Abstract
Mutations in the polymerase basic 2 (PB2) gene of avian influenza viruses are important signatures for their adaptation to mammalian hosts. Various adaptive mutations have been identified around the 627 and nuclear localization sequence (NLS) domains of PB2 protein, and these mutations contribute to the replicative ability of avian influenza viruses. However, few studies have focused on adaptive mutations in other regions of PB2. In this study, we investigated the functional roles of the D253N mutation in PB2 in an H9N2 virus. This mutation was found to affect an amino acid residue in the middle domain of the PB2 protein. The virus with the D253N mutation showed higher polymerase activity and transiently increased viral replication in human cells. However, the mutant did not show significant differences in viral replication in the respiratory tract of mice upon infection. Our results supported that the D253N mutation in the middle domain of PB2, similar to mutations at the 627 and NLS domains, specifically contributed to the replication of avian influenza viruses in human cells.
Keywords: Avian influenza virus, Mammalian adaptation, D253N, Polymerase basic 2 (PB2), H9N2
Introduction
Outbreaks of pathogenic avian influenza virus in humans are associated with high fatality rates and have become a serious public health issue. These viruses can be directly transmitted from avian species to humans and are associated with mortality rates of around 30%–50% among infected patients (Li and Cao 2017). Most of these patients exhibit primary viral pneumonia, with some progressing to acute respiratory distress syndrome (Liem et al.2009; Gao et al.2013). Although the pathogenic mechanisms of avian influenza viruses in human are still not clear, these viruses have been found to replicate efficiently in the lower respiratory tract, as supported by the results of clinical, in vitro, ex vivo, and in vivo studies (de Jong et al.2006; Mok et al.2013a, b; Chan et al.2013; Zhou et al.2013). Moreover, hyperinduction of pro-inflammatory cytokines, which is a hallmark of the infection, is frequently found in patients or animals infected by mammalian-adapted avian influenza viruses (de Jong et al. 2006; Mok et al.2013a, b; Zhou et al.2013; Perrone et al.2008). Lung epithelial cells and macrophages have also been shown to play key roles in the pathogenesis of avian influenza viruses because they are both primary cell targets of these viruses and are the main sources of many pro-inflammatory cytokines upon infection (Cheung et al.2002; Chan et al.2005).
Avian influenza viruses, such as H5N1, H7N9, and H9N2, have been frequently isolated from virus-infected patients. Genetic analyses have shown that specific mutations in the polymerase basic 2 (PB2) gene of these viruses are important signatures of their adaptation to mammalian hosts. Various studies have demonstrated that these mutations, which have mostly been identified around the 627 and nuclear localization sequence (NLS) domains of PB2 protein, contribute to the virulence of the viruses by increasing viral replication efficiency or overwhelming immune responses (Mok et al.2009, 2013a, b; Hatta et al.2001, 2007). In addition to two well-studied adaptive mutations, E627K and D701N, our previous study identified two mutations, D253N and Q591K, which occurred simultaneously in the PB2 gene of an H9N2 virus after serial passages in a mammalian Madin-Darby Canine Kidney (MDCK) cell line (Mok et al.2011). Although we and others have shown that the Q591K mutation (also located in the 627 domain) in this virus contributes to pathogenesis in our in vitro and in vivo models, no systemic investigations have described the roles of the PB2 D253N mutation (located in the middle protein domain) (Hatta et al.2007; Wang et al.2016).
Accordingly, in this study, we further investigated the role of the D253N mutation in PB2 in mammalian hosts when introduced into the H9N2 virus.
Materials and Methods
Cells
To obtain primary human macrophages, blood mononuclear cells from healthy donors (Hong Kong Red Cross Blood Transfusion Service) were separated by Ficoll-Paque centrifugation. Monocytes were purified by the adherence method and differentiated into macrophages as previously described (Mok et al.2009; Wang et al.2016). Normal human bronchial epithelial (NHBE) cells were purchased from Lonza. The cells were seeded at 1 × 105 cells/well in 24-well plates and differentiated for 21 days with an air–liquid interface, as previously described (Wang et al.2016). Human embryonic kidney 293T and MDCK cells were maintained in Eagle’s minimal essential medium containing 10% fetal calf serum and antibiotics.
Polymerase Activity Assay
A single mutation at D253N was introduced into the PB2 plasmid of the A/Duck/Hong Kong/Y280/97 (H9N2/Y280) virus using a point mutation kit (Roche, USA). The PB2 plasmid was transferred into 293T cells monolayers together with PB1, PA, and NP plasmids of the H9N2/Y280 virus as well as the luciferase reporter plasmid (pluci) and the internal control plasmid (phRL-CMV). After 24 h of incubation, cell extracts were prepared in 500 μL lysis buffer. The luciferase levels were assayed with a Luciferase Assay System (Promega, Madison, WI, USA) and detected using a luminometer.
Generation of Recombinant Viruses
Eight plasmids containing the full genome of the H9N2/Y280 virus were transfected into 293T cells using TransIT for 48 h. Supernatants were then inoculated into embryonic eggs for 48 h. The titer of the viruses was determined by plaque forming assays in MDCK cells.
In Vitro Experiments
Primary human macrophages or primary human bronchial epithelial cells were infected with H9N2/Y280 or the PB2 mutant at a multiplicity of infection (MOI) of 2 or 0.01 according to the indicated conditions. Total RNA or supernatants were collected at 3, 6, 24, 48, and 72 h postinfection. Evidence of viral replication from the supernatants of the infected cells was determined according to the tissue culture infectious dose 50 (TCID50) in MDCK cells. Induction of cytokine mRNAs and proteins was determined by quantitative reverse transcription-polymerase chain reaction (PCR) and bead-based enzyme-linked immunosorbent assay (ELISA; Biolegend), respectively.
Animal Experiments
Eight 6- to 8-week-old BALB/c mice per group were inoculated intranasally with recombinant H9N2 viruses with or without the D253N mutation in PB2. Lethality and weight loss in the infected mice were recorded for 14 days. In the next experiment, three mice per group infected by either the wild-type or mutant PB2 were sacrificed on days 3 and 6 after infection to compare cytokine induction and viral replication in the lungs. The brain, liver, and lungs were isolated and homogenized with 1 mL phosphate-buffered saline (PBS). The supernatant was collected after centrifugation, and the levels of cytokines and viral replication were determined by bead-based ELISA (Biolegend) and TCID50 assays, respectively. Nasal wash was collected by inoculating 200 μL PBS into the noses of the mice.
This study protocol was carried out in strict accordance with the recommendations and approval of the Committee on the Use of Live Animals in Teaching and Research of the University of Hong Kong (approval no.: CULATR 2270-10). Humane endpoints for animal experiments and methods were undertaken to minimize potential pain and distress. All animals were euthanized using pentobarbital (200 mg/kg, intravenous injection) at the end of the experiments or once they were severely sick and showed more than one of the following signs (score > 1, as one sign = 1): loss of weight more than 30%, respiratory signs, depression, diarrhea, cyanosis of the exposed skin, edema of the face and head, and neurological signs. No mice fulfilled the above criteria or were euthanized before the end of the experiment.
Quantitative Analysis of Cytokine Levels
Expression levels of tumor necrosis factor (TNF)-α, macrophage inflammatory protein (MIP)-1α, MIP-1β, monocyte chemotactic protein (MCP)-1, MCP-3, and interferon-γ-induced protein-10 (IP-10) from the samples were quantitatively determined by flow cytometry-based immunoassay (Biolegend). Twenty-five microliters of each sample was processed according to the manufacturer’s protocol. The amounts of cytokines (pg/mL) in the samples were determined using a BD LSRII (BD Bioscience) and was calculated using the software provided by Biolegend.
Statistical Analysis
The statistical significance of differences between experimental groups was determined using unpaired, parametric Student’s t tests. Differences with P values less than 0.05 were considered significant.
Results
The H9N2 Virus with the D253N Mutation in PB2 Showed Enhanced Polymerase Activity and Transiently Increased Viral Replication in Human Cells
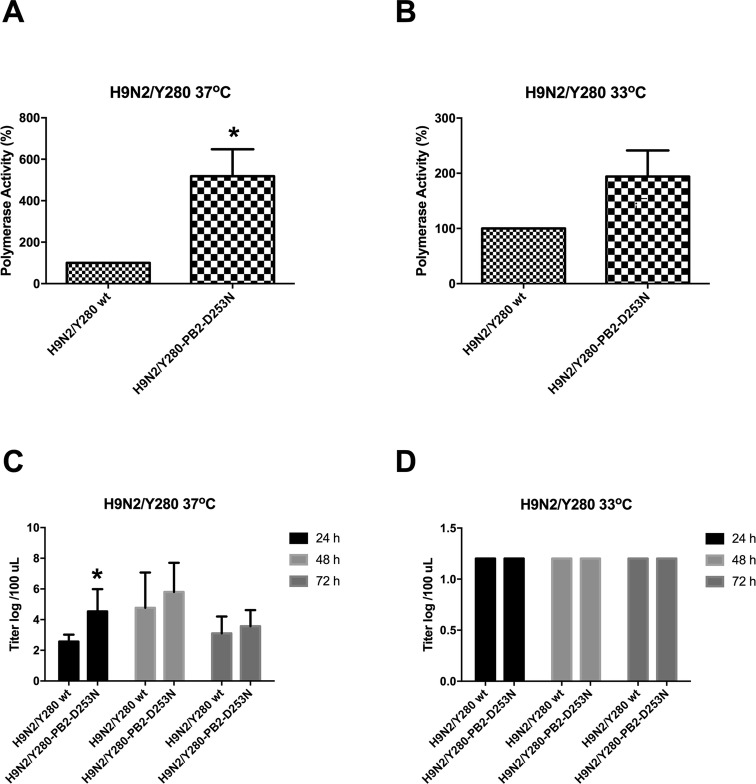
Our previous studies showed that the amino acid substitution D253N in the PB2 gene of an avian influenza virus H9N2/G1 resulted in increased polymerase activity and transiently higher viral replication in human cells (Mok et al.2011). To determine whether these observations were strain-specific and occurred in one virus strain, we further examined the functions of this mutation in another virus, H9N2/Y280, which belongs to another representative lineage of the H9N2 subtype. We compared the activity of the polymerase complex between wild-type H9N2/Y280 (wt) virus and the mutant virus harboring the D253N substitution in PB2 in a minigenome reporter assay using human 293T cells. Compared with the polymerase activity of the wt H9N2/Y280, the D253N mutant PB2 showed higher polymerase activity at 37 °C (temperature of the lower respiratory tract) but not at 33 °C (temperature of the upper respiratory tract) in 293T cells (Fig. 1A, 1B).
Fig. 1.
The polymerase activity and virus replication of H9N2/Y280 is enhanced by the D253N mutant in PB2. 293T cells were transfected with plasmids containing H9N2/Y280 PB2, PB1, PA, and NP genes plus a control luciferase reporter plasmid and a viral untranslated region-driven luciferase reporter plasmid. Transfected cells were cultured at A 37 °C and B 33 °C for 24 h, and luciferase activity was assayed in cell extracts. Results are the averages of three experiments. The values were statistical analyzed by two-tailed, paired t-tests. *P < 0.05. Primary NHBE cells were infected at an MOI of 0.01 with H9N2/Y280 and the PB2 mutant. The viral titers were measured from the supernatants at 24, 48, and 72 h postinfection using TCID50 assays. C 37 °C, D 33 °C. The data are shown as means of three independent experiments. *P < 0.05.
Initial infection and onward transmission of an influenza virus depends on efficient viral replication in the human respiratory tract. Primary NHBE cells were infected with H9N2/Y280 or the D253N mutant PB2 at an MOI of 0.01 and incubated at either 33 °C or 37 °C. Higher viral replication was found from the supernatant collected from cells infected with the PB2 mutant at 24 h postinfection but not at 48 or 72 h postinfection at 37 °C compared with those infected by the wt control. No significant differences in viral replication were found between cells infected by the wt and mutant PB2 at 33 °C (Fig. 1C, 1D).
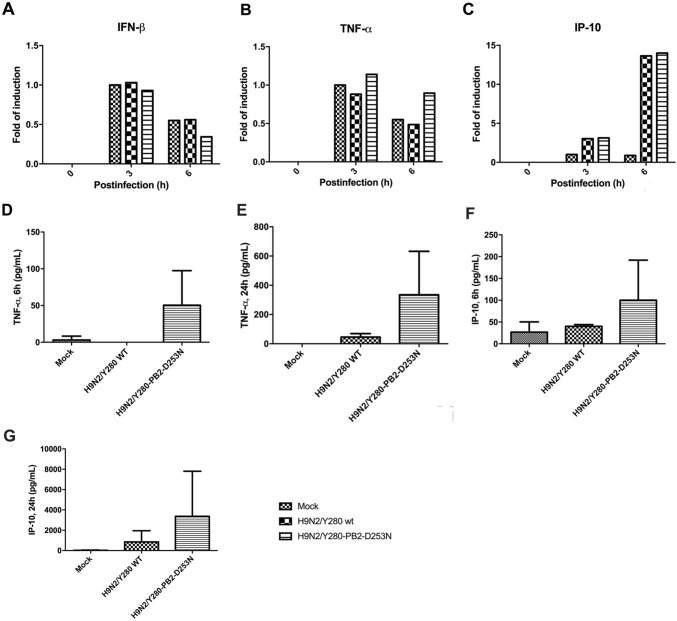
The D253N Mutant in PB2 Did Not Contribute to Cytokine Induction in Primary Human Macrophages
Human macrophages are a primary target of avian influenza virus. Secretion of pro-inflammatory cytokines has been shown to play a role in virus pathogenesis during the infection. Moreover, we previously showed that the single mutation of D253N in PB2 in the H9N2/G1 virus did not enhance cytokine induction in primary human monocyte-derived macrophages. To further confirm this phenomenon, we compared the level of cytokine induction in macrophages infected with the H9N2/Y280 virus or the D253N mutant. Total mRNA and culture supernatants of virus-infected primary human macrophages were collected and tested using quantitative PCR and ELISA. No significant differences in the mRNA and protein levels of interferon-β (IFN-β), IP-10, and TNF-α were found between the wt H9N2/Y280 and PB2 mutant (Fig. 2). Taken together, these findings demonstrated that the D253N mutation in PB2 did not contribute to the upregulation of pro-inflammatory cytokines in primary human macrophages.
Fig. 2.
Induction of cytokine mRNA and protein in primary human macrophages does not enhanced by the PB2-D253N mutation of H9N2/Y280 virus. Primary human macrophages were infected at an MOI of 2 with the H9N2/Y280 virus or the PB2 variant. mRNA was collected at 0, 3, and 6 h postinfection. The gene copy numbers of (A) IFN-β, (B) TNF-α, and (C) IP-10 were analyzed by real-time PCR. The data are shown based on the results of a representative donor. Mock: uninfected cells. GAPDH was used as internal control. The supernatants were collected at 6 and 24 h postinfection. The protein levels of (D) TNF-α (6 h), (E) TNF-α (24 h), (F) IP-10 (6 h), and (G) IP-10 (24 h) were analyzed using an ELISA-based cytomix detection kit. Results are expressed as means ± standard deviations of three independent experiments. Mock: uninfected cells.
The D253N Mutation in PB2 in the H9N2 Virus Did Not Contribute to Viral Replication in the Respiratory Tract of Mice
We next examined the pathogenicity of the D253N mutant in PB2 using an in vivo model of healthy female BALB/c mice (Fig. 3). No mice died or showed virus dissemination to the brain and liver in both groups (data not shown). Mice infected with the wt H9N2/Y280 virus showed weight loss of approximately 10%–12% within 14 days postinfection. Additionally, mice infected with the D253N mutant showed slightly more weight loss than mice infected with wt virus on days 4–7 (18%–20%) (Fig. 3A). However, mice infected with the D253N mutant in PB2 showed no significant differences in viral replication in nasal wash and lung homogenates compared with that in the wt control (Fig. 3B, 3C).
Fig. 3.
The PB2-D253N mutation of H9N2/Y280 virus causes more weight loss but not virus replication in mice. Female BALB/c mice were infected intranasally with 1 × 105 PFU of H9N2/Y280 or the D253N mutant in PB2. A The weights of the infected mice were expressed as the percentage change compared with the weight at day 0. Results from each time point are expressed as means ± standard deviations of eight mice. B The nasal wash and C lungs were harvested for viral titration at 3 and 6 days postinoculation. Lungs were homogenized in 1 mL PBS, and 200 μL PBS was used as a nasal wash. The results from each group are represented by the average titers of three mice. The viral titers were determined by calculating the TCID50 in MDCK cells. Results from each time point were expressed as means ± standard deviations. *P < 0.05.
We also compared the induction of pro-inflammatory cytokines in the lungs of mice infected with the wt H9N2/Y280 virus and the D253N mutant. All wt and PB2 mutants produce higher level of cytokines than uninfected controls. However, mice infected with the PB2 mutant showed comparable levels of pro-inflammatory cytokines (IP-10, MCP-3, MCP-1, MIP-1α, MIP-1β, and TNF-α) in the lungs compared with those in wt controls (Fig. 4A–4F; P > 0.05).
Fig. 4.
Cytokine expression in the lungs of mice infected with H9N2/Y280 and the D253N mutant in PB2. The lungs of virus-infected mice were collected from days 3 and 6 postinoculation, and A IP-10, B MCP-3, C MCP-1, D MIP-1α, E MIP-1β, and F TNF-α levels were measured by bead ELISAs. Results from each time point are expressed as means ± standard deviations of three infected mice (n = 3).
Discussion
In our previous study, we showed that the D253N mutation occurred together with the Q591K mutation in the PB2 gene (Mok et al.2011). We and others subsequently demonstrated that the Q591K mutation is an important factor determining the pathogeneses of different avian influenza viruses in mammalian hosts (Wang et al.2016; Yamada et al.2010). In this study, we showed that the D253N mutation in PB2 contributed to increased polymerase activity and viral replication in human cells at 37 °C using another H9N2 lineage different from that in our previous studies. These results were consistent with the temperatures at which we identified this mutation in MDCK cells, suggesting that the D253N mutation may be responsible for adaptation of the virus at 37 °C, i.e., the temperature of the lower respiratory tract. However, unlike the Q591K mutation, H9N2 virus with only the D253N mutation in PB2 did not showed enhanced viral replication or cytokine induction in mice, despite causing more weight loss on days 4–7 compared with that in the wt control. These results explained why we could not find the D253N mutation in human isolates because this mutation did not provide a long-lasting advantage for selection among all other mutations for mammalian adaptation, similar to the wt isotype.
Although the H9N2 virus with the D253N mutation in PB2 caused significantly more weight loss in the mice compared with the wt control, the viral loads and cytokine levels in the lungs were not correlated with differences in pathogenesis. We previously showed that the E627K and Q591K mutations in PB2 could increase the influx of neutrophils compared with the wt H9N2 virus (Wang et al.2016). Human neutrophils are the most abundant leukocytes in the body and rapidly respond to both bacterial and viral infections (Németh and Mócsai 2016; Parkos 2016). However, excessive infiltration of neutrophils into the lungs is also correlated with lung inflammation and immunopathology, which causes pneumonia (De Filippo et al.2014; Sugamata et al.2012). Whether the virus with the D253N mutation in PB2 attracts more neutrophils or other immune cells will need to be further investigated.
PB2 is a viral polymerase with 759 amino acid residues composed of an N-terminal domain, a middle domain, a cap-binding domain, a 627 domain, and an NLS domain (Lo et al.2018). Residue 253 is located in the middle domain, which has four intertwining helices spanning amino acid residues 251–316. The structural analysis showed that this residue is not in close contact with PB1 or PA but is involved in a global domain reorientation, suggesting that this domain serves as a bridge to link the N-terminal domain and other domains (Lo et al.2018). Our results further showed that the D253N mutation in this region enhanced the replication of the avian influenza virus through adaptation in mammalian cells. We did not show that changing the aspartic acid residue to asparagine would alter the structure of the middle domain. However, switching of the charge from negative (D) to neutral (N) may affect the interaction of PB2 with other components of ribonucleoprotein complex (RNP), which could increase viral transcription; additional studies are needed to support this hypothesis. Moreover, similar results were observed for two other H9N2 viruses: A/Quail/Hong Kong/G1/97 (H9N2/G1) and A/Duck/Hong Kong/Y280/97 (H9N2/Y280), in which their genetic backgrounds were highly distinct, suggesting that this functional role was not strain specific (Mok et al.2011). Interestingly, another study showed that the T271A mutation in PB2, which is also located in the middle domain, enhances polymerase activity but not virulence in mice (Bussey et al.2010).
Taken together, our results supported that the D253N mutation in the middle domain of PB2, similar to mutations at the 627 and NLS domains, specifically contributed to the replication process of avian influenza viruses in human cells.
Acknowledgements
This project was supported by the Science Research Project of the Guangdong Province (Grant no. 2016A050503047), Health and Medical Research Fund (Grant No. 12111832), Guangzhou Medical University High Level University Construction Project Funding and Research Grants Council of the Hong Kong Special Administrative Region, China, through the Theme Based Research Scheme (Ref: T11-705/14N).
Author Contributions
JZ, RS, and CKPM designed the study, HA and CKPM performed the experiments; JZ, XJ, RJ, and YW analyzed the data, JZ and CKPM wrote the main manuscript. All authors read and approved the final manuscript.
Conflict of interest
We declare that no authors have conflict interests.
Animal and Human Rights Statement
All institutional and national guidelines for the care and use of laboratory animals were followed.
References
- Bussey KA, Bousse TL, Desmet EA, Kim B, Takimoto T. PB2 residue 271 plays a key role in enhanced polymerase activity of influenza A viruses in mammalian host cells. J Virol. 2010;84:4395–4406. doi: 10.1128/JVI.02642-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan MC, Cheung CY, Chui WH, Tsao SW, Nicholls JM, Chan YO, Chan RW, Long HT, Poon LL, Guan Y, Peiris JS. Proinflammatory cytokine responses induced by influenza A (H5N1) viruses in primary human alveolar and bronchial epithelial cells. Respir Res. 2005;6:135. doi: 10.1186/1465-9921-6-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan MC, Chan RW, Chan LL, Mok CK, Hui KP, Fong JH, Tao KP, Poon LL, Nicholls JM, Guan Y, Peiris JS. Tropism and innate host responses of a novel avian influenza A H7N9 virus: an analysis of ex vivo and in vitro cultures of the human respiratory tract. Lancet Respir Med. 2013;1:534–542. doi: 10.1016/S2213-2600(13)70138-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung CY, Poon LL, Lau AS, Luk W, Lau YL, Shortridge KF, Gordon S, Guan Y, Peiris JS. Induction of proinflammatory cytokines in human macrophages by influenza A (H5N1) viruses: a mechanism for the unusual severity of human disease? Lancet. 2002;360:1831–1837. doi: 10.1016/S0140-6736(02)11772-7. [DOI] [PubMed] [Google Scholar]
- De Filippo K, Neill DR, Mathies M, Bangert M, McNeill E, Kadioglu A, Hogg N. A new protective role for S100A9 in regulation of neutrophil recruitment during invasive pneumococcal pneumonia. FASEB J. 2014;28:3600–3608. doi: 10.1096/fj.13-247460. [DOI] [PubMed] [Google Scholar]
- de Jong MD, Simmons CP, Thanh TT, Hien VM, Smith GJ, Chau TN, Hoang DM, Chau NV, Khanh TH, Dong VC, Qui PT, Cam BV, Ha do Q, Guan Y, Peiris JS, Chinh NT, Hien TT, Farrar J. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat Med. 2006;12:1203–1207. doi: 10.1038/nm1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao HN, Lu HZ, Cao B, Du B, Shang H, Gan JH, Lu SH, Yang YD, Fang Q, Shen YZ, et al. Clinical findings in 111 cases of influenza A (H7N9) virus infection. N Engl J Med. 2013;368:2277–2285. doi: 10.1056/NEJMoa1305584. [DOI] [PubMed] [Google Scholar]
- Hatta M, Gao P, Halfmann P, Kawaoka Y. Molecular basis for high virulence of Hong Kong H5N1 influenza A viruses. Science. 2001;293:1840–1842. doi: 10.1126/science.1062882. [DOI] [PubMed] [Google Scholar]
- Hatta M, Hatta Y, Kim JH, Watanabe S, Shinya K, Nguyen T, Lien PS, Le QM, Kawaoka Y. Growth of H5N1 influenza A viruses in the upper respiratory tracts of mice. PLoS Pathog. 2007;3:1374–1379. doi: 10.1371/journal.ppat.0030133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Cao B. Pandemic and avian influenza A viruses in humans: epidemiology, virology, clinical characteristics, and treatment strategy. Clin Chest Med. 2017;38:59–70. doi: 10.1016/j.ccm.2016.11.005. [DOI] [PubMed] [Google Scholar]
- Liem NT, Tung CV, Hien ND, Hien TT, Chau NQ, Long HT, Hien NT, le Mai Q, Taylor WR, Wertheim H, Farrar J, Khang DD, Horby P. Clinical features of human influenza A (H5N1) infection in Vietnam: 2004–2006. Clin Infect Dis. 2009;48:1639–1646. doi: 10.1086/599031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo CY, Tang YS, Shaw PC. Structure and function of influenza virus ribonucleoprotein. Subcell Biochem. 2018;88:95–128. doi: 10.1007/978-981-10-8456-0_5. [DOI] [PubMed] [Google Scholar]
- Mok KP, Wong CH, Cheung CY, Chan MC, Lee SM, Nicholls JM, Guan Y, Peiris JS. Viral genetic determinants of H5N1 influenza viruses that contribute to cytokine dysregulation. J Infect Dis. 2009;200:1104–1112. doi: 10.1086/605606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok CK, Yen HL, Yu MY, Yuen KM, Sia SF, Chan MC, Qin G, Tu WW, Peiris JS. Amino acid residues 253 and 591 of the PB2 protein of avian influenza virus A H9N2 contribute to mammalian pathogenesis. J Virol. 2011;85:9641–9645. doi: 10.1128/JVI.00702-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok CK, Lee HH, Chan MC, Sia SF, Lestra M, Nicholls JM, Zhu H, Guan Y, Peiris JM. Pathogenicity of the novel A/H7N9 influenza virus in mice. MBio. 2013;pii:e00362–13. doi: 10.1128/mBio.00362-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok CKP, Lee HHY, Lestra M, Nicholls JM, Chan MCW, Sia SF, Zhu H, Poon LLM, Guan Y, Peiris JSM. Amino-acid substitutions in polymerase basic protein 2 gene contributes to the pathogenicity of the novel A/H7N9 influenza virus in mammalian hosts. J Virol. 2013;88:3568–3576. doi: 10.1128/JVI.02740-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Németh T, Mócsai A. Feedback amplification of neutrophil function. Trends Immunol. 2016;37:412–424. doi: 10.1016/j.it.2016.04.002. [DOI] [PubMed] [Google Scholar]
- Parkos CA. Neutrophil-epithelial interactions: a double-edged sword. Am J Pathol. 2016;186:1404–1416. doi: 10.1016/j.ajpath.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrone LA, Plowden JK, García-Sastre A, Katz JM, Tumpey TM. H5N1 and 1918 pandemic influenza virus infection results in early and excessive infiltration of macrophages and neutrophils in the lungs of mice. PLoS Pathog. 2008;4:e1000115. doi: 10.1371/journal.ppat.1000115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugamata R, Dobashi H, Nagao T, Yamamoto K, Nakajima N, Sato Y, Aratani Y, Oshima M, Sata T, Kobayashi K, Kawachi S, Nakayama T, Suzuki K. Contribution of neutrophil-derived myeloperoxidase in the early phase of fulminant acute respiratory distress syndrome induced by influenza virus infection. Microbiol Immunol. 2012;56:171–182. doi: 10.1111/j.1348-0421.2011.00424.x. [DOI] [PubMed] [Google Scholar]
- Wang C, Lee HHY, Yang ZF, Mok CKP, Zhang Z. PB2-Q591K mutation determines the pathogenicity of avian H9N2 influenza viruses for mammlian species. PLoS ONE. 2016;11:e0162163. doi: 10.1371/journal.pone.0162163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada S, Hatta M, Staker BL, Watanabe S, Imai M, Shinya K, Sakai-Tagawa Y, Ito M, Ozawa M, Watanabe T, Sakabe S, Li C, Kim JH, Myler PJ, Phan I, Raymond A, Smith E, Stacy R, Nidom CA, Lank SM, Wiseman RW, Bimber BN, O’Connor DH, Neumann G, Stewart LJ, Kawaoka Y. Biological and structural characterization of a host-adapting amino acid in influenza virus. PLoS Pathog. 2010;6(8):e1001034. doi: 10.1371/journal.ppat.1001034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Wang D, Gao R, Zhao B, Song J, Qi X, Zhang Y, Shi Y, Yang L, Zhu W, et al. Biological features of novel avian influenza A (H7N9) virus. Nature. 2013;499:500–503. doi: 10.1038/nature12379. [DOI] [PubMed] [Google Scholar]