Abstract
Purpose
To identify the genetic variation in two unrelated probands with congenital cataract and to perform functional analysis of the detected variants.
Methods
Clinical examination and phenotyping, segregation, and functional analysis were performed for the two studied pedigrees.
Results
A novel OCRL gene variant (c.1964A>T, p. (Asp655Val)) was identified. This variant causes defects in OCRL protein folding and mislocalization to the cytoplasm. In addition, the variant’s location close to the Rab binding site is likely to be associated with membrane targeting abnormalities.
Conclusions
The results highlight the importance of early genetic diagnosis in infants with congenital cataract and show that mutations in the OCRL gene can present as apparently isolated congenital cataract.
Introduction
Mutations in the oculocerebrorenal syndrome of Lowe (OCRL; OMIM 300535) gene have been described in oculocerebrorenal syndrome of Lowe [1] and Dent disease type 2 [2]. Both conditions can cause progressive renal dysfunction leading to renal failure through impairment of proximal renal tubular cells (selective proximal tubulopathy) that leads to hypercalciuria, aminoaciduria, low molecular weight proteinuria, and renal tubular acidosis [3,4]. In addition, both conditions have ocular and central nervous system manifestations. The conditions are associated with congenital cataract, glaucoma, learning disability, and increased susceptibility to seizures [3,5]. Dent disease caused by mutations in the CLCN5 gene (OMIM 300008) has a more severe phenotype than the 15% of patients with Dent disease type 2 caused by mutations in the OCRL gene [3]. In one series, two of 28 patients with Dent disease type 2 had mild, asymptomatic cataract, and none had glaucoma [3]. In contrast, the classical Lowe syndrome phenotype includes symptomatic cataract, severe learning difficulties, and behavioral disturbance in addition to the proximal tubulopathy [6].
The OCRL gene is located at Xq25–26 and spans 52,278 bp. It contains 24 exons; exons 1–23 are the coding regions [7]. A variant protein with eight additional amino acids is created by alternative splicing of exon 18a, and in contrast to most tissues where both alternatively spliced forms are expressed, the variant is the sole OCRL variant in the brain [7,8]. Both OCRL isoforms localize to various cellular compartments, including the trans-Golgi network and endosomes, with the longer isoform displaying a stronger association with clathrin-coated pits due to better binding to the clathrin coat protein [9]. More than 200 variants of the OCRL gene associated with Lowe syndrome or Dent disease type 2 have been identified. However, in one series, no variant was identified in about 20% of patients [10].
Other variant types of unknown significance have been described throughout the OCRL gene and are not related to the Lowe syndrome or Dent disease type 2 phenotypes [11]. No clear correlation between the class of variant and clinical severity of the phenotype has been found. Additionally, the same missense variants (e.g., p.Arg318Cys) have been associated with the classic Lowe syndrome phenotype and the mild Dent disease type 2 phenotype [10]. A possible explanation for this variability may be the presence of compensatory factors that differ between patients as the expression of these factors are likely to be determined by the patient’s genetic background [10].
Methods
We report two pedigrees with a novel OCRL gene variant presenting with variable, but hypomorphic, Lowe syndrome phenotypes, including, and importantly, apparent isolated congenital cataract. Written informed consents were obtained from the participants and their families. This work was conducted according to the tenets of the declaration of Helsinki. Ethics approval was not required as this work not done as a research project. All the collected clinical data and images were consented for. Proband 1 is a child who was diagnosed with visually significant mixed nuclear and cortical bilateral (but asymmetric) congenital cataract shortly after birth. He underwent left lens extraction (lensectomy) at the age of 6 months, and his right cataract was monitored thereafter. A contact lens and patching were prescribed to treat left amblyopia. After the child was referred to the oculogenetic clinic for further investigation, full prenatal and neonatal histories were taken from his parents and were unremarkable. He was born at 40 weeks’ gestation with a birthweight of 2.8 kg. Gross motor milestones were within normal range. He sat unsupported at 6 months, crawled at 9 months, and walked unaided at 15 months of age. He had normal hearing. However, he had delayed speech. He was also being seen by the dermatology department for congenital melanocytic nevus syndrome. He had no other medical conditions. His family history included a maternal uncle and a maternal great-uncle known to have had cataract surgery before the age of 20 years, but no other information was available. The proband had one younger sibling with no relevant medical history who has good vision but bilateral peripheral cortical cataracts. The proband’s mother and maternal grandmother did not have a significant medical history or ocular phenotype, and no family members had raised intraocular pressure or additional features of glaucoma.
Proband 1’s DNA sample was screened for 114 genes associated with congenital cataract via the United Kingdom Genetic Testing Network (UKGTN) registered, Manchester Retinal Gene Panel. Next-generation sequencing was performed, using a custom-designed, Sure Select targeting kit (Agilent, Santa Clara, CA), and 100% of the targeted regions were captured and sequenced.
Materials and antibodies
The following antibodies were used: mouse anti-EEA1 (BD Biosciences, San Jose, CA, 610456), sheep anti-GFP [12], rabbit anti-GORAB (Proteintech, Manchester, UK, 17798), sheep anti-GST [13], sheep anti-Pacsin 2 [14], and sheep anti-TGN46 (Vas Ponnambalam, University of Leeds, Leeds, UK). Cy3- and Cy5-conjugated secondary antibodies were from Jackson ImmunoLabs (West Grove, PA). Horseradish peroxidase (HRP)-conjugated secondary antibodies were from Santa Cruz Biotechnology (Santa Clara, CA) or Tago Immunochemicals (Camarillo, CA).
Molecular biology
IPIP27A, OCRL, and Rab6 constructs have been described previously [9,15,16]. The NCBI RefSeq accession numbers for each are NM_001177997.1 (IPIP27A), NM_000276.3 (OCRL), and NM_198896.1 (Rab6). GFP-OCRL D655V was generated with PCR using the Quikchange method (Agilent Technologies). PCR reactions were performed with Pfu Turbo Agilent Technologies (Santa Clara, CA) using the following primers (F: GGG AGA AGA TAA GAT TGA AGT TAT TCT CGT CCT TCA CCT GG; R: CCA GGT GAA GGA CGA GAA TAA CTT CAA TCT TAT CTT CTC CC) and PCR conditions: 5 min 95 ˚C, followed by 16 cycles of 1 min 95 ˚C, 30 sec 55 ˚C, 18 min 68 ˚C, and a final incubation at 68 ˚C for 18 min
Cell culture and transfection
HeLaM cells were cultured at 37 °C and 5% CO2 in Dulbecco’s modified eagle medium (DMEM; Sigma-Aldrich, St Louis, MO), supplemented with 10% Hyclone fetal bovine serum (Thermo Fisher Scientific; Waltham, MA), 1% penicillin/streptomycin (100 units/ml), and 1 mM L-glutamine. DNA transfections were performed with Fugene HD (Promega; Fitchburg, WI)) according to the manufacturer’s instructions. A total of 2 µg DNA per six wells was used. Cells were assayed between 18 and 24 h after transfection.
Fluorescence microscopy
Cells were grown on coverslips and fixed in paraformaldehyde (3% w/v in PBS 1X; 8.1 mM Na2HPO4, 1.47 mM KH2PO4, 27 mM KCl, 137 mM NaCl, pH 7.4) for 20 min at room temperature (RT) and then washed 3X in PBS. Cells were permeabilized by incubating in 0.1% Triton X-100 in PBS for 4 min and then washed 3X in PBS. Coverslips were incubated for 20 min at RT with primary antibodies. Then fluorophore-conjugated secondary antibodies were diluted in PBS containing 0.5 mg/ml bovine serum albumin and 100 ng/ml Hoechst 33,342 before mounting in Mowiol. Slides were viewed using an Olympus (Bolton, UK) BX60 upright microscope with a 60X objective, and images were taken using a MicroMax-cooled, slow-scan charge-coupled device (CCD) camera (Roper Scientific, Tucson, AZ) controlled by MetaVue software (MDS Analytical Technologies, San Jose, CA). Images were processed using ImageJ (Bethesda, MD) and Adobe Photoshop Creative Suite 5 (San Jose, CA). Figures were assembled in Adobe Illustrator Creative Suite 5.
Protein binding experiments
Cells were lysed on ice for 20 min in lysis buffer (20 mM HEPES, pH 7.4, 5 mM MgCl2, 0.1 M NaCl2, 0.5% (wt/vol) Triton X-100, and complete, EDTA-free Protease Inhibitor Cocktail (Roche Diagnostics) using 1 ml per 10 cm dish. Lysates were clarified with centrifugation at full speed in a microfuge for 10 min at 4 °C. GST-tagged bait protein (80–400 µg) coupled to 10 µl glutathione-Sepharose was then incubated with 300 µl cell lysate for 4 h at 4 °C. After washing in lysis, bound proteins were eluted in sodium dodecyl sulfate (SDS) sample buffer and analyzed with sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) and western blotting or Coomassie blue staining.
Analysis of the effect of the OCRL D655V mutation on Rab binding
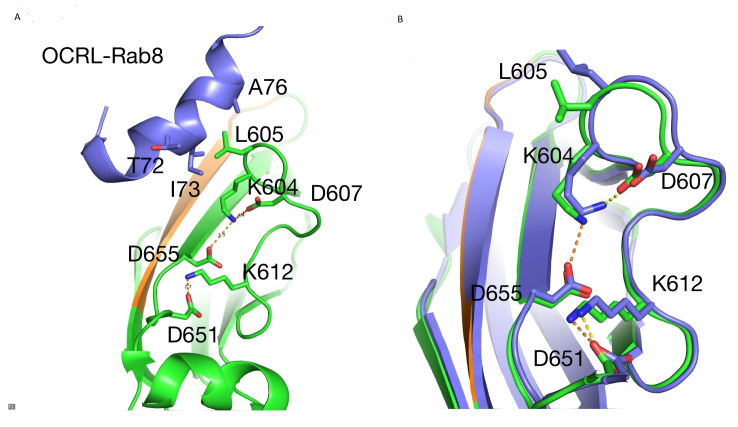
The crystal structures of OCRL with the Rab8a [17] and the complex of OCRL with Ipip27A/SES1 [18] were analyzed. OCRL Asp655 is located at the N-terminus of β5 of the ASH domain in a region rich in charged residues (K604, D607, K612, and D651), most of which are located in a loop connecting β5 and β6 (Figure 1A). In the Rab8a complex, L605 in this loop forms hydrophobic interactions with T72, I73, and A76 in the switch II region of Rab8a (Figure 1A), thus contributing to the overall interface between OCRL and Rab8. In this complex, D655 forms a hydrogen bond with K604 (Figure 1A). Interestingly, in the complex of OCRL and Ipip27A/SES1, there are minor structural rearrangements affecting the Rab-binding loop. The hydrogen bond between D655 and K604 is lost, and this results in a change in the position of L605, which is critical for Rab8a binding (Figure 1B). Therefore, it is likely that the role of D655 is to help stabilize the loop conformation required to engage in hydrophobic interactions with Rab8a. In the D655V mutation, the interaction with K604 will be lost, thus favoring a conformation for the β5–β6 loop that is incompatible with Rab binding.
Figure 1.
The effect of the OCRL D655V mutation on Rab binding. A: Structure of ORCL-Ash domain (green) with Rab8a (purple). L605 is critical for interaction with Rab8a. Residues involved in hydrogen bond interactions within the β5-β6 loop are shown as sticks, including D655. Intercations are shown in dashed lines. B. superposition of the structures of OCRL-Rab8a (3QBT in green) and OCRL-SES1 (3QIS in purple), showing the change in the position of K604 and L605.
Results
A novel hemizygous OCRL variant, c.1964A>T, p. (Asp655Val), was identified (OMIM 300535). Segregation analysis showed that the proband’s mother and grandmother were heterozygous for the variant and his younger brother was also hemizygous. A renal profile in the proband revealed no significant abnormalities (serum sodium 141 mmol/l, serum potassium 4.1 mmol/l, serum urea 4.8 mmol/l, and creatinine 50 mmol/l, which is slightly raised). This raised the possibility that this novel variant was causative for the hypomorphic Lowe/Dent-2 phenotype observed in the male hemizygous proband and his brother. The phenotype of the isolated cataract in this proband and his younger brother is novel for both of the known OCRL-related conditions (Lowe syndrome and Dent disease type 2).
Subsequently, through the Manchester clinical laboratory, a second proband with the same variant was identified in an unrelated family from Northern Ireland. He was diagnosed with bilateral congenital cataract shortly after birth. He had cataracts removed at 2 weeks of age. He was being monitored afterward in the pediatric ophthalmology clinic and developed secondary glaucoma. He was diagnosed with hypodontia with 11 missing teeth. He was tested for Nance-Horan syndrome (dental cataract syndrome) at the age of 15 years when he was referred to local genetics services with developmental delay and mild facial dysmorphism. The National Health Service (NHS) gene mutation analysis was reported as normal ruling out the possibility of Nance-Horan syndrome. However, array comparative genomic hybridization (CGH) identified maternally inherited 8p23 duplication of 187 kb. The duplication interval contains the genes NEIL2 (OMIM 608933), CTSB (OMIM 116810), and FDFT1 (OMIM 184420) and the OMIM morbid gene GATA4 (OMIM 600576). The duplication noted in him was smaller than, but located within, the region commonly found to be duplicated in 8p23.1 duplication syndrome. This syndrome is caused by a partial duplication of the short arm of chromosome 8 and is associated with speech delay, developmental delay, mild facial dysmorphism, and congenital heart disease (but not congenital cataract or renal disease). He presented with a generalized tonic clonic seizure at the age of 19 years, and investigations suggested that the proband had stage 3 chronic kidney disease (proteinuria, urine pH 5.5, creatinine 156 μmol/l, estimated glomerular filtration rate (eGFR) 54 ml/min with normal electrolyte levels and blood pressure). Other blood tests such as anti-neutrophil cytoplasmic antibody (ANCA), anti-glomerular basement membrane (Anti-GBM), compliment component 3 (C3) and compliment component 4 (C4) were normal. Computed tomography (CT) brain and electroencephalogram (EEG) were also normal. In view of his evolved clinical phenotype, he was investigated for Lowe’s syndrome at the age of 19 years, and OCRL1 gene (OMIM 300535) mutation analysis was requested. He also had the same OCRL variant as described in proband 1 (hemizygous OCRL change, c.1964A>T, p. (Asp655Val). Segregation analysis confirmed that the variant was present in proband 2’s mother, who did not have any ocular abnormalities. The variant was not found in the proband’s sister or in an unaffected maternal uncle. However, the family reported that another maternal uncle had died of renal failure. Proband 2 had phosphatidylinositol bisphosphate phosphatase enzyme analysis on skin fibroblasts in view of the uncertain significance of the OCRL1 variant. The enzyme level was found to be very low suggesting his affected status.
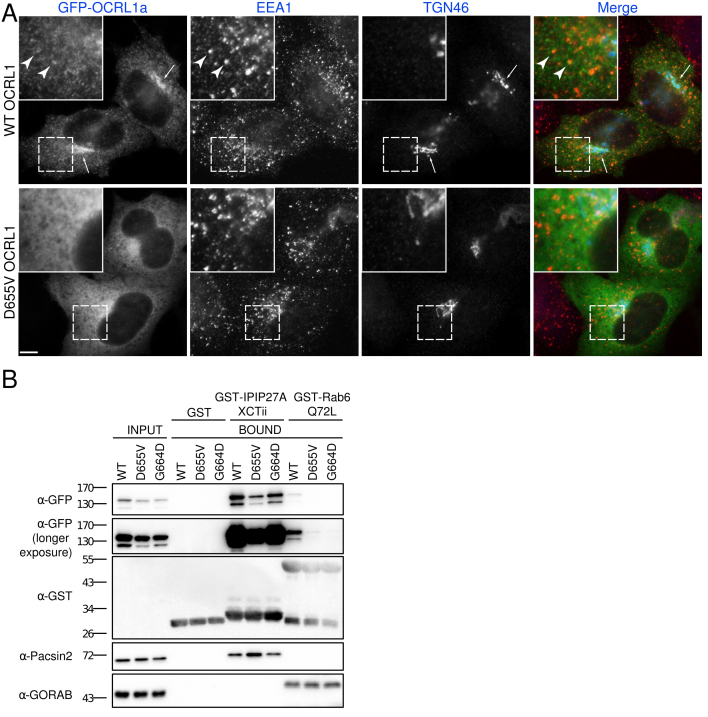
Due to the novel phenotypes described here and the novelty of the c.1964A>T, p. (Asp655Val) variant, we performed studies to determine the cellular consequences of this novel OCRL variant. Specifically, localization and protein binding studies with the D655V variant were performed. The D655V variant protein and a wild-type control were tagged with GFP and expressed in human tissue culture cells. In comparison to the wild-type protein, which localized to the perinuclear Golgi apparatus and punctate endosomal structures as expected, the D655V variant was mislocalized to the cytoplasm (Figure 2A). To explain this loss of membrane targeting, binding of OCRL to two binding partners known to function in this process, IPIP27A and members of the Rab GTP family that includes Rab6, was analyzed [13,15,19,20]. As shown in Figure 2B, the D655V mutation completely abrogated binding to Rab6, giving a similar loss of binding to the previously identified Rab binding deficient mutant G664D [13]. In contrast, binding to IPIP27A was only marginally affected (Figure 1B). Thus, the loss of membrane targeting of D655V can be explained by a loss of interaction with Rabs.
Figure 2.
The D655V mutation compromises the membrane localization and interaction of OCRL with Rab6. A: HeLaM cells transiently transfected with wild-type (WT) or D655V green fluorescent protein (GFP-OCRL) were fixed, immunolabeled with antibodies raised against the early endosome marker EEA1 (red) and trans-Golgi network protein TGN46 (blue), and examined with immunofluorescence microscopy. Scale bar, 10 µm. B: Glutathione S-transferase tagged (GST-tagged) bait proteins were incubated with extracts from HeLaM cells expressing GFP-tagged WT, D655V, or G664D OCRL, and binding was assessed with western blotting with anti-GFP antibodies. Anti-Pacsin2 and anti-GORAB antibodies were used as positive controls for binding to GST-IPIP27A and GST-Rab6 Q72L, respectively.
Discussion
We report a novel variant in the OCRL gene in two unrelated pedigrees associated with a variable, hypomorphic Lowe syndrome phenotype. Support for the pathogenicity of this variant comes from segregation within the family in addition to functional analysis identifying defects in protein folding and mislocalization to the cytoplasm, explained by perturbation of interaction with Rab GTPases that mediate membrane targeting. However, the authors appreciate that due to the variation in phenotypes presented here, it is possible that other genetic factors, such as an unidentified gene deletion or the interaction of other modifying genes, may play a role in the disease phenotypes presented here. These findings have important clinical implications for infants with seemingly isolated congenital cataract. The findings support the argument for early genetic diagnosis in these patients who may develop further features of Lowe syndrome or Dent disease type 2, including significant ocular (glaucoma) and systemic (renal) disease later in life, requiring early treatment.
References
- 1.Attree O, Olivos IM, Okabe I, Bailey LC, Nelson DL, Lewis RA, McInnes RR, Nussbaum RL. The Lowe’s oculocerebrorenal syndrome gene encodes a protein highly homologous to inositol polyphosphate-5-phosphatase. Nature. 1992;358:239–42. doi: 10.1038/358239a0. [DOI] [PubMed] [Google Scholar]
- 2.Hoopes RR, Jr, Shrimpton AE, Knohl SJ, Hueber P, Hoppe B, Matyus J, Simckes A, Tasic V, Toenshoff B, Suchy SF, Nussbaum RL, Scheinman SJ. Dent Disease with mutations in OCRL1. Am J Hum Genet. 2005;76:260–7. doi: 10.1086/427887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bokenkamp A, Bockenhauer D, Cheong HI, Hoppe B, Tasic V, Unwin R, Ludwig M. Dent-2 disease: a mild variant of Lowe syndrome. J Pediatr. 2009;155:94–9. doi: 10.1016/j.jpeds.2009.01.049. [DOI] [PubMed] [Google Scholar]
- 4.Bockenhauer D, Bokenkamp A, van’t Hoff W, Levtchenko E, Kist-van Holthe JE, Tasic V, Ludwig M. Renal phenotype in Lowe Syndrome: a selective proximal tubular dysfunction. Clin J Am Soc Nephrol. 2008;3:1430–6. doi: 10.2215/CJN.00520108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Loi M. Lowe syndrome. Orphanet J Rare Dis. 2006;1:16. doi: 10.1186/1750-1172-1-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kleta R. Fanconi or not Fanconi? Lowe syndrome revisited. Clin J Am Soc Nephrol. 2008;3:1244–5. doi: 10.2215/CJN.02880608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nussbaum RL, Orrison BM, Janne PA, Charnas L, Chinault AC. Physical mapping and genomic structure of the Lowe syndrome gene OCRL1. Hum Genet. 1997;99:145–50. doi: 10.1007/s004390050329. [DOI] [PubMed] [Google Scholar]
- 8.Johnson JM, Castle J, Garrett-Engele P, Kan Z, Loerch PM, Armour CD, Santos R, Schadt EE, Stoughton R, Shoemaker DD. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–4. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- 9.Choudhury R, Noakes CJ, McKenzie E, Kox C, Lowe M. Differential clathrin binding and subcellular localization of OCRL1 splice isoforms. J Biol Chem. 2009;284:9965–73. doi: 10.1074/jbc.M807442200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hichri H, Rendu J, Monnier N, Coutton C, Dorseuil O, Poussou RV, Baujat G, Blanchard A, Nobili F, Ranchin B, Remesy M, Salomon R, Satre V, Lunardi J. From Lowe syndrome to Dent disease: correlations between mutations of the OCRL1 gene and clinical and biochemical phenotypes. Hum Mutat. 2011;32:379–88. doi: 10.1002/humu.21391. [DOI] [PubMed] [Google Scholar]
- 11.Bokenkamp A, Ludwig M. The oculocerebrorenal syndrome of Lowe: an update. Pediatr Nephrol. 2016;31:2201–12. doi: 10.1007/s00467-016-3343-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Noakes CJ, Lee G, Lowe M. The PH domain proteins IPIP27A and B link OCRL1 to receptor recycling in the endocytic pathway. Mol Biol Cell. 2011;22:606–23. doi: 10.1091/mbc.E10-08-0730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hyvola N, Diao A, McKenzie E, Skippen A, Cockcroft S, Lowe M. Membrane targeting and activation of the Lowe syndrome protein OCRL1 by rab GTPases. EMBO J. 2006;25:3750–61. doi: 10.1038/sj.emboj.7601274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Swan LE, Tomasini L, Pirruccello M, Lunardi J, De Camilli P. Two closely related endocytic proteins that share a common OCRL-binding motif with APPL1. Proc Natl Acad Sci USA. 2010;107:3511–6. doi: 10.1073/pnas.0914658107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diao A, Rahman D, Pappin DJ, Lucocq J, Lowe M. The coiled-coil membrane protein golgin-84 is a novel rab effector required for Golgi ribbon formation. J Cell Biol. 2003;160:201–12. doi: 10.1083/jcb.200207045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fukuda M, Kanno E, Ishibashi K, Itoh T. Large scale screening for novel rab effectors reveals unexpected broad Rab binding specificity. Molecular & cellular proteomics. MCP. 2008;7:1031–42. doi: 10.1074/mcp.M700569-MCP200. [DOI] [PubMed] [Google Scholar]
- 17.Hou X, Hagemann N, Schoebel S, Blankenfeldt W, Goody RS, Erdmann KS, Itzen A. A structural basis for Lowe syndrome caused by mutations in the Rab-binding domain of OCRL1. EMBO J. 2011;30:1659–70. doi: 10.1038/emboj.2011.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pirruccello M, Swan LE, Folta-Stogniew E, De Camilli P. Recognition of the F&H motif by the Lowe syndrome protein OCRL. Nat Struct Mol Biol. 2011;18:789–95. doi: 10.1038/nsmb.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Billcliff PG, Noakes CJ, Mehta ZB, Yan G, Mak L, Woscholski R, Lowe M. OCRL1 engages with the F-BAR protein pacsin 2 to promote biogenesis of membrane-trafficking intermediates. Mol Biol Cell. 2016;27:90–107. doi: 10.1091/mbc.E15-06-0329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choudhury R, Diao A, Zhang F, Eisenberg E, Saint-Pol A, Williams C, Konstantakopoulos A, Lucocq J, Johannes L, Rabouille C, Greene LE, Lowe M. Lowe syndrome protein OCRL1 interacts with clathrin and regulates protein trafficking between endosomes and the trans-Golgi network. Mol Biol Cell. 2005;16:3467–79. doi: 10.1091/mbc.E05-02-0120. [DOI] [PMC free article] [PubMed] [Google Scholar]