aa), 2-(prop-2-yn-1-yloxy)ethyl 4-methylbenzenesulfonate (3ab) were prepared according to reference(Cserép, et al., 2014; McConnell, et al., 2010). To a solution of 2-(prop-2-yn-1-yloxy)ethyl 4-methylbenzenesulfonate. (3ab) (5400 mg, 21 mmol) in MeOH (30 mL) ethylenediamine was added (20 mL, 300 mmol) dropwise at 0 °C. After the addition of ethylenedi amine, the reaction mixture was stirred overnight. Then, 10% K2CO3 was added to the reaction mixture and extracted with DCM. The combined organic layers were dried over Na2SO4 and concentrated in vacuo. The residue was purified by distillation (85 °C/ < 1 mmHg) to affor d 1.6 g of 3a (53% yield), a colorless oil. 1H NMR(400 MHz, CDCl3) δ 4.17–4.16 (m, 2H), 3.65 (t, J = 5.1 Hz, 2H), 2.85–2.80 (m, 4H), 2.69 (t, J = 7.8, 2H), 2.44–2.43 (m, 1H) 1.34 (br s, 3H). 13C NMR(100 MHz, CDCl3) δ 41.8, 49.1, 52.5, 58.4, 69.5, 74.4, 79.7. MS m/z (ESI); calculated for C7H15N2O (M + H)+ 143.1; found 143.3
Synthesis of Mitoxantrone Derivative 3b:
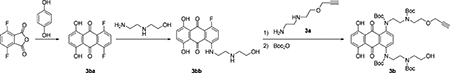
The compounds 1,4-difluoro-5,8-dihydroxyanthracene-9,10-dione (3ba) and 1-fluoro-5,8-dihydroxy-4-((2-((2-hydroxyethyl)amino)ethyl)amino)anthracene-9,10-dione (3bb) were prepared according to Liu et al. and Mansour et al. (Liu, et al., 2009; Mansour, et al., 2010). To a solution of 3bb (50 mg, 0.14 mmol) in pyridine (4 mL) was added 3a (36 mg, 0.25 mmol). Then the mixture was stirred at 100 °C under argon for 2 h. The reaction mixture was cooled to room temperature. Boc2O (283 mg, 1.3 mmol) was added, and the reaction mixture was stirred overnight. The reaction mixture was evaporated and the residue was purified by silica gel column chromatography (15–60% ethyl acetate in hexane) to afford 50 mg of 3b (43% yield) as a dark blue powder. 1H NMR(CD3OD, 700 MHz) δ 7.51–7.49 (m, 1H), 7.46–7.40 (m, 2H), 7.38–7.27 (m, 1H), 4.21–4.19 (m, 2H), 3.68–3.62 (m, 8H), 3.50–3.47 (m, 6H), 3.38–3.36 (m, 2H), 2.88 (s, 1H), 1.58 (s, 18H), 1.48 (s, 9H), 1.40–1.39 (m, 9H). 13C NMR(175 MHz, CD3OD) δ 181.9, 157.3, 157.2, 157.1, 152.8, 149.3, 147.3, 147.2, 147.1, 129.2, 128.7, 125.2, 125.0, 124.9, 124.8, 111.1, 111.0, 84.6, 81.6, 81.5, 81.4, 81.3, 80.7, 76.2, 69.8, 69.6, 61.5, 61.3, 59.2, 52.1, 51.3, 41.8, 41.3, 28.8, 28.7, 28.2. HRMS m/z (ESI): calculated for C45H63N4O14 (M + H)+ 883.4341, found 883.4375.
Synthesis of the Peptoid backbone (3c):
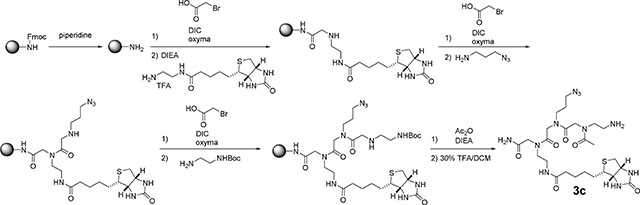
Rink amide resin (400 mg, 0.23 mmol) was swollen in DMF at room temperature for 10 min and then deprotected with a solution of 20% piperidine in DMF (5 mL, 2 × 20 min). The resin was washed with DMF (3 × 5 mL). To the resin were added 1.1 mL of 1.0 M bromoacetic acid in DMF, DIC (0.3 mL, 1.2 mmol) and oxyma (163 mg, 1.1 mmol) and the mixture was shaken at room temperature for 2 h. The resin was washed with DCM (5 × 6 mL) and DMF (5 × 6 mL). Then to the resin was added a solution of N-(2-aminoethyl) biotinamide (384 mg, 1.0 mmol) in DMF (2 mL) and DIPEA (0.2 mL, 1.2 mmol) and the mixture was shaken at room temperature for 1.5 h. The resin was washed with DCM (5 × 6 mL) and DMF (5 × 6 mL). Then to the resin were added 1.1 mL of 1.0 M bromoacetic acid in DMF, DIC (0.3 mL, 1.2 mmol) and oxyma (163 mg, 1.1 mmol) and the mixture was shaken at room temperature for 2 h. The resin was washed with DCM (5 × 6 mL) and DMF (5 × 6 mL). Then to the resin were added DMF (2 mL) and 3-azidopropan-1-amine (230 mg, 2.3 mmol). The mixture was shaken at room temperature overnight. The mixture was washed with DCM (5 × 6 mL) and DMF (5 × 6 mL). Then to the resin were added 1.1 mL of 1.0 M bromoacetic acid in DMF, DIC (0.3 mL, 1.2 mmol) and oxyma (163 mg, 1.1 mmol) and the mixture was shaken at room temperature for 2 h. The resin was washed with DCM (5 × 6 mL) and DMF (5 × 6 mL). Then to the resin were added DMF (2 mL), N-Boc-ethylenediamine (184 mg, 1.2 mmol) and DIEA (0.2 mL, 1.2 mmol) and the mixture was shaken at room temperature for 1 h. The beads were washed with DCM (5 × 6 mL) and DMF (5 × 6 mL) followed by addition of DMF (4 mL), acetic anhydride (0.12 mL 1.3 mmol) and DIEA (0.4 mL, 2.3 mmol). The mixture was shaken at room temperature for 1 h followed by washing the resin with DCM (5 × 6 mL) and DMF (5 × 6 mL). Then to the resin was added 30% TFA in DCM (2 mL), and the mixture was shaken at room temperature for 30 min. The mixture was filtered, and the filtrate was evaporated. The crude product was purified by preparative HPLC (linear gradient of 0% to 100% CH3OH in H2O with 0.1% (v/v) TFA over 60 min) to afford compound 3c as a colorless oil (8 mg, 4.8% yield). HRMS m/z (ESI): calculated for C25H44N11O6S (M + H)+ 626.3197, found 626.3216.
Synthesis of 3-CA-Biotin (10)
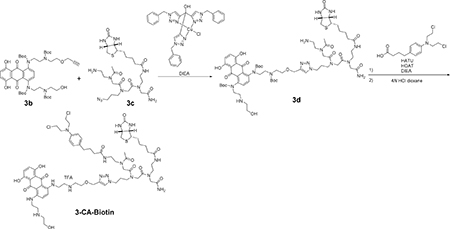
To a solution of 3b (57 mg, 0.065 mmol) and 3c (51 mg, 0.082 mmol) in DMSO (0.6 mL) were added Cu(I)-catalyst (7.5 mg, 0.0048 mmol) and DIEA (0.5 mL, 2.9 mmol). The reaction mixture was stirred at 60 °C overnight. Then saturated NaH CO3 was added, the aqueous layer was extracted with DCM, and the organic layer was evaporated. The residue was purified by silica gel column chromatography (0–5% MeOH with 1% NH3 aq in DCM) to give 70 mg of 3d (partially purified).
To a solution of 3d (24 mg, 0.016 mmol) in DCM were added chlorambucil (42 mg, 0.14 mmol), HOAt (19 mg, 0.14 mmol), HATU (53 mg, 0.14 mmol) and DIEA (40 uL, 0.23 mmol). The reaction mixture was stirred at room temperature overnight and then evaporated. To the residue was added 4N HCl dioxane (3 mL) and the mixture was stirred at room temperature for 2 h followed by evaporation of the suspension. The residue was diluted with water (3 mL) and the aqueous layer was washed with DCM and ethyl acetate. The aqueous layer was directly purified by preparative HPLC with a linear gradient of 0–100% acetonitrile in H2O with 0.1% TFA over 60 min. Fractions containing the compound were concentrated in vacuo and the residue was dissolved in 200 μL of DMSO. The concentration of the DMSO stock solution (1.69 mM, 200 μL, 2.1% yield) was determined with 10 mM Tris-HCl (pH 7.4) and molecular extinction coefficient of mitoxantrone hydrochloride (19200 M−1 cm−1 at 608 nm)(Hajihassan and Rabbani-Chadegani, 2011). Purity was analyzed on an analytical HPLC with a linear gradient of 0–100% acetonitrile in water with 0.1% TFA (see Figure S6). HRMS m/z (ESI): calculated for C64H91Cl2N16O13S (M + H)+ 1393.6049, found 1393.6036.
Quantification and Statistical Analysis
All plots show means with error bars representing S.E.M., unless dictated otherwise. Experiments were completed at least in triplicate. Data were plotted using Graphpad’s Prism 7.
Data and Software Availability
The Inforna software is free for academic use with a software license agreement. Please see: https://disney.florida.scripps.edu/software/.
Key Resources Table
A Key Resources Table is provided and contains detailed information about the sources and identifiers of antibodies, chemicals, oligonucleotides (siRNAs), recombinant DNA, and software and algorithms.
Supplementary Material
Key Resource table:
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-phospho-Histone-H2A.X (Ser139) | EMD Millipore | clone JBW301 |
| Goat anti-Mouse IgG Secondary Antibody, DyLight 488 | Thermo Fisher | Catalog #:35502 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| 1 | Cayman Chemicals | Catalog# 15007 |
| 2 | Biotang, Inc. | Catalog# RS023 |
| 3 | Biotang, Inc. | Catalog# RM11 |
| NIH Clinical Collection (NIH-CC) | Scripps Molecular Screening Center | NA |
| SYNLibrary-95 | Synkinase | Catalog# SYN-2103 |
| pre-mRNA splicing regulators | California Institute Biomedical for Research | N/A |
| RNA-focused compound library | Scripps Molecular Screening Center | N/A |
| Critical Commercial Assays | ||
| Topoisomerase II Drug Screening Kit | TopoGEN, Inc. | Catalog# TG1009 |
| Oligonucleotides | ||
| Primers used for RT-qPCR, see Table S2 | This paper | N/A |
| hsa-miR-21–5p, miRCURY LNA Power microRNA inhibitor | Exiqon (now a part of Qiagen) | Catalog# 4100689101 |
| Negative control A, miRCURY LNA Power microRNA inhibitor control | Exiqon (now a part of Qiagen) | Catalog# 199006–102 |
| Recombinant DNA | ||
| pGL3-PTEN-3'UTR | Addgene | Plasmid # 21326 |
| pcDNA3-miR21 | Addgene | Plasmid # 21114 |
| pRL Renilla Luciferase Control | Promega | Plasmid # E223A |
| Software and Algorithms | ||
| Olympus Fluoview software version 3.0 | Olympus | N/A |
| QuantityOne | BioRad | N/A |
| Graphpad Prism 7 | GraphPad Prism Software, Inc. | N/A |
| Inforna | Disney Laboratory | https://disney.florida.scripps.edu/software/ |
Highlights.
Broad classes of small molecule medicines bind RNA
Sequence-based design shows that approved medicines target oncogenic microRNA-21
The medicines target microRNA-21 in cells, reversing phenotype in cancer cells
Oncogenic non-coding RNAs could be an established drug target all along
SIGNIFICANCE.
Herein, we have developed a microarray-based method dubbed AbsorbArray to identify the preferred RNA motifs for unmodified small molecules. That is, the compounds do not require installation of a functional group for immobilization. AbsorbArray was applied to the NIH Clinical Collection, a library of RNA-focused small molecules, RNA splicing modulators, and a library of kinase inhibitors. Indeed, these drugs, in particular topoisomerase inhibitors, kinase inhibitors, and RNA splicing modulators bind to RNAs. 2DCS and HiT-STARTS identified privileged motifs for each small molecule, and overlap was identified with oncogenic miR-21. One topoisomerase inhibitor, 3, selectively inhibited miR-21 biogenesis, de-repressed a downstream protein, and reversed an invasive phenotype in MDA-MB-231 cells. These studies suggest that many known drugs have affinity for RNA and RNA may indeed be one of their cellular targets. Further, they lay the foundation for the rational re-purposing of known drugs.
ACKNOWLEDGMENTS
This work was funded by the Scheller Graduate Student Fellowship to M.G.C., a Swiss National Science Foundation Early Postdoc Mobility Fellowship to B.R.V., and the National Institutes of Health (5R01GM097455) to M.D.D.
Footnotes
DECLARATION OF INTERESTS
A patent application has been submitted on the Inforna software: Disney & Velagapudi, U.S. Patent Application 14/911,032.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Binaschi M, Zunino F, and Capranico G (1995). Mechanism of action of DNA topoisomerase inhibitors. Stem Cells. 13, 369–379. [DOI] [PubMed] [Google Scholar]
- Blount KF, and Breaker RR (2006). Riboswitches as antibacterial drug targets. Nat. Biotechnol. 24, 1558–1564. [DOI] [PubMed] [Google Scholar]
- Bose D, Jayaraj G, Suryawanshi H, Agarwala P, Pore SK, Banerjee R, and Maiti S (2012). The tuberculosis drug streptomycin as a potential cancer therapeutic: inhibition of miR-21 function by directly targeting its precursor. Angew. Chem. Int. Ed 51, 1019–1023. [DOI] [PubMed] [Google Scholar]
- Calin GA, and Croce CM (2006). MicroRNA-cancer connection: the beginning of a new tale. Cancer Res. 66, 7390–7394. [DOI] [PubMed] [Google Scholar]
- Carter AP, Clemons WM, Brodersen DE, Morgan-Warren RJ, Wimberly BT, and Ramakrishnan V (2000). Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature 407, 340–348. [DOI] [PubMed] [Google Scholar]
- Chan JA, Krichevsky AM, and Kosik KS (2005). MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 65, 6029–6033. [DOI] [PubMed] [Google Scholar]
- Childs-Disney JL, Wu M, Pushechnikov A, Aminova O, and Disney MD (2007). A small molecule microarray platform to select RNA internal loop-ligand interactions. ACS Chem. Biol 2, 745–754. [DOI] [PubMed] [Google Scholar]
- Childs-Disney JL, Yildirim I, Park H, Lohman JR, Guan L, Tran T, Sarkar P, Schatz GC, and Disney MD (2014). Structure of the myotonic dystrophy type 2 RNA and designed small molecules that reduce toxicity. ACS Chem. Biol 9, 538–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirayil S, Wu Q, Amezcua C, Luebke KJ (2014). NMR Characterization of an Oligonucleotide Model of the MIR-21 Pre-ELement. PLOS ONE 9, e108231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly CM, Boer RE, Moon MH, Gareiss P, and Schneekloth JS Jr. (2017). Discovery of Inhibitors of MicroRNA-21 Processing Using Small Molecule Microarrays. ACS Chem. Biol 12, 435–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper TA, Wan L, and Dreyfuss G (2009). RNA and disease. Cell 136, 777–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cserép GB, Baranyai Z, Komáromy D, Horváti K, Bősze S, and Kele P (2014). Fluorogenic tagging of peptides via Cys residues using thiol-specific vinyl sulfone affinity tags. Tetrahedron 70, 5961–5965. [Google Scholar]
- Dibrov SM, Parsons J, Carnevali M, Zhou S, Rynearson KD, Ding K, Garcia Sega E, Brunn ND, Boerneke MA, Castaldi MP, et al. (2014). Hepatitis C virus translation inhibitors targeting the internal ribosomal entry site. J. Med. Chem 57, 1694–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Disney MD, Labuda LP, Paul DJ, Poplawski SG, Pushechnikov A, Tran T, Velagapudi SP, Wu M, and Childs-Disney JL (2008). Two-dimensional combinatorial screening identifies specific aminoglycoside-RNA internal loop partners. J. Am. Chem. Soc 130, 11185–11194. [DOI] [PubMed] [Google Scholar]
- du Rieu MC, Torrisani J, Selves J, Al Saati T, Souque A, Dufresne M, Tsongalis GJ, Suriawinata AA, Carrere N, Buscail L, et al. (2010). MicroRNA-21 is induced early in pancreatic ductal adenocarcinoma precursor lesions. Clin. Chem 56, 603–612. [DOI] [PubMed] [Google Scholar]
- Feofanov A, Sharonov S, Kudelina I, Fleury F, and Nabiev I (1997). Localization and molecular interactions of mitoxantrone within living K562 cells as probed by confocal spectral imaging analysis. Biophys. J 73, 3317–3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazier KS (2015). Antisense oligonucleotide therapies: the promise and the challenges from a toxicologic pathologist’s perspective. Toxicol. Pathol 43, 78–89. [DOI] [PubMed] [Google Scholar]
- Gareiss PC, Sobczak K, McNaughton BR, Palde PB, Thornton CA, and Miller BL (2008). Dynamic combinatorial selection of molecules capable of inhibiting the (CUG) repeat RNA-MBNL1 interaction in vitro: Discovery of lead compounds targeting myotonic dystrophy (DM1). J. Am. Chem. Soc 130, 16254–16261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan L, and Disney MD (2013). Covalent small molecule-RNA complex formation enables cellular profiling of small molecule-RNA interactions. Angew. Chem. Int. Ed 52, 10010–10013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumireddy K, Young DD, Xiong X, Hogenesch JB, Huang Q, and Deiters A (2008). Small-molecule inhibitors of microrna miR-21 function. Angew. Chem. Int. Ed 47, 7482–7484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajihassan Z, and Rabbani-Chadegani A (2011). The effect of mitoxantrone as an anticancer drug on hepatocytes nuclei and chromatin: Selective release of histone proteins. Indian J.ournal of pharmacology 43, 187–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampf M, and Gossen M (2006). A protocol for combined Photinus and Renilla luciferase quantification compatible with protein assays. Anal. Biochem 356, 94–99. [DOI] [PubMed] [Google Scholar]
- Hergenrother PJ, Depew KM, and Schreiber SL (2000). Small-molecule microarrays: Covalent attachment and screening of alcohol-containing small molecules on glass slides. J. Am. Chem. Soc 122, 7849–7850. [Google Scholar]
- Huang TH, Wu F, Loeb GB, Hsu R, Heidersbach A, Brincat A, Horiuchi D, Lebbink RJ, Mo YY, Goga A, et al. (2009). Up-regulation of miR-21 by HER2/neu signaling promotes cell invasion. J. Biol. Chem 284, 18515–18524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov AI, Christodoulou J, Parkinson JA, Barnham KJ, Tucker A, Woodrow J, and Sadler PJ (1998). Cisplatin Binding Sites on Human Albumin. Journal of Biological Chemistry 273, 14721–14730. [DOI] [PubMed] [Google Scholar]
- Jahromi AH, Nguyen L, Fu Y, Miller KA, Baranger AM, and Zimmerman SC (2013). A novel CUG(exp).MBNL1 inhibitor with therapeutic potential for myotonic dystrophy type 1. ACS Chem. Biol 8, 1037–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Parkesh R, Sznajder LJ, Childs-Disney JL, Sobczak K, and Disney MD (2012). Chemical correction of pre-mRNA splicing defects associated with sequestration of muscleblind-like 1 protein by expanded r(CAG)-containing transcripts. ACS Chem. Biol 7, 496–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee ER, Blount KF, and Breaker RR (2009). Roseoflavin is a natural antibacterial compound that binds to FMN riboswitches and regulates gene expression. RNA Biol. 6, 187–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MK, Bottini A, Kim M, Bardaro MF Jr., Zhang Z, Pellecchia M, Choi BS, and Varani G (2014). A novel small-molecule binds to the influenza A virus RNA promoter and inhibits viral replication. Chem. Commun 50, 368–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang SI, Puc J, Miliaresis C, Rodgers L, McCombie R, et al. (1997). PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science 275, 1943–1947. [DOI] [PubMed] [Google Scholar]
- Liang X. h., Sun H, Shen W, and Crooke ST (2015). Identification and characterization of intracellular proteins that bind oligonucleotides with phosphorothioate linkages. Nucleic Acids Res 43, 2927–2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Childs-Disney JL, Znosko BM, Wang D, Fallahi M, Gallo SM, and Disney MD (2016). Analysis of secondary structural elements in human microRNA hairpin precursors. BMC Bioinf. 17, 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Peacey E, Dickson J, Donahue CP, Zheng S, Varani G, and Wolfe MS (2009). Mitoxantrone analogues as ligands for a stem-loop structure of tau pre-mRNA. J. Med. Chem 52, 6523–6526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lown JW, Hanstock CC, Bradley RD, and Scraba DG (1984). Interactions of the antitumor agents mitoxantrone and bisantrene with deoxyribonucleic acids studied by electron microscopy. Mol. Pharmacol 25, 178–184. [PubMed] [Google Scholar]
- MacBeath G, Koehler AN, and Schreiber SL (1999). Printing small molecules as microarrays and detecting protein- ligand interactions en masse. J. Am. Chem. Soc 121, 79677968. [Google Scholar]
- MacBeath G, and Schreiber SL (2000). Printing proteins as microarrays for high-throughput function determination. Science 289, 1760–1763. [DOI] [PubMed] [Google Scholar]
- Mansour OC, Evison BJ, Sleebs BE, Watson KG, Nudelman A, Rephaeli A, Buck DP, Collins JG, Bilardi RA, Phillips DR, et al. (2010). New anthracenedione derivatives with improved biological activity by virtue of stable drug-DNA adduct formation. J. Med. Chem 53, 6851–6866. [DOI] [PubMed] [Google Scholar]
- Marcheschi RJ, Mouzakis KD, and Butcher SE (2009). Selection and Characterization of Small Molecules That Bind the HIV-1 Frameshift Site RNA. ACS Chem. Biol 4, 844–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell AJ, Serpell CJ, Thompson AL, Allan DR, and Beer PD (2010). Calix[4]arene-based rotaxane host systems for anion recognition. Chemistry. 16, 1256–1264. [DOI] [PubMed] [Google Scholar]
- Mei H-Y, Galan AA, Halim NS, Mack DP, Moreland DW, Sanders KB, Hoa NT, and Czarnik AW (1995). Inhibition of an HIV-1 Tat-derived peptide binding to TAR RNA by aminoglycoside antibiotics. Bioorg. Med. Chem. Lett 5, 2755–2760. [Google Scholar]
- Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, and Patel T (2007). MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology 133, 647–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon MH, Hilimire TA, Sanders AM, and Schneekloth JS Jr. (2018). Measuring RNA-Ligand Interactions with Microscale Thermophoresis. Biochemistry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow T (2017). New Therapy for Spinal Muscular Atrophy Offers Modest Bang for Pharamaceutical Buck. Managed care 26, 36–37. [PubMed] [Google Scholar]
- Naro Y, Thomas M, Stephens MD, Connelly CM, and Deiters A (2015). Aryl amide small-molecule inhibitors of microRNA miR-21 function. Bioorg. Med. Chem. Lett 25, 4793–4796. [DOI] [PubMed] [Google Scholar]
- Naryshkin NA, Weetall M, Dakka A, Narasimhan J, Zhao X, Feng Z, Ling KK, Karp GM, Qi H, Woll MG, et al. (2014). Motor neuron disease. SMN2 splicing modifiers improve motor function and longevity in mice with spinal muscular atrophy. Science 345, 688–693. [DOI] [PubMed] [Google Scholar]
- O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, and Mendell JT (2005). c-Myc-regulated microRNAs modulate E2F1 expression. Nature 435, 839–843. [DOI] [PubMed] [Google Scholar]
- Palacino J, Swalley SE, Song C, Cheung AK, Shu L, Zhang X, Van Hoosear M, Shin Y, Chin DN, Keller CG, et al. (2015). SMN2 splice modulators enhance U1-pre-mRNA association and rescue SMA mice. Nat. Chem. Biol 11, 511–517. [DOI] [PubMed] [Google Scholar]
- Panwar B, Omenn GS, and Guan Y (2017). miRmine: a database of human miRNA expression profiles. Bioinformatics 33, 1554–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilch DR, Sedelnikova OA, Redon C, Celeste A, Nussenzweig A, and Bonner WM (2003). Characteristics of gamma-H2AX foci at DNA double-strand breaks sites. Biochem. Cell Biol 81, 123–129. [DOI] [PubMed] [Google Scholar]
- Poehlsgaard J, and Douthwaite S (2005). The bacterial ribosome as a target for antibiotics. Nat. Rev. Microbiol 3, 870–881. [DOI] [PubMed] [Google Scholar]
- Pushechnikov A, Lee MM, Childs-Disney JL, Sobczak K, French JM, Thornton CA, and Disney MD (2009). Rational design of ligands targeting triplet repeating transcripts that cause RNA dominant disease: application to myotonic muscular dystrophy type 1 and spinocerebellar ataxia type 3. J. Am. Chem. Soc 131, 9767–9779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou EP, Boon C, Redon C, and Bonner WM (1999). Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol 146, 905–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rzuczek SG, Colgan LA, Nakai Y, Cameron MD, Furling D, Yasuda R, and Disney MD (2017). Precise small-molecule recognition of a toxic CUG RNA repeat expansion. Nat. Chem. Biol 13, 188–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rzuczek SG, Southern MR, and Disney MD (2015). Studying a Drug-like, RNA-Focused Small Molecule Library Identifies Compounds That Inhibit RNA Toxicity in Myotonic Dystrophy. ACS Chem. Biol 10, 2706–2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel SAI, Dijkman PM, Lea WA, van den Bogaart G, Jerabek-Willemsen M, Lazic A, Joseph JS, Srinivasan P, Baaske P, Simeonov A, et al. (2013). Microscale thermophoresis quantifies biomolecular interactions under previously challenging conditions. Methods 59, 301–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seike M, Goto A, Okano T, Bowman ED, Schetter AJ, Horikawa I, Mathe EA, Jen J, Yang P, Sugimura H, et al. (2009). MiR-21 is an EGFR-regulated anti-apoptotic factor in lung cancer in never-smokers. Proc. Natl. Acad. Sci. U.S.A 106, 12085–12090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaul P, Frenkel M, Goldstein EB, Mittelman L, Grunwald A, Ebenstein Y, Tsarfaty I, and Fridman M (2013). The Structure of Anthracycline Derivatives Determines Their Subcellular Localization and Cytotoxic Activity. ACS Med. Chem. Lett 4, 323–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shortridge MD, and Varani G (2015). Structure based approaches for targeting non-coding RNAs with small molecules. Curr. Opin. Struct. Biol 30, 79–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shortridge MD, Walker MJ, Pavelitz T, Chen Y, Yang W, and Varani G (2017). A Macrocyclic Peptide Ligand Binds the Oncogenic MicroRNA-21 Precursor and Suppresses Dicer Processing. ACS Chem. Biol 12, 1611–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Z, Zhang Y, Gendron TF, Bauer PO, Chew J, Yang WY, Fostvedt E, Jansen-West K, Belzil VV, Desaro P, et al. (2014). Discovery of a Biomarker and Lead Small Molecules to Target r(GGGGCC)-Associated Defects in c9FTD/ALS. Neuron 84, 239. [DOI] [PubMed] [Google Scholar]
- Swift LP, Rephaeli A, Nudelman A, Phillips DR, and Cutts SM (2006). Doxorubicin-DNA adducts induce a non-topoisomerase II-mediated form of cell death. Cancer Res. 66, 4863–4871. [DOI] [PubMed] [Google Scholar]
- Tenson T, and Mankin A (2006). Antibiotics and the ribosome. Mol. Microbiol 59, 1664–1677. [DOI] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. (2012). An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velagapudi SP, Gallo SM, and Disney MD (2014). Sequence-based design of bioactive small molecules that target precursor microRNAs. Nat. Chem. Biol 10, 291–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velagapudi SP, Luo Y, Tran T, Haniff HS, Nakai Y, Fallahi M, Martinez GJ, Childs-Disney JL, and Disney MD (2017). Defining RNA-small molecule affinity landscapes enables design of a small molecule inhibitor of an oncogenic noncoding RNA. ACS Cent. Sci 3, 205–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et al. (2006). A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. U.S.A 103, 2257–2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan LX, Huang XF, Shao Q, Huang MY, Deng L, Wu QL, Zeng YX, and Shao JY (2008). MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA. 14, 2348–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WY, He F, Strack RL, Oh SY, Frazer M, Jaffrey SR, Todd PK, and Disney MD (2016). Small Molecule Recognition and Tools to Study Modulation of r(CGG)(exp) in Fragile X-Associated Tremor Ataxia Syndrome. ACS Chem. Biol 11, 2456–2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WY, Wilson HD, Velagapudi SP, and Disney MD (2015). Inhibition of Non-ATG Translational Events in Cells via Covalent Small Molecules Targeting RNA. J. Am. Chem. Soc 137, 5336–5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapp ML, Stern S, and Green MR (1993). Small molecules that selectively block RNA binding of HIV-1 Rev protein inhibit Rev function and viral production. Cell 74, 969–978. [DOI] [PubMed] [Google Scholar]
- Zheng S, Chen Y, Donahue CP, Wolfe MS, and Varani G (2009). Structural Basis for Stabilization of the Tau Pre-mRNA Splicing Regulatory Element by Novantrone (Mitoxantrone). Chem. Biol 16, 557–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Wu H, Wu F, Nie D, Sheng S, and Mo YY (2008). MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res. 18, 350–359. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Inforna software is free for academic use with a software license agreement. Please see: https://disney.florida.scripps.edu/software/.