SUMMARY
Objective:
This study examined the oral epithelial immunotranscriptome response patterns modulated by oral bacterial planktonic or biofilm challenge.
Methods:
We assessed gene expression patterns when epithelial cells were challenged with a multispecies biofilm composed of S. gordonii, F. nucleatum, and P. gingivalis representing a type of periodontopathic biofilm compared to challenge with the same species of planktonic bacteria.
Results:
Of the 579 human immunology genes, a substantial signal of the epithelial cells was observed to 181 genes. Biofilm challenged stimulated significant elevations compared to planktonic bacteria for IL32, IL8, CD44, B2M, TGFBI, NFKBIA, IL1B, CD59, IL1A, CCL20 representing the top 10 signals comprising 55% of the overall signal for the epithelial cell responses. Levels of PLAU, CD9, IFITM1, PLAUR, CD24, TNFSF10, and IL1RN were all elevated by each of the planktonic bacterial challenge versus the biofilm responses. While the biofilms upregulated 123/579 genes (>2-fold), fewer genes were increased by the planktonic species [36 (S. gordonii), 30 (F. nucleatum), 44 (P. gingivalis)].
Conclusions:
A wide array of immune genes were regulated by oral bacterial challenge of epithelial cells that would be linked to the local activity of innate and adaptive immune response components in the gingival tissues. Incorporating bacterial species into a structured biofilm dramatically altered the number and level of genes expressed. Additionally a specific set of genes were significantly decreased with the multispecies biofilms suggesting that some epithelial cell biologic pathways are down-regulated when in contact with this type of pathogenic biofilm.
INTRODUCTION
Recent evidence has demonstrated the critical nature of the human gut microbiome as a component of the maintenance of health at this mucosal surface. This feature of the host-bacterial interactions has been shown to interact with and regulate epithelial cell functions in the gut. Additionally, these interactions have been documented to be a significant component of the characteristics and maturation of the host immune system both locally and systemically 1–3. However, while clear data delineate the relationship of selected gut bacteria in driving this communication with the immune system, similar data from other mucosal tissues, including the oral cavity is much more limited.
Historically the epithelium and epithelial cells were viewed more as a mechanical barrier in innate immunity, including the routine sloughing of the cells removing the bound bacteria from the mucosal surface. However, more recent information has provided insights into the numerous biological functions of epithelial cells. These include the production of a constitutive and induced profile of antimicrobial peptides 4–6, various cell growth and communication factors to help maintain the integrity of the epithelium 7–9, and an array of pro- and anti-inflammatory chemokines and cytokines to inform the cells of the immune system concerning the epithelial cell interactions with the juxtaposed microbiome 7,8,10.
Previous results have demonstrated a limited response profile of oral epithelial cells following challenge with various planktonic oral bacteria 11–13. We and others have also provided novel data regarding the influence of various mono- and multispecies biofilms on these response profiles with results demonstrating significant differences in planktonic versus biofilm challenge within a bacteria species, and a different profile of responses to the multispecies biofilms that varied from simply a summation of the individual bacterial components 14–16. This report describes our studies focusing on the capabilities of the epithelial cells to activate an array of immune response pathway signals that would prime the gingival tissues for responses to the oral microbiome. We assessed these gene expression patterns related to challenge of the epithelial cells with a multispecies biofilm composed of Streptococcus gordonii, Fusobacterium nucleatum, and Porphyromonas gingivalis representing a type of periodontopathic biofilm 16,17. These response profiles were compared to challenge with the same species of planktonic bacteria reflecting the ongoing process of detachment and dispersal that would occur with oral bacteria throughout the oral cavity to establish biofilms at other sites 18–20.
MATERIALS and METHODS
Growth of bacteria and multispecies biofilms
F. nucleatum ATCC 25586, S. gordonii ATCC 10558, and P. gingivalis FDC381) were grown in Brain Heart Infusion (Becton Dickinson, Sparks, MD) medium supplemented with 5 µg hemin ml−1 and 1 µg menadione ml−1 under anaerobic conditions (85% N2, 10% H2, 5% CO2) at 37oC as we have described previously 15,17 . Biofilms were grown on Rigid Gas Permeable Lenses (RGPL) (Advanced Vision Technologies, Golden, CO), 10.5 mm in diameter in a single well of a 48-well plate, which allows the RGPLs to cover the entire surface of the well. Prior to biofilm formation, RGPLs were coated with 1% fetal bovine serum (Invitrogen) and monospecies planktonic cultures of the 3 bacteria were mixed and used to create the biofilms, with bacterial input from 1-11×108 21. Our previous studies of these biofilms have shown an approximate composition of the final biofilms at 3.4×109 with 92% S. gordonii, 2% F. nucleatum, and 6% P. gingivalis.
Oral epithelial cell culture model
An immortalized epithelial cell line OKF6 22 was cultured in standard KFM media to form a confluent monolayer 17. Planktonic bacteria and biofilm challenge, and control treatments were each carried out in 6 wells in 1ml/well fresh media seeded with 5×104 OKF6 cells, and continuously incubated for 12 h under anaerobic conditions (85% N2, 5% CO2, and 10% H2). The results of gene expression levels in the epithelial cells that were challenged with the planktonic bacteria at an MOI of 1:50-1:100 were combined since no significant differences were generally noted in response profiles with these 2 doses 23. Three day old biofilms grown on contact lenses were overlaid with the biofilm surface juxtaposed to the epithelial cells. OKF6 cells with or without overlaid RGPL were used as controls and maintain high viability (XTT conversion and level of housekeeping gene expression23) and function for the 24 hr. experimental interval 15,24. Based upon estimated calculations of the area of the biofilms on the RGPL 23 and the surface area of an epithelial cell, we estimated that the direct interaction of the biofilm surface with the epithelial cells would approximate an MOI of 10:1 to 50:1 bacterial cells on the surface of the biofilms were in contact with an individual epithelial cell.
NanoString analysis
Gene expression profiles of the oral epithelial cells exposed to the biofilms and bacteria were assessed using the n Counter Human Immunology Kit panel (NanoString, Seattle, WA; https://www.nanostring.com/products/gene-expression-panels/ncounter-immunology-panels) containing a set of 579 genes representing pathways that cover an array of inflammatory, and innate and adaptive immune responses. After exposure of cell cultures to the bacteria, media only or RGPL, total mRNA was isolated using the Pure Link RNA Mini (Life Technologies, NY, USA) kit following the manufacturer’s instructions. RNA (100ng) with integrity numbers of 9-10 from each sample was hybridized with the reporter code set beads 25 in a final volume of 30 μl at 65°C for 12 hours and processed using the NanoString Cell Prep Station. Data normalized to total RNA levels was collected using the NanoString Digital Analyzer (NanoString Technologies, Seattle, WA, USA) through the Microarray Core facility at the University of Kentucky.
Statistical analysis:
The mean ± standard deviation of the bacteria/biofilm stimulation of OKF6 were compared using an ANOVA on ranks test with Dunn’s test for multiple comparisons to evaluate the data from stimulated cells compared with unchallenged cells or RGPL overlaid OKF6 cells (Sigma Stat 3.5; Systat Software, Inc., Chicago, IL).
RESULTS
Comparison of gene expression profiles of oral epithelial cells to planktonic bacteria and biofilms
Figure 1 provides an overview of the primary response profiles of the oral epithelial cells to challenge with each of the three planktonic bacteria and the biofilms organized based upon the magnitude of gene expression with the biofilm. Of the 579 genes in the NanoString human immunology portfolio, substantial signal (>20 copies under one or more conditions) of the epithelial cells was observed to 181 genes. Highlighted on the figure are genes that were most greatly affected (fold differences >2) by the bacterial challenge. Summation of the mRNA signal across the 181 genes showed that this set of response genes accounted for 98.2% of total biofilm mRNA signal, and 98.9%, 98.8%, and 98.3% of the mRNA signals from the epithelial cells stimulated with S. gordonii, F. nucleatum, and P. gingivalis, respectively. Many of these host response genes were at an increased or decreased signal level following challenge with S. gordonii, F. nucleatum, or P. gingivalis as planktonic bacteria. Of note was the high gene expression of IL32 following biofilm challenge, and the substantially lower level of IL8, IL1B, CCL20, PML, LTB4R2, LIF, SOCS3, and MBP induced by all the planktonic bacteria versus the biofilm. In contrast, levels of PLAU, CD9, IFITM1, PLAUR, CD24, TNFSF10, and IL1RN were all elevated by each of the planktonic bacterial challenge versus the biofilm responses.
Figure 1:
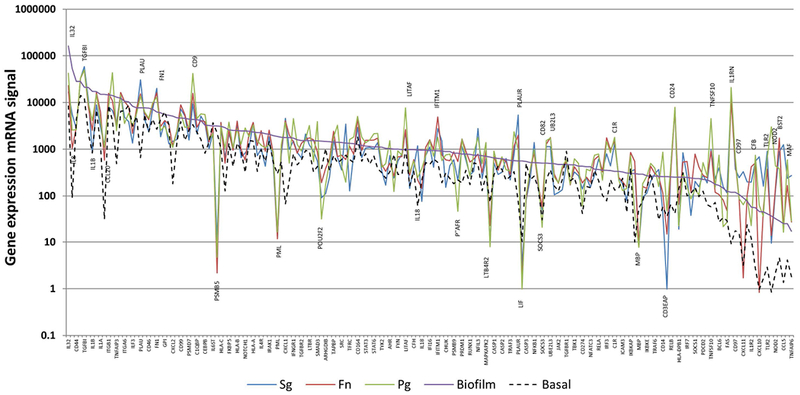
Gene expression mRNA signal of genes expressed by oral epithelial cells with signal >100 to biofilm and/or planktonic bacterial challenge. The genes are ordered based upon the magnitude of signal with the biofilm challenge. The highlighted genes are those expressed by planktonic challenge with all the species that resulted in substantially (>4-fold) increased or decreased levels compared to the biofilm signal. Values denote the mean of values from 5 independent cell culture wells for the biofilms and basal levels (cells only), and duplicate cell culture wells for the planktonic bacteria.
Figure 2 displays the fold changes in response patterns of the oral epithelial cells following interaction with serum coated or biofilms on rigid gas permeable lens (RGPL). Generally the RGPL interaction resulted in minimal gene expression differences compared to the media control cell cultures. Of note was the large array of genes (those elevated by ≥16-fold highlighted) that were elevated following the biofilm challenge representing various pathways of host response in which the epithelial cells may be participating to help maintain homeostasis and communicate with immune system cells.
Figure 2:
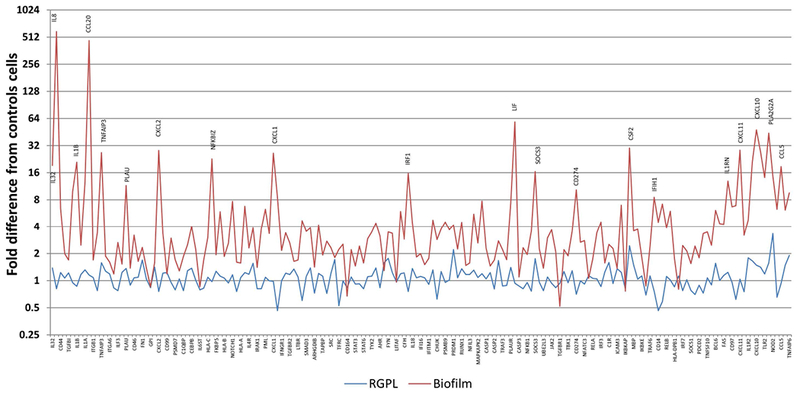
Comparison of fold difference in gene expression with biofilm or RGPL interaction with the oral epithelial cells. Genes are ordered based upon the magnitude of signal expression following biofilm challenge. Control cells represented basal production in media. Values denote the mean of 5 values for the biofilms and 3 for the RGPL.
Figure 3 summarizes the differences in gene expression profiles of the multispecies biofilm with each of the individual planktonic microorganisms used to challenge the epithelial cells. Fig. 3A highlights major gene differences between the biofilm and challenge with S. gordonii. Beyond these targeted genes, the profile demonstrates an upregulation of >2-fold with 123 of the 181 highly expressed genes (>20 copies) with the biofilm and only 36 with S. gordonii. Of these elevated responses, only IL8, CCL20, CXCL2, PSMB5, CXCl1, ICAM1, MBP, IL23A, CXCL11, CXCL10, and CCL5 were increased >8-fold. Nineteen genes showed expression levels decreased by >2-fold following challenge with S. gordonii. Fig. 3B provides a similar profile comparing the biofilm responses to those elicited by challenge with F. nucleatum. We observed 30 genes with expression levels >2-fold increased to F. nucleatum with only MBP, IL23A, IL1R2, and TRAF1 being substantially increased and NOTCH1, and MAF being decreased by >2-fold compared to control cells. Interestingly, Fig. 3C presents patterns comparing the biofilm challenge to responses following infection with planktonic P. gingivalis. Few genes were elevated beyond 16-fold with P. gingivalis and included IL23A, CXCL11, CXCL10, and TRAF1. As observed previously 26 the planktonic bacteria increased PLA2G2A by over 3000-fold that appeared to be attenuated when P. gingivalis was in a biofilm milieu. The expression of 11 genes was decreased by >2-fold compared to control cells. Moreover, of the genes that were decreased by the planktonic bacteria, only MX1, STAT1, and POU2F2 were down-regulated by both S. gordonii and P. gingivalis.
Figure 3A-C:
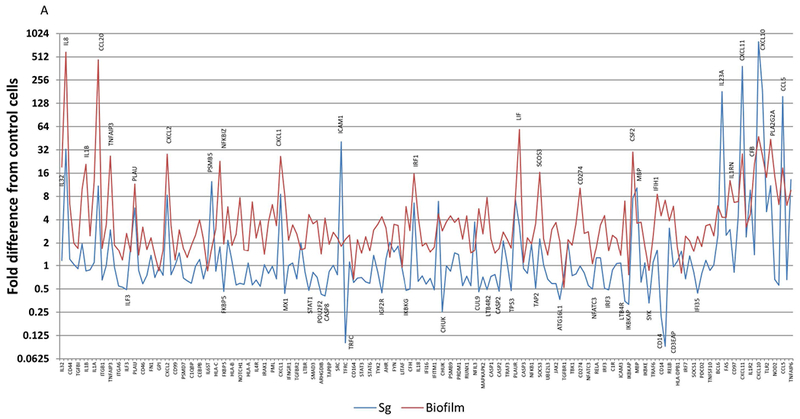
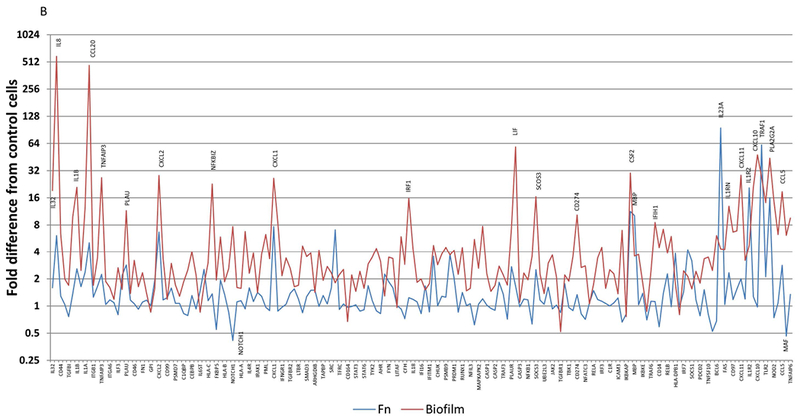
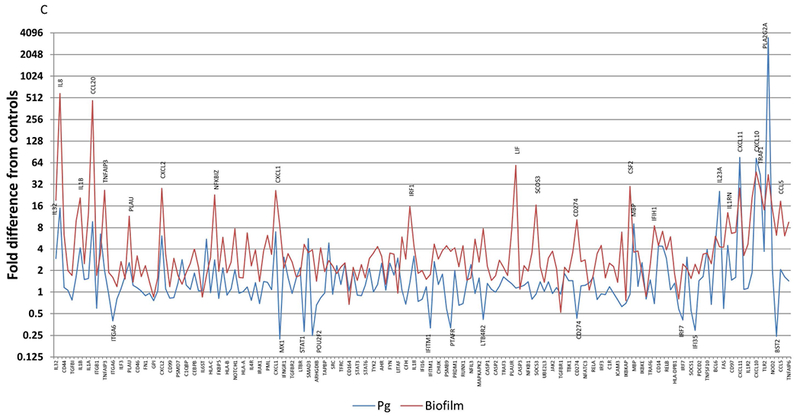
Comparison of fold differences in gene expression with biofilms compared to individual planktonic bacteria, S. gordonii (A), F. nucleatum (B), and P. gingivalis (C). Highlighted genes are those increased by >8-fold or decreased by >2-fold. Values denote the mean of 5 values for the biofilms and duplicates for the planktonic bacteria.
Figure 4 summarizes the relationship of the up-regulated genes between the biofilm and the individual planktonic species. Fig. 4A compares the unique and overlapping genes between across the bacterial challenge conditions. The data shows that the majority of the genes that were up-regulated by the individual planktonic bacteria were also increased by the biofilm, albeit, nearly 60% of the gene up-regulated by the biofilms were not represented across the planktonic species. Fig. 4B depicts the features of altered gene expression across the planktonic species. Only 1 gene (ICAM1) was up-regulated with all species, which was lost when the bacteria were in a biofilm. Additionally, P. gingivalis demonstrated a feature of a larger set of unique genes that were increased versus the other species (eg. IL32, CTSC, NOTCH1, C3, CD14, NOD2).
Figure 4:
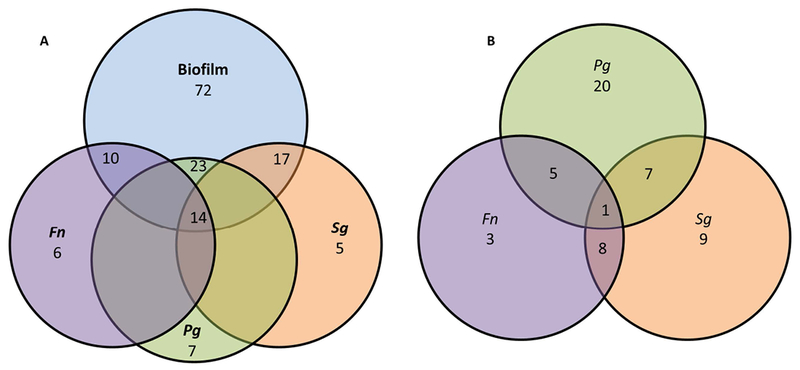
Venn diagram depicting the number of genes upregulated by (A) biofilm or planktonic bacteria and identifying overlap or unique alteration of the expression levels. (B) Provides similar comparison of gene IDs upregulated among the planktonic bacteria only.
Target genes affected by planktonic bacteria and biofilms
Figure 5A-C focuses on specific gene profiles that were elevated by at least 2 times with the biofilms compared to any of the planktonic bacteria driven gene expression. As such, 72/579 of the human immunology genes were specifically elevated by the multispecies biofilm challenge compared to all planktonic challenges. The Protein ANalysis THrough Evolutionary Relationships [PANTHER; 27] classification system was used to assess the gene ontology of these profiles. The pathways that were significantly over-represented are shown in Table 1. These included an array of pathways directly involved in communicating the cellular response to infection to immune system components. Additionally, this periodontopathic biofilm surrogate elicited various pathways controlling cell survival and apoptotic processes.
Figure 5A-C:
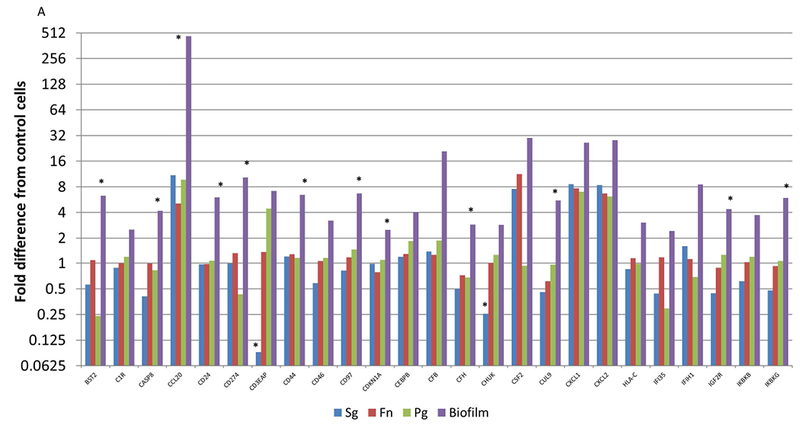
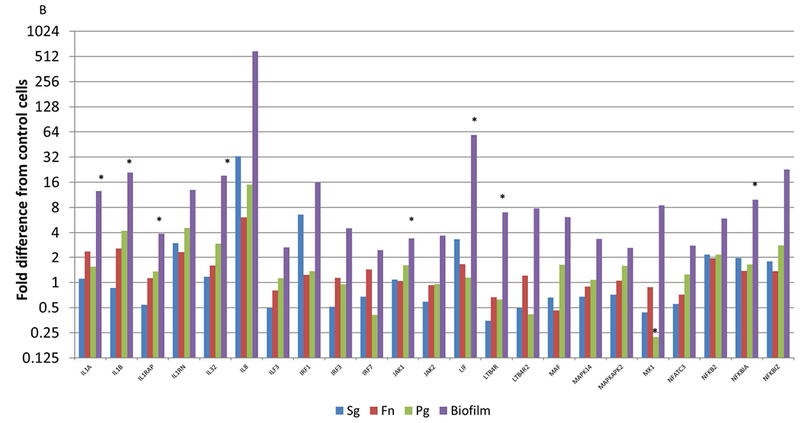
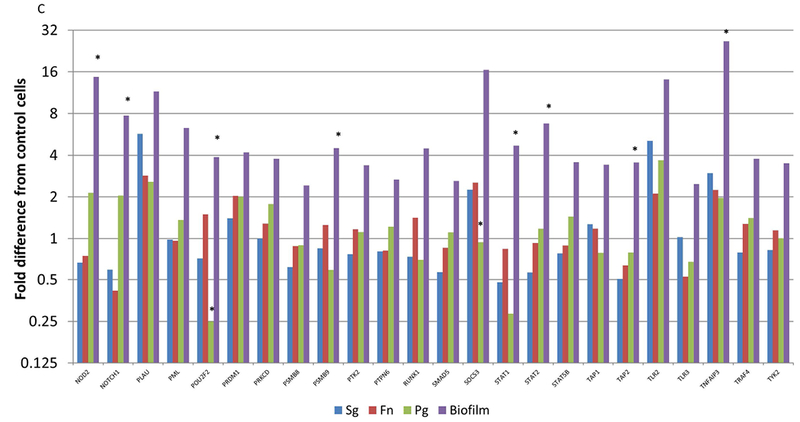
Identification of genes with at least 2-fold increased expression induced by biofilms compared to all of the planktonic species. Values denote the mean of 5 values for the biofilms and duplicates for the planktonic bacteria. Asterisk (*) denotes at least p<0.01 from the other conditions.
Table 1:
Major pathways of immunology gene upregulation by bacterial challenge of oral epithelial cells.
| PANTHER Pathways | H. sapiens Genome # | Biofilms # | Expected | Fold Enrichment | Raw P-value | FDR |
|---|---|---|---|---|---|---|
| Biofilm | ||||||
| JAK/STAT signaling pathway | 17 | 6 | .05 | >100 | 6.22E-11 | 2.03E-09 |
| Interferon-gamma signaling pathway | 34 | 6 | .10 | 57.59 | 2.27E-09 | 5.29E-08 |
| Toll receptor signaling pathway | 64 | 11 | .20 | 56.09 | 3.74E-16 | 3.04E-14 |
| B cell activation | 69 | 8 | .21 | 37.84 | 8.66E-11 | 2.35E-09 |
| Interleukin signaling pathway | 94 | 10 | .29 | 34.72 | 7.16E-13 | 2.92E-11 |
| p53 pathway feedback loops 2 | 49 | 3 | .15 | 19.98 | 5.40E-04 | 5.87E-03 |
| VEGF signaling pathway | 68 | 4 | .21 | 19.20 | 6.98E-05 | 8.75E-04 |
| T cell activation | 88 | 5 | .27 | 18.54 | 9.61E-06 | 1.31E-04 |
| PDGF signaling pathway | 142 | 8 | .44 | 18.39 | 1.82E-08 | 3.70E-07 |
| Apoptosis signaling pathway | 129 | 7 | .40 | 17.71 | 1.91E-07 | 2.83E-06 |
| Inflammation mediated by chemokine/cytokine signaling pathway | 286 | 15 | .88 | 17.12 | 1.41E-14 | 7.66E-13 |
| Angiogenesis | 169 | 8 | .52 | 15.45 | 6.60E-08 | 1.19E-06 |
| CCKR signaling map | 180 | 8 | .55 | 14.50 | 1.05E-07 | 1.71E-06 |
| RAS pathway | 76 | 3 | .23 | 12.88 | 1.82E-03 | 1.75E-02 |
| EGF receptor signaling pathway | 153 | 5 | .47 | 10.66 | 1.22E-04 | 1.42E-03 |
| Planktonic | ||||||
| Inflammation mediated by chemokine/cytokine signaling pathway | 286 | 5 | .22 | 23.18 | 1.81E-06 | 2.94E-04 |
Figure 6 provides a depiction of the group of genes (n=17) in which the response to one or more of the individual planktonic bacteria was greater than the biofilm challenge, or in many cases multiple planktonic bacteria elicited elevated responses that appeared to be decreased when the bacteria were in the biofilm structure. Table 1 provides a pathway analysis of the over-represented responses, which were limited to inflammation mediated chemokine/cytokine signaling pathways. This could be interpreted that these bacteria in biofilms, while upregulating a number of chemokines/cytokines appeared to “block” the increase in the expression of these genes, for example CCL5, CXCL10, CXCL11, ICAM1, IL23A, PLA2G2A, and PSMB5.
Figure 6:
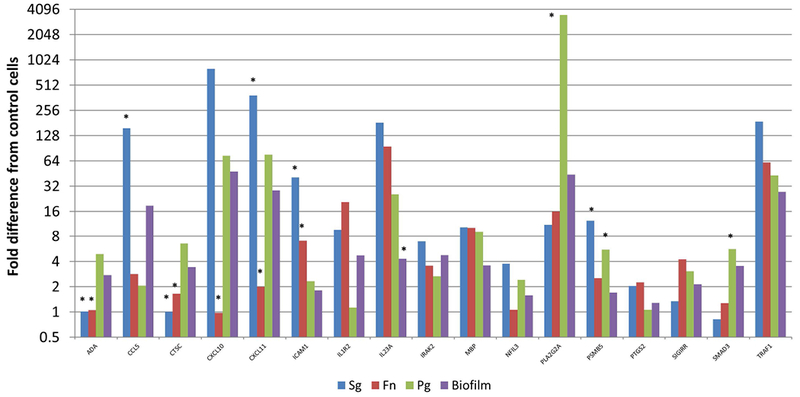
Identification of genes with at least 2-fold increased expression induced by one or more planktonic bacteria compared to the multispecies biofilm. Values denote the mean of 5 values for the biofilms and duplicates for the planktonic bacteria. Asterisk (*) denotes at least p<0.01 from the other conditions.
DISCUSSION
Epithelial cells that provide a mechanical barrier at mucosal surfaces are gaining a stronger recognition regarding their ability to respond to both the commensal microbiome and deleterious microbial insults, and are involved in regulating mucosal inflammatory and immune responses. In this study, we evaluated gene expression profiles of oral epithelial cells, specifically targeting genes that could be more directly involved in communicating regulatory signals to the inflammatory, innate, and adaptive immune system in the periodontium. The specific hypothesis to be tested was that distinct differences would be observed in response profiles of oral epithelial cells reacting to challenge with planktonic bacteria, representing the characteristics of detached and dispersed microorganisms throughout the oral cavity, and the response patterns to these species when in organized biofilms.
Numerous reports have documented responses of oral epithelial cells to bacterial challenge generally with a biased sampling of a very limited number of host response biomolecules 10,28–30. This study examined a broad array of genes and gene pathways that describe the capacity of epithelial cells to defend and communicate against a noxious microbial challenge. We identified that >98% of the basal mRNA signal within this array of 579 genes was accounted for by 181 genes. Interestingly, the message signal for basal epithelial cell responses and control reactions to the biofilm carrier (RPGL) within this immunology array showed that the top 20 gene signals (TGFB1, B2M, IL32, ITGB1, APP, CD59, ITGA6, CTNNB1, ITGA5, CD44, GPI, PSMB7, CD99, CD81, ETS1, FN1, BCAP31, CD9, CTSC, IL6ST) comprised >60% of the overall message for both conditions. In contrast, these 20 genes only accounted for 31%, 27%, 47%, and 55% of the total response message to the biofilm, S. gordonii, F. nucleatum, and P. gingivalis, respectively. This supports that the bacterial challenge was not only substantially increasing the level of selected genes, but triggering a much broader array of responses from the epithelial cells that could be engaged in host responses to the infection. Also of interest was the identification of some of these major responses, for example IL32, amyloid beta precursor protein (APP), PSMB7 (Proteasome Subunit Beta 7; component of the 20S core proteasome complex), ETS1 (ETS Proto-Oncogene 1, Transcription Factor), and IL6ST (interleukin 6 signal transducer) that have not been well described in the oral epithelium.
Biofilm challenge of the oral epithelial cells induced up regulation of a wide array of the immune response-related genes with >16-fold up regulation of cytokines/chemokines (IL32, IL8, IL1B, CCL20, CXCL2, CXCL1, CXCL11, CXCL10, CCL5, LIF). Interestingly, IL32 and LIF (leukemia inhibitory factor) have not been previously described in oral epithelial cell biology. IL32 has been implicated in the pathogenesis and progression of various chronic inflammatory disorders 31–33 and is induced by microbial ligands via TLR pathways 34. The IL32 gene leads to 9 splice variants and isoforms that have been shown to have varied activities under different cellular conditions 32,35,36. This molecule has been described as a pro-inflammatory cytokine that induces differentiation of monocytes to macrophages 37, as well as up-regulation of other pro-inflammatory cytokines in these cells 38. Recently, it was shown that IL-32 levels were elevated in saliva and GCF in periodontitis 39 and is consistent with elevated levels of IL-32 in gingival tissues in periodontitis 40. LIF (Leukemia inhibitory factor) regulates cellular differentiation, growth and inflammation 41. It is a member of the IL-6 family of cytokines with pro-inflammatory capabilities 42. This molecule has been shown to be produced by the epithelial cells in the lung and protect against bacterial pneumonia infections 42,43. However, the same ligand has been shown to induce the production of an array of pro-inflammatory molecules (eg. IL-1, IL-6, IL-8, inflammatory lipids) by gut 44, uterine 45, corneal 46, nasal airway and bronchial epithelial cells 47, and keratinocytes 41,48. This activity appears to occur through the LIF receptor and JAK1-STAT3 signaling pathway 45,46. Literature on its role in the oral mucosa is more limited with decreased levels in periodontitis GCF 49, and production by gingival fibroblasts in response to host pro-inflammatory signals 50. Thus, this report of the substantial transcription of the LIF gene by oral epithelial cells in response to biofilm challenge provides new information on this potentially important molecule in gingival tissue responses.
IL-23 activates STAT4 inducing IFNγ, preferentially targeting memory CD4+ T cells and promoting the production of various proinflammatory cytokines 51. This cytokine is directly linked to Th17 T cell functions and production of IL-17 52, and the IL-23/IL-17 axis has been reported as a major host response abnormality in leukocyte adhesion deficiency type 1 53 with a clear impact on expression of early onset severe periodontal disease 54. Information on this cytokine in periodontitis has been accumulating and is clearly interconnected with the importance of IL-17 in the inflammatory changes in periodontitis 55–58. Additional studies have shown that antigen-presenting cells (eg. macrophages, dendritic cells) challenged with P. gingivalis or Prevotella spp. demonstrated increases in IL-23 within the broader repertoire of inflammatory mediator responses 59–61, and P. endodontalis LPS elicited IL-23 linked to enhanced osteoclastogenesis 62, the cytokine cascade of granuloma tissues 63 and in responses of periodontal ligament cells to LPS 62. However, IL-23 in responses of oral epithelial cells is sparse. A single report describes IL-23 elevations in immortalized gingival keratinocytes following infection with P. gingivalis 61. The interesting findings with this important regulatory cytokine were its elevated expression with each of the planktonic species that was decreased substantially with the combined biofilms.
An interesting profile of the cytokine family of genes was also noted to be significantly elevated in the infected epithelial cells. IL8 was a major gene induced by the biofilms, albeit its constitutive production was rather low 24,64. CCL20 (MIP3a) is a chemokine for dendritic cells, and can recruit both Th17 and Treg cells to sites of inflammation. Previous studies have shown increased CCL20 expression by epithelial cells stimulated with oral bacteria 13,59,65 , and is elevated in periodontitis tissues 66,67 . CXCL1 (Groα) together with IL-8 are major chemotaxins for neutrophils. This chemokine is upregulated in gingival epithelial cells 68, and Ramage et al. 14 have shown that both CXCL1 and CXCL2 are expressed in the junctional epithelium potentially contributing to attempts to maintain homeostasis 69. Gingival fibroblasts were reported to produce CXCL11 (SCYB11; I-TAC) in response to challenge with muramydipeptide 70, while macrophages stimulated with P. gingivalis also upregulated this chemokine. Thus, with each of these cytokines/chemokines/signaling factors that were elevated in the oral epithelial cells there is a biological consideration regarding roles in the gingival communication of innate immune system processes.
CSF2 (colony stimulating factor 2; GMCSF) is a cytokine associated with functions of granulocytes and macrophages 71. It has been shown to be elevated in gingival tissues of periodontitis 72 and was suggested to drive MMP-12 production in diseased tissues 73. Interestingly, CSF2 was overexpressed by gingival epithelial cells following challenge with A. actinomycetemcomitans 74 and was one of the target cytokines that were increased in oral epithelial cell cultures treated with mono- and multi-species biofilms 14, particularly associated with a mixed species biofilm containing F. nucleatum and P. gingivalis. In this study, the biofilms upregulated this gene by 30-fold; however, of interest was that only S. gordonii (7.6-fold) and F. nucleatum (11.3-fold) in planktonic form stimulated expression of this important cell communication factor compared with P. gingivalis (−1.07-fold). Thus, the biofilms appear to be reflecting a synergistic stimulation of CSF2 when these species are organized into this type of structure.
Additionally a number of altered genes following biofilm challenge were directly related to control and regulation of the NF-κB pathway and as an activator of genes involved in both innate and acquired immune responses by binding to an interferon-stimulated response element (ISRE) in their promoters., including: TNFAIP3 [A20; 75], NFKBIZ (Iκbζ) 76–78, SOCS3 (suppressor of cytokine signaling 3) 79 and IRF1 [Interferon Regulatory Factor 1; 80]. While there exists some information on certain of these regulatory factors in periodontitis, their combined expression profiles by epithelial cells in response to biofilm challenge has not be described. TNFAIP3 has not been reported related to oral epithelial cell biology, some recent studies suggest a role in dampening osteoclastogenesis 81, and elevations in gingival tissue related to decreased periodontitis coupled with TLR9 activity 82. NFKBIZ regulates antimicrobial peptide production by epithelial cells 77,78 and has recently been confirmed as a major factor in epithelial cell functions, controlling communication signals with inflammatory/immune cells 83. In recent studies IRF1 showed a decreased expression in chronic periodontitis 84,85, although at the cellular level, how epithelial cells were affected has not be described. Finally, there is a robust literature on SOCS3 in periodontal disease, including the identification of this regulatory molecule in human periodontal tissues 86 and rodent models of periodontal disease 87–89, as well as responses of periodontal ligament cells 90 and osteoblasts 91. However, minimal data is available regarding the role of the inflammation regulatory molecule in the oral epithelium.
Comparison of the biofilm challenge for gene expression profiles with the individual planktonic bacteria provided a somewhat differential pattern with each bacterial species. Substantial overlap in up-regulated genes was noted between the biofilm and S. gordonii challenge. However, PSMB5, ICAM1, MBP, IL23A, CXCL11, CXCL10 and CCL5 were significantly increased with the S. gordonii challenge versus the biofilm containing this species. In addition, CXCL10 and CXCL11 were also increased by P. gingivalis infection. IL23A was the only gene that was substantially increased by all the planktonic bacteria compared to the biofilms. This cassette of genes that appears to be affected most by challenge with the planktonic bacteria comprises some related activities in control of local immune response. MBP (myelin basic peptide) is involved in signaling pathways in T-cells and can induce T-cell proliferation. This is coupled with CXCL11 that is chemotactic for interleukin-activated T-cells, CCL5 that is a chemo attractant for monocytes and memory T-helper cells, CXCL10 that binds to CXCR3 and stimulates monocytes, natural killer and T-cell migration, and modulation of adhesion molecule expression, such as ICAM1. ICAM1 is expressed on endothelial and immune system cells engaging integrins of the CD11/CD18 type that are implicated in interactions with monocytes, macrophages and granulocytes, as well as binding to the iC3b fragment of the third complement component. Finally, PSMB5 is a component of the core proteasome complex involved in the proteolytic degradation of most intracellular proteins. This type of proteolysis is necessary for generation of a subset of MHC class I-presented antigenic peptides in adaptive immune responses. Thus, it appears that this group of genes depicts the potential for this commensal bacterium to communicate to both innate and adaptive immune mechanisms to potentially regulate the symbiotic relationship with the host.
Also of interest was the distribution of genes whose levels were substantially decreased by treatment of the epithelial cells with the planktonic or biofilm bacteria. S. gordonii challenged decreased 24 genes by 2-fold or greater versus the basal cell levels, with P. gingivalis down-regulating only 11 genes and F. nucleatum only 2 genes. Interestingly, there was minimal overlap in this gene expression inhibition across the planktonic species. In contrast, of these 579 immune response associated genes, only TGFBR1 was down-regulated by the biofilms at approximately 2-fold.
This study provided a robust assessment of major gene expression patterns of host response system biomolecules associated with the biology of the epithelium. Importantly, the breadth of gene expression and up-regulation following challenge with the individual planktonic species and the multispecies biofilms emphasized a critical role for epithelial cell responses in the periodontium enabling both direct interactions with the microbial insult, as well as a sophisticated communication and regulatory system for the inflammatory and immune infiltrate to reestablish homeostasis. More generally, the pathway analysis did provide some insight into the features of the pathogenic biofilm stimulation of a breadth of epithelial cell responses that signal the immune system. These pathways included chemokines/cytokines that would communicate effective host responses to T cells, B cells, and endothelial cells, as well as controlling cell behavior including survival and replication. Nevertheless, limitations of these types of in vitro studies are the capacity to truly model the complex microbial biofilms that occur in situ, as well as fully understanding the dynamics of interactions between the bacterial biofilms and individual epithelial cells that are critical for maintaining homeostasis. The character of these cellular responses in health, and the changes that occur with disease initiation still remain to be fully elucidated. The data also identified some unique gene profiles for the oral epithelial cells including IL32 and LIF as cytokines that have not been linked to major responses of these cells to microbial challenge. Thus, the potential for a unique role of these host response factors to pathogenic biofilms may provide additional insights into the underlying biologic mechanisms of the chronic inflammation in periodontitis.
ACKNOWLEDGEMENTS
We want to acknowledge support from U.S.P.H.S. grant P30 GM103538 from the National Institute of General Medical Sciences and the Center for Oral Health Research in the University of Kentucky College of Dentistry. We also thank Dr. Kuey Chen in the Department of Pharmacology, College of Medicine, University of Kentucky for support with the NanoString analyses. The authors explicitly state no conflicts of interest in connection with this report.
REFERENCES
- 1.Takiishi T, Fenero CIM, Camara NOS. Intestinal barrier and gut microbiota: Shaping our immune responses throughout life. Tissue Barriers. 2017;5(4):e1373208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Partida-Rodriguez O, Serrano-Vazquez A, Nieves-Ramirez ME, et al. Human Intestinal Microbiota: Interaction Between Parasites and the Host Immune Response. Arch Med Res. 2017;48(8):690–700. [DOI] [PubMed] [Google Scholar]
- 3.Ahluwalia B, Magnusson MK, Ohman L. Mucosal immune system of the gastrointestinal tract: maintaining balance between the good and the bad. Scand J Gastroenterol. 2017;52(11):1185–1193. [DOI] [PubMed] [Google Scholar]
- 4.Ghosh SK, Feng Z, Fujioka H, Lux R, McCormick TS, Weinberg A. Conceptual Perspectives: Bacterial Antimicrobial Peptide Induction as a Novel Strategy for Symbiosis with the Human Host. Frontiers in microbiology. 2018;9:302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McCormick TS, Weinberg A. Epithelial cell-derived antimicrobial peptides are multifunctional agents that bridge innate and adaptive immunity. Periodontol 2000. 2010;54(1):195–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diamond G, Beckloff N, Weinberg A, Kisich KO. The roles of antimicrobial peptides in innate host defense. Current pharmaceutical design. 2009;15(21):2377–2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pardo-Camacho C, Gonzalez-Castro AM, Rodino-Janeiro BK, Pigrau M, Vicario M. Epithelial immunity: priming defensive responses in the intestinal mucosa. American journal of physiology Gastrointestinal and liver physiology. 2018;314(2):G247–G255. [DOI] [PubMed] [Google Scholar]
- 8.Andrews C, McLean MH, Durum SK. Cytokine Tuning of Intestinal Epithelial Function. Frontiers in immunology. 2018;9:1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buckley KM, Rast JP. An Organismal Model for Gene Regulatory Networks in the Gut-Associated Immune Response Frontiers in immunology. 2017;8:1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ji S, Choi Y. Innate immune response to oral bacteria and the immune evasive characteristics of periodontal pathogens. J Periodontal Implant Sci. 2013;43(1):3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Montreekachon P, Nongparn S, Sastraruji T, et al. Favorable interleukin-8 induction in human gingival epithelial cells by the antimicrobial peptide LL-37. Asian Pacific journal of allergy and immunology / launched by the Allergy and Immunology Society of Thailand. 2014;32(3):251–260. [DOI] [PubMed] [Google Scholar]
- 12.Dickinson BC, Moffatt CE, Hagerty D, et al. Interaction of oral bacteria with gingival epithelial cell multilayers. Molecular oral microbiology. 2011;26(3):210–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Milward MR, Chapple IL, Wright HJ, Millard JL, Matthews JB, Cooper PR. Differential activation of NF-kappaB and gene expression in oral epithelial cells by periodontal pathogens. Clin Exp Immunol. 2007;148(2):307–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramage G, Lappin DF, Millhouse E, et al. The epithelial cell response to health and disease associated oral biofilm models. J Periodontal Res. 2017;52(3):325–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peyyala R, Kirakodu SS, Novak KF, Ebersole JL. Oral epithelial cell responses to multispecies microbial biofilms. J Dent Res. 2013;92(3):235–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peyyala R, Ebersole JL. Multispecies biofilms and host responses: “discriminating the trees from the forest”. Cytokine. 2013;61(1):15–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peyyala R, Kirakodu SS, Ebersole JL, Novak KF. Novel model for multispecies biofilms that uses rigid gas-permeable lenses. Applied and environmental microbiology. 2011;77(10):3413–3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guilhen C, Forestier C, Balestrino D. Biofilm dispersal: multiple elaborate strategies for dissemination of bacteria with unique properties. Molecular microbiology. 2017;105(2):188–210. [DOI] [PubMed] [Google Scholar]
- 19.Barraud N, Kjelleberg S, Rice SA. Dispersal from Microbial Biofilms. Microbiol Spectr. 2015;3(6). [DOI] [PubMed] [Google Scholar]
- 20.Kostakioti M, Hadjifrangiskou M, Hultgren SJ. Bacterial biofilms: development, dispersal, and therapeutic strategies in the dawn of the postantibiotic era. Cold Spring Harbor perspectives in medicine. 2013;3(4):a010306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peyyala R, Kirakodu SS, Ebersole JL, Novak KF. Novel model for multispecies biofilms that uses rigid gas-permeable lenses. Applied and environmental microbiology. 2011;77(10):3413–3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rheinwald JG, Hahn WC, Ramsey MR, et al. A Two-Stage, p16INK4A- and p53-Dependent Keratinocyte Senescence Mechanism That Limits Replicative Potential Independent of Telomere Status. Mol Cell Biol. 2002;22(14):5157–5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peyyala R, Kirakodu S, Novak KF, Ebersole JL. Epithelial interleukin-8 responses to oral bacterial biofilms. Clinical and vaccine immunology : CVI. 2011;18(10):1770–1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peyyala R, Kirakodu S, Novak K, Ebersole JL. Epithelial interleukin-8 responses to oral bacterial biofilms. Clinical and Vaccine Immunology. 2011;18(10):1770–1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Al-Attar A, Alimova Y, Kirakodu S, et al. Activation of Notch-1 in oral epithelial cells by P. gingivalis triggers the expression of the antimicrobial protein PLA2-IIA. Mucosal Immunol. 2018;11(4):1047–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Al-Attar A, Alimova Y, Kirakodu S, et al. Activation of Notch-1 in oral epithelial cells by P. gingivalis triggers the expression of the antimicrobial protein PLA2-IIA. Mucosal Immunol. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thomas PD, Campbell MJ, Kejariwal A, et al. PANTHER: a library of protein families and subfamilies indexed by function. Genome research. 2003;13(9):2129–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benakanakere M, Kinane DF. Innate cellular responses to the periodontal biofilm. Frontiers of oral biology. 2012;15:41–55. [DOI] [PubMed] [Google Scholar]
- 29.Andrian E, Grenier D, Rouabhia M. Porphyromonas gingivalis-epithelial cell interactions in periodontitis. J Dent Res. 2006;85(5):392–403. [DOI] [PubMed] [Google Scholar]
- 30.Amano A Molecular interaction of Porphyromonas gingivalis with host cells: implication for the microbial pathogenesis of periodontal disease. J Periodontol. 2003;74(1):90–96. [DOI] [PubMed] [Google Scholar]
- 31.Damen MS, Popa CD, Netea MG, Dinarello CA, Joosten LA. Interleukin-32 in chronic inflammatory conditions is associated with a higher risk of cardiovascular diseases. Atherosclerosis. 2017. [DOI] [PubMed] [Google Scholar]
- 32.Khawar MB, Abbasi MH, Sheikh N. IL-32: A Novel Pluripotent Inflammatory Interleukin, towards Gastric Inflammation, Gastric Cancer, and Chronic Rhino Sinusitis. Mediators Inflamm. 2016;2016:8413768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Joosten LA, Heinhuis B, Netea MG, Dinarello CA. Novel insights into the biology of interleukin-32. Cellular and molecular life sciences : CMLS. 2013;70(20):3883–3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang L, Che C, Lin J, Liu K, Li DQ, Zhao G. TLR-mediated induction of proinflammatory cytokine IL-32 in corneal epithelium. Current eye research. 2013;38(6):630–638. [DOI] [PubMed] [Google Scholar]
- 35.Jeong HJ, Shin SY, Oh HA, Kim MH, Cho JS, Kim HM. IL-32 up-regulation is associated with inflammatory cytokine production in allergic rhinitis. The Journal of pathology. 2011;224(4):553–563. [DOI] [PubMed] [Google Scholar]
- 36.Imaeda H, Andoh A, Aomatsu T, et al. A new isoform of interleukin-32 suppresses IL-8 mRNA expression in the intestinal epithelial cell line HT-29. Molecular medicine reports. 2011;4(3):483–487. [DOI] [PubMed] [Google Scholar]
- 37.Netea MG, Lewis EC, Azam T, et al. Interleukin-32 induces the differentiation of monocytes into macrophage-like cells. Proc Natl Acad Sci U S A. 2008;105(9):3515–3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hong J, Bae S, Kang Y, et al. Suppressing IL-32 in monocytes impairs the induction of the proinflammatory cytokines TNFalpha and IL-1beta. Cytokine. 2010;49(2):171–176. [DOI] [PubMed] [Google Scholar]
- 39.Ongoz Dede F, Balli U, Bozkurt Dogan S, Guven B. Interleukin-32 levels in gingival crevicular fluid and saliva of patients with chronic periodontitis after periodontal treatment. J Periodontal Res. 2017;52(3):397–407. [DOI] [PubMed] [Google Scholar]
- 40.Ouhara K, Kawai T, Silva MJ, et al. Expression levels of novel cytokine IL-32 in periodontitis and its role in the suppression of IL-8 production by human gingival fibroblasts stimulated with Porphyromonas gingivalis. Journal of oral microbiology. 2012;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paglia D, Kondo S, Ng KM, Sauder DN, McKenzie RC. Leukaemia inhibitory factor is expressed by normal human keratinocytes in vitro and in vivo. The British journal of dermatology. 1996;134(5):817–823. [PubMed] [Google Scholar]
- 42.Quinton LJ, Mizgerd JP, Hilliard KL, Jones MR, Kwon CY, Allen E. Leukemia inhibitory factor signaling is required for lung protection during pneumonia. J Immunol. 2012;188(12):6300–6308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Traber KE, Symer EM, Allen E, et al. Myeloid-epithelial cross talk coordinates synthesis of the tissue-protective cytokine leukemia inhibitory factor during pneumonia. Am J Physiol Lung Cell Mol Physiol. 2017;313(3):L548–L558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guimbaud R, Abitbol V, Bertrand V, et al. Leukemia inhibitory factor involvement in human ulcerative colitis and its potential role in malignant course. Eur Cytokine Netw. 1998;9(4):607–612. [PubMed] [Google Scholar]
- 45.Pawar S, Starosvetsky E, Orvis GD, Behringer RR, Bagchi IC, Bagchi MK. STAT3 regulates uterine epithelial remodeling and epithelial-stromal crosstalk during implantation. Mol Endocrinol. 2013;27(12):1996–2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu X, Tseng SC, Zhang MC, et al. LIF-JAK1-STAT3 signaling delays contact inhibition of human corneal endothelial cells. Cell cycle. 2015;14(8):1197–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fayon M, Lacoste-Rodrigues A, Barat P, et al. Nasal airway epithelial cell IL-6 and FKBP51 gene expression and steroid sensitivity in asthmatic children. PLoS One. 2017;12(5):e0177051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parodi A, Sanguineti R, Catalano M, et al. A comparative study of leukaemia inhibitory factor and interleukin-1alpha intracellular content in a human keratinocyte cell line after exposure to cosmetic fragrances and sodium dodecyl sulphate. Toxicology letters. 2010;192(2):101–107. [DOI] [PubMed] [Google Scholar]
- 49.Becerik S, Ozturk VO, Atmaca H, Atilla G, Emingil G. Gingival crevicular fluid and plasma acute-phase cytokine levels in different periodontal diseases. J Periodontol. 2012;83(10):1304–1313. [DOI] [PubMed] [Google Scholar]
- 50.Souza PP, Palmqvist P, Lundberg P, et al. Interleukin-4 and interleukin-13 inhibit the expression of leukemia inhibitory factor and interleukin-11 in fibroblasts. Molecular immunology. 2012;49(4):601–610. [DOI] [PubMed] [Google Scholar]
- 51.Floss DM, Schröder J, Franke M, Scheller J. Insights into IL-23 biology: from structure to function. Cytokine & growth factor reviews. 2015;26(5):569–578. [DOI] [PubMed] [Google Scholar]
- 52.Jain R, Chen Y, Kanno Y, et al. Interleukin-23-Induced Transcription Factor Blimp-1 Promotes Pathogenicity of T Helper 17 Cells. Immunity. 2016;44(1):131–142. [DOI] [PubMed] [Google Scholar]
- 53.Moutsopoulos NM, Zerbe CS, Wild T, et al. Interleukin-12 and Interleukin-23 Blockade in Leukocyte Adhesion Deficiency Type 1. N Engl J Med. 2017;376(12):1141–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hajishengallis G, Moutsopoulos NM. Role of bacteria in leukocyte adhesion deficiency-associated periodontitis. Microbial pathogenesis. 2016;94:21–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sadeghi R, Sattari M, Dehghan F, Akbari S. Interleukin-17 and interleukin-23 levels in gingival crevicular fluid of patients with chronic and aggressive periodontitis. Cent Eur J Immunol. 2018;43(1):76–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cifcibasi E, Koyuncuoglu C, Ciblak M, et al. Evaluation of Local and Systemic Levels of Interleukin-17, Interleukin-23, and Myeloperoxidase in Response to Periodontal Therapy in Patients with Generalized Aggressive Periodontitis. Inflammation. 2015;38(5):1959–1968. [DOI] [PubMed] [Google Scholar]
- 57.Menegat JS, Lira-Junior R, Siqueira MA, et al. Cytokine expression in gingival and intestinal tissues of patients with periodontitis and inflammatory bowel disease: An exploratory study. Arch Oral Biol. 2016;66:141–146. [DOI] [PubMed] [Google Scholar]
- 58.Liukkonen J, Gursoy UK, Pussinen PJ, Suominen AL, Kononen E. Salivary Concentrations of Interleukin (IL)-1beta, IL-17A, and IL-23 Vary in Relation to Periodontal Status. J Periodontol. 2016;87(12):1484–1491. [DOI] [PubMed] [Google Scholar]
- 59.Larsen JM. The immune response to Prevotella bacteria in chronic inflammatory disease. Immunology. 2017;151(4):363–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hou Y, Yu H, Liu X, et al. Gingipain of Porphyromonas gingivalis manipulates M1 macrophage polarization through C5a pathway. In Vitro Cell Dev Biol Anim. 2017;53(7):593–603. [DOI] [PubMed] [Google Scholar]
- 61.Glowczyk I, Wong A, Potempa B, et al. Inactive Gingipains from P. gingivalis Selectively Skews T Cells toward a Th17 Phenotype in an IL-6 Dependent Manner. Frontiers in cellular and infection microbiology. 2017;7:140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ma N, Yang D, Okamura H, et al. Involvement of interleukin23 induced by Porphyromonas endodontalis lipopolysaccharide in osteoclastogenesis. Molecular medicine reports. 2017;15(2):559–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Araujo-Pires AC, Francisconi CF, Biguetti CC, et al. Simultaneous analysis of T helper subsets (Th1, Th2, Th9, Th17, Th22, Tfh, Tr1 and Tregs) markers expression in periapical lesions reveals multiple cytokine clusters accountable for lesions activity and inactivity status. Journal of applied oral science : revista FOB. 2014;22(4):336–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Peyyala R, Kirakodu SS, Novak KF, Ebersole JL. Oral microbial biofilm stimulation of epithelial cell responses. Cytokine. 2012;58(1):65–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huynh J, Scholz GM, Aw J, et al. IRF6 Regulates the Expression of IL-36gamma by Human Oral Epithelial Cells in Response to Porphyromonas gingivalis. J Immunol. 2016;196(5):2230–2238. [DOI] [PubMed] [Google Scholar]
- 66.Souto GR, Queiroz CM Jr, Costa FO, Mesquita RA. Relationship between chemokines and dendritic cells in human chronic periodontitis. J Periodontol. 2014;85(10):1416–1423. [DOI] [PubMed] [Google Scholar]
- 67.Luo Z, Wang H, Chen J, Kang J, Sun Z, Wu Y. Overexpression and Potential Regulatory Role of IL-17F in Pathogenesis of Chronic Periodontitis. Inflammation. 2015;38(3):978–986. [DOI] [PubMed] [Google Scholar]
- 68.Chen SC, Constantinides C, Kebschull M, Papapanou PN. MicroRNAs Regulate Cytokine Responses in Gingival Epithelial Cells. Infect Immun. 2016;84(12):3282–3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tsukamoto Y, Usui M, Yamamoto G, et al. Role of the junctional epithelium in periodontal innate defense and homeostasis. J Periodontal Res. 2012;47(6):750–757. [DOI] [PubMed] [Google Scholar]
- 70.Hosokawa Y, Hosokawa I, Ozaki K, Nakae H, Matsuo T. TNFSF14 coordinately enhances CXCL10 and CXCL11 productions from IFN-gamma-stimulated human gingival fibroblasts. Molecular immunology. 2010;47(4):666–670. [DOI] [PubMed] [Google Scholar]
- 71.Fjell CD, Thair S, Hsu JL, Walley KR, Russell JA, Boyd J. Cytokines and signaling molecules predict clinical outcomes in sepsis. PLoS One. 2013;8(11):e79207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sun Y, Guo QM, Liu DL, Zhang MZ, Shu R. In vivo expression of Toll-like receptor 2, Toll-like receptor 4, CSF2 and LY64 in Chinese chronic periodontitis patients. Oral Dis. 2010;16(4):343–350. [DOI] [PubMed] [Google Scholar]
- 73.Bjornfot Holmstrom S, Clark R, Zwicker S, et al. Gingival Tissue Inflammation Promotes Increased Matrix Metalloproteinase-12 Production by CD200R(low) Monocyte-Derived Cells in Periodontitis. J Immunol. 2017;199(12):4023–4035. [DOI] [PubMed] [Google Scholar]
- 74.Umeda JE, Demuth DR, Ando ES, Faveri M, Mayer MP. Signaling transduction analysis in gingival epithelial cells after infection with Aggregatibacter actinomycetemcomitans. Mol Oral Microbiol. 2012;27(1):23–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dang RJ, Yang YM, Zhang L, et al. A20 plays a critical role in the immunoregulatory function of mesenchymal stem cells. Journal of cellular and molecular medicine. 2016;20(8):1550–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.MaruYama T, Kobayashi S, Ogasawara K, Yoshimura A, Chen W, Muta T. Control of IFN-gamma production and regulatory function by the inducible nuclear protein IkappaB-zeta in T cells. Journal of leukocyte biology. 2015;98(3):385–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cowland JB, Muta T, Borregaard N. IL-1beta-specific up-regulation of neutrophil gelatinase-associated lipocalin is controlled by IkappaB-zeta. J Immunol. 2006;176(9):5559–5566. [DOI] [PubMed] [Google Scholar]
- 78.Kao CY, Kim C, Huang F, Wu R. Requirements for two proximal NF-kappaB binding sites and IkappaB-zeta in IL-17A-induced human beta-defensin 2 expression by conducting airway epithelium. J Biol Chem. 2008;283(22):15309–15318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Carow B, Rottenberg ME. SOCS3, a Major Regulator of Infection and Inflammation. Frontiers in immunology. 2014;5:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yarilina A, Park-Min KH, Antoniv T, Hu X, Ivashkiv LB. TNF activates an IRF1-dependent autocrine loop leading to sustained expression of chemokines and STAT1-dependent type I interferon-response genes. Nat Immunol. 2008;9(4):378–387. [DOI] [PubMed] [Google Scholar]
- 81.Hong JY, Bae WJ, Yi JK, Kim GT, Kim EC. Anti-inflammatory and anti-osteoclastogenic effects of zinc finger protein A20 overexpression in human periodontal ligament cells. J Periodontal Res. 2016;51(4):529–539. [DOI] [PubMed] [Google Scholar]
- 82.Crump KE, Oakley JC, Xia-Juan X, et al. Interplay of Toll-Like Receptor 9, Myeloid Cells, and Deubiquitinase A20 in Periodontal Inflammation. Infect Immun. 2017;85(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sundaram K, Rahman MA, Mitra S, et al. IkappaBzeta Regulates Human Monocyte Pro-Inflammatory Responses Induced by Streptococcus pneumoniae. PLoS One. 2016;11(9):e0161931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tanaka MH, Giro EM, Cavalcante LB, et al. Expression of interferon-gamma, interferon-alpha and related genes in individuals with Down syndrome and periodontitis. Cytokine. 2012;60(3):875–881. [DOI] [PubMed] [Google Scholar]
- 85.Nepomuceno R, Villela BS, Corbi SC, et al. Dyslipidemia rather than Type 2 Diabetes Mellitus or Chronic Periodontitis Affects the Systemic Expression of Pro- and Anti-Inflammatory Genes. Mediators Inflamm. 2017;2017:1491405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Menezes R, Garlet TP, Trombone AP, et al. The potential role of suppressors of cytokine signaling in the attenuation of inflammatory reaction and alveolar bone loss associated with apical periodontitis. J Endod. 2008;34(12):1480–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.de Souza JA, Nogueira AV, de Souza PP, Cirelli JA, Garlet GP, Rossa C Jr Expression of suppressor of cytokine signaling 1 and 3 in ligature-induced periodontitis in rats. Arch Oral Biol. 2011;56(10):1120–1128. [DOI] [PubMed] [Google Scholar]
- 88.Papathanasiou E, Kantarci A, Konstantinidis A, Gao H, Van Dyke TE. SOCS-3 Regulates Alveolar Bone Loss in Experimental Periodontitis. J Dent Res. 2016;95(9):1018–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.de Vries TJ, Andreotta S, Loos BG, Nicu EA. Genes Critical for Developing Periodontitis: Lessons from Mouse Models. Frontiers in immunology. 2017;8:1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fukushima A, Kajiya H, Izumi T, Shigeyama C, Okabe K, Anan H. Pro-inflammatory cytokines induce suppressor of cytokine signaling-3 in human periodontal ligament cells. J Endod. 2010;36(6):1004–1008. [DOI] [PubMed] [Google Scholar]
- 91.Gao A, Kantarci A, Herrera BS, Gao H, Van Dyke TE. A critical role for suppressors of cytokine signaling 3 in regulating LPS-induced transcriptional activation of matrix metalloproteinase-13 in osteoblasts. PeerJ. 2013;1:e51. [DOI] [PMC free article] [PubMed] [Google Scholar]


