Abstract
FLT3-ITD and FLT3-TKD are the most frequent tyrosine kinase mutations in acute myeloid leukemia (AML), with the former conferring a poor prognosis. We have recently revealed that FLT3-ITD confers resistance to the PI3K/AKT pathway inhibitors by protecting the mTORC1/4EBP1/Mcl-1 pathway through Pim kinases induced by STAT5 activation in AML. The proteasome inhibitor bortezomib has recently been reported as a promising agent for treatment of AML. Here, we show that the proteasome inhibitor bortezomib as well as carfilzomib induces apoptosis through the intrinsic pathway more conspicuously in cells transformed by FLT3-TKD than FLT3-ITD. Mechanistically, bortezomib upregulated the stress-regulated protein REDD1 and induced downregulation of the mTORC1 pathway more distinctively in cells transformed by FLT3-TKD than FLT-ITD, while overexpression of Pim-1 partly prevented this downregulation and apoptosis in FLT3-TKD–transformed cells. Genetic enhancement of the REDD1 induction or pharmacological inhibition of STAT5, Pim kinases, mTORC1, or S6K by specific inhibitors, such as pimozide, AZD1208, PIM447, rapamycin, and PF-4708671, accelerated the downregulation of mTORC1/Mcl-1 pathway to enhance bortezomib-induced apoptosis in FLT3-ITD–expressing cells, including primary AML cells, while overexpression of Mcl-1 prevented induction of apoptosis. Thus, FLT3-ITD confers a resistance to the proteasome inhibitors on AML cells by protecting the mTORC1/Mcl-1 pathway through the STAT5/Pim axis, and inhibition of these signaling events remarkably enhances the therapeutic efficacy.
Introduction
FLT3 is a receptor-tyrosine kinase expressed on hematopoietic progenitor cells and plays important roles in regulation of progenitor cell proliferation, survival, and differentiation [1], [2]. Internal tandem duplication (ITD) mutations in the juxtamembrane domain of FLT3 (FLT3-ITDs) are the most frequent mutations in acute myeloid leukemia (AML) and occur in 25%-30% of cases, while point mutations within the tyrosine kinase domain (FLT3-TKDs), such as the most frequent D835Y mutation, are found in 5%-10% of patients with AML. It is well established that FLT3-ITD but probably not FLT3-TKD confers a poor prognosis because of intrinsic therapy resistance with lower complete response rates and higher relapse rates, resulting in inferior disease-free and overall survivals [3], [4]. On the other hand, clinical trials with specific FLT3 tyrosine kinase inhibitors alone have so far shown only transient responses because of emergence of resistance mutations and through other various mechanisms in the case of FLT3-specific inhibitor quizartinib (AC-220) [5], [6].
FLT3-ITD as well as FLT3-TKD constitutively stimulates the various signaling pathways, such as the PI3K/Akt/mTOR and MEK/ERK pathways, thus leading to survival and proliferation of hematopoietic progenitor cells [1], [2]. Importantly, FLT3-ITD but not FLT3-TKD strongly activates STAT5, which contributes to enhance transforming potentials of FLT3-ITD as compared with FLT3-TKD [7], [8], [9]. The serine/threonine kinase mTOR is mainly activated downstream of the PI3K/Akt pathway forming two multiprotein complexes, mTORC1 and mTORC2, to regulate various cellular events, such as proliferation, apoptosis, and autophagy [10], [11]. On the other hand, mTOR is downregulated in response to nutrient depletion or a variety of cellular stressors, such as hypoxia and cellular damage. REDD1, also known as DDIT4 or RTP801, has been identified as a key stress-regulated protein acting as a potent inhibitor of mTORC1 [12]. Notably, mTORC1 plays a critical role in regulation of cap-dependent translation by phosphorylating 4EBP1 to release it from the mRNA m7-GTP cap-binding protein eIF4E, which interacts with the scaffolding protein eIF4G to initiate the formation of the translation-initiating complex eIF4F. This factor is required for the translation of mRNAs containing long 5′-UTRs, which are highly structured and have a high G + C content, such as those for c-Myc, Mcl-1, and cyclin D1 [13], [14], [15]. In addition, mTORC1 activates S6K, which phosphorylates eIF4B, as well as S6RP, to enhance cap-dependent translation by the eIF4F complex [16]. Mcl-1 is a highly unstable antiapoptotic Bcl-2 family member playing a crucial role in survival of hematopoietic progenitor cells and various malignant hematopoietic cells including AML cells [17]. We have previously found that FLT3-ITD confers resistance to the PI3K/Akt pathway inhibitors through the robust STAT5 activation to induce expression of Pim kinases, which protected the mTORC1 pathway to maintain the expression level of Mcl-1 [18], [19].
Proteasome inhibitors, such as bortezomib and carfilzomib, have been widely used for treatment of multiple myeloma and have shown excellent efficacies [20]. However, although a promising result has been reported for bortezomib combined with the standard combination chemotherapy for AML, bortezomib used alone has shown only modest effects in various studies [21]. Thus, many studies are currently investigating effects of bortezomib in combination with various agents, including tyrosine kinase inhibitors, histone deacetylase inhibitors, and hypomethylating agents, as well as chemotherapeutic agents. As for the mechanisms of action, inhibition of the transcription factor NFκB by bortezomib was previously implicated in its cellular effects in AML as well as myeloma cells [21]. More recently, bortezomib was shown to inhibit mTORC1, thus leading to induction of autophagy and degradation of FLT3-ITD [22]. However, mechanisms involved in inhibition of mTORC1 and effects on downstream events other than autophagy have remained to be known.
In the present study, we examine the responses of FLT3-ITD–transformed cells to proteasome inhibitors and the molecular mechanisms underlying the responses. The results obtained shed light on mechanisms involved in apoptosis induced by bortezomib in FLT3-ITD–positive AML cells and suggest that the STAT5/Pim pathway and the downstream mTORC1/Mcl-1 pathway would provide promising targets to enhance the effectiveness of therapies with proteasome inhibitors against this type of AML with poor prognosis.
Materials and Methods
Cells and Reagents
Murine IL-3–dependent 32Dcl3 cells, Ton.32D/FLT3-ITD (32D/ITD) or Ton.32D/FLT3-D835Y (32D/TKD) inducibly expressing FLT3-ITD or FLT3-D835Y, respectively, when cultured with doxycycline as well as 32D/TKD cells expressing the constitutively activated STAT5 mutant STAT5A1*6 (32D/TKD/STAT5A1*6), Pim1 (32D/TKD/Pim-1) or 32D/ITD cells overexpressing Mcl-1 (32D/ITD/Mcl-1) and their vector control cells (32D/TKD/pMXs, 32D/TKD/pMXs-IG, 32D/ITD/pMXs) have been described previously [18], [19], [23] and maintained in RPMI 1640 medium supplemented with 10% fetal calf serum (FCS) and 5 U/ml recombinant mIL-3 (PeproTech, Rocky Hill, NJ). Before analyses, these cells were cultured in medium containing doxycycline without IL-3 to proliferate dependent on FLT3-ITD or FLT3-TKD and independent of mIL-3. A clone of murine IL-3–dependent BaF3 cells expressing the reverse tet-transactivator Ton.BaF.1 was kindly provided by Dr. George Daley [24] and cultured in RPMI 1640 containing 10% FCS and 5 U/ml recombinant mIL-3. MV4-11 cells were purchased from ATCC and cultured in Iscove's modified Dulbecco medium containing 10% FCS. PLAT-A, an amphotropic virus packaging cell line, was kindly provided by Dr. Toshio Kitamura and maintained in DMEM supplemented with 10% FCS.
The proteasomal inhibitors bortezomib and carfilzomib as well as the FLT3 inhibitor quizartinib were purchased from LC laboratories (Woburn, MA). The Pim kinase inhibitor AZD1208 and the FLT3 inhibitor gilteritinib were from Active Biochem (Kowloon, Hong Kong). The Pim kinase inhibitor PIM447 was from Selleckchem (Houston, TX). The mTOR inhibitor PP242 was from Wako Pure Chemical Industries (Osaka, Japan). The S6K inhibitor PF-4708671 was from Adooq Bioscience (Irvine, CA). The pan-caspase inhibitor Boc-D-FMK was from Bio Vision (Milpitas, CA). DiOC6 was from Invitrogen (Carlsbad, CA). Doxycycline and propidium iodide were from Sigma Aldrich (St Louis, MO). Anti-Bax (TACS-2281) and anti-Bak (AM03) monoclonal antibodies were from Trevigen (Gaithersburg, MD) and Merck Millipore (Darmstadt, Germany), respectively. An APC-conjugated anti-mouse IgG antibody was from Biotech R&D systems (Minneapolis, MN). Anti–Bcl-xL (610211) was from BD Biosciences (San Jose, CA). An anti-REDD1 (PGI 10638) antibody was from Proteintech Group, Inc. (Rosemont, IL). The STAT5 inhibitor pimozide and antibodies against FLT3 (SC-479), STAT5A (SC-1081), and HSP-90 (SC-13119) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Antibodies against Caspase-3 (CS-9662), cleaved Caspase-3 (CS-9661), cleaved Caspase-9 (CS-9509), eIF4E (CS-2067), eIF4G (CS-1469), eIF4B (CS-3592), phospho-S422-eIF4BP (CS-3591), phospho-S406-eIF4BP (CS-8151), LC3B (CS-2775), phospho-Y694-STAT5 (CS-9359), 4EBP1 (CS-9644), non-phospho-T46-4EBP1 (CS-4923), phospho-T37/46-4EBP1 (CS-2855), phospho-S65-4EBP1 (CS-9451), phospho-T389-S6K (CS-9234), phospho-T308-Akt (CS-9275), Pim-2 (CS-4730), phospho-S235/S236-S6RP (CS4858), and Mcl-1 (CS-5453) were purchased from Cell Signaling (Beverly, MA). An anti-β-actin (A1978) antibody was from Sigma Aldrich.
Expression plasmids, transfection, and infection
Retroviral expression plasmids for FLT3-ITD and Mcl-1, pMXs-IG-FLT3-ITD and pMXs-puro-Mcl-1, respectively, were described previously [18], [23]. For construction of a retroviral expression plasmid for inducible expression of REDD1, pRevTRE-REDD1, the EcoRI/SalI fragment of pCMS-eGFP-RTP801 (Addgene, #65057) was first subcloned into the EcoRI/ XhoI site of pMXs-IG to give pMXs-IG-REDD1. The BamHI/SalI region of this plasmid coding REDD1 was then subcloned into the BamHI/HpaI site of pRevTRE (Clontech; Palo Alto, CA) to give pRevTRE-REDD1.
Ton.BaF3/pRevTRE-REDD1 (BaF3/REDD1) and Ton.BaF3/pRevTRE (BaF3/Cont) cells were obtained by infection of Ton.BaF3.1 cells with pRevTRE-REDD1 or pRevTRE, essentially as described previously [23], followed by selection with hygromycin. Ton.BaF3/pRevTRE-REDD1/pMXsIG-FLT3-ITD (BaF3/ITD/REDD1) and Ton.BaF3/pRevTRE/pMXsIG-FLT3-ITD (BaF3/ITD/Cont) cells were obtained by infection of BaF3/REDD1 or BaF3/Cont cells with the recombinant retrovirus obtained from PLAT-A transfected with pMXsIG-FLT3-ITD followed by sorting for GFP-expressing cells using the BD Biosciences Arial II flow cytometer (Mountain View, CA). MV4-11 cells overexpressing Mcl-1 were obtained by infection with pMXs-puro-Mcl-1 or pMXs-puro, as a control, followed by selection with puromycin.
Analyses of Cell Proliferation and Viability
Cell proliferation and viability were assessed by counting viable and nonviable cell numbers by the trypan blue-dye exclusion method. Cell viability was calculated by dividing number of viable cells by that of total cells. Viable cell numbers were also assessed by the sodium 3-[1-(phenylaminocarbonyl)-3,4-tetrazolium]-bis (4-methoxy-6-nitro) benzene sulfonic acid hydrate (XTT) colorimetric assay using the Cell Proliferation Kit II (Roche Molecular Biochemicals, Mannheim, Germany) according to the manufacturer's instructions. Viable cell numbers were also assessed by the sodium 3-[1-(phenylaminocarbonyl)-3,4-tetrazolium]-bis (4-methoxy-6-nitro) benzene sulfonic acid hydrate (XTT) colorimetric assay using the Cell Proliferation Kit II (Roche Molecular Biochemicals, Mannheim, Germany) according to the manufacturer's instructions.
Flow Cytometric Analyses
Flow cytometric analyses for cell cycle and apoptosis, mitochondrial membrane potential, conformational changes of Bax and Bak, and cleavage of Caspase-3 were performed essentially as described previously [18], [25]. Flow cytometric analysis of phosphorylation of S6RP was performed in the same manner using anti-phospho-S240/S244-S6RP (CS5367) and PE-conjugated anti-rabbit IgG (SB4052-09; Southern Biotech, Birmingham, AL). The surface expression of FLT3-ITD was analyzed by using APC-conjugated anti-FLT3 (17-1357-42; eBioscience Inc., San Diego, CA) and APC-conjugated anti-mouse IgG1 kappa (17-4714-81; eBioscience Inc.), as an isotype control, without fixation and permeabilization of cells.
Immunoblotting and Cap-Binding Assays
For immunoblotting experiments, cells were lysed and subjected to immunoblot analysis essentially as described previously [26]. Cap-binding assays were performed as described previously [18].
Analyses of Primary AML Cells
Mononuclear cells were isolated and subjected to immunoblot analysis and viability assays as described previously [18]. Primary AML cases #1 and #2 correspond to those described in our previous report [19]. The study was approved by the ethical committee of Tokyo Medical and Dental University. Written informed consent was obtained from the patients in compliance with the Declaration of Helsinki.
Results
FLT3-ITD Confers Resistance to Bortezomib by Inhibiting Activation of the Mitochondria-Mediated Intrinsic Apoptotic Pathway Involving Activation of Bax and Caspases
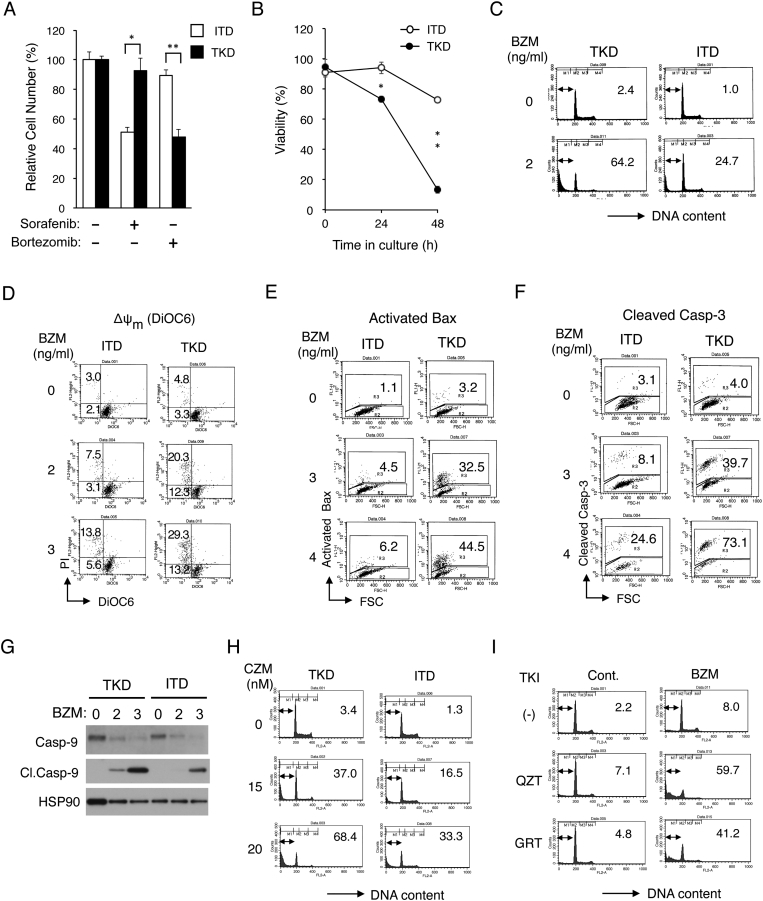
We first examined the sensitivities of 32D cells transformed by FLT3-ITD or FLT-TKD to bortezomib comparatively by the XTT proliferation assay. As shown in Figure 1A, bortezomib affected proliferation of 32D/TKD much more prominently than that of 32D/ITD, while sorafenib inhibited proliferation of 32D/ITD much more remarkably than 32D/TKD, as expected [23]. Furthermore, bortezomib reduced viability of 32D/TKD much more remarkably than that of 32D/ITD by inducing apoptosis more prominently as judged by increases in cells with sub-G1 cellular DNA content, which is a hallmark of apoptotic cells (Figure 1, B and C). In accordance with this, bortezomib induced decline in mitochondrial membrane potential (Δψm), activation of Bax, and cleavage of Caspase-3 as well as Caspase-9 much more prominently in 32D/TKD cells than in 32D/ITD cells (Figure 1, D-G). Carfilzomib, the other proteasome inhibitor in clinical use, also dose-dependently induced apoptosis more prominently in 32D/TKD cells than in 32D/ITD cells (Figure 1H). Finally, it was found that a modest apoptosis induced by a low concentration of bortezomib in the FLT3-ITD–positive AML cell line MV4-11 was drastically enhanced by a low concentration of the clinically relevant FLT3 inhibitor quizartinib or gilteritinib (Figure 1I). These results indicate that, as compared with FLT3-TKD, FLT3-ITD confers resistance to bortezomib by inhibiting activation of the mitochondria-mediated intrinsic pathway involving activation of Bax and Caspases.
Figure 1.
FLT3-ITD confers resistance to bortezomib by inhibiting activation of the mitochondria-mediated intrinsic apoptotic pathway involving activation of Bax and caspases. (A) 32D/ITD (ITD) or 32D/TKD (TKD) cells were cultured for 48 hours with or without 50 nM sorafenib or 3 ng/ml bortezomib, as indicated. The means of relative viable cell numbers from triplicate measurements are plotted as percentages of control cells without inhibitors, with error bars indicating standard errors. The asterisks indicate a statistically significant difference determined by Student's t test (*P < .05, *P < .005). (B) Cells indicated were cultured with 3 ng/ml bortezomib for indicated times, and viable cell numbers were counted after trypan blue staining. The means of percentages of viable cells from triplicate measurements are shown, with error bars indicating standard errors (*P < .001, *P < .0001). (C) Cells indicated were treated for 48 hours with or without 2 ng/ml bortezomib (BZM), as indicated, and analyzed for the cellular DNA content by flow cytometry. Percentages of apoptotic cells with sub-G1 DNA content are indicated. (D) Cells indicated were cultured for 24 hours with BZM, as indicated, and analyzed for the mitochondrial membrane potential (∆ψm) using DiOC6 by flow cytometry. Percentages of cells with reduced ∆ψm stained with or without PI are indicated. (E, F) Cells indicated were cultured for 24 hours with BZM, as indicated, and analyzed for activated Bax (E) and cleaved Caspase-3 (F) by flow cytometry. Percentages of cells with activated Bax or cleaved Caspase-3 are indicated. FSC: forward scatter. (G) Cells indicated were treated with indicated concentrations (ng/ml) of BZM for 42 hours and subjected to Western blot analysis with antibodies against indicated proteins. Abbreviations: Casp-9, Caspase-9; Cl.Casp-9, cleaved Caspase-9. HSP90 was used for the loading control. (H) Cells indicated were treated for 48 hours with indicated concentrations of carfilzomib (CZM) and analyzed. (I) 32D/ITD cells were treated for 48 hours with or without 1 ng/ml BZM in the presence or absence of 0.5 nM quizartinib (QZT) or 5 nM gilteritinib (GRT), as indicated, and analyzed.
FLT3-ITD Confers the Resistance to Bortezomib Through Activation of STAT5
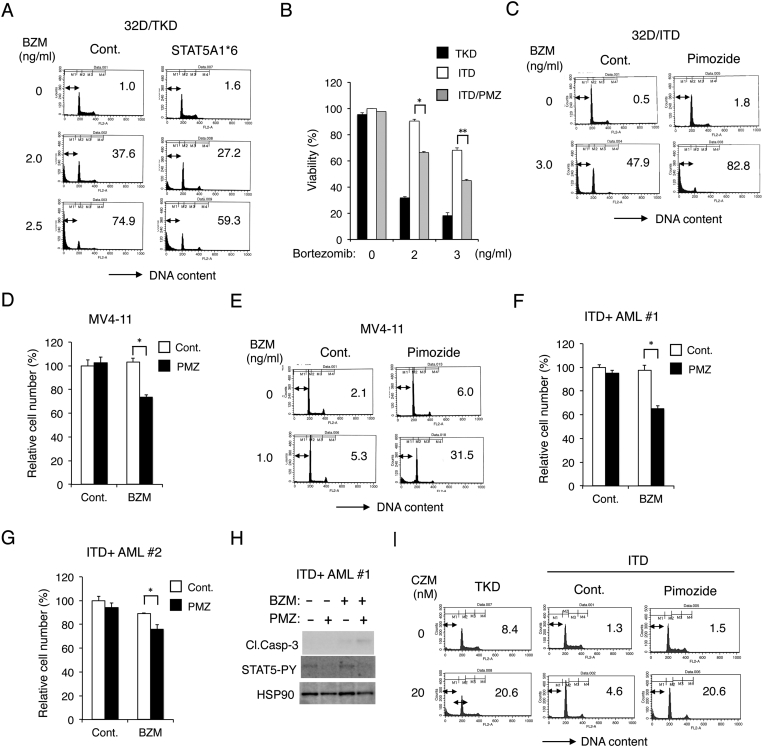
To address the possibility that STAT5 may be involved in conferring the resistance to bortezomib by FLT3-ITD, we examined the 32D/TKD cells expressing the constitutively activated STAT5 mutant STAT5A1*6 [18], [27]. Bortezomib induced apoptosis to lesser extents in STAT5A1*6-expressing cells than in vector control cells, thus suggesting that the robust activation of STAT5 may confer resistance to bortezomib (Figure 2A). We next examined effects of the STAT5 inhibitor pimozide, which we had confirmed to reduce the activation-specific phosphorylation of STAT5 on Y694 in 32D/ITD or MV4-11 cells previously [18]. Pimozide at 2.5 μM did not significantly affect the viability of 32D/ITD cells, but significantly augmented the bortezominb-induced decrease in viability of cells (Figure 2B). Furthermore, pimozide remarkably enhanced apoptosis induced by bortezomib in 32D/ITD cells (Figure 2C). As shown in Figure 2D, pimozide at 1 μM did not show any inhibitory effect on proliferation of MV4-11 cells but significantly reduced their proliferation in combination with bortezomib at 0.1 ng/ml, which also did not show any inhibitory effect when used alone. Moreover, pimozide and bortezomib at higher concentrations cooperatively induced apoptosis in MV4-11 cells (Figure 2E). Bortezomib also reduced viable cell numbers of primary leukemic cells from the two patients with FLT3-ITD–positive AML we described previously [19] more distinctively in the presence of pimozide than its absence (Figure 2, F and G). Furthermore, bortezomib induced activation of Caspase-3 more distinctively in the presence of pimozide than in its absence in one of the primary AML samples we could examine (Figure 2H). Finally, pimozide enhanced apoptosis induced by carfilzomib in 32D/ITD to the similar extent with that induced in 32D/TKD (Figure 2I). Together, these data indicate that FLT3-ITD may confer resistance to the proteasome inhibitors through the robust activation of STAT5.
Figure 2.
Activation of STAT5 mediates the resistance of FLT3-ITD–expressing cells including primary AML cells to bortezomib. (A) 32D/TKD cells transduced with STAT5A1*6 or vector control cells, as indicated, were cultured for 48 hours with indicated concentrations of bortezomib (BZM) and analyzed for the cellular DNA content by flow cytometry. Percentages of apoptotic cells with sub-G1 DNA content are indicated. (B) 32D/ITD cells in the absence (ITD) or presence (ITD/PMZ) of 2.5 μM pimozide as well as 32D/TKD (TKD) cells were cultured with indicated concentrations of bortezomib for 48 hours, and viable cell numbers were counted after trypan blue staining. The means of percentages of viable cells from triplicate measurements are shown, with error bars indicating standard errors. The asterisks indicate a statistically significant difference determined by Student's t test (*P < .0001, **P < .0005). (C) 32D/ITD cells were cultured for 48 hours with or without BZM or 5 μM pimozide, as indicated, and analyzed. (D) MV4-11 cells were cultured for 48 hours with or without 0.1 ng/ml BZM or 1 μM pimozide (PMZ), as indicated. The means of relative viable cell numbers from triplicate measurements expressed as percentages of control cells are plotted, with error bars indicating standard errors (*P < .01). (E) MV4-11 cells were cultured for 48 hours with BZM or 10 μM pimozide, as indicated, and analyzed. (F) Primary AML cells from a FLT3-ITD–positive patient (ITD+ AML #1) were culture for 48 hours with 0.5 ng/ml BZM or 2 μM pimozide PMZ, as indicated, and analyzed (*P < .01). (G) Primary AML cells from a FLT3-ITD–positive patient (ITD+ AML #2) were culture for 24 hours with 1 ng/ml BZM or 10 μM PMZ, as indicated, and analyzed (*P < .05). (H) Cells indicated were cultured for 24 hours with 0.5 ng/ml BZM or 2 μM PMZ, as indicated, and subjected to Western blot analysis with antibodies against indicated proteins. Abbreviations: Cl.Casp-3, cleaved Caspase-3; STAT5-PY, phospho-Y694-STAT5. (I) 32D/ITD (ITD) or 32D/TKD (TKD) cells were cultured with indicated concentrations of carfilzomib (CZM) with or without 10 μM pimozide for 48 hours and analyzed.
FLT3-ITD May Protect the mTORC1 Pathway Through the Robust STAT5 Activation in Bortezomib-Treated Cells to Enhance Cell Survival
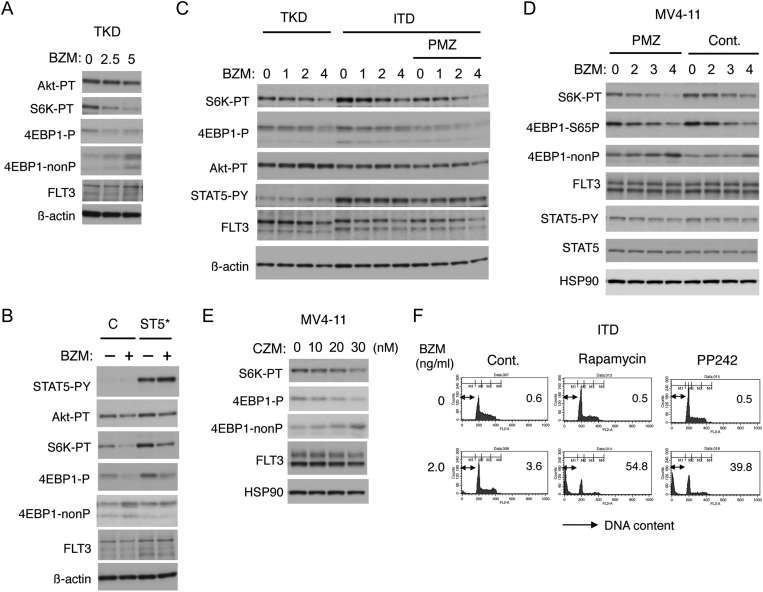
We previously revealed that the robust activation of STAT5 by FLT3-ITD conferred the resistance to the PI3K/Akt pathway inhibitors by protecting the mTORC1 pathway [18]. To examine the possibility that the resistance to proteasome inhibitors is mediated by similar mechanisms, we first examined the effect of bortezomib on the Akt/mTORC1 pathway. As shown in Figure 3A, bortezomib dose-dependently decreased phosphorylation of the mTORC1 substrates S6K and 4EPB1 without affecting the expression level of FLT3-TKD or activation-specific phosphorylation of Akt. Furthermore, the bortezomib-induced downregulation of mTORC1 was mitigated by STAT5A1*6 in 32D/TKD cells, observed less significantly in 32D/ITD than 32D/TKD cells, and augmented by treatment with the STAT5 inhibitor pimozide in 32D/ITD as well as in MV4-11 cells (Figure 3, B-D). Carfilzomib also inhibited the mTORC1 pathway without affecting the FLT3-ITD level in MV4-11 cells (Figure 3E). Finally, inhibition of mTORC1 by rapamycin or PP242 abrogated the resistance of 32D/ITD and drastically enhanced the bortezomib-induced apoptosis, whereas these inhibitors did not induce apoptosis when used alone. These data suggest that protection of the mTORC1 pathway through the robust STAT5 activation in FLT3-ITD–expressing cells may play an important role in acquisition of the resistance to proteasome inhibitors.
Figure 3.
FLT3-ITD protects the mTORC1 pathway through the robust STAT5 activation in bortezomib-treated cells to enhance cell survival. (A) 32D/TKD (TKD) cells were treated with indicated concentrations (ng/ml) of bortezomib (BZM) for 6 hours and subjected to Western blot analysis. Abbreviations: Akt-PT, phospho-T308-Akt; S6K-PT, phospho-T389-S6K; 4EBP1-P, phospho-T37/46-4EBP1; 4EBP1-nonP, non-phospho-T46-4EBP1. (B) 32D/TKD cells transduced with STAT5A1*6 (ST5*) or vector control cells, as indicated, were treated with or without 5 ng/ml BZM for 6 hours and analyzed. (C) 32D/ITD (ITD) or 32D/TKD (TKD) cells were treated with indicated concentrations (ng/ml) of BZM with or without 5 μM pimozide (PMZ), as indicated, for 6 hours and analyzed. STAT5-PY: phospho-Y694-STAT5. (D) MV4-11 cells were cultured for 3 hours with or without 10 μM PMZ, as indicated, and then treated with indicated concentrations (ng/ml) of BZM for 6 hours and analyzed. 4EBP1-S65P: phospho-S65-4EBP1. (E) MV4-11 cells were treated with indicated concentrations of carfilzomib (CZM) for 6 hours and analyzed. (F) 32D/ITD (ITD) cells were cultured with indicated concentrations of BZM with or without 50 nM rapamycin or 0.2 mM PP242, as indicated, for 48 hours and analyzed for the cellular DNA content by flow cytometry. Percentages of apoptotic cells with sub-G1 DNA content are indicated.
Bortezomib May Downregulate the mTORC1 Pathway at Least Partly by Inducing REDD1 to Decrease the FLT3-ITD Expression Level
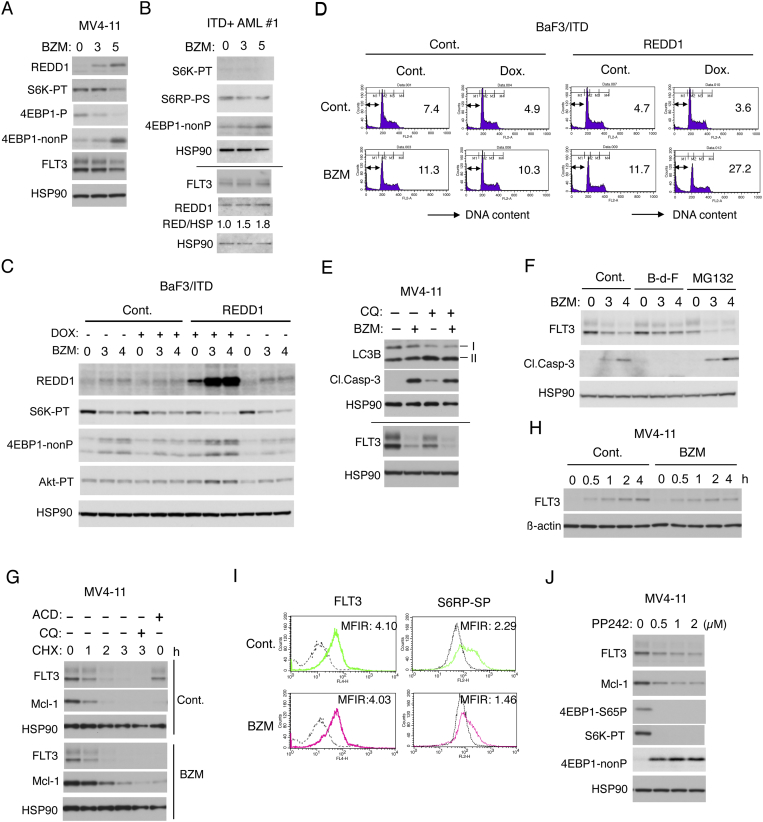
To explore the mechanisms involved in downregulation of the mTORC1 pathway by bortezomib, we examined the possible involvement of REDD1, known to be involved in inhibition of this pathway in response to various cellular stresses [12]. Bortezomib increased the expression level of REDD1 while downregulating the mTORC1 pathway in MV4-11 and primary FLT3-ITD–positive AML cells (Figure 4, A and B). Furthermore, in FLT3-ITD–transformed murine hematopoietic BaF3 cells inducibly expressing REDD1 when cultured with doxycycline, the bortezomib-induced increases in REDD1 expression correlated with downregulation of the mTORC1 pathway, while the activation specific phosphorylation of Akt correlated with the expression level of REDD1 for an unknown reason (Figure 4C). As expected, apoptosis induced by bortezomib in these cells was remarkably increased under the conditions in which induction of REDD1 expression was enhanced (Figure 4D). These results suggest that bortezomib may downregulate the mTORC1 pathway at the level downstream of Akt at least partly through induction of REDD1 to induce apoptosis.
Figure 4.
Bortezomib induces REDD1 expression to downregulate the mTORC1 pathway and subsequently the FLT3-ITD expression. (A) MV4-11 cells were treated with indicated concentrations (ng/ml) of bortezomib (BZM) for 6 hours and subjected to Western blot analysis with antibodies against indicated proteins. Abbreviations: S6K-PT, phospho-T389-S6K; 4EBP1-P, phospho-T37/46-4EBP1; 4EBP1-nonP, non-phospho-T46-4EBP1. (B) Primary AML cells from FLT3-ITD–positive patients (ITD+ AML #1) were treated with indicated concentrations (ng/ml) of BZM for 6 hours and lysed. Cell lysates were run on duplicate gels and analyzed. Relative expression levels of REDD1 analyzed by densitometry and normalized by that of HSP90 (RED/HSP) are indicated. S6RP-SP: phospho-S240/244-S6RP. (C) BaF3/ITD cells inducibly overexpressing REDD1 (REDD1) or vector control cells (Cont.) cultured with or without doxycycline (DOX), as indicated, were treated with indicated concentrations (ng/ml) of BZM for 6 hours and analyzed. Akt-PT: phospho-T308-Akt. (D) Cells indicated were treated with 2 ng/ml BZM for 48 hours and analyzed for the cellular DNA content by flow cytometry. Percentages of apoptotic cells with sub-G1 DNA content are indicated. (E) MV4-11 cells were treated with or without 5 ng/ml BZM and 50 μM chloroquine (CQ), as indicated, for 8 hours and analyzed. Positions of LC3B-I and -II are indicated. Cl.Casp-3: cleaved Caspase-3. (F) MV4-11 cells were treated with indicated concentrations (ng/ml) of BZM for 8 hours simultaneously with 100 μM Boc-d-FMK (B-d-F) or with 5 μM MG132 added for the last 2 hours, as indicated, and analyzed. (G) MV4-11 cells were left untreated as control (Cont.) or treated with 5 ng/ml BZM, as indicated, for 5 hours. Cells were then treated with 50 μg/ml cycloheximide (CHX) for indicated times along with 100 μM CQ where indicated or treated with 5 μg/ml actinomycin D (ACD) for 4 hours, as indicated, and lysed for Western blot analysis. (H) MV4-11 cells were cultured for 4 hours with 50 μg/ml CHX in the absence (Cont.) or presence of 5 ng/ml of BZM. Cells were then washed three times and cultured with or without BZM, as indicated, for indicated times and analyzed. (I) MV4-11 cells were treated for 6 hours with 4 ng/ml BZM or left untreated as control (Cont.), as indicated, and subjected to flow cytometric analysis with an antibody against FLT3 or phospho-S240/244-S6RP (S6RP-SP), as indicated. Gray lines represent isotype controls. MFIR: mean fluorescent intensity ratio. (J) MV4-11 cells were treated with indicated concentrations of PP242 for 8 hours and analyzed.
It has been reported that bortezomib downregulated mTORC1 to induce degradation of FLT3-ITD through autophagy in AML cells, including MV4-11 cells [22]. Concordantly, the expression level of FLT3-ITD was remarkably downregulated after treatment with 5 ng/ml bortezomib for 8 h (Figure 4E), a longer period of time than in experiments shown in Figure 3 or Figure 4, A and B. However, bortezomib failed to increase the LC3B-II levels in the presence or absence of the lysosome inhibitor chloroquine, which indicates that the autophagic flow was not increased by bortezomib under these conditions (Figure 4E). Furthermore, the decrease in FLT3-ITD expression was partly inhibited by the pan-caspase inhibitor Boc-d-FMK but not by chloroquine (Figure 4, E and F). On the other hand, co-treatment with the proteasomal inhibitor MG132 enhanced the bortezomib-induced downregulation of FLT3-ITD expression as well as activation of Caspase-3 in MV4-11 cells. These results indicate that bortezomib may reduce the expression level of FLT3-ITD in MV4-11 cells at least partly through activation of caspases but not through the autophagic, lysosomal, or proteasomal degradation under these conditions.
To further explore the mechanisms involved in downregulation of FLT3-ITD, we next examined the effects of the transcription inhibitor actinomycin D and the translation inhibitor cycloheximide. Bortezomib drastically or only modestly accelerated the decrease in FLT3-ITD expression when transcription or translation was shut down, respectively (Figure 4G). Furthermore, bortezomib retarded the increase in FLT3-ITD expression in MV4-11 cells released from the inhibition of translation by cycloheximide (Figure 4H). These results suggest that bortezomib may decrease the expression level of FLT3-ITD posttranscriptionally but not mainly through posttranslational mechanisms and strongly implicate the translational downregulation as the most plausible mechanism.
To address the possibility that the inhibition of mTORC1, which plays a critical role in regulation of translation mechanisms, may play a role in downregulation of FLT3-ITD in cells treated with proteasome inhibitors, we first performed flow cytometric analyses to confirm the data shown by Western blot analyses that the downregulation of mTORC1 preceded that of FLT3-ITD. As shown in Figure 4I, treatment with bortezomib at 4 ng/ml for 6 h barely affected the surface expression of FLT3-ITD but remarkably decreased phosphorylation of S6RP, a downstream substrate of the mTORC1/S6K pathway. Furthermore, the mTOR inhibitor PP242 inhibited the mTORC1 activity and reduced the expression level of FLT3-ITD as well as Mcl-1 in MV4-11 cells (Figure 4J). Taken together, these results suggest that bortezomib inhibits the mTORC1 pathway at least partly by inducing REDD1 to reduce FLT3-ITD expression through translational mechanisms and later by cleavage by caspases.
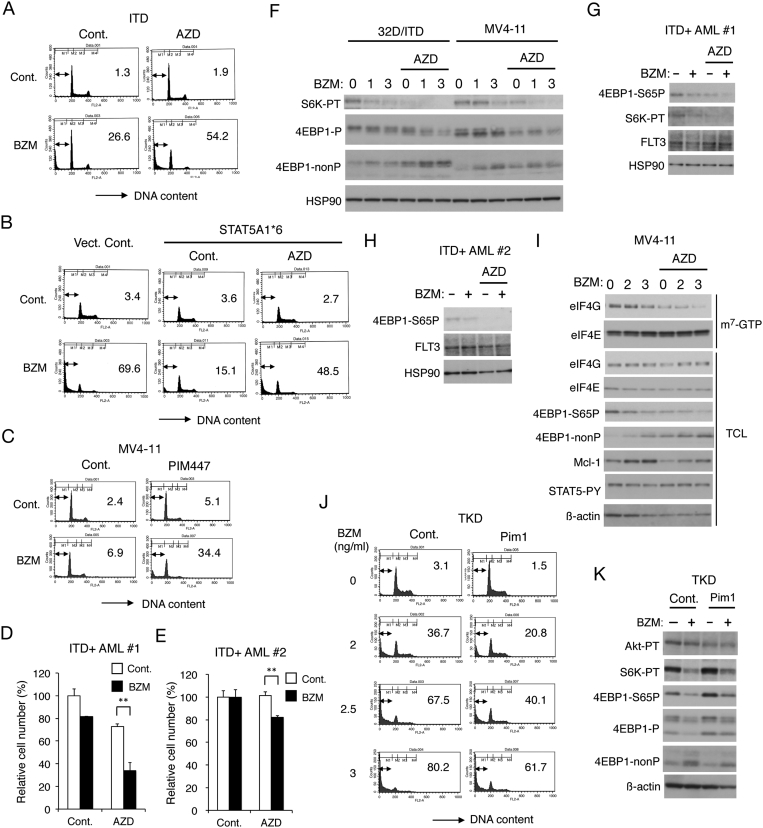
Pim Kinases Play a Key Role in STAT5-Mediated Acquisition of Bortezomib Resistance by Sustaining the mTORC1 Pathway in FLT3-ITD–Expressing Cells
We have recently revealed that the Pim kinases, which are STAT5 target gene products, play a role in acquisition of resistance to the PI3K/Akt inhibitors by FLT3-ITD–expressing cells [19]. To address the possibility that Pim kinases may also play a role in acquisition of resistance to bortezomib by these cells, we first examined the effects of the pan-Pim inhibitor AZD1208. AZD1208 remarkably enhanced apoptosis induced by bortezomib in 32D/ITD cells to a similar level with that induced by bortezomib in 32D/TKD cells (Figure 5A and data not shown), in which Pim-1 is expressed at a much lower level than in 32D/ITD cells [19]. As expected, AZD1208 also conquered the resistance of STAT5A1*6-expressing 32D/TKD cells (Figure 5B), which express Pim-1 at a higher level than in 32D/TKD [19]. The pan-Pim inhibitor AZD1208 or PIM447 also remarkably enhanced apoptosis induced by bortezomib in MV4-11 cells (Figure 5C and supplementary Figure S1A), in which Pim-2 is expressed at least partly through STAT5 activation by FLT3-ITD [19]. AZD1028 also significantly enhanced the decrease in viable cell numbers induced by bortezomib in primary leukemic cells from the two patients with FLT3-ITD–positive AML we could examine (Figure 5, D and E). Furthermore, AZD1208 cooperatively reduced the mTORC1 activity with bortezomib in 32D/ITD as well as MV4-11 cells and also in primary AML cells without affecting the expression level of FLT3-ITD (Figure 5, F-H). We then examined the effect of bortezomib and AZD1208 on formation of the eIF4E-eIF4G complex, which plays a critical role in cap-dependent translation [14], by the pull-down assays using m7-GTP beads. As shown in Figure 5I, bortezomib and AZD1208 cooperatively reduced the amount of eIF4G pulled down with eIF4E bound to m7-GTP as well as that of 4EBP1 phosphorylated in MV4-11 cells. Under these conditions, bortezomib increased the expression level of Mcl-1, most likely by inhibiting its rapid degradation through the ubiquitin/proteasome system. However, AZD1208 partly prevented the increase of Mcl-1, which is produced mostly by the cap-dependent translation [14]. We next analyzed 32D/TKD cells transduced with and overexpressing Pim-1 [19] and found that apoptosis induced by bortezomib as well as carfilzomib was mitigated by overexpression of Pim-1 (Figure 5J and supplementary Figure S1B). In these cells overexpressing Pim-1, phosphorylation of S6K and 4EBP1, but not that of Akt, was increased in accordance with our previous report [19] and was less distinctively downregulated by bortezomib (Figure 5K). Taken together, these results suggest that Pim kinases expressed downstream of STAT5 may play a role in acquisition of bortezomib resistance in FLT3-ITD–expressing cells by sustaining the mTORC1 signaling pathway.
Figure 5.
Pim kinases play a key role in STAT5-mediated acquisition of bortezomib resistance by sustaining the mTORC1 pathway in FLT3-ITD–expressing cells. (A) 32D/ITD cells were treated for 48 hours with or without 1 ng/ml bortezomib (BZM) and 1 μM AZD1208 (AZD), as indicated, and analyzed for the cellular DNA content by flow cytometry. Percentages of apoptotic cells with sub-G1 DNA content are indicated. (B) 32D/TKD cells transduced with STAT5A1*6 or vector control cells (Vect. Cont.) were cultured for 48 hours with 2 ng/ml BZM and 1 μM AZD, as indicated, and analyzed. (C) MV4-11 cells were treated for 48 hours with or without 0.75 ng/ml BZM and 2 μM PIM447, as indicated, and analyzed. (D) Primary AML cells from a FLT3-ITD–positive patient (ITD+ AML #1) were cultured for 48 hours with 0.5 ng/ml BZM and 0.5 μM AZD, as indicated. The means of relative viable cell numbers from triplicate measurements expressed as percentages of control cells are plotted, with error bars indicating standard errors. The asterisks indicate statistically significant differences determined by Student's t test (**P < .01). (E) Primary AML cells from FLT3-ITD–positive patients (ITD+ AML #2) were cultured for 24 hours with 0.3 ng/ml BZM and 0.25 μM AZD, as indicated, and analyzed. (F) 32D/ITD or MV4-11 cells were treated with indicated concentrations (ng/ml) of BZM with or without 0.5 μM AZD, as indicated, for 6 hours and subjected to Western blot analysis with antibodies against indicated proteins. Abbreviations: S6K-PT, phospho-T389-S6K; 4EBP1-P, phospho-T37/46-4EBP1; 4EBP1-nonP, non-phospho-T46-4EBP1. (G) Cells indicated were treated for 8 hours with 3 ng/ml BZM and 0.5 μM AZD, as indicated, and analyzed. 4EBP1-S65P: phospho-S65-4EBP1. (H) Cells indicated were treated for 8 hours with 4 ng/ml BZM and 0.3 μM AZD, as indicated, and analyzed. (I) MV4-11 cells were treated with indicated concentrations (ng/ml) of BZM and 1 μM AZD for 6 hours and subjected to the cap-binding assay to analyze eIF4E-eIF4G complex formation. Proteins bound to m7-GTP-sepharose (m7-GTP) as well as total cell lysates (TCL) were analyzed by immunoblotting with indicated antibodies. STAT5-PY: phospho-Y694-STAT5. (J) 32D/TKD cells transduced with Pim-1 or vector control cells, as indicated, were treated with indicated concentrations of BZM for 48 hours and analyzed. (K) Cells indicated were treated with or without 4 ng/ml BZM, as indicated, for 6 hours and analyzed.
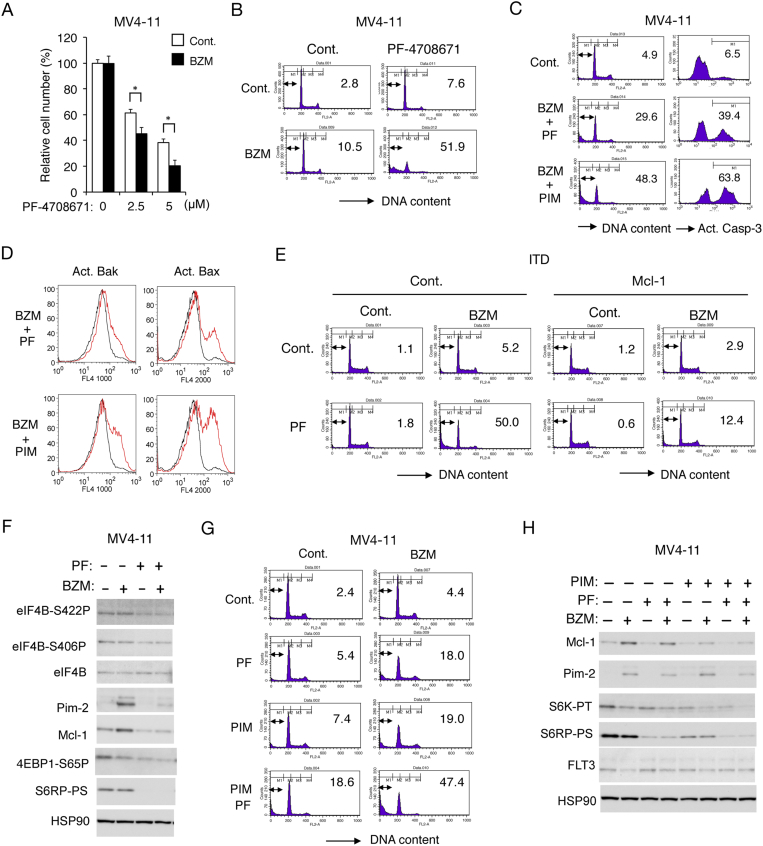
S6K Also Plays an Essential Role in Acquisition of the Bortezomib Resistance by FLT3-ITD–Expressing Leukemic Cells
We finally examined the possible involvement of S6K activated downstream of mTORC1 in acquisition of the bortezomib resistance by FLT3-ITD–expressing leukemic cells. As shown in Figure 6A, the S6K inhibitor PF-4708671 not only dose-dependently reduced viable cell numbers of MV4-11 but also significantly enhanced the ability of bortezomib to reduce them. Furthermore, PF-4708671 drastically enhanced apoptosis induced by bortezomib as well as carfilzomib (Figure 6B and supplementary Figure S2A). In accordance with this, inhibition of S6K as well as Pim kinases remarkably induced activation of Bak as well as Bax and Caspase-3 in combination with bortezomib (Figure 6, C and D). Although the pan-caspase inhibitor Boc-d-FMK prevented apoptosis in MV4-11 cells induced by the combination treatment with bortezomib and PF-4708671 or PIM447, it only modestly inhibited activation of Bak or Bax, thus indicating that these proapoptotic Bcl-2 family effector molecules were activated upstream of caspase activation (Supplementary Figure S2B, C). PF-4708671 and bortezomib synergistically induced apoptosis also in 32D/ITD cells, which was mostly prevented by overexpression of Mcl-1 in these cells as well as in MV4-11 (Figure 6E and Supplementary Figure S2D). Interestingly, PF-4708671 reduced not only phosphorylation of S6RP on S240/244 but also that of eIF4B on S422 and S406 (Figure 6F), which is known to play a role in enhancement of cap-dependent translation through modulation of the eIF4A helicase activity [16]. Accordingly, PF-4708671 reduced the expression levels of Mcl-1 and Pim-2, which are regulated through both cap-dependent translation and proteasome-dependent degradation, and mostly prevented their upregulation by bortezomib (Figure 6F). Moreover, inhibition of S6K and Pim kinases cooperatively induced apoptosis in MV4-11 cells, which was remarkably enhanced by bortezomib (Figure 6G). Consistent with this, PF-4708671 and PIM447 cooperatively downregulated the mTORC1 pathway and expression levels of Mcl-1 in MV4-11 cells treated with or without bortezomib (Figure 6H). Together, these results imply that S6K activated downstream of mTORC1 may also play a role in acquisition of the bortezomib resistance possibly and at least partly through phosphorylation of eIF4B to protect the cap-dependent translation of Mcl-1 and other proteins in FLT3-ITD–positive leukemic cells.
Figure 6.
S6K also plays an essential role in acquisition of the bortezomib resistance by FLT3-ITD–expressing leukemic cells. (A) MV4-11 cells were cultured for 48 hours with or without 0.75 ng/ml bortezomib (BZM) and indicated concentrations of PF-4708671. The means of relative viable cell numbers from triplicate measurements expressed as percentages of cells cultured without PF-4708671 are plotted, with error bars indicating standard errors. The asterisks indicate statistically significant differences determined by Student's t test (*P < .05). (B) MV4-11 cells were treated for 48 hours with or without 0.75 ng/ml BZM and 10 μM PF-4708671, as indicated, and analyzed for the cellular DNA content by flow cytometry. Percentages of apoptotic cells with sub-G1 DNA content are indicated. (C) MV4-11 cells were treated for 36 hours with or without 0.75 ng/ml BZM and 5 μM PF-4708671 (PF) or 3 μM PIM447 (PIM), as indicated, and analyzed. (D) MV4-11 cells were treated as described for C and analyzed for activation of Bak and Bax by flow cytometry. Red and black lines represent cells treated with or without inhibitors. (E) 32D/ITD cells transduced with Mcl-1 or vector control cells were treated with or without 1.5 ng/ml bortezomib BZM and 10 μM PF, as indicated, for 48 hours and analyzed. (F) MV4-11 cells were treated with or without 2 ng/ml BZM and 10 μM PF, as indicated, for 8 hours and subjected to Western blot analysis with antibodies against indicated proteins. Abbreviations: eIF4B-S422P, phospho-S422-eIF4B; eIF4B-S406P, phospho-S406-eIF4B; 4EBP1-S65P, phospho-S65-4EBP1; S6RP-PS: phospho-S240/244-S6RP. (G) MV4-11 cells were treated for 48 hours with or without 0.75 ng/ml BZM, 7.5 μM PF, or 2 μM PIM, as indicated, and analyzed. (H) MV4-11 cells were treated for 6 hours with or without 3 ng/ml BZM, 10 μM PF, or 3 μM PIM, as indicated, and analyzed.
Discussion
The present study indicates that apoptosis induced by proteasome inhibitors may be mainly mediated by downregulation of the mTORC1 pathway, which is partly prevented through the STAT5/Pim kinase axis by FLT3-ITD. This is because the induction of apoptosis closely correlated with downregulation of mTORC1 under a variety of conditions in various cell types, including primary AML cells, and was synergistically enhanced by inhibition of mTORC1. On the other hand, previous studies have shown that bortezomib-induced downregulation of the mTORC1 pathway played protective roles in multiple myeloma and prostate cancer cells [28], [29]. This protective role could be explained by the decrease in translation or induction of autophagy to reduce accumulation of misfolded proteins causing lethal stress responses. On the other hand, cap-dependent translation regulated by phosphorylation of 4EBP1 downstream of mTORC1 has been reported to contribute to resistance of multiple myeloma cells to bortezomib [30]. The present study is in line with this and further suggests that S6K activated by mTORC1 may also contribute to resistance to bortezomib because its inhibition drastically enhanced apoptosis induced by bortezomib in a synergistic manner. In this regard, it should be noted that S6K upregulates cap-dependent as well as global translation of mRNAs by phosphorylating eIF4B to enhance eIF4A helicase activity [16]. The present study also revealed that Mcl-1 may mediate a protective role downstream of mTORC1, which regulates cap-dependent translation, because its exogenous overexpression showed a potent protective effect on cells treated with combination of bortezomib and the S6K inhibitor (Figure 6E and Supplementary Figure 2D). Previous studies have also shown that upregulation of Mcl-1 may mediate resistance to bortezomib or carfilzomib in AML [21]. Thus, reduction of bortezomib-induced upregulation of Mcl-1 by downregulation of cap-dependent translation in cells cotreated with the PIM or S6K inhibitor likely plays an important role in increasing sensitivity to bortezomib.
The present study suggests that upregulation of REDD1 expression induced by bortezomib may play a role in downregulation of the mTORC1 pathway because genetic enhancement of REDD1 induction distinctively augmented the bortezomib-induced downregulation of this pathway (Figure 4C). It has been previously reported that bortezomib increased the expression level of REDD1 also in multiple myeloma and prostate cancer cells [28], [29], [31]. Although the increase in REDD1 transcript level was previously implicated in multiple myeloma cells [29], it was later reported that bortezomib induced REDD1 expression not through transcriptional upregulation but through stabilization in prostate cancer cells [28]. In accordance with this, bortezomib upregulated the expression levels of both endogenous and exogenous REDD1 in the present study, thus implicating the stabilization of this highly unstable protein rapidly degraded by the proteasome [32], rather than the transcriptional upregulation of its gene as the underlying major mechanism. REDD1 has been reported to inhibit the mTORC1 pathway by various molecular mechanisms, including inhibition of the upstream Akt kinase [12], [33], [34]. In the present study, Akt was rather activated by overexpression of REDD1, possibly through a negative feedback mechanism, implying that it affected signaling events downstream of Akt. In contrast to the present study, the bortezomib-induced REDD1 expression was implicated in protection of cell survival through downregulation of mTORC1 pathway to enhance autophagy in the previous reports [28], [29]. Thus, REDD1 induced by bortezomib may play different roles in regulation of cell survival in different cellular contexts. Future studies are warranted to explore the possibility that various pharmacological agents inducing cellular stresses to induce REDD1 expression may promote the effects of bortezomib on AML with or without FLT3-ITD.
The present study has shown that bortezomib downregulated the expression level of FLT3-ITD in AML cells, which is in agreement with previous reports [22], [35]. However, different mechanisms have been described to induce the downregulation. While Blum et al. [35] reported that bortezomib inhibited the FLT3-ITD expression at the transcriptional level in MV4-11 as well as primary AML cells, Larrue et al. [22] more recently reported that bortezomib inhibited the mTORC1 pathway to induce autophagic degradation in these cells, which was not prevented by caspase inhibitors. In the present study, however, the decrease in FLT3-ITD expression was observed under the conditions where autophagic flux was not increased and was at least partly prevented by inhibition of caspases but not autophagy (Figure 4, E and F). Moreover, bortezomib only modestly or drastically accelerated the decrease in FLT3-ITD expression when translation or transcription was shut down, respectively, thus implicating translational downregulation as the major underlying mechanism (Figure 4G). In accordance with this, bortezomib retarded the recovery of FLT3-ITD expression after reinitiation of translation in MV4-11 cells (Figure 4H). Furthermore, inhibition of mTORC1 downregulated FLT3-ITD (Figure 4J). Thus, our results strongly suggest that bortezomib reduced the FLT3-ITD expression by the translation mechanisms regulated by mTORC1 and subsequently by the caspase-mediated mechanisms in cells undergoing apoptosis. The discrepancy with the previous reports might have been caused at least partly by the differences in experimental conditions. For instance, Larrue et al. examined the effect of bortezomib on autophagy in MV4-11 cells treated with 10 nM bortezomib for 24 hours, while we examined the earlier (8 hours) effects of a slightly higher concentration (5 ng/ml or 13.0 nM) of bortezomib on FLT3-ITD and autophagy in these cells. Together, these data suggest that proteasome inhibitors may reduce the expression of FLT3-ITD through various mechanisms dependent on cellular and experimental conditions. Irrespective of the underlying mechanisms, the decrease in FLT3-ITD expression may play a significant role in enhancement of long-term cytotoxic effects of proteasome inhibitors on FLT3-ITD–positive AML cells, which may explain the report that primary AML samples bearing FLT3-ITD mutations are more sensitive to proteasome inhibitors than wild-type samples [22].
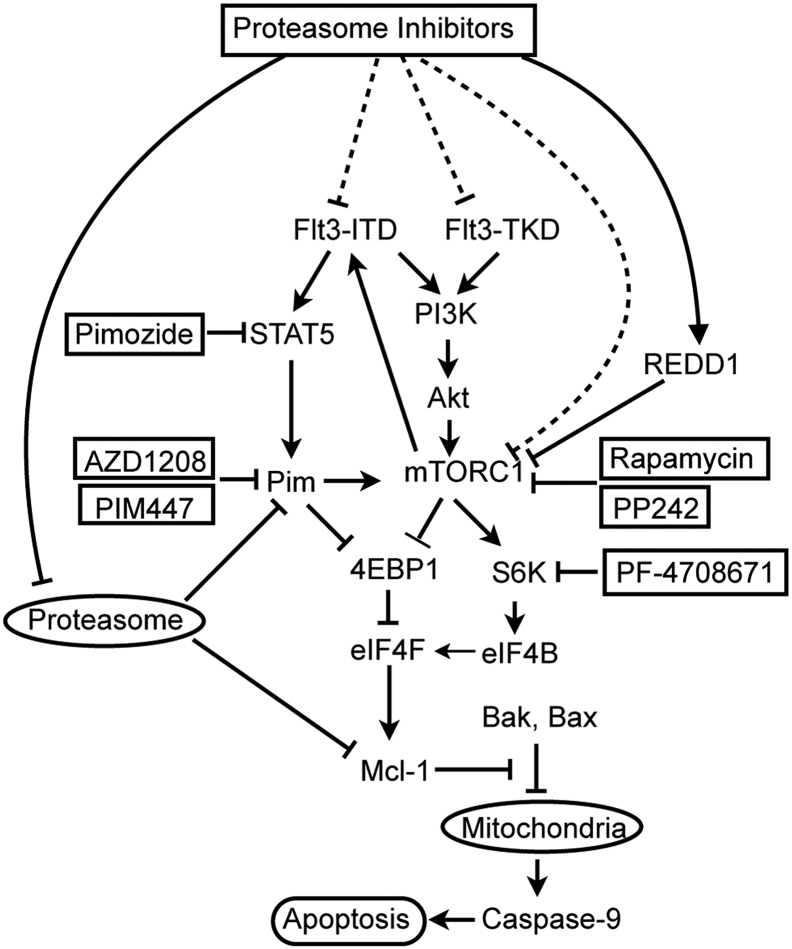
In summary, we have demonstrated that the proteasome inhibitors bortezomib and carfilzomib downregulated the mTORC1 pathway to induce apoptosis mediated through the intrinsic mitochondrial pathway in hematopoietic cells including the FLT3-ITD–positive MV4-11 and primary AML cells. FLT3-ITD as compared with FLT3-TKD diminished the cytotoxic effect of proteasome inhibitors by partly sustaining the mTORC1 pathway through activation of the STAT5/Pim kinase axis. Induction of REDD1 by bortezomib was implicated in reducing the mTORC1 activity critical for translation of mRNAs, which led to decrease in FLT3-ITD expression as well as decrease in induction of Mcl-1 expression by proteasome inhibitors to result in induction of apoptosis. Because inhibition of the STAT5/Pim kinase pathway as well as that of the mTORC1 pathway remarkably enhanced apoptosis induced by the proteasome inhibitors, various molecules in these pathways are supposed to be promising targets to augment the therapeutic effects of proteasome inhibitors against therapy-resistant FLT3-ITD–positive AML (Figure 7).
Figure 7.
A schematic model of intracellular signaling mechanisms regulating proteasome inhibitor-induced apoptosis by FLT3-ITD involving the STAT5/Pim axis and the mTORC1/eIF4F/Mcl-1 pathway.
Acknowledgements
We thank Maho Kawakami and Cheng Chen for excellent experimental and technical supports.
This work was supported in part by grants from Ministry of Education, Culture, Sports, Science and Technology of Japan (grant numbers 15K09467, and 18K08349).
Footnotes
Conflict of Interest: The authors declare that they have no conflict of interest.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.tranon.2018.11.001.
Appendix A. Supplementary Data
Supplementary figures
References
- 1.Hospital MA, Green AS, Maciel TT, Moura IC, Leung AY, Bouscary D, Tamburini J. FLT3 inhibitors: clinical potential in acute myeloid leukemia. OncoTargets Ther. 2017;10:607–615. doi: 10.2147/OTT.S103790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meshinchi S, Appelbaum FR. Structural and functional alterations of FLT3 in acute myeloid leukemia. Clin Cancer Res. 2009;15:4263–4269. doi: 10.1158/1078-0432.CCR-08-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schlenk RF, Kayser S, Bullinger L, Kobbe G, Casper J, Ringhoffer M, Held G, Brossart P, Lubbert M, Salih HR. Differential impact of allelic ratio and insertion site in FLT3-ITD positive AML with respect to allogeneic hematopoietic stem cell transplantation. Blood. 2014;124:3441–3449. doi: 10.1182/blood-2014-05-578070. [DOI] [PubMed] [Google Scholar]
- 4.Schnittger S, Bacher U, Haferlach C, Alpermann T, Kern W, Haferlach T. Diversity of the juxtamembrane and TKD1 mutations (exons 13-15) in the FLT3 gene with regards to mutant load, sequence, length, localization, and correlation with biological data. Genes Chromosomes Cancer. 2012;51:910–924. doi: 10.1002/gcc.21975. [DOI] [PubMed] [Google Scholar]
- 5.Smith CC, Wang Q, Chin CS, Salerno S, Damon LE, Levis MJ, Perl AE, Travers KJ, Wang S, Hunt JP. Validation of ITD mutations in FLT3 as a therapeutic target in human acute myeloid leukaemia. Nature. 2012;485:260–263. doi: 10.1038/nature11016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith CC, Paguirigan A, Jeschke GR, Lin KC, Massi E, Tarver T, Chin CS, Asthana S, Olshen A, Travers KJ. Heterogeneous resistance to quizartinib in acute myeloid leukemia revealed by single-cell analysis. Blood. 2017;130:48–58. doi: 10.1182/blood-2016-04-711820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choudhary C, Brandts C, Schwable J, Tickenbrock L, Sargin B, Ueker A, Böhmer FD, Berdel WE, Müller-Tidow C, Serve H. Activation mechanisms of STAT5 by oncogenic Flt3-ITD. Blood. 2007;110:370–374. doi: 10.1182/blood-2006-05-024018. [DOI] [PubMed] [Google Scholar]
- 8.Hayakawa F, Towatari M, Kiyoi H, Tanimoto M, Kitamura T, Saito H, Naoe T. Tandem-duplicated Flt3 constitutively activates STAT5 and MAP kinase and introduces autonomous cell growth in IL-3-dependent cell lines. Oncogene. 2000;19:624–631. doi: 10.1038/sj.onc.1203354. [DOI] [PubMed] [Google Scholar]
- 9.Janke H, Pastore F, Schumacher D, Herold T, Hopfner KP, Schneider S, Berdel WE, Büchner T, Woermann BJ, Subklewe M. Activating FLT3 mutants show distinct gain-of-function phenotypes in vitro and a characteristic signaling pathway profile associated with prognosis in acute myeloid leukemia. PLoS One. 2014;9 doi: 10.1371/journal.pone.0089560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Altman JK, Sassano A, Platanias LC. Targeting mTOR for the treatment of AML. New agents and new directions. Oncotarget. 2011;2:510–517. doi: 10.18632/oncotarget.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martelli AM, Evangelisti C, Chiarini F, McCubrey JA. The phosphatidylinositol 3-kinase/Akt/mTOR signaling network as a therapeutic target in acute myelogenous leukemia patients. Oncotarget. 2010;1:89–103. doi: 10.18632/oncotarget.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lipina C, Hundal HS. Is REDD1 a metabolic eminence grise? Trends Endocrinol Metab. 2016;27:868–880. doi: 10.1016/j.tem.2016.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsieh AC, Costa M, Zollo O, Davis C, Feldman ME, Testa JR, Meyuhas O, Shokat KM, Ruggero D. Genetic dissection of the oncogenic mTOR pathway reveals druggable addiction to translational control via 4EBP-eIF4E. Cancer Cell. 2010;17:249–261. doi: 10.1016/j.ccr.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mills JR, Hippo Y, Robert F, Chen SM, Malina A, Lin CJ, Trojahn U, Wendel HG, Charest A, Bronson RT. mTORC1 promotes survival through translational control of Mcl-1. Proc Natl Acad Sci U S A. 2008;105:10853–10858. doi: 10.1073/pnas.0804821105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nawijn MC, Alendar A, Berns A. For better or for worse: the role of Pim oncogenes in tumorigenesis. Nat Rev Cancer. 2011;11:23–34. doi: 10.1038/nrc2986. [DOI] [PubMed] [Google Scholar]
- 16.Magnuson B, Ekim B, Fingar DC. Regulation and function of ribosomal protein S6 kinase (S6K) within mTOR signalling networks. Biochem J. 2012;441:1–21. doi: 10.1042/BJ20110892. [DOI] [PubMed] [Google Scholar]
- 17.Gores GJ, Kaufmann SH. Selectively targeting Mcl-1 for the treatment of acute myelogenous leukemia and solid tumors. Genes Dev. 2012;26:305–311. doi: 10.1101/gad.186189.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nogami A, Oshikawa G, Okada K, Fukutake S, Umezawa Y, Nagao T, Kurosu T, Miura O. FLT3-ITD confers resistance to the PI3K/Akt pathway inhibitors by protecting the mTOR/4EBP1/Mcl-1 pathway through STAT5 activation in acute myeloid leukemia. Oncotarget. 2015;6:9189–9205. doi: 10.18632/oncotarget.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okada K, Nogami A, Ishida S, Akiyama H, Chen C, Umezawa Y, Miura O. FLT3-ITD induces expression of Pim kinases through STAT5 to confer resistance to the PI3K/Akt pathway inhibitors on leukemic cells by enhancing the mTORC1/Mcl-1 pathway. Oncotarget. 2018;9:8870–8886. doi: 10.18632/oncotarget.22926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crawford LJ, Irvine AE. Targeting the ubiquitin proteasome system in haematological malignancies. Blood Rev. 2013;27:297–304. doi: 10.1016/j.blre.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 21.Csizmar CM, Kim DH, Sachs Z. The role of the proteasome in AML. Blood Cancer J. 2016;6:e503. doi: 10.1038/bcj.2016.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Larrue C, Saland E, Boutzen H, Vergez F, David M, Joffre C, Hospital MA, Tamburini J, Delabesse E, Manenti S. Proteasome inhibitors induce FLT3-ITD degradation through autophagy in AML cells. Blood. 2016;127:882–892. doi: 10.1182/blood-2015-05-646497. [DOI] [PubMed] [Google Scholar]
- 23.Oshikawa G, Nagao T, Wu N, Kurosu T, Miura O. c-Cbl and Cbl-b ligases mediate 17-allylaminodemethoxygeldanamycin-induced degradation of autophosphorylated Flt3 kinase with internal tandem duplication through the ubiquitin proteasome pathway. J Biol Chem. 2011;286:30263–30273. doi: 10.1074/jbc.M111.232348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klucher KM, Lopez DV, Daley GQ. Secondary mutation maintains the transformed state in BaF3 cells with inducible BCR/ABL expression. Blood. 1998;91:3927–3934. [PubMed] [Google Scholar]
- 25.Kurosu T, Ohki M, Wu N, Kagechika H, Miura O. Sorafenib induces apoptosis specifically in cells expressing BCR/ABL by inhibiting its kinase activity to activate the intrinsic mitochondrial pathway. Cancer Res. 2009;69:3927–3936. doi: 10.1158/0008-5472.CAN-08-2978. [DOI] [PubMed] [Google Scholar]
- 26.Miura O, Cleveland JL, Ihle JN. Inactivation of erythropoietin receptor function by point mutations in a region having homology with other cytokine receptors. Mol Cell Biol. 1993;13:1788–1795. doi: 10.1128/mcb.13.3.1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Onishi M, Nosaka T, Misawa K, Mui AL, Gorman D, McMahon M, Miyajima A, Kitamura T. Identification and characterization of a constitutively active STAT5 mutant that promotes cell proliferation. Mol Cell Biol. 1998;18:3871–3879. doi: 10.1128/mcb.18.7.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barakat DJ, Mendonca J, Barberi T, Zhang J, Kachhap SK, Paz-Priel I, Friedman AD. C/EBPbeta regulates sensitivity to bortezomib in prostate cancer cells by inducing REDD1 and autophagosome-lysosome fusion. Cancer Lett. 2016;375:152–161. doi: 10.1016/j.canlet.2016.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Decaux O, Clement M, Magrangeas F, Gouraud W, Charbonnel C, Campion L, Loiseau HA, Minvielle S. Inhibition of mTORC1 activity by REDD1 induction in myeloma cells resistant to bortezomib cytotoxicity. Cancer Sci. 2010;101:889–897. doi: 10.1111/j.1349-7006.2009.01467.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mancino M, Grosso S, Terragna C, Borsi E, Cavo M, Biffo S. Cap dependent translation contributes to resistance of myeloma cells to bortezomib. Translation. 2013;1 doi: 10.4161/trla.27245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gu Y, Kaufman JL, Bernal L, Torre C, Matulis SM, Harvey RD, Chen J, Sun SY, Boise LH, Lonial S. MLN4924, an NAE inhibitor, suppresses AKT and mTOR signaling via upregulation of REDD1 in human myeloma cells. Blood. 2014;123:3269–3276. doi: 10.1182/blood-2013-08-521914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Katiyar S, Liu E, Knutzen CA, Lang ES, Lombardo CR, Sankar S, Toth JI, Petroski MD, Ronai Z, Chiang GG. REDD1, an inhibitor of mTOR signalling, is regulated by the CUL4A-DDB1 ubiquitin ligase. EMBO Rep. 2009;10:866–872. doi: 10.1038/embor.2009.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.DeYoung MP, Horak P, Sofer A, Sgroi D, Ellisen LW. Hypoxia regulates TSC1/2-mTOR signaling and tumor suppression through REDD1-mediated 14-3-3 shuttling. Genes Dev. 2008;22:239–251. doi: 10.1101/gad.1617608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dennis MD, Coleman CS, Berg A, Jefferson LS, Kimball SR. REDD1 enhances protein phosphatase 2A-mediated dephosphorylation of Akt to repress mTORC1 signaling. Sci Signal. 2014;7:ra68. doi: 10.1126/scisignal.2005103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blum W, Schwind S, Tarighat SS, Geyer S, Eisfeld AK, Whitman S, Walker A, Klisovic R, Byrd JC, Santhanam R. Clinical and pharmacodynamic activity of bortezomib and decitabine in acute myeloid leukemia. Blood. 2012;119:6025–6031. doi: 10.1182/blood-2012-03-413898. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures