Abstract
Bacterial growth and cell division requires precise spatiotemporal regulation of the synthesis and remodelling of the peptidoglycan layer that surrounds the cytoplasmic membrane. GpsB is a cytosolic protein that affects cell wall synthesis by binding cytoplasmic mini-domains of peptidoglycan synthases to ensure their correct subcellular localisation. Here, we describe critical structural features for the interaction of GpsB with peptidoglycan synthases from three bacterial species (Bacillus subtilis, Listeria monocytogenes and Streptococcus pneumoniae) and suggest their importance for cell wall growth and viability in L. monocytogenes and S. pneumoniae. We use these structural motifs to identify novel partners of GpsB in B. subtilis and extend the members of the GpsB interactome in all three bacterial species. Our results support that GpsB functions as an adaptor protein that mediates the interaction between membrane proteins, scaffolding proteins, signalling proteins and enzymes to generate larger protein complexes at specific sites in a bacterial cell cycle-dependent manner.
GpsB is a cytosolic protein that modulates bacterial cell wall synthesis by interacting with cytoplasmic domains of peptidoglycan synthases. Here, Cleverley et al. describe structural features that are important for these interactions, and identify new interacting partners of GpsB in three bacterial species.
Introduction
Peptidoglycan (PG), a network of glycan strands connected by short peptides, forms the essential cell envelope that maintains cell shape and protects bacteria from osmotic stresses1. High molecular weight (HMW) bi-functional penicillin binding proteins (class A PBPs) are PG synthases that catalyse glycan strand polymerisation and peptide crosslinking, whereas HMW class B mono-functional PBPs only have transpeptidase functions2. The PG layer needs remodelling to enable normal cell growth and division and thus the bacterial cell cycle requires the extracellular activities of PBPs3 and PG hydrolases4 to be co-ordinated. The outer membrane-anchored LpoA/B lipoproteins activate their cognate PBP1A/1B PG synthases in the synthesis of the thin, periplasmic PG layer in the Gram-negative paradigm Escherichia coli5,6. By contrast, Gram-positive bacteria have a much thicker PG layer that is complemented with other anionic cell wall polymers. PG synthesis regulation in Gram-positive bacteria involves protein phosphorylation by orthologues of the serine/threonine kinase PknB7/StkP8, and dedicated cell cycle scaffolding proteins including DivIVA9, EzrA10 and GpsB11,12. However, the molecular mechanisms that modulate PG synthesis in Gram-positive bacteria are virtually unknown.
GpsB has emerged as a major regulator of PG biosynthesis in low G+C Gram-positive bacteria, and its homologues (DivIVA/Wag31/antigen 84) in Actinobacteria play essential roles in hyphal growth and branching13–15. GpsB was initially characterised in Bacillus subtilis where severe cell division and growth defects were observed when both gpsB and ezrA11 or gpsB and ftsA12 were deleted. Both EzrA and FtsA play roles in the dynamics and membrane anchoring of the FtsZ Z-ring, the constriction of which is fundamental to cell division16. The Z-ring also recruits downstream proteins, including PBPs17,18, to complete the process. Deletion of gpsB alone in Listeria monocytogenes caused marked growth and division defects at 37 °C and was lethal at 42 °C19. Moreover, gpsB deletion in L. monocytogenes also resulted in enhanced susceptibility to β-lactam19 and fosfomycin20 antibiotics, reduced virulence in an insect infection model19, and caused alterations to PG structure21. Mutations in gpsB that affected binding to the PG synthase PBPA1 also showed a lethal phenotype in L. monocytogenes at 42 °C19. The gpsB gene is essential in the Streptococcus pneumoniae D39 progenitor strain as well as in some of its laboratory derivatives and its inactivation resulted in elongated cells unable to divide22–24. In addition, a genome-wide association study of S. pneumoniae clinical isolates revealed that the presence of gpsB variants was correlated significantly to β-lactam resistance25, suggesting that GpsB may have fitness and pleiotropic roles in maintaining cell wall integrity during antibiotic stress. The gpsB gene has also been described as essential in the spherical bacterium Staphylococcus aureus, but the biochemical properties and physiological functions of S. aureus GpsB in this recent report26 differ to what has been described in B. subtilis11,12,18,19,27 L. monocytogenes19,27 and S. pneumoniae22–24.
In both B. subtilis11 and L. monocytogenes19, the cytosolic GpsB localises to the lateral side walls of newborn, growing cells and to the septum of dividing cells, the same localisation pattern as that of B. subtilis PBP111. In S. pneumoniae, GpsB localises to mid-cell22, the only region of active PG synthesis for both peripheral (side-wall) elongation and cell division in this bacterium. The localisation of GpsB at regions of active PG synthesis allows for the interaction of GpsB with the poorly characterised cytoplasmic mini-domains of PG synthases11,19,27,28. S. pneumoniae GpsB (SpGpsB) has been found to co-immunoprecipitate with SpPBP2a, SpPBP2b and SpMreC24, suggesting these proteins form a complex that is regulated by SpGpsB24.
To gain molecular understanding of GpsB function, we report three crystal structures of PBP cytoplasmic mini-domains in complex with GpsB, the first structures of a PG synthase in complex with a cytoplasmic cell cycle regulator. Despite a marked absence of sequence and structural homology, we find that the PBP domains interact with equivalent surfaces in GpsB using an arginine that is conserved in the respective orthologues of the PBPs. The visualisation of each complex permits a comprehensive mutagenesis strategy and functional study to rationalise the role of each interfacial amino acid in the PBP:GpsB pairs. We uncover a sequence motif used by the B. subtilis PG synthase to interact with GpsB. We identify two new members of the GpsB interactome in this organism by querying the B. subtilis proteome with this motif for potential new partners of GpsB, and provide evidence for their connection to other, established proteins in growth and division. We also identify extensive GpsB interactomes in B. subtilis, L. monocytogenes and S. pneumoniae by bacterial two-hybrid assays (BACTH). Therefore, the role of GpsB in the bacterial cell cycle is that of an adaptor29–31, docking PG synthases to other cell wall enzymes, scaffolds and shape determinants into protein complexes for division (the divisome) and peripheral growth (the elongasome).
Results
The first 16 residues of BsPBP1 dictate the BsGpsB interaction
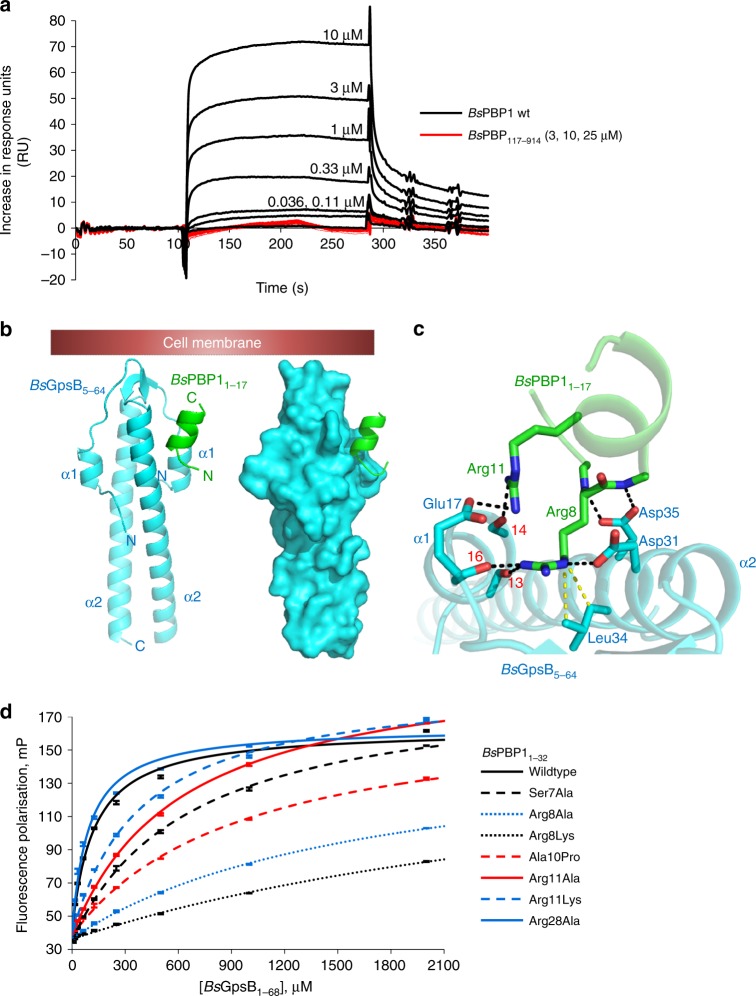
GpsB is an influential cell cycle regulator in low G+C Gram-positive bacteria and we set out to establish the common rules by which GpsB interacts with major PG synthases in three important bacteria—one model species (B. subtilis) and two pathogens (L. monocytogenes and S. pneumoniae). It has been determined previously by us by SPR that the first 31 amino acids of the cytoplasmic mini-domain of B. subtilis PBP1 (BsPBP1) were critical for binding of BsGpsB19. Using the same technique the binding site was further mapped to the first 16 amino acids of BsPBP1 by comparing the interaction of BsGpsB with SPR chips coated with full-length BsPBP1 and a PBP1 deletion mutant, BsPBP117-914, where the codons for the first 16 amino acids were removed genetically. While submicromolar concentrations of BsGpsB bound to chips coated with the wild-type BsPBP1, there was no interaction even when 25 µM BsGpsB was injected over equivalent chips coated with BsPBP117-914 (Fig. 1a).
Fig. 1.
BsGpsB:BsPBP1 interactions require conserved arginines in the α-helical cytoplasmic minidomain of BsPBP1. a BsGpsB interacts with the first 16 amino acids of BsPBP1. SPR sensorgrams of full-length BsGpsB against immobilised full-length BsPBP1 (black) and BsPBP117-914 (red). BsGpsB does not interact with the BsPBP117-914 coated chip, even when 25 μM GpsB is injected. b Cartoon (left) and surface representation (right) of the crystal structure of the BsGpsB5-64:BsPBP11-17 complex. BsGpsB5-64 is coloured cyan and BsPBP11-17 is coloured green and the likely plane of the bacterial membrane is shown as a red box. c The BsGpsB5-64:BsPBP11-17 complex is dependent upon a conserved SRxxR(R/K) motif in BsPBP1. Key interfacial residues in the BsGpsB5-64:BsPBP11-17 complex are shown as sticks and coloured (and labelled) blue and green, respectively. The carbonyl oxygens of BsGpsBIle13, BsGpsBLeu14 and BsGpsBLys16 are labelled with their respective red numerals. Hydrogen bonds are shown as black dashed lines and the van der Waals’ interactions between BsGpsBLeu34 and BsPBP1Arg8 are in yellow. d Mutation of key BsPBP1 interfacial residues in the structure of BsGpsB5-64:BsPBP11-17 leads to a loss of binding of position 16 TAMRA-labelled BsPBP11-32 variants to BsGpsB1-68 as measured by fluorescence polarisation. When the BsPBP11-32 peptide was labelled with fluorescein at position 31 the measured affinity was the same as if the fluorophore was at position 16, indicating that the fluorophore itself or its position in the peptide has no impact on the binding interaction. The calculated dissociation constants can be found in Supplementary Table 1
We subsequently solved the crystal structure of the complex between the N-terminal domain of BsGpsB whose termini were truncated slightly to expedite crystallisation, BsGpsB5-64, with the first 17 amino acids of BsPBP1, BsPBP11-17 (Fig. 1b, c). The BsPBP11-17 α-helix is stabilised by an intramolecular salt bridge between BsPBP11-17Glu9 and BsPBP11-17Arg12 and by a hydrogen bond between the sidechain of BsPBP11-17Ser7 and the backbone amide of BsPBP11-17Ala10. A prominent feature of the complex is the deep penetration of the sidechain of BsPBP1Arg8 into the groove between BsGpsB5-64 α-helices 1 and 2, contacting the mainchain carbonyl oxygens of BsGpsBIle13, BsGpsBLeu14 and BsGpsBLys16 and forming a salt bridge with BsGpsBAsp31 (Fig. 1c), which in turn is tethered in place by hydrogen bonds to the hydroxyl of BsGpsBTyr25. The backbone amides of BsPBP1Arg8 and BsPBP1Glu9 interact with BsGpsBAsp35, mimicking the mainchain interactions in successive turns in an α-helix. In a longer α-helix, the backbone amides of BsPBP1Arg8 and BsPBP1Glu9 would not be available to interact with BsGpsBAsp35 because of intra-helical hydrogen bonds with the mainchain carbonyls of BsPBP1Phe5 and BsPBP1Asn6. The sidechain of BsPBP1Arg11 forms hydrogen bonds with the carbonyl oxygen of BsGpsBLeu14 and a salt bridge with BsGpsBGlu17. Van der Waals’ interactions connect BsPBP1Arg8 to BsGpsBLeu34 (Fig. 1c) and BsPBP1Glu9 with BsGpsBLys32. The BsPBP11-17 peptide binds to a groove between α-helices 1 and 2 in only one molecule of BsGpsB5-64 in the crystallographic asymmetric unit as the second BsGpsB-binding site is blocked by crystal contacts. The key interactions are mapped onto GpsB and PBP1 sequences in Supplementary Figure 1A.
To rationalise the features of PBP cytoplasmic domains important for determining recognition by GpsB, the interactions described above were analysed further by fluorescence polarisation (FP) and circular dichroism (CD). The BsGpsB1-68Glu17Ala, BsGpsBTyr25Phe, BsGpsB1-68Asp31Ala and BsGpsB1-68Asp35Ala mutations had little impact on protein stability (Supplementary Figure 1B and Supplementary Note 1) and each reduced the affinity for BsPBP11-32 by more than 8-fold (Supplementary Figure 1C, Supplementary Table 1). BsPBP1Arg8Lys, BsPBP1Arg8Ala and BsPBP1Arg11Ala mutations each resulted in reduced affinities for BsGpsB1-68 by at least 5-fold (Fig. 1d, Supplementary Table 1). BsPBP11-32Arg28Ala had no effect on binding (Fig. 1d, Supplementary Table 1), suggesting that the overall positive charge of the peptide is not the primary determinant of BsGpsB:BsPBP1 interactions, rather the unique physicochemical characteristics of an arginine at position 8 in BsPBP1 is essential. The BsPBP11-32Ser7Ala and BsPBP11-32Ala10Pro mutations each reduced the affinity for BsGpsB1-68 by at least 6-fold by affecting the α-helix of BsPBP11-32 (Fig. 1d, Supplementary Figure 1D, Supplementary Table 1 and Supplementary Note 1), and the reduction in affinity was the same irrespective of the position of the fluorophore in the peptide (Supplementary Figure 1E, Supplementary Table 1). BsPBP11-32Ser7 acts as the helix N-cap, a role that is performed preferentially by Ser, Asn and Thr32, and alanine and proline substitutions in helical positions equivalent to BsPBP11-32Ser7 and BsPBP11-32Ala10, respectively, destabilise model peptides32,33. Finally, the importance of BsPBP11-32Ser7, BsPBP11-32Arg8 and BsPBP11-32Arg11 to GpsB binding is highlighted because these are the most well conserved residues in an alignment of the cytoplasmic mini-domains of Bacillaceae PBP1 PG synthases (Supplementary Figure 1F).
L. monocytogenes GpsB interacts with PBPA1 by a TRxxYR motif
The deletion of gpsB alone in B. subtilis has no readily apparent phenotype until combined with deletions in ezrA11 or ftsA12; by contrast, the deletion of gpsB in L. monocytogenes is lethal when grown at 42 °C19. Since GpsB in both species interact with class A PG synthases, we next determined whether the rules established above for the BsGpsB:BsPBP1 interaction could be applied directly to LmGpsB:LmPBPA1. The cytoplasmic mini-domain of LmPBPA1, LmPBPA11-29, has an abundance of positively charged residues, but lacks an exact copy of the SRxxR(R/K) motif of Bacillaceae PBP1 (Supplementary Figure 1F, 2A); the closest equivalent is TRxxYR. In FP, the first 20 amino acids of LmPBPA1, LmPBPA11-20, bound to the N-terminal domain of LmGpsB, LmGpsB1-73, with an affinity similar to that of BsPBPA11-32 for BsGpsB1-68 (Supplementary Figure 2B, Supplementary Table 1), but we were unable to co-crystallise LmGpsB constructs with LmPBPA1 peptides to visualise these interactions and to compare them to BsGpsB5-64:BsPBP11-17. Consequently, and to expedite crystallisation, we solved the structure of the first 15 amino acids of LmPBPA1, LmPBPA11-15, bound to BsGpsB5-64Lys32Glu, which is a surrogate for LmGpsB1-73 because (i) all the residues within 8 Å of the BsGpsB5-64:BsPBP11-17 interface are conserved in LmGpsB except for Lys32, which is glutamate in LmGpsB, and thus all peptide-contacting residues are maintained—as well as bystander residues that help indirectly to shape the PBP binding site; (ii) LmGpsB and BsGpsB use overlapping PBP binding sites19; (iii) the Kds of BsGpsB1-68Lys32Glu and LmGpsB1-73 for the first 20 amino acids of LmPBPA1, LmPBPA11-20, are almost identical (Supplementary Figure 2B, Supplementary Table 1). Therefore, the K32E variant of the N-terminal domain of BsGpsB is as close a surrogate for the equivalent LmGpsB domain that could be obtained.
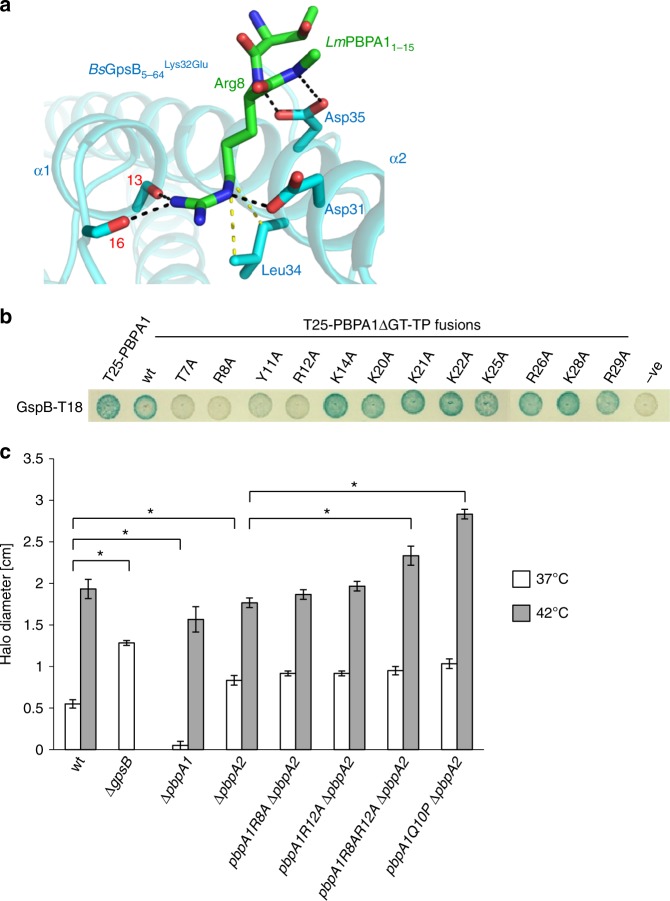
The subsequent structure of the BsGpsB5-64Lys32Glu:LmPBPA11-15 complex revealed that, in contrast to the BsGpsB5-64:BsPBP11-17 complex, the majority of the LmPBPA1 peptide was disordered except for LmPBPA11-15Arg8 and the immediately-adjacent mainchain atoms. LmPBPA11-15Arg8 adopts the same orientation and makes the same interactions as described above for BsPBP11-17Arg8 in the BsGpsB5-64:BsPBP11-17 complex (Figs. 1c, 2a). The bidentate interaction of Glu35 of BsGpsB5-64Lys32Glu with the backbone amides of LmPBPA1Arg8 and LmPBPA1Ser9 are maintained just as in the BsGpsB5-64:BsPBP11-17 complex and, as the backbone torsion angles of LmPBPA11-15Arg8 are also α-helical, it suggests that the role of the conserved LmPBPA1Thr7 is to N-cap this helix32. The interaction of BsGpsB5-64Lys32Glu with LmPBPA11-15 seemingly centres almost entirely on a single arginine. How LmGpsB discerns LmPBPA1Arg8 over other positively charged residues in the arginine- and lysine-rich cytoplasmic domain of LmPBPA1 (Supplementary Figure 2A) was determined by FP and BACTH. LmPBPA11-20Arg8Ala and LmPBPA11-20Arg12Ala mutations reduced the affinity for BsGpsB1-68Lys32Glu by >15- and ~4-fold, respectively (Supplementary Figure 2B, Supplementary Table 1), and had significant negative impact on the LmGpsB:LmPBPA1 interaction by BACTH (Fig. 2b). Alanine substitution of LmPBPA1Tyr11 had a comparatively milder impact in BACTH (Fig. 2b) and in FP (Supplementary Figure 2B, Supplementary Table 1). Reintroducing positive charge into LmPBPA11-20Arg8Ala did not restore wild-type binding affinity in FP as LmPBPA11-20Arg8Ala,Ser16Arg still bound to BsGpsB1-68Lys32Glu with an affinity at least 10-fold weaker than wild-type (Supplementary Figure 2B, Supplementary Table 1). BACTH also supports the central importance of LmPBPA1Thr7, which presumably plays a structural role in the LmPBPA11-15 peptide possibly by acting as an N-cap and positioning LmPBPA1Arg8 at the start of an α-helix at the. Alanine substitution of the other positively charged residues downstream of residue 14 had no impact in BACTH (Fig. 2b), consistent with the particularly stringent specificity of LmGpsB for LmPBPA1Arg8 in comparison to other positively charged residues.
Fig. 2.
The LmGpsB:LmPBPA1 interactions are also governed by a conserved arginine. a The structure of the BsGpsB5-64Lys32Glu:LmPBPA11-15 complex reveals that only Arg8 and adjacent backbone atoms of the LmPBPA11-15 peptide are ordered. In this cartoon, BsGpsB5-64Lys32Glu is coloured cyan and selected sidechains are drawn as stick with cyan carbons, whereas the LmPBPA11-15 peptide is represented in stick form, with green carbons. The carbonyl oxygens of BsGpsBIle13 and BsGpsBLys16 are denoted by respective red numerals. Hydrogen bonds are shown as black dashed lines and the van der Waals’ interactions between BsGpsBLeu34 and LmPBPA1Arg8 are in yellow. Only one PBP-binding site is occupied by peptide in these crystals because the second site is blocked by crystal contacts. b Mutation of conserved LmPBPA1 residues results in a loss of interaction by BACTH. The removal of residues 92–827, correlating to the glycosyltransferase and transpeptidase domains of LmPBPA1, results in the PBPA1ΔGT-TP peptide. Empty pKT25 (−) was used as a negative control. Agar plates were photographed after 24 h at 30 °C. c Effect of N-terminal pbpA1 mutations on fosfomycin sensitivity of a ΔpbpA2 mutant. Wild-type and mutant L. monocytogenes EGD-e strains were grown as confluent layers on BHI agar plates at 37 °C and 42 °C and halo diameters around fosfomycin-containing filter discs were measured and corrected for the disc diameter. The experiment was performed in triplicate, and average values and standard deviations are shown. Asterisks indicate statistically significant differences (P < 0.01, t-test)
LmPBPA1Arg8 and BsPBP1Arg8 are equivalent in their interactions with GpsB. Of the other GpsB-binding determinants of BsPBP1, LmPBPA1 lacks an analogous BsPBP1Arg11. The sequential equivalent is LmPBPA1Tyr11, but this residue is completely disordered and its mutation to alanine reduced the affinity for BsGpsB1-68Lys32Glu only by 2-fold (Supplementary Figure 2B). To further decipher the reason for the specificity for LmPBPA1Arg8, the importance of α-helix formation in LmPBPA11-15 for GpsB binding was analysed with Q10P-mutated LmPBPA1 peptides. A Q10P mutation caused a >7-fold reduction in binding affinity (Supplementary Figure 2B, Supplementary Table 1) and CD confirmed a significant impact of the Q10P mutation on the α-helicity of LmPBPA11-15 peptides (Supplementary Figure 2C). Finally, the effects of mutations to the crucial LmGpsB-interacting residues in LmPBPA1 were also probed in Listeria using fosfomycin sensitivity as a reporter since ΔgpsB mutants are more susceptible to this antibiotic at 37 °C than wild-type L. monocytogenes20. Fosfomycin inhibits the first enzyme in the biosynthetic pathway of PG, MurA, and the ΔgpsB mutant is hypersensitive to it probably because of unproductive consumption of PG precursors due to mis-regulated LmPBPA120. By contrast, removal of pbpA1 reduces substrate turnover in PG biosynthesis and thus the cells become more resistant to fosfomycin, and a pbpA1 pbpA2 double mutant is not viable34. Effects on fosfomycin sensitivity were apparent in mutants carrying the pbpA1Arg8Ala,Arg12Ala and pbpA1Gln10Pro alleles but only when LmPBPA2, the LmPBPA1 paralogue, was also absent (Fig. 2c). Synthetic lethality with pbpA2 and a growth defect at 42 °C is characteristic of the L. monocytogenes null gpsB mutant19, suggesting that the observed effects partially phenocopy ΔgpsB. However, no pbpA1 mutation phenocopied the ΔgpsB mutant completely (Supplementary Figure 2D).
Taken together, our data highlight the importance of a conserved arginine and α-helicity in class A PG synthases for interacting with GpsB in two species. In both cases the arginine is adjacent to a conserved residue with high propensity to act as a helix N-cap, implying that positioning of the arginine at the start of the helix is critical. Furthermore, since pbpA1 does not phenocopy gpsB in L. monocytogenes, and gpsB deletion on its own in B. subtilis has no clear phenotype, GpsB must have additional functions in both bacteria.
Extending the B. subtilis and L. monocytogenes GpsB interactomes
The data presented above describe features critical for interactions involving BsGpsB, which include a helical SRxxR(R/K) motif in close proximity to the membrane. To identify hitherto unidentified BsGpsB-interacting proteins, the B. subtilis proteome was queried with the SRxxR(R/K) motif. Two previously uncharacterised ORFs, BsYpbE and BsYrrS, conform to all the features described above. BsYpbE is a membrane protein with a 56-residue cytoplasmic domain that encodes a SRVERR motif. The extracellular region, residues 80–240, contains a LysM (lysin motif) domain between residues 189 and 235; LysM domains are ~40-residue, degenerate PG- and chitin-binding modules widespread in bacteria and eukaryotes. yrrS is found in a bicistronic operon widely conserved in the Bacillaceae with the gene (yrrR) encoding a class B PBP, PBP4b35, suggesting these genes have a linked function in cell wall homoeostasis36. BsYrrS comprises an 18-residue cytoplasmic domain with two potential, overlapping BsGpsB-binding motifs SRYENR and NRDKRR and an extracellular domain that belongs to the widespread and currently uncharacterised DUF1510 family.
LysM domains are frequently found as tandem repeats within bacterial proteins37 and the individual domains can act co-operatively to bind PG38,39. BsYpbE contains one LysM domain hence oligomerization of BsYpbE may enhance PG binding, with the oligomerisation of the extracellular LysM domain of BsYpbE presumably controlled by cytoplasmic, hexameric BsGpsB27, the essential form of the protein in bacteria19. In the absence of purified, full-length BsYpbE to test this hypothesis directly, monomeric and dimeric forms of BsYpbE130-240, which encompasses the sole extracellular LysM domain, were generated instead. Dimeric BsYpbE130-240 was prepared by mutation of Ser132 to cysteine to enable disulphide-linked BsYpbE130-240Ser132Cys dimers to be purified. In pulldown assays, the binding of BsYpbE130-240Ser132Cys dimers to PG was enhanced considerably relative to the monomeric, cysteine-free version of BsYpbE130-240 (Supplementary Figure 3A). Therefore, the binding of YpbE to PG in B. subtilis will be stimulated by YpbE multimerisation, induced by hexameric GpsB.
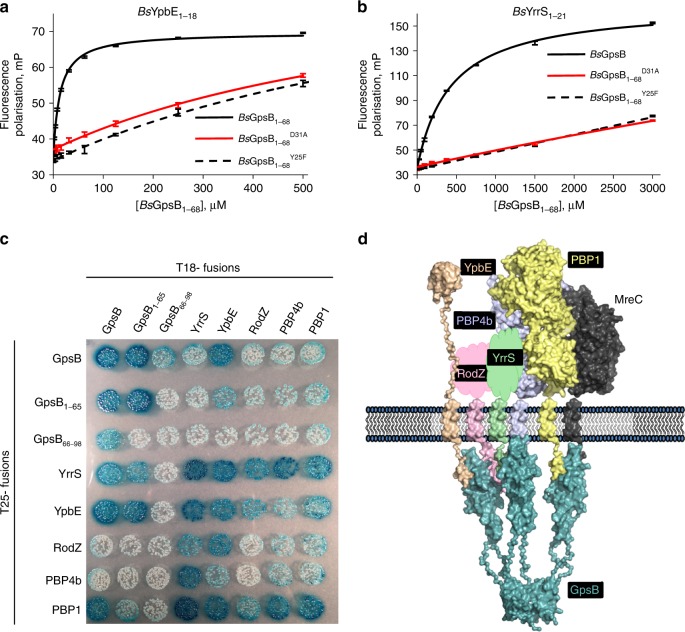
The interaction of BsGpsB1-68 with BsYrrS and BsYpbE was assessed by FP and BACTH. BsGpsB1-68 bound to a 21-residue fragment of the cytoplasmic domain of BsYpbE, BsYpbE1-21 that encompasses the SRVERR motif, and the entire cytoplasmic mini-domain of BsYrrS, BsYrrS1-18, with Kd values of 13 μM (Fig. 3a) and 430 μM (Fig. 3b), respectively. The modest affinity of the BsYrrS1-18 peptide for BsGpsB1-68 (Supplementary Table 1) probably translates to a substantially tighter affinity in bacterial cells, because of an avidity effect resulting from BsGpsB and BsYrrS associating with membranes, effectively increasing the local concentration of each significantly. The specificity of these interactions measured by FP was consistent with the impact of BsGpsBAsp31Ala and BsGpsBTyr25Phe mutations, each of which reduced the affinities for BsYrrS1-18 and BsYpbE1-21 by 7- and ~40-fold, respectively (Fig. 3a, b), and in-line with the roles of BsGpsBAsp31 and BsGpsBTyr25 in defining the BsPBP1 binding site. Interactions of BsGpsB with BsYrrS and BsYpbE were also detected by BACTH, with the interactions mapping to the N-terminal domain of BsGpsB in both cases (Fig. 3c). GpsB is only conditionally essential in B. subtilis11,12, and perhaps it is not surprising that no obvious cell growth or division phenotypes were identified in our hands or previously40 on deleting the genes encoding BsYrrS, BsPBP4b or BsYpbE. BACTH was used to confirm that BsYrrS interacted with BsPBP4b, BsPBP1 and BsRodZ; the latter two proteins have established roles in cell division, growth and morphogenesis41,42. The interaction between BsPBP1 and a fragment of BsYrrS lacking residues 13-16, BsYrrSΔ13-16 (which reduced non-specific binding to the BsPBP1-immobilised SPR chip and did not affect the SRYENR motif) was quantified by SPR, and BsYrrS Δ13-16 bound to BsPBP1 with a Kd of 20 nM (Supplementary Figure 3B). Therefore, these gene products are capable of forming a network of interactions (Fig. 3d) that may be nucleated by the formation of a BsPBP1:BsYrrS complex.
Fig. 3.
The SRxxR(R/K) motif identifies BsYpbE and BsYrrS as new BsGpsB binding partners. a, b BsYpbE1-18 and BsYrrS1-21 bind to BsGpsB1-68 at the same site as BsPBP1. Fluorescence polarisation of BsGpsB1-68 binding to fluorescein-labelled BsYpbE1-18 (a) and fluorescein-labelled BsYrrS1-21 (b). The interaction of wild-type proteins is depicted by black curves, whereas the red curves and dashed black lines correspond to BsGpsB1-68Asp31Ala and BsGpsB1-68Tyr25Phe mutants, respectively. c BACTH reveals a new BsGpsB interaction network involving proteins encoding the SRxxR(R/K) motif. The panel shows pairwise combinations of proteins expressed as N-terminal fusions to both halves of adenylate cyclase in the BACTH host strain. Their presence in complexes containing BsRodZ, BsPBP4b and BsPBP1 imply roles for BsYrrS and BsYpbE in sidewall synthesis during cell growth. The validity of the observed interactions is supported by the behaviour of the T18-GpsB66-98 fusion, which does not interact with any other partner except T25-GpsB, and therefore acts as an internal control. The T18-BsGpsB66-98:T25-BsGpsB interaction is consistent with the hexameric nature of BsGpsB19,26. The absence of interactions between BsGpsB66-98 and BsYrrS or BsYpbE is consistent with data in b that shows that the N-terminal domain of GpsB interacts with the SRxxR(R/K) motif of YpbE and YrrS. d A model recapitulates interactions between BsGpsB and partners. Surface representations of the SAXS structure of LmGpsB26 (coloured teal) and the closest homologues in the PDB by sequence (BsPBP1, yellow, PDBid 2OLV; BsPBP4b, pale blue, 4L0L; BsRodZ, pink, 2WUS; BsMreC, dark grey, 2J5U; BsYpbE, salmon, 2MKX and the linker represents the disordered region, residues 131-188) are used as models for the B. subtilis proteins. Amorphous blobs, the surface area of which are scaled proportional to molecular weight, are used for BsYrrS (green) and the extracellular domain of BsRodZ (pink) where there is no structural information. The N-terminal domain of each membrane protein is cytoplasmic and 22-amino acid model helices represent each TM helix. The TMpred-predicted TM boundaries are: BsPBP1 (38-60); BsPBP4b (9-31); BsYrrS (19-41); BsYpbE (57-79); BsRodZ (89-111) and BsMreC (7-24). The GpsB-interacting domains of BsYpbE, BsYrrS and BsPBP1 are based on the BsPBP11-17 structure
Homologues of YpbE do not exist in L. monocytogenes and the YrrS homologue (Lmo1495) does not contain a signature BsGpsB-binding motif and neither protein is found in S. pneumoniae. No strong potential GpsB-interacting candidates were identified when the L. monocytogenes proteome was searched with either TRxxYR or SRxxR(R/K) as the query. BACTH was thus used to uncover additional potential LmGpsB functions in L. monocytogenes using a bank of 27 listerial components from the known elongation and division machineries in bacteria (Supplementary Note 2). Twelve proteins were shown to interact with LmGpsB and only these are shown in Supplementary Figure 3C. There is no consensus motif shared by these proteins, though all have at least one arginine present in their cytoplasmic regions that is conserved in their respective orthologues. Two classes of hits were identified in the BACTH screen; class I hits (LmPBPA1, LmMreC and LmSepF, and LmGpsB self-interactions) turned blue after one day of incubation (Supplementary Figure 3C). Class II hits turned blue after 2 days incubation at 30 °C, including LmZapA, LmEzrA, LmDivIB, LmDivIC, LmMreC, LmMreBH and the other HMW LmPBPs (Supplementary Figure 3C). All of these interactions, except for the GpsB self-interactions, required the LmGpsB N-terminal domain (Supplementary Figure 3C). In good agreement with the absence of a TRxxYR motif in LmMreC, LmSepF and LmZapA, interactions with these proteins did not require key residues in the known PBP-binding groove in LmGpsB (Supplementary Figure 3D) and reciprocal tests validated the LmGpsB:LmPBPA1 interactions21. It would thus seem that LmPBPA1 represents the only GpsB binding partner that employs the TRxxYR motif in L. monocytogenes.
Two arginines dictate S. pneumoniae GpsB:PBP2a molecular recognition
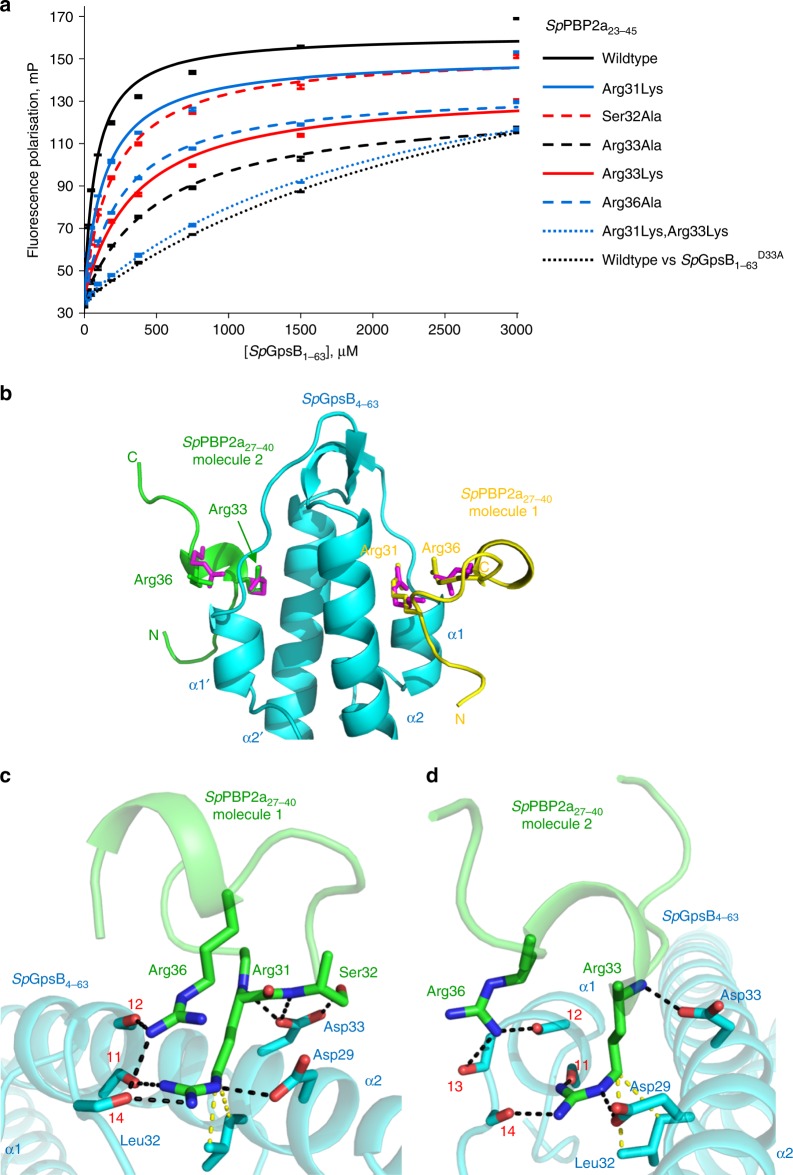
S. pneumoniae, more distantly related to either Bacillus or Listeria, is an ovoid-shaped Gram-positive coccus in which GpsB is essential22–24. SpGpsB co-immunoprecipitated with SpPBP2a (one of three pneumococcal class A PBPs), SpMreC and other proteins, suggesting they interact at some point in the pneumococcal cell cycle24. Synthetic lethality studies in pneumococcal ΔgpsB suppressor mutants revealed that pbp1a, and not pbp2a, became essential in the absence of gpsB indicating that SpPBP2a is the class A PBP regulated by SpGpsB in S. pneumoniae24. We found that the cytoplasmic mini-domain of SpPBP2a and many of its orthologues contain the consensus sequence (S/R)RS(R/G)(K/S)xR (Supplementary Figure 4A) that resembles the Bacillaceae PBP1 SRxxR(R/K) motif (Supplementary Figure 1F). A 22-residue peptide of SpPBP2a that encompasses this region, SpPBP2a23-45, was found by FP to bind to the N-terminal domain of SpGpsB, SpGpsB1-63, with a Kd of 80 μM whereas SpGpsB1-63Asp33Ala (equivalent to BsGpsBAsp35Ala) had a ~40-fold reduced affinity for SpPBP2a23-45 (Fig. 4a, Supplementary Table 1). The crystal structure of a slightly truncated form of the N-terminal domain of SpGpsB, SpGpsB4-63 (to expedite crystallisation), was solved in the presence of a 14-residue peptide of SpPBP2a, SpPBP2a27-40, which includes the (S/R)RS(R/G)(K/S)xR motif. In this instance, each subunit of the SpGpsB dimer is peptide-bound (Fig. 4b). Peptide binding principally involves two arginines but each SpGpsB subunit recognises the peptide differently. In SpGpsB4-63 molecule 1, SpPBP2a27-40 recognition centres on SpPBP2aArg31 and SpPBP2aArg36 (Fig. 4c), whereas molecule 2 involves SpPBP2aArg33 and SpPBP2aArg36 (Fig. 4d). The arginine pairs occupy the same positions as BsPBP1Arg11 and BsPBP1Arg8 in the BsGpsB5-64:BsPBP11-17 complex (Fig. 1c); SpPBP2aArg36 is equivalent to BsPBP1Arg11whereas SpPBP2aArg31 and SpPBP2aArg33 are equivalent to BsPBP1Arg8.
Fig. 4.
The SpPBP2a minidomain is not α-helical but still interacts with SpGpsB through conserved arginines. a Arginine residues of SpPBP2a play a key role in binding to SpGpsB. Unless otherwise indicated, the fluorescence polarisation binding curves represent the interaction of TAMRA-labelled SpPBP2a23-45 peptides with wildtype SpGpsB1-63. The relevant dissociation constants are listed in Supplementary Table 1. b The structure of the SpGpsB4-63:SpPBP2a27-40 complex reveals the critical role of SpPBP2a arginines for the interaction with SpGpsB. In this cartoon, SpGpsB4-63 is coloured cyan, and the SpPBP2a27-40 peptide is coloured yellow (molecule 1) and green (molecule 2). The sidechains of Arg8 and Arg11 from the BsGpsB5-64:BsPBP11-17 complex are shown as red sticks after a global superimposition of equivalent GpsB atoms. In molecule 1, SpPBP2aArg31 and SpPBP2aArg36 superimpose with BsGpsB5-64Arg8 and BsGpsB5-64Arg11 whereas molecule 2 accommodates SpPBP2aArg33 and SpPBP2aArg36. c, d Close-up view of the interactions of SpPBP2a from molecule 1 (c) and 2 (d) with SpGpsB4-63. Key interfacial sidechains and backbone atoms are represented in stick format; SpGpsB4-63 is coloured cyan and SpPBP2A27-40 is coloured green. The van der Waals’ interactions between SpGpsBLeu32 and SpPBP1Arg31 (molecule 1) and SpPBP1Arg33 (molecule 2) are in yellow. The carbonyl oxygens of SpGpsBIle11, SpGpsBPhe12, SpGpsBGlu13 and SpGpsBGln14 are denoted by respective red numerals
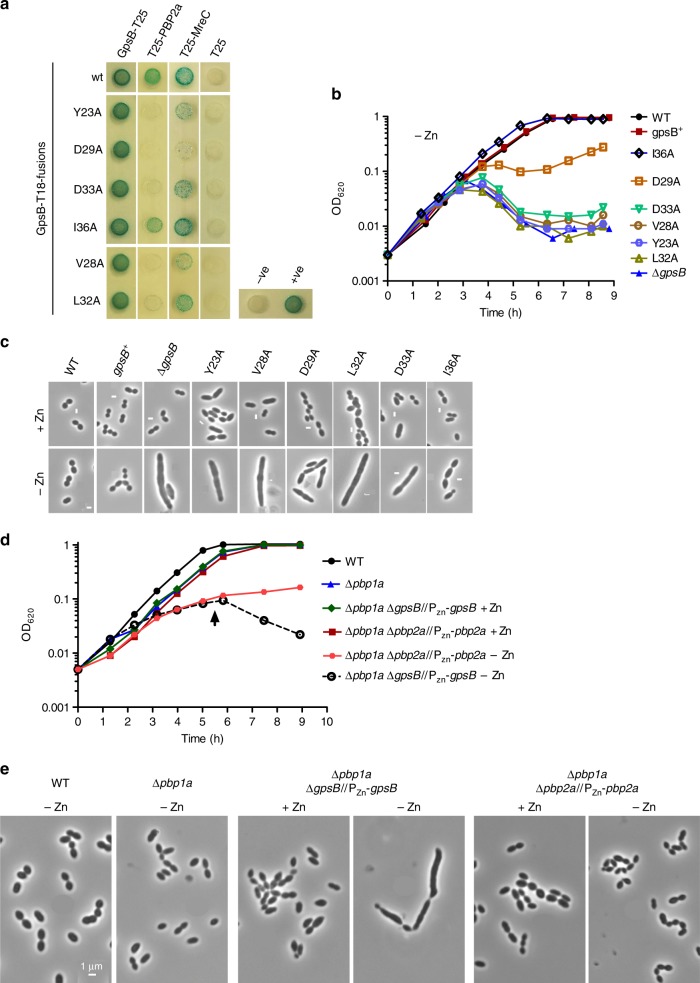
The SpGpsB:SpPBP2a interaction was confirmed by BACTH (Supplementary Figure 4B). The interaction was lost completely with SpGpsBTyr23Ala, SpGpsBVal28Ala, SpGpsBAsp29Ala, SpGpsBLeu32Ala and SpGpsBAsp33Ala mutated proteins and reduced with SpGpsBIle36Ala (Fig. 5a). All the SpGpsB variants retained the ability to self-interact and to interact with wild-type SpGpsB (Fig. 5a, Supplementary Figure 4C); the impact of the mutations on interactions with SpPBP2a thus does not reflect impaired expression of the relevant fusion proteins. Moreover, all the SpGpsB variants, except SpGpsBAsp29Ala, retained some ability to interact with SpMreC, which was also confirmed to interact with SpGpsB by BACTH (Fig. 5a, Supplementary Figure 4B).
Fig. 5.
Depletion of SpPBP2a does not phenocopy depletion of SpGpsB. a Mutations in SpGpsB differentially affect interactions with SpPBP2a and SpMreC. BACTH analysis of the interactions of SpGpsB-T18 variants with wild-type SpGpsB, SpPBP2a and SpMreC. pKT25/pUT18C and pKT25-zip/pUT18C-zip plasmid pairs were used as negative (−ve) and positive (+ve) controls, respectively. The agar plates were photographed after 40h incubation at 30 °C. b, c SpGpsB variants that have lost SpPBP2a binding have a gpsB null growth and morphology phenotype. Representative growth curve of S. pneumoniae strains with ectopic expression of gpsB+ under a Zn2+-dependent promoter. GpsB variants of Y23A, V28A, L32A and D33A showed a gpsB null growth phenotype (b) and elongated cell morphology (c) on gpsB depletion. The D29A variant showed an intermediate growth phenotype, which was also obtained with an independent isogenic isolate and with a gpsBD29A-FLAG labelled strain. The I36A strain has a reduced elongation phenotype. All phase‐contrast micrographs are at the same magnification (scale bar = 1 µm). d, e SpPBP2a depletion does not phenocopy SpGpsB. Representative growth curves (d) and phase-contrast micrographs (e) of parent IU1824 (WT, D39 ∆cps rpsL1), IU13444 (∆pbp1a), IU14381 (∆pbp2a//ΔbgaA::PZn-pbp2a+ ∆pbp1a) and IU14383 (∆gpsB//ΔbgaA::PZn-gpsB+ ∆pbp1a). Similar to the depletion of SpGpsB in S. pneumoniae pbp1a+ strains (see panel b), depletion of SpGpsB in IU14383 leads to extremely elongated cells, a growth cessation and lysis phenotype. By contrast, depletion of SpPBP2a in the ∆pbp1a background (right hand panels) leads to small but mostly ovococcal cells that do not lyse during the time course examined. All phase-contrast micrographs were taken at OD620 ≈ 0.15 or at the time point marked by arrows in a for IU14381 and IU14383 under zinc depletion and are at the same magnification (scale bar = 1 µm)
Despite differences in the secondary structures of the two independent SpPBP2a peptides bound to the SpGpsB4-63 dimer (Fig. 4b), the two arginines form a similar network of interactions with SpGpsB as described for BsGpsB and LmGpsB (Figs. 1c, 2a) with additional sidechain contacts in molecule 1 between SpPBP2aSer32 and SpGpsBAsp33 (Fig. 4c), and SpPBP2aArg31 and SpGpsBTyr23. The importance of SpPBP2aArg31 and SpPBP2aArg33 is further supported by their sequence conservation (Supplementary Figure 4A) and FP (Fig. 4a, Supplementary Table 1). Although SpPBP2aArg31Lys had only a 2-fold reduced affinity, which probably reflects the ability of SpPBP2aArg33 to compensate for the loss of SpPBP2aArg31, the binding affinity of SpPBP2aArg31Lys,Arg33Lys was reduced >25-fold relative to wild type. The importance of the SpGpsB residues involved in the interactions with SpPBP2a is also consistent with the phenotype because of the severe growth (Fig. 5b) and morphological defects (Fig. 5c) of S. pneumoniae strains harbouring the SpGpsBTyr23Ala, SpGpsBVal28Ala, SpGpsBAsp29Ala, SpGpsBLeu32Ala and SpGpsBAsp33Ala alleles even though the mutated proteins were still capable of self-interactions (Supplementary Figure 4C) and were expressed at wild-type levels (Supplementary Figure 4D). However, no obvious phenotype was observed in S. pneumoniae strains carrying the corresponding SpPBP2aArg31Ala, SpPBP2aArg33Ala or SpPBP2aArg31Ala,Ser32Ala,Arg36Ala alleles, even when pbp1a was deleted to decouple the effects of mutations in SpPBP2a from SpPBP1a activity (Supplementary Table 2). Nevertheless, SpPBP2a mutants in which amino acids 32–37 or 27–38 or 26–45 were deleted in a Δpbp1a background showed progressively reduced growth rates in the three deletion strains and pronounced morphological defects in the two strains with larger deletions (Supplementary Figure 5A, 5B), despite wild-type levels of protein expression at the respective expected molecular masses (Supplementary Figure 5C). At least eight residues, including two arginines and one lysine, were retained in all the deletion constructs before the predicted start of the SpPBP2a TM helix (residue 54), providing the necessary charge to satisfy the inside-positive rule43. Consistent with the pbp2a deletion mutant phenotypes, BACTH results for the correspondent truncated SpPBP2a variants showed reduced interactions with SpGpsB in comparison to the wildtype but not with SpMreC (Supplementary Figure 5D, 5E).
Together, these results support a critical role of the (S/R)RS(R/G)(K/S)xR motif between SpPBP2a residues 30 and 36 for mediating protein–protein interactions with SpGpsB and the importance of its reduction or loss in S. pneumoniae. However, the observation that all three Δpbp1a pbp2a deletion mutants were viable and both growth and morphology phenotypes were different between S. pneumoniae Δpbp1a strains depleted for gpsB (Fig. 5d) and Δpbp1a strains depleted for pbp2a (Fig. 5e) implies that SpPBP2a binding is also just one function for SpGpsB. It is possible that the SpPBP2a:GpsB interaction is only part of the S. pneumoniae ΔgpsB phenotype and this interaction per se is not essential, unless the interaction of GpsB with an additional partner is also lost. Alternatively, since deleting the GpsB binding motif of SpPBP2a did not abolish the interaction with GpsB completely, the deletion might reveal, or even generate, an unpredicted GpsB binding site, perhaps in the juxta-membrane region untouched by the deletions.
Since the pbp2a mutant did not phenocopy gpsB depletion, and given the complex network of GpsB interactions in B. subtilis and L. monocytogenes, we also sought to extend the pneumococcal GpsB interactome starting from those proteins that were reported to form a complex with SpGpsB by co-immunoprecipitation (co-IP)24. Besides the SpGpsB interactions detected with SpPBP2a and SpMreC by BACTH, we also detected an interaction between SpGpsB and the essential class B SpPBP2x44 (Supplementary Figure 6A). SpPBP2x, which contains a cytoplasmic domain abundant with arginines, also interacted with SpGpsB in FP experiments (Supplementary Figure 6B). However, a SpGpsB:SpPBP2x interaction was not detected by co-IP either using SpGpsB-L-FLAG3 as the bait and an anti-HA antibody to detect HA-tagged SpPBP2x24 or an anti-SpPBP2x for native SpPBP2x immnodetection, or SpPBP2x-FLAG3 as bait and an anti-SpGpsB antibody for immunodetection of SpGpsB (Supplementary Figure 6C). These results suggest that SpGpsB:SpPBP2x complexes may be insufficiently abundant, or present only transiently, to be detected by co-IP in pneumococcal cells. SpGpsB also interacted in BACTH with the class A SpPBP1a (Supplementary Figure 6A), which contains a LRLIKY motif in a putative helical region in its small cytoplasmic domain of 18 amino acids. On the other hand, no direct interaction was detected between SpGpsB and SpPBP2b either by BACTH (Supplementary Figure 6A) or by FP (Supplementary Figure 6B), consistent with the lack of any candidate GpsB-binding sequence motif in SpPBP2b. The SpPBP2b:SpGpsB interaction detected previously by co-IP24 (Supplementary Figure 6C) is therefore likely to be indirect, presumably involving a bridging protein. The SpGpsBAsp33Ala allele that resulted in a complete loss of binding to SpPBP2a by FP (Supplementary Table 1) also abrogated interactions in BACTH with a markedly greater impact on interactions with SpPBP2a compared to SpMreC (Supplementary Figure 6A). SpGpsBTyr23Ala and SpGpsBAsp29Ala mutations had a greater impact on SpMreC interactions than SpGpsBAsp33Ala; this pattern mirrors the impact of the equivalent mutations (LmGpsBTyr27Ala, LmGpsBAsp33Ala and LmGpsBAsp37Ala) on MreC interactions in L. monocytogenes (Supplementary Figure 3D) and thus substantiates the evidence that different surfaces are used by GpsB to bind a specific class A PBP and MreC. Whilst the interactions of SpGpsB with SpEzrA and SpStkP were confirmed by BACTH, and observed with the reciprocal hybrid pairs, they were unaffected by any of the SpGpsB alleles mentioned above (Supplementary Figure 6A). The interactions of GpsB in the pneumococcus, as detected by BACTH, FP and co-IP, are summarised in Supplementary Figure 6D.
Taken as a whole, our data on three important bacterial systems agree that GpsB is an adaptor protein29–31 that connects a major class A PG synthase with other cell wall and cell cycle proteins, and to cell shape determinants such as MreC. The identity and mode of interaction of the GpsB-binding partners varies from species to species and may reflect the different physiologies of each bacterium and their modes of growth and division.
Discussion
Bacterial cell growth and division necessitates tight co-ordination between the replication and segregation of the chromosome, the fission of the cell membrane and the remodelling of the PG. Consequently proteins and their complexes with major functions on either side of the membrane must co-ordinate their activities. One potential mechanism involves the interactions of major PG synthases with their intracellular regulators. Herein we present the first structures of the cell cycle adaptor, GpsB, in complex with the cytoplasmic mini-domains of PG synthases from three different bacteria, the rod-shaped B. subtilis and L. monocytogenes, in which gpsB is conditionally essential11,12,19 and the ovococcal S. pneumoniae in which gpsB is essential22–24. In common with mammalian adaptors GGA31 and 14-3-345 proteins, the primary binding surface of GpsB is restricted to a conserved groove between α-helices The cytoplasmic mini-domains of the three PG synthases in the three organisms have little in common except that each utilises a conserved arginine in their respective sequences to interact with the cognate GpsB. The PG synthase arginine finger pokes into a negatively charged cavity situated between α-helices 1 and 2 of GpsB and is fixed in the same orientation in all structures, just as the phosphoryl group defines the binding orientation of peptides to 14-3-345. The arginine complements the cavity best when the mainchain amide protons of it and its downstream residue are accessible to form hydrogen bonds with BsGpsBAsp35, LmGspBAsp37 or SpGspBAsp33. This scenario can occur when the arginine is either at the start of an α-helix, such as BsPBP1Arg8, or at the i + 1 position in a type I β turn, such as SpPBP2aArg31, which explains why free l-arginine does not displace pre-bound PBP peptides from GpsB even when present at 100-fold molar excess (Supplementary Figure 2E). Similarly, contact to the backbone amide at the i + 2 position in 14-3-3 ligands is essential for binding45. Despite a lack of strong sequence and structural homology in the PG synthase cytoplasmic mini-domains, their binding is dependent upon an identical subset of GpsB residues including BsGpsBTyr25, BsGpsBAsp31, BsGpsBAsp35 (Fig. 1c) and their structural equivalents LmGspBTyr27, LmGspBAsp33, LmGspBAsp37 (Fig. 2a), and SpGpsBTyr23, SpGpsBAsp29 and SpGspBAsp33 (Fig. 4c, d, Supplementary Figure 1A). Mutations at these positions in GpsB from S. pneumoniae and L. monocytgenes are lethal or have marked growth defects (Fig. 5b, c)19 and do not interact with their cognate peptidoglycan synthase by FP (Supplementary Table 1)19 or by BACTH (Fig. 5a)19. A phenotype analysis of equivalent mutations in B. subtilis to assess their importance is not straightforward to determine for two reasons. First, a phenotype for the gpsB null mutant is only observed synthetically, when gpsB is mutated alongside either ftsA12 or ezrA11. Second, a B. subtilis mutant lacking all class A PBPs is viable46, precluding an unequivocal assessment of the precise importance of the BsGpsB:BsPBP1 interaction. Nonetheless, the convergence of all our experimental data on three bacterial systems suggests strongly that the tyrosine and aspartate dyad also have important roles in B. subtilis. Furthermore, these amino acids are also conserved in the DivIVA/Wag31/antigen 84 actinobacterial homologues of GpsB, suggesting a role for them in recruiting cell wall synthesis enzymes to the hyphal tip and future branch sites13,14, regions that require nascent PG synthesis in filamentous bacteria.
We originally set out to establish the common rules by which GpsB interacts with major PG synthases. Other than the arginine finger mode of GpsB recognition, we discovered how GpsB interacts with at least one class A PBP in each species; that LmGpsB interacts with both the cell shape determinant, MreC47, and a regulator of Z-ring dynamics, EzrA10, while in S. pneumoniae we have confirmed the SpGpsB:MreC interaction and newly identified interactions with the class A SpPBP1a and with the essential class B SpPBP2x44. These new data complement what was previously known about these interactions in B. subtilis11 and S. pneumoniae23,24. How GpsB can interact with such disparate targets remains unknown but BsGpsBAsp31, LmGpsBAsp33 and SpGpsBAsp29 are important for interactions with PBPs and other proteins, including MreC, while LmGpsBAsp37 and SpGpsBAsp33 only interact with PBPs. There must be at least one other surface that is used by GpsB to form complexes with other proteins in its function as an adaptor.
The GpsB:PBP interaction interface notably requires no more than three sidechains from any PBP to complex with GpsB. Protein:peptide contacts involving less well-conserved exosites that flank a small core, conserved peptide motif can contribute significantly to the affinity of protein:peptide interactions48,49, and the contribution of exosites to affinity, such as the juxta-membrane region, may explain why point mutations in LmPBPA1 and SpPBP2a have a significant impact using peptide fragments in vitro, but have reduced impact in bacterial cells. For instance, the LmPBPA1Arg8Ala mutation had negligible effect (Fig. 2c, Supplementary Figure 2D) yet it reduced binding by > 15-fold (Supplementary Table 1). Similarly SpPBP2aArg31Lys,Arg33Lys had a > 25-fold impact on binding (Supplementary Table 1) yet growth phenotypes were not evident (Supplementary Table 2) until significant stretches of the cytoplasmic mini-domain were deleted (Supplementary Figure 5B).
We also found some differences in GpsB interactions between the species that may be related to GpsB species-specific function. In B. subtilis, we discovered a critical motif, SRxxR(R/K), found in close proximity to the membrane that could be used to predict novel GpsB partners. We used this information to identify an interaction network involving cell envelope binding and modifying proteins that most likely is underpinned by the GpsB hexamer. An RSxxxR motif was identified in class A PBPs from most streptococci and sequence features that could dictate GpsB-binding can be found in class A PBPs in other Gram-positive organisms such as the lactococci, Leuconostocaceae and enterococci, including the ESKAPE pathogen E. faecium (Supplementary Figure 7). However, sequence-based searches alone will not identify complete GpsB interactomes because the local structure of the sequence and its proximity to the membrane are also key parameters of GpsB binding. SpPBP2a contains a partially conserved sequence RSxxxR (Supplementary Figure 4A) that resembles the BsPBP1 signature motif and the SpPBP2a mini-domain interacts with each subunit of the SpGpsB dimer in a different way (Fig. 4b–d). We have observed here that mutation or deletion of the cytoplasmic mini-domain of SpPBP2a does not phenocopy deletion or depletion of gpsB (Fig. 5c, e). Similarly, a ΔpbpA1 strain did not phenocopy gpsB deletion in L. monocytogenes19. Taken together these results imply that there must be at least one other critical GpsB interaction partner, beyond respective class A PG synthases, that dictates its conditional essentiality in L. monocytogenes and B. subtilis, and essentiality under normal growth conditions in S. pneumoniae.
In every Firmicute (and Actinobacterium) tested thus far, apart from S. aureus26, GpsB (or the homologous DivIVA/Wag31/antigen 84) acts as an adaptor to co-ordinate PG synthase activity with other processes depending on the physiology of the cell. GpsB hexamerisation can bridge the interaction of multiple binding partners, a function GpsB shares with 14-3-3 proteins that can form ternary complexes with BCR and Raf-1 by 14-3-3 dimerisation50. In bacilli, BsGpsB plays a role in shuttling between the side wall during elongation and the septum during division11 and, given that BsPBP4b is regulated by σ factors E[35] and F[51], complexes of BsPBP4b and BsYrrS, bridged by BsGpsB (Fig. 3c, d), presumably play a role in the asymmetric cell division characteristic of endospore-forming bacilli. In listeria, which is closely related to bacilli and shares with them a rod-like morphology, GpsB appears to connect several PBPs with proteins with known roles in cytokinesis, including Z-ring polymerisation modulators (ZapA, EzrA, SepF), late division proteins (DivIB, DivIC) and the elongasome (MreC, MreBH) (Supplementary Figure 3C,D), all of which except SepF and MreBH have also been tested in B. subtilis and found not to interact with BsGpsB11. By contrast, S. aureus GpsB appears to modulate Z-ring assembly26, however, no interaction between FtsZ and GpsB has been identified in B. subtilis11, S. pneumoniae23,24 or L. monocytogenes (Supplementary Figure 3C).
The pneumococci have an ovoid cell shape and lack key components such as the MinCD system for cell division site selection52, and MreB-like proteins required for side wall synthesis53. Presumably SpGpsB interacts with one or more pneumococcal-specific proteins, the loss of which may be related to the lethal phenotype. Furthermore, SpGpsB affects both StkP autophosphorylation23,24 and the StkP-catalysed phosphorylation of SpDivIVA23,24, SpMapZ/LocZ24,54, SpJag/EloR/KhpB55–57 and SpMacP58. It is not yet clear how the complexes formed by these proteins are affected by their phosphorylation, except that SpPBP2a activity is dependent upon phosphorylated SpMacP58, at least in the presence of functional SpStkP, or what the impact is of potential cross-talk to two-component signalling systems59.
Finally, the different phenotypic outcomes associated with gpsB deletion or depletion in the three systems studied herein may reflect the presence of redundant systems in the large genome (4.2 Mbp) of the bacilli, partial redundancy in listeria (2.9 Mbp), and a relative absence of redundancy in the stripped-down genome (2.1 Mbp) of the pneumococci. The relative affinities and cellular concentrations of GpsB partners probably dictate which protein(s) is bound by GpsB at any point in a cell cycle-dependent manner; simultaneous interactions with multiple target proteins is likely to lead to an increase in avidity of GpsB19 as commonly found in antibody:antigen interactions. However, the intricate networks involving GpsB will only be uncovered by validating the full GpsB interactome.
Methods
Bacterial strains and growth conditions
Supplementary Table 3 lists the bacterial strains used in this study. L. monocytogenes EGD-e-derived strains were routinely cultivated in brain heart infusion (BHI) broth or on BHI agar plates at 37 °C or 42 °C, where indicated. If required, erythromycin (5 µg−1mL−1) and X-Gal (40–100 µg−1mL−1) were added. All L. monocytogenes growth experiments were repeated three times and average values and standard deviations are shown. All S. pneumoniae strains were derived from unencapsulated serotype 2 D39 strains IU1824 (D39 Δcps rpsL1) or IU1945 (D39 Δcps)60. S. pneumoniae strains were grown on Petri dishes containing the appropriate antibiotic, modified trypticase soy agar II (Becton-Dickinson) and 5% (vol per vol) defibrinated sheep blood (TSAII-BA), and the plates were incubated at 37 °C in 5% CO2. Bacteria were cultured statically in BHI (Becton-Dickinson) broth at 37 °C in an atmosphere of 5% CO2, and growth was monitored by OD620. Escherichia coli TOP10 and DH5α (lab stocks) were used as standard hosts for all cloning procedures61, and BL21(DE3) strains (lab stocks) were used for recombinant protein production.
General methods, manipulation of DNA and oligonucleotide primers
E. coli transformation and isolation of plasmid DNA (the plasmids used are listed in Supplementary Table 4) was performed according to standard protocols61. L. monocytogenes strains were transformed by electroporation. Enzymatic modification of plasmid DNA was carried out as described by the instructions given by the manufacturers. Quikchange mutagenesis was employed for restriction-free modification of plasmids62. DNA sequences of oligonucleotide primers are listed in Supplementary Tables 5–7. All deletions and insertions were confirmed by PCR, and all constructs and clones were verified by Sanger DNA sequencing.
Construction of L. monocytogenes mutant strains
Plasmid pSH497 was first constructed to facilitate mutagenesis of the chromosomal copy of pbpA1. A DNA fragment comprising recU and the first 2063 bp of pbpA1 was amplified from chromosomal DNA using primer pair SHW773/SHW774 and cloned into pMAD using BamHI/NcoI. An unwanted mutation in front of recU resulted in a correction of this plasmid by Quikchange with primer pair SHW777/SHW778. The corrected pSH497 was used as template in further Quikchange reactions to introduce Thr7Ala (SHW744/SHW745, pSH504), Arg8Ala (SHW746/SHW747, pSH505), Tyr11Ala (SHW755/SHW756, pSH506), Gln10Pro (SHW787/SHW788, pSH509) and Arg12Ala mutations (SHW748/SHW749, pSH507) into pbpA1, or to remove pbpA1 nucleotides 4-1250 (SHW775/SHW776, pSH503). pSH508 (pbpA1Arg8Ala/Arg12Ala) was obtained by Quikchange with primers SHW750/SHW751 and plasmid pSH505 as the template. An N-terminal fragment of pbpA1 was excised from the L. monocytogenes chromosome using plasmid pSH503 and the insertion/excision protocol for construction of clean deletions, resulting in strain LMS211. The mutated pbpA1 alleles present on pSH504-pSH509 were then reintroduced into LMS211 following the same protocol.
L. monocytogenes fosfomycin susceptibility assays
Fosfomycin susceptibility was recorded using filter discs (Ø 6 mm) soaked with 10 µL of a 10 mg−1mL−1 fosfomycin solution. L. monocytogenes colonies, grown on BHI agar plates, were resuspended in BHI broth and used to swab-inoculate BHI agar plates. Filter discs soaked with fosfomycin were placed on top of the agar surface and the plates were incubated at 37 °C overnight. Diameter of growth inhibition zones was measured and corrected for the filter disc diameter. All experiments were performed three times and average values and standard deviations were calculated. Significance was determined using the t-test and differences were considered to be significant when P < 0.01.
L. monocytogenes and B. subtilis BACTH assay
The BACTH system63 was used for analysis of protein:protein interactions. First, LmGpsB was screened against LmPBPA1 (lmo1892) variants to identify residues in LmPBPA1 that are important for binding LmGpsB. To facilitate screening, we used a T25 fragment of Bordetella pertussis adenylate cyclase that had been fused to the N-terminal 91 amino acids of LmPBPA1 to create a T25-LmPBPA1 fusion containing the N-terminal cytoplasmic mini-domain and the transmembrane helix, but lacking lacking the extracellular glycosyltransferase and transpeptidase domains (T25-PBPA1ΔGT-TP) in plasmid pSH43721. LmPBPA1 residues that corresponded to the BsGpsB binding motif of BsPBP1 (Thr7, Arg8, Tyr11, Arg12) and additional positively charged amino acids (Lys14, Lys20, Lys21, Lys22, Lys25, Arg26, Lys28 and Arg29) in T25-PBP A1ΔGT-TP were replaced with alanine prior to screening against GpsB-T18. The pSH437 plasmid was mutated by Quikchange mutagenesis using the primer pairs SHW744/SHW754 (T7A, pSH485), SHW746/SHW747 (R8A, pSH486), SHW755/SHW756 (Y11A, pSH487), SHW748/SHW749 (R12A, pSH488), SHW757/SHW758 (K14A, pSH489), SHW759/SHW760 (K20A, pSH490), SHW761/SHW762 (K21A, pSH491), SHW763/SHW764 (K22A, pSH492), SHW765/SHW766 (K25A, pSH493), SHW767/SHW768 (R26A, pSH494), SHW769/SHW770 (K28A, pSH495) and SHW771/SHW772 (R29A, pSH496).
pJR164 and pJR169 were constructed by cloning sepF (primers JR334/JR335 and XbaI/KpnI digestion) into pKT25 and p25-N, respectively. pJR165 and pJR170 were generated by cloning ezrA into pKT25 and p25-N, respectively, using primers JR339/JR340 and XbaI/KpnI for digestion. pJR172 and pJR174 were obtained by cloning zapA into pKT25 and p25-N, respectively, using primers JR337/JR338 and PstI/KpnI. pJR213 was constructed by amplification of divIB with primers JR353/JR354 and cloning into pKT25 using XbaI/KpnI. pJR214 was generated by PCR amplification of divIC with primers JR355/JR356 and cloning into pKT25 using XbaI/KpnI. mreBH was amplified by PCR using oligonucleotides JR368/JR369 and cloned into pKT25 and p25-N using XbaI/KpnI, resulting in pJR233 and pJR236, respectively. pJR242 and pJR243 were obtained by cloning mreC into pKT25 and p25-N, respectively, using primers JR364/JR365 and PstI/KpnI digestion. pbpA2 was cloned into pKT25 using oligonucleotides SHW153/SHW154 and XbaI/KpnI to create pJR250. pbpB1 was amplified with primers SHW157/SHW158 and cloned with SmaI/EcoRI into pKT25, yielding pSH236. pbpB2 was amplified with primers SHW155/SHW156 and cloned into pKT25 after PstI/BamHI digestion, yielding plasmid pSH235. pbpB3 was amplified using primers SHW159/160 and cloned by PstI/BamHI digestion into pKT25, resulting in pSH237. Finally, pSH484 was obtained by removing the N-terminal domain from gpsB present on plasmid pSH226 in a PCR with the primer pair SHW752/SHW753.
Plasmids encoding the respective genes fused to the N- or C-termini of the T18- or the T25-fragment of the Bordetella pertussis adenylate cyclase were co-transformed into E. coli BTH101. Transformants were selected on LB agar plates containing ampicillin (100 µg−1mL−1), kanamycin (50 µg−1mL−1), X-Gal (40 µg−1mL−1) and IPTG (0.1 mM) and photographs were taken after 24 and 48h growth at 30 °C. The BACTH experiments in Fig. 2b and Supplementary Figure 3C, D were repeated at least twice.
The same procedures outlined above were followed for the BACTH using B. subtilis GpsB except that co-transformants were grown on nutrient agar plates11, not LB, before imaging. These experiments were replicated reproducibly at least three times and a representative image is provided in Fig. 3c. Reciprocal tests for some interactions do not show the same results presumably because of the different expression levels for the individual fusion constructs as pKT25 is a low copy number plasmid whereas pUT18C is a high copy number plasmid.
S. pneumoniae strain construction
Mutant strains containing antibiotic resistance markers were constructed by transformation of CSP-1 induced competent pneumococcal cells with linear DNA amplicons synthesised by overlapping fusion PCR64. Mutant constructs were confirmed by PCR and DNA sequencing of chromosomal regions corresponding to the amplicon region used for transformation.
Ectopic expression of wildtype gpsB with a Zn2+-inducible promoter (IU11286) was achieved by insertion into bgaA to create a ΔbgaA::tet-PZn-gpsB+ construct using PCR-fused fragments. To investigate whether GpsB with allele changes of Y23A, V28A, D29A L32A D33A or I36A were functional, and given that gpsB is essential for the growth of S. pneumoniae D39, a merodiploid D39 Δcps ΔgpsB<>aad9//ΔbgaA::PZn-gpsB+ strain (IU11388) was constructed by transforming ΔgpsB<>aad9 into IU11286 in the presence of 0.5 mM ZnCl2 + 0.05 mM MnSO4, which is required in the media to counteract the Zn2+ toxicity57,64. For selection of gpsB alleles, a Pc-erm cassette was fused to the 3ʹ end of gpsB with the desired mutations in the native chromosomal locus. The gpsB allele change-Pc-erm amplicon was transformed into IU11388 in the presence of erythromycin and 0.5 mM ZnCl2 + 0.05 mM MnSO4. The Pc-erm cassette inserted at the 3ʹ end of gpsB+ imparted no discernible growth phenotype compared to a wild-type strain (Fig. 5b). The growth of merodiploid strain IU11388, and strains containing gpsB with allele changes complemented with ectopic expression of WT gpsB (IU12361, IU12363, IU12440, IU12612, IU12615, IU13121), or gpsB with allele changes linked to a FLAG tag complemented with ectopic expression of WT gpsB (IU13141, IU13364, IU13366, IU13368, IU13370, IU13372 and IU13374) were performed in the presence of 0.5 mM ZnCl2 + 0.05 mM MnSO4. Transformation of ∆pbp1a::Pc-erm into IU11388 (ΔgpsB<>aad9//ΔbgaA::tet-PZn-gpsB+) and IU14365 (∆pbp2a markerless//ΔbgaA::kan-PZn-pbp2a+) was performed in the presence of 0.4 mM ZnCl2 + 0.04 mM MnSO4. Strains containing markerless pbp2a alleles in the native chromosomal locus were constructed using Janus cassette allele replacement65 via an rpsL1 strain containing a ∆pbp2a::Pc-[kan-rpsL+] construct (IU7853).
Transformation of a Δpbp1a amplicon into S. pneumoniae strains
The function of the mutated pbp2a alleles was evaluated by transformations of 30 ng of a ∆pbp1a::Pc-erm amplicon, obtained from strain E177 and containing ≈1 kb of flanking chromosomal DNA, into strains harbouring pbp2a mutations. Transformants were visualised after 24h incubation at 37 °C in 5% CO2. The numbers of colonies were normalised to 1 mL of transformation mixture.
Growth and microscopy of S. pneumoniae strains
Strains IU1945 (wild-type parent) and IU11286 (gpsB+//ΔbgaA::PZn-gpsB+) were inoculated from frozen glycerol stocks into BHI broth, serially diluted, and incubated for <13h statically at 37 °C in an atmosphere of 5% CO2. For IU11388 (∆gpsB//ΔbgaA::PZn-gpsB+) and all gpsB allele exchange strains complemented with bgaA::PZn-gpsB+, 0.5 mM ZnCl2 + 0.05 mM MnSO4 (+Zn) was added to BHI broth cultures to enable the ectopic expression of gpsB64. The next day, cultures at OD620 ≈ 0.1–0.4 were diluted to OD620 ≈ 0.003 in BHI broth lacking (−Zn) or containing (+Zn) 0.5 mM ZnCl2 + 0.05 mM MnSO4 and cultured under the same conditions. Growth was monitored turbidimetrically every 45 min to 1 h with a Genesys 2 spectrophotometer (Thermo Scientific). At 3.75 h after dilution into BHI broth with or without Zn/Mn supplement, cell pellets were obtained from 1 mL of culture, and resuspended in 100 µL of 4% paraformaldehyde. Cells were fixed for 15 min at RT and the tubes with fixed cells were left on ice until microscopy. For microscopic analyses, samples (1.5 μL) were taken and examined using a Nikon E-400 epifluorescence phase-contrast microscope. Growth and microscopy of strains containing pbp2a alleles were performed as above, but with no Zn/Mn addition to the BHI broth. Microscopy was performed at OD620 ≈ 0.15.
Growth and microscopy experiments of IU14381 (∆pbp2a//PZn-pbp2a+ ∆pbp1a) and IU14383 (ΔgpsB//PZn-gpsB+ ∆pbp1a) were performed as for IU11388 (∆gpsB//PZn-gpsB+) except that overnight growths were carried out in BHI broth containing 0.2 mM ZnCl2 + 0.02 mM MnSO4 for IU14381, and 0.4 mM ZnCl2 + 0.04 mM MnSO4 for IU14383. Cultures were diluted into the same media as the overnight media for +Zn condition and into BHI broth without Zn/Mn for the −Zn condition. All growth measurements were reproducible and were repeated at least three times. The mean doubling time and the standard error of the mean ( ± ) are reported in Supplementary Figure 5B.
Western blotting and immunodetection of S. pneumoniae strains
To confirm the expression of gpsB with Y23A, V28A, D29A, L32A, D33A or I36A allele exchanges, western blot analyses were performed of strains constructed with gpsB-FLAG or gpsB variant-FLAG at the native site complemented by ectopic Zn-dependent expression of WT gpsB. 0.5 mM ZnCl2 + 0.05 mM MnSO4 (+Zn) was added to all BHI broth cultures to enable the ectopic expression of gpsB, and were diluted to OD620 ≈ 0.003 and cultured under the same conditions to OD620 ≈ 0.15 to 0.2. Under the +Zn condition, all FLAG-tagged gpsB allele exchange strains grew identically to WT gpsB+ strain and to the untagged gpsB allele exchange strains. A parallel set of cultures was performed using BHI broth without addition of ZnCl2 and MnSO4 to confirm that FLAG-tagged gpsB allele strains grew identically to their untagged counterparts. Aliquots of 1.8 mL were centrifuged (5 min, 16,000 × g at 4 °C), and cell pellets were washed once with PBS. The supernatant was discarded and the pellets were frozen on dry ice for 5 min. The pellets were then thawed for 5 min at RT, resuspended in 80 µL of prewarmed 37 °C SEDS lysis buffer (0.1% deoxycholate, 150 mM NaCl, 0.2% SDS, 15 mM EDTA pH 8.0)24. Samples were incubated on a 37 °C shaking block at 300 rpm for 15 min, and were vortexed vigorously twice during the 15 min incubation. Total protein concentrations were determined using a Bio-Rad DCTM protein assay kit. 4 µg of total protein was loaded per lane on a 4–15% precast gradient SDS-PAGE gel (Bio-Rad), subjected to electrophoresis and transferred to a nitrocellulose membrane. FLAG-tagged GpsB proteins were detected with an anti‐FLAG rabbit polyclonal antibody (Sigma, F7425, 1:1400 dilution) as primary antibodies, and ECL anti-rabbit IgG horseradish peroxidase linked whole antibody as secondary (dilution 1:10,000). Chemiluminescent signals in protein bands were detected with an IVIS imaging system. MreC was detected with anti-MreC antibodies66 by relabelling the nitrocellulose membrane after detection with anti-FLAG.
Western detection of PBP2a in various PBP2a variant strains was performed essentially as above with exponentially growing cultures in BHI broth. 4 µg of total protein was loaded per lane onto a 10% precast SDS-PAGE gel (Bio-Rad). Electrophoresis was carried out for 1.5 h to allow separation of SpPBP2a from a non-specific band. Anti-SpPBP2a serum67 was used at a dilution of 1:5000.
The anti-serum against purified, recombinant SpGpsB1-63 was characterised by preparing lysates from gpsB+ divIVA+ WT (strain IU1945), ΔgpsB (IU6442) or ΔdivIVA (IU8496) strains as described above. 4 μg of total protein was loaded per lane on a 4–20% precast gradient SDS-PAGE gel (Bio-Rad), and anti-GpsB serum from rabbit Ab-1432 was used at a dilution of 1:2000.
Co-immunoprecipitation (Co-IP) of S. pneumoniae strains
Co-IP experiments of S. pneumoniae FLAG-tagged strains were performed with the use of anti-FLAG magnetic beads24. Lysates were obtained from cultures grown exponentially at 37 °C in an atmosphere of 5% CO2 in 400 mL of BHI to OD620 ≈ 0.25–0.4024. Cell pellets were washed once with 30 mL of 1X PBS (4 °C) and resuspended in 19.8 mL 1× PBS (4 °C). About 200 μL of 10% (vol per vol) paraformaldehyde solution (EMS) were added for crosslinking to a final concentration of 0.1% (vol per vol). Mixtures were incubated at 37 °C in an air incubator for 1 h. Cross‐linking reactions were quenched by the addition of 4 mL 1.0 M glycine followed by incubation at 25 °C for 10 min. Cells were collected by centrifugation (16,500 × g for 5 min at 4 °C). Pellets were washed once with 20 mL cold 1× PBS (4 °C) and resuspended in 2 mL of cold lysis buffer (50 mM Tris‐HCl pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton X100 (vol per vol)) with 1 tablet of protease inhibitor (ThermoFisher Scientific, 78429) freshly added per 10 mL of lysis buffer. The suspension was transferred into two lysing matrix B tubes (MP Biomedicals) with 1 mL in each tube. The tubes were shaken ten times in a FastPrep homogenizer (4×, 5 min on ice, 3×, 5 min on ice and 3×) with 6.0 M s−1 for 40 s each at 4 °C. Cell debris and lysing matrix from tubes were removed by centrifugation at 16,000 × g for 5 min at 4°C. The protein concentration of each sample was determined by Bio‐Rad DCTM protein assay (Bio‐Rad). About 1 mL of lysate with similar amounts of total protein (5–7 mg−1mL−1) was added to tubes with 50 μL of anti‐FLAG magnetic beads (Sigma, M8823). The same amount of protein was loaded onto the beads for strains expressing FLAG‐tagged proteins and the corresponding control strains lacking FLAG‐tagged proteins in each experiment. The tubes were rotated for 2 h at 4 °C. The beads were washed three times with 1 mL of lysis buffer (4 °C) with 10 min incubation at 4 °C each time. FLAG‐tagged proteins were eluted from the beads by incubation with 100 μL of FLAG elution solution (150 ng 3X FLAG peptide per μL) (Sigma, F4799) for 30 min at 4 °C. A volume of 100 μL of the elution and of the original lysate added to magnetic beads (input) were separately mixed with 100 μL 2× Laemmli sample buffer (Bio‐Rad) containing 5% (vol per vol) β‐mercaptoethanol (Sigma) and heated at 95 °C for 1 h to break the cross‐links. A volume of 40 µL of each elution sample mixed with 2× sample buffer was loaded on each lane of a 4–15% precast SDS-PAGE gel and subjected to western blotting using affinity-purified SpPBP2x polyclonal antibodies (dilution 1:10,000)67 or an anti-GpsB antibody as the primary antibodies.
S. pneumoniae BACTH assay
The target genes were amplified by the PCR from S. pneumoniae D39 chromosomal DNA. PCR fragments for pbp2a, mreC, pbp1a, pbp2x and pbp2b were purified, digested with appropriate restriction enzymes and cloned into the corresponding sites of the pKT25/pUT18C vectors to generate plasmids encoding the corresponding hybrid proteins fused at the C-terminal ends of the T25 and T18 fragments, respectively. Plasmids pKNT25-gpsB/pUT18-gpsB, pKNT25-stkP/pUT18-stkP and pKNT25-ezrA/pUT18-ezrA were already constructed24. The mutated and truncated gpsB and pbp2a alleles were amplified from their respective DNA templates and the corresponding PCR products cloned into the corresponding sites of the BACTH vectors as described above for the wild-type alleles. E. coli DH5α transformants were selected on LB agar plates containing ampicillin (100 μg−1mL−1) or kanamycin (50 μg−1mL−1) and 0.4% glucose to repress leaky expression63.
BACTH experiments, each pair of plasmids was co-transformed into the E. coli cya- strain BTH101 and co-transformation mixtures were spotted onto LB agar plates supplemented with ampicillin (100 mg−1mL−1), kanamycin (50 mg−1mL−1) and X-Gal (40 μg−1mL−1), followed by incubation at 30 °C. Plates were inspected and photographed after 24 and 40 h. Plasmid pairs pKNT25/pUT18 and pKT25-zip/pUT18C-zip were used as negative and positive controls, respectively. All experiments were repeated at least twice.
Plasmid construction for recombinant protein and peptide work
(i) GpsB: All mutagenesis was undertaken by the Quikchange protocol; where mutagenesis reactions failed to generate a DNA fragment of the expected size, a modification of the Quikchange protocol with two separate PCR steps was used instead69. Glu17Ala, Asp31Ala and Asp35Ala mutations were introduced to the BsGpsB1-68 expression construct plasmid19 by Quikchange. The BsGpsB5-64 construct was prepared by PCR amplification of gpsB from B. subtilis 168 genomic DNA using primers BsGpsB5start and BsGpsB64stop. The PCR product was ligated between the NdeI and XhoI sites of modified pET15b (M26L DJ-1 TEV site pET15b; gift from Mark Wilson, Addgene) with the nucleotides encoding the thrombin cleavage site replaced by a sequence encoding a TEV cleavage site. The SpGpsB1-63 expression construct (pET28SpGpsB1-63) was prepared by PCR amplification of gpsB from S. pneumoniae R6 genomic DNA with primers SpGpsB5ndeI and SpGpsB3XhoI. This PCR product was ligated between the NdeI and XhoI sites of pET28a, and a stop codon was introduced by Quikchange mutagenesis in place of the codon for Pro64. The SpGpsB4-63 expression construct was generated in two steps; first the ORF encoding residues 1–63 of SpGpsB was amplified with primers SpGpsBM11NcoI and SpGpsBM11XhoI using plasmid pET28SpGpsB1-63 as the template. This DNA fragment was then ligated between the NcoI and XhoI sites of pET11M70 before the codons for residues 1–3 of SpGpsB were deleted by Quikchange mutagenesis with primers SpGpsB1to3del5 and SpGpsB1to3del3.
(ii) PBP peptides; Ser16Cys, Ser7Ala, Arg8Ala, Ala10Pro, Arg11Ala and Arg28Ala mutations in BsPBP11-32 fused to maltose binding protein (MBP)19 were introduced by Quikchange mutagenesis. Plasmids expressing LmPBPA11-20 and SpPBP2a23-45 peptides fused to MBP were prepared by PCR amplifying the relevant ORFs from L. monocytogenes strain EGD-e and S. pneumoniae R6 genomic DNA with primers LmPBPA1ncoI5, LmPBPA1xhoI3, SpPBP2AncoI5 and SpPBP2AxhoI3, respectively. The subsequent PCR fragments were digested with NcoI and XhoI for ligating into similarly restricted pMAT11, a modified version of pHAT470. Ser19 and Lys21 in LmPBPA1 were mutated in a single step by Quikchange to cysteine and stop codons, respectively; similarly Quikchange was used to change SpPBP2a Gly43 and Arg46 to cysteine and stop codons, respectively. Further mutations in LmPBPA1 were also made by Quikchange; the Arg8AlaSer16Arg double mutant was prepared in two successive steps with the Arg8Ala mutation made first, followed by Ser16Arg.
The SpPBP2x1-29 construct was prepared in several steps. First, nucleotides 1–1165 of pbp2x were PCR amplified in two steps: an initial fragment generated from S. pneumoniae R6 genomic DNA with primers SpPBP2x5/SpPBP2x3st1 was used as a template for further PCR amplification with SpPBP2x5/SpPBP2x3st2 primers; this fragment was then ligated into the NcoI and XhoI sites of pMAT11. Second, cysteine, serine and stop codons were introduced by Quikchange in place of residues 30–32 with primers SpPBP2xS30CL31SL32STOP5 and SpPBP2xS30CL31SL32STOP3. A second Quikchange step (primers SpPBP2xinsgly5, SpPBP2xinsgly3) introduced extra codons encoding a GSG sequence after the TEV cleavage to improve cleavage of the MBP-PBP2x1-29 fusion protein.
(iii) YpbE and YrrS; The construct for expressing YpbE130-240 was prepared in two steps; first, the ORF encoding residues 80-240 of YpbE was PCR amplified from genomic DNA and ligated between the NdeI and XhoI sites of pET28a. Second, the ORF encoding the N-terminal His-tag and residues 80–129 of YpbE was subsequently deleted by PCR amplification of the entire plasmid with primers YpbEtruncatencoi5 and YpbEtruncatencoi3, followed by NcoI restriction digestion and religation of the DNA with T4 DNA ligase. The construct for expressing YrrSΔ13-16 was prepared by amplifying the yrrS gene from B. subtilis genomic DNA with flanking NdeI and XhoI sites and then ligating the PCR fragment between the same restriction sites in pET28a. The region coding for residues 13–16 of YrrS was subsequently deleted by Quikchange mutagenesis.
Recombinant protein purification
(i) GpsB; BsGpsB1-68 proteins used for FP assays were purified by Ni-NTA affinity chromatography followed by proteolytic removal of the His6-tag by thrombin and subsequent size exclusion chromatography19. The same protocol was used for SpGpsB1-63 except that an ammonium sulphate precipitation step was added after thrombin removal of the His6-tag. Ammonium sulphate was added to the thrombin-cleaved protein to a final concentration of 2.34 M (60% saturation at 0 °C) by adding a stock solution of 100% saturated ammonium sulphate that was prepared by adding solid ammonium sulphate directly to 50 mM Tris.HCl (pH 8), 300 mM NaCl, 10 mM imidazole. After stirring for 30 min at 4 °C the mixture was centrifuged at 19,000 × g for 20 min and solid ammonium sulphate then added to the supernatant to a final concentration of 2.8 M. After stirring at 4 °C for another 30 min the mixture was again centrifuged 19,000 × g for 20 min and further ammonium sulphate added to a final concentration of 3.5 M. After 30 min stirring the mixture was again centrifuged at 19,000 × g and the pellet resuspended in 10 mM Tris.HCl (pH 8), 250 mM NaCl before further purification by gel filtration19. BsGpsB5-64, BsGpsB5-64Lys32Glu and SpGpsB4-63 were expressed and purified by a similar protocol except that TEV protease, rather than thrombin, was used to remove the N-terminal His6-tag by overnight cleavage with TEV (1:50 ratio of TEV:GpsB) at 4 °C in a buffer of 50 mM Tris.HCl (pH 8.0), 300 mM NaCl, 250 mM imidazole, 1 mM DTT. The cleaved protein was then dialysed against 20 mM Tris.HCl (pH 8.0), 200 mM NaCl, 10 mM imidazole and manually passed over a 5 mL Ni-NTA superflow cartridge (Qiagen) to remove TEV and uncleaved proteins. The flow through from the column was concentrated and loaded onto a Superdex 75 XK16/60 (GE Healthcare) gel filtration column equilibrated in 10 mM HEPES.NaOH (pH 8.0), 100 mM NaCl.
(ii) PBP peptides; The PBP peptides, generated as His-tagged MBP-fusion proteins, were purified by standard Ni-NTA affinity chromatography procedures19. The purified MBP-PBP fusions at 5 mg−1mL−1 were fluorescently labelled in a buffer of 50 mM Tris.HCl (pH 7.0), 300 mM NaCl, 250 mM imidazole by mixing with either TAMRA-maleimide (Santa Cruz Biotechnology) or with fluorescein maleimide (Vector laboratories) to final concentrations of 625 μM and 1 mM from 25 and 40 mM stocks made in DMSO, respectively. After overnight incubation at 4 °C the labelling reaction was exchanged into a buffer of 10 mM Tris.HCl (pH 8.0), 250 mM NaCl, cleaved with TEV protease (1:50) and the labelled peptide separated from the MBP fusion partner by passing through a 3 kDa molecular mass cutoff centrifugal ultrafiltration device19.
The BsPBP11-17, LmPBPA11-15 and SpPBP2a27-40 peptides used for crystallisation and circular dichroism were synthesised chemically (Protein and Peptide Research Ltd, UK and Severn Biotech, UK). The fluorescein-labelled BsYpbE1-21 and SpPBP2b1-17 peptides used in FP assays were also synthesised chemically (Protein and Peptide Research Ltd); the SpPBP2b1-17 peptide encompasses the first seventeen amino acids of SpPBP2b from strain R6, based on the annotated sequence in the NCBI database (accession NP_359110).
(iii) BsPBP1 proteins; Recombinant BsPBP1 proteins were purified from E. coli membrane pellets19 resuspended in a buffer of 50 mM HEPES.NaOH pH 7.5, 500 mM NaCl, 3 mM MgCl2, 0.3 mM DTT supplemented with Roche complete EDTA-free protease inhibitor cocktail, lysozyme and DNAase. The lysate was clarified by centrifugation at 10,000 × g for 20 min and the supernatant centrifuged further for 1 h at 4 °C at 100,000 × g. The membrane pellet was resuspended in lysis buffer supplemented with 15% glycerol, 0.2 mM AEBSF, 0.5 mM DTT and Roche complete EDTA-free protease inhibitor. The resuspended membranes were ultracentrifuged for 1 h at 4 °C at 100,000 × g and the membrane pellet resuspended in resuspension buffer (50 mM HEPES.NaOH pH 7.5, 500 mM NaCl, 3 mM MgCl2, 2% reduced Triton X-100, 15% glycerol, 10 mM β-mercaptoethanol). After 2 h stirring at 4 °C the extract was ultracentrifuged at 100,000 × g for 30 min at 4 °C. The supernatant was supplemented with imidazole to 5 mM then incubated for 2 h at 4 °C with Sigma His select resin. This mixture was then passed through a gravity column and the resin washed with resuspension buffer supplemented with 5 mM imidazole and then BsPBP1 proteins were eluted with resuspension buffer containing 250 mM imidazole. The His6-tag was removed by thrombin treatment, before the digest was purified by Superdex S200 size exclusion chromatography in a buffer of 10 mM HEPES.NaOH pH 7.5, 500 mM NaCl, 3 mM MgCl2, 0.1% reduced Triton X-100, 2% glycerol. Residual uncleaved BsPBP1 proteins were removed by incubation with His select resin before concentraing and snap-freezing in liquid nitrogen.
(iv) YpbE; The YpbE130-240 protein and its S132C variant were expressed in BL21(DE3) cells by induction overnight at 17 °C with 0.5 mM IPTG when the culture had reached an optical density at 600 nm of 0.6. The cell pellet from a 2 L culture was resuspended in 70 mL of 50 mM Tris.HCl (pH 8), 10 mM NaCl, supplemented with 1 mL of 25× working concentration of Roche Complete EDTA free protease inhibitor cocktail and lysed by sonication. The lysate was then passed over a XK16 column packed with a 25 mL bed volume of Fast Flow Q-Sepharose and the flow through from the column then mixed with an equal volume of lysis buffer supplemented with saturated ammonium sulphate at room temperature. After 30 min stirring at 4 °C the mixture was centrifuged at 19,000 × g for 20 min and the supernatant mixed with saturated ammonium sulphate to a final concentration of 60%. This mixture was stirred for 30 min at 4 °C then centrifuged for 20 min at 19,000 × g. The supernatant was mixed further with ammonium sulphate to a final concentration of 80%. After 30 min stirring at 4 °C the mixture was centrifuged for 30 min at 19,000 × g then the pellet resuspended in 5 mL 10 mM MES buffer pH 6. The resuspended pellet was then loaded onto a 5 mL Fast Flow HiTrap SP column (GE Healthcare) equilibrated in 25 mM MES.NaOH pH 6 and the protein eluted with a linear gradient of 0–500 mM NaCl. Fractions containing YpbE130-240 were pooled, concentrated and loaded onto a Superdex 75 XK16/60 gel filtration column (GE Healthcare) pre-equilibrated in PBS.
(v) YrrS; YrrSΔ13-16 was expressed in BL21(DE3) cells by overnight induction at 20 °C with 0.4 mM IPTG. The pellet from a 2 L culture was resuspended in 25 mL 50 mM Tris.HCl (pH 8), 500 mM NaCl supplemented with 0.5 mg−1mL−1 lysozyme, 2 µg−1mL−1 DNAase and Roche Complete protease inhibitor cocktail. The cell suspension was lysed by sonication, and centrifuged at 10,000 × g for 20 min at 4 °C to remove intact cells. The supernatant was centrifuged at 100,000 × g at 4 °C for 1 h and the membrane pellet was resuspended in 15 mL 50 mM Tris.HCl (pH 8), 500 mM NaCl 10% (vol per vol) glycerol and centrifuged again for 1 hr at 100,000 × g at 4 °C. The membrane pellet was frozen in liquid nitrogen and stored at −80 °C. The membrane pellet was resuspended in 10 mL of 50 mM Tris.HCl (pH 8), 500 mM NaCl, 2% reduced Triton X-100, 5 mM imidazole supplemented with Roche Complete EDTA free protease inhibitor cocktail. After 3 h gentle agitation at 4 °C the membrane suspension was centrifuged at 4 °C at 100,000 × g for 30 min. A 0.4 mL bed volume of Ni-NTA Sepharose (Qiagen) was added to the supernatant and mixed overnight at 4 °C in a tube roller before filtering through an empty gravity column. The Ni-NTA beads were washed three times with 4 mL of 50 mM Tris.HCl (pH 8), 500 mM NaCl, 0.1% reduced Triton X-100, 5 mM imidazole and YrrSΔ13-16 was eluted from the column with 4 mL of the same buffer supplemented with 250 mM imidazole. The eluate was concentrated to 0.5 mL and loaded onto a Superdex 200 HR10/300 gel filtration column (GE Healthcare) equilibrated in 10 mM Tris.HCl (pH 8), 250 mM NaCl, 0.1% reduced Triton X-100.
All recombinant proteins or peptides were concentrated and flash frozen in small aliquots in liquid nitrogen and stored at −80 °C.
Crystallisation and structure determination
Successful crystallisation of the N-terminal domains of GpsB proteins was improved by truncation of a few amino acids from both N- and C-termini, which are too far from the PBP binding site (25 and 45 Å, respectively) to affect binding and correspond to the minimum ordered portion of GpsB19. Co-crystallisation of BsGpsB5-64:BsPBP11-17, BsGpsB5-64Lys32Glu:LmPBPA11-15 and SpGpsB4-63:SpPBP2a27-40 followed the same procedure. Equal volumes of GpsB protein and PBP peptide, both in a buffer of 10 mM HEPES.NaOH (pH 8.0) and 100 mM NaCl, were mixed at final concentrations of 20 and 25 mg−1 mL−1, respectively. This mixture, corresponding to a 1:5 molar ratio of protein:peptide, was subjected to sparse matrix crystallisation screening at room temperature with 100 nL drops dispensed by a Mosquito (TTP Labtech) liquid handling robot into 96 well MRC crystallisation plates. Unbound SpGpsB4-63 was crystallised by the same procedure, at the same protein concentration, except for the absence of added peptide. Crystals of BsGpsB5-64:BsPBP11-17 grew from 0.1 M HEPES/MOPS pH 7.5, 12.5% MPD, 12.5% PEG 1000 12.5% PEG 3350, 0.03 M MgCl2, 0.03 M CaCl2 and were mounted in rayon loops and frozen directly in liquid nitrogen. Crystals of BsGpsB5-64Lys32Glu:LmPBPA11-15 grew from 0.1 M imidazole pH 8, 0.2 M zinc acetate, 20% PEG 3000 were transferred to a cryoprotectant comprising the well solution supplemented with 20% vol per vol PEG 300 and 2.5 mg−1 mL−1 of the LmPBPA11-15 peptide; after 3 min the crystals were mounted in rayon loops and frozen in liquid nitrogen. The SpGpsB4-63:SpPBP2a27-40 complex crystallised in 0.1 M Tris.HCl pH 8.5, 0.2 M lithium sulphate, 40% PEG400; crystals were mounted after 1 μL of the crystallisation mother liquor containing 2.5 mg−1mL−1 SpPBP2a27-40 peptide was layered over the crystallisation drop immediately prior to freezing directly in liquid nitrogen. Crystals of unbound SpGpsB4-63 were obtained from 0.1 M Tris.HCl pH 8.5, 0.2 M magnesium chloride, 20% PEG 8000 and were cryoprotected by direct transfer to a solution of the mother liquor supplemented with 20% PEG400 before mounting and freezing as above.
X-ray data were collected from crystals of unbound SpGpsB4-63 in house with a gallium METALJETTM (Bruker AXS GmbH) as X-ray source at a wavelength of 1.34 Å. All other diffraction data were collected at the DIAMOND synchrotron: for BsGpsB5-64:BsPBP11-17 data were collected on beamline I24 at a wavelength of 0.9686 Å; for BsGpsB5-64Lys32Glu:LmPBPA11-15 data were collected on beamline I04 at a wavelength of 0.9795 Å and for SpGpsB4-63:SpPBP2a27-40 data were collected on beamline I03 at a wavelength of 0.9762 Å. For BsGpsB5-64:BsPBP11-17 and SpGpsB4-63:SpPBP2a27-40 the diffraction images were indexed and integrated with XDS71 and scaled with AIMLESS72. For BsGpsB5-64Lys32Glu:LmPBPA11-15 the diffraction images were indexed and integrated with DIALS73, scaled in XDS71 and merged with AIMLESS72. For unbound SpGpsB4-63 the images were indexed, integrated, scaled and merged in Proteum 3 (Bruker AXS GmbH). For the three different GpsB:PBP complexes and the unbound SpGpsB4-63 the initial phases were obtained by molecular replacement (MR) in PHASER74. For BsGpsB5-64:BsPBP11-17 residues 5-64 from the structure of dimeric GpsB1-68 (PDBid 4UG319) were used as the search model for MR whereas for BsGpsB5-64Lys32Glu:LmPBPA11-15 the MR search model was the co-ordinates of the BsGpsB5-64:BsPBP11-17 complex with the peptide removed. For the unbound SpGpsB4-63 the search model for MR was the N-terminal domain of LmGpsB (PDB code 4UG119). For the SpGpsB4-63:SpPBP2a27-40 complex, the co-ordinates of the unbound SpGpsB4-63 protein were used as a search model for MR. In all cases the molecular replacement solutions were visualised and rebuilt in COOT75. The BsGpsB5-64Lys32Glu:LmPBPA11-15 and SpGpsB4-63:SpPBP2a27-40 complexes were refined in PHENIX.REFINE76 while the BsGpsB5-64:BsPBP11-17 complex and unbound SpGpsB4-63 were refined using REFMAC77. The Ramachandran plot statistics (favoured/allowed) are: BsGpsB5-64:BsPBP11-17 100%/0%; BsGpsB5-64Lys32Glu:LmPBPA11-15 99.2%/0.8%; SpGpsB4-63:SpPBP2a27-40 98.6%/1.4%, and unbound SpGpsB4-63 99.6%/0.4% allowed. Statistics for data collection and for the final refined models can be found in Table 1. Representative electron density maps for the peptide-bound structures are displayed in stereographic mode in Supplementary Figure 8.
Table 1.
Data collection and refinement statistics
| BsGpsB5-64:BsPBP11-17 | BsGpsB5-64Lys32Glu:LmPBPA11-15 | Unbound SpGpsB4-63 | SpGpsB4-63:PBP2a27-40 | |
|---|---|---|---|---|
| Data collection | ||||
| Space group | P 21 21 21 | P 1 21 1 | P 1 21 1 | C 1 2 1 |
| a, b, c (Å) | 31.5, 53.8, 85.9 | 26.6, 31.4, 81.0 | 39.1, 54.5, 61.6 | 83.1, 26.4, 65.9 |
| α, β, γ (°) | 90, 90, 90 | 90, 92.4, 90 | 90, 106.1, 90 | 90, 106.4, 90 |
| Resolution (Å) | 45.57–1.95 (2.00–1.95)a | 31.42–1.60 (1.63–1.60)a | 27.22–1.90 (1.94–1.90)a | 39.85–1.80 (1.84–1.80)a |
| Rpim | 0.059 (0.473) | 0.044 (0.386) | 0.041 (0.311) | 0.051 (0.269) |
| CC (1/2) (%) | 99.4 (72.0) | 99.7 (74.3) | 99.8 (78.0) | 97.6 (88.3) |
| I/σI | 8.8 (2.1) | 8.4 (1.8) | 13.9 (4.0) | 9.0 (2.2) |
| Completeness (%) | 100.0 (100.0) | 99.8 (99.9) | 100 (100) | 99.8 (99.9) |
| No. of observations | 78,385 (5459) | 63,406 (3019) | 497,944 (28,050) | 46,904 (2454) |
| No. of unique reflections | 11,206 (768) | 17,944 (902) | 19,776 (1353) | 13,052 (763) |
| Redundancy | 7.0 (7.1) | 3.5 (3.3) | 25.2 (20.7) | 3.6 (3.2) |
| Refinement | ||||
| Resolution (Å) | 45.57–1.95 | 29.29–1.60 | 26.01–1.90 | 39.85–1.80 |
| No. of reflections | 11,162 | 17,912 | 19,738 | 13,007 |
| Rwork/Rfree | 0.187/0.214 | 0.173/0.204 | 0.188/0.249 | 0.178/0.225 |
| No. of atoms | ||||
| Protein | 951 | 986 | 1919 | 999 |
| Peptide | 94 | 20 | — | 220 |
| Ligand/ion | 1 | 5/8 | — | 30/1 |
| Water | 48 | 106 | 260 | 87 |
| B-factors (Å2) | ||||
| Protein | 34.0 | 25.2 | 24.8 | 18.7 |
| Peptide | 39.3 | 36.0 | — | 32.5 |
| Ligand/ion | 23.7 | 30.9/36.5 | — | 57.9/12.1 |
| Water | 36.4 | 35.6 | 30.6 | 28.1 |
| R.m.s. deviations | ||||
| Bonds (Å) | 0.006 | 0.016 | 0.017 | 0.015 |
| Angles (°) | 0.673 | 1.372 | 1.399 | 1.393 |
aValues in parentheses refer to the highest resolution shell
FP assays
FP experiments were undertaken in a buffer of 10 mM Tris.HCl (pH 8.0), 250 mM NaCl, 0.1% reduced Triton X-100. The excitation wavelength was 540 nm and fluorescence emission was recorded above 590 nm for TAMRA-labelled peptides. The N-terminal domains of GpsB proteins BsGpsB1-68, LmGpsB1-73, SpGpsB1-63 were used in all experiments in preference to the full-length GpsB proteins due to the high solubility of the former, which facilitated achieving the high protein concentrations necessary to saturate peptide binding. The validity of the N-terminal domain as a substitute for full-length GpsB proteins in these experiments is supported by the comparable affinity of BsGpsB and BsGpsB1-68 for a labelled BsPBP1A1-32 peptide in FP experiments (Supplementary Table 1). Kd values ± the standard error, generated after simultaneously fitting all the binding data to a 1:1 interaction model are reported and there are three measurements at each data point in a titration.
SPR
All SPR experiments used a running buffer of 10 mM Tris.HCl (pH 8.0), 250 mM NaCl, 0.1% reduced Triton X-100. BsPBP1 and BsPBP117-914 were immobilised on the surface of a CM5 chip (GE Healthcare). For the BsPBP1/YrrSΔ13-16 titration 800 RUs of BsPBP1 were immobilised on the chip surface; for the BsPBP1/BsGpsB titration 1200 RUs of BsPBP1 were immobilised. YrrSΔ13-16, which lacks residues 13–16 in the cytoplasmic domain, was used in these experiments to minimise non-specific interactions with the chip matrix; binding of YrrSΔ13-16 to the reference surface was negligible and the deletion of residues 13–16 leaves the GpsB-binding motif intact. The YrrSΔ13-16 titration was carried out in single-cycle mode, without regenerating the surface between successive injections of protein, because harsh reagents were necessary for full surface regeneration. The fit of the single-cycle mode data to a 1:1 binding model was achieved with a U-value of 2 and a corresponding χ2 of 46.1.
CD analysis
CD spectra were recorded on a JASCO J-810 spectropolarimeter with a PTC-4235 Peltier temperature controller using 1 mm path length quartz cuvettes. For full wavelength scans a scan speed of 10 nm−1min−1 and a response time of 4 s were used, the final spectra were the average of at least four measurements. BsPBP11-32 CD spectra were recorded at 4 °C in a buffer of 20 mM sodium phosphate, pH 7.3 at a peptide concentration of 25 μM. Peptide helical contents were estimated from molar ellipticities at 222 nm using the equation helix = [θ]222 nm / (39,500(1–2.57/n))78 where n is the number of residues in the peptide. LmPBPA11-15 CD spectra were analysed at 40 μM in 20 mM sodium phosphate buffer, pH 7.8, for samples containing 0, 40 and 60% trifluoroethanol (TFE). For samples containing 80% TFE, 5 mM sodium phosphate buffer, pH 7.8, was used. The CD spectra of BsGpsB1-68 proteins were measured at a concentration of 4.5 μM in a buffer of 20 mM sodium phosphate (pH 7.8) 150 mM NaCl. For thermal melt experiments, a response time of 8 s and scan rate of 1 °C−1min−1 were used. All thermal melts were reversible as CD spectra recorded before and after the melts in the wavelength range 200–240 nm could be superimposed.
Peptidoglycan pulldown assay
PG pulldown assays were carried out in PBS buffer. B. subtilis PG (SigmaAldrich) was prepared as a 10 mg−1 mL−1 stock solution in PBS containing 0.02% sodium azide. 25 µg of protein were added to a 100 µL sample containing 66 µg of PG and the mixture incubated with gentle agitation at room temperature for 30 min. The mixture was centrifuged at 16,000 × g for 7 min, the supernatant removed and the pellet resuspended in 1 mL of ice-cold PBS and immediately centrifuged for 2 min at 16,000 × g. The pellet was resuspended in 1 mL of the appropriate buffer and centrifuged for 2 min at 16,000 × g. Finally the pellet was boiled for 3 min in 60 μL SDS-PAGE loading buffer before analysis by SDS-PAGE; the SDS-PAGE loading buffer lacked reducing agents for YpbE130-240Ser132Cys samples.
Electronic supplementary material
Acknowledgements
This work was supported by grants from the UK BBSRC (BB/M001180/1 to R.J.L.), the German DFG (HA 6830/1-1 and HA 6830/1-2 to S.H.), the American NIH (RO1GM113172, RO1GM114315 and RO1GM127715 to M.E.W). O.M. is funded by a Contribution to Basic Research (LR7/2007) from the Autonomous Region of Sardinia. F.C. is funded by the European Commission (International Training Network Train2Target, No. 721484) and Z.R. is funded by a UK BBSRC DTP studentship (BB/M011186/1). We thank the Diamond synchrotron light source for access to its beamlines and thank its staff for support during data collection. We are indebted to Waldemar Vollmer for critical reading of the manuscript and for supplying S. pneumoniae genomic DNA. We thank Helen Waller for technical assistance with SPR experiments, Dalia Denapaite, Regine Hakenbeck and Reinhold Bruckner for the supply of affinity-purified anti-SpPBP2x antibodies, Jiaqi Zheng and Amilcar J. Perez for co-IP analysis with the anti-SpPBP2x and anti-SpGpsB antibodies, and Simon Thorpe at the University of Sheffield for mass spectroscopy analysis of peptide samples.
Author contributions
R.M.C. and Z.J.R. conducted the biochemical and X-ray crystallography experiments and R.M.C., Z.J.R. and R.J.L. analysed these data. R.M.C. performed bioinformatics analyses. J.R. and S.H. conducted the Listeria BACTH screen and the fosfomycin resistance assays and analysed both these data. F.A.A. conducted the Bacillus BACTH screen, and F.A.A. and R.A.D. analysed these data. F.C. conducted the Streptococcus BACTH screen and F.C. and O.M. analysed these data. H.-C.T.T. conducted the Streptococcus phenotype and growth analyses and performed the Western experiments and H.-C.T.T., O.M. and M.W.E. analysed these data. R.M.C., H.-C.T.T, S.H., O.M. and R.J.L. wrote the first draft of the paper and all authors contributed to the final submitted manuscript.
Data availability
The crystallographic data that support the findings of this study are available in the PDB with the identifiers: 6GP7 for BsGpsB5-64:BsPBP11-17; 6GPZ for BsGpsB5-64Lys32Glu:LmPBPA11-15; 6GQN for SpGpsB4-63:SpPBP2a27-40; and 6GQA for unbound SpGpsB4-63. All the other data that support the findings of this study are available from the corresponding author upon request.
Competing interests
The authors declare no competing interests.
Footnotes
Journal peer review information: Nature Communications thanks the anonymous reviewers for their contributions to the peer review of this work. Peer Reviewer Reports are available.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary Information accompanies this paper at 10.1038/s41467-018-08056-2.
References
- 1.Vollmer W, Blanot D, de Pedro MA. Peptidoglycan structure and architecture. FEMS Microbiol. Rev. 2008;32:149–167. doi: 10.1111/j.1574-6976.2007.00094.x. [DOI] [PubMed] [Google Scholar]
- 2.Sauvage E, Kerff F, Terrak M, Ayala JA, Charlier P. The penicillin-binding proteins: structure and role in peptidoglycan biosynthesis. FEMS Microbiol. Rev. 2008;32:234–258. doi: 10.1111/j.1574-6976.2008.00105.x. [DOI] [PubMed] [Google Scholar]
- 3.Egan AJ, Vollmer W. The physiology of bacterial cell division. Ann. N. Y. Acad. Sci. 2013;1277:8–28. doi: 10.1111/j.1749-6632.2012.06818.x. [DOI] [PubMed] [Google Scholar]
- 4.Uehara T, Bernhardt TG. More than just lysins: peptidoglycan hydrolases tailor the cell wall. Curr. Opin. Microbiol. 2011;14:698–703. doi: 10.1016/j.mib.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Typas A, et al. Regulation of peptidoglycan synthesis by outer-membrane proteins. Cell. 2010;143:1097–1109. doi: 10.1016/j.cell.2010.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paradis-Bleau C, et al. Lipoprotein cofactors located in the outer membrane activate bacterial cell wall polymerases. Cell. 2010;143:1110–1120. doi: 10.1016/j.cell.2010.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fernandez P, et al. The Ser/Thr protein kinase PknB is essential for sustaining mycobacterial growth. J. Bacteriol. 2006;188:7778–7784. doi: 10.1128/JB.00963-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beilharz K, et al. Control of cell division in Streptococcus pneumoniae by the conserved Ser/Thr protein kinase StkP. Proc. Natl Acad. Sci. USA. 2012;109:E905–E113. doi: 10.1073/pnas.1119172109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cha JH, Stewart GC. The divIVA minicell locus of Bacillus subtilis. J. Bacteriol. 1997;179:1671–1683. doi: 10.1128/jb.179.5.1671-1683.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levin PA, Kurtser IG, Grossman AD. Identification and characterization of a negative regulator of FtsZ ring formation in Bacillus subtilis. Proc. Natl Acad. Sci. USA. 1999;96:9642–9647. doi: 10.1073/pnas.96.17.9642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Claessen D, et al. Control of the cell elongation-division cycle by shuttling of PBP1 protein in Bacillus subtilis. Mol. Microbiol. 2008;68:1029–1046. doi: 10.1111/j.1365-2958.2008.06210.x. [DOI] [PubMed] [Google Scholar]
- 12.Tavares JR, de Souza RF, Meira GL, Gueiros-Filho FJ. Cytological characterization of YpsB, a novel component of the Bacillus subtilis divisome. J. Bacteriol. 2008;190:7096–7197. doi: 10.1128/JB.00064-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flärdh K. Essential role of DivIVA in polar growth and morphogenesis in Streptomyces coelicolor A3(2) Mol. Microbiol. 2003;49:1523–1536. doi: 10.1046/j.1365-2958.2003.03660.x. [DOI] [PubMed] [Google Scholar]
- 14.Hempel AM, Wang SB, Letek M, Gil JA, Flärdh K. Assemblies of DivIVA mark sites for hyphal branching and can establish new zones of cell wall growth in Streptomyces coelicolor. J. Bacteriol. 2008;190:7579–7583. doi: 10.1128/JB.00839-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Santos-Beneit F, Roberts DM, Cantlay S, McCormick JR, Errington J. A mechanism for FtsZ-independent proliferation in Streptomyces. Nat. Commun. 2017;8:1378. doi: 10.1038/s41467-017-01596-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma X, Ehrhardt DW, Margolin W. Colocalization of cell division proteins FtsZ and FtsA to cytoskeletal structures in living Escherichia coli cells by using green fluorescent protein. Proc. Natl Acad. Sci. USA. 1996;93:12998–13003. doi: 10.1073/pnas.93.23.12998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bisson-Filho AW, et al. Treadmilling by FtsZ filaments drives peptidoglycan synthesis and bacterial cell division. Science. 2017;355:739–743. doi: 10.1126/science.aak9973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gamba P, Veening JW, Saunders NJ, Hamoen LW, Daniel RA. Two-step assembly dynamics of the Bacillus subtilis divisome. J. Bacteriol. 2009;191:4186–4194. doi: 10.1128/JB.01758-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rismondo J, et al. Structure of the bacterial cell division determinant GpsB and its interaction with penicillin binding proteins. Mol. Microbiol. 2016;99:978–998. doi: 10.1111/mmi.13279. [DOI] [PubMed] [Google Scholar]
- 20.Rismondo J, Bender JK, Halbedel S. Suppressor mutations linking gpsB with the first committed step of peptidoglycan biosynthesis in Listeria monocytogenes. J. Bacteriol. 2017;199:e00393–16. doi: 10.1128/JB.00393-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rismondo J, Wamp S, Aldridge C, Vollmer W, Halbedel S. Stimulation of PgdA-dependent peptidoglycan N-deacetylation by GpsB-PBP A1 in Listeria monocytogenes. Mol. Microbiol. 2018;107:472–487. doi: 10.1111/mmi.13893. [DOI] [PubMed] [Google Scholar]
- 22.Land AD, et al. Requirement of essential Pbp2x and GpsB for septal ring closure in Streptococcus pneumoniae D39. Mol. Microbiol. 2013;90:939–955. doi: 10.1111/mmi.12408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fleurie A, et al. Interplay of the serine/threonine-kinase StkP and the paralogs DivIVA and GpsB in pneumococcal cell elongation and division. PLoS Genet. 2014;10:e1004275. doi: 10.1371/journal.pgen.1004275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rued BE, et al. Suppression and synthetic-lethal genetic relationships of ΔgpsB mutations indicate that GpsB mediates protein phosphorylation and penicillin-binding protein interactions in Streptococcus pneumoniae D39. Mol. Microbiol. 2017;103:931–957. doi: 10.1111/mmi.13613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mobegi FM, et al. Deciphering the distance to antibiotic resistance for the pneumococcus using genome sequencing data. Sci. Rep. 2017;7:42808. doi: 10.1038/srep42808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eswara PJ, et al. An essential Staphylococcus aureus cell division protein directly regulates FtsZ dynamics. Elife. 2018;7:e38856. doi: 10.7554/eLife.38856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cleverley RM, et al. Subunit arrangement in GpsB, a regulator of cell wall biosynthesis. Microb. Drug Resist. 2016;22:446–460. doi: 10.1089/mdr.2016.0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berg KH, Straume D, Håvarstein LS. The function of the transmembrane and cytoplasmic domains of pneumococcal penicillin-binding proteins 2x and 2b extends beyond that of simple anchoring devices. Microbiology. 2014;160:1585–1598. doi: 10.1099/mic.0.078535-0. [DOI] [PubMed] [Google Scholar]
- 29.Morrison D. 14-3-3: modulators of signaling proteins? Science. 1994;266:56–57. doi: 10.1126/science.7939645. [DOI] [PubMed] [Google Scholar]
- 30.Pawson T, Scott JD. Signaling through scaffold, anchoring, and adaptor proteins. Science. 1997;278:2075–2080. doi: 10.1126/science.278.5346.2075. [DOI] [PubMed] [Google Scholar]
- 31.Bonifacino JS. The GGA proteins: adaptors on the move. Nat. Rev. Mol. Cell. Biol. 2004;5:23–32. doi: 10.1038/nrm1279. [DOI] [PubMed] [Google Scholar]
- 32.Doig AJ, Baldwin RL. N- and C-capping preferences for all 20 amino acids in alpha-helical peptides. Prot. Sci. 1995;4:1325–1336. doi: 10.1002/pro.5560040708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iqbalsyah TM, Doig AJ. Effect of the N3 residue on the stability of the alpha-helix. Prot. Sci. 2004;13:32–39. doi: 10.1110/ps.03341804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rismondo J, et al. Discrete and overlapping functions of peptidoglycan synthases in growth, cell division and virulence of Listeria monocytogenes. Mol. Microbiol. 2015;95:332–351. doi: 10.1111/mmi.12873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wei Y, McPherson DC, Popham DL. A mother cell-specific class B penicillin-binding protein, PBP4b, in Bacillus subtilis. J. Bacteriol. 2004;186:258–261. doi: 10.1128/JB.186.1.258-261.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Snel B, Bork P, Huynen MA. The identification of functional modules from the genomic association of genes. Proc. Natl Acad. Sci. USA. 2002;99:5890–5895. doi: 10.1073/pnas.092632599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Buist G, Steen A, Kok J, Kuipers OP. LysM, a widely distributed protein motif for binding to (peptido)glycans. Mol. Microbiol. 2008;68:838–847. doi: 10.1111/j.1365-2958.2008.06211.x. [DOI] [PubMed] [Google Scholar]
- 38.Wong JE, et al. Cooperative binding of LysM domains determines the carbohydrate affinity of a bacterial endopeptidase protein. FEBS J. 2014;281:1196–1208. doi: 10.1111/febs.12698. [DOI] [PubMed] [Google Scholar]
- 39.Mesnage S, et al. Molecular basis for bacterial peptidoglycan recognition by LysM domains. Nat. Commun. 2014;5:4269. doi: 10.1038/ncomms5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koo BM, et al. Construction and analysis of two genome-scale deletion libraries for Bacillus subtilis. Cell Syst. 2017;4:291–305. doi: 10.1016/j.cels.2016.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scheffers D, Errington J. PBP1 is a component of the Bacillus subtilis cell division machinery. J. Bacteriol. 2004;186:5153–5156. doi: 10.1128/JB.186.15.5153-5156.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alyahya SA, et al. RodZ, a component of the bacterial core morphogenic apparatus. Proc. Natl Acad. Sci. USA. 2009;106:1239–1244. doi: 10.1073/pnas.0810794106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.von Heijne G. Control of topology and mode of assembly of a polytopic membrane protein by positively charged residues. Nature. 1989;341:456–458. doi: 10.1038/341456a0. [DOI] [PubMed] [Google Scholar]
- 44.Kell CM, et al. Deletion analysis of the essentiality of penicillin-binding proteins 1A, 2B and 2X of Streptococcus pneumoniae. FEMS Microbiol. Lett. 1993;106:171–175. doi: 10.1111/j.1574-6968.1993.tb05954.x. [DOI] [PubMed] [Google Scholar]
- 45.Yang. X, et al. Structural basis for protein-protein interactions in the 14-3-3 protein family. Proc. Natl Acad. Sci. USA. 2006;103:17237–17242. doi: 10.1073/pnas.0605779103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Popham DL, Setlow P. Phenotypes of Bacillus subtilis mutants lacking multiple class A high-molecular-weight penicllin-binding proteins. J. Bacteriol. 1996;178:2079–2085. doi: 10.1128/jb.178.7.2079-2085.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leaver M, Errington J. Roles for MreC and MreD proteins in helical growth of the cylindrical cell wall in Bacillus subtilis. Mol. Microbiol. 2005;57:1196–1209. doi: 10.1111/j.1365-2958.2005.04736.x. [DOI] [PubMed] [Google Scholar]
- 48.Stein A, Aloy P. Contextual specificity in peptide-mediated protein interactions. PLoS ONE. 2008;3:e2524. doi: 10.1371/journal.pone.0002524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Frappier V, Duran M, Keating AE. PixelDB: protein-peptide complexes annotated with structural conservation of the peptide binding mode. Protein Sci. 2018;27:276–285. doi: 10.1002/pro.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Braselmann S, McCormick F. Bcr and Raf form a complex in vivo via 14-3-3 proteins. EMBO J. 1995;14:4839–4848. doi: 10.1002/j.1460-2075.1995.tb00165.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Steil L, Serrano M, Henriques AO, Völker U. Genome-wide analysis of temporally regulated and compartment-specific gene expression in sporulating cells of Bacillus subtilis. Microbiology. 2005;151:399–420. doi: 10.1099/mic.0.27493-0. [DOI] [PubMed] [Google Scholar]
- 52.de Boer PA, Crossley RE, Rothfield LI. A division inhibitor and a topological specificity factor coded for by the minicell locus determine proper placement of the division septum in E. coli. Cell. 1989;56:641–649. doi: 10.1016/0092-8674(89)90586-2. [DOI] [PubMed] [Google Scholar]
- 53.Figge RM, Divakaruni AV, Gober JW. MreB, the cell shape-determining bacterial actin homologue, co-ordinates cellwall morphogenesis in Caulobacter crescentus. Mol. Microbiol. 2004;51:1321–1332. doi: 10.1111/j.1365-2958.2003.03936.x. [DOI] [PubMed] [Google Scholar]
- 54.Fleurie A, et al. MapZ marks the division sites and positions FtsZ rings in Streptococcus pneumoniae. Nature. 2014;516:259–262. doi: 10.1038/nature13966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ulrych A, et al. Characterization of pneumococcal Ser/Thr protein phosphatase phpP mutant and identification of a novel PhpP substrate, putative RNA binding protein Jag. BMC Microbiol. 2016;16:247. doi: 10.1186/s12866-016-0865-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stamsås GA, et al. Identification of EloR (Spr1851) as a regulator of cell elongation in Streptococcus pneumoniae. Mol. Microbiol. 2017;105:954–967. doi: 10.1111/mmi.13748. [DOI] [PubMed] [Google Scholar]
- 57.Zheng JJ, Perez AJ, Tsui HT, Massidda O, Winkler ME. Absence of the KhpA and KhpB (JAG/EloR) RNA-binding proteins suppresses the requirement for PBP2b by overproduction of FtsA in Streptococcus pneumoniae D39. Mol. Microbiol. 2017;106:793–814. doi: 10.1111/mmi.13847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fenton AK, et al. Phosphorylation-dependent activation of the cell wall synthase PBP2a in Streptococcus pneumoniae by MacP. Proc. Natl Acad. Sci. USA. 2018;115:2812–2817. doi: 10.1073/pnas.1715218115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kellogg, S. L. & Kristich, C. J. Convergence of PASTA kinase and two-component signaling in response to cell wall stress in Enterococcus faecalis. J. Bacteriol. 200, e00086-18 (2018). [DOI] [PMC free article] [PubMed]
- 60.Lanie JA, et al. Genome sequence of Avery’s virulent serotype 2 strain D39 of Streptococcus pneumoniae and comparison with that of unencapsulated laboratory strain R6. J. Bacteriol. 2007;189:38–51. doi: 10.1128/JB.01148-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sambrook, J., Fritsch, E. F. & Maniatis, T. Molecular Cloning: a Laboratory Manual (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, 1989).
- 62.Zheng L, Baumann U, Reymond JL. An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res. 2004;32:3115. doi: 10.1093/nar/gkh635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Karimova G, Pidoux J, Ullmann A, Ladant D. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc. Natl Acad. Sci. USA. 1998;95:5752–5756. doi: 10.1073/pnas.95.10.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tsui HC, et al. Suppression of a deletion mutation in the gene encoding essential PBP2b reveals a new lytic transglycosylase involved in peripheral peptidoglycan synthesis in Streptococcus pneumoniae D39. Mol. Microbiol. 2016;100:1039–1065. doi: 10.1111/mmi.13366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sung CK, Li H, Claverys JP, Morrison DA. An rpsL cassette, janus, for gene replacement through negative selection in Streptococcus pneumoniae. Appl. Environ. Microbiol. 2001;67:5190–5196. doi: 10.1128/AEM.67.11.5190-5196.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Land AD, Winkler ME. The requirement for pneumococcal MreC and MreD is relieved by inactivation of the gene encoding PBP1a. J. Bacteriol. 2011;193:4166–4179. doi: 10.1128/JB.05245-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhao G, Meier TI, Hoskins J, McAllister KA. Identification and characterization of the penicillin-binding protein 2a of Streptococcus pneumoniae and its possible role in resistance to β-lactam antibiotics. Antimicrob. Agents Chemother. 2000;44:1745–1748. doi: 10.1128/AAC.44.6.1745-1748.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Maurer PB, et al. Penicillin-binding protein 2x of Streptococcus pneumoniae: three new mutational pathways for remodelling an essential enzyme into a resistance determinant. J. Mol. Biol. 2008;376:1403–1416. doi: 10.1016/j.jmb.2007.12.058. [DOI] [PubMed] [Google Scholar]
- 69.Wang W, Malcolm BA. Two-stage PCR protocol allowing introduction of multiple mutations, deletions and insertions using QuikChange site-directed mutagenesis. BioTechniques. 1999;26:680–682. doi: 10.2144/99264st03. [DOI] [PubMed] [Google Scholar]
- 70.Peränen J, Rikkonen M, Hyvönen M, Kääriäinen L. T7 vectors with modified T7lac promoter for expression of proteins in Escherichia coli. Anal. Biochem. 1996;236:371–373. doi: 10.1006/abio.1996.0187. [DOI] [PubMed] [Google Scholar]
- 71.Kabsch, W. XDS. Acta Crystallogr.D66, 125–132 (2010). [DOI] [PMC free article] [PubMed]
- 72.Evans PR, Murshudov GN. How good are my data and what is the resolution? Acta Crystallogr. 2013;D69:1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Waterman DG, et al. Diffraction-geometry refinement in the DIALS framework. Acta Crystallogr. 2016;D72:558–575. doi: 10.1107/S2059798316002187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.McCoy A, et al. Phaser crystallographic software. J. Appl. Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. 2010;D66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Adams, P. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr.D66, 213–221 (2010). [DOI] [PMC free article] [PubMed]
- 77.Murshudov GN, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. 2011;D67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen YH, Yang JT, Chau KH. Determination of the helix and beta form of proteins in aqueous solution by circular dichroism. Biochemistry. 1974;13:3350–3359. doi: 10.1021/bi00713a027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The crystallographic data that support the findings of this study are available in the PDB with the identifiers: 6GP7 for BsGpsB5-64:BsPBP11-17; 6GPZ for BsGpsB5-64Lys32Glu:LmPBPA11-15; 6GQN for SpGpsB4-63:SpPBP2a27-40; and 6GQA for unbound SpGpsB4-63. All the other data that support the findings of this study are available from the corresponding author upon request.