DELLA proteins were first identified as inhibitors of gibberellin (GA) signalling but have since been shown to be involved in regulation of many growth responses. GA causes responses by triggering the destruction of DELLA proteins (Willige et al., 2007). These proteins contain a conserved DELLA domain, which is followed by a GRAS domain. The GRAS domain which defines a family of proteins, including the DELLA proteins, is a conserved domain named after the first three family members: GAI (gibberellic‐acid insensitive), RGA (Repressor of ga1‐3), and SCR (Scarecrow). The DELLA domain encompasses the DELLA, LExLE and TVHYNP motifs. These motifs are important for binding to the GA‐bound GA receptor GID1, which is one of the initial steps in targeting the DELLA proteins for destruction by the 26S proteasome (Dill et al., 2001). The GA‐GID1‐DELLA complex interacts with the SLY1/GID2 SCF complex, which marks it for destruction by modifying it with ubiquitin. In this way, the suppression of GA signalling by DELLA is released and GA responses are activated. A number of mutants with mutations affecting the DELLA domain that increase the abundance of DELLA by weakening the interaction with the GA‐GID1 complex are known (Ueguchi‐Tanaka et al., 2007; Willige et al., 2007). Since these mutant DELLA proteins retain the ability to inhibit GA responses, the mutants are dwarfed.
Tomato has one DELLA protein called PROCERA (PRO). There is a well‐studied partial loss‐of‐function mutant, pro, which has a single amino acid substitution, valine to glutamate, in the VHVID motif of its GRAS domain (Bassel et al., 2008; Jupe et al., 1988). Strong recessive pro loss‐of‐function alleles, pro TALEN and pro ∆GRAS, were generated using transcription activator‐like effector nucleases (TALENs) and an Ac/Ds system, respectively (Livne et al., 2015; Lor et al., 2014). Consistent with these mutations fully activating GA signalling, homozygous mutants are extremely tall, have light green leaves with smoother leaf margins, and do not respond to the GA biosynthesis inhibitor paclobutrazol (PAC) or GA treatment. Recently, a gain‐of‐function allele affected in the DELLA motif that causes mild dwarfing and reduced GA sensitivity was reported (Tomlinson et al., 2018) but strong gain‐of‐function alleles have not been reported. Here, we report the generation and characterization of strong dwarfing alleles produced using the clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR‐associated protein9 (Cas9) system to induce intragenic suppressor mutations of pro TALEN alleles.
In the course of experiments to identify regenerating shoots in which the pro TALEN loss‐of‐function mutation has been repaired due to CRISPR/cas9 stimulated homologous recombination, we recovered several dwarf plants with dark green serrated leaves (Figure 1a, b). The regenerated T0 plants were dwarfs and the dwarfing alleles PRO GF8 and PRO GF9 from plants #8 and #9, respectively, were heritable. Sequencing confirmed that the PRO GF8 and PRO GF9 are intragenic suppressors of pro TALEN1 and pro TALEN7 respectively and that each encodes a full‐length mutant protein (Figure 1c, d). The protein encoded by PRO GF8 is predicted to have a 12 amino acid insertion and 2 amino acid substitution affecting the LExLE motif, while the protein encoded by PRO GF9 has a three amino acid deletion and an E to R substitution that affects the LExLE motif. PRO GF8 was characterized further. F1 seedlings from a cross between plant #8 and wild‐type (M82) were either extreme dwarfs with dark green leaves or wild‐type in appearance (Figure 1e). Only the dwarf plants had inherited the PRO GF8 allele indicating that it was dominant or semidominant.
Figure 1.
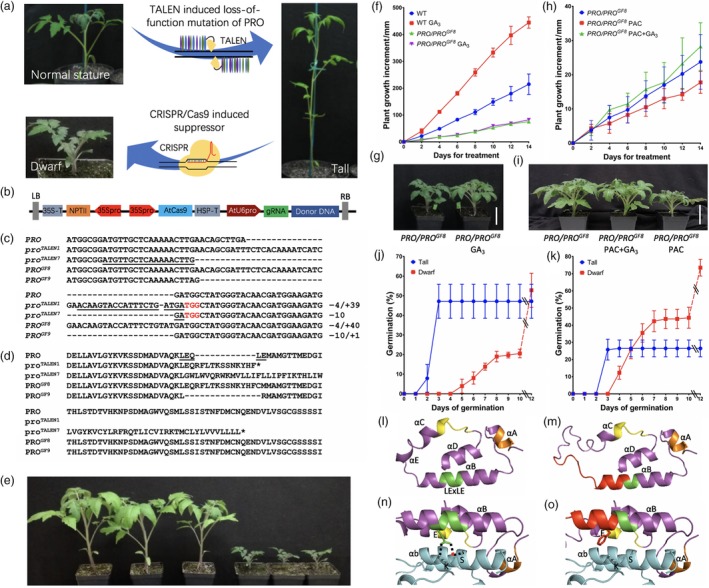
(a) Strategy for generating pro TALEN loss‐of‐function alleles and subsequent generation of suppressor mutations. (b) T‐DNA construct used to edit pro TALEN mutation sites were constructed in pTRANS220 as described by previously (Čermák et al., 2017). The gRNAs targeting pro TALEN 1 and pro TALEN 7 were constructed in pMOD_B2515 and the resulting module (B) was assembled together with modules from pMOD_A0101 (module A), and pMOD_C0000 (module C). Module C consisted of a fragment of genomic DNA spanning the pro TALEN mutation site. (c) DNA and (d) protein sequence of CRISPR/Cas9 induced alleles. The gRNA binding site is underlined and PAM site is highlighted in red in the pro TALEN 1 and pro TALEN 7 sequences. The size of insertion (+) and deletion (−) compared with wild‐type is indicated to the right of sequences. The conserved LExLE motif of DELLA protein is underlined in the wild‐type sequence. (e) Four‐week old F1 seedlings from a cross between T0 plant #8 and M82. Left to right, three tall plants that lack PROGF 8 and three PRO/ PROGF 8 plants. (f, h) Plants were spayed to runoff every other day with 0.07% ethanol (control treatment), 50 μm GA 3, 100 μm Paclobutrazol (PAC) or GA 3 plus PAC and the height was measured every other day for 2 weeks. After 14 days of treatment, the height of control and GA 3‐treated PRO/ PROGF 8 plants were similar. After 14 days, PAC treated PRO/ PROGF 8 were significantly shorter than GA 3‐treated PRO/ PROGF 8 plants based on an ANOVA followed by t‐test (P value 0.0014). (g) Seven‐week old PRO/ PROGF 8 plants after 2 weeks of treatment as in panel f. (i) Seven‐week old PRO/ PROGF 8 plants after 2 weeks of treatment as in panel h. Scale bar, 50 mm. (j) and (k), Time course of germination of seeds giving rise to tall and dwarf seedlings from (j) 4 month‐old F1 seeds from crossing T0 plant #8 with wild‐type pollen and (k) seeds immediately after harvest from a selfed PRO/pro GF 8 plant. Seeds that had not germinated after 10 days were scarified by removing the endosperm and seed coat adjacent to the root tip. Dash lines indicate the germination after scarification, which was scored at day 12. At least 40 seeds are used for each test, which was repeated three times. (l‐o) 3D models of PROCERA (l) and PROGF 8 (m) based on template 2ZSH (GA 3‐GID1A‐GAI). Close‐up view of the hydrogen bonds between LExLE motif of PROCERA and αb of GID1 (n) LExLE motif of proGF8 and αb of GID1 (o) based on template. The DELLA motif is highlighted in orange and TVHYNP motif in yellow. The LExLE motif in PRO and PROGF 8 is highlighted in green; the insertion of PROGF 8 is highlighted in red. Hydrogen bonds are indicated as dot lines. A water molecule bridging E and S is showed as a red sphere.
We tested the effect of PRO GF8 on GA responsiveness. In contrast to wild‐type M82 plants, which grew more rapidly when sprayed every 2 days with 50 μM GA3, PRO/PRO GF8 were unaffected (Figure 1f, g). Because DELLA proteins promote GA synthesis, endogenous GA levels are highly elevated in plants carrying DELLA gain‐of‐function alleles (Talón et al., 1990). To determine if the apparent GA insensitivity of PRO/PRO GF8 plants is because they contain saturating levels of endogenous bioactive GA, we treated PRO/PRO GF8 (Figure 1h, i) and wild‐type (not shown) plants with the GA biosynthesis inhibitor paclobutrazol (PAC) or a combination of PAC and GA3. PAC treatment reduced the height of both PRO/PRO GF8 and wild‐type plants and the GA3 treatment reversed this effect. The effects of the treatments were much smaller for PRO/PRO GF8, indicating that, while PRO/PRO GF8 responds to GA, it is nearly insensitive to it.
While germination tests found that initially only one half of the newly harvested F1 seeds from a cross between the T0 plant #8 and M82 germinated, the non‐germinated seeds germinated after they were scarified. The seedlings from seeds that did not require scarification were tall and did not carry the PRO GF allele while all of the seedlings from seeds that required scarification were PRO/PRO GF8 and dwarf (not shown). When we examined the germination kinetics after 4 months of storage, seeds that germinated by day 3 gave rise to tall plants whereas all seeds that germinated after day 3 produced dwarf seedlings (Figure 1j). After 10 days, 30% of the seeds had not germinated. Following scarification, these ungerminated seeds germinated and gave rise to dwarf seedlings.
We also tested germination of fresh seeds harvested from selfed PRO/PRO GF8 (Figure 1k). After 3 days, all of the seeds that produced wild‐type stature seedlings had germinated (26%). The seeds that had not germinated could be divided into two groups. One group comprising 44% of the seeds germinated by day 10 and produced dwarf seedlings. The remaining seed required scarification and also produced dwarf seedlings. The observed ratios for the tall seedlings, dwarf seedlings from seed that germinated without scarification, and dwarf from seeds requiring scarification [Tall vs. Dwarf (non‐scarified) vs. Dwarf (scarified)] fit a 1 : 2 : 1 ratio (three trials Chi‐square range from 0.458 to 3.471, P value from 0.176 to 0.795), which suggested that the PRO GF8 allele is semidominant. Consistent with this hypothesis, when genotyped, 16 out of 16 dwarf plants from seeds that germinated without scarification were heterozygous and 10 of 11 dwarf seedlings from seeds requiring scarification were homozygous for PRO GF8 and the remaining plant was heterozygous. Plants from seeds that require scarification to germinate were shorter than the dwarf plants from seeds that germinated without scarification (not shown) indicating that pro GF8 is also semidominant with respect to plant stature.
Molecular modelling of PRO and PROGF8 predicts that PROGF8 protein has a longer disordered region between the LExLE and TVHYNP motifs and the intervening alpha helix structure(s) are also affected (Figure 1l–o). The changes in PROGF8 are predicted to weaken its interaction with the GA‐bound GID1 because the stabilizing interaction between the second glutamic acid in LExLE motif and arginine and lysine in GID1 are disrupted. Weakening the interaction with GID1 likely increases the abundance of PROGF8.
Conflict of interest
The authors declare that there is no conflict of interest.
Acknowledgements
We thank Kathryn Fajardo for assistance with growing and maintain plants and Kristin Grandt for comments on the manuscript. This work was supported by a grant from the US Israel Binational Agriculture Research and Development fund to N.O. and D.W. (grant no. US‐4813‐15C). Zhiguo Zhu was supported by a State Scholarship Fund from China Scholarship Council (no. 201606050088).
References
- Bassel, G.W. , Mullen, R.T. and Bewley, J.D. (2008) Procera is a putative DELLA mutant in tomato (Solanum lycopersicum): effects on the seed and vegetative plant. J. Exp. Bot. 59, 585–593. [DOI] [PubMed] [Google Scholar]
- Čermák, T. , Curtin, S.J. , Gil‐Humanes, J. , Čegan, R. , Kono, T.J.Y. , Konečná, E. , Belanto, J.J. et al (2017) A multipurpose toolkit to enable advanced genome engineering in plants. Plant Cell, 29, 1196–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill, A. , Jung, H.‐S. and Sun, T.‐P. (2001) The DELLA motif is essential for gibberellin‐induced degradation of RGA. Proc. Natl Acad. Sci. 98, 14162–14167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jupe, S. , Causton, D. and Scott, I. (1988) Cellular basis of the effects of gibberellin and the pro gene on stem growth in tomato. Planta, 174, 106–111. [DOI] [PubMed] [Google Scholar]
- Livne, S. , Lor, V.S. , Nir, I. , Eliaz, N. , Aharoni, A. , Olszewski, N.E. , Eshed, Y. et al (2015) Uncovering DELLA‐independent gibberellin responses by characterizing new tomato procera mutants. Plant Cell, 27, 1579–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lor, V.S. , Starker, C.G. , Voytas, D.F. , Weiss, D. and Olszewski, N.E. (2014) Targeted mutagenesis of the tomato PROCERA gene using transcription activator‐like effector nucleases. Plant Physiol. 166, 1288–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talón, M. , Koornneef, M. and Zeevaart, J.A. (1990) Accumulation of C19‐gibberellins in the gibberellin‐insensitive dwarf mutant gai of Arabidopsis thaliana (L.) Heynh. Planta, 182, 501–505. [DOI] [PubMed] [Google Scholar]
- Tomlinson, L. , Yang, Y. , Emenecker, R. , Smoker, M. , Taylor, J. , Perkins, S. , Smith, J. et al (2018) Using CRISPR/Cas9 genome editing in tomato to create a gibberellin‐responsive dominant dwarf DELLA allele. Plant Biotechnol J, 10.1111/pbi.12952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi‐Tanaka, M. , Nakajima, M. , Katoh, E. , Ohmiya, H. , Asano, K. , Saji, S. , Hongyu, X. et al (2007) Molecular interactions of a soluble gibberellin receptor, GID1, with a rice DELLA protein, SLR1, and gibberellin. Plant Cell, 19, 2140–2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willige, B.C. , Ghosh, S. , Nill, C. , Zourelidou, M. , Dohmann, E.M.N. , Maier, A. and Schwechheimer, C. (2007) The DELLA domain of GA INSENSITIVE mediates the interaction with the GA INSENSITIVE DWARF1A gibberellin receptor of Arabidopsis . Plant Cell, 19, 1209–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]


