Abstract
Objective: Published studies have demonstrated a closer association between vitamin D receptor (VDR) gene polymorphisms and polycystic ovary syndrome (PCOS) risk, but the results were inconsistent. We therefore performed this meta-analysis to explore the precise associations between VDR gene polymorphisms and PCOS risk.
Methods: Five online electronic databases (PubMed, Embase, SCI index, CNKI and Wanfang) were searched. Odds ratios (ORs) with 95% confidence interval (CIs) were calculated to assess the association between VDR Fok I C/T (rs10735810), BsmI A/G (rs1544410), ApaI A/C (rs7975232), and TaqI T/C (rs731236) polymorphisms and PCOS risk. In addition, heterogeneity, accumulative/sensitivity analysis and publication bias were conducted to check the statistical power.
Results: Overall, 10 publications (31 independent case-control studies) involving 1,531 patients and 1,174 controls were identified. We found that the C mutation of ApaI A/C was a risk factor for PCOS (C vs. A: OR = 1.20, 95%CI = 1.06–1.35, P < 0.01, I2 = 29.7%; CC vs. AA: OR = 1.49, 95%CI = 1.17–1.91, P < 0.01, I2 = 0%; CC vs. AA+AC: OR = 1.36, 95%CI = 1.09–1.69, P = 0.01, I2 = 12.8%). Moreover, the BsmI A/G polymorphism also showed a dangerous risk for PCOS in Asian population (G vs. A: OR = 1.62, 95%CI = 1.24–2.11, P < 0.01, I2 = 0%; AG vs. AA: OR = 2.08, 95%CI = 1.26–3.20, P < 0.01, I2 = 0%; GG vs. AA: OR = 2.21, 95%CI = 1.29–3.77, P < 0.01, I2 = 0%; AG+GG vs. AA: OR = 2.12, 95%CI = 1.42–3.16, P < 0.01, I2 = 0%). In addition, no significant association of Fok I C/T, and TaqI T/C polymorphisms was observed.
Conclusions: In summary, our meta-analysis suggested that VDR gene polymorphisms contribute to PCOS development, especially in Asian populations.
Keywords: vitamin D receptor, polycystic ovary syndrome, polymorphism, meta-analysis, risk
Introduction
Polycystic ovary syndrome (PCOS), characterized by clinical features including menstrual disorder, persistent anovulation, and polycystic ovaries, is one of the most common reproductive, endocrine, and metabolic disorder syndromes among women of reproductive age (Sirmans and Pate, 2013). Polycystic ovary syndrome (PCOS) has become a highly prevalent disorder that affects women in their reproductive age and contributes to multiple complications. According to the NIH 1990 criteria and/or Rotterdam 2003 criteria, the cumulative prevalence of PCOS was ~4–21% worldwide (Knochenhauer et al., 1998; Asuncion et al., 2000; Azziz et al., 2004). High prevalence and elevated risk of the development of type 2 diabetes mellitus (T2DM) and cardiovascular disease (CVD) were reported in women with PCOS (Repaci et al., 2011; Ollila et al., 2017). Moreover, long-term complications including the mental dysfunctions, such as mood and sleeping disorders, are also found. However, the precise etiology and underlying pathological mechanism of PCOS remain unclear.
Vitamin D, a steroid hormone, plays an important role in maintaining calcium homeostasis and promoting bone mineralization (Shen et al., 2013). Beyond these fundamental relationships, accumulating evidence indicates a close association of vitamin D status with the pathogenesis, signs and symptoms of PCOS (Wehr et al., 2009; Krul-Poel et al., 2013). A recent meta-analysis found significant differences in serum 25-hydroxyvitamin D, serum insulin, total cholesterol, triglycerides, and low-density lipoprotein cholesterol in patients with PCOS compared with that in healthy controls (Bacopoulou et al., 2017).
Vitamin D receptor (VDR) is widely distributed in several tissues of the female reproductive system (Kato, 2000). Vitamin D receptor (VDR) could mediate the biological responses of the 1α,25(OH)2D3 hormone, through generating a signal transduction complex with a heterodimer of 1α,25(OH)2D3-liganded VDR and unoccupied retinoid X receptor (RXR). Then, this transcriptional unit combines with the vitamin D response element (VDRE) in the promoter region of genes and regulates its actions through altering the transcriptional expression of target genes (Haussler et al., 2011). Single nucleotide polymorphisms (SNPs) are the most frequent nucleotide variations in the human genome. The VDR gene is located on chromosome 12q13.11, includes eight protein coding exons and one untranslated exon, and encodes a 427-amino-acid protein (Baker et al., 1988). To date, four most common VDR polymorphisms of FokI (rs10735810 C>T), BsmI (rs1544410 G>A), ApaI (rs7975232 G>T), and TaqI (rs731236 T>C) have been investigated to explore the association between VDR and PCOS susceptibility. However, the results were conflicting and inconclusive owing to the small sample size and limited statistical power. We therefore conducted this comprehensive meta-analysis to evaluate the association between the above polymorphisms and PCOS susceptibility precisely.
Materials and Methods
This meta-analysis was conducted according to the guideline of Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) statement (Moher et al., 2009). All included data were based on published studies, and no ethical issues were involved.
Search Strategy
Five online electronic databases (PubMed, Embase, SCI index, CNKI, and Wanfang) were searched with the following terms from their inception up to March 20, 2018: “vitamin D receptor,” “VDR,” “rs10735810,” “rs1544410,” “rs7975232,” “rs731236,” “polymorphism,” “variant,” “mutation” “polycystic ovary syndrome,” and “PCOS.” Some relevant references cited within retrieved articles were reviewed with manual searched.
The following search strategy was used:
#1 vitamin D receptor
#2 VDR
#3 rs10735810
#4 rs1544410
#5 rs7975232
#6 rs731236
#7 #1 OR #2 OR #3 OR #4 OR #5 OR #6
#8 polymorphism
#9 variant
#10 mutation
#11 #8 OR #9 OR #10
#12 polycystic ovary syndrome
#13 PCOS
#14 #12 OR #13
# 15 #7 AND #11 AND #14
Eligibility Criteria
Studies were selected when they met the following criteria by two independent investigators (NYM and HYY): (1) the study followed a case-control design; (2) at least one polymorphisms of VDR gene was reported; (3) sufficient information about the distribution frequency of different polymorphism loci could be extracted to calculate the odds ratios (ORs) and 95% confidence intervals (CIs); (4) the most recent or largest sample sizes were selected when multiple publications were repeatedly reported with same team; and (5) the articles were written in English and Chinese.
Data Extraction
All included studies were reviewed and extracted by two independent investigators (NYM and WYD). Disagreements and compared results were settled through discussion. The following information and data were extracted from included studies: the first author of each study, published year, study country or region where the study was conducted, ethnicity of research population, the source of the controls, the sample sizes of patients with PCOS and healthy controls, data of the frequency genotype of distribution, and the genotyping method.
Risk Assessment of Bias Within Studies
All included studies in this meta-analysis were subject to make risk assessment of bias by two independent authors (JGB and BG) via the modified Newcastle-Ottawa Quality Assessment Scale (Niu et al., 2015). The score was based on five parameters (representativeness of cases, source of controls, Hardy-Weinberg equilibrium (HWE) in controls, genotyping examination and association assessment), with a maximum score of 11 points. Studies of at least a score of 8 were identified with a high quality (Table 1).
Table 1.
Scale for quality evaluation.
| Criteria | Score |
|---|---|
| Representativeness of cases | |
| Consecutive/randomly selected cases with clearly defined sampling frame Not consecutive/randomly selected case or without clearly defined sampling frame Not described |
2 1 0 |
| Source of controls | |
| Population-based Hospital-bases or healthy-bases Not described |
2 1 0 |
| Hardy-weinberg equilibrium in controls | |
| Hardy-weinberg equilibrium Hardy-weinberg disequilibrium Not available |
2 1 0 |
| Genotyping examination | |
| Genotyping done under “blinded” condition and repeated again Genotyping done under “blinded” condition or repeated again Unblinded done or not mentioned and unrepeated |
2 1 0 |
| Subjects | |
| Number ≥300 Number < 300 |
1 0 |
| Association assessment | |
| Assess association between genotypes and PCOS with appropriate statistics and adjustment for confounders Assess association between genotypes and PCOS with appropriate statistics and without adjustment for confounders Inappropriate statistics used |
2 1 0 |
Statistical Analysis
Crude ORs with 95% CIs were calculated to examine the statistical power of the association between the VDR gene polymorphisms and PCOS risk. For example, four genetic models of Fok I C/T polymorphisms were calculated: allele contrast (T vs. C), co-dominant models (heterozygote model: CT vs. CC, homozygote model: TT vs. CC), dominant model (CT+TT vs. CC), and recessive model (TT vs. CC+CT) (Minelli et al., 2005; Lewis and Knight, 2012). Similar genetic models were also calculated with the others [BsmI A/G (rs1544410), ApaI A/C (rs7975232), TaqI T/C (rs731236)]. Subgroup analysis based on HWE status, ethnicity difference, control design and genotyping methods were performed to clarify the potential risk. Heterogeneity was investigated by I2 index which describes the percentage of variation among the included studies in the pooled analysis (Huedo-Medina et al., 2006). The fixed-effect model (Mantel-Haenszel method) was used when the I2 value was < 40% (Mantel and Haenszel, 1959). Otherwise, a random-effects model (DerSimonian and Laird method) was adopted (DerSimonian, 1996). Cumulative analyses were conducted to explore the tendency of the results whit the published studies were added. Sensitivity analyses were performed to investigate the stability of the results when each study was removed one at a time. Publication bias was assessed with the Egger's bias test and Begg's funnel plots (Begg and Mazumdar, 1994; Egger et al., 1997). Data analysis was conducted using STATA version 14.0 (Stata Corporation, College Station, TX, USA). P < 0.05 indicated a statistically significant difference.
Results
Study Characteristics
At first, 120 publications were identified through the systematic literature search. Three important steps according to the eligibility criteria were conducted to screen the selected studies were as follows: duplicate check, title and abstract check and text review. The selection of screening is presented in Figure 1. Finally, 10 studies were included in the meta-analysis with 1,531 patients with PCOS and 1,174 control individuals (Mahmoudi, 2009; Wehr et al., 2011; Bagheri et al., 2012, 2013; El-Shal et al., 2013; Dasgupta et al., 2015; Jedrzejuk et al., 2015; Mahmoudi et al., 2015; Cao and Tu, 2016; Siddamalla et al., 2018). The studies comprised seven case-control studies on FokI C/T (Mahmoudi, 2009; Wehr et al., 2011; Bagheri et al., 2012; Dasgupta et al., 2015; Jedrzejuk et al., 2015; Mahmoudi et al., 2015; Cao and Tu, 2016), seven case-control studies on BsmI A/G (Mahmoudi, 2009; Wehr et al., 2011; Bagheri et al., 2012; Jedrzejuk et al., 2015; Mahmoudi et al., 2015; Siddamalla et al., 2018), eight case-control studies on ApaI A/C (Mahmoudi, 2009; Wehr et al., 2011; El-Shal et al., 2013; Dasgupta et al., 2015; Jedrzejuk et al., 2015; Mahmoudi et al., 2015; Cao and Tu, 2016; Siddamalla et al., 2018), and nine case-control studies on TaqI T/C (Mahmoudi, 2009; Wehr et al., 2011; Bagheri et al., 2013; El-Shal et al., 2013; Dasgupta et al., 2015; Jedrzejuk et al., 2015; Mahmoudi et al., 2015; Cao and Tu, 2016; Siddamalla et al., 2018), respectively. Furthermore, three publications involved the Asians (Dasgupta et al., 2015; Cao and Tu, 2016; Siddamalla et al., 2018), and seven studies involved Caucasians (Mahmoudi, 2009; Wehr et al., 2011; Bagheri et al., 2012, 2013; El-Shal et al., 2013; Jedrzejuk et al., 2015; Mahmoudi et al., 2015). In the control groups, there are two case-control studies in BsmI A/G (Mahmoudi, 2009; Siddamalla et al., 2018), three case-control studies in ApaI A/C (Wehr et al., 2011; Dasgupta et al., 2015; Siddamalla et al., 2018) and two case-control studies in TaqI T/C (Cao and Tu, 2016; Siddamalla et al., 2018) polymorphisms deviated from the HWE. The main characteristics of the selected studies are shown in Table 2.
Figure 1.
Flow diagram of the study selection process.
Table 2.
Characteristics of included studies on vitamin D receptor gene polymorphisms and polycystic ovary syndrome risk.
| References | Country/region | Racial | Source of controls | Case | Control | Genotype distribution | Genotyping methods | P for HWE | MAF in control | NOS | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case | Control | ||||||||||||||
| Fok I C/T (rs10735810) | |||||||||||||||
| CC | CT | TT | CC | CT | TT | ||||||||||
| Mahmoudi, 2009 | Iran | Caucasian | Hospital-B | 162 | 162 | 83 | 67 | 12 | 96 | 59 | 7 | PCR-RFLP | 0.58 | 0.23 | 8 |
| Wehr et al., 2011 | Austria | Caucasian | Hospital-B | 538 | 135 | 215 | 241 | 82 | 53 | 60 | 22 | Genotyping assay | 0.47 | 0.39 | 8 |
| Bagheri et al., 2012 | Iran | Caucasian | Healthy-B | 46 | 46 | 22 | 20 | 4 | 29 | 15 | 2 | PCR-RFLP | 0.97 | 0.21 | 7 |
| Jedrzejuk et al., 2015 | Poland | Caucasian | Healthy-B | 90 | 98 | 28 | 51 | 11 | 23 | 50 | 25 | PCR- SNaPshot | 0.84 | 0.51 | 7 |
| Dasgupta et al., 2015 | India | Asian | Healthy-B | 250 | 250 | 155 | 85 | 10 | 153 | 82 | 15 | PCR-RFLP | 0.37 | 0.22 | 8 |
| Mahmoudi et al., 2015 | Iran | Caucasian | Healthy-B | 35 | 35 | 16 | 17 | 2 | 24 | 10 | 1 | PCR-RFLP | 0.97 | 0.17 | 8 |
| Cao and Tu, 2016 | China | Asian | Hospital-B | 120 | 120 | 70 | 40 | 10 | 65 | 45 | 10 | PCR-RFLP | 0.60 | 0.27 | 7 |
| BsmI A/G (rs1544410) | |||||||||||||||
| AA | AG | GG | AA | AG | GG | ||||||||||
| Mahmoudi, 2009 | Iran | Caucasian | Hospital-B | 162 | 162 | 24 | 85 | 53 | 18 | 91 | 53 | PCR-RFLP | 0.02 | 0.61 | 8 |
| Wehr et al., 2011 | Austria | Caucasian | Hospital-B | 537 | 137 | 77 | 244 | 216 | 22 | 66 | 49 | Genotyping assay | 0.98 | 0.60 | 8 |
| Bagheri et al., 2012 | Iran | Caucasian | Healthy-B | 46 | 46 | 15 | 27 | 4 | 20 | 24 | 2 | PCR-RFLP | 0.12 | 0.30 | 7 |
| Jedrzejuk et al., 2015 | Poland | Caucasian | Healthy-B | 90 | 98 | 14 | 45 | 31 | 13 | 42 | 43 | PCR- SNaPshot | 0.59 | 0.65 | 7 |
| Mahmoudi et al., 2015 | Iran | Caucasian | Healthy-B | 35 | 35 | 10 | 12 | 13 | 5 | 23 | 7 | PCR-RFLP | 0.06 | 0.53 | 8 |
| Cao and Tu, 2016 | China | Asian | Hospital-B | 120 | 120 | 23 | 60 | 37 | 40 | 55 | 25 | PCR-RFLP | 0.45 | 0.44 | 7 |
| Siddamalla et al., 2018 | India | Asian | Hospital-B | 95 | 130 | 35 | 45 | 15 | 72 | 41 | 17 | PCR-RFLP | 0.01 | 0.29 | 7 |
| ApaI A/C (rs7975232) | |||||||||||||||
| AA | AC | CC | AA | AC | CC | ||||||||||
| Mahmoudi, 2009 | Iran | Caucasian | Hospital-B | 162 | 162 | 58 | 68 | 36 | 49 | 90 | 23 | PCR-RFLP | 0.07 | 0.42 | 8 |
| Wehr et al., 2011 | Austria | Caucasian | Hospital-B | 543 | 145 | 142 | 274 | 127 | 48 | 60 | 37 | Genotyping assay | 0.04 | 0.46 | 7 |
| El-Shal et al., 2013 | Egypt | Caucasian | Healthy-B | 150 | 150 | 63 | 65 | 22 | 68 | 64 | 18 | PCR-RFLP | 0.62 | 0.33 | 8 |
| Jedrzejuk et al., 2015 | Poland | Caucasian | Healthy-B | 90 | 98 | 19 | 52 | 19 | 32 | 49 | 17 | PCR- SNaPshot | 0.81 | 0.42 | 7 |
| Dasgupta et al., 2015 | India | Asian | Healthy-B | 250 | 250 | 117 | 120 | 13 | 122 | 116 | 12 | PCR-RFLP | 0.02 | 0.28 | 7 |
| Mahmoudi et al., 2015 | Iran | Caucasian | Healthy-B | 35 | 35 | 15 | 11 | 9 | 8 | 21 | 6 | PCR-RFLP | 0.23 | 0.47 | 8 |
| Cao and Tu, 2016 | China | Asian | Hospital-B | 120 | 120 | 22 | 58 | 40 | 39 | 55 | 26 | PCR-RFLP | 0.43 | 0.45 | 7 |
| Siddamalla et al., 2018 | India | Asian | Hospital-B | 95 | 130 | 42 | 21 | 32 | 70 | 35 | 25 | PCR-RFLP | < 0.01 | 0.33 | 7 |
| TaqI T/C (rs731236) | |||||||||||||||
| TT | TC | CC | TT | TC | CC | ||||||||||
| Mahmoudi, 2009 | Iran | Caucasian | Hospital-B | 162 | 162 | 71 | 71 | 20 | 72 | 76 | 14 | PCR-RFLP | 0.33 | 0.32 | 8 |
| Wehr et al., 2011 | Austria | Caucasian | Hospital-B | 536 | 137 | 226 | 238 | 72 | 49 | 65 | 23 | Genotyping assay | 0.85 | 0.41 | 8 |
| Bagheri et al., 2013 | Iran | Caucasian | Healthy-B | 38 | 38 | 16 | 14 | 8 | 17 | 19 | 2 | PCR-RFLP | 0.26 | 0.30 | 7 |
| El-Shal et al., 2013 | Egypt | Caucasian | Healthy-B | 150 | 150 | 40 | 74 | 36 | 69 | 61 | 20 | PCR-RFLP | 0.27 | 0.34 | 8 |
| Jedrzejuk et al., 2015 | Poland | Caucasian | Healthy-B | 90 | 98 | 37 | 45 | 8 | 49 | 37 | 12 | PCR- SNaPshot | 0.24 | 0.31 | 7 |
| Dasgupta et al., 2015 | India | Asian | Healthy-B | 250 | 250 | 112 | 91 | 47 | 109 | 104 | 37 | PCR-RFLP | 0.14 | 0.36 | 8 |
| Mahmoudi et al., 2015 | Iran | Caucasian | Healthy-B | 35 | 35 | 15 | 14 | 6 | 15 | 16 | 4 | PCR-RFLP | 0.93 | 0.34 | 8 |
| Cao and Tu, 2016 | China | Asian | Hospital-B | 120 | 120 | 57 | 52 | 11 | 40 | 72 | 8 | PCR-RFLP | < 0.01 | 0.37 | 6 |
| Siddamalla et al., 2018 | India | Asian | Hospital-B | 95 | 130 | 40 | 31 | 24 | 71 | 42 | 17 | PCR-RFLP | 0.01 | 0.29 | 7 |
HWE in control. MAF, Minor allele frequency in control group; Hospital-B, Hospital-based control; Healthy-B, Healthy-based control.
Quantitative Analysis
Fok I C/T Locus and PCOS Risk
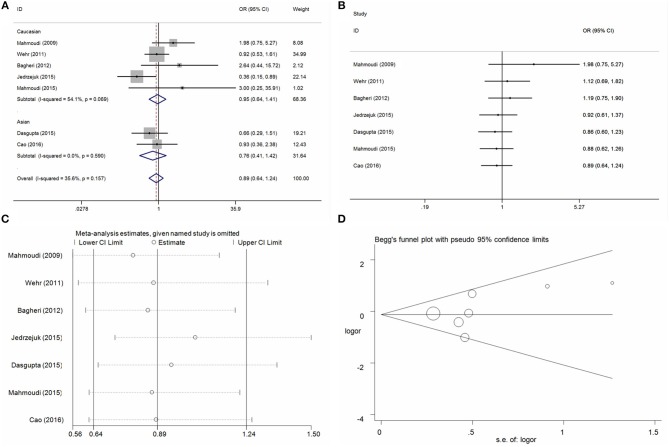
Seven case-control studies with 1,241 PCOS cases and 846 control individuals were identified with regard to the association between Fok I C/T locus and PCOS risk. Overall, the pool analysis did not find any significant association between this locus on PCOS risk in five genetic models (T vs. C: OR = 1.04, 95%CI = 0.83–1.30, P = 0.77, I2 = 53.2%; CT vs. CC: OR = 1.08, 95%CI = 0.89–1.32, P = 0.40, I2 = 7.0%; TT vs. CC: OR = 0.89, 95%CI = 0.64–1.25, P = 0.50, I2 = 35.6%; CT+TT vs. CC: OR = 1.06, 95%CI = 0.88–1.27, P = 0.56, I2 = 36.4%; TT vs. CC+CT: OR = 0.86, 95%CI = 0.63–1.18, P = 0.34, I2 = 23.6%) (Table 3, Figure 2A for TT vs. CC model). Heterogeneity was only indentified in allele contrast and the meta-regression analyses did not find any distinct factors that contributed to the heterogeneity. Furthermore, no significant association was identified in stratified analysis of HWE status, ethnicity difference, and control design and genotyping methods (Table 3). Cumulative analyses by publication date showing the negative results according to the new studies were added (Figure 2B for TT vs. CC model). Sensitivity analysis presented a consistent tendency of negative results without any apparent changes (Figure 2C for TT vs. CC model). Publication bias was assessed using the Egger's bias test and Begg's funnel plot tests, and no significant asymmetrical evidence was found (T vs. C: P = 0.23; CT vs. CC: P = 0.20; TT vs. CC: P = 0.33; CT+TT vs. CC: P = 0. 27; TT vs. CC+CT: P = 0.37) (Figure 2D for TT vs. CC model).
Table 3.
Summary ORs and 95% CI of vitamin D receptor gene polymorphisms and polycystic ovary syndrome risk.
| Locus | N* | OR | 95% CI | P | I2 | OR | 95% CI | P | I2 | OR | 95% CI | P | I2 | OR | 95% CI | P | I2 | OR | 95% CI | P | I2 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Fok I C/T (rs10735810) | T vs. C | CT vs. CC | TT vs. CC | CT+TT vs. CC | TT vs. CC+CT | ||||||||||||||||
| Total | 7 | 1.04 | 0.83–1.30 | 0.77 | 53.2 | 1.08 | 0.89–1.32 | 0.42 | 7.0 | 0.89 | 0.64–1.25 | 0.50 | 35.6 | 1.06 | 0.88–1.27 | 0.56 | 36.4 | 0.86 | 0.63–1.18 | 0.34 | 23.6 |
| ETHNICITY | |||||||||||||||||||||
| Caucasian | 5 | 1.14 | 0.80–1.60 | 0.47 | 66.6 | 1.18 | 0.92–1.53 | 0.20 | 19.3 | 1.07 | 0.52–2.19 | 0.86 | 54.1 | 1.22 | 0.84–1.77 | 0.31 | 49.5 | 0.96 | 0.53–1.76 | 0.91 | 45.0 |
| Asian | 2 | 0.91 | 0.72–1.16 | 0.46 | 0 | 0.95 | 0.70–1.30 | 0.77 | 0 | 0.77 | 0.41–1.43 | 0.40 | 0 | 0.92 | 0.69–1.24 | 0.60 | 0 | 0.79 | 0.43–1.45 | 0.44 | 0 |
| DESIGN | |||||||||||||||||||||
| Hospital-B | 3 | 1.04 | 0.86–1.27 | 0.65 | 28.1 | 1.04 | 0.80–1.36 | 0.75 | 0.0 | 1.07 | 0.70–1.56 | 0.76 | 0 | 1.05 | 0.82–1.36 | 0.68 | 15.9 | 1.06 | 0.71–1.59 | 0.78 | 0 |
| Healthy-B | 4 | 1.08 | .0.69–1.69 | 0.72 | 68.7 | 1.14 | 0.85–1.52 | 0.39 | 33.8 | 0.78 | 0.33–1.83 | 0.56 | 45.1 | 1.19 | 0.72–1.97 | 0.50 | 57.5 | 0.62 | 0.37–1.03 | 0.07 | 23.7 |
| GENOTYPING | |||||||||||||||||||||
| PCR-RFLP | 5 | 1.17 | 0.89–1.54 | 0.26 | 45.9 | 1.15 | 0.91–1.46 | 0.27 | 26.6 | 1.14 | 0.70–1.84 | 0.59 | 11.6 | 1.22 | 0.88–1.68 | 0.24 | 42.3 | 1.09 | 0.68–1.75 | 0.72 | 0 |
| Other | 2 | 0.82 | 0.56–1.19 | 0.30 | 57.7 | 0.95 | 0.67–1.35 | 0.76 | 0 | 0.62 | 0.25–1.53 | 0.30 | 66.6 | 0.88 | 0.63–1.23 | 0.47 | 0 | 0.65 | 0.29–1.44 | 0.28 | 66.5 |
| BsmI A/G (rs1544410) | G vs. A | AG vs. AA | GG vs. AA | AG+GG vs. AA | GG vs. AA+AG | ||||||||||||||||
| Total | 7 | 1.17 | 0.95–1.45 | 0.14 | 49.6 | 1.15 | 0.75–1.78 | 0.52 | 59.9 | 1.28 | 0.95–1.74 | 0.11 | 35.7 | 1.22 | 0.82–1.81 | 0.34 | 57.1 | 1.16 | 0.93–1.45 | 0.18 | 19.8 |
| HWE-yes | 5 | 1.17 | 0.90–1.51 | 0.24 | 45.2 | 1.10 | 0.67–1.82 | 0.70 | 50.8 | 1.38 | 0.96–2.00 | 0.09 | 36.9 | 1.19 | 0.76–1.89 | 0.48 | 47.5 | 1.26 | 0.84–1.89 | 0.27 | 42.2 |
| HWE-no | 2 | 1.20 | 0.70–2.07 | 0.50 | 78.2 | 1.27 | 0.40–4.01 | 0.68 | 84.7 | 1.15 | 0.48–2.72 | 0.76 | 61.2 | 1.26 | 0.43–3.64 | 0.67 | 84.1 | 1.06 | 0.72–1.58 | 0.16 | 0 |
| ETHNICITY | |||||||||||||||||||||
| Caucasian | 5 | 1.02 | 0.86–1.21 | 0.84 | 0.0 | 0.90 | 0.64–1.26 | 0.54 | 31.4 | 0.99 | 0.68–1.44 | 0.95 | 0.0 | 0.94 | 0.68–1.30 | 0.72 | 11.7 | 1.08 | 0.84–1.39 | 0.57 | 27.2 |
| Asian | 2 | 1.62 | 1.24–2.11 | < 0.01 | 0.0 | 2.08 | 1.26–3.20 | < 0.01 | 0 | 2.21 | 1.29–3.77 | < 0.01 | 0.0 | 2.12 | 1.42–3.16 | < 0.01 | 0 | 1.51 | 0.95–2.39 | 0.08 | 0 |
| DESIGN | |||||||||||||||||||||
| Hospital-B | 4 | 1.26 | 0.97–1.64 | 0.08 | 60.1 | 1.35 | 0.81–2.24 | 0.25 | 64.5 | 1.43 | 0.87–2.35 | 0.16 | 51.3 | 1.41 | 0.86–2.29 | 0.17 | 65.5 | 1.22 | 0.95–1.57 | 0.12 | 0 |
| Healthy-B | 3 | 0.96 | 0.71–1.31 | 0.82 | 20.8 | 0.82 | 0.33–2.00 | 0.66 | 59.8 | 0.90 | 0.45–1.78 | 0.76 | 0 | 0.90 | 0.45–1.80 | 0.76 | 40.0 | 1.28 | 0.49–3.30 | 0.62 | 58.9 |
| GENOTYPING | |||||||||||||||||||||
| PCR-RFLP | 5 | 1.29 | 0.99–1.99 | 0.06 | 46.3 | 1.17 | 0.62–2.20 | 0.63 | 71.6 | 1.49 | 0.85–2.60 | 0.16 | 40.7 | 1.29 | 0.74–2.25 | 0.37 | 66.9 | 1.30 | 0.97–1.80 | 0.07 | 0 |
| Other | 2 | 0.98 | 0.68–1.41 | 0.89 | 55.3 | 1.04 | 0.65–1.65 | 0.87 | 0 | 1.04 | 0.65–1.69 | 0.86 | 28.1 | 1.04 | 0.67–1.61 | 0.85 | 0 | 0.94 | 0.53–1.66 | 0.84 | 62.2 |
| ApaI A/C (rs7975232) | C vs. A | AC vs. AA | CC vs. AA | AC+CC vs. AA | CC vs. AA+AC | ||||||||||||||||
| Total | 8 | 1.20 | 1.06–1.35 | < 0.01 | 29.7 | 1.10 | 0.80–1.49 | 0.56 | 59.7 | 1.49 | 1.17–1.91 | < 0.01 | 0 | 1.21 | 0.94–1.57 | 0.15 | 51.3 | 1.36 | 1.09–1.69 | 0.01 | 12.8 |
| HWE-yes | 5 | 1.22 | 1.03–1.45 | 0.02 | 32.6 | 1.00 | 0.58–1.72 | 0.99 | 72.6 | 1.61 | 1.13–2.27 | 0.01 | 0 | 1.14 | 0.70–1.83 | 0.60 | 68.8 | 1.55 | 1.15–2.10 | < 0.01 | 0 |
| HWE-no | 3 | 1.21 | 0.95–1.54 | 0.13 | 49.0 | 1.21 | 0.94–1.56 | 0.14 | 0 | 1.38 | 0.97–1.97 | 0.07 | 18.1 | 1.26 | 1.00–1.59 | 0.06 | 0 | 1.25 | 0.71–2.20 | 0.44 | 62.7 |
| ETHNICITY | |||||||||||||||||||||
| Caucasian | 5 | 1.11 | 0.95–1.30 | 0.18 | 0 | 0.98 | 0.60–1.62 | 0.95 | 72.6 | 1.28 | 0.94–1.74 | 0.12 | 0 | 1.07 | 0.74–1.57 | 0.71 | 58.0 | 1.20 | 0.91–1.57 | 0.20 | 0 |
| Asian | 3 | 1.41 | 1.02–1.95 | 0.04 | 63.9 | 1.19 | 0.90–1.57 | 0.23 | 18.9 | 1.97 | 1.30–2.97 | < 0.01 | 22.8 | 1.42 | 0.96–2.10 | 0.08 | 48.6 | 1.72 | 1.19–2.50 | < 0.01 | 0 |
| DESIGN | |||||||||||||||||||||
| Hospital-B | 4 | 1.31 | 1.02–1.67 | 0.03 | 55.8 | 1.16 | 0.71–1.89 | 0.56 | 68.9 | 1.59 | 1.17–2.15 | < 0.01 | 38.1 | 1.33 | 0.90–1.97 | 0.16 | 61.1 | 1.50 | 0.98–2.30 | 0.06 | 59.9 |
| Healthy-B | 4 | 1.11 | 0.93–1.34 | 0.24 | 0 | 1.03 | 0.64–1.64 | 0.91 | 60.5 | 1.31 | 0.85–2.02 | 0.22 | 0 | 1.10 | 0.74–1.62 | 0.64 | 49.9 | 1.26 | 0.85–1.87 | 0.25 | 0 |
| GENOTYPING | |||||||||||||||||||||
| PCR-RFLP | 5 | 1.22 | 1.00–1.49 | 0.05 | 45.4 | 0.95 | 0.66–1.36 | 0.78 | 58.5 | 1.60 | 1.18–2.16 | < 0.01 | 3.8 | 1.10 | 0.80–1.54 | 0.52 | 57.8 | 1.63 | 1.24–2.15 | < 0.01 | 0 |
| Other | 2 | 1.17 | 0.94–1.46 | 0.16 | 0 | 1.61 | 1.12–2.32 | 0.01 | 0 | 1.30 | 0.85–2.00 | 0.22 | 0 | 1.50 | 1.07–2.10 | 0.02 | 0 | 0.98 | 0.68–1.41 | 0.89 | 0 |
| TaqI T/C (rs731236) | C vs. T | TC vs. TT | CC vs. TT | TC+ CC vs. TT | CC vs. TT+ TC | ||||||||||||||||
| Total | 9 | 1.16 | 0.94–1.44 | 0.18 | 66.5 | 1.00 | 0.75–1.34 | 0.99 | 58.5 | 1.44 | 0.97–2.15 | 0.07 | 54.2 | 1.10 | 0.82–1.48 | 0.52 | 65.4 | 1.37 | 1.09–1.74 | 0.01 | 38.3 |
| HWE-yes | 7 | 1.16 | 0.93–1.46 | 0.19 | 61.3 | 1.07 | 0.79–1.45 | 0.66 | 51.2 | 1.39 | 0.87–2.22 | 0.17 | 57.8 | 1.15 | 0.85–1.56 | 0.38 | 58.1 | 1.31 | 0.90–1.90 | 0.16 | 43.4 |
| HWE-no | 2 | 1.15 | 0.52–2.53 | 0.73 | 88.0 | 0.81 | 0.32–2.05 | 0.65 | 81.1 | 1.65 | 0.65–4.18 | 0.29 | 56.3 | 0.95 | 0.33–2.80 | 0.93 | 87.9 | 1.91 | 1.09–3.33 | 0.02 | 0 |
| ETHNICITY | |||||||||||||||||||||
| Caucasian | 6 | 1.20 | 0.89–1.60 | 0.23 | 67.2 | 1.13 | 0.79–1.63 | 0.50 | 54.2 | 1.46 | 0.79–2.71 | 0.23 | 64.8 | 1.20 | 0.82–1.76 | 0.34 | 62.8 | 1.33 | 0.81–2.17 | 0.26 | 52.5 |
| Asian | 3 | 1.11 | 0.74–1.67 | 0.60 | 76.4 | 0.82 | 0.51–1.32 | 0.41 | 63.0 | 1.45 | 0.99–2.12 | 0.06 | 36.9 | 0.95 | 0.55–1.65 | 0.86 | 75.9 | 1.55 | 1.08–2.22 | 0.02 | 0 |
| DESIGN | |||||||||||||||||||||
| Hospital-B | 4 | 1.03 | 0.74–1.44 | 0.85 | 74.5 | 0.83 | 0.59–1.17 | 0.29 | 48.0 | 1.23 | 0.66–2.27 | 0.52 | 63.5 | 0.91 | 0.60–1.38 | 0.66 | 68.0 | 1.33 | 0.79–2.22 | 0.28 | 54.0 |
| Healthy-B | 5 | 1.31 | 1.00–1.70 | 0.05 | 48.0 | 1.21 | 0.78–1.87 | 0.40 | 57.9 | 1.70 | 0.98–2.96 | 0.06 | 46.0 | 1.33 | 0.89–1.98 | 0.16 | 56.4 | 1.51 | 1.09–2.08 | 0.01 | 30.8 |
| GENOTYPING | |||||||||||||||||||||
| PCR-RFLP | 7 | 1.25 | 0.97–1.60 | 0.08 | 64.5 | 0.98 | 0.69–1.40 | 0.91 | 61.8 | 1.74 | 1.30–2.33 | < 0.01 | 27.3 | 1.14 | 0.79–1.63 | 0.50 | 68.5 | 1.68 | 1.28–2.22 | < 0.01 | 0 |
| Other | 2 | 0.89 | 0.71–1.13 | 0.34 | 38.9 | 1.09 | 0.55–2.17 | 0.81 | 71.8 | 0.73 | 0.44–1.19 | 0.20 | 0 | 1.01 | 0.55–1.86 | 0.98 | 68.1 | 0.75 | 0.48–1.18 | 0.22 | 0 |
Numbers of comparisons.
Hospital-B, Hospital-based control; Healthy-B, Healthy-based control; RT-PCR, Real-time PCR.
Figure 2.
Statistical analysis of the association between VDR Fok I C/T polymorphism and PCOS risk in the TT vs. CC model. (A) ORs and 95% CIs; (B) cumulative analysis; (C) sensitivity analysis; (D) publication bias.
BsmI A/G Locus and PCOS Risk
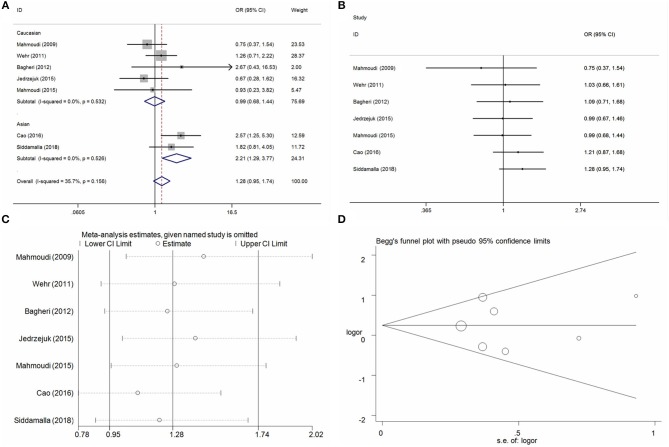
Seven case-control studies with 1,085 PCOS cases and 728 control individuals were identified on the association between BsmI A/G locus and PCOS risk. Overall, the pool analysis did not find any significant association between this locus on PCOS risk in five genetic models (G vs. A: OR = 1.17, 95%CI = 0.95–1.45, P = 0.14, I2 = 49.6%; AG vs. AA: OR = 1.15, 95%CI = 0.75–1.78, P = 0.52, I2 = 59.9%; GG vs. AA: OR = 1.28, 95%CI = 0.95–1.74, P = 0.11, I2 = 35.7%; AG+GG vs. AA: OR = 1.22, 95%CI = 0.82–1.81, P = 0.34, I2 = 57.1%; GG vs. AA+AG: OR = 1.16, 95%CI = 0.93–1.45, P = 0.18, I2 = 19.8%) (Table 3, Figure 3A for GG vs. AA model). Heterogeneity was observed in allele contrast, heterozygote model and dominant model. Meta-regression analyses were conducted, and the results indicated that the ethnicity diversity maybe the critical factors contributing to the existed heterogeneity (G vs. A: Pethnicity = 0.04; AG vs. AA: Pethnicity = 0.04; AG+GG vs. AA: Pethnicity = 0.03). In addition, the subgroup of ethnicity proved that the heterogeneity was alleviated in the Asian and Caucasian subgroups apparently. Furthermore, the subgroup analyses based on ethnicity presented an increased risk in Asian populations in some genetic models (G vs. A: OR = 1.62, 95%CI = 1.24–2.11, P < 0.01, I2 = 0%; AG vs. AA: OR = 2.08, 95%CI = 1.26–3.20, P < 0.01, I2 = 0%; GG vs. AA: OR = 2.21, 95%CI = 1.29–3.77, P < 0.01, I2 = 0%; AG+GG vs. AA: OR = 2.12, 95%CI = 1.42–3.16, P < 0.01, I2 = 0%). Cumulative analyses by publication date were conducted and indicated apparent consistence and stability of pool results (Figure 3B for GG vs. AA model). Sensitivity analysis was conducted and indicated some changes of results in allele contrast, homozygote, and recessive models without the publication by Jedrzejuk et al. (2015) (Figure 3C for GG vs. AA model). Publication bias was assessed using the Egger bias test and a Begg funnel plot test, and no significant asymmetrical evidence was found (T vs. C: P = 0.82; CT vs. CC: P = 0.17; TT vs. CC: P = 0.94; CT+TT vs. CC: P = 0.19; TT vs. CC+CT: P = 0.36) (Figure 3D for GG vs. AA model).
Figure 3.
Statistical analysis of the association between VDR BsmI A/G polymorphism and PCOS risk in the GG vs. AA model. (A) ORs and 95% CIs; (B) cumulative analysis; (C) sensitivity analysis; (D) publication bias.
ApaI A/C Locus and PCOS Risk
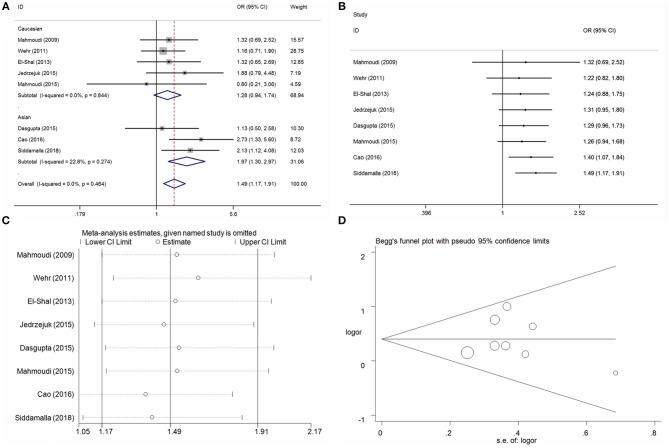
Eight case-control studies with 1,445 cases and 1,090 controls individuals were identified on the association between ApaI A/C locus and PCOS risk. Overall, significant association of increased risk were observed in three genetic models (C vs. A: OR = 1.20, 95%CI = 1.06–1.35, P = 0.01, I2 = 29.7%; CC vs. AA: OR = 1.49, 95%CI = 1.17–1.91, P < 0.01, I2 = 0%; CC vs. AA+AC: OR = 1.36, 95%CI = 1.09–1.69, P = 0.01, I2 = 12.8%) (Table 3, Figure 4A for CC vs. AA model). Heterogeneity was observed in heterozygote model (AC vs. AA) and dominant model (AC+CC vs. AA), and the meta-regression analyses did not find any distinct factors that contributed to the heterogeneity. Subgroup analyses by ethnicity presented an increased risk in Asian populations in the genetic models mentioned (C vs. A: OR = 1.22, 95%CI = 1.03–1.45, P = 0.02, I2 = 32.6%; CC vs. AA: OR = 1.61, 95%CI = 1.13–2.27, P < 0.01, I2 = 0%; CC vs. AA+AC: OR = 1.55, 95%CI = 1.15–2.10, P = 0.01, I2 = 0%). Moreover, the same significant PCOS risk was observed in some genetic models in these subgroups of HWE-yes, hospital control and genotyping groups (Table 3). Cumulative analyses demonstrated a significant alteration when the study of Cao and Tu (2016) was added in 2016 (Table 2, Figure 4B for CC vs. AA model). Sensitivity analysis was conducted in every genetic model and did not indicate some apparent changes except for the dominant modes (Figure 4C for CC vs. AA model). Publication bias was assessed using the Egger bias test and a Begg's funnel plot test, and no significant asymmetrical evidence was found (C vs. A: P = 0.74; AC vs. AA: P = 0.55; CC vs. AA: P = 0.97; AC+CC vs. AA: P = 0.86; CC vs. AA+AC: P = 0.37) (Figure 4D for CC vs. AA model).
Figure 4.
Statistical analysis of the association between VDR ApaI A/C polymorphism and PCOS risk in the CC vs. AA model. (A) ORs and 95% CIs; (B) cumulative analysis; (C) sensitivity analysis; (D) publication bias.
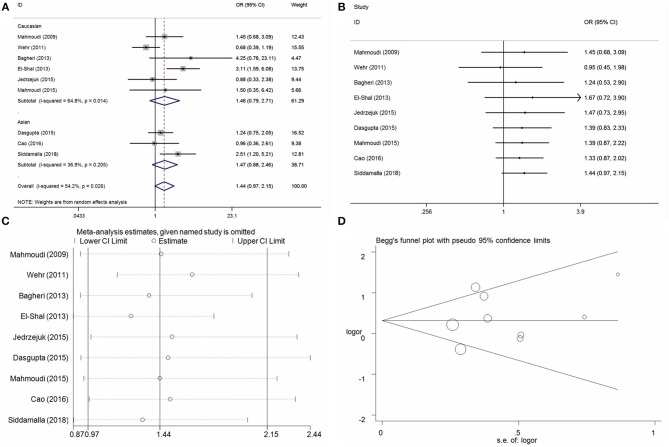
TaqI T/C Locus and PCOS Risk
Nine case-control studies with 1,476 cases and 1,120 controls individuals were identified on the association between TaqI T/C locus and PCOS risk. Overall, the increased risk was observed only in the recessive model (CC vs. TT+TC: OR = 1.37, 95%CI = 1.09-1.74, P = 0.01, I2 = 38.3%) (Table 3, Figure 5A for CC vs. TT model). Heterogeneities were identified in the allele contrast (C vs. T), heterozygote model (TC vs. TT), homozygote (CC vs. TT), and dominant model (TC+CC vs. TT). Meta-regression analyses only found that the genotyping methods contributed to the existing heterogeneity in the dominant model, but not in other models. Subgroup analyses revealed an increased PCOS risks in Asian populations (CC vs. TT+TC: OR = 1.55, 95%CI = 1.08-2.22, P = 0.02, I2 = 0%) and other subgroups in the recessive model (Table 3). Cumulative analyses by publication date demonstrated a negative association except for the recessive model (Figure 5B for CC vs. TT model). Sensitivity analysis indicated some slight alterations when the studies of Wehr et al. (2011) and El-Shal et al. (2013) were deleted in the homozygote and recessive models, respectively (Figure 5C for CC vs. TT model). Publication bias was assessed using the Egger bias test and a Begg funnel plot test, and no significant asymmetrical evidence was found (C vs. T: P = 0.48; TC vs. TT: P = 0.74; CC vs. TT: P = 0.49; TC+CC vs. TT: P = 0.62; CC vs. TT+TC: P = 0.38) (Figure 5D for CC vs. TT model).
Figure 5.
Statistical analysis of the association between VDR TaqI T/C polymorphism and PCOS risk in the CC vs. TT model. (A) ORs and 95% CIs; (B) cumulative analysis; (C) sensitivity analysis; (D) publication bias.
Discussion
To date, the pathogenesis and etiology of PCOS have remained unknown. The complex gene-gene and gene-environment interactions have been reported to be an important risk for PCOS development. Consistent epidemiologic evidence demonstrated that PCOS always suffered a series of complications, comprising hyperandrogenism, oligo-anovulation, insulin resistance and associated metabolic abnormalities.
Many studies proved that a dysregulated vitamin D serum level is closely related to PCOS occurrence. In addition, the vitamin D supplementation therapy would decreases fasting plasma glucose, serum insulin concentrations, and homeostasis model assessment-estimated insulin resistance (HOMA-IR) (Asemi et al., 2015; Foroozanfard et al., 2017). All these evidences suggested that vitamin D disorder is associated with multiple metabolic risks in women with PCOS.
Noteworthily, the 1α,25(OH)2D3 is an important active form of vitamin D, it is mediated by the vitamin D receptor [1α,25(OH)2D3 receptor, VDR] (Yoshizawa et al., 1997). Vitamin D receptor (VDR) is a DNA-binding transcription factor, combined with a heterodimer of the 1a,25(OH)2D3-ligand VDR and unoccupied RXR to generate an active signal transduction complex (Haussler et al., 2011). To date, several functional SNP loci reported in these polymorphisms presented an increased susceptibility of various diseases (Valdivielso and Fernandez, 2006), such as multiple cancers (Vidigal et al., 2017), diabetes mellitus (Yu et al., 2017), rheumatoid arthritis (Tizaoui et al., 2014), and cardiocerebrovascular disease (Moradi et al., 2017).
In 2009, Mahmoudi et al. published the first case-control study to explore the association between the above four polymorphisms and PCOS susceptibility in the Iranian population, and the results suggested that these individuals with an CC genotype have an increased risk for PCOS compared with the AA genotype. Since then, a series of case-control studies was conducted to evaluate the association between the vitamin D polymorphisms and PCOS susceptibility, but some controversies arose and bewildered us completely. In 2017, Reis et al. published a system review on vitamin D polymorphisms and PCOS with most literature on this theme (Reis et al., 2017). However, the synthesis and calculation of all selected data were not conducted. We also made a comprehensive understanding of all the studies but failed to draw a clear conclusion. Thus, we conducted the meta-analysis to investigate the precise relationships between VDR Fok I C/T, BsmI A/G, ApaI A/C, and TaqI T/C polymorphisms and PCOS risk based on 10 published case-control studies.
In the current meta-analysis, the pooled results indicated some significant association between ApaI A/C polymorphism and PCOS susceptibility in allele contrast, homozygote genotype and recessive models, presenting 1.20-, 1.49- and 1.36-fold high risk for PCOS. Furthermore, an increased PCOS risk was observed in the subgroup analysis of the HWE-yes group and hospital based group, especially in the Asian group. In BsmI A/G polymorphism, only some increased PCOS risks were observed in the Asians based on ethnic diversity. These pieces of evidence demonstrated that the ethnicity differences may play an important role, contributing to the varying PCOS susceptibility among the Asian and Caucasian races. In addition, no signification association was observed in TaqI T/C and Fok I C/T polymorphisms for PCOS risk, except for a few scattered cases of increased PCOS risk in the former in the recessive models.
The restriction fragment length polymorphism sties of BsmI and ApaI are located in the intron (between exons 8 and 9), and the TaqI polymorphism was located in exon 9 (Zmuda et al., 2000). They are all located near the 3'-untranslated region of the VDR gene, which was suggested to be involved in the regulation of gene expression by modulating mRNA stability (Zmuda et al., 2000; Ogunkolade et al., 2002). In this meta-analysis, some increased and significant risks were observed in the above three polymorphism, indicating that the potential synergism among these polymorphisms would play an important role for PCOS occurrence. Regrettably, this hypothesis could not be verified without valid haplotype data to assess interaction between the adjacent polymorphism loci with all included studies. FokI polymorphism located in exon 2, resulting in a incorporation VDR protein production in the NH2 terminal, which was suggested to influence the transcriptional activity of VDR gene combined with the modulation of transcription factor IIB (Jurutka et al., 2000; Whitfield et al., 2001). Some publications indicated that this polymorphism would regulate the expression of mRNA and contribute to susceptibility to various diseases (Arai et al., 1997; Colombini et al., 2014), but no significant association between FokI polymorphism and PCOS was found based on the current meta-analysis. So, all these evidences indicated that there were a causation between the mutation of the above SNPs located in VDR gene and PCOS occurrence (Hill, 1965).
To our knowledge, this is the first meta-analysis to assess the association between VDR polymorphisms and PCOS risk. Some advantages were presented in this meta-analysis compared with the published case-control studies: First, all case-control studies published on the four polymorphisms were considered, and the risk assessment of bias within studies would enhance the statistical power and help understand the association between VDR polymorphisms and PCOS risk. Second, a stratified analysis based on ethnic diversity, control design, and genotyping methods was conducted to explore the potential relationships that were modulated under these subgroup biologic factors. Third, a scientific retrieval strategy and rigorous methodology were used, including cumulative analyses and sensitivity analyses. Publication bias was also used to guarantee the stability and credibility of the conclusions of the analysis.
However, there were some limitations of this study, which should be pointed out. First, only 10 publications were included in this present meta-analysis. The studies and sample size of each polymorphic locus were limited, and the pooled results and subgroup analysis could not reveal the reality association between VDR polymorphisms and PCOS susceptibility. Second, interactive risk factors, such as living habits, diet, age, and family history were not adjusted in this meta-analysis due to data deficiency. Third, included studies were written only in English and Chinese, and the included subjects were mostly Asian and Caucasian populations. Therefore, the results of this meta-analysis cannot represent all ethnic populations, and the application of the conclusions was restricted. Fourth, all examined polymorphisms were assessed separately, and the gene-gene interactions especially the haplotype analyses were not assessed due to the insufficient data.
In conclusion, this meta-analysis suggests that VDR gene polymorphisms play an important role in PCOS development, especially on the ApaI A/C and BsmI A/G among the Asian populations. Further case-control studies on various ethnic populations with a larger sample size are need to verify the current conclusions in the future.
Author Contributions
Y-MN, Y-DW, and Y-YH conceived the study. Y-MN and Y-DW searched the databases and extracted the data. X-FL, G-BJ, and GB analyzed the data. Y-MN, Y-YH, and H-BC wrote the draft of the paper. Y-DW and MS reviewed the manuscript. All the authors approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This study was supported by the National Natural Science Foundation of China (81400535), the Foundations of the Natural Science Foundation of Jiangsu Province (BK20140912) and Hubei Province (No. 2016CFB567), the Hubei Province health and family planning scientific research project (No. WJ2017F069, WJ2015Q041) and Taihe Hospital (2016BSQD02).
References
- Arai H., Miyamoto K., Taketani Y., Yamamoto H., Iemori Y., Morita K., et al. (1997). A vitamin D receptor gene polymorphism in the translation initiation codon: effect on protein activity and relation to bone mineral density in Japanese women. J. Bone Miner. Res. 12, 915–921. 10.1359/jbmr.1997.12.6.915 [DOI] [PubMed] [Google Scholar]
- Asemi Z., Foroozanfard F., Hashemi T., Bahmani F., Jamilian M., Esmaillzadeh A. (2015). Calcium plus vitamin D supplementation affects glucose metabolism and lipid concentrations in overweight and obese vitamin D deficient women with polycystic ovary syndrome. Clin. Nutr. 34, 586–592. 10.1016/j.clnu.2014.09.015 [DOI] [PubMed] [Google Scholar]
- Asuncion M., Calvo R. M., San Millan J. L., Sancho J., Avila S., Escobar-Morreale H. F. (2000). A prospective study of the prevalence of the polycystic ovary syndrome in unselected Caucasian women from Spain. J. Clin. Endocrinol. Metab. 85, 2434–2438. 10.1210/jc.85.7.2434 [DOI] [PubMed] [Google Scholar]
- Azziz R., Woods K. S., Reyna R., Key T. J., Knochenhauer E. S., Yildiz B. O. (2004). The prevalence and features of the polycystic ovary syndrome in an unselected population. J. Clin. Endocrinol. Metab. 89, 2745–2749. 10.1210/jc.2003-032046 [DOI] [PubMed] [Google Scholar]
- Bacopoulou F., Kolias E., Efthymiou V., Antonopoulos C. N., Charmandari E. (2017). Vitamin D predictors in polycystic ovary syndrome: a meta-analysis. Eur. J. Clin. Invest. 47, 746–755. 10.1111/eci.12800 [DOI] [PubMed] [Google Scholar]
- Bagheri M., Abdi Rad I., Hosseini Jazani N., Nanbakhsh F. (2013). Vitamin D receptor TaqI gene variant in exon 9 and polycystic ovary syndrome risk. Int. J. Fertil. Steril. 7, 116–121. [PMC free article] [PubMed] [Google Scholar]
- Bagheri M., Rad I. A., Jazani N. H., Nanbakhsh F. (2012). Lack of association of vitamin D receptor FokI (rs10735810) (C/T) and BsmI (rs1544410) (A/G) genetic variations with polycystic ovary syndrome risk: a case-control study from Iranian Azeri Turkish women. Maedica 7, 303–308. [PMC free article] [PubMed] [Google Scholar]
- Baker A. R., McDonnell D. P., Hughes M., Crisp T. M., Mangelsdorf D. J., Haussler M. R., et al. (1988). Cloning and expression of full-length cDNA encoding human vitamin D receptor. Proc. Natl. Acad. Sci. U. S. A. 85, 3294–3298. 10.1073/pnas.85.10.3294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begg C. B., Mazumdar M. (1994). Operating characteristics of a rank correlation test for publication bias. Biometrics 50, 1088–1101. 10.2307/2533446 [DOI] [PubMed] [Google Scholar]
- Cao H. B., Tu L. (2016). Association between Vitamin D receptor gene polymorphism and polycystic ovary syndrome (Chinese). Pract. Clin. Med. 17, 40–42. 10.13764/j.cnki.lcsy.2016.02.017 [DOI] [Google Scholar]
- Colombini A., Brayda-Bruno M., Lombardi G., Croiset S. J., Vrech V., Maione V., et al. (2014). FokI polymorphism in the vitamin D receptor gene (VDR) and its association with lumbar spine pathologies in the Italian population: a case-control study. PLoS ONE 9:e97027. 10.1371/journal.pone.0097027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta S., Dutta J., Annamaneni S., Kudugunti N., Battini M. R. (2015). Association of vitamin D receptor gene polymorphisms with polycystic ovary syndrome among Indian women. Indian J. Med. Res. 142, 276–285. 10.4103/0971-5916.166587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DerSimonian R. (1996). Meta-analysis in the design and monitoring of clinical trials. Stat. Med. 15, 1237–1248. [DOI] [PubMed] [Google Scholar]
- Egger M., Davey Smith G., Schneider M., Minder C. (1997). Bias in meta-analysis detected by a simple, graphical test. BMJ 315, 629–634. 10.1136/bmj.315.7109.629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Shal A. S., Shalaby S. M., Aly N. M., Rashad N. M., Abdelaziz A. M. (2013). Genetic variation in the vitamin D receptor gene and vitamin D serum levels in Egyptian women with polycystic ovary syndrome. Mol. Biol. Rep. 40, 6063–6073. 10.1007/s11033-013-2716-y [DOI] [PubMed] [Google Scholar]
- Foroozanfard F., Talebi M., Samimi M., Mehrabi S., Badehnoosh B., Jamilian M., et al. (2017). Effect of two different doses of vitamin D supplementation on metabolic profiles of insulin-resistant patients with polycystic ovary syndrome: a randomized, double-blind, placebo-controlled trial. Horm. Metab. Res. 49, 612–617. 10.1055/s-0043-112346 [DOI] [PubMed] [Google Scholar]
- Haussler M. R., Jurutka P. W., Mizwicki M., Norman A. W. (2011). Vitamin D receptor (VDR)-mediated actions of 1alpha,25(OH)(2)vitamin D(3): genomic and non-genomic mechanisms. Best Pract. Res. Clin. Endocrinol. Metab. 25, 543–559. 10.1016/j.beem.2011.05.010 [DOI] [PubMed] [Google Scholar]
- Hill A. B. (1965). The environment and disease: association or causation? Proc. R. Soc. Med. 58, 295–300. [PMC free article] [PubMed] [Google Scholar]
- Huedo-Medina T. B., Sanchez-Meca J., Marin-Martinez F., Botella J. (2006). Assessing heterogeneity in meta-analysis: Q statistic or I2 index? Psychol. Methods 11, 193–206. 10.1037/1082-989X.11.2.193 [DOI] [PubMed] [Google Scholar]
- Jedrzejuk D., Laczmanski L., Milewicz A., Kuliczkowska-Plaksej J., Lenarcik-Kabza A., Hirnle L., et al. (2015). Classic PCOS phenotype is not associated with deficiency of endogenous vitamin D and VDR gene polymorphisms rs731236 (TaqI), rs7975232 (ApaI), rs1544410 (BsmI), rs10735810 (FokI): a case-control study of lower Silesian women. Gynecol. Endocrinol. 31, 976–979. 10.3109/09513590.2015.1062865 [DOI] [PubMed] [Google Scholar]
- Jurutka P. W., Remus L. S., Whitfield G. K., Thompson P. D., Hsieh J. C., Zitzer H., et al. (2000). The polymorphic N terminus in human vitamin D receptor isoforms influences transcriptional activity by modulating interaction with transcription factor IIB. Mol. Endocrinol. 14, 401–420. 10.1210/mend.14.3.0435 [DOI] [PubMed] [Google Scholar]
- Kato S. (2000). The function of vitamin D receptor in vitamin D action. J. Biochem. 127, 717–722. 10.1093/oxfordjournals.jbchem.a022662 [DOI] [PubMed] [Google Scholar]
- Knochenhauer E. S., Key T. J., Kahsar-Miller M., Waggoner W., Boots L. R., Azziz R. (1998). Prevalence of the polycystic ovary syndrome in unselected black and white women of the southeastern United States: a prospective study. J. Clin. Endocrinol. Metab. 83, 3078–3082. 10.1210/jc.83.9.3078 [DOI] [PubMed] [Google Scholar]
- Krul-Poel Y. H., Snackey C., Louwers Y., Lips P., Lambalk C. B., Laven J. S., et al. (2013). The role of vitamin D in metabolic disturbances in polycystic ovary syndrome: a systematic review. Eur. J. Endocrinol. 169, 853–865. 10.1530/EJE-13-0617 [DOI] [PubMed] [Google Scholar]
- Lewis C. M., Knight J. (2012). Introduction to genetic association studies. Cold Spring Harb. Protoc. 2012, 297–306. 10.1101/pdb.top068163 [DOI] [PubMed] [Google Scholar]
- Mahmoudi T. (2009). Genetic variation in the vitamin D receptor and polycystic ovary syndrome risk. Fertil. Steril. 92, 1381–1383. 10.1016/j.fertnstert.2009.05.002 [DOI] [PubMed] [Google Scholar]
- Mahmoudi T., Majidzadeh A. K., Farahani H., Mirakhorli M., Dabiri R., Nobakht H., et al. (2015). Association of vitamin D receptor gene variants with polycystic ovary syndrome: a case control study. Int. J. Reprod. Biomed. 13, 793–800. 10.29252/ijrm.13.12.793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantel N., Haenszel W. (1959). Statistical aspects of the analysis of data from retrospective studies of disease. J. Natl. Cancer Inst. 22, 719–748. [PubMed] [Google Scholar]
- Minelli C., Thompson J. R., Abrams K. R., Thakkinstian A., Attia J. (2005). The choice of a genetic model in the meta-analysis of molecular association studies. Int. J. Epidemiol. 34, 1319–1328. 10.1093/ije/dyi169 [DOI] [PubMed] [Google Scholar]
- Moher D., Liberati A., Tetzlaff J., Altman D. G. (2009). Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. J. Clin. Epidemiol. 62, 1006–1012. 10.1016/j.jclinepi.2009.06.005 [DOI] [PubMed] [Google Scholar]
- Moradi N., Fadaei R., Ahmadi R., Mohammad M. H., Shahmohamadnejad S., Tavakoli-Yaraki M., et al. (2017). Role of serum MMP-9 levels and vitamin D receptor polymorphisms in the susceptibility to coronary artery disease: an association study in Iranian population. Gene 628, 295–300. 10.1016/j.gene.2017.07.060 [DOI] [PubMed] [Google Scholar]
- Niu Y. M., Weng H., Zhang C., Yuan R. X., Yan J. Z., Meng X. Y., et al. (2015). Systematic review by multivariate meta-analyses on the possible role of tumor necrosis factor-alpha gene polymorphisms in association with ischemic stroke. Neuromol. Med. 17, 373–384. 10.1007/s12017-015-8365-7 [DOI] [PubMed] [Google Scholar]
- Ogunkolade B. W., Boucher B. J., Prahl J. M., Bustin S. A., Burrin J. M., Noonan K., et al. (2002). Vitamin D receptor (VDR) mRNA and VDR protein levels in relation to vitamin D status, insulin secretory capacity, and VDR genotype in Bangladeshi Asians. Diabetes 51, 2294–2300. 10.2337/diabetes.51.7.2294 [DOI] [PubMed] [Google Scholar]
- Ollila M. E., West S., Keinanen-Kiukaanniemi S., Jokelainen J., Auvinen J., Puukka K., et al. (2017). Overweight and obese but not normal weight women with PCOS are at increased risk of Type 2 diabetes mellitus-a prospective, population-based cohort study. Hum. Reprod. 32, 423–431. 10.1093/humrep/dew329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis G. V., Gontijo N. A., Rodrigues K. F., Alves M. T., Ferreira C. N., Gomes K. B. (2017). Vitamin D receptor polymorphisms and the polycystic ovary syndrome: a systematic review. J. Obstet. Gynaecol. Res. 43, 436–446. 10.1111/jog.13250 [DOI] [PubMed] [Google Scholar]
- Repaci A., Gambineri A., Pasquali R. (2011). The role of low-grade inflammation in the polycystic ovary syndrome. Mol. Cell. Endocrinol. 335, 30–41. 10.1016/j.mce.2010.08.002 [DOI] [PubMed] [Google Scholar]
- Shen M., Luo Y., Niu Y., Chen L., Yuan X., Goltzman D., et al. (2013). 1,25(OH)2D deficiency induces temporomandibular joint osteoarthritis via secretion of senescence-associated inflammatory cytokines. Bone 55, 400–409. 10.1016/j.bone.2013.04.015 [DOI] [PubMed] [Google Scholar]
- Siddamalla S., Reddy T. V., Govatati S., Erram N., Deenadayal M., Shivaji S., et al. (2018). Vitamin D receptor gene polymorphisms and risk of polycystic ovary syndrome in South Indian women. Gynecol. Endocrinol. 34, 161–165. 10.1080/09513590.2017.1371128 [DOI] [PubMed] [Google Scholar]
- Sirmans S. M., Pate K. A. (2013). Epidemiology, diagnosis, and management of polycystic ovary syndrome. Clin. Epidemiol. 6, 1–13. 10.2147/CLEP.S37559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tizaoui K., Kaabachi W., Ouled Salah M., Ben Amor A., Hamzaoui A., Hamzaoui K. (2014). Vitamin D receptor TaqI and ApaI polymorphisms: a comparative study in patients with Behcet's disease and Rheumatoid arthritis in Tunisian population. Cell. Immunol. 290, 66–71. 10.1016/j.cellimm.2014.05.002 [DOI] [PubMed] [Google Scholar]
- Valdivielso J. M., Fernandez E. (2006). Vitamin D receptor polymorphisms and diseases. Clin. Chim. Acta 371, 1–12. 10.1016/j.cca.2006.02.016 [DOI] [PubMed] [Google Scholar]
- Vidigal V. M., Silva T. D., de Oliveira J., Pimenta C. A. M., Felipe A. V., Forones N. M. (2017). Genetic polymorphisms of vitamin D receptor (VDR), CYP27B1 and CYP24A1 genes and the risk of colorectal cancer. Int. J. Biol. Markers 32, e224–e230. 10.5301/jbm.5000248 [DOI] [PubMed] [Google Scholar]
- Wehr E., Pilz S., Schweighofer N., Giuliani A., Kopera D., Pieber T. R., et al. (2009). Association of hypovitaminosis D with metabolic disturbances in polycystic ovary syndrome. Eur. J. Endocrinol. 161, 575–582. 10.1530/EJE-09-0432 [DOI] [PubMed] [Google Scholar]
- Wehr E., Trummer O., Giuliani A., Gruber H. J., Pieber T. R., Obermayer-Pietsch B. (2011). Vitamin D-associated polymorphisms are related to insulin resistance and vitamin D deficiency in polycystic ovary syndrome. Eur. J. Endocrinol. 164, 741–749. 10.1530/EJE-11-0134 [DOI] [PubMed] [Google Scholar]
- Whitfield G. K., Remus L. S., Jurutka P. W., Zitzer H., Oza A. K., Dang H. T., et al. (2001). Functionally relevant polymorphisms in the human nuclear vitamin D receptor gene. Mol. Cell. Endocrinol. 177, 145–159. 10.1016/S0303-7207(01)00406-3 [DOI] [PubMed] [Google Scholar]
- Yoshizawa T., Handa Y., Uematsu Y., Takeda S., Sekine K., Yoshihara Y., et al. (1997). Mice lacking the vitamin D receptor exhibit impaired bone formation, uterine hypoplasia and growth retardation after weaning. Nat. Genet. 16, 391–396. 10.1038/ng0897-391 [DOI] [PubMed] [Google Scholar]
- Yu F., Wang C., Wang L., Jiang H., Ba Y., Cui L., et al. (2017). Study and evaluation the impact of vitamin D receptor variants on the risk of type 2 diabetes mellitus in Han Chinese. J. Diabetes 9, 275–284. 10.1111/1753-0407.12413 [DOI] [PubMed] [Google Scholar]
- Zmuda J. M., Cauley J. A., Ferrell R. E. (2000). Molecular epidemiology of vitamin D receptor gene variants. Epidemiol. Rev. 22, 203–217. 10.1093/oxfordjournals.epirev.a018033 [DOI] [PubMed] [Google Scholar]