Summary
Heading date is an important agronomic trait affecting crop yield. The GRAS protein family is a plant‐specific super family extensively involved in plant growth and signal transduction. However, GRAS proteins are rarely reported have a role in regulating rice heading date. Here, we report a GRAS protein DHD1 (Delayed Heading Date1) delays heading and enhances yield in rice. Biochemical assays showed DHD1 physically interacts with OsHAP5C/D both in vitro and in vivo. DHD1 and OsHAP5C/D located in the nucleus and showed that rhythmic expression. Both DHD1 and OsHAP5C/D affect heading date by regulating expression of Ehd1. We propose that DHD1 interacts with OsHAP5C/D to delay heading date by inhibiting expression of Ehd1.
Keywords: rice, heading date, GRAS, Ehd1, OsHAP5C/D, yield
Introduction
Rice is one of the most important food crops in the world. Heading date has a significant impact on agronomic traits associated with yield (Jung and Muller, 2009). Photoperiod is the most important environmental factor affecting flowering time in rice. Many flowering time‐related genes have been cloned in rice. Heading date 3a (Hd3a) and RICE FLOWERING LOCUS T1 (RFT1) are florigen proteins in rice (Chardon and Damerval, 2005; Komiya et al., 2009; Tsuji et al., 2013). Hd3a is activated under SD and inhibited under LD, whereas RFT1 expression is increased at later developmental stages to promote flowering under LD condition (Komiya et al., 2008, 2009). Hd3a and RFT1 expression is activated by a rice‐specific B‐type response factor encoded by Early heading date 1 (Ehd1) under both SD and LD (Doi et al., 2004). Ehd1 is up‐regulated by some proteins such as Ehd2/OsINDETERMINATE 1 (OsID1)/Rice INDETERMINATE 1 (RID1) (Matsubara et al., 2008; Park et al., 2008; Wu et al., 2008), Ehd3 (Matsubara et al., 2011), Ehd4 (Gao et al., 2013), VERNALIZATION INSENSITIVE 3‐LIKE 2 (OsVIL2) (Wang et al., 2013), SET domain group protein 724 (SDG724) (Sun et al., 2012a), OsMADS50 (Ryu et al., 2009) and OsMADS51 (Kim et al., 2007). In contrast, multiple proteins also act as negative regulators of Ehd1, including Grain yield and heading date 7 (Ghd7) (Xue et al., 2008), LEC2 and FUSCA3‐Like 1 (OsLFL1) (Peng et al., 2007), CONSTANS‐Like 4 (OsCOL4) (Lee et al., 2010), OsCOL10 (Tan et al., 2016), OsCOL13 (Sheng et al., 2016), Date To Heading on chromosome 7 (DTH7) (Gao et al., 2014), DTH8 (Wei et al., 2010), Heading date 16 (Hd16)/Early Flowering 1 (EL1) (Dai and Xue, 2010; Hori et al., 2013), SUPERNUMERARY BRACT (SNB), OsINDETERMINATE SPIKELET 1 (OsIDS1) (Lee et al., 2014), OsMADS56 (Ryu et al., 2009), ABA responsive element binding factor 1 (OsABF1) (Zhang et al., 2016) and Heme Activator Protein like 1 (OsHAPL1) (Zhu et al., 2017). Heading date 1 (Hd1) also activates Ehd1 expression under SD, but functions as a repressor of Ehd1 through interaction with Ghd7 under LD (Nemoto et al., 2016).
The plant‐specific GRAS (GAI, RGA and SCR) protein family, contains five conserved motifs named LHRI, VHIID, LHRII, PFYRE and SAW in their C‐termini (Bolle, 2004; Sun et al., 2011). This family can be divided into 10 subfamilies according to several independent phylogenetic analyses based on the relatively conserved motifs in the N‐terminal domains (Sun et al., 2011). GRAS proteins have been extensively studied in plants and are known involved in gibberellin (GA) signaling, root development, auxin response, stress responses, phytochrome (Phy) signaling, brassinosteroid (BR) signaling and control of tillering (Bolle, 2004; Sun et al., 2011, 2012b). SLENDER RICE1 (SLR1) protein is a negative regulator of GA signaling and belongs to the DELLA subfamily of GRAS (Ikeda et al., 2001). Tillering in rice is controlled by MONOCULM 1 (MOC1), which belongs to the Arabidopsis LATERAL SUPPRESSOR (AtLAS) subfamily (Li et al., 2003). DWARF AND LOW‐TILLERING (DLT), the DLT subfamily of GRAS, plays positive roles in brassinosteroid signaling (Tong et al., 2009). Rice homologs of SHORTROOT (SHR) and SCARECROW (SCR) have an evolutionarily conserved mechanism that defines a single endodermal layer similar to Arabidopsis thaliana (Cui et al., 2007). Two Arabidopsis Phytochrome A signal transduction (AtPAT) subfamily members, Chitin‐Inducible Gibberellin‐Responsive 1 (CIGR1) and CIGR2, are responsive to N‐acetyl‐chitooligosaccharide elicitor and GA (Day et al., 2004). Recently, OsGRAS23 was reported to be involved in drought stress response by regulating expression of stress‐responsive genes (Xu et al., 2015). However, GRAS proteins have not been reported in connection with heading date in rice.
Here, we identified a new GRAS protein DHD1 that delays heading date by down‐regulating expression of Ehd1, Hd3a and RFT1, and also enhances yield. DHD1 physically interacts with OsHAP5C and OsHAP5D both in vitro and in vivo. OsHAP5C/D also delays heading date by suppressing Ehd1. Both DHD1 and OsHAP5C/D have rhythmic expression modes. Our findings reveal functional roles of DHD1 in rice flowering time and rice yield.
Results
DHD1 delays heading date in rice
Approximately 1,685 cDNAs of transcription factors from rice were fused to the VP64 activation domain, driven by the maize (Zea mays) ubiquitin (pUbi) promoter (Zhao et al., 2015). By analyzing the phenotypes of 57 751 independent transgenic plants of a Japonica rice variety Kitaake, we observed 30 independent lines, containing a VP64‐LOC_Os11 g47920 construct, showed a delayed heading compared to wild type (WT, Figure S1). And then, we focused on the new gene (LOC_Os11 g47920) with unknown function, namely DHD1 (Delayed Heading Date1).
To eliminate potential effects of VP64 activation activities and further confirm the role of DHD1, we fused DHD1 to a 3 × Flag‐tag and overexpressed the fusion under control of pUbi. We obtained 13 independent transgenic lines, and found that all these independent transgenic lines had the delayed heading phenotype compared to WT. We randomly chose three independent transgenic lines (OE18, OE19 and OE26) and used their homozygous offsprings for further study. Under natural long day condition (NLD), flowering time of all three lines was delayed by about 1 month, and panicle size was significantly increased compared to WT (Figure 1a–d). Under controlled long day (LD, 14 h light/10 h darkness) and short day (SD, 10 h light/14 h darkness) conditions flowering time of OE19 plants was delayed by about 3 weeks relative to WT (Figure 1e). The leaf emergence rates of WT and OE19 were investigated until the heading stage. The leaf emergence rates were similar to WT, indicating that the delayed flowering time in the transgenic plants was not due to retarded growth (Figure 1f,g). Plant height, panicle length, primary branching and secondary branching of OE lines were significantly increased while tiller number and thousand grain weight were not changed (Table 1). These results suggest that DHD1 may have potential to increase yield in breeding programs.
Figure 1.
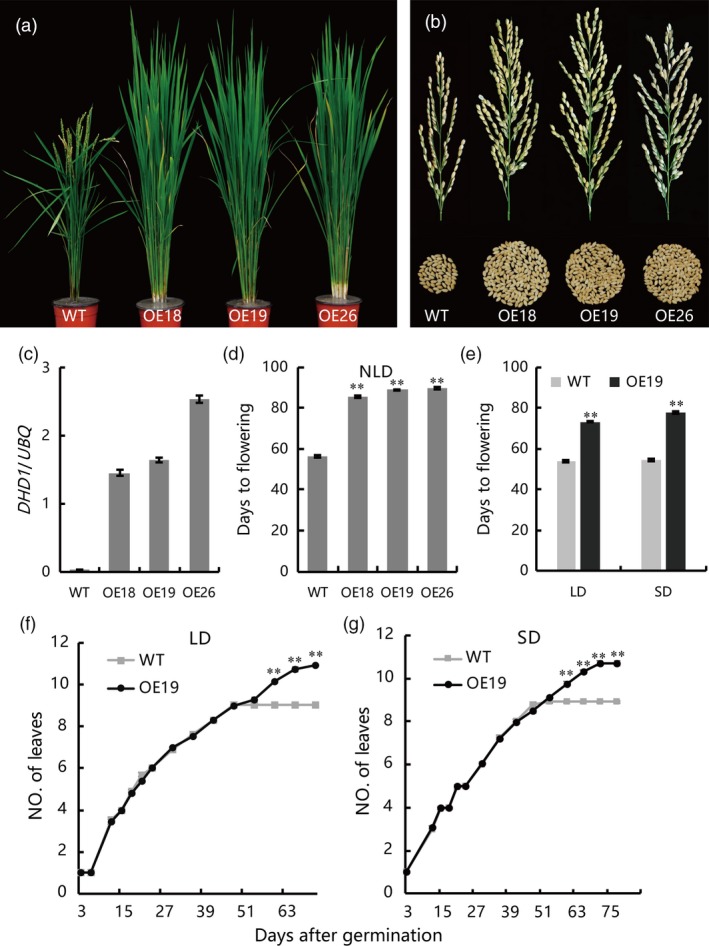
Phenotypes of DHD1‐Flag overexpression lines and wild type (WT) plants under natural LD, control LD and control SD conditions. (a) Phenotypes of overexpression and WT plants. Plants were grown under natural LD conditions for 70 days. OE18, OE19 and OE26, independent overexpression lines of DHD1‐Flag. (b) Main panicle size and grains per panicle of overexpression lines and WT. (c) Expression of DHD1 in overexpression lines and WT. Means ± SE (n = 3). (d) Heading dates of overexpression lines and WT. Means ± SE (n > 15). (e) Flowering time of OE19 and WT under LD and SD conditions. Means ± SE (n > 15). (f) Leaf emergence rate of OE19 and WT under LD conditions. Means ± SE (n > 15). (g) Leaf emergence rate of OE19 and WT under SD conditions. Means ± SE (n > 15), **P ≤ 0.01.
Table 1.
Agronomic traits of wild type Kitaake and overexpression lines when planted in the field under NLD conditions
| Trait | Kitaake | OE18 | OE19 | OE26 |
|---|---|---|---|---|
| Plant height (cm) | 65.66 ± 1.02 | 76.30 ± 0.84** | 78.00 ± 1.07** | 78.31 ± 0.98** |
| No. of tillers | 19.00 ± 0.74 | 18.67 ± 1.15 | 16.80 ± 1.31 | 17.53 ± 0.94 |
| Panicle length (cm) | 13.03 ± 0.35 | 17.97 ± 0.22** | 19.31 ± 0.61** | 19.08 ± 0.43** |
| Primary branches, No./panicle | 6.30 ± 0.15 | 16.40 ± 0.40** | 14.90 ± 0.41** | 16.30 ± 0.50** |
| Secondary branches, No./panicle | 9.10 ± 0.53 | 24.40 ± 1.29** | 26.60 ± 1.86** | 26.90 ± 0.67** |
| Grains/panicle | 58.40 ± 2.01 | 163.70 ± 6.16** | 163.60 ± 7.16** | 174.00 ± 2.51** |
| Thousand grain weight (g) | 26.35 ± 0.12 | 25.23 ± 0.47 | 24.93 ± 0.64 | 24.92 ± 0.42 |
**P ≤ 0.01.
To further confirm the function of DHD1 in plants, we created DHD1 mutants by CRISPR‐Cas9 technology in Kitaake, but there was no difference in flowering time between the mutants and Kitaake (Figure S2). Considering that Kitaake is an early flowering and photoperiod‐insensitive variety, we chose Nipponbare, a late flowering and photoperiod‐sensitivity variety, for further study to see if there is background effect. We created DHD1‐RNAi plants in Nipponbare background. There was no change in heading date when DHD1‐RNAi plants were planted in the field (Figure S5a–c). A genome‐wide search of rice revealed a homolog, DHD1L, with an amino acid sequence highly similar to DHD1 (Figure S3). We overexpressed DHD1L in Kitaake, and the heading date of the overexpression lines was also delayed (Figure S4). To eliminate the redundancy of DHD1L, we created a double mutant dhd1 dhd1 l in Nipponbare. We compared DHD1‐RNAi, dhd1 and dhd1 dhd1 l phenotypes in the field, but found no significant difference in heading date compared to the WT (Figure S5). In addition, the agronomic traits such as plant height and panicle size of dhd1 and dhd1 dhd1 l were decreased significantly (Table 2).
Table 2.
Agronomic traits of wild type Nipponbare, dhd1 and dhd1 dhd1 l when planted in the field under NLD conditions
| Trait | Nip | dhd1 | dhd1 dhd1 l |
|---|---|---|---|
| Plant height (cm) | 100.54 ± 0.57 | 87.93 ± 0.58** | 78.87 ± 0.82** |
| No. of tillers | 16.70 ± 0.84 | 13.47 ± 0.39** | 10.96 ± 0.41** |
| Panicle length (cm) | 22.75 ± 0.24 | 20.12 ± 0.25** | 19.37 ± 0.19** |
| Primary branches, No./panicle | 13.11 ± 0.14 | 12.00 ± 0.21** | 10.95 ± 0.26** |
| Secondary branches, No./panicle | 30.28 ± 0.93 | 27.76 ± 0.72** | 22.20 ± 0.71** |
| Grains/panicle | 164.94 ± 3.10 | 151.52 ± 2.78** | 121.60 ± 2.36** |
**P ≤ 0.01.
DHD1 has a constitutive and rhythmic expression pattern and DHD1 is located in the nucleus
In order to examine the expression pattern of DHD1, quantitative real‐time PCR (qRT‐PCR) experiments were performed with different tissues (Figure 2a). Although DHD1 was expressed in all tissues, expression levels were higher in leaves, leaf sheaths and panicles (Figure 2b). To investigate whether expression of DHD1 was rhythmic, we assayed the expression every 4 h in a 24 h time course under LD and SD conditions. The transcription level of DHD1 peaked at dawn and then slowly decreased until evening under both LD and SD conditions (Figure 2c,d). Our rice protoplast transient assays showed that DHD1‐GFP protein was mostly located in the nucleus, overlapping with the nuclear marker D53‐mCherry (Zhou et al., 2013) (Figure 2e).
Figure 2.
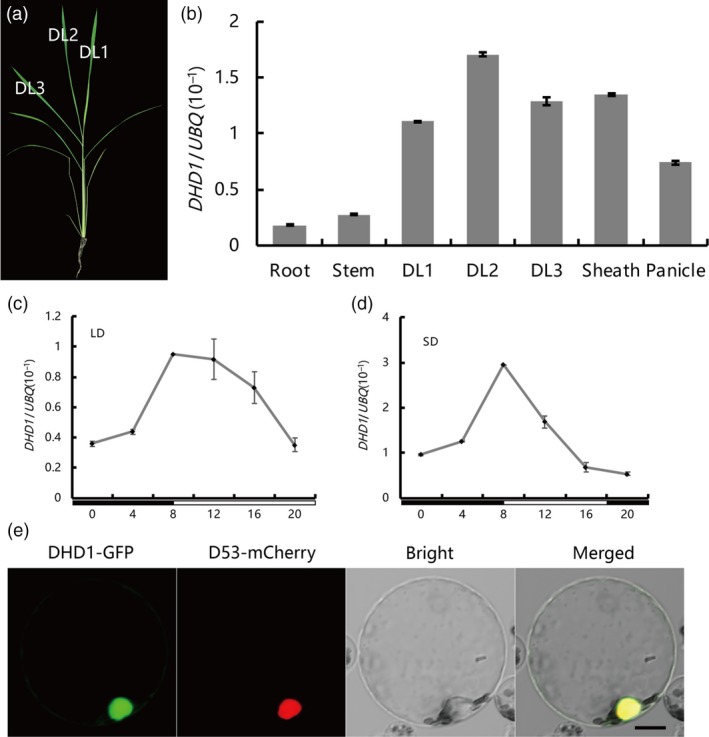
DHD1 expression patterns, subcellular localization of protein and transcriptional activity. (a) Leaf samples at different developmental stages were obtained from 30 day old wild type plants. DL, developed leaf. (b) The relative expression levels of DHD1 in different plant tissues. (c) Rhythmic expression pattern of DHD1 under LD conditions. Black and white boxes denote dark and light periods respectively. (d) Rhythmic expression pattern of DHD1 under SD conditions. Black and white boxes denote dark and light periods respectively. (e) Subcellular localization of DHD1‐GFP in rice protoplasts. Bar, 10 μm.
DHD1 interacts with OsHAP5C and OsHAP5D in vitro and in vivo
To investigate the action mode of DHD1, yeast two‐hybrid library screening with DHD1 as bait was performed and 73 positive clones were selected. By further confirmation of yeast‐two‐hybrid assay, we confirmed that OsHAP5C (LOC_Os03 g14669) and OsHAP5D (LOC_Os08 g38780) can interact with DHD1 in yeast (Figure 3a). A pull‐down assay was performed to confirm the physical interactions between DHD1 and OsHAP5C/D in vitro (Figure 3b).
Figure 3.
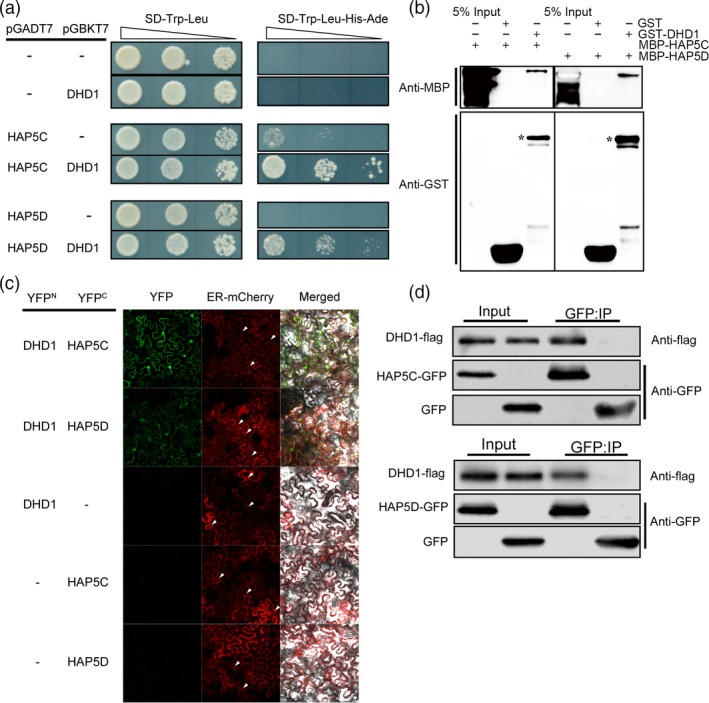
DHD1 interacts with OsHAP5C and OsHAP5D in vitro and in vivo. (a) Interactions of DHD1 with OsHAP5C/D in yeast strains. Triangle denotes 10‐fold dilution. (b) In vitro pull‐down analysis validates interactions of DHD1 with OsHAP5C/D. (c) BiFC analysis validates the interactions between DHD1 and OsHAP5C/D; the small triangles indicate the nucleus. (d) Co‐IP analysis in rice protoplasts verified interactions of DHD1 with OsHAP5C/D.
BiFC and Co‐IP experiments were performed to further confirm the interactions in vivo. In the BiFC assay, YFP fluorescence was produced in tobacco leaves co‐infected by Agrobacterium containing DHD1‐YFPN and OsHAP5C‐YFPC or OsHAP5D‐YFPC. YFP fluorescence was not generated when the tobacco was co‐infected by DHD1‐YFPN and empty YFPC. OsHAP5C‐YFPC and OsHAP5D‐YFPC with an empty YFPN also did not result in YFP fluorescence (Figure 3c). The Co‐IP assay was performed in rice protoplasts. The vector containing OsHAP5C‐GFP or OsHAP5D‐GFP was co‐transformed with a vector containing DHD1‐Flag into rice protoplasts. Both OsHAP5C‐GFP and OsHAP5D‐GFP proteins co‐immunoprecipitated with the DHD1‐Flag protein, whereas GFP protein alone did not (Figure 3d). These results indicated that OsHAP5C and OsHAP5D could interact with DHD1 in tobacco and rice.
OsHAP5C/D delayed heading date in rice
By qRT‐PCR and subcellular localization experiments, we found that OsHAP5C and OsHAP5D were subject to rhythmic expression and the proteins were mainly located in the nucleus (Figure S6). To investigate whether OsHAP5C/D are involved in heading date, we created OsHAP5C and OsHAP5D overexpression plants. These plants showed later flowering than WT (Kitaake) under natural LD, control LD and control SD conditions (Figure 4a,b). qRT‐PCR showed that expression of Ehd1, Hd3a and RFT1 was down‐regulated in OsHAP5C/D overexpressing plants (Figure 4c,d). These results indicated that OsHAP5C/D control heading date by regulating expression of Ehd1, Hd3a and RFT1.
Figure 4.
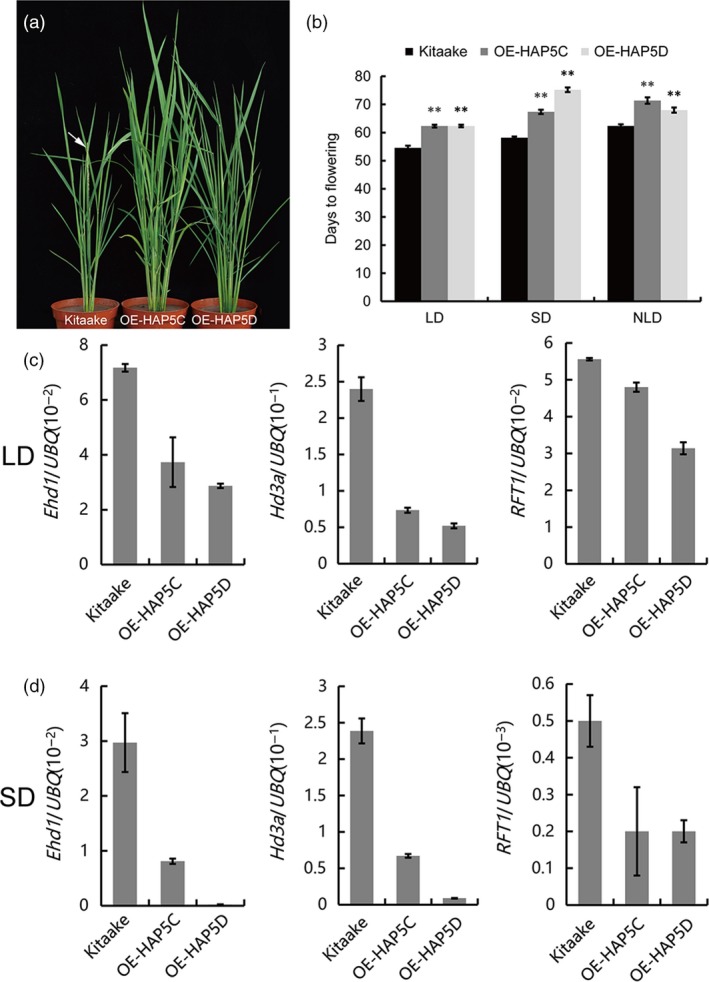
OsHAP5C/D overexpression delayed the heading date in rice. (a) Phenotypes of WT and OsHAP5C/D overexpression lines under NLD conditions. Arrow indicates the panicle. (b) Flowering time of WT and OsHAP5C/D overexpression lines under LD, SD and NLD conditions. Means ± SE (n > 12), **P ≤ 0.01. (c) Expression levels of Ehd1, Hd3a and RFT1 in WT and OsHAP5C/D overexpression lines under LD conditions. Means ± SE. (d) Expression levels of Ehd1, Hd3a and RFT1 in WT and OsHAP5C/D overexpression lines under SD conditions. Means ± SE.
DHD1 delays heading date by down‐regulating Ehd1, Hd3a and RFT1
In order to investigate downstream genes regulated by DHD1, we examined the expression of key genes involved in flowering in WT and OE‐DHD1‐19 under LD and SD conditions. qRT‐PCR data showed that expression of Ehd1, Hd3a and RFT1 was greatly decreased under both LD and SD conditions in OE‐DHD1‐19 compared to WT, particularly, at the time of peak expression in WT. Expression of OsMADS14, a downstream gene of Hd3a and RFT1, was also significantly decreased (Figure 5). This suggests that DHD1 delays flowering in rice by suppressing expression of Ehd1, Hd3a and RFT1.
Figure 5.
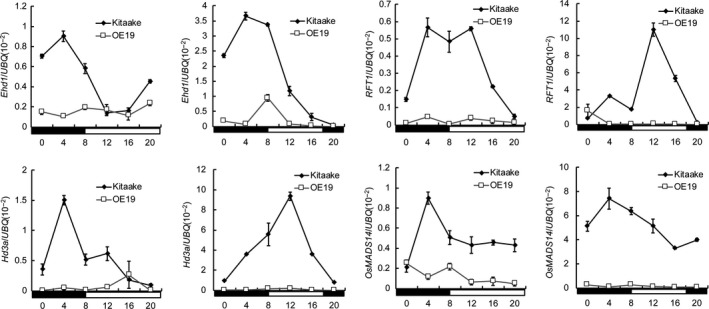
DHD1 inhibited expression of downstream genes Ehd1, Hd3a,RFT1 and OsMADS14 under LD and SD conditions. Means ± SE. The black and white boxes denote dark and light periods, respectively.
We examined expression of some other heading‐related genes, including OsPhyA, OsPhyB, OsPhyC, PHOTOPERIOD SENSITIVITY5 (SE5), OsGIGANTEA (OsGI), OsMADS50, OsMADS51, OsMADS56, DTH2, DTH8, OsCOL4, RICE FLORICULA/LEAFY (RFL), Ehd2, Ehd3, Ehd4 and Hd1, but detected no significant changes (Figures S7 and S8). We also analyzed the expression of DHD1 in mutants or near isogenic lines affecting heading date, in order to identify upstream genes that regulate DHD1. The results showed no changes of DHD1 expression between WT and these mutants or near isogenic lines (Figures S9 and S10).
Discussion
DHD1 could be involved in GA and ABA‐related pathways of flowering
No change in the heading date of DHD1‐RNAi, dhd1 or dhd1 dhd1 l plants when planted in the field. DHD1 and DHD1L may be involved in other special rice flowering pathways. The homologous of DHD1, DELLA proteins, are involved in the GA flowering pathway in Arabidopsis thaliana (Wang et al., 2016). DELLA protein REPRESSOR of ga1‐3 (RGA) can be degraded by GA (Willige et al., 2007) while ABA promotes its accumulation (Guo et al., 2014). Since DHD1 and DELLA proteins have conserved GRAS domains, we speculated that DHD1 has a similar function as DELLA. DHD1 was expressed transiently in the rice protoplasts, and treated with hormones GA, ABA and BR. We found that accumulation of DHD1 was increased under ABA treatment and decreased under GA treatment (Figure S11). Together, we speculated that DHD1 was involved in GA and ABA‐related pathways of flowering.
Salt stress delays flowering in Arabidopsis, which relies on the DELLA proteins that negatively regulate GA signaling and delay flowering by inhibiting downstream CONSTANS (CO) and FLOWERING LOCUS T (FT) expression (Achard et al., 2006; Li et al., 2007). Drought induces expression of OsABF1, which directly binds to the downstream promoter of OsWARKY104 to activate its expression, which in turn, inhibits expression of Ehd1 and delays flowering (Zhang et al., 2016). Previous studies have shown that the Lilium longiflorum SCR‐like (LISCL) subfamily proteins may be involved in the stress response, and recently, OsGRAS23, a member of the LISCL subfamily of rice, has been reported to be involved in drought response (Xu et al., 2015). We observed that the dhd1 dhd1 l double mutant flowered earlier than WT and the dhd1 single mutant under drought and salt treatment. There was no significant difference in flowering time under normal conditions (Figure S12). We therefore hypothesized that DHD1 may be involved in the flowering regulation of stress, which remains to be further investigated.
DHD1 interacts with OsHAP5C/D and participates in regulating heading date
Heme activator protein (HAP) proteins, also known as NUCLEAR FACTOR Y (NF‐Y) proteins, are a class of regulatory proteins widely found in fungi, animals and plants (Mantovani, 1999). In Arabidopsis, NF‐Y proteins bind CCAAT elements to form long‐distance chromatin loops on the FT promoter and initiate expression of FT by recruiting CO (Cao et al., 2014). The NF‐Y complex also partially modulates trimethylated H3K27 levels of the SUPPRESOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1) promoter by interacting with a H3K27 demethylase, RELATIVE OF EARLY FLOWERING 6 (REF6) (Hou et al., 2014). In rice, DTH8/OsHAP3H delays heading by decreasing the expression of Ehd1 and Hd3a under LD (Wei et al., 2010). Li et al. (2016) overexpressed a series of HAP family genes and found that OsHAP3D, OsHAP3E, OsHAP5A and OsHAP5B delayed flowering under LD conditions. Furthermore, an OsHAPL1‐DTH8‐Hd1 complex was reported to function as a repressor of heading (Zhu et al., 2017). Yeast two‐hybrid library screening showed that DHD1 can interact with OsHAP5C and OsHAP5D. Furthermore, yeast two‐hybrid experiments and pull‐down experiments demonstrated that DHD1 physically interacts with OsHAP5C and OsHAP5D in vitro (Figure 3a,b). BiFC experiments in tobacco and the Co‐IP experiments in rice protoplasts verified these interactions in vivo (Figure 3c,d). Overexpression of OsHAP5C/D also delays heading in Kitaake under LD, SD and NLD conditions (Figure 4a,b), as well as reducing expression of Ehd1, Hd3a and RFT1 (Figure 4c,d), in a similar manner to lines overexpressing DHD1. These results suggest that DHD1 and OsHAP5C/D may play roles in the same flowering pathway. We also created single and double mutants of OsHAP5C and OsHAP5D, but these had no effect on heading date relative to WT when grown in the field (Figure S13). Considering there are 11 HAP5 subunits in rice (Li et al., 2016), it is likely that many other HAP5 genes play redundant roles.
DHD1‐OsHAP5C/5D inhibits flowering by repressing Ehd1 expression
It is generally considered that there are two flowering regulation pathways in rice: the Hd1‐mediated pathway and the Ehd1‐mediated pathway, which modulate expression of the florigen genes Hd3a and RFT1 to control heading date (Sun et al., 2014; Tsuji et al., 2011). Additional genes directly regulate the expression of Hd3a and RFT1, such as DTH2 (Wu et al., 2013), OsCO3 (Kim et al., 2008) and RFL (Rao et al., 2008). It was reported that Hd1 interacts with Ghd7 and binds directly to a cis‐regulatory region in Ehd1 to repress its expression (Nemoto et al., 2016). Drought, GA and low temperature also affect expression of Ehd1 to control the flowering time (Cho et al., 2017). Thus, Ehd1 may be an important integrator in regulating flowering time in rice. DHD1 delayed heading by inhibiting Ehd1 expression (Figure 5). NF‐YA, or HAP2, binds to CCAAT elements and recruits other HAP subunit proteins to bind the promoter (Gnesutta et al., 2017). There are several CCAAT elements in the promoter of Ehd1. We hypothesize that it is possible that other HAP2 proteins bind to the Ehd1 promoter and recruit HAP5s and DHD1 to form a complex that regulates flowering. The relationships between DHD1 and HAP proteins and Ehd1 remain to be further studied.
DHD1 regulates heading date and has potential to increase yield in rice
The GRAS proteins are involved in many aspects of plant development and signal transduction in rice (Bolle, 2004; Sun et al., 2012b). However, the relationship between GRAS proteins and heading date is less reported. Here, we describe a GRAS family protein encoded by DHD1 in rice. Overexpression of DHD1 in Kitaake, a short‐season variety, delayed heading date and caused increases in a number of agronomic traits such as panicle length, primary branch number, secondary branch number and grain number per panicle, without a change number of tillers per plant. Thus, we speculated DHD1 could be utilized to improve agronomic traits including yield in rice breeding.
Experimental procedures
Plant materials and growth conditions
The genotypes used for transformation were ssp. japonica varieties Kitaake and Nipponbare. All plants were grown in paddy fields at Beijing (116°13′E, 39°54′N) during summer, representing natural long day conditions. Plants were also grown in climate chambers with controlled LD (10 h darkness, 25 °C/14 h light, 30 °C) and SD (14 h darkness, 25 °C/10 h light, 30 °C) conditions with about 800 μmol/m2/s light intensity and 70% relative humidity.
Vector construction and rice transformation
To generate DHD1, DHD1L, OsHAP5C and OsHAP5D overexpressing plants, full‐length CDSs of DHD1, DHD1L, OsHAP5C and OsHAP5D were amplified, and PCR products were subcloned into binary vector pCAMBIA1390 using an In‐Fusion Advantage PCR Cloning Kit (Clontech, Beijing, China). To knockdown the DHD1 gene, construct pCUbi1390‐ΔFAD2 (Ubi promoter and a FAD2 intron inserted into pCAMBIA1390) was used as an RNAi vector (Tan et al., 2014). Both antisense and sense versions of a specific 172 bp fragment from the coding region were amplified (primer pairs DHD1‐RNAi‐1 and DHD1‐RNAi‐2; Table S1), and inserted into pCUbi1390‐ΔFAD2 to form the RNAi construct pUbi‐dsRNAi‐DHD1. To knockout the DHD1 gene, 20 bp gene‐specific spacer sequence of the target gene (Table S1) was cloned into the entry vector pOs‐sgRNA and then subcloned into the destination vector containing the CAS9 expression cassette using the Gateway LR Clonase II Enzyme mix [Invitrogen, Shanghai, China; (Miao et al., 2013)]. To develop dhd1 dhd1 l double mutant, we constructed a CRISPR‐Cas9 vector containing guide RNAs targeting DHD1 and DHD1L respectively. Mutants of hap5c/d were created in the same way. The resulting plasmids were transformed into Agrobacterium tumefaciens strain EHA105 and then introduced into rice variety Kitaake or Nipponbare as described previously (Hiei et al., 1994). Primer sequences for construction of these vectors are listed in Table S1.
Total RNA was isolated from frozen tissues using a ZR Plant RNA MiniPrep Kit (ZYMO Research, Beijing, China) and reverse transcribed using a QuantiTect reverse transcription kit (Qiagen, Shanghai, China) according to the manufacturer's protocol. qRT‐PCR was performed with a SYBR premix Ex Taq Kit (TaKaRa, Dalian, China) according to the operation manual and amplified in an ABI 7500 using primers listed in Table S1. Data from three biological replicates were analyzed following the relative quantification method (Livak and Schmittgen, 2001). Primer sequences for qRT‐PCR are listed in Table S1.
Phenotype measurement of agronomic traits
Various agronomic traits, including days to heading, plant height, tillers number, panicle length, primary branches number, secondary branches number, grain number per panicle and thousand grains weight (1000‐grain weight), were manually measured. Days to heading were counted from seed sowing to first panicle heading about 1–2 cm of each plant. The reproductive tillers having panicles with filled grains were counted for tillers number. Thousand grain weight was determined by measuring the weight of harvested seeds that were air‐dried in a glasshouse and oven‐dried at 50 °C until they reached ~14.0% moisture content. Panicle‐related traits containing panicle length, primary branches number, secondary branches number and grain number per panicle were measured from main tillers of 15 plants in WT and transgenic lines respectively (Kim et al., 2018). All data are analyzed by the LSD‐test.
Subcellular localization
The full length cDNAs of DHD1, OsHAP5C and OsHAP5D were fused at the C‐terminal with GFP in the pAN580 vector, and then transiently expressed the resulting construct in rice leaf protoplasts to explore the subcellular localization (Table S1). The method for rice leaf protoplast transformation is available in a previous report (Zhang et al., 2011). Fluorescence signals were detected by a ZESS LSM880 confocal microscope.
Yeast two‐hybrid assay
The full length DHD1 cDNA was amplified, inserted into the pGBKT7 vector (Table S1), and then transformed into the yeast strain AH109. BD‐DHD1 was used for yeast two‐hybrid library screening experiment (Yeastmaker™ Yeast Transformation Manual User, Clontech). The complete coding sequences of the OsHAP5C and OsHAP5D genes were inserted into the pGADT7 vector and co‐transformed into yeast with pGBKT7‐DHD1 as described above (Table S1).
In vitro pull‐down assay
The coding region of DHD1 was inserted into the prokaryotic expression vector pGEX4T‐1 for fusing the GST‐tag. The CDSs of OsHAP5C and OsHAP5D were inserted into pMAL‐c2X for fusing the MBP‐tag (Table S1). The vectors were transformed into the expression strain Escherichia coli BL21. Fusion proteins GST‐DHD1 and MBP‐OsHAP5C/D were induced with 0.5 mm IPTG at 16 °C for 12 h. Pull‐down assays were performed as previously reported (Miernyk and Thelen, 2008). MBP and GST antibodies with horseradish peroxidase (MBL) were used in western blotting experiments.
Bimolecular fluorescence complementation (BiFC) analysis
The coding region of DHD1 was inserted into the pSPYNE173 vector and OsHAP5C/D were inserted into the pSPYCE (M) (Table S1). Fusion proteins DHD1‐YNE and OsHAP5C/D‐YCE were transiently expressed in 5‐6‐week‐old Nicotiana benthamiana leaves. The method for infiltrating N. benthamiana leaves with Agrobacterium strain EHA105 followed a previous report (Waadt and Kudla, 2008). Fluorescence signals were detected by a ZESS LSM880 confocal microscope 48–72 h after infiltration.
In vivo co‐immunoprecipitation (Co‐IP) assay
The CDS of DHD1 with a 3 × Flag‐tag was inserted into the pAN580 vector. OsHAP5C/D were also inserted into pAN580 vector for fusing GFP at the C‐terminal (Table S1). The vectors were co‐transformed into rice protoplasts as described above. The Co‐IP assay was performed as previously reported (Yang et al., 2014). Western blotting was detected by the Flag and GFP antibodies with horseradish peroxidase (MBL).
Hormone treatment experiment
The pAN580‐DHD1‐Flag vector was transformed into rice protoplast as described above. The protoplasts were divided into four groups, three of which were separately treated with 10 μm GA, ABA and BR. Protein was extracted from each group after 4 h and subjected to western blotting.
Supporting information
Figure S1 VP64‐DHD1 overexpression plants flowered later than the wild type Kitaake.
Figure S2 Heading date of dhd1 mutant in Kitaake background under NLD, LD and SD conditions.
Figure S3 Phylogenetic tree and protein sequence alignment of DHD1 and DHD1L
Figure S4 DHD1L overexpression plants flowered later than the wild type Kitaake.
Figure S5 Phenotypes of wild type Nipponbare, DHD1‐RNAi, dhd1 and dhd1 dhd1 l plants grown in the field under NLD.
Figure S6 Rhythmic expression pattern of OsHAP5C/D and subcellular localization of proteins.
Figure S7 Relative expression levels of heading date‐related genes in wild type Kitaake and OE19 overexpression lines.
Figure S8 Relative expression levels of heading date‐related genes in wild type Kitaake and OE19 overexpression lines.
Figure S9 The relative expression levels of DHD1 in heading date‐related mutants or near‐isogenic lines.
Figure S10 The relative expression levels of DHD1 in the heading date‐related mutants or near‐isogenic lines.
Figure S11 Accumulation of DHD1‐Flag protein in rice protoplasts subjected to ABA, GA and BR hormone treatments.
Figure S12 Flowering time of NIP, dhd1 and dhd1 dhd1 l under normal, salt and drought conditions in SD climate chamber.
Figure S13 Heading date of hap5c, hap5d and hap5c hap5d grown in the field under NLD condition.
Table S1 Primers used in the current work.
Acknowledgements
This work was supported by the National Natural Science Foundation (31771764), the National Key R&D Program of China (2016YFD0100600 and 2017YFD0100501) and the National Transgenic Major Program (2016ZX08009‐003). This research was also supported by the Key Laboratory of Biology, Genetics and Breeding of Japonica Rice in Mid‐lower Yangtze River, Ministry of Agriculture, P. R. China, and Jiangsu Collaborative Innovation Center for Modern Crop Production. All authors have approved this manuscript and declare no conflict of interests.
References
- Achard, P. , Cheng, H. , De Grauwe, L. , Decat, J. , Schoutteten, H. , Moritz, T. , Van Der Straeten, D. et al (2006) Integration of plant responses to environmentally activated phytohormonal signals. Science, 311, 91–94. [DOI] [PubMed] [Google Scholar]
- Bolle, C. (2004) The role of GRAS proteins in plant signal transduction and development. Planta, 218, 683–692. [DOI] [PubMed] [Google Scholar]
- Cao, S. , Kumimoto, R.W. , Gnesutta, N. , Calogero, A.M. , Mantovani, R. and Holt, B.F. III (2014) A distal CCAAT/NUCLEAR FACTOR Y complex promotes chromatin looping at the FLOWERING LOCUS T promoter and regulates the timing of flowering in Arabidopsis. Plant Cell, 26, 1009–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chardon, F. and Damerval, C. (2005) Phylogenomic analysis of the PEBP gene family in cereals. J. Mol. Evol. 61, 579–590. [DOI] [PubMed] [Google Scholar]
- Cho, L.H. , Yoon, J. and An, G. (2017) The control of flowering time by environmental factors. Plant J. 90, 708–719. [DOI] [PubMed] [Google Scholar]
- Cui, H. , Levesque, M.P. , Vernoux, T. , Jung, J.W. , Paquette, A.J. , Gallagher, K.L. , Wang, J.Y. et al (2007) An evolutionarily conserved mechanism delimiting SHR movement defines a single layer of endodermis in plants. Science, 316, 421–425. [DOI] [PubMed] [Google Scholar]
- Dai, C. and Xue, H.W. (2010) Rice early flowering1, a CKI, phosphorylates DELLA protein SLR1 to negatively regulate gibberellin signalling. EMBO J. 29, 1916–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day, R.B. , Tanabe, S. , Koshioka, M. , Mitsui, T. , Itoh, H. , Ueguchi‐Tanaka, M. , Matsuoka, M. et al (2004) Two rice GRAS family genes responsive to N ‐acetylchitooligosaccharide elicitor are induced by phytoactive gibberellins: evidence for cross‐talk between elicitor and gibberellin signaling in rice cells. Plant Mol. Biol. 54, 261–272. [DOI] [PubMed] [Google Scholar]
- Doi, K. , Izawa, T. , Fuse, T. , Yamanouchi, U. , Kubo, T. , Shimatani, Z. , Yano, M. et al (2004) Ehd1, a B‐type response regulator in rice, confers short‐day promotion of flowering and controls FT‐like gene expression independently of Hd1. Genes Dev. 18, 926–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, H. , Zheng, X.M. , Fei, G. , Chen, J. , Jin, M. , Ren, Y. , Wu, W. et al (2013) Ehd4 encodes a novel and Oryza‐genus‐specific regulator of photoperiodic flowering in rice. PLoS Genet. 9, e1003281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, H. , Jin, M. , Zheng, X.M. , Chen, J. , Yuan, D. , Xin, Y. , Wang, M. et al (2014) Days to heading 7, a major quantitative locus determining photoperiod sensitivity and regional adaptation in rice. Proc. Natl Acad. Sci. USA, 111, 16337–16342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnesutta, N. , Kumimoto, R.W. , Swain, S. , Chiara, M. , Siriwardana, C. , Horner, D.S. , Holt, B.F.I.I.I. et al (2017) CONSTANS imparts DNA sequence specificity to the histone fold NF‐YB/NF‐YC dimer. Plant Cell, 29, 1516–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, W. , Cong, Y. , Hussain, N. , Wang, Y. , Liu, Z. , Jiang, L. , Liang, Z. et al (2014) The remodeling of seedling development in response to long‐term magnesium toxicity and regulation by ABA‐DELLA signaling in Arabidopsis. Plant Cell Physiol. 55, 1713–1726. [DOI] [PubMed] [Google Scholar]
- Hiei, Y. , Ohta, S. , Komari, T. and Kumashiro, T. (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T‐DNA. Plant J. 6, 271–282. [DOI] [PubMed] [Google Scholar]
- Hori, K. , Ogiso‐Tanaka, E. , Matsubara, K. , Yamanouchi, U. , Ebana, K. and Yano, M. (2013) Hd16, a gene for casein kinase I, is involved in the control of rice flowering time by modulating the day‐length response. Plant J. 76, 36–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou, X. , Zhou, J. , Liu, C. , Liu, L. , Shen, L. and Yu, H. (2014) Nuclear factor Y‐mediated H3K27me3 demethylation of the SOC1 locus orchestrates flowering responses of Arabidopsis. Nat. Commun. 5, 4601. [DOI] [PubMed] [Google Scholar]
- Ikeda, A. , Ueguchi‐Tanaka, M. , Sonoda, Y. , Kitano, H. , Koshioka, M. , Futsuhara, Y. , Matsuoka, M. et al (2001) Slender rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height‐regulating gene GAI/RGA/RHT/D8. Plant Cell, 13, 999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung, C. and Muller, A.E. (2009) Flowering time control and applications in plant breeding. Trends Plant Sci. 14, 563–573. [DOI] [PubMed] [Google Scholar]
- Kim, S.L. , Lee, S. , Kim, H.J. , Nam, H.G. and An, G. (2007) OsMADS51 is a short‐day flowering promoter that functions upstream of Ehd1, OsMADS14, and Hd3a. Plant Physiol. 145, 1484–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S.K. , Yun, C.H. , Lee, J.H. , Jang, Y.H. , Park, H.Y. and Kim, J.K. (2008) OsCO3, a CONSTANS‐LIKE gene, controls flowering by negatively regulating the expression of FT‐like genes under SD conditions in rice. Planta, 228, 355–365. [DOI] [PubMed] [Google Scholar]
- Kim, S.R. , Ramos, J.M. , Hizon, R.J.M. , Ashikari, M. , Virk, P.S. , Torres, E.A. , Nissila, E. et al (2018) Introgression of a functional epigenetic OsSPL14(WFP) allele into elite indica rice genomes greatly improved panicle traits and grain yield. Sci. Rep. 8, 3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komiya, R. , Ikegami, A. , Tamaki, S. , Yokoi, S. and Shimamoto, K. (2008) Hd3a and RFT1 are essential for flowering in rice. Development, 135, 767–774. [DOI] [PubMed] [Google Scholar]
- Komiya, R. , Yokoi, S. and Shimamoto, K. (2009) A gene network for long‐day flowering activates RFT1 encoding a mobile flowering signal in rice. Development, 136, 3443–3450. [DOI] [PubMed] [Google Scholar]
- Lee, Y.S. , Jeong, D.H. , Lee, D.Y. , Yi, J. , Ryu, C.H. , Kim, S.L. , Jeong, H.J. et al (2010) OsCOL4 is a constitutive flowering repressor upstream of Ehd1 and downstream of OsphyB. Plant J. 63, 18–30. [DOI] [PubMed] [Google Scholar]
- Lee, Y.S. , Lee, D.Y. , Cho, L.H. and An, G. (2014) Rice miR172 induces flowering by suppressing OsIDS1 and SNB, two AP2 genes that negatively regulate expression of Ehd1 and florigens. Rice, 7, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X. , Qian, Q. , Fu, Z. , Wang, Y. , Xiong, G. , Zeng, D. , Wang, X. et al (2003) Control of tillering in rice. Nature, 422, 618–621. [DOI] [PubMed] [Google Scholar]
- Li, K. , Wang, Y. , Han, C. , Zhang, W. , Jia, H. and Li, X. (2007) GA signaling and CO/FT regulatory module mediate salt‐induced late flowering in Arabidopsis thaliana. Plant Growth Regul. 53, 195–206. [Google Scholar]
- Li, Q.P. , Yan, W.H. , Chen, H.X. , Tan, C. , Han, Z.M. , Yao, W. , Li, G.W. et al (2016) Duplication of OsHAP family genes and their association with heading date in rice. J. Exp. Bot. 67, 1759–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak, K.J. and Schmittgen, T.D. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2(T)(‐Delta Delta C) method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Mantovani, R. (1999) The molecular biology of the CCAAT‐binding factor NF‐Y. Gene, 239, 15–27. [DOI] [PubMed] [Google Scholar]
- Matsubara, K. , Yamanouchi, U. , Wang, Z.X. , Minobe, Y. , Izawa, T. and Yano, M. (2008) Ehd2, a rice ortholog of the maize INDETERMINATE1 gene, promotes flowering by up‐regulating Ehd1. Plant Physiol. 148, 1425–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubara, K. , Yamanouchi, U. , Nonoue, Y. , Sugimoto, K. , Wang, Z.X. , Minobe, Y. and Yano, M. (2011) Ehd3, encoding a plant homeodomain finger‐containing protein, is a critical promoter of rice flowering. Plant J. 66, 603–612. [DOI] [PubMed] [Google Scholar]
- Miao, J. , Guo, D.S. , Zhang, J.Z. , Huang, Q.P. , Qin, G.J. , Zhang, X. , Wan, J.M. et al (2013) Targeted mutagenesis in rice using CRISPR‐Cas system. Cell Res. 23, 1233–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miernyk, J.A. and Thelen, J.J. (2008) Biochemical approaches for discovering protein‐protein interactions. Plant J. 53, 597–609. [DOI] [PubMed] [Google Scholar]
- Nemoto, Y. , Nonoue, Y. , Yano, M. and Izawa, T. (2016) Hd1, a CONSTANS ortholog in rice, functions as an Ehd1 repressor through interaction with monocot‐specific CCT‐domain protein Ghd7. Plant J. 86, 221–233. [DOI] [PubMed] [Google Scholar]
- Park, S.J. , Kim, S.L. , Lee, S. , Je, B.I. , Piao, H.L. , Park, S.H. , Kim, C.M. et al (2008) Rice Indeterminate 1 (OsId1) is necessary for the expression of Ehd1 (Early heading date 1) regardless of photoperiod. Plant J. 56, 1018–1029. [DOI] [PubMed] [Google Scholar]
- Peng, L.T. , Shi, Z.Y. , Li, L. , Shen, G.Z. and Zhang, J.L. (2007) Ectopic expression of OsLFL1 in rice represses Ehd1 by binding on its promoter. Biochem. Biophys. Res. Commun. 360, 251–256. [DOI] [PubMed] [Google Scholar]
- Rao, N.N. , Prasad, K. , Kumar, P.R. and Vijayraghavan, U. (2008) Distinct regulatory role for RFL, the rice LFY homolog, in determining flowering time and plant architecture. Proc. Natl Acad. Sci. USA, 105, 3646–3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu, C.H. , Lee, S. , Cho, L.H. , Kim, S.L. , Lee, Y.S. , Choi, S.C. , Jeong, H.J. et al (2009) OsMADS50 and OsMADS56 function antagonistically in regulating long day (LD)‐dependent flowering in rice. Plant, Cell Environ. 32, 1412–1427. [DOI] [PubMed] [Google Scholar]
- Sheng, P. , Wu, F. , Tan, J. , Zhang, H. , Ma, W. , Chen, L. , Wang, J. et al (2016) A CONSTANS‐like transcriptional activator, OsCOL13, functions as a negative regulator of flowering downstream of OsphyB and upstream of Ehd1 in rice. Plant Mol. Biol. 92, 209–222. [DOI] [PubMed] [Google Scholar]
- Sun, X. , Xue, B. , Jones, W.T. , Rikkerink, E. , Dunker, A.K. and Uversky, V.N. (2011) A functionally required unfoldome from the plant kingdom: intrinsically disordered N‐terminal domains of GRAS proteins are involved in molecular recognition during plant development. Plant Mol. Biol. 77, 205–223. [DOI] [PubMed] [Google Scholar]
- Sun, C. , Fang, J. , Zhao, T. , Xu, B. , Zhang, F. , Liu, L. , Tang, J. et al (2012a) The histone methyltransferase SDG724 mediates H3K36me2/3 deposition at MADS50 and RFT1 and promotes flowering in rice. Plant Cell, 24, 3235–3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, X. , Jones, W.T. and Rikkerink, E.H. (2012b) GRAS proteins: the versatile roles of intrinsically disordered proteins in plant signalling. Biochem J. 442, 1–12. [DOI] [PubMed] [Google Scholar]
- Sun, C. , Chen, D. , Fang, J. , Wang, P. , Deng, X. and Chu, C. (2014) Understanding the genetic and epigenetic architecture in complex network of rice flowering pathways. Protein Cell, 5, 889–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, J. , Tan, Z. , Wu, F. , Sheng, P. , Heng, Y. , Wang, X. , Ren, Y. et al (2014) A novel chloroplast‐localized pentatricopeptide repeat protein involved in splicing affects chloroplast development and abiotic stress response in rice. Mol. Plant, 7, 1329–1349. [DOI] [PubMed] [Google Scholar]
- Tan, J. , Jin, M. , Wang, J. , Wu, F. , Sheng, P. , Cheng, Z. , Wang, J. et al (2016) OsCOL10, a CONSTANS‐like gene, functions as a flowering time repressor downstream of Ghd7 in rice. Plant Cell Physiol. 57, 798–812. [DOI] [PubMed] [Google Scholar]
- Tong, H. , Jin, Y. , Liu, W. , Li, F. , Fang, J. , Yin, Y. , Qian, Q. et al (2009) DWARF AND LOW‐TILLERING, a new member of the GRAS family, plays positive roles in brassinosteroid signaling in rice. Plant J. 58, 803–816. [DOI] [PubMed] [Google Scholar]
- Tsuji, H. , Taoka, K. and Shimamoto, K. (2011) Regulation of flowering in rice: two florigen genes, a complex gene network, and natural variation. Curr. Opin. Plant Biol. 14, 45–52. [DOI] [PubMed] [Google Scholar]
- Tsuji, H. , Taoka, K. and Shimamoto, K. (2013) Florigen in rice: complex gene network for florigen transcription, florigen activation complex, and multiple functions. Curr. Opin. Plant Biol. 16, 228–235. [DOI] [PubMed] [Google Scholar]
- Waadt, R. and Kudla, J. (2008) In planta visualization of protein interactions using bimolecular fluorescence complementation (BiFC). CSH Protoc. 2008, 10.1101/pdb.prot4995 [DOI] [PubMed] [Google Scholar]
- Wang, J. , Hu, J. , Qian, Q. and Xue, H.W. (2013) LC2 and OsVIL2 promote rice flowering by photoperoid‐induced epigenetic silencing of OsLF. Mol. Plant, 6, 514–527. [DOI] [PubMed] [Google Scholar]
- Wang, H. , Pan, J. , Li, Y. , Lou, D. , Hu, Y. and Yu, D. (2016) The DELLA‐CONSTANS transcription factor cascade integrates gibberellic acid and photoperiod signaling to regulate flowering. Plant Physiol. 172, 479–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, X. , Xu, J. , Guo, H. , Jiang, L. , Chen, S. , Yu, C. , Zhou, Z. et al (2010) DTH8 suppresses flowering in rice, influencing plant height and yield potential simultaneously. Plant Physiol. 153, 1747–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willige, B.C. , Ghosh, S. , Nill, C. , Zourelidou, M. , Dohmann, E.M. , Maier, A. and Schwechheimer, C. (2007) The DELLA domain of GA INSENSITIVE mediates the interaction with the GA INSENSITIVE DWARF1A gibberellin receptor of Arabidopsis. Plant Cell, 19, 1209–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, C. , You, C. , Li, C. , Long, T. , Chen, G. , Byrne, M.E. and Zhang, Q. (2008) RID1, encoding a Cys2/His2‐type zinc finger transcription factor, acts as a master switch from vegetative to floral development in rice. Proc. Natl Acad. Sci. USA, 105, 12915–12920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, W.X. , Zheng, X.M. , Lu, G.W. , Zhong, Z.Z. , Gao, H. , Chen, L.P. , Wu, C.Y. et al (2013) Association of functional nucleotide polymorphisms at DTH2 with the northward expansion of rice cultivation in Asia. Proc. Natl Acad. Sci. USA, 110, 2775–2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, K. , Chen, S. , Li, T. , Ma, X. , Liang, X. , Ding, X. , Liu, H. et al (2015) OsGRAS23, a rice GRAS transcription factor gene, is involved in drought stress response through regulating expression of stress‐responsive genes. BMC Plant Biol. 15, 141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue, W. , Xing, Y. , Weng, X. , Zhao, Y. , Tang, W. , Wang, L. , Zhou, H. et al (2008) Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat. Genet. 40, 761–767. [DOI] [PubMed] [Google Scholar]
- Yang, J.W. , Fu, J.X. , Li, J. , Cheng, X.L. , Li, F. , Dong, J.F. , Liu, Z.L. et al (2014) A novel Co‐immunoprecipitation protocol based on protoplast transient gene expression for studying protein‐protein interactions in rice. Plant Mol. Biol. Rep. 32, 153–161. [Google Scholar]
- Zhang, Y. , Su, J. , Duan, S. , Ao, Y. , Dai, J. , Liu, J. , Wang, P. et al (2011) A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast‐related processes. Plant Methods, 7, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, C. , Liu, J. , Zhao, T. , Gomez, A. , Li, C. , Yu, C. , Li, H. et al (2016) A drought‐inducible transcription factor delays reproductive timing in rice. Plant Physiol. 171, 334–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, T. , Liu, J. , Li, H.Y. , Lin, J.Z. , Bian, M.D. , Zhang, C.Y. , Zhang, Y.X. et al (2015) Using hybrid transcription factors to study gene function in rice. Sci. China Life Sci. 58, 1160–1162. [DOI] [PubMed] [Google Scholar]
- Zhou, F. , Lin, Q. , Zhu, L. , Ren, Y. , Zhou, K. , Shabek, N. , Wu, F. et al (2013) D14‐SCF(D3)‐dependent degradation of D53 regulates strigolactone signalling. Nature, 504, 406–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, S. , Wang, J. , Cai, M. , Zhang, H. , Wu, F. , Xu, Y. , Li, C. et al (2017) The OsHAPL1‐DTH8‐Hd1 complex functions as the transcription regulator to repress heading date in rice. J. Exp. Bot. 68, 553–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 VP64‐DHD1 overexpression plants flowered later than the wild type Kitaake.
Figure S2 Heading date of dhd1 mutant in Kitaake background under NLD, LD and SD conditions.
Figure S3 Phylogenetic tree and protein sequence alignment of DHD1 and DHD1L
Figure S4 DHD1L overexpression plants flowered later than the wild type Kitaake.
Figure S5 Phenotypes of wild type Nipponbare, DHD1‐RNAi, dhd1 and dhd1 dhd1 l plants grown in the field under NLD.
Figure S6 Rhythmic expression pattern of OsHAP5C/D and subcellular localization of proteins.
Figure S7 Relative expression levels of heading date‐related genes in wild type Kitaake and OE19 overexpression lines.
Figure S8 Relative expression levels of heading date‐related genes in wild type Kitaake and OE19 overexpression lines.
Figure S9 The relative expression levels of DHD1 in heading date‐related mutants or near‐isogenic lines.
Figure S10 The relative expression levels of DHD1 in the heading date‐related mutants or near‐isogenic lines.
Figure S11 Accumulation of DHD1‐Flag protein in rice protoplasts subjected to ABA, GA and BR hormone treatments.
Figure S12 Flowering time of NIP, dhd1 and dhd1 dhd1 l under normal, salt and drought conditions in SD climate chamber.
Figure S13 Heading date of hap5c, hap5d and hap5c hap5d grown in the field under NLD condition.
Table S1 Primers used in the current work.


