Abstract
Enhancers function as DNA logic gates and may control specialized functions of billions of neurons. Here we show a tailored program of noncoding genome elements active in situ in physiologically distinct dopamine neurons of the human brain. We found 71,022 transcribed noncoding elements, many of which were consistent with active enhancers and with regulatory mechanisms in zebrafish and mouse brains. Genetic variants associated with schizophrenia, addiction, and Parkinson’s dis- ease were enriched in these elements. Expression quantitative trait locus analysis revealed that Parkinson’s disease-associated variants on chromosome 17q21 cis-regulate the expression of an enhancer RNA in dopamine neurons. This study shows that enhancers in dopamine neurons link genetic variation to neuropsychiatric traits.
To date the majority of disease- and trait-associated variants emerging from genome-wide association studies (GWAS) of neurologic and psychiatric diseases lie within non-protein coding sequence. Several lines of evidence suggest the involvement of a proportion of such variants in transcriptional regulatory mechanisms, including modulation of enhancer elements1. Many regulatory elements are cell-type preferential2,3 and therefore sequence variants with functional consequences are expected to manifest their effects more strongly in cell type most relevant to a specific disease phenotype.
Here we focused on systematically identifying all noncoding regulatory elements transcriptionally active in a morphologically, functionally, and biochemically distinct neuronal archetype --- dopamine neurons of the substantia nigra pars compacta in human midbrain. We hypothesized that genetic variation associated with diseases involving dopaminergic neurotransmission exert their effects through modulation of enhancers functionally active in this particular type of neurons. Perturbations of the dopaminergic system are important in the pathogenesis and treatment responses of many increasingly prevalent complex genetic diseases, including 0.5 million people with Parkinson’s disease (PD)4, 2.2 million with schizophrenia5, and 23.5 million people with addiction6 in the United States alone. In healthy people, these dopaminergic neurons shape how we conduct our everyday lives, encoding activities related to motivation and reward. Signals from these neurons to the striatum have a profound impact on action learning and automatic movements, while projections to hippocampus and prefrontal cortex influence memories and behavior7.
Our analysis is powered by an integrated hardware-software solution for comprehensively detecting noncoding transcription in one single and minuscule RNA sample, and mapping the variation in noncoding transcription to genetic variation within dopamine neurons across multiple individuals. This method combines the base-pair resolution and a comprehensive genome-wide view afforded by ultra-deep, total RNA sequencing, with the positional and cytoarchitectural precision afforded by traditional light microscopy.
Results
Identification of noncoding elements actively transcribed in dopamine neurons of human brain
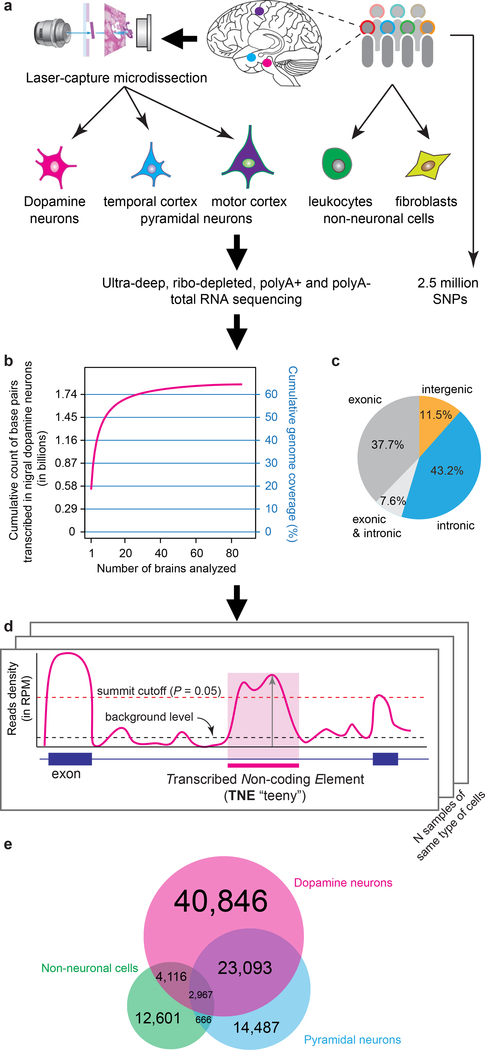
To systematically identify noncoding elements actively transcribed in dopamine neurons of human brain we used laser-capture microdissection total RNA sequencing (lcRNAseq). Beyond traditional messenger (mRNA) sequencing, all polyadenylated and non-polyadenylated transcripts were ultra-deeply sequenced using ribo-depleted RNAs from ~40,400 neurons laser-captured from 99 human post-mortem brains and seven non-neuronal cell-type samples with an average 178 million reads per sample yielding 2.0 × 1010 pair-ended RNA-seq reads (Supplementary Table 1). Melanized neurons from the midbrain substantia nigra pars compacta (SNpc) of 86 high-quality human brains (dopamine neurons), pyramidal neurons from layers V/VI of the middle temporal cortex of ten brains and from the primary motor cortex of three brains (pyramidal neurons) were laser-captured as described Ref.8–10 (Fig. 1a). Human fibroblasts from four and peripheral blood mononuclear white cells (non-neuronal cells) from three individuals were analyzed in the same pipeline (Supplementary Fig. 1a, 2; Supplementary Table 2). Cumulatively, we found that at least 64.4% of the human genome is transcribed to produce detectable RNAs in dopamine neurons of the human brain (Fig. 1b, Online Methods) consistent with observations from ENCODE in cultured cells11. More than half of these reads (54.7%) mapped to intergenic or intronic regions (Fig. 1c) indicating a massive hidden layer of active non-coding transcription in human brain neurons.
Figure 1. Identification of noncoding elements actively transcribed in dopamine neurons of human brain.
a, Laser-capture microdissection total RNA sequencing (lcRNAseq) was used to systematically identify noncoding elements transcribed in dopamine neurons and pyramidal neurons of human brains. Dopamine neurons of the substantia nigra pars compacta from 89 high-quality autopsy brains were analyzed. Pyramidal neurons from temporal cortex from ten and motor cortex from three brains were evaluated. Moreover, fibroblasts from four and peripheral blood mononuclear white cells (non-neuronal cells) from three individuals were analyzed. b, Cumulatively, 64.4% of the human genome were transcribed in dopamine neurons of human brain. c, 54.7% of reads mapped to intergenic or intronic genome sequence. d, Schematic of the method for identifying transcribed noncoding elements (TNEs): a stringent six-step filter was applied to the RNA-seq reads aggregated from dopamine neurons, pyramidal neurons, and non-neuronal cells, respectively. Briefly, a putative TNE site was defined as a genomic region with RNA-seq reads density higher than the background transcriptional level (black dashed line) and the “summit” RPM (vertical arrow) achieving a local detection P value ≤ 0.05. TNE were required to exceed 100 bp; known genes and splice junctions were excluded. Finally, TNE were required to achieve Bonferroni-corrected expression P value ≤ 0.05 across all samples of one cell type (indicated by multiple copies of the schematic) compared to length-matched, randomly selected background regions using a binomial distribution. See text, Online Methods, Supplementary Fig. 14 for detail. e, Venn diagram with TNE detected in dopamine neurons, pyramidal neurons, and non-neuronal cells.
Enhancer RNA (eRNA) expression is a marker for active enhancers12–14 and can be used to estimate enhancers active in a particular cell type at given time13. Genetic enhancer elements control the cell type-specific activation of gene expression. We designed a sophisticated method for the systematic identification of noncoding elements including known and novel candidate enhancers that are significantly expressed in dopamine neurons, pyramidal neurons, and non-neuronal cells, respectively using a stringent six-step filter (Fig. 1d, see Online Methods for detail). We required aggregated reads for each cell type to achieve local peak read densities (“summit”) with detection P-value of 0.05 compared to randomly sampled background; without overlap with exons from annotated genes and TSS-proximal regions; with a minimal element length of 100 bp; without splicing junction reads (to avoid multi-exon non-coding RNAs (ncRNA)). We then rigorously determined the statistical significance of each of these candidate transcribed noncoding elements across multiple independent samples of the same cell type (e.g. across 86 independent samples for dopamine neurons) with a family-wise adjusted P-value less or equal than 0.05 taken as evidence of statistical significant expression.
We discovered 71,022, 37,007, and 19,690 transcribed non-coding elements (TNEs) in dopamine neurons, pyramidal neurons, or non-neuronal cells, respectively, with detection P values equal or better than the Bonferroni-corrected significance thresholds of 7.0 × 10−7, 5.1 × 10−7, 6.6 × 10−7 for each of the three cell types, respectively (Supplementary Table 3). The length distribution of TNEs peaked around 400 bp (Supplementary Fig. 3a), consistent with that of the enhancer RNAs previously reported by FANTOM513 and of activity-regulated enhancer RNAs found in mouse cortical neurons12. Unlike promoter regions, TNEs showed a GC content distribution similar to random genomic background regions and this is inconsistent with PCR bias (Supplementary Fig. 3b). The vast majority of TNEs (92%) localized to intronic regions (Supplementary Fig. 3c) and they tended to be positionally biased towards the 5′ end of gene body; a pattern opposite to that of partial RNA degradation, which preferentially degrades 5′ ends (Supplementary Fig. 3d). TNEs accounted for 31.42% and 32.35% of reads transcribed in dopamine and pyramidal neurons, respectively, compared to 21.08% in peripheral cells. 26.38% of dopamine neuron TNEs were also presented in pyramidal neurons (Fig. 1e; Fisher’s exact test P < 2.2 × 10−16, odds ratio = 3.22), but only 7.85% in peripheral cells. Subprograms of protein-coding mRNAs and non-coding RNAs (ncRNAs) expressed in dopamine neurons, pyramidal neurons, and non-neuronal cells were also characterized (see Supplementary Fig. 4, Supplementary Table 4, and Online Methods for detail).
Transcribed noncoding elements (TNEs) identify putative enhancers active in dopamine neurons
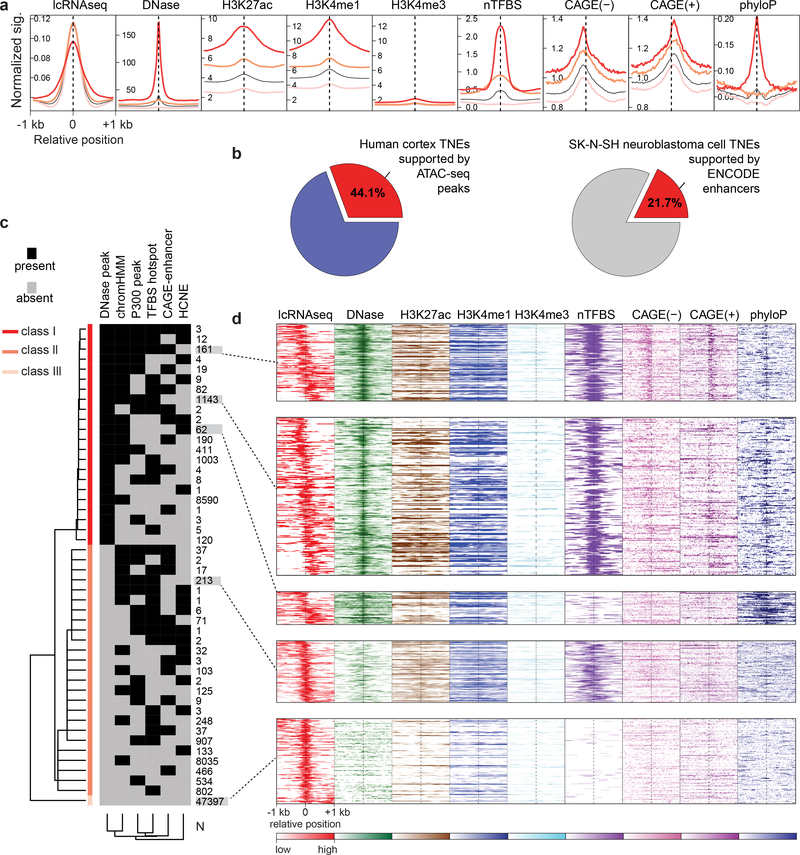
23,625 of 71,022 (33%) TNEs active in dopamine neurons coincided with enhancers defined by one or more genomic or epigenetic features (Fig. 2; see Online Methods). These features included DNase I hypersensitivity sites (DHS)15, characteristic histone modifications (such as high H3K27ac, high H3K4me1, and low H3K4me3)16, capped analysis of gene expression (CAGE)13-defined enhancers, transcriptional coactivator P30017 binding sites, transcription factor ‘hotspots’18, and sequence conservation19. 20,505 of 71,022 TNEs coincided with chromatin state-defined putative active enhancers from Roadmap Epigenomics20 and 1,212 TNEs coincided with CAGE-defined putative active enhancers13. The overlap was significantly higher than expected by chance alone with P < 2.2 × 10−16 by permutation test (Supplementary Table 5).
Figure 2. Transcribed noncoding elements (TNEs) identify putative enhancers active in dopamine neurons.
a, TNEs in dopamine neurons are, in part, supported by epigenetic and genetic features of active enhancers. The aggregation plot visualizes normalized signals for DNase I hypersensitivity sites (DHS), histone modification marks (H3K27ac, H3K4me1, and H3K4me3), transcription factor binding sites ‘hotspots’ (nTFBS), CAGE signals (forward and reverse strand), and sequence conservation score (phyloP) in the [−1000, +1000]bp regions centered on the DHS peak (if any) or middle position of TNEs. TNE with multiple supportive enhancer features are shown as red line (class I); TNE with one supportive feature are shown with a pink line (class II), and TNE previously not identified by published histone- or CAGE-enhancers are shown light pink (class III). The black line indicates the aggregation plot for all three TNE classes combined. b, TNEs significantly overlapped with putative enhancers predicted by other methods applied to the same human brain samples and to the same human dopaminergic SK-N-SH neuroblastoma cell line (P < 2.2 × 10−16 by permutation test). c, 71,022 dopamine neuron-TNEs were clustered into three classes (rows) according to the presence or absence of supporting features of active enhancers (columns) such as DHS, P300 peak, enhancer chromatin state (chromHMM), transcription factor binding sites (TFBS) hotspot, CAGE-enhancer, sequence conservation from external databases as described above. d, The heatmaps visualize TNEs with distinct combinations of epigenetic and genetic enhancer features. The relative abundance of TNEs (rows) in dopamine neurons as measured by lcRNAseq (red, relative abundance is color-coded) and physically co-localizing signals of supportive epigenetic and genetic enhancer features (the relative abundance of each supportive enhancer features is color-coded) is shown for the same [−1000, +1000]bp window as above.
We performed two experiments to directly benchmark TNE to putative enhancers predicted by two other methods applied to the same source (Fig. 2b). 44.1% of TNE (14,904 of the 33,762 TNE) called by our pipeline in the human cortex data set from PsychENCODE21 overlapped with a strong ATAC-seq peak (which maps chromatin accessibility22) identified in the same samples (Fig. 2b). This was a significantly higher than expected by chance with P < 2.2 × 10−16 by permutation test. In SK-N-SH cells, 21.7% of called TNEs (11,465 of 52,733) overlapped with putative enhancer features (Fig. 2b; e.g. H3K27ac, H3K4me3, transcriptional regulator CCCTC-binding factor (CTCF) chromatin immunoprecipitation sequencing (ChIP-seq), P300 ChIP-Seq, DNase I hypersensitivity, and transcription factor hotspots) delineated by ENCODE in this cell line with P < 2.2 × 10−16 by permutation test, similar to the 25% overlap previously reported between CAGE-defined and chromatin state-predicted putative enhancers13.
We grouped 71,022 dopamine TNEs into three classes according to the presence or absence of supporting features (see Online Methods). Specifically, 11,835 TNEs coincided with multiple supportive features (designated class I TNE), that is a known DHS site plus at least one of five additional external features (chromHMM, CAGE-enhancer, P300 peak, transcription factor binding sites (TFBS) hotspot, highly conserved noncoding elements (HCNEs) between human and zebrafish; Fig. 2c,d). A second set of 11,790 TNEs was supported by at least one of the five external features, but lacked additional DHS evidence (designated class II TNEs; Fig. 2c,d). A third set of 47,397 TNEs had no previously reported supporting external features (termed class III TNE; Fig. 2c,d). Bi-directional transcription of select dopamine TNEs was seen using CAGE in substantia nigra of four of the same brains used for lcRNAseq (Supplementary Fig. 5a). Moreover, transcription factor binding sites were enriched in TNE sites based on in silico analysis of ChIP-seq peaks and motif scanning (Supplementary Fig. 5b–d, Supplementary Note).
Replication of TNEs in independent cohorts.
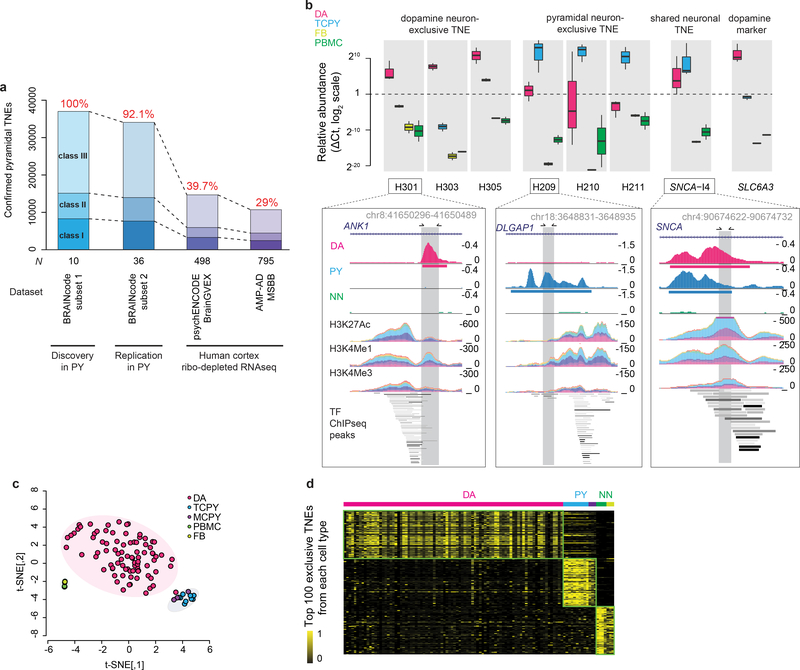
We replicated pyramidal neuron-TNE in three independent cohorts representing 36, 498, and 795 human brain samples, respectively (Fig. 3a), and additionally confirmed select TNE with two secondary methods (Fig. 3b and Supplementary Fig. 5a). Out of the 37,007 pyramidal neuron-TNE discovered, 34,077 (92.1%) were replicated in an independent cohort of pyramidal neurons laser-captured from layer V/VI of 36 new human autopsy brains (Fig. 3a, BRAINcode-replication subset). 14,679 (39.7%) and 10,718 (29%) of 37,007 pyramidal neuron-TNEs were identified from ribo-depleted total RNA-seq data of frontal cortex (PsychENCODE21) and four cortex areas (Accelerating Medicines Partnership–Alzheimer’s Disease Consortium (AMP-AD)), respectively Fig. 3a). Select brain cell type-specific TNE were confirmed with a secondary method, qPCR, in laser-captured dopamine neurons (Fig. 3b). As expected, qPCR analysis of control samples lacking template or reverse transcriptase showed no expression of TNE. Finally, we confirmed a subset of dopamine neuron TNE by performing CAGE on four substantia nigra homogenate samples (Supplementary Fig. 5a, see Online Methods).
Figure 3. TNEs signatures accurately cluster dopamine and pyramidal neurons.
a, Of the 37,007 pyramidal neuron TNEs discovered, 34,077 (92.1%) were replicated in a second cohort of pyramidal neurons (PY) laser-captured from layer V/VI of 36 new human autopsy brains that had not been used for the discovery study. Moreover, using our TNE detection pipeline, we deconvoluted expression of 14,679 (39.7%) and 10,718 (29%) of pyramidal neuron TNEs in two ribodepleted total RNA-seq datasets representing cortex homogenates from 498 and 795 individuals, respectively. AMP-AD, Accelerating Medicines Partnership–Alzheimer’s Disease Consortium; MSBB, Mount Sinai VA Medical Center Brain Bank; psychENCODE BrainGVEX (https://www.synapse.org/#!Synapse:syn4590909); b, Cell type preferential expression of three dopamine neuron-exclusive TNEs, three pyramidal neuron-exclusive TNEs, and one TNE common to both dopamine and pyramidal neurons (in intron 4 of the PD gene SNCA; see also Supplementary Fig. 6a) was confirmed additionally by qPCR (box plots, top panel) in addition to lcRNAseq (pile graphs, bottom panels; and Supplementary Fig. 6b). Relative mRNA abundance of a classical dopamine-neuron marker, the dopamine transporter gene SLC6A3 (DAT), was assayed as positive control. Dopamine neuron samples (DA), N = 3; temporal cortex pyramidal neurons (TCPY), N = 3; primary human fibroblasts (FB), N = 2; human peripheral blood mononuclear white cells (PBMC), N =2. The pile graph tracks show RNA-seq read densities in dopamine neurons (DA), pyramidal neurons (PY), and non-neuronal cells (NN), as well as corresponding histone enhancer marks and transcription factor (TF) ChIP-seq peaks from ENCODE25. Box plots indicate the median (bold line), the 25th and 75th percentiles (box edges), and the most extreme data point which is no more than 1.5 times the interquartile range from the box (whiskers). c, A signature based on cell type-exclusive TNE clustered 106 individual samples with 99.1% accuracy by t-SNE. d, The heatmap visualizes normalized counts for the 100 most abundant TNEs exclusively expressed in dopamine neuron (DA), pyramidal neuron (PY), and non-neuronal cells (NN).
TNEs signatures accurately cluster dopamine and pyramidal neurons
57.5% (40,846 of 71,022) of the detected TNEs were exclusively expressed in human dopamine neurons. They were not detected in pyramidal neurons or non-neuronal cells. 39% (14,487 of 37,007) of pyramidal neuron-TNEs were exclusive to this cell type; 64% (12,601 of 19,690) of non-neuronal TNEs were exclusively expressed in non-neuronal cells (Fig. 1e). A signature based on cell type-exclusive TNE clustered 106 individual samples with 99.1% accuracy (Fig. 3c) — similar to the classification accuracy afforded by mRNAs and ncRNAs (Supplementary Fig. 4). Normalized counts for the 100 most abundant exclusive TNEs in each cell type are visualized in Fig. 3d. Cell type preferential expression of three dopamine neuron-exclusive TNEs, three pyramidal neuron-exclusive TNEs, and one TNE common to both dopamine and pyramidal neurons (in intron 4 of the PD gene SNCA23,24, Supplementary Fig. 6a), was confirmed by qPCR in addition to lcRNAseq (Fig. 3b and Supplementary Fig. 6b). These TNEs were in close proximity to histone marks typical of active enhancers25 as well as multiple transcription factor occupancy sites25 (Fig. 3b, lower panels).
In vivo validation of TNE enhancer activity in zebrafish, mice, and neuronal cells (Fig. 4)
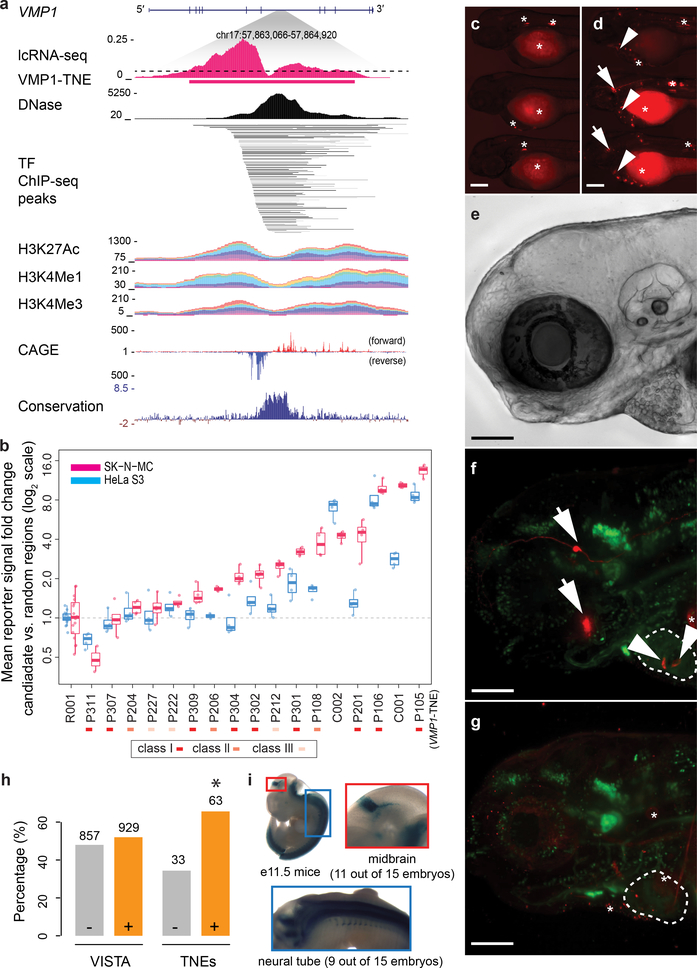
Figure 4. In vivo validation of TNE enhancer activity in zebrafish, mice, and neuronal cells.
a, VMP1-TNE located in intron 7 of the human VMP1 gene. This evolutionary conserved TNE is supported by the classical epigenetic features of a putative active enhancer including open chromatin (DNase)20, high levels of H3K4me1 and H3K27ac25, and bidirectional CAGE signal13.
b, Enhancer reporter assays for TNE-defined putative enhancers in HeLa S3 (cyan) and SK-N-MC neuroblastoma cells (magenta). TNE regions are labeled Pxxx (see Supplement for detail); C001 and C002, denote enhancers from FANTOM5 (positive controls)13; R001, random genomic background region (negative control). N = 4 transfections were independently performed and analyzed for each TNE in HeLa S3; another N = 4 independent transfections in SK-N-MC cells; *, P < 0.05; **, P < 0.01; ***, P < 0.001; two-tailed Student’s t-test. Box plots as in Fig. 3.
c-g, Group view of embryos injected with control:gata2:mRuby2 and VMP1-TNE:gata2:mRuby2 respectively. Enhanced reporter activity was observed in the embryos injected with VMP1-TNE containing reporter construct (e) compared to the control element (d). In addition, embryos injected VMP1-TNE:gata2:mRuby2 reporter construct (e) show tissue specific reporter expression in a group of telencephalic neurons in proximity to the eye (arrows) and atrioventricular canal (arrowheads). Background (ectopic) activity (marked by stars) predominantly in skin yolk muscle and auto-fluorescence from blood and eye pigmentation (stars) was observed in both, VMP1-TNE:gata2:mRuby2 and control:gata2:mRuby2 injected embryos.
f,g, High magnification view on the head and heart region of ETvmat2:gfp transgenic embryos injected with control:gata2:mRuby2 and VMP1-TNE:gata2:mRuby2 respectively. The expression of the GFP reporter in this transgenic was utilized as marker for the heart ventricle (dashed line). Expression in the telencephalic neurons (arrows) and atrioventricular canal (arrow heads) in VMP1-TNE:gata2:mRuby2 injected embryos can be seen. Stars mark ectopic activity. e, A bright filed reference image of the corresponding embryo region shown on f-g. Scale bars equal to 200 μm on c-d and 100 μm on e-g. The experiment was repeated independently with similar results four times with 478 and 408 embryos screened in total, for VMP1-TNE and control constructs, respectively (see Supplemental Table 7 for details).
h, Putative enhancers evaluated in mice by VISTA28. 929 (52%) out of all 1786 putative enhancers (left two bars) tested in mice by VISTA were active in mice. By contrast, 63 (66%) of 96 TNE-defined putative enhancers were found to be active enhancers in mice (*, indicates P = 3.91 × 10−3, Hypergeometric test).
i, Reporter activity of a neuron-specific intronic TNE located in the AUTS2 gene is seen in the midbrain (red insert) and neuronal tube (blue insert) of mouse e11.5 embryos by VISTA28. Embryos have an average crown-rump length of 6 mm.
To determine if TNEs can function as enhancers we tested 15 TNEs (Supplementary Table 6) in vitro in human SK-N-MC neuroblastoma cells and non-neuronal HeLa cells. TNE sequences were inserted into a modified pGL4.10 vector (as in ref.13), e.g. upstream of an EF1a basal promoter separated by a synthetic polyA signal/transcriptional pause site in order to avoid promoter effects. 11 of the 15 TNEs (73%) significantly increased reporter activity in neuronal cells compared to control inserts representing random background sites (Fig. 4b). Eight TNEs induced more than a twofold increase in reporter signal (Fig. 4b) and all but one TNE exhibited considerably higher enhancer activity in the neuronal cells compared to HeLa cells (Fig. 4b). VMP1-TNE (chr17_57863430_57864538) is located in intron 7 of the human VMP1 gene, a key regulator of autophagy. The VMP1-TNE site is evolutionary conserved among vertebrates and actively transcribed in human brain dopamine neurons, pyramidal neurons, and non-neuronal cells. VMP1-TNE was a class I TNE with a bimodal distribution of RNA-seq reads (centered on the DHS peak; Fig. 4a), bidirectional CAGE signal (Fig. 4a), occupancy by 90 TFs (Fig. 4a), high levels of H3K4me1 and H3K27ac (Fig. 4a), and was predicted as putative enhancer by ChromHMM26 in Roadmap Epigenomics20. It was highly active in neuroblastoma and HeLa cells in culture (P105 in Fig. 4b). To assess the activity of VMP1-TNE in vivo, transient transgenic reporter assays were carried out in zebrafish embryos. The PCR-amplified sequence was cloned upstream of zebrafish gata227 minimal promoter, linked to an mRuby2 reporter gene in a modified pDB896 vector. A similarly sized sequence amplified from a nonconserved intergenic region with very low or no signal for enhancer marks was used to generate a control construct. Embryos injected with Has.VMP1-TNE:gata2:mRuby2 (Fig. 4c–g) reporter construct showed reproducible enrichment of enhancer activity in a specific subset of telencephalic neurons near the eyes and in cardiac cells in proximity of the atrioventricular canal compared to embryos carrying control construct (Has.control:gata2:mRuby2; Fig. 4c–g; Supplementary Table 7) consistent with the expression pattern of miR-21 (www.zfin.org), the putative target gene in the synteny block as suggested by comparative genomics (Supplementary Fig. 7).
The VISTA consortium has established one of the largest repositories of in vivo enhancer screens during mouse development28. Sequences overlapping with 96 dopamine neuron TNEs were evaluated by VISTA28, 63 (65.6%) of which were positive enhancers in vivo in mice, considerably more than expected by chance alone with P = 3.91 × 10−3 by Fisher’s exact test (Fig. 4h). The enrichment for VISTA- validated enhancers was similar for class I and III TNEs (Supplementary Fig. 6c). Interestingly, 35 of these 63 (55.6%) VISTA-validated TNEs drove reporter gene expression in neuronal tissues, particularly midbrain, hindbrain, and the neural tube. For example, a neuron-specific TNE located in the intron of autism susceptibility candidate 2 gene (AUTS2) enhanced reporter activity specifically in the midbrain in 11 out of 15 mouse embryos tested28 (Supplementary Fig. 6d, Fig. 4i). In comparison, of 31 exclusively non-neuronal TNE evaluated by VISTA, 14 (45%) were positive enhancers, and only 9 (29%) were active in neuronal tissues. Collectively, these test cases show that select TNE sites enhance reporter gene expression in human neuronal cells and in neurons in the brain of zebrafish and in mouse.
Variants associated with diseases of the dopamine system are over-represented in TNE actively transcribed in dopamine neurons
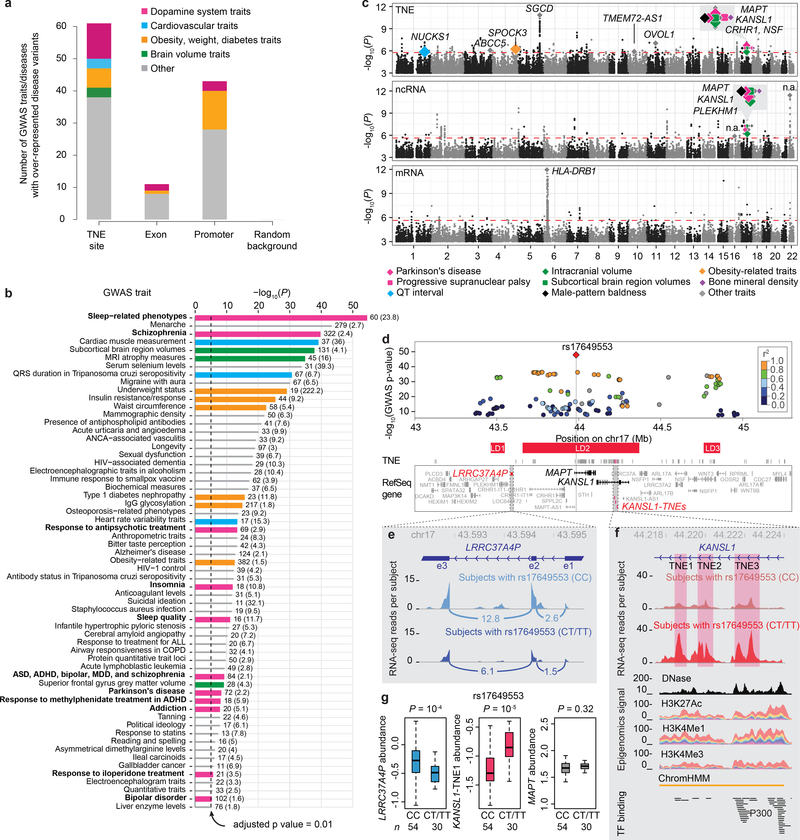
GWAS variants for 61 diseases and traits were significantly enriched within noncoding elements functional in dopamine neurons with P values below the Bonferroni-corrected significance threshold of 9.64 × 10−6 by Fisher’s exact test (e.g. 0.01 divided by 1037, the total number of traits in the NHGRI GWAS catalog29) compared to random background (Fig. 5a; Online Methods). By contrast, only 43 traits were significantly enriched in promoters, 11 in exons (Fig. 5a). Consistent with our hypothesis, variants associated with eleven diseases and medications perturbing the dopamine system were significantly enriched in dopamine neuron-TNE sites (strawberry color in Fig. 5a,b). These included variants associated with schizophrenia with P = 1.75 × 10−40, PD with P = 5.05 × 10−9, addiction with P = 1.33 × 10−8, and bipolar disorder with P = 5.05 × 10−6. Moreover, pharmacogenetic variants associated with response to antipsychotics were enriched in these TNE sites with P = 4.39 × 10−14. Classical antipsychotics are dopamine receptor antagonists that are the standard treatment for schizophrenia. Variants associated with response to iloperidone, a specific antipsychotic medication for schizophrenia were also enriched with P = 1.94 × 10−6. Variants associated with response to the dopamine reuptake inhibitor methylphenidate (used to treat attention deficit hyperactivity disorder) were enriched in dopamine neuron-TNE sites with P = 8.74 × 10−9. By contrast, none of these trait variants were enriched in promoters or exons. Interestingly, traits relating to sleep phenotypes, which are modulated by dopamine neurons (e.g. ref. 30,31) and perturbed in PD32, were highly enriched in these TNE sites with P = 2.6 × 10−55 (Fig. 5a,b). Surprisingly, cardiovascular traits (blue in Fig. 5a,b); diseases and traits clustering around obesity, weight and diabetes (orange in Fig. 5a,b); and brain volume related traits (green in Fig. 5a,b) were also over-represented in dopamine neuron-TNEs compared to random genomic background. The enrichment density for dopamine system traits was similar for each of the three TNE classes (Supplementary Fig. 8a).
Figure 5. Putative enhancers active in dopamine neurons link genetic variation to neuropsychiatric disease.
a, GWAS diseases and traits with variants significantly enriched in dopamine TNEs, exons, promoters, and random background regions. Variants for 61 diseases and traits were enriched within TNE-defined putative active enhancers in dopamine neurons with P values below the Bonferroni-corrected significance threshold of 9.64 × 10−6 by one-sided Fisher’s exact test compared to 71,022 random genomic background regions (see Online Methods). The largest share of traits (N = 11) enriched within putative active enhancers clustered around perturbations of the dopamine system (strawberry color). By contrast, only 43 traits were enriched in promoters (including two involving the dopamine system), 11 in exons, and none in random background regions.
b, Diseases and traits significantly enriched in TNE-defined putative enhancers in dopamine neurons. Variants associated with eleven diseases or medications perturbing the dopamine system (horizontal strawberry colored bars) were dramatically over-localized in dopamine neuron TNE. The number of disease-variants colocalizing to dopamine TNEs for each trait as well as odds ratios (in parenthesis) are shown next to each bar. x-axis, P-values by one-sided Fisher’s exact test (-log10 scale); y-axis, diseases and traits. ADHD, attention deficit hyperactivity disorder; ASD, autism spectrum disorder; MDD, major depressive disorder; ANCA, antineutrophil cytoplasmic antibody; ALL, acute lymphoblastic leukemia; COPD, chronic obstructive pulmonary disorder; QRS duration, a feature on an electrocardiogram.
c, Expression quantitative trait locus analysis reveals transcribed noncoding elements in synapse genes as main cell-autonomous effector of cis-acting genetic variation in human brain dopamine neurons. Manhattan plots for TNE-eQTLs (top), ncRNA-eQTLs (middle), and mRNAs (bottom) are shown. Diamonds indicate eSNPs with RTC scores ≥ 0.85, colors indicate different groups of diseases. Gene symbols of the host loci for these eSNPs are shown (n.a., intergenic regions). Y-axis, P-values of eSNP-transcript associations from Matrix-eQTL linear regression model (N = 84). A false discovery rate (FDR) of 0.05 was considered significant (indicated by the red dashed line); QT interval, another feature on an electrocardiogram.
d, Parkinson’s disease-associated variants cis-regulate a noncoding element in the KANSL1 gene in dopamine neurons. The locus plot visualizes P values (y-axis) and chromosomal location (x-axis) of genetic variants associated with susceptibility for PD in the chromosome 17q21 GWAS peak35 (chr17:43,000,000–45,300,000 in hg19). A red diamond visualizes the lead susceptibility variant rs17649553; other PD-associated variants in the locus are represented by circles (r2 with the lead SNP is color-coded). 45 Refseq genes are physically localized under this GWAS peak (box). Linkage disequilibrium (LD) blocks are shown as red horizontal bars. P-values extracted from www.pdgene.org for GWAS meta-analysis of 13,708 PD cases and 95,282 controls are shown.
e-f, Increased expression of KANSL1-TNE1 (right panel, red) and decreased expression of LRRC37A4P (left panel, blue) is seen in dopamine neurons of individuals carrying one or two copies of the risk allele (CT/TT) compared to individuals without the risk allele (CC). e, Pile graphs of average RNA-seq reads density aligning to the exon 3–2 and exon 2–1 junctions of LRRC37A4P are visualized for individuals not carrying (cyan) or carrying the risk allele (navy). f, Genomic landscape of KANSL1-TNE1 expression in non-carriers (red-brown) and carriers (red) of the risk allele. KANSL1-TNE1 is expressed from intron 2 of the KANSL1 gene (chr17_44218414_44219042). Note, nearby KANSL1-TNE2 and -TNE3 showing similar expression patterns. KANSL1-TNE1 is expressed from a chromatin state-defined enhancer. Lower tracks display DNase uniformed signal (Roadmap20), stacked H3K27ac, H3K4me1, and H3K4me3 signals (ENCODE25), chromHMM predicted enhancer based on histone modifications (Roadmap20), as well as transcription factor ChIP-seq peak clusters (ENCODE25).
g, Boxplots representing the eQTL relation between the lead PD-associated rs17649553 variant and transcript abundance of KANSL1-TNE1, LRRC37A4P, and MAPT in dopamine neurons. Box plots as in Fig. 3b. P values from Matrix-eQTL linear regression model, N = 84 biologically independent samples.
Dopamine neuron-TNEs harbor a higher density of GWAS variants linked to traits of the dopamine system than enhancer predictions without cell-type specificity
GWAS SNP density analyses showed a higher density of GWAS variants for dopamine system traits in TNE active in midbrain dopamine neurons compared to FANTOM5-predicted and ChromHMM-predicted putative enhancers, exons, promoters, introns, intergenic regions, and length-matched random regions (Supplementary Fig. 9).
Expression quantitative trait locus analysis reveals transcribed noncoding elements in synapse genes as main cell-autonomous effectors of cis-acting genetic variation
Expression quantitative trait locus (eQTL) analysis for TNE, ncRNAs, and mRNAs was --- for the first time --- performed across cell type-specific transcriptomes from 84 human brains (Fig. 5c). 4,283,750 SNPs were measured or imputed and associated with normalized TNE expression using Matrix eQTL33 (see Methods). 8,676 cis-acting TNE-eQTLs achieved a false discovery rate (FDR) ≤ 0.05, comprising 3,461 unique expression-associated SNPs (eSNPs) and 151 unique TNEs (Fig. 5c, top panel). On average 23 eSNPs associated with expression changes in one TNE. Furthermore, 3,381 ncRNA-eQTLs were significant (FDR ≤ 0.05), comprising combinations of 3,320 unique eSNPs and 52 unique expressed ncRNA genes (Fig. 5c, middle panel; Supplementary Fig. 10). By contrast only 1,150 mRNA-eQTLs reached statistical significance (FDR ≤ 0.05), comprising combinations of 676 unique eSNPs and 46 unique associated expressed protein-coding genes (Fig. 5c, bottom panel; Supplementary Fig. 10).
These 151 cis-regulated TNEs physically localized to introns of 102 host genes. These host genes were highly enriched in Gene Ontology terms related to synapse function (with P values less than 4.79 × 10−7 by enrichment analysis using the hypergeometric test; see Supplementary Table 8 for full results and Online Methods) and in MeSH terms for brain disorders with P = 5.1 × 10−10 (Supplementary Table 9). Mutations of several of these synapse-related host genes can cause abnormal brain development and function (Supplementary Fig. 10; Supplementary Note). Taken together this gene-regulatory analysis indicates that genetic variation is linked to variation in the activity of putative enhancers in synapse genes, including several loci linked to Mendelian brain disorders.
Parkinson’s disease-associated variants cis-regulate a noncoding element in the KANSL1 gene
Leveraging 495,085 SNPs associated with one or more of 1,037 human diseases or traits (19,188 disease-associated SNPs from NHGRI-EBI GWAS catalog29, extended via imputation of proxy SNPs with r2 ≥ 0.8), we identified 1,989 disease-associated SNPs that influence expression of 19 TNEs, 4 ncRNAs, and 5 mRNAs in cis. To distinguish coincidental co-localizations of GWAS and eQTL associations, we used regulatory trait concordance (RTC) scores34, which assess whether a cis-eQTL and a trait association are tagging the same underlying functional effect. Applying a stringent RTC threshold of 0.85, we identified 23 disease-associated TNE-eQTLs for which the trait and TNE expression associations may be tagging the same effect in dopamine neurons (Fig. 5c; Supplementary Table 10). 17 and 1 disease-associated eQTLs were identified for ncRNAs and mRNAs, respectively.
Eight of these 23 TNE-eQTLs linked PD-associated variants to a putative eRNA expressed from intron 2 of the KANSL1 gene with P values as low as 1.57 × 10−7 (Supplementary Table 10). The corresponding RTC scores were 0.91–1.00, indicating that the GWAS-derived disease variants explain the eQTL observation. Six of the eight PD-associated eSNPs mapped to the exact same 712,000 bp-long LD block on chromosome 17q21 (termed here LD2; Fig. 5d) and were significantly associated with up-regulation of the same KANSL1-TNE1 in carriers of risk alleles (6.46 × 10−7). Two additional eSNPs mapped to a nearby LD block (LD3; Fig. 5d). Conditional eQTL analysis adjusting for the lead GWAS variant rs17649553 suggested that some eSNPs in LD2 and one in LD3 might carry an independent signal (Online Methods, Supplementary Fig. 11, and Supplementary Table 11). Chromosome 17q21 is the second most important GWAS peak for sporadic PD (after SNCA) and unequivocally associated with susceptibility for PD with P values as low as 2.23 × 10−48 in a meta-GWAS of more than 100,000 cases and controls35. There is precedent that copy number variation in the KANSL1 locus causally impacts brain function as microdeletions of the locus cause Koolen de Vries syndrome, a neurologic disease with severe learning disability and developmental delay. In addition to up-regulating the KANSL1-TNE, the same PD-associated variants in LD2 (but not those localized to LD3) were associated with down-regulation of an expressed pseudogene, LRRC37A4P with P = 2.36 × 10−07 (Supplementary Table 10). LRRC37A4P is localized near the KANSL1 gene under the chromosome 17q21 GWAS peak. By contrast, eQTL associations for MAPT mRNA, a biological candidate in this region, did not reach genome-wide significance (Fig. 5e–g). The inverse eQTL relation between the lead GWAS-derived SNP rs17649553 and KANSL1-TNE1 and LRRC37A4P, respectively, was confirmed by a second method, cell type-specific qPCR (Supplementary Fig. 12a). Moreover, this association was independently replicated in a second cohort of neurons laser-captured from 31 high-quality control brains (Supplementary Fig. 12b, Supplementary Table 12). Third, the rs17649553-LRRC37A4P eQTL association was further confirmed in 56 substantial nigra and 96 frontal cortex samples from GTEx (Supplementary Fig. 12c,d), which used a polyA+ selecting protocol that does not allow for assaying KANSL1-TNE1 RNA.
Discussion
eRNA expression is a feature of active enhancers12–14 and can be used as a marker to estimate their activity in a particular cell type13. Genome elements with enhancer chromatin marks that are transcribed into eRNAs have significantly higher validation rates in in vitro enhancer assays than enhancers defined exclusively by chromatin-states13. Moreover, in transgenic mouse reporter assays, over half of putative enhancers identified on the basis of deep RNA sequencing functioned as enhancers with reproducible activity in the predicted tissue36. Many chromatin-defined enhancers are not regulatory active in a particular cellular state, but may be active in other cells37 or are pre-marked for fast regulatory activity upon stimulation38.
We show a highly specific program of enhancer elements that are actively transcribed in physiologically and morphologically distinct, disease-relevant dopamine and pyramidal neurons --- in situ, in human brains. 64.4% of the genome were cumulatively transcribed in dopamine neurons, including 71,022 noncoding elements many of which consistent with histone-state and CAGE-defined active enhancers, and with in vivo regulatory functions in zebrafish and mouse neurons. We provided mechanistic evidence that some of these elements function as enhancers of transcription in zebrafish brain, in the midbrain of mice, and in human cultured neuronal cells using genetics and reporter assays. Moreover, multiple independent lines of evidence—including chromatin state, CAGE expression, and transcription factor binding analyses— supporting the view that these transcribed noncoding elements are putative enhancers specifically active in dopamine neurons.
Variants associated with eleven diseases or medications perturbing the dopamine system were enriched in dopamine neuron-specific TNE sites --- much more so than in promoters and or exons (Fig. 5a,b). Risk alleles associated with major disorders of dopaminergic neurotransmission, schizophrenia, PD, addiction as well as with bipolar disorder over-localized to active TNE sites. Compellingly, even pharmacogenetic variants linked to treatment response were enriched in active enhancers. These observations suggest that GWAS variants might modulate enhancers active in dopamine neurons and thereby regulate the transcriptional program underlying susceptibility for these neuropsychiatric diseases. Finally, risk alleles associated with sleep-related phenotypes were enriched in TNE sites with P = 2.6 × 10−55. Indeed, dopamine neurons have a role in sleep regulation30,31 and REM sleep behavior disorder is an early sign of PD32.
eQTL analysis for putative eRNAs was for the first time performed across cell type-specific transcriptomes from 84 human brains (Fig. 5c). It uncovered transcribed noncoding elements in synapse genes as a main cell-autonomous effector of cis-acting genetic variation in dopamine neurons. Importantly, the number of TNE-eQTLs greatly surpassed the number of mRNA-eQTLs and ncRNA-eQTLs identified.
The second most significant GWAS locus for sporadic PD is located on chromosome 17q21. This locus shows unequivocal evidence for association with PD. The regulated gene has not been established, but MAPT has been commonly assumed to be the prime candidate. Using expression quantitative trait locus analysis we provide surprising evidence pointing at regulation of a putative enhancer RNA expressed from intron 2 of the KANSL1 gene as a novel gene-regulatory mechanism for this susceptibility locus. The KANSL1 locus is important for normal brain function. Microdeletions cause Koolen de Vries syndrome, a neurologic disease with severe learning disability and developmental delay39. The KANSL1-TNE1 eQTL association was confirmed by cell type-specific qPCR and replicated in an independent cohort. By contrast, eQTL associations for MAPT did not reach statistical significance in dopamine neurons (P = 0.32). Long-read sequencing and larger data sets will be required to comprehensively illuminate the relation between structural variation and transcriptional function in this complex locus.
The KANSL1-TNE1 eQTL appears to be a “super-eQTL” of variants associated with eight dopaminergic, radiographic, pulmonary, and dermatologic traits all localized to the same LD2 block on chromosome 17q21 and all associated with KANSL1-TNE1 up-regulation (Fig. 5c, LD2). Six of these seemingly disparate traits are clinically implicated in multisystem features of PD. Progressive supranuclear palsy (trait 2), leads to neurodegeneration of dopamine neurons (Fig. 5c and Supplementary Table 10). Men with early-onset male pattern baldness (trait 3) have a 28% higher risk of developing PD40. Genetic variants for intracranial volume (traits 4 & 5) are related to PD41 and PD patients are prone for reduced bone mineral density (trait 6)42,43. Thus, PD and seven clinically related traits with variants localizing to an LD block on chromosome 17q21 are associated with KANSL1-TNE1 expression through a uniform gene-regulatory mechanism.
This study is powered by innovations both in wet and dry lab methods and provides a valuable online resource of mRNAs, ncRNAs, and TNE expression in dopamine and pyramidal neurons as well as dopamine neuron-specific mRNA-, ncRNA-, and TNE-eQTLs (www.humanbraincode.org). The method allows detecting the full complement of mRNAs, ncRNAs, and active enhancers in one single and minuscule RNA sample and combines the base-pair resolution and a comprehensive genome-wide view afforded by ultra-deep, total RNA sequencing, with the positional and cytoarchitectural information afforded by traditional light microscopy. It can be transferred to other morphologically or regionally defined brain and peripheral cells of critical relevance to health and disease. Moreover, the three-in-one approach (detecting three types of RNAs: TNEs, mRNAs, and ncRNAs) offers simplicity and noise-reduction compared to approaches relying on separate methodologies, experiments, and source material for assaying enhancers and mRNAs. LcRNAseq offers advantages to RNA sequencing of brain region homogenates (a suspension of all types of glial, neuronal, immune, and vascular cells resident in a tissue block) or of sorted nuclei without precise information on the 3-D origins in human brain and morphological features44,45. Conversely, FISSEQ and other in situ hybridization-based methods preserve valuable positional information but the number of transcripts probed has been limited46.
This analysis shows that putative enhancers active in dopamine neurons link genetic variation to neuropsychiatric traits. It has clear applications for the genetics of more than twenty million patients in the United States alone with perturbed dopamine systems, to narrow the search window for functional associations and therapeutic nodes, and for defining the regulatory networks that underpin this archetype of a human brain neuron.
Online Methods
Sample Collection and Processing
We started with 107 high-quality, frozen postmortem human control brain samples identified from Banner Sun Health Institute, Brain Tissue Center at Massachusetts General Hospital, Harvard Brain Tissue Resource Center at McLean Hospital, University of Kentucky ADC Tissue Bank, University of Maryland Brain and Tissue Bank, Pacific Northwest Dementia and Aging Neuropathology Group (PANDA) at University of Washington Medicine Center, and Neurological Foundation of New Zealand Human Brain Bank. Detailed quality measures and demographic characteristics of these high-quality, frozen postmortem samples are shown in Supplementary Table 1. Median RNA integrity numbers (RIN) were 7.8, 7.8, and 7.2 for substantia nigra samples (used to laser-capture dopamine neurons), temporal cortex (used to laser-capture temporal cortex pyramidal neurons), and motor cortex samples (used to laser-capture motor cortex pyramidal neurons) indicating high RNA quality. Median post-mortem intervals were exceptionally short with 3 hours for substantia nigra, 3 hours for temporal cortex, and 13 hours for motor cortex samples further consistent with highest sample quality (Supplementary Table 1).
The 107 brain samples represented 93 subjects without a clinico-pathological diagnosis of a neurodegenerative disease meeting the following stringent inclusion and exclusion criteria. Inclusion criteria: (1) absence of clinical or neuropathological diagnosis of a neurodegenerative disease e.g. Parkinson’s disease according to the UKPDBB criteria47, Alzheimer’s disease according to NIA-Reagan criteria48, dementia with Lewy bodies by revised consensus criteria49. For the purpose of this analysis incidental Lewy body cases (not meeting clinico-pathological diagnostic criteria for PD or other neurodegenerative disease) were accepted for inclusion. (2) PMI ≤ 48 hours; (3) RIN50 ≥ 6.0 by Agilent Bioanalyzer (good RNA integrity); (4) visible ribosomal peaks on the electropherogram. Exclusion criteria were: (1) a primary intracerebral event as the cause of death; (2) brain tumor (except incidental meningiomas); (3) systemic disorders likely to cause chronic brain damage. We also included eight non-brain tissue samples as controls, including five samples of peripheral blood mononuclear cell (PBMC) and three fibroblasts (FB), provided by Harvard Biomarker Study and Coriell Institute. This study was approved by the Institutional Review Board of Brigham and Women’s Hospital.
We then performed Laser-capture Microdissection (LCM) on the brain samples to extract neurons from different brain regions. LCM was performed similar to what we and others reported8,51–53. For each substantia nigra sample, 300–800 dopamine neurons, readily visualized in HistoGene-stained frozen sections based on hallmark neuromelanin granules were laser-captured using the Arcturus Veritas Microdissection System (Applied Biosystems). For each temporal cortex (middle gyrus) or motor cortex sample, about 300 pyramidal neurons were outlined in layers V/VI by their characteristic size, shape, and location in HistoGene-stained frozen sections and laser-captured using the Arcturus Veritas Microdissection System (Applied Biosystems). Total RNA was isolated and treated with DNase (Qiagen) using the Arcturus Picopure method (Applied Biosystems), yielding approximately 7–8 ng RNA per subject. Total RNA was linearly amplified into 5~10 μg of double-stranded cDNA using the validated, precise, isothermal RNA amplification method implemented in the Ovation RNA-seq System V2 (NuGen)54,55. Unlike PCR-based methods that exponentially replicate original transcript and copies, with this method only the original transcripts are linearly replicated54,55, and amplification is initiated at the 3’ end as well as randomly thus allowing for amplification of both mRNA and non-polyadenylated transcripts54,55. Sequencing libraries were generated from 500 ng of the double-stranded (ds) cDNA using the TruSeq RNA Library Prep Kit v2 (Illumina) according to the manufacturer’s protocol. The cDNA was fragmented, and end repair, A-tailing, adapter ligation were performed for library construction. Sequencing library quality and quantity control was performed using the Agilent DNA High Sensitivity Chip and qPCR quantification, respectively. Libraries were sequenced (50 or 75 cycles, paired-end) on a Illumina HiSeq 2000 and 2500 at the Harvard Partners Core.
Genotyping and Imputation
Each sample was genotyped using the Infinium Omni2.5Exome-8 BeadChips (Illumina), which includes more than 2.5 million tag SNPs from the HapMap and 1000 Genomes Project. The total 98 samples from 93 subjects were genotyped in three batches, with technical replicates for 5 subjects. We computed the pairwise IBD of genotypes between replicates using PLINK2, and reach 0.9991 proportion IBD averagely. Thus, we kept unique sample and replicates in batch 1 for further quality control analysis.
We applied PLINK256 (v1.9beta) and in-house scripts to perform rigorous subject and SNP quality control (QC) (Supplementary Fig. 13a) that includes (1) SNP GC score filtering, (2) subjects call rate, (3) gender misidentification, (4) genotype call rate (5), Hardy-Weinberg Equilibrium testing, (6) Test-mishap, (7) heterozygosity outlier, and (8) IBS/IBD filtering. In total, we excluded 5,249 SNPs with GC score < 0.25, 1,955 SNPs not in the genome assembly we used (hg19) and 20,049 SNPs with call rate < 95%, 57 SNPs with Hardy-Weinberg equilibrium p-value < 10−6, 1,295,546 SNPs with MAF < 0.05, and finally 2 subjects with IBS/IBD PI_HAT > 0.9. In total, 91 subjects with 1,235,673 SNPs passed QC.
We employed SHAPEIT257 (v2.5) to perform pre-phasing and then IMPUTE257 (v2.3.1) to impute the post-QC genotyped markers in autosomal chromosomes using reference Haplotype panels from the 1000 Genomes Project (Phase 3) which includes a total 77.8 million SNPs in 2,504 individual samples. For genotyped markers in chromosome X, we used the 1000 Genomes Project Phase I Integrated Release Version 3 as reference Haplotype in 1,092 individuals. The genotyped calls of imputed genotypes with posterior probability <0.9 were marked as missing and we kept biallelic genotypes for further analysis. After genotype imputation, we filtered out imputed SNPs with MAF < 0.05 and info metric < 0.5 that has been compared in previous review58, which resulted in 4,889,047 imputed SNPs. In total 6,124,720 SNPs are passed to downstream eQTL analysis.
RNA sequencing data analysis pipeline
RNA-seq raw files in FASTQ format were processed in a customized pipeline. For each sample, we first filtered out reads that failed vendor check or are too short (<15nt) after removing the low-quality ends or possible adaptor contamination by using fastq-mcf with options of “-t 0 –x 10 –l 15 –w 4 –q 10 –u”. We then checked the quality using FastQC and generated k-mer profile using kpal59 for the remaining reads. Reads were then mapped to the human genome (GRCh37/hg19) using Tophat60 (v2.0.8) by allowing up to 2 mismatches and 100 multiple hits. Reads mapped to ribosomal RNAs or to the mitochondrial genome were excluded from downstream analysis. Gene expression levels were quantified using FPKM (Fragments Per Kilobase of transcript per Million mapped reads). Only uniquely mapped reads were used to estimate FPKM. To calculate normalized FPKM, we first ran Cuffquant61 (v2.2.1) with default arguments for genes annotated in GENCODE (v19), and then ran Cuffnorm with parameters of “-total-hits-norm –library-norm-method quartile” on the CBX files generated from Cuffquant.
Sample QC based on RNA-seq data
We performed sample QC similar to ‘t Hoen PA et al.62. In brief, we ran k-mer profiling for filtered reads using kpal59 and calculated the median profile distance for each sample. Samples with distances clearly different from the rest samples were marked as outliers (Supplementary Fig. 1c). We also calculated pair-wise Spearman correlations of gene expression quantification across samples and measured the median correlation (D-statistics) for each sample (Supplementary Fig. 1b,d). Samples with D-statistics markedly different from the rest of samples were deemed outliers. Moreover, we tested for concordance between reported clinical sex and sex indicated by the expression of female-specific XIST gene and male-specific Y-chromosome gene (Supplementary Fig. 1e). Samples from the first batch with a relative low sequencing depth were also excluded. In addition to these samples used for cell type-specific transcriptome analyses, various additional control samples were analyzed (e.g. amplification controls, tissue homogenate), and technical replicates (Supplementary Fig. 1f–h). At the end, 106 out of 115 samples passed QC and are used for downstream analysis (Supplementary Fig. 1a).
Defining the cumulative transcribed region by RNA-seq
Previously, ENCODE reported that in cell lines cumulatively 62.1% of the genome was transcribed with at least five mapped reads (Supplementary Table 11 of ref. 11). In our study, we rigorously accounted for sequencing depth and thus considered a genomic sequence as transcribed only if it had a read coverage of more than 0.05 RPM (unique reads per million). This approximately corresponds to 10 mapped reads (considering that for each sample we had on average 178 million mapped reads). With this rigorous definition, we showed that the cumulative coverage of transcribed regions in the dopamine neuron samples is 64.4%.
Defining catalogs of expressed ncRNAs and mRNAs
Normalized expression values of 106 samples passed QC were used as input. We first excluded genes with FPKM of zero in all 106 samples. Next, surrogate variable analysis and batch adjustment was performed using the sva63 and ComBat64 packages in R. In brief, the FPKM values were log10-transformed after adding a pseudocount of 0.0001. FPKM values within each group were adjusted for age, sex, and RIN as well as hidden covariates using frozen surrogate variable analysis (sva63). ComBat64 was used to adjust for batch effects. Median expression values for each gene were calculated for each cell type. To rigorously exclude low abundance genes, genes with median adjusted FPKM values < 0.01 in a cell type were not considered expressed in that cell type. GENCODE genes meeting these criteria were used to create a detailed catalog of mRNAs and ncRNAs expressed in a cell type.
Genes “exclusive” to dopamine neurons, pyramidal neurons, or non-neuronal cells, respectively, were defined as those, which achieved a median adjusted FPKM ≥ 0.01 in only one of these three cell types (with adjusted FPKMs < 0.01 in the other two cell types). We used the t-SNE package in R for t-Distributed Stochastic Neighbor Embedding analysis and the heatmap2 package for clustering and visualization purposes of cell type-exclusive ncRNAs and mRNAs.
Definition of TNE regions
A schematic of the TNE identification pipeline is shown in Fig. 1d and a flow chart in Supplementary Fig. 14a. TNE identification analysis was performed separately for each of the three cell types of dopamine neurons, pyramidal neurons, and non-neuronal cells, respectively. We first calculated the reads density values (in RPMs) at each genomic nucleotide position for all samples. We then calculated the aggregation signal for each cell type by computing the trimmed mean (e.g. trim the 10% of highest and lowest data points) of RPMs across the total N samples from the cell type of interest for each nucleotide position. We then scanned this aggregation signal in UCSC BegGraph format with a six-step filter:
-
(1)
scan each nucleotide position to filter for (keep for analysis) genomic regions with RPMs higher than the background level. The background level is defined as the average reads density across the nuclear genome (i.e. sum of all RPMs in a cell type divided by the total number of base-pairs comprising the nuclear genome (e.g. 3,095,677,412 for hg19). The borders of the selected genomic regions for each candidate TNE site are thus defined by the first and the last nucleotide for each TNE site that meets the RPM cutoff;
-
(2)
for each candidate region from step #1, require the summit RPM (i.e. maximal RPM in the region) to achieve a detection P value ≤ 0.05 compared to transcriptional background noise. The transcriptional background was defined by randomly selecting 1,000,000 single nucleotide positions outside of the EXCLUSION region (see Method) and calculating the distribution of their RPMs. The background signal was fitted to a normal distribution using the fitdist(x,’norm’) function in R. See Supplementary Fig. 14b for the distribution of background signals. Neighboring regions were merged into one region if the genomic distance between them was less than 100 bp;
-
(3)
exclude any regions overlapping with the EXCLUSION regions defined below (e.g. known genes, CAGE-defined promoters, and genomic gap regions);
-
(4)
require candidate regions to be longer than 100 bp;
-
(5)
exclude candidate regions containing junction sites that are supported by more than ten spliced reads in each of at least five samples. Junction sites were combined from the junctions.bed files of Tophat output.
-
(6)
For candidate regions meeting these criteria, we then required statistically significant expression across samples. We first computed the mean RPM values of each candidate region and then estimate the significance (P-value) compared to expression noise observed in random background regions of the sample. The P-value were computed by comparing the expression levels to the random background distribution of each sample, e.g. P = 1 - Fn(x), where Fn(x) is the empirical cumulative distribution function of expression levels of the same number of background regions with matched length randomly picked up beyond the EXCLUSION regions. Then for each candidate region, we computed the number of samples ‘called’ with a P-value ≤ 0.05 and calculated the probability of observing this number of ‘called’ samples by chance alone using a binomial distribution with the population probability set at 0.05. Finally, we rigorously corrected the binomial P-values for each candidate region for the total number of tests performed using Bonferroni correction. Candidate regions with Bonferroni-corrected P-values ≤ 0.05 were considered significantly expressed in the given cell type.
Regions excluded from the construction of random background regions
We defined “EXCLUSION” as a set of regions to exclude when constructing the random background regions. The EXCLUSION regions included any known transcribed regions (i.e. [−500, +500] bp of annotated exons from GENCODE (v19)65, UCSC known genes, lincRNA from NONCODE (v4)66, and rRNA from repeatMasker), FANTOM5 CAGE-defined promoters (i.e. [−500, +500] bp regions flanking the CAGE-predicted TSS), and genomic gap regions in the UCSC hg19 assembly.
The N of background regions picked for the analysis of dopamine neurons, pyramidal neurons, or non-neuronal cells equaled the N of 71,022, 37,007, and 19,690 TNEs detected in each of the three cell types, respectively. Background regions were randomly picked from the human genome (without the EXCLUSION regions) with length distributions matched to each TNE set, respectively.
Defining exclusive vs. shared TNEs
TNEs were further annotated into “shared” and “exclusive” classes depending on whether they overlap (i.e. at least 1nt) with TNEs detected in the other cell types. Cell type-exclusive TNEs were exclusively detected in one cell type. They do not overlap with TNEs detected in another cell type. TNEs detected in more than one cell type were termed “shared”. Infrequently, a dopamine neuron TNE overlapped with more than one pyramidal neuron TNE. Thus, in Fig. 1e the intersections between dopamine neuron TNEs and pyramidal neuron TNEs (or dopamine neuron TNE and non-neuronal TNE) shows the number of dopamine neuron TNEs that physically overlap with any pyramidal neuron TNE (or non-neuronal TNE). Similarly, the intersection between pyramidal neuron TNEs and non-neuronal TNEs (not shared with dopamine neurons) shows the number of pyramidal neuron TNEs that physically overlap with any non-neuronal TNE. The area-proportional Venn diagram was generated using eulerAPE67.
Characterization of TNEs using regulatory annotations
To explore the possible role of TNEs in gene regulation, we characterized TNEs with various known regulatory data in human brain (if available) or cell lines. For example, we used chromHMM ‘enhancer’ states in any of the ten human brain tissues in the Roadmap Epigenomics Project for histone-defined enhancers20,26. Enhancer is marked as the E6, E7 or E12 state from the 15-state chromHMM segmentation defined by five core marks, e.g. H3K4me3, H3K4me1, H3K36me3, H3K27me3, and H3K9me3. The ten brain tissues are Hippocampus Middle, Substantia Nigra, Anterior Caudate, Cingulate Gyrus, Inferior Temporal Lobe, Angular Gyrus, Dorsolateral Prefrontal Cortex, Germinal Matrix, Fetal Brain Female, and Fetal Brain Male. We used DNase-seq peak called in fetal brain of the Roadmap Epigenomics Project20 for DNase hypersensitivity sites. For TF binding, we used the TF ChIP-seq peak clusters (wgEncodeRegTfbsClusteredV3 from UCSC Genome Browser) from the ENCODE project25,68, which contains the most comprehensive TF ChIP-seq repository (so far). Other regulatory data include EP300 binding peaks from the ENCODE project23, CAGE-defined enhancers from the FANTOM5 project13, and sequence conservation score (phyloP) based on 100 vertebrate genomes comparison69.
By converting these features into binary codes (1 or 0) according to their presence or absence in TNE regions, we further built a simple classifier using these binary codes. For example, we defined a presence of TF binding hotspot if at least 5 distinct TFs ChIP-seq peaks found in a region. Epigenomic enhancer is presented if either of chromHMM ‘enhancer’ states (E6|E7|E12) is overlapped with the region. For conservation, we overlapped TNEs with HCNEs (highly conserved noncoding elements) defined in Ancora19 and defined “being conserved” if a TNE overlaps with a HCNE between human and zebrafish with at least 70% similarity and 50nt in length. We built the weighted classifier with relative 2-fold higher weight for DNase signal and implemented in R using the function daisy().
GWAS SNP enrichment analysis
We first downloaded the GWAS-associated SNPs from the NHGRI-EBI GWAS catalog70 (v1.0, downloaded on November 4, 2015), which includes 19,188 SNP-disease/trait associations after successfully lifting over back to the hg19 assembly. We then extended this set to 495,085 autosomal associations by including proxy SNPs imputed from the 1000 Genomes project. Proxy SNPs were extracted using SNAP71 from either of three populations in the 1000 Genomes Pilot 1 dataset with distance limit of 250kb and linkage disequilibrium (LD) r2 threshold of 0.8. Non-associated SNPs were extracted from dbSNP (build 137). We calculated the number of trait-associated and non-associated SNPs that physically localized (or did not localize) to TNE, promoters (unique locations of all GENCODE v19 protein-coding gene TSSs ± 200 bp), exons (unique locations of all GENCODE v19 protein-coding gene transcript inner exons), or random regions (100,000 genomic regions of 400 bp randomly selected beyond the TNEs, FANTOM5 permissive enhancers and EXCLUSION regions defined above), respectively. Only diseases/traits with more than three associated SNPs localizing to TNEs were considered for this analysis. For each genomic feature associated with a disease/traits with an odds ratio > 1, we performed a Fisher’s exact test. P values equal or below 9.64 ×10−6 (i.e. 0.01 divided by 1037, the total number of diseases/traits tested in NHGRI-EBI GWAS catalog as of November 4, 2015) were considered statistically significant.
Validating enhancer activity in HeLa S3 and neuroblastoma cells
PCR primers for the amplification of TNE-defined enhancer candidates and control regions from genomic DNA were designed using the Primer3web (v4.0.0)72, restriction sites of SalI and BamHI were separately added to 5′ end of the sense and antisense primer. Combined primer sequences were pre-validated with UCSC In-Silico PCR web tool, and synthesized by Thermo Fisher Scientific. All primers sequences are listed in Supplementary Table 6.
The modified vector of pGL4.10_mod3_EF1α was kindly provided by RIKEN and its structure was also described as Supplementary Figure 9d in their publication13. In brief, an EF1a basal promoter fragment was inserted into HindIII and NheI sites of the promoter-less pGL4.10 (Promega) to construct the pGL4.10EF1a vector, then the BamHI and SalI containing fragment (as the enhancer insertion site) was removed and re-inserted at the SpeI site located upstream of the synthetic poly(A) signal/transcriptional pause site to generate modified versions of pGL4.10EF1a vector. The introduction of the poly(A) site between the enhancers insertion site and the basal promoter is to avoid read-through from the enhancer, since we expect that many of our test elements are transcribed.
The PCR reaction was performed in 50 μl reaction to amplify each sequence of interest from 100 ng of human cerebellum tissue gDNA using One Taq DNA polymerase Kit (New England Biolabs). The PCR product was digested with BamHI and SalI (New England Biolabs), the restriction DNA fragment (insert) was isolated using agarose gel electrophoresis and purified by the MinElute Gel Extraction Kit (Qiagen). The pGL4.10_mod3_EF1α vector was also digested with BamHI and SalI, the double digested DNA (vector) was isolated and purified in the same way as insert. 100 ng of insert and 20 ng of vector were ligated in 10 μl reaction using T4 DNA Ligase (New England Biolabs). 1 μl of ligation reaction was transformed to 100 μl of DH5α competent cells (Invitrogen). Positive colonies were selected by colony PCR and correct insertion in the plasmid was confirmed by sequencing. Cloned plasmids for transfections were purified using the QIAamp DNA Midi Kit (Qiagen).
HeLa-S3 cells were cultured in MEM (Gibco) supplemented with 10% FBS (Gibco), 100 U/ml penicillin and streptomycin (Gibco). SK-N-MC neuroblastoma cells were cultured in DMEM (Gibco) supplemented with 10% FBS (Nichirei Bioscience Inc.), and MEM (WAKO) supplemented with 10% FBS (Gibco), 100 U/ml penicillin and streptomycin (Gibco). 7.5 × 103 cells per well of HeLa-S3 cells and 4 × 104 cells per well SK-N-MC were seeded in 96 well plates 24 hours before transfection.
190 ng of plasmids inserted with the PCR products and 10 ng of pGL4.73 Renilla luciferase plasmid (Promega) were co-transfected into HeLa-S3 and SK-N-MC cells respectively using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instruction. Each transfection was independently performed three times. After 24 hours, the luciferase activities were measured by Gen5 Microplate Reader (BioTek) using the Dual-glo luciferase assay system (Promega) according to the manufacturer’s instruction.
Validating enhancer activity in zebrafish
Selected TNEs with potential enhancer activity and one negative control element (non-conserved intergenic sequence region with very low or no signal for enhancer marks, e.g. DNase I hypersensitivity, H3K4me1 and H3K27ac) were amplified from human genomic DNA using primers (Supplementary Table 6). PCR products were purified using NucleoSpin Gel and PCR Clean-up Kit (Macherey-Nagel) and cloned upstream of zebrafish gata2 promoter73 linked to mRuby2 reporter gene into modified pDB896 vector (a gift from D. Balciunas, Temple University). The cloning procedures were performed using In-Fusion HD Cloning Kit (Clontech) according the manufacture instructions into BamHI linearized vector. Plasmid DNA was purified using QIAGEN-tip 20 miniprep kit (QIAGEN) and verified by restriction digest and sequencing.
Zebrafish stocks (Danio rerio) were kept and used according to Home Office regulations (UK) at the University of Birmingham. For these experiments the enhancer trap transgenic line ETvmat2:GFP74 was used. Adults were crossed pairwise and eggs were collected and injected within 20 min after fertilization. Microinjection solutions contained 20 ng/μl of plasmid DNA, 0.1% of phenol red (Sigma). Injections were performed through the chorion and into the cytoplasm of zygotes using an analogue microinjector MINJ-1 (Tritech Research). About 150–200 eggs were injected per construct and experiments were replicated at least three times. Embryos were kept in E3 Medium containing 50 μg/ml gentamicin (Thermo Fisher Scientific) and 0.003% phenylthiourea (Sigma) at 28.5 °C.
Injected embryos were screened for expression during the first 5d postfertilization and group images were taken on Zeiss Axio Zoom V16 stereo microscope. Selected embryos showing specific expression pattern were imaged at the relevant developmental stage on a Zeiss Lightsheet Z1 microscope with 20× objective and 0.5 optical zoom. Stacks containing 250–300 slices with 2-μm thickness were acquired, and maximum intensity projections were made using Zeiss ZEN Black Software.
eQTL analysis pipeline
The eQTL analysis was performed for both GENCODE genes and TNEs using the 84 subjects for which lcRNAseq data from dopamine neurons as well as genotyping data were available. For genes, we first filtered for genes with >0.05 FPKM in at least 10 individuals, then transformed FPKM to rank normalized gene expression. In brief, the FPKM values were log10-transformed (adding a pseudocount of 0.01). The measurements for each gene were transformed into normally distributed while preserving relative rankings (quantile normalization) and the mean and standard deviation of the original measurement. For TNEs, the expression distribution is close to a normal distribution and thus quantile normalization was not indicated. Moreover, our TNE identification method already selects for TNE pervasively expressed across multiple individuals. We then performed surrogate variable analysis (SVA) with the sva R package63 to adjust the effects of known covariates, including batch, age, gender, RIN, PMI, and reads length. Adjusted expression values extracted from fsva() function were used for downstream eQTL analysis. We used RLE (relative log expression) plots to visually inspect the effects of covariates adjustment. We also filtered out SNPs with missing values or with MAF ≤ 0.05 in the 84 subjects. Matrix-eQTL33 was applied for cis-eQTL analysis, with the cis window defined as 1 megabase between the SNP and the nearest end of a gene or TNE. Nominal P-values were generated for SNP-gene pairs in linear regression mode. See Supplementary Fig. 13b for detail.
TNE-host gene function enrichment analysis
151 cis-regulated TNEs physically localized to introns of 102 host genes. Gene set enrichment analysis was performed using the C5 gene sets (GO terms) implemented in the MSigDB database using the hypergeometic test. Each gene set contains genes annotated to the same GO term. For each gene set, the hypergeometric test was performed for k-1, K, N - K, n; where k is the number of TNE host genes genes that are part of a GO term gene set; K is the total number of genes annotated to the same GO term gene set; N is the total number of all known human genes; and n is the number of genes in the query set. The top 50 GO terms enriched in these TNE host genes are shown in Supplementary Table 8 (all with a FDR q value < 0.05).
We also evaluated whether there is a specific enrichment among cis-regulated TNEs in genes associated with brain disorders. We used diseases in MeSH C10 (Nervous System Diseases) or F03 (Mental Disorders) for brain disorders, and associated disease to genes using GenDisNet database. The disease-gene association was extracted from DisGeNet76 (http://www.disgenet.org/) filtered with GDA score > 0.1. For all annotated protein-coding genes, we performed Fisher’s exact test based on whether a gene is associated with brain disorder and a gene hosts a cis-eQTL TNE (Supplementary Table 9).
TF binding motif enrichment analysis
For TFs with ENCODE ChIP-Seq, we extracted their peak coordinates from the wgEncodeRegTfbsClusteredV3 file downloaded from UCSC Genome Browser, which contains 4,380,444 TF binding peaks from 161 TFs in total. We also downloaded 579 non-redundant TF motifs in vertebrate from JASPAR (version 2018)77 and then scanned the whole genome with the motifs using the program FIMO78 (default parameters with P value < 10−4) to get 418,034,884 putative binding sites. For each TF, Fisher’s exact test was performed to see if observed occurrences of TF peaks (for ENCODE ChIP-Seq) or binding sites (for JASPAR motifs) in TNEs are significantly enriched than expected. In brief, for each TF in JASPAR, we assigned the full set of putative binding regions of JASPAR motifs to one cell of the 2×2 table, according to if a region is bound by the TF or not, and if it’s overlapped with TNE or not. So, each TF has a 2×2 table for Fisher’s exact test. This is similarly done for ENCODE TF ChIP-seq peaks.
We also tested the TF motif enrichment against random genomic sequences that are GC- and length-matched. We first extracted the GC- and length- matched random genomic background regions using the GC_compo (http://opossum.cisreg.ca/GC_compo/) and then tested motif enrichment using the AME program in MEME suite for all 579 non-redundant TF motifs in vertebrate from JASPAR CORE 2018.
Causality analysis for TNE-, ncRNA-, and mRNA-eQTLs
We used the Relative Trait Concordance (RTC) method to integrate QTL and GWAS data to detect potential disease-causing cis-regulatory effects according to the method described in Ref. 34. Using this method, an RTC score of 1 or near 1 indicates a potentially causal cis-regulatory effect.
In order to reduce the redundancy in the output of the RTC analysis due to SNPs in strong LD, we pruned the result using the following rules. If multiple eSNPs shared the same LD block with a GWAS SNP, we only took the eSNP with best RTC score for each GWAS variant-transcript pair. If multiple eSNPs achieved the exact same top RTC score and they included the GWAS-derived variant itself, we selected the GWAS variant as top eSNP. If multiple variants achieved the exact same top RTC score (but did not include the GWAS-derived variant itself), then we arbitrarily picked one of these top-scoring eSNPs as representative eSNP. The pruned result is shown in Supplementary Table 10.
Three haplotype blocks were defined for the chr17q21 locus by plink2 (plink --blocks --blocks-max-kb 1000) using the CEU subpopulation (n = 99) in the 1000 Genome Project.
Conditional eQTL analysis was performed for the chr17q21 locus by including the rs17649553 genotype as an additional covariate. All eQTL pairs for genes/TNEs and SNPs in the locus (chr17:43,000,000–45,300,000 in hg19) are displayed in Supplementary Fig. 11. Majority of significant eQTL SNPs become insignificant after conditional analysis of rs17649553, except 31 SNPs in the KANSL1 gene (green dots on the top-right corner) and one SNP in NSF gene (red dot on the top-right corner). The 31 SNPs are in the same LD block as rs17649553.
Confirming TNE and mRNA expression by qPCR
Quantitative PCR was performed using SYBR Green Master Mix (Life Technologies) on an ABI 7900HT instrument (Applied Biosystems). Primer sequences are shown in Supplementary Table 6. In order to confirm the expression of lcRNAseq-derived TNE and mRNAs in dopamine neurons and pyramidal neurons, relative abundances of target TNE or mRNAs were evaluated by qPCR in linearly amplified laser-captured, microdissected samples from human substantia nigra or temporal cortex, as well as in linearly amplified human fibroblast and PBMC samples (shown in Fig. 3b). TNE and mRNA expression was further confirmed in SK-N-MC human neuroblastoma cells and Human Universal Reference RNA (not shown). The human reference gene GUSB was used to normalize for RNA loading. Control samples lacking template and those lacking reverse transcriptase showed virtually no expression of these target TNEs and mRNAs indicating that primer dimers or DNA contamination did not materially influence results. Expression values were analyzed using the comparative threshold cycle method24. Equal amplification efficiencies for target and reference transcripts were confirmed using melting curve analysis.
Evaluation of the chromosome 17q21 eQTL in a second, independent cohort of 31 individuals by qPCR
Postmortem brain samples from 31 individuals were analyzed. These individuals were without a clinical or neuropathological diagnosis of neurodegenerative disease and met the inclusion and exclusion criteria described in the "Sample collection and processing" section. These new brain samples were obtained from Banner Sun Health Institute, Brain Tissue Center at Massachusetts General Hospital, and University of Kentucky ADC Tissue Bank. Pyramidal neurons were laser-captured from the middle temporal gyrus of each of the 31 individuals and linearly amplified as described in the "Sample collection and processing" section.x These samples showed exceptional quality as documented by a median RNA integrity number 7.7 and a median post-mortem interval of 2.9 hours (Supplementary Table 12). Relative expression abundances of the two target transcripts, KANSL1-TNE1 and LRRC37A4P, were assayed using SYBR Green qPCR (Life Technologies). The geometric mean of two reference genes, EIF4A2 and RPL13, was used to control for RNA loading. Control samples lacking template and those lacking reverse transcriptase showed virtually no detectable expression. Relative expression abundance of each of the target genes was compared in subjects carrying one or two risk alleles (CT or TT) and those without risk allele (CC) at the rs17649553. A two-tailed Student’s homoscedastic t-test was used to determine statistical significance. Data were visualized in Supplementary Fig. 12.
Technical confirmation of lcRNAseq eQTL results in laser-captured dopamine neurons by qPCR
We confirmed the lcRNAseq-based dopamine neuron eQTL for KANSL1-TNE1 and LRRC37A4P, respectively, using SYBR Green qPCR (Life Technologies). The geometric mean of two reference genes, EIF4A2 and RPL13, was used to control for RNA loading. For this confirmatory experiment, laser-captured dopamine neuron samples from 35 substantia nigra samples (also used for lcRNAseq) were analyzed. Data were visualized in Supplementary Fig. 12.
Post-mortem brain CAGE methods
Four human post-mortem brains (healthy controls) were obtained from University of Maryland, University of Washington, and McLean Hospital, with the same inclusion/exclusion criteria as described above. Substantia nigra tissue samples were utilized for Cap Analysis Gene Expression (CAGE). 5 μg of total RNA was exacted from each sample using the RNeasy RNA Kit (Qiagen) with an RNA integrity number (RIN) > 6. Use of postmortem samples for expression analysis was approved by the IRB of Brigham & Women’s Hospital.
Libraries were constructed using a published CAGEseq protocol adapted for next-generation sequencing79. Briefly, complementary DNA (cDNA) was synthesized from total RNA using random primers, and this process was carried out at high temperature in the presence of trehalose and sorbitol to extend cDNA synthesis through GC-rich regions in 5′ untranslated regions. The 5′ ends of messenger RNA within RNA-DNA hybrids were selected by the cap-trapper method and ligated to a linker so that an EcoP15I recognition site was placed adjacent to the start of the cDNA, corresponding to the 5′ end of the original messenger RNA. This linker was used to prime second-strand cDNA synthesis. Subsequent EcoP15I digestion released the 27-base pair (bp) CAGEseq reads. After ligation of a second linker, CAGEseq tags were polymerase chain reaction amplified, purified, and sequenced on the HiSeq 2000 (Illumina) using standard protocol for 50 bp single end runs.
CAGEseq data were filtered for CAGEseq artifacts using TagDust80 (version 1.12), removal of reads mapping to known ribosomal RNA genes and low quality reads, mapping to the human genome (hg19) using Burrows-Wheeler Aligner (version 0.5.9) for short reads. Reads mapping to autosomes were used to minimize gender and normalization biases for subsequent analysis. Normalization was done based on the amount of reads per million sequence reads.
Data collection, statistical analysis and data presentation
Sample sizes were based on the total number of available high-quality brain samples that met inclusion and exclusion criteria. No statistical methods were used to pre-determine sample sizes but our sample sizes are consistent with those recommended by the Genotype-Tissue Expression Consortium81. No randomization of data collection was performed in this study. Brains were selected based on pre-defined inclusion and exclusion criteria (see above). Sample outliers were rationally identified as described in the Section on Sample QC based on RNA-seq data. TNE were defined in a rigorous six-step process as detailed in the Section Definition of TNE regions. Data were not excluded based on arbitrary post-hoc considerations. Data collection and analysis were not performed blind to the conditions of the experiments. Data distribution was assumed to be normal but this was not formally tested, except that the normality of transcriptional background signal was checked by visual inspection.
R (The R Foundation for Statistical Computing, Vienna, Austria) was used for other statistical tests. Box plots were used to present multi-groups comparison. In all box plots, center line represents the median value; box limits, first and third quartiles; whiskers, the most extreme data point which is no more than 1.5 times the interquartile range from the box.
Statistical tests used in each figure: Fig. 4b, two-tailed Student’s t-test; Fig. 4h, hypergeometric test; Fig. 5b, one-sided Fisher’s exact test; Fig. 5c, linear regression model in Matrix-eQTL; Fig. 5d, meta-GWAS from www.pdgene.org. Fig. 5g, two-sided Student’s t-test; Supplementary Fig. 5b–d, one-sided Fisher’s exact test; Supplementary Fig. 6c, hypergeometric test; Supplementary Fig. 10a, linear regression model in Matrix-eQTL; Supplementary Fig. 10b, linear regression model in Matrix-eQTL (for 3 groups comparison) or two-sided Student’s t-test (for two groups comparison); Supplementary Fig. 11, linear regression model in Matrix-eQTL; Supplementary Fig. 12, two-sided Student’s t-test.
Reporting Summary
Further information on experimental design is available in the Nature Research Reporting Summary linked to this article.
Code availability
Custom code associated with this study is available upon reasonable request.
Data availability
RNA-seq and genotyping raw data have been deposited in dbGAP under accession number phs001556.v1.p1. The proceeded data and eQTL result for BRAINcode project can be queried at http://www.humanbraincode.org through a user-friendly interface. Other data supporting the findings of this study are available upon reasonable request.
Supplementary Material
Acknowledgements
We thank H. Suzuki and T. Suzuki of RIKEN for providing the modified pGL4.10_mod3_EF1α vector and consultation. We are grateful to C. Vanderburg of the Advanced Tissue Resource Center, Massachusetts General Hospital, for his expertise and support. We thank Z. Weng at the University of Massachusetts Medical School for sharing additional data from the ENCODE consortium. We thank A. Sandelin and R. Andersson, both from Copenhagen University; A. Regev, Broad Institute; and M. Feany, Brigham & Women's Hospital, for insightful comments and guidance. We thank C Liu, A Shieh, and T Goodman for assisting to extract the RNA-Seq and ATAC-seq data in BrainGVEX dataset. We gratefully acknowledge the Banner Sun Health Institute, Massachusetts Alzheimer’s Disease Research Center at Massachusetts General Hospital, Harvard Brain Tissue Resource Center at McLean Hospital, University of Kentucky ADC Tissue Bank, University of Maryland Brain and Tissue Bank, Pacific Northwest Dementia and Aging Neuropathology Group at University of Washington Medicine Center, and Neurological Foundation of New Zealand for providing human brain tissue.
This study was funded in part by NIH grant U01 NS082157 and the U.S. Department of Defense (to C.R.S.); NIH R01AG057331 (to C.R.S.) funded RNAseq of pyramidal neurons; with additional support from the Michael J. Fox Foundation (MJFF) (to C.R.S. and C.H.A., respectively); the Australia NHMRC GNT1067350 (to A.C. and J.M.); NIA P30 AG028383 (to P.T.N.); U.K. Wellcome Trust Investigator award (to F.M.); NINDS U24 NS072026 National Brain and Tissue Resource for Parkinson’s Disease and Related Disorders (to T.G.B. and C.H.A.); NIA P50 AG005134 (to M.P.F.).
The MSBB data were generated as part of the AMP-AD Consortium from postmortem brain tissue collected through the Mount Sinai VA Medical Center Brain Bank and were provided by Dr. Eric Schadt from Mount Sinai School of Medicine.
PsychENCODE data were generated as part of the PsychENCODE Consortium, supported by: U01MH103339, U01MH103365, U01MH103392, U01MH103340, U01MH103346, R01MH105472, R01MH094714, R01MH105898, R21MH102791, R21MH105881, R21MH103877, and P50MH106934 awarded to: Schahram Akbarian (Icahn School of Medicine at Mount Sinai), Gregory Crawford (Duke), Stella Dracheva (Icahn School of Medicine at Mount Sinai), Peggy Farnham (USC), Mark Gerstein (Yale), Daniel Geschwind (UCLA), Thomas M. Hyde (LIBD), Andrew Jaffe (LIBD), James A. Knowles (USC), Chunyu Liu (UIC), Dalila Pinto (Icahn School of Medicine at Mount Sinai), Nenad Sestan (Yale), Pamela Sklar (Icahn School of Medicine at Mount Sinai), Matthew State (UCSF), Patrick Sullivan (UNC), Flora Vaccarino (Yale), Sherman Weissman (Yale), Kevin White (UChicago) and Peter Zandi (JHU).
Footnotes
Competing Interests
C.R.S. has collaborated with Pfizer, Sanofi; has consulted for Sanofi; has served as Advisor to the Michael J. Fox Foundation, NIH, Department of Defense; is on the Scientific Advisory Board of the American Parkinson Disease Association; has received funding from the NIH, the U.S. Department of Defense, the Michael J. Fox Foundation, and the American Parkinson Disease Association. C.R.S. is named as co-inventor on two US patent applications on biomarkers for PD held in part by Brigham & Women’s Hospital. B.G. is the founder of Pacific Analytics PTY LTD, Australia; a founding member of the International Cerebral Palsy Genetics Consortium; a member of the Australian Genomics Health Alliance and is on the Scientific Advisory Board of Iggy Get Out!, Australia. T.G.B provides consultancies to Prothena and GSK; is on the Advisory Board of Vivid Genomics; has contracted research with Avid Radiopharmaceuticals, Navidea Biopharmaceuticals, and Aprinoia Therapeutics. The other authors declare no competing financial interests.
References
- 1.Cookson W, Liang L, Abecasis G, Moffatt M & Lathrop M Mapping complex disease traits with global gene expression. Nat Rev Genet 10, 184–194 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.ENCODE Project Consortium et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447, 799–816 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heintzman ND et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459, 108–12 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kowal SL, Dall TM, Chakrabarti R, Storm MV & Jain A The current and projected economic burden of Parkinson’s disease in the United States. Mov. Disord 28, 311–318 (2013). [DOI] [PubMed] [Google Scholar]
- 5.Cloutier M. et al. The economic burden of schizophrenia in the United States in 2013. J. Clin. Psychiatry 77, 764–771 (2016). [DOI] [PubMed] [Google Scholar]
- 6.National Institute of Drug Abuse. Treatment Statistics. (2011). at <https://www.drugabuse.gov/publications/drugfacts/treatment-statistics>
- 7.Hassan A & Benarroch EE Heterogeneity of the midbrain dopamine system. Neurology 85, 1795–1805 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Zheng B et al. PGC-1α, a potential therapeutic target for early intervention in Parkinson’s disease. Sci. Transl. Med 2, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liang WS et al. Neuronal gene expression in non-demented individuals with intermediate Alzheimer’s Disease neuropathology. Neurobiol. Aging 31, 549–66 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elstner M et al. Neuromelanin, neurotransmitter status and brainstem location determine the differential vulnerability of catecholaminergic neurons to mitochondrial DNA deletions. Mol Brain 4, 43 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Djebali S et al. Landscape of transcription in human cells. Nature 489, 101–108 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim T-K et al. Widespread transcription at neuronal activity-regulated enhancers. Nature 465, 182–187 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andersson R et al. An atlas of active enhancers across human cell types and tissues. Nature 507, 455–61 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Core LJ et al. Analysis of nascent RNA identifies a unified architecture of initiation regions at mammalian promoters and enhancers. Nat. Genet 46, 1311–1320 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thurman RE et al. The accessible chromatin landscape of the human genome. Nature 489, 75–82 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heintzman ND et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet 39, 311–318 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Visel A et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature 457, 854–8 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yip KY et al. Classification of human genomic regions based on experimentally determined binding sites of more than 100 transcription-related factors. Genome Biol. 13, R48 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Engström PG, Fredman D & Lenhard B Ancora: a web resource for exploring highly conserved noncoding elements and their association with developmental regulatory genes. Genome Biol. 9, R34 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Consortium RE et al. Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Akbarian S et al. The PsychENCODE project. Nature Neuroscience 18, 1707–1712 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buenrostro JD, Wu B, Chang HY & Greenleaf WJ ATAC-seq: A method for assaying chromatin accessibility genome-wide. Curr. Protoc. Mol. Biol 2015, 21.29.1–21.29.9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mittal S et al. β2-Adrenoreceptor is a regulator of the α-synuclein gene driving risk of Parkinson’s disease. Science 357, 891–898 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scherzer CR et al. GATA transcription factors directly regulate the Parkinson’s disease-linked gene alpha-synuclein. Proc. Natl. Acad. Sci. U. S. A 105, 10907–12 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ernst J & Kellis M ChromHMM: automating chromatin-state discovery and characterization. Nat. Methods 9, 215–216 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ellingsen S et al. Large-scale enhancer detection in the zebrafish genome. Development 132, 3799–811 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Visel A, Minovitsky S, Dubchak I & Pennacchio LA VISTA Enhancer Browser--a database of tissue-specific human enhancers. Nucleic Acids Res. 35, D88–D92 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Welter D et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 42, D1001–6 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang Y et al. A Genetic Screen To Assess Dopamine Receptor (DopR1) Dependent Sleep Regulation in Drosophila. G3 (Bethesda). 6, 4217–4226 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.González S et al. Circadian-Related Heteromerization of Adrenergic and Dopamine D4 Receptors Modulates Melatonin Synthesis and Release in the Pineal Gland. PLoS Biol. 10, e1001347 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Breen DP et al. Sleep and circadian rhythm regulation in early Parkinson disease. JAMA Neurol. 71, 589–95 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shabalin AA Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinformatics 28, 1353–1358 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nica AC et al. Candidate causal regulatory effects by integration of expression QTLs with complex trait genetic associations. PLoS Genet. 6, e1000895 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nalls M. a et al. Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson’s disease. Nat. Genet 46, 989–93 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu H et al. Tissue-Specific RNA Expression Marks Distant-Acting Developmental Enhancers. PLoS Genet. 10, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mercer EM et al. Multilineage Priming of Enhancer Repertoires Precedes Commitment to the B and Myeloid Cell Lineages in Hematopoietic Progenitors. Immunity 35, 413–425 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ostuni R et al. Latent enhancers activated by stimulation in differentiated cells. Cell 152, 157–171 (2013). [DOI] [PubMed] [Google Scholar]
- 39.Koolen D a et al. Mutations in the chromatin modifier gene KANSL1 cause the 17q21.31 microdeletion syndrome. Nat. Genet 44, 639–641 (2012). [DOI] [PubMed] [Google Scholar]
- 40.Li R et al. Six Novel Susceptibility Loci for Early-Onset Androgenetic Alopecia and Their Unexpected Association with Common Diseases. PLoS Genet. 8, e1002746 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Adams HHH et al. Novel genetic loci underlying human intracranial volume identified through genome-wide association. Nat. Neurosci 19, 1569–1582 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Torsney KM et al. Bone health in Parkinson’s disease: a systematic review and meta-analysis. J Neurol Neurosurg Psychiatry 85, 1159–1166 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ding H et al. Unrecognized vitamin D3 deficiency is common in Parkinson disease: Harvard Biomarker Study. Neurology 81, 1531–1537 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saliba AE, Westermann AJ, Gorski SA & Vogel J Single-cell RNA-seq: Advances and future challenges. Nucleic Acids Research 42, 8845–8860 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lake BB et al. Neuronal subtypes and diversity revealed by single-nucleus RNA sequencing of the human brain. Science 352, 1586–1590 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Je A et al. Fluorescent in situ sequencing (FISSEQ) of RNA for gene expression profiling in intact cells and tissues. Nat. Protoc 10, 442–58 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only References
- 47.Hughes AJ, Daniel SE, Kilford L & Lees AJ Accuracy of clinical diagnosis of idiopathic Parkinson’s disease: a clinico-pathological study of 100 cases. J. Neurol. Neurosurg. Psychiatry 55, 181–4 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.The National Institute on Aging and Reagan Institute Working Group on Diagnostic Criteria for the Neuropathological Assessment of Alzheimer’s Disease. Consensus Recommendations for the Postmortem Diagnosis of Alzheimer’s Disease. Neurobiol. Aging 18, S1–S2 (1997). [PubMed] [Google Scholar]
- 49.Bonanni L, Thomas A, Onofrj M & McKeith IG Diagnosis and management of dementia with Lewy bodies: Third report of the DLB Consortium. Neurology 66, 1455–1455 (2006). [DOI] [PubMed] [Google Scholar]
- 50.Schroeder A et al. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol. Biol 7, 3 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Unni VK, Ebrahimi-Fakhari D, Vanderburg CR, McLean PJ & Hyman BT Studying protein degradation pathways in vivo using a cranial window-based approach. Methods 53, 194–200 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ingelsson M et al. No alteration in tau exon 10 alternative splicing in tangle-bearing neurons of the Alzheimer’s disease brain. Acta Neuropathol. 112, 439–449 (2006). [DOI] [PubMed] [Google Scholar]
- 53.Liu G et al. Metal exposure and Alzheimer’s pathogenesis. J. Struct. Biol 155, 45–51 (2006). [DOI] [PubMed] [Google Scholar]
- 54.Kurn N Novel Isothermal, Linear Nucleic Acid Amplification Systems for Highly Multiplexed Applications. Clin. Chem 51, 1973–1981 (2005). [DOI] [PubMed] [Google Scholar]
- 55.Faherty SL, Campbell CR, Larsen PA & Yoder AD Evaluating whole transcriptome amplification for gene profiling experiments using RNA-Seq. BMC Biotechnol. 15, 65 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chang CC et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4, 7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Delaneau O, Zagury J-F & Marchini J Improved whole-chromosome phasing for disease and population genetic studies. Nat. Methods 10, 5–6 (2013). [DOI] [PubMed] [Google Scholar]
- 58.Marchini J & Howie B Genotype imputation for genome-wide association studies. Nat. Rev. Genet 11, 499–511 (2010). [DOI] [PubMed] [Google Scholar]
- 59.Anvar S et al. Determining the quality and complexity of next-generation sequencing data without a reference genome. Genome Biol. 15, 555 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Trapnell C, Pachter L & Salzberg SL TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Trapnell C et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–78 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.‘t Hoen PAC et al. Reproducibility of high-throughput mRNA and small RNA sequencing across laboratories. Nat. Biotechnol. 31, 1015–22 (2013). [DOI] [PubMed] [Google Scholar]
- 63.Leek JT & Storey JD Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet. 3, 1724–1735 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Johnson WE, Li C & Rabinovic A Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8, 118–127 (2007). [DOI] [PubMed] [Google Scholar]
- 65.Harrow J et al. GENCODE: The reference human genome annotation for the ENCODE project. Genome Res. 22, 1760–1774 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhao Y et al. NONCODE 2016: An informative and valuable data source of long non-coding RNAs. Nucleic Acids Res. 44, D203–D208 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Micallef L & Rodgers P eulerAPE: Drawing Area-Proportional 3-Venn Diagrams Using Ellipses. PLoS One 9, e101717 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang J et al. Factorbook. org: a Wiki-based database for transcription factor-binding data generated by the ENCODE consortium. Nucleic Acids Res. (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pollard KS, Hubisz MJ, Rosenbloom KR & Siepel A Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 20, 110–121 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.MacArthur J et al. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res. 45, D896–D901 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Johnson AD et al. SNAP: A web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics 24, 2938–2939 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Untergasser A et al. Primer3—new capabilities and interfaces. Nucleic Acids Res. 40, e115–e115 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Meng a, Tang H, Ong B. a, Farrell MJ & Lin S Promoter analysis in living zebrafish embryos identifies a cis-acting motif required for neuronal expression of GATA-2. Proc. Natl. Acad. Sci. U. S. A 94, 6267–6272 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wen L et al. Visualization of monoaminergic neurons and neurotoxicity of MPTP in live transgenic zebrafish. Dev. Biol. 314, 84–92 (2008). [DOI] [PubMed] [Google Scholar]
- 75.Forster B, Van De Ville D, Berent J, Sage D & Unser M Complex wavelets for extended depth-of-field: A new method for the fusion of multichannel microscopy images. Microsc. Res. Tech 65, 33–42 (2004). [DOI] [PubMed] [Google Scholar]
- 76.Piñero J et al. DisGeNET: A comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 45, D833–D839 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mathelier A et al. JASPAR 2016: A major expansion and update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 44, D110–D115 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Grant CE, Bailey TL & Noble WS FIMO: Scanning for occurrences of a given motif. Bioinformatics 27, 1017–1018 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Takahashi H, Lassmann T, Murata M & Carninci P 5’ end-centered expression profiling using cap-analysis gene expression and next-generation sequencing. Nat. Protoc 7, 542–61 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lassmann T, Hayashizaki Y & Daub CO TagDust - A program to eliminate artifacts from next generation sequencing data. Bioinformatics 25, 2839–2840 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Aguet F et al. Genetic effects on gene expression across human tissues. Nature (2017). doi: 10.1038/nature24277 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq and genotyping raw data have been deposited in dbGAP under accession number phs001556.v1.p1. The proceeded data and eQTL result for BRAINcode project can be queried at http://www.humanbraincode.org through a user-friendly interface. Other data supporting the findings of this study are available upon reasonable request.