Abstract
Reprogramming of mouse and human somatic cells can be achieved by ectopic expression of transcription factors, but with low efficiencies. We report that DNA methyltransferase and histone deacetylase (HDAC) inhibitors improve reprogramming efficiency. In particular, valproic acid (VPA), an HDAC inhibitor, improves reprogramming efficiency by more than 100 fold, using Oct4-GFP as a reporter. VPA also enables efficient induction of pluripotent stem cells without introduction of the oncogene c-Myc.
Patient specific stem cells may be created by reprogramming somatic cells to a pluripotent state. Recently, pioneering work by Yamanaka and colleagues showed that the forced expression of just four transcription factors, Oct4, Klf4, Sox2 and c-Myc, reprograms mouse embryonic fibroblasts (MEFs) into induced pluripotent stem (iPS) cells that closely resemble embryonic stem (ES) cells1–4. Reprogramming human somatic cells has now been achieved through similar means5–8, suggesting that the mechanism of reprogramming is conserved between human and the mouse. However, reprogramming by viral infection is a slow and inefficient process. In addition, the genetic transformation with exogenous genes, in particular oncogenes such as c-Myc and Klf4, and the use of viral delivery systems handicap this method in terms of human therapeutic applications.
Previous studies have shown histone deacetylase (HDAC) inhibitors and DNA demethylation have a modest effect (2–5 fold) on the efficiency of reprogramming mediated by somatic cell nuclear transfer (SCNT)9–11. We speculated that reprogramming by defined factors may share common mechanisms with SCNT. Using an Oct4-GFP transgenic reporter12, we tested whether small molecules involved in chromatin modification have any effect on reprogramming (Supplementary Fig. 1). Retroviral expression of four transcription factors, Oct4, Sox2, Klf4 and c-Myc, in MEFs hemizygous for the Oct4-GFP transgene (Oct4-GFP/+) induced 0.03% ± 0.02% (mean ± standard deviation) of the cells to become GFP+, starting at 7 days post-infection, and the percentage of GFP+ cells remained at similar levels between 7 and 13 days post-infection (Supplementary Fig. 2). Treating 4-factor infected MEFs with 2 μM 5’-azacytidine (5’-azaC), a DNA methyltransferase inhibitor, increased the percentage of GFP+ cells by ~10 fold to 0.50% ± 0.06% (mean ± standard deviation) (Fig. 1a, b). 5’-azaC promoted reprogramming efficiency in a dose-dependant manner, with an EC50 of ~2.4 μM (Supplementary Fig. 3a). Dexamethasone (1 μM), a synthetic glucocorticoid, improved the effect of 5’-azaC by 2.6 fold when used in combination, although dexamethasone alone had no significant effect. Three known HDAC inhibitors, suberoylanilide hydroxamic acid (SAHA), trichostatin A (TSA) and valproic acid (VPA) also greatly improved reprogramming efficiency. VPA was the most potent of the three. Treating 4-factor infected MEFs with 2 mM VPA for a week induced approximately 11.8% ± 2.2% GFP+ cells which amounts to a more than 100 fold improvement over the control (Fig. 1a, b). This reprogramming efficiency approached the estimated 13–41% viral co-transduction rate, arguing that most if not all cells infected with all four factors can be reprogrammed. VPA promoted reprogramming efficiency in a dose-dependant manner, with an EC50 of ~1.9 mM (Supplementary Fig. 3b). The effect of VPA is much stronger than 5’-azaC and other HDAC inhibitors tested. This could be due to toxicity of other chemicals at higher dosages. Alternatively, VPA may have additional activities, beyond inhibition of HDACs.
Figure 1. Chemicals that promote reprogramming efficiency.
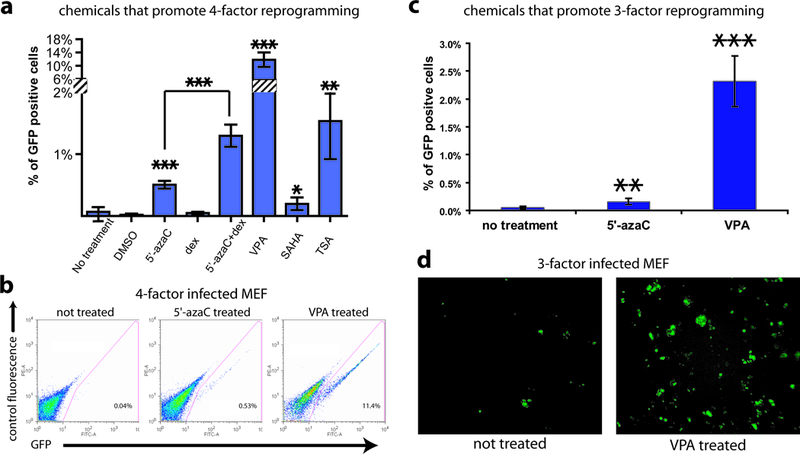
(a) The percentages of GFP+ cells induced in 4-factor (Oct4, Sox2, Klf4 and c-Myc) infected Oct4-GFP/+ MEFs treated with chemicals. Chemical treatments were: 5’-azaC (2 μM), dexamethasone (dex, 1 μM), 5’-azaC and dexamethasone, VPA (2 mM), SAHA (5 μM) and TSA (20 nM). The controls were infected MEFs without chemical treatment or treated with DMSO (the solvent for dexamethasone, SAHA and TSA). The y axis is truncated to accommodate the high percentage from the VPA treatment. For all figures in this study, standard deviations are indicated by error bars, and P values by two-tailed student t-test smaller than 0.05, 0.01 and 0.001 are indicated by one, two and three asterisks respectively. (b) Representative FACS plots from 4-factor infected MEFs treated with 5’-azaC and VPA compared to the control infected MEFs without treatment. (c) MEFs infected with the 3 factors (Oct4, Sox2, Klf4, but not c-Myc) were treated with 5’-azaC or VPA for a week and the percentage of Oct4-GFP+ cells induced was measured by FACS analysis at 10 days post-infection, and compared to 3-factor infected MEFs without chemical treatment. (d) Representative pictures at 16 days post-infection in 3-factor infected MEFs with VPA treatment compared to the control infected MEFs without VPA treatment.
Chemical treatment induced GFP+ iPS colonies in greater numbers, consistent with the FACS data. 8 days post-infection, an average of 10 and 241 colonies were observed in 5’-azaC and VPA treated MEF culture (out of 270,000 cells seeded) respectively. No GFP+ colonies were observed 8 days post-infection without chemical treatment, though some do emerge after 10 days post-infection in untreated cells. The dramatic difference in colony numbers was maintained as more GFP+ iPS colonies emerged in both the chemical treated and non-treated MEF cultures during the following days; more than 8 and 40 fold difference in colony number was observed of 5’-azaC and VPA treatment at two weeks post-infection, respectively.
In addition to improving reprogramming efficiency on 4-factor infected MEFs, chemical treatment allowed efficient induction of iPS cells without the oncogene c-Myc, the use of which could cause tumorigenicity in cells derived from the iPS cells2. Although reprogramming is possible with three factors (Oct4, Sox2 and Klf4) without c-Myc, the efficiency is extremely low and the appearance of iPS colonies significantly delayed compared to reprogramming with 4 factors. Nakagawa et al. found that fewer than 1 iPS colony was formed from 100,000 human dermal fibroblasts infected (<0.001%)13, an efficiency that can make it difficult to derive patient-specific iPS cells from a small starting population of cells. Similar low efficiency was also reported for induction of iPS cells from mouse fibroblasts without c-Myc14. We tested whether treating the cells with 5’-azaC or VPA could improve the efficiency of iPS colony formation without the need for c-Myc. Oct4-GFP/+ MEFs were first infected with Oct4, Sox2 and Klf4, then treated with 5’-azaC or VPA for a week starting 1 day post-infection. FACS analysis 10 days post-infection showed that treatment with 5’-azaC increased reprogramming efficiency by 3 fold, a small improvement (Fig. 1c). Treatment with VPA improved reprogramming efficiency by 50 fold (Fig. 1c). Notably, this reprogramming efficiency is superior to that achieved when MEFs were infected with all four factors, without VPA treatment. Consistent with the FACS data, a 30–40 fold increase of GFP+ colonies was observed compared to control infected MEFs without treatment (Fig. 1d). This allowed for picking of iPS colonies within two weeks post-infection, sooner than the typical ~30 days post-infection or later without chemical treatment13, 14.
To examine whether VPA treatment changes the type of iPS cells generated, we established multiple iPS cell lines from 3-factor infected MEFs. These iPS cells closely resemble mouse ES cells. They have typical ES cell morphology, stain for alkaline phosphatase, and express pluripotent marker genes (Fig. 2a, b, Supplementary Fig. 4). They were readily cultured without further chemical treatment, and passaged more than 10 times, while maintaining ES cell morphology. Global gene expression profiling of iPS cells, MEFs and mouse ES cells shows that iPS cells induced with VPA treatment are distinct from MEFs, and most similar to mouse ES cells (Fig. 2c). The r2 value (square of linear correlation coefficient) between iPS cells and mouse ES cells is 0.94–0.97 (Supplementary Table 1), comparable to previous reports3. In contrast, the r2 value between iPS cells (or mouse ES cells) and MEFs is only 0.62–0.66. Like ES cells, iPS cells induced by the 3 factors with VPA treatment develop teratomas in three to five weeks, and differentiate into tissues representing all three germ layers (Fig. 2d).
Figure 2. c-Myc-free iPS cells induced by VPA treatment resemble ES cells in gene expression and pluripotency.
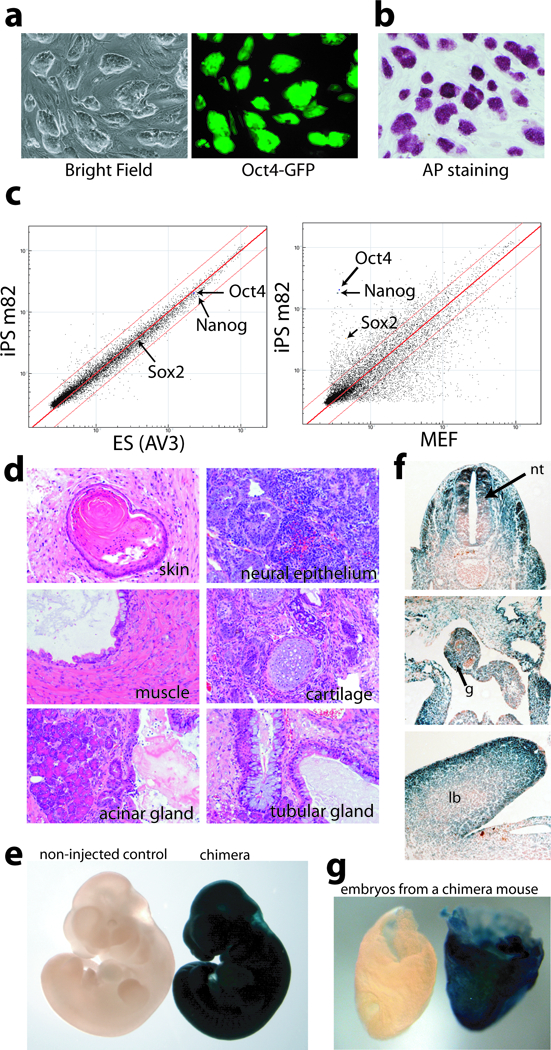
(a) iPS colonies exhibited typical ES cell morphology and expressed Oct4-GFP homogeneously. (b) iPS colonies exhibited high alkaline phosphatase activities. (c) Scatter plots comparing global gene expression patterns between iPS cells and ES cells, and iPS cells and MEFs. Red lines indicate the linear equivalent and two fold changes in gene expression levels between the samples. (d) Hematoxylin and eosin staining of teratoma sections showed differentiation of iPS cells to various tissues. (e) lacZ staining of a midgestation chimeric embryo from donor iPS cells carrying the Rosa26-lacZ allele, compared to the non-injected control. (f) Sections of chimeric embryos showed contribution of donor iPS cells to tissues derived from all three germ layers, including the neural tube (nt, ectoderm derivative), gut endoderm (g) and limb bud (lb, mesoderm derivative). (g) Shown here is a lacZ positive e8.5 embryo with a littermate control on the left from a mating between a wild type female and a chimera from blastocyst injection of iPS cells. Both embryos have yolk sacs attached, and are oriented with the anterior to the left.
To further evaluate the pluripotency of the iPS cells induced by VPA treatment, MEFs were derived from mouse embryos carrying both the Oct4-GFP transgenic allele and the Rosa26-lacZ knock-in allele. iPS cell lines were derived from these MEFs infected with Oct4, Sox2 and Klf4. β-galactosidase staining showed that high-contribution chimeras were obtained from all four iPS cell lines tested, with extensive contribution of the iPS cell derivatives to all three germ layers (Fig. 2e, f). A number of chimeras developed into healthy adults. Germline transmission was confirmed by positive staining of β-galactosidase activity in embryos from matings between chimeric males and wild type females (Fig. 2g). Therefore, the iPS cells induced with VPA treatment fulfill stringent criteria for pluripotency and closely resemble ES cells in all aspects examined.
The dramatic effect of VPA suggests that it may control a rate-limiting step in reprogramming. VPA treatment of uninfected MEFs does not induce Oct4- GFP+ cells, indicating that VPA treatment alone is insufficient to reprogram MEFs. Nor does VPA treatment cause genetic changes when examined at the level of chromosomal abnormalities (Supplementary Table 2). Microarray analysis on uninfected MEFs treated with VPA for a week showed that VPA treatment in uninfected MEFs induced a transcriptional program that can be described as leaning toward an ES-like pattern. Among 968 genes (out of 18,918 total genes) that are up-regulated by more than ten fold in ES cells, compared to untreated MEFs, 66% are up-regulated by more than two fold in VPA treated MEFs. Only 4.5% of these genes are down-regulated by more than two fold (Fig. 3). For example, Rex3 and Zfp7, two genes expressed in undifferentiated ES cells, but not in untreated MEFs, are up-regulated by more than 20 fold in MEFs treated with VPA (Supplementary Fig. 5). Likewise, among the 214 genes down-regulated by more than 10 fold in ES cells, compared to untreated MEFs, 55% are down-regulated by more than two fold in VPA treated MEFs, whereas only 6.2% were up-regulated by more than two fold (Fig. 3). For example, Aspn and Meox2, two genes expressed in MEFs but not in ES cells, were both down-regulated by more than 20 fold in VPA treated MEFs (Supplementary Fig. 5). Therefore, the effect of VPA on reprogramming may be due to the collective effects of up-regulation of ES-specific genes and down-regulation of MEF-specific genes.
Figure 3. The effect of VPA treatment on uninfected MEFs.
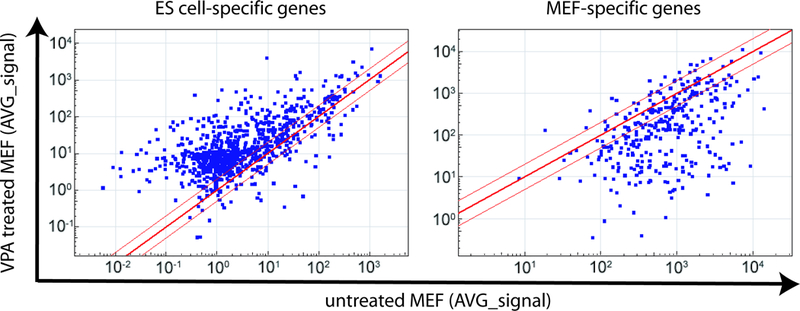
Microarray data were obtained from ES cells, iPS cells, untreated MEFs and MEFs treated with VPA. Genes that were specifically expressed in ES cells and MEFs (>10 fold difference) were selected, and scatter plots were generated to visualize the effect of VPA treatment on the expression of these genes. Red lines indicate the linear equivalent and two fold changes in gene expression levels.
Our findings provide proof of principle that chemicals can increase reprogramming efficiency and may be used to replace one or more factors used for reprogramming. The demonstration that both DNA methyltransferase and HDAC inhibitors improve reprogramming efficiency suggests that chromatin modification is a key step in reprogramming fibroblasts to pluripotent cells. Given that the reprogramming factors are conserved between human and the mouse1–5, 7, 8, these findings will likely apply to human cells. This encourages one to explore high-throughput screening of small molecule libraries to achieve reprogramming through pure chemical means, making therapeutic use of reprogrammed cells safer and more practical.
Supplementary Material
Acknowledgement
D.A.M. is a Howard Hughes Medical Institute Investigator. D.H. is funded by the Helen Hay Whitney Foundation and Novartis Institutes for BioMedical Research. W.G. is funded by the Susan G. Komen Breast Cancer Foundation. A.E.C. is supported by the Jane Coffin Childs Memorial Fund for Medical Research and Merck Research Laboratories. The authors would like to thank Julia Lamenzo and Anastasie Kweudjeu for assistance with micoarray analysis, Shuibing Chen for insightful discussions and critical review of the manuscript. We would also like to thank Dr. Robert Weinberg for support of this study. Some monoclonal antibodies were obtained from the Developmental Studies Hybridoma Bank, which was developed under the auspices of the National Institute of Child Health and Human Development and is maintained by The University of Iowa, Department of Biological Sciences.
Reference:
- 1.Takahashi K & Yamanaka S Cell 126, 663–676 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Okita K, Ichisaka T & Yamanaka S Nature 448, 313–317 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Maherali N et al. Cell Stem Cell 1, 55–70 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Wernig M et al. Nature 448, 318–324 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Takahashi K et al. Cell 131, 861–872 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Yu J et al. Science 318, 1917–1920 (2007). [DOI] [PubMed] [Google Scholar]
- 7.Park IH et al. Nature 451, 141–146 (2008). [DOI] [PubMed] [Google Scholar]
- 8.Lowry WE et al. Proc Natl Acad Sci U S A 105, 2883–2888 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kishigami S et al. Biochem Biophys Res Commun 340, 183–189 (2006). [DOI] [PubMed] [Google Scholar]
- 10.Rybouchkin A, Kato Y & Tsunoda Y Biol Reprod 74, 1083–1089 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Blelloch R et al. Stem Cells 24, 2007–2013 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Szabo PE, Hubner K, Scholer H & Mann JR Mech. Dev. 115, 157–160 (2002). [DOI] [PubMed] [Google Scholar]
- 13.Nakagawa M et al. Nat Biotechnol 26, 101–106 (2008). [DOI] [PubMed] [Google Scholar]
- 14.Wernig M, Meissner A, Cassady JP & Jaenisch R Cell Stem Cell 2, 10–12 (2008). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


