Summary
Tuberous sclerosis complex (TSC) is a rare neurodevelopmental disorder resulting from autosomal dominant mutations in the TSC1 or TSC2 genes, leading to a hyperactivated mammalian target of rapamycin (mTOR) pathway, and gray and white matter defects in the brain. To study the involvement of neuron-glia interactions in TSC phenotypes, we generated TSC patient induced pluripotent stem cell (iPSC)-derived cortical neuronal and oligodendrocyte (OL) cultures. TSC neuron mono-cultures showed increased network activity, as measured by calcium transients and action potential firing, and increased dendritic branching. However, in co-cultures with OLs, neuronal defects became more apparent, showing cellular hypertrophy and increased axonal density. In addition, TSC neuron-OL co-cultures showed increased OL cell proliferation and decreased OL maturation. Pharmacological intervention with the mTOR regulator rapamycin suppressed these defects. Our patient iPSC-based model, therefore, shows a complex cellular TSC phenotype arising from the interaction of neuronal and glial cells and provides a platform for TSC disease modeling and drug development.
Keywords: tuberous sclerosis complex, autism, iPSC, co-culture, glia, oligodendrocyte, neuron, in vitro model, neuronal activity, myelin
Highlights
-
•
TSC neuron mono-cultures show an increase in network activity and dendritic branching
-
•
TSC co-cultures show hypertrophy and an increase in axonal length and OL proliferation
-
•
mTOR regulators normalize TSC neuronal and glial phenotypes
Nadadhur et al. generated TSC disease models using patient iPSCs. While neuron mono-cultures showed an increase in network activity and dendritic branching, only when co-cultured with oligodendrocytes (OLs), hypertrophy and axonal abnormalities were observed. Neuron-OL interactions, modulated by mTOR regulators, support use of mixed cultures for TSC disease modeling and drug development.
Introduction
Tuberous sclerosis complex (TSC) is a rare genetic, neurodevelopmental disorder, which affects multiple organs (Leung and Robson, 2007), causing benign tumors in the brain, eyes, heart, lung, liver, kidney, and skin. The neuropathological features in the brain involve cortical tubers and subependymal nodules (Curatolo et al., 2002). Many studies also indicate glial-associated phenotypes, including giant astrocytoma (Beaumont et al., 2015) and white matter abnormalities (Curatolo et al., 2002, Marti-Bonmati et al., 2000). TSC is caused by autosomal dominant mutations in the genes TSC1 and TSC2, resulting in a mammalian target of rapamycin (mTOR) pathway hyperactivity (Curatolo and Moavero, 2012). The mTOR pathway plays key roles in multiple cellular processes, such as cell growth (Huang and Manning, 2008), and is differentially regulated by factors, such as rapamycin (Rapa) (Curatolo et al., 2016), insulin-like growth factor 1 (IGF-1) (Min et al., 2012), and guanabenz (Guana) (Jiang et al., 2016) depending on cell type and maturation state, as shown in cellular systems and in vivo models (Oh and Jacinto, 2011, Watanabe et al., 2011). Clinical studies showed encouraging improvements in TSC patients after Rapa treatment (Sadowski et al., 2016). However, to develop improved treatment regimens, we lack a detailed understanding of how TSC disease mechanisms affect human neuronal and glial cell types.
TSC has been studied in different rodent model systems, which showed neuronal defects, such as distorted synaptic balance, neuronal hyperactivity (Bateup et al., 2013), and axonal defects (Choi et al., 2008), as well as glial defects, such as reduced myelin (Meikle et al., 2007), increased oligodendrocyte (OL) progenitor proliferation, and increased OL death (Carson et al., 2015, Ercan et al., 2017, Jiang et al., 2016). In human pluripotent stem cell-based assays, homozygous mutations for TSC2 in human embryonic stem cell line showed a hyperactive mTOR pathway, increased soma size, neuronal hyperactivity, and increased dendritic branching (Costa et al., 2016), while a heterozygous gene disruption of TSC2 presented with mild neuronal phenotypic defects (Costa et al., 2016). Two TSC patient induced pluripotent stem cell (iPSC)-based studies have been presented before, where one showed increased astrocyte proliferation, and increased soma size and decreased neurite length in iPSC-derived neuronal cultures (Li et al., 2017) and the other showed delayed neuronal differentiation related to phosphatidylinositol 3-kinase/AKT (Zucco et al., 2018). As TSC neuropathological features implicate both neuronal and glial cells, and proper neural development involve complex interactions between different neural cell types, mixed neuron-glia systems would be valuable to study the modulatory actions of mTOR pathway inhibitors on TSC disease phenotypes.
Here we present a TSC patient iPSC-based model involving neuron-OL co-cultures. While increased neuronal activity was quantifiable in neuronal mono-cultures, morphological changes, such as increased axonal density and hypertrophy, only became apparent after the TSC neurons were co-cultured with human OLs. Treatment of known mTOR regulators Rapa, IGF-1, and Guana confirmed modulating effects in both neurons and OLs and validated our TSC model for future drug screens to design treatments for TSC patients.
Results
TSC Patient iPSCs Show Normal Stem Cell Properties and a Decrease in TSC Protein Levels
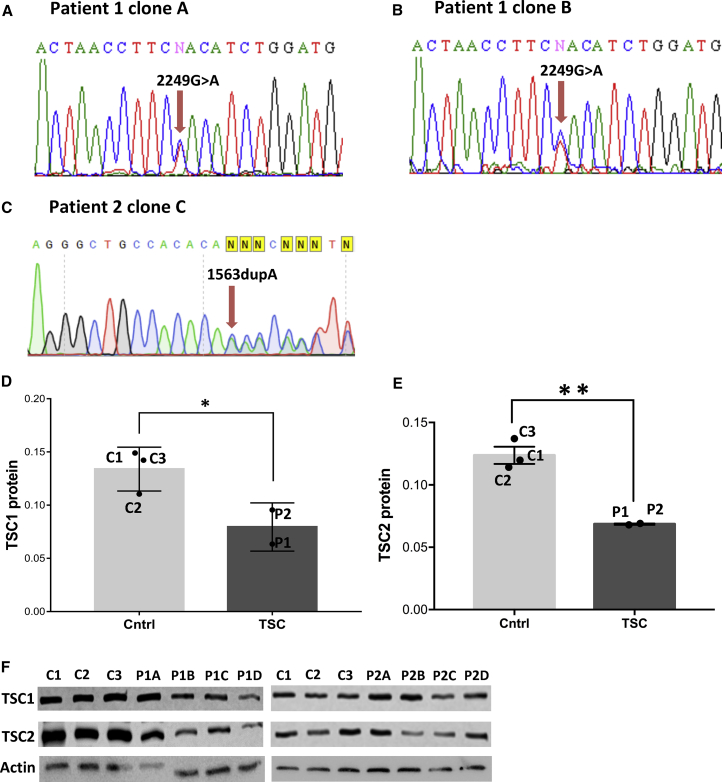
To generate iPSC-based models for TSC, we selected fibroblasts from teenage patients. Patient 1 (P1) carried a heterozygous mutation 2249G > A in the TSC1 gene, resulting in a de novo mutation W750X reported earlier (Jones et al., 1999); Patient 2 (P2) had a single nucleotide duplication (1563dupA in TSC2 gene resulting in frameshift mutation H522T). Control iPSC lines were generated from one infant (C1) and two teenagers (C2, C3). Several iPSC clones were obtained from each patient and control fibroblast line and verified by sequencing (Figures 1A–1C). To characterize TSC patient and control iPSCs for pluripotency, we performed standard assays, including immunocytochemistry for markers OCT3&4, SSEA4, TRA-1-60, and TRA-1-80, and expression of alkaline phosphatase (Figures S1A–S1F).
Figure 1.
Mutations in TSC Genes Lead to Decreased TSC Protein Levels in Patient iPSCs
(A–C) Sequencing of patient iPSC line (A) P1A, (B) P1B, and (C) P2.
(D and E) Western blot quantifications of (D) TSC1 and (E) TSC2 protein expression in controls and patients iPSC cells. Individual points indicate the average values of the two patients (P1 and P2) and controls (C1, C2, and C3). P1 and P2 represent four iPSC clones each. A significant difference was obtained in the mean TSC1 and TSC2 protein expression level between the control and patient groups (p < 0.05, unpaired one-tailed t test).
(F) Western blots presenting labeled TSC1, TSC2, and actin protein of four iPSC clones of TSC patient P1 and P2, and three control iPSCs (means ± SEM).
See also Figure S1.
To investigate how the patient TSC mutations change TSC1 and TSC2 protein expression, we performed western blot analysis (Figures 1D and 1E). The stoichiometric ratio of TSC1 and TSC2 proteins are known to be tightly regulated in the cell, so changes in the level of one TSC protein could be expected to lead to changes of the other TSC protein level. Indeed when we investigated the TSC1 and TSC2 proteins of both patients we found this to be the case. The level of one TSC patient was obtained by averaging the levels of different iPSC clones of that patient: four clones for both P1 and P2 (Figures 1D–1F). By comparing the average levels of the patients (P1 and P2) to the average levels of the controls (C1–C3), we found a significant reduction in both TSC1 (p < 0.03) and TSC2 (p < 0.004) in TSC iPSCs (Figures 1D and 1E). The TSC protein levels are controlled for loading and blotting, by measuring the total protein content on the blot, as described by previous studies (Gurtler et al., 2013, Rivero-Gutierrez et al., 2014), although a protein control actin is shown in Figure 1F. We selected two iPSC clones hVS-233 (P1A) and hVS-248 (P1B) from P1, one iPSC clone hVS-417 (P2C) from P2, and control iPSC clones hVS-88 (C1), hVS-228 (C2), and hVS-421 (C3) for the following experiments. Overall, the TSC patient iPSCs showed a significant reduction in both TSC1 and TSC2 proteins compared with the controls, but expressed normal stem cell properties, justifying a combined study of both TSC1 and TSC2 patients.
Increased Neuronal Activity in TSC Patient iPSC-Derived Neuron Mono-cultures
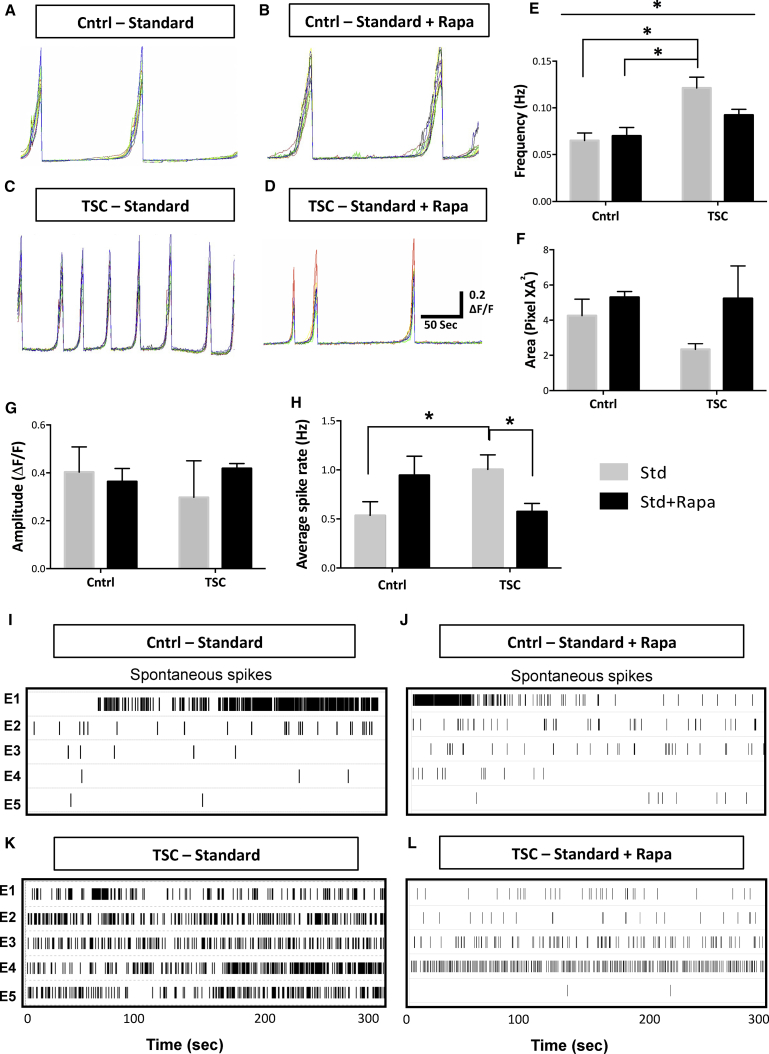
Network hyperexcitability has been reported in several models of TSC (Bateup et al., 2013, Lasarge and Danzer, 2014, Wang et al., 2007). To investigate neuronal activity in TSC patients, we generated neuronal cultures from controls C1, C2, and C3, and patient lines P1A, P1B, and P2, according to established protocols (Nadadhur et al., 2017). At day 56, we measured spontaneous calcium transients in the neuronal cultures (standard [Std] conditions; Figures 2A and 2C). Under Std conditions, the spontaneous calcium influx frequency in the networks was significantly higher (p < 0.01, post hoc test) in the TSC neurons (0.12 ± 0.01 Hz; Figures 2C and 2E) compared with the control neuron cultures (0.065 ± 0.008 Hz; Figures 2A and 2E). We further studied whether mTOR pathway inhibition, by administration of Rapa (Std + Rapa) or IGF-1 (Std + IGF-1), modulated the spontaneous calcium transients (Figures 2B and 2D). The frequency of calcium transients in TSC neuron cultures treated with Rapa (Figure 2D) or IGF-1 (Figures S2G and S2L) reduced to around 0.09 ± 0.006 and 0.089 ± 0.01 Hz, respectively, whereas the frequency in control neurons after Rapa and IGF-1 treatment remained around 0.069 ± 0.009 and 0.07 ± 0.008, respectively (Figures 2B and S2F). The reduction in the total event area in the TSC cultures (2.3 ± 0.3) compared with control cultures (4.2 ± 0.9) under Std conditions showed a trend toward statistical significance (p = 0.08; Figures 2F and S2M). Lastly, event amplitudes between TSC and control neurons were similar (Figures 2G and S2N). Overall, the increased event frequency in the TSC neurons suggests an increase in neuronal activity, i.e., spontaneous firing, which was diminished by treatment with Rapa or IGF-1.
Figure 2.
Increased Network Activity in TSC Neuron Mono-cultures Can Be Modulated by Rapa
(A–D) Representative traces of calcium influx of neuronal cultures generated from control and TSC patients, under Std (A and C) and Std + Rapa (B and D) culture conditions. The multicolor traces in (A–D) represent activity in 12 regions of interest per FOV.
(E) Average calcium event frequency for the patient (P1 and P2) and control (C1, C2, and C3) neurons under Std and Std + Rapa (means ± SEM). Average for P1 was obtained from P1A and P1B. The average calcium event frequency of every iPSC line is calculated by averaging the analysis of two to four differentiations. Asterisks in (E) indicate statistical differences found in Tukey's post hoc test, ∗p < 0.05.
(F and G) Calcium event amplitude (F) and event area (G) in control and patient groups under Std and Std + Rapa conditions.
(H–L) Average spike rate (Hz) at 50 days post plating on MEAs under Std and Std + Rapa condition from C1, C2, P1A, and P2 (H) (n = 6–8 independent experiments; means ± SEM). Asterisks in (H) indicate unpaired t test, ∗p < 0.05. A representative raster plot showing the activity in control neurons under; (I) Std and (J) Std + Rapa conditions and TSC neurons under; (K) Std and (L) Std + Rapa conditions.
See also Figure S2.
To further assess activity, we measured spontaneous action potential firing of neuronal cultures from controls C1, C2, patient lines P1A and P2, differentiated and plated on multi-electrode arrays (MEAs). At day 106 under Std conditions, the neuronal firing (measured as spike rate) was significantly higher in TSC neurons (p < 0.03, unpaired t test; 1.09 ± 0.14 Hz; Figure 2K) compared with the control cells (0.53 ± 0.14 Hz; Figure 2I), which significantly reduced to control levels, i.e., 0.52 ± 0.08 Hz upon Rapa treatment (p < 0.02, unpaired t test; Figures 2L and 2H). ANOVA analysis between the four groups did not show a statistically significant difference. Altogether, we found higher spontaneous neuronal firing in TSC neuron mono-cultures suggesting an increased neuronal activity, which was modulated to control levels after Rapa treatment.
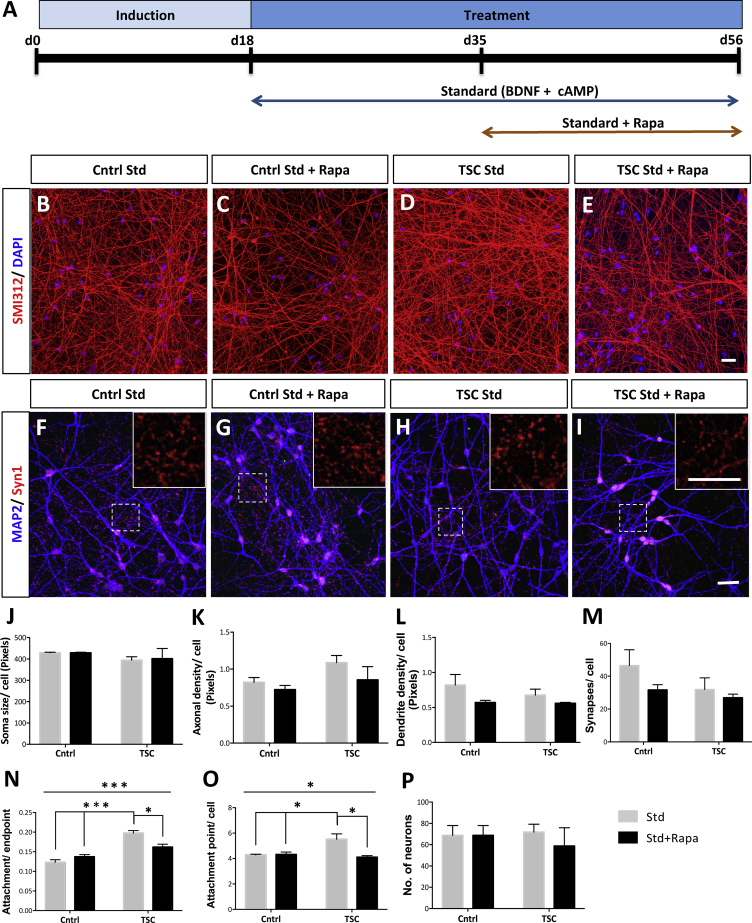
Morphological Analysis Only Reveals Increased Dendritic Arborization in TSC Neuron Mono-cultures
To investigate whether TSC patient iPSC-derived neuron cultures show morphological changes, we measured the axonal density in the cultures under Std, Std + Rapa, and Std + IGF-1 conditions at day 56 (Figures 3A and S2A). No significant changes in axonal densities (SMI312-positive pixels/DAPI-positive cell) were observed in the TSC (Figure 3D) compared with the control neuron mono-cultures (Figures 3B–3E). As hypertrophy is a known phenotype of TSC (Ruppe et al., 2014), we further measured neuronal soma size (Nissl-positive pixels/DAPI-positive cell). We did not observe differences in the soma size in the TSC compared with the control neuron cultures, under Std or Std + Rapa (Figure 3J), or under Std + IGF-1 conditions (Figure S2H). To assess differences in dendritic and synaptic density between TSC patient and control neuron cultures, we performed staining for dendritic marker MAP2 and pre-synaptic marker Synaptophysin1 (Figures 3F–3I, S2D, and S2E). No significant changes in the synaptic (Figure 3M) and the dendritic densities (Figure 3L) were found between the TSC and control neuron mono-cultures under Std, Std + Rapa, or Std + IGF-1 conditions (Figures S2K and S2J). To study changes in basal dendritic branching, prominent in TSC (Jaworski and Sheng, 2006), we utilized NeurphologyJ, an ImageJ plugin, to measure the number of dendritic branches per neuron (i.e., attachment points per cell; Figures 3O and S2P), and the ratio between number of branches per neuron and the dendritic ends (i.e., attachment points/endpoints; Figures 3N and S2O). Interestingly, the TSC cultures showed a significant increase in the dendritic branching, as measured by attachment points per cell (p < 0.02, ANOVA: p < 0.02; Tukey's post hoc test), and attachment points/endpoints (p < 0.0002, ANOVA: p < 0.0002, Tukey's post hoc test) compared with control neuron cultures, which was normalized after Rapa treatment (Figures 3N and 3O). The total number of neurons in the cultures (Figures 3P and S2Q) remained around 50 to 65 per field of view (FOV) for all groups. Altogether, although neurite densities did not change and hypertrophy was not observant, the TSC neurons do exhibit an increased basal dendritic branching compared with control neuron mono-cultures.
Figure 3.
Increased Basal Dendritic Branching in TSC Patient Neuronal Mono-cultures
(A) Schematic of neuronal differentiation protocol under the Std and Std + Rapa conditions.
(B–I) Immunochemistry of axonal marker SMI312, nuclear marker DAPI in control Std (B), control Std + Rapa (C), TSC Std (D), TSC Std + Rapa (E), and dendritic marker MAP2 and pre-synaptic marker Synaptophysin1 in control Std (F), control Std + Rapa (G), TSC Std (H) and TSC Std + Rapa (I) at day 56 for C1, C2, C3, P1, and P2 (n = 6–8 independent differentiations; means ± SEM). P1 includes P1A and P1B.
(J) Soma size is measured as the mean number of Nissl-positive pixels per soma.
(K) Total axonal density per soma is calculated as the number of SMI312-positive pixels per DAPI-positive soma.
(L) Total dendritic density per soma is calculated as the number of MAP2-positive pixels per DAPI-positive soma.
(M) Number of synapses is expressed as the number of Synaptophysin1-positive puncta per soma.
(N–P) Ratio of dendritic attachment points/endpoints of dendrites (N), number of dendrites (i.e., attachment points) per soma (O), and total number of neurons per field of view (FOV) measured by number of Nissl- and DAPI-double-positive soma (P).
Scale bars, 25 μm. Asterisks in (N) and (O) indicate statistical significant differences found using one-way ANOVA and Tukey's post hoc test, ∗p < 0.05, ∗∗∗p < 0.0005. While several differences were found using Tukey's post hoc, only the most important one's are represented. See also Figure S2.
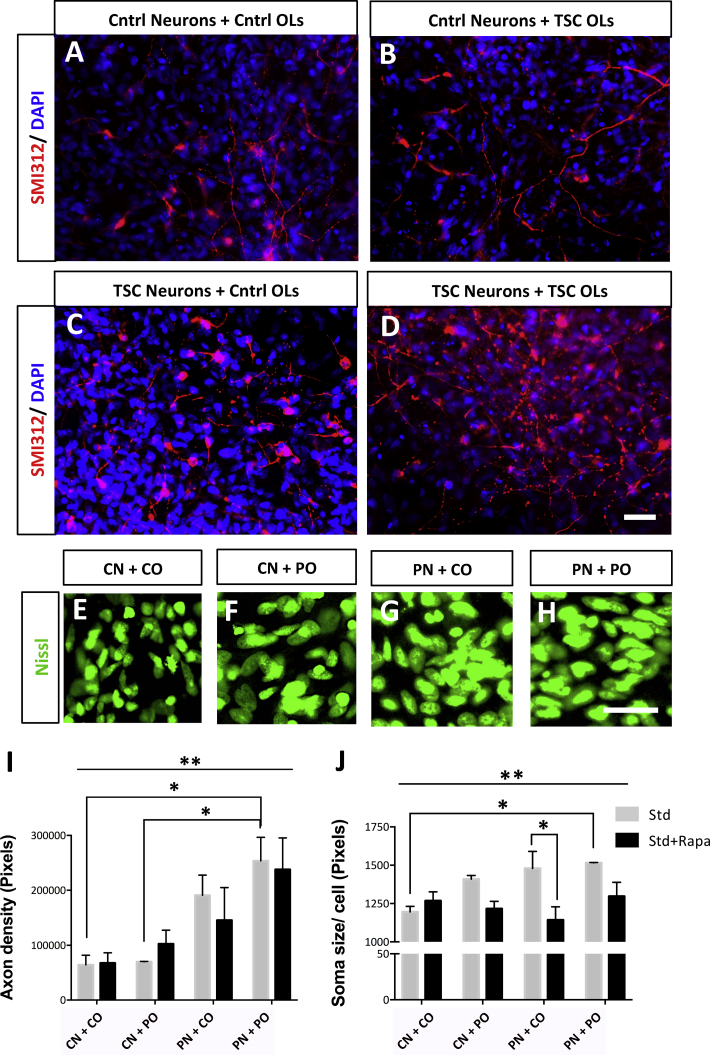
TSC Neuron-OL Co-cultures Show Hypertrophy and an Increased Neurite Density
Axonal changes and hypertrophy have been reported in brain tissue of TSC patients (Ruppe et al., 2014) and TSC mouse models (Meikle et al., 2007). Considering the white matter defects in TSC (Ruppe et al., 2014) and the mTOR pathway involvement in OL development and maturation (Meikle et al., 2007, Wahl et al., 2014), we examined whether neuronal abnormalities increase through complex interactions with OLs. To measure morphological changes in neuron-glia cultures, we grew co-cultures of neurons and OLs from TSC and control cells (CN + CO, CN + PO, PN + CO, and PN + PO). We first generated neurons and OLs separately and later co-cultured neurons (day 37) and OLs (day 65) under Std conditions for another 28 days and included treatments with Rapa (Std + Rapa) or Guana (Std + Guana) from day 7 of co-culture on (Figures 4A and S4A). Since IGF- administration could affect OL generation, we used Rapa and Guana as mTOR pathway modulators. The axonal density significantly changed between the different co-cultures (p < 0.004, ANOVA; Figures 4A–4D and 4I). The PN + PO culture showed a statistically significant 4-fold increase in axonal density compared with the CN + CO (p < 0.01, Tukey's post hoc test). CN + PO cultures appeared to be similar to control co-cultures and statistically different from PN + PO (p < 0.03, Tukey's post hoc test) cultures (Figures 4A–4D and 4I). This suggested that the increased axonal density did not increase just through elevated signaling from TSC OLs. Consistent with this hypothesis, PN + CO cultures showed a 3-fold increase in axonal density compared with CN + CO and CN + PO cultures, although not significant with a post hoc test. Treatments with Rapa or Guana did not suppress the increased axonal density (Figures 4I and S4B). The total number of cells in the co-cultures under Std and treated conditions remained the same (Figure S4F). Therefore, when co-cultures contained both TSC neurons and TSC OLs, the axonal density significantly increased compared with cultures containing control neurons, which was unaffected by treatment with Rapa or Guana.
Figure 4.
Hypertrophy and Increased Axonal Density in TSC iPSC-Derived Neuron-OL Co-cultures
(A–D) Immunocytochemistry of co-cultures, showing SMI312 and DAPI expression in CN + CO (A), CN + PO (B), PN + CO (C), and PN + PO (D) groups.
(E–H) Nissl soma staining in co-cultures of CN + CO (E), CN + PO (F), PN + CO (G), and PN + PO (H) groups.
(I and J) Total axonal pixel density (I) and average soma size (J) per cell in co-cultures measured at day 28 of co-culturing. Mean per group (means ± SEM) was obtained in total from n = 4–9 independent experiments including C1-, C3-, P1B-, and P2-derived neurons and OLs. Scale bars, 25 μm. Asterisks in (I) and (J) indicate statistically significant differences found in one-way ANOVA and Tukey's post hoc test, ∗p < 0.05, ∗∗p < 0.005, only the most important differences are represented in the graph.
See also Figures S4 and S5.
The soma size also significantly changed between the different co-cultures (p < 0.007, ANOVA; Figures 4E–4H and 4J). There was a statistically significant difference in soma size between PN + PO and CN + CO cultures (p < 0.02, Tukey's post hoc test), and between the Std and the Std + Rapa condition in the PN + CO culture (p < 0.04, Tukey's post hoc test; Figure 4J). The decrease between the Std and the Std + Rapa condition in the CN + PO and PN + PO cultures was not statistically significant (Figure 4J). Treatments with Guana did not normalize increased soma size (Figure S4C). Altogether, when co-cultures contained both TSC neurons and TSC OLs, the soma size significantly increased compared with cultures containing control neurons and control OLs. Rapa showed regulating effects on soma size when co-cultures contained TSC neurons and/or TSC OLs.
To study how interactions between TSC neurons and TSC OLs affect the density of neuronal processes after long periods of time, we transplanted control (CN + CO) and TSC (PN + PO) neuron-OL populations into the brain of immunocompromised RAG2null mice. After transplanting neuronal populations, we observed an abundant incorporation of human (HN-positive) cells in the mouse brain at 2.5 months post-transplantation (Figures S5A and S5B). To identify the human neurons in the mixed neuron-OL grafts, they were labeled with Synapsin-GFP viral construct before transplantation of mixed neuron-OL populations. HN-positive and GFP-positive cells were spread over several brain regions including the cerebral cortex (Figures S5C–S5F), the caudate putamen, and the olfactory bulb. While the total number of grafted control and patient neurons remained the same (Figure S5G), the average neurite length (GFP pixel density per HN-positive soma) in the PN + PO grafts (7,349 ± 1,247) was significantly higher (p < 0.02) compared with the CN + CO grafts (3,504 ± 233; Figure S5H). Therefore, mixed TSC neuron-OL populations showed increased neurite densities compared with control neuron-OL grafts at 2.5 months post-transplantation. Overall, changes in neurite density and hypertrophy in TSC patient cells can be measured in mixed neuron-OL populations both in vitro and in vivo, which can be partially altered by mTOR regulators.
Altered OL Properties in Mixed TSC Neuron-OL Populations
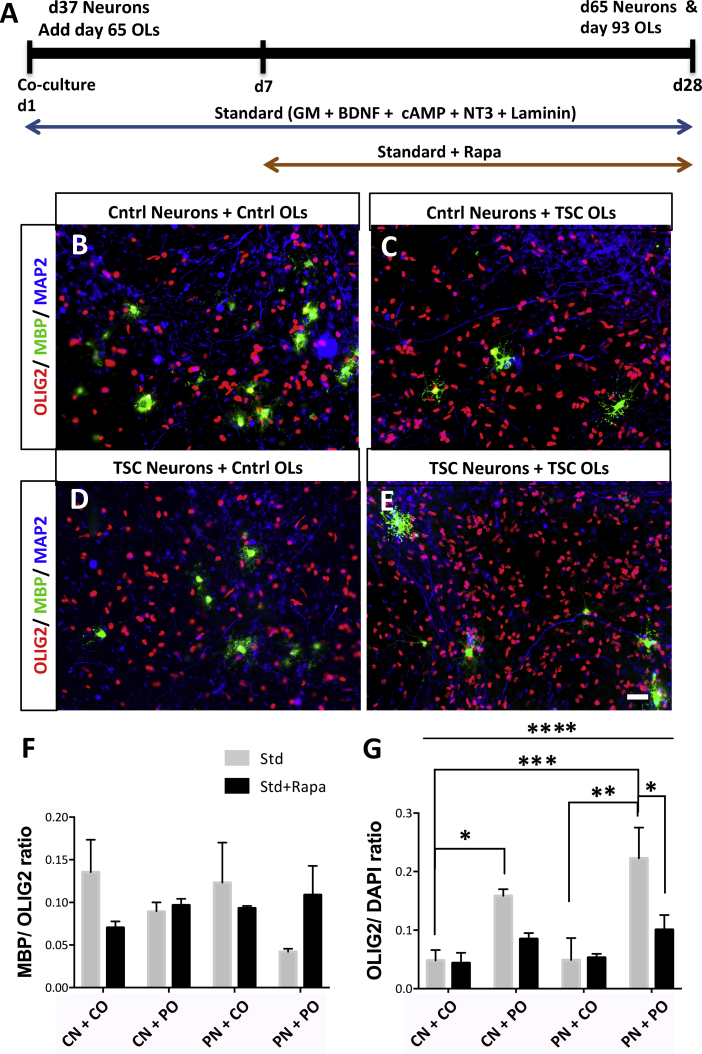
As TSC patients show glial dysfunctions, we further investigated whether TSC neuron-OL co-cultures show OL abnormalities. To study whether TSC neuron-OL co-cultures affected OL maturation, we analyzed the ratio of MBP/OLIG2-positive cells in the different co-cultures with and without Rapa or Guana treatment (Figures 5B–5F and S4D). While ANOVA analysis did not show significant changes (Figure 5F), t test analysis showed a significant decrease in OL maturation in PN + PO (0.04 ± 0.003) compared with CN + CO (0.13 ± 0.03; p < 0.03, t test) cultures, which improved after treatment with Rapa and Guana (0.10 ± 0.03 and 0.07 ± 0.004, respectively; Figures 5F and S4D). Overall, TSC neuron-OL co-cultures showed reduced maturation, which was improved by Rapa and Guana treatment.
Figure 5.
Differential Proliferation and Maturation of TSC OLs in Co-cultures
(A) Schematic of neuron-OL co-culture protocol in the Std and Std + Rapa conditions.
(B–E) Immunocytochemical images of co-cultures showing OL precursor marker OLIG2- and OL maturity marker MBP-positive cells in the CN + CO (B), CN + PO (C), PN + CO (D), and PN + PO (E) groups.
(F and G) Ratio of MBP/OLIG2- (F) and OLIG2/DAPI-positive cells (G) in the co-cultures at day 28. Mean per group (means ± SEM) was obtained in total from n = 4–9 independent experiments including C1-, C3-, P1B-, and P2-derived neurons and OLs.
Scale bars, 25 μm. Asterisks in (G) indicate statistically significant differences found in one-way ANOVA and Tukey's post hoc test. While several differences were found using Tukey's post hoc only important differences are represented in the graph, ∗p < 0.05, ∗∗p < 0.005, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. See also Figures S3–S5.
To study whether co-cultures with TSC neurons and/or OLs affected OL proliferation, we measured the number of OLs in mono- and co-cultures. In mono-cultures, we found a significant 2-fold increase in the number of OLIG2-positive cells in TSC (OLIG2/DAPI cells; p < 0.02, unpaired t test, Figures S3A–S3C) compared with control. We further found an increase in the number of the bromodeoxyuridine (BrdU)-positive cells 2 hr post addition in the TSC cultures (Figures S3D–S3F), confirming increased proliferation. However, there were no changes in the number of BrdU-positive cells 1 week post BrdU labeling (Figures S3G–S3I), suggestive of cell death in TSC cultures, which was further confirmed by an increase in the cleaved caspase 3-positive cells in TSC glial cultures (Figures S3J–S3L). In co-cultures, we also found significant changes in the number of OLs (OLIG2/DAPI cells) between the different groups (one-way ANOVA; p < 0.0001). Post hoc analysis showed a 4-fold significant increase (p < 0.0002, Tukey's post hoc test) in the number of OLIG2-positive cells in the PN + PO (0.22 ± 0.05; Figures 5E and 5G) compared with the CN + CO (0.048 ± 0.01; Figures 5B and 5G) cultures. In addition, when the TSC OLs were co-cultured with control neurons (CN + PO; 0.15 ± 0.01; Figures 5C and 5G), a three-times significant increase (p < 0.01, Tukey's post hoc test) in the OLIG2/DAPI-positive cells compared with CN + CO cultures was observed. The PN + CO cultures (Figure 5D) showed no significant changes in the OLIG2/DAPI-positive cells compared with CN + CO cultures (Figure 5G), indicating that TSC patient neurons do not affect proliferation levels of control OLs. Interestingly, PN + PO cultures under Std + Rapa conditions were significantly reduced compared with PN + PO cultures under Std conditions (p < 0.04; Tukey's post hoc test; Figure 5G). Treatment with Guana did not normalize OL numbers (Figure S4E). Hence, TSC patient OL cultures show increased proliferation, which increased in co-culture with control and TSC neurons, which was modulated by Rapa.
To study proliferation of TSC OLs in vivo, transplanted control (CN + CO) and TSC (PN + PO) neuron-OL populations were studied 2.5 months post-transplantation. Human OL somas were traced by OLIG2 and HN double labeling. The numbers of TSC OLs in the PN + PO grafts (0.33 ± 0.02) were significantly increased (p < 0.02) compared with OL numbers in the CN + CO grafts (0.12 ± 0.03; Figure S5I). So, after in vivo transplantation, the number of OLs increased in the grafted mixed TSC neuron-OL populations.
Altogether, TSC patient OLs showed a significantly increased proliferation when co-cultured with control neurons, which further increased by the addition of TSC patient neurons both in vitro and in vivo, and could be normalized by Rapa treatment.
Discussion
TSC neuropathology is known to involve defects in both neuronal and glial cell populations; however, the inter-relationships between each neural cell type during disease progression is unclear, hampering the development of treatment strategies. Here we present evidence for an interaction between neurons and OLs leading to changes in axonal density, cellular hypertrophy, and OL cell proliferation, using TSC patient iPSC-based neuron-OL co-cultures. However, increased basal dendritic branching and enhanced neuronal activity appeared also in neuron mono-cultures in the absence of OLs. Administration of mTOR pathway regulators reduced the increase in network activity and partly normalized the cellular defects. Overall, we present TSC patient iPSC-derived models involving TSC1 and TSC2 mutations, showing several known TSC-associated neuronal and OL phenotypes.
Here, we generated TSC patient-derived iPSC models that mimic some of the known effects of TSC mutations in the CNS. We substantially expand on previous observations reported by Li et al. (2017), who investigated iPSC-derived neural stem cell (NSC), neuronal, and astrocyte morphological properties from a single patient carrying a c.1444-2A > C mutation in TSC2. The authors showed an increase in NSC and astrocyte proliferation, hypertrophy, and neurite abnormalities in patient cultures (Li et al., 2017). While we did find an increase in the number of basal dendrites, we observed no significant changes in cell soma size in TSC patient neuron mono-cultures in contrast to Li et al. (2017). This might underlie differences in the culture set up, such as the generation of different neuronal subtypes, or the supplementation of rodent astrocytes (Nadadhur et al., 2017). In the current study, we did measure changes in neurite density and hypertrophy when neurons were co-cultured with OLs, next to increased OL cell proliferation and decreased OL maturation, indicating that interactions between neurons and OLs are of utmost importance for the development of TSC-associated phenotypes. In our TSC neuron mono-cultures, we further showed an increase in spontaneous calcium event frequency, consistent with the network hyperactivity seen in the TSC mouse model (Bateup et al., 2013), and an altered inhibition reported in TSC patient tissue (Talos et al., 2012). The MEA data showed that an increase in activity is also present at the level of action potential generation, which is the output of the summation of calcium activity. Lastly, our patient-derived iPSC models involved either TSC1 or TSC2 dominant mutations. TSC2 gene loss often causes a more severe phenotype (Kothare et al., 2014), as also seen in mouse studies (Onda et al., 1999, Zeng et al., 2011). As specific genetic variations in the TSC genes lead to variable disease symptoms and severity in patients (Jentarra et al., 2011), there is a need for model systems that involve either TSC1 or TSC2 mutations.
Interestingly, while we did not measure a difference in axonal density in the neuron mono-cultures, the presence of TSC patient neurons in the co-cultures seems to be the most important determinant to allow axonal differences. OLs have critical roles in regulating neuronal survival, the stability of the axon (Morrison et al., 2013), and neuronal assembly (Simons and Trajkovic, 2006). Furthermore, it is suggested that myelinated axons need OLs for metabolic support, i.e., maintenance of ATP homeostasis (Philips and Rothstein, 2017). To ensure firing of action potentials and activity of Na+ and K+ channels, neurons need a significant amount of ATP. This might further explain why neuron-OL co-cultures (or mixed neural organoid cultures) can maintain neuronal health for longer periods of time, while neuron mono-cultures degenerate at certain stages. So this could also suggest that the neurons in mono-cultures versus the neurons in co-cultures had different maturation states and that only at more mature stages do axonal differences in TSC became apparent. Together, co-culture model systems give the opportunity to study morphological defects in TSC neuronal populations that might be absent in neuronal mono-cultures, and thereby supports the use of mixed neural cultures for future screens.
Administration of mTOR pathway inhibitors to the neuron-OL co-cultures, showed different modulating effects. While Rapa treatment normalized the enhanced OL numbers and increase in soma size, the axonal density remained unchanged. Earlier reports showed that the mTOR pathway is differentially regulated in different neural cell types (Edinger et al., 2003, Li et al., 2014, Oh and Jacinto, 2011, Watanabe et al., 2011), and that dose-dependent Rapa treatment suppresses mTOR hyperactivation and rescues TSC neuronal phenotypes and epilepsy in rodent models (Yasuda et al., 2014, Zeng et al., 2008). Furthermore, Rapa can enhance neurite outgrowth in primary control neurons (Lyons et al., 1994) and TSC plays a role in neuronal polarization (Choi et al., 2008). Rodent models also showed that the mTOR pathway regulates OL proliferation and maturation (Wahl et al., 2014), as well as glial abnormalities (Carson et al., 2012). Therefore, our neuron-OL models confirm that both neuronal and OL cell types are affected by TSC mutations and that the mTOR pathway regulators have differential effects on these cell types.
An increased amount of OLIG2-positive cells was present in the TSC cultures, which was highest when TSC OLs were co-cultured with TSC neurons. This is in line with earlier studies showing axonal influence on OL precursor proliferation, maturation, and myelination (Barres and Raff, 1999, Simons and Trajkovic, 2006), such as neuron-induced secretion of neurotransmitters and neurotrophins (Leferink and Heine, 2017). Interestingly, a recent study showed that TSC expression in neurons regulates OL differentiation via neuronal connective tissue growth factor (Ercan et al., 2017). The increased OL proliferation in patient neuron-OL cultures was suppressed by Rapa treatment. While these results support Rapa treatment to normalize OL defects in patients, the treatment regimen need further study, as earlier reports showed that both increased and decreased mTOR pathway activation negatively affects OL maturation (Lebrun-Julien et al., 2014, Wahl et al., 2014). Similarly, a recent study showed that TSC1 deletion in OL precursors accelerates remyelination (McLane et al., 2017), thereby illustrating the different roles of TSC proteins in OL development and myelin health. Guana, an antihypertensive drug, was shown to prevent OL cell death and to induce partial rescue of myelination defects by altering mTOR-related endoplasmic reticulum stress via inhibiting Gadd34-PP1 phosphatase (Jiang et al., 2016). However, we did not find significant changes after Guana treatment. As TSC phenotypes are also measurable in neuron-OL grafts in vivo, chimeric mouse models could function to test treatment dose and strategies. Hence, mixed neuron-OL populations represent TSC phenotypes in OL populations, thereby providing physiological models to study the therapeutic effects of mTOR regulators.
In summary, we have used a TSC iPSC-based cell models to show: (1) common neuronal defects caused by both autosomal dominant TSC1 and TSC2 mutations in a patient genetic background; (2) complex neuron-OL interactions in TSC phenotypes; and (3) therapeutic effects of compounds such as mTOR regulator Rapa, IGF-1, and Guana. This model opens future prospects for the development of disease-relevant cell-based assays that capture the complex brain interactions underlying TSC and other mTOR-mediated neuropathies.
Experimental Procedures
iPSCs and Neuronal Cultures
TSC patient (P1, GM06149; P2, GM03958) and control fibroblasts (C3, GM23973) were obtained from the Coriell Biorepository (all teenage donors), and controls C1 (infant) and C2 (teenage) were obtained from anonymous control donors. The selected TSC patients were reported with mental retardation and seizures along with other TSC-related abnormalities. iPSC reprogramming was done based on polycistronic construct with OCT3/4, c-MYC, SOX2, and KLF4 (Warlich et al., 2011). All the iPSC lines were cultured in Essential 8 medium (Gibco) on Geltrex LDEV-free (Gibco). Generation of in-direct contact iPSC-neuronal mono-cultures was performed as described earlier (Nadadhur et al., 2017) with slight modifications. Neuronal mono-cultures with a density of 62.5 K/2.0 cm2 in the Std condition medium contained Neurobasal composition with brain-derived neurotrophic factor (BDNF) (20 ng/mL; Peprotech) and cAMP (1 μM; Sigma). The Std + IGF-1 and Std + Rapa conditions contained Std medium with IGF-1 (10 ng/mL; Peprotech) or Rapa (30 nM; Sigma), respectively. For detailed culturing methods and culture conditions for MEA experiments please see Supplemental Experimental Procedures.
Glial Cells and Co-cultures
OLs were obtained as described previously (Izrael et al., 2007). Before neuron-OL co-cultures were generated, we first separately created neuron mono-cultures (62.5 K/2.0 cm2 density) until day 37 (Nadadhur et al., 2017) and OL progenitors until day 65. To generate neuron-OL co-cultures, the neuronal coverslips were removed from rat astrocyte feeder plates and OL progenitors were passaged with Accutase (Millipore) and plated at a density of 200 K/2.0 cm2 on neuronal coverslips. The co-cultures were maintained in Std co-culture medium (DMEM/F12 and Neurobasal [1:1; both Gibco], 1% N1 supplement [in-house], 2% B27 supplement [Gibco], 1% NEAA [Gibco], 60 ng/mL T3 [Sigma], 100 ng/mL Biotin [Sigma] medium with NT3 [10 ng/mL; Peprotech], mouse laminin [1 μg/mL; Sigma], BDNF [20 ng/mL], and cAMP [1 μM]). From day 1 to 7 of co-culture, all cells were refreshed with Std medium. From day 8 to 28 of co-culture, cells were refreshed with either Std, Std + Guana (5 μM; Sigma) or Std + Rapa (10 nM) medium twice a week. At day 28 of co-culture, the cell culture coverslips were processed for immunocytochemistry.
Calcium Imaging
At day 56, neuronal mono-culture coverslips were incubated with Fluo-5-AM ester 1 μM in Neurobasal medium without growth factors for 5 min at 37°C and imaged under constant perfusion at 0.5 mL/min rate with Tyrode's solution (119 NaCl. 2.5 KCl, 2 CaCl2, 2 MgCl2, 25 HEPES, 30 glucose; all in mM) in an Axiovert 200M inverted microscope (Zeiss) using a 40× oil objective (numerical aperture 1.3; Zeiss), a Polychrome IV light source (TILL Photonics), a 488/6 nm excitation filter (Semrock), a 525/50-nm emission filter (Semrock), and a 500-nm beam splitter (Semrock). Images (1 image/s) were acquired using a Cascade 512 EM-CCD camera (Roper Scientific) and Metamorph 6.2 software (Molecular Devices). The total length of 4.5 min was recorded per FOV, and a minimum of five FOVs per coverslip was recorded each from two to four differentiations for three controls and three TSC lines. For analysis, image series were stacked using z stack projection in ImageJ, and regions of interest were placed on a minimum of 12 neurites per FOV. Calcium traces were detected using “event detection analysis (EVA)” with previously described methods in MATLAB (Hjorth et al., 2016).
MEA
60MEA200/30iR-Tigr MEAs were used. Spontaneous activity was recorded, for a period of 10 min, every 10 days at 37°C using MC Rack at a sample rate of 25,000 Hz. Electrode data were filtered online with a 200 Hz high pass and a 5,000 Hz low pass filter (both second-order Butterworth). Data were stored as .mcd files, which contain the filtered continuous traces for each electrode together with the spikes detected using the online threshold detection built into MC Rack. After selecting electrodes for analysis, chosen data were converted to asci files for offline analysis using the MC Data tool. Offline analysis was achieved with custom scripts written in MATLAB. Spikes were detected from filtered data using an automatic threshold-based method set at −5.5×σ, where σ is an estimate of the noise of each electrode. Spike timestamps were analyzed to provide statistics on the general excitability of cultures. The average spike rate (Hz) represents the median of the average spike rate for all electrodes (AvgSpkRateE1-n). The average spike rate for each electrode (AvgSpkRateE) was calculated as AvgSpkRateE = number of spikes/tmax, where tmax is the total length of the recording in seconds. This is used to control for particularly high or low firing rates.
Immunofluorescence
For immunocytochemistry, neuronal mono-cultures and neuron-OL co-culture coverslips were fixed with 4% PFA for 20 min and processed for staining. For immunohistochemistry, slides were washed with PBS (6 times for 5 min each in PBS 1×), optionally treated for 2 min with citrate buffer (0.01 M [pH 6]) for antigen retrieval, followed by washing in PBS (5 min for 1 time). All the coverslips/slides were blocked for 1 hr at room temperature (RT) in blocking buffer (5% NGS, 0.1% BSA, 0.3% Triton X-100 in PBS 1×), incubated with primary antibody in blocking buffer overnight at 4°C. Next day, after washing again with PBS (6 times for 5 min each), the coverslips/slides were incubated with secondary antibody solution for 2 hr, washed again (6× for 5 min), and then mounted with Fluoromount G (Southern Biotech) on glass slides (VWR; Super frost plus)/glass coverslips. Images were acquired from stained slides using either a Carl Zeiss 510 Meta confocal microscope with 40× (1.2 numerical aperture) oil objective or a Leica DM6000B fluorescent microscope with 10×, 20×, and 40× magnification. For details of primary antibodies used please see Supplemental Experimental Procedures.
Cell Property Analysis
Cell properties such as axonal density, dendritic density, soma size, and the number of synapses; OLIG2- and MBP-positive cells were measured using Columbus 2.5 online software. Precisely, images acquired with confocal and fluorescent microscope were analyzed with algorithms for morphology, soma recognition, and co-localization tools using pixel density measurement and the number of objects present, respectively. To quantify the soma sizes and morphological parameters, pixel densities were measured in each FOV (6–10 per condition/coverslip) with images of 20× or 40× magnification. To quantify dendritic branching, the NeurophologyJ tool was used in ImageJ.
Western Blot Analysis
Cell lysates were collected from feeder-free iPSCs at day 3 or 4 after passage and from neuroepithelial stem cells between P1 and P5. SDS-PAGE was run on Bolt 4%–12% Bis-Tris Plus Gels (Thermo Fisher Scientific). The PVDF membranes were blocked with 5% non-fat dry milk in TBS-0.05% Tween, followed by primary antibody incubations at 1:1,000 concentration in 5% BSA in TBS-0.05% Tween and incubated ON at 4°C (primary antibodies are listed in Supplement Information). The blots were then washed thrice for 10 min with TBS-Tween and incubated with secondary antibody in milk-TBS-Tween for 1 hr at RT. After washing thrice again, bands were visualized using SuperSignal West Femto substrate (Thermo Scientific) on Odyssey Fc imaging system (LI-COR). Primary antibodies against TSC1 (no. 6935) and TSC2 (no. 4308; both from Cell Signaling Technology) were used at 1:1,000 concentrations. The secondary antibody used was horseradish peroxidase rabbit antibody (1:1,000; no. 7074S, Cell Signaling Technology).
Statistical Analysis
All statistical analysis was performed using Prism 7 software. All the raw data were tested for normal distribution and then analyzed using unpaired t test or one-way ANOVA, depending on group numbers. In cases where significant differences were obtained in the ANOVA (p < 0.05), a Tukey's post hoc test was performed.
Author Contributions
A.G.N. and V.M.H. conceptualized and designed the experiments. V.M.H. provided iPSC/study material and financial support. A.G.N. performed, collected, and analyzed all the cell culture, immunofluorescence, and calcium imaging data. M.A. and A.J.H. performed the MEA analysis. L.G. supported the cell culture experiments and provided administrative support. P.C.S., E.v.H., and A.G.N. performed and analyzed the western blot experiments. S.D., V.M.H., and A.G.N. collected all transplantation data. A.G.N. and V.M.H. wrote the manuscript with inputs from all authors.
Acknowledgments
We thank Prof. Eleonora Aronica for providing TSC patient material. We thank Gerbren Jacobs and Frank den Oudsten for help with iPSC and rat astrocyte cultures. We thank Joke Wortel and Nina Straat for performing transplantation and Carola Berkel for performing sequencing experiments. We thank Jurjen Broeke for valuable suggestions on calcium imaging experiments. We thank Nicole Breeuwsma and Jorik Methorst for help with immunohistochemistry and western blot. This study was supported and funded by Amsterdam Neuroscience and EU MSCA-ITN CognitionNet (FP7-PEOPLE-2013-ITN 607508). M.A. and A.J.H. are supported by DEFINE, a Wellcome Trust Strategic Award (100202). V.M.H. is supported by ZonMw VIDI Research grant (91712343), E-Rare Joint Call project (9003037601) and a European Leukodystrophy Association (ELA) Research Grant (2014-012L1).
Published: December 20, 2018
Footnotes
Supplemental Information includes Supplemental Experimental Procedures and five figures and can be found with this article online at https://doi.org/10.1016/j.stemcr.2018.11.019.
Supplemental Information
References
- Barres B.A., Raff M.C. Axonal control of oligodendrocyte development. J. Cell Biol. 1999;147:1123–1128. doi: 10.1083/jcb.147.6.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateup H.S., Johnson C.A., Denefrio C.L., Saulnier J.L., Kornacker K., Sabatini B.L. Excitatory/inhibitory synaptic imbalance leads to hippocampal hyperexcitability in mouse models of tuberous sclerosis. Neuron. 2013;78:510–522. doi: 10.1016/j.neuron.2013.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaumont T.L., Godzik J., Dahiya S., Smyth M.D. Subependymal giant cell astrocytoma in the absence of tuberous sclerosis complex: case report. J. Neurosurg. Pediatr. 2015;16:134–137. doi: 10.3171/2015.1.PEDS13146. [DOI] [PubMed] [Google Scholar]
- Carson R.P., Kelm N.D., West K.L., Does M.D., Fu C., Weaver G., McBrier E., Parker B., Grier M.D., Ess K.C. Hypomyelination following deletion of Tsc2 in oligodendrocyte precursors. Ann. Clin. Transl. Neurol. 2015;2:1041–1054. doi: 10.1002/acn3.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson R.P., Van Nielen D.L., Winzenburger P.A., Ess K.C. Neuronal and glia abnormalities in Tsc1-deficient forebrain and partial rescue by rapamycin. Neurobiol. Dis. 2012;45:369–380. doi: 10.1016/j.nbd.2011.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y.J., Di Nardo A., Kramvis I., Meikle L., Kwiatkowski D.J., Sahin M., He X. Tuberous sclerosis complex proteins control axon formation. Genes Dev. 2008;22:2485–2495. doi: 10.1101/gad.1685008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa V., Aigner S., Vukcevic M., Sauter E., Behr K., Ebeling M., Dunkley T., Friedlein A., Zoffmann S., Meyer C.A. mTORC1 inhibition corrects neurodevelopmental and synaptic alterations in a human stem cell model of tuberous sclerosis. Cell Rep. 2016;15:86–95. doi: 10.1016/j.celrep.2016.02.090. [DOI] [PubMed] [Google Scholar]
- Curatolo P., Bjornvold M., Dill P.E., Ferreira J.C., Feucht M., Hertzberg C., Jansen A., Jozwiak S., Kingswood J.C., Kotulska K. The role of mTOR inhibitors in the treatment of patients with tuberous sclerosis complex: evidence-based and expert opinions. Drugs. 2016;76:551–565. doi: 10.1007/s40265-016-0552-9. [DOI] [PubMed] [Google Scholar]
- Curatolo P., Moavero R. mTOR inhibitors in tuberous sclerosis complex. Curr. Neuropharmacol. 2012;10:404–415. doi: 10.2174/157015912804143595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curatolo P., Verdecchia M., Bombardieri R. Tuberous sclerosis complex: a review of neurological aspects. Eur. J. Paediatr. Neurol. 2002;6:15–23. doi: 10.1053/ejpn.2001.0538. [DOI] [PubMed] [Google Scholar]
- Edinger A.L., Linardic C.M., Chiang G.G., Thompson C.B., Abraham R.T. Differential effects of rapamycin on mammalian target of rapamycin signaling functions in mammalian cells. Cancer Res. 2003;63:8451–8460. [PubMed] [Google Scholar]
- Ercan E., Han J.M., Di Nardo A., Winden K., Han M.J., Hoyo L., Saffari A., Leask A., Geschwind D.H., Sahin M. Neuronal CTGF/CCN2 negatively regulates myelination in a mouse model of tuberous sclerosis complex. J. Exp. Med. 2017;214:681–697. doi: 10.1084/jem.20160446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurtler A., Kunz N., Gomolka M., Hornhardt S., Friedl A.A., McDonald K., Kohn J.E., Posch A. Stain-free technology as a normalization tool in Western blot analysis. Anal. Biochem. 2013;433:105–111. doi: 10.1016/j.ab.2012.10.010. [DOI] [PubMed] [Google Scholar]
- Hjorth J.J., Dawitz J., Kroon T., Pires J., Dassen V.J., Berkhout J.A., Emperador Melero J., Nadadhur A.G., Alevra M., Toonen R.F. Detection of silent cells, synchronization and modulatory activity in developing cellular networks. Dev. Neurobiol. 2016;76:357–374. doi: 10.1002/dneu.22319. [DOI] [PubMed] [Google Scholar]
- Huang J., Manning B.D. The TSC1-TSC2 complex: a molecular switchboard controlling cell growth. Biochem. J. 2008;412:179–190. doi: 10.1042/BJ20080281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izrael M., Zhang P., Kaufman R., Shinder V., Ella R., Amit M., Itskovitz-Eldor J., Chebath J., Revel M. Human oligodendrocytes derived from embryonic stem cells: effect of noggin on phenotypic differentiation in vitro and on myelination in vivo. Mol. Cell. Neurosci. 2007;34:310–323. doi: 10.1016/j.mcn.2006.11.008. [DOI] [PubMed] [Google Scholar]
- Jaworski J., Sheng M. The growing role of mTOR in neuronal development and plasticity. Mol. Neurobiol. 2006;34:205–219. doi: 10.1385/MN:34:3:205. [DOI] [PubMed] [Google Scholar]
- Jentarra G.M., Rice S.G., Olfers S., Saffen D., Narayanan V. Evidence for population variation in TSC1 and TSC2 gene expression. BMC Med. Genet. 2011;12:29. doi: 10.1186/1471-2350-12-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M., Liu L., He X., Wang H., Lin W., Wang H., Yoon S.O., Wood T.L., Lu Q.R. Regulation of PERK-eIF2alpha signalling by tuberous sclerosis complex-1 controls homoeostasis and survival of myelinating oligodendrocytes. Nat. Commun. 2016;7:12185. doi: 10.1038/ncomms12185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones A.C., Shyamsundar M.M., Thomas M.W., Maynard J., Idziaszczyk S., Tomkins S., Sampson J.R., Cheadle J.P. Comprehensive mutation analysis of TSC1 and TSC2-and phenotypic correlations in 150 families with tuberous sclerosis. Am. J. Hum. Genet. 1999;64:1305–1315. doi: 10.1086/302381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kothare S.V., Singh K., Chalifoux J.R., Staley B.A., Weiner H.L., Menzer K., Devinsky O. Severity of manifestations in tuberous sclerosis complex in relation to genotype. Epilepsia. 2014;55:1025–1029. doi: 10.1111/epi.12680. [DOI] [PubMed] [Google Scholar]
- Lasarge C.L., Danzer S.C. Mechanisms regulating neuronal excitability and seizure development following mTOR pathway hyperactivation. Front. Mol. Neurosci. 2014;7:18. doi: 10.3389/fnmol.2014.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebrun-Julien F., Bachmann L., Norrmen C., Trotzmuller M., Kofeler H., Ruegg M.A., Hall M.N., Suter U. Balanced mTORC1 activity in oligodendrocytes is required for accurate CNS myelination. J. Neurosci. 2014;34:8432–8448. doi: 10.1523/JNEUROSCI.1105-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leferink P.S., Heine V.M. The healthy and diseased microenvironment regulate oligodendrocyte properties: implications for regenerative medicine. Am. J. Pathol. 2017;188:39–52. doi: 10.1016/j.ajpath.2017.08.030. [DOI] [PubMed] [Google Scholar]
- Leung A.K., Robson W.L. Tuberous sclerosis complex: a review. J. Pediatr. Health Care. 2007;21:108–114. doi: 10.1016/j.pedhc.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Li J., Kim S.G., Blenis J. Rapamycin: one drug, many effects. Cell Metab. 2014;19:373–379. doi: 10.1016/j.cmet.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Cao J., Chen M., Li J., Sun Y., Zhang Y., Zhu Y., Wang L., Zhang C. Abnormal neural progenitor cells differentiated from induced pluripotent stem cells partially mimicked development of TSC2 neurological abnormalities. Stem Cell Reports. 2017;8:883–893. doi: 10.1016/j.stemcr.2017.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons W.E., George E.B., Dawson T.M., Steiner J.P., Snyder S.H. Immunosuppressant FK506 promotes neurite outgrowth in cultures of PC12 cells and sensory ganglia. Proc. Natl. Acad. Sci. U S A. 1994;91:3191–3195. doi: 10.1073/pnas.91.8.3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marti-Bonmati L., Menor F., Dosda R. Tuberous sclerosis: differences between cerebral and cerebellar cortical tubers in a pediatric population. AJNR Am. J. Neuroradiol. 2000;21:557–560. [PMC free article] [PubMed] [Google Scholar]
- McLane L.E., Bourne J.N., Evangelou A.V., Khandker L., Macklin W.B., Wood T.L. Loss of tuberous sclerosis complex1 in adult oligodendrocyte progenitor cells enhances axon remyelination and increases myelin thickness after a focal demyelination. J. Neurosci. 2017;37:7534–7546. doi: 10.1523/JNEUROSCI.3454-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meikle L., Talos D.M., Onda H., Pollizzi K., Rotenberg A., Sahin M., Jensen F.E., Kwiatkowski D.J. A mouse model of tuberous sclerosis: neuronal loss of Tsc1 causes dysplastic and ectopic neurons, reduced myelination, seizure activity, and limited survival. J. Neurosci. 2007;27:5546–5558. doi: 10.1523/JNEUROSCI.5540-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min J., Singh S., Fitzgerald-Bocarsly P., Wood T.L. Insulin-like growth factor I regulates G2/M progression through mammalian target of rapamycin signaling in oligodendrocyte progenitors. Glia. 2012;60:1684–1695. doi: 10.1002/glia.22387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison B.M., Lee Y., Rothstein J.D. Oligodendroglia: metabolic supporters of axons. Trends Cell Biol. 2013;23:644–651. doi: 10.1016/j.tcb.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadadhur A.G., Emperador Melero J., Meijer M., Schut D., Jacobs G., Li K.W., Hjorth J.J.J., Meredith R.M., Toonen R.F., Van Kesteren R.E. Multi-level characterization of balanced inhibitory-excitatory cortical neuron network derived from human pluripotent stem cells. PLoS One. 2017;12:e0178533. doi: 10.1371/journal.pone.0178533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh W.J., Jacinto E. mTOR complex 2 signaling and functions. Cell Cycle. 2011;10:2305–2316. doi: 10.4161/cc.10.14.16586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onda H., Lueck A., Marks P.W., Warren H.B., Kwiatkowski D.J. Tsc2(+/−) mice develop tumors in multiple sites that express gelsolin and are influenced by genetic background. J. Clin. Invest. 1999;104:687–695. doi: 10.1172/JCI7319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philips T., Rothstein J.D. Oligodendroglia: metabolic supporters of neurons. J. Clin. Invest. 2017;127:3271–3280. doi: 10.1172/JCI90610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivero-Gutierrez B., Anzola A., Martinez-Augustin O., de Medina F.S. Stain-free detection as loading control alternative to Ponceau and housekeeping protein immunodetection in Western blotting. Anal. Biochem. 2014;467:1–3. doi: 10.1016/j.ab.2014.08.027. [DOI] [PubMed] [Google Scholar]
- Ruppe V., Dilsiz P., Reiss C.S., Carlson C., Devinsky O., Zagzag D., Weiner H.L., Talos D.M. Developmental brain abnormalities in tuberous sclerosis complex: a comparative tissue analysis of cortical tubers and perituberal cortex. Epilepsia. 2014;55:539–550. doi: 10.1111/epi.12545. [DOI] [PubMed] [Google Scholar]
- Sadowski K., Kotulska K., Schwartz R.A., Jozwiak S. Systemic effects of treatment with mTOR inhibitors in tuberous sclerosis complex: a comprehensive review. J. Eur. Acad. Dermatol. Venereol. 2016;30:586–594. doi: 10.1111/jdv.13356. [DOI] [PubMed] [Google Scholar]
- Simons M., Trajkovic K. Neuron-glia communication in the control of oligodendrocyte function and myelin biogenesis. J. Cell Sci. 2006;119:4381–4389. doi: 10.1242/jcs.03242. [DOI] [PubMed] [Google Scholar]
- Talos D.M., Sun H., Kosaras B., Joseph A., Folkerth R.D., Poduri A., Madsen J.R., Black P.M., Jensen F.E. Altered inhibition in tuberous sclerosis and type IIb cortical dysplasia. Ann. Neurol. 2012;71:539–551. doi: 10.1002/ana.22696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl S.E., McLane L.E., Bercury K.K., Macklin W.B., Wood T.L. Mammalian target of rapamycin promotes oligodendrocyte differentiation, initiation and extent of CNS myelination. J. Neurosci. 2014;34:4453–4465. doi: 10.1523/JNEUROSCI.4311-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Greenwood J.S., Calcagnotto M.E., Kirsch H.E., Barbaro N.M., Baraban S.C. Neocortical hyperexcitability in a human case of tuberous sclerosis complex and mice lacking neuronal expression of TSC1. Ann. Neurol. 2007;61:139–152. doi: 10.1002/ana.21058. [DOI] [PubMed] [Google Scholar]
- Warlich E., Kuehle J., Cantz T., Brugman M.H., Maetzig T., Galla M., Filipczyk A.A., Halle S., Klump H., Scholer H.R. Lentiviral vector design and imaging approaches to visualize the early stages of cellular reprogramming. Mol. Ther. 2011;19:782–789. doi: 10.1038/mt.2010.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe R., Wei L., Huang J. mTOR signaling, function, novel inhibitors, and therapeutic targets. J. Nucl. Med. 2011;52:497–500. doi: 10.2967/jnumed.111.089623. [DOI] [PubMed] [Google Scholar]
- Yasuda S., Sugiura H., Katsurabayashi S., Shimada T., Tanaka H., Takasaki K., Iwasaki K., Kobayashi T., Hino O., Yamagata K. Activation of Rheb, but not of mTORC1, impairs spine synapse morphogenesis in tuberous sclerosis complex. Sci. Rep. 2014;4:5155. doi: 10.1038/srep05155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L.H., Rensing N.R., Zhang B., Gutmann D.H., Gambello M.J., Wong M. Tsc2 gene inactivation causes a more severe epilepsy phenotype than Tsc1 inactivation in a mouse model of tuberous sclerosis complex. Hum. Mol. Genet. 2011;20:445–454. doi: 10.1093/hmg/ddq491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L.H., Xu L., Gutmann D.H., Wong M. Rapamycin prevents epilepsy in a mouse model of tuberous sclerosis complex. Ann. Neurol. 2008;63:444–453. doi: 10.1002/ana.21331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zucco A.J., Pozzo V.D., Afinogenova A., Hart R.P., Devinsky O., D'Arcangelo G. Neural progenitors derived from tuberous sclerosis complex patients exhibit attenuated PI3K/AKT signaling and delayed neuronal differentiation. Mol. Cell. Neurosci. 2018;92:149–163. doi: 10.1016/j.mcn.2018.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.