Abstract
The cellular prion protein (PrPC) is a cell surface glycoprotein attached to the membrane by a glycosylphosphatidylinositol (GPI)-anchor and plays a critical role in transmissible, neurodegenerative and fatal prion diseases. Alterations in membrane attachment influence PrPC-associated signaling, and the development of prion disease, yet our knowledge of the role of the GPI-anchor in localization, processing, and function of PrPC in vivo is limited We exchanged the PrPC GPI-anchor signal sequence of for that of Thy-1 (PrPCGPIThy-1) in cells and mice. We show that this modifies the GPI-anchor composition, which then lacks sialic acid, and that PrPCGPIThy-1 is preferentially localized in axons and is less prone to proteolytic shedding when compared to PrPC. Interestingly, after prion infection, mice expressing PrPCGPIThy-1 show a significant delay to terminal disease, a decrease of microglia/astrocyte activation, and altered MAPK signaling when compared to wild-type mice. Our results are the first to demonstrate in vivo, that the GPI-anchor signal sequence plays a fundamental role in the GPI-anchor composition, dictating the subcellular localization of a given protein and, in the case of PrPC, influencing the development of prion disease.
Author summary
The prion protein (PrPC) is a glycoprotein attached to the neuronal surface via a GPI-anchor. When misfolded to PrPSc, it leads to fatal neurodegenerative diseases which propagates from host to host. PrPSc is the principal component of the infectious agent of prion diseases, the “prion”. Misfolding occurs at the plasma membrane, and when PrPC lacks the GPI-anchor, neuropathology and incubation time of prion disease are strongly modified. Moreover, the composition of the PrPC GPI-anchor impacts on the conversion process. To study the role of the GPI-anchor in the pathophysiology of prion diseases in vivo, we have generated transgenic mice where the PrPC GPI-signal sequence (GPI-SS) is replaced for the one of Thy-1, a neuronal protein with a distinct GPI-anchor and membrane localization. We found that the resulting protein, PrPCGPIThy-1, shows a different GPI-anchor composition, increased axonal localization, and reduced enzymatic shedding. After prion infection, disease progression is significantly delayed, and the neuropathology and cellular signaling are changed.
The present work demonstrates that the GPI-SS per se determines the GPI-anchor composition and localization of a given protein and it stresses the importance of PrPC membrane anchorage in prion disease.
Introduction
The cellular prion protein (PrPC) is a cell surface GPI-anchored protein (GPI-AP) with two putative N-glycosylation sites [1, 2] targeted to detergent-resistant membranes (DRMs; or lipid rafts)[3]. All GPI-APs share a common GPI-anchor core structure which is highly conserved among species and consists of ethanolamine phosphate linked through an amide group to the carboxyl terminus of the protein, three mannose residues, glucosamine, and a phosphatidylinositol (PI) group. During biosynthesis, the signal sequence for a GPI-anchor (GPI-SS) is rapidly removed after ER translocation, and a common GPI-anchor core is attached to the protein via a GPI-transamidase. Once attached to the protein, this core undergoes several modification steps during ER and Golgi transport involving both elimination/addition of side branching sugars to the glycan moiety together with fatty acid remodeling [4, 5]. How these side chains and lipid moieties are chosen, and how this, in turn, affects the trafficking of GPI-APs is unclear, but it is probably cell- and species-specific and may depend on the functional context [6]. It could also be, that the GPI-SS itself influences GPI-anchor remodeling [7] which could then affect its intracellular sorting [3, 4]. In polarized Madin-Darby canine kidney (MDCK) cells, N-glycosylated GPI-APs are mostly apically sorted, suggesting that the GPI-anchor is an apical sorting signal [8], yet PrPC is an exception in this regard being basolaterally sorted in these cells [9, 10]. When the GPI-SS of PrPC is replaced by the one of the DRM-resident Thy-1, PrPC partially relocates from the basolateral to the apical side in MDCK cells [11]. Likewise, GPI-SS-dependent relocalization of EGFP-tagged proteins occurs in cultured cells [12].
Most recently, Bate et al. demonstrated that sialylation of the PrPC GPI-anchor plays a role in its synaptic targeting [13]. Although it has already been shown that the GPI-SS influences intracellular sorting in vitro, how differences in the GPI-SS impact on the GPI-anchor composition in neuronal cells or in the brain, has not been investigated yet.
A key event in the pathogenesis of prion diseases is a (templated) conformational change of PrPC to its misfolded isoform (PrPSc), the critical component of prion infectivity [14]. Neuropathological characteristics of prion diseases include vacuolization of neuropil and white matter, astro- and microgliosis, neuronal loss and PrPSc deposition [15]. Of outstanding importance for the pathogenesis of prion diseases are the membrane attachment and lipid raft localization of PrPC [16]. Thus, cells expressing GPI-anchorless PrP cannot be infected with prions [17], and cholesterol depletion or expression of a transmembrane PrP-CD4 fusion protein (shifting PrP-CD4 out of DRMs) in cells, interferes with prion propagation [18]. Furthermore, when cell membranes are treated with analogs of GPI-anchors, such as glucosamine-PI, the membrane composition is altered, and the formation of PrPSc is reduced, probably by displacing PrPC from lipid rafts [19]. Along the same line, enzymatic elimination of one GPI-anchor acyl chain in cells removes PrPC from DRMs and reduces PrPSc amounts [20]. Also, mice expressing GPI-anchorless PrPC show delayed clinical onset of prion disease together with altered clinical and neuropathological presentation [21, 22].
Interestingly, the GPI-anchor of PrPC bears a sialic acid, a rare modification for a mammalian GPI-AP [23]. PrPC lacking sialic acid in its GPI-anchor cannot convert to PrPSc in vitro [13, 24]. Sialo and asialo GPI-anchored PrPSc are equally present in infected mice [25]. Therefore, the contribution of the GPI-anchor sialic acid modification to the conversion of PrPC to PrPSc clearly needs further investigations.
In the present study and based on our previous results [11], we assessed how the substitution of the GPI-SS of PrPC for that of Thy-1 influences the biology of the resulting chimeric PrPCGPIThy-1 in vivo, and how this impacts on the pathophysiology of prion disease. We provide evidence that this replacement alters the resulting GPI-anchor composition regarding sialic acid content and leads to relocalization of PrPCGPIThy-1, increasing its presence in axons when compared to wild-type PrPC (WTPrPC). This is accompanied by decreased proteolytic shedding of the protein. After intracerebral challenge with mouse-adapted prions, incubation time to clinical prion disease is extended, correlating with decreased activation of the mitogen-activated protein kinase ERK and reduced glial activation.
Results
Characterization of transgenic PrPCGPIThy-1 mice
We generated four lines of transgenic mice (PrPCGPIThy-1) expressing PrPC with the GPI-SS of Thy-1 (Fig 1A) which were backcrossed into PrP knockout (Prnp0/0 in C57/Bl6 background) mice. Two of the lines, L27 and L16, had the 3F4 tag [26]. However, since this modification can increase prion disease incubation time [27], we also generated two other lines, L150 and L159, without the 3F4 tag, of which L150 was used for prion inoculation experiments. A WTPrPC line was generated with littermates of the founders, which did not contain the transgene but had an identical genetic background. WTPrPC mice were backcrossed with C57/Bl6 mice to avoid differences in prion incubation times depending on the genetic background. All lines described here developed normally for the period observed (>350 days) and did not present any obvious phenotypic alterations.
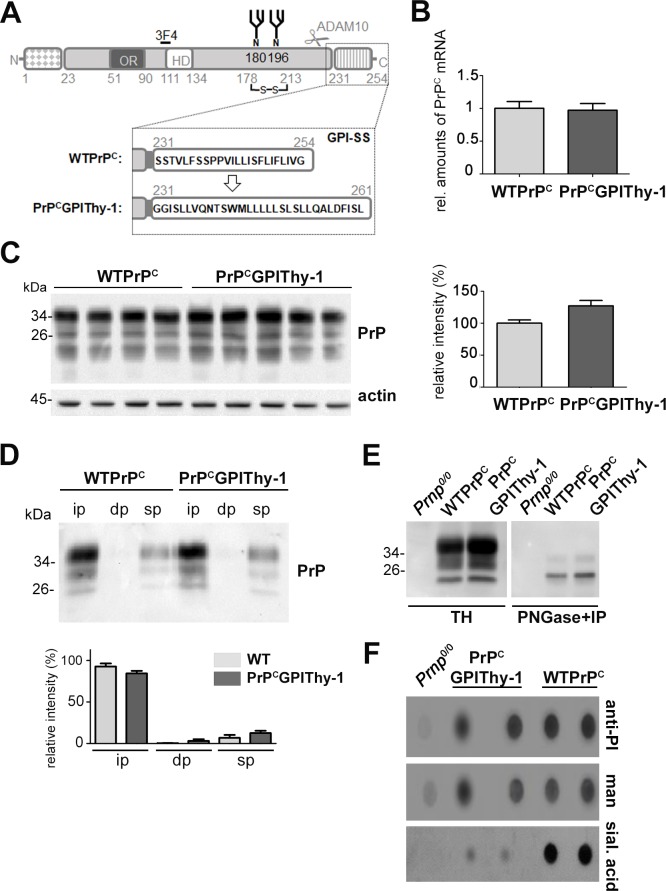
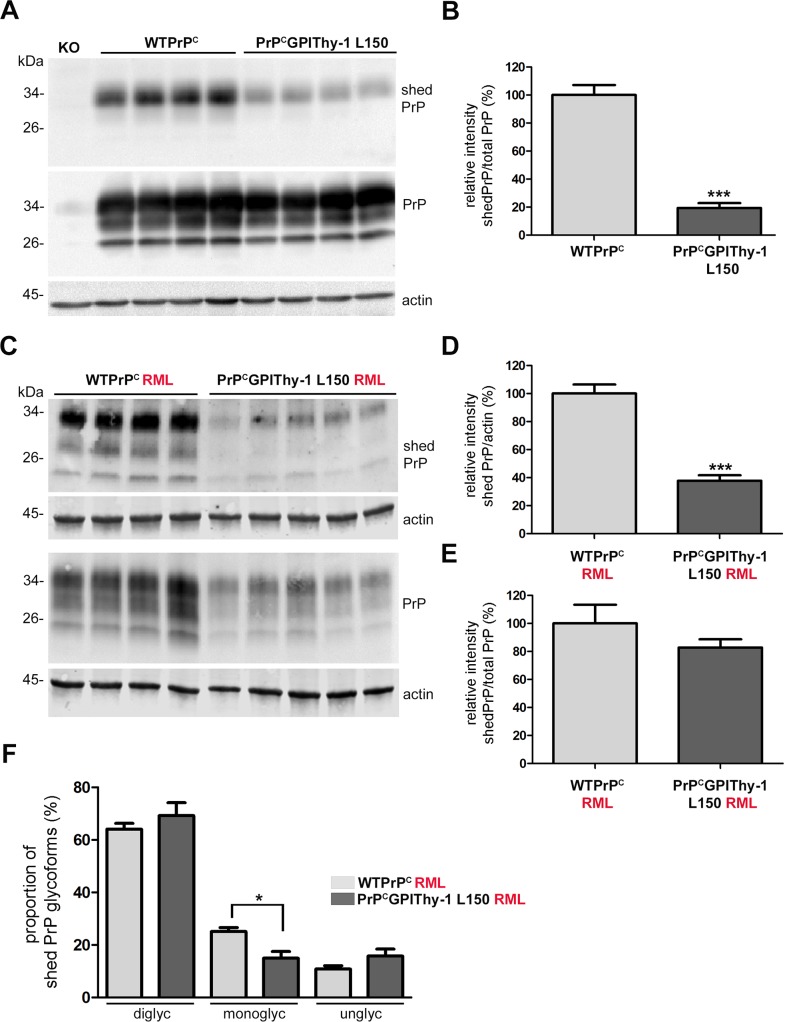
Fig 1. Biochemical characterization of PrPCGPIThy-1 mice.
(A) Schematic representation of the PrPCGPIThy-1 fusion protein. PrPC comprises an N-terminal and a C-terminal (GPI-anchor) signal sequence, both of them not being present in the mature protein. Positions of an octameric repeat region (OR), a hydrophobic domain (HD), two N-glycans (N), a disulfide bridge, the 3F4 tag and epitope (present in mouse lines L27 and L16) and of the cleavage site for the ADAM10-mediated shedding (scissors) are indicated. The substitution of the GPI-SS of PrPC for the one of Thy-1 is shown in the dotted box. The substitution of the GPI-SS of PrPC for the one of Thy-1 is indicated. (B) Relative amount of PrPC mRNA from WTPrPC (n = 5) and PrPCGPIThy-1 (n = 5) measured by RT-qPCR. WTPrPC mRNA is set to one. Error bars are SEM. (C) Representative western blot showing PrPCGPIThy-1 protein expression in brain compared to WTPrPC. Bar chart shows the mean of PrP relative intensity related to actin intensity (used as a loading control). WTPrPC is set to 100%. Error bars are SEM. (D) Triton X-114 phase partitioning assay. Both, WTPrPC and PrPCGPIThy-1 were mainly found in the insoluble phase (ip), indicating that both carry a GPI-anchor as described [28] (dp: detergent phase; sp: soluble phase). Diagram shows the relative signal intensity of PrP from 3 independent experiments. WTPrPC ip is set to 100%. Error bars are SEM. (E) Representative western blot showing total homogenates (TH) from PrP KO (Prnp0/0, used as a negative control), WTPrPC and PrPCGPIThy-1 brains used for PNGase treatment (to eliminate N-glycans) followed by PrP immunoprecipitation (IP), shown in the adjacent western blot. PNGase+IP treated samples were then used to isolate the GPI-anchors, showed in (F). (F) Dot blot analysis of the GPI-anchors from deglycosylated, immunoprecipitated PrP and PK digested samples from mouse brain. Phosphatidylinositol (PI), mannose (man) and sialic acid (sial. acid) were detected as described in Methods. Note that the amounts of PI and mannose are similar between PrPCGPIThy-1 and WTPrPC whereas sialic acid is almost absent in PrPCGPIThy-1. Sialic acid background signal in PrPCGPIThy-1 could be explained by deglycosylation not being 100% achieved as shown in blot in (E).
Analyses of PrPC expression by RT-qPCR and western blot (Fig 1B and 1C) showed similar PrPC-levels for line L27 and a two-fold increase of PrPC levels for line L16 (S1A Fig) when compared to wild-type mice (WTPrPC). Both lines of transgenic mice also showed a similar pattern of expression in different organs as WTPrPC (S1B Fig). L27 was used for the detailed biochemical characterization of PrPCGPIThy1 in mice (Figs 1 and 2).
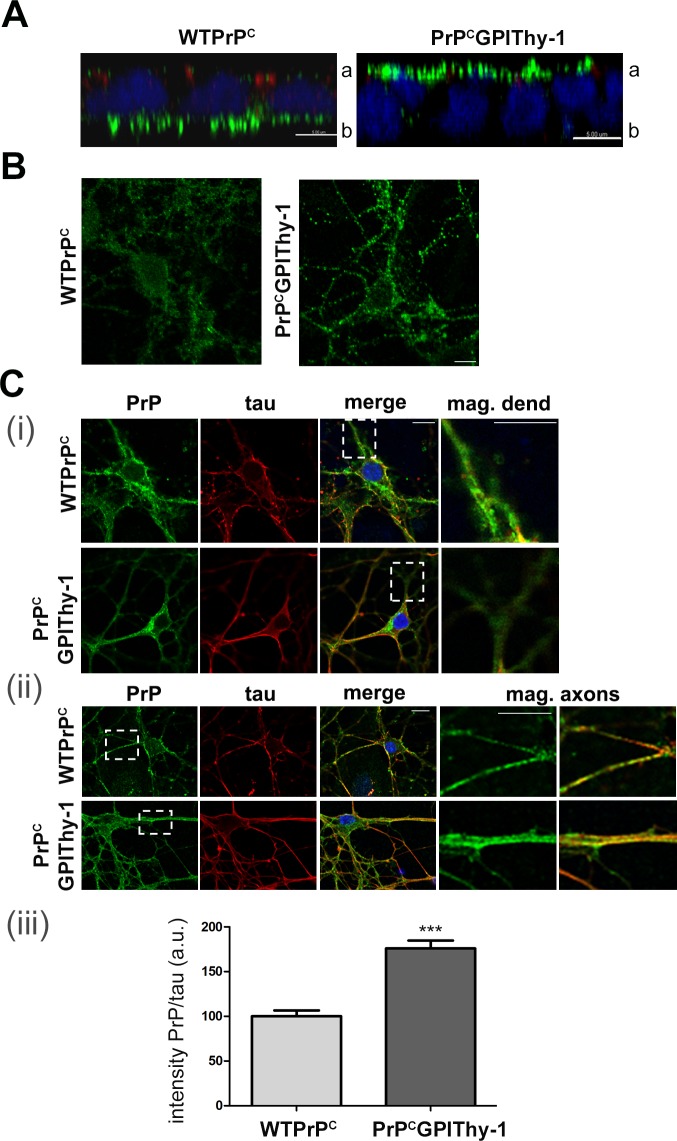
Fig 2. PrPCGPIThy-1 shows altered sorting.
(A) Confocal microscopy showing expression of WTPrPC and PrPCGPIThy-1 (green) in fully polarized MDCK epithelial cells. ZO1 (red) is expressed at the tight junction and delimitates the apical (a)/basolateral (b) side. Note that PrPCGPIThy-1 relocalizes to the apical side compared to WTPrPC, which is predominantly basolaterally located (scale bar is 5 μm). (B) Confocal microscopy of primary neuronal cultures stained with an antibody against PrP (POM1; green) under non-permeabilizing conditions. Both WTPrPC and PrPCGPIThy-1 are expressed at the plasma membrane (scale bars are 10 μm). (C) Representative confocal microscopy pictures of primary neuronal cultures stained with antibodies against PrP (POM1; green) and tau (red). (i) The squares indicate selected areas of the dendrites (where tau is absent) showing decreased PrPCGPIThy-1 amounts whereas staining is present in WTPrPC. (ii) Same staining as in (i) but focusing on tau-positive axons (the squares indicate magnifications where the relative increase in PrP staining at the axons in PrPCGPIThy-1 neurons can be observed). (iii) Bar diagram of a semi-automated quantification showing that the amount of PrPCGPIThy-1 present in tau-positive axons is significantly increased compared to WTPrPC (***p = 0.0001).
To assess whether a proper GPI-anchor was added, we performed a Triton X-114 phase separation assay [28]. We found that PrPCGPIThy-1, like WTPrPC, is mainly present in the insoluble pellet, indicating GPI-anchorage (Fig 1D). Thy-1 and PrPC are both DRM residents, but share different lipid domains at the plasma membrane that can, in principle, be isolated by differential detergent solubilization [29]. We hypothesized that the exchange of the GPI-SS of PrPC for the one of Thy-1 would alter the lipid subdomain localization of the protein. We isolated lipid rafts from frontal cortex using Brij 96 (0.5%) and sodium deoxycholate (NaDOC, 0.5%), as these detergents were described to discriminate between lipid subdomains [30]. As shown in S2A Fig, both WTPrPC and PrPCGPIThy-1 were mainly solubilized and present at the bottom of the gradient (fractions 11 and 12), whereas flotillin, a marker of lipid rafts, and Thy-1 were found in the upper fractions of the gradient under these conditions (S2B Fig). We myelin-depleted the sample before detergent incubation, as myelin interferes with proper detergent solubilization. After solubilization with either a Brij 96 (0.5%)/NaDOC (0.5%) mixture or with Brij 98 (1%) as previously described [31], we obtained similar results, with the majority of WTPrPC and PrPCGPIThy-1 solubilized and present at the bottom of the gradient (S2C Fig).
Since the PrPC GPI-anchor contains sialic acid and the Thy-1 GPI-anchor does not, we assessed sialic acid content of both GPI-anchors. WTPrPC and PrPCGPIThy-1 brain homogenates were first deglycosylated (as the N-glycans of PrPC also contain sialic acid), immunoprecipitated with the PrPC-directed POM1 antibody (Fig 1E) and, after proteinase K (PK) digestion, their GPI-anchors were dot-blotted. As shown in Fig 1F, the amounts of phosphatidylinositol (PI) and mannose (man) as controls did not differ substantially between samples, whereas the amount of sialic acid was drastically reduced in PrPCGPIThy-1, showing a GPI-anchor composition similar to Thy-1 (S3 Fig). Thus, although the sublipid domain occupancy was not altered, the composition of the GPI-anchor was indeed changed. This suggests that the GPI-SS dictates the composition of the resulting GPI-anchor.
Altered sorting of PrPCGPIThy-1 compared to WTPrPC
As we have described [11], PrPCGPIThy-1, in contrast to WTPrPC, is mainly sorted to the apical compartment when expressed in a polarized model of epithelial cells, thus behaving like Thy-1 (Fig 2A). In neurons, PrPC is mainly present in the somatodendritic compartment whereas Thy-1 is more uniformly present in cell bodies and axons [29].
We could observe that, under non-permeabilizing conditions, PrPCGPIThy-1 was found at the plasma membrane similarly to WTPrPC, indicating that GPI-SS does not act on overall plasma membrane localization (Fig 2B). Next, we evaluated the presence of PrPC in axons of primary hippocampal neurons isolated from PrPCGPIThy-1 and WTPrPC mice (Fig 2C). PrPCGPIThy-1 shows a significantly higher degree of localization in tau-positive axons than WTPrPC when assessed in primary neurons (100 ± 6.5% for WTPrPC vs. 176 ± 8.8% for PrPCGPIThy-1; ***p = 0.0001; unpaired t-test).
PrPCGPIThy-1 mice challenged with prions show delayed clinical presentation and different neuropathology when compared to WTPrPC mice
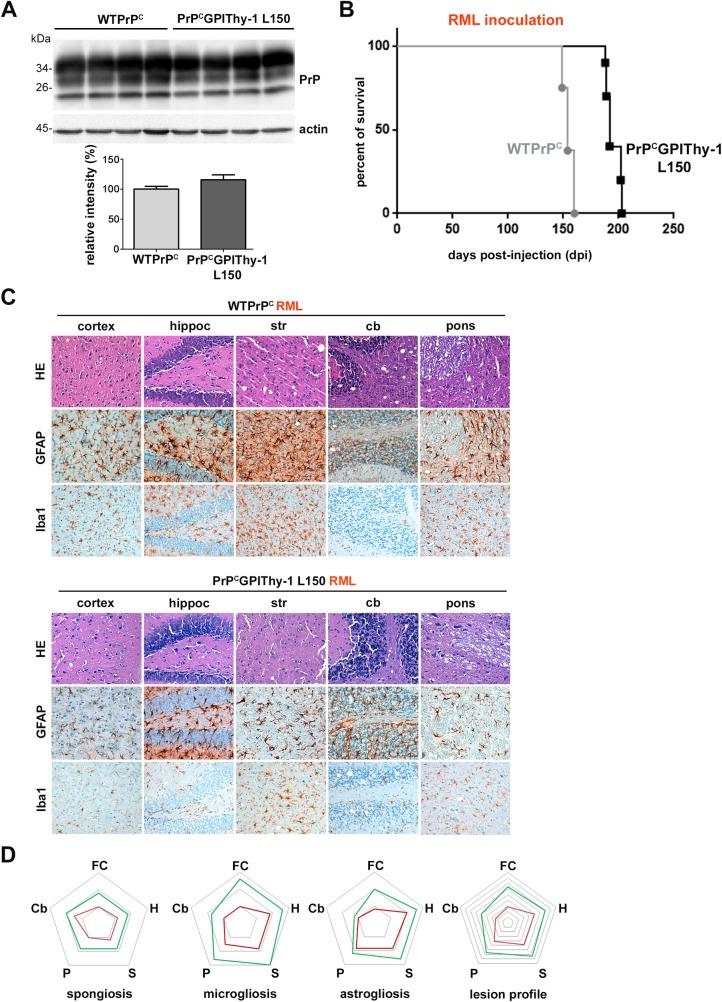
As stated in material and methods, for prion inoculation experiments we generated new lines of transgenic mice lacking the 3F4 tag (S1 Table). As shown in Fig 3A and S4 Fig, we obtained two lines, PrPCGPIThy-1 L150 and PrPCGPIThy-1 L159, with different expression levels of the transgene. We chose L150 for prion infection as these mice expressed amounts of PrPCGPIThy-1 equal to endogenous PrPC levels in WTPrPC mice (Fig 3A and S4A Fig). As shown in Fig 3B, terminal prion disease in transgenic animals inoculated with mouse-adapted RML prions occurred at 195 ± 2 days post-infection (dpi; SEM; n = 10), thus showing a significant delay compared to WTPrPC mice (155 ± 1.6 dpi (SEM; n = 8); Log Rank (Mantel-Cox) ****p<0.0001). The delay was independent of the prion strain as PrPCGPIThy-1 L150 mice inoculated with another strain (22L; S5 Fig), also presented with a significant delay in incubation time (156 ± 3 dpi; (n = 5) compared to 144 dpi in WTPrPC (n = 5); Log Rank (Mantel-Cox) **p = 0.003). Interestingly, transgenic RML-infected mice, although showing a delay to terminal disease, presented a more rapid disease progression after clinical onset than the WT mice (duration of clinical phase: 20 ± 2 dpi in PrPCGPIThy-1 L150, compared to 40 ± 2 dpi in WT [32]).
Fig 3. PrPCGPIThy-1 L150 mice show a delay to terminal disease and altered neuropathology upon prion infection.
(A) Representative blot of PrPGPIThy-1 L150 (generated for infection experiments) and WTPrPC mouse brain homogenates showing no relevant differences in the amount of PrPCGPIThy-1 and WTPrPC in the brain. Chart underneath shows mean relative intensity of PrP referred to actin where WTPrPC samples are set to 100% as a reference (error bars are SEM; n = 5 for each genotype). (B) Kaplan-Meier survival curve. Mice were inoculated intracerebrally (i.c.) with RML prions and sacrificed at terminal disease. Note the significant delay (****p<0.0001) between PrPCGPIThy-1 L150 (n = 10) and WTPrPC mice (n = 8) incubation time. (C) Neuropathological characterization of terminally diseased mice. Note that PrPCGPIThy-1 L150 mouse brains show a general decrease in spongiosis as observed with HE staining. Gliosis is also decreased in the transgenic mice as manifested by reduced GFAP (astrocytes) and Iba1 staining (microglia) in all the studied regions. (D) Lesion profile after semiquantitative assessment (n = 3 or n = 4 for each genotype) of spongiosis and gliosis (FC: frontal cortex; H: hippocampus; S: striatum; P: pons; Cb: cerebellum). Note the general decrease in lesion severity for the PrPCGPIThy-1 L150 mice (red line), compared to WTPrPC profile (green line).
Sagittal brain sections of terminally prion-diseased WTPrPC and PrPCGPIThy-1 L150 mice were neuropathologically examined. As shown in Fig 3C, RML prion-infected PrPCGPIThy-1 L150 mice presented with decreased spongiosis and less severe astro- and microgliosis when compared to WTPrPC. By analyzing the lesion profile (as described elsewhere [33], Fig 3D), we found an overall decrease in the severity of prion-associated lesions, namely less spongiosis in all the studied areas, reduced microgliosis (less pronounced in the cerebellum) and astrogliosis (although with similar amounts in pons).
In another set of experiments, we also inoculated PrPCGPIThy-1 L16 mice with RML and 22L mouse-adapted prions. RML-inoculated transgenic mice reached terminal disease after 400 ± 56 dpi (n = 10), whereas WTPrPC mice became terminally sick at 155 ± 1.6 dpi (n = 8), thus also showing a highly significant delay for PrPCGPIThy-1-expressing mice (Log Rank (Mantel-Cox)****p<0.0001; S6A Fig). Upon inoculation with 22L prions, PrPCGPIThy-1 L16 mice again presented with a significant delay (200 ± 23 dpi; n = 5) compared to controls (144 ± 1 dpi; n = 5; Log Rank (Mantel-Cox) **p<0.003; S6A Fig). Although this drastic delay to terminal disease in PrPCGPIThy-1 L16 mice may partially be explained by the presence of the 3F4 tag, decreased lesion severity in these mice was conspicuously similar to the infected PrPCGPIThy-1 L150 mice lacking the 3F4 tag, with an overall decrease in spongiosis and glial activation (S6B Fig).
The PrPSc pattern differs between PrPCGPIThy-1 and WTPrPC mice
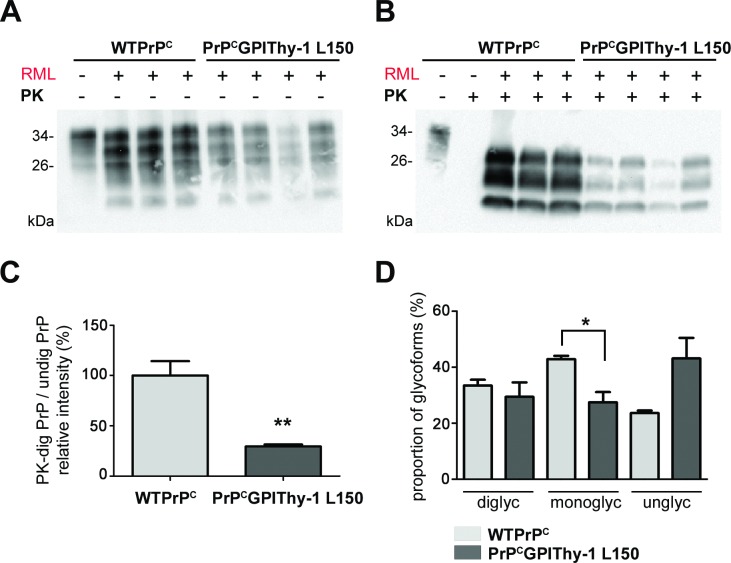
To investigate the amount and type of PrPSc, we performed biochemical analyses of brain samples from terminally prion-diseased mice. Interestingly, PrPCGPIThy-1 L150 mice infected with RML showed decreased amounts of total PrP (Fig 4A) with significantly less PrPSc compared to WTPrPC mice (100 ± 14.26% for WTPrPC mice vs. 29.4 ± 2% for PrPCGPIThy-1 L150 mice; Fig 4B and 4C, **p = 0.0021; unpaired t-test). Likewise, RML-infected PrPCGPIThy-1 L16 mice also had significantly less PK-resistant PrPSc (S6C Fig). The reduction in total PrP was not due to an age-dependent decrease in PrPC expression as we did not observe significant differences between 20 and 40 weeks-old PrPCGPIThy-1 L150 mice (S7A Fig). Moreover, no significant differences in total PrP levels were observed between terminally prion-diseased (around 30 weeks old) and non-infected PrPCGPIThy-1 L150 mice (40 weeks old; S7B(ii) Fig). Taken together, these findings suggest that the relative decrease of total PrP after infection in PrPCGPIThy-1 is due to less efficient conversion to PrPSc in these mice compared to WTPrPC. In addition, the PrPSc glycopattern was also changed in prion-diseased PrPCGPIThy-1 L150 mice, which showed significantly less monoglycosylated PrPSc (Fig 4D, 42.9 ± 1.9% in WTPrPC vs. 27.5 ± 3.6% in PrPCGPIThy-1 L150; *p = 0.0173, unpaired t-test).
Fig 4. The amount of PK-resistant PrP is decreased, and the glycopattern is changed in PrPCGPIThy-1 L150 mouse brains after RML prion infection.
Representative blots of total brain homogenates from terminally sick WTPrPC and PrPCGPIThy-1 L150 mice infected with RML prions, without (A) and after (B) PK digestion at 37°C and detected with POM1 antibody. Note that there is a significant overall decrease in the total amounts of PrPCGPIThy-1 in the infected brains (n = 4) which is more conspicuous after digestion with PK (B). (C) Bar chart representing the mean relative intensity of PK-digested samples related to the undigested amounts. Note the significant decrease in the amount of resistant PrPSc for PrPCGPIThy-1 compared to WTPrPC (**p = 0.0021). (D) Quantification of the glycoform banding pattern of PrPSc. The intensity of each band in the PK-digested samples was measured and referred to the total amount of PrP as a percentage. The glycopattern is significantly changed in PrPCGPIThy-1 L150 after RML infection, where the monoglycosylated isoform is less PK-resistant compared to WTPrPC (*p = 0.019).
Proteolytic shedding of PrPCGPIThy-1 is reduced compared to WTPrPC
PrPC undergoes several physiological cleavages which are highly conserved through evolution, indicating essential functions [34]. We have previously shown that the metalloprotease ADAM10 is the responsible protease for the shedding of PrPC in vivo [35] and that impaired shedding has significant consequences for prion diseases [32]. Thus, to investigate the possible mechanisms implicated in the clinical delay and the altered neuropathology, we decided to analyze the shedding of PrPCGPIThy-1. We have recently generated an antibody (sPrPG228) that specifically recognizes shed PrP (as the antibody is directed against the carboxy terminus Gly228 only exposed upon ADAM10-mediated cleavage) allowing for the direct detection of shed PrP in mouse brain homogenates [36]. By using this antibody, we found a significantly decreased shedding in PrPCGPIThy-1 L150 and PrPCGPIThy-1 L159 mice compared to WTPrPC (Fig 5A and 5B; 100 ± 72% in WTPrPC vs. 19.37 ± 3.5% in PrPCGPIThy-1 L150; ***p = 0.0005; S8A and S8B Fig, 100 ± 14.8% in WTPrPC vs. 37.5 ± 3.9% in PrPCGPIThy-1 L159; *p = 0.015; unpaired t-test). This indicates that the altered GPI-anchor and/or localization of PrPCGPIThy-1 interferes with the ADAM10-mediated release from the plasma membrane. Remarkably, in terminally diseased RML-infected PrPCGPIThy-1 mice, even though total PrP shedding is decreased compared to WTPrPC (Fig 5C and 5D, 100 ± 6.4% in WTPrPC vs. 37.77 ± 3.9% in PrPCGPIThy-1 L150; ***p<0.0001; unpaired t-test), it does not show differences when it is referred to total PrP amounts (Fig 5E). This indicates a relative increase of PrPCGPIThy-1shedding upon infection, which was further confirmed by comparing levels of shed PrPCGPIThy-1 between non-infected and infected mice (S7B(i) Fig; 100 ± 11.8% in PrPCGPIThy-1 L150 vs. 272 ± 21% in PrPCGPIThy-1 L150 RML infected; ***p = 0.0004; unpaired t-test). We also observed that in brains of both, RML-infected WTPrPC and PrPCGPIThy-1 L150 mice, all PrP glycoforms could be shed (Fig 5C), contrasting with the almost exclusive shedding of diglycosylated PrPC in non-infected samples (Fig 5A and S7 Fig, also characterized in [36]). In PrPCGPIThy-1 L150 mice, the monoglycosylated isoform is significantly less shed compared to WTPrPC (Fig 5F, 25.08 ± 1.6% in WTPrPC vs. 15 ± 2.5% in PrPCGPIThy-1 L150; *p = 0.0158; unpaired t-test), possibly reflecting its decreased presence in these infected brains as shown in Fig 4D. In conclusion, ADAM10-mediated shedding is significantly altered for PrPCGPIThy-1 compared to WTPrPC under both physiological and pathological conditions. In order to properly interpret these results, one should keep in mind that shed PrP (made visible by the highly sensitive sPrPG228 antibody) represents only a minor fraction of the total PrP pool (repeated own observations and [37]).
Fig 5. Shedding of PrP is decreased in PrPCGPIThy-1 L150 mice but increases after RML infection.
(A) Representative western blot showing shed PrP (detected with our new antibody described in [36]) and total PrP levels (detected with POM1). The blots for shed PrP and total homogenates were run in parallel (n = 4 for each genotype). The diglycosylated band is preferentially shed in both WTPrPC and PrPCGPIThy-1 L150 brains, but in the latter, shedding is reduced to 27% (***p = 0.0005) as shown in the bar chart (B). To quantify, each blot (shed and total PrP) was first referred to actin prior to the relative quantification of shed PrP referred to total PrP. WTPrPC is set to 100% and error bars are SEM. (C) Representative western blot of shed PrP and total PrP from RML infected brain homogenates of terminally sick mice (n = 4 for WTPrPC and n = 5 for PrPCGPIThy-1 L150). Note that in the infected brains all the isoforms are shed, in both WTPrPC and PrPCGPIThy-1 L150. (D) Bar chart showing the quantification of shed PrP referred to actin. WTPrPC is set to 100%. Note that shedding in PrPCGPIThy-1 L150 is still decreased compared to WTPrPC (***p = 0.0004). (E) Bar chart showing that when shed PrP is referred to the total PrP in RML infected mice, there are no significant differences in the relative amount of shedding, implying a relatively increased shedding in infected PrPCGPIThy-1 L150 brains. Blots were run in parallel and first referred to its actin. WTPrPC is set to 100%. (F) Quantification of the shed PrP glycopattern. Each band intensity was referred to the total amount of shed PrP. In infected brains, the monoglycosylated isoform is significantly less shed in PrPCGPIThy-1 L150 mice (*p = 0.0158).
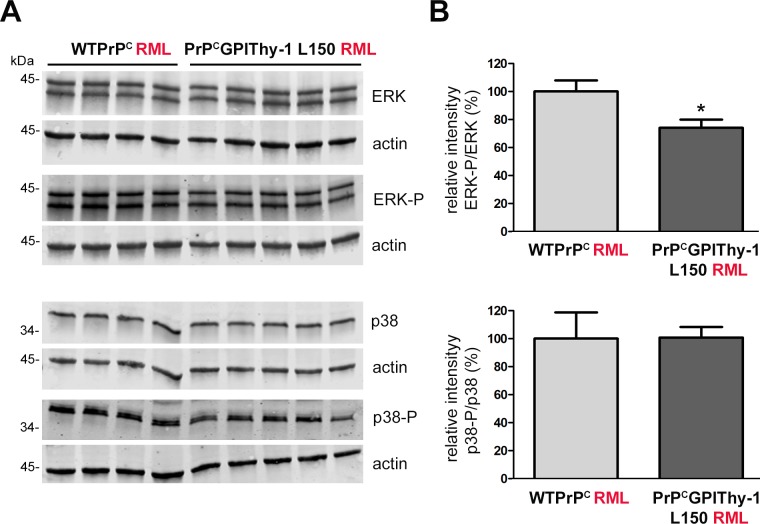
Decreased ERK activation in brains of PrPCGPIThy-1 L150 mice after RML infection
Activation of ERK1/2, a member of the mitogen-activated protein kinase (MAPK) family, can promote the formation of PrPSc in a cell model of prion infection [38]. In the same study, it was demonstrated that activation of other MAPKs, such as p38 or JNK, has an adverse effect on the formation of PrPSc. We have recently shown that a C-terminal deletion retains PrP in the secretory pathway, leading to p38 activation and neuronal death [39]. Since in the present study RML-infected PrPCGPIThy-1 L150 mice showed decreased PrPSc formation and increased survival, we performed western blot analyses to assess the status of ERK1/2 and p38 MAPK phosphorylation. As shown in Fig 6A and 6B, clinically terminal PrPCGPIThy-1 L150 mice presented a significant decrease of phosphorylated ERK1/2 (*p = 0.031) when compared to WTPrPC. Phosphorylation of p38 instead was not significantly changed at terminal disease.
Fig 6. Phosphorylated ERK (ERK-P) is decreased in terminally prion-diseased PrPCGPIThy-1 L150 mice.
(A) Representative western blots of total brain homogenates from prion diseased mice infected with RML prions, incubated with antibodies against total ERK and ERK-P (upper part) as well as total p38 and p38-P (lower part). (B) Quantifications of the relative intensity of ERK-P signal related to total ERK signal and p38-P signal (upper part) related to the total p38 signal (lower part). (WTPrPC, n = 4; PrPCGPIThy-1, n = 5). For the quantification, each signal was first related to the corresponding actin signal. Note that there is a significant decrease in ERK-P signal (*p = 0.031) in PrPCGPIThy-1 RML infected mice compared to WTPrPC mice, whereas no changes are observed for p38-P.
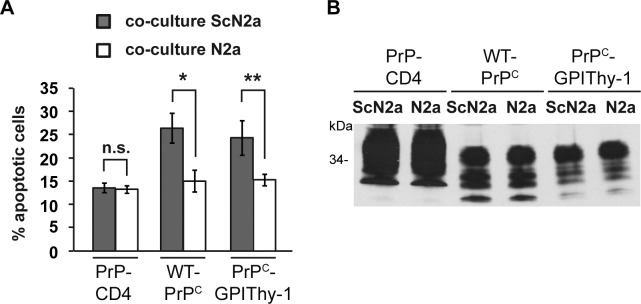
PrPCGPIThy-1, like WTPrPC, mediates toxic PrPSc-associated signaling
Since MAPK signaling was changed, and to assess whether PrPCGPIThy-1 is capable of mediating pro-apoptotic signaling induced by PrPSc, we employed a cell culture model [40]. This assay is based on the co-cultivation of PrPC-expressing cells with chronically prion-infected cells that release PrPSc into the cell culture medium. As illustrated in Fig 7A, co-cultivation of SH-SY5Y cells expressing PrPC with prion-infected mouse neuroblastoma (ScN2a) cells increased apoptotic cell death, as determined by activation of caspase-3. In contrast, SH-SY5Y cells expressing a PrP mutant containing a heterologous C-terminal transmembrane domain instead of the GPI-anchor (PrP-CD4) could be co-cultured with ScN2a cells without signs of apoptosis. Notably, PrP-CD4 also inhibits PrPSc-formation in scrapie-infected neuroblastoma cells [18]. However, SH-SY5Y cells expressing PrPCGPIThy-1 transduce PrPSc-mediated toxicity similar to WTPrPC. This supports that, despite an altered GPI-anchor composition and localization at the cell surface, PrPCGPIThy-1 is still able to contribute to the formation of signaling-competent complexes relevant to prion diseases.
Fig 7. PrPCGPIThy-1 can induce prion-associated apoptosis in cells.
(A) SH-SY5Y cells expressing the indicated PrP constructs (PrP-CD4 is a transmembrane version of PrP and served as a negative control) were co-cultured with ScN2a or N2a cells for 16 h. For quantification of apoptotic cell death, SH-SY5Y cells were fixed, permeabilized and stained for active caspase-3. Quantifications are based on triplicates of at least three independent experiments. (B) Comparable expression of PrP versions was confirmed by western blot using the anti-PrP antibody 3F4.
Discussion
The GPI-anchor is a complex structure for the attachment of proteins to the outer leaflet of the plasma membrane that involves more than 20 proteins in its production [5]. It bestows the capacity to localize in lipid rafts (determining compartmentalization), and although it does not reach the intracellular space, it can confer the ability of signal transduction through transmembrane spanning partners [41, 42]. The signal sequence for GPI-anchor attachment has a wide range of sequence diversity and its participation in determining the final composition of the GPI-anchor itself is not known. It is assumed that remodeling of the core GPI-anchor depends on the protein it is attached to, and on the cell type by which it is synthesized [42]. In the present study, we have made several novel observations important not only for prion diseases but the biology of GPI-APs. We could show in a newly generated transgenic mouse model that (i) by changing the amino acids of the GPI-SS, the composition of the GPI-anchor is modified, resulting in a differential sorting and proteolytic processing of the protein. Upon infection with prions, these alterations (ii) are associated with significantly prolonged prion disease incubation time, and (iii) influence the conversion to PrPSc as well as the neuropathological presentation including decreased gliosis and spongiosis. Fittingly, we also found (iv) reduced activation of the ERK signal cascade during prion disease (please refer to Fig 8 for a graphical summary of the principal findings). To our knowledge, this study is the first to demonstrate the importance of the GPI-SS in determining a GPI-anchor composition and sorting of a GPI-AP in a mouse model. In the specific case of PrPC, this has a direct link to the pathophysiology of prion disease.
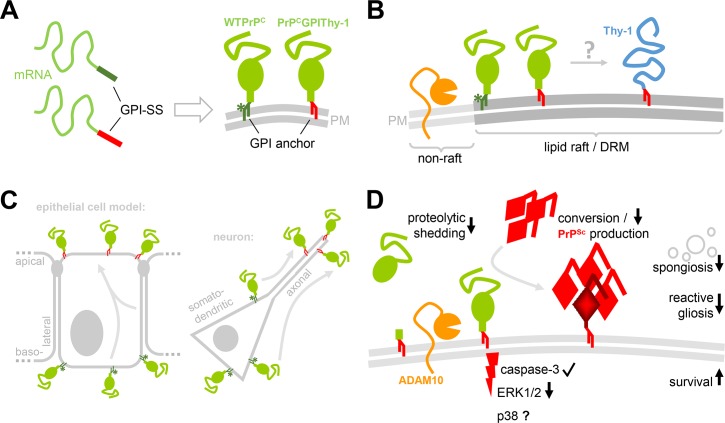
Fig 8. Graphical summary of research aim and key findings.
(A) To study the role of the GPI-anchor signal sequence (GPI-SS) in determining localization and biology of a protein, the GPI-SS of PrPC (dark green) was exchanged for the one of Thy-1 (red). Both proteins, WTPrPC and the PrPCGPIThy-1 mutant, are expressed at the cell surface and comprise a GPI-anchor. A sialic acid modification (green asterisk) typical for the GPI-anchor of PrPC is lacking in the GPI-anchor of PrPCGPIThy-1 (note its red GPI-anchor referring to the GPI-anchor of Thy-1 [in B]). This suggests that an altered GPI-SS in the mRNA results in a different GPI-anchor of the mature protein. (B) PrPC (green) and Thy-1 (blue) are both known residents of lipid rafts/DRMs. While PrPC is located in their periphery and able to leave and re-enter these membrane subdomains, Thy-1 has been shown to occupy more central regions therein. Despite several biochemical methods applied in our study, we were unable to demonstrate a relative re-distribution of PrPCGPIThy-1 towards Thy-1 compared to WTPrPC. (C) Nevertheless, the overall cellular sorting is altered with PrPCGPIThy-1 being relatively more transported towards the apical compartment compared to WTPrPC with its predominant basolateral sorting in a polarized epithelial cell model such as MDCK cells. This also holds true in our transgenic mice and translates to an increased axonal sorting of PrPCGPIThy-1 in primary neurons compared to a mainly somatodendritic presence of PrPC in wild-type neurons. (D) Altered GPI-anchor composition and sorting of PrPCGPIThy-1 results in different biological consequences (in comparison to WTPrPC): (i) Endogenous proteolytic shedding by the metalloprotease ADAM10 (orange) at the cell surface is reduced. (ii) Although PrPCGPIThy-1, in principle, is able to transduce PrPSc-associated toxic signaling (e.g. via cleaved caspase-3), signaling via the MAP kinase ERK1/2 is reduced upon prion infection. Though not investigated here, p38 signaling may be reduced at early time points, contributing to delay to terminal disease (showed as p38?). Key hallmarks of prion-associated neuropathology are also altered in the transgenic mice including (iii) decreased PrPSc production and deposition, (iv) reduced vacuolization (spongiosis) of the brain parenchyma, and (v) reduced induction of astrocytes and microglia (reactive gliosis). These changes are accompanied with prolonged survival of the PrPCGPIThy-1 mice and support a relevant impact of the GPI-SS on a GPI-AP`s biology.
Our previous observation in a model of polarized epithelial cells already indicated that the GPI-SS plays a role in protein sorting. We observed that, by changing the GPI-SS of PrPC for the one of Thy-1, PrPCGPIThy-1 was partially, yet significantly, relocalized from the basolateral to the apical side [11], confirming previous observations from others [12]. In the present work, we extended our observations to transgenic mice expressing PrPCGPIThy-1 on a Prnp knock-out background. PrPCGPIThy-1 is correctly expressed at the plasma membrane, showing identical N-glycosylation as WTPrPC, and is located in lipid rafts. In primary hippocampal neurons, we showed a 1.8-fold increase of PrPCGPIThy-1 in axons when compared to controls, despite equal PrPC expression levels. Notably, we showed that by changing the GPI-SS, the biochemical composition of the GPI-anchor is altered, presenting with a loss of the sialic acid usually present in the GPI-anchor of PrPC. It has recently been shown that the sialic acid in the GPI-anchor of PrPC has a role in the synaptic targeting of PrPC in cultured cells [13]. Thus, when sialic acid from exogenously administered GPI-anchored PrP is depleted, or the lipid moiety is changed, PrPC still partitions in DRMs but is no longer targeted to synapses. In vivo, we also observed that PrPCGPIThy-1 lacking sialic acid is differently sorted, but we observed an increase in axonal targeting. The fact, that not only the sialic acid is missing but also the GPI-SS was changed for PrPCGPIThy-1, may account for these differences.
The fact that in the GPI-anchor of PrPCGPIThy-1 not only the sialic acid is missing but also the SS-GPI anchor is changed, can account for these differences. Our mouse model provides the opportunity to address this on the molecular level.
In mammals, the sorting of GPI-APs is complex, and in epithelial cell models it appears to depend on (i) the presence of the GPI-anchor itself, (ii) its remodelling in the trans-Golgi network (TGN), (iii) its partitioning in DRMs, and (iv) the capacity to oligomerize in the Golgi, which in turn depends on the clustering of DRMs among others [7, 43]. In neurons, the signals implicated in the targeting of proteins either exclusively to dendrites/axons or to both sides, are not clear. For GPI-APs, such as Thy-1, it has been suggested that early interaction with DRM components (especially with sphingomyelin and cholesterol) is necessary for axonal delivery of Thy-1 in mature neurons [44]. According to our data and recent findings of others, it could be hypothesized that the GPI-SS itself directs the protein to different lipid environments, which then influence how the GPI-anchor will be remodeled in the TGN, thereby affecting its oligomerization capacity and, hence, its further sorting [7, 45]. Actually, PrPC and Thy-1 are both located in DRMs but occupy different subdomains therein [30]. However, even after using different detergents previously reported to allow for discrimination between PrPC- and Thy-1-enriched DRM subdomains [29, 31], we were unable to observe differences in the distribution between PrPCGPIThy-1 and WTPrPC.
Nonetheless, support for a relocalization of PrPCGPIThy-1 to a different subdomain comes from our finding of reduced ADAM10-mediated shedding. This proteolytic cleavage at the cell surface is likely to occur at the interface between raft and non-raft regions, where–supported by the ability of PrPC to leave and re-enter DRMs [46]–protease and substrate are thought to interact [34]. In that regard, the in vivo data presented here complement our recent finding of significantly decreased shedding of PrPCGPIThy-1 in N2a cells [36]. Relocalization of the prion protein from the periphery of rafts towards more central regions (where Thy-1 resides [29]) could, therefore, explain the reduced shedding in PrPCGPIThy-1 mice. It is interesting that, despite reduced shedding compared to WTPrPC under normal conditions, upon prion infection this cleavage is relatively increased in PrPCGPIThy-1 mice. We and others have shown that shedding interferes with PrPSc formation and beneficially influences prion disease incubation times [32, 47, 48]. Thus, upregulation of shedding during prion disease could represent a protective feedback mechanism to lower PrPSc production and prolong survival in our transgenic mice.
Our further aim was to elucidate how the differential localization of PrPCGPIThy-1 impacts on prion disease. We intracerebrally inoculated our transgenic and WTPrPC mice with prions. Upon RML-infection, PrPCGPIThy-1 mice showed (i) delayed onset of terminal disease, (ii) different neuropathological presentation with decreased spongiosis, gliosis, reduced PrPSc amounts, and an altered PrPSc-glycotype pattern, (iii) relatively increased shedding, and (iv) a decrease in ERK phosphorylation. Although, as demonstrated by others [27, 49, 50] and now confirmed by us, the 3F4 tag prolongs prion disease incubation times, we included these mice (PrPCGPIThy-1 L16) as they showed several neuropathological characteristics that were likewise present in the non-3F4-tagged transgenic mice (PrPCGPIThy-1 L150), thus strengthening our overall results.
It is known that not only the presence of PrPC but also its type of membrane anchorage is fundamental for the pathophysiology of prion disease. Briefly, (i) presence of extracellular PrPSc does not lead to prion-induced neurodegeneration in the absence of PrPC on neurons [51, 52], (ii) cells expressing either anchorless PrP or PrP with a transmembrane domain instead of the GPI-anchor are resistant to prion infection [17, 18, 53], and (iii) prion-infected transgenic mice expressing low amounts of anchorless PrP do not show clinical symptoms of prion disease despite high titers of infectivity and high levels of PrPSc [21, 54]. When the expression of anchorless PrP is increased, this leads to delayed onset of disease and generation of a new prion strain [22, 55]. Remarkably, the type of GPI-anchor also affects PrPSc formation. Thus, when amino acids in the C-terminus or within the GPI-SS of the murine sequence are replaced by their cunicular homologs (with rabbits being naturally resistant to prion infection), this modified PrPC cannot be converted to PrPSc in a cell culture model [56].
Moreover, the presence of sialic acid in the GPI-anchor can influence conversion as PrP with a desialylated GPI-anchor cannot be converted to PrPSc and can even stop an ongoing infection in vitro [57]. Thus, a desialylated GPI-anchor may modify the lipid environment leading to inhibition of the signal transduction associated with PrPSc neurotoxicity. Fitting to this, our model with a PrP mutant lacking sialic acid in its GPI-anchor also shows delay to terminal disease as well as altered signal transduction when compared to WTPrPC, highlighting the importance of the sialic acid modification of the GPI-anchor in prion disease. However, in vivo we found conversion of PrPCGPIThy-1 to be reduced, not completely abolished.
As stated above, the sialylation status of the PrPC GPI-anchor modifies the lipid environment in vitro [58]. PrPC and Thy-1 share different lipid environments, and it has been shown that the lipid environment composition is fundamental for conversion of PrPC to PrPSc [59, 60]. PrPC-enriched lipid rafts isolated from rat brain have a significant increase in cholesterol compared to Thy-1-containing lipid rafts [30]. In vitro, desialylated PrPC is found in lipid rafts with increased cholesterol content, probably stabilizing PrPC in lipid rafts and increasing its half-life at the plasma membrane [58]. Although, by performing differential detergent extraction, we could not detect a differential distribution of PrPC/PrPCGPIThy-1 in lipid rafts, we cannot rule out that the lipid environment of PrPCGPIThy-1 is altered in a way that impairs the conversion to PrPSc.
In prion-infected mice both, sialo and asialo GPI-anchored PrPC forms, can be converted to PrPSc and are present in infected brains and spleens [61, 62]. Our results, where delay to clinical disease is associated with a significant decrease in the amount of PK-resistant PrPSc, suggest that asialo forms can be converted in vivo but with comparably low efficiency. Interestingly, Katorcha et al. [63] have found that a decrease in PrP N-glycan sialylation leads to a different glycoform pattern after prion infection. They also observed that when prion-infected desialylated brain homogenates where inoculated to Syrian hamsters, there was a drop in infectivity and PK-resistance. Because in their experiments they used a sialidase that can also eliminate the sialic acid of the GPI-anchor [25], and in view of our present results, it would be interesting to study if the sialic acid of the PrP GPI-anchor also partially contributes to the observations of Katorcha et al.
Several other aspects may also contribute to reduced PrPSc conversion and prolonged survival of our PrPCGPIThy-1 mice. On the one hand, Nemoto et al. [64] have recently described that the behavior of Thy-1 and PrPC at the plasma membrane is different, with PrPC showing a slower membrane diffusion compared to Thy-1. This may increase the probability of homophilic interactions for WTPrPC but not for PrPCGPIThy-1. Since the conversion of PrPC to PrPSc occurs at the plasma membrane [65], this effect could favor interactions between WTPrPC and critical PrPSc seeds, thus enhancing the propagation of PrPSc. In contrast, the lateral diffusion behavior of PrPCGPIThy-1 may be more similar to that of Thy-1, thus decreasing the conversion rate. On the other hand, the GPI-anchor itself influences the structure of a given protein [41]. In the case of PrPC, this could result in structural hindrance and, thus, less efficient PrPC to PrPSc conversion.
PrPCGPIThy-1 mice presented with significantly increased incubation times with two different prion strains, RML and 22L (with longest delay to terminal disease with RML prions). Several factors may account for that. On the one hand, each prion strain has different cellular tropism [66] and 22L is mainly associated to astrocytes where it mediates indirect neuronal damage. On the other hand, the fact that RML and 22L prions use different endocytic pathways to infect the cell [67], could indicate that PrPCGPIThy-1 follows an altered endocytic pathway that facilitates 22L over RML infection.
Intriguingly, after prion infection, we observed a decrease in the glial response in all PrPCGPIThy-1 lines compared to controls. Prion diseases, like other neurodegenerative diseases, are accompanied by an increase in microglia with a phagocytic phenotype and by reactive astrocytes. Microglia, the resident immune cells of the central nervous system, may act beneficial or detrimental in prion diseases [68, 69]. In the present study, a decreased amount of microglia in PrPCGPIThy-1 mice coincides with a delay to the terminal stage of prion disease, but further experiments would be needed to establish a direct correlation. Of note, a recent study has functionally linked ADAM10-mediated shedding of PrPC to inflammatory responses and monocyte recruitment to the brain [70]. So, it is conceivable that reduced levels of shed PrP (as a potential chemoattractant and activating factor) account for the impaired glial response in our mice. With regard to astrogliosis, it has been shown that astrocytes (i) accumulate PrPSc early in the disease [71], (ii) can rapidly internalize, traffic, and spread PrPSc in cell culture [72, 73], and (iii) that astrocytic PrPC supports the development of prion disease in mice [74]. Since astrogliosis is decreased in our transgenic mice, this could contribute to both, delay to terminal disease and reduced PrPSc deposition. Moreover, because destruction of the extracellular matrix due to factors released by microglia and astrocytes may lead to the vacuolization observed in prion diseases [75, 76], decreased gliosis in PrPCGPIThy-1 mice could also explain their low degree of spongiosis. Interestingly, lack of vacuolization of the gray matter was also observed in prion-infected mice expressing GPI-anchorless PrP [22].
The GPI-anchor is necessary for toxic signaling associated to PrP [40] and it seems plausible that an altered GPI-anchor and localization of PrPC affect its association with signaling-competent, membrane-spanning binding partners and, thus, signaling outcome [77]. We observed that, in vitro, PrPCGPIThy-1 is as able as WTPrPC to transduce neurotoxic signals, and, in vivo, we showed decreased ERK1/2 phosphorylation whereas p38 MAPK was unchanged at a terminal stage of disease. Using an in vitro model of prion infection, Fang et al. [78] recently showed that phosphorylation of p38 is an early event in synaptotoxicity and that PrPSc specifically targets post-synaptic PrPC. In our transgenic mice, PrPCGPIThy-1 is partially depleted from the dendritic compartment, and we observed a delay in the clinical onset. We did not detected a decrease in p38 phosphorylation but we cannot rule out the possibility that, at an earlier disease state, p38 activation in PrPCGPIThy-1 mice might be lower than in controls, thus contributing to delayed disease onset.
An increase in ERK1/2 signaling has been consistently observed in cellular [79–82], mouse [83] and hamster models of prion infection [84], and is probably related to neurodegeneration and cell death. Activated ERK1/2 also participates in the conversion of PrPC to PrPSc in vitro [38]. Accordingly, it could be hypothesized that the decrease in ERK phosphorylation in our model delays clinical disease and reduces PrPSc deposition. How exactly PrPCGPIThy-1 affects MAPK signaling deserves further studies, but it is interesting that interfering with this signaling cascade is considered a potential therapeutic option [80, 81]. However, since astrocytes represent the main cell population that increases ERK phosphorylation after prion infection [83], it is also possible that the observed reduction is due to reduced astrogliosis in our transgenic mice.
Although PrPCGPIThy-1 mice present with extended survival and delay to clinical disease, we observed a faster progression to terminal disease compared to controls. Several factors can account for this. On the one hand, the different localization of PrPCGPIThy-1 may induce alternative signaling pathways not investigated in this study and these could follow a kinetic leading to rapid disease development once a critical threshold is reached [85]. On the other hand, glia is less activated (maybe due to decreased levels of PrPSc), which may prolong survival (given less inflammatory brain injury) but could also contribute to toxicity as non-functional cells and PrPSc may not be efficiently eliminated. The final sum up of positive and negative factors may influence the complex findings observed in PrPCGPIThy-1 mice.
In conclusion, we showed that the GPI-SS influences GPI-anchor composition and localization of a GPI-AP in vivo. This study attributes a novel function to the GPI-SS in subcellular trafficking in vivo and sheds light on several molecular events underlying prion-associated neurodegeneration.
Materials and methods
Ethics statement
Animal experiments were approved by the Behörde für Gesundheit und Verbraucherschutz of the Freie und Hansestadt Hamburg (permit numbers 80/08, 38/07 and 84/13). All the procedures were performed under the guidelines of the animal facility of the University Medical Center Hamburg-Eppendorf and in compliance with the Guide for the Care and Use of Laboratory Animals. Mice used for prion infection were anesthetized with a mixture of Xylazin hydrochloride and Ketamine hydrochloride in 0.9% NaCl prior to intracerebral prion inoculation. To sacrifice the mice, first they were anesthetized with halothane followed by neck dislocation.
Generation of transgenic PrPCGPIThy-1 mice
The generation of the PrPCGPIThy-1 construct was already described elsewhere [11]. To insert PrPCGPIThy-1 in the half-genomic expression vector (mPrPHGC, a generous gift from M. Groschup, Institute for Novel and Emerging Infectious Diseases at the Friedrich-Loeffler-Institut, Greifswald-Insel Riems, Germany)[86], a PmlI restriction site was inserted after the stop codon of PrPCGPIThy-1 DNA by using the following primers: F:5´-CCCAAGGAGAAACACGTGCCCTCGAGGTCCTTC-3´; R: 5´-GAAGGACCTCGAGGGCACGTGTTTCTCCTTGGG-3´ (PmlI restriction site is in bold), with the QuickChange Lightning mutagenesis kit (Stratagene). PrPC was excised from mPrPHG by AgeI and PmlI (Fast Digest, Fermentas). PrPCGPIThy-1 was cut with AgeI and PmlI and ligated into the mPrPHG. For pronuclear injection, the mPrPHG vector was cut with SalI and NotI and separated in an agarose gel. The pronuclear injection was performed at the Transgenic Mouse Facility (ZMNH, Hamburg). Positive animals for the transgene were selected by PCR with the following primers: F: 5´-ATGTGGACTGATGTCGGCCT-3´; R: 5’-CTTGGAGGAGGGAGAGGGAA-3’. Lines L27 and L16 were established, and animals were backcrossed to PrP0/0 mice (on a C57/Bl6 background). To generate the control line, a littermate not presenting the transgene was backcrossed with C57/Bl6 mice with the same backcrossing scheme as the transgenic mice.
To eliminate the 3F4 tag we used the PrPCGPIThy-1 in mPrPHGC as a template and we change the two methionine residues at position 108 and 111 for leucine and valine respectively by using QuickChange Lightning (Stratagene) and the following primers: F: 5´-CAAACCAAAAACCAACCTCAAGCATGTGGCAGGGGCTGCGGCAGC-3´; R: 5´-GCTGCCGCAGCCCCTGCCACATGCTTGAGGTTGGTTTTTGGTTTG-3´ (mutations are in bold).
Quantitative real time PCR
RNA was extracted from mouse brain tissue using Precellys Lysing Kit (Bertin Technologies) and precooled QIAzol (Qiagen). Tissue (n = 7 and n = 5 for WTPrPC; n = 6 for PrPCGPIThy-1 L16; n = 5 for PrPCGPIThy-1 L27; n = 5 for PrPCGPIThy-1 L150) was homogenized for 30 s at 2,000 rpm in a dismembrator and subsequently centrifuged at 2,000xg for 2 min at room temperature. The supernatant was mixed with 200 μl chloroform and incubated at room temperature for 3 min following centrifugation for 15 min at 12,000xg. Total RNA was purified from the upper phase using RNeasy Mini Kit (Qiagen) according to the manufacturer’s instructions. RNA concentration and purity were determined using the NanoDrop system (Thermo Fisher Scientific). First strand cDNA was synthesized using 1 μg of total DNase-treated RNA using RevertAid H Minus First Strand cDNA Synthesis kit (Thermo Fisher Scientific). Real-time PCR reactions were performed in a volume of 10 μl, consisting of 10 ng cDNA, 2XSYBR GreenPCRMasterMix (Applied Biosystems), and 0.2 μM of each primer in Rotor Gene Q (Qiagen). For the detection of murine Prnp gene, the following primer pairs were used: 5´GGCCAAGGAGGGGGTACCCATAAT 3´ and 5´TAGTAGCGGTCCTCCCAGTCGTTGC 3´. The RPL gene (F: 5´CGGAATGGCATGATACTGAAGCC 3´; R: 5´TTGGTGTGGTATCTCACTGTAGG 3‘) was used as a reference to calculate relative expression levels of Prnp using ΔCT values.
Inoculation of mice with RML and 22L mouse-adapted prions
Briefly, 8–10 weeks old WTPrPC (n = 8), PrPCGPIThy-1 L16 (n = 10) and PrPCGPIThy-1 L150 mice (n = 10) mice were anasthesized and intracerebrally inoculated with 30 μl (corresponding to 3x105 log LD50) of RML 5.0 prion inoculum. Moreover, WTPrPC (n = 5), PrPCGPIThy-1 L16 (n = 5) and PrPCGPIThy-1 L150 (n = 5) mice were inoculated with 30 μl of 10% 22L prion inoculum. Mice were checked two to three times per week for clinical signs of prion disease and daily once clinical symptoms appeared.
Western blot analysis
Frontal cortex samples were homogenized as a 10% in RIPA buffer (50 mM Tris-HCl pH8, 150 mM NaCl, 1% NP40, 0.5% Na-Deoxycholate, 0.1% SDS) containing a cocktail of protease and phosphatase inhibitors (Roche), left for 10 min on ice, and centrifuged for 5 min at 12,000xg. The protein content of the supernatant was assessed by colorimetric analysis (QuickStart Bradford 1x Dye, Biorad) following the instructions of the supplier. Samples were standardized to 1 μg/μl in 4x loading buffer (250mM Tris-HCl, 8% SDS, 40% glycerol, 20% β-mercaptoethanol, 0.008% Bromophenol Blue, pH 6.8), boiled for 5 min at 95°C and subjected to electrophoresis (20 μg per sample). Proteins were then transferred to nitrocellulose membranes (Biorad) and incubated with mouse monoclonal antibodies against PrPC (POM1, 1:2.500; A. Aguzzi, Zurich, Switzerland) and actin (1:2,000; Millipore), ERK (1:1,000), ERK-P (1:1,000) p38 (1:1,000), p38-P (1:1,000) all from Cell Signaling Technologies; and rabbit polyclonal specific for shed PrP (1:1,000 [36]). Membranes were incubated with appropriate secondary antibodies and then developed either with Pierce ECL Western Blotting Substrate or Pierce Femto (Thermo Scientific) in a CD camera imaging system (BioRad) or with Odyssey Image system (Licor) and quantified with Image Studio software (Licor).
Immunohistochemistry
Processing of samples, including cutting and staining of paraffin sections with HE, was performed as published [39]. For immunohistochemistry, all sections were stained using the Ventana Benchmark XT machine (Ventana, Tuscon, Arizona, USA). Deparaffinised sections were boiled for 30–60 min in CC1 solution (Ventana, Tuscon, Arizona, USA) for antigen retrieval. Primary antibodies were diluted in 5% goat serum (Dianova), 45% Tris-buffered saline pH 7.6 (TBS) and 0.1% Triton X-100 in antibody diluent solution (Zytomed, Berlin, Germany). Sections were then incubated with primary antibody POM1 (1:100), Iba1 (Dako, 1:2.000) or GFAP (Dako, 1:400) for one hour. Anti-mouse or anti-rabbit histofine Simple Stain MAX PO Universal immunoperoxidase polymer (Nichirei Biosciences) was used as secondary antibody. Detection of secondary antibodies was performed with an ultraview universal DAB detection kit from Ventana with appropriate counterstaining and sections were cover-slipped using TissueTek glove mounting media (Sakura Finetek).
The neuropathological assessment (n = 3 or n = 4 depending on the genotype) was conducted by three independent investigators in a blinded fashion by grading the samples 1 (mild) to 3 (severe) depending on the staining intensity or spongiosis.
Triton X-114 assay
The assay was performed as previously described by Hooper et al. [28] which is a modification of the method described earlier by Pryde and Philips [87]. All the buffers contained a protease inhibitor cocktail (Roche). Briefly, frontal cortex samples (n = 3 for each genotype) were homogenized in 10 vol. of 0.32M sucrose in 50mM HEPES/NaOH pH 7.4, centrifuged for 15 min at 8,000xg and the supernatant further centrifuged at 26,000xg for 2h. The resulting pellet was resuspended in H buffer (10mM HEPES/NaOH pH 7.4) with the addition of 2% of pre-condensed Triton X-114 (Sigma-Aldrich) in a total volume of 200 μl (final concentration of 2 μg/μl). Samples were vortex mixed for 1–2 s, let on ice for 5 min and centrifuged again at 8,880xg at 4°C in a fixed angle rotor. The resulting pellet was washed with 0.2 ml H buffer, centrifuged at 8,800xg for 10 min at 4°C and the resulting pellet was resuspended in 180 μl of H buffer. This was kept as the Insoluble Pellet. The supernatant was layered over a 0.3 ml of 6% sucrose cushion in T buffer (10mM Tris-HCl pH 7.4, 0.15M NaCl) and 0.06% precondensed Triton X-114, incubated at 30°C for 3 min and further centrifuged at 3,000xg for 3 min in a swinging bucket rotor. The sucrose cushion was removed from the pellet (Detergent Phase), and the latter was resuspended in 180 μl of H buffer.
The supernatant (upper aqueous phase) was mixed with 0.5%(v/v) pre-condensed Triton X-114, vortexed 1–2 s, kept 5 min on ice, and further layered over a 0.3 ml 6% sucrose cushion in T buffer, incubated at 30°C for 3 min and centrifuged again at 3,000xg for 3 min in a swinging bucket rotor. The pellet was discarded, and the upper phase mixed again with 2% (v/v) pre-condensed Triton X-114 and processed again as described in the step before but without the sucrose cushion. After the last centrifugation, the supernatant was kept as final aqueous phase.
An equal amount of sample was then mixed with 4x loading buffer, and 30 μl of sample was subjected to gel electrophoresis and western blot as described above.
The procedure was repeated with three times with three different samples for quantification.
Assessment of sialic acid modifications in mouse brain homogenates
GPI anchors were isolated as previously described, with few modifications [88]. Briefly, 10% mouse brain homogenates were prepared with RIPA buffer plus protease inhibitor cocktail (Roche) and protein content was determined as described before. As sialic acids in N-glycans would interfere with the analysis of GPI-anchor modifications, samples containing 100 μg of total protein were first subjected to deglycosylation for 4 h at 37°C using the PNGase F kit (New England Biolabs) according to the manufacturer`s protocol. To only yield PrPC and avoid presence of interfering IgGs in eluates, deglycosylated samples were subsequently used for immunoprecipitation using monoclonal antibody POM1 that had been covalently linked to beads following the instructions of the Pierce Co-Immunoprecipitation Kit (Thermo Scientific) before. Thy-1 was immunoprecipitated with the rat anti-mouse Thy-1 (MCA1474, Serotech). The eluted samples containing purified deglycosylated PrPC were digested with 100 μg/ml proteinase K, at 37°C for 24 hours, resulting in GPI anchors attached to the terminal amino acid. The released GPIs were extracted with water-saturated butanol, washed with water 5 times and loaded onto C18 columns. GPIs were eluted under a gradient of propanol and water. The presence of GPIs was detected by ELISA. Maxisorb immunoplates were coated with 0.5 μg/ml concanavalin A (binds mannose) and blocked with 5% milk powder. Samples were added and any bound GPI was detected by the addition of the phosphatidylinositol-reactive mAb 5AB3-11, followed by a biotinylated anti-mouse IgM (Sigma), extravidin-alkaline phosphatase and 1mg/ml 4-nitrophenyl phosphate.
The presence of phosphatidylinositol in GPI anchors was identified using mAb (5AB3-11) and specific glycans were detected with biotinylated lectins. Isolated GPI anchors were bound to nitrocellulose membranes by dot blot and blocked with 5% milk powder. Samples were incubated with mAb 5AB3-11, biotinylated SNA (detects terminal sialic acid residues bound α-2,6 or α-2,3 to galactose), biotinylated concanavalin A (detects mannose) or biotinylated RCA I (detects terminal galactose) (Vector Labs). Bound lectins were visualised using extravidin peroxidase and enhanced chemiluminescence. The mAb was visualised by incubation with a horseradish peroxidase conjugated anti-murine-IgG and chemiluminescence.
Primary neurons
For preparation of primary hippocampal neurons, we used postnatal P0-P2 mice. Briefly, pups were killed by decapitation, and after removing skull and meninges, the hippocampus was dissected and collected in 10mM glucose in PBS containing 0.5 mg/ml papain (Sigma-Aldrich) and 10μg/ml DNAse (Roche). Hippocampi of PrPCGPIThy-1 mice were collected individually, and the tails were used for genotyping. After 30 min incubation at 37°C, samples were washed 4 times with plating medium (MEM 1X (Gibco), 20mM glucose (Sigma), 10% Horse serum (PAA Laboratories) and 3% of NaHCO3 7.5% (Gibco)) and carefully pipetted up and down several times in order to mince the tissue. Cells were plated in 6-well plates containing coverslips previously treated with 0.5 mg/ml of Poly-L-Lys and incubated at 37°C in a 5% CO2 cell culture incubator. After 4 hours, the media was changed to Neurobasal A medium (Gibco) containing 2% of B27 serum, Glutamax (Gibco) and penicillin/streptomycin (PAA Laboratories). Next day, AraC (Sigma-Aldrich) was added to kill proliferating cells. Half of the media was changed every three days.
Confocal immunofluorescence microscopy of primary neurons
After seven days in culture, coverslips were washed in PBS (Sigma-Aldrich), fixed (4% paraformaldehyde in PBS, 15 min room temperature (RT)), washed (PBS) and permeabilized with PBS containing 0.1% bovine serum albumin (BSA) and 0.3% Triton-X-100 for 1h at RT. Cells were then incubated with primary antibody (POM1 antibody at 1:250 and anti-tau antibody (Synaptic Systems, 1:500) diluted in PBS/0.1% BSA) for 1h at RT, washed (PBS), incubated with secondary antibody (donkey anti-mouse antibody AlexaFluor 488 and anti-guinea pig AlexaFluor 555 (Invitrogen), diluted in PBS/0.1% BSA, 1 h at RT) and washed again with PBS. DAPI (Roche) was added to the last wash and samples were then mounted with Fluoromount G media (SouthernBiotech). Consecutive Z-stacks (between 30–35 Z-stacks per picture) were taken with Leica Laser Scanner Confocal Microscope TCS SP5 (Leica), and the reconstructed 3D images were further processed with the IMARIS Software to quantify colocalization. For this, 12 pictures for the controls (from 5 animals) and 7 pictures for PrPCGPIThy-1 (from 5 animals) were analyzed. Out of every picture, ten regions of interest (ROI) were selected, and the total intensity of PrP and tau within these ROIs was measured. The ratio of PrP/tau intensity of 10 ROIs was then added to make a total mean. Total mean of all the pictures for WTPrPC and the ones from PrPCGPIThy-1 neurons were then added and subjected to statistical analysis (unpaired Student's t-Test).
DRM isolation
DRMs were isolated as previously described [29]. Briefly, about 0.2 mg of frontal cortex was homogenized in 10 vol. of homogenization buffer (10mM Tris-HCl pH 8.2, 0.02% sodium azide and 0.32M sucrose) on ice. After centrifugation at 500xg for 5 min to pellet nuclei and cell debris, the supernatant was further centrifuged for 40 min at 18,000xg to obtain a membrane-enriched fraction. The amount of protein from this fraction was quantified, and 1–2 mg of protein were diluted with 2x detergent buffer (10mM Tris-HCl pH 8.2, 1% Brij 96 (Sigma-Aldrich), 1% sodium deoxycholate (Sigma-Aldrich)) that had previously been mixed at least for 16h. Samples were then incubated for 30 min at 4°C and mixed at a ratio of 1:1 with 80% sucrose in the detergent buffer. 35% of sucrose (8 ml) and 5% of sucrose (1 ml) in detergent buffer were layered on top. After 18 h of centrifugation at 200,000xg, 12 fractions were taken and subjected to electrophoresis and western blot as previously described. Primary antibodies for DMR characterization used in the western blot were Flotillin (1:1,000; Cell Signaling Technologies) and calnexin (1:1,000; BD Transduction). For DRM isolation of samples depleted of myelin, a modification of the protocol from Chen et al. was used [31]. Briefly, samples were homogenized with ISB buffer (10mM HEPES/KOH pH 7.6; 200mM sucrose, 50mM K acetate, 1mM Mg acetate, 1mM EGTA, 1mM DTT and protease inhibitors) and centrifuged at 5000xg for 5 min. The resulting supernatant was further centrifuged at 22.000xg for 60 min to obtain a pellet with a membrane enriched fraction. This pellet was resuspended in ½ of the ISB buffer starting volume and centrifuged at 20.000xg for 60 min again. The resulting pellet was resuspended in 1ml ISB buffer and layered on a top of a 0.83M sucrose cushion in ISB buffer, and further centrifuged at 75.000xg for 35 min. After centrifugation, a myelin sheath band was visible at the border of the 0.83M sucrose. All the sample under the myelin layer was further diluted to 0.2M sucrose and centrifuged 30.000xg for 40 min. The resulting pellet was resuspended in 200 μl of ISB buffer without sucrose, the amount of protein quantified, and equal amounts of protein were further incubated either with 2X detergent (1% Brij 96/1% sodium deoxycholate in ISB buffer without sucrose) for 30 min at 4°C or with Brij 98 (Sigma-Aldrich) for 5 min at 37°C. After incubation, samples were mixed at a ratio of 1:1 with 80% sucrose in ISB buffer and proceeded with the sucrose gradient and ultracentrifugation as described above.
Proteinase K (PK) digestion
Frontal cortex brain samples (n = 3 for WTPrPC; n = 4 for PrPCGPIThy-1) were homogenized 1:10 in RIPA buffer and digested with 20 μgrs/ml of PK (Roche) at 37°C for 1 h. The reaction was stopped as before. All samples were subjected to electrophoresis and western blot analysis.
Cell culture, transfection, and co-cultivation
Cells were cultivated and transfected as described [89]. The human SH-SY5Y cell line is a human neuroblastoma cell line (DSMZ number ACC 209). Co-cultivation experiments were done as described previously [40, 90]. In brief, SH-SY5Y cells were grown on glass cover slips and transfected with Lipofectamine (Invitrogen). 2 h after transfection cover slips were transferred into dishes containing a 90% confluent cell layer of either N2a (immortalized neuroblastoma cell line, ATCC No. Ccl 131) or chronically infected ScN2a cells (established by infecting N2a cells with an enriched preparation of prions isolated from the brains of mice infected with RML prions [91]). After 16 h of co-cultivation, apoptotic cell death was analyzed (see below).
Apoptosis assay
16 h after co-cultivation, SH-SY5Y cells were fixed on glass cover slips with 3.7% paraformaldehyde for 20 min, washed and permeabilized with 0.2% Triton X-100 in PBS for 10 min at room temperature. Fixed cells were incubated with an anti-active caspase 3 antibodies overnight at 4°C, followed by incubation with the fluorescently labeled secondary antibody Alexa Fluor 555 for one hour at room temperature. Cells were then mounted onto glass slides and examined by fluorescence microscopy using a Zeiss Axiovert 200M microscope (Carl Zeiss). The numbers of cells positive for activated caspase-3 out of at least 1,000 transfected cells were determined in a blinded manner. All quantifications were based on at least three independent experiments.
Statistical analysis
IBM SPSS Statistics 22 and GraphPad Prism 5 statistic software programs were used in the statistical analysis. To assess differences between Kaplan-Meier survival curves, the Breslow test was used. For comparison between the groups in western blots and neuronal countings, unpaired Student's t-test was used. Statistical significance was considered when p-values were as follows: *p < 0.05, **p < 0.005, ***p < 0.001, ****p<0.0001. The exact p value is also given.
Supporting information
(A) Bar chart of PrPC mRNA levels measured by RT-qPCR. PrPCGPIThy-1 L16 (n = 6) presents a doubled amount of PrP mRNA levels compared to WTPrPC (n = 7). (B) Representative western blots showing PrPCGPIThy-1 protein expression in different organs in the two transgenic mice lines compared to WTPrPC. (B: brain; Cb: cerebellum; SC: spinal cord; Sp: spleen; M: muscle). POM1 antibody was used to detect PrP.
(TIF)
(A) DRMs isolation of WTPrPC and PrPCGPIThy-1 mouse brain (n = 3 for each genotype) with a mixture of 0.5% Brij96 and 0.5% sodium deoxycholate. After extraction at 4°C and overnight centrifugation on a sucrose density gradient, twelve fractions were loaded on a gel. Flotillin is used as a marker for DRMs, whereas calnexin, a chaperone resident in the ER, is used as a marker of non-DRM fractions. Note that there no difference was found between controls and transgenic mice in the PrP solubilization pattern (quantifications are shown below). (B) Under the same conditions as in (A), Thy-1 stays in the DRMs (mainly in fractions 2 and 3). (C) Because myelin could impair the proper solubilization of the DRMs, in another set of experiments we depleted the samples from myelin and used the same procedure as in (A). Again, no differences in the solubilization pattern were seen by incubating with 0.5% Brij 96 and 0.5% sodium deoxycholate. With another detergent (Brij 98 at 37°C), although the isoform solubilization pattern differs between WTPrPC and PrPCGPIThy-1, no differences in the fraction distribution were observed.
(TIF)
Dot blot analysis of PrPC and Thy-1 GPI-anchors from WT mouse brain and PrP knock-out mice (Prnp0/0 mice). Phosphatidylinositol (PI), mannose (man) and sialic acid (sial. acid) were detected as described in the methods section. Note that the amounts of PI and mannose are similar between Thy-1 and WTPrPC, whereas sialic acid is only present in WTPrPC.
(TIF)
(A) Bar chart showing relative amounts of PrPC mRNA extracted from brains of WTPrPC and PrPCGPIThy-1 L150 mice showing no differences. WTPrPC is set to 1. (B) Representative western blot of total brain homogenates from PrPCGPIThy-1L159 and quantification of the signal showing that these animals present around 50% of the transgene compared to amounts of PrP in WTPrPC mice (n = 3 for each genotype; **p = 0.0011).
(TIF)
Kaplan-Meier survival curve of WTPrPC and PrPCGPIThy-1 L150 mice inoculated with 22L prions. PrPCGPIThy-1 L150 mice (n = 5; black line) reached terminal disease and were sacrificed at day 156 ± 3 dpi, compared to 144 dpi for WTPrPC (n = 5; grey line); Log Rank (Mantel-Cox) **p<0.003).
(TIF)
(A) Kaplan-Meier survival curve of PrPCGPIThy-1 L16 mice infected either with RML or 22L prions. Note the substantial delay to terminal disease for the transgenic mice (RML: 400 ± 56 dpi (n = 10); 22L: 200 ± 23 dpi (n = 5); black line) compared to WTPrPC mice (RML: 157 ± 6 dpi (n = 8); 22L: 144 ± 1 dpi (n = 5); grey line; Log Rank (Mantel-Cox) RML: ****p<0.0001; 22L: **p = 0.003). (B) Neuropathological analysis of terminally diseased PrPCGPIThy-1 L16 mice infected with RML. In PrPCGPIThy-1 L16 brains a general decrease in spongiosis is observed with HE staining. Gliosis is also decreased as observed with antibodies against astrocytes (GFAP) and microglia (Iba1). (C) Representative blot of WTPrPC and PrPCGPIThy-1 L16 brain homogenates infected with RML prions and digested with PK. Decreased amounts of PK-resistant PrPSc were observed for PrPCGPIThy-1 L16 brain homogenates, despite of the two-fold expression of the transgene (shown in S1A Fig).
(TIF)
(A) Western blot analysis of PrP expression in PrPCGPIThy-1 L150 mice at 20 weeks (20W, n = 3) and 40 weeks of age (40W, n = 4). No changes in expression were observed between the two groups (bar chart shows the mean of PrP relative intensity related to actin used as a loading control; WTPrPC is set to 100%). (B) (i) Representative western blot showing expression of shed PrP in terminally RML-infected mice (n = 4) compared with non-infected PrPCGPIThy-1 mice (n = 4). Note that shed PrP increases about 3-fold in RML-infected animals. Bar chart shows relative intensity of shed PrP related to the total amount of PrP (shown in the re-probed blot in (ii); PrPCGPIThy-1 in uninfected mice is set to 100%). (ii) Representative western blot showing that the amounts of total PrP do not change between RML-infected (n = 4) and uninfected PrPCGPIThy-1 mice (n = 4). The bar chart shows relative intensity of PrP developed with POM1 related to actin used as a loading control. All error bars are SEM.
(TIF)
(A) Representative blot of total brain homogenates from WTPrPC and PrPCGPIThy-1 L159 mice detected with the antibody against shed PrP. (B) Although this line of transgenic mice expresses per se less PrPCGPThy-1 levels than WTPrPC mice (as shown in S3 Fig), when the corresponding signal of shed PrP is referred to the signal of total PrP, there is a significant decrease (*p = 0.015) in shed PrP in PrPCGPIThy-1 L159 brains (n = 3) compared to WTPrPC (n = 3).
(TIF)
(DOCX)
Acknowledgments
We thank Dr. S. Krasemann and K. Hartmann from the Mouse Pathology Facility, University Medical Center Hamburg-Eppendorf, and Dr. I. Hermans-Borgmeyer from the Transgenic Mouse Facility, Center for Molecular Neurobiology Hamburg (ZMNH). We also want to thank Dr. V. Failla from the Microscopic Imaging Facility (UMIF), University Medical Center Hamburg-Eppendorf. We also thank Beata Szalay for excellent technical assistance.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
BP, HCA and LL were supported by the Werner Otto Stifftung. HCA received funding from the Creutzfeldt-Jakob Disease Foundation. MG received funding from DFG (FG885, GRK1459, SFB877 project A12) JT received funding from DFG (TA 167/6) The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript
References
- 1.Haraguchi T, Fisher S, Olofsson S, Endo T, Groth D, Tarentino A, et al. Asparagine-linked glycosylation of the scrapie and cellular prion proteins. Arch Biochem Biophys. 1989;274(1):1–13. . [DOI] [PubMed] [Google Scholar]
- 2.Stahl N, Borchelt DR, Hsiao K, Prusiner SB. Scrapie prion protein contains a phosphatidylinositol glycolipid. Cell. 1987;51(2):229–40. 10.1016/0092-8674(87)90150-4. [DOI] [PubMed] [Google Scholar]
- 3.Mayor S, Riezman H. Sorting GPI-anchored proteins. Nat Rev Mol Cell Biol. 2004;5(2):110–20. 10.1038/nrm1309 . [DOI] [PubMed] [Google Scholar]
- 4.Fujita M, Kinoshita T. GPI-anchor remodeling: potential functions of GPI-anchors in intracellular trafficking and membrane dynamics. Biochim Biophys Acta. 2012;1821(8):1050–8. 10.1016/j.bbalip.2012.01.004 . [DOI] [PubMed] [Google Scholar]
- 5.Fujita M, Kinoshita T. Structural remodeling of GPI anchors during biosynthesis and after attachment to proteins. FEBS Lett. 2010;584(9):1670–7. 10.1016/j.febslet.2009.10.079 . [DOI] [PubMed] [Google Scholar]
- 6.Saha S, Anilkumar AA, Mayor S. GPI-anchored protein organization and dynamics at the cell surface. J Lipid Res. 2016;57(2):159–75. 10.1194/jlr.R062885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zurzolo C, Simons K. Glycosylphosphatidylinositol-anchored proteins: Membrane organization and transport. Biochim Biophys Acta. 2016;1858(4):632–9. 10.1016/j.bbamem.2015.12.018 . [DOI] [PubMed] [Google Scholar]
- 8.Lisanti MP, Rodriguez-Boulan E. Glycophospholipid membrane anchoring provides clues to the mechanism of protein sorting in polarized epithelial cells. Trends Biochem Sci. 1990;15(3):113–8. . [DOI] [PubMed] [Google Scholar]
- 9.Sarnataro D, Paladino S, Campana V, Grassi J, Nitsch L, Zurzolo C. PrPC is sorted to the basolateral membrane of epithelial cells independently of its association with rafts. Traffic. 2002;3(11):810–21. . [DOI] [PubMed] [Google Scholar]
- 10.Uelhoff A, Tatzelt J, Aguzzi A, Winklhofer KF, Haass C. A pathogenic PrP mutation and doppel interfere with polarized sorting of the prion protein. J Biol Chem. 2005;280(7):5137–40. 10.1074/jbc.C400560200 . [DOI] [PubMed] [Google Scholar]
- 11.Puig B, Altmeppen HC, Thurm D, Geissen M, Conrad C, Braulke T, et al. N-glycans and glycosylphosphatidylinositol-anchor act on polarized sorting of mouse PrP(C) in Madin-Darby canine kidney cells. PLoS One. 2011;6(9):e24624 10.1371/journal.pone.0024624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paladino S, Lebreton S, Tivodar S, Campana V, Tempre R, Zurzolo C. Different GPI-attachment signals affect the oligomerisation of GPI-anchored proteins and their apical sorting. J Cell Sci. 2008;121(Pt 24):4001–7. 10.1242/jcs.036038 . [DOI] [PubMed] [Google Scholar]
- 13.Bate C, Nolan W, McHale-Owen H, Williams A. Sialic Acid within the Glycosylphosphatidylinositol Anchor Targets the Cellular Prion Protein to Synapses. Journal of Biological Chemistry. 2016;291(33):17093–101. 10.1074/jbc.M116.731117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prusiner SB. Prions. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(23):13363–83. PMC33918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeffrey M, McGovern G, Siso S, Gonzalez L. Cellular and sub-cellular pathology of animal prion diseases: relationship between morphological changes, accumulation of abnormal prion protein and clinical disease. Acta Neuropathol. 2011;121(1):113–34. 10.1007/s00401-010-0700-3 . [DOI] [PubMed] [Google Scholar]
- 16.Taylor DR, Hooper NM. The prion protein and lipid rafts. Mol Membr Biol. 2006;23(1):89–99. 10.1080/09687860500449994 . [DOI] [PubMed] [Google Scholar]
- 17.McNally KL, Ward AE, Priola SA. Cells expressing anchorless prion protein are resistant to scrapie infection. J Virol. 2009;83(9):4469–75. 10.1128/JVI.02412-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taraboulos A, Scott M, Semenov A, Avrahami D, Laszlo L, Prusiner SB. Cholesterol depletion and modification of COOH-terminal targeting sequence of the prion protein inhibit formation of the scrapie isoform. J Cell Biol. 1995;129(1):121–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bate C, Tayebi M, Williams A. A glycosylphosphatidylinositol analogue reduced prion-derived peptide mediated activation of cytoplasmic phospholipase A2, synapse degeneration and neuronal death. Neuropharmacology. 2010;59(1–2):93–9. 10.1016/j.neuropharm.2010.04.002 . [DOI] [PubMed] [Google Scholar]
- 20.Bate C, Williams A. The cellular prion protein with a monoacylated glycosylphosphatidylinositol anchor modifies cell membranes, inhibits cell signaling and reduces prion formation. Prion. 2011;5(2):65–8. 10.4161/pri.5.2.16095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stohr J, Elfrink K, Weinmann N, Wille H, Willbold D, Birkmann E, et al. In vitro conversion and seeded fibrillization of posttranslationally modified prion protein. Biol Chem. 2011;392(5):415–21. 10.1515/BC.2011.048 . [DOI] [PubMed] [Google Scholar]
- 22.Chesebro B, Race B, Meade-White K, Lacasse R, Race R, Klingeborn M, et al. Fatal transmissible amyloid encephalopathy: a new type of prion disease associated with lack of prion protein membrane anchoring. PLoS Pathog. 2010;6(3):e1000800 10.1371/journal.ppat.1000800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stahl N, Baldwin MA, Hecker R, Pan KM, Burlingame AL, Prusiner SB. Glycosylinositol phospholipid anchors of the scrapie and cellular prion proteins contain sialic acid. Biochemistry. 1992;31(21):5043–53. . [DOI] [PubMed] [Google Scholar]
- 24.Bate C, Nolan W, Williams A. Glycosylphosphatidylinositols: More than just an anchor? Communicative & Integrative Biology. 2016;9(2):e1149671 10.1080/19420889.2016.1149671 PMC4857774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Katorcha E, Srivastava S, Klimova N, Baskakov IV. Sialylation of Glycosylphosphatidylinositol (GPI) Anchors of Mammalian Prions Is Regulated in a Host-, Tissue-, and Cell-specific Manner. J Biol Chem. 2016;291(33):17009–19. 10.1074/jbc.M116.732040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kascsak RJ, Rubenstein R, Merz PA, Tonna-DeMasi M, Fersko R, Carp RI, et al. Mouse polyclonal and monoclonal antibody to scrapie-associated fibril proteins. J Virol. 1987;61(12):3688–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Supattapone S, Muramoto T, Legname G, Mehlhorn I, Cohen FE, DeArmond SJ, et al. Identification of two prion protein regions that modify scrapie incubation time. J Virol. 2001;75(3):1408–13. 10.1128/JVI.75.3.1408-1413.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hooper NM, Bashir A. Glycosyl-phosphatidylinositol-anchored membrane proteins can be distinguished from transmembrane polypeptide-anchored proteins by differential solubilization and temperature-induced phase separation in Triton X-114. Biochem J. 1991;280 (Pt 3):745–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Madore N, Smith KL, Graham CH, Jen A, Brady K, Hall S, et al. Functionally different GPI proteins are organized in different domains on the neuronal surface. EMBO J. 1999;18(24):6917–26. 10.1093/emboj/18.24.6917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brugger B, Graham C, Leibrecht I, Mombelli E, Jen A, Wieland F, et al. The membrane domains occupied by glycosylphosphatidylinositol-anchored prion protein and Thy-1 differ in lipid composition. J Biol Chem. 2004;279(9):7530–6. 10.1074/jbc.M310207200 . [DOI] [PubMed] [Google Scholar]
- 31.Chen X, Jen A, Warley A, Lawrence MJ, Quinn Peter J, Morris Roger J. Isolation at physiological temperature of detergent-resistant membranes with properties expected of lipid rafts: the influence of buffer composition. Biochemical Journal. 2009;417(2):525–33. 10.1042/BJ20081385 [DOI] [PubMed] [Google Scholar]
- 32.Altmeppen HC, Prox J, Krasemann S, Puig B, Kruszewski K, Dohler F, et al. The sheddase ADAM10 is a potent modulator of prion disease. Elife. 2015;4 10.7554/eLife.04260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haybaeck J, Heikenwalder M, Klevenz B, Schwarz P, Margalith I, Bridel C, et al. Aerosols transmit prions to immunocompetent and immunodeficient mice. PLoS Pathog. 2011;7(1):e1001257 10.1371/journal.ppat.1001257 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.Linsenmeier L, Altmeppen HC, Wetzel S, Mohammadi B, Saftig P, Glatzel M. Diverse functions of the prion protein–Does proteolytic processing hold the key? Biochimica et Biophysica Acta (BBA)—Molecular Cell Research. 2017. 10.1016/j.bbamcr.2017.06.022. [DOI] [PubMed] [Google Scholar]
- 35.Altmeppen HC, Prox J, Puig B, Dohler F, Falker C, Krasemann S, et al. Roles of endoproteolytic alpha-cleavage and shedding of the prion protein in neurodegeneration. FEBS J. 2013;280(18):4338–47. 10.1111/febs.12196 . [DOI] [PubMed] [Google Scholar]
- 36.Linsenmeier L, Mohammadi B, Wetzel S, Puig B, Jackson WS, Hartmann A, et al. Structural and mechanistic aspects influencing the ADAM10-mediated shedding of the prion protein. Mol Neurodegener. 2018;13(1):18 10.1186/s13024-018-0248-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Borchelt DR, Rogers M, Stahl N, Telling G, Prusiner SB. Release of the cellular prion protein from cultured cells after loss of its glycoinositol phospholipid anchor. Glycobiology. 1993;3(4):319–29. 10.1093/glycob/3.4.319 [DOI] [PubMed] [Google Scholar]
- 38.Nordstrom E, Fisone G, Kristensson K. Opposing effects of ERK and p38-JNK MAP kinase pathways on formation of prions in GT1-1 cells. FASEB J. 2009;23(2):613–22. 10.1096/fj.08-115360 . [DOI] [PubMed] [Google Scholar]
- 39.Puig B, Altmeppen HC, Ulbrich S, Linsenmeier L, Krasemann S, Chakroun K, et al. Secretory pathway retention of mutant prion protein induces p38-MAPK activation and lethal disease in mice. Scientific Reports. 2016;6:24970 10.1038/srep24970 https://www.nature.com/articles/srep24970#supplementary-information. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rambold AS, Muller V, Ron U, Ben-Tal N, Winklhofer KF, Tatzelt J. Stress-protective signalling of prion protein is corrupted by scrapie prions. EMBO J. 2008;27(14):1974–84. 10.1038/emboj.2008.122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paulick MG, Bertozzi CR. The Glycosylphosphatidylinositol Anchor: A Complex Membrane- Anchoring Structure for Proteins. Biochemistry. 2008;47(27):6991–7000. 10.1021/bi8006324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ferguson MAJ, Hart GW, Kinoshita T. Glycosylphosphatidylinositol Anchors In: Varki A, Cummings RD, Esko JD, Stanley P, Hart GW, Aebi M, et al. , editors. Essentials of Glycobiology. 3rd ed Cold Spring Harbor; (NY)2015. 10.1093/glycob/cwv091 [DOI] [Google Scholar]
- 43.Brown DA, Rose JK. Sorting of GPI-anchored proteins to glycolipid-enriched membrane subdomains during transport to the apical cell surface. Cell. 1992;68(3):533–44. . [DOI] [PubMed] [Google Scholar]
- 44.Ledesma MD, Brugger B, Bunning C, Wieland FT, Dotti CG. Maturation of the axonal plasma membrane requires upregulation of sphingomyelin synthesis and formation of protein-lipid complexes. EMBO J. 1999;18(7):1761–71. 10.1093/emboj/18.7.1761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Puig B, Altmeppen H, Glatzel M. The GPI-anchoring of PrP: implications in sorting and pathogenesis. Prion. 2014;8(1):11–8. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sunyach C, Jen A, Deng J, Fitzgerald KT, Frobert Y, Grassi J, et al. The mechanism of internalization of glycosylphosphatidylinositol-anchored prion protein. EMBO J. 2003;22(14):3591–601. 10.1093/emboj/cdg344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Endres K, Mitteregger G, Kojro E, Kretzschmar H, Fahrenholz F. Influence of ADAM10 on prion protein processing and scrapie infectiosity in vivo. Neurobiol Dis. 2009;36(2):233–41. 10.1016/j.nbd.2009.07.015 . [DOI] [PubMed] [Google Scholar]
- 48.Glatzel M, Linsenmeier L, Dohler F, Krasemann S, Puig B, Altmeppen HC. Shedding light on prion disease. Prion. 2015;9(4):244–56. 10.1080/19336896.2015.1065371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bouybayoune I, Mantovani S, Del Gallo F, Bertani I, Restelli E, Comerio L, et al. Transgenic fatal familial insomnia mice indicate prion infectivity-independent mechanisms of pathogenesis and phenotypic expression of disease. PLoS Pathog. 2015;11(4):e1004796 10.1371/journal.ppat.1004796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jackson WS, Borkowski AW, Faas H, Steele AD, King OD, Watson N, et al. Spontaneous generation of prion infectivity in fatal familial insomnia knockin mice. Neuron. 2009;63(4):438–50. 10.1016/j.neuron.2009.07.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bueler H, Aguzzi A, Sailer A, Greiner RA, Autenried P, Aguet M, et al. Mice devoid of PrP are resistant to scrapie. Cell. 1993;73(7):1339–47. . [DOI] [PubMed] [Google Scholar]
- 52.Brandner S, Isenmann S, Raeber A, Fischer M, Sailer A, Kobayashi Y, et al. Normal host prion protein necessary for scrapie-induced neurotoxicity. Nature. 1996;379:339 10.1038/379339a0 [DOI] [PubMed] [Google Scholar]
- 53.Marshall KE, Hughson A, Vascellari S, Priola SA, Sakudo A, Onodera T, et al. PrP Knockout Cells Expressing Transmembrane PrP Resist Prion Infection. Journal of virology. 2017;91(2):e01686–16. 10.1128/JVI.01686-16 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chesebro B, Trifilo M, Race R, Meade-White K, Teng C, LaCasse R, et al. Anchorless prion protein results in infectious amyloid disease without clinical scrapie. Science. 2005;308(5727):1435–9. 10.1126/science.1110837 . [DOI] [PubMed] [Google Scholar]
- 55.Raymond GJ, Race B, Hollister JR, Offerdahl DK, Moore RA, Kodali R, et al. Isolation of novel synthetic prion strains by amplification in transgenic mice coexpressing wild-type and anchorless prion proteins. J Virol. 2012;86(21):11763–78. 10.1128/JVI.01353-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nisbet RM, Harrison CF, Lawson VA, Masters CL, Cappai R, Hill AF. Residues surrounding the glycosylphosphatidylinositol anchor attachment site of PrP modulate prion infection: insight from the resistance of rabbits to prion disease. J Virol. 2010;84(13):6678–86. 10.1128/JVI.02709-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bate C, Nolan W, Williams A. Sialic Acid on the Glycosylphosphatidylinositol Anchor Regulates PrP-mediated Cell Signaling and Prion Formation. J Biol Chem. 2016;291(1):160–70. 10.1074/jbc.M115.672394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bate C, Nolan W, Williams A. Does the tail wag the dog? How the structure of a glycosylphosphatidylinositol anchor affects prion formation. Prion. 2016;10(2):127–30. 10.1080/19336896.2016.1148237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang F, Yang F, Hu Y, Wang X, Wang X, Jin C, et al. Lipid Interaction Converts Prion Protein to a PrPSc-like Proteinase K-Resistant Conformation under Physiological Conditions. Biochemistry. 2007;46(23):7045–53. 10.1021/bi700299h [DOI] [PubMed] [Google Scholar]
- 60.Ma J. Prion Protein Conversion and Lipids In: Zou W-Q, Gambetti P, editors. Prions and Diseases: Volume 1, Physiology and Pathophysiology. New York, NY: Springer; New York; 2013. p. 107–19. [Google Scholar]
- 61.Katorcha E, Daus ML, Gonzalez-Montalban N, Makarava N, Lasch P, Beekes M, et al. Reversible off and on switching of prion infectivity via removing and reinstalling prion sialylation. Sci Rep. 2016;6:33119 10.1038/srep33119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Baskakov IV, Katorcha E. Multifaceted Role of Sialylation in Prion Diseases. Front Neurosci. 2016;10:358 10.3389/fnins.2016.00358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Katorcha E, Makarava N, Savtchenko R, D'Azzo A, Baskakov IV. Sialylation of prion protein controls the rate of prion amplification, the cross-species barrier, the ratio of PrPSc glycoform and prion infectivity. PLoS pathogens. 2014;10(9):e1004366–e. 10.1371/journal.ppat.1004366 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nemoto YL, Morris RJ, Hijikata H, Tsunoyama TA, Shibata ACE, Kasai RS, et al. Dynamic Meso-Scale Anchorage of GPI-Anchored Receptors in the Plasma Membrane: Prion Protein vs. Thy1. Cell Biochemistry and Biophysics. 2017. 10.1007/s12013-017-0808-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Goold R, Rabbanian S, Sutton L, Andre R, Arora P, Moonga J, et al. Rapid cell-surface prion protein conversion revealed using a novel cell system. Nat Commun. 2011;2:281 10.1038/ncomms1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Carroll JA, Striebel JF, Rangel A, Woods T, Phillips K, Peterson KE, et al. Prion Strain Differences in Accumulation of PrPSc on Neurons and Glia Are Associated with Similar Expression Profiles of Neuroinflammatory Genes: Comparison of Three Prion Strains. PLoS pathogens. 2016;12(4):e1005551–e. 10.1371/journal.ppat.1005551 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fehlinger A, Wolf H, Hossinger A, Duernberger Y, Pleschka C, Riemschoss K, et al. Prion strains depend on different endocytic routes for productive infection. Scientific Reports. 2017;7:6923 10.1038/s41598-017-07260-2 PMC5537368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Aguzzi A, Zhu C. Microglia in prion diseases. J Clin Invest. 2017;127(9):3230–9. 10.1172/JCI90605 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Obst J, Simon E, Mancuso R, Gomez-Nicola D. The Role of Microglia in Prion Diseases: A Paradigm of Functional Diversity. Front Aging Neurosci. 2017;9:207 10.3389/fnagi.2017.00207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Megra BW, Eugenin EA, Berman JW. The Role of Shed PrPc in the Neuropathogenesis of HIV Infection. J Immunol. 2017;199(1):224–32. 10.4049/jimmunol.1601041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Diedrich JF, Bendheim PE, Kim YS, Carp RI, Haase AT. Scrapie-associated prion protein accumulates in astrocytes during scrapie infection. Proc Natl Acad Sci U S A. 1991;88(2):375–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hollister JR, Lee KS, Dorward DW, Baron GS. Efficient uptake and dissemination of scrapie prion protein by astrocytes and fibroblasts from adult hamster brain. PLoS One. 2015;10(1):e0115351 10.1371/journal.pone.0115351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Victoria GS, Arkhipenko A, Zhu S, Syan S, Zurzolo C. Astrocyte-to-neuron intercellular prion transfer is mediated by cell-cell contact. Sci Rep. 2016;6:20762 10.1038/srep20762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Raeber AJ, Race RE, Brandner S, Priola SA, Sailer A, Bessen RA, et al. Astrocyte-specific expression of hamster prion protein (PrP) renders PrP knockout mice susceptible to hamster scrapie. EMBO J. 1997;16(20):6057–65. 10.1093/emboj/16.20.6057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Betmouni S, Clements J, Perry VH. Vacuolation in murine prion disease: an informative artifact. Current Biology. 1999;9(18):R677–R9. 10.1016/S0960-9822(99)80437-0. [DOI] [PubMed] [Google Scholar]
- 76.Rezaie P, Lantos PL. Microglia and the pathogenesis of spongiform encephalopathies. Brain Research Reviews. 2001;35(1):55–72. 10.1016/S0165-0173(01)00042-X. [DOI] [PubMed] [Google Scholar]
- 77.Linden R. The Biological Function of the Prion Protein: A Cell Surface Scaffold of Signaling Modules. Frontiers in Molecular Neuroscience. 2017;10(77). 10.3389/fnmol.2017.00077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fang C, Wu B, Le NTT, Imberdis T, Mercer RCC, Harris DA. Prions activate a p38 MAPK synaptotoxic signaling pathway. PLoS pathogens. 2018;14(9):e1007283–e. 10.1371/journal.ppat.1007283 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Marella M, Gaggioli C, Batoz M, Deckert M, Tartare-Deckert S, Chabry J. Pathological prion protein exposure switches on neuronal mitogen-activated protein kinase pathway resulting in microglia recruitment. J Biol Chem. 2005;280(2):1529–34. 10.1074/jbc.M410966200 . [DOI] [PubMed] [Google Scholar]
- 80.Didonna A, Legname G. Aberrant ERK 1/2 complex activation and localization in scrapie- infected GT1-1 cells. Mol Neurodegener. 2010;5:29 10.1186/1750-1326-5-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nordstrom EK, Luhr KM, Ibanez C, Kristensson K. Inhibitors of the mitogen-activated protein kinase kinase 1/2 signaling pathway clear prion-infected cells from PrPSc. J Neurosci. 2005;25(37):8451–6. 10.1523/JNEUROSCI.2349-05.2005 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pradines E, Hernandez-Rapp J, Villa-Diaz A, Dakowski C, Ardila-Osorio H, Haik S, et al. Pathogenic prions deviate PrP(C) signaling in neuronal cells and impair A-beta clearance. Cell Death Dis. 2013;4:e456 10.1038/cddis.2012.195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.LaCasse RA, Striebel JF, Favara C, Kercher L, Chesebro B. Role of Erk1/2 activation in prion disease pathogenesis: absence of CCR1 leads to increased Erk1/2 activation and accelerated disease progression. J Neuroimmunol. 2008;196(1–2):16–26. 10.1016/j.jneuroim.2008.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lee HP, Jun YC, Choi JK, Kim JI, Carp RI, Kim YS. Activation of mitogen-activated protein kinases in hamster brains infected with 263K scrapie agent. J Neurochem. 2005;95(2):584–93. 10.1111/j.1471-4159.2005.03429.x . [DOI] [PubMed] [Google Scholar]
- 85.Zinkle A, Mohammadi M. A threshold model for receptor tyrosine kinase signaling specificity and cell fate determination. F1000Research. 2018;7:F1000 Faculty Rev-872. 10.12688/f1000research.14143.1 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fischer M, Rulicke T, Raeber A, Sailer A, Moser M, Oesch B, et al. Prion protein (PrP) with amino-proximal deletions restoring susceptibility of PrP knockout mice to scrapie. EMBO J. 1996;15(6):1255–64. [PMC free article] [PubMed] [Google Scholar]
- 87.Pryde JG, Phillips JH. Fractionation of membrane proteins by temperature-induced phase separation in Triton X-114. Application to subcellular fractions of the adrenal medulla. Biochem J. 1986;233(2):525–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bate C, Williams A. Neurodegeneration Induced by Clustering of Sialylated Glycosylphosphatidylinositols of Prion Proteins. Journal of Biological Chemistry. 2012;287(11):7935–44. 10.1074/jbc.M111.275743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Winklhofer KF, Heller U, Reintjes A, Tatzelt J. Inhibition of complex glycosylation increases the formation of PrPsc. Traffic. 2003;4(5):313–22. . [DOI] [PubMed] [Google Scholar]
- 90.Resenberger UK, Harmeier A, Woerner AC, Goodman JL, Muller V, Krishnan R, et al. The cellular prion protein mediates neurotoxic signalling of beta-sheet-rich conformers independent of prion replication. EMBO J. 2011;30(10):2057–70. 10.1038/emboj.2011.86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Winklhofer KF, Tatzelt J. Cationic Lipopolyamines Induce Degradation of PrPSc in Scrapie- Infected Mouse Neuroblastoma Cells. Biological Chemistry 2000. p. 463 10.1515/BC.2000.061 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Bar chart of PrPC mRNA levels measured by RT-qPCR. PrPCGPIThy-1 L16 (n = 6) presents a doubled amount of PrP mRNA levels compared to WTPrPC (n = 7). (B) Representative western blots showing PrPCGPIThy-1 protein expression in different organs in the two transgenic mice lines compared to WTPrPC. (B: brain; Cb: cerebellum; SC: spinal cord; Sp: spleen; M: muscle). POM1 antibody was used to detect PrP.
(TIF)
(A) DRMs isolation of WTPrPC and PrPCGPIThy-1 mouse brain (n = 3 for each genotype) with a mixture of 0.5% Brij96 and 0.5% sodium deoxycholate. After extraction at 4°C and overnight centrifugation on a sucrose density gradient, twelve fractions were loaded on a gel. Flotillin is used as a marker for DRMs, whereas calnexin, a chaperone resident in the ER, is used as a marker of non-DRM fractions. Note that there no difference was found between controls and transgenic mice in the PrP solubilization pattern (quantifications are shown below). (B) Under the same conditions as in (A), Thy-1 stays in the DRMs (mainly in fractions 2 and 3). (C) Because myelin could impair the proper solubilization of the DRMs, in another set of experiments we depleted the samples from myelin and used the same procedure as in (A). Again, no differences in the solubilization pattern were seen by incubating with 0.5% Brij 96 and 0.5% sodium deoxycholate. With another detergent (Brij 98 at 37°C), although the isoform solubilization pattern differs between WTPrPC and PrPCGPIThy-1, no differences in the fraction distribution were observed.
(TIF)
Dot blot analysis of PrPC and Thy-1 GPI-anchors from WT mouse brain and PrP knock-out mice (Prnp0/0 mice). Phosphatidylinositol (PI), mannose (man) and sialic acid (sial. acid) were detected as described in the methods section. Note that the amounts of PI and mannose are similar between Thy-1 and WTPrPC, whereas sialic acid is only present in WTPrPC.
(TIF)
(A) Bar chart showing relative amounts of PrPC mRNA extracted from brains of WTPrPC and PrPCGPIThy-1 L150 mice showing no differences. WTPrPC is set to 1. (B) Representative western blot of total brain homogenates from PrPCGPIThy-1L159 and quantification of the signal showing that these animals present around 50% of the transgene compared to amounts of PrP in WTPrPC mice (n = 3 for each genotype; **p = 0.0011).
(TIF)
Kaplan-Meier survival curve of WTPrPC and PrPCGPIThy-1 L150 mice inoculated with 22L prions. PrPCGPIThy-1 L150 mice (n = 5; black line) reached terminal disease and were sacrificed at day 156 ± 3 dpi, compared to 144 dpi for WTPrPC (n = 5; grey line); Log Rank (Mantel-Cox) **p<0.003).
(TIF)
(A) Kaplan-Meier survival curve of PrPCGPIThy-1 L16 mice infected either with RML or 22L prions. Note the substantial delay to terminal disease for the transgenic mice (RML: 400 ± 56 dpi (n = 10); 22L: 200 ± 23 dpi (n = 5); black line) compared to WTPrPC mice (RML: 157 ± 6 dpi (n = 8); 22L: 144 ± 1 dpi (n = 5); grey line; Log Rank (Mantel-Cox) RML: ****p<0.0001; 22L: **p = 0.003). (B) Neuropathological analysis of terminally diseased PrPCGPIThy-1 L16 mice infected with RML. In PrPCGPIThy-1 L16 brains a general decrease in spongiosis is observed with HE staining. Gliosis is also decreased as observed with antibodies against astrocytes (GFAP) and microglia (Iba1). (C) Representative blot of WTPrPC and PrPCGPIThy-1 L16 brain homogenates infected with RML prions and digested with PK. Decreased amounts of PK-resistant PrPSc were observed for PrPCGPIThy-1 L16 brain homogenates, despite of the two-fold expression of the transgene (shown in S1A Fig).
(TIF)
(A) Western blot analysis of PrP expression in PrPCGPIThy-1 L150 mice at 20 weeks (20W, n = 3) and 40 weeks of age (40W, n = 4). No changes in expression were observed between the two groups (bar chart shows the mean of PrP relative intensity related to actin used as a loading control; WTPrPC is set to 100%). (B) (i) Representative western blot showing expression of shed PrP in terminally RML-infected mice (n = 4) compared with non-infected PrPCGPIThy-1 mice (n = 4). Note that shed PrP increases about 3-fold in RML-infected animals. Bar chart shows relative intensity of shed PrP related to the total amount of PrP (shown in the re-probed blot in (ii); PrPCGPIThy-1 in uninfected mice is set to 100%). (ii) Representative western blot showing that the amounts of total PrP do not change between RML-infected (n = 4) and uninfected PrPCGPIThy-1 mice (n = 4). The bar chart shows relative intensity of PrP developed with POM1 related to actin used as a loading control. All error bars are SEM.
(TIF)
(A) Representative blot of total brain homogenates from WTPrPC and PrPCGPIThy-1 L159 mice detected with the antibody against shed PrP. (B) Although this line of transgenic mice expresses per se less PrPCGPThy-1 levels than WTPrPC mice (as shown in S3 Fig), when the corresponding signal of shed PrP is referred to the signal of total PrP, there is a significant decrease (*p = 0.015) in shed PrP in PrPCGPIThy-1 L159 brains (n = 3) compared to WTPrPC (n = 3).
(TIF)
(DOCX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.