Abstract
Cell fate specification is mediated primarily through the expression of cell-type-specific genes. The regulatory pathway that governs the sperm/egg decision in the hermaphrodite germ line of Caenorhabditis elegans has been well characterized, but the transcription factors that drive these developmental programs remain unknown. We report the identification of ELT-1, a GATA transcription factor that specifies hypodermal fate in the embryo, as a regulator of sperm-specific transcription in the germ line. Computational analysis identified a conserved bipartite sequence element that is found almost exclusively in the promoters of a number of sperm genes. ELT-1 was recovered in a yeast one-hybrid screen for factors that bind to that sperm consensus site. In vitro assays defined the sperm consensus sequence as an optimal binding site for ELT-1. We determined that expression of elt-1 is elevated in the sperm-producing germ line, and that ELT-1 is required for sperm function. Deletion of the ELT-1 binding site from a sperm promoter abrogates sperm-specific expression of a reporter transgene. This work demonstrates a role for the ELT-1 transcription factor in sperm, and provides a critical link between the germ line sex determination program and gamete-specific gene expression.
Keywords: Germ line, Spermatogenesis, GATA factor, Gene expression
Introduction
A recurring theme in development is the specification of cell fate through the activities of transcription factors that regulate cell-type-specific gene expression. This fundamental mechanism of control extends from relatively simple systems, such as mating-type determination in the unicellular Saccharomyces cerevisiae by proteins encoded by the MAT locus, through the interplay of a variety of transcription factors that pattern the developing Drosophila embryo, to the less-well-characterized complexity of mammalian development. In some instances (e.g., MyoD) (Tapscott et al., 1988) expression of a single transcription factor is capable of directing a particular developmental program and specifying a single cell type from among multiple potential fates. Such master switch genes can directly regulate expression of additional transcription factors (as well as cell-type structural genes) that further promote cell-type-specific gene expression (including other transcription factors and structural genes). This transcriptional cascade produces a unique combination of regulatory proteins, and combinatorial control of gene expression provides a mechanism whereby the same transcription factor can be employed in different tissues or at different times of development to govern different sets of targets.
Genome-scale microarray screens of the nematode Caenorhabditis elegans have demonstrated that transcriptional regulation also underlies the determination of cell type in the germ line of this hermaphroditic organism. A population of mitotically dividing stem cells gives rise to first male and then female gametes. Comparisons of transcriptional profiles during spermatogenesis or oogenesis have identified hundreds of genes that are differentially regulated (Reinke et al., 2000, 2004). Recent work indicates that, for sperm genes, transcriptional control is the primary mechanism of regulation (Merritt et al., 2008). Because the C. elegans germ line stem cell is restricted to one of the two choices, it provides a simple model for the investigation of metazoan cell fate specification.
The sex determination program that governs male vs. female sexual fate in C. elegans has been well studied (reviewed in Zarkower, 2006). The ratio of X chromosomes to autosomes controls a signaling pathway that governs the activity of TRA-1, which shares homology with the Drosophila cubitus interruptus and mammalian GLI family of zinc finger transcription factors (Hodgkin, 1993; Zarkower and Hodgkin, 1992). In the soma, TRA-1 is the terminal regulator of sexual identity and acts as a master switch to promote the female fate while inhibiting the male fate. On the basis of homology and its sequence-specific DNA binding activity (Zarkower and Hodgkin, 1993), TRA-1 was proposed to activate transcription of genes required for female development and/or repress transcription of genes needed for male development. This model was supported by the identification of mab-3, which encodes a doublesex homolog required for male somatic development, as a putative target of TRA-1-mediated regulation (Raymond et al., 1998; Shen and Hodgkin, 1988; Yi et al., 2000). Subsequent targets of TRA-1 likewise exhibit roles in sexually dimorphic programs of neuronal cell death (egl-1, ceh-30) or tail development (dmd-3) (Conradt and Horvitz, 1999; Mason et al., 2008; Peden et al., 2007; Schwartz and Horvitz, 2007).
As the only predicted transcription factor in the sex determination pathway, TRA-1 is an attractive candidate as a direct regulator of gamete-type-specific gene expression. However, several lines of evidence suggest that other transcription factors must mediate sperm or oocyte-specific transcription. First, animals with tra-1 null alleles are capable of producing both sperm and oocytes (Hodgkin, 1987; Schedl et al., 1989), demonstrating that specification and cell-type-specific gene expression can occur independently of TRA-1. Second, in contrast to the soma, TRA-1 is not the final determinant of gamete identity in the germ line sex determination pathway. Rather, a number of genes that do not encode transcription factors are nonetheless epistatic to tra-1 in the specification of gamete cell type. The FEM proteins, which form part of a CUL-2 ubiquitin ligase complex (Starostina et al., 2007), function downstream of TRA-1 to promote spermatogenesis in the germ line (Doniach and Hodgkin, 1984; Hodgkin, 1986). FOG-1, a CPEB-related RNA-binding protein, and FOG-3, a Tob/BTG homolog, also act subsequently to TRA-1 to specify sperm cell fate (Barton and Kimble, 1990; Chen et al., 2000; Ellis and Kimble, 1995; Luitjens et al., 2000). Finally, molecular data indicate that TRA-1 may act directly on the fog-3 promoter both as an activator and repressor (Chen and Ellis, 2000), a function at odds with its predicted role as solely a repressor of male (i.e., sperm) gene expression.
One might predict that genetic screens for sperm-specific sterile mutations would recover alleles of transcriptional regulators responsible for sperm gene expression. However, none of the 20 spe or fer genes cloned to date encodes a transcription factor homolog (see review by L’Hernault, 2006). In lieu of genetics, molecular data has been used to suggest at least one potential candidate for that role. A multigene family that encodes the major sperm protein (MSP) is expressed solely in sperm (Klass et al., 1982). Alignment of 5′ flanking sequences identified a conserved motif (AGATCTN7WGATAA) among a subset of the MSP genes (Klass et al., 1988). The last portion of that motif matches the canonical WGATAR sequence that is recognized by GATA transcription factors (Yamamoto et al., 1990). A C. elegans GATA factor homolog encoded by the elt-1 gene was recovered via a degenerate oligonucleotide strategy (Spieth et al., 1991). Shim et al. demonstrated that ELT-1 is able to promote expression from a yeast reporter plasmid containing concatamers of GATA-containing elements or combinations of GATA and GATC sequences (Shim et al., 1995). Subsequent work demonstrated that the conserved MSP 5′ motif likewise functions in yeast as a target for ELT-1, and differential Northern blot analysis indicated that elt-1 is expressed in the germ line of C. elegans (Shim, 1999). On the basis of those data, it was proposed that ELT-1 might function as a transcriptional regulator of MSP genes. However, the sole mutation in elt-1 available at that time resulted in embryonic lethality (Page et al., 1997), which precluded genetic characterization of an in vivo role in sperm development.
The current work extends the analysis of ELT-1 and its hypothesized role in sperm gene transcription. A computational screen of sperm promoter sequences recovered the MSP 5′ motif, which is also found upstream of a number of other sperm-enriched genes. We demonstrate that the motif is a preferred binding site for ELT-1 in vitro, and is required for sperm-specific transgene expression in vivo. We find that elt-1 is expressed in the sperm-producing germ line, and inactivation of elt-1 produces sperm-specific sterility and cytological defects in sperm. Taken together, the data support a direct role for ELT-1 in the activation of sperm gene expression.
Materials and methods
C. elegans strains were derived from wild-type isolate N2 (Bristol) and contained one of the following mutations: fem-1(hc17)IV, fem-3 (q20)IV, fem-3(q23)IV, elt-1(ok1002)IV, and pha-1(e2123)III. Strains were maintained at 15 °C unless otherwise noted. Genetic manipulations and media were according to Brenner (1974).
Computational analysis
Promoter sequences, defined as the 500 basepairs of sequence immediately preceding the predicted initiation codon for each gene, were downloaded from WormBase release 190 (www.wormbase.org). The size limit was selected on the basis of transgenic rescue studies for a variety of spermatogenesis-defective (Spe) mutations: of the eight Spe genes for which deletion constructs were employed to define a minimally sufficient rescuing fragment, none of the promoters was longer than 500 basepairs (Supplemental Table S1). Also, sperm-specific expression of GFP transgenes has been observed with promoters of this size range (spe-11, 272 basepairs; msp-56, 513 basepairs; Merritt et al., 2008). Promoters containing repetitive elements were removed from the dataset. An oligomer-counting algorithm (Jacobs Anderson and Parker, 2000) was modified to identify pairs of co-represented sequence elements within a restricted interval, in the hope of isolating bipartite binding sites. Promoters were divided into two groups on the basis of sperm-enriched expression (Reinke et al., 2000, 2004). For each promoter, the occurrence of every pair of 5-mer sequences separated by a defined distance (from 0 to 9 basepairs) was counted. For each group (sperm and non-sperm), the number of occurrences for each of the 410 possible pairs of 5-mers was summed and divided by the total number of promoters counted to calculate the relative frequency. 5-mer pairs that were most over-represented among sperm promoters were identified by dividing the sperm frequency by the non-sperm frequency and placing in rank order. Thresholds were set at the mean (for non-sperm) and mean plus three standard deviations (for sperm) to control for differences that might occur by chance among sequences of low frequency. Overlapping elements were identified by visual inspection and assembled into longer sequences when appropriate. The position-weighted alignment in Fig. 1B was generated using WebLogo (weblogo.berkeley.edu; Schneider and Stephens, 1990; Crooks et al., 2004).
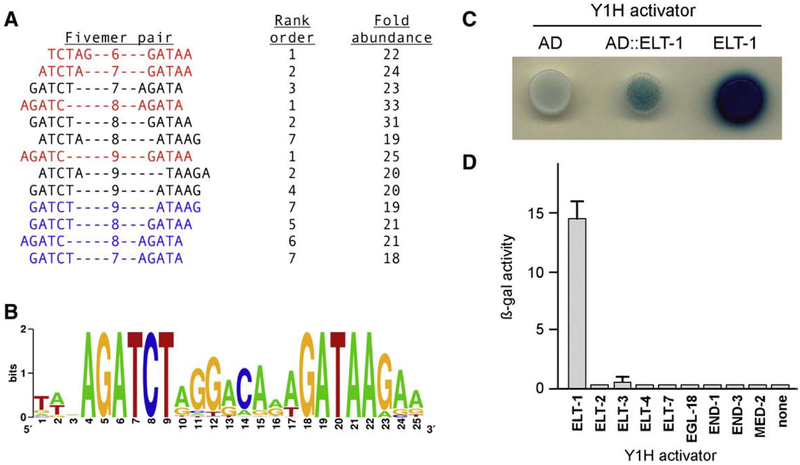
Fig. 1.
Transcriptional activation of the sperm consensus site by ELT-1. (A) Computational analysis of promoters. Shown are the 5-mer pairs and intervening gaps, their rank order among all over-represented 5-mer pairs with the indicated gap, and their relative abundance compared to the non-sperm promoters. (B) Weighted sequence alignment of the 48 promoters that contain the sperm consensus site; the individual sequences are listed in Supplemental Table S2. (C) Yeast one-hybrid assay for activation. Expression of the P2X-SPE::lacZ reporter gene in yeast transformed with a plasmid expressing the B42 transcriptional activation domain alone (AD), B42 fused to ELT-1 at residue 98 (AD::ELT-1), or full-length ELT-1 alone (ELT-1). β-galactosidase activity was detected on X-Gal indicator plates. (D) Yeast one-hybrid assay of C. elegans GATA transcription factors. Expression of the P1X-SPE::lacZ reporter gene in yeast transformed with a plasmid expressing the indicated full-length GATA factor fused to the GAL4 transcriptional activation domain. β-galactosidase activity was quantified by ONPG assay. Assays were performed in triplicate for each plasmid.
Yeast one-hybrid cDNA library construction and screening
Poly(A)+ RNA was isolated from fem-3(q23gf) young adult hermaphrodites reared at 25 °C as described previously (Reinke et al., 2000). cDNA was synthesized, ligated to EcoRI adaptors, phosphorylated, and size-fractionated according to the manufacturer’s protocol (Clontech). The cDNA library was ligated into the yeast activation domain (AD) plasmid pJG4–5 (Gyuris et al., 1993) digested with EcoRI, then transformed into E. coli strain DH10B by electroporation. The library contained 3×106 independent clones, with an average insert size of 600 basepairs. Amplified library DNA was recovered by Qiagen maxiprep.
Yeast media and manipulations followed standard protocols (Rose et al., 1990). One-hybrid lacZ reporter plasmids contained one (P1X-SPE::lacZ) or two (P2X-SPE::lacZ) copies of the sperm consensus site AGATCTAGGACAGAGATAA (all sequences shown 5′→3′) inserted as annealed oligonucleotides into plasmid pLacZi (Clontech). Each reporter plasmid was digested with NcoI, transformed into yeast strain EGY48 (Golemis and Khazak, 1997), and plated on SC-Ura selective medium. Integration at the URA3 locus was confirmed by Southern blot. The P2X-SPE::lacZ-bearing yeast strain was transformed with the AD-cDNA fusion library, plated onto SC-TrpUra selective medium, then replica-plated onto X-Gal indicator plates (SC-TrpUra containing 1% galactose + 2% raffinose to induce AD-cDNA expression) to detect β-galactosidase activity. Plasmid DNA was recovered from lacZ-expressing (i.e., blue) colonies, retransformed into yeast containing the P2X-SPE::lacZ reporter or pLacZi with no binding site, and rescreened on X-Gal indicator plates. Plasmids that retested for P2X-SPE–dependent lacZ expression were sequenced with plasmidspecific primers to identify the cDNA insert.
The full-length elt-1 cDNA absent the heterologous activation domain was amplified by PCR from a C. elegans cDNA library with gene-specific primers containing appropriate restriction sites, digested, and ligated into a derivative of pJG4–5 from which the activation domain had been deleted. DNA sequencing confirmed the integrity of elt-1. The plasmid was transformed into the P2X-SPE::lacZ-bearing strain and assessed on X-Gal indicator plates as above. Comparison of C. elegans GATA factor binding shown in Fig. 1D employed the AD-TF minilibrary (Deplancke et al., 2004). Two genes (med-1, absent; elt-6, incorrect construct) were not available from this collection. The remaining nine individual GATA factor clones were transformed in the P1X-SPE::lacZ-bearing strain and plated onto SC-TrpUra selective medium. Colonies were grown in SC-TrpUra liquid medium, cultures harvested by centrifugation at mid-log (OD600=0.5±0.1), and β-galactosidase activity quantified in chloroform-sodium dodecyl sulfate-permeabilized cells (Stern et al., 1984).
In vitro binding assays
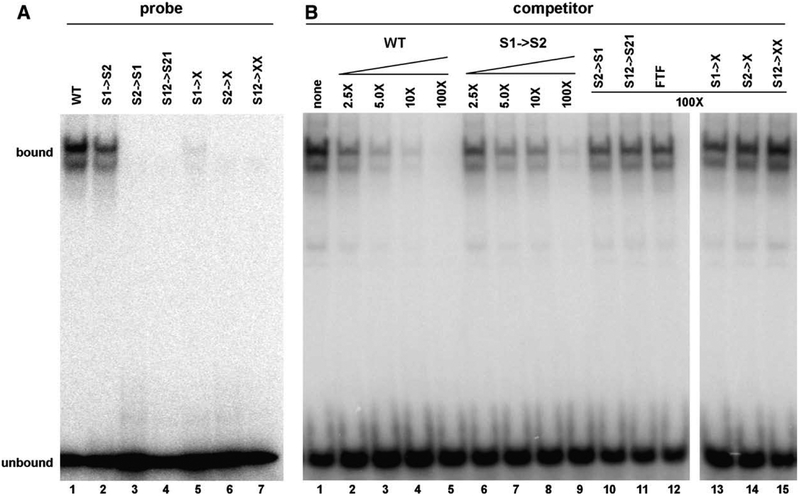
Probe sequences are listed in Table 1. Protein was synthesized from elt-1 cDNAs encoding wild-type, F1, or F2 zinc finger mutations using the TNT T7-coupled rabbit reticulocyte lysate system according to the manufacturer’s protocol (Promega). Binding reactions included 50 mM KCl, 20 mM HEPES, pH 7.9, 0.2 mM EDTA, 0.5 mM DTT,3.0 mM MgCl2, 4% (w/v) Ficoll, 1.0 μg poly(dI–dC), 1 μl translated protein diluted in 3 μl Dignam buffer [20 mM HEPES, pH 7.4, 20% (v/v) glycerol, 100 mM KCl, 0.2 mM EDTA, 0.5 mM DTT, 1×protease inhibitor cocktail (Sigma P2714)], and 2000-fold molar excess of non-target single-stranded DNA in a final volume of 20 μl. After pre-cooling on ice, 320 fmol of the indicated 32P-labeled DNA probe (~2×104 cpm; all probes adjusted to same specific activity) was added to the reaction. After incubation for 20 min on ice, 2 μl of 0.02% (w/v) bromophenol blue was added and samples were loaded onto a 4.5% (w/v) polyacrylamide gel in 0.5×TBE and subjected to electrophoresis for 2 1/2 h at 150 V, 4 °C. Gels were dried, exposed, and developed using a Storm 800 Phosphorimager (Molecular Dynamics). For competitive binding experiments shown in Fig. 2B, the indicated fold excess of cold competitor was added to each reaction and incubated an additional 10 min on ice prior to loading.
Table 1.
Probe sequences for in vitro binding assays.
| Designation | Sequencea |
|---|---|
| WT | AGCTCAAGATCTAGGACAGAGATAAGA |
| SW→S2 | AGCTCAAGATAAAGGACAGAGATAAGA |
| S2→S1 | AGCTCAAGATAAGGACAGAGATCTGA |
| S12→S21 | AGCTCAAGATAAAGGACAGAGATCTGA |
| Sl→X | AGCTCAAGAGGTAGGACAGAGATAAGA |
| S2→X | AGCTCAAGATCTAGGACAGAGAGGAGA |
| S12→XX | AGCTCAAGAGGTAGGACAGAGAGGAGA |
| FTF | AGCTCAGAGCAAGGTCCAAGGGCATGGG |
The 5′ and 3′ sequence elements are indicated in bold in the WT sequence. Nucleotide changes from WT are underlined.
Fig. 2.
In vitro DNA binding activity of ELT-1. (A) Binding to the sperm consensus site or sequence variants. Electrophoretic mobility shift assay of in vitro-generated ELT-1 protein with 32P-labeled oligonucleotide probes. See text for details. (B) Competitive binding assay. All samples contain equal amounts of ELT-1 protein plus 32P-labeled probe containing the sperm consensus site. Unlabeled probes were added at the indicated fold excess relative to the 32P-labeled probe to assess their relative binding affinities. See text for details.
In situ hybridization
Gonads were dissected from young adult hermaphrodites homo-zygous for the fem-3(q20ts) gain-of-function allele or fem-1(hc17ts) loss-of-function allele. Gonads were fixed by treatment with paraformaldehyde plus glutaraldehyde and permeabilized with proteinase K treatment according to published protocols (Lee and Schedl, 2006). Strand-specific digoxigenin-labeled probes were amplified linearly from an elt-1 cDNA template according to the manufacturer’s protocol (Roche). Probe synthesis was assessed by denaturing agarose gel electrophoresis and quantified by dot blot comparison to a standard of known concentration. Following hybrization to fixed gonads, the probe was detected by colorimetric assay with alkaline phosphatase-conjugated anti-digoxigenin antibody and NBT/BCIP substrate. After probe detection, gonads were stained with DAPI, mounted onto agarose pads, and visualized on Olympus BX51 microscope equipped for DIC Nomarski and fluorescent imaging.
RNAi and transgenic assays
RNAi experiments employed the double T7 promoter vector L4440 (Timmons and Fire, 1998) that contained or lacked a 1.4 kbp fragment of the elt-1 gene. Production of dsRNA in E. coli host strain HT115 (DE3) was induced on RNAi feeding plates (NGM medium containing carbenicillin plus IPTG). To bypass the embryonic requirement for elt-1, untreated embryos were obtained by bleaching gravid hermaphrodites. Those embryos were hatched in the absence of food to obtain L1 larvae, then transferred to RNAi feeding plates in bulk. L4 larvae were picked onto individual fresh RNAi feeding plates to assess self-fertility. Sterile adult hermaphrodites were transferred to fresh NGM plates populated with four wild-type adult males, allowed to mate for 24 h, then transferred to fresh NGM plates to assess cross-fertility. Sperm morphology was visualized by microdissection of gonads from young adult hermaphrodites into a drop of SM medium on poly-L-lysine-coated slides (Shakes and Ward, 1989).
Rescued transgenic lines of elt-1(ok1002) for mosaic analysis were obtained by germ line microinjection of heterozygous hermaphrodites with an 11 kbp genomic fragment of elt-1 plus the dominant rol-6 (su1006) marker (Mello et al., 1991). C. elegans genomic DNA was included to facilitate transgene expression in the germ line (Kelly et al., 1997). Transgenic F2 progeny were identified by the roller phenotype. Homozygous lines were identified by the lack of viable non-roller progeny in subsequent generations, indicating a requirement for the transgene to complement the elt-1(ok1002) embryonic lethality. L4 hermaphrodites were picked to individual plates and assessed for self and cross-fertility as above.
Transgenic lines for sperm-specific GFP expression were obtained by biolistic transformation (Praitis et al., 2001). GFP transgenes contained the 0.1 kbp msp-64 promoter (wild-type or deleted for the elt-1 consensus site), 1.0 kbp of msp-142 3′ sequence, and 6.6 kbp pha-1 gene as a selectable marker (Granato et al., 1994). Worms containing the temperature-sensitive pha-1(e2123) mutation (Schnabel and Schnabel, 1990) were age-synchronized by bleach treatment, hatched without food, then shifted to bacterial lawns at 25 °C. Worms were bombarded as young adults and maintained at 25 °C to identify progeny rescued for pha-1 lethality. Integration was confirmed by Mendelian segregation, and copy number estimated by Southern blotting.
Results
Identification of a sperm promoter element and its cognate binding factor
Prior results from genome-scale microarray screening identified 1343 genes (7.5% of the 18,010 genes assayed) that exhibited elevated expression during sperm development in C. elegans (Reinke et al., 2000, 2004). That list of sperm-enriched genes was the basis for an unbiased computational approach to identify potential binding sites for transcription factors that regulate sperm gene expression. We constructed a database of putative promoter sequences from the 20,177 coding genes in the C. elegans genome, and divided them into sperm-enriched and non-enriched groups. For each group, we determined the frequency of every pair of 5-mers separated by a defined interval (see Materials and methods for details). By comparing the relative frequencies in the sperm and non-sperm groups, we identified 5-mer pairs that were over-represented among the sperm-enriched group.
The most significant differences detected between the sperm and non-sperm classes were 5-mer pairs that overlapped the sequence AGATCT coupled with the sequence GATAA. Comparisons with gaps of six, seven, eight, or nine intervening nucleotides between 5-mers identified matching sequences as the most (6, 8, or 9 nucleotide gap) or second-most (7 nucleotide gap) over-represented pairs in the sperm-enriched group (Fig. 1A, in red). Additional 5-mer pairs that partially overlapped these elements were also among the ten most over-represented sequences, as were overlapping 5-mer pairs on the opposite strand (Fig. 1A, blue). All were 18- to 33-fold more abundant among the sperm group relative to the non-sperm group. Alignment of all the sperm promoters that contained both of the AGATCT and GATAA elements defined a putative sperm consensus site AGATCT(N)8GATAA. We screened the promoters of all coding genes in the C. elegans genome for this consensus site. A weighted sequence alignment of all the matching promoters indicates that, in addition to the bipartite consensus, preferred nucleotides are detected in the intervening and downstream sequences as well (Fig. 1B).
We found the distribution of the sperm consensus site to be decidedly non-random between the sperm and non-sperm groups of promoters. The consensus is found upstream of only 48 genes present on the microarray (Supplemental Table S2). Forty-five of those genes fall in the sperm-enriched category, vs. the four genes (48 × 7.5%) predicted by chance. Furthermore, those 45 genes exhibit high expression ratios (mean value = 39.5, vs. 14.6 for all sperm-enriched genes in the microarray screen), suggesting that this consensus site may act as a potent promoter of transcription during spermatogenesis. Targets include 22 of the 47 genes or pseudogenes that encode the major sperm protein (MSP) family; this protein is present only in sperm and is required for cell motility and signaling to the oocyte (Klass et al., 1982; Miller et al., 2001; Smith, 2006). The presence of this conserved sequence in the promoters of MSP genes had been noted previously (Klass et al., 1988; Shim, 1999). Other targets within this class include members of the group D family of nematode-specific peptides, protein kinases, and a number of novel sperm-enriched genes.
We used the putative sperm consensus site in a yeast one-hybrid screen (Li and Herskowitz, 1993) to determine the relevant binding factor(s) for this target sequence. We constructed an activation domain-cDNA fusion library enriched in sperm transcripts by isolating mRNA from fem-3(gf) adult hermaphrodites, which make only sperm. The library was screened against a lacZ reporter gene containing two tandem copies of the consensus site (designated P2X-SPE::lacZ). Only three plasmids out of 2×106 total transformants reproducibly yielded blue colonies on X-Gal indicator plates. All contained the same in-frame fusion of the activation domain to the elt-1 gene beginning at codon 98 (Fig. 1C). We then constructed a yeast expression vector for the full-length elt-1 gene absent the heterologous activation domain and observed even stronger expression of lacZ (Fig. 1C), which indicates that ELT-1 functions as a potent activator of transcription. Stimulation of lacZ expression by ELT-1 was specific for the consensus site, as LacZ activity was undetectable from control reporters lacking the element or containing an unrelated binding site (data not shown). These results are consistent with an earlier study of ELT-1 transcriptional activity in yeast (Shim et al., 1995).
The elt-1 gene encodes a GATA transcription factor, a class of zinc finger proteins with binding specificity for the sequence WGATAR (Yamamoto et al., 1990). The C. elegans genome encodes a total of 11 GATA factor homologs, so we tested the nine available from a C. elegans transcription factor library (Deplancke et al., 2004) for the ability to activate transcription from the sperm consensus site. We constructed a lacZ reporter that contained a single copy of the consensus site (designated P1X-SPE::lacZ), which replicates more faithfully the in vivo structure of sperm promoters, and also utilized fusions of each GATA factor to the same heterologous activation domain, in order to minimize potential differences in strength for the endogenous activation domains. Some of these gene products, such as the likely non-functional ELT-4 (Fukushige et al., 2003) and the atypical binding factor MED-2 (Broitman-Maduro et al., 2005) were not expected to exhibit activity. However, the results clearly indicate that only the ELT-1 fusion protein significantly stimulates transcription from the sperm consensus site (Fig. 1D), thereby demonstrating the specificity of this particular binding site for ELT-1 vs. the other GATA factors.
In vitro characterization of ELT-1 binding
ELT-1 is unique among the C. elegans GATA factor homologs in that it contains two zinc finger domains (the others possess only a single zinc finger). In that regard, ELT-1 is more similar to the vertebrate family of GATA factors that typically have two zinc fingers (Gillis et al., 2008). Studies of those proteins demonstrate different functional roles for the two fingers: the carboxy-terminal finger binds with high affinity (in the nanomolar range) to the WGATAR consensus site, while the amino-terminal finger stabilizes the interaction with DNA (Martin and Orkin, 1990; Omichinski et al., 1993). For some GATA factors, the amino-terminal finger exhibits preferential binding for sequences containing GATC instead of GATA (Newton et al., 2001; Pedone et al.,1997). Similarly, studies of ELT-1 in yeast suggest that the two fingers might also exhibit different binding specificities, with the amino-terminal finger being required for activation through GATC sequences (Shim et al., 1995). Therefore, we sought to determine directly the binding preferences of ELT-1 through DNA binding assays with in vitro-translated protein.
We first characterized the sequence requirements for the DNA target site. We observed efficient binding of ELT-1 to the consensus site derived from sperm promoters (Fig. 2A, WT; probe sequences listed in Table 1). The gel shift produces a doublet, which has been observed for other GATA factors (e.g., see Merika and Orkin, 1993). Binding required functional ELT-1 protein (see below) and was specific for this sequence, as no mobility shift was observed with an unrelated sequence (data not shown). We then assessed whether the two elements that define this bipartite consensus site were functionally equivalent or not. Note that the 3′ element AGATAA matches the canonical WGATAR site, while the 5′ element AGATCT is divergent. Conversion of the 5′ element to the 3′ element (designated S1→S2 in Fig. 2A) reduced the binding affinity of ELT-1. The converse replacement of the 3′ element with the 5′ element (Fig. 2A, S2→S1) completely abolished ELT-1 binding, consistent with a requirement for at least one copy of the canonical WGATAR site. Swapping the order of the two elements (Fig. 2A, S12→S21) also disrupted binding of ELT-1, indicating that the relative orientation of the elements contributes to binding specificity. Similar results were obtained when the sequence elements were mutated to non-GATA target sites. Mutation of the 5′ element (Fig. 2A, S1→X) substantially reduced binding, while mutation of the 3′ element singly or in combination with the 5′ element (Fig. 2A, S2→X or S12→XX, respectively) abrogated ELT-1 binding.
We confirmed these results by comparing the relative affinity of ELT-1 for each of these sequences in binding competition assays between a constant concentration of labeled probe containing the sperm consensus site and increasing amounts of unlabeled competitor DNA (Fig. 2B). The only mutant probe that competed for binding was the one with two canonical WGATAR sites (S1→S2), and even that binding was less robust than the sperm consensus sequence (WT in Fig. 2B; compare lanes 2–5 to lanes 6–9). All of the other mutant variants of the sperm consensus site were no more effective competitors than a completely unrelated sequence from the FTF promoter (Fig. 2B, lanes 10–11 and 13–15 compared to 12), even at 100-fold excess. Taken together, the data indicate that the two elements of the bipartite consensus site are functionally distinct, that the canonical 3′ site is essential for ELT-1 binding, and that the relative order of these two elements is also necessary for optimal binding.
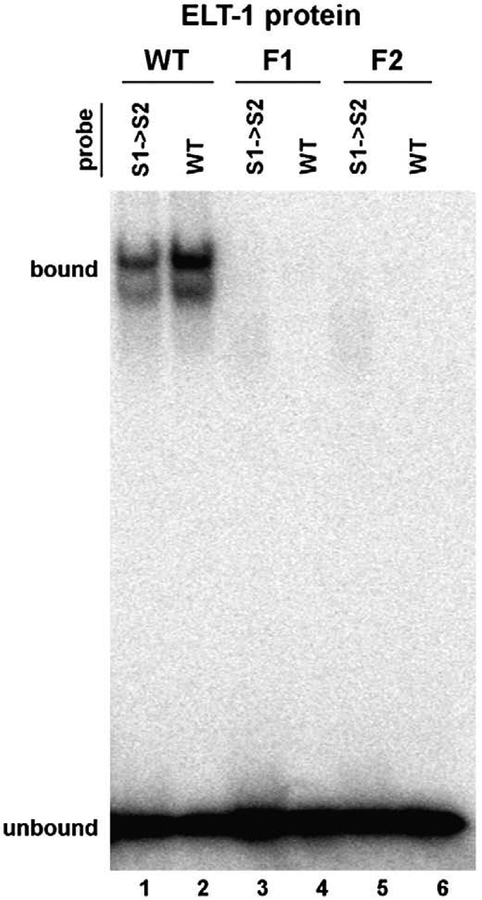
We next assessed the binding characteristics of the individual zinc fingers of ELT-1 by generating site-directed mutations to disrupt the function of one or the other binding domain. Each mutation converted a pair of cysteine residues to serine residues within the first or second zinc finger (F1 or F2, respectively). These mutations are predicted to preclude the coordination of zinc that is required for proper conformation of the DNA binding domain; the same mutations have been shown to abrogate zinc finger binding activity in other GATA factors (Yang and Evans, 1992). We compared binding to the sperm consensus site as well as the S1→S2 variant, which contains two copies of the canonical GATA site and exhibits binding (albeit reduced) by ELT-1. In contrast to the wild-type ELT-1 protein, neither F1 nor F2 yielded detectable binding activity for either probe (Fig. 3, compare lanes 1–2 to 3–6). Protein labeling controls indicated that equal amounts of soluble protein were produced from the wild-type and mutant genes (data not shown), suggesting that the overall conformation of the F1 or F2 protein is not adversely affected by the mutation. Instead, the data indicate that both zinc fingers of ELT-1 are essential for DNA binding activity. This conclusion stands in contrast to prior work, which suggested that only the carboxy-terminal finger is needed to bind DNA (Shim et al., 1995). That result was based on transcriptional activation in yeast from concatemers of WGATAA elements by ELT-1 deletion derivatives, so the discrepancy likely reflects differences between the two assays.
Fig. 3.
Loss of DNA binding by ELT-1 zinc finger mutations. Electrophoretic mobility shift assay of in vitro-generated ELT-1 protein (WT) or variants with mutations in the first (F1) or second (F2) zinc finger. 32P-labeled oligonucleotide probes contain the sperm consensus site (WT) or two copies of the canonical GATA element (S1→S2). Samples were run on the same gel as Fig. 2A; lanes 1 and 2 are lanes 2 and 1, respectively, in Fig. 2A.
In vivo role for ELT-1 in sperm
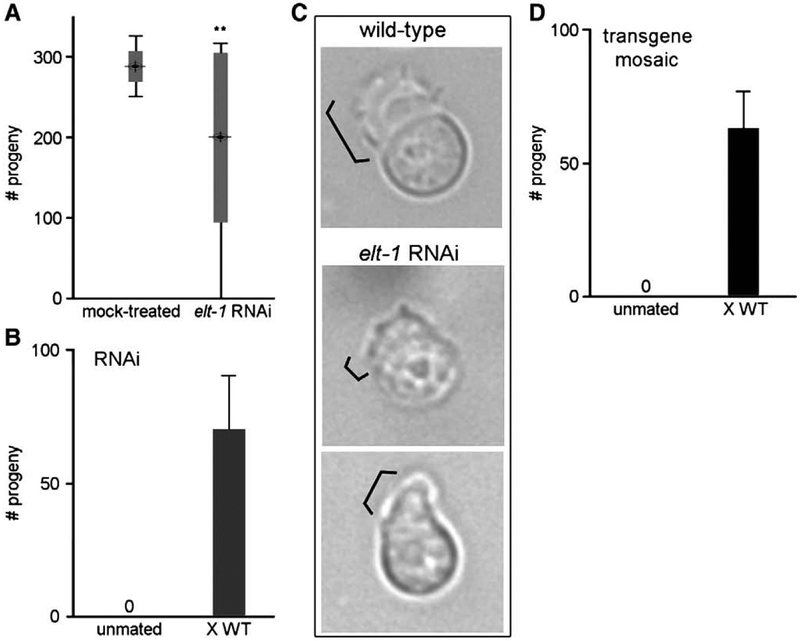
Functional roles for elt-1 have been demonstrated at multiple stages of development in C. elegans. The elt-1 gene product is required to specify the majority of hypodermal cell lineages in the early embryo, and mutation of elt-1 results in embryonic arrest (Page et al., 1997). ELT-1 is also necessary for maintaining seam cell fates throughout larval development; elt-1 RNA interference (RNAi) causes loss of seam cells and produces a ruptured vulva at the onset of egg-laying in adult hermaphrodites (Smith et al., 2005). The same study also reported roles for elt-1 in dauer formation and locomotion. We sought to determine if ELT-1 activity might also be required for proper sperm development. We performed elt-1 RNAi by the bacterial feeding technique (Timmons and Fire, 1998) during larval development. We observed the ruptured vulva phenotype in the majority (>95%) of treated hermaphrodites upon reaching adulthood, which prevented characterization of sperm function in those animals. Among the non-ruptured animals, we observed a significant reduction in the production of progeny (Fig. 4A). Individual fecundity varied widely. Approximately half of the animals (44%) were fully fertile, and presumably unaffected by elt-1 RNAi treatment. 21% produced a brood size 20–40% smaller than normal. 15% were half-sterile and laid a mixture of embryos and unfertilized oocytes simultaneously, consistent with a fertility defect in one of the two gonad arms. The remaining 21% were completely sterile and laid only oocytes. No sterile or half-sterile animals were observed among 1210 mock-treated controls.
Fig. 4.
Sperm sterility and motility defects caused by loss of elt-1. (A) Reduction in fertility by elt-1 RNAi. The total number of fertilized embryos was quantified for individual hermaphrodites following mock treatment or elt-1 RNAi (N =39 for each). Bull’s eye indicates mean progeny number; wide gray bar indicates ± standard deviation (S.D.); narrow bar indicates range of progeny number. **, p < 0.0001. (B) Restoration of fertility by mating. Sterile elt-1 RNAi hermaphrodites were individually mated to wild-type males for 24 h; shown is the mean number of total fertilized embryos (N =7, ±S.D.). (C) Pseudopod defect. Individual spermatozoa dissected from wild-type (top panel) or elt-1 RNAi-treated (middle and bottom) hermaphrodites, visualized by DIC Nomarski. Bracket indicates pseudopod. (D) Rescue of transgene mosaic sterility by mating. Sterile hermaphrodites were individually mated to wild-type males for 24 h; shown is the mean number of total fertilized embryos (N =6, ±S.D.).
Because sperm are the limiting gamete for reproduction in C. elegans, we tested if the introduction of wild-type sperm could restore fertility to the fully sterile hermaphrodites. Embryo production was observed following mating with wild-type males (Fig. 4B), demonstrating that the oocytes are competent for fertilization. Therefore, the sterility is a result of a defect in sperm fertility. We attempted to characterize sperm function of elt-1 RNAi-treated males; however, treatment resulted in a severe morphological defect in the male tail copulatory structures (Supplemental Fig. S1). This tail defect blocked the transfer of sperm during mating and thereby precluded any assessment of sperm fertility in males.
We repeated the screen and dissected the gonads from fully sterile hermaphrodites to examine the sperm for morphological defects. Wild-type sperm possess an extended pseudopod and are capable of directed motility on a poly-lysine-coated microscope slide (Fig. 4C, top panel). In contrast, the pseudopod of sperm from elt-1 RNAi-treated animals is either absent or short and aberrantly shaped (Fig. 4C, bottom panels) and the cells are unable to crawl. The sperm motility defect is also detectable within the reproductive tract of intact hermaphrodites; whereas wild-type sperm are localized to the spermathecae, where fertilization occurs, sperm from treated animals have been displaced by passing oocytes and are found primarily in the uterus (data not shown). Since motility is understood to be a necessary prerequisite for fertilization, the sperm motility defect is likely responsible for the observed sterility.
The variable penetrance of sperm sterility by RNAi was not unanticipated, since sperm genes as a group appear particularly refractory to inactivation by this technique. Both large-scale and targeted RNAi screens have failed to recapitulate the sperm-specific sterility produced by bona fide mutations in approximately two dozen known Spe or Fer genes (Fraser et al., 2000; our unpublished results). Therefore, we tried an alternative approach, mosaicism of transgenic rescue, to evaluate further the role of elt-1 in sperm. We first rescued the embryonic lethality associated with the elt-1(ok1002) deletion mutation by microinjection of a wild-type elt-1 transgene plus rol-6 (su1006) as a morphological marker (Mello et al., 1991), then assessed individual progeny for potential defects in fertility. We anticipated that loss of the elt-1 transgene in the primordial germ line stem cells Z2 and/or Z3 during development, or silencing of the transgene in the germ line (a commonly reported phenomenon; Kelly et al. 1997; Kelly and Fire, 1998; Jedrusik and Schulze, 2001), might result in sterility. Much like the elt-1 RNAi experiments, we observed a number of completely sterile hermaphrodites (6 of 1213 animals, 0.5%), plus a similar fraction (10, or 0.8%) of half-sterile animals. Fertility of the completely sterile animals was restored by mating with wild-type males (Fig. 4D), which indicates that the sterility is sperm-specific. We assessed the outcross progeny for phenotypic expression of the rol-6 (su1006) coinjection marker; the lack of rollers confirmed that the transgenic array had been lost or silenced in the germ line in each of the sterile parental hermaphrodites. Therefore, both the RNAi and transgene mosaic experiments indicate a functional role for elt-1 in sperm.
Expression studies for elt-1 have reported its presence in a variety of cells, consistent with its functional roles. The elt-1 gene is first expressed in hypodermal precursors in the early embryo, and remains detectable in the lateral seam cells (Page et al., 1997). At later stages of development, elt-1 expression is observed in a number of neuronal cells, in the vulval muscles of hermaphrodites, and in the sensory ray precursors of the male tail (Smith et al., 2005). Expression of elt-1 was also inferred from differential Northern blots to be present in the germ line during both oogenesis and spermatogenesis (Shim, 1999). We directly evaluated the expression of elt-1 in the germ lines of hermaphrodites by in situ hybridization of dissected gonads.
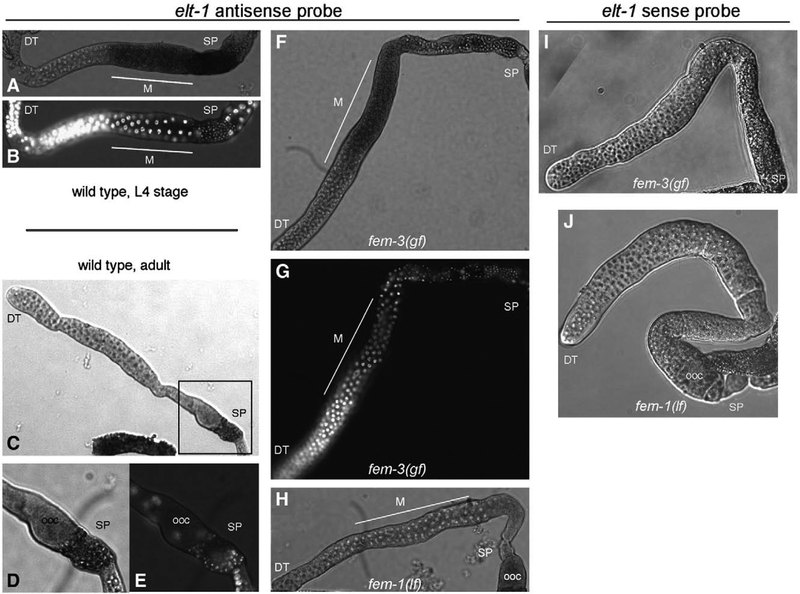
With an elt-1 antisense probe, expression was readily detectable in the germ line of wild-type hermaphrodites during the L4 larval stage, when spermatogenesis occurs (Fig. 5A). Co-staining with DAPI indicated that expression begins in pachytene nuclei during the first meiotic division, the developmental stage at which gamete-type-specific genes first become expressed, and is detected through the formation of haploid spermatids at the completion of meiosis (Fig. 5B). The expression pattern differed in wild-type adults, in which gametogenesis has switched from sperm to oocyte production. Staining was restricted to the sperm but was not detectable in the oocyte-producing germ line (Figs. 5C–E). To determine whether the observed elt-1 expression pattern is a consequence of developmental age or restricted by gamete type, we compared expression in age-synchronized adults that make only sperm (due to fem-3 gain-of-function mutation) or only oocytes (from fem-1 loss of function). Expression of elt-1 was detectable during spermatogenesis in fem-3 adults beginning at pachytene and extending through the formation of haploid spermatids (Figs. 5F, G), the same pattern that was observed in wild-type L4 animals (compare to Figs. 5A, B). In contrast, elt-1 was not detectably expressed in the germ line of fem-1 adults undergoing oogenesis (Fig. 5H; additional images available in Supplemental Fig. S2). These results indicate that elt-1 expression in the germ line is limited to sperm development. No signal was present when using the sense-stranded probe as a control in either fem-3 or fem-1 mutant animals (Figs. 5I, J). Our results are consistent with the microarray data, which indicate that elt-1 expression is elevated during spermatogenesis compared to oogenesis (Reinke et al., 2000). Therefore, elt-1 is expressed in the appropriate tissue and at the appropriate stage to play a functional role during sperm development.
Fig. 5.
Expression of elt-1 in the sperm-producing germ line. (A–H) In situ hybridizations of elt-1 antisense probe to dissected gonads. (A) Wild-type hermaphrodite, L4 stage during spermatogenesis. (B) The same gonad stained with DAPI to visual nuclear morphology. Highly condensed sperm nuclei are visible in the spermatheca (SP) at right. (C) Wild-type adult hermaphrodite, during oogenesis. (D) Enlargement of boxed region in C. (E) DAPI staining of boxed region in C. (F) Adult fem-3(gf) hermaphrodite. (G) DAPI staining of the same gonad. (H) Adult fem-1(lf) hermaphrodite. The weak oocyte (ooc) staining is variable and often seen using the sense strand control. (I, J) In situ hybridizations with elt-1 sense strand control of fem-3 (I) and fem-1 (J) adult hermaphrodites. A minimum of 25 gonads was examined for each genotype/probe combination; shown are representative images. DT, distal tip; SP, spermatheca; M, meiotic zone of pachytene nuclei; ooc, oocyte.
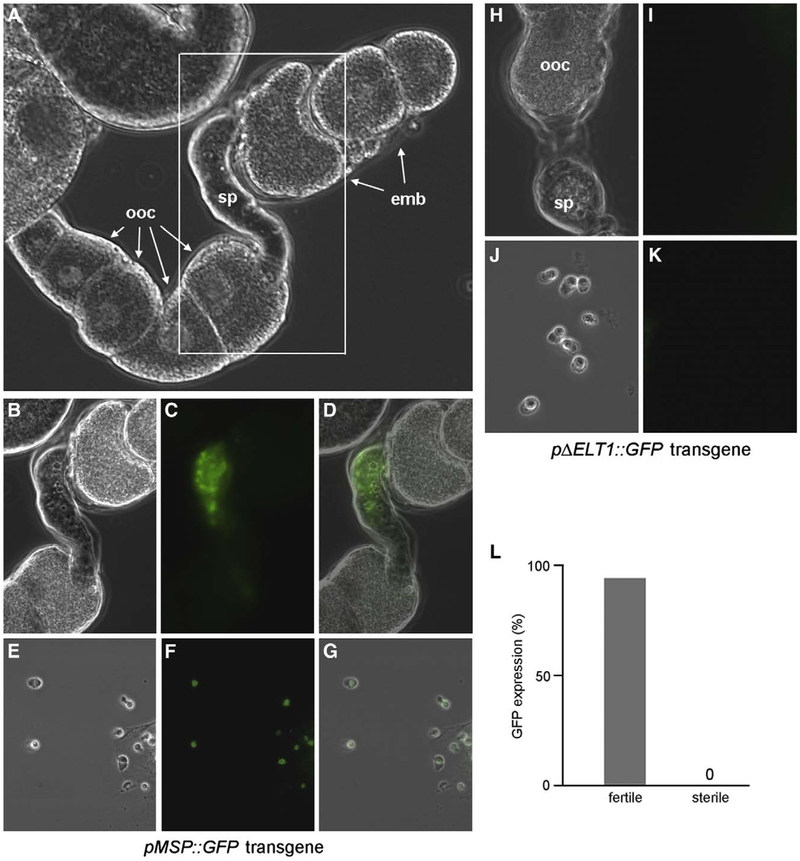
We assessed the ability of a sperm promoter containing the ELT-1 bipartite consensus site to drive expression of GFP in the sperm-producing germ line. We constructed a transgene, pMSP::GFP, with the GFP reporter expressed from the msp-64 promoter. The introduction of transgenes into C. elegans by microinjection produces highly repetitive extrachromosomal arrays that are typically silenced in the germ line (Kelly et al., 1997). Therefore, we utilized biolistic transformation to generate low-copy-number integrated transgenes that might escape germ line silencing (Praitis et al., 2001). We recovered a total of six independent integrated transgenic lines, all of which exhibited the same pattern of GFP expression. GFP fluorescence was observed exclusively in spermatozoa stored within the spermatheca in the germ line of adult hermaphrodites (Figs. 6A–G). GFP was not observed in oocytes or in any other tissues throughout development, demonstrating that this promoter confers sperm-specific gene expression. To ascertain the importance of the bipartite ELT-1 binding site, we constructed a GFP transgene in which the site was deleted (pΔELT1::GFP). A total of 11 transgenic lines were obtained by biolistic transformation as above; none of these animals exhibited GFP expression at detectable levels (Figs. 6H–K). Therefore, the ELT-1 binding site is essential for expression of the GFP transgene in sperm.
Fig. 6.
Expression of an ELT-1 target transgene in sperm. (A) DIC image of dissected gonad from pMSP::GFP transgenic hermaphrodite. (B–D) Higher magnification of boxed region in panel A showing DIC (B), GFP (C), and composite (D) images. (E–G) Spermatozoa from dissected spermatheca of pMSP::GFP transgenic hermaphrodite by DIC (E), GFP (F), and composite (G). (H–K) Dissection of pΔELT1::GFP transgenic hermaphrodites showing gonad (H, I) or spermatozoa (J, K) by DIC or GFP fluorescence, respectively. (L) Loss of GFP expression in sterile hermaphrodites. 1440 homozygous elt-1(ok1002); pMSP::GFP hermaphrodites containing the elt-1+rol-6(su1006) extrachromosomal array were individually screened for sterility then visualized for GFP in sperm (fertile, N=200; sterile, N=8). ooc, oocytes in the proximal arm of the uterus; sp, spermatheca; emb, embryos in the uterus.
We attempted to ascertain whether knockdown of elt-1 expression by RNAi adversely affected expression of the pMSP::GFP reporter. However, we observed the absence of GFP in a significant fraction (4–11%, depending upon the particular transgenic line) of untreated or control RNAi animals, presumably due to a residual level of germ line transgene silencing. Given the low penetrance of sperm-specific sterility by elt-1 RNAi, we were unable to detect a statistically significant increase in the percentage of GFP-negative animals following treatment. Instead, we employed mosaicism of transgene rescue as above to assess the role of elt-1 in reporter gene expression. The integrated pMSP::GFP reporter was crossed into the elt-1(ok1002) strain containing the rescuing elt-1 + rol6(su1006) extrachromosomal array. Homozygous elt-1(ok1002); pMSP::GFP lines were then screened for sterility (due to germ line loss or silencing of the array) and GFP expression. GFP was not observed in any of the sterile hermaphrodites, whereas sperm-specific GFP expression was visible in 94% of the fertile hermaphrodites (Fig. 6L). The strong correlation between sterility and loss of GFP demonstrates a role for elt-1 in sperm-specific gene expression.
Discussion
We have identified ELT-1, a GATA transcription factor previously shown to function in the embryo to specify hypodermal cell lineages, as an activator of sperm gene expression in the germ line. A computational approach identified a bipartite consensus site present almost exclusively in the promoters of sperm genes. In vivo and in vitro analyses demonstrated preferential binding of ELT-1 to this sperm consensus site. The elt-1 gene is expressed in the sperm-producing germ line, and functional studies indicated a requirement for ELT-1 in sperm fertility and sperm-specific gene expression. Our work is the first to demonstrate direct regulation of sperm gene expression in C. elegans, and fills a critical gap in the pathway linking germ line sex determination to its ultimate output in cell-type-specific gene expression.
GATA factors have been implicated in the transcriptional regulation of sperm development in other systems as well. Vertebrates possess six GATA factors, all of which (like ELT-1) contain two zinc finger motifs (Gillis et al., 2008). These genes are expressed in a partially overlapping variety of tissues and, given the similarities of their binding preferences, target specificity is conferred in partnership with different cofactors (e.g., FOG; reviewed in Cantor and Orkin, 2005). Three (GATA-1, −4, and −6) are expressed in somatic cells of the developing or adult testis of mammals (Ito et al., 1993; Viger et al., 1998; Ketola et al., 1999). Transcriptional targets involved in male gonadogenesis or steroidogenesis have been identified for all three (reviewed in Viger et al., 2004). The best evidence for a functional role in testis differentiation exists for GATA-4. In vivo disruption of its interaction with FOG cofactors impairs murine testicular development during embryogenesis and dramatically reduces expression of the sex-determining gene Sry in the developing gonad (Tevosian et al., 2002). In mice and humans, GATA-4 partners with the WT1 transcription factor to promote expression directly through the Sry promoter (Miyamoto et al., 2008). Also, GATA-4 is expressed in human male germ cells as well as the somatic testis, suggestive of a role in gametespecific gene expression subsequent to sex determination (Ketola et al., 2000).
Our data indicate that control of sperm-specific transcription by ELT-1 is mediated by a conserved binding site found only in the promoters of a subset of sperm genes. This bipartite site consists of a non-canonical element with a GATC core followed by a canonical GATA element. In this regard, ELT-1 seems most similar to GATA-1 from mouse (Newton et al., 2001) and GATA-2 and GATA-3 from chicken (Pedone et al., 1997). Those homologs possess an N-terminal zinc finger that binds preferentially to motifs containing GATC, and a C-terminal finger that exhibits high affinity for GATA-containing sequences. ELT-1 is the only GATA factor in C. elegans with two zinc fingers, and prior work suggested that the two fingers of ELT-1 demonstrate different binding specificities (Shim et al., 1995). Our one-hybrid assay of C. elegans GATA factors indicates that only ELT-1 exhibits detectable binding to the bipartite sperm consensus site, thereby providing a mechanism for selective activation of sperm genes by ELT-1.
ELT-1 activity is required in different tissues at different stages of development. In addition to sperm, functional and expression studies demonstrate roles in hypodermal precursor cells in the embryo, lateral seam cells during larval development, dauer formation, locomotory neurons, and morphogenesis of the male tail (Page et al., 1997; Smith et al., 2005; our Supplemental Fig. S1). How is its activity restricted to the appropriate target genes? An earlier model invoked different levels of ELT-1 (low in the germ line and high in the embryo) as the primary mechanism of differential regulation (Shim, 1999). However, additional modes of regulation must exist to explain the observed patterns of gene expression. Microarray data indicate that the sperm gene targets of ELT-1 are not expressed during embryogenesis or larval development (Baugh et al., 2003; Jiang et al., 2001), despite a functional requirement for ELT-1 in hypodermal cells at those stages. Expression of the pMSP::GFP transgene is likewise restricted to the sperm-producing germ line. Repression of ELT-1 sperm genes by regional chromatin modification seems unlikely, at least for the GFP reporter: the transgene contained minimal flanking sequences and, despite the construction of multiple independent lines integrated at random loci, expression in all instances was restricted to the sperm. There-ore, it seems likely that one or more additional binding factors (either repressor in hypodermal cells and/or activator in sperm) are necessary for the observed pattern of GFP expression. Alternatively, sequences present in both the native MSP and reporter transgene transcripts could be targeted for silencing in hypodermal cells by an endogenous RNAi mechanism.
For hypodermal target genes, cell-type-specific expression by ELT-1 might also require different regulatory components, such as a repressor in sperm. However, it is possible that the postulated difference in the concentration of ELT-1 protein might suffice to confer differential regulation of hypodermal gene expression. Hypodermal genes lack the consensus ELT-1 binding site, which is found only in the promoters of sperm genes. Instead, control by ELT-1 is understood to be mediated by the presence of multiple copies of the canonical GATA site in the promoters of hypodermal targets. In the best-characterized example, a 4 kbp enhancer from lin-26 can direct GFP expression in hypodermal precursors and seam cells, and expression is dependent on ELT-1 (Landmann et al., 2004). This lin-26 enhancer contains four GATA sites that are evolutionarily conserved with C. briggsae, which suggests a likely functional role for these elements. Multiple, evolutionarily conserved copies of the canonical GATA site are also present in the promoters of the ELT-1 hypodermal targets elt-3 and nhr-25 (Yanai et al., 2008). Our in vitro studies clearly indicate that ELT-1 binds preferentially to the sperm consensus site in comparison to two GATA elements (WT vs. S1→S2 in Fig. 2). Expression from hypodermal promoters might be predicted to require higher concentrations of ELT-1 protein than expression from the sperm consensus site. Additional studies of ELT-1 protein levels in vivo, in combination with reporter transgenes bearing appropriate binding sites, will be necessary to address this possibility.
Although ELT-1 is clearly required for expression of some sperm genes, it is unlikely to act as the sole (or even primary) regulator of transcription during spermatogenesis. Only a small subset of sperm genes (45 of 1343 identified by microarray) contains the consensus ELT-1 site AGATCT(N)8GATAA, and at least one promoter that lacks this consensus is capable of driving sperm-specific GFP expression (spe-11; Merritt et al., 2008). We acknowledge that limiting the list of candidate sperm promoters to those that perfectly match this site is overly restrictive and undoubtedly excludes bona fide ELT-1 binding sites that diverge from this consensus. Although ELT-1 clearly exhibits preferential binding to the consensus in comparison to the other sequences tested, we have not determined the full constellation of possible ELT-1 binding sites. However, we demonstrate the absolute requirement for at least one copy of the canonical GATA element for ELT-1 binding (Fig. 2), and that sequence is not found in the majority of sperm promoters, which strongly suggests that those genes are not directly regulated by ELT-1. Among the sperm promoters that lack the ELT-1 consensus site, we have identified several additional sperm-enriched sequence elements unrelated to the ELT-1 site that might serve as binding sites for other, as-yet-unidentified transcriptional regulators.
It is also improbable that ELT-1 functions as a master switch at the apex of a sperm-specific transcriptional cascade, since none of the putative ELT-1 sperm target genes is predicted to encode additional transcription factors that would be necessary for expression of non-ELT-1 targets. The requirement for ELT-1 in hypodermal gene expression (as well as in other cell types) also argues against the role of a master switch that specifies sperm cell fate. Rather, we believe that ELT-1 lies nearer the end of the pathway and, in conjunction with other regulatory proteins, directly mediates the expression of a subset of sperm structural genes, such as those encoding MSP.
One unanswered question is how expression of elt-1 itself is controlled in the germ line by the sex determination pathway. The microarray data and in situ hybridizations clearly demonstrate that ELT-1 is governed, at least in part, at the level of transcription. Direct regulation by the transcription factor TRA-1 seems implausible. The elt-1 promoter does not contain sequences that match the TRA-1 consensus binding site. Furthermore, elt-1 is differentially expressed in the fem-1 vs. fem-3gf mutants (Fig. 5), and epistasis analysis indicates that these gene products act subsequent to TRA-1 in the germ line sex determination program. The FEM proteins are part of an E3 ubiquitin ligase complex (Starostina et al., 2007), so one possibility is ubiquitin-mediated degradation of a repressor of elt-1 during spermatogenesis. Our recent identification of the E1 ubiquitin-activating enzyme encoded by uba-1 as essential for sperm-specific fertility (Kulkarni and Smith, 2008) is also consistent with this model. Alternatively, several of the sperm-enriched genes identified by microarray screening are predicted to encode transcriptional regulators; one (or more) of those proteins is an attractive candidate for sperm-specific control of elt-1 expression. Ongoing efforts are directed at identifying the mechanism(s) that govern elt-1 transcription in the germ line and contribute to the specification of sperm cell fate.
Supplementary Material
Acknowledgments
We wish to thank the Caenorhabditis Genetics Center and the C. elegans Gene Knockout Consortium for providing strains and plasmids, Bronessa Fernandes for assistance with the yeast one-hybrid assays, one anonymous reviewer for suggesting the mosaic/GFP experiment, and members of the lab and the Baltimore/Washington worm community for fruitful discussions. This work was supported in part by National Science Foundation grant number 0445684 to H.E.S.
Footnotes
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.ydbio.2009.06.044.
References
- Barton MK, Kimble J, 1990. fog-1, a regulatory gene required for specification of spermatogenesis in the germ line of Caenorhabditis elegans. Genetics 125, 29–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baugh LR, Hill AA, Slonim DK, Brown EL, Hunter CP, 2003. Composition and dynamics of the Caenorhabditis elegans early embryonic transcriptome. Development 130, 889–900. [DOI] [PubMed] [Google Scholar]
- Brenner S, 1974. The genetics of Caenorhabditis elegans. Genetics 77, 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broitman-Maduro G, Maduro MF, Rothman JH, 2005. The noncanonical binding site of the MED-1 GATA factor defines differentially regulated target genes in the C. elegans mesendoderm. Dev. Cell 8, 427–433. [DOI] [PubMed] [Google Scholar]
- Cantor AB, Orkin SH, 2005. Coregulation of GATA factors by the Friend of GATA (FOG) family of multitype zinc finger proteins. Semin. Cell Dev. Biol 16, 117–128. [DOI] [PubMed] [Google Scholar]
- Chen P, Ellis RE, 2000. TRA-1A regulates transcription of fog-3, which controls germ cell fate in C. elegans. Development 127, 3119–3129. [DOI] [PubMed] [Google Scholar]
- Chen PJ, Singal A, Kimble J, Ellis RE, 2000. A novel member of the tob family of proteins controls sexual fate in Caenorhabditis elegans germ cells. Dev. Biol 217, 77–90. [DOI] [PubMed] [Google Scholar]
- Conradt B, Horvitz HR, 1999. The TRA-1A sex determination protein of C. elegans regulates sexually dimorphic cell deaths by repressing the egl-1 cell death activator gene. Cell 98, 317–327. [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE, 2004. WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deplancke B, Dupuy D, Vidal M, Walhout AJ, 2004. A Gateway-compatible yeast one-hybrid system. Genome Res. 14, 2093–2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doniach T, Hodgkin J, 1984. A sex-determining gene, fem-1, required for both male and hermaphrodite development in Caenorhabditis elegans. Dev. Biol 106, 223–235. [DOI] [PubMed] [Google Scholar]
- Ellis RE, Kimble J, 1995. The fog-3 gene and regulation of cell fate in the germ line of Caenorhabditis elegans. Genetics 139, 561–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser AG, Kamath RS, Zipperlen P, Martinez-Campos M, Sohrmann M, Ahringer J, 2000. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature 408, 325–330. [DOI] [PubMed] [Google Scholar]
- Fukushige T, Goszczynski B, Tian H, McGhee JD, 2003. The evolutionary duplication and probable demise of an endodermal GATA factor in Caenorhabditis elegans. Genetics 165, 575–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillis WQ, Bowerman BA, Schneider SQ, 2008. The evolution of protostome GATA factors: molecular phylogenetics, synteny, and intron/exon structure reveal orthologous relationships. BMC Evol. Biol 8, 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golemis EA, Khazak V, 1997. Alternative yeast two-hybrid systems. The interaction trap and interaction mating. Methods Mol. Biol 63, 197–218. [DOI] [PubMed] [Google Scholar]
- Granato M, Schnabel H, Schnabel R,1994. pha-1, a selectable marker for gene transfer in C. elegans. Nucleic Acids Res. 22, 1762–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyuris J, Golemis E, Chertkov H, Brent R, 1993. Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell 75, 791–803. [DOI] [PubMed] [Google Scholar]
- Hodgkin J, 1986. Sex determination in the nematode C. elegans: analysis of tra-3 suppressors and characterization of fem genes. Genetics 114, 15–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin J, 1987. A genetic analysis of the sex-determining gene, tra-1, in the nematodeCaenorhabditis elegans. Genes Dev. 1, 731–745. [DOI] [PubMed] [Google Scholar]
- Hodgkin J, 1993. Molecular cloning and duplication of the nematode sex-determining gene tra-1. Genetics 133, 543–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito E, Toki T, Ishihara H, Ohtani H, Gu L, Yokohama M, Engel JD, Yamamoto M, 1993. Erythroid transcription factor GATA-1 is abundantly transcribed in mouse testis. Nature 362, 466–468. [DOI] [PubMed] [Google Scholar]
- Jacobs Anderson JS, Parker R, 2000. Computational identification of cis-acting elements affecting post-transcriptional control of gene expression in Saccharomyces cerevisiae. Nucleic Acids Res. 28, 1604–1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jedrusik MA, Schulze E, 2001. A single histone H1 isoform (H1.1) is essential for chromatin silencing and germline development in Caenorhabditis elegans. Development 128, 1069–1080. [DOI] [PubMed] [Google Scholar]
- Jiang M, Ryu J, Kiraly M, Duke K, Reinke V, Kim SK, 2001. Genome-wide analysis of developmental and sex-regulated gene expression profiles in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 98, 218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly WG, Fire A, 1998. Chromatin silencing and the maintenance of a functional germline in Caenorhabditis elegans. Development 125, 2451–2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly WG, Xu S, Montgomery MK, Fire A, 1997. Distinct requirements for somatic and germline expression of a generally expressed Caernorhabditis elegans gene. Genetics 146, 227–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketola I, Pentikainen V, Vaskivua T, Ilvesmaki V, Herva R, Dunkel L, Tapanainen JS, Toppari J, Heikinheimo M, 2000. Expression of transcription factor GATA-4 during human testicular development and disease. J. Clin. Endocrinol. Metab 85, 3925–3931. [DOI] [PubMed] [Google Scholar]
- Ketola I, Rahman N, Toppari J, Bielinska M, Porter-Tinge SB, Tapainen JS, Huhtaniemi IT, Wilson DB, Heikinheimo M, 1999. Expression and regulation of transcription factors GATA-4 and GATA-6 in developing mouse testis. Endocrinology 140, 1470–1480. [DOI] [PubMed] [Google Scholar]
- Klass M, Dow B, Herndon M, 1982. Cell-specific transcriptional regulation of the major sperm protein in Caenorhabditis elegans. Dev. Biol 93, 152–164. [DOI] [PubMed] [Google Scholar]
- Klass M, Ammons D, Ward S, 1988. Conservation in the 5′ flanking sequences of transcribed members of the Caenorhabditis elegans major sperm protein gene family. J. Mol. Biol 199, 15–22. [DOI] [PubMed] [Google Scholar]
- Kulkarni M, Smith HE, 2008. E1 ubiquitin-activating enzyme UBA-1 plays multiple roles throughout C. elegans development. PLoS Genet. 4, e1000131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landmann F, Quintin S, Labouesse M, 2004. Multiple regulatory elements with spatially and temporally distinct activities control the expression of the epithelial differentiation gene lin-26 in C. elegans. Dev. Biol 265, 478–490. [DOI] [PubMed] [Google Scholar]
- Lee MH, Schedl T, 2006. RNA in situ hybridization of dissected gonads WormBook, pp. 1–7. June 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- L’Hernault SW, 2006. Spermatogenesis WormBook, pp. 1–14. February 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JJ, Herskowitz I, 1993. Isolation of ORC6, a component of the yeast origin recognition complex by a one-hybrid system. Science 262, 1870–1874. [DOI] [PubMed] [Google Scholar]
- Luitjens C, Gallegos M, Kraemer B, Kimble J, Wickens M, 2000. CPEB proteins control two key steps in spermatogenesis in C. elegans. Genes Dev. 14, 2596–2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DI, Orkin SH, 1990. Transcriptional activation and DNA binding by the erythroid factor GF-1/NF-E1/Eryf 1. Genes Dev. 4, 1886–1898. [DOI] [PubMed] [Google Scholar]
- Mason DA, Rabinowitz JS, Portman DS, 2008. dmd-3, a doublesex-related gene regulated by tra-1, governs sex-specific morphogenesis in C. elegans. Development 135, 2373–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello CC, Kramer JM, Stinchcomb D, Ambros V, 1991. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10, 3959–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merika M, Orkin SH, 1993. DNA-binding specificity of GATA family transcription factors. Mol. Cell. Biol 13, 3999–4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt C, Rasoloson D, Ko D, Seydoux G, 2008. 3′ UTRs are the primary regulators of gene expression in the C. elegans germline. Curr. Biol 18, 1476–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MA, Nguyen VQ, Lee MH, Kosinski M, Schedl T, Caprioli RM, Greenstein D, 2001. A sperm cytoskeletal protein that signals oocyte meiotic maturation and ovulation. Science 291, 2144–2147. [DOI] [PubMed] [Google Scholar]
- Miyamoto Y, Taniguchi H, Hamel F, Silversides DW, Viger RS, 2008. A GATA4/WT1 Cooperation Regulates Transcription of Genes Required for Mammalian Sex Determination and Differentiation. . [DOI] [PMC free article] [PubMed]
- Newton A, Mackay J, Crossley M, 2001. The N-terminal zinc finger of the erythroid transcription factor GATA-1 binds GATC motifs in DNA. J. Biol. Chem 276, 35794–35801. [DOI] [PubMed] [Google Scholar]
- Omichinski JG, Trainor C, Evans T, Gronenborn AM, Clore GM, Felsenfeld G, 1993. A small single-”finger” peptide from the erythroid transcription factor GATA-1 binds specifically to DNA as a zinc or iron complex. Proc. Natl. Acad. Sci. U.S.A. 90, 1676–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page BD, Zhang W, Steward K, Blumenthal T, Priess JR, 1997. ELT-1, a GATA-like transcription factor, is required for epidermal cell fates in Caenorhabditis elegans embryos. Genes Dev. 11, 1651–1661. [DOI] [PubMed] [Google Scholar]
- Peden E, Kimberly E, Gengyo-Ando K, Mitani S, Xue D, 2007. Control of sex-specific apoptosis in C. elegans by the BarH homeodomain protein CEH-30 and the transcriptional repressor UNC-37/Groucho. Genes Dev. 21, 3195–3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedone PV, Omichinski JG, Nony P, Trainor C, Gronenborn AM, Clore GM, Felsenfeld G, 1997. The N-terminal fingers of chicken GATA-2 and GATA-3 are independent sequence-specific DNA binding domains. EMBO J. 16, 2874–2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praitis V, Casey E, Collar D, Austin J, 2001. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics 157, 1217–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond CS, Shamu CE, Shen MM, Seifert KJ, Hirsch B, Hodgkin J, Zarkower D, 1998. Evidence for evolutionary conservation of sex-determining genes. Nature 391, 691–695. [DOI] [PubMed] [Google Scholar]
- Reinke V, Gil IS, Ward S, Kazmer K, 2004. Genome-wide germline-enriched and sex-biased expression profiles in Caenorhabditis elegans. Development 131, 311–323. [DOI] [PubMed] [Google Scholar]
- Reinke V, Smith HE, Nance J, Wnag J, Van Doren C, Begley R, Jones SJ, Davis EB, Scherer S, Ward S, Kim SK, 2000. A global profile of germline gene expression inC. elegans. Mol. Cell 6, 605–616. [DOI] [PubMed] [Google Scholar]
- Rose MD, Winston F, Hieter P, 1990. Methods in Yeast Genetics: a Laboratory CourseManual. CSHL Press, Cold Spring Harbor, NY. [Google Scholar]
- Schedl T, Graham PL, Barton MK, Kimble J, 1989. Analysis of the role of tra-1 in germline sex determination in the nematode Caenorhabditis elegans. Genetics 123, 755–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnabel H, Schnabel R, 1990. An organ-specific differentiation gene, pha-1, fromCaenorhabditis elegans. Science 250, 686–688. [DOI] [PubMed] [Google Scholar]
- Schneider TD, Stephens RM, 1990. Sequence logos: a new way to display consensus sequences. Nucl. Acids Res. 18, 6097–6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz HT, Horvitz HR, 2007. The C. elegans protein CEH-30 protects male-specific neurons from apoptosis independently of the Bcl-2 homolog CED-9. Genes Dev. 21, 3181–3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakes DC, Ward S,1989. Initiation of spermiogenesis in C. elegans: a pharmacological and genetic analysis. Dev. Biol 134, 189–200. [DOI] [PubMed] [Google Scholar]
- Shen MM, Hodgkin J, 1988. mab-3, a gene required for sex-specific yolk protein expression and a male-specific lineage in C. elegans. Cell 54, 1019–1031. [DOI] [PubMed] [Google Scholar]
- Shim YH, 1999. elt-1, a gene encoding a Caenorhabditis elegans GATA transcription factor, is highly expressed in the germ lines with msp genes as the potential targets. Mol. Cells 9, 535–541. [PubMed] [Google Scholar]
- Shim YH, Bonner JJ, Blumenthal T, 1995. Activity of a C. elegans GATA transcription factor, ELT-1, expressed in yeast. J. Mol. Biol 253, 665–676. [DOI] [PubMed] [Google Scholar]
- Smith HE, 2006. Sperm motility and MSP WormBook, pp. 1–8. February 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JA, McGarr P, Gilleard JS, 2005. The Caenorhabditis elegans GATA factor elt-1 is essential for differentiation and maintenance of hypodermal seam cells and for normal locomotion. J. Cell Sci. 118, 5709–5719. [DOI] [PubMed] [Google Scholar]
- Spieth J, Shim YH, Lea K, Conrad R, Blumenthal T, 1991. elt-1, an embryonically expressed Caenorhabditis elegans gene homologous to the GATA transcription factor family. Mol. Cell. Biol 11, 4651–4659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starostina NG, Lim JM, Schvarzstein M, Wells L, Spence AM, Kipreos ET, 2007. A CUL-2 ubiquitin ligase containing three FEM proteins degrades TRA-1 to regulate C. elegans sex determination. Dev. Cell 13, 127–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern M, Jensen R, Herskowitz I, 1984. Five SWI genes are required for expression of the HO gene in yeast. J. Mol. Biol 178, 853–868. [DOI] [PubMed] [Google Scholar]
- Tapscott SJ, Davis RL, Thayer MJ, Cheng PF, Weintraub H, Lassar AB, 1988. MyoD1: a nuclear phosphoprotein requiring a Myc homology region to convert fibroblasts to myoblasts. Science 242, 405–411. [DOI] [PubMed] [Google Scholar]
- Tevosian SG, Albrecht KH, Crispino JD, Fujiwara Y, Eicher EM, Orkin SH, 2002. Gonadal differentiation, sex determination and normal Sry expression in mice require direct interaction between transcription partners GATA4 and FOG2. Development 129, 4627–4634. [DOI] [PubMed] [Google Scholar]
- Timmons L, Fire A, 1998. Specific interference by ingested dsRNA. Nature 395, 854. [DOI] [PubMed] [Google Scholar]
- Viger RS, Mertineit C, Trasler JM, Nemer M, 1998. Transcription factor GATA-4 is expressed in a sexually dimorphic pattern during mouse gonadal development and is a potent activator of the Mullerian inhibiting substance promoter. Development 125, 2665–2675. [DOI] [PubMed] [Google Scholar]
- Viger RS, Taniguchi H, Robert NM, Tremblay JJ, 2004. Role of the GATA family of transcription factors in andrology. J. Andrology 25, 441–452. [DOI] [PubMed] [Google Scholar]
- Yamamoto M, Ko LJ, Leonard MW, Beug H, Orkin SH, Engel JD, 1990. Activity and tissue-specific expression of the transcription factor NF-E1 multigene family. Genes Dev. 4, 1650–1662. [DOI] [PubMed] [Google Scholar]
- Yanai I, Baugh LR, Smith JJ, Roehrig C, Shen-Orr SS, Claggett JM, Hill AA, Slomin DK, Hunter CP, 2008. Pairing of competitive and topologically distinct regulatory modules enhances patterned gene expression. Mol. Syst. Biol 4, 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HY, Evans T, 1992. Distinct roles for the two cGATA-1 finger domains. Mol. Cell Biol. 12, 4562–4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi W, Ross JM, Zarkower D, 2000. Mab-3 is a direct tra-1 target gene regulating diverse aspects of C. elegans male sexual development and behavior. Development 127, 4469–4480. [DOI] [PubMed] [Google Scholar]
- Zarkower D, 2006. Somatic sex determination WormBook, pp. 1–12. February 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarkower D, Hodgkin J, 1992. Molecular analysis of the C. elegans sex-determining gene tra-1: a gene encoding two zinc finger proteins. Cell 70, 237–249. [DOI] [PubMed] [Google Scholar]
- Zarkower D, Hodgkin J, 1993. Zinc fingers in sex determination: only one of the two C. elegans Tra-1 proteins binds DNA in vitro. Nucleic Acids Res. 21, 3691–3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.