Abstract
Burkholderia cenocepacia H111 is an opportunistic pathogen associated with chronic lung infections in cystic fibrosis patients. Biofilm formation, motility and virulence of B. cenocepacia are regulated by the second messenger cyclic di-guanosine monophosphate (c-di-GMP). In the present study, we analyzed the role of all 25 putative c-di-GMP metabolizing proteins of B. cenocepacia H111 with respect to motility, colony morphology, pellicle formation, biofilm formation, and virulence. We found that RpfR is a key regulator of c-di-GMP signaling in B. cenocepacia, affecting a broad spectrum of phenotypes under various environmental conditions. In addition, we identified Bcal2449 as a regulator of B. cenocepacia virulence in Galleria mellonella larvae. While Bcal2449 consists of protein domains that may catalyze both c-di-GMP synthesis and degradation, only the latter was essential for larvae killing, suggesting that a decreased c-di-GMP level mediated by the Bcal2449 protein is required for virulence of B. cenocepacia. Finally, our work suggests that some individual proteins play a role in regulating exclusively motility (CdpA), biofilm formation (Bcam1160) or both (Bcam2836).
Keywords: Cyclic-di-GMP, Burkholderia cenocepacia, biofilm formation, motility, RpfR, Bcal2449, GGDEF EAL domain proteins
Introduction
Cyclic di-guanosine monophosphate (c-di-GMP) is an ubiquitous intracellular second messenger, regulating motility, biofilm formation, cell differentiation and virulence in numerous bacteria and some Dictyostelia (Chen and Schaap, 2012; Römling et al., 2013). It is synthesized out of two molecules of GTP by diguanylate cyclases (DGCs) with conserved GGDEF-domains as enzymatically active sites. Phosphodiesterases (PDEs) with conserved EAL- or HD-GYP-domains are required for converting c-di-GMP into pGpG, while further hydrolysis into GMP is catalyzed by some PDEs or by additional oligoribonucleases (Hengge, 2009; Römling et al., 2013; Orr et al., 2015; Jenal et al., 2017). Cyclic-di-GMP metabolizing domains are often coupled with sensor domains, enabling the cell to react to intracellular or environmental changes with modified c-di-GMP levels through modulation of DGC or PDE activities (Galperin, 2004). Once synthesized, c-di-GMP binds to a number of receptor molecules, e.g., enzymes, transcription factors and riboswitches, or it stabilizes multi-protein-complexes (Hengge, 2009; Ryan et al., 2012; Römling et al., 2013). Equally diverse are the regulated target molecules and phenotypic outputs. High c-di-GMP levels are known to decrease single-cell motility through inhibition of flagellar gene expression and rotation, and also down-regulate synthesis of acute virulence factors. Moreover, high c-di-GMP levels stimulate the production of extracellular matrix substances, and thereby positively regulate biofilm formation (Ryjenkov et al., 2006; Hickman and Harwood, 2008; Boehm et al., 2010; Baraquet et al., 2012; Hall and Lee, 2018). Biofilms in the host can often resist antibiotic treatments and serve as permanent pathogen reservoirs, often leading to chronic infections (Parsek and Singh, 2003; Ciofu et al., 2015). Due to the functional redundancy of c-di-GMP-forming, -binding and -degrading proteins, a theory of multiple c-di-GMP regulation modules acting in parallel or on subsequent hierarchic levels has been proposed recently (Kader et al., 2006; Povolotsky and Hengge, 2012; Sarenko et al., 2017; Dahlstrom et al., 2018). Within those modules, only one or a few DGCs and PDEs regulate a specific effector and target molecule, thereby affecting only one cellular output. For example, some c-di-GMP-metabolizing proteins, such as AdrA and DgcC in Salmonella and Escherichia coli or FimX in P. aeruginosa, regulate a single specific phenotype, in this case cellulose biosynthesis and twitching motility, respectively (Zogaj et al., 2001; Brombacher et al., 2003; Kazmierczak et al., 2006). In contrast, global regulators, such as PdeR in E. coli, BldD in Streptomyces or PleD in C. crescentus, affect multiple c-di-GMP-controlled processes (Paul et al., 2004; Lindenberg et al., 2013; Tschowri et al., 2014).
The Burkholderia cepacia complex (Bcc) consists of 22 species (Vanlaere et al., 2009; De Smet et al., 2015; Depoorter et al., 2016; Bach et al., 2017; Martina et al., 2018), often associated with infections in plants, animals and humans. Besides the Bcc, the Burkholderia genus also includes B. pseudomallei and B. mallei, the causative agents of melioidosis and glanders in humans and animals, respectively. Over the past decades, the Bcc member B. cenocepacia has emerged as an important opportunistic human pathogen for immunocompromised individuals and patients suffering from cystic fibrosis (CF) (Mahenthiralingam et al., 2005; Chiarini et al., 2006). Within the CF lung, B. cenocepacia infections cause severe complications, known as the cepacia-syndrome, including decline in lung function and increased mortality (Drevinek and Mahenthiralingam, 2010; Bragonzi et al., 2012). B. cenocepacia can produce several biofilm matrix components, including the exopolysaccharides Bep, cellulose, cepacian and poly-β-1,6-N-acetylglucosamine, proteinaceous compounds, such as BapA, lectins and type-1 fimbria, and extracellular DNA (Conway et al., 2004; Chiarini et al., 2006; McCarthy et al., 2010; Fazli et al., 2011, 2012; Yakandawala et al., 2011; Inhulsen et al., 2012; Messiaen et al., 2014).
Similar to many other Gram-negative bacteria, the production of extracellular matrix components and the resulting formation of pellicles and flow-cell biofilms in Burkholderia sp. are shown to be positively regulated by c-di-GMP; whereas motility is negatively regulated by the second messenger (Lee et al., 2010; Fazli et al., 2011, 2012, 2014; Plumley et al., 2017; Kumar et al., 2018). However, in contrast to the situation in, e.g., P. aeruginosa, where c-di-GMP stimulates biofilm formation in microtiter plates (Hickman et al., 2005m et al., 2015.), its role in this specific form of biofilm formation in the members of the genus Burkholderia is unclear, and contradicting results have been reported (Lee et al., 2010; Deng et al., 2012; Jung et al., 2017; Plumley et al., 2017; Schmid et al., 2017). Prior work in our group has suggested that synthesis of the exopolysaccharide Bep is positively regulated by c-di-GMP and the two transcriptional regulators BerA and BerB (Fazli et al., 2011, 2012, 2017). Moreover, it is known that the cis-2-dodecenoic acid-based quorum sensing system (referred to as the Burkholderia diffusible signal factor, BDSF, in Burkholderia spp.) stimulates c-di-GMP degradation via RpfR, a BDSF-binding GGDEF-EAL domain protein, and thereby regulates virulence in B. cenocepacia (Deng et al., 2012, 2013; Schmid et al., 2017; Yang et al., 2017). Point mutations in the BDSF-binding domain of RpfR have been reported in B. multivorans strains isolated from patients with chronic lung infections (Silva et al., 2016), highlighting the role of c-di-GMP in chronic infections not only in B. cenocepacia, but also in other related Burkholderia spp.
In the present study, we investigated individual mutants of all 25 GGDEF-/EAL-/HD-GYP-domain protein encoding genes present in B. cenocepacia H111 for their roles in the regulation of multiple c-di-GMP-dependent phenotypes. We found that RpfR acts as a key regulator in various c-di-GMP signaling-dependent processes, we present evidence that the Bcal2449 protein is a regulator of virulence, and we identified network-components with specific effects on a limited number of c-di-GMP regulated processes.
Materials and Methods
Bacterial Strains and Growth Conditions
Bacterial strains and plasmids used in this study are listed in Tables 1, 2, respectively. Unless stated otherwise, B. cenocepacia and E. coli strains were incubated at 37°C. Lysogeny Broth (LB) was used for overnight cultivation, and it was supplemented with antibiotics when necessary at the following concentrations: 25 μg/ml gentamicin (Gm), 50 μg/ml kanamycin (Km), 80 μg/ml (liquid medium) or 120 μg/ml (solid medium) tetracycline (Tet), 100 μg/ml trimethoprim (Tp) for B. cenocepacia strains; 100 μg/ml ampicillin (Amp), 6 μg/ml chloramphenicol (Cm), 10 μg/ml Gm, 50 μg/ml Km, 20 μg /ml Tet, 50 μg/ml Tp for E. coli strains. After conjugations, bacteria were incubated on LB medium supplemented with appropriate antibiotics and Amp or on AB medium (Clark and Maaløe, 1967) supplemented with 10 mM Na-citrate as carbon source to inhibit E. coli growth and select for B. cenocepacia transconjugants. When indicated, salt-free medium (LBnoNaCl or ABnoNaCl) was used for biofilm analysis.
Table 1.
Bacterial strains used in this study.
| Bacterial strain | Characteristics | Reference |
|---|---|---|
| B. cenocepacia H111 | Clinical isolate from a cystic fibrosis patient | Gotschlich et al., 2001; Carlier et al., 2014 |
| B. cenocepacia H111 ΔrpfR | rpfR gene deletion mutant | Deng et al., 2012 |
| B. cenocepacia H111 cdpA | Insertional mutant with cdpA interrupted by pEX18Gm, Gmr | This study |
| B. cenocepacia H111 Δbcal0430 | bcal0430 gene deletion mutant | |
| B. cenocepacia H111 Δbcal1635 | bcal1635 gene deletion mutant | |
| B. cenocepacia H111 Δbcal1975 | bcal1975 gene deletion mutant | |
| B. cenocepacia H111 Δbcal2852 | bcal2852 gene deletion mutant | |
| B. cenocepacia H111 Δbcam0748 | bcam0748 gene deletion mutant | |
| B. cenocepacia H111 Δbcam1161 | bcam1161 gene deletion mutant | |
| B. cenocepacia H111 Δbcam1554 | bcam1554 gene deletion mutant | |
| B. cenocepacia H111 Δbcam1670 | bcam1670 gene deletion mutant | |
| B. cenocepacia H111 Δbcam2256 | bcam2256 gene deletion mutant | |
| B. cenocepacia H111 Δbcam2822 | bcam2822 gene deletion mutant | |
| B. cenocepacia H111 Δbcam2836 | bcam2836 gene deletion mutant | |
| B. cenocepacia H111 Δbcas0398 | bcas0398 gene deletion mutant | |
| B. cenocepacia H111 Δbcal0621 | bcal0621 gene deletion mutant | |
| B. cenocepacia H111 Δbcal2449 | bcal2449 gene deletion mutant | |
| B. cenocepacia H111 Δbcam1160 | bcam1160 gene deletion mutant | |
| B. cenocepacia H111 Δbcal0652 | bcal0652 gene deletion mutant | |
| B. cenocepacia H111 Δbcal1100 | bcal1100 gene deletion mutant | |
| B. cenocepacia H111 Δbcal2749 | bcal2749 gene deletion mutant | |
| B. cenocepacia H111 Δbcal3188 | bcal3188 gene deletion mutant | |
| B. cenocepacia H111 Δbcam0158 | bcam0158 gene deletion mutant | |
| B. cenocepacia H111 Δbcam2426 | bcam2426 gene deletion mutant | |
| B. cenocepacia H111 Δbcas0263 | bcas0263 gene deletion mutant | |
| B. cenocepacia H111 Δbcas0378 | bcas0378 gene deletion mutant | |
| E. coli DH5α | Used for standard DNA manipulations | Invitrogen |
| E. coli DB3.1 | Host for Gateway-compatible gene replacement vectors | Invitrogen |
| E. coli S17-1-βpir | Host of the mini-Tn7-kan-gfp delivery vector | Simon et al., 1983 |
Table 2.
Plasmids used in this study.
| Plasmid | Characteristics | Reference |
|---|---|---|
| pEX18Gm | Suicide vector for cdpA mutant construction | Hoang et al., 1998 |
| Pmini-Tn7-kan-gfp | Delivery vector for mini-Tn7-kan-gfp, Kmr | Norris et al., 2010 |
| pRK600 | Helper plasmid in tri- and four-parental matings; ori-ColE1 RK-mob+ RK-tra+; Cmr | Kessler et al., 1992 |
| pUX-BF13 | Helper plasmid providing the Tn7 transposition functions in trans; mob+ ori-R6K; Ampr | Bao et al., 1991 |
| pDAI-SceI-pheS | Cloning vector containing the I-SceI endonuclease and pheS; Tetr | Fazli et al., 2015 |
| pDONRPEX18Tp-SceI-pheS | Gateway compatible gene replacement vector based on SceI and pheS; Tpr | Fazli et al., 2015 |
| pDONRPEX18Gm-SceI-pheS | Gateway compatible gene replacement vector based on SceI and pheS; Gmr | Fazli et al., 2015 |
| pENTRPEX18Tp-SceI-pheS-bcal0430 | Gene replacement vector containing the bcal0430 deletion allele, Tpr | This study |
| pENTRPEX18Gm-SceI-pheS-bcal1635 | Gene replacement vector containing the bcal1635 deletion allele, Gmr | |
| pENTRPEX18Tp-SceI-pheS-bcal1975 | Gene replacement vector containing the bcal1975 deletion allele, Tpr | |
| pENTRPEX18Gm-SceI-pheS-bcal2852 | Gene replacement vector containing the bcal2852 deletion allele, Gmr | |
| pENTRPEX18Tp-SceI-pheS-bcam0748 | Gene replacement vector containing the bcam0748 deletion allele, Tpr | |
| pENTRPEX18Tp-SceI-pheS-bcam1161 | Gene replacement vector containing the bcam1161 deletion allele, Tpr | |
| pENTRPEX18Tp-SceI-pheS-bcam1554 | Gene replacement vector containing the bcam1554 deletion allele, Tpr | |
| pENTRPEX18Tp-SceI-pheS-bcam1670 | Gene replacement vector containing the bcam1670 deletion allele, Tpr | |
| pENTRPEX18Tp-SceI-pheS-bcam2256 | Gene replacement vector containing the bcam2256 deletion allele, Tpr | |
| pENTRPEX18Tp-SceI-pheS-bcam2822 | Gene replacement vector containing the bcam2822 deletion allele, Tpr | |
| pENTRPEX18Tp-SceI-pheS-bcam2836 | Gene replacement vector containing the bcam2836 deletion allele, Tpr | |
| pENTRPEX18Tp-SceI-pheS-bcas0398 | Gene replacement vector containing the bcas0398 deletion allele, Tpr | |
| pENTRPEX18Tp-SceI-pheS-bcal0621 | Gene replacement vector containing the bcal0621 deletion allele, Tpr | |
| pENTRPEX18Tp-SceI-pheS-bcal2449 | Gene replacement vector containing the bcal2449 deletion allele, Tpr | |
| pENTRPEX18Tp-SceI-pheS-bcam1160 | Gene replacement vector containing the bcam1160 deletion allele, Tpr | |
| pENTRPEX18Tp-SceI-pheS-bcal0652 | Gene replacement vector containing the bcal0652 deletion allele, Tpr | |
| pENTRPEX18Tp-SceI-pheS-bcal1100 | Gene replacement vector containing the bcal1100 deletion allele, Tpr | |
| pENTRPEX18Tp-SceI-pheS-bcal2749 | Gene replacement vector containing the bcal2749 deletion allele, Tpr | |
| pENTRPEX18Tp-SceI-pheS-bcal3188 | Gene replacement vector containing the bcal3188 deletion allele, Tpr | |
| pENTRPEX18Tp-SceI-pheS-bcam0158 | Gene replacement vector containing the bcam0158 deletion allele, Tpr | |
| pENTRPEX18Tp-SceI-pheS-bcam2426 | Gene replacement vector containing the bcam2426 deletion allele, Tpr | |
| pENTRPEX18Tp-SceI-pheS-bcas0263 | Gene replacement vector containing the bcas0263 deletion allele, Tpr | |
| pENTRPEX18Tp-SceI-pheS-bcas0378 | Gene replacement vector containing the bcas0378 deletion allele, Tpr |
Construction of B. cenocepacia H111 Mutants and gfp-Fusions
Deletion of 23 of the PDE/DGC genes in B. cenocepacia was carried out using a protocol based on an allelic exchange system described previously (Fazli et al., 2015). Sequences of the oligonucleotides used for the construction of the gene replacement vectors are available on request. The rpfR (bcam0580) mutant used in this study has been described previously (Deng et al., 2012). The cdpA (bcal1069) mutant was constructed as follows. An internal fragment of cdpA was amplified by PCR using the primers 5′-ggatccCGTACGAAATGCACCATGAC and 5′-aagcttCTGCCCGTGGTACACGTAG and inserted as a BamHI/HindIII fragment into the respective sites of the suicide plasmid pEX18Gm (Hoang et al., 1998). The resulting plasmid was transferred by triparental conjugation to B. cenocepacia H111 wild type to generate a cdpA mutant. The introduction of a chromosomal gfp-tag into the B. cenocepacia strains for confocal laser scanning microscopy (CLSM) was carried out as described previously (Fazli et al., 2012).
Motility Assays
Swimming motility was tested on plates containing 0.5% tryptone, 0.5% sodium chloride and 0.3% agar (Pesavento et al., 2008). Three μl of an overnight culture (adjusted to OD600 4.0) was inoculated into the swimming plates and cells were allowed to swim for 10 h at the temperature indicated. Assessment of swarming motility was performed using AB medium plates containing 0.5% glucose, 0.5% casamino acids and 0.6% agar as described previously (Givskov et al., 1998). Three μl of an overnight culture (adjusted to OD600 4.0) was spotted on top of the agar medium and cells were allowed to swarm for up to 48 h. Twitching motility was tested using LB plates containing 1% agar. Cell material from overnight grown colonies was stab inoculated through the LB agar medium, ensuring that bacteria grew and moved between the inner bottom of the petri dish and the agar medium. After 48 h incubation at 37°C, the agar medium was removed carefully and bacteria attached to the petri dish were stained with 0.1% crystal violet (CV) solution to visualize the area covered with bacteria.
Crystal Violet Biofilm Assay
Attachment to abiotic surfaces was assayed as described in Dahlstrom et al. (2018) using BIOLOG PM-1 nutrient plates for phenotypic microarrays with minor modifications. LB overnight cultures were adjusted to OD600 0.716 in ABnoNaCl, mixed 1.5:100 with ABnoNaCl. Hundred-twenty five μl of the inoculated medium was pipetted up and down in the BIOLOG plate and transferred to a 96-well polystyrene microtiter plate (Thermo Fisher, Cat. Nr. 269787) covered with a lid, and the microtiter plate was incubated for 24 h at 37°C to allow biofilm formation. The following day, attached bacteria were quantified using a standard biofilm protocol (O′Toole and Kolter, 1998) with minor modifications as follows. Bacterial growth was measured at OD600, the medium was discarded, and the wells were washed twice with distilled water. To avoid biofilm disruption during the following washing steps, the attached cells were fixed by drying the plates overnight. The wells were stained with 175 μl of 0.1% CV at room temperature for 20 min and subsequently washed twice with distilled water. To quantify biofilm formation, the cells were de-stained with 200 μl 30% acetic acid and absorbance of the CV-acetic acid solution was measured at 590 nm. Based on the results obtained from the BIOLOG plates, 18 substances were selected for further investigation, and biofilm assays were repeated at least three times with 4 replicate wells per strain (the substance concentrations used were: 5 mM citric acid; 10 mM L-arabinose/ L-asparagine/ D-fructose/ L-fucose/ L-glutamine/ L-lactic acid/ D-mannitol/ L-proline; 20 mM galactose/ glycolic acid/ glyoxylic acid/ D-mannose; 50 mM L-glutamic acid/ α-D-lactose/ L-serine/ tyramine; 5 mg/ml mucin from porcine stomach). In addition, ABnoNaCl containing glucose, glycerol or Na-citrate (all at 10 mM) as well as LBnoNaCl was included. CV assays were performed similar to the ones with the BIOLOG plates, however, each well contained 150 μl of medium inoculated to an OD600 of 0.01. Upon transfer to 37°C, the plates were covered with a 96-pegs polystyrene lid (Thermo Scientific; Cat. Nr. 445497), referred to here as the “Calgary Biofilm Device” (Ceri et al., 1999), which allows bacterial cells to attach to the microtiter wells and the pegs. Bound CV and therefore biofilm formation on the pegs was quantified and analyzed separately in a second microtiter plate containing 30% acetic acid to de-stain the pegs-biofilms for subsequent absorbance measurements.
Macrocolony Formation
Five μl of an overnight culture grown in LB was spotted on ABnoNaCl medium supplemented with 1.5% agar. After 5 days of incubation at 37°C photos of the macrocolonies were acquired using a Nikon D3300 SLR camera equipped with a 52 mm Nikon DX SWM Micro 1:1 objective.
Pellicle Formation
Pellicle formation was assayed as described previously (Fazli et al., 2011) with minor modifications. Three ml ABnoNaCl supplemented with carbon sources as indicated was inoculated 1:100 with overnight-grown B. cenocepacia culture, and incubated for up to 5 days at 23°C or 30°C. Pellicle formation was photographed on each day.
Flow-Cell Biofilm Formation and CLSM Analysis
Cultivation of flow-cell biofilms and optical visualization using CLSM were performed as described previously (Fazli et al., 2011, 2012). gfp-tagged strains were used in order to visualize biofilms in flow-cell chambers via CLSM. Microscopic observation of biofilms was performed after 24, 48, and 72 h of growth at 37°C using a Zeiss LSM710 confocal laser scanning microscope equipped with a 63×/1.4 objective and an argon laser. Images were acquired with an AxioCam MRm camera and processed using ZEN (Zeiss, Germany) and IMARIS (Bitplane, Zürich, Switzerland) software. Quantification of biofilm biomass was performed by using COMSTAT software (Heydorn et al., 2000). For COMSTAT analysis, a total of 6 CLSM image stacks were acquired randomly for each strain from flow-cell biofilms. Flow-cell experiments were performed at least twice for each mutant strain along with a wild-type strain control in each individual experiment.
Virulence Assay
Virulence assays were performed using larvae of the Greater Wax Moth (Galleria mellonella), purchased from BioSystems Technologies (Exeter, United Kingdom), and experiments were performed as described previously (Agnoli et al., 2014) with minor modifications. Briefly, LB medium supplemented with antibiotics, when necessary, was inoculated 1:100 with bacterial overnight cultures and incubated with shaking at 37°C for 3–3.5 h to an OD600 of 0.4–0.7. Thereafter, 250 μl of bacterial culture was harvested by centrifugation, and the cell pellet was resuspended in 10 mM MgSO4 to obtain an OD600 of 0.125, corresponding to 4∗107 CFU/ml. The cell suspension was further diluted 1:10 in 10 mM MgSO4 supplemented with Amp, and 5 μl of the cell suspension (10.000–20.000 CFU) was injected into the second last right proleg of G. mellonella larvae using a Hamilton syringe. The syringe was washed with 2.6% NaClO, 70% ethanol, and MgSO4 prior to continuing with the next strain. Eight-to-10 larvae were used for each strain tested and experiments were performed at least 3 times per strain with a wild-type control in each experiment. Upon arrival, un-infected larvae were kept at 12°C for up to 10 days and were transferred to room temperature 4 h prior to infection. After injection of the bacteria the larvae were transferred with a small tissue paper into petri dishes and incubated in a plastic box containing wet paper towels at 30°C for up to 4 days. The larvae were observed daily for viability. In parallel, serial dilutions of the remaining inocula were spread on LB-agar plates for CFU counts to verify that each larva was injected with a similar amount of bacterial cells for each strain tested.
Results
The B. cenocepacia H111 Genome Codes for 25 Putative c-di-GMP-Metabolizing Proteins
Comparative BLAST analysis using the GGDEF domain of E. coli YdaM [aa241-410], the EAL domain of E. coli YjcC [aa275-510] and the HD-GYP domain of Xanthomonas campestris pv. Campestris RpfG [aa202-319] revealed that the B. cenocepacia H111 genome codes for twenty-five GGDEF, EAL, or HD-GYP domain containing proteins (Figure 1). Based on the presence of amino acid residues known to be required for enzymatic activities and co-factor binding (Rao et al., 2008; Tchigvintsev et al., 2010; Römling et al., 2013), we predicted possible c-di-GMP metabolizing activities. Twelve genes encode proteins with conserved GGDEF domains, and therefore being potential DGCs. Eight genes encode proteins with EAL or HD-GYP domains, four (Bcal0652, Bcal1100, Bcam0158, and Bcam2426) of which have conserved domains and therefore being potential PDEs. Five genes encode composite proteins with both GGDEF and EAL domains. Among the composite proteins two (Bcal0621 and CdpA) contain only one conserved domain, indicating that these proteins most likely function as DGC and PDE, respectively; whereas the three other proteins (Bcal2449, RpfR, and Bcam1160) contain intact GGDEF and EAL domains, indicating that these proteins might be bifunctionally active. The chromosome 1 of B. cenocepacia J2315 contains a 57-kb duplication (from bcal0969 to bcal1026 and from bcal2901 to bcal2846), resulting in a gene duplication of bcal1020 (with the paralog bcal2852) (Holden et al., 2009) encoding putative DGCs. However, the H111 genome does not have this duplication (Carlier et al., 2014), hence the corresponding H111 homolog is termed bcal2852 here.
FIGURE 1.
Domain structure of GGDEF/EAL/HD-GYP proteins of B. cenocepacia H111. Proteins were identified by comparative BLAST analysis of well described GGDEF-, EAL- and HDY-GYP domains from E. coli and X. campestris (see text for details). Protein sequences were analyzed with Pfam and NCBI-CDD web tools. Transmembrane domains (gray squares) were verified with the TMHMM web tool. GGDEF domains are highlighted in red, EAL domains in blue and HD-GYP domains in violet, sensor domains in yellow or green. If a protein name has been assigned, it is also shown here.
Besides their c-di-GMP-metabolizing domains, twenty proteins have additional sensor domains, such as phospho-receiver domains (REC) and domains associated with nucleotide (GAF) or ligand binding (PAS, CBS) (Ponting and Aravind, 1997; Ho et al., 2000; Hoch, 2000; Ereno-Orbea et al., 2013). Eleven proteins are predicted to be integrated into the cytoplasmic membrane and possess membrane-associated sensor domains (MASE) formed by multiple transmembrane helices, periplasmic ligand-binding domains (CACHE, CHASE), or domains necessary for formation of disulfide bonds (CSS) (Anantharaman and Aravind, 2000; Galperin et al., 2001; Nikolskaya et al., 2003; Hengge et al., 2015), whereas intramolecular signal transduction from, e.g., periplasmic sensor domains to cytoplasmic output domains can be facilitated through HAMP domains (Aravind and Ponting, 1999). Although the Bcal0652, Bcal1100, and Bcal2749 protein sequences are longer than solitary EAL domains, we failed to identify additional domains. However, sensor domains are most likely present here, too, and might be identified with future domain database versions of Pfam or CDD. In contrast to E. coli (e.g., PdeH), Salmonella enterica (e.g., YdiV and YhjH), and Klebsiella pneumoniae (e.g., KPN_03274), the B. cenocepacia genome does not code for a protein consisting of a solitary, conserved ∼250 AAs long EAL domain only, suggesting that none of the c-di-GMP metabolizing proteins underlies transcriptional and translational regulation only. Besides the composite proteins Bcal0621 and CdpA, which have in addition to their degenerated EAL- or GGDEF- domains, respectively, a second conserved c-di-GMP associated domain and are therefore most likely enzymatically active, the solitary proteins Bcal2749 and Bcal3188 harbor strongly degenerated EAL domains (Figure 1). However, a regulatory function as it has been described for BluF and CsrD in E. coli or LapD in Pseudomonas fluorescens and P. putida (Gjermansen et al., 2005, 2010; Suzuki et al., 2006; Newell et al., 2009; Tschowri et al., 2009) seems likely for Bcal2749 and Bcal3188, particularly as both are conserved among different Burkholderia species (Plumley et al., 2017).
Using a recently published method developed by our group (Fazli et al., 2015), we constructed a mutant library with individual gene deletions of the GGDEF/EAL/HD-GYP domain encoding genes in B. cenocepacia H111, and investigated the resulting strains for c-di-GMP dependent phenotypes. Several studies have suggested that the lack of a single GGDEF/EAL/HD-GYP domain protein does not necessarily change global intracellular levels of c-di-GMP, indicating that most enzymes interact with specific effectors and affect distinct c-di-GMP-regulated phenotypes without affecting the total cellular c-di-GMP pool (Merritt et al., 2010; Massie et al., 2012; Sarenko et al., 2017). Recent studies have suggested that molecular regulation takes place at a local level via direct protein-protein interactions or gradients and closed pools providing locally defined c-di-GMP concentrations (Abel et al., 2011; Lindenberg et al., 2013; Dahlstrom et al., 2015). In addition, these studies have provided evidence that c-di-GMP regulated phenotypes, such as colony morphology, congo red binding and motility, are not necessarily correlated with changes in global cellular c-di-GMP levels (Dahlstrom et al., 2015; Sarenko et al., 2017). Therefore, in the present study we focused on assessing the phenotypic changes of mutants defective of putative c-di-GMP metabolizing proteins in B. cenocepacia, rather than determining c-di-GMP concentrations of whole-cell extracts of our mutants.
The Swimming Phenotype of Distinct DGC and PDE Mutants Suggests That c-di-GMP Is a Negative Regulator of Motility in B. cenocepacia
In many bacteria, high intracellular levels of c-di-GMP has been shown to inhibit motility, by either negatively regulating flagellar gene expression or binding to the molecular break, YcgR, hence interfering with flagellar rotation (Ryjenkov et al., 2006; Boehm et al., 2010; Römling et al., 2013). Recent findings in B. pseudomallei (Plumley et al., 2017), B. lata (Jung et al., 2017), B. cepacia (Ferreira et al., 2013), and B. cenocepacia (Deng et al., 2012; Kumar et al., 2018) suggest that the modulation of c-di-GMP levels in Burkholderia sp. affects swimming motility in semi-solid agar.
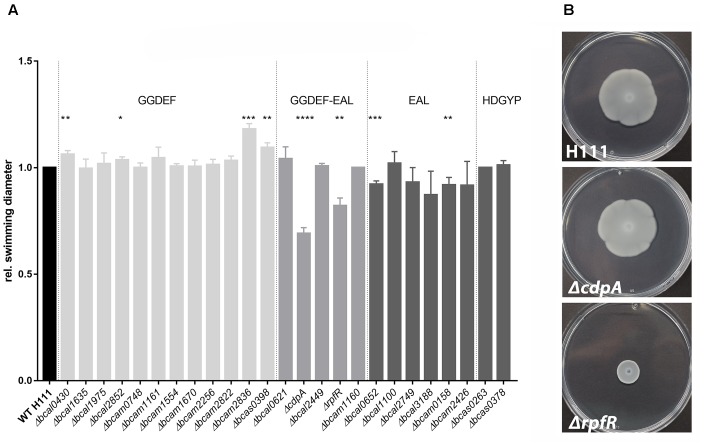
In the present study, we provide evidence for a negative effect of elevated c-di-GMP levels on motility in B. cenocepacia H111. While the deletion of the GGDEF domain encoding genes resulted in increased swimming motility in semi-solid medium up to 18% (Δbcam2836), the deletion of several EAL domain genes (e.g., Δbcal0652 and Δbcam0158) caused a slightly impaired motility phenotype (Figure 2A). However, the deletion of rpfR (bcam0580) or cdpA (bcal1069) resulted in the strongest reduction in swimming diameter (18 and 31% compared to the wild type level) (Figure 2A). Similar results were reported for a bcal1069 mutant in B. cenocepacia K56-2 in synthetic CF sputum medium and homolog gene mutations in B. pseudomallei, leading to renaming of bcal1069 to cdpA (Lee et al., 2010; Kumar et al., 2018). In contrast to swimming motility in semi-solid medium, swarming motility can be observed in medium with higher viscosity and requires hyper-flagellation as well as production of extracellular products, such as surfactants, proteins, or polysaccharides (Harshey, 1994). However, CdpA′s stimulating effect on motility seems to be restricted to swimming motility, as swarming motility was only affected in the rpfR mutant under the conditions tested (Figure 2B). The DGC/PDE mutants showed wild-type levels of pilus-dependent twitching motility under the conditions tested in the present study (data not shown).
FIGURE 2.
Motility of B. cenocepacia H111 and PDE/DGC mutants in semi-solid medium. (A) Swimming motility. Overnight cultures of B. cenocepacia H111 and mutant derivatives were adjusted to OD600 4 and inoculated into swim plates. Plates were incubated at 37°C and the relative swimming diameter was measured after 10 h. Strains with disruption of genes encoding GGDEF-only proteins are highlighted in light gray, composite GGDEF-EAL protein mutants in gray, EAL-only and HD-GYP-domain protein mutants in dark gray, respectively. Data was normalized to wild type results and shows mean swimming diameter and standard derivations of three individual experiments. Asterisks indicate P < 0.05 with unpaired t-test. (B) Swarming motility. Overnight cultures of B. cenocepacia H111 and mutant derivatives were adjusted to OD600 4.0 and inoculated on top of a swarm plate. Plates were incubated at 37°C and pictures were captured after 24 h. Data shown here are representatives of three independent experiments.
Taken together, our data showed that the deletion of specific genes encoding GGDEF- or EAL-domain proteins resulted in altered swimming motility of B. cenocepacia H111. Increased swimming motility of the bcam2836 mutant, which is deleted for a gene that has only a GGDEF domain, and reduced swimming motility of the cdpA and rpfR mutants, for which increased c-di-GMP levels have been reported in B. cenocepacia and B. pseudomallei (Lee et al., 2010; Deng et al., 2012; Plumley et al., 2017; Kumar et al., 2018), suggest that swimming motility is negatively affected by c-di-GMP in B. cenocepacia. However, in contrast to what has been shown for B. pseudomallei, where swimming motility was completely abolished by the cdpA mutation (Lee et al., 2010), none of our B. cenocepacia DGC/PDE mutants displayed a total loss of swimming motility.
rpfR Deletion Stimulates Pellicle Formation by B. cenocepacia Under Multiple Conditions
While bacterial motility is negatively regulated by high intracellular levels of c-di-GMP, the synthesis of extracellular matrix substances resulting in biofilm formation is usually stimulated by the second messenger. In B. cenocepacia, the Bep polysaccharide, synthesized by a multi-protein complex encoded by the bepA-L locus, is positively regulated by c-di-GMP and functions as an extracellular matrix component in flow-cell biofilms, pellicles, and macrocolonies, especially in cells with artificially increased c-di-GMP levels (Fazli et al., 2011, 2012, 2017). While the appearance of flow-cell biofilms and macrocolonies depends also on additional matrix components (Huber et al., 2002; Conway et al., 2004; Yakandawala et al., 2011; Inhulsen et al., 2012), the formation of pellicles at the air-liquid interface of liquid cultures was shown to be a Bep-specific phenotype (Fazli et al., 2011, 2012).
As Bep expression is stimulated by high levels of c-di-GMP, the question arose which DGC stimulates its synthesis through c-di-GMP production and which PDE represses its synthesis through degradation of the signal molecule. Therefore, we performed pellicle biofilm formation assays with all 25 DGC/PDE mutants and the wild type strain under different medium and temperature conditions. None of the twelve GGDEF gene deletion mutants were impaired in pellicle formation (data not shown), excluding the existence of a Bep-specific DGC. Furthermore, the EAL and HD-GYP gene deletion mutants and four out of five GGDEF-EAL mutants did not show altered pellicle formation. However, the rpfR mutant developed a pellicle already after 48 h of incubation at 30°C in minimal medium, which is significantly faster than the wild type. Prolonged incubation led to pellicle formation by the wild type, too, but the pellicle formed by the rpfR mutant was significantly thicker than the wild type pellicle after 72 h incubation (Figure 3). When incubated at room temperature in LBnoNaCl, pellicle formation after 120 h was increased in the rpfR mutant, too (data not shown). Therefore, the composite protein RpfR appears to be a negative regulator of the formation of pellicle biofilms. However, we did not identify a single DGC as responsible for delivering c-di-GMP to this process.
FIGURE 3.
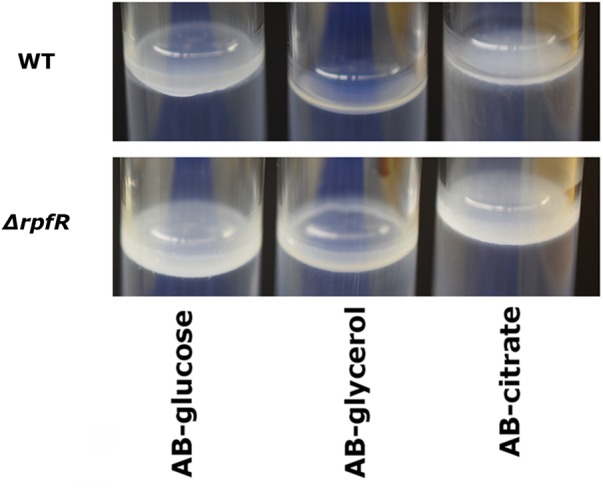
Pellicle formation of B. cenocepacia H111 and the rpfR mutant in static batch cultures. Wild type and mutants were grown at 30°C in ABnoNaCl supplemented with 10 mM glucose, 10 mM glycerol or 10 mM citrate, respectively, in glass tubes and sealed with parafilm to diminish evaporation but allow air exchange. Pictures of pellicles were captured after 72 h.
RpfR and Bcal2449 Modulate the Morphology of B. cenocepacia Macrocolonies
Burkholderia spp. in biofilms were shown to produce multiple extracellular polysaccharides (e.g., Bep, cellulose and cepacian) and protein-based matrix components (e.g., BapA, fimbria, lectins) (Ferreira et al., 2011; Yakandawala et al., 2011; Fazli et al., 2012; Inhulsen et al., 2012; Cescutti et al., 2013). A correlation between colony morphology and the production of extracellular matrix components has been reported for several bacterial species, including B. cenocepacia (e.g., Fazli et al., 2012). To investigate whether some of our DGC/PDE mutants displayed changed matrix production, we used solid agar plates spot inoculated with cell culture and allowed them to develop into macrocolonies for 5 days at 37°C. In contrast to the wild type and the majority of the GGDEF/EAL/HD-GYP mutants, the ΔrpfR and Δbcal2449 mutants developed dry and structured colonies, suggesting that these mutants secrete increased amounts of extracellular matrix material or that it has an altered composition (Figure 4 and Supplementary Figure S1). We note that unlike the ΔrpfR mutant the Δbcal2449 mutant did not display increased pellicle formation, indicating that the matrix produced by the Δbcal2449 mutant differ from that produced by the ΔrpfR mutant. The RpfR and Bcal2449 proteins have conserved GGDEF and EAL domains and therefore can potentially act as DGC or PDE, depending on intra-molecular regulation or interaction with protein partners. Our data suggest that under the conditions tested RpfR and Bcal2449 both have a negative effect on the production of one or more matrix components.
FIGURE 4.
Macrocolony formation of the B. cenocepacia wild type, and rpfR and bcal2449 mutants on solid agar. B. cenocepacia H111 and mutant derivatives were grown on ABnoNaCl agar supplemented with 1% glucose and incubated 5 days, after which images were acquired.
RpfR Regulates B. cenocepacia Biofilm Formation in Microtiter Plates
Growth of bacteria in the wells of microtiter plates followed by staining of attached cells with CV serves as an established method for quantification of biofilm formation. BIOLOG phenotypic microarray plates (BIOLOG, Inc.) allow growth of bacteria under 95 different medium conditions in parallel, and was used recently to elucidate the effect of environmental conditions on the regulation of c-di-GMP dependent biofilm formation in P. fluorescens (Dahlstrom et al., 2018). In addition, recent publications have shown a medium and temperature dependent motility and biofilm phenotype for c-di-GMP mutants in B. cenocepacia, B. pseudomallei, and P. fluorescens (Plumley et al., 2017; Dahlstrom et al., 2018; Kumar et al., 2018). Besides transcriptional and translational regulation, DGCs and PDEs can be regulated at the post-translational level, and environmental stimuli may be perceived by sensor domains. Therefore, individual DGCs and PDEs might respond individually to different environmental signals, such as carbon sources.
Using BIOLOG PM-1 phenotype microarray plates, we assessed the effect of 95 different carbon sources on the biofilm formation properties of our GGDEF/EAL/HD-GYP mutants. After 24 h, detectable cell growth (OD600 > 0.2) for the B. cenocepacia H111 wild type and mutants was observed for 65 carbon sources, while CV binding was measurable for the wild type and mutants for 90 carbon sources (OD590 > 0.25). In total, 52 microtiter wells showed both bacterial growth and visible CV binding after medium and planktonic cells had been discarded. Among these, we focused on the carbon sources which altered CV values by at least 20 percent when compared to the wild type in at least one mutant and, in addition, included the medium used for macrocolony and pellicle experiments, resulting in 22 different media. We performed standard biofilm assays in microtiter plates and quantified cell attachment on microtiter wells as well as pegs dipped into the wells.
Compared to the wild type, the ΔrpfR mutant showed altered biofilm formation under most culture conditions (Figure 5 and Supplementary Figure S8). However, while ΔrpfR biofilm formation on the pegs was increased, attachment to the microtiter wells was decreased when grown with asparagine, citric acid, fructose, galactose, glucose, lactic acid, mannitol, mannose, and mucin as the carbon source. In contrast, growth with fucose, glutamine or proline altered only one form of biofilm formation, and growth on glutamic acid enhanced attachment to both the pegs and the wells. Among the remaining 24 strains tested, the GGDEF mutants Δbcam2836 and Δbcas0398 showed slightly enhanced attachment under several conditions to both the pegs and the wells (e.g., Δbcas0398 in fucose media), and for the GGDEF-EAL mutant Δbcam1160 reduced attachment to the wells was observed in medium supplemented with glycolic acid and glyoxylic acid (Supplementary Figures S2–S7). These two compounds did not change attachment patterns of the rfpR mutant. In contrast to our initial results from the BIOLOG plates, cell growth and detectable amounts of CV binding could not be observed in medium supplemented with lactose, serine and tyramine, and the results with these media are therefore not shown.
FIGURE 5.
Biofilm formation in microtiter plates by the B. cenocepacia rpfR mutant under various medium conditions. Cells were grown in ABnoNaCl supplemented with carbon sources as indicated (see material and methods for concentrations) under static conditions at 37°C for 24 h. Subsequently, the amount of biofilm on the wells and on the pegs, respectively, was determined via a CV staining assay. Graphs show OD590nm values of CV bound by the ΔrpfR mutant normalized to values obtained from the wild type. Error bars indicate standard deviations of 4 replicates per strain, and data shown here are representatives of three independent experiments.
Taken together, our results suggest that RpfR is the master regulator of biofilm formation in microtiter plates under most conditions, although Bcam2836, Bcas0398, and Bcam1160 seem to affect biofilm formation under some conditions. Our results suggest that B. cenocepacia biofilm formation on pegs is positively regulated by c-di-GMP, whereas biofilm formation on wells (the “standard” microtiter plate assay) is negatively regulated by c-di-GMP, suggesting that these two forms of biofilm formation differ, and are under different regulatory mechanisms in B. cenocepacia.
RpfR Regulates B. cenocepacia Biofilm Formation in Flow-Cells
Similar to microtiter plates, flow-cells provide an abiotic surface for cell attachment and formation of biofilms. However, in contrast to microtiter plates, flow-cell incubation guarantees constant nutrient supply due to a continuous flow of fresh medium as well as removal of metabolic waste products, used medium and cell debris. In addition, the maturing biofilm is exposed to mild shear forces, without the risk of desiccation due to medium evaporation. Prior work in our group has demonstrated that B. cenocepacia biofilm formation in flow-cells is positively regulated by c-di-GMP (Fazli et al., 2011, 2012).
To assess the effect of individual GGDEF, EAL, or HD-GYP deletions, we tested biofilm formation phenotypes of our 25 GGDEF/EAL/HD-GYP mutants and the corresponding wild type in flow-cells over a time frame of 3 days. Most of the mutants did not display altered biofilm formation in the flow-cells compared to the wild type (Supplementary Figure S9). However, similar to the results obtained from biofilm assays in microtiter plates, the rpfR mutant displayed an altered biofilm formation phenotype (Figure 6 and Supplementary Figure S9). The rpfR mutant formed compact, spherical microcolonies after 24 h, which were absent in the biofilms formed by the wild type and the other DGC/PDE mutants. After 48 h the rpfR mutant cells had colonized the entire substratum with a thick matt-like biofilm, which appeared smooth at the surface, but showed denser structures in underlying layers. Notably, the biomass of the ΔrpfR biofilms was increased in comparison to the biomass of the wild type biofilms at all time points (Figure 6B). In contrast to the case of macrocolony formation (Figure 4), the bcal2449 mutant did not display altered biofilm formation in flow-cells (Supplementary Figure S9).
FIGURE 6.
Biofilm formation in flow-cells by B. cenocepacia H111 and the rpfR and bcal2449 mutants. (A) Confocal laser scanning micrographs of 3-day-old biofilms formed by gfp-expressing B. cenocepacia strains. The images show top-down easy3D views (B) Comstat analysis of biomass for 1, 2 and 3 day old biofilms of wild type and mutants. Figure shows mean and standard derivations of six CLSM images per strain and time point.
Taken together, our results suggest that RpfR negatively regulates the formation of microcolonies in young biofilms and development into mature biofilms during prolonged incubation in flow-cells. We did not identify a single DGC as responsible for delivering c-di-GMP to stimulate biofilm formation in flow-cells.
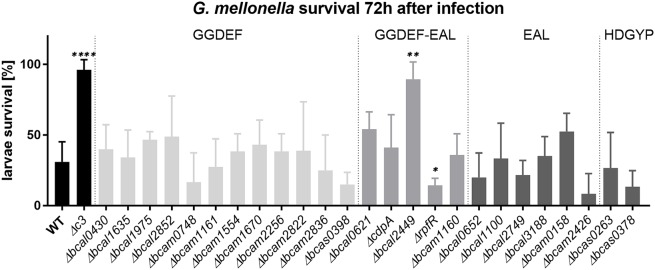
Bcal2449 Is a Major Regulator of B. cenocepacia Virulence
Besides motility and biofilm formation, c-di-GMP is known to regulate virulence in a number of bacteria (Römling et al., 2013; Jenal et al., 2017; Hall and Lee, 2018), including B. cenocepacia, where high c-di-GMP levels appear to be associated with impaired virulence (Schmid et al., 2017). We determined the virulence phenotype of each individual GGDEF, EAL, or HD-GYP mutant in larvae of the Greater Wax Moth G. mellonella (Figure 7). Interestingly, we found that the deletion of the GGDEF-EALxR-containing Bcal2449 protein almost abolished B. cenocepacia virulence (Figure 7). The virulence was restored when bcal2449 was expressed in trans from the plasmid pBBR1-bcal2449 (Figure 8). Larvae killing was strictly dependent on a conserved EAL-motif, as the complementation of the bcal2449 mutant strain with a mutated EAL domain (Δbcal2449/pBBR1-bcal2449-AAL and Δbcal2449/pBBR1-bcal2449-GGAAF-AAL) failed to restore the virulence (Figure 8), suggesting that a decreased c-di-GMP level mediated by the Bcal2449 protein is required for virulence. In contrast, bacteria expressing a bcal2449-variant with a mutated GGDEF domain (Δbcal2449/pBBR1-bcal2449-GGAAF) were as virulent as the complemented mutant (Figure 8). Besides, our screen suggested that infection with the rpfR or bcam2426 mutants resulted in reduced larval survival in comparison to the wild type (Figure 7), although the results for the rpfR and bcam2426 mutants are not as pronounced as the result obtained for the bcal2449 mutant. An attenuated virulence phenotype of ΔcdpA was not observed in the present study. Therefore, we suggest that CdpA only acts on motility in B. cenocepacia and does not play a major role in other c-di-GMP regulated processes, in contrast to its homolog in B. pseudomallei (Lee et al., 2010).
FIGURE 7.
Virulence of B. cenocepacia H111 and PDE/DGC mutants in G. mellonella. Ten larvae were infected with 10.000–20.000 B. cenocepacia wild type or mutants, and killing was observed over 3 days. Data presented here show mean and standard deviations of larvae survival of at least 3 independent experiments per strain 72 h after infection. Knockout mutants of genes coding for GGDEF-only proteins are highlighted in light gray, those encoding composite GGDEF-EAL proteins in gray, EAL-only proteins and HD-GYP-domain proteins in dark gray, wild type and Δc3 control in black, respectively. Asterisks indicate P < 0.05 with unpaired t-test with wild type control.
FIGURE 8.

Effect of mutations in the EAL and GGDEF domain of Bcal2449 on virulence of B. cenocepacia in G. mellonella. Ten larvae were infected with 10.000–20.000 B. cenocepacia wild type or mutants harboring pBBR1 plasmid derivatives as indicated, and larvae killing was observed over 3 days. Data presented here show mean and standard deviations of larvae survival of at least 3 independent experiments per strain and the corresponding wild type control 72 h after infection. Asterisks indicate P < 0.05 with unpaired t-test with wild type control.
Taken together, our results suggest a major role for the composite protein Bcal2449 in B. cenocepacia virulence, which is dependent on a functional EAL domain, suggesting that a local decrease in c-di-GMP mediated by the Bcal2449 protein is required for virulence in the G. mellonella larvae model.
Discussion
The intracellular signaling molecule c-di-GMP is synthesized and degraded by GGDEF- and EAL-/HD-GYP-domain proteins, respectively, and regulates a variety of cellular processes in a wide range of bacteria, including B. cenocepacia. This study represents the first detailed analysis of the function of all 25 GGDEF-/ EAL- and HD-GYP-domain proteins encoded by the B. cenocepacia H111 genome, focusing on their individual effects on motility, biofilm formation and virulence.
RpfR, a multi-domain protein consisting of a BDSF-binding PAS-, a GGDEF- and an EAL-domain, was previously shown to regulate motility, protease activity, biofilm formation and virulence of B. cenocepacia (Deng et al., 2012; Jung et al., 2017; Schmid et al., 2017). Our data presented in this report suggest that RpfR is a key regulator of c-di-GMP signaling in B. cenocepacia. Previous studies revealed a reduced ability of biofilm formation in the wells of microtiter plates for rpfR mutants (Deng et al., 2012; Suppiger et al., 2016; Yang et al., 2017). Striking results, considering that deletion of rpfR also resulted in reduced motility and increased intracellular c-di-GMP levels. Our analysis verified decreased biofilm formation of the rpfR mutant in the wells of microtiter plates, however, we found that biofilm formation on polystyrene pegs, in flow-cells and in pellicles was increased for the rpfR mutant, suggesting that high levels of c-di-GMP, due to a lack of this key-PDE, positively regulate biofilm formation by B. cenocepacia under various conditions.
The CepIR quorum sensing system appears to control biosurfactant production in B. cenocepacia (Huber et al., 2001) and is, in turn, modulated by the RpfRF system (Schmid et al., 2012). Hence, RpfR affects both flagellar gene expression (BDSF regulated) and indirectly biosurfactant production (AHL regulated) and therefore swimming and swarming motility (Lee et al., 2010). Accordingly, we observed reduced swimming motility for the rpfR mutant and, in addition, also for the cdpA mutant. In B. pseudomallei swimming motility is completely abolished in a cdpA mutant, and the cells are aflagellate and do not express fliC (Lee et al., 2010). In agreement with our results, reduced swimming motility for a B. cenocepacia cpdA mutant has also been described by Kumar et al. (2018), and this was found not to be the result of altered flagellin expression or flagellation patterns, suggesting that cellular targets of CdpA-dependent regulation differ among these species. While motility in B. cenocepacia is regulated by CdpA in response to arginine and glutamate (Kumar et al., 2018), biofilm formation is not affected under the conditions tested. However, we show that attachment to abiotic surfaces in the presence of glutamate is regulated by RpfR, suggesting that one extracellular stimulus can regulate different phenotypic outputs through independent c-di-GMP cascades.
To analyze the effect of environmental conditions on biofilm formation, we performed biofilm assays in media supplemented with different carbon sources, which we chose with a focus on, inter alia, the conditions likely prevailing in the natural habitats of B. cenocepacia, such as soil and water, plant rhizosphere and CF sputum, or as a component of Bcc extracellular polysaccharides (Mahenthiralingam et al., 2005; Palmer et al., 2007; Cuzzi et al., 2014; Mongkolrob et al., 2015). We found that RpfR regulates biofilm formation under multiple nutrient conditions, many of them associated with components present in sputum (galactose, glutamine, lactic acid, mucin, proline) (Palmer et al., 2007). In contrast to the rpfR mutant, the bcam1160 mutant showed reduced biofilm formation in response to plant-associated components, such as glycolic and glyoxylic acid, emphasizing the importance of adjacent sensor domains for signal specificity of individual DGCs and PDEs.
In this study, we simultaneously quantified biofilm formation on the surface of microtiter plate wells and biofilm formation on pegs dipped into the wells. We found that biofilm formation of the rpfR mutant was increased on the pegs but decreased on the surface of the wells under various conditions. We also assessed biofilm formation of the wild-type and mutants on the surface of the wells in microtiter plates without pegs, and found no difference compared to when pegs were present (data not shown). These results argue against the possibility that the rpfR mutant attached and grew better on pegs, and therefore fewer cells grew attached to the surface of the microtiter wells, resulting in simply a redistribution of biomass. Moreover, given that the RpfR protein has been reported to act as a PDE under the conditions of the microtiter tray assay (Deng et al., 2012), our data for biofilm formation on the surface of microtiter wells agrees with former studies, where a correlation between high c-di-GMP levels (either through PDE mutation or plasmid-borne DGC expression) and reduced biofilm formation in microtiter plates has been observed (Deng et al., 2012; Plumley et al., 2017; Schmid et al., 2017; Yang et al., 2017).
Our analysis of macrocolony morphology of the B. cenocepacia wild type and DGC/PDE mutants showed that the rpfR and bcal2449 mutants formed more structured colonies than the wild type, indicating that the RpfR and Bcal2449 proteins act as PDEs at 37°C. During long-term infection, strains with mutations within the N-terminal PAS domain of RpfR were isolated (Silva et al., 2016). As mutations affected amino acid residues essential for BDSF-binding, the mutated protein-variant is most likely unable to bind BDSF, resulting in a constantly inactive or unregulated PDE, which may lead to increased biofilm formation in the lungs of the patient. RpfR was also identified as a key player in an evolutionary biofilm model that allows long-term selection for adherence to and dispersal from a plastic bead in a test tube, emphasizing the central role of this regulator in biofilm formation (Traverse et al., 2013).
Strikingly, RpfR, while being a key component in the c-di-GMP network of B. cenocepacia, is not fully conserved within the genus Burkholderia, raising the question about a homolog PDE with a comparable broad regulon in these species. The genes bcal2449, bcal2749 and bcal3188 are conserved among B. cenocepacia, B. glumae, B. mallei, B. pseudomallei, and B. thailandensis (Plumley et al., 2017). The genes bcal2749 and bcal3188 code for proteins with solitary, degenerated EAL domains, which most likely do not take direct action in c-di-GMP degradation. However, their wide distribution as well as their mutated active site within the EAL domain (HASxR and GYLxx, respectively) within the genus Burkholderia suggests a prominent role in cellular regulation. Moreover, the two domains of Bcal2449 (GGDEF and EAL) are highly conserved. Our study indicates a major role for Bcal2449 in the regulation of virulence, which is dependent on its EAL domain and therefore most likely its PDE activity. This is in agreement with prior studies, indicating that virulence of B. cenocepacia is stimulated at low c-di-GMP levels obtained by plasmid-borne expression of a PDE (Schmid et al., 2017). As this effect is restricted to Bcal2449 only and was not observed for other PDEs such as RpfR or CdpA, which have been shown to regulate global intracellular c-di-GMP levels (Lee et al., 2010; Deng et al., 2012), we propose a local mode of regulation on virulence factors mediated by Bcal2449. Further studies will identify these Bcal2449-controled virulence factors, and the molecular regulation and environmental signals modulating Bcal2449 activity presumably via its periplasmic Cache domain.
Our study identified individual c-di-GMP-metabolizing proteins, which regulate a specific c-di-GMP-dependent phenotype. Our mutant analysis indicated that the putative DGC Bcam2836 and the PDE CdpA have opposite effects on motility, suggesting that locally high c-di-GMP levels inhibit motility of B. cenocepacia. Consistent with a recent publication (Plumley et al., 2017), we found that Bcam2836 regulates biofilm formation under some conditions. In addition, the GGDEF-EAL domain proteins Bcam1160 and Bcal2449 regulate only some c-di-GMP-dependent phenotypes (macrocolony and microtiter plate biofilm formation/ macrocolony biofilms and virulence), suggesting a high specificity within the c-di-GMP network. In contrast, the quorum sensing regulator RpfR modulates multiple c-di-GMP phenotypes and therefore represents an important key-regulator of multiple phenotypes in B. cenocepacia.
Collectively, our study demonstrates that c-di-GMP inhibits motility and stimulates biofilm formation by B. cenocepacia under many conditions similar to what has been found for many other bacterial species. RpfR seems to be a master regulator of biofilm formation under various conditions, whereas a few of the other DGC/PDEs were shown to affect biofilm formation under specific conditions. In addition, we present evidence that Bcal2449 is a regulator of B. cenocepacia virulence, which is dependent on its EAL domain, presumably lowering the level of c-di-GMP and thereby stimulating virulence. However, the virulence factors and extracellular matrix components controlled by Bcal2449 remain to be elucidated. The role of the remaining GGDEF-, EAL-, and HD-GYP domain proteins as well as environmental clues regulating them has to be addressed in future studies.
Author Contributions
AR and TT-N designed the experiments. AR, MF, NS, RS, and AS performed the experiments. AR, TT-N, LE, and MG interpreted the data. AR wrote the manuscript with input from all authors.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was supported by a Postdoc research grant from the Deutsche Forschungsgemeinschaft (RI 2747/1-1) to AR, and grants from the Lundbeck Foundation (R198-2015-486) to TT-N and MG, and a grant from the Swiss National Science Foundation (SNSF; Project 31003A_169307) to LE.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.03286/full#supplementary-material
References
- Abel S., Chien P., Wassmann P., Schirmer T., Kaever V., Laub M. T., et al. (2011). Regulatory cohesion of cell cycle and cell differentiation through interlinked phosphorylation and second messenger networks. Mol. Cell 43 550–560. 10.1016/j.molcel.2011.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agnoli K., Frauenknecht C., Freitag R., Schwager S., Jenul C., Vergunst A., et al. (2014). The third replicon of members of the Burkholderia cepacia Complex, plasmid pC3, plays a role in stress tolerance. Appl. Environ. Microbiol. 80 1340–1348. 10.1128/AEM.03330-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anantharaman V., Aravind L. (2000). Cache – a signaling domain common to animal Ca(2+)-channel subunits and a class of prokaryotic chemotaxis receptors. Trends Biochem. Sci. 25 535–537. 10.1016/S0968-0004(00)01672-8 [DOI] [PubMed] [Google Scholar]
- Aravind L., Ponting C. P. (1999). The cytoplasmic helical linker domain of receptor histidine kinase and methyl-accepting proteins is common to many prokaryotic signalling proteins. FEMS Microbiol. Lett. 176 111–116. 10.1111/j.1574-6968.1999.tb13650.x [DOI] [PubMed] [Google Scholar]
- Bach E., Sant’Anna F. H., Magrich Dos Passos J. F., Balsanelli E., de Baura V. A., Pedrosa F. O., et al. (2017). Detection of misidentifications of species from the Burkholderia cepacia complex and description of a new member, the soil bacterium Burkholderia catarinensis sp. nov. Pathog. Dis. 75:ftx076. 10.1093/femspd/ftx076 [DOI] [PubMed] [Google Scholar]
- Bao Y., Lies D. P., Fu H., Roberts G. P. (1991). An improved Tn7-based system for the single-copy insertion of cloned genes into chromosomes of gram-negative bacteria. Gene 109 167–168. 10.1016/0378-1119(91)90604-A [DOI] [PubMed] [Google Scholar]
- Baraquet C., Murakami K., Parsek M. R., Harwood C. S. (2012). The FleQ protein from Pseudomonas aeruginosa functions as both a repressor and an activator to control gene expression from the pel operon promoter in response to c-di-GMP. Nucleic Acids Res. 40 7207–7218. 10.1093/nar/gks384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm A., Kaiser M., Li H., Spangler C., Kasper C. A., Ackermann M., et al. (2010). Second messenger-mediated adjustment of bacterial swimming velocity. Cell 141 107–116. 10.1016/j.cell.2010.01.018 [DOI] [PubMed] [Google Scholar]
- Bragonzi A., Farulla I., Paroni M., Twomey K. B., Pirone L., Lore N. I., et al. (2012). Modelling co-infection of the cystic fibrosis lung by Pseudomonas aeruginosa and Burkholderia cenocepacia reveals influences on biofilm formation and host response. PLoS One 7:e52330. 10.1371/journal.pone.0052330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brombacher E., Dorel C., Zehnder A. J., Landini P. (2003). The curli biosynthesis regulator CsgD co-ordinates the expression of both positive and negative determinants for biofilm formation in Escherichia coli. Microbiology 149(Pt 10), 2847–2857. 10.1099/mic.0.26306-0 [DOI] [PubMed] [Google Scholar]
- Carlier A., Agnoli K., Pessi G., Suppiger A., Jenul C., Schmid N., et al. (2014). Genome sequence of Burkholderia cenocepacia H111, a cystic fibrosis airway isolate. Genome Announc. 2:e00298-14. 10.1128/genomeA.00298-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceri H., Olson M. E., Stremick C., Read R. R., Morck D., Buret A. (1999). The calgary biofilm device: new technology for rapid determination of antibiotic susceptibilities of bacterial biofilms. J. Clin. Microbiol. 37 1771–1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cescutti P., Cuzzi B., Herasimenka Y., Rizzo R. (2013). Structure of a novel exopolysaccharide produced by Burkholderia vietnamiensis, a cystic fibrosis opportunistic pathogen. Carbohydr. Polym. 94 253–260. 10.1016/j.carbpol.2013.01.047 [DOI] [PubMed] [Google Scholar]
- Chen Z. H., Schaap P. (2012). The prokaryote messenger c-di-GMP triggers stalk cell differentiation in Dictyostelium. Nature 488 680–683. 10.1038/nature11313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiarini L., Bevivino A., Dalmastri C., Tabacchioni S., Visca P. (2006). Burkholderia cepacia complex species: health hazards and biotechnological potential. Trends Microbiol. 14 277–286. 10.1016/j.tim.2006.04.006 [DOI] [PubMed] [Google Scholar]
- Ciofu O., Tolker-Nielsen T., Jensen P. O., Wang H., Hoiby N. (2015). Antimicrobial resistance, respiratory tract infections and role of biofilms in lung infections in cystic fibrosis patients. Adv. Drug Deliv. Rev. 85 7–23. 10.1016/j.addr.2014.11.017 [DOI] [PubMed] [Google Scholar]
- Clark D. J., Maaløe O. (1967). DNA replication and the division cycle in Escherichia coli. J. Mol. Biol. 23 99–112. 10.1016/S0022-2836(67)80070-6 4578860 [DOI] [Google Scholar]
- Conway B. A., Chu K. K., Bylund J., Altman E., Speert D. P. (2004). Production of exopolysaccharide by Burkholderia cenocepacia results in altered cell-surface interactions and altered bacterial clearance in mice. J. Infect. Dis. 190 957–966. 10.1086/423141 [DOI] [PubMed] [Google Scholar]
- Cuzzi B., Herasimenka Y., Silipo A., Lanzetta R., Liut G., Rizzo R., et al. (2014). Versatility of the Burkholderia cepacia complex for the biosynthesis of exopolysaccharides: a comparative structural investigation. PLoS One 9:e94372. 10.1371/journal.pone.0094372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlstrom K. M., Collins A. J., Doing G., Taroni J. N., Gauvin T. J., Greene C. S., et al. (2018). A multimodal strategy used by a large c-di-GMP network. J. Bacteriol. 200:e00703-17. 10.1128/JB.00703-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlstrom K. M., Giglio K. M., Collins A. J., Sondermann H., O’Toole G. A. (2015). Contribution of physical interactions to signaling specificity between a diguanylate cyclase and its effector. MBio 6:e01978-15. 10.1128/mBio.01978-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet B., Mayo M., Peeters C., Zlosnik J. E., Spilker T., Hird T. J., et al. (2015). Burkholderia stagnalis sp. nov. and Burkholderia territorii sp. nov., two novel Burkholderia cepacia complex species from environmental and human sources. Int. J. Syst. Evol. Microbiol. 65 2265–2271. 10.1099/ijs.0.000251 [DOI] [PubMed] [Google Scholar]
- Deng Y., Lim A., Wang J., Zhou T., Chen S., Lee J., et al. (2013). Cis-2-dodecenoic acid quorum sensing system modulates N-acyl homoserine lactone production through RpfR and cyclic di-GMP turnover in Burkholderia cenocepacia. BMC Microbiol. 13:148. 10.1186/1471-2180-13-148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y., Schmid N., Wang C., Wang J., Pessi G., Wu D., et al. (2012). Cis-2-dodecenoic acid receptor RpfR links quorum-sensing signal perception with regulation of virulence through cyclic dimeric guanosine monophosphate turnover. Proc. Natl. Acad. Sci. U.S.A. 109 15479–15484. 10.1073/pnas.1205037109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depoorter E., Bull M. J., Peeters C., Coenye T., Vandamme P., Mahenthiralingam E. (2016). Burkholderia: an update on taxonomy and biotechnological potential as antibiotic producers. Appl. Microbiol. Biotechnol. 100 5215–5229. 10.1007/s00253-016-7520-x [DOI] [PubMed] [Google Scholar]
- Drevinek P., Mahenthiralingam E. (2010). Burkholderia cenocepacia in cystic fibrosis: epidemiology and molecular mechanisms of virulence. Clin. Microbiol. Infect. 16 821–830. 10.1111/j.1469-0691.2010.03237.x [DOI] [PubMed] [Google Scholar]
- Ereno-Orbea J., Oyenarte I., Martinez-Cruz L. A. (2013). CBS domains: ligand binding sites and conformational variability. Arch. Biochem. Biophys. 540 70–81. 10.1016/j.abb.2013.10.008 [DOI] [PubMed] [Google Scholar]
- Fazli M., Almblad H., Rybtke M. L., Givskov M., Eberl L., Tolker-Nielsen T. (2014). Regulation of biofilm formation in Pseudomonas and Burkholderia species. Environ. Microbiol. 16 1961–1981. 10.1111/1462-2920.12448 [DOI] [PubMed] [Google Scholar]
- Fazli M., Harrison J. J., Gambino M., Givskov M., Tolker-Nielsen T. (2015). In-frame and unmarked gene deletions in Burkholderia cenocepacia via an allelic exchange system compatible with gateway technology. Appl. Environ. Microbiol. 81 3623–3630. 10.1128/AEM.03909-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazli M., McCarthy Y., Givskov M., Ryan R. P., Tolker-Nielsen T. (2012). The exopolysaccharide gene cluster Bcam1330-Bcam1341 is involved in Burkholderia cenocepacia biofilm formation, and its expression is regulated by c-di-GMP and Bcam1349. Microbiologyopen 2 105–122. 10.1002/mbo3.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazli M., O’Connell A., Nilsson M., Niehaus K., Dow J. M., Givskov M., et al. (2011). The CRP/FNR family protein Bcam1349 is a c-di-GMP effector that regulates biofilm formation in the respiratory pathogen Burkholderia cenocepacia. Mol. Microbiol. 82 327–341. 10.1111/j.1365-2958.2011.07814.x [DOI] [PubMed] [Google Scholar]
- Fazli M., Rybtke M., Steiner E., Weidel E., Berthelsen J., Groizeleau J., et al. (2017). Regulation of Burkholderia cenocepacia biofilm formation by RpoN and the c-di-GMP effector BerB. Microbiologyopen 6:e00480. 10.1002/mbo3.480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira A. S., Silva I. N., Oliveira V. H., Becker J. D., Givskov M., Ryan R. P., et al. (2013). Comparative transcriptomic analysis of the Burkholderia cepacia tyrosine kinase bceF mutant reveals a role in tolerance to stress, biofilm formation, and virulence. Appl. Environ. Microbiol. 79 3009–3020. 10.1128/AEM.00222-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira A. S., Silva I. N., Oliveira V. H., Cunha R., Moreira L. M. (2011). Insights into the role of extracellular polysaccharides in Burkholderia adaptation to different environments. Front. Cell Infect. Microbiol. 1:16. 10.3389/fcimb.2011.00016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin M. Y. (2004). Bacterial signal transduction network in a genomic perspective. Environ. Microbiol. 6 552–567. 10.1111/j.1462-2920.2004.00633.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin M. Y., Nikolskaya A. N., Koonin E. V. (2001). Novel domains of the prokaryotic two-component signal transduction systems. FEMS Microbiol. Lett. 203 11–21. 10.1111/j.1574-6968.2001.tb10814.x [DOI] [PubMed] [Google Scholar]
- Givskov M., Ostling J., Eberl L., Lindum P. W., Christensen A. B., Christiansen G., et al. (1998). Two separate regulatory systems participate in control of swarming motility of Serratia liquefaciens MG1. J. Bacteriol. 180 742–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gjermansen M., Nilsson M., Yang L., Tolker-Nielsen T. (2010). Characterization of starvation-induced dispersion in Pseudomonas putida biofilms: genetic elements and molecular mechanisms. Mol. Microbiol. 75 815–826. 10.1111/j.1365-2958.2009.06793.x [DOI] [PubMed] [Google Scholar]
- Gjermansen M., Ragas P., Sternberg C., Molin S., Tolker-Nielsen T. (2005). Characterization of starvation-induced dispersion in Pseudomonas putida biofilms. Environ. Microbiol. 7 894–906. 10.1111/j.1462-2920.2005.00775.x [DOI] [PubMed] [Google Scholar]
- Gotschlich A., Huber B., Geisenberger O., Togl A., Steidle A., Riedel K., et al. (2001). Synthesis of multiple N-acylhomoserine lactones is wide-spread among the members of the Burkholderia cepacia complex. Syst. Appl. Microbiol. 24 1–14. 10.1078/0723-2020-00013 [DOI] [PubMed] [Google Scholar]
- Hall C. L., Lee V. T. (2018). Cyclic-di-GMP regulation of virulence in bacterial pathogens. Wiley Interdiscip Rev. RNA 9:e1454. 10.1002/wrna.1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harshey R. M. (1994). Bees aren’t the only ones: swarming in gram-negative bacteria. Mol. Microbiol. 13 389–394. 10.1111/j.1365-2958.1994.tb00433.x [DOI] [PubMed] [Google Scholar]
- Hengge R. (2009). Principles of c-di-GMP signalling in bacteria. Nat. Rev. Microbiol. 7 263–273. 10.1038/nrmicro2109 [DOI] [PubMed] [Google Scholar]
- Hengge R., Galperin M. Y., Ghigo J. M., Gomelsky M., Green J., Hughes K. T., et al. (2015). Systematic nomenclature for GGDEF and EAL domain-containing c-di-GMP turnover proteins of Escherichia coli. J. Bacteriol. 198 7–11. 10.1128/JB.00424-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heydorn A., Nielsen A. T., Hentzer M., Sternberg C., Givskov M., Ersboll B. K., et al. (2000). Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146(Pt 10), 2395–2407. 10.1099/00221287-146-10-2395 [DOI] [PubMed] [Google Scholar]
- Hickman J. W., Harwood C. S. (2008). Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Mol. Microbiol. 69 376–389. 10.1111/j.1365-2958.2008.06281.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman J. W., Tifrea D. F., Harwood C. S. (2005). A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc. Natl. Acad. Sci. U.S.A. 102 14422–14427. 10.1073/pnas.0507170102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Y. S., Burden L. M., Hurley J. H. (2000). Structure of the GAF domain, a ubiquitous signaling motif and a new class of cyclic GMP receptor. EMBO J. 19 5288–5299. 10.1093/emboj/19.20.5288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang T. T., Karkhoff-Schweizer R. R., Kutchma A. J., Schweizer H. P. (1998). A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212 77–86. 10.1016/S0378-1119(98)00130-9 [DOI] [PubMed] [Google Scholar]
- Hoch J. A. (2000). Two-component and phosphorelay signal transduction. Curr. Opin. Microbiol. 3 165–170. 10.1016/S1369-5274(00)00070-9 [DOI] [PubMed] [Google Scholar]
- Holden M. T., Seth-Smith H. M., Crossman L. C., Sebaihia M., Bentley S. D., Cerdeno-Tarraga A. M., et al. (2009). The genome of Burkholderia cenocepacia J2315, an epidemic pathogen of cystic fibrosis patients. J. Bacteriol. 191 261–277. 10.1128/JB.01230-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber B., Riedel K., Hentzer M., Heydorn A., Gotschlich A., Givskov M., et al. (2001). The cep quorum-sensing system of Burkholderia cepacia H111 controls biofilm formation and swarming motility. Microbiology 147(Pt 9), 2517–2528. 10.1099/00221287-147-9-2517 [DOI] [PubMed] [Google Scholar]
- Huber B., Riedel K., Kothe M., Givskov M., Molin S., Eberl L. (2002). Genetic analysis of functions involved in the late stages of biofilm development in Burkholderia cepacia H111. Mol. Microbiol. 46 411–426. 10.1046/j.1365-2958.2002.03182.x [DOI] [PubMed] [Google Scholar]
- Inhulsen S., Aguilar C., Schmid N., Suppiger A., Riedel K., Eberl L. (2012). Identification of functions linking quorum sensing with biofilm formation in Burkholderia cenocepacia H111. Microbiologyopen 1 225–242. 10.1002/mbo3.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenal U., Reinders A., Lori C. (2017). Cyclic di-GMP: second messenger extraordinaire. Nat. Rev. Microbiol. 15 271–284. 10.1038/nrmicro.2016.190 [DOI] [PubMed] [Google Scholar]
- Jung H. I., Kim Y. J., Lee Y. J., Lee H. S., Lee J. K., Kim S. K. (2017). Mutation of the cyclic di-GMP phosphodiesterase gene in Burkholderia lata SK875 attenuates virulence and enhances biofilm formation. J. Microbiol. 55 800–808. 10.1007/s12275-017-7374-7 [DOI] [PubMed] [Google Scholar]
- Kader A., Simm R., Gerstel U., Morr M., Römling U. (2006). Hierarchical involvement of various GGDEF domain proteins in rdar morphotype development of Salmonella enterica serovar Typhimurium. Mol. Microbiol. 60 602–616. 10.1111/j.1365-2958.2006.05123.x [DOI] [PubMed] [Google Scholar]
- Kazmierczak B. I., Lebron M. B., Murray T. S. (2006). Analysis of FimX, a phosphodiesterase that governs twitching motility in Pseudomonas aeruginosa. Mol. Microbiol. 60 1026–1043. 10.1111/j.1365-2958.2006.05156.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler B., de Lorenzo V., Timmis K. N. (1992). A general system to integrate lacZ fusions into the chromosomes of gram-negative eubacteria: regulation of the Pm promoter of the TOL plasmid studied with all controlling elements in monocopy. Mol. Gen. Genet. 233 293–301. 10.1007/BF00587591 [DOI] [PubMed] [Google Scholar]
- Kumar B., Sorensen J. L., Cardona S. T. (2018). A c-di-GMP-modulating protein regulates swimming motility of Burkholderia cenocepacia in response to arginine and glutamate. Front. Cell Infect. Microbiol. 8:56. 10.3389/fcimb.2018.00056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. S., Gu F., Ching S. M., Lam Y., Chua K. L. (2010). CdpA is a Burkholderia pseudomallei cyclic di-GMP phosphodiesterase involved in autoaggregation, flagellum synthesis, motility, biofilm formation, cell invasion, and cytotoxicity. Infect. Immun. 78 1832–1840. 10.1128/IAI.00446-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenberg S., Klauck G., Pesavento C., Klauck E., Hengge R. (2013). The EAL domain protein YciR acts as a trigger enzyme in a c-di-GMP signalling cascade in E. coli biofilm control. EMBO J. 32 2001–2014. 10.1038/emboj.2013.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahenthiralingam E., Urban T. A., Goldberg J. B. (2005). The multifarious, multireplicon Burkholderia cepacia complex. Nat. Rev. Microbiol. 3 144–156. 10.1038/nrmicro1085 [DOI] [PubMed] [Google Scholar]
- Martina P., Leguizamon M., Prieto C. I., Sousa S. A., Montanaro P., Draghi W. O., et al. (2018). Burkholderia puraquae sp. nov., a novel species of the Burkholderia cepacia complex isolated from hospital settings and agricultural soils. Int. J. Syst. Evol. Microbiol. 68 14–20. 10.1099/ijsem.0.002293 [DOI] [PubMed] [Google Scholar]
- Massie J. P., Reynolds E. L., Koestler B. J., Cong J. P., Agostoni M., Waters C. M. (2012). Quantification of high-specificity cyclic diguanylate signaling. Proc. Natl. Acad. Sci. U.S.A. 109 12746–12751. 10.1073/pnas.1115663109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy Y., Yang L., Twomey K. B., Sass A., Tolker-Nielsen T., Mahenthiralingam E., et al. (2010). A sensor kinase recognizing the cell-cell signal BDSF (cis-2-dodecenoic acid) regulates virulence in Burkholderia cenocepacia. Mol. Microbiol. 77 1220–1236. 10.1111/j.1365-2958.2010.07285.x [DOI] [PubMed] [Google Scholar]
- Merritt J. H., Ha D. G., Cowles K. N., Lu W., Morales D. K., Rabinowitz J., et al. (2010). Specific control of Pseudomonas aeruginosa surface-associated behaviors by two c-di-GMP diguanylate cyclases. MBio 1:e00183-10. 10.1128/mBio.00183-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messiaen A. S., Nelis H., Coenye T. (2014). Investigating the role of matrix components in protection of Burkholderia cepacia complex biofilms against tobramycin. J. Cyst. Fibros 13 56–62. 10.1016/j.jcf.2013.07.004 [DOI] [PubMed] [Google Scholar]
- Mongkolrob R., Taweechaisupapong S., Tungpradabkul S. (2015). Correlation between biofilm production, antibiotic susceptibility and exopolysaccharide composition in Burkholderia pseudomallei bpsI, ppk, and rpoS mutant strains. Microbiol. Immunol. 59 653–663. 10.1111/1348-0421.12331 [DOI] [PubMed] [Google Scholar]
- Newell P. D., Monds R. D., O’Toole G. A. (2009). LapD is a bis-(3’,5’)-cyclic dimeric GMP-binding protein that regulates surface attachment by Pseudomonas fluorescens Pf0-1. Proc. Natl. Acad. Sci. U.S.A. 106 3461–3466. 10.1073/pnas.0808933106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolskaya A. N., Mulkidjanian A. Y., Beech I. B., Galperin M. Y. (2003). MASE1 and MASE2: two novel integral membrane sensory domains. J. Mol. Microbiol. Biotechnol. 5 11–16. 10.1159/000068720 [DOI] [PubMed] [Google Scholar]
- Norris M. H., Kang Y., Wilcox B., Hoang T. T. (2010). Stable, site-specific fluorescent tagging constructs optimized for burkholderia species. Appl. Environ. Microbiol. 76 7635–7640. 10.1128/AEM.01188-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr M. W., Donaldson G. P., Severin G. B., Wang J., Sintim H. O., Waters C. M., et al. (2015). Oligoribonuclease is the primary degradative enzyme for pGpG in Pseudomonas aeruginosa that is required for cyclic-di-GMP turnover. Proc. Natl. Acad. Sci. U.S.A. 112 E5048–E5057. 10.1073/pnas.1507245112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O′Toole G. A., Kolter R. (1998). Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple pathways convergent signalling: a genetic analysis. Mol. Microbiol. 28 449–461. 10.1046/j.1365-2958.1998.00797.x [DOI] [PubMed] [Google Scholar]
- Palmer K. L., Aye L. M., Whiteley M. (2007). Nutritional cues control Pseudomonas aeruginosa multicellular behavior in cystic fibrosis sputum. J. Bacteriol. 189 8079–8087. 10.1128/JB.01138-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsek M. R., Singh P. K. (2003). Bacterial biofilms: an emerging link to disease pathogenesis. Annu. Rev. Microbiol. 57 677–701. 10.1146/annurev.micro.57.030502.090720 [DOI] [PubMed] [Google Scholar]
- Paul R., Weiser S., Amiot N. C., Chan C., Schirmer T., Giese B., et al. (2004). Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes Dev. 18 715–727. 10.1101/gad.289504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesavento C., Becker G., Sommerfeldt N., Possling A., Tschowri N., Mehlis A., et al. (2008). Inverse regulatory coordination of motility and curli-mediated adhesion in Escherichia coli. Genes Dev. 22 2434–2446. 10.1101/gad.475808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plumley B. A., Martin K. H., Borlee G. I., Marlenee N. L., Burtnick M. N., Brett P. J., et al. (2017). Thermoregulation of biofilm formation in Burkholderia pseudomallei is disrupted by mutation of a putative diguanylate cyclase. J. Bacteriol. 199:e00780-16. 10.1128/JB.00780-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting C. P., Aravind L. (1997). PAS: a multifunctional domain family comes to light. Curr. Biol. 7 R674–R677. 10.1016/S0960-9822(06)00352-6 [DOI] [PubMed] [Google Scholar]
- Povolotsky T. L., Hengge R. (2012). ‘Life-style’ control networks in Escherichia coli: signaling by the second messenger c-di-GMP. J. Biotechnol. 160 10–16. 10.1016/j.jbiotec.2011.12.024 [DOI] [PubMed] [Google Scholar]
- Rao F., Yang Y., Qi Y., Liang Z. X. (2008). Catalytic machanism of c-di-GMP specific phosphodiesterase: a study of the EAL domain-containing RocR from Pseudomonas aeruginosa. J. Bacteriol. 190 3622–3631. 10.1128/JB.00165-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römling U., Galperin M. Y., Gomelsky M. (2013). Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol. Mol. Biol. Rev. 77 1–52. 10.1128/MMBR.00043-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan R. P., Tolker-Nielsen T., Dow J. M. (2012). When the PilZ don’t work: effectors for cyclic di-GMP action in bacteria. Trends Microbiol. 20 235–242. 10.1016/j.tim.2012.02.008 [DOI] [PubMed] [Google Scholar]
- Ryjenkov D. A., Simm R., Römling U., Gomelsky M. (2006). The PilZ domain is a receptor for the second messenger c-di-GMP: the PilZ domain protein YcgR controls motility in enterobacteria. J. Biol. Chem. 281 30310–30314. 10.1074/jbc.C600179200 [DOI] [PubMed] [Google Scholar]
- Sarenko O., Klauck G., Wilke F. M., Pfiffer V., Richter A. M., Herbst S., et al. (2017). More than enzymes that make or break cyclic Di-GMP-local signaling in the interactome of GGDEF/EAL domain proteins of Escherichia coli. mBio 8:e01639-17. 10.1128/mBio.01639-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid N., Pessi G., Deng Y., Aguilar C., Carlier A. L., Grunau A., et al. (2012). The AHL- and BDSF-dependent quorum sensing systems control specific and overlapping sets of genes in Burkholderia cenocepacia H111. PLoS One 7:e49966. 10.1371/journal.pone.0049966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid N., Suppiger A., Steiner E., Pessi G., Kaever V., Fazli M., et al. (2017). High intracellular c-di-GMP levels antagonize quorum sensing and virulence gene expression in Burkholderia cenocepacia H111. Microbiology 163 754–764. 10.1099/mic.0.000452 [DOI] [PubMed] [Google Scholar]
- Silva I. N., Santos P. M., Santos M. R., Zlosnik J. E., Speert D. P., Buskirk S. W., et al. (2016). Long-term evolution of Burkholderia multivorans during a chronic cystic fibrosis infection reveals shifting forces of selection. mSystems 1 e29–e16. 10.1128/mSystems.00029-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon R., Priefer U., Pühler A. (1983). A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Nat. Technol. 1:784–791. 10.1038/nbt1183-784 [DOI] [Google Scholar]
- Suppiger A., Eshwar A. K., Stephan R., Kaever V., Eberl L., Lehner A. (2016). The DSF type quorum sensing signalling system RpfF/R regulates diverse phenotypes in the opportunistic pathogen Cronobacter. Sci. Rep. 6:18753. 10.1038/srep18753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K., Babitzke P., Kushner S. R., Romeo T. (2006). Identification of a novel regulatory protein (CsrD) that targets the global regulatory RNAs CsrB and CsrC for degradation by RNase E. Genes Dev. 20 2605–2617. 10.1101/gad.1461606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchigvintsev A., Xu X., Singer A., Chang C., Brown G., Proudfoot M., et al. (2010). Structural insight into the mechanism of c-di-GMP hydrolysis by EAL domain phosphodiesterases. J. Mol. Biol. 402 524–538. 10.1016/j.jmb.2010.07.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traverse C. C., Mayo-Smith L. M., Poltak S. R., Cooper V. S. (2013). Tangled bank of experimentally evolved Burkholderia biofilms reflects selection during chronic infections. Proc. Natl. Acad. Sci. U.S.A. 110 E250–E259. 10.1073/pnas.1207025110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschowri N., Busse S., Hengge R. (2009). The BLUF-EAL protein YcgF acts as a direct anti-repressor in a blue-light response of Escherichia coli. Genes Dev. 23 522–534. 10.1101/gad.499409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschowri N., Schumacher M. A., Schlimpert S., Chinnam N. B., Findlay K. C., Brennan R. G., et al. (2014). Tetrameric c-di-GMP mediates effective transcription factor dimerization to control streptomyces development. Cell 158 1136–1147. 10.1016/j.cell.2014.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanlaere E., Baldwin A., Gevers D., Henry D., De Brandt E., LiPuma J. J., et al. (2009). Taxon K, a complex within the Burkholderia cepacia complex, comprises at least two novel species, Burkholderia contaminans sp. nov. and Burkholderia lata sp. nov. Int. J. Syst. Evol. Microbiol. 59 102–111. 10.1099/ijs.0.001123-0 [DOI] [PubMed] [Google Scholar]
- Yakandawala N., Gawande P. V., LoVetri K., Cardona S. T., Romeo T., Nitz M., et al. (2011). Characterization of the poly-beta-1,6-N-acetylglucosamine polysaccharide component of Burkholderia biofilms. Appl. Environ. Microbiol. 77 8303–8309. 10.1128/AEM.05814-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C., Cui C., Ye Q., Kan J., Fu S., Song S., et al. (2017). Burkholderia cenocepacia integrates cis-2-dodecenoic acid and cyclic dimeric guanosine monophosphate signals to control virulence. Proc. Natl. Acad. Sci. U.S.A. 114 13006–13011. 10.1073/pnas.1709048114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zogaj X., Nimtz M., Rohde M., Bokranz W., Römling U. (2001). The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol. Microbiol. 39 1452–1463. 10.1046/j.1365-2958.2001.02337.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.