Abstract
Sulfur oxidation is an essential component of the earth’s sulfur cycle. Acidithiobacillus spp. can oxidize various reduced inorganic sulfur compounds (RISCs) with high efficiency to obtain electrons for their autotrophic growth. Strains in this genus have been widely applied in bioleaching and biological desulfurization. Diverse sulfur-metabolic pathways and corresponding regulatory systems have been discovered in these acidophilic sulfur-oxidizing bacteria. The sulfur-metabolic enzymes in Acidithiobacillus spp. can be categorized as elemental sulfur oxidation enzymes (sulfur dioxygenase, sulfur oxygenase reductase, and Hdr-like complex), enzymes in thiosulfate oxidation pathways (tetrathionate intermediate thiosulfate oxidation (S4I) pathway, the sulfur oxidizing enzyme (Sox) system and thiosulfate dehydrogenase), sulfide oxidation enzymes (sulfide:quinone oxidoreductase) and sulfite oxidation pathways/enzymes. The two-component systems (TCSs) are the typical regulation elements for periplasmic thiosulfate metabolism in these autotrophic sulfur-oxidizing bacteria. Examples are RsrS/RsrR responsible for S4I pathway regulation and TspS/TspR for Sox system regulation. The proposal of sulfur metabolic and regulatory models provide new insights and overall understanding of the sulfur-metabolic processes in Acidithiobacillus spp. The future research directions and existing barriers in the bacterial sulfur metabolism are also emphasized here and the breakthroughs in these areas will accelerate the research on the sulfur oxidation in Acidithiobacillus spp. and other sulfur oxidizers.
Keywords: Acidithiobacillus, sulfur oxidation, two-component system, elemental sulfur oxidation, thiosulfate oxidation pathways, sulfide oxidation, sulfite oxidation
Introduction
Acidithiobacillus, the gram-negative sulfur-oxidizing chemolithotrophic bacteria in the proteobacterial class Acidithiobacillia, formerly belonged to the genus “Thiobacillus” (Vishniac and Santer, 1957; Kelly and Wood, 2000; Williams and Kelly, 2013). However, due to their higher acid-tolerance and relatively closer evolutionary relationships with each other compared to other species in the genus Thiobacillus, they were reclassified as a new genus “Acidithiobacillus” in 2000 (Kelly and Wood, 2000). Members of this genus have the remarkable capability of oxidizing various reduced inorganic sulfur compounds (RISCs) to obtain electrons for carbon dioxide fixation, and some of them also have ferrous iron oxidation ability (Harrison, 1984). On the basis of physiological characters and 16S rRNA gene sequence comparisons, the genus Acidithiobacillus has been classified into seven different species (Table 1; Waksman and Joffe, 1922; Temple and Colmer, 1951; Hallberg and Lindstrom, 1994; Xia et al., 2007; Liljeqvist et al., 2012; Falagan and Johnson, 2016; Nunez et al., 2017). Based on the differences in the energy-substrates, species in Acidithiobacillus can be divided into two groups: the sulfur-oxidizing-only species, including Acidithiobacillus thiooxidans, Acidithiobacillus caldus and Acidithiobacillus albertensis, and the sulfur- and ferrous- oxidizing species, including Acidithiobacillus ferrooxidans, Acidithiobacillus ferrivorans, Acidithiobacillus ferriphilus, and Acidithiobacillus ferridurans (Table 1). Acidithiobacillus strains are widely distributed in acidic sulfur-containing environments on land or in the sea, including soil, sediments, hot springs, iron-sulfur mineral deposits and acid mine drainage (AMD), where these bacteria participate in the global element cycles of sulfur and iron, promoting the oxidation of RISCs to sulfate and the conversions between ferrous and ferric ions (London and Rittenberg, 1964; Nielsen and Beck, 1972; Taylor et al., 1984; Schrenk et al., 1998; Jones et al., 2012; Hua et al., 2015; Sharmin et al., 2016).
Table 1.
Taxonomic traits of seven species in the genus of Acidithiobacillus.
| Trait | A. ferrooxidans | A. ferrivorans | A. ferriphilus | A. ferridurans | A. thiooxidans | A. caldus | A. albertensis |
|---|---|---|---|---|---|---|---|
| Gram stain | - | - | - | - | - | - | - |
| Cell size (μm) | 1.0 × 0.5 | 2.4 × 0.5 | 1–2 | 1–2 | 1.0–2.0 × 0.5 | 1.2–1.9 × 0.7 | 1–2 × 0.4–0.6 |
| Motility | +/- | + | + | + | + | + | + |
| Growth pH (optimum) | 1.3–4.5 (2.0–2.5) | 1.9–3.4 (2.5) | 1.5 (2.0) | 1.4–3.0 (2.1) | 0.5–5.5 (2.0–3.0) | 1.0–3.5 (2.0–2.5) | 0.5-6.0 (3.5–4.0) |
| Growth T/°C (optimum) | 10–37 (30–35) | 4–37 (28–33) | 5–33 (30) | 10–37 (29) | 10–37 (28–30) | 32–52 (40–45) | 10–40 (25–30) |
| Oxidation of S0, S4O62-, S2O32- | + | + | + | + | + | + | + |
| Oxidation of Fe2+ | + | + | + | + | - | - | - |
| Growth on sulfide minerals | + | + | + | + | - | - | - |
| Growth on hydrogen | + | (+) | - | + | - | + | NR |
| Anaerobic growth with Fe3+ | + | + | + | + | - | - | - |
| N2 fixation | + | + | NR | NR | - | - | - |
| Mol% G+C | 58–59 | 55–56 | 57.4 | 58.4 | 52 | 63–64 | 61.5 |
| Thiosulfate-metabolic pathways | TSD enzyme; S4I pathway | Sox system; TSD enzyme; S4I pathway | NR | TSD enzyme; S4I pathway | Sox system; S4I pathway | Sox system; S4I pathway | Sox system; S4I pathway |
| Reference | Janiczek et al., 2007; Valdés et al., 2008a,b; Hallberg et al., 2010; Hedrich and Johnson, 2013b; Kikumoto et al., 2013 | Hallberg et al., 2010; Hedrich and Johnson, 2013b; Christel et al., 2016 | Falagan and Johnson, 2016 | Hedrich and Johnson, 2013a,b; Miyauchi et al., 2018 | Valdés et al., 2008b; Hallberg et al., 2010; Yin et al., 2014 | Valdés et al., 2008b; Hallberg et al., 2010; Mangold et al., 2011; Hedrich and Johnson, 2013b | Bryant et al., 1983; Xia et al., 2007; Castro et al., 2017 |
+, positive; -, negative; +/-, the positive or negative result from different reports; (+), some strains have the ability to oxidize hydrogen; NR, not reported; Tm, temperature.
Acidithiobacillus spp. are prevalent in acid mines due to their capabilities of utilizing the sulfur and iron in ores and adapting to extremely acidic environments. As a consequence, Acidithiobacillus spp. have become the most active bacteria used in the biohydrometallurgy industry in bioleaching or biomining, whereby metals are extracted from ores through microbial oxidation (Rawlings et al., 1999; Rawlings, 2005). Three species, A. ferrooxidans A. thiooxidans, and A. caldus, have been studied extensively and applied widely in bioleaching for mineral extraction from ores (Valdés et al., 2008b). In addition, the ability of heavy metal leaching has expanded the application of Acidithiobacillus spp. from hydrometallurgy to the treatment of wastes containing heavy metals, such as sewage sludge, spent household batteries, mine tailings, and printed circuit boards (Pathak et al., 2009; Bayat and Sari, 2010; Arshadi and Mousavi, 2014; Ijadi Bajestani et al., 2014; Nguyen et al., 2015; Rastegar et al., 2015). Moreover, these bacteria have been widely studied in microbial desulfurization of coal and gas (Azizan et al., 2000; He et al., 2012; Charnnok et al., 2013). Taken together, Acidithiobacillus spp. have shown their great value of applications not only in metal leaching (copper, uranium, gold and so on) from mineral ores, but also in solving environmental pollution problems caused by heavy metals and inorganic sulfur compounds.
Sulfur oxidation, as the essential physiological feature of Acidithiobacillus spp. and the important character for their application, has attracted extensive attention (Suzuki and Werkman, 1959; London, 1963; London and Rittenberg, 1964; Suzuki et al., 1992; Hallberg et al., 1996; Quatrini et al., 2009; Chen et al., 2012; Yin et al., 2014; Wang et al., 2016; Nunez et al., 2017; Zhang et al., 2018). The element sulfur can exist in various oxidation states ranging from -2 to +6, which results in a variety of RISCs including tetrathionate (S4O62-), thiosulfate (S2O32-), sulfite (SO32-), sulfide (S2-), and elemental sulfur (S0). A variety of enzymes and proteins involved in the oxidation of RISCs were discovered, including sulfur-oxidizing enzymes, sulfur transferases and sulfur carrier proteins. The sulfur-metabolic enzymes in Acidithiobacillus spp., based on their substrates, can be categorized as elemental sulfur oxidation enzymes, enzymes in thiosulfate oxidation pathways, sulfide oxidation enzymes, and sulfite oxidation enzymes. These enzymes work cooperatively to oxidize the RISCs to the final product sulfate. Therefore, the identification of novel sulfur-metabolic proteins and investigation of the metabolic and regulatory mechanisms of these known sulfur oxidation proteins have been the main subject of researches on sulfur oxidation in Acidithiobacillus spp. Significant research progress was made over the last decades in these areas. Here, the key points are summarized to provide an overall picture of sulfur oxidation in Acidithiobacillus spp.
Elemental Sulfur Oxidation
Sulfur Dioxygenase (SDO)
Elemental sulfur (S0), mainly present in the form of insoluble homocyclic S8, is hydrophobic, metastable, and almost insoluble in water. Elemental sulfur oxidation activity was first detected in A. thiooxidans as early as 1959 (Suzuki and Werkman, 1959). The purified enzyme that is associated with this activity was named sulfur dioxygenase (SDO, EC 1.13.11.18) and reduced glutathione (GSH) was necessary for activity detection in in vitro assays. It was reported that Sulfur dioxygenase (SDO) is composed of a 21- and a 26- kDa protein in A. thiooxidans or two 23 kDa subunits in A. ferrooxidans (Suzuki, 1965; Silver and Lundgren, 1968a; Sugio et al., 1987a). Assays in vitro revealed that the actual substrate for SDO-catalyzed reaction was the sulfane sulfur atom of glutathione persulfide (GSSH) and its homologs (GSSnH, n > 1) (Rohwerder and Sand, 2003, 2008), but the amino acid sequences of SDO were not identified at that time.
In recent years, SDO activity was also detected in the mitochondria of plants and animals and in heterotrophic bacteria, and protein sequences were identified in these organisms. This enzyme was originally named as SDO in Acidithiobacillus spp. Based on the further studies, two new names, ETHE1 and persulfide dioxygenase (PDO), were proposed for SDO homologs in mitochondria and heterotrophic bacteria, respectively (Jackson et al., 2012; Kabil and Banerjee, 2012; Liu H. et al., 2014; Sattler et al., 2015). To avoid any confusion, here we use SDO throughout this paper to refer to this enzyme in Acidithiobacillus spp. The mitochondrial ETHE1s identified in human and Arabidopsis both belong to the metallo-β-lactamase superfamily, and their metal-binding active sites consist of an aspartate and two histidine residues (Tiranti et al., 2009; Holdorf et al., 2012). Based on phylogenetic analysis, heterotrophic bacterial PDOs were classified into three subgroups (Liu H. et al., 2014). Structural analyses indicated that the PDOs in heterotrophic bacteria contained the conserved amino acid residues, and differences in the GS-moiety binding sites of the key amino acid residues supported grouping of PDOs (Sattler et al., 2015). The major physiological function of SDOs in mitochondria or in heterotrophic bacteria has been proposed: ETHE1s/ PDOs works cooperatively with sulfide:quinone oxidoreductase (SQR) to oxidize H2S, relieving the toxic effect of H2S to the cells (Guimaraes et al., 2011; Kabil and Banerjee, 2012; Liu H. et al., 2014).
Sulfur dioxygenase in Acidithiobacillus spp. was previously proposed to serve as the first enzyme for extracellular elemental sulfur oxidation. This was concluded from a SDO-dependent sulfur oxidation model: extracellular elemental sulfur was first activated by thiol-containing outer-membrane proteins to generate persulfide sulfane sulfur, and this product was further oxidized by periplasmic SDO to produce sulfite (Rohwerder and Sand, 2003). The nucleotide and amino acid sequences of SDO in Acidithiobacillus spp. were not identified until 2014 (Wang et al., 2014). Two homologs of human ETHE1, AFE_0269 in A. ferrooxidans ATCC 23270 and A5904_0790 in A. caldus MTH-04, exhibited a remarkable GSH-dependent SDO activity in in vitro assays (Wang et al., 2014). Recently, a second SDO (A5904_0421, termed SDO1), with 33% amino acid identity to previously identified Ac-SDO (A5904_0790, termed SDO2), was identified in A. caldus MTH-04. The enzymatic activity of SDO1 is much lower than that of SDO2 (Wu et al., 2017). The latest study on ETHE1-like SDO (SDO2 homolog) from A. caldus C-SH12 suggested that AcSDO is a homotetramer containing a mononuclear iron site with a 2-His-1-carboxylate facial triad in the active site. The key amino acid residues of this protein were described recently (Ruhl et al., 2018).
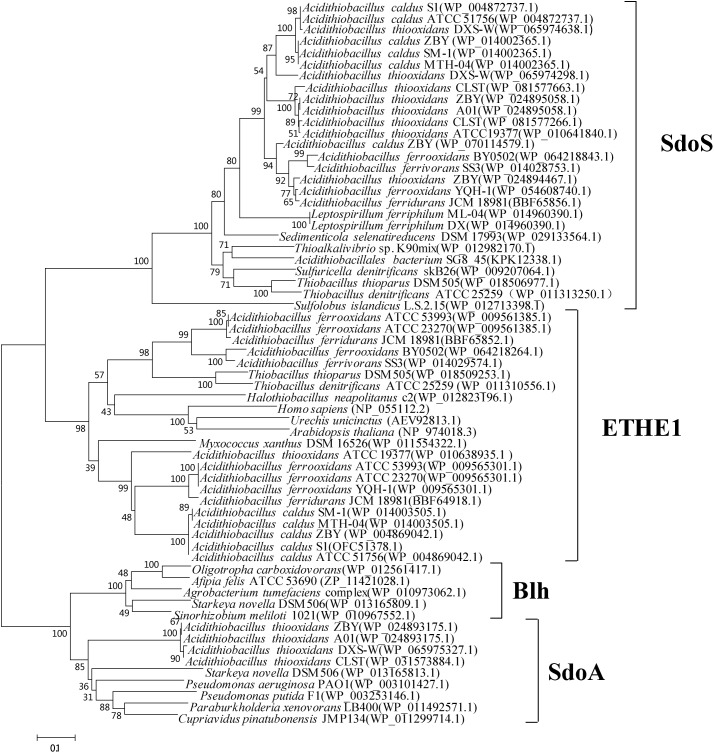
There are two to three copies of SDO paralogs in different species of Acidithiobacillus (Figure 1; Wu et al., 2017). A. caldus SDO1 and its homologs form a new subgroup distinct from the well described subgroups (ETHE1, Blh, SdoA), named as SdoS, whereas the SDO2 of A. caldus and its homologs belong to the ETHE1 subgroup (Figure 1; Wu et al., 2017). Phylogenetic analysis also revealed that some potential SDOs from A. thiooxidans belong to the SdoA subgroup (Figure 1). Studies on the SDOs indicated the ubiquitous existence of this elemental sulfur-oxidizing enzyme in these chemoautotrophic sulfur-oxidizing bacteria.
FIGURE 1.
Phylogenetic analysis of SDOs in Acidithiobacillus spp. and other typical prokaryotes. MEGA 5.0 with the bootstrap test (1000 replicates) were used to construct NJ-tree. The protein ID or locus_tag of each SDO is present in parentheses. The predicted functional domains of proteins whose sequences identities are higher than 30% are selected for this phylogenetic analysis.
Recently, further phylogenetic analysis on bacterial PDOs suggested PDOs can be reclassified into three types (Xia et al., 2017). The subgroups of SdoA and Blh were categorized into the type II PDOs, and ETHE1 homologous proteins are grouped as type I PDOs. The type III PDOs exhibited low sequence identities with the type I and type II PDOs. According to the new classification of PDOs, the SDOs in the SdoS subgroup belong to the type III category. Studies on SDOs in SdoS and ETHE1 subgroups found that there are obvious differences in the key amino acid residues of the substrate binding regions (unpublished data), indicating different functions of SDO1 and SDO2 in the process of sulfur oxidation in Acidithiobacillus spp.
The role of SDOs in sulfur oxidation in Acidithiobacillus was studied using sdo deletion and overexpression strains. When grown in liquid S0-medium, the sdo (AFE_0269) mutant of A. ferrooxidans ATCC 23270 grew much slower than the wild type. Moreover, cell extracts of the mutant still maintained the SDO activity when cultivated in S0- or Fe2+- medium (Wang et al., 2014). No significant differences in growth were observed among the three sdo mutants of A. caldus MTH-04 (Δsdo1, Δsdo2, and Δsdo1&2) and the wild type strain in S0-media, and the SDO activities of cell extracts from these mutants were not lower than that from wild type (Wu et al., 2017). All these results indicated that the absence of SDOs in A. ferrooxidans and A. caldus neither impaired their elemental sulfur oxidation activities nor caused lethal effects on their growth rates in S0-media. It was suggested that SDO1 in A. caldus is involved in the S4O62- metabolic process, because when cultivated in tetrathionate at a concentration of 2.27 g/L, strains Δsdo1 and Δsdo1&2 did not exhibit any OD increase in growth experiments (Wu et al., 2017). Transcriptional analysis on sdo deletion and overexpression strains of A. caldus showed that the transcription levels of sdo1 and sdo2 had close linkages to those of tetH (encoding a tetrathionate hydrolase) and sqr (encoding a sulfide:quinone oxidoreductase), respectively (Wu et al., 2017). In contrary to the previous hypothesis that SDO functioned in the oxidation of persulfide sulfane sulfur in periplasm, it is now believed that all the SDO homologs in Acidithiobacillus spp. are cytoplasmic proteins due to the lack of signal peptides and transmembrane regions (Wu et al., 2017). Therefore, based on the current knowledge, SDOs in Acidithiobacillus spp. are believed to be involved in cytoplasmic elemental sulfur oxidation, and different subgroups of SDOs are probably responsible for the oxidation of elemental sulfur generated by different pathways: the ETHE1-subgroup of SDOs are involved in the H2S-oxidation pathway and the SdoS-subgroup of SDOs are related to the S4O62--decomposition pathway (Wu et al., 2017).
Sulfur oxygenase reductase (SOR) is another elemental sulfur oxidizing enzyme found in Acidithiobacillus spp. This enzyme, first reported in several acidophilic and thermophilic archaea, can catalyze the disproportionation of cytoplasmic elemental sulfur and generate thiosulfate, sulfite, and sulfide (Kletzin, 1989; Kletzin et al., 2004; Ghosh and Dam, 2009). The reaction is dioxygen (O2)-dependent with no external cofactors or electron donors required, and the oxidation is coupled with neither electron transfer nor substrate-level phosphorylation (Kletzin, 1989; Urich et al., 2004). The SORs from archaeal and bacterial species are large hollow spheres consisting of 24 identical subunits. Each SOR monomer has a catalytic pocket containing an indispensable cysteine and a low-potential non-heme iron site (Urich et al., 2006; Li et al., 2008).
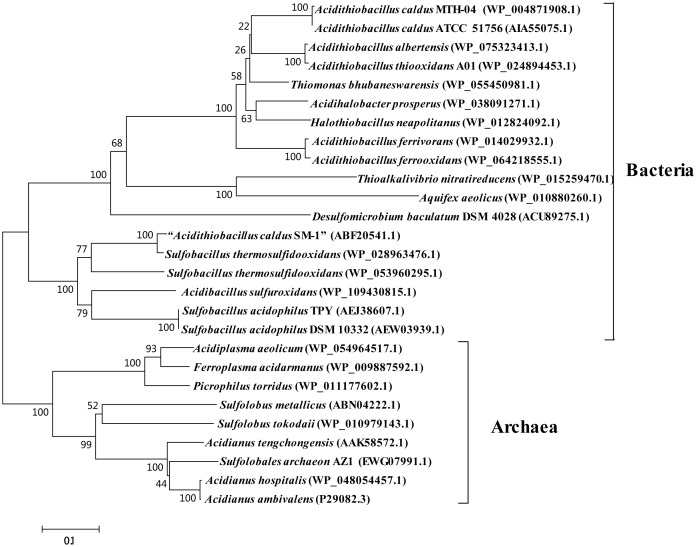
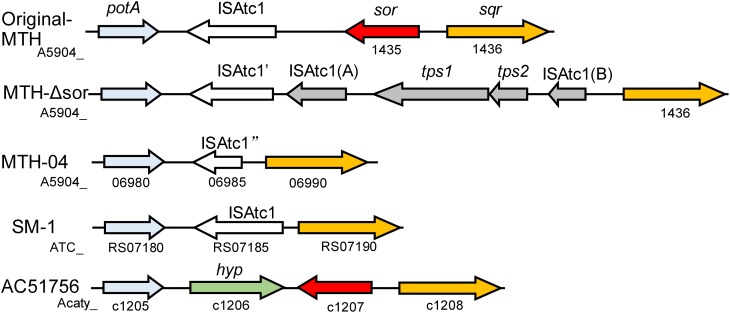
Although SOR activity from A. caldus SM-1 cultivated in bioreactors treating gold-bearing concentrates was reported (Chen et al., 2007), a subsequent study indicated that the sor gene was absent in the complete genome sequence of this strain (You et al., 2011). Sequence analysis of SOR proteins indicated that the SOR ascribed to A. caldus SM-1, was actually isolated from Sulfobacillus contaminated sample (Figure 2; Janosch et al., 2015). The SOR homolog was found in our laboratorial strain A. caldus MTH-04, and the overexpression of sor in A. caldus MTH-04 increased SOR activity by 22.2% and meanwhile resulted in a growth advantage after the mid-log phase (unpublished data). SOR was defined as a cytoplasmic S0-oxidizing enzyme based on studies on a sor deficient mutant of A. caldus MTH-04 (Chen et al., 2012). SOR homologs have been found in some, but not all, strains of A. thiooxidans, A. ferrooxidans, A. ferrivorans, A. albertensis (Figure 2; Valdés et al., 2008b; Yin et al., 2014; Christel et al., 2016). Phylogenetic analysis indicated that the SORs in these Acidithiobacillus spp. strains were probably acquired from sulfur-oxidizing archaea via horizontal gene transfer (Figure 2). Moreover, the sor gene could be eliminated via transposition in strain MTH-04 of A. caldus (Figure 3; Chen et al., 2012). For our laboratory strain MTH-04, both the electrotransformation of suicide plasmid specific for sor gene mutagenesis and the continuous passage in S0-media in the laboratory cultivation environment resulted in the loss of sor gene (Figure 3). The tendency of eliminating the sor gene in A. caldus, the sporadic distribution and the relatively low retention rate of the sor gene in Acidithiobacillus spp. indicate that SOR is supplementary (You et al., 2011; Chen et al., 2012; Janosch et al., 2015; Christel et al., 2016), but not necessary, for cytoplasmic elemental sulfur oxidation in these sulfur-oxidizing bacteria.
FIGURE 2.
Phylogenetic analysis of SORs in some Acidithiobacillus spp. and other typical prokaryotes. MEGA 5.0 with the bootstrap test (1000 replicates) were used to construct NJ-tree. The protein ID or locus_tag of each SOR is present in parentheses. The predicted functional domains of proteins whose sequences identities are higher than 30% are selected for this phylogenetic analysis. The SOR reported from Acidithiobacillus caldus SM-1 was actually SOR from Sulfobacillus spp. Thus, it was marked as “A. caldus SM-1” in the phylogenetic tree (Chen et al., 2007; Janosch et al., 2015).
FIGURE 3.
The loci of the sor gene on the genomes of different A. caldus strains. Original-MTH, A. caldus MTH-04 wild type harboring sor gene (the sor-sqr gene cluster sequence GenBank: MK165447); MTH-Δsor, A. caldus MTH-04 sor mutant generated by electroporation of the suicide plasmid (the sqr gene cluster sequence GenBank: MK165449); MTH-04, A. caldus MTH-04 wild type lacking sor gene (GenBank: CP026328.1); SM-1, A. caldus SM-1 (GenBank: CP002573.1); ACA, A. caldus ATCC 51765 (GenBank:CP005986.1); potA, ABC transporter ATP-binding protein; ISAtc1, IS elements; tps1&2, transposase; sor, sulfur oxygenase reductase; sqr, sulfide:quinone oxidoreductase; hyp, hypothetical protein. The gene locus on the chromosome was shown below the corresponding gene.
Heterodisulfide Reductase (Hdr)-Like System
A Hdr-like complex is proposed to serve as an elemental sulfur oxidation enzyme in the cytoplasmic space of Acidithiobacillus and many other sulfur-oxidizing bacteria and archaea (Quatrini et al., 2009; Mangold et al., 2011; Chen et al., 2012; Inskeep et al., 2013; Yin et al., 2014; Liu L.J. et al., 2014; Dahl, 2015; Christel et al., 2016). Proteins of the Hdr-like system from bacterial and archaeal sulfur oxidizers are homologous to those in the HdrABC complex from methanogenic archaea, sulfate-reducing archaea and sulfate-reducing bacteria (Hedderich et al., 2005). HdrABC is a unique disulfide reductase that catalyzes the reversible reduction of the disulfide bond X-S-S-X coupled with the energy conservation (Thauer et al., 2008; Kaster et al., 2011; Wagner et al., 2017). Studies have shown that the HdrABC complex is composed of three subunits: HdrA carrying a typical FAD binding motif and four binding motifs for [4Fe-4S] clusters, HdrB harboring two identical non-cubane [4Fe-4S] clusters and both of these clusters are consisted of [3Fe-4S] and [2Fe-2S] subclusters, and the ferredoxin-like HdrC containing two binding motifs for [4Fe-4S] clusters (Hamann et al., 2007; Wagner et al., 2017). In methanogenic and sulfate-reducing archaea, HdrA receives the electrons from a hydrogenase and transfers them through HdrC to the heterodisulfide reductase catalytic site on HdrB (Mander et al., 2004). Unlike the HdrABC complex, the Hdr-like system in sulfur-oxidizing bacteria and archaea is encoded by a hdrC1B1A-hyp-hdrC2B2 gene cluster and consists of at least five subunits (HdrA, HdrB1, HdrB2, HdrC1, and HdrC2) (Boughanemi et al., 2016).
Hdr-like systems have been discovered in different species of Acidithiobacillus, and the function of this complex was proposed to be oxidation of disulfide intermediates (most likely sulfane sulfur as in GSSH or other sulfur carriers) to sulfite in the cytoplasm (Quatrini et al., 2009; Mangold et al., 2011; Chen et al., 2012; Yin et al., 2014; Christel et al., 2016; Koch and Dahl, 2018.) The transcriptional levels of hdr genes were upregulated when A. ferrooxidans or A. thiooxidans were cultivated in sulfur-containing media (Ehrenfeld et al., 2013). HdrC from A. ferrooxidans was heterologously expressed and the recombinant protein harbored [4Fe–4S] clusters (Ossa Henao et al., 2011). However, biochemical evidence confirming the function of Hdr-like systems is still absent. Recently, an indirect genetic study indicated that a Hdr-like complex functions in the oxidation of thiosulfate to sulfite in Hyphomicrobium denitrificans (Koch and Dahl, 2018). Moreover, a lipoate-binding protein (LbpA) was identified as a necessary component for the Hdr-like sulfur-oxidizing system in H. denitrificans (Cao et al., 2018). Thus, a sulfur oxidation pathway was proposed in which the Hdr-like complex oxidizes the sulfane sulfur delivered by the sulfur carrier TusA to sulfite, and the released electrons in the reaction might be transferred via LbpA to generate NADH (Cao et al., 2018). The confirmation of the sulfur-oxidizing ability of Hdr-like complex and the sulfur-metabolizing process catalyzed by this system in H. denitrificans, would undoubtedly provide new insights in the elemental sulfur oxidation in the cytoplasm of Acidithiobacillus spp., and promote research on the Hdr-like complex in these sulfur-oxidizers.
The presence of SDO, SOR, and Hdr-like complex in Acidithiobacillus spp. indicates the diversity and complexity of elemental sulfur oxidation in these acidophilic bacteria. The triple sor-sdo1-sdo2 mutant of A. caldus MTH-04 exhibited an increased elemental sulfur oxidation activity, indicating the existence of undetermined elemental sulfur oxidation enzymes in Acidithiobacillus spp. (Wu et al., 2017). Direct and conclusive evidences are needed to confirm the function of Hdr-like complex in these Acidithiobacillus strains. In addition, the distinct roles, the potential cooperative effects and regulation modes of the three cytoplasmic elemental sulfur oxidation enzymes in Acidithiobacillus spp. could be the emphasis of future researches.
Sulfur Trafficking in Acidithiobacillus
The sulfur carrier proteins TusA and DsrE exist in many sulfur oxidation bacteria and archaea (Liu L.J. et al., 2014; Dahl, 2015). The rhd-tusA-dsrE genes were reported in A. caldus and A. ferrooxidans, and these genes, together with the Hdr-like system gene cluster (hdrC1B1A-hyp-hdrC2B2) and sulfite-oxidation-enzyme genes (soeABC), exist in one gene cluster in A. caldus SM-1 (soeABC-mogA-rhd-tusA-dsrE-hdrC1-hdrB1-hdrA-hyp-hdrC2- hdrB2, Atc_2359-2347) (Liu L.J. et al., 2014). Similar gene clusters can also be found in other species of Acidithiobacillus (unpublished data). The unique gene arrangement character suggests potential functional connections among elemental sulfur oxidation (Hdr-like complex), Rhd-DsrE-TusA-mediated sulfur transfer and sulfite oxidation in the cytoplasm of Acidithiobacillus spp. Studies on the sulfur-trafficking proteins from Metallosphaera cuprina and Allochromatium vinosum indicated that inorganic sulfur compounds were successively transferred by Rhd, DsrE, and TusA to form sulfane sulfur at the cysteine of TusA (Liu L.J. et al., 2014; Stockdreher et al., 2014; Dahl, 2015). TusA serves as a central component of cytoplasmic sulfur trafficking in sulfur-oxidizing prokaryotes (Dahl, 2015), and might deliver the sulfane sulfur to the Hdr-like sulfur-oxidizing system (Cao et al., 2018). Thus, TusA can be considered as a joint between sulfur trafficking mediated by Rhd/DsrE/TusA and the sulfur oxidation catalyzed by Hdr-like complex in these sulfur-oxidizers. The discovery of conserved genetic clusters in several Acidithiobacillus species suggested that similar sulfur trafficking and oxidation pathways might also work in the cytoplasm of these sulfur-oxidizers. Thus, it would be interesting to perform further protein characterization assays in vitro and gene function studies in vivo to confirm the function of these proteins in the process of sulfur trafficking and oxidation in Acidithiobacillus spp.
Rhodanese (Rhd) might be an important functional enzyme in sulfur trafficking and sulfur oxidation of Acidithiobacillus (Quatrini et al., 2009; Chen et al., 2012; Liu L.J. et al., 2014; Yin et al., 2014; Koch and Dahl, 2018). Rhd belongs to the sulfurtransferase family, which is found in organisms from all three domains of life and involved in various cellular processes (Smit and Urbanska, 1986; Aminlari and Gilanpour, 1991; Cipollone et al., 2007). This enzyme is a thiosulfate:cyanide sulfur transferase (TST), which cleaves the S–S bond present in thiosulfate, producing sulfur and sulfite. Rhd activities were detected in crude enzyme extracts of A. ferrooxidans, A. thiooxidans and A. caldus (Tabita et al., 1969; Gardner and Rawlings, 2000). Genomic sequences revealed that there are multiple copies of putative rhd genes in the genomes of Acidithiobacillus (Valdés et al., 2008a, 2011; Liljeqvist et al., 2011; Yin et al., 2014). A 21-kDa rhodanese-like protein (P21) of A. ferrooxidans was induced when cells were grown on metal sulfides and different sulfur compounds, but the purified recombinant P21 protein did not show Rhd activity in vitro (Ramírez et al., 2002). Furtherly, eight rhodanese-like proteins from A. ferrooxidans were cloned and expressed in E. coli. Some of the recombinant proteins had the Rhd activities, and the others including the P21 did not show this activity, indicating the potential different physiological roles of these rhodanese-like proteins in this bacterium (Acosta et al., 2005). Rhd probably plays an essential role in sulfur oxidation in A. caldus since we had no success in the deletion of the rhd gene in the hdr-rhd-tusA-dsrE gene cluster (Zhang et al., 2014). Further bioinformatics analysis of these Rhd proteins is needed to reveal the sequence similarities and potential protein localizations in these autotrophic sulfur-oxidizing bacteria.
Thiosulfate Oxidation
Thiosulfate (S2O32-) plays an important role in the biogeochemical sulfur cycle. It is a common substrate and a key sulfur-metabolic intermediate oxidized by almost all sulfur-oxidizing microorganisms, thus thiosulfate metabolism is essential for these sulfur-oxidizers (Dahl and Prange, 2006; Ghosh and Dam, 2009). In acidic environments (pH < 4.0), thiosulfate can decompose to sulfur and sulfite chemically (Johnston and McAmish, 1973), and the generated sulfur exists as colloidal sulfur that cannot be used by Acidithiobacillus spp. To efficiently metabolize thiosulfate, all the species in the genus Acidithiobacillus possess complicated thiosulfate-oxidizing multi-enzyme systems to immediately oxidize thiosulfate and sulfur-transfer enzymes to transform the thiosulfate to other forms of sulfur-substrates (Valdés et al., 2008b).
S4I Pathway
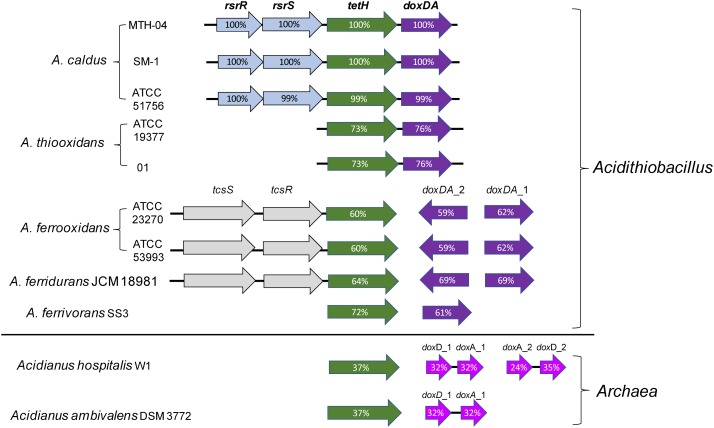
A thiosulfate oxidation pathway via formation of tetrathionate as an intermediate (S4I) is widely found in β- and γ- proteobacteria, particularly in obligately chemolithotrophic genera including Acidithiobacillus, Thermithiobacillus, Halothiobacillus, and Tetrathiobacter (Dam et al., 2007; Ghosh and Dam, 2009). The S4I pathway is made up of a thiosulfate:quinol oxidoreductase (TQO or DoxDA) and a tetrathionate hydrolase (TetH or TTH). TQO oxidizes thiosulfate to tetrathionate while TetH hydrolyzes tetrathionate to thiosulfate and other products. The fact that the predicted tetH and doxDA genes are found in all the published genomes of Acidithiobacillus (Table 1), indicates the universality and importance of the S4I pathway in these acidophilic chemotrophic bacteria (Rzhepishevska et al., 2007; Wang et al., 2016). As shown in Figure 4, tetH and doxDA are arranged in a cluster in A. caldus and A. thiooxidans, while they are located separately in the genomes of A. ferrooxidans, A. ferrivorans, and A. ferridurans. Two copies of doxDA genes are located separately in the genomes of A. ferrooxidans and A. ferridurans. There are two-component systems (TCSs) located upstream of the tetH genes in A. caldus, A. ferrooxidans, and A. ferridurans. Moreover, the transcription of tetH and doxDA is influenced significantly by different sulfur-substrates in the media (Mangold et al., 2011; Chen et al., 2012; Yin et al., 2014; Christel et al., 2016), indicating that Acidithiobacillus spp. can modulate the S4I pathway at the transcriptional level in response to the various sulfur-metabolites in the growth environment.
FIGURE 4.
Arrangement of genes of S4I pathways in Acidithiobacillus spp. and sulfur-oxidizing archaea. The percentages of similarities between protein sequences are indicated by the values marked in the gene. Accession numbers (GenBank) for these proteins are: A. caldus MTH-04, RsrR (ANJ65973.1), RsrS (ANJ65974.1), TetH (OAN03451.1), DoxDA (OAN03452.1) (GenBank: MK165448); A. caldus SM-1, RsrR (AEK59530.1), RsrS (AEK58242.1), TetH (AEK58243.1), DoxDA (AEK58244.1); A. caldus ATCC 51756, RsrR (ABP38227.1), RsrS (ABP38226.1), TetH (ABP38225.1), DoxDA (ABP38224.1); Acidithiobacillus ferrooxidans ATCC 23270, TcsS (ACK79489.1), TcsR (ACK79259.1), TetH (ACK80599.1), DoxDA_2 (ACK79881.1), DoxDA_1 (ACK78481.1); A. ferrooxidans ATCC 53993, TcsS (ACH82290.1), TscR (ACH82291.1), TetH (ACH82292.1), DoxDA_2(ACH82311.1), DoxDA_1(ACH82307.1); A.thiooxidans ATCC 19377, TetH (WP_029316048.1), DoxDA (WP_010638552.1); A.thiooxidans A01, TetH (WP_024894935.1), DoxDA (WP_024894934.1); Acidithiobacillus ferridurans JCM 18981, TcsS (BBF65177.1), TcsR (BBF65176.1), TetH (BBF65175.1),DoxDA_2 (BBF65156.1),DoxDA_1 (BBF65160.1); Acidithiobacillus ferrivorans SS3, TetH (AEM46280.1), DoxDA (AEM47534.1); Acidianus hospitalis W1, TetH (AEE94548.1), DoxD_1 (AEE93006.1), DoxA_1 (AEE93005.1), DoxA_2 (AEE93131.1), DoxD_2 (AEE93130.1); Acidianus ambivalens DSM 3772, TetH (CBY66038.1), DoxD (CAA69986.1), DoxA(CAA69987.1).
Thiosulfate:quinol oxidoreductase (TQO), first discovered in Acidianus ambivalens (A. ambivalens), is composed of two 28-kDa DoxA and two 16-kDa DoxD subunits that formed a α2β2-tetramer (Muller et al., 2004). The membrane-bound TQO oxidizes thiosulfate to tetrathionate, and ferricyanide or decylubiquinone (DQ) takes the electrons generated from this reaction to the electron transport chain (Muller et al., 2004). Phylogenetic analysis indicated that the subunits DoxD and DoxA are fused into one protein in Acidithiobacillus spp. (Wang et al., 2016; Figure 4), indicating differences of TQOs between archaeal and bacterial species. Until now, the catalytic mechanism of TQO in Acidithiobacillus spp. is still unclear, and the functional role of TQO in sulfur oxidation needs to be further confirmed experimentally.
The hydrolysis of tetrathionate by tetrathionate hydrolase has been studied extensively in Acidithiobacillus, including enzymatic properties, protein localization and function in the sulfur-metabolic network (Ralf et al., 1987; de Jong et al., 1997; Bugaytsova and Lindstrom, 2004; Kanao et al., 2010, 2018; Beard et al., 2011; Yu et al., 2014). The fact that ΔtetH strains of A. ferrooxidans and A. caldus could not survive in tetrathionate-medium but could grow on other sulfur-substrates, indicates that tetrathionate-metabolism in these sulfur-oxidizing bacteria is TetH-dependent (van Zyl et al., 2008; Yu et al., 2014). In addition to its role in tetrathionate hydrolysis, a ferric reductase activity associated with TetH was reported for A. ferrooxidans as well (Sugio et al., 2009).
TetH proteins purified from A. thiooxidans, A. ferrooxidans, and A. caldus are all homodimers and the optimal activities are detected under acidic conditions (pH 3.0–4.0) (Hazeu et al., 1988; Sugio et al., 1996; Tano et al., 1996; Bugaytsova and Lindstrom, 2004; Kanao et al., 2007, 2018; Beard et al., 2011). Studies on the products of tetrathionate hydrolysis catalyzed by TetH proteins from different species of Acidithiobacillus yielded conflicting results. When suspensions of A. ferrooxidans were used in the assays, polythionates (up to S13O62-) and sulfur rings (S0, containing 98% S8, and small amounts of S6, S7, S9, and S12) were detected in tetrathionate hydrolysates (Ralf et al., 1987). However, using the pure enzyme from A. ferrooxidans, the end products were thiosulfate, sulfur and sulfate (de Jong et al., 1997). TetH assays for A. thiooxidans showed that thiosulfate was a main product of tetrathionate decomposition and the thiosulfate probably was further decomposed to generate elemental sulfur (Tano et al., 1996). In contrast, for A. caldus, thiosulfate and pentathionate were detected after TetH assays (Bugaytsova and Lindstrom, 2004). These different results are probably due to the differences in the detection methods or the protein components used in the assays. It is possible that the final products of TetH-mediated reactions in Acidithiobacillus spp. are thiosulfate, sulfur and sulfate, even though the intermediate metabolites may exist.
Meanwhile, the catalytic mechanism of TetH is still unclear. Crystallization and preliminary X-ray diffraction analysis of TetH from A. ferrooxidans was published (Kanao et al., 2013). The crystal of recombinant Af-TetH was a hexagonal cylinder with dimensions of 0.2 mm × 0.05 mm × 0.05 mm, and the crystal diffracted to 2.15 Å resolution. However, the three-dimensional structure was not obtained until now. It was reported that the sole cysteine residue (Cys301) in Af-TetH was involved in neither the tetrathionate hydrolysis reaction nor the subunit assembly, indicating a novel cysteine-independent reaction mechanism for this enzyme (Kanao et al., 2014).
Different speculations on the localization of TetH in Acidithiobacillus spp. were made based on the results of indirect biochemical assays. Both the low optimal pH (∼4) for TetH activities and the purification of TetH proteins from the soluble fraction implied that TetH proteins from A. ferrooxidans and A. thiooxidans were periplasmic proteins (Tano et al., 1996; de Jong et al., 1997; Kanao et al., 2007). The TetH activity analysis on different fractionations of A. caldus cells indicated TetH was a potential periplasmic protein (Bugaytsova and Lindstrom, 2004). However, the requirement of an acidic environment in the in vitro refolding experiments on recombinant Af-TetH indicated TetH probably localized in the outer membrane of A. ferrooxidans (Kanao et al., 2010). Results based on marine A. thiooxidans strain SH also suggested that TetH locates on the outer membrane of the cell (Kanao et al., 2018). Another research group proposed that TetH in A. ferrooxidans was probably secreted to the extracellular space (Beard et al., 2011). Signal peptides of TetHs from different strains of Acidithiobacillus spp. were predicted in bioinformatics analysis (unpublished data), implying TetH was secreted into the periplasm of these sulfur-oxidizing bacteria. Further immunocytochemical experiments and micro-imaging technologies are required to obtain conclusive evidence for the localization of this enzyme.
Sox System
The sulfur oxidizing enzyme (Sox) system, a typical periplasmic multi-enzyme system, was first found in lithoautotrophic Paracoccus pantotrophus (Friedrich et al., 2000, 2005). This complex has been thought to be widely distributed among the various phylogenetic groups of photo- and chemo-lithotrophic sulfur-oxidizing prokaryotes (summarized by Ghosh and Dam, 2009; Frigaard and Dahl, 2009). The Sox system in P. pantotrophus is made up of four components: SoxXA, SoxYZ, SoxB, and Sox(CD)2 (Friedrich et al., 2005). This typical Sox system has the capability of oxidizing thiosulfate, sulfide, sulfite, and elemental sulfur to sulfate as the final product (summarized by Ghosh and Dam, 2009). Thiosulfate oxidation is the core of this oxidation pathway: first, SoxXA catalyzes the reaction between the sulfane sulfur of thiosulfate and the SoxY-cysteine-sulfhydryl group of the SoxYZ complex, forming a cysteine S-thiosulfonate derivative (SoxYZ-S-S-SO3-); second, SoxB hydrolyzes sulfate (SO42-) from the terminal sulfone (-SO3-) group of SoxYZ complex; third, Sox(CD)2 oxidizes the sulfane sulfur (–S-) of the residual SoxY-cysteine persulfide (SoxYZ-S-S-) to cysteine-S-sulfate (SoxYZ-S-SO3-); eventually, the sulfonate moiety (-SO3-) is hydrolyzed again by SoxB, regenerating SoxYZ (summarized by Ghosh and Dam, 2009; Welte et al., 2009; Zander et al., 2011). However, in contrast to the current Sox metabolic models, a new discovery on the intermediates of the Sox pathway was reported: it was proposed that instead of cysteine S-thiosulfonate (SoxYZ-S-(S)-SO3-), SoxYZ that conjugates with multiple sulfane atoms (SoxYZ-S-(S)n-SO3-, n ≥ 2) is the true carrier species in the Sox pathway (Grabarczyk and Berks, 2017). In addition, Sox(CD)2 is absent in a wide range of the Sox-system-dependent sulfur-oxidizing bacteria. In the Sox pathway that lacks Sox(CD)2 (so-called truncated Sox pathway), the sulfur atom of the sulfane intermediate (SoxYZ-S-S-) is probably fed into other Sox pathways or transformed into storage forms of sulfur (summarized by Ghosh and Dam, 2009; Welte et al., 2009). In recent years, structural and biological studies on Sox proteins, including the analysis of their three-dimensional structures and the identification of active sites, have provided more information regarding the catalytic mechanism of the Sox system at atomic-level resolution (Bamford et al., 2002; Sauve et al., 2007, 2009; Bradley et al., 2012; Grabarczyk et al., 2015).
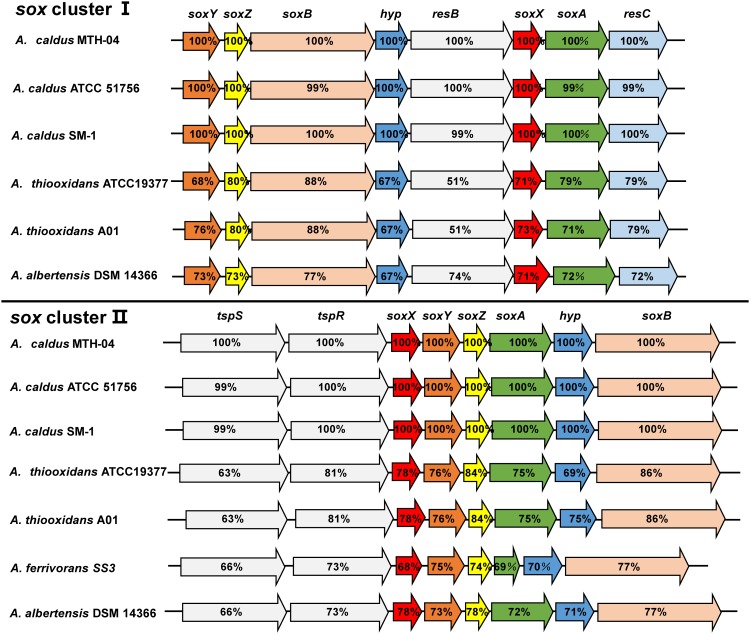
Comparative analysis of the published genomic sequences from species of the genus Acidithiobacillus indicated that sox clusters without soxCD genes are present in A. caldus, A. thiooxidans, A. albertensis and A. ferrivorans, but not found in A. ferrooxidans strains (Figure 5 and Table 1; Valdés et al., 2008b; Talla et al., 2014; Christel et al., 2016; Castro et al., 2017). Two separate sox clusters are present in A. caldus, A. thiooxidans and A. albertensis. One cluster is arranged in the order soxXYZA-hyp-soxB that is located downstream of a sigma54-dependent two component system tspS-tspR (termed as sox-II cluster), the other one was soxYZB-hyp-resB-soxAX-resC (termed as sox-I cluster). Only the sox-II-like cluster was found in A. ferrivorans (Figure 5).
FIGURE 5.
The sox clusters in A. caldus and A. thiooxidans. The percentages of similarities between protein sequences are indicated by the values marked in the gene. A. caldus MTH-04: sox I (Gene ID: A5904_10510-10475), sox II (A5904_11270- 11305); A. caldus ATCC 51756: sox I (Acaty_c2059- c2052), sox II (Acaty_c2206-c2213); A. caldus SM-1: sox I (Atc_2217-2209), sox II (Atc_2363-2370); A. thiooxidans ATCC19377: sox I (ATHIO_RS0100375-RS0100340), sox II (ATHIO_RS0101665-RS0101630); A. thiooxidans A01: sox I (X795_RS0118310-RS0118345), sox II (RS0104295-RS0104260); A.albertensis DSM 14366: sox I (BLW97_RS03815-RS03850), sox II (BLW97_RS11430-RS11465); A. ferrivorans SS3: sox II (Acife_2487–2494).
Due to the absence of Sox(CD)2 protein in the Sox system of Acidithiobacillus, two possible ways for SoxYZ regeneration in bacteria with the truncated Sox pathway were proposed, including the cleavage of sulfur atom of the sulfane intermediate (SoxYZ–S–S-) and the oxidation of SoxYZ–S–S- by the predicted sulfur dioxygenase (SDO) (Chen et al., 2012; Yin et al., 2014). Since it is hypothetized that SDO is located in cytoplasm where it oxidizes the elemental sulfur to sulfite (Wu et al., 2017), it is inferred that the sulfur atom of the sulfane intermediate (SoxYZ-S-S-) should be cleaved to form elemental sulfur via an unknown mechanism, so that SoxYZ is regenerated. As a consequence, studies on SoxYZ regeneration and sulfur globule accumulation are important for uncovering the catalytic process of the truncated Sox system in Acidithiobacillus spp.
Thiosulfate Dehydrogenase (TSD)
The first report on the thiosulfate dehydrogenase in Acidithiobacillus was released by Silver and Lundgren (1968b), that the thiosulfate-oxidizing enzyme purified from A. ferrooxidans oxidizes one mole of thiosulfate to produce 0.5 mole of tetrathionate. The optimal pH for this thiosulfate-oxidizing enzyme was 5.0 and no cofactor was required for this reaction (Silver and Lundgren, 1968b). Janiczek et al. (2007) described that the purified TSD from A. ferrooxidans was a tetramer consisting of four identical subunits of 45 kDa and its optimal activity was observed at pH 3.0. It was not until 2013, that the gene (AFE_0042) encoding thiosulfate dehydrogenase (∼25 kDa) was identified by Kikumoto et al. (2013) Maximum enzyme activity appeared at pH 2.5 and 70°C, but this enzyme could reduce neither ubiquinone nor horse heart cytochrome c. The low pH optimum of TSD indicated that this enzyme probably metabolized thiosulfate in the periplasmic space. It was reported that different TSDs were purified from A. thiooxidans (Nakamura et al., 2001), but the corresponding gene sequences are unidentified.
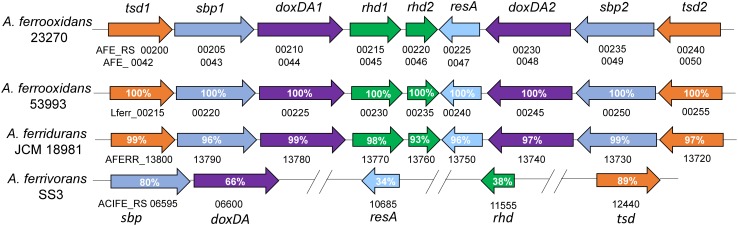
Using the protein sequence of AFE_0042 in A. ferrooxidans as the reference sequence, another potential TSD (AFE_0050) was discovered in A. ferrooxidans with 76% identities to the protein sequence of AFE_0042. These two copies of tsd genes, together with other thiosulfate-oxidation-related genes (doxDA and rhd), are arranged in a unique thiosulfate-metabolic gene cluster in the genome of A. ferrooxidans (Figure 6). This thiosulfate gene cluster was also discovered in A. ferridurans, but does not exist in A. ferrivorans that contains TSD (Figure 6). However, no TSD homologs are found in A. caldus or A. thiooxidans (Table 1). The conflict between the absence of TSD homologs and the reported TSD activity in A. thiooxidans should be clarified in further investigations (Nakamura et al., 2001). Taken together, these results suggest that the existence of the TSD-dependent thiosulfate-oxidation pathway is probably a typical character for sulfur- and ferrous-oxidizing bacteria (A. ferrooxidans, A. ferridurans, and A. ferrivorans).
FIGURE 6.
Schematic map of the thiosulfate-metabolic gene clusters in A. ferrooxidans and other Acidithiobacillus sp. tsd, thiosulfate dehydrogenase; sbp, sulfate/molybdate binding protein; doxDA, thiosulfate:quinol oxidoreductase; rhd, rhodanese-like domain-containing protein; resA, thiol-disulfide oxidoreductase. The numbers after tsd, sbp, doxDA and rhd were there to differentiate the two copies of these genes in the cluster. The identities of TSD1 and TSD2, SBP1 and SBP2, DoxDA1 and DoxDA2 in A. ferrooxidans are 76, 74, and 71%, respectively.
The most intensively studied tetrathionate-forming thiosulfate dehydrogenase (TsdA) was identified and investigated in the purple sulfur bacterium A. vinosum (Denkmann et al., 2012). This enzyme is a periplasmic, monomeric 25.8 kDa c-type cytochrome, most active at pH 4.0. The catalytic mechanism of TsdA has been elucidated (Liu et al., 2013; Brito et al., 2015; Kurth et al., 2016). Homologous proteins are not encoded in Acidithiobacillus genomes, indicating potential differences in thiosulfate dehydrogenases (Kikumoto et al., 2013).
The TSD in Acidithiobacillus and TsdA in Allochromatium can be considered as two different kinds of thiosulfate dehydrogenase based on protein similarity, enzymatic features and their existence in different sulfur-oxidizing bacteria. Thus, investigations on the distribution of TSD and TsdA homologs in different sulfur-oxidizing prokaryotes will be essential for illustrating the differences in thiosulfate metabolism during the evolution of these sulfur-oxidizing species. The unusual gene arrangement of the tsd-doxDA clusters in A. ferrooxidans, A. ferridurans and A. ferrivorans implies a unique thiosulfate-metabolic mode in these sulfur- and ferrous-oxidizing Acidithiobacillus spp. Studies on gene function and regulation mechanism of this cluster would provide new insights in thiosulfate metabolism in A. ferrooxidans, A. ferridurans, and A. ferrivorans. Meanwhile, any further knowledge on the protein structure and catalytic mechanism of TSD would be helpful for understanding the TSD-catalyzed thiosulfate-metabolic process in these sulfur-oxidizing bacteria.
Sulfide Oxidation
Sulfide is an important sulfur-substrate and metabolic-intermediate during elemental sulfur oxidation in Acidithiobacillus. The sulfide-oxidizing enzyme SQR is present in different domains of life from prokaryotes to animals (Griesbeck et al., 2000). It has been proved that sulfide can be oxidized to zero-valent sulfur by the membrane-bound protein SQR and at the same time, electrons are generated and fed into the membrane quinone pool (Griesbeck et al., 2000). SQR activity was detected in A. ferrooxidans NAsF-1. When cells were cultured in S0 medium, the activity was 17 times higher than in Fe2+-medium (Wakai et al., 2004). The crystal structure of A. ferrooxidans SQR suggested the potential oxidation mechanism: when a sulfide ion interacts with the Sγ of Cys356, two electrons are acquired by FAD and the sulfur atom is attached to the polysulfide bridge (Cherney et al., 2010). The active sites of A. ferrooxidans SQR includes two cysteines (Cys160, Cys356) involved in the transfer of electrons to FAD and the formation of the polysulfide bridge, a third cysteine (Cys128) related to the release of the polysulfur product, and two histidine residues (His132, His198) that are essential for its function (Cherney et al., 2010; Zhang and Weiner, 2014). Multiple copies of putative sqr genes were annotated in the genome of Acidithiobacillus strains, but their roles in the sulfur oxidation are still unclear. SQRs have been classified into several groups (Pham et al., 2008; Marcia et al., 2009, 2010; Sousa et al., 2018), and the potential SQRs in a single species of Acidithiobacillus may belong to different groups. The different catalytic properties and distinct functions of SQRs in Acidithiobacillus spp. could be investigated in future researches to reveal their different roles in the sulfur oxidation of these bacteria.
Sulfite Oxidation
Sulfite needs to be quickly oxidized or transformed in the organism due to its harmful effect on the cell. Enzyme-mediated sulfite oxidation can occur in the periplasm and the cytoplasm in different bacteria. Besides, sulfite also can be converted to sulfate, thiosulfate or glutathione S-sulfonate chemically with the help of Fe3+ or sulfur (Sugio et al., 1987b; Suzuki et al., 1992; Harahuc and Suzuki, 2001).
Sulfite oxidase activity from A. ferrooxidans strains was reported (Vestal and Lundgren, 1971; Sugio et al., 1988; Sugio et al., 1992; Suzuki, 1994), but the gene sequence was still unidentified (Valdés et al., 2008a). Until now, the knowledge on sulfite oxidation in Acidithiobacillus spp. was mainly acquired based on the prediction of potential genes and inference of protein functions. Three pathways/enzymes are proposed to participate in the sulfite oxidation process in Acidithiobacillus: the Sox system that oxidizes the periplasmic sulfite, an adenosine-5’-phosphosulfate (APS) pathway involving two different enzymes that catalyze the cytoplasmic sulfite oxidation and a predicted SoeABC complex.
In fact, similar APS pathways associated with cytoplasmic sulfite oxidation were reported in A. ferrooxidans, A. caldus, A. thiooxidans and A. ferrivorans (Quatrini et al., 2009; Chen et al., 2012; Yin et al., 2014; Christel et al., 2016). The APS pathway consists of an APS reductase (AprBA) and an ATP sulfurylase (SAT). These enzymes are involved in the dissimilatory sulfate reduction pathway in sulfate-reducing prokaryotes. SAT utilizes ATP and sulfate to generate APS which is further converted to AMP and sulfite by AprBA. In some phototrophic and chemotrophic sulfur-oxidizing bacteria, this pathway was postulated to work in the reverse direction: sulfite is oxidized to sulfate; meanwhile, substrate level phosphorylation is achieved (Hipp et al., 1997; Meyer and Kuever, 2007; Meyer and Kuever, 2008). The genomic sequence analysis showed that putative sat genes encoding enzymes catalyzing turnover of APS to sulfate and ATP are present in A. ferrooxidans and A. caldus, but AprBA homologs that oxidize sulfite to APS are not encoded in these strains (Quatrini et al., 2009; Chen et al., 2012). In A. thiooxidans A01, phosphoadenosine phosphosulfate (PAPS) reductase and adenylylsulfate kinase genes were discovered, while the sat gene was not detected (Yin et al., 2014). To elucidate APS-mediated sulfite oxidation process in these sulfur-oxidizing bacteria, more information on the essential genes and their functions is required.
SoeABC, a heterotrimeric membrane-bound complex, was first reported to be involved in sulfite oxidation in the cytoplasm of the chemotrophic Ruegeria pomeroyi (Lehmann et al., 2012). The complex is formed by three subunits: an NrfD/PsrC like membrane protein (SoeC), an iron–sulfur protein (SoeB), and a molybdoprotein (SoeA) (Lehmann et al., 2012; Dahl et al., 2013). The membrane-bound iron-sulfur molybdoprotein SoeABC has been identified as a major direct sulfite-oxidizing enzyme in the cytoplasm of the purple sulfur bacterium A. vinosum (Dahl et al., 2013). The genes encoding SoeABC were reported in the genome of A. caldus SM-1 (Liu L.J. et al., 2014) and SoeABC homologs are also found in A. ferrivorans, A. ferrooxidans, A. thiooxidans and A. albertensis (unpublished data), implying the presence of a direct cytoplasmic sulfite-oxidizing pathway in these chemoautotrophic sulfur-oxidizers.
Sulfur Oxidation Network in Acidithiobacillus Spp.
With the identification of new enzymes, application of omics technologies and gene functional studies based on gene knockout techniques, different models of the sulfur-metabolic networks have been proposed to elucidate the sulfur-oxidizing processes in Acidithiobacillus species (Hallberg et al., 1996; Valdés et al., 2008a; Mangold et al., 2011; Chen et al., 2012; Bobadilla Fazzini et al., 2013; Yin et al., 2014). Based on the knowledge regarding discovered sulfur-oxidizing enzymes and the previous models, sulfur oxidation in Acidithiobacillus spp. can be classified into two modes: one is the Sox-pathway-dependent sulfur oxidation networks in A. caldus, A. thiooxidans, A. albertensis, and A. ferrivorans, and the other is the Sox-pathway-independent sulfur-metabolic network in A. ferrooxidans (Table 1). The Sox system is present in both the sulfur-oxidizing-only species and the sulfur- and ferrous-oxidizing species A. ferrivorans. Results from our laboratory indicate that the Sox-containing strains of A. caldus and A. thiooxidans have higher sulfur oxidation capacities (higher elemental-sulfur-oxidizing rate and growth density in S0-medium) compared with a Sox- deficient A. ferreooxidans strain. Although A. ferrooxidans has thiosulfate dehydrogenase (TSD), the differences in the abilities of obtaining electrons from TSD and Sox system probably contribute to the different sulfur oxidation capacities of these Acidithiobacillus strains. Interestingly, A. ferrivorans possesses a single copy of both the sox cluster and tsd gene while in other species double copies of these genes are found. Based on the important role of periplasmic thiosulfate metabolism in cell growth and the observed differences in thiosulfate-metabolic pathways in Acidithiobacillus spp., it may be suggested that the variations in thiosulfate-metabolic pathways/enzymes should be a key difference of the sulfur oxidation process in different species of Acidithiobacillus.
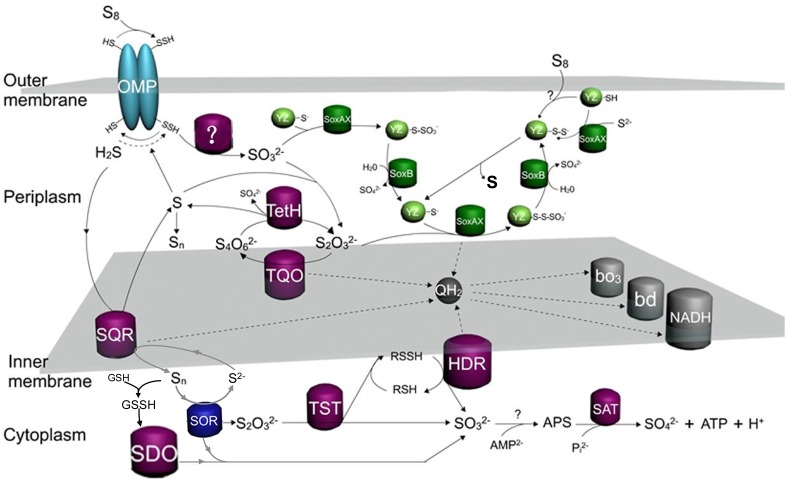
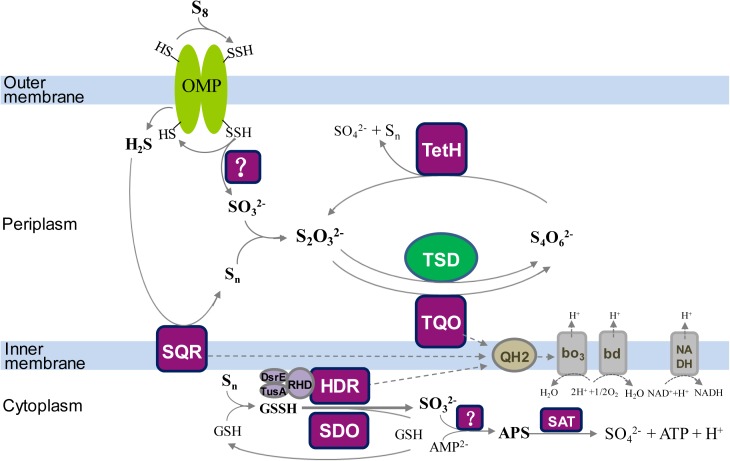
To better understand the Sox-pathway-dependent and Sox-pathway-independent sulfur-metabolic processes in Acidithiobacillus, modified sulfur oxidation models for A. caldus and A. ferrooxidans were proposed based on cutting-edge research progresses (Valdés et al., 2008a; Quatrini et al., 2009; Mangold et al., 2011; Chen et al., 2012). In A. caldus (Figure 7), extracellular elemental sulfur (S8) is activated and transported by special outer-membrane proteins (OMP) into the periplasm where persulfide sulfane sulfur is oxidized by an unknown enzyme; the resulting sulfite can directly enter the Sox pathway or form S2O32- via a non-enzymatic reaction between SO32- and a sulfur atom; the periplasmic thiosulfate is then processed by the truncated Sox pathway, producing sulfate and elemental sulfur, or be catalyzed by TQO to generate S4O62- that is further hydrolyzed by TetH; the hydrogen sulfide generated in the activation of S8 is oxidized by SQR located in the inner membrane; some sulfur-metabolic processes, including the truncated Sox pathway mediated thiosulfate metabolism, tetrathionate hydrolysis and sulfide oxidation, can produce elemental sulfur that may be re-activated and -oxidized at the outer membrane region, or be mobilized into the cytoplasm where it is oxidized by SDO and SOR; the products formed by SDO and SOR can activate the cytoplasmic sulfur-metabolic pathways including the pathway of S2O32- utilization catalyzed by rhodanese (TST) and the Hdr-like complex (HDR), the degradation of SO32- via the APS pathway and the oxidation of S2- by SQR. Sulfur oxidation in A. ferrooxidans differs from that of A. caldus in its absence of the Sox pathway, however, it possesses TSD that could be an alternative thiosulfate-metabolic pathway in the periplasm (Figure 8).
FIGURE 7.
The updated model of sulfur oxidation in A. caldus. OMP, outer-membrane proteins; TQO, thiosulfate quinone oxidoreductase; TetH, tetrathionate hydrolase; SQR, sulfide:quinone oxidoreductase; SDO, sulfur dioxygenase; SOR, Sulfur oxygenase reductase; TST, rhodanese; HDR, Hdr-like complex; SAT, ATP sulfurylase; bd, bo3, terminal oxidases; QH2, quinol pool; NADH, NADH dehydrogenase complex I.
FIGURE 8.
The updated model of sulfur oxidation in A. ferrooxidans. OMP, outer-membrane proteins; TQO, thiosulfate quinone oxidoreductase; TSD, thiosulfate dehydrogenase; TetH, tetrathionate hydrolase; SQR, sulfide:quinone oxidoreductase; SDO, sulfur dioxygenase; HDR, Hdr-like complex; SAT, ATP sulfurylase; bd, bo3, terminal oxidases; QH2, quinol pool; NADH, NADH dehydrogenase complex I.
The discovery of a TSD homolog in Sox-system-containing A. ferrivorans indicates the existence of a hybrid thiosulfate-metabolic mode in the periplasm of this species of Acidithiobacillus. Some of the sulfur-oxidizing pathways/enzymes, including TQO in the S4I pathway and the Hdr-like complex and SQR, are proposed to feed electrons via the quinol pool (QH2) in the inner membrane to the terminal oxidases bd or bo3 for producing ATP or to NADH dehydrogenase (complex I) for generating NADH (Figures 7, 8; Valdés et al., 2008a; Quatrini et al., 2009; Mangold et al., 2011; Chen et al., 2012; Yin et al., 2014; Christel et al., 2016).
The sulfur-metabolic networks in Acidithiobacillus spp. differ from those of the sulfur-oxidizing archaea (Kletzin et al., 2004; Liu et al., 2012). Regarding the thiosulfate metabolism, the S4I pathway exists in sulfur-oxidizing archaea such as Acidianus ambivalens (Muller et al., 2004; Wang et al., 2016), but there is no report on the Sox system and thiosulfate dehydrogenase (TSD) in these archaeal sulfur-oxidizers. At least two of the three thiosulfate metabolic pathways/enzymes (Sox system, S4I pathway and TSD enzyme) are employed in each species of Acidithiobacillus. The complexity of thiosulfate metabolism could be considered as a difference between Acidithiobacillus and sulfur-oxidizing archaea. As for elemental sulfur oxidation, SOR plays a critical role in sulfur-oxidizing archaea (Kletzin et al., 2004), but it is not an indispensable enzyme in Acidithiobacillus spp. (You et al., 2011; Chen et al., 2012). In these chemoautotrophic sulfur-oxidizing bacteria, SDO is one of the important cytoplasmic elemental-sulfur-oxidizing enzymes (Figure 1; Wang et al., 2014; Wu et al., 2017). Meanwhile, there are sulfur-oxidizing genes possessed by both sulfur-oxidizing archaea and Acidithiobacillus, such as dsrE-tusA, soeABC, and hdrC1B1A-hyp-hdrC2B2 genes (Liu L.J. et al., 2014).
Sulfur oxidation in Acidithiobacillus spp. is a sophisticated process that involves various enzymes/proteins and sulfur compounds in different cellular compartments. The proposal of sulfur oxidation models provides an overall and systematic understanding of the sulfur-metabolic process in these bacteria. However, there are still some questions and doubts regarding these models: (i) The localization of these sulfur-oxidizing enzymes in the cell. For example, SQR is proposed to be located at the periplasmic surface of the cytoplasmic membrane in the published models of Acidithiobacillus spp. (Valdés et al., 2008a; Quatrini et al., 2009; Mangold et al., 2011; Chen et al., 2012; Yin et al., 2014; Christel et al., 2016). Similar localization of SQR was reported for Rhodobacter capsulatus (Schutz et al., 1999), while results for heterotrophic Cupriavidus pinatubonensis indicated that SQR located on the cytoplasmic side of the membrane and the soluble cytoplasmic PDO was in the vicinity of the membrane (Gao et al., 2017). The close location of SQR and PDO in the cytoplasm increases the efficiency of sulfide oxidation in heterotrophic bacteria (Gao et al., 2017). However, whether the SQR in Acidithiobacillus spp. works in the same way as that of heterotrophic bacteria remains unknown. (ii) The actual sulfur-intermediates in the metabolic process. Various reduced sulfur compounds are produced during the oxidation of elemental sulfur to the final product sulfate, and some of them (thiosulfate, tetrathionate, sulfane sulfur and so on) are hypothesized to be important sulfur-substrates for sulfur-oxidizing pathways in Acidithiobacillus spp. models (Valdés et al., 2008a; Quatrini et al., 2009; Mangold et al., 2011; Chen et al., 2012; Yin et al., 2014; Christel et al., 2016). However, experimental confirmation on the existence of these sulfur-intermediates in vivo is lacking, thus the actual intracellular transformation of sulfur compounds is obscure. (iii) Electron transport chain. The quinol pool was proposed to mediate the electron transport in Acidithiobacillus spp. In published models, it was proposed that the electrons generated by the Sox complex were transferred via the quinol pool (Valdés et al., 2008a; Quatrini et al., 2009; Mangold et al., 2011; Chen et al., 2012; Yin et al., 2014; Christel et al., 2016). However, because of the presence of cytochrome c (SoxXA) in the Sox system (Mangold et al., 2011; Chen et al., 2012; Yin et al., 2014; Christel et al., 2016), the electrons obtained from Sox complex could be transferred directly to cytochrome c oxidase via SoxXA. Thus, which is the actual electron transport for the Sox system in Acidithiobacillus spp. remains to be verified.
Regulation of Thiosulfate-Metabolic Pathways in Acidithiobacillus Spp.
How to sense the sulfur-metabolites and regulate their sulfur-metabolic pathways is fundamental for these sulfur-oxidizers to catalyze various RISCs. The TCS, including a membrane-bound sensor histidine kinase HK and a cognate response regulator RR, is a predominant regulatory mechanism for prokaryotic microorganisms to initiate specific adaptive responses in response to environmental stimuli (Capra and Laub, 2012). Acidithiobacillus spp. have evolved a set of TCSs to regulate the thiosulfate metabolism in the periplasm. TCS genes upstream of the tetH gene of S4I pathway were found in A. caldus, A. ferrooxidans and A. ferridurans, but not in other species of Acidithiobacillus (Rzhepishevska et al., 2007; Wang et al., 2016). Phylogenetic analysis indicated that RsrS/RsrR in A. caldus was an EnvZ/OmpR-like TCS, whereas the TcsS/TcsR in A. ferrooxidans and A. ferridurans was a σ54-denpendent TCS similar to ZraS/ZraR-like systems (Rzhepishevska et al., 2007; Wang et al., 2016). EnvZ/OmpR mediates osmotic stress response in various gram-negative bacteria, and ZraS/ZraR (HydH/HydG) is found responding to high concentrations of zinc or lead in the medium (Leonhartsberger et al., 2001; Cai and Inouye, 2002). The TspS/TspR that regulate the Sox pathway was also a σ54-dependent TCS, located upstream of sox-II cluster in both A. caldus and A. thiooxidans (Li et al., 2017). In A. ferrooxidans, the global redox responding TCS RegB/RegA could regulate the ferrous iron and RISC oxidation pathways (Sandoval Ponce et al., 2012; Moinier et al., 2017).
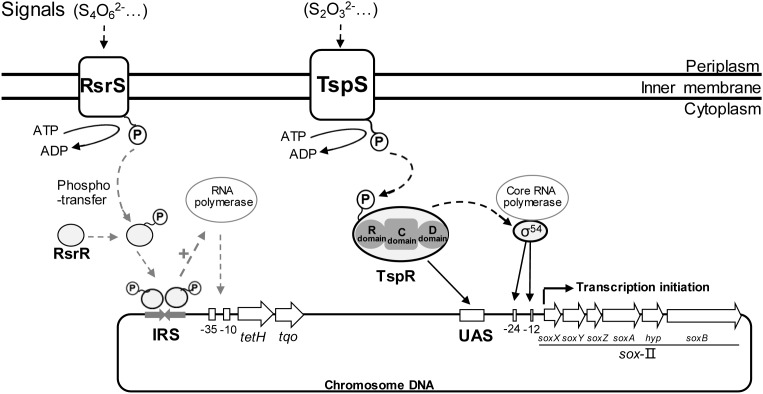
Based on previous results on RsrS/RsrR and TspS/TspR (Wang et al., 2016; Li et al., 2017), a new regulation model (shown in Figure 9) is summarized to better illustrate the significance of TCSs in the regulation of the periplasmic thiosulfate-metabolism in A. caldus. In the model, S2O32-, the potential signal molecule, is sensed by TspS and the activated TspS transmits the signal to TspR; the phosphorylated TspR binds to the upstream activator sequence (UAS; TGTCCCAAATGGGACA), initiating the transcription of sox-II gene cluster to express the Sox system for the metabolism of thiosulfate; S2O32- can also be oxidized by TQO to generate S4O62- that is later sensed by RsrS; the signal is transferred to RsrR, generating the active dimer. The RsrR dimer combines to a 19bp-inverted-repeat-sequence (IRS, AACACCTGTTACACCTGTT), stimulating the transcription of tetH and tqo, thus tetrathionate is decomposed to regenerate thiosulfate. Therefore, both the regulation of TspS/TspR of the Sox system and RsrS/RsrR of the S4I pathway are important for maintaining the balance between the oxidation and the conversion of S2O32-. The existence of TCSs that are associated with thiosulfate metabolism regulation in the periplasm of A. caldus and other acidophilic sulfur-oxidizers, allows these sulfur-oxidizing microbes to detect the generated S2O32- in the periplasm, and then modulate thiosulfate-metabolic pathways to quickly oxidize the instable S2O32- or convert it to the acid-stable S4O62-. The discovery of the regulation of TCSs on the S4I and Sox pathways also indicates the influence of these thiosulfate-metabolic pathways on the sulfur-metabolic network of Acidithiobacillus spp.
FIGURE 9.
Regulation model of periplasmic thiosulfate metabolism in A. caldus. RsrS/RsrR and TspS/TspR regulates the transcription of tetH and sox-II gene cluster, respectively.
Conclusion and Future Directions
Sulfur oxidation in chemoautotrophic Acidithiobacillus is an important component of microbial sulfur metabolism in the global sulfur cycle. Research on sulfur oxidation of Acidithiobacillus has made remarkable progresses in multiple aspects over the past decades, from enzymological studies and gene identification to protein structure and catalytic mechanism. The establishment of sulfur oxidation models for Acidithiobacillus spp. provides overall understanding of the sulfur-metabolic process. Sulfur metabolism in Acidithiobacillus spp. involves various sulfur-oxidizing pathways and enzymes located in different compartments of the cell, indicating the complexity and diversity of sulfur oxidation in these acidophilic autotrophic bacteria. Gene regulation systems including TCSs and other undetected regulatory mechanisms, also contribute to the remarkable sulfur-oxidizing abilities of Acidithiobacillus spp. However, detailed and in-depth studies on gene function and enzymatic properties involved in sulfur oxidation are insufficient or absent, resulting in the ambiguous conclusions or questions without answers in certain aspects of sulfur metabolism in Acidithiobacillus. (i) The reason for the existence of multiple copies of sulfur-oxidizing genes (rhd, dsrE, sox and so on) and their different roles in sulfur oxidation; (ii) the enzymatic properties of some proteins in Acidithiobacillus are speculated but not experimentally confirmed, such as the Hdr-like complex, SoeABC and the APS-sulfite-oxidizing pathway (Quatrini et al., 2009; Liu L.J. et al., 2014); (iii) structural analysis and catalytic mechanism of some sulfur-oxidizing enzymes (TSD, TetH, and TQO) remains to be clarified. Moreover, the utilization of various omics technologies at DNA, RNA and protein levels to study these sulfur-metabolic genes would facilitate the discovery of new sulfur-oxidizing proteins and improve the understanding of the sulfur-metabolic networks in Acidithiobacillus strains. Besides, the developments of novel methods or techniques, such as methods for visualization of protein localization and sulfur-metabolite detection techniques in vivo and in vitro, are needed to facilitate the studies of the enzymatic function and catalytic process in Acidithiobacillus spp. and other sulfur-oxidizing microorganisms.
Author Contributions
RW, J-QuL, and L-XC designed and composed the manuscript. J-QiL, X-ML, and XP helped in revising the manuscript. C-JZ, X-YG, C-ML, Y-QL, and YL prepared the figures. C-LY analyzed the gene and protein sequences.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We appreciate the support from the National Natural Science Foundation of China (31570036, 31370138, 31400093, 31570041, 31872621, and 30800011).
References
- Acosta M., Beard S., Ponce J., Vera M., Mobarec J. C., Jerez C. A. (2005). Identification of putative sulfurtransferase genes in the extremophilic Acidithiobacillus ferrooxidans ATCC 23270 genome: structural and functional characterization of the proteins. Omics 9 13–29. 10.1089/omi.2005.9.13 [DOI] [PubMed] [Google Scholar]
- Aminlari M., Gilanpour H. (1991). Comparative studies on the distribution of rhodanese in different tissues of domestic animals. Comp. Biochem. Physiol. B 99 673–677. 10.1016/0305-0491(91)90353-F [DOI] [PubMed] [Google Scholar]
- Arshadi M., Mousavi S. M. (2014). Simultaneous recovery of Ni and Cu from computer-printed circuit boards using bioleaching: statistical evaluation and optimization. Bioresour. Technol. 174 233–242. 10.1016/j.biortech.2014.09.140 [DOI] [PubMed] [Google Scholar]
- Azizan A., Najafpour G., Harun A. (2000). “Microbial desulfurization of coal with mixed cultures of Thiobacillus ferrooxidans and Thiobacillus thiooxidans,” in Proceedings of the 2nd International Conference on Advances in Strategic Technologies-ICAST 2000, (Putrajaya: Faculty of Engineering; ),1457–1462. [Google Scholar]
- Bamford V. A., Bruno S., Rasmussen T., Appia-Ayme C., Cheesman M. R., Berks B. C., et al. (2002). Structural basis for the oxidation of thiosulfate by a sulfur cycle enzyme. EMBO J. 21 5599–5610. 10.1093/emboj/cdf566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayat B., Sari B. (2010). Comparative evaluation of microbial and chemical leaching processes for heavy metal removal from dewatered metal plating sludge. J. Hazard. Mater. 174 763–769. 10.1016/j.jhazmat.2009.09.117 [DOI] [PubMed] [Google Scholar]
- Beard S., Paradela A., Albar J. P., Jerez C. A. (2011). Growth of Acidithiobacillus ferrooxidans ATCC 23270 in thiosulfate uUnder oxygen-limiting conditions generates extracellular sulfur globules by means of a secreted tetrathionate hydrolase. Front. Microbiol. 2:79. 10.3389/fmicb.2011.00079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobadilla Fazzini R. A., Cortes M. P., Padilla L., Maturana D., Budinich M., Maass A., et al. (2013). Stoichiometric modeling of oxidation of reduced inorganic sulfur compounds (Riscs) in Acidithiobacillus thiooxidans. Biotechnol. Bioeng. 110 2242–2251. 10.1002/bit.24875 [DOI] [PubMed] [Google Scholar]
- Boughanemi S., Lyonnet J., Infossi P., Bauzan M., Kosta A., Lignon S., et al. (2016). Microbial oxidative sulfur metabolism: biochemical evidence of the membrane-bound heterodisulfide reductase-like complex of the bacterium Aquifex aeolicus. FEMS Microbiol. Lett. 363:fnw156. 10.1093/femsle/fnw156 [DOI] [PubMed] [Google Scholar]
- Bradley J. M., Marritt S. J., Kihlken M. A., Haynes K., Hemmings A. M., Berks B. C., et al. (2012). Redox and chemical activities of the hemes in the sulfur oxidation pathway enzyme SoxAX. J. Biol. Chem. 287 40350–40359. 10.1074/jbc.M112.396192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brito J. A., Denkmann K., Pereira I. A., Archer M., Dahl C. (2015). Thiosulfate dehydrogenase (TsdA) from Allochromatium vinosum: structural and functional insights into thiosulfate oxidation. J. Biol. Chem. 290 9222–9238. 10.1074/jbc.M114.623397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant R. D., Mcgroarty K. M., Costerton J. W., Laishley E. J. (1983). Isolation and characterization of a new acidophilic Thiobacillus species (T. albertis). Rev. Can. Microbiol. 29 1159–1170. 10.1139/m83-178 [DOI] [Google Scholar]
- Bugaytsova Z., Lindstrom E. B. (2004). Localization, purification and properties of a tetrathionate hydrolase from Acidithiobacillus caldus. Eur. J. Biochem. 271 272–280. 10.1046/j.1432-1033.2003.03926.x [DOI] [PubMed] [Google Scholar]
- Cai S. J., Inouye M. (2002). EnvZ-OmpR interaction and osmoregulation in Escherichia coli. J. Biol. Chem. 277 24155–24161. 10.1074/jbc.M110715200 [DOI] [PubMed] [Google Scholar]
- Cao X., Koch T., Steffens L., Finkensieper J., Zigann R., Cronan J. E., et al. (2018). Lipoate-binding proteins and specific lipoate-protein ligases in microbial sulfur oxidation reveal an atpyical role for an old cofactor. eLife 7:e37439. 10.7554/eLife.37439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capra E. J., Laub M. T. (2012). Evolution of two-component signal transduction systems. Annu. Rev. Microbiol. 66 325–347. 10.1146/annurev-micro-092611-150039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro M., Moya-Beltran A., Covarrubias P. C., Gonzalez M., Cardenas J. P., Issotta F., et al. (2017). Draft genome sequence of the type strain of the sulfur-oxidizing acidophile, Acidithiobacillus albertensis. Stand. Genomic Sci. 12:77. 10.1186/s40793-017-0282-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charnnok B., Suksaroj T., Boonswang P., Chaiprapat S. (2013). Oxidation of hydrogen sulfide in biogas using dissolved oxygen in the extreme acidic biofiltration operation. Bioresour. Technol. 131 492–499. 10.1016/j.biortech.2012.12.114 [DOI] [PubMed] [Google Scholar]
- Chen L., Ren Y., Lin J., Liu X., Pang X., Lin J. (2012). Acidithiobacillus caldus sulfur oxidation model based on transcriptome analysis between the wild type and sulfur oxygenase reductase defective mutant. PLoS One 7:e39470. 10.1371/journal.pone.0039470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z. W., Liu Y. Y., Wu J. F., She Q., Jiang C. Y., Liu S. J. (2007). Novel bacterial sulfur oxygenase reductases from bioreactors treating gold-bearing concentrates. Appl. Microbiol. Biotechnol. 74 688–698. 10.1007/s00253-006-0691-0 [DOI] [PubMed] [Google Scholar]
- Cherney M. M., Zhang Y., Solomonson M., Weiner J. H., James M. N. (2010). Crystal structure of sulfide:quinone oxidoreductase from Acidithiobacillus ferrooxidans: insights into sulfidotrophic respiration and detoxification. J. Mol. Biol. 398 292–305. 10.1016/j.jmb.2010.03.018 [DOI] [PubMed] [Google Scholar]
- Christel S., Fridlund J., Buetti-Dinh A., Buck M., Watkin E. L., Dopson M. (2016). RNA transcript sequencing reveals inorganic sulfur compound oxidation pathways in the acidophile Acidithiobacillus ferrivorans. FEMS Microbiol. Lett. 363:fnw057. 10.1093/femsle/fnw057 [DOI] [PubMed] [Google Scholar]
- Cipollone R., Frangipani E., Tiburzi F., Imperi F., Ascenzi P., Visca P. (2007). Involvement of Pseudomonas aeruginosa rhodanese in protection from cyanide toxicity. Appl. Environ. Microbiol. 73 390–398. 10.1128/aem.02143-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl C. (2015). Cytoplasmic sulfur trafficking in sulfur-oxidizing prokaryotes. IUBMB Life 67 268–274. 10.1002/iub.1371 [DOI] [PubMed] [Google Scholar]
- Dahl C., Franz B., Hensen D., Kesselheim A., Zigann R. (2013). Sulfite oxidation in the purple sulfur bacterium Allochromatium vinosum: identification of SoeABC as a major player and relevance of SoxYZ in the process. Microbiology 159(Pt 12), 2626–2638. 10.1099/mic.0.071019-0 [DOI] [PubMed] [Google Scholar]
- Dahl C., Prange A. (2006). “Bacterial sulfur globules: occurrence, structure and metabolism,” in Inclusions in Prokaryotes, ed. Shively J. (Berlin: Springer; ), 21–51. [Google Scholar]
- Dam B., Mandal S., Ghosh W., Das Gupta S. K., Roy P. (2007). The S4-intermediate pathway for the oxidation of thiosulfate by the chemolithoautotroph Tetrathiobacter kashmirensis and inhibition of tetrathionate oxidation by sulfite. Res. Microbiol. 158 330–338. 10.1016/j.resmic.2006.12.013 [DOI] [PubMed] [Google Scholar]
- de Jong G. A. H., Hazeu W., Bos P., Kuenen J. G. (1997). Polythionate degradation by tetrathionate hydrolase of Thiobacillus ferrooxidans. Microbiology 143 499–504. 10.1099/00221287-143-2-499 [DOI] [PubMed] [Google Scholar]
- Denkmann K., Grein F., Zigann R., Siemen A., Bergmann J., van Helmont S., et al. (2012). Thiosulfate dehydrogenase: a widespread unusual acidophilic c-type cytochrome. Environ. Microbiol. 14 2673–2688. 10.1111/j.1462-2920.2012.02820.x [DOI] [PubMed] [Google Scholar]
- Ehrenfeld N., Levican G., Parada P. (2013). Heterodisulfide reductase from Acidithiobacilli is a key component involved in metabolism of reduced inorganic sulfur compounds. Adv. Mater. Res. 825 194–197. 10.4028/www.scientific.net/AMR.825.194 [DOI] [Google Scholar]
- Falagan C., Johnson D. B. (2016). Acidithiobacillus ferriphilus sp. nov., a facultatively anaerobic iron- and sulfur-metabolizing extreme acidophile. Int. J. Syst. Evol. Microbiol. 66 206–211. 10.1099/ijsem.0.000698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich C. G., Bardischewsky F., Rother D., Quentmeier A., Fischer J. (2005). Prokaryotic sulfur oxidation. Curr. Opin. Microbiol. 8 253–259. 10.1016/j.mib.2005.04.005 [DOI] [PubMed] [Google Scholar]
- Friedrich C. G., Quentmeier A., Bardischewsky F., Rother D., Kraft R., Kostka S., et al. (2000). Novel genes coding for lithotrophic sulfur oxidation of Paracoccus pantotrophus GB17. J. Bacteriol. 182 4677–4687. 10.1128/JB.182.17.4677-4687.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frigaard N. U., Dahl C. (2009). Sulfur metabolism in phototrophic sulfur bacteria. Adv. Microb. Physiol. 54 103–200. 10.1016/s0065-2911(08)00002-7 [DOI] [PubMed] [Google Scholar]
- Gao R., Liu H., Xun L. (2017). Cytoplasmic localization of sulfide:quinone oxidoreductase and persulfide dioxygenase of Cupriavidus pinatubonensis JMP134. Appl. Environ. Microbiol. 83:e1820–17. 10.1128/aem.01820-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner M. N., Rawlings D. E. (2000). Production of rhodanese by bacteria present in bio-oxidation plants used to recover gold from arsenopyrite concentrates. J. Appl. Microbiol. 89 185–190. 10.1046/j.1365-2672.2000.01117.x [DOI] [PubMed] [Google Scholar]
- Ghosh W., Dam B. (2009). Biochemistry and molecular biology of lithotrophic sulfur oxidation by taxonomically and ecologically diverse bacteria and archaea. FEMS Microbiol. Rev. 33 999–1043. 10.1111/j.1574-6976.2009.00187.x [DOI] [PubMed] [Google Scholar]
- Grabarczyk D. B., Berks B. C. (2017). Intermediates in the sox sulfur oxidation pathway are bound to a sulfane conjugate of the carrier protein SoxYZ. PLoS One 12:e0173395. 10.1371/journal.pone.0173395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabarczyk D. B., Chappell P. E., Johnson S., Stelzl L. S., Lea S. M., Berks B. C. (2015). Structural basis for specificity and promiscuity in a carrier protein/enzyme system from the sulfur cycle. Proc. Natl. Acad. Sci. U.S.A. 112 E7166–E7175. 10.1073/pnas.1506386112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griesbeck C., Hauska G., Schütz M. (2000). Biological sulfide oxidation: sulfide-quinone reductase (SQR), the primary reaction. Recent Res. Dev. Microbiol. 4 179–203. [Google Scholar]
- Guimaraes B. G., Barbosa R. L., Soprano A. S., Campos B. M., de Souza T. A., Tonoli C. C., et al. (2011). Plant pathogenic bacteria utilize biofilm growth-associated repressor (BigR), a novel winged-helix redox switch, to control hydrogen sulfide detoxification under hypoxia. J. Biol. Chem. 286 26148–26157. 10.1074/jbc.M111.234039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallberg K. B., Dopson M., Lindstrom E. B. (1996). Reduced sulfur compound oxidation by Thiobacillus caldus. J. Bacteriol. 178 6–11. 10.1128/jb.178.1.6-11.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallberg K. B., Gonzalez-Toril E., Johnson D. B. (2010). Acidithiobacillus ferrivorans, sp. nov.; facultatively anaerobic, psychrotolerant iron-, and sulfur-oxidizing acidophiles isolated from metal mine-impacted environments. Extremophiles 14 9–19. 10.1007/s00792-009-0282-y [DOI] [PubMed] [Google Scholar]
- Hallberg K. B., Lindstrom E. B. (1994). Characterization of Thiobacillus caldus sp. nov., a moderately thermophilic acidophile. Microbiology 140( Pt 12), 3451–3456. 10.1099/13500872-140-12-3451 [DOI] [PubMed] [Google Scholar]
- Hamann N., Mander G. J., Shokes J. E., Scott R. A., Bennati M., Hedderich R. (2007). A cysteine-rich CCG domain contains a novel [4Fe-4S] cluster binding motif as deduced from studies with subunit B of heterodisulfide reductase from Methanothermobacter marburgensis. Biochemistry 46 12875–12885. 10.1021/bi700679u [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harahuc L., Suzuki I. (2001). Sulfite oxidation by iron-grown cells of Thiobacillus ferrooxidans at pH 3 possibly involves free radicals, iron, and cytochrome oxidase. Can. J. Microbiol. 47 424–430. 10.1139/w01-024 [DOI] [PubMed] [Google Scholar]
- Harrison A. P., Jr (1984). The acidophilic thiobacilli and other acidophilic bacteria that share their habitat. Annu. Rev. Microbiol. 38 265–292. 10.1146/annurev.mi.38.100184.001405 [DOI] [PubMed] [Google Scholar]
- Hazeu W., Batenburg-van der Vegte W. H., Bos P., van der Pas R. K., Kuenen J. G. (1988). The production and utilization of intermediary elemental sulfur during the oxidation of reduced sulfur compounds by Thiobacillus ferrooxidans. Arch. Microbiol. 150 574–579. 10.1007/bf00408252 [DOI] [Google Scholar]
- He H., Hong F.-F., Tao X.-X., Li L., Ma C.-Y., Zhao Y.-D. (2012). Biodesulfurization of coal with Acidithiobacillus caldus and analysis of the interfacial interaction between cells and pyrite. Fuel Process. Technol. 101 73–77. 10.1016/j.fuproc.2012.04.006 [DOI] [Google Scholar]
- Hedderich R., Hamann N., Bennati M. (2005). Heterodisulfide reductase from methanogenic archaea: a new catalytic role for an iron-sulfur cluster. Biol. Chem. 386 961–970. 10.1515/bc.2005.112 [DOI] [PubMed] [Google Scholar]
- Hedrich S., Johnson D. B. (2013a). Acidithiobacillus ferridurans sp. nov., an acidophilic iron-, sulfur- and hydrogen-metabolizing chemolithotrophic gammaproteobacterium. Int. J. Syst. Evol. Microbiol. 63(Pt 11), 4018–4025. 10.1099/ijs.0.049759-0 [DOI] [PubMed] [Google Scholar]
- Hedrich S., Johnson D. B. (2013b). Aerobic and anaerobic oxidation of hydrogen by acidophilic bacteria. FEMS Microbiol. Lett. 349 40–45. 10.1111/1574-6968.12290 [DOI] [PubMed] [Google Scholar]
- Hipp W. M., Pott A. S., Thum-Schmitz N., Faath I., Dahl C., Truper H. G. (1997). Towards the phylogeny of APS reductases and sirohaem sulfite reductases in sulfate-reducing and sulfur-oxidizing prokaryotes. Microbiology 143( Pt 9), 2891–2902. 10.1099/00221287-143-9-2891 [DOI] [PubMed] [Google Scholar]
- Holdorf M. M., Owen H. A., Lieber S. R., Yuan L., Adams N., Dabney-Smith C., et al. (2012). Arabidopsis ETHE1 encodes a sulfur dioxygenase that is essential for embryo and endosperm development. Plant Physiol. 160 226–236. 10.1104/pp.112.201855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Z. S., Han Y. J., Chen L. X., Liu J., Hu M., Li S. J., et al. (2015). Ecological roles of dominant and rare prokaryotes in acid mine drainage revealed by metagenomics and metatranscriptomics. ISME J. 9 1280–1294. 10.1038/ismej.2014.212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ijadi Bajestani M., Mousavi S. M., Shojaosadati S. A. (2014). Bioleaching of heavy metals from spent household batteries using Acidithiobacillus ferrooxidans: statistical evaluation and optimization. Sep. Purif. Technol. 132 309–316. 10.1016/j.seppur.2014.05.023 [DOI] [Google Scholar]
- Inskeep W. P., Jay Z. J., Herrgard M. J., Kozubal M. A., Rusch D. B., Tringe S. G., et al. (2013). Phylogenetic and functional analysis of metagenome sequence from high-temperature archaeal habitats demonstrate linkages between metabolic potential and geochemistry. Front. Microbiol. 4:95 10.3389/fmicb.2013.00095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson M. R., Melideo S. L., Jorns M. S. (2012). Human sulfide:quinone oxidoreductase catalyzes the first step in hydrogen sulfide metabolism and produces a sulfane sulfur metabolite. Biochemistry 51 6804–6815. 10.1021/bi300778t [DOI] [PubMed] [Google Scholar]
- Janiczek O., Zemanova J., Mandl M. (2007). Purification and some properties of thiosulfate dehydrogenase from Acidithiobacillus ferrooxidans. Prep. Biochem. Biotechnol. 37 101–111. 10.1080/10826060701199015 [DOI] [PubMed] [Google Scholar]
- Janosch C., Remonsellez F., Sand W., Vera M. (2015). Sulfur oxygenase reductase (Sor) in the moderately thermoacidophilic leaching bacteria: studies in Sulfobacillus thermosulfidooxidans and Acidithiobacillus caldus. Microorganisms 3 707–724. 10.3390/microorganisms3040707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston F., McAmish L. (1973). A study of the rates of sulfur production in acid thiosulfate solutions using S-35. J. Colloid Interface Sci. 42 112–119. 10.1016/0021-9797(73)90013-1 [DOI] [Google Scholar]
- Jones D. S., Albrecht H. L., Dawson K. S., Schaperdoth I., Freeman K. H., Pi Y., et al. (2012). Community genomic analysis of an extremely acidophilic sulfur-oxidizing biofilm. ISME J. 6 158–170. 10.1038/ismej.2011.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabil O., Banerjee R. (2012). Characterization of patient mutations in human persulfide dioxygenase (ETHE1) involved in H2S catabolism. J. Biol. Chem. 287 44561–44567. 10.1074/jbc.M112.407411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanao T., Kamimura K., Sugio T. (2007). Identification of a gene encoding a tetrathionate hydrolase in Acidithiobacillus ferrooxidans. J. Biotechnol. 132 16–22. 10.1016/j.jbiotec.2007.08.030 [DOI] [PubMed] [Google Scholar]
- Kanao T., Kosaka M., Yoshida K., Nakayama H., Tamada T., Kuroki R., et al. (2013). Crystallization and preliminary X-ray diffraction analysis of tetrathionate hydrolase from Acidithiobacillus ferrooxidans. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 69(Pt 6), 692–694. 10.1107/s1744309113013419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanao T., Matsumoto C., Shiraga K., Yoshida K., Takada J., Kamimura K. (2010). Recombinant tetrathionate hydrolase from Acidithiobacillus ferrooxidans requires exposure to acidic conditions for proper folding. FEMS Microbiol. Lett. 309 43–47. 10.1111/j.1574-6968.2010.02019.x [DOI] [PubMed] [Google Scholar]
- Kanao T., Nakayama H., Kato M., Kamimura K. (2014). The sole cysteine residue (Cys301) of tetrathionate hydrolase from Acidithiobacillus ferrooxidans does not play a role in enzyme activity. Biosci. Biotechnol. Biochem. 78 2030–2035. 10.1080/09168451.2014.948374 [DOI] [PubMed] [Google Scholar]
- Kanao T., Onishi M., Kajitani Y., Hashimoto Y., Toge T., Kikukawa H., et al. (2018). Characterization of tetrathionate hydrolase from the marine acidophilic sulfur-oxidizing bacterium, Acidithiobacillus thiooxidans strain SH. Biosci. Biotechnol. Biochem. 82 152–160. 10.1080/09168451.2017.1415128 [DOI] [PubMed] [Google Scholar]
- Kaster A. K., Moll J., Parey K., Thauer R. K. (2011). Coupling of ferredoxin and heterodisulfide reduction via electron bifurcation in hydrogenotrophic methanogenic archaea. Proc. Natl. Acad. Sci. U.S.A. 108 2981–2986. 10.1073/pnas.1016761108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly D. P., Wood A. P. (2000). Reclassification of some species of Thiobacillus to the newly designated genera Acidithiobacillus gen. nov., Halothiobacillus gen. nov. and Thermithiobacillus gen. nov. Int. J. Syst. Evol. Microbiol. 50(Pt 2), 511–516. 10.1099/00207713-50-2-511 [DOI] [PubMed] [Google Scholar]
- Kikumoto M., Nogami S., Kanao T., Takada J., Kamimura K. (2013). Tetrathionate-forming thiosulfate dehydrogenase from the acidophilic, chemolithoautotrophic bacterium Acidithiobacillus ferrooxidans. Appl. Environ. Microbiol. 79 113–120. 10.1128/aem.02251-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kletzin A. (1989). Coupled enzymatic production of sulfite, thiosulfate, and hydrogen sulfide from sulfur: purification and properties of a sulfur oxygenase reductase from the facultatively anaerobic archaebacterium Desulfurolobus ambivalens. J. Bacteriol. 171 1638–1643. 10.1128/jb.171.3.1638-1643.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kletzin A., Urich T., Muller F., Bandeiras T. M., Gomes C. M. (2004). Dissimilatory oxidation and reduction of elemental sulfur in thermophilic archaea. J. Bioenerg. Biomembr. 36 77–91. 10.1023/B:JOBB.0000019600.36757.8c [DOI] [PubMed] [Google Scholar]
- Koch T., Dahl C. (2018). A novel bacterial sulfur oxidation pathway provides a new link between the cycles of organic and inorganic sulfur compounds. ISME J. 12 2479–2491. 10.1038/s41396-018-0209-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurth J. M., Brito J. A., Reuter J., Flegler A., Koch T., Franke T., et al. (2016). Electron accepting units of the diheme cytochrome c TsdA, a bifunctional thiosulfate dehydrogenase/tetrathionate reductase. J. Biol. Chem. 291 24804–24818. 10.1074/jbc.M116.753863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann S., Johnston A. W. B., Curson A. R. J., Todd J. D., Cook A. M. (2012). “SoeABC, a novel sulfite dehydrogenase in the roseobacters?,” in Poster at the Programme and Abstract Book EMBO Workshop on Microbial Sulfur Metabolism, Noordwijkerhout, 29. [Google Scholar]
- Leonhartsberger S., Huber A., Lottspeich F., Bock A. (2001). The hydH/G genes from Escherichia coli code for a zinc and lead responsive two-component regulatory system. J. Mol. Biol. 307 93–105. 10.1006/jmbi.2000.4451 [DOI] [PubMed] [Google Scholar]
- Li L. F., Fu L. J., Lin J. Q., Pang X., Liu X. M., Wang R., et al. (2017). The sigma(54)-dependent two-component system regulating sulfur oxidization (Sox) system in Acidithiobacillus caldus and some chemolithotrophic bacteria. Appl. Microbiol. Biotechnol. 101 2079–2092. 10.1007/s00253-016-8026-2 [DOI] [PubMed] [Google Scholar]
- Li M., Chen Z., Zhang P., Pan X., Jiang C., An X., et al. (2008). Crystal structure studies on sulfur oxygenase reductase from Acidianus tengchongensis. Biochem. Biophys. Res. Commun. 369 919–923. 10.1016/j.bbrc.2008.02.131 [DOI] [PubMed] [Google Scholar]
- Liljeqvist M., Rzhepishevska O. I., Dopson M. (2012). Gene identification and substrate regulation provides insights into sulfur accumulation during bioleaching with the psychrotolerant Acidithiobacillus ferrivorans. Appl. Environ. Microbiol. 71 951–957. 10.1128/aem.02989-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liljeqvist M., Valdes J., Holmes D. S., Dopson M. (2011). Draft genome of the psychrotolerant acidophile Acidithiobacillus ferrivorans SS3. J. Bacteriol. 193 4304–4305. 10.1128/jb.05373-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Xin Y., Xun L. (2014). Distribution, diversity, and activities of sulfur dioxygenases in heterotrophic bacteria. Appl. Environ. Microbiol. 80 1799–1806. 10.1128/aem.03281-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L. J., Stockdreher Y., Koch T., Sun S. T., Fan Z., Josten M., et al. (2014). Thiosulfate transfer mediated by DsrE/TusA homologs from acidothermophilic sulfur-oxidizing archaeon Metallosphaera cuprina. J. Biol. Chem. 289 26949–26959. 10.1074/jbc.M114.591669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Beer L. L., Whitman W. B. (2012). Sulfur metabolism in archaea reveals novel processes. Environ. Microbiol. 14 2632–2644. 10.1111/j.1462-2920.2012.02783.x [DOI] [PubMed] [Google Scholar]
- Liu Y. W., Denkmann K., Kosciow K., Dahl C., Kelly D. J. (2013). Tetrathionate stimulated growth of Campylobacter jejuni identifies a new type of bi-functional tetrathionate reductase (TsdA) that is widely distributed in bacteria. Mol. Microbiol. 88 173–188. 10.1111/mmi.12176 [DOI] [PubMed] [Google Scholar]
- London J. (1963). Cytochrome in Thiobacillus thiooxidans. Science 140 409–410. 10.1126/science.140.3565.409 [DOI] [PubMed] [Google Scholar]
- London J., Rittenberg S. C. (1964). Path of sulfur in sulfide and thiosulfate oxidation by Thiobacilli. Proc. Natl. Acad. Sci. U.S.A. 52 1183–1190. 10.1073/pnas.52.5.1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mander G. J., Pierik A. J., Huber H., Hedderich R. (2004). Two distinct heterodisulfide reductase-like enzymes in the sulfate-reducing archaeon Archaeoglobus profundus. Eur. J. Biochem. 271 1106–1116. 10.1111/j.1432-1033.2004.04013.x [DOI] [PubMed] [Google Scholar]
- Mangold S., Valdes J., Holmes D. S., Dopson M. (2011). Sulfur metabolism in the extreme acidophile Acidithiobacillus caldus. Front. Microbiol. 2:17 10.3389/fmicb.2011.00017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcia M., Ermler U., Peng G., Michel H. (2009). The structure of Aquifex aeolicus sulfide:quinone oxidoreductase, a basis to understand sulfide detoxification and respiration. Proc. Natl. Acad. Sci. U.S.A. 106 9625–9630. 10.1073/pnas.0904165106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcia M., Ermler U., Peng G., Michel H. (2010). A new structure-based classification of sulfide:quinone oxidoreductases. Proteins 78 1073–1083. 10.1002/prot.22665 [DOI] [PubMed] [Google Scholar]
- Meyer B., Kuever J. (2007). Molecular analysis of the distribution and phylogeny of dissimilatory adenosine-5’-phosphosulfate reductase-encoding genes (aprBA) among sulfur-oxidizing prokaryotes. Microbiology 153(Pt 10), 3478–3498. 10.1099/mic.0.2007/008250-0 [DOI] [PubMed] [Google Scholar]
- Meyer B., Kuever J. (2008). Homology modeling of dissimilatory APS reductases (AprBA) of sulfur-oxidizing and sulfate-reducing prokaryotes. PLoS One 3:e1514. 10.1371/journal.pone.0001514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyauchi T., Kouzuma A., Abe T., Watanabe K. (2018). Complete genome sequence of Acidithiobacillus ferridurans JCM 18981. Microbiol. Resour. Announc. 7:e01028–18. 10.1128/mra.01028-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moinier D., Byrne D., Amouric A., Bonnefoy V. (2017). The global redox responding RegB/RegA signal transduction system regulates the genes involved in ferrous iron and inorganic sulfur compound oxidation of the acidophilic Acidithiobacillus ferrooxidans. Front. Microbiol. 8:1277. 10.3389/fmicb.2017.01277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller F. H., Bandeiras T. M., Urich T., Teixeira M., Gomes C. M., Kletzin A. (2004). Coupling of the pathway of sulphur oxidation to dioxygen reduction: characterization of a novel membrane-bound thiosulphate:quinone oxidoreductase. Mol. Microbiol. 53 1147–1160. 10.1111/j.1365-2958.2004.04193.x [DOI] [PubMed] [Google Scholar]
- Nakamura K., Nakamura M., Yoshikawa H., Amano Y. (2001). Purification and properties of thiosulfate dehydrogenase from Acidithiobacillus thiooxidans JCM7814. Biosci. Biotechnol. Biochem. 65 102–108. 10.1271/bbb.65.102 [DOI] [PubMed] [Google Scholar]
- Nguyen V. K., Lee M. H., Park H. J., Lee J.-U. (2015). Bioleaching of arsenic and heavy metals from mine tailings by pure and mixed cultures of Acidithiobacillus spp. J. Ind. Eng. Chem. 21 451–458. 10.1016/j.jiec.2014.03.004 [DOI] [Google Scholar]
- Nielsen A. M., Beck J. V. (1972). Chalcocite oxidation and coupled carbon dioxide fixation by Thiobacillus ferrooxidans. Science 175 1124–1126. 10.1126/science.175.4026.1124 [DOI] [PubMed] [Google Scholar]
- Nunez H., Moya-Beltran A., Covarrubias P. C., Issotta F., Cardenas J. P., Gonzalez M., et al. (2017). Molecular systematics of the genus Acidithiobacillus: insights into the phylogenetic structure and diversification of the taxon. Front. Microbiol. 8:30 10.3389/fmicb.2017.00030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossa Henao D., Oliveira R. R., Murakami M., Vicentini R., Costa-Filho A., Alexandrino F., et al. (2011). Expression, purification and spectroscopic analysis of an HdrC: an iron–sulfur cluster-containing protein from Acidithiobacillus ferrooxidans. Process Biochem. 46 1335–1341. 10.1016/j.procbio.2011.03.001 [DOI] [Google Scholar]
- Pathak A., Dastidar M. G., Sreekrishnan T. R. (2009). Bioleaching of heavy metals from sewage sludge: a review. J. Environ. Manage. 90 2343–2353. 10.1016/j.jenvman.2008.11.005 [DOI] [PubMed] [Google Scholar]
- Pham V. H., Yong J. J., Park S. J., Yoon D. N., Chung W. H., Rhee S. K. (2008). Molecular analysis of the diversity of the sulfide : quinone reductase (sqr) gene in sediment environments. Microbiology 154(Pt 10), 3112–3121. 10.1099/mic.0.2008/018580-0 [DOI] [PubMed] [Google Scholar]
- Quatrini R., Appia-Ayme C., Denis Y., Jedlicki E., Holmes D. S., Bonnefoy V. (2009). Extending the models for iron and sulfur oxidation in the extreme acidophile Acidithiobacillus ferrooxidans. BMC Genomics 10:394. 10.1186/1471-2164-10-394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralf S., Gabriele H., Thomas G., Willem H. (1987). Chromatographic separation of higher polythionates SnO (n = 3…22) and their detection in cultures of Thiobacillus ferroxidans; molecular composition of bacterial sulfur secretions. Angew. Chem. Int. Ed. Engl. 26 151–153. 10.1002/anie.198701511 [DOI] [Google Scholar]
- Ramírez P., Toledo H., Guiliani N., Jerez C. A. (2002). An exported rhodanese-like protein is induced during growth of Acidithiobacillus ferrooxidans in metal sulfides and different sulfur compounds. Appl. Environ. Microbiol. 68 1837–1845. 10.1128/AEM.68.4.1837-1845.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastegar S. O., Mousavi S. M., Shojaosadati S. A., Sarraf Mamoory R. (2015). Bioleaching of V, Ni, and Cu from residual produced in oil fired furnaces using Acidithiobacillus ferrooxidans. Hydrometallurgy 157 50–59. 10.1016/j.hydromet.2015.07.006 [DOI] [Google Scholar]
- Rawlings D. E. (2005). Characteristics and adaptability of iron- and sulfur-oxidizing microorganisms used for the recovery of metals from minerals and their concentrates. Microb. Cell Fact. 4:13. 10.1186/1475-2859-4-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings D. E., Tributsch H., Hansford G. S. (1999). Reasons why ‘Leptospirillum’-like species rather than Thiobacillus ferrooxidans are the dominant iron-oxidizing bacteria in many commercial processes for the biooxidation of pyrite and related ores. Microbiology 145(Pt 1), 5–13. 10.1099/13500872-145-1-5 [DOI] [PubMed] [Google Scholar]
- Rohwerder T., Sand W. (2003). The sulfane sulfur of persulfides is the actual substrate of the sulfur-oxidizing enzymes from Acidithiobacillus and Acidiphilium spp. Microbiology 149(Pt 7), 1699–1710. 10.1099/mic.0.26212-0 [DOI] [PubMed] [Google Scholar]
- Rohwerder T., Sand W. (2008). Properties of thiols required for sulfur dioxygenase activity at acidic pH. J. Sulfur Chem. 29 293–302. 10.1080/17415990802146972 [DOI] [Google Scholar]
- Ruhl P., Haas P., Seipel D., Becker J., Kletzin A. (2018). Persulfide dioxygenase from Acidithiobacillus caldus: variable roles of cysteine residues and hydrogen bond networks of the active site. Front. Microbiol. 9:1610. 10.3389/fmicb.2018.01610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rzhepishevska O. I., Valdes J., Marcinkeviciene L., Gallardo C. A., Meskys R., Bonnefoy V., et al. (2007). Regulation of a novel Acidithiobacillus caldus gene cluster involved in metabolism of reduced inorganic sulfur compounds. Appl. Environ. Microbiol. 73 7367–7372. 10.1128/aem.01497-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoval Ponce J., Moinier D., Byrne D., Amouric A., Bonnefoy V. (2012). Acidithiobacillus ferrooxidans oxidizes ferrous iron before sulfur likely through transcriptional regulation by the global redox responding RegBA signal transducing system. Hydrometallurgy 127–128, 187–194. 10.1016/j.hydromet.2012.07.016 [DOI] [Google Scholar]
- Sattler S. A., Wang X., Lewis K. M., DeHan P. J., Park C. M., Xin Y., et al. (2015). Characterizations of two bacterial persulfide dioxygenases of the metallo-beta-lactamase superfamily. J. Biol. Chem. 290 18914–18923. 10.1074/jbc.M115.652537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauve V., Bruno S., Berks B. C., Hemmings A. M. (2007). The SoxYZ complex carries sulfur cycle intermediates on a peptide swinging arm. J. Biol. Chem. 282 23194–23204. 10.1074/jbc.M701602200 [DOI] [PubMed] [Google Scholar]
- Sauve V., Roversi P., Leath K. J., Garman E. F., Antrobus R., Lea S. M., et al. (2009). Mechanism for the hydrolysis of a sulfur-sulfur bond based on the crystal structure of the thiosulfohydrolase SoxB. J. Biol. Chem. 284 21707–21718. 10.1074/jbc.M109.002709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrenk M. O., Edwards K. J., Goodman R. M., Hamers R. J., Banfield J. F. (1998). Distribution of Thiobacillus ferrooxidans and Leptospirillum ferrooxidans: implications for generation of acid mine drainage. Science 279 1519–1522. 10.1126/science.279.5356.1519 [DOI] [PubMed] [Google Scholar]
- Schutz M., Maldener I., Griesbeck C., Hauska G. (1999). Sulfide-quinone reductase from Rhodobacter capsulatus: requirement for growth, periplasmic localization, and extension of gene sequence analysis. J. Bacteriol. 181 6516–6523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharmin S., Yoshino E., Kanao T., Kamimura K. (2016). Characterization of a novel thiosulfate dehydrogenase from a marine acidophilic sulfur-oxidizing bacterium, Acidithiobacillus thiooxidans strain SH. Biosci. Biotechnol. Biochem. 80 273–278. 10.1080/09168451.2015.1088377 [DOI] [PubMed] [Google Scholar]
- Silver M., Lundgren D. G. (1968a). Sulfur-oxidizing enzyme of Ferrobacillus ferrooxidans (Thiobacillus ferrooxidans). Can. J. Biochem. 46 457–461. 10.1139/o68-069 [DOI] [PubMed] [Google Scholar]
- Silver M., Lundgren D. G. (1968b). The thiosulfate-oxidizing enzyme of Ferrobacillus ferrooxidans (Thiobacillus ferrooxidans). Can. J. Biochem. 46 1215–1220. 10.1139/o68-181 [DOI] [PubMed] [Google Scholar]
- Smit J. D. G., Urbanska K. M. (1986). Rhodanese activity in Lotus corniculatus s.l. J. Nat. Hist. 20 1467–1476. 10.1080/00222938600770991 [DOI] [Google Scholar]
- Sousa F. M., Pereira J. G., Marreiros B. C., Pereira M. M. (2018). Taxonomic distribution, structure/function relationship and metabolic context of the two families of sulfide dehydrogenases: SQR and FCSD. Biochim. Biophys. Acta Bioenerg. 1859 742–753. 10.1016/j.bbabio.2018.04.004 [DOI] [PubMed] [Google Scholar]
- Stockdreher Y., Sturm M., Josten M., Sahl H. G., Dobler N., Zigann R., et al. (2014). New proteins involved in sulfur trafficking in the cytoplasm of Allochromatium vinosum. J. Biol. Chem. 289 12390–12403. 10.1074/jbc.M113.536425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugio T., Hirose T., Ye L. Z., Tano T. (1992). Purification and some properties of sulfite:ferric ion oxidoreductase from Thiobacillus ferrooxidans. J. Bacteriol. 174 4189–4192. 10.1128/jb.174.12.4189-4192.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugio T., Kanao T., Furukawa H., Nagasawa T., Blake R. C. (1996). Isolation and identification of an iron-oxidizing bacterium which can grow on tetrathionate medium and the properties of a tetrathionate-decomposing enzyme isolated from the bacterium. J. Ferment. Bioeng. 82 233–238. 10.1016/0922-338X(96)88813-1 [DOI] [Google Scholar]
- Sugio T., Katagiri T., Moriyama M., Zhen Y. L., Inagaki K., Tano T. (1988). Existence of a new type of sulfite oxidase which utilizes ferric ions as an electron acceptor in Thiobacillus ferrooxidans. Appl. Environ. Microbiol. 54 153–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugio T., Mizunashi W., Inagaki K., Tano T. (1987a). Purification and some properties of sulfur:ferric ion oxidoreductase from Thiobacillus ferrooxidans. J. Bacteriol. 169 4916–4922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugio T., Noguchi M., Tano T. (1987b). Detoxification of sulfite produced during the oxidation of elemental sulfur by Thiobacillus ferrooxidans. Agric. Biol. Chem. 51 1431–1433. 10.1271/bbb1961.51.1431 [DOI] [Google Scholar]
- Sugio T., Taha T. M., Takeuchi F. (2009). Ferrous iron production mediated by tetrathionate hydrolase in tetrathionate-, sulfur-, and iron-grown Acidithiobacillus ferrooxidans ATCC 23270 cells. Biosci. Biotechnol. Biochem. 73 1381–1386. 10.1271/bbb.90036 [DOI] [PubMed] [Google Scholar]
- Suzuki I. (1965). Oxidation of elemental sulfur by an enzyme system of Thiobacillus thiooxidans. Biochim. Biophys. Acta 104 359–371. 10.1016/0304-4165(65)90341-7 [DOI] [PubMed] [Google Scholar]
- Suzuki I. (1994). Sulfite: Cytochrome c Oxidoreductase of Thiobacilli Methods in Enzymology. Cambridge, MA: Academic Press, 447–454. [Google Scholar]
- Suzuki I., Chan C. W., Takeuchi T. L. (1992). Oxidation of elemental sulfur to sulfite by Thiobacillus thiooxidans cells. Appl. Environ. Microbiol. 58 3767–3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki I., Werkman C. H. (1959). Glutathione and sulfur oxidation by Thiobacillus thiooxidans. Proc. Natl. Acad. Sci. U.S.A. 45 239–244. 10.1073/pnas.45.2.239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabita R., Silver M., Lundgren D. G. (1969). The rhodanese enzyme of Ferrobacillus ferrooxidans (Thiobacillus ferrooxidans). Can. J. Biochem. 47 1141–1145. 10.1139/o69-184 [DOI] [PubMed] [Google Scholar]
- Talla E., Hedrich S., Mangenot S., Ji B., Johnson D. B., Barbe V., et al. (2014). Insights into the pathways of iron- and sulfur-oxidation, and biofilm formation from the chemolithotrophic acidophile Acidithiobacillus ferrivorans CF27. Res. Microbiol. 165 753–760. 10.1016/j.resmic.2014.08.002 [DOI] [PubMed] [Google Scholar]
- Tano T., Kitaguchi H., Harada M., Nagasawa T., Sugio T. (1996). Purification and some properties of a tetrathionate decomposing enzyme from Thiobacillus thiooxidans. Biosci. Biotechnol. Biochem. 60 224–227. 10.1271/bbb.60.224 [DOI] [PubMed] [Google Scholar]
- Taylor B. E., Wheeler M. C., Nordstrom D. K. (1984). Isotope composition of sulphate in acid mine drainage as measure of bacterial oxidation. Nature 308 538–541. 10.1038/308538a0 [DOI] [Google Scholar]
- Temple K. L., Colmer A. R. (1951). The autotrophic oxidation of iron by a new bacterium, Thiobacillus ferrooxidans. J. Bacteriol. 62 605–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thauer R. K., Kaster A. K., Seedorf H., Buckel W., Hedderich R. (2008). Methanogenic archaea: ecologically relevant differences in energy conservation. Nat. Rev. Microbiol. 6 579–591. 10.1038/nrmicro1931 [DOI] [PubMed] [Google Scholar]
- Tiranti V., Viscomi C., Hildebrandt T., Di Meo I., Mineri R., Tiveron C., et al. (2009). Loss of ETHE1, a mitochondrial dioxygenase, causes fatal sulfide toxicity in ethylmalonic encephalopathy. Nat. Med. 15 200–205. 10.1038/nm.1907 [DOI] [PubMed] [Google Scholar]
- Urich T., Bandeiras T. M., Leal S. S., Rachel R., Albrecht T., Zimmermann P., et al. (2004). The sulphur oxygenase reductase from Acidianus ambivalens is a multimeric protein containing a low-potential mononuclear non-haem iron centre. Biochem. J. 381(Pt 1), 137–146. 10.1042/bj20040003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urich T., Gomes C. M., Kletzin A., Frazao C. (2006). X-ray structure of a self-compartmentalizing sulfur cycle metalloenzyme. Science 311 996–1000. 10.1126/science.1120306 [DOI] [PubMed] [Google Scholar]
- Valdés J., Ossandon F., Quatrini R., Dopson M., Holmes D. S. (2011). Draft genome sequence of the extremely acidophilic biomining bacterium Acidithiobacillus thiooxidans ATCC 19377 provides insights into the evolution of the Acidithiobacillus genus. J. Bacteriol. 193 7003–7004. 10.1128/jb.06281-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdés J., Pedroso I., Quatrini R., Dodson R. J., Tettelin H., Blake R., et al. (2008a). Acidithiobacillus ferrooxidans metabolism: from genome sequence to industrial applications. BMC Genomics 9:597. 10.1186/1471-2164-9-597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdés J., Pedroso I., Quatrini R., Holmes D. S. (2008b). Comparative genome analysis of Acidithiobacillus ferrooxidans, A. thiooxidans and A. caldus: insights into their metabolism and ecophysiology. Hydrometallurgy 94 180–184. 10.1016/j.hydromet.2008.05.039 [DOI] [Google Scholar]
- van Zyl L. J., van Munster J. M., Rawlings D. E. (2008). Construction of arsB and tetH mutants of the sulfur-oxidizing bacterium Acidithiobacillus caldus by marker exchange. Appl. Environ. Microbiol. 74 5686–5694. 10.1128/aem.01235-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vestal J. R., Lundgren D. G. (1971). The sulfite oxidase of Thiobacillus ferrooxidans (Ferrobacillus ferrooxidans). Can. J. Biochem. 49 1125–1130. 10.1139/o71-162 [DOI] [PubMed] [Google Scholar]
- Vishniac W., Santer M. (1957). The thiobacilli. Bacteriol. Rev. 21 195–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner T., Koch J., Ermler U., Shima S. (2017). Methanogenic heterodisulfide reductase (HdrABC-MvhAGD) uses two noncubane [4Fe-4S] clusters for reduction. Science 357 699–703. 10.1126/science.aan0425 [DOI] [PubMed] [Google Scholar]
- Wakai S., Kikumoto M., Kanao T., Kamimura K. (2004). Involvement of sulfide:quinone oxidoreductase in sulfur oxidation of an acidophilic iron-oxidizing bacterium, Acidithiobacillus ferrooxidans NASF-1. Biosci. Biotechnol. Biochem. 68 2519–2528. 10.1271/bbb.68.2519 [DOI] [PubMed] [Google Scholar]
- Waksman S. A., Joffe J. S. (1922). Microorganisms concerned in the oxidation of sulfur in the soil: II. Thiobacillus thiooxidans, a new sulfur-oxidizing organism isolated from the soil. J. Bacteriol. 7 239–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Liu S., Liu X., Li X., Wen Q., Lin J. (2014). Identification and characterization of an ETHE1-like sulfur dioxygenase in extremely acidophilic Acidithiobacillus spp. Appl. Microbiol. Biotechnol. 98 7511–7522. 10.1007/s00253-014-5830-4 [DOI] [PubMed] [Google Scholar]
- Wang Z. B., Li Y. Q., Lin J. Q., Pang X., Liu X. M., Liu B. Q., et al. (2016). The two-component system RsrS-RsrR regulates the tetrathionate intermediate pathway for thiosulfate oxidation in Acidithiobacillus caldus. Front. Microbiol. 7:1755. 10.3389/fmicb.2016.01755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welte C., Hafner S., Kratzer C., Quentmeier A., Friedrich C. G., Dahl C. (2009). Interaction between sox proteins of two physiologically distinct bacteria and a new protein involved in thiosulfate oxidation. FEBS Lett. 583 1281–1286. 10.1016/j.febslet.2009.03.020 [DOI] [PubMed] [Google Scholar]
- Williams K. P., Kelly D. P. (2013). Proposal for a new class within the phylum Proteobacteria, Acidithiobacillia classis nov., with the type order Acidithiobacillales, and emended description of the class Gammaproteobacteria. Int. J. Syst. Evol. Microbiol. 63(Pt 8), 2901–2906. 10.1099/ijs.0.049270-0 [DOI] [PubMed] [Google Scholar]
- Wu W., Pang X., Lin J., Liu X., Wang R., Lin J., et al. (2017). Discovery of a new subgroup of sulfur dioxygenases and characterization of sulfur dioxygenases in the sulfur metabolic network of Acidithiobacillus caldus. PLoS One 12:e0183668. 10.1371/journal.pone.0183668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J.-L., Peng A.-A., He H., Yang Y., Liu X.-D., Qiu G.-Z. (2007). A new strain Acidithiobacillus albertensis BY-05 for bioleaching of metal sulfides ores. Trans. Nonferrous Metal. Soc. China 17 168–175. 10.1016/S1003-6326(07)60067-3 [DOI] [Google Scholar]
- Xia Y., Lu C., Hou N., Xin Y., Liu J., Liu H., et al. (2017). Sulfide production and oxidation by heterotrophic bacteria under aerobic conditions. ISME J. 11 2754–2766. 10.1038/ismej.2017.125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H., Zhang X., Li X., He Z., Liang Y., Guo X., et al. (2014). Whole-genome sequencing reveals novel insights into sulfur oxidation in the extremophile Acidithiobacillus thiooxidans. BMC Microbiol. 14:179. 10.1186/1471-2180-14-179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- You X. Y., Guo X., Zheng H. J., Zhang M. J., Liu L. J., Zhu Y. Q., et al. (2011). Unraveling the Acidithiobacillus caldus complete genome and its central metabolisms for carbon assimilation. J. Genet. Genomics 38 243–252. 10.1016/j.jgg.2011.04.006 [DOI] [PubMed] [Google Scholar]
- Yu Y., Liu X., Wang H., Li X., Lin J. (2014). Construction and characterization of tetH overexpression and knockout strains of Acidithiobacillus ferrooxidans. J. Bacteriol. 196 2255–2264. 10.1128/jb.01472-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zander U., Faust A., Klink B. U., de Sanctis D., Panjikar S., Quentmeier A., et al. (2011). Structural basis for the oxidation of protein-bound sulfur by the sulfur cycle molybdohemo-enzyme sulfane dehydrogenase SoxCD. J. Biol. Chem. 286 8349–8360. 10.1074/jbc.M110.193631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Chen L., Dong N., Lin H., Lin J., Lin J. (2014). “Markerless deletion of a rhodanese in Acidithiobacillus caldus MTH-04” in Proceedings of the International Conference on Human Health and Medical Engineering, (Hoboken, NJ: WileyPublishing company; ), 745–752. 10.2495/HHME131002 [DOI] [Google Scholar]
- Zhang X., Liu Z., Wei G., Yang F., Liu X. (2018). In silico genome-wide analysis reveals the potential links between core genome of Acidithiobacillus thiooxidans and its autotrophic lifestyle. Front. Microbiol. 9:1255. 10.3389/fmicb.2018.01255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Weiner J. H. (2014). Characterization of the kinetics and electron paramagnetic resonance spectroscopic properties of Acidithiobacillus ferrooxidans sulfide:quinone oxidoreductase (SQR). Arch. Biochem. Biophys. 564 110–119. 10.1016/j.abb.2014.Vesta09.016 [DOI] [PubMed] [Google Scholar]