Abstract
Autophagy delivers cytoplasmic constituents to autolysosomes and has been linked to both innate and adaptive immunity. Toll-Like Receptor 4 (TLR4) signaling induces autophagy and recruits Beclin 1 to the receptor complex. Here we show that Traf6-mediated lysine (K) 63-linked ubiquitination of Beclin 1 is critical for TLR4 triggered autophagy in macrophages. Two Traf6 binding motifs in Beclin 1 facilitated Traf6 binding and its ubiquitination. Beclin 1 K117, strategically located in the Beclin 1 BH3 domain is a major site for K63-linked ubiquitination. A20, a known deubiquitinating enzyme, reduced Beclin 1 K63-linked ubiquitination, and limited the induction of autophagy following TLR signaling. Treatment of macrophages with either interferon-γ or IL-1 also triggered K63-linked ubiquitination of Beclin 1 and autophagosome formation. These results indicate that the status of Beclin 1 K63-linked ubiquitination plays a key role in regulating autophagy during inflammatory responses.
Keywords: Toll-like receptor, interferon, macrophage, autophagy, signal transduction, ubiquitination
INTRODUCTION
Toll-like receptors (TLRs) contain leucine-rich repeat regions that recognize specific molecular patterns present in microbial components. TLR stimulation leads to the activation of innate immunity and helps to instruct the development of antigen-specific acquired immunity (1, 2). TLRs contain intracellular Toll/IL-1 receptor (TIR) domains that trigger signals via homotypic interactions with proximal adaptor proteins such as MyD88. Upon ligand activation, MyD88 recruits IL-1R-associated kinase (IRAK)-1 and IRAK-4. IRAK-1 is phosphorylated resulting in the recruitment of TNF receptor-associated factor 6 (Traf6). This is followed by Traf6 autoubiquitination with K63-linked ubiquitin chains and K63-linked IRAK-1 ubiquitination. Ubiquitin modified Traf6 activates the TGF-ß-activated protein kinase 1 (TAK1)/TAK-1-binding protein complex, which phosphorylates the inhibitor of κB (IκΒ) kinase (IKK) complex. This results in IκB phosphorylation and degradation, which promotes the translocation of NF-κB into nuclei and the induction of inflammatory cytokines production as well as A20 expression. A20 functions both as a deubiquitinating enzyme and as an E3 ligase (3). A20 helps terminate the activation of NF-κB by deubiquitinating Traf6. A20-deficient mice exhibit spontaneous inflammation, cachexia and premature death that are TLR signaling dependent (4, 5).
TLRs also use MyD88 and the adaptor TRIF to trigger autophagy in macrophages (6–8). Autophagy is a cellular response to starvation as well as a quality control system that can remove damaged organelles and long-lived proteins from the cytoplasm. Autophagy plays a role in programmed cell death; the promotion and prevention of cancer; neurodegeneration; major histocompatibility complex class II antigen presentation; and cellular defense by removing intracellular pathogens (9). Numerous lines of evidence indicate that autophagy is involved in both innate and adaptive immunity against intracellular protozoa, bacteria and viruses (10–13). Engagement of TLR4 recruits not only MyD88, but also Beclin 1 into the TLR4-signaling complex (7). Beclin 1 is the mammalian homologue of yeast Atg6, a key component of a Class III phosphatidylinositol 3-kinase complex (PI3KC3) that initiates autophagosome formation by helping to localize other autophagy proteins to the pre-autophagosomal membrane (14). Oligomerization of Beclin 1 may allow it to serve as scaffold for other molecules known to associate with Beclin 1. Exposure of macrophages to TLR4 ligands reduces the association of Beclin-1 with Bcl2, an anti-apoptotic protein known to bind Beclin 1 and to reduce autophagy (15). The mechanism by which prosurvival proteins such as Bcl2 reduce autophagy is still not understood. It is known that the interaction of Bcl2 with Beclin 1 interferes with complex formation between PI3KC3 and Beclin 1. However, the PI3KC3 binding site and the BH3 domain of Beclin 1, which binds Bcl2 proteins, do not overlap (16, 17). One possibility is that Bcl2 like proteins inhibit Beclin 1 oligomerization, which normally facilitates the binding of interacting proteins such as PI3KC3.
Previously, we had noted that immunoprecipitates prepared from macrophages using an agonistic TLR4 antibody contained Beclin 1 that had a higher than expected molecular mass and a smeared appearance on SDS-PAGE. In this study we tested whether Beclin 1 undergoes ubiquitination, examined whether the ubiquitination is K63-or K48-linked, and determined the physiologic significance of this modification. This has led us to the identification of a key ubiquitination site on Beclin 1. We have also shown that similar to their role in the regulation of NF-κB activation, TRAF6 functions as an E3 ligase to ubiquinate Beclin 1 and that A20 negatively regulates Beclin 1 ubiquitination to control the level of autophagy elicited by TLR4 signaling.
RESULTS
TLR4 signaling triggers K63-linked ubiquitination of Beclin 1 via a mechanism that depends upon Traf6
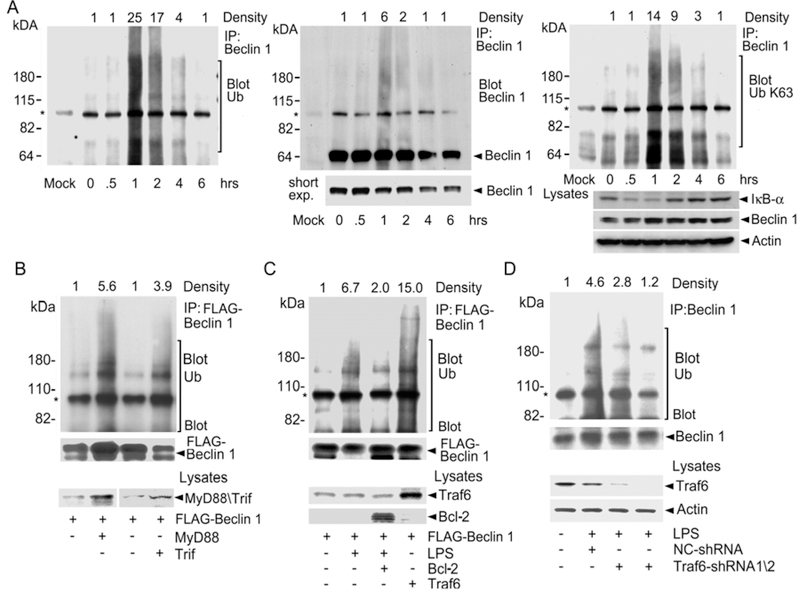
To determine whether Beclin 1 underwent ubiquitination, we stimulated the murine macrophage cell line Raw 264.7 for various durations with the TLR4 agonist LPS. We lysed the cells and dissociated any protein-protein interactions prior to immunoprecipitating Beclin 1 and immunoblotting for ubiquitin (Fig. 1A, left panel). The immunoblot revealed the induction of a smear of ubiquitinated proteins, most prominent in the range of 100–200 kDa at 1 hr after exposure, which disappeared by 6 hours after stimulation. Stripping and re-blotting for Beclin 1 disclosed a prominent Beclin 1 band at the expected molecular mass and a less intense smear of Beclin 1 in a pattern similar to the ubiquitin immunoblot (Fig. 1A, middle panel). These results indicate that Beclin 1 was ubiquitinated following LPS stimulation. Next, we examined whether Beclin 1 ubiquitination occurred via a K63-linkage. To do so we stripped and re-probed the immunoblot with an antibody specific for K63-linked ubiquitin (Fig. 1A, right panel). The resultant immunoblot appeared similar to the previous ubiquitin immunoblot indicating that much of the Beclin 1 ubiquitination occurred by a K63-linkage. We also found that treating primary human monocytes with LPS resulted in Beclin 1 K63-linked ubiquitination (Fig. 1S). Since TLRs and the interleukin-1 receptor (IL-1R) signaling pathways share some common elements including usage of the E3 ligase Traf6, we checked whether IL-1 stimulation of Raw 264.7 cells triggered Beclin 1 ubiquitination. We found that IL-1R signaling resulted in an even more prominent K63-linked ubiquitination of Beclin 1 than did LPS. Like LPS stimulation, IL-1R signaling also induced autophagosome formation in these cells (Fig. 2S).
Fig. 1.
TLR4 engagement, MyD88, Trif, and Traf6 promote the ubiquitination of Beclin 1. (A) Ubiquitination of Beclin 1. LPS was used to stimulate Raw 267.4 cells for various durations. Shown is a Beclin 1 immunoprecipitation immunoblotted for ubiquitin (Ub, left panel), Beclin 1 (middle panel), and K63-linked ubiquitin (right panel). After each immunoblotting the membrane was stripped and the efficacy of the stripping verified. The major non-specific band (*) results from the secondary antibody recognizing the immunoprecipitating antibody. Lysates were immunoblotted as indicated. LPS stimulation reduced IκB levels in the cell lysates as expected. (B) Overexpression of MyD88 or Trif enhances Beclin 1 ubiquitination. FLAG immunoprecipitates from cells expressing MyD88 or Trif with FLAG-Beclin 1 were immunoblotted for ubiquitin, stripped and re-blotted for Beclin 1. Lysates immunoblotted as indicated. (C) Bcl-2 attenuates while Traf6 overexpression enhances Beclin 1 ubiquitination. FLAG immunoprecipitates from Raw 264.7 cells expressing Bcl-2 or Traf6 with FLAG-Beclin 1 were immunoblotted for ubiquitin, stripped, and re-blotted for Beclin 1. Lysates were immunoblotted as indicated. (D) Reduced Traf6 expression impairs Beclin 1 ubiquitination. Beclin 1 immunoprecipitates from Raw 264.7 cells expressing control or shRNAs targeted at Traf6 were immunoblotted for ubiquitin, stripped, and re-blotted for Beclin 1. The lysates were immunoblotted as indicated.
Since both MyD88 and Trif overexpression had led to the recruitment of Beclin 1, we tested whether their overexpression enhanced the ubiquitination of co-expressed FLAG-Beclin 1 (Fig. 1B). We observed that co-expression of MyD88 or Trif with Flag-Beclin 1 led to an increase in the amount of ubiquitin in the FLAG immunoprecipitates. Next, we tested Traf6’s involvement in Beclin 1 ubiquitination. When over-expressed along with FLAG-Beclin 1, the amount of ubiquitin detected in the FLAG immunoprecipitates increased as compared to those from cells not over-expressing Traf6. In contrast, Bcl-2 overexpression, which reduces LPS induced autophagosome formation (7), had the opposite effect decreasing the LPS-induced increase in ubiquitin that we detected in the FLAG-Beclin 1 immunoprecipitates (Fig. 1C). Confirming the importance of Traf6, expression of either of two different Traf6 shRNAs in Raw 267.4 cells decreased in the amount of ubiquitin associated with endogenous Beclin 1 following LPS treatment (Fig. 1D).
Beclin 1 binds Traf6 and purified Traf6 catalyzes Beclin 1 ubiquitination
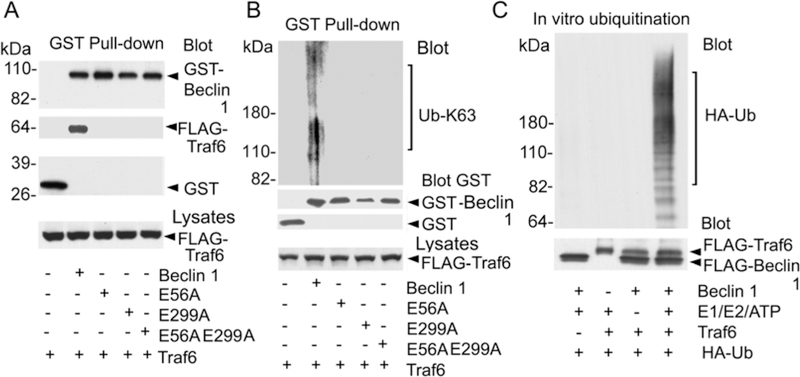
Inspection of the human Beclin 1 amino acid sequence revealed two potential Traf6 binding sites (P-X-E-X-X-aromatic/acidic) (18) located at amino acids 54–58 and 297–301. The first motif is conserved between mouse and human Beclin 1, but not in chicken or zebra fish, while the second is conserved in all four species. To determine whether these sites functioned in the recruitment of Traf6 to Beclin 1, we mutated the conserved glutamic acid residue to alanine in each potential binding site, individually and together. A Beclin 1 GST pull-down assay revealed that both sites contributed to the interaction between Traf6 and Beclin 1 (Fig. 2A). The Beclin 1 Traf6 binding mutants also exhibited much less K63-linked ubiquitination following Traf6 overexpression (Fig. 2B). To provide evidence that Traf6 directly ubiquitinated Beclin 1, we reconstituted the reaction in vitro using the indicated combinations of purified FLAG-Traf6, purified FLAG-Beclin 1, recombinant E1/E2, ATP, and recombinant HA-ubiquitin. Following the ubiquitination reaction, we dissociated the proteins, immunoprecipitated Beclin 1, and immunoblotted for HA-ubiquitin. The results showed that purified Traf6 catalyzed the in vitro ubiquitination of Beclin 1 as we observed a smear of HA-ubiquitin in the Beclin 1 immunoprecipitates that depended upon the presence of Beclin 1, Traf6, HA-ubiquitin, and E1/E2/ATP (Fig. 2C).
Fig. 2.
Traf6 binds Beclin 1 and triggers in vitro ubiquitination of Beclin 1. Traf6 binding sites identified in Beclin 1. FLAG-Traf6 was expressed along with various GST Beclin 1 fusion proteins. GST pull-downs were immunoblotted for GST, GST-Beclin 1, various GST-Beclin 1 mutants, and FLAG-Traf6. Lysates were blotted for Traf6. (B) Traf6 binding mutants of Beclin 1 undergo less ubiquitination. Following stringent washing the indicated GST pull-downs were immunoblotted for K63-linked ubiquitin. The blot was stripped and re-blotted for GST to detect GST-Beclin 1 and the various GST-Beclin 1 mutant proteins. Lysates were blotted for FLAG. (C) FLAG-Traf6 catalyzes FLAG-Beclin 1 ubiquitination in vitro. Purified proteins along with the indicated reaction components were combined and incubated. After which Beclin 1 immunoprecipitates from each reaction were analyzed by immunoblotting them for HA-ubiquitin (HA-Ub). Saved portions of each of the reactions were immunoblotted for FLAG to detect Beclin 1 and Traf6.
A20 negatively regulates the ubiquitination of Beclin 1
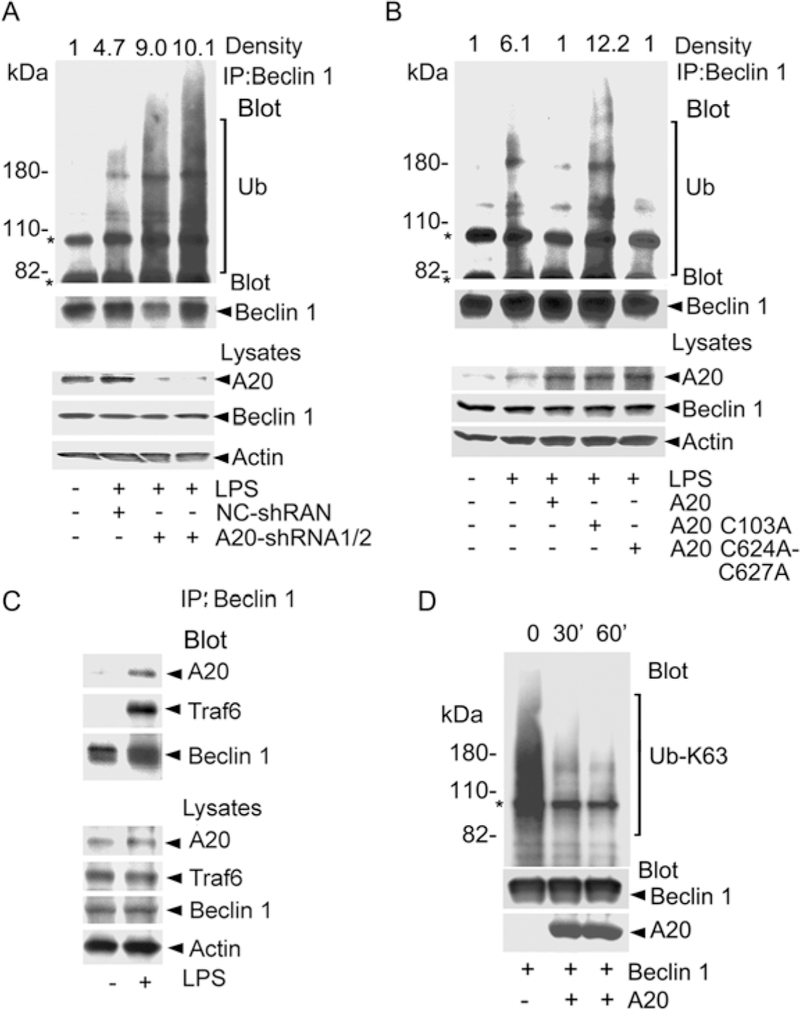
If Traf6 functions to ubiquitinate Beclin 1, then A20 by virtue of its deubiquitination activity for Traf6 (3) should counter it. We tested this hypothesis by silencing A20 expression in Raw 264.7 cells and examining endogenous Beclin 1 ubiquitination following LPS treatment. Two different A20 shRNAs reduced A20 expression and significantly increased the amount of ubiquitin detected in the Beclin 1 immunoprecipitation. Reducing A20 expression did not affect the Beclin 1 protein levels (Fig. 3A). Next, we made use of two A20 mutant proteins; A20 C103A, which is incapable of deubiquitinating Traf6, and A20 C624A/C627A, which lacks K48 E3 ligase activity (3). The results showed that A20 and the K48 E3 ligase dead form both markedly attenuated LPS-induced ubiquitination of Beclin 1, whereas the deubiquitination dead form did not (Fig. 3B). To determine whether the deubiquitinase activity of A20 might directly target ubiquitinated Beclin 1, we first examined whether A20 associates with Beclin 1. Prior to LPS stimulation A20 did not immunoprecipitate with Beclin 1, not unexpectedly since A20 is poorly expressed prior to LPS stimulation. Following LPS stimulation, the Beclin 1 immunoprecipitates contained A20, but also Traf6, which did not help clarify whether A20 directly targeted ubiquitinated Beclin 1 (Fig. 3C). To examine the effect of A20 on Beclin 1 ubiquitination in the absence of Traf6, we performed an in vitro deubiquitination assay using recombinant A20 (1–371), which lacks ubiquitin ligase activity, and purified Beclin 1. As a consequence of the overexpression some of the purified Beclin 1 is ubiquitinated. We found that following the addition of A20 the amount of K63-linked ubiquitinated Beclin 1 declined indicating that Beclin 1 is a direct A20 target (Fig. 3D). Thus, A20 can limit Beclin 1 ubiquitination not only by controlling the E3 ligase activity of Traf6, but also by directly deubiquitinating Beclin 1.
Fig. 3.
A20 regulates the ubiquitination of Beclin 1. (A) Reduced A20 expression promotes LPS-induced ubiquitination of Beclin 1. Beclin 1 immunoprecipitates from Raw 264.7 cells expressing indicated shRNAs were blotted for ubiquitin (Ub), stripped and re-blotted for Beclin 1. The major non-specific bands (*) are indicated. (B) A20 and its E3 ligase mutant, but not the deubiquitination dead form attenuate LPS-induced ubiquitination of Beclin 1. Cell lysates and Beclin 1 immunoprecipitates from cells expressing A20 or various mutants were immunoblotted for as indicated. (C) Interaction between Beclin 1 and A20. Cell lysates and Beclin 1 immunoprecipitates from Raw 264.7 cells stimulated with LPS (1 hr) or not were immunoblotted as indicated. (D) A20 deubiquitinates Beclin 1. Purified FLAG-Beclin 1 was incubated with recombinant A20 (1–371) and the reactions immunoblotted as indicated.
Beclin 1 has ubiquitin binding domain(s) that preferentially Interacts with K63-linked ubiquitin
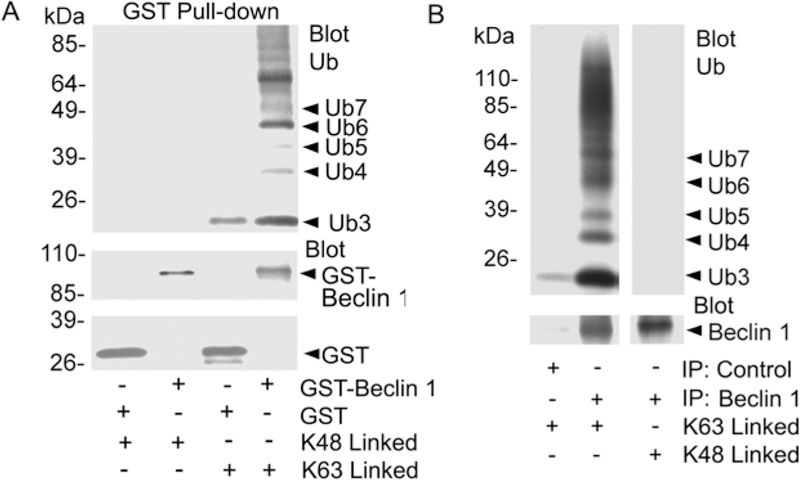
Some proteins that undergo ubiquitination also bind ubiquitin. To date 11 families of ubiquitin binding domains (UBDs) have been identified (19). While most UBDs show little chain specificity, some are capable of selecting distinct linkages from an ubiquitin chain mixture. The NEMO UBAN (ubiquitin binding in ABIN and NEMO) motif of NEMO binds linear ubiquitin chains, while the protein kinase TAK1 relies on K63 chains, which are specifically recognized by its adaptor TAB2 (20). To determine whether Beclin can bind K48-linked and/or K63-linked ubiquitin, we expressed GST or GST-Beclin 1 in Raw 267.4 cells and purified the proteins. The purified proteins were incubated with either K48-linked or K63-linked ubiquitin peptides. We found that GST-Beclin 1 exhibited a strong preference for K63-linked versus K48-linked ubiquitin peptides while GST failed to bind either (Fig. 4A). To confirm this result we examined endogenous Beclin 1 immunoprecipitated from Raw 264.7 cells. Similar to the GST-Beclin 1 endogenous Beclin 1 bound K63-linked ubiquitin peptides better than K48-linked peptides (Fig. 4B). These data provide a mechanism by which Beclin 1 can interact with K-63 linked ubiquitinated proteins.
Fig. 4.
Beclin 1 binds ubiquitin. (A) GST-Beclin 1 preferentially binds K63-linked ubiquitin. GST and GST-Beclin 1 pull-downs were incubated with K63- or K48-linked peptides, washed, and analyzed by immunoblotting for Ub. The blot was stripped and re-probed for GST to identify GST or GST-Beclin 1. (B). Endogenous Beclin 1 preferentially binds K63-linked ubiquitin. Beclin 1 immunoprecipitates from Raw 264.7 cells were similarly evaluated for K63- and K48-linked Ub peptide binding. The initial blot was stripped and re-immunoblotted for Beclin 1.
Lysine 117 of Beclin 1 is a major ubiquitination site
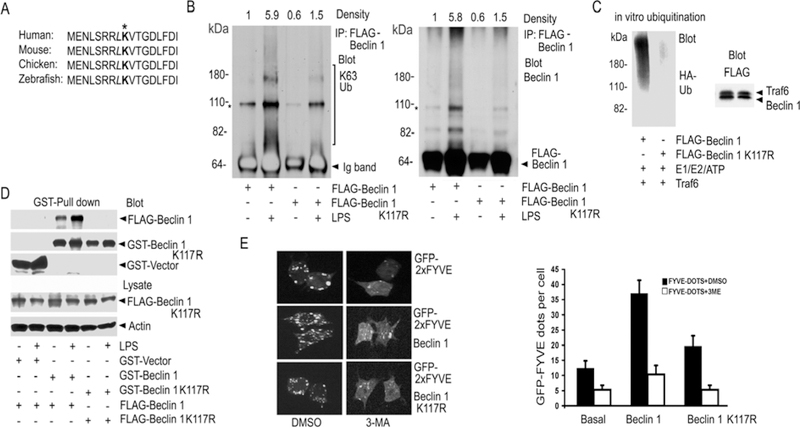
Beclin 1 contains a conserved BH3 domain required for binding Bcl-2 (15, 16). Shown is an alignment of the amino acid residues in the BH3 domain of Beclin 1 from various species (Fig. 5A). A L116A mutation in the BH3 domain of Beclin 1 attenuates Bcl-2 binding and enhances Beclin 1 induced autophagy (15). Adjacent to this residue is a conserved lysine, which we viewed as a prime candidate to link ubiquitin chains. To test that possibility, we stimulated Raw 264.7 cells expressing a FLAG-K117R mutant of Beclin 1 and analyzed its ubiquitination. We found that FLAG immunoprecipitates from the cells expressing Beclin 1 K117R had little associated K63-linked ubiquitin as compared to the FLAG immunoprecipitates from the cells expressing wild type Beclin 1 (Fig. 5B). Next, we compared wild type and Beclin 1 K117R in an in vitro ubiquitination reaction. While Traf6 again catalyzed the addition of HA-ubiquitin to wild type Beclin 1 the amount of HA-Beclin added to Beclin 1 K117R was greatly reduced (Fig. 5C). To begin to address the functional role of LPS-induced ubiquitination of Beclin 1, we tested whether K63-linked polyubiquitination might facilitate Beclin 1-Beclin 1 interactions. To do so we expressed GST-Beclin 1 and FLAG-Beclin 1 or GST-Beclin 1 K117R and FLAG-Beclin 1 K117R in Raw 264.7 cells. In the absence of LPS stimulation the GST-Beclin 1 pull-downs contained some FLAG-Beclin 1, while stimulation enhanced the amount. In contrast, the GST-Beclin 1 K117R pull-downs contained little FLAG-Beclin 1 K117R and LPS stimulation had no effect (Fig. 5D). Next, we check whether the ubiquitination of Beclin 1 might affect the lipid kinase activity of PI3KC3 by expressing either Beclin 1 or its K117R mutant in Raw 264.7 cells along with a phosphatidylinositol 3-phosphate (PI3P) sensor (21, 22). We found that PI3P production was reduced about 50 percent in Beclin 1 K117R expressing cells as compared to Beclin 1 expressing cells (Fig. 5E). These results suggest that K117 ubiquitination of Beclin 1 promotes Beclin 1 multimerization and affects the activity of PI3KC3.
Fig. 5.
Mutation of Lysine 117 in Beclin 1 reduces Beclin 1 ubiquitination. (A) Amino acid alignment of a portion of the BH3 domains of different species. (B) Contrast between LPS-induced ubiquitination of Beclin 1 and its K117R mutant. FLAG immunoprecipitates of Raw 264.7 cells expressing FLAG-Beclin 1 or FLAG-K117R Beclin 1 were immunoblotted for K63 linked ubiquitin (K63 Ub), stripped, and re-blotted for Beclin 1. The major non-specific bands (*) are indicated. The heavy chain of the FLAG antibody was detected by the secondary antibody in the left panel and is labeled Ig band. The density of each lane over the indicated area is shown. (C) Beclin 1 K117R is poorly ubiquitinated in vitro. Purified FLAG-Beclin 1 or FLAG-Beclin 1 K117R was incubated with eluted FLAG-Traf6 along with reaction components. The reactions were immunoblotted for HA-Ub. Similar amounts of Traf6 and the FLAG-tagged proteins were in the reaction. (D) Beclin 1 ubiquitination enhances Beclin 1 oligomerization. GST-pull downs from Raw 264.7 cells expressing GST-Beclin 1 and FLAG-Beclin 1 or GST-Beclin 1 K117R and FLAG-Beclin 1 K117R stimulated with LPS, or not, were immunoblotted for FLAG. (E) Beclin 1, but not its K117R mutant, enhances the activity of PI3KC3 in Raw 264.7 cells. GFP-2xFYVE dots were imaged. Beclin 1 or Beclin 1 K117R was expressed in Raw 264.7 cells along with GFP-2xFYVE. The cells were imaged and then treated with 5mM 3-MA or DMSO for 2 h. The cells were then re-imaged. Representative cells are shown (top panel). The average number of dots in 100 cells from the indicated each sample was determined. Data displayed as mean +/− SD of the number of dots/cell (bottom panel). Similar results in 3 experiments performed.
Ubiquitination of Beclin 1 amplifies LPS-induced autophagy
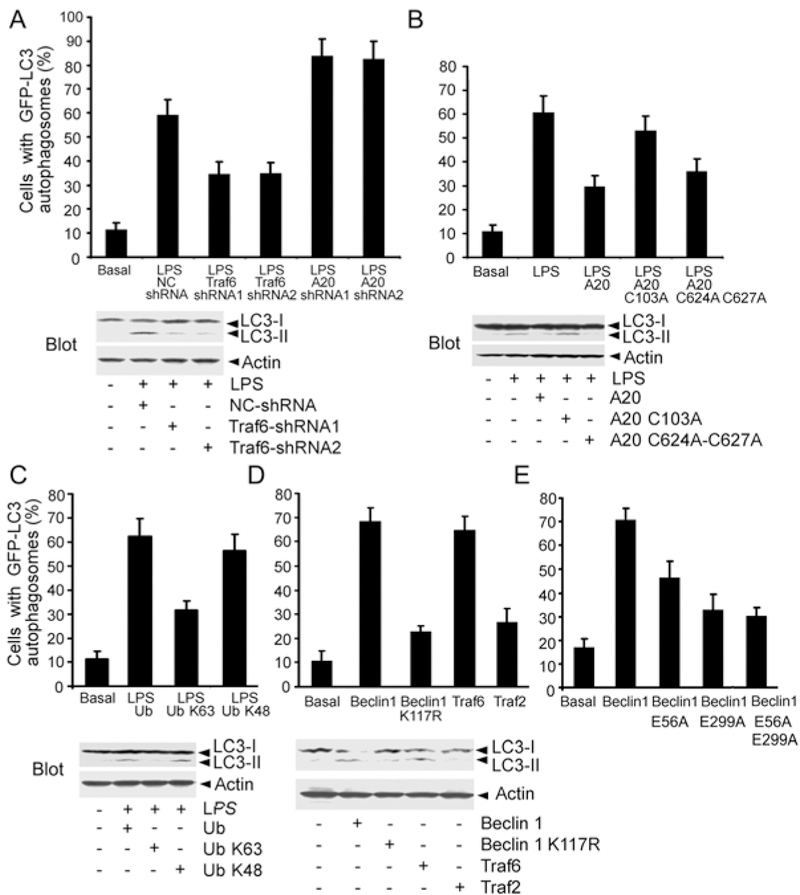
Next, we investigated whether the ubiquitination of Beclin 1 is important for LPS-induced autophagy. To detect autophagosome formation, we used GFP fused to LC3 (GFP-LC3) as a surrogate marker (23). When autophagosomes form, GFP-LC3 is processed and recruited to the autophagosome membrane, where it can be imaged by confocal microscopy. As another assay of autophagosome formation, the ratio between LC3-I and LC3-II can be monitored by western blotting (24). We first verified that LPS stimulation of Raw 267.4 cells increased autophagosome formation, by assessing LC3-II levels in the presence of lysosomal inhibitors (25). The results of this analysis are consistent with increased autophagosome synthesis. Similar results were found following Traf6 overexpression (Fig. 3S). We then examined LPS induced autophagosome formation in Traf6 and A20 knock-down Raw 264.7 cells. Reducing Traf6 expression decreased the percentage of cells with LPS-induced GFP-LC3 puncta formation by 40%, whereas reducing A20 expression increased the percentage of cells with LPS-induced GFP-LC3 puncta formation by a similar amount. Decreasing Traf6 expression also impaired the LPS-induced increases in the LC3-II/LC3-I ratio (Fig. 6A). Expression of A20 or the K48 E3 ligase dead form attenuated the percentage of cells with GFP-LC3 puncta formation by 50% and 40%, respectively, while the expression of the deubiquitination dead form had little effect. Monitoring the LC3-II/LC3–1 ratio gave similar results (Fig. 6B). To test the role of K63-linked polyubiquitination in autophagosome formation, we transfected Raw 264.7 macrophages with wild type, K63R, or K48R ubiquitin together with the GFP-LC3 marker. The K48R and K63R ubiquitin mutants impede the addition of K48- or K63-linked ubiquitin chains, respectively. We noted that the disturbed addition of K63-linked ubiquitination chains attenuated the percentage of cells with LPS-induced GFP-LC3 puncta formation by 45%, while interfering with K48-linked ubiquitination had little effect. We again confirmed these results by checking the endogenous LC3-II/LC3-I ratio (Fig. 6C). Since Beclin 1 overexpression is known to trigger autophagosome formation (7), we compared autophagosome formation following overexpression of wild type or Beclin 1 K117R. The GFP-LC3 assay and the LC3-II/LC3–1 ratio showed a marked difference between the two constructs indicating that K117 is a key residue in Beclin 1 for autophagosome formation (Fig. 6D). We found no role for Traf2, another E3 ligase that mediates Tumor Necrosis Factor induced c-Jun N-terminal protein kinase activation (26) (Fig. 6D). Showing the importance of the Traf6/Beclin 1 interaction for Beclin 1 overexpression induced autophagosome formation Beclin 1 lacking both Traf6 binding sites minimally increased the percentage of cells with GFP-LC3 puncta above the background. The E299A mutation in Beclin 1 had a greater effect than the E56A mutation (Fig. 6E).
Fig. 6.
The ubiquitination of Beclin 1 facilitates LPS-induced autophagy. (A) Modulation of LPS-induced autophagy by decreased Traf6 or A20 expression. Raw 264.7 cells expressing GFP-LC3 along with various shRNAs were treated with LPS. The % GFP-LC3 positive cells with GFP-LC3 dots were counted (top). Immunoblots of endogenous LC3-I and LC3-II and actin in similar cells is shown (bottom). (B) A20 deubiquitinating activity attenuates LPS-induced autophagy. Raw 264.7 cells expressing A20 or various mutants and GFP-LC3 were treated with LPS or not. The % GFP-LC3 positive cells with GFP-LC3 dots were counted (top). Similar experiment but the levels of endogenous LC3-I and LC3-II were measured by immunoblotting (bottom). (C) Blocking the addition of K63-linked Ub chains impairs LPS-induced autophagy. The indicated proteins were expressed in Raw 264.7 cells. Autophagy was detected with GFP-LC3 marker and immunoblots of endogenous LC3. (D) Induction of autophagy by expressing Beclin 1 or Traf6, but not by Beclin 1 K117R or Traf2. Raw 267.4 cells expressing Beclin 1, Traf6, K117R Beclin 1, or Traf2 along with the GFP-LC3 marker to monitor autophagy or similar experiment where the levels of autophagy were assessed by LC3 immunoblotting. (E) Beclin 1 mutants that are defective in Traf6 binding poorly induce autophagosome formation. GST or GST fusion proteins of Beclin 1 or its mutants were expressed in Raw 264.7 cells along with the GFP-LC3 marker to monitor autophagy. Data shown in all the panels as mean +/− SD.
Finally to determine if other signals known to trigger autophagy might also trigger Beclin 1 ubiquitination we examined the effects of interferon-γ (IFN-γ) and amino acid starvation on Raw 264.7 cells. IFN-γ-induced-autophagy is known to help eliminates M. tuberculosis in macrophages (27). We observed that similar to LPS and IL-1, IFN-γ treatment triggered the ubiquitination of endogenous Beclin 1 in Raw 264.7 cells (Fig. 4S). Furthermore, the K117R mutant of Beclin 1 was significantly less ubiquitinated than wild type Beclin 1 following IFN-γ treatment (Fig. 4S). In addition, amino acid starvation, which also induces autophagosome formation in Raw 267.4 cells (27), caused a rapid although transient increase in K63-linked ubiquitination of Beclin 1 (Fig. 4S). To compare wild type Beclin and the K117R mutant of Beclin 1 in amino acid starvation induced autophagy, we reduced the endogenous level of mouse Beclin 1 in Raw 267.4 cells using one of the Beclin shRNAs, expressed GFP-LC3, and introduced either wild type or human Beclin 1 K117R. The day following the transfection we starved the cells for 2 hours and then examined the percentage of cells with GFP-LC3 puncta above background. We found that expression of wild type Beclin 1 significantly increased the % of cells with GFP-LC3 puncta above that of the cells not reconstituted while the addition of Beclin 1 K117R had no effect (Fig. 4S). Immunoblotting verified similar levels of Beclin 1. Thus, like TLR and IL-1R signaling, IFN-γ and amino acid starvation also induce the ubiquitination of Beclin 1, which contributes to the induction of autophagy.
DISCUSSION
Our studies have provided several insights into the regulation TLR-induced autophagy in macrophages. First, we showed that the engagement of TLR4 results in the modification of Beclin 1 by the addition of K63-linked ubiquitin chains. Second, we provided evidence that Traf6 functions as an ubiquitin ligase in this reaction and that A20 is a critical enzyme for regulating the deubiquitination of Beclin 1. Third, we mapped two Traf6 binding sites in Beclin 1 that contribute to the ubiquitination of Beclin 1. Fourth, we identified K117 as a key site in Beclin 1 for linking ubiquitin chains. Fifth, the ubiquitination of Beclin 1 on K117 enhances its multimerization and may augment Beclin 1 mediated increases in PI3KC3 enzymatic activity. Finally, we have provided evidence that the ubiquitination of Beclin 1 amplifies TLR-induced autophagy and that A20 plays an important counter-regulatory role. These results indicate that an intimate association exists between the TLR-induced activation of the NF-κB pathway and the induction of autophagy in macrophages.
The transcription factor NF-κB has a pivotal role in inflammation and in adaptive immunity. Its translocation to the nucleus triggers the expression of a wide variety of genes implicated in the initiation and propagation of the host innate and adaptive immune responses (28). Pathogen-associated molecular patterns from bacteria, viruses, fungi or parasites are recognized by TLRs on immune cells. Engagement of TLRs triggers NF-κB activation, which critically depends on the E3 ligase activity of Traf6 (29). TLR signaling induces A20 expression, which functions to limit NF-κB activation by deubiquitinating Traf6 (3). Our findings indicate that the Traf6 and A20 function in an analogous fashion to control the induction and termination of TLR-induced autophagy in macrophages. In this signaling pathway Traf6 likely directly recruits Beclin 1 resulting in the ubiquitination of Beclin 1. Ubiquitination of Beclin 1 enhances its multimerization and augments the associated PI3KC3 activity. TLR signaling induces NF-κB translocation to the nucleus and A20 transcriptional activation leading to a reduction in Beclin 1 ubiquitination, which acts to limits autophagy in macrophages.
A20 deficient mice are cachexic, exhibit severe tissue damage in multiple organs, are hypersensitive to LPS and tumor necrosis factor (TNF), and suffer neonatal lethality (4). Studies of mice doubly deficient for A20 and TNF or for A20 and TNF receptor 1 indicate that A20 is required to terminate TLR-induced NF-κB activation and proinflammatory gene expression in macrophages (30). Interestingly, A20 and MyD88 double deficient mice have reduced inflammation and are protected from the premature lethality observed in A20 deficient mice (5). These observations indicate that A20 functions in a critical negative feedback loop to regulate TLR signaling. Our findings indicate that A20 restricted LPS-induced autophagy by reducing the ubiquitination of Beclin 1 acting perhaps on both Traf6 and Beclin 1. Besides its deubiquitinating activity A20 also functions of as an E3 ligase to trigger K48-linked ubiquitination of a targeted protein, which leads to its degradation. For example A20 is known to function as an E3 ligase for RIP1 (3). However, we did not detect any degradation of Beclin 1 in LPS stimulated macrophages arguing that A20 does not function as an E3 ligase for Beclin 1.
Human TRAF6 has been previously linked to the induction of autophagy in macrophages. Signaling through the cell surface molecule CD40 activates macrophages to kill Toxoplasma gondii through a Beclin 1 dependent process that leads to the recruitment of autophagosomes around the parasitophorous vacuole. This causes the lysosomal degradation of the parasite (31). The intracytoplasmic tail of CD40 has a TRAF6 binding site that has been shown to be important in this signaling pathway (32). In this model TRAF6 signaling enhances autocrine Tumor Necrosis Factor-α production, which synergizes with downstream TRAF6 signals to recruit autophagosome markers to the parasitophorous vacuole (32). Based on the studies reported here we would suggest that TRAF6 and A20 would also regulate the level of Beclin 1 ubiquitination following CD40 stimulation of macrophages.
The antiapoptotic protein Bcl2, binds to BH3 domain of Beclin 1 and attenuates autophagy (15, 16). TLR stimulation of Raw 267.4 cells reduces the association between Bcl2 and Beclin 1 (7). This could arise as a consequence of the ubiquitination of Beclin 1 at lysine 117, which is located in the BH3 domain of Beclin 1 and adjacent to a critical amino acid for the Bcl-2/Beclin 1 interaction. The interpretation of preliminary experiments to address this issue has been complicated by the presence of endogenous Beclin 1 in Raw 267.4 cells and the tendency of Beclin 1 to multimerize following LPS stimulation. However, they do suggest that the decrease in the Beclin 1/Bcl-2 interaction that occurs upon LPS activation does not happen when with the K117R mutation is present in Beclin 1. Starvation can lead to c-Jun N-terminal kinase 1 (JNK1) mediated phosphorylation of Bcl2, which reduces the association between Bcl2 and Beclin 1, and enhances autophagy (33). In our study starvation also led to the ubiquitination of Beclin 1, which could further reduce the interaction between Bcl-2 and Beclin 1 contributing to starvation-induced autophagy.
Besides undergoing K63-linked ubiquitination, Beclin 1 also possesses a UBD. Numerous cellular processes depend upon the interaction between ubiquitinated proteins and intracellular proteins with a UBD. Nonproteolytic functions of ubiquitin rely on the recognition of monomeric ubiquitin or ubiquitin chains branched from lysines other than K48 and constitute an intracellular signaling network. Despite marked structural differences between the various characterized UBDs they share a general mode of interaction with ubiquitin (19). A hydrophobic surface on the UBD contacts a hydrophobic pocket centered on ubiquitin isoleucine 44. Although many UBD share little specificity for the different types of ubiquitin chains some do (20). In our study Beclin 1 exhibited a strong preference for K63-versus K48-linked ubiquitin peptides. The basis of this selectivity is currently unknown. Little structural information is available to make a prediction on which region in Beclin 1 serves as a UBD. Besides its BH3-like domain, Beclin 1 has a central coiled-coil domain, and an evolutionarily conserved domain, which is essential for PI3KC3 binding. It lacks the NZF fingers (Npl4 zinc finger) that Trabid utilizes to bind K63-linked ubiquitin chains (34). We are attempting to map the UBD in Beclin 1 by expressing various truncated proteins.
Although our investigations have shown that LPS trigger, IFN-γ, and starvation trigger the ubiquitination of Beclin 1 and autophagy, whether Traf6 and A20 regulate Beclin 1 ubiquitination following starvation or IFN-γ treatment is currently unknown. We are testing whether modifying Traf6 and A20 expression affects IFN-γ or starvation induced Beclin 1 ubiquitination and autophagy. If Traf6 and A20 do not, other ubiquitin modifying enzymes must exist that regulate IFN-γ- and starvation-induced autophagy. Recently it has been reported that the IFN-γ-induced autoantigen Ro52 is an E3 ligase that ubiquitinates IRF-8 resulting in enhanced cytokine expression in macrophages (35). As such it is a candidate to have a role in IFN-γ-induced ubiquitination of Beclin 1.
In conclusion, in TLR4-induced autophagy in macrophages Traf6 and A20 function in an analogous fashion to their defined roles in controlling NF-κB activation. The Traf6/A20 axis by controlling the level of Beclin 1 ubiquitination acts as a rheostat to up- or down-regulate autophagy. IFN-γ exposure and amino acid starvation also cause the ubiquitination of Beclin 1 suggesting that K63-linked ubiquitination of Beclin 1 may be a shared mechanism that different inductive stimuli use to trigger autophagosome formation.
MATERIAL AND METHODS
Cell Culture, Plasmids, and Reagents
Raw 264.7 cells were obtained from the ATCC (American Type Culture Collection, Rockville, MD) and were maintained in DMEM 10% FBS. For amino acid and serum starvation Raw 267.4 were washed twice with PBS and then cultured in Earle’s balanced salts solution at 37 °C for 30 minutes or 1 hour. Human monocytes were obtained by elutriation from healthy donors after they provided informed consent approved by the institutional review board. The ubiquitin expression vectors were previously described (Shi and Kehrl, 2003). The LC3 cDNA was a gift from Dr. N. Mizushima (Tokyo Medical and Dental University). Full length Flag-MyD88 was kindly provided by Dr. A. Ding (Cornell University). The Trif expression vector was a gift from Dr. D. Golenbock (University of Massachusetts Medical School). The Beclin 1 construct was kindly provided by Dr. B. Levine (Baylor College of Medicine). The A20 expression vector was a kind gift Dr. K. T. Jeang (Laboratory of Molecular Microbiology, NIAID, NIH). The constructs expressing C103A A20 and C624A-C627A A20 were made with a QuickChange Site-Directed Mutagenesis kit (Stratagene) following the manufacturer’s protocol. The GFP-2xFYVE construct was kindly provided by Dr. H. Stenmark (Institute for Cancer Research, The Norwegian Radium Hospital). The Traf6 and Traf2 expression vectors were provided by Dr. U. Siebenlist (Laboratory of Immunoregulation, NIAID, NIH). The GST-Beclin 1 was made by subcloning the coding region of Beclin 1 into the GST fusion vector pEGG. That plasmid was used to make GST-Beclin 1 K117R, GST-Beclin 1 E56A, GST-Beclin 1 E299A, and GST-Beclin 1 E56AE299A using the above mutagenesis kit. The lipopolysaccharide (Alexis) for the stimulation of Raw 264.7 cells was used at a concentration of 200–500 ng/ml. The mouse IFN-γ (PeproTech) was used at a final concentration of 200 ng/ml. 3-methyladenine (3-MA, Sigma-Aldrich) was used at a final concentration of 5 mM. IL-1α and IL-1β (PeproTech) were used at a final concentration of 200 ng/ml.
RNA Interference
Software provided by the Clontech shRNA website was used to generate a list of potential 19-mer sequences for use as shRNAs targeted against the mRNA of interest. Each shRNA was verified not to have non-specific targets by database searches. The shRNA sequences of the two different targeted sites in mouse A20, Traf6, and Beclin 1 were as follows: GCTGGAGATGTTCAGAACA and CAGCAACATCCTCAGAAGA for A20, GAGACATCTCGAGGATCAT and TCCACACAATGCAAGGAGA for Traf6, TGGACACGAGCTTCAAGAT and TGGAGATCCTAGAGCAGAT for Beclin 1. Annealed inserts were cloned into the RNAi-Ready pSIREN-RetroQ vector (Clontech) that had been previously digested with EcoRI-BamHI. The negative control shRNAs were purchased from Clontech. The shRNAs for silencing A20, Traf6, or Beclin 1 mRNAs or a negative control were transfected into RAW 264.7 cells twice separated by 24 hours using FuGene HD Transfection Reagent (Roche Applied Science). The cells were treated with LPS for 4–7 hours.
GFP-LC3 and GFP-2xFYVE Assays
A Perkin-Elmer Ultraview spinning wheel confocal system mounted on a Zeiss Axiovert 200 microscope and equipped with an argon/krypton laser, an Orca-ERII CCD camera (Hamamatsu), and filters suitable for the visualization of both FITC and red dyes was used for the quantization of cells transfected with GFP-LC3 and GFP-2xFVYE. The assays were performed as detailed previously (7, 21). For the GFP-LC3 assay a minimum of 50–100 GFP positive cells per sample were counted and the number of GFP-LC3 dots enumerated. Cells were scored as positive if they had more than 3 large GFP-LC3 dots and the data was presented as percent of the total GFP positive cells.
Western Blot Analysis and Immunoprecipitations
Monoclonal mouse Flag M2 antibodies and their conjugated agarose were purchased from Sigma. The monoclonal mouse anti-ubiquitin antibodies (P4D1) and the antibodies directed against MyD88, A20, Traf6, Bcl2, and actin were from Santa Cruz. Rabbit anti-Beclin 1 antibodies were from Cell Signaling. Monoclonal mouse anti-Beclin 1 antibodies were from BD Transduction Laboratories. Monoclonal mouse anti-K63 poly-ubiquitin chain antibodies (HWA4C4) were from eBiosciences. Monoclonal mouse anti-HA tag antibodies (16B12) were from Covance. Antibodies directed at Trif (Imgenex), and PI3KC3 (Cell Signaling Technology), and LC3 (MBL International) were also used. For immunoblotting cell lysates Raw 264.7 cells were lysed in a buffer containing 2% Triton X-100 in PBS with a protease inhibitor mixture (Roche Applied Science). The cell lysates were centrifuged, and equivalent amounts of protein were loaded onto 4–20% or 10% Tris/glycine/SDS-polyacrylamide gels (Invitrogen). After electrophoresis, the proteins were transferred to nitrocellulose membranes. The specific antibodies as indicated were used for immunoblots. For the co-immunoprecipitation assays, the cells were lysed in a buffer that contained 20 mM Hepes pH 7.4, 50 mM β-glycerophosphate, 1 mM Na3VO4, 0.5 % Triton X-100, 0.5 % CHAPS and 10% glycerol with a protease inhibitor mixture (Roche Applied Science) was used. The lysates were incubated with the appropriate antibodies for 2 hours at 4 °C, and then Protein G PLUS-Agarose was added and incubated for 1 hour at 4 °C. The immunoprecipitates were collected, washed 8 times with lysis buffer, and analyzed by SDS-Page and immunoblotting. To detect ubiquitin and K63 linked-ubiquitin modified Beclin 1 or FLAG-Beclin 1, we first lysed the cells using a buffer (20 mM HEPES pH 7.4, 2mM EGTA, 50 mM β-glycerophosphate, 0.5 % Triton X-100, 0.5% CHAPS, 10% Glycerol) containing complete protease inhibitor cocktail (Roch) by shaking 30 minutes on 4 °C. After which 1% SDS (v/v) was added and the lysates heated at 90 °C for 5 minutes to dissociate protein-protein interactions. Then the samples were diluted 10 fold with the same buffer and the FLAG-agarose or the Beclin 1 antibody was added. The immunoprecipitations were performed at 4 °C for 4 hours while the samples were rotated. The immunoprecipitates were collected and the same lysis buffer was used to wash them 8 times. The samples were analyzed by SDS-PAGE followed by immunoblotting. In general the first immunoblot was performed with the ubiquitin or K63-linked ubiquitin antibody. To do so the membrane was first incubated in TBST buffer (Tris-buffered saline-Tween, 20 mM Tris-HCL pH 7.6, 133 mM NaCl, 0.1 Tween) with 5% bovine albumin or 10% milk for 1 hour. The membrane was washed once with same buffer, and the first antibody was added to membrane in TBST buffer plus milk or albumin over-night at 4 °C. The membrane was washed in 4 × 10 minutes with TBST buffer, incubated with HRP-conjugated secondary antibodies for 2 hours, washed in 6 × 10 minutes in TBST buffer, and subjected to enhanced chemiluminence, which was detected using HyBlot CL film (Denville Scientific Incorporation). The immunoblot was stripped and the membrane re-probed with a rabbit antibody against Beclin 1 antibody. To strip the membrane it was submerged in stripping buffer (100 mM 2-Mercaptoethanol, 2% SDS, 62.5 mM Tris-HCL pH 6.7) at 55–60 °C for 60 minutes while shaking. The membrane was washed 6 times (10 minutes each wash) in 100 ml of TBST at room temperature while rapidly shaking. The HRP-second antibodies were used to check the efficacy of the stripping prior to re-immunoblotting. Immunoblots were scanned and imported into Photoshop as unmodified TIFF files (Adobe Systems). The amount of signal in different lanes was determined using Photoshop and normalized to the level in the control lane. All experiments were repeated a minimum of three times with similar results.
In Vitro Ubiquitination Assay
FLAG-TRAF6, FLAG-Beclin 1, or FLAG-Beclin 1 K117R mutant were transfected into HEK293T cells for 24 hours. The proteins were purified essentially as previously described (Wertz et al., 2004). The cells were washed once with PBS, and then lysed with a buffer (20 mM HEPES pH 7.4, 2mM EGTA, 50 mM β-glycerophosphate, 0.5 % Triton X-100, 0.5% CHAPS, 10% Glycerol) containing a complete protease inhibitor cocktail (Roche) by shaking for 30 minutes on 4 °C. Lysates were cleared by centrifugation and immunoprecipitated with anti-FLAG coupled agarose (Sigma) overnight. The immunoprecipitates were washed once for 30 minutes with buffer (20 mM HEPES pH 7.9, 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA containing 25% glycerol and the protease inhibitor cocktail and four times (each for 30 minutes) with a second buffer (20 mM Tris pH 7.4, 20% glycerol, 0.2 mM EDTA, 300 mM NaCl, 0.1% NP-40, and protease inhibitor cocktail) while rotating. The purified proteins were eluted with 300 μg/ml FLAG peptide (Sigma) according to the manufacturer’s instructions. The eluted proteins were concentrated using a Microcon (Millipore) with a 5–10 kDa molecular weight cut-off. The assays were performed as previously described (Wertz et al., 2004). The assays were carried out in a 20 μl reaction volume and that contained all or some of the following components: UBE1 0.2 μg (Boston Biochem), UbeH13/Uev1a heterodimer complex 0.5 μg (Boston Biochem), HA-Ubiquitin 100 μM (Boston Biochem), 2 μl of a 10x reaction buffer (Boston Biochem) and 2 μl of a Mg+2-ATP solution (Boston Biochem), and 0.5 μg purified FLAG-Beclin 1 and 0.5 μg FLAG-Traf6 proteins. The reaction was carried at 30 °C for 1 hour with agitation. After the reaction a portion was saved for immunoblotting and the remaining proteins were dissociated by heating at 90 °C in 1% SDS (v/v) for 5 minutes, diluted 1:10 in lysis buffer, and Beclin 1 immunoprecipitated with the Beclin 1 antibody as above. The immunoprecipitates were prepared for immunoblotting with the HA-antibody to detect HA-ubiquitin.
In Vitro De-ubiquitination Assay
Immunoprecipitated FLAG-Beclin 1 prepared from 293T cells. The beads with Flag-Beclin 1 were treated with dissociated washes by the above mentioned methods. The FLAG-Beclin 1 protein was mixed with His6-A20 catalytic domain recombinant protein (2.5 μM, Boston Biochem) in a 20 μl reaction with the buffer (50 mM HEPES pH 8.0, 0.01% Brij, and 3 mM DTT). The reaction was performed at 30 °C with agitation for 30 or 60 minutes.
In vitro Ubiquitin Binding Assay
The binding assay was done as has been previously described (Tran et al., 2009). Raw cells 267.4 cells were transfected with constructs expressing GST-Beclin 1 or GST. GST or GST-Beclin 1 was purified (see below) or endogenous Beclin 1 immunoprecipitated. After washing with lysis buffer 8 times, K63- or K48-linked ubiquitin chain peptides (BIOMOL) 0.5 μg for each sample were added to the samples, and then the samples were incubated by rotating at 4 °C for 1 hour. Afterwards, the samples were washed 4 times with same buffer and analyzed by SDS-PAGE followed by immunoblotting for ubiquitin.
Beclin 1 GST Pull-down Assay
Raw 264.7 were transfected with the constructs to express GST-Beclin 1 or its various mutants in the presence of FLAG-Traf6 or FLAG-Beclin 1. Whole cell lysates were prepared using a lysis buffer contains 20 mM Hepes pH 7.4, 50 mM β-glycerophosphate, 1 mM Na3VO4, 0.5 % Triton X-100, 0.5 % CHAPS and 10% glycerol with a protease inhibitor mixture (Roche Applied Science, Basel, Switzerland) was used. The GST-fusion proteins were purified on Glutathione-Sepharose 4B beads as instructed by the manufacturer (Amersham Pharmacia). The beads were washed with 0.5 ml of the same buffer 8 times. The pulled down proteins were subjected to SDS-PAGE gel electrophoresis and detected by immunoblot analysis as described above.
Supplementary Material
ACKNOWLEDGMENTS
We thank M. Rust for her excellent editorial assistance, and Dr. A. Fauci for his continued support. This research was supported by the intramural program of the National Institutes of Allergy and Infectious Diseases.
This research was supported by the Intramural Research Program of NIAID, NIH.
REFERENCES AND NOTES
- 1.Akira S, Takeda K, Kaisho T, Toll-like receptors: critical proteins linking innate and acquired immunity. Nat Immunol 2, 675–680 (2001). [DOI] [PubMed] [Google Scholar]
- 2.Trinchieri G, Sher A, Cooperation of Toll-like receptor signals in innate immune defence. Nat Rev Immunol 7, 179–190 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Wertz IE, O’Rourke KM, Zhou H, Eby M, Aravind L, Seshagiri S, WU P, Wiesmann C, Baker R, Boone DL et al. , De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature 430, 694–699 (2004). [DOI] [PubMed] [Google Scholar]
- 4.Lee EG, Boone DL, Chai S, Libby SL, Chien M, Lodolce JP, Ma A, Failure to regulate TNF-induced NF-kappaB and cell death responses in A20-deficient mice. Science 289, 2350–2354 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Turer EE, Tavares RM, Mortier E, O Hitotsumatsu, Advincula R, Lee B, Shifrin N, Malynn BA, Ma A, Homeostatic MyD88-dependent signals cause lethal inflamMation in the absence of A20. J Exp Med 205, 451–464 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delgado MA, Elmaoued RA, Davis AS, Kyei G, Deretic V, Toll-like receptors control autophagy. EMBO J 27, 1110–1121 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shi CS, Kehrl JH, J Biol Chem 283, 33175–33182 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu Y, Jagannath C, Liu XD, Sharafkhaneh A, Kolodziejska KE, Eissa NT, Toll-like receptor 4 is a sensor for autophagy associated with innate immunity. Immunity 27, 135–144 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mizushima N, Levine B, Cuervo AM, Klionsky DJ, Nature 451, 1069–1075 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deretic V, Autophagy in innate and adaptive immunity. Trends Immunol 26, 523–528 (2005). [DOI] [PubMed] [Google Scholar]
- 11.Levine B, Deretic V, Unveiling the roles of autophagy in innate and adaptive immunity. Nat Rev Immunol 7, 767–777 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmid D, Munz C, Innate and adaptive immunity through autophagy. Immunity 27, 11–21 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Virgin HW, Levine B, Autophagy genes in immunity. Nat Immunol 10, 461–470 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xie Z, Klionsky DJ, Autophagosome formation: core machinery and adaptations. Nat Cell Biol 9, 1102–1109 (2007). [DOI] [PubMed] [Google Scholar]
- 15.Pattingre S et al. , Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell 122, 927–939 (2005). [DOI] [PubMed] [Google Scholar]
- 16.Maiuri MC, Le Toumelin G, Criollo A, Rain JC, Gautier F, Julin P, Tasdemir E, Pierron G, Troulinaki K, Tavernarakis N, et al. , Functional and physical interaction between Bcl-X(L) and a BH3-like domain in Beclin-1. EMBO J 26, 2527–2539 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oberstein A, Jeffrey PD, Shi Y, Crystal structure of the Bcl-XL-Beclin 1 peptide complex: Beclin 1 is a novel BH3-only protein. J Biol Chem 282, 13123–3132 (2007). [DOI] [PubMed] [Google Scholar]
- 18.Ye H, Arron JR, Lamothe B, Cirilli M, Kobayashi T, Shevde NK, Segal D, Dzivenu OK, Vologodskaia M, Yim M et al. , Distinct molecular mechanism for initiating TRAF6 signalling. Nature 418, 443–447 (July 25, 2002). [DOI] [PubMed] [Google Scholar]
- 19.Hicke L, Schubert HL, Hill CP, Ubiquitin-binding domains. Nat Rev Mol Cell Biol 6, 610–621 (2005). [DOI] [PubMed] [Google Scholar]
- 20.Komander D, Reyes-Turcu F, Licchesi JD, Odenwaelder P, Wilkinson KD, Barford D, Molecular discrimination of structurally equivalent Lys 63-linked and linear polyubiquitin chains. EMBO Rep 10, 466–473 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gillooly DJ, Morrow IC, Lindsay M, gould R, Bryant NJ, Gaullier JM, parton RG, Stenmark H, Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J 19, 4577–4588 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Obara K, Noda T, Niimi K, Ohsumi Y, Transport of phosphatidylinositol 3-phosphate into the vacuole via autophagic membranes in Saccharomyces cerevisiae. Genes Cells 13, 537–547 (2008). [DOI] [PubMed] [Google Scholar]
- 23.Mizushima N, Methods for monitoring autophagy. Int J Biochem Cell Biol 36, 2491–2502 (2004). [DOI] [PubMed] [Google Scholar]
- 24.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T et al. , LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J 19, 5720–5728 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rubinsztein DC, Cuervo AM, Ravikumar B, Sarkar S, Korochuk V, Kaushik S, Klionsky DJ, In search of an “autophagomometer”. Autophagy 5, 585–589 (2009). [DOI] [PubMed] [Google Scholar]
- 26.Shi CS, Kehrl JH, Tumor necrosis factor (TNF)-induced germinal center kinase-related (GCKR) and stress-activated protein kinase (SAPK) activation depends upon the E2/E3 complex Ubc13-Uev1A/TNF receptor-associated factor 2 (TRAF2). J Biol Chem 278, 15429–15434 (April 25, 2003). [DOI] [PubMed] [Google Scholar]
- 27.Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V, Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell 119, 753–766 (December 17, 2004). [DOI] [PubMed] [Google Scholar]
- 28.Hayden MS, West AP, Ghosh S, NF-kappaB and the immune response. Oncogene 25, 6758–6780 (2006). [DOI] [PubMed] [Google Scholar]
- 29.Kawai T, Akira S, TLR signaling. Cell Death Differ 13, 816–825 (2006). [DOI] [PubMed] [Google Scholar]
- 30.Boone DL, Turer EE, lee EG, Ahmad RC, Wheeler MT, Tsui C, Hurley P, Chien M, Chai S, Hitotsumatsu O et al. , The ubiquitin-modifying enzyme A20 is required for termination of Toll-like receptor responses. Nat Immunol 5, 1052–1060 (2004). [DOI] [PubMed] [Google Scholar]
- 31.Andrade RM, Wessendarp M, Gubbels M-J, Striepen B, Subauste CS, CD40 induces macrophage anti-Toxoplasma gondii activity by triggering autophagy-dependent fusion of pathogen-containing vacuoles and lysosomes. J Clin Invest 116, 2366–2377, (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Subauste CS, Andrade RM, Wessendarp M, CD40-TRAF6 and autophagy-dependent anti-microbial activity in Marcrophages. Autophagy 3, 245–248 (2007). [DOI] [PubMed] [Google Scholar]
- 33.Wei Y, Pattingre S, Sinha S, Bassik M, Levine B, JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol Cell 30, 678–688 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tran H, Hamada F, Schwarz-Romond T, Bienz M, Trabid, a new positive regulator of Wnt-induced transcription with preference for binding and cleaving K63-linked ubiquitin chains. Genes Dev 22, 528–542 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kong HJ, Anderson D.e., Lee CH, Jang MK, Tamura T, Tailor P, Cho HK, Cheong J, Xiong H, Morse HC, Ozato K et al. , Cutting edge: autoantigen Ro52 is an interferon inducible E3 ligase that ubiquitinates IRF-8 and enhances cytokine expression in macrophages. J Immunol 179, 26–30 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.