Abstract
During exocytosis, vesicles fuse with the plasma membrane and release their contents. The fusion pore is the initial, nanometer-sized connection between the plasma membrane and the cargo-laden vesicle. A growing body of evidence points towards the fusion pore being a regulator of exocytosis, but the shortcomings of current experimental techniques to investigate single fusion pores make it difficult to study factors governing pore behavior. Here we describe an assay that fuses v-SNARE-reconstituted nanodiscs with cells ectopically expressing “flipped” t-SNAREs to monitor dynamics of single fusion pores in a biochemically defined system using electrical recordings. We also describe a fluorescence microscopy based approach to monitor nanodisc-cell fusion that is much simpler to employ, but cannot resolve single pores.
Keywords: membrane fusion, exocytosis, nanodisc, SNAREs, fusion pore, electrophysiology
Introduction
1.1. Exocytosis and the fusion pore
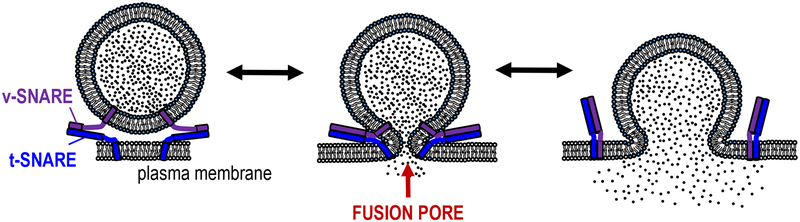
Membrane fusion is an important biological process that involves making two membranes into one continuous membrane1–3. The initial connection between these two membranes is called the fusion pore (Figure 1). It is a 1–3 nm wide dynamic structure that can fluctuate in size, flicker open-closed, and ultimately dilate fully or reseal altogether4,5. In calcium-triggered exocytosis, a cargo-loaded vesicle fuses its membrane with that of the plasma membrane in order to release its soluble cargo into the extracellular space. This is especially important for communication between cells either locally (in the case of neurons), or distally (in the case of hormone secreting cells).
Figure 1.
The fusion pore is a key intermediate during exocytosis. Left: complex formation between v-SNARE proteins on the vesicle and t-SNARE proteins on the plasma membrane drive fusion between the membranes. This results in the formation of a fusion pore (middle). The initial pore is only 1–3 nm wide and can fluctuate in size, flicker open-closed multiple times, then either reseal or dilate irreversibly (right).
Exocytosis is a highly regulated process that involves a myriad of different proteins2, but the core drivers of membrane fusion are the SNARE proteins2,3,6 (Figure 1). Vesicular v-SNAREs complex with target-membrane t-SNAREs on the plasma membrane in a highly exergonic reaction that brings the two membranes together and drives fusion3,7. For neuronal exocytosis, there are two t-SNARE proteins (SNAP25 and syntaxin1) and one v-SNARE protein (VAMP2) involved in this process2,3.
Exocytosis is tightly regulated at various stages, including after the formation of the initial fusion pore. Studies in neuroendocrine cells have firmly established that the fusion pore can control the amount of cargo released, the kinetics of cargo release, and the mode and kinetics of vesicle recycling4,5,8–11. However, while transient fusion and/or flickering pores have been observed or deduced for several synapses12–16, there is still no consensus on whether or not fusion pore dynamics contribute significantly to the control of neuronal exocytosis. This is mostly due to the fact that probing single fusion pores during synaptic vesicle release in neurons is much harder than probing release in neuroendocrine cells.
1.2. Current approaches to study membrane fusion and single fusion pores
Most of our knowledge regarding fusion pores comes from studies of exocytosis in neuroendocrine cells using time-resolved capacitance or amperometry8,17. Time-resolved capacitance measurements directly detect membrane area changes due to exocytosis (membrane addition) or endocytosis (membrane removal). The same approach allows estimation of fusion pore conductance, and so it is a direct way of assessing pore size4,8. Amperometry detects single exocytosis events using a carbon fiber electrode placed close to a secretory cell17. When certain cargo molecules reach the electrode surface, they are oxidized and an oxidation current can be recorded with excellent time resolution. The oxidation profile contains information about pore dynamics. Finally, fluorescence methods can be employed to measure influx or efflux of probes through the pore during exocytosis16. Although these approaches to detect fusion pores in live cells are crucial to characterize pore properties in native settings, they are limited in their power in revealing molecular mechanisms that control pore properties. Such information is best obtained using complementary biochemical reconstitution of the fusion reaction.
Indeed, starting with the first successful reconstitution of SNARE-mediated fusion in a bulk fluorescence based assay6, much of our mechanistic understanding about exocytosis has come from similar bulk fluorescence monitoring of fusion among proteoliposomes. However, such assays cannot probe fusion pores directly. More recently, assays monitoring fusion of single-liposomes with other liposomes immobilized on a surface18–20, or with a supported bilayer have been developed21–25. Stratton et al. recently were able to deduce pore properties from the kinetics of lipid mixing monitored with ~15 ms time resolution25. Nevertheless, such approaches deduce fusion pores only indirectly and do not yet have the sub-millisecond time resolution required to detect individual pore flickers.
Clearly, a limitation of previous approaches is the inability to combine single-pore sensitivity, sub-millisecond time resolution, and biochemical reconstitution. We have recently developed an assay that combines these three requirements. This has allowed us, for the first time, to start characterizing pore properties in detail in biochemically defined settings. This protocol describes how to carry out such measurements.
1.3. Nanodisc-cell fusion
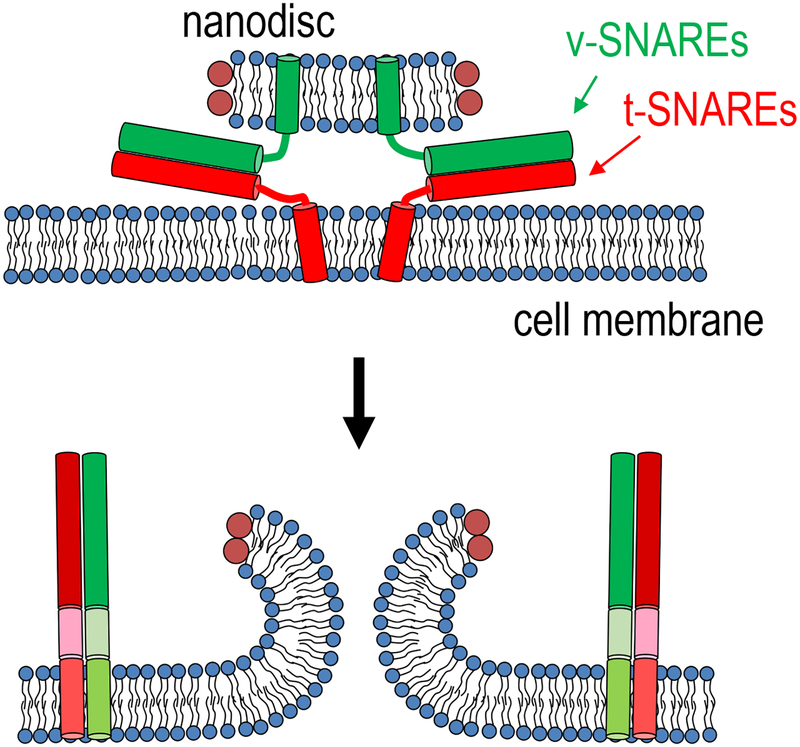
Our single-pore assay fuses nanodiscs reconstituted with neuronal/exocytotic v-SNAREs with engineered cells ectopically expressing the cognate t-SNAREs in a “flipped” configuration26,27. This approach builds upon previous work mostly carried out in the Rothman laboratory, where nanodiscs were employed for the first time in membrane fusion studies as fusion partners with proteoliposomes in a bulk assay28. We fuse these v-SNARE reconstituted nanodiscs to cells that are genetically engineered to express t-SNAREs “flipped” to be on the outside of the cell (t-cells). Flipped t-SNARE cells were previously shown to fuse with cells expressing the cognate, flipped v-SNAREs on their surfaces, also in the Rothman lab29. We asked if the v-SNARE reconstituted nanodiscs and the flipped t-SNARE cells, developed for distinct assays, could be used as fusion partners. We expected that when a v-disc encountered t-SNAREs on the plasma membrane of a t-cell, the v- and t-SNAREs would interact to drive the opening of a fusion pore that would connect the cytoplasm of the cell to the extracellular solution (Figure 2). This direct connection, in the form of a nanometer sized pore, should in principle allow us to use sensitive electrophysiological methods developed for measuring very small currents passing through single ion channels30.
Figure 2.
Fusion of a nanodisc with a cell creates a pore that connects the cytosol to the extracellular medium. Nanodiscs containing v-SNAREs fuse with the membrane of a cell containing flipped t-SNAREs facing outward. A fusion pore is formed that connects the nanodisc membrane to that of the cell. Figure modified from26.
A very simple approach to assess fusion between nanodiscs and cells is to load a t-cell with a calcium indicator dye26,27. When v-SNARE nanodiscs are added into the bath, if they fuse with the cells, extracellular calcium enters the cytoplasm through fusion pores and the fluorescence of the cytoplasmic calcium indicator increases. While this nanodisc-cell calcium-influx assay (which we describe in section 3.2) is useful in its own right, we also suggest using it as a way to test nanodisc and cell fusion competency before embarking upon the more technically-challenging nanodisc-cell single fusion pore assay (described in section 3.3). Note that variations of this calcium-influx assay include use of probes other than calcium to asses average pore size, e.g. passage of neurotransmitter-like fluorescent probes through the pores26.
1.4. Detecting single pores during nanodisc-cell fusion
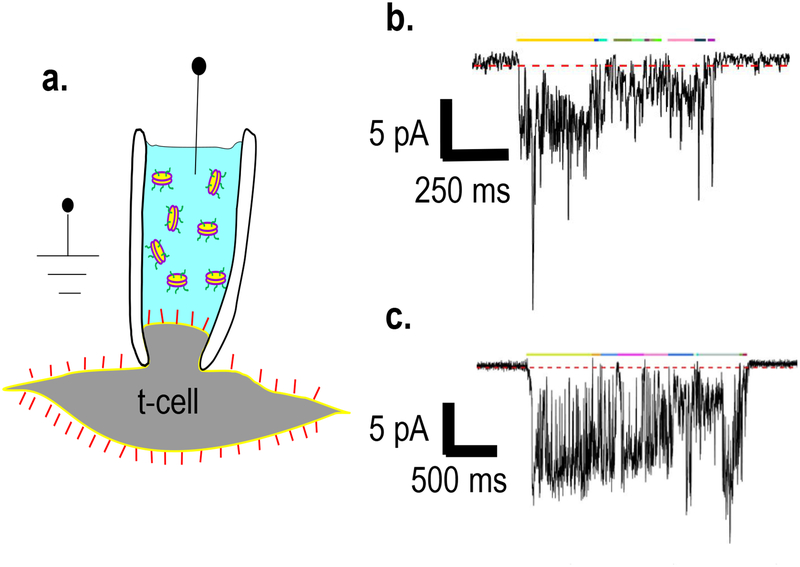
In order to detect single fusion pores during nanodisc-cell fusion, we employ on-cell voltage clamp recordings using t-cells, with the patch pipette filled with nanodiscs containing a defined number of v-SNAREs (Figure 3a). When a v-SNARE disc fuses with the plasma membrane, ions can then move through the fusion pore down their electrochemical gradient, resulting in a current. Finally, using a dilute concentration of nanodiscs such that pores appear infrequently, ensures that single-pores are probed26,27.
Figure 3.
Nanodisc-cell single fusion pore assay. a) A pipette containing v-SNARE nanodiscs is used to establish an on-cell patch on a flipped t-SNARE cell. Discs that are initially layered on top of a disc-free solution in the pipette diffuse to the cell surface. When a disc that reaches the cell surface fuses with it, a fusion pore is formed. (Figure from26). b) An example of a fusion pore current burst (using 16 v-SNARE copies per disc). The threshold for detecting open-pore sub-periods is indicated as the dashed red line just below the baseline. Current must be <−0.25 pA for 60 ms or longer to be considered due to an open pore. Burst open sub-states are indicated by colored lines above the pore. c) a second example of a fusion pore current burst, as in b.
Materials
2.1. Cell Culture
HeLa cells genetically engineered to express t-SNAREs (SNAP25 and syntaxin1) ectopically on the outside29 (see Note 1).
Sodium citrate detaching solution: 5.5 g KCl, 2.2 g sodium citrate, add deionized H2O to 500 ml. Filter using a 0.22 μm vacuum filter/storage system (Corning) to ensure the solution is sterile.
Growth medium: DMEM (with 4500 mg/L glucose, sodium pyruvate and sodium bicarbonate), 10% (v/v) Fetal Bovine Serum, 1% Penicillin/Streptomycin.
10 cm culture dishes
35 mm culture dishes
15 ml conical
Centrifuge
Hemocytometer
2.2. Nanodisc-cell fusion: calcium influx
Cell permeable acetoxymethyl ester (AM) conjugated calcium-sensitive fluorophore Fluo-4 (Life Technologies, New York, NY, USA).
PBS −/−: Phosphate-buffered saline without Ca2+ and Mg2+
PBS +/+: Phosphate-buffered saline containing both 2 mM Ca2+ and 1 mM Mg2+
A Nikon Eclipse Ti spinning disc confocal microscope (see Note 2).
2.3. Nanodisc-cell single fusion pore assay:
Pipette solution: 125 mM NaCl, 4 mM KCl, 1 mM MgCl2, 10 mM HEPES, 30 mM TEA-Cl. Adjust pH to 7.2 with NaOH. Store at −20°C in 1 ml aliquots.
Extracellular Solution: 125 mM NaCl, 4 mM KCl, 2 mM CaCl2-2H2O, 1 mM MgCl2, 10 mM HEPES, 10 mM glucose. Adjust pH to 7.2 with NaOH. Store at 4°C.
Nanodiscs: lipid composition 82% palmitoyl-2-oleoyl phosphatidylcholine (POPC) (Avanti Polar Lipids, Alabaster, AL, USA), 15% 1,2-dioleoyl phosphatidylserine (DOPS) (Avanti), 1.5% N-(7-nitro 2,1,3-benzoxadiazole-4-yl)-1,2-dipalmitoyl phosphatidylethanolamine (NBD-PE) (Avanti), and 1.5% N-(Lissamine rhodamine B sulfonyl)-1,2-dipalmitoyl phosphatidylethanolamine (LR-PE) (Avanti), using Apolipoprotein E as a scaffold, and with an average disc diameter of 24 nm. We follow a protocol developed in the Rothman lab for making these discs, described in ref.31, and Stroeva and Krishnakumar [32].
- Equipment: Any low-noise patch clamp setup should be suitable. We use the following equipment in our lab.
- Inverted microscope (Olympus IX71, Olympus Corporation, Tokyo, Japan.) equipped with an EMCCD camera (DU-885K, Andor Technology Ltd., UK) controlled by Solis software (Andor).
- A HEKA EPC10 Double USB amplifier (HEKA Electronics, Inc., Quebec, Canada), controlled by the Patchmaster software (HEKA).
- Temperature-controlled stage (Thermo Plate, Tokai Hit, Shizuoka, Japan)
- Pipette puller: model P-1000 pipette puller (Sutter Instruments, Novato, CA, USA)
Glass capillaries with filament, borosilicate glass (Sutter Instruments)
Two plastic syringes (1 mL volume), sterile
Scissors
Bunsen burner
Computer
Analysis software. Minimally, a software capable of baseline subtraction, digital filtering, and peak detection is required. These features are typically found in software packages used for analysis of electrophysiological recordings. We have not extensively tested the suitability of various off-the-shelf software packages, but wrote our own package (available upon request) using Matlab.
Methods
3.1. Cell culture
Grow cells in 10 cm dish, in Growth Medium in incubator at 37°C with 5.0% CO2, to 80–85% confluency (see Note 3).
Remove cells from incubator and handle in a biological hood.
Aspirate media from the cells.
Add 5 ml Detaching Solution (see Note 4).
Return dish to the incubator for 2 minutes.
Transfer cells from the dish into 15 ml conical by pipetting against the back of the dish to detach cells.
Spin conical in a small tabletop centrifuge for 2.5 minutes at 0.4 rcf.
Remove conical from centrifuge, aspirate supernatant, and replace with 4–6 ml Growth Medium.
Pipette cells up and down (~15 times) until cells are well dispersed.
Determine cell density by counting cells using a hemocytometer.
Plate cells into 35 mm dishes at a density of 30,000 to 60,000 cells per dish, depending on when they will be used (see Note 5).
Return cells to incubator and allow them to recover for 12–24 hours before use (see Note 6).
3.2. Nanodisc-cell fusion: Calcium influx assay
Use flipped t-SNARE cells plated in a 35 mm dish coated in Poly-D-Lysine.
Wash cells three times with PBS −/−.
Add 5–7 μM cell permeable Fluo-4 AM (a cell permeable calcium-sensitive fluorophore) to the cells. Incubate for 30 minutes at room temperature. This step loads the Fluo-4 dye into the cells. From this step onwards, protect the cells from light by shielding with aluminum foil and minimizing time spent in light whenever possible.
After incubation, wash cells twice with PBS −/− and incubate in the same PBS −/− buffer for an additional 10 minutes to allow the cells to recover.
During the incubation, dilute 15 μl nanodisc stock into 85 μl PBS +/+, or so that the final concentration of lipid in step 8 will be 36 μM.
Mount cells on the stage of a Nikon Eclipse Ti confocal microscope and keep the cells at 37°C.
Remove the PBS −/− media and add 900 μl of PBS +/+.
Add 100 μl diluted nanodisc solution dropwise to the cells and immediately begin imaging.
Acquire time-lapse images of Fluo-4 signals every 5 seconds for 20 minutes.
Use ImageJ software to analyze fluorescence intensity over time. Normalize intensity relative to the initial fluorescence value as a function of time. With 80% confluency, one can simply measure the mean pixel value over the entire frame and plot it against frame number (or time).
3.3.1. Nanodisc-cell single fusion pore assay: Electrophysiology
Aliquot ~20–50 ml extracellular solution and warm in 37°C water bath for ~10 minutes.
Take out 1 ml aliquot of pipette solution from −20°C and allow to thaw. Keep on ice.
Using a pipette puller and glass capillaries, freshly pull pipettes to a resistance of 5–10MΩ in NaCl-based solution. This is about 1–2 μm wide at the tip (see Note 7).
Using a Bunsen burner, heat the tip of a 1 ml plastic, sterile syringe. When it begins to melt and is close to dripping, remove from heat, allow the first drip to fall, and then carefully pull from the thin, dripped area by hand until the syringe tip is ~0.5 mm in diameter (see Note 8). Blow on the tip to cool and harden it. Use scissors to cut off the excess plastic. Repeat for a second syringe (see Note 9).
In a new 1.5 ml Eppendorf tube, dilute nanodiscs in pipette solution to a concentration of 30 μM lipid (see Note 10). The final volume should be between 50–500 μl.
Turn on the electrophysiology setup. Allow heated stage to reach 37°C.
Retrieve t-cells in 35 mm dish, and warmed extracellular solution. Replace culture media with 2.5 ml extracellular solution. Place dish on heated stage of microscope.
Add nanodisc-free pipette solution to the pipette tip: Using the plastic syringe with the smallest tip, fill the tip of the pipette with pipette solution. Flick to get solution into tip. Then use the second pulled syringe to add 30 μM lipid nanodisc solution on top of the solution in the tip. Fill to about 0.5 cm from the tip, or to the height you would for other electrophysiological recordings (see Note 11).
Attach the filled pipette to the head stage. Choose a t-cell that is separated from others, and patch onto this cell, creating a gigaseal (see Note 12).
Do an on-cell patch clamp recording. Use a holding potential of −40 mV relative to the bath for t-cells with a resting potential of ~−56 mV26 (see Note 13). Use a gain of 20mV/pA or 50mV/pA. Record for 10–15 minutes, using a sampling frequency of 20 kHz (see Note 14).
3.3.2. Nanodisc-cell single fusion pore assay: Fusion Pore Analysis
Open a recording using the Patchmaster program (HEKA) that is also used for data acquisition. Go through different traces, using a digital filter (100 Hz). Pick a candidate pore current burst that is well-isolated in time from other currents, lasts at least 250 ms, and has an (absolute) amplitude larger than around 2 pA (assuming a transmembrane voltage of −16 mV, and a gain of 20 mV/pA). This increases the likelihood that the current burst is caused by a single pore and not due to short-lived, small spikes caused by artifacts. Export traces which contain candidate pore currents in Matlab format (or whatever analysis software will be used). For exporting, select “trace time relative to series” option such that the timing of every candidate pore current is maintained relative to the start of the recording (i.e. the beginning of the series).
Open an exported trace in Matlab. It is convenient to convert the current values from A to pA. Apply a low-pass filter on the data (see Note 15).
If needed, apply a notch filter to the data to remove noise with specific frequencies, such as those resulting from line voltage (see Note 16).
Average data points in blocks. We typically use a block size of 40 or 80 depending on sampling frequency (10 or 20 kHz, respectively) to achieve 4 ms final separation between successive points. This reduces noise and ensures a normal distribution of noise around the mean (see26 for details)
Fit the baseline to a polynomial (usually a 1st to 3rd degree). Subtract the fitted baseline from the trace.
Look at the background-corrected trace and visually identify regions that are good candidates for currents that may be due to a fusion pore.
Detect regions in the trace where the current is lower (more negative) than a threshold for a minimum amount of time. Our program marks such regions in colored bars above the trace (Figure 3b,c). Since pore currents fluctuate significantly, we choose to set a threshold close to the baseline such that low-conductance values are not missed in the analysis. This however, results in occasional, random crossings of the threshold by baseline currents. To avoid including such random fluctuations as fusion pore currents, we additionally impose a minimum crossing time over the threshold26 (see Notes 17 and 18). Analyze the current further by determining the baseline noise on either side of the pore. We typically average ~500 ms of baseline (125 points with 4 ms separation between successive points) on either side of the pore. If the baseline root mean squared noise is >1 pA the current burst is not analyzed further.
Analyze the pore current further to characterize pore properties, such as burst lifetime, open sub-states (Figure 3b,c), average pore current for the open sub-states, distribution of point-by-point conductance values and the mean conductance (for the open sub-states), the fraction of time the pore was open during the burst, the number of pore flickers, and fluctuations of pore currents relative to mean.
Assuming that a pore is a 15 nm long cylinder33, the conductance distribution can be converted to a distribution of pore radii26,27.
Once a large number of individual pores have been analyzed, pore characteristics can be analyzed across the sample. For example, we find pore burst lifetimes are well described by an exponential distribution, with a characteristic lifetime of 5–10 s. Due to the large inherent variability in pore properties (such as an exponential distribution of pore lifetimes), we recommend analyzing 40–100 pores per condition.
Acknowledgement
We thank all members of the Karatekin laboratory for stimulating discussions, D. Zenisek and F. Sigworth (Cellular and Molecular Physiology, Yale University) for expert advice and discusssions, Oscar Bello, Shyam Krishnakumar, and other members of the Rothman laboratory (Cell Biology, Yale University) for critical advice and introducing us to the use of nanodiscs. This work was supported by the National Institute of General Medical Sciences (grant R01GM108954), and a Kavli Foundation Neuroscience Scholar Award (to EK).
Notes
We noticed t-SNARE expression can be lost from these cells over time. To regain higher t-SNARE expression, we used the cytoplasmic domain of VAMP2 (CDV) tagged with a fluorescent label, such as AlexaFluor 647 (CDV-647) and sorted using FACS.
Any good-quality fluorescence microscope equipped with the correct filter set should work, including wide-field image acquisition.
Cells must be out of tetracycline and doxycycline for at least 5 days prior to use. It is possible to continuously culture these cells in tetracycline- and doxycycline-free media.
Do not use trypsin. Sodium citrate is used to keep SNAREs on the outside of the cell intact.
Cells must be separated from other cells, so do not plate too densely.
Insufficient recovery time can result in bad or noisy recordings.
Fire polishing can be used, but in our hands is not necessary to obtain a GΩ seal.
The very small diameter is very important for the first pipette because it needs to be able to get down to the very bottom of the glass pipettes that were pulled.
Off-the-shelf filling syringes are also acceptable (ex. MicroFil from World Precision Instruments, Sarasota, FL, USA).
Up to 60 μM lipid (assuming 24 nm diameter discs) is okay, but a concentration that is too high will run the risk of the nanodiscs clogging the pipette tip.
It is important to fill the tip with nanodisc-free solution before filling with nanodiscs. Otherwise, establishing a gigaseal patch will be difficult, presumably because the nanodiscs will stick to the pipette walls.
If it is usually difficult to patch, it is possible that the concentration of discs is too high, or the time between nanodisc addition to the pipette and patching onto the cell was too long.
There is some voltage-dependence to the currents that are observed. Using a holding potential of −40 mV is empirically found to be a good compromise between the signal amplitude and noise.
About 7–9 minutes are needed for the discs to diffuse to the tip (counting from the time they are overlaid onto the disc-free solution in the pipette) and produce fusion pore currents. As time increases, more and more discs will be fusing and the seal will be weaker. Because it is difficult to distinguish between these processes, recordings beyond ~17–18 minutes (from the time the discs are overlaid onto the disc-free solution in the pipette) are usually unusable.
We use a passband of 280 or 480 Hz, depending on the gain (20 mV/pA or 50 mV/pA, respectively).
To identify the specific frequencies to filter, look at the power spectrum of the baseline.
For a transmembrane potential of −16 mV and gain 20 mV/pA, we use a threshold of −0.25 pA, and a minimum crossing time of 60 ms to define pore open sub-states in a current burst. These values were found empirically, after analyzing hundreds of pores26.
Overlapping pores become more likely towards the end of a recording, as the concentration of the nanodiscs near the plasma membrane, hence the fusion rate, increases over time. It is difficult to tell the difference between overlapping pore currents and a patch seal that is becoming loose. If the origin of a current burst is questionable, discard it from analysis. We also discard currents that occur while the baseline is not stable (usually during the first few minutes of recording).
References
- 1.Chernomordik LV & Kozlov MM Mechanics of membrane fusion. Nat Struct Mol Biol 15, 675–683 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jahn R & Fasshauer D Molecular machines governing exocytosis of synaptic vesicles. Nature 490, 201–207, doi: 10.1038/nature11320 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sudhof TC & Rothman JE Membrane fusion: grappling with SNARE and SM proteins. Science 323, 474–477, doi: 10.1126/science.1161748 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lindau M & de Toledo GA The fusion pore. Bba-Mol Cell Res 1641, 167–173 (2003). [DOI] [PubMed] [Google Scholar]
- 5.Jackson MB & Chapman ER The fusion pores of Ca2+ -triggered exocytosis. Nat Struct Mol Biol 15, 684–689, doi: 10.1038/nsmb.1449 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weber T et al. SNAREpins: Minimal machinery for membrane fusion. Cell 92, 759–772 (1998). [DOI] [PubMed] [Google Scholar]
- 7.Gao Y et al. Single reconstituted neuronal SNARE complexes zipper in three distinct stages. Science 337, 1340–1343, doi: 10.1126/science.1224492 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lindau M High resolution electrophysiological techniques for the study of calcium-activated exocytosis. Bba-Gen Subjects 1820, 1234–1242 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fulop T, Radabaugh S & Smith C Activity-dependent differential transmitter release in mouse adrenal chromaffin cells. J Neurosci 25, 7324–7332 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hastoy B, Clark A, Rorsman P & Lang J Fusion pore in exocytosis: More than an exit gate? A beta-cell perspective. Cell calcium 68, 45–61, doi: 10.1016/j.ceca.2017.10.005 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Collins SC et al. (2016) Increased expression of the diabetes gene SOX4 reduces insulin secretion by impaired fusion pore expansion. Diabetes 65, 1952–1961. 10.2337/db15-1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Staal RGW, Mosharov EV & Sulzer D Dopamine neurons release transmitter via a flickering fusion pore. Nat Neurosci 7, 341–346 (2004). [DOI] [PubMed] [Google Scholar]
- 13.Pawlu C, DiAntonio A & Heckmann M Postfusional control of quantal current shape. Neuron 42, 607–618 (2004). [DOI] [PubMed] [Google Scholar]
- 14.Chapochnikov NM et al. Uniquantal release through a dynamic fusion pore is a candidate mechanism of hair cell exocytosis. Neuron 83, 1389–1403, doi: 10.1016/j.neuron.2014.08.003 (2014). [DOI] [PubMed] [Google Scholar]
- 15.He LM, Wu XS, Mohan R & Wu LG Two modes of fusion pore opening revealed by cell-attached recordings at a synapse. Nature 444, 102–105 (2006). [DOI] [PubMed] [Google Scholar]
- 16.Alabi AA & Tsien RW Perspectives on kiss-and-run: role in exocytosis, endocytosis, and neurotransmission. Annual review of physiology 75, 393–422, doi: 10.1146/annurev-physiol-020911-153305 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Travis ER & Wightman RM Spatio-temporal resolution of exocytosis from individual cells. Annual review of biophysics and biomolecular structure 27, 77–103, doi: 10.1146/annurev.biophys.27.1.77 (1998). [DOI] [PubMed] [Google Scholar]
- 18.Kyoung M, Zhang Y, Diao J, Chu S & Brunger AT Studying calcium-triggered vesicle fusion in a single vesicle-vesicle content and lipid-mixing system. Nature Protocols 8, 1–16, doi:nprot.2012.134 [pii] 10.1038/nprot.2012.134 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoon TY, Okumus B, Zhang F, Shin YK & Ha T Multiple intermediates in SNARE-induced membrane fusion. P Natl Acad Sci USA 103, 19731–19736 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lai Y et al. Fusion pore formation and expansion induced by Ca2+ and synaptotagmin 1. Proc Natl Acad Sci U S A 110, 1333–1338, doi: 10.1073/pnas.1218818110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kiessling V, Liang B, Kreutzberger AJ & Tamm LK Planar Supported Membranes with Mobile SNARE Proteins and Quantitative Fluorescence Microscopy Assays to Study Synaptic Vesicle Fusion. Frontiers in molecular neuroscience 10, 72, doi: 10.3389/fnmol.2017.00072 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karatekin E et al. A fast, single-vesicle fusion assay mimics physiological SNARE requirements. Proc Natl Acad Sci U S A 107, 3517–3521, doi: 10.1073/pnas.0914723107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karatekin E & Rothman JE Fusion of single proteoliposomes with planar, cushioned bilayers in microfluidic flow cells. Nat Protoc 7, 903–920, doi: 10.1038/nprot.2012.019 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith MB et al. Interactive, computer-assisted tracking of speckle trajectories in fluorescence microscopy: application to actin polymerization and membrane fusion. Biophysical journal 101, 1794–1804, doi: 10.1016/j.bpj.2011.09.007 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stratton BS et al. Cholesterol Increases the Openness of SNARE-Mediated Flickering Fusion Pores. Biophysical journal 110, 1538–1550, doi: 10.1016/j.bpj.2016.02.019 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu Z et al. Nanodisc-cell fusion: control of fusion pore nucleation and lifetimes by SNARE protein transmembrane domains. Scientific reports 6, 27287, doi: 10.1038/srep27287 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu Z et al. Dilation of fusion pores by crowding of SNARE proteins. eLife 6, doi: 10.7554/eLife.22964 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi L et al. SNARE Proteins: One to Fuse and Three to Keep the Nascent Fusion Pore Open. Science 335, 1355–1359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu C et al. Fusion of cells by flipped SNAREs. Science 300, 1745–1749 (2003). [DOI] [PubMed] [Google Scholar]
- 30.Sakmann B & Neher E Single-channel recording. 2nd edn, (Springer, 2009). [Google Scholar]
- 31.Bello OD, Auclair SM, Rothman JE & Krishnakumar SS Using ApoE Nanolipoprotein Particles To Analyze SNARE-Induced Fusion Pores. Langmuir : the ACS journal of surfaces and colloids 32, 3015–3023, doi: 10.1021/acs.langmuir.6b00245 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stroeva E, Krishnakumar SS (2018) Using nanodiscs to probe Ca2+-dependent membrane interaction of Synaptotagmin-1 In: Fratti R (ed) SNAREs, Methods and protocols. Springer, New York: [DOI] [PubMed] [Google Scholar]
- 33.Breckenridge LJ & Almers W Currents through the Fusion Pore That Forms during Exocytosis of a Secretory Vesicle. Nature 328, 814–817 (1987). [DOI] [PubMed] [Google Scholar]