Abstract
Background
Microscopic evaluation of urine is inconsistently performed in veterinary clinics. The IDEXX SediVue Dx® Urine Sediment Analyzer (SediVue) recently was introduced for automated analysis of canine and feline urine and may facilitate performance of urinalyses in practice.
Objective
Compare the performance of the SediVue with manual microscopy for detecting clinically relevant numbers of cells and 2 crystal types.
Samples
Five‐hundred thirty urine samples (82% canine, 18% feline).
Methods
For SediVue analysis (software versions [SW] 1.0.0.0 and 1.0.1.3), uncentrifuged urine was pipetted into a cartridge. Images were captured and processed using a convolutional neural network algorithm. For manual microscopy, urine was centrifuged to obtain sediment. To determine sensitivity and specificity of the SediVue compared with manual microscopy, thresholds were set at ≥5/high power field (hpf) for red blood cells (RBC) and white blood cells (WBC) and ≥1/hpf for squamous epithelial cells (sqEPI), non‐squamous epithelial cells (nsEPI), struvite crystals (STR), and calcium oxalate dihydrate crystals (CaOx Di).
Results
The sensitivity of the SediVue (SW1.0.1.3) was 85%‐90% for the detection of RBC, WBC, and STR; 75% for CaOx Di; 71% for nsEPI; and 33% for sqEPI. Specificity was 99% for sqEPI and CaOx Di; 87%‐90% for RBC, WBC, and nsEPI; and 84% for STR. Compared to SW1.0.0.0, SW1.0.1.3 had increased sensitivity but decreased specificity. Performance was similar for canine versus feline and fresh versus stored urine samples.
Conclusions and Clinical Importance
The SediVue exhibits good agreement with manual microscopy for the detection of most formed elements evaluated, but improvement is needed for epithelial cells.
Keywords: automated analyzer, cat, dog, urinalysis, urine formed elements, urine sediment
Abbreviations
- CaOx Di
calcium oxalate dihydrate crystals
- CNN
convolutional neural network
- CRYu
unclassified crystals
- CV
coefficient of variation
- hpf
high power field
- nsEPI
non‐squamous epithelial cells
- QCM
quality control material
- RBC
red blood cells
- ROC
receiver operating characteristic
- SediVue
IDEXX SediVue Dx® Urine Sediment Analyzer
- sqEPI
squamous epithelial cells
- STR
struvite crystals
- SW
software version
- WBC
white blood cells
1. INTRODUCTION
Urinalysis is a key component in the evaluation of patients with urologic disease. Urinalysis also is an important part of the minimum laboratory database, as it may provide valuable information, even in patients without clinical signs of urinary tract disease. A complete urinalysis consists of macroscopic examination (including urine specific gravity), biochemical analysis, and microscopic urine sediment examination. Of these, sediment examination is the most technically difficult. Microscopic urine sediment examination is time‐consuming and, based on studies performed in human medical laboratories, is associated with high interobserver variability.1, 2, 3 In our experience, many veterinarians and veterinary technicians have limited training in urine sediment analysis, and urine sediment evaluation in veterinary practices suffers from similar interobserver variability as documented in human medicine. Unfamiliarity with the identification of formed elements in urine as well as the microscope adjustments required for effective examination of urine are some of the reasons urine sediment evaluation may be inconsistently performed in veterinary practices.
In human medicine, several instruments have been introduced in an attempt to automate urine sediment analysis. The UF series (eg, UF‐100; Sysmex Corporation, Kobe, Japan) classifies particles in urine based on flow cytometric analysis, whereas the iQ200 (Iris Diagnostics, Chatsworth, California) and UriSed (77 Elektronika, Budapest, Hungary) use imaging flow cytometry or a camera‐microscope system, respectively, to record images of the urine sediment, followed by the identification of formed elements using image recognition software. Overall, these instruments demonstrate good agreement with manual microscopy for the detection of red blood cells (RBC), white blood cells (WBC), and squamous epithelial cells (sqEPI), although performance for non‐squamous epithelial cells (nsEPI), casts, crystals, bacteria, and yeast is less reliable.4, 5, 6
Automated urine sediment analyzers can provide advantages over manual microscopy, including higher intra‐assay precision,4, 7, 8, 9, 10 faster turnaround time,6, 9, 11, 12 and the requirement of a small sample volume. A combination of automated sediment analysis and dipstick test results has been utilized in human medical laboratories as a screening method to decrease the number of samples requiring manual microscopic review.4, 6, 13, 14, 15 These instruments do not completely eliminate the need for manual microscopy or captured image analysis, because pathological samples often are flagged for technician review.7, 15, 16
The recently introduced IDEXX SediVue Dx® Urine Sediment Analyzer (SediVue) is the first instrument designed to perform automated urine sediment analysis for veterinary patients. This instrument is closely modeled after the UriSed, but more images are captured, and the image recognition software has been adapted for canine and feline urine. Our objective was to compare the performance of the SediVue with manual microscopy for the detection of clinically relevant numbers of RBC, WBC, sqEPI, nsEPI, struvite crystals (STR), and calcium oxalate dihydrate crystals (CaOx Di) in canine and feline urine sediments.
2. MATERIALS AND METHODS
2.1. Urine samples
Prospectively, residual urine (urine leftover after routine analysis) was obtained using samples from client‐owned dogs and cats presented to the Texas A&M University Veterinary Medical Teaching Hospital (n = 258). Additionally, residual canine and feline urine submitted to the IDEXX Reference Laboratory in North Grafton, Massachusetts (n = 303) was included in the study. All samples were obtained between August 2015 and February 2017. For study inclusion, ≥1.1 mL was required for centrifugation to obtain sediment for manual microscopy, in addition to approximately 200 μL uncentrifuged urine for SediVue analysis. Urine samples from healthy patients, as well as patients with urologic or non‐urologic disease, were permitted. Multiple samples from the same patient were allowed, as long as >12 hours had passed between sample submissions. Both fresh (analyzed within 24 hours of submission to the laboratory, typically <8 hours) and stored (analyzed >24 hours post‐collection) samples were included. Each sample was evaluated by the SediVue (software versions 1.0.0.0 and 1.0.1.3) and by manual microscopy.
2.2. SediVue analysis
For SediVue analysis, 165 μL of well‐mixed, uncentrifuged urine was manually pipetted into a disposable cartridge. After a 10‐second, on‐board centrifugation period, 70 high‐quality grayscale images of the sediment (resolution limit, 1.2 μm) were captured by a built‐in camera‐microscope system (Figure 1). Together, these images covered an area equivalent to approximately 45 high power fields (×400, hpf). The instrument analyzed the images using a veterinary‐specific convolutional neural network (CNN) algorithm (software version [SW] 1.0.0.0) performed by an on‐board computer to identify and quantify formed elements. Cells identified by the SediVue included RBC, WBC, sqEPI, and nsEPI. Crystals first were labeled as unclassified crystals (CRYu), and if they met appropriate criteria, they were relabeled as STR or CaOx Di. The SediVue used a formula to convert the number of cells per image to elements per hpf or low power field, providing semiquantitative and quantitative results for each element (only semiquantitative results are currently provided to veterinary practices). After collection of data for the study, an updated software version (SW1.0.1.3) was introduced. Comparison of this version with SW1.0.0.0 was possible upon reanalysis of captured images.
Figure 1.

Example image of unstained urine sediment from a dog taken by the SediVue. Each image represents approximately 66% of a typical microscopic ×400 field. (scale bar = 50 μM)
2.3. Manual microscopic examination
Within 1 hour of SediVue analysis, each sample also was reviewed by manual bright‐field microscopy (Olympus BX43; Olympus Corporation of the Americas, Scientific Solutions Group, Waltham, Massachusetts). A minimum of 1.1 mL (range, 1.1‐4.2 mL) of urine was centrifuged at 1228 × g for 5 minutes. The sediment was processed using a KOVA system (KOVA Tubes and KOVA Petters; KOVA International Inc, Garden Grove, California) according to the manufacturer's instructions and loaded into a DeciSlide (Fisherbrand UriSystem DeciSlide 10‐Test Slides; Fisher Healthcare, Houston, Texas). The same DeciSlide preparation was reviewed separately by 2 individuals skilled in interpretation of urine sediments from a group of clinical pathology residents, clinical pathologists, and laboratory personnel (veterinary technicians and medical technologists). A maximum of 1 hour was allowed between the first and second review, and both observers were blinded to the results of SediVue analysis. The 2 reviewers independently assigned elements to a semiquantitative category and came to a consensus. If an agreement could not be reached, a third observer was consulted to reach a majority decision. One observer also performed a quantitative assessment by counting the number of RBC, WBC, sqEPI, and nsEPI per 10 hpf. The average number of each element per hpf was recorded. When necessary, acid or base was added to the urine sediment to aid in determination of crystal types. For documentation, 1 observer captured a minimum of 5 representative images of each urine sediment using a digital microscopy camera (Moticam 5 digital microscopy camera; Motic, China Group Co Ltd, Hong Kong, China).
2.4. Intra‐ and inter‐assay precision for RBC and WBC
The intra‐ and inter‐assay precision of the SediVue for the detection of RBC and WBC were assessed using commercial quality control material (QCM) containing at least some human‐derived components (IDEXX SediVue QC fluid, IDEXX Laboratories Inc, Westbrook, Maine). For intra‐assay precision, 2 levels of QCM (normal and abnormal) were analyzed 10 consecutive times. For inter‐assay precision, 2 levels of QCM were analyzed once daily for 5 consecutive days.
Intra‐assay precision also was assessed using residual fresh urine samples. One to 6 samples were included for each of 6 categories: RBC‐low, RBC‐medium, RBC‐high, WBC‐low, WBC‐medium, and WBC‐high. Each sample was analyzed 8‐10 consecutive times, depending on the available volume of urine. A single sample could fulfill 2 categories if it contained both RBC and WBC.
2.5. Dilutional linearity for RBC
To determine dilutional linearity, 3 samples with high numbers of RBC/hpf were selected (sample 1: approximately 200 RBC/hpf; sample 2: approximately 500 RBC/hpf; and sample 3: approximately 1400 RBC/hpf, based on SediVue analysis). Each sample was diluted serially from 1:2 to 1:64 using sterile saline (0.9% NaCl). After gentle mixing, each dilution was immediately analyzed in duplicate before making the next dilution. The mean value of the 2 measurements and the expected value for each dilution were calculated. Additionally, a sample with low numbers of RBC (approximately 1 RBC/hpf based on SediVue analysis) was used to make a solution with approximately 1500 RBC/hpf (by adding 4 μL EDTA whole blood to 2 mL urine) and another with approximately 500 RBC/hpf (by diluting the first solution 1:4 with urine). The 2 solutions were mixed in different ratios as follows: 200, 150, 100, and 50 μL of the first solution spiked with 50, 100, 150, and 200 μL of the second solution, respectively. Measured and expected values for RBC were calculated.
2.6. Statistical analyses
All statistical analyses were performed using commercially available computer software (Microsoft Excel 2013; Microsoft Corporation, Redmond, Washington). We calculated the sensitivity and specificity of the SediVue (SW1.0.0.0 and SW1.0.1.3) in comparison to manual microscopy for the detection of clinically relevant numbers of each formed element, along with 95% Clopper‐Pearson confidence intervals. For these calculations, thresholds for clinical relevance for both manual microscopy and SediVue analysis were defined as ≥5/hpf for RBC and WBC and ≥1/hpf for sqEPI, nsEPI, STR, and CaOx Di. The following scale was used to rate sensitivity and specificity: excellent (95.0%‐100.0%), good (85.0%‐94.9%), moderate (70.0%‐84.9%), fair (60.0%‐69.9%), and poor (≤59.9%). Cohen's kappa coefficient also was calculated to determine the level of agreement between manual microscopy and SediVue analysis, using the following scale for classification: excellent (0.81‐1.00), substantial (0.61‐0.80), moderate (0.41‐0.60), fair (0.21‐0.40), and slight (0.0‐0.20).17 Receiver operating characteristic (ROC) curve analysis was performed to determine the sensitivity and specificity of the SediVue for the detection of each formed element at various thresholds, holding the manual threshold constant. The SediVue thresholds with maximal sensitivity and specificity for each element were determined by calculating the Youden index (sensitivity + specificity − 1).
3. RESULTS
3.1. Urine samples
Of 561 urine samples evaluated, 31 were excluded for the following reasons: instrument hardware problem at the time of analysis (n = 13), sample mislabeling (n = 5), excessive time between SediVue analysis and manual microscopic review (n = 5), unknown volume of urine centrifuged (n = 3), inappropriate numerical rounding during data recording (n = 3), and the presence of glass shards in the urine used for SediVue analysis (n = 2). Therefore, 530 samples were included (432 canine and 98 feline). The number of samples positive for each element on manual microscopy and SediVue analysis is listed in Table 1. The centrifuged volume of urine was between 1.1 and 1.4 mL for 3%, 1.5 and 1.9 mL for 8%, 2.0 and 2.4 mL for 13%, 2.5 and 2.9 mL for 18%, 3.0 mL for 57%, and 3.1 and 4.5 mL for 1% of samples.
Table 1.
Number of urine samples positive for each element on manual microscopy and SediVue analysis
| Total | Canine | Feline | Positive threshold | ||||
|---|---|---|---|---|---|---|---|
| Manual | SediVue | Manual | SediVue | Manual | SediVue | ||
| RBC | 171/530 (32%) | 187/530 (35%) | 121/432 (28%) | 134/432 (31%) | 50/98 (51%) | 53/98 (54%) | ≥5/hpf |
| WBC | 126/530 (24%) | 154/530 (29%) | 114/432 (26%) | 140/432 (32%) | 12/98 (12%) | 14/98 (14%) | ≥5/hpf |
| sqEPI | 24/530 (5%) | 11/530 (2%) | 23/432 (5%) | 10/432 (2%) | 1/98 (1%) | 1/98 (1%) | ≥1/hpf |
| nsEPI | 56/530 (11%) | 100/530 (19%) | 49/432 (11%) | 91/432 (21%) | 7/98 (7%) | 9/98 (9%) | ≥1/hpf |
| STR | 127/530 (24%) | 178/530 (34%) | 102/432 (24%) | 147/432 (34%) | 25/98 (26%) | 31/98 (32%) | ≥1/hpf |
| CaOx Di | 52/530 (10%) | 43/530 (8%) | 49/432 (11%) | 41/432 (9%) | 3/98 (3%) | 2/98 (2%) | ≥1/hpf |
Abbreviations: CaOx Di, calcium oxalate dihydrate crystals; hpf, high power field; nsEPI, non‐squamous epithelial cells; RBC, red blood cells; sqEPI, squamous epithelial cells; STR, struvite crystals; WBC, white blood cells.
3.2. Comparison of SediVue detection of formed elements to manual microscopy using set thresholds
Compared to the older software version (SW1.0.0.0), the newer version (SW1.0.1.3) exhibited increased sensitivity for the detection of all elements tested, but confidence intervals overlapped for all elements except STR. The sensitivity of the SediVue (SW1.0.1.3) was good for the detection of RBC, WBC, and STR; moderate for nsEPI and CaOx Di; and poor for sqEPI (Table 2). In contrast, specificity decreased when elements were evaluated with SW1.0.1.3 compared to the older software version (except for sqEPI), although confidence intervals overlapped for all elements except WBC and STR. The specificity of the SediVue (SW1.0.1.3) was excellent for the detection of sqEPI and CaOx Di; good for RBC, WBC, and nsEPI; and moderate for STR (Table 2). Based on Cohen's kappa coefficient, agreement between the SediVue and manual microscopy for formed element detection was substantial for RBC, WBC, STR, and CaOx Di, and moderate for sqEPI and nsEPI. Agreement classification based on the coefficients was unchanged between software versions for all elements except sqEPI and nsEPI, which improved from fair to moderate in the newer software version (Table 2).
Table 2.
Sensitivities and specificities of the SediVue in comparison to manual microscopy for the detection of formed elements in urine using thresholds of ≥5/hpf for RBC and WBC and ≥1/hpf for sqEPI, nsEPI, STR, and CaOx Di. Kappa coefficients were also calculated to determine the level of agreement between the SediVue and manual microscopy for detection of each element.
| Sensitivity (95% CI) | Specificity (95% CI) | Kappa coefficient | ||||
|---|---|---|---|---|---|---|
| SW1.0.0.0 | SW1.0.1.3 | SW1.0.0.0 | SW1.0.1.3 | SW1.0.0.0 | SW1.0.1.3 | |
| RBC | 84.2% (77.9%‐89.3%) | 88.9% (83.2%‐93.2%) | 93.3% (90.2%‐95.7%) | 90.3% (86.7%‐93.1%) | 0.78 | 0.77 |
| WBC | 76.2% (67.8%‐83.3%) | 85.7% (78.4%‐91.3%) | 94.3%a (91.6%‐96.4%) | 88.6%a (85.1%‐91.5%) | 0.72 | 0.69 |
| sqEPI | 25.0% (9.8%‐46.7%) | 33.3% (15.6%‐55.3%) | 99.0% (97.7%‐99.7%) | 99.4% (98.3%‐99.9%) | 0.32 | 0.44 |
| nsEPI | 57.1% (43.2%‐70.3%) | 71.4% (57.8%‐82.7%) | 89.9% (86.8%‐92.4%) | 87.3% (84.0%‐90.2%) | 0.40 | 0.44 |
| STR | 71.7%a (63.0%‐79.3%) | 90.6%a (84.1%‐95.0%) | 93.1%a (90.1%‐95.3%) | 84.4%a (80.5%‐87.8%) | 0.66 | 0.66 |
| CaOx Di | 63.5% (49.0%‐76.4%) | 75.0% (61.1%‐86.0%) | 99.8% (98.8%‐100.0%) | 99.2% (97.9%‐99.8%) | 0.75 | 0.80 |
Abbreviations: CaOx Di, calcium oxalate dihydrate crystals; CI, confidence intervals; RBC, red blood cells; nsEPI, non‐squamous epithelial cells; sqEPI, squamous epithelial cells; STR, struvite crystals; WBC, white blood cells.
Comparisons in which confidence intervals did not overlap.
3.3. Evaluation of false‐negative and false‐positive results for selected elements
The sensitivity of the SediVue for the detection of sqEPI, nsEPI, and CaOx Di was relatively low (<80%) compared with other elements. Therefore, images tagged using SW1.0.1.3 were reviewed for samples with false‐negative results for these elements.
Sixty‐seven percent of samples positive for sqEPI on manual microscopy (16/24) had false‐negative results on SediVue analysis. For these samples, the average number of sqEPI detected by manual microscopy was 1.8/hpf (range, 1.0‐3.7/hpf) versus 0.3/hpf (range, 0‐0.9/hpf) for the SediVue. Mislabeling of sqEPI as nsEPI was noted as a possible cause of false‐negative results (Figure 2A).
Figure 2.
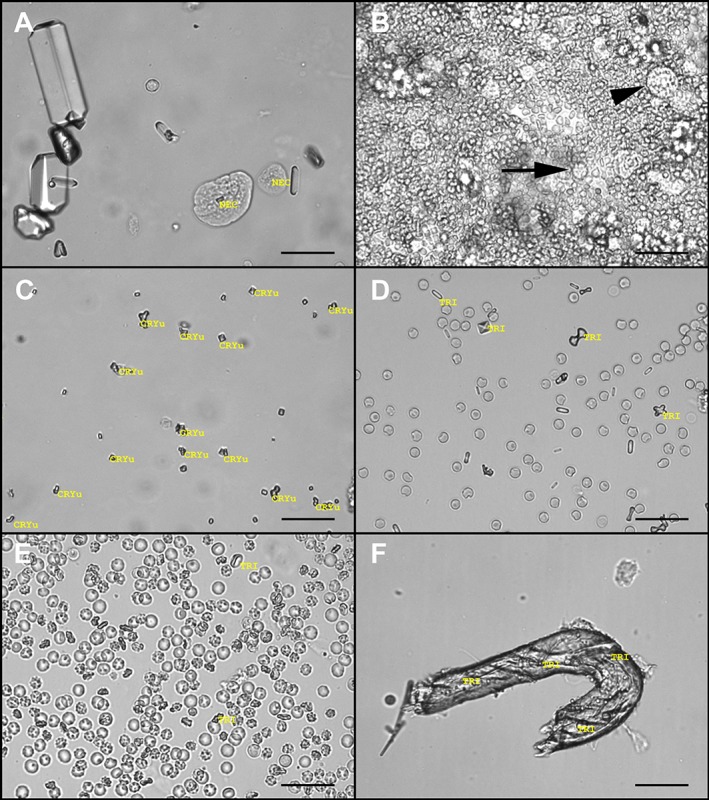
Images of unstained urine sediments showing examples of improper identification of formed elements by the SediVue. A, Urine sediment from a dog. A non‐squamous epithelial cell (NEC*) is on the right and a probable squamous epithelial cell labeled as an NEC is on the left. B, Densely cellular urine sediment from a cat containing numerous red blood cells (RBC), with fewer white blood cells (WBC) (arrow) and NEC (arrowhead). In this case, the SediVue reported false‐negative results for RBC and WBC caused by the extremely crowded nature of the sample; <1 NEC per high power field was observed on both manual microscopy and SediVue analysis. In clinics, this sample would likely be flagged for image review with no results displayed. C, Urine sediment from a dog containing many small calcium oxalate dihydrate crystals (CaOxd*), which were labeled as unidentified crystals (CRYu). D, Urine sediment from a dog containing many calcium oxalate monohydrate crystals that were labeled as struvite crystals (TRI*). A CaOxd was also labeled as TRI. E, Urine sediment from a dog containing many RBC. Occasional RBCs viewed on an angle are identified as TRI. F, Urine sediment from a dog containing debris that was labeled multiple times as TRI (scale bars = 50 μM). *In the version of the SediVue used for the study, non‐squamous epithelial cells were labeled as NEC, calcium oxalate dihydrate crystals were labeled as CaOxd, and struvite crystals were labeled as TRI in the generated images
For nsEPI, 29% of samples positive on manual microscopy (16/56) had false‐negative results on SediVue analysis. Although nsEPI typically were observed in low numbers upon image review, these samples frequently were densely cellular, containing >100 RBC/hpf, >100 WBC/hpf, or both (Figure 2B). Other samples were crowded with large amounts of other elements, such as bacteria or amorphous crystals.
For CaOx Di, 25% of samples positive on manual microscopy (13/52) had false‐negative results on SediVue analysis. In some samples, the crystals were near the limit of resolution for both manual microscopy (×400) and the SediVue, and therefore were too small to visualize the typical calcium oxalate dihydrate structure. In other samples, CaOx Di were recognized as crystals but were labeled as either CRYu or STR by the SediVue (Figure 2C,D).
Specificity for the detection of STR was the lowest of all elements (84%), with 16% of samples negative for STR on manual microscopy (63/403) displaying false‐positive results on SediVue analysis. Image review indicated that the SediVue sometimes misclassified other crystal types (eg, CaOx Di, calcium oxalate monohydrate, amorphous crystals, ammonium biurates), other formed elements (eg, RBC, WBC, nsEPI, when viewed on an angle or in densely crowded samples), and various types of debris or contaminants as STR (Figure 2D‐F). In several samples, STR were present and correctly identified in SediVue images, but, on manual microscopy, only rare STR were observed that were unevenly distributed and did not exceed the required threshold for a positive result.
3.4. Receiver operating characteristic analysis for determination of optimal thresholds
The ROC curves for each element are displayed in Figure 3, comparing both software versions. The ability of the SediVue to detect nsEPI, STR, and CaOx Di visually improved in SW1.0.1.3, but visual improvement was difficult to discern for the detection of RBC, WBC, and sqEPI. The SediVue (SW1.0.1.3) thresholds with optimal sensitivity and specificity were similar to the original thresholds used for RBC, WBC, nsEPI, and STR, but they were lower for sqEPI and CaOx Di (Table 3).
Figure 3.
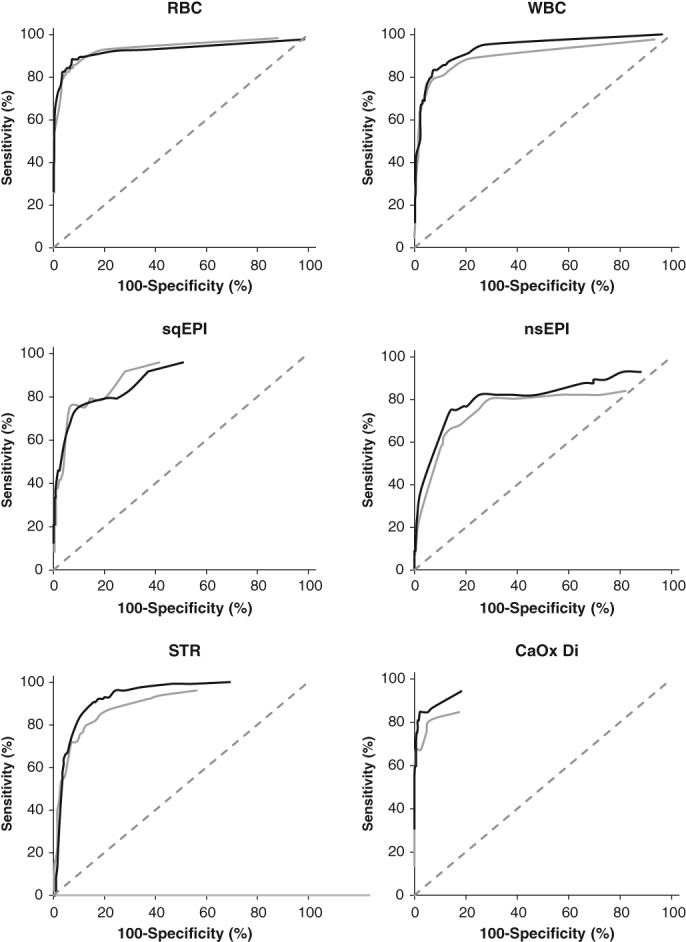
Receiver operating characteristic curves for detection of red blood cells (RBC), white blood cells (WBC), squamous epithelial cells (sqEPI), non‐squamous epithelial cells (nsEPI), struvite crystals (STR), and calcium oxalate dihydrate crystals (CaOx Di) by the SediVue compared with manual microscopy. Software version 1.0.0.0 (gray line) was compared with version 1.0.1.3 (black line)
Table 3.
Optimal SediVue (SW1.0.1.3) thresholds for the detection of formed elements in urine based on ROC analysis with corresponding sensitivities and specificities, as compared to manual microscopy
| Optimal threshold | Sensitivity (95% CI) | Specificity (95% CI) | |
|---|---|---|---|
| RBC | 6.5/hpf | 88.3% (82.5%‐92.7%) | 92.8% (89.6%‐95.2%) |
| WBC | 6.5/hpf | 83.3% (75.7%‐89.4%) | 92.6% (89.6%‐94.9%) |
| sqEPI | 0.2/hpf | 75.0% (53.3%‐90.2%) | 90.3% (87.4%‐92.8%) |
| nsEPI | 0.9/hpf | 75.0% (61.6%‐85.6%) | 85.9% (82.4%‐88.9%) |
| STR | 1.0/hpf | 90.6% (84.1%‐95.0%) | 84.4% (80.5%‐87.8%) |
| CaOx Di | 0.3/hpf | 84.6% (71.9%‐93.1%) | 97.9% (96.2%‐99.0%) |
Abbreviations: CaOx Di, calcium oxalate dihydrate crystals; CI, confidence intervals; hpf, high power field; nsEPI, non‐squamous epithelial cells; RBC, red blood cells; sqEPI, squamous epithelial cells; STR, struvite crystals; WBC, white blood cells.
3.5. Comparison between canine and feline urine samples
The sensitivity of the SediVue (SW1.0.1.3) compared with manual microscopy was similar for the detection of RBC and STR between canine and feline urine samples. Sensitivity for the detection of WBC in feline urine samples was lower than in canine urine samples, although confidence intervals overlapped (Table 4). Four of 5 feline urine samples with false‐negative results for WBC contained >100 RBC/hpf, whereas 1 sample contained many amorphous crystals. The number of feline urine samples positive on manual microscopy for sqEPI, nsEPI, and CaOx Di was too low (n < 10 for each element) to compare sensitivity of the SediVue between species. For all elements except nsEPI, specificity of the SediVue was similar between canine and feline urine samples.
Table 4.
Sensitivity and specificity of the SediVue (SW1.0.1.3) in comparison to manual microscopy for the detection of formed elements in canine (n = 432) versus feline (n = 98) urine using thresholds of ≥5/hpf for RBC and WBC and ≥1/hpf for sqEPI, nsEPI, STR, and CaOx Di
| Sensitivity (95% CI) | Specificity (95% CI) | |||
|---|---|---|---|---|
| Canine | Feline | Canine | Feline | |
| RBC | 87.6% (80.4%‐92.9%) | 92.0% (80.8%‐97.8%) | 91.0% (87.3%‐93.9%) | 85.4% (72.2%‐93.9%) |
| WBC | 88.6% (81.3%‐93.8%) | 58.3% (27.7%‐84.8%) | 87.7% (83.6%‐91.1%) | 91.9% (84.0%‐96.7%) |
| sqEPI | 30.4% (13.2%‐52.9%) | N/A (2.5%‐100.0%) | 99.3% (97.9%‐99.9%) | 100.0% (96.3%‐100.0%) |
| nsEPI | 71.4% (56.7%‐83.4%) | N/A (29.0%‐96.3%) | 85.4%a (81.4%‐88.8%) | 95.6%a (89.1%‐98.8%) |
| STR | 90.2% (82.7%‐95.2%) | 92.0% (74.0%‐99.0%) | 83.3% (78.9%‐87.2%) | 89.0% (79.5%‐95.2%) |
| CaOx Di | 79.6% (65.7%‐89.8%) | N/A (0.0%‐70.8%) | 99.5% (98.1%‐99.9%) | 97.9% (92.6%‐99.7%) |
Abbreviations: CaOx Di, calcium oxalate dihydrate crystals; CI, confidence intervals; hpf, high power field; nsEPI, non‐squamous epithelial cells; N/A, not applicable because of wide confidence interval; RBC, red blood cells; sqEPI, squamous epithelial cells; STR, struvite crystals; WBC, white blood cells.
Comparisons in which confidence intervals did not overlap.
3.6. Comparison of fresh and stored urine samples
Comparison of the SediVue (SW1.0.1.3) with manual microscopy for fresh (n = 227) versus stored (n = 303) urine samples disclosed higher sensitivity for STR and higher specificity for RBC in stored urine (Table 5), but an overlap in confidence intervals was observed for both comparisons. The number of fresh urine samples positive on manual microscopy for sqEPI and CaOx Di was too low (n < 10 for each element) to compare sensitivities.
Table 5.
Sensitivity and specificity of the SediVue (SW1.0.1.3) in comparison to manual microscopy for the detection of formed elements in fresh (n = 227) versus stored (n = 303) urine using thresholds of ≥5/hpf for RBC and WBC and ≥1/hpf for sqEPI, nsEPI, STR, and CaOx Di
| Sensitivity (95% CI) | Specificity (95% CI) | |||
|---|---|---|---|---|
| Fresh | Stored | Fresh | Stored | |
| RBC | 89.0% (79.5%‐95.2%) | 88.8% (80.8%‐94.3%) | 85.1% (78.4%‐90.3%) | 94.2% (90.0%‐96.9%) |
| WBC | 89.5% (75.2%‐97.1%) | 84.1% (74.8%‐91.0%) | 87.8% (82.3%‐92.1%) | 89.3% (84.4%‐93.1%) |
| sqEPI | N/A (0.0%‐84.2%) | 36.4% (17.2%‐59.3%) | 100.0% (98.4%‐100.0%) | 98.9% (96.9%‐99.8%) |
| nsEPI | 72.2% (46.5%‐90.3%) | 71.1% (54.1%‐84.6%) | 90.9% (86.2%‐94.4%) | 84.5% (79.6%‐88.7%) |
| STR | 75.0% (42.8%‐94.5%) | 92.2% (85.7%‐96.4%) | 87.0% (81.7%‐91.2%) | 81.4% (75.1%‐86.7%) |
| CaOx Di | N/A (0.0%‐70.8%) | 79.6% (65.7%‐89.8%) | 99.1% (96.8%‐99.9%) | 99.2% (97.2%‐99.9%) |
Abbreviations: CaOx Di, calcium oxalate dihydrate crystals; CI, confidence intervals; hpf, high power field; nsEPI, non‐squamous epithelial cells; N/A, not applicable because of wide confidence interval; RBC, red blood cells; sqEPI, squamous epithelial cells; STR, struvite crystals; WBC, white blood cells.
3.7. Volume comparison
Urine samples were compared to determine if the variation in volume centrifuged for manual microscopy impacted sensitivity or specificity for formed element detection (Table 6). Increased sensitivity with increasing volume centrifuged was observed for most elements, but confidence intervals overlapped.
Table 6.
Sensitivity and specificity of the SediVue (SW1.0.1.3) in comparison to manual microscopy for the detection of formed elements in urine samples based on different volumes centrifuged for the manual method (n = 163 for ≤2.5 mL, n = 367 for >2.5 mL)
| Sensitivity (95% CI) | Specificity (95% CI) | |||
|---|---|---|---|---|
| ≤2.5 mL | >2.5 mL | ≤2.5 mL | >2.5 mL | |
| RBC | 84.6% (71.9%‐93.1%) | 90.8% (84.1%‐95.3%) | 87.4% (79.7%‐92.9%) | 91.5% (87.4%‐94.7%) |
| WBC | 80.0% (61.4%‐92.3%) | 87.5% (79.2%‐93.4%) | 88.7% (82.1%‐93.6%) | 88.6% (84.2%‐92.1%) |
| sqEPI | 60.0% (14.7%‐94.7%) | 26.3% (9.2%‐51.2%) | 99.4% (96.5%‐100.0%) | 99.4% (97.9%‐99.9%) |
| nsEPI | 68.8% (41.3%‐89.0%) | 72.5% (56.1%‐85.4%) | 87.8% (81.3%‐92.6%) | 87.2% (83.0%‐90.6%) |
| STR | 89.5% (75.2%‐97.1%) | 91.0% (83.1%‐96.0%) | 80.0% (71.9%‐86.6%) | 86.3% (81.7%‐90.1%) |
| CaOx Di | 61.1% (35.8%‐82.7%) | 82.4% (65.5%‐93.2%) | 100.0% (97.5%‐100.0%) | 98.8% (97.0%‐99.7%) |
Abbreviations: CaOx Di, calcium oxalate dihydrate crystals; CI, confidence intervals; nsEPI, non‐squamous epithelial cells; RBC, red blood cells; sqEPI, squamous epithelial cells; STR, struvite crystals; WBC white blood cells.
3.8. Intra‐ and inter‐assay precision for RBC and WBC
Intra‐ and inter‐assay precision for QCM (Table 7) and intra‐assay precision for patient samples (Table 8) indicated that average coefficients of variation (CVs) for each category were <20%.
Table 7.
Intra‐ and inter‐assay precision of the SediVue (SW1.0.1.3) using quality control material
| Intra‐assay | Inter‐assay | ||||
|---|---|---|---|---|---|
| Number of cells/hpf, mean (range) | %CV | Number of cells/hpf, mean (range) | %CV | ||
| RBC | Normal | N/Aa | N/Aa | N/Aa | N/Aa |
| Abnormal | 33.6 (30.8‐35.9) | 4.6 | 34.7 (30.8‐37.1) | 7.6 | |
| WBC | Normal | 2.6 (1.9‐3.3) | 15.6 | 2.6 (2.6‐2.7) | 1.3 |
| Abnormal | 32.1 (27.7‐39.5) | 10.6 | 33.0 (29.1‐35.0) | 7.1 | |
Abbreviations: CV, coefficient of variation; hpf, high power field; N/A, not applicable; RBC, red blood cells; WBC, white blood cells.
The CV could not be calculated for RBC in “normal” QCM, as the numbers of RBC present were too low (<1/hpf).
Table 8.
Intra‐assay precision of the SediVue (SW1.0.1.3) using patient samples
| Number of samples | Number of cells/hpf, mean (range) | %CV, mean (range) | ||
|---|---|---|---|---|
| RBC | Low | 5 | 2.9 (1.1‐5.4) | 19.3 (11.9‐25.4) |
| Medium | 4 | 13.2 (5.6‐19.2) | 9.9 (6.9‐14.7) | |
| High | 1 | 26.4 (24.9‐28.1) | 4.5 | |
| WBC | Low | 3 | 2.1 (0.8‐3.8) | 10.6 (7.9‐12.0) |
| Medium | 6 | 11.6 (5.5‐18.6) | 11.9 (8.0‐16.6) | |
| High | 3 | 41.7 (28.6‐54.4) | 10.3 (7.3‐15.9) |
Abbreviations: CV, coefficient of variation; hpf, high power field; RBC, red blood cells; WBC, white blood cells.
3.9. Dilutional linearity for RBC
Mean percentages of recovery obtained by the linearity under dilution test in samples 1, 2, and 3 were 114%, 89%, and 179%, respectively. Samples 1 and 2 yielded recovery values between 80% and 120%, except for the 1:16 dilution (132%) for sample 1 and 1:64 dilution (25%) for sample 2. Recovery values for sample 3 ranged from 152% to 207% when expected values were based on the original, undiluted sample. However, recoveries ranged from 112% to 136% when expected values were based on the 1:2 dilution.
Recovery values ranged from 91% to 102% (mean, 95%) for the high‐RBC solutions mixed in various ratios.
4. DISCUSSION
We evaluated the performance of the SediVue as compared to manual microscopy for the detection of cells and 2 common crystal types in urine. Overall, the ability of the SediVue (SW1.0.1.3) to detect clinically relevant numbers of formed elements was considered acceptable for RBC, WBC, STR, and CaOx Di (ie, both sensitivity and specificity ranged from moderate to excellent), whereas improvement is needed for accurate detection of sqEPI and nsEPI. Review of the provided images was crucial in determining the reasons for false‐positive and false‐negative results and supports the opportunity for improved SediVue results when complemented with image reviews.
Sensitivity of the SediVue was lowest for sqEPI, nsEPI, and CaOx Di. The poor sensitivity for sqEPI (33%) partly could be because of the low number of samples positive for sqEPI on manual microscopy. Although many samples contained sqEPI scattered throughout, only 5% contained enough sqEPI to exceed the positive threshold (≥1/hpf), resulting in a wide confidence interval. Regardless, the upper confidence limit (55%) was poor, supporting the need for improvement in sqEPI detection. In the false‐negative samples, the SediVue detected an average of 0.3 versus 1.8 sqEPI/hpf on manual microscopy. This small difference should not impact clinical decision‐making, because squamous epithelial cells in urine typically are considered an incidental finding. However, several of these samples had false‐positive results for nsEPI, and a review of the SediVue images confirmed mislabeling of sqEPI as nsEPI in some samples (Figure 2A). This mislabeling could have a clinical impact, because misclassification of sqEPI as nsEPI may raise suspicion for a pathological process (eg, transitional cell carcinoma). Because epithelial cell types can be difficult to distinguish even for human observers,18 some reference laboratories do not routinely distinguish between sqEPI and nsEPI, reporting both types as “epithelial cells.” Therefore, in some samples, the SediVue may have correctly labeled cells as nsEPI that were incorrectly labeled as sqEPI on manual microscopy.
Several causes are suspected for the relatively low sensitivities for nsEPI and CaOx Di. For nsEPI, the low sensitivity partially was caused by overshadowing of relatively low numbers of nsEPI by large numbers of WBC and RBC (Figure 2B), and dilution might have enhanced nsEPI detection in these samples. For CaOx Di, several false‐negative samples contained CaOx Di that were extremely small and difficult to distinguish from background debris on manual microscopy. In other samples, CaOx Di were labeled as CRYu, indicating they were identified as crystals but were not subtyped as CaOx Di (Figure 2C). Occasional mislabeling of CaOx Di as STR also supports the need for improvement in CaOx Di detection (Figure 2D).
Overall, the specificity of the SediVue for the detection of formed elements was good to excellent. The lowest specificity was for STR. In some samples with false‐positive results, scattered STR were observed on manual microscopy, but their number was too small to classify as a positive result. The higher number of STR/hpf detected by the SediVue in some samples compared to manual microscopy may indicate increased sedimentation of formed elements by the SediVue or more even distribution of elements in SediVue preparations. Additionally, both software versions evaluated often labeled a single STR multiple times (ie, opposite ends of the same crystal), contributing to the overestimation of STR. In some samples, other crystal types, debris, and environmental contaminants were mislabeled as STR (Figure 2F), supporting the need for review of images as well as continued enhancement in specificity for STR detection.
Several software iterations have been developed to improve the detection of formed elements, and one of the latest versions (SW1.0.1.3) was compared with one of the earliest versions (SW1.0.0.0). The sensitivity of the SediVue for the detection of all formed elements increased with the later version, at the expense of a typically mild decrease in specificity.
Furthermore, ROC analysis was utilized to determine optimal thresholds for the detection of each element when compared with the originally defined thresholds for manual microscopy. This substantiated the use of the original thresholds for most elements. However, optimal thresholds for sqEPI and CaOx Di were notably lower, and using the optimal threshold resulted in a substantial increase in sensitivity, albeit at the expense of a mild to moderate decrease in specificity. To determine whether threshold adjustments for these 2 elements should be considered, further evaluation is needed because of the low number of positive samples in our study.
The apparent lower sensitivity for WBC detection in feline urine samples compared to canine urine samples partially could be because of the low number of feline urine samples that contained ≥5 WBC/hpf on manual microscopy (n = 12), which resulted in a wide confidence interval for sensitivity. Additionally, feline urine samples with false‐negative results contained large numbers of RBC or amorphous crystals, interfering with the identification of WBC by the SediVue. Dilution of such samples might have increased sensitivity for WBC. Analysis of additional pyuric feline urine samples would help to more accurately determine sensitivity.
Urine samples used in our study included fresh and stored samples. Overall, performance was similar between methods for each sample type. However, in stored samples, the SediVue demonstrated apparently higher sensitivity for STR and higher specificity for RBC. The majority of samples positive for STR on manual microscopy were stored before analysis (n = 115), whereas substantially fewer fresh samples were positive (n = 12). This difference resulted in a wide 95% confidence interval for sensitivity for fresh urine samples, and the sensitivity in stored urine samples likely better reflects the true sensitivity for STR detection. The reason for higher specificity for RBC in stored urine samples is unclear but might be partially because of the larger number of stored urine samples that were negative for RBC on manual microscopy (n = 205 for stored urine samples versus n = 154 for fresh urine samples).
The SediVue displayed good precision for detecting RBC and WBC, especially compared to that reported for manual microscopy. In several studies, the imprecision of manual microscopy is consistently higher than that reported for automated methods. For manual microscopy, CVs as low as 8.5% have been reported for urine samples with large numbers of cells, but CVs typically exceed 40% in urine samples of low cellularity (Chase J, Hammond J, Bilbrough G, DeNicola DB. Examination of imprecision and effectiveness of different centrifugation and uncentrifugation methods for urine sediment microscopic evaluation (abstract). Vet Clin Pathol. 2015;44(4):E13).4,7‐10 In our study, imprecision of the SediVue also tended to be greater, but still typically <20%, in samples with low cell counts. Intra‐assay precision based on QCM and patient urine samples were comparable, supporting sufficient repeatability for patient urine samples that are reanalyzed within a short period of time (ie, 1 hour). However, this is not true for urine samples reanalyzed within 1 or several days, because cells and other formed elements degenerate, whereas crystals can proliferate, obscuring other elements (personal observation).
Manual microscopic examination was used as the gold standard in our study, but it has several limitations. It is time‐consuming, veterinarians and technicians may not feel comfortable identifying formed elements, and manual preparation of urine sediments is poorly standardized. Variables such as volume of urine centrifuged, force of centrifugation, and method used for sediment examination can vary markedly among clinics. Substantial inter‐operator variation may exist during urine sediment preparation. Even if performed by a single person, variation may occur in the amount of supernatant used for sediment resuspension or the volume of sediment examined. Manual microscopy also has high interobserver variability regarding the identification and quantification of formed elements, further contributing to imprecision.1, 2, 3 Lastly, standard centrifugation of urine may destroy fragile elements, creating inaccuracies.2
Automated urine sediment analysis by the SediVue addresses many of the limitations of manual microscopy. It is quick and simple to perform and requires only a small volume of uncentrifuged urine, both of which might increase the number of complete urinalyses that are performed in practice. Urine does not need to be centrifuged before analysis, decreasing the number of steps involved and eliminating several sources of variability. Additionally, the volume of urine and force of centrifugation are standardized. The SediVue utilizes a short and gentle centrifugation technique, which could minimize destruction of fragile formed elements. Furthermore, our study supports that precision is higher for the SediVue compared to that reported for human observers for the detection and quantification of RBC and WBC in urine sediments.1, 2, 3 Notably, the ability of the SediVue to capture and display images of urine sediment is a major benefit that could minimize the need for personnel to examine urine under the microscope. Future studies should evaluate the utility of the provided images in decreasing the need for manual microscopy.
Despite these advantages, there are several limitations associated with this instrument. Densely cellular samples will likely need to be diluted for the SediVue to accurately identify and quantify formed elements. Samples were not diluted in our study for consistency in the experimental protocol. Results from densely cellular samples (Figure 2B) often were inaccurate and likely had a negative impact on the reported sensitivities and specificities. In the commercial version of the SediVue, results are not reported if a sample is too crowded to accurately identify and quantify formed elements. Instead, the run is flagged, and the instrument prompts the operator to dilute the sample. Based on the results of our study, dilution of samples with large numbers of RBCs produces linear results, particularly when the starting concentration is ≤1000‐1500 RBC/hpf. Additional studies are warranted to evaluate the detection of other formed elements after dilution of densely cellular samples.
Another limitation of the SediVue is the impact of lipid droplets on performance. Lipid droplets are common in urine samples from veterinary species. Because lipids reside in the plane above other formed elements, the SediVue camera sometimes focuses on the lipid plane rather than on the cellular plane, producing out‐of‐focus images, particularly in lowly cellular samples with many lipid droplets (Figure 4). It is possible that the centrifugation time of 10 seconds used in our study was inadequate for the separation of lipids from other formed elements for some samples. The current configuration of the SediVue has an increased centrifugation time, which could improve SediVue performance for general formed element identification. Further studies investigating the impact of this higher centrifugation time on lipid interference are needed.
Figure 4.
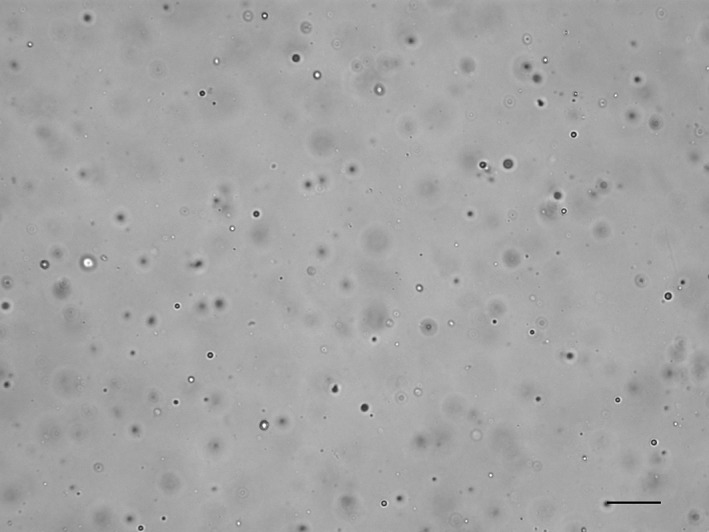
SediVue image from a lowly cellular, unstained urine sediment from a dog containing many lipid droplets, where the image is focused on the lipid plane rather than on the few cells present. On manual microscopy, red and white blood cells were <1/high power field, and no epithelial cells or crystals were observed (scale bar = 50 μM)
Our study had several limitations. In an attempt to minimize inter‐operator variation regarding the identification and quantification of formed elements, at least 2 observers evaluated each sediment and reached a majority decision regarding the type and semiquantitative category of elements present. However, minor differences in operator performance, including the steps involved in urine sediment preparation, cannot be completely excluded as a source of variation. Additionally, definitive identification of elements in urine sediments can be challenging even for experienced technicians and pathologists.
In our study, a single method for urine sediment preparation and evaluation was used. Although doing so helped standardize the manual preparation of urine, it limited the ability to apply the results to other laboratories. KOVA tubes and DeciSlides were selected, because these are the standard materials used by our clinical pathology laboratory for routine processing of urine samples. Many reference laboratories and clinics use other preparation methods (eg, slide and coverslip) for microscopic examination. In our experience, when comparing the DeciSlide and slide‐and‐coverslip methods, urine sediments will appear more concentrated on the DeciSlide preparation. Therefore, the method used in our study does not necessarily reflect the variety of methods utilized by veterinarians in practice.
The volume of urine centrifuged was not standardized in our study in order to maximize the number of samples available for analysis. Although it is preferable to standardize the volume centrifuged, a standard volume of urine is not always submitted to the laboratory. For the majority of samples in our study (88%), 2.0‐3.0 mL urine was centrifuged. Our experience (Chase J, Hammond J, Bilbrough G, DeNicola DB. Examination of imprecision and effectiveness of different centrifugation and uncentrifugation methods for urine sediment microscopic evaluation (abstract). Vet Clin Pathol. 2015;44(4):E13.) and results from our study support the contention that the volume of urine centrifuged does not substantially impact sediment results when comparing small differences in centrifuged volumes. In fact, decreased sensitivity would be expected for the SediVue when higher volumes were centrifuged. However, we observed the opposite, likely because of the lower number of samples with ≤2.5 mL centrifuged and therefore wider confidence intervals in this group.
Our study protocol emphasized the need to thoroughly mix urine immediately before SediVue analysis, but a similar emphasis was not placed on mixing urine samples for manual microscopic examination before centrifugation. Although mixing urine samples before aliquoting for centrifugation should be standard practice in the laboratory, it is possible that the lack of emphasis on this aspect of urine sediment preparation could have resulted in inconsistencies that affected study results. Because formed elements in urine may settle in as little as 15 seconds (Hammond J, Ericson C, Myrick C, et al. Impact of urine formed element settling and sample aspiration location on microscopic urinalysis (abstract). ACVP/ASVCP Concurrent Annual Meeting; New Orleans, Louisiana; December 3‐7, 2016.), proper mixing of urine is essential to obtain representative results not only for SediVue analysis but also for preparation of urine sediments if more urine is collected than centrifuged for manual microscopy.19
Despite the large number of urine samples included in our study, the number of samples positive on manual microscopy for certain elements was low (particularly sqEPI, nsEPI, and CaOx Di), resulting in wide 95% confidence intervals for sensitivity. This was especially a problem for feline urine samples, because they composed a minority of samples. In the future, collection of additional samples would help to more accurately determine the sensitivity of the SediVue for the detection of these elements, particularly in feline urine. For specificity, the 95% confidence intervals generally were much narrower because of the relatively large number of samples considered negative for each element. Therefore, the specificities observed in our study are more likely to be representative of the SediVue for all elements.
Lastly, although our study determined the ability of the SediVue to accurately detect the presence or absence of clinically relevant numbers of cells and crystals, correlation of the number of elements identified per hpf between the 2 methods was not evaluated. For example, a sample with 10 RBC/hpf on manual microscopy and 50 RBC/hpf on SediVue analysis would be considered a true positive (both methods exceeded ≥5/hpf), despite the lack of a close correlation between numerical results.
In summary, our results support that clinical interpretation would be similar between the SediVue and manual microscopy in the majority of urine samples for the formed elements evaluated, but further improvement is needed for the identification of epithelial cells. Additionally, the images provided by the SediVue may decrease the need for manual microscopy in many situations. The extent to which these images may increase the efficiency and ease of performing complete urinalyses should be evaluated in future studies. Evaluation of future software versions will be necessary to determine if updates to the CNN algorithm continue to enhance the accuracy of the detection of formed elements.
CONFLICT OF INTEREST DECLARATION
This study was supported by IDEXX Laboratories, Inc. S. Edwards is employed by and G.E.A. Bilbrough, D.B. DeNicola, C. Myrick, and J.M. Hammond are employed by and have stock or stock options with IDEXX Laboratories, Inc. M.B. Nabity has served on an advisory board and received travel reimbursement and speaker honoraria from IDEXX Laboratories, Inc. within the past 3 years. A.M. Hernandez and A.N. Myers received reimbursement for travel expenses incurred as a result of this study. A.M. Hernandez was employed by IDEXX Laboratories, Inc after study completion.
OFF‐LABEL ANTIMICROBIAL DECLARATION
Authors declare no off‐label use of antimicrobials.
INSTITUTIONAL ANIMAL CARE AND USE COMMITTEE (IACUC) OR OTHER APPROVAL DECLARATION
Authors declare no IACUC or other approval was needed.
HUMAN ETHICS APPROVAL DECLARATION
Authors declare human ethics approval was not needed for this study.
ACKNOWLEDGMENTS
The authors acknowledge the laboratory personnel at Texas A&M University and the IDEXX Reference Laboratory in North Grafton, MA. Work was performed in the Department of Veterinary Pathobiology at Texas A&M University College of Veterinary Medicine and Biomedical Sciences, College Station, TX, and at IDEXX Laboratories, North Grafton, MA. The open access publishing fees for this article have been covered by the Texas A&M University Open Access to Knowledge Fund (OAKFund), supported by the University Libraries and the Office of the Vice President for Research. The study was presented in part at the 2016 ACVP and ASVCP Concurrent Annual Meeting, New Orleans, LA, and at the 2017 Annual ESVCP/ECVCP Congress, London, United Kingdom.
Hernandez AM, Bilbrough GEA, DeNicola DB, et al. Comparison of the performance of the IDEXX SediVue Dx® with manual microscopy for the detection of cells and 2 crystal types in canine and feline urine. J Vet Intern Med. 2019;33:167–177. 10.1111/jvim.15341
Funding information: IDEXX Laboratories, Inc
REFERENCES
- 1. Winkel P, Statland BE, Jorgensen K. Urine microscopy, an ill‐defined method, examined by a multifactorial technique. Clin Chem. 1974;20:436‐439. [PubMed] [Google Scholar]
- 2. Carlson DA, Statland BE. Automated urinalysis. Clin Lab Med. 1988;8:449‐461. [PubMed] [Google Scholar]
- 3. Wald R, Bell CM, Nisenbaum R, et al. Interobserver reliability of urine sediment interpretation. Clin J Am Soc Nephrol. 2009;4:567‐571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fenili D, Pirovano B. The automation of sediment urinalysis using a new urine flow cytometer (UF‐100). Clin Chem Lab Med. 1998;36:909‐917. [DOI] [PubMed] [Google Scholar]
- 5. Linko S, Kouri TT, Toivonen E, Ranta PH, Chapoulaud E, Lalla M. Analytical performance of the Iris iQ200 automated urine microscopy analyzer. Clin Chim Acta. 2006;372:54‐64. [DOI] [PubMed] [Google Scholar]
- 6. Zaman Z, Fogazzi GB, Garigali G, Croci MD, Bayer G, Kránicz T. Urine sediment analysis: analytical and diagnostic performance of sediMAX—a new automated microscopy image‐based urine sediment analyser. Clin Chim Acta. 2010;411:147‐154. [DOI] [PubMed] [Google Scholar]
- 7. Akin OK, Serdar MA, Cizmeci Z, Genc O, Aydin S. Comparison of LabUMat‐with‐UriSed and iQ200 fully automatic urine sediment analysers with manual urine analysis. Biotechnol Appl Biochem. 2009;53:139‐144. [DOI] [PubMed] [Google Scholar]
- 8. Chien TI, Kao JT, Liu HL, et al. Urine sediment examination: a comparison of automated urinalysis systems and manual microscopy. Clin Chim Acta. 2007;384:28‐34. [DOI] [PubMed] [Google Scholar]
- 9. Mayo S, Acevedo D, Quinones‐Torrelo C, et al. Clinical laboratory automated urinalysis: comparison among automated microscopy, flow cytometry, two test strips analyzers, and manual microscopic examination of the urine sediments. J Clin Lab Anal. 2008;22:262‐270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yuksel H, Kilic E, Ekinci A, et al. Comparison of fully automated urine sediment analyzers H800‐FUS100 and LabUMat‐UriSed with manual microscopy. J Clin Lab Anal. 2013;27:312‐316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Alves L, Ballester F, Camps J, Joven J. Preliminary evaluation of the Iris IQ 200 automated urine analyser. Clin Chem Lab Med. 2005;43:967‐970. [DOI] [PubMed] [Google Scholar]
- 12. Bottini PV, Martinez MH, Garlipp CR. Urinalysis: comparison between microscopic analysis and a new automated microscopy image‐based urine sediment instrument. Clin Lab. 2014;60:693‐697. [DOI] [PubMed] [Google Scholar]
- 13. Hannemann‐Pohl K, Kampf SC. Automation of urine sediment examination: a comparison of the Sysmex UF‐100 automated flow cytometer with routine manual diagnosis (microscopy, test strips, and bacterial culture). Clin Chem Lab Med. 1999;37:753‐764. [DOI] [PubMed] [Google Scholar]
- 14. Roggeman S, Zaman Z. Safely reducing manual urine microscopy analyses by combining urine flow cytometer and strip results. Am J Clin Pathol. 2001;116:872‐878. [DOI] [PubMed] [Google Scholar]
- 15. Shayanfar N, Tobler U, von Eckardstein A, Bestmann L. Automated urinalysis: first experiences and a comparison between the iris iQ200 urine microscopy system, the Sysmex UF‐100 flow cytometer and manual microscopic particle counting. Clin Chem Lab Med. 2007;45:1251‐1256. [DOI] [PubMed] [Google Scholar]
- 16. Bogaert L, Peeters B, Billen J. Evaluation of a new automated microscopy urine sediment analyser—sediMAX conTRUST(R). Acta Clin Belg. 2017;72:91‐94. [DOI] [PubMed] [Google Scholar]
- 17. Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics. 1977;33:159‐174. [PubMed] [Google Scholar]
- 18. Stockham SL, Scott MA, System U. In: Stockham SL, Scott MA, eds. Fundamentals of Veterinary Clinical Pathology. 2nd ed. Ames, IA: Blackwell Publishing; 2008:473‐474. [Google Scholar]
- 19. Osborne CA, SJB. Urine sediment: under the microscope In: Osborne CA, Stevens JB, eds. Urinalysis: A Clinical Guide to Compassionate Patient Care. Shawnee Mission, KS: Bayer Corporation; 1999:127. [Google Scholar]


