Summary
Relapse of acute myeloid leukemia (AML) remains a significant clinical challenge due to limited therapeutic options and poor prognosis. Leukemic stem cells (LSCs) are the cellular units responsible for relapse in AML, and strategies that target LSCs are thus critical. One proposed potential strategy to this end is to break the quiescent state of LSCs, thereby sensitizing LSCs to conventional cytostatics. The hypoxia-inducible factor (HIF) pathway is a main driver of cellular quiescence and a potential therapeutic target, with precedence from both solid cancers and leukemias. Here, we used a conditional knockout Hif-1α mouse model together with a standard chemotherapy regimen to evaluate LSC targeting in AML. Contrary to expectation, our studies revealed that Hif-1α-deleted-leukemias displayed a faster disease progression after chemotherapy. Our studies thereby challenge the general notion of cancer stem cell sensitization by inhibition of the HIF pathway, and warrant caution when applying HIF inhibition in combination with chemotherapy in AML.
Keywords: hypoxia, HIF-1α, acute myeloid leukemia, single-cell transcriptional analysis, chemotherapy, mouse model
Graphical Abstract
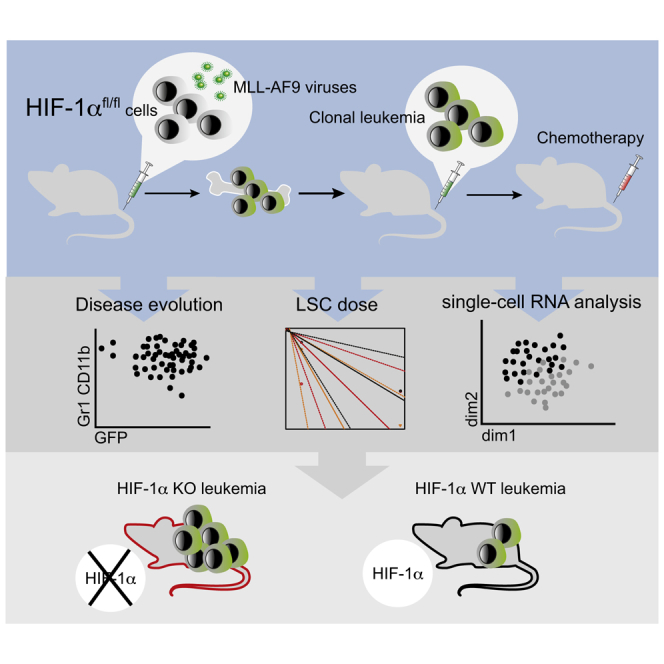
Highlights
-
•
Deletion of Hif-1α accelerates the progression of chemotherapy-treated MLL-AF9-AML
-
•
Deletion of Hif-1α does not decrease LSC frequency after chemotherapy
-
•
Chemotherapy targets more mature cells indicated by transcriptional analysis
-
•
Hif-1α deletion affects few transcriptional pathways in AML cells
In this article, Bryder, Velasco-Hernandez and colleagues show that the combination of chemotherapy and Hif-1α deletion in a murine model of MLL-AF9-AML leads to a faster progression of the disease without altering LSCs frequency. Single-cell transcriptional analysis indicates that Hif-1α deletion leads to few transcriptional modifications.
Introduction
Acute myeloid leukemia (AML) is a clonal disorder characterized by a rapid accumulation of differentiation-arrested myeloid blasts. Remission can be achieved in most patients by a combination of cytarabine (AraC) and anthracycline therapy. However, few treatment improvements have arisen over the past decades (Rowe and Tallman, 2010) and 40%–60% of patients still relapse (Burnett et al., 2011, Yanada et al., 2008), which is associated in general with limited treatment options (Dohner et al., 2010).
Relapse is attributed to a minor subpopulation of cells referred to as leukemic stem cells (LSCs). LSCs are resilient to cytotoxic effects of chemotherapy, via mechanisms that include resistance to apoptosis, increased capacities to efflux drugs, and relative quiescence (Thomas and Majeti, 2017). This quiescence in turn underlies resistance to compounds that target energy metabolism (Essers and Trumpp, 2010).
Cellular quiescence is typically related to activation of hypoxia-signaling pathway, driven by hypoxia-inducible factor (HIF) complex, which constitutes a family of 3 heterodimeric transcription factors HIF-1, HIF-2, and HIF-3 (Flamme et al., 1997, Makino et al., 2002, Semenza and Wang, 1992). The oxygen-dependent regulation of HIFs depends on stabilization of an associated α subunit. Hence, at oxygen levels above 5%, the α subunit is proteasomally degraded. In contrast, under hypoxic conditions the α subunit is stabilized, dimerizes with constitutively expressed β subunit, and promotes transcription of target genes regulating key cellular processes such as angiogenesis, proliferation, metabolism, and apoptosis (Semenza, 2003).
The most primitive hematopoietic stem cells (HSCs) reside in hypoxic niches in the bone marrow (BM) (Morrison and Scadden, 2014). Thereby, HSCs are kept dormant and free of genetic changes that mostly occur during DNA replication. However, while HIF-1α signaling was reported to critically regulate HSC maintenance (Takubo et al., 2010), other studies showed that depletion of HIF-1α/HIF-2α had no such effect (Guitart et al., 2013). The HIF pathway has also been proposed to play important roles in AML, a notion supported by the preferential localization of chemotherapy-resistant AML cells to the hypoxic endosteal niche in the BM (Ishikawa et al., 2007). Accordingly, several studies have shown that loss of HIF-1/2 leads to abrogation of LSCs (Rouault-Pierre et al., 2013, Wang et al., 2011, Zhang et al., 2012). Contrasting these results are studies demonstrating that loss of HIF-1/2 does not impact on mouse models of AML, or alternatively can give rise to an even more severe leukemic phenotype (Velasco-Hernandez et al., 2014, Vukovic et al., 2015). Nonetheless, targeting hypoxia and HIFs has been considered a key anticancer approach (Frolova et al., 2012, Rouault-Pierre et al., 2013, Wang et al., 2011).
Here, we investigated the possibility to sensitize LSCs from a mouse model of AML to a clinically relevant chemotherapy regimen by targeting the hypoxia pathway via Hif-1α. Contrary to expectation, our work suggests a more aggressive disease outcome upon Hif-1α deletion, which challenges the general use of hypoxia targeting for therapeutic benefit in AML.
Results
Deletion of Hif-1α Accelerates the Progression of Chemotherapy-Treated Leukemia
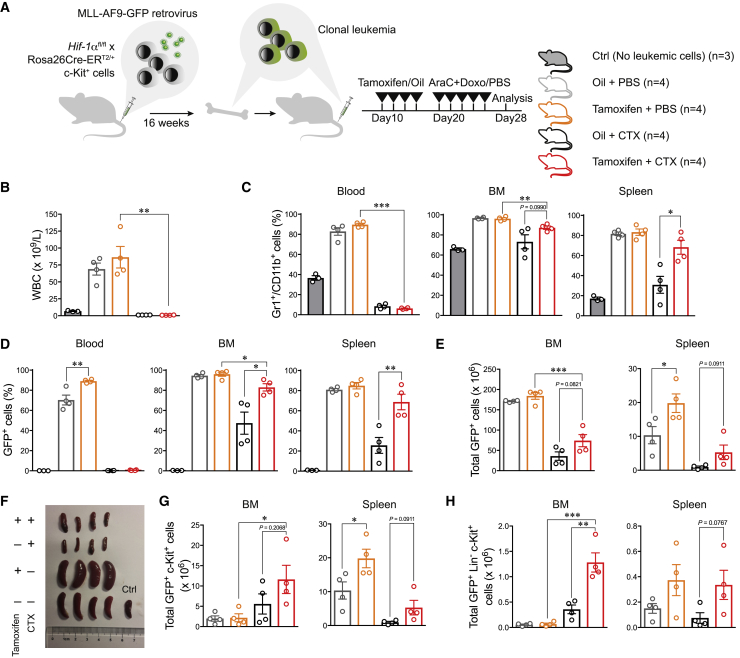
To study effects of Hif-1α on the sensitivity of LSCs to chemotherapy, we bred Hif-1αfl mice (Ryan et al., 2000) with Rosa26Cre-ERT2 mice (Ventura et al., 2007), generating a model that allows for Hif-1α deletion upon tamoxifen treatment (Figure S1A). c-Kit+ cells from Hif-1αfl/fl;Rosa26Cre-ERT2/+ mice were transduced with MLL-AF9-GFP retrovirus and transplanted into mice, which led to development of MLL-AF9-driven AML. Primary leukemias were next used to study AML progression in secondary hosts.
After comparing two different chemotherapy protocols (Figure S1B), we used the one described by Zuber et al. (2009), which resulted in severely decreased white blood cells, erythrocytes, and platelets, akin to what is observed in human patients. We then titrated the latency of the disease using different leukemic cell doses (Figure S1C) and chose a dose of 104 cells, which ensured a sufficiently long latency for posterior procedures. Cells from individual MLL-AF9-leukemic mice were transplanted into new recipients and subjected to Hif-1α deletion 10 days after transplantation, and to a standard chemotherapeutic protocol at day 20. As a control, we included mice injected with both vehicles but receiving no leukemic cells (Figure 1A).
Figure 1.
Deletion of Hif-1α Leads to a Faster Progression of AML after Chemotherapy Treatment
(A–H) Experimental design for the generation of leukemic clones, deletion of Hif-1α, and applied chemotherapy protocol (A). At day 28 after transplantation, mice were sacrificed and disease evolution was examined according to different parameters: total white blood cell count in PB (B), myeloid cells (Gr1+-CD11b+) (C), GFP+ cells in PB, BM, and spleen (D and E), splenomegaly (F), GFP+ c-Kit+ cells in BM and spleen (G), and GFP+ Lin– c-Kit+ in BM and spleen (H). Results are from 1 of 2 experiments and depict results from 4 mice per group (3 for wild-type control mice). Plots represent mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.01. AraC, cytarabine; Dox, doxorubicin; CTX, chemotherapy; BM, bone marrow; Ctrl, control.
See also Figures S1 and S2.
Three days after the last injection, we started to observe mortality in the PBS group (Figure S2A). We collected peripheral blood (PB), BM, spleen, and liver samples from half of the cohort and analyzed multiple disease parameters (Figures 1B–1F and S2). As expected, disease progressed more slowly in the chemotherapy-treated (CTX) mice (Figures 1B–1F). More intriguingly, we observed a faster disease evolution upon Hif-1α deletion. This was seen in both CTX and PBS groups (Figures 1C–1F). When we assessed phenotypically more primitive GFP+ c-Kit+ or GFP+ Lin– c-Kit+ cells, we observed a higher abundance in the Hif-1α-deleted (Hif-1α–/–) than in the Hif-1α-intact (Hif-1α+/+) group after chemotherapy (Figures 1G, 1H, and S2). While these populations were very infrequent in BM of PBS groups, with no clear differences between Hif-1α–/– and Hif-1α+/+ cells, a higher number of such cells upon Hif-1α deletion were observed in the spleens of the same animals. Taken together, these results demonstrate an increment of potential LSCs (c-Kit+) upon Hif-1α–/– deletion, which was particularly significant after chemotherapy.
Deletion of Hif-1α Does Not Decrease the Frequency of LSCs after Chemotherapy Treatment
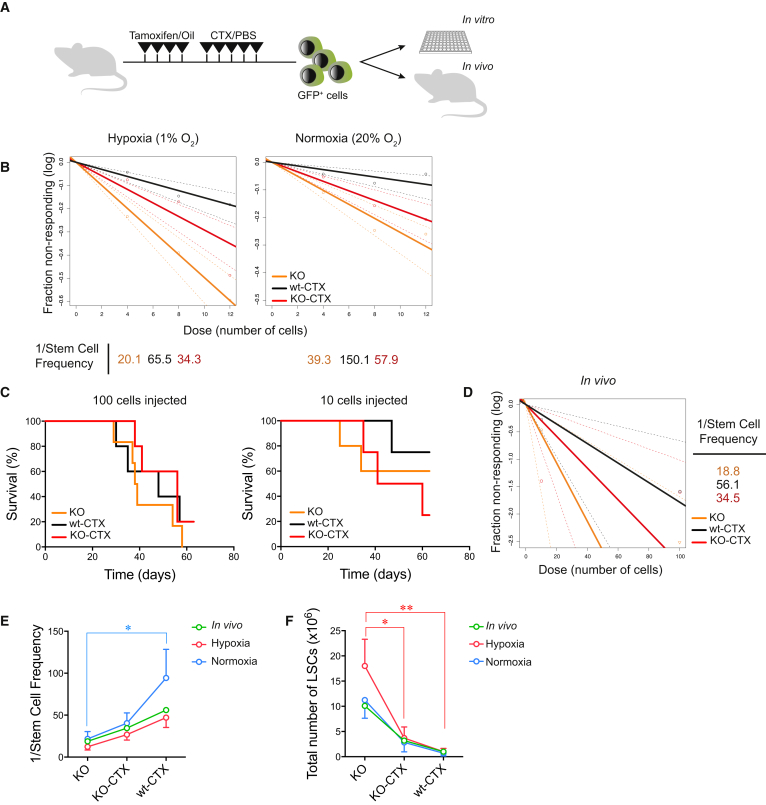
To evaluate the impact of chemotherapy and Hif-1α deletion on LSCs more directly, we performed limiting dilution analysis (LDA) (Figure 2A).
Figure 2.
Deletion of Hif-1α Does Not Decrease LSC Frequency after Chemotherapy
(A) Experimental design for the analysis of LSC frequency.
(B) Limiting dilution analysis (LDA) from in vitro data. Variable numbers of GFP+ cells (4, 8, or 12 cells) from the 3 experimental groups were cultured in vitro under normoxic or hypoxic conditions and proliferative capacity was evaluated after 10 days. LSC frequencies were calculated using ELDA software. Plots show 1 representative of 3 independent experiments (cells from 3 different donors) with 96 replicates per group and condition.
(C) Survival curves of mice transplanted with 10 or 100 GFP+ cells from mice treated with the combination of tamoxifen + chemotherapy (KO-CTX), tamoxifen + PBS (KO-PBS), or oil + chemotherapy (wt-CTX) (n = 4–6 mice per group from 1 experiment).
(D) LDA from in vivo data. Recipient mice were injected intravenously with variable numbers of GFP+ cells from the 3 experimental groups (KO-CTX, KO-PBS, and wt-CTX). Mice survival was evaluated 2 months after transplantation, and frequency of LSCs was calculated using ELDA software.
(E) Stem cell frequency of LSCs from the different experiments: hypoxia in vitro (red, n = 3), normoxia in vitro (blue, n = 3), and in vivo (green, n = 1) experiments. ∗p < 0.05, two-way ANOVA.
(F) LSC total numbers from the different experiments: hypoxia in vitro (red, n = 3), normoxia in vitro (blue, n = 3), and in vivo (green, n = 1) experiments. Plot shows mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, two-way ANOVA. CTX, chemotherapy; KO, Hif-1α–/–; wt, Hif-1α+/+.
For an in vitro approach, we sorted 4, 8, or 12 GFP+ leukemic BM cells from each group into individual wells. Proliferation was next evaluated in both normoxic (20% oxygen) and hypoxic (1% oxygen) conditions (Figures 2B and 2E). We obtained the highest LSC frequency from PBS-Hif-1α–/– cells (1/12.3 and 1/21.7 in hypoxia or normoxia, respectively, means from 3 independent experiments). Within CTX-treated cells, Hif-1α–/– samples contained a higher LSC frequency compared with Hif-1α+/+ cells (1/26.8 versus 1/47.1 in hypoxia, and 1/40.4 versus 1/94.4 in normoxia, respectively, means from 3 independent experiments), although this failed to reach statistical significance.
We next assessed LSC frequencies in vivo, injecting 10, 102, 103, or 104 GFP+ BM cells from each group into new recipients (Figure 2C). While transplantation of 103 and 104 cells resulted in 100% mortality, lower doses of 10 and 102 cells indicated an LSC frequency highest in the PBS-Hif-1α–/– group (1/18.8). Among CTX-treated samples, we observed a higher LSC frequency in the Hif-1α–/– group (1/34.5) than in the Hif-1α+/+ group (1/56.1) (Figures 2D and 2E).
Estimation of total number of LSCs in the BM (Figure 2F) demonstrated that deletion of Hif-1α fails to reduce LSC frequencies/numbers after chemotherapy.
Hif-1α Deletion Affects Transcriptional Expression of Replication, Transcription, and Translation-Related Genes
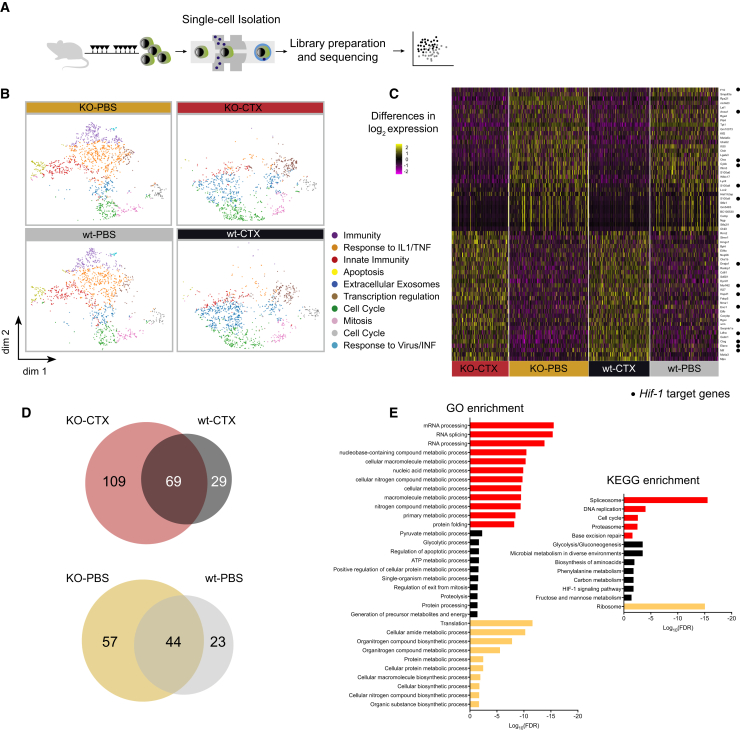
Our data indicated that Hif-1α deletion contributed to a more rapid disease progression (Figure 1). To tease out a potential mechanism, we therefore conducted single-cell RNA sequencing on leukemic cells from the 4 evaluated settings (Hif-1α–/–/Hif-1α+/+ ± chemotherapy) (Figure 3A). Aggregated data were subjected to dimensionality reduction using the t-distributed stochastic neighbor embedding (t-SNE) method (Figure 3B), revealing dramatically different transcriptional profiles of cells following chemotherapy, but less different according to Hif-1α status. This indicates either a modification of transcription upon chemotherapy or, more likely, a selective survival regardless of Hif-1α status.
Figure 3.
Single-Cell RNA Sequencing of Leukemic Cells after Chemotherapy Treatment
(A) Experimental design for single-cell RNA-sequencing analysis. After treatment with tamoxifen/oil and chemotherapy/PBS, GFP+ cells were collected at day 28 after transplantation and subjected to RNA processing.
(B) t-SNE plots representing gene expression profiles of all individual cells analyzed: KO-CTX (n = 1,217 cells), KO-PBS (n = 1,696 cells), wt-CTX (n = 1,301 cells), and wt-PBS (n = 1,450 cells). Each dot represents one cell. K-means clustering groups cells into 10 clusters, which are represented by different colors. Differentially expressed genes in each cluster were correlated with the main biological function indicated at the right side of the plots.
(C) Heatmap depicting significantly differentially expressed genes in single cells. Heatmap with the full set of genes can be found in Figure S3. Hif-1α target genes are indicated by black dots.
(D) Venn diagrams showing the distribution among groups of differentially expressed genes (adjusted p value <0.05). Of note, there are no overlapping genes between chemotherapy- and PBS-treated groups among this set of differentially expressed genes.
(E) Gene ontology (GO) and KEGG pathway enrichment analysis from uniquely differentially expressed genes in each group indicating the associated biological process.
CTX, chemotherapy; IL1/TNF, interleukin-1/tumor necrosis factor; INF, interferon; dim, dimension; FDR, false discovery rate.
See also Figures S3 and S4.
By K-means clustering, we divided the entire dataset into 10 different clusters. We observed that clusters with functions related to more differentiated cells (e.g., immune system process, response to interleukin-1/tumor necrosis factor, innate immunity or response to virus/interferon) were predominant in PBS samples, while others (e.g., mitotic cell cycle, cellular response to DNA damage, and transcriptional regulation) were associated with CTX samples. Multiple genes related to myeloid differentiation were predominantly expressed in PBS samples (data not shown), strongly suggesting effective chemotherapy targeting of more mature leukemia-associated cells.
Pairwise sample comparisons produced a collection of 331 differentially expressed genes (Figures S3 and 3C). When comparing significantly overexpressed genes among groups (adjusted p value <0.05), we found a very different distribution of cells between CTX and PBS samples, but only a small subset of genes uniquely expressed in either Hif-1α–/– or Hif-1α+/+ cells (Figures 3D and S4). In single-cell analysis, this indicates that there are differences in transcriptional regulation or that analyzed cells from each sample are different.
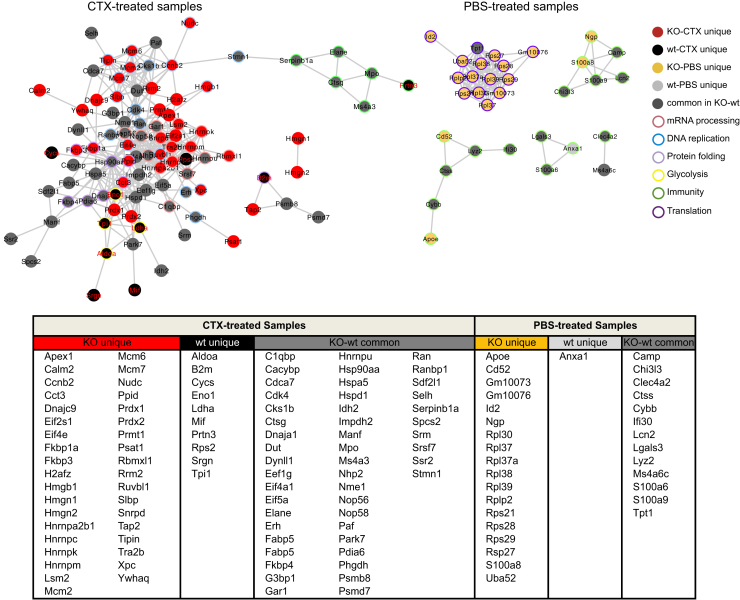
To identify functional protein association networks among differentially expressed genes, we used the STRING database (Figures 3E and 4). For the 4 sets of differentially expressed genes (log fold change >0.4, adjusted p value <0.05), we obtained a significant protein-protein interaction (PPI) enrichment score, indicating that there are more interactions among these proteins than a random set of proteins from the genome (PPI enrichment p values: Hif-1α–/–-CTX = 0; Hif-1α+/+-CTX = 1.03 × 10−6; Hif-1α–/–-PBS = 0; Hif-1α+/+-PBS = 0.0251). This indicates that genes in each set are at least partially biologically connected. To correlate these genes with a biological function, we performed gene ontology and KEGG pathway enrichment analyses (Figure 3E). This revealed that mRNA processing, DNA replication, and protein folding were significantly overrepresented in the Hif-1α–/–-CTX group, while the Hif-1α+/+-CTX group was associated with the HIF-1α pathway, glycolysis, and other carbon metabolism processes. In PBS groups, translation was the most represented pathway associated with Hif-1α–/– cells, while no significant pathways could be assigned to Hif-1α+/+ cells.
Figure 4.
Protein Association Networks Affected by Chemotherapy
Protein association network analysis of differentially expressed set of genes (average log fold change >0.4, adjusted p value <0.05). Uniquely expressed genes from each group are indicated by its assigned color. Common genes among KO and wt samples, within each specific set (CTX- or PBS-treated), are shown in dark gray. Associated biological function to each gene is indicated, with the color of the border showing the main processes regulated by these genes. Interaction enrichment score >0.5. CTX, chemotherapy.
Discussion
Relapse of AML remains a major therapeutic challenge, not least because of the few therapeutic options for many of these patients, which in most cases is restricted to palliative care.
One potential strategy for eradicating LSCs, responsible for relapse in AML, is to sensitize them to chemotherapy. We here explored whether targeting the HIF pathway could be used for this using an MLL-AF9 murine AML model, which was subjected to a standard and clinically relevant chemotherapy protocol. In our work, Hif-1α–/– cells displayed a faster evolution of the disease compared with Hif-1α+/+ cells following chemotherapy. While the results are consistent with our previous work (Velasco-Hernandez et al., 2014) and recently published data (Vukovic et al., 2015), they appear in sharp contrast to previous studies (Wang et al., 2011, Zhang et al., 2012) in which Hif-1α was critical for LSCs maintenance.
In our work, we used the Rosa26Cre-ERT2 rather than the more commonly used Mx1-Cre model, which is associated with significant leakiness (Velasco-Hernandez et al., 2016). This allowed us to study the effect of Hif-1α inhibition specifically in AML cells of the same founder clone, which circumvents the risk that different leukemic clones have intrinsically different behavior. Inducing the deletion in Rosa26Cre-ERT2 mice requires injection of tamoxifen that might itself affect the initiation of MLL-AF9 leukemia (Sanchez-Aguilera et al., 2014). However, as the described effect of tamoxifen is a regression of the disease, the isolated effect of knocking out Hif-1α could be even more pronounced than the one we report.
The standard chemotherapy regimen for AML is a combination of AraC and an anthracycline (such as doxorubicin). Doxorubicin has itself been attributed Hif-inhibiting properties (Lee et al., 2009). Different disease progression in CTX-Hif-1α–/– and Hif-1α+/+ samples indicates that even if doxorubicin is inhibiting HIF function, this effect is not complete and can be further increased by alternative means.
There is evidence that c-Kit deregulation and overexpression could be factors contributing to the chemotherapy resistance in AML (Advani et al., 2008). The higher numbers of c-Kit+ cells in the Hif-1α–/– chemotherapy group might therefore be an indicator of higher drug resistance of these cells, with c-Kit being a well-described stem cell marker of murine MLL-AF9-driven LSCs (Krivtsov et al., 2006). However, our c-Kit expression data correlated poorly with LSC frequencies measured by LDA. Previously it was thought that chemotherapy promoted LSC enrichment (Ishikawa et al., 2007, Saito et al., 2010). However, although cytarabine decreases the frequency of LSCs in human AML models (Griessinger et al., 2014), it fails to promote enrichment either of quiescent cells or more primitive CD34+CD38– cells, emphasizing that functional assessments are critical. We did not observe an increment in LSC frequencies measured by LDA but observed a substantial reduction (around 3-fold) of the total bulk of leukemic cells, indicating an overall enrichment of LSCs following chemotherapy. In addition, we observed an increased frequency of c-Kit+ cells by Hif-1α deletion, which further supports the increased aggressive phenotype of Hif-1α–/– cells.
Hif-1α is rapidly degraded in normoxia and Hif-1α+/+ cells should thus theoretically behave as Hif-1α–/– cells, which would suggest similar LSC frequencies between Hif-1α–/– and Hif-1α+/+ cells in normoxia. Somewhat surprisingly, we observed a higher LSC frequency of Hif-1α–/– cells, although not significant (p = 0.1079). One possibility is that this indicates the involvement of one or more Hif-1α-oxygen-independent mechanisms. For instance, it is well established that Hif-1α can be induced by the PTEN/PI3K/AKT axis (Zundel et al., 2000), a pathway that appears to extend also to MLL-AF9 leukemia (Hoshii et al., 2012). Regardless, even if hypoxia in general promotes a quiescent state of cells, such conditions seem to promote AML cell proliferation, which is also in line with previous results on human AML (Griessinger et al., 2014).
After chemotherapy, we observed a more aggressive phenotype as a consequence of Hif-1α deletion. We hypothesized that this faster development of disease could be explained by two scenarios: (1) leukemic Hif-1α–/– cells are less sensitive to the chemotherapy, which would be in contrast to our original expectation; and/or (2) leukemic Hif-1α–/– cells expand faster than Hif-1α+/+ cells after chemotherapy. To discriminate between these two options and unveil possible mechanisms leading to this effect, we conducted a single-cell transcriptome analysis.
We found that chemotherapy-treated cells displayed a distinct transcriptomic profile when compared with PBS-treated cells, but with fewer differences when comparing Hif-1α–/– and Hif-1α+/+ cells. Still, of the significantly overexpressed genes specific for each group, we identified an increment in genes involved in translation (PBS-Hif-1α–/– cells) and in mRNA processing, DNA replication, and protein folding (CTX-Hif-1α–/– cells). Overall, this is in line with a more active proliferative status of Hif-1α–/– cells and suggests that deletion of Hif-1α is transcriptionally deregulating these aforementioned specific pathways. Chemotherapy-depleted cells with an immune-related phenotype represent the cells expanded in leukemia. As expected, Hif-1α–/– cells were depleted in expression of Hif-1α targets, and Hif-1α deletion leads to an enhanced transcription of genes involved mainly in replication, mRNA processing, and translation.
These results are in accordance with the described effect of Hif-1α, inhibiting cell cycle and promoting quiescence, actions attributed to blockage of c-MYC and enhancement of cyclin-dependent kinase inhibitors such as p21/CDKN1A (Koshiji et al., 2004). Since we are deleting only one member of a multigene family, it remains a possibility that other members, i.e., Hif-2α, could have a compensatory effect. Future work could benefit from an increased understanding of both gene redundancy and potential hypoxia-independent effects of Hif-1α.
Overall, our data suggest that Hif-1α inhibition failed to improve the outcome of chemotherapy in MLL-AF9-driven AML and, in contrast, led to a faster progression of disease upon withdrawal of the treatment. Further investigations are needed to extend these conclusions to other genetic subtypes of AML.
Deletion of Hif-1α in our setting was permanent, which would be a different scenario than in a clinical setting where administration of Hif-1α inhibitors would be transient. Our results nevertheless emphasize that the effects of HIF inhibition have to be further investigated before this strategy can be applied in a clinical setting.
Experimental Procedures
Mice
Hif-1αlox mice (Ryan et al., 2000) (JAX 007561) were crossed with the tamoxifen-inducible Rosa26Cre-ERT2 mice (Ventura et al., 2007) (JAX 008463) to generate a combined conditional knockout (KO) model. Mice were maintained at the animal facility of the Biomedical Center at Lund University (Sweden) and all experiments were performed with consent from a local ethics committee.
Statistical Analysis
All data are expressed as the mean ± SEM. Differences between groups were assessed using unpaired Student's t tests. All analyses were performed with Prism software, version 7.0 (GraphPad Software).
Detailed descriptions of experiments can be found in Supplemental Experimental Procedures.
Author Contributions
T.V.-H. designed the study, performed experiments, analyzed data, and wrote the manuscript; S.S. performed bioinformatics analysis; I.H. and E.E. performed experiments; and J.C and D.B. designed the study and wrote the manuscript. All authors revised the manuscript and approved the final version of this study.
Acknowledgments
We thank Dr. Göran Karlsson and Parashar Dhapola for helpful discussions on data analysis. This work was supported with a project grant to T.V.-H. from the Sigurd och Elsa Goljes Minne Memorial Foundation, to J.C. from The Swedish Cancer Society (Cancerfonden) and Barncancerfonden, and to D.B. from The Swedish Cancer Society, The Swedish Medical Research Council, ERC (Leukemiabarrier 615068), The Knut and Alice Wallenberg Foundation, and the Tobias Foundation.
Published: December 27, 2018
Footnotes
Supplemental Information includes Supplemental Experimental Procedures and four figures and can be found with this article online at https://doi.org/10.1016/j.stemcr.2018.11.023.
Contributor Information
Talia Velasco-Hernandez, Email: tvelasco@carrerasresearch.org.
David Bryder, Email: david.bryder@med.lu.se.
Accession Numbers
The accession number for transcriptomic data is GEO: GSE119484.
Supplemental Information
References
- Advani A.S., Rodriguez C., Jin T., Jawde R.A., Saber W., Baz R., Kalaycio M., Sobecks R., Sekeres M., Tripp B. Increased C-kit intensity is a poor prognostic factor for progression-free and overall survival in patients with newly diagnosed AML. Leuk. Res. 2008;32:913–918. doi: 10.1016/j.leukres.2007.08.019. [DOI] [PubMed] [Google Scholar]
- Burnett A., Wetzler M., Lowenberg B. Therapeutic advances in acute myeloid leukemia. J. Clin. Oncol. 2011;29:487–494. doi: 10.1200/JCO.2010.30.1820. [DOI] [PubMed] [Google Scholar]
- Dohner H., Estey E.H., Amadori S., Appelbaum F.R., Buchner T., Burnett A.K., Dombret H., Fenaux P., Grimwade D., Larson R.A. Diagnosis and management of acute myeloid leukemia in adults: recommendations from an international expert panel, on behalf of the European Leukemianet. Blood. 2010;115:453–474. doi: 10.1182/blood-2009-07-235358. [DOI] [PubMed] [Google Scholar]
- Essers M.A., Trumpp A. Targeting leukemic stem cells by breaking their dormancy. Mol. Oncol. 2010;4:443–450. doi: 10.1016/j.molonc.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flamme I., Frohlich T., von Reutern M., Kappel A., Damert A., Risau W. HRF, a putative basic helix-loop-helix-PAS-domain transcription factor is closely related to hypoxia-inducible factor-1 alpha and developmentally expressed in blood vessels. Mech. Dev. 1997;63:51–60. doi: 10.1016/s0925-4773(97)00674-6. [DOI] [PubMed] [Google Scholar]
- Frolova O., Samudio I., Benito J.M., Jacamo R., Kornblau S.M., Markovic A., Schober W., Lu H., Qiu Y.H., Buglio D. Regulation of HIF-1alpha signaling and chemoresistance in acute lymphocytic leukemia under hypoxic conditions of the bone marrow microenvironment. Cancer Biol. Ther. 2012;13:858–870. doi: 10.4161/cbt.20838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griessinger E., Anjos-Afonso F., Pizzitola I., Rouault-Pierre K., Vargaftig J., Taussig D., Gribben J., Lassailly F., Bonnet D. A niche-like culture system allowing the maintenance of primary human acute myeloid leukemia-initiating cells: a new tool to decipher their chemoresistance and self-renewal mechanisms. Stem Cells Transl. Med. 2014;3:520–529. doi: 10.5966/sctm.2013-0166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guitart A.V., Subramani C., Armesilla-Diaz A., Smith G., Sepulveda C., Gezer D., Vukovic M., Dunn K., Pollard P., Holyoake T.L. Hif-2alpha is not essential for cell-autonomous hematopoietic stem cell maintenance. Blood. 2013;122:1741–1745. doi: 10.1182/blood-2013-02-484923. [DOI] [PubMed] [Google Scholar]
- Hoshii T., Tadokoro Y., Naka K., Ooshio T., Muraguchi T., Sugiyama N., Soga T., Araki K., Yamamura K., Hirao A. mTORC1 is essential for leukemia propagation but not stem cell self-renewal. J. Clin. Invest. 2012;122:2114–2129. doi: 10.1172/JCI62279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa F., Yoshida S., Saito Y., Hijikata A., Kitamura H., Tanaka S., Nakamura R., Tanaka T., Tomiyama H., Saito N. Chemotherapy-resistant human AML stem cells home to and engraft within the bone-marrow endosteal region. Nat. Biotechnol. 2007;25:1315–1321. doi: 10.1038/nbt1350. [DOI] [PubMed] [Google Scholar]
- Koshiji M., Kageyama Y., Pete E.A., Horikawa I., Barrett J.C., Huang L.E. HIF-1alpha induces cell cycle arrest by functionally counteracting Myc. EMBO J. 2004;23:1949–1956. doi: 10.1038/sj.emboj.7600196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krivtsov A.V., Twomey D., Feng Z., Stubbs M.C., Wang Y., Faber J., Levine J.E., Wang J., Hahn W.C., Gilliland D.G. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 2006;442:818–822. doi: 10.1038/nature04980. [DOI] [PubMed] [Google Scholar]
- Lee K., Qian D.Z., Rey S., Wei H., Liu J.O., Semenza G.L. Anthracycline chemotherapy inhibits HIF-1 transcriptional activity and tumor-induced mobilization of circulating angiogenic cells. Proc. Natl. Acad. Sci. U S A. 2009;106:2353–2358. doi: 10.1073/pnas.0812801106. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Makino Y., Kanopka A., Wilson W.J., Tanaka H., Poellinger L. Inhibitory PAS domain protein (IPAS) is a hypoxia-inducible splicing variant of the hypoxia-inducible factor-3alpha locus. J. Biol. Chem. 2002;277:32405–32408. doi: 10.1074/jbc.C200328200. [DOI] [PubMed] [Google Scholar]
- Morrison S.J., Scadden D.T. The bone marrow niche for haematopoietic stem cells. Nature. 2014;505:327–334. doi: 10.1038/nature12984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouault-Pierre K., Lopez-Onieva L., Foster K., Anjos-Afonso F., Lamrissi-Garcia I., Serrano-Sanchez M., Mitter R., Ivanovic Z., de Verneuil H., Gribben J. HIF-2alpha protects human hematopoietic stem/progenitors and acute myeloid leukemic cells from apoptosis induced by endoplasmic reticulum stress. Cell Stem Cell. 2013;13:549–563. doi: 10.1016/j.stem.2013.08.011. [DOI] [PubMed] [Google Scholar]
- Rowe J.M., Tallman M.S. How I treat acute myeloid leukemia. Blood. 2010;116:3147–3156. doi: 10.1182/blood-2010-05-260117. [DOI] [PubMed] [Google Scholar]
- Ryan H.E., Poloni M., McNulty W., Elson D., Gassmann M., Arbeit J.M., Johnson R.S. Hypoxia-inducible factor-1alpha is a positive factor in solid tumor growth. Cancer Res. 2000;60:4010–4015. [PubMed] [Google Scholar]
- Saito Y., Uchida N., Tanaka S., Suzuki N., Tomizawa-Murasawa M., Sone A., Najima Y., Takagi S., Aoki Y., Wake A. Induction of cell cycle entry eliminates human leukemia stem cells in a mouse model of AML. Nat. Biotechnol. 2010;28:275–280. doi: 10.1038/nbt.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Aguilera A., Arranz L., Martin-Perez D., Garcia-Garcia A., Stavropoulou V., Kubovcakova L., Isern J., Martin-Salamanca S., Langa X., Skoda R.C. Estrogen signaling selectively induces apoptosis of hematopoietic progenitors and myeloid neoplasms without harming steady-state hematopoiesis. Cell Stem Cell. 2014;15:791–804. doi: 10.1016/j.stem.2014.11.002. [DOI] [PubMed] [Google Scholar]
- Semenza G.L. Targeting HIF-1 for cancer therapy. Nat. Rev. Cancer. 2003;3:721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- Semenza G.L., Wang G.L. A nuclear factor induced by hypoxia via de novo protein synthesis binds to the human erythropoietin gene enhancer at a site required for transcriptional activation. Mol. Cell. Biol. 1992;12:5447–5454. doi: 10.1128/mcb.12.12.5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takubo K., Goda N., Yamada W., Iriuchishima H., Ikeda E., Kubota Y., Shima H., Johnson R.S., Hirao A., Suematsu M. Regulation of the HIF-1alpha level is essential for hematopoietic stem cells. Cell Stem Cell. 2010;7:391–402. doi: 10.1016/j.stem.2010.06.020. [DOI] [PubMed] [Google Scholar]
- Thomas D., Majeti R. Biology and relevance of human acute myeloid leukemia stem cells. Blood. 2017;129:1577–1585. doi: 10.1182/blood-2016-10-696054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco-Hernandez T., Hyrenius-Wittsten A., Rehn M., Bryder D., Cammenga J. HIF-1alpha can act as a tumor suppressor gene in murine acute myeloid leukemia. Blood. 2014;124:3597–3607. doi: 10.1182/blood-2014-04-567065. [DOI] [PubMed] [Google Scholar]
- Velasco-Hernandez T., Sawen P., Bryder D., Cammenga J. Potential pitfalls of the Mx1-Cre system: implications for experimental modeling of normal and malignant hematopoiesis. Stem Cell Reports. 2016;7:11–18. doi: 10.1016/j.stemcr.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura A., Kirsch D.G., McLaughlin M.E., Tuveson D.A., Grimm J., Lintault L., Newman J., Reczek E.E., Weissleder R., Jacks T. Restoration of p53 function leads to tumour regression in vivo. Nature. 2007;445:661–665. doi: 10.1038/nature05541. [DOI] [PubMed] [Google Scholar]
- Vukovic M., Guitart A.V., Sepulveda C., Villacreces A., O'Duibhir E., Panagopoulou T.I., Ivens A., Menendez-Gonzalez J., Iglesias J.M., Allen L. Hif-1alpha and Hif-2alpha synergize to suppress AML development but are dispensable for disease maintenance. J. Exp. Med. 2015;212:2223–2234. doi: 10.1084/jem.20150452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Liu Y., Malek S.N., Zheng P., Yang L. Targeting HIF1alpha eliminates cancer stem cells in hematological malignancies. Cell Stem Cell. 2011;8:399–411. doi: 10.1016/j.stem.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanada M., Garcia-Manero G., Borthakur G., Ravandi F., Kantarjian H., Estey E. Relapse and death during first remission in acute myeloid leukemia. Haematologica. 2008;93:633–634. doi: 10.3324/haematol.12366. [DOI] [PubMed] [Google Scholar]
- Zhang H., Li H., Xi H.S., Li S. HIF1alpha is required for survival maintenance of chronic myeloid leukemia stem cells. Blood. 2012;119:2595–2607. doi: 10.1182/blood-2011-10-387381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber J., Radtke I., Pardee T.S., Zhao Z., Rappaport A.R., Luo W., McCurrach M.E., Yang M.M., Dolan M.E., Kogan S.C. Mouse models of human AML accurately predict chemotherapy response. Genes Dev. 2009;23:877–889. doi: 10.1101/gad.1771409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zundel W., Schindler C., Haas-Kogan D., Koong A., Kaper F., Chen E., Gottschalk A.R., Ryan H.E., Johnson R.S., Jefferson A.B. Loss of PTEN facilitates HIF-1-mediated gene expression. Genes Dev. 2000;14:391–396. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.