Summary
Heterozygous de novo mutations in GATA6 are the most frequent cause of pancreatic agenesis in humans. In mice, however, a similar phenotype requires the biallelic loss of Gata6 and its paralog Gata4. To elaborate the human-specific requirements for GATA6, we chose to model GATA6 loss in vitro by combining both gene-edited and patient-derived pluripotent stem cells (hPSCs) and directed differentiation toward β-like cells. We find that GATA6 heterozygous hPSCs show a modest reduction in definitive endoderm (DE) formation, while GATA6-null hPSCs fail to enter the DE lineage. Consistent with these results, genome-wide studies show that GATA6 binds and cooperates with EOMES/SMAD2/3 to regulate the expression of cardinal endoderm genes. The early deficit in DE is accompanied by a significant reduction in PDX1+ pancreatic progenitors and C-PEPTIDE+ β-like cells. Taken together, our data position GATA6 as a gatekeeper to early human, but not murine, pancreatic ontogeny.
Keywords: GATA6, definitive endoderm, pancreatic agenesis, human pluripotent stem cells, disease modeling
Highlights
-
•
Heterozygous loss of human GATA6 results in pancreatic agenesis
-
•
GATA6+/− hPSCs form fewer pancreatic progenitors and β cells in vitro
-
•
GATA6−/− hPSCs entirely fail to form the definitive endoderm in vitro
-
•
GATA6 binds to and cooperates with EOMES/SMAD2/3 to regulate cardinal endodermal genes
In this article, Chia et al. model human pancreatic agenesis in vitro that results from heterozygous loss of the GATA6 gene. Using both gene-edited and patient-derived hPSCs, they demonstrate that GATA6 serves as a gatekeeper to early human, but not murine, pancreatic ontogeny. Genome-wide studies further show that GATA6 binds to and cooperates with EOMES/SMAD2/3 to regulate the expression of cardinal endoderm genes.
Introduction
Pancreatic agenesis is an extremely rare human condition resulting from the impaired formation of the pancreas during embryonic development. Clinically, patients can entirely lack the pancreas or present with only a partially formed organ (hypoplasia). The majority of patients have complete absence of a functioning pancreas, resulting in intrauterine growth retardation, neonatal diabetes, and exocrine pancreatic failure, and thus require insulin and exocrine enzyme replacement therapy. Less commonly, less severely affected patients can display a reduction in total islet number or insulin-secreting β cells and present diabetic symptoms during adolescence or adulthood.
The vast majority of human pancreatic agenesis cases owe their genetic origins to mutations in a small handful of pancreatic regulatory genes. The first described is Pancreatic and Duodenal Homeobox 1 (Pdx1) (Schwitzgebel, 2014, Stoffers et al., 1997). In mice, Pdx1 transcripts label the incipient pancreatic primordium—two epithelial buds that are situated dorsally and ventrally on opposite sides of the posterior foregut around embryonic day 9.5 (Jørgensen et al., 2007, Pan and Wright, 2011). In Pdx1-null mutant mice, these buds initially form but quickly regress, resulting in complete pancreatic agenesis, severe hyperglycemia, and death within a few days of birth (Ahlgren et al., 1996, Jonsson et al., 1994, Offield et al., 1996). PDX1 similarly labels the human embryonic dorsal and ventral foregut around Carnegie stage 12 (29–31 days post conception) (Jennings et al., 2013). Significantly, the pathology of human patients with homozygous or compound heterozygous mutations in PDX1 mirrors the agenesis phenotype observed in Pdx1-deficient mice (Schwitzgebel et al., 2003, Stoffers et al., 1997).
The most common cause of pancreatic agenesis in humans is heterozygous mutations in the GATA6 gene (De Franco et al., 2013, Lango Allen et al., 2011). GATA6 encodes a highly conserved zinc-finger transcription factor that recognizes and binds the (A/T)GATA(A/G) regulatory motif, two of which are located in the mouse Pdx1 and human PDX1 promoters (Carrasco et al., 2012, Lentjes et al., 2016, Patient and McGhee, 2002, Viger et al., 2008, Xuan et al., 2012). GATA6, along with its five other family members (GATA1–5), functions in diverse cellular contexts, from coordinating morphogenesis during embryonic development to the maintenance of lineage-specific gene expression in adult hematopoietic stem cells (Lentjes et al., 2016, Viger et al., 2008). Gata6 is expressed in the definitive endoderm (DE) that emerges during gastrulation, as well as its derivative the gut tube epithelium and the early pancreas primordium (Freyer et al., 2015, Morrisey et al., 1996). Gata6 expression persists as the pancreas undergoes branching morphogenesis, becoming restricted in later development to the ductal epithelial compartment and a subset of endocrine cells (Decker et al., 2006, Ketola et al., 2004).
In contrast to PDX1, GATA6 mutations that result in pancreatic agenesis are heterozygous and predominantly de novo (Chao et al., 2015, De Franco et al., 2013, Lango Allen et al., 2011, Stanescu et al., 2015, Suzuki et al., 2014). The majority of cases have full pancreatic agenesis, but there are some associated with incomplete penetrance, resulting in a broad spectrum of clinical manifestations (De Franco et al., 2013). At the extreme, family members with the same inherited GATA6 allele can present with markedly different phenotypes (Bonnefond et al., 2012, Yau et al., 2017, Yorifuji et al., 2012). In addition, GATA6 patients usually display a number of extrapancreatic abnormalities, including congenital heart defects, as well as several whose origins are endodermal—hepatobiliary malformations, gall bladder agenesis, and gut herniation (Chao et al., 2015, De Franco et al., 2013, Lango Allen et al., 2011).
Given the observations that haploinsufficiency results in severe pancreatic and non-pancreatic anomalies in humans, it is surprising that Gata6 heterozygous null mice are viable and fertile, with no reported abnormalities (Koutsourakis et al., 1999, Morrisey et al., 1998). In a recent study, Schrode et al. (2014) showed that the specification of the extraembryonic primitive endoderm entirely fails in Gata6 homozygous embryos at the blastocyst stage, while in a series of older reports Gata6-null mutant embryos were recovered at post-implantation stages with defects in the cardiac mesoderm and visceral endoderm (Koutsourakis et al., 1999, Morrisey et al., 1998). Interestingly, tetraploid complementation experiments between wild-type embryos and Gata6-deficient embryonic stem cells, a technique that overcomes the early lethality resulting from the absence of Gata6 in the extraembryonic lineages, reveal that Gata6-deficient cells can indeed contribute descendants to the DE in chimeric embryos (Zhao et al., 2005). Moreover, conditional loss of Gata6 specifically in Pdx1+ pancreatic progenitors has no impact on pancreatic morphogenesis. Only when a closely related gene, Gata4, is simultaneously deleted is an agenesis phenotype recovered that resembles GATA6 heterozygous human patients (Carrasco et al., 2012, Xuan et al., 2012).
The striking discrepancy between the mouse and the human phenotypes and the complex genetic landscape of GATA6 agenesis patients led us to model GATA6 deficiency in vitro using human pluripotent stem cells (hPSCs). We generated a large panel of heterozygous, homozygous, and compound heterozygous GATA6 mutations by performing genome editing in human embryonic stem cells (hESCs) and human induced pluripotent stem cells (hiPSCs). We additionally derived hiPSCs from two GATA6 heterozygous pancreatic agenesis patients. Subjecting these GATA6 heterozygous hPSCs to directed differentiation into the pancreatic lineage unexpectedly revealed a modest requirement for wild-type GATA6 gene dosage for robust formation of the DE. In contrast to the mouse, complete loss of GATA6 abrogates DE production. Consistent with these results, genome-wide studies show that GATA6 binds and cooperates with EOMES/SMAD2/3 to regulate the expression of cardinal endoderm genes. In addition, GATA6 haploinsufficiency diminishes the ability of those DE cells that form to become PDX1+ pancreatic progenitors and to further mature into C-PEPTIDE-containing β-like cells. These findings show that in humans, the formation of DE and acquisition of pancreatic fate are exquisitely sensitive to GATA6 gene dosage.
Results
GATA6 Expression during Directed Differentiation of hPSCs into the Endocrine Lineage
Consistent with Gata6 expression in the mouse embryo, we previously showed that GATA6 is activated during the early differentiation of hESCs into the DE lineage (Teo et al., 2015, Vallier et al., 2009). We next determined the precise expression kinetics of GATA6 during extended differentiation into the pancreatic lineage using the well-characterized hESC line H9 and a slightly revised version of an 18-day chemically defined protocol previously published by our group (Figure S1A and see Experimental Procedures for complete details) (Cho et al., 2012). GATA6 transcripts are not detected in undifferentiated hESCs, but are abundant by day 3, a time point characterized by the expression of canonical DE markers (SOX17, GATA4, FOXA2, and HNF4A) (Figure S1B). Roughly, ∼75% and ∼98% of cells on day 3 are SOX17+ and GATA6+, respectively (Figure S1D). GATA6 expression persists from day 6 onward, coinciding with the activation of the signature pancreatic lineage marker PDX1 (Figure S1B). By day 12, GATA6 is co-expressed with genes associated with endocrine commitment (NGN3 and NKX6-1), with approximately 76% and 88% of the differentiated cells PDX1+ or GATA6+, respectively (Figures S1B and S1E). The expression of islet hormone genes (INSULIN, GLUCAGON, and SOMATOSTATIN) increases from day 12 (Figure S1B). Importantly, immunofluorescence (IF) staining reveals co-localization of SOX17 and GATA6 in day 3 DE as well as PDX1 and GATA6 in day 12 pancreatic endoderm (PE) (Figure S1C). These data were confirmed in a healthy hiPSC line, FSPS13.B, hereafter designated 13.B (Figures S2A–S2C). Taken together, these findings establish developmental windows where GATA6 insufficiency can result in the pancreatic hypoplasia observed in human GATA6 heterozygous patients.
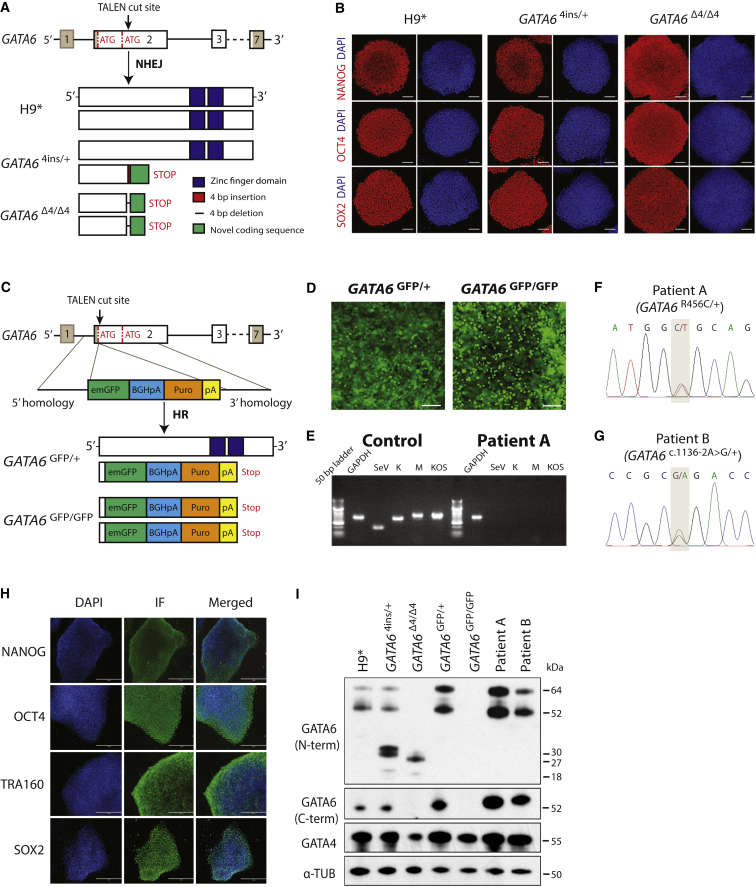
Generation of GATA6 Mutant Alleles Using TALENs and Derivation of hiPSCs from Two Independent GATA6 Heterozygous Patients
To pinpoint the precise role of GATA6 in the human pancreatic lineage, we performed genome-editing in hPSCs as well as isolated patient-derived hiPSCs to generate a panel of GATA6 mutant alleles to model pancreatic agenesis in vitro. The human GATA6 gene is transcribed from two distinct promoter regions, contains two initiation codons in exon 2 (a second at Met147), and consequently encodes two GATA6 protein isoforms, with masses of 60 and 45 kDa, respectively (Figure 1A; Brewer et al., 1999). We initially targeted both H9 and 13.B at a TALEN cut site immediately 3′ of the first ATG in GATA6. Despite the introduction of frameshift mutations that result in premature stop codons, translation still initiated at the second ATG, producing the shorter GATA6 isoform at wild-type levels (data not shown). Thus, in subsequent experiments, we targeted GATA6 at a second TALEN cut site 3′ of the second ATG (Figure 1A) and successfully recovered GATA6 heterozygous (GATA6c.618_619insTGCA/+, hereafter GATA64ins/+) and homozygous (GATA6c.611_614delACCT/c.611_614delACCT, hereafter GATA6Δ4/Δ4) mutations in H9 cells. We generated similar insertion or deletion alleles, both heterozygous (GATA6c.del614_627TGCAGGGGTCGGGC/+, hereafter GATA6Δ14/+) and compound heterozygous (GATA6c.del614_627TGCAGGGGTCGGGC/c.del613_623CTGCAGGGGTC, hereafter GATA6Δ14/Δ11), in 13.B. In parallel, we inserted an emerald GFP (emGFP) reporter in-frame with the first GATA6 ATG via homologous recombination (GATA6GFP/+) in H9 cells (Figures 1C and 1D) and 13.B cells. An H9 GATA6GFP homozygous clone was also recovered (GATA6GFP/GFP) (Figure 1C). Unfortunately, no 13.B GATA6GFP homozygous clone was recovered despite numerous attempts. Control TALEN-targeted lines that harbor no mutations in GATA6 (designated H9∗ or 13.B∗) served as wild-type, isogenic positive controls for differentiation experiments involving genome-edited hPSCs. Last, we obtained fibroblasts from two GATA6 heterozygous patients, whose mutations were previously described (De Franco et al., 2013, Yu et al., 2014). Patient A contains a missense mutation (c.1366C>T) at a highly conserved amino acid within the second zinc-finger DNA-binding domain (Arg456Cys) (Figure 1F), while patient B contains a splice acceptor mutation in exon 3 (GATA6c.1136–2A>G/+) (Figure 1G). Three independent hiPSC clones were isolated for each patient line. All hESC and hiPSC lines were found to have a normal karyotype by multiplex fluorescence in situ hybridization (see Supplemental Experimental Procedures) and assayed by immunohistochemistry to confirm their pluripotency (Figures 1B and 1H and data not shown). hiPSC lines were also monitored for absence of the Sendai virus (Figure 1E).
Figure 1.
Derivation and Characterization of GATA6 Mutant Lines
(A) Schematic of the GATA6 locus. Gray shading highlights the 5′ and 3′ untranslated regions. The TALEN cut site lies downstream of the second start ATG in exon 2. Successful gene editing in H9 cells yielded a GATA6 heterozygous line containing a 4-bp insertion (GATA64ins/+) and a homozygous line with an identical 4-bp deletion on each chromosome (GATA6Δ4/Δ4). Each mutation results in the addition of novel coding sequence (green) and a premature stop. H9∗ cells were subjected to gene editing and selection, but have no mutation in GATA6.
(B) OCT4, SOX2, and NANOG immunofluorescence in H9∗, GATA64ins/+, and GATA6Δ4/Δ4 lines confirms pluripotency in gene-edited clones. Scale bars, 100 μm.
(C) A second TALEN cut site downstream of the first ATG in exon 2 of GATA6 is depicted. Cartoon schematic of the “knockin” vector that introduces an emerald GFP (emGFP) reporter in-frame and a puromycin-resistance cassette. Successful homologous recombination resulted in both heterozygous (GATA6GFP/+) and homozygous (GATA6GFP/GFP) mutant cells
(D) Immunofluorescence showing emGFP-expressing heterozygous GATA6GFP/+ and homozygous GATA6GFP/GFP mutant cells on day 3 of differentiation. Scale bars, 100 μm.
(E) PCR showing loss of transgenes in a patient A mutant hiPSC line, clone 1, compared with positive controls. Data are representative of three independent clones derived from either patient A or patient B.
(F and G) Genotype confirmation by Sanger sequencing of two GATA6 patient-derived hiPSC lines: (F) patient A, GATA6R465C/+, and (G) patient B, GATA6c.1136–2A>G/+.
(H) Immunofluorescence confirming the successful reprogramming and pluripotency of one patient A-derived (GATA6R465C/+) mutant line. Scale bars, 200 μm. Images are representative of three independent clones derived from either patient A or patient B (GATA6c.1136–2A>G/+).
(I) Western blot analysis of GATA6 and GATA4 protein levels in undifferentiated H9∗, GATA64ins/+, GATA6Δ4/Δ4, GATA6GFP/+, and GATA6GFP/GFP mutant cells, as well as the two patient-derived mutant lines: patient A, GATA6R465C/+, and patient B, GATA6c.1136–2A>G/+. α-tubulin was used as a loading control. Long and short isoforms of wild-type GATA6 are 60 and 45 kDa, respectively; the partial protein products for GATA64ins/+ are 30 and 18 kDa for the long and short isoforms, respectively; the partial protein products for GATA6Δ4/Δ4 are 27 and 15 kDa for the long and short isoforms, respectively. No GATA6 protein was present for the GATA6GFP/GFP mutant.
Differentiation of GATA6 Mutant hPSC Lines into the Definitive Endoderm Lineage
Mutant lines were next differentiated to the DE stage and GATA6 protein levels determined by western blot using anti-N- and anti-C-terminal GATA6 antibodies (Figure 1I). In H9∗ DE cells, both GATA6 isoforms are detected by the N-terminal antibody, whereas the C-terminal antibody predominantly recognizes the short isoform (Figure 1I) (Brewer et al., 1999). GATA64ins/+ and GATA6Δ4/Δ4 contain frameshift mutations that result in truncated partial protein products predicted to contain 205 and 203 N-terminal amino acids, respectively, of the longer GATA6 isoform as well as additional novel C-terminal sequences (Figures 1A and 1I) that terminate before the two C-terminal zinc-finger DNA-binding domains. The insertion of the GFP reporter and puromycin-resistance cassettes in GATA6 exon 2 generates a loss-of-function allele, since neither wild-type GATA6 isoform nor novel partial protein products were observed in GATA6GFP/GFP knockin H9 cells (Figures 1C and 1I).
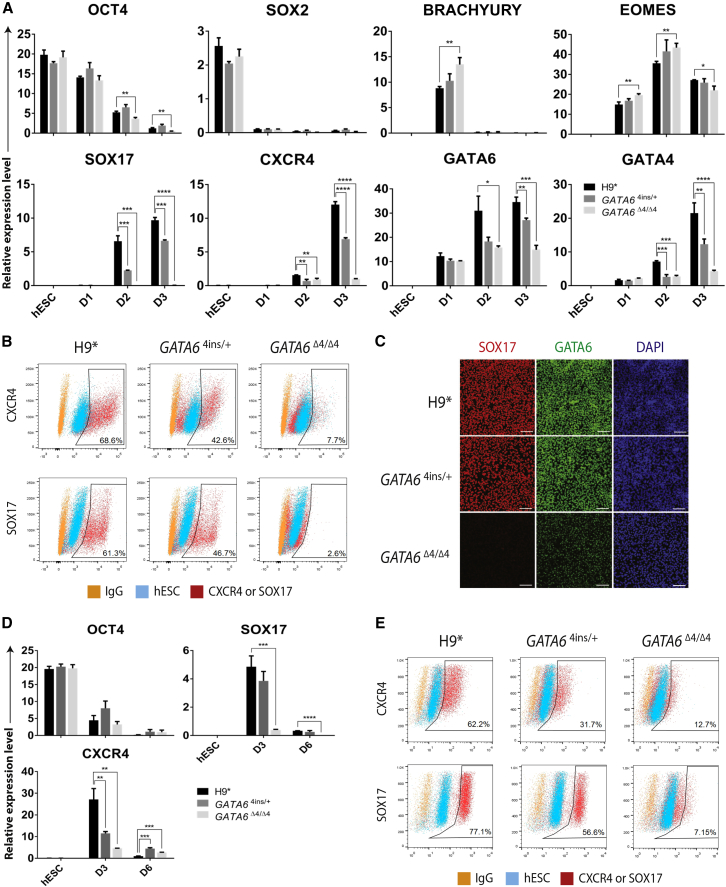
Using the H9-derived GATA64ins/+ and GATA6Δ4/Δ4 lines, we next asked whether reduced levels of GATA6 have an impact on early mesendoderm (corresponding to days 1 and 2) to DE (day 3) differentiation. qRT-PCR analyses show that in H9∗, GATA64ins/+, and GATA6Δ4/Δ4 cells, the levels of the pluripotency markers OCT4 and SOX2 were comparable in undifferentiated cells and expectedly declined during differentiation (Figure 2A). The expression of primitive streak (BRACHYURY) and mesendoderm (EOMESODERMIN [EOMES]) markers was also relatively unchanged across the control H9∗ and GATA6 mutant lines, suggesting that early mesendoderm formation was not affected by either single or biallelic loss of GATA6 (Figure 2A). Key DE markers SOX17 and CXCR4 were, however, modestly downregulated beginning on day 2 in GATA64ins/+ cells (Figure 2A), and on day 3, GATA64ins/+ differentiations yielded roughly 25% fewer SOX17+ cells by fluorescence-activated cell sorting (FACS) and IF compared with wild-type H9∗ (Figures 2B and 2C). This heterozygous effect on SOX17 transcription was also observed to varying degrees in H9-GATA6GFP/+ and 13.B-derived GATA6Δ14/+ as well as in patients A (GATA6R456C/+) and B (GATA6c.1136–2A>G/+) (Figures S2D–S2F). Interestingly, this heterozygous effect was not observed in 13.B-derived GATA6GFP/+ (Figure S2E). Further depleting GATA6 with homozygous (GATA6Δ4/Δ4 or GATA6GFP/GFP) or compound heterozygous (13.B-GATA6Δ14/Δ11) allelic combinations dramatically affects DE formation, yielding ∼3% SOX17+ cells on day 3 (Figure 2B and data not shown). We further validated these results using the commercially available STEMdiff pancreatic progenitor kit from STEMCELL Technologies. Using this differentiation platform, H9∗, GATA64ins/+, and GATA6Δ4/Δ4 formed DE at efficiencies indistinguishable from the results obtained with the protocol outlined in Figure 1A (cf. Figures 2A and 2B with 2D and 2E). Taken together, these findings show that diminished levels of GATA6 compromise early DE formation, and complete loss of GATA6 significantly perturbs the gene regulatory network (GRN) governing human DE specification.
Figure 2.
GATA64ins/+ and GATA6Δ4/Δ4 Mutant hESC Lines Display Impaired DE Formation
(A) Expression of pluripotency (OCT4, SOX2), primitive streak (BRACHYURY), mesendoderm (EOMES), and definitive endoderm (CXCR4, SOX17, GATA4) markers, as well as GATA6 itself, in H9∗ and H9-derived GATA64ins/+ and GATA6Δ4/Δ4 mutant cells over 3 days of differentiation (Figure S1A).
(B) Differentiation efficiency measured by FACS analysis of CXCR4 and SOX17 at day 3 DE in H9∗ and H9-derived GATA64ins/+ and GATA6Δ4/Δ4 mutant cells.
(C) Immunofluorescence analyses for the key DE markers GATA6 with SOX17 in H9∗ and H9-derived GATA64ins/+ and GATA6Δ4/Δ4 mutant cells. DAPI, 4′,6-diamidino-2-phenylindole. Scale bars, 100 μm.
(D) Expression of pluripotency (OCT4) and definitive endoderm (SOX17, CXCR4) markers in H9∗ and H9-derived GATA64ins/+ and GATA6Δ4/Δ4 mutant cells on days 3 and 6 of differentiation with the STEMdiff pancreatic progenitor kit.
(E) Differentiation efficiency measured by FACS analysis of CXCR4 and SOX17 at day 3 DE in H9∗ and H9-derived GATA64ins/+ and GATA6Δ4/Δ4 mutant cells differentiated using the STEMdiff pancreatic progenitor kit.
(A and D) Error bars represent the SE of three independent experiments. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
(B and E) Undifferentiated hESCs stained with the respective primary and secondary antibodies and secondary antibody only (IgG) were both used as controls. Gates were set according to an hESC control.
Establishing the GATA6 Gene Regulatory Network
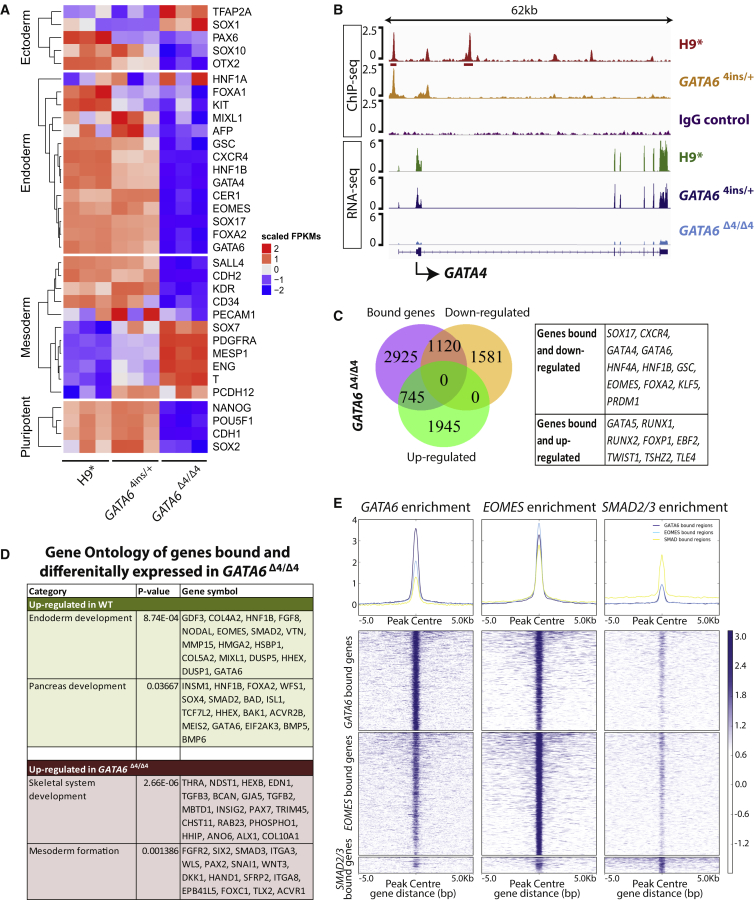
To establish comprehensively how GATA6 mutations alter the DE transcriptional network, we performed RNA sequencing (RNA-seq) for H9∗, GATA64ins/+ and GATA6Δ4/Δ4, and patient A cells on day 3 of differentiation. Comparative analyses revealed 7,472 genes that are differentially expressed (adjusted p ≤ 0.01; fold change ≥2) between H9∗ and GATA6Δ4/Δ4, 2,898 genes between H9∗ and GATA64ins/+, and 6,977 genes between H9∗ and hiPSC clones 1 to 3 from patient A (Table S1). We observed that, consistent with our qRT-PCR data in Figure 2, GATA6Δ4/Δ4 mutant cells show significantly decreased expression of cardinal endoderm markers (e.g., SOX17, CXCR4, HNF1B, and FOXA2) (Figure 3A). Similar results were observed when wild-type H9∗ was compared with GATA64ins/+ and hiPSC clones 1 to 3 from patient A (Figures 3A and S3A).
Figure 3.
GATA6 Is a Key Regulator of the DE Transcriptional Network
(A) Heatmap illustrating differential gene expression of key germ layer markers via RNA-seq between H9∗ cells and H9-derived GATA64ins/+ and GATA6Δ4/Δ4 mutant cells at the DE stage. n = 3 biological replicates for each cell line.
(B) ChIP-seq binding profiles of H9∗ and GATA64ins/+ showing GATA6 enrichment near GATA4, and GATA4 expression by RNA-seq in H9∗ and H9-derived GATA64ins/+ and GATA6Δ4/Δ4 mutant cells at the DE stage. The input control profile (IgG control) is included for comparison. The ChIP-seq binding profile is derived from merging two biological replicates.
(C) Venn diagram indicating the overlap of GATA6-bound genes from ChIP-seq at the DE stage with downregulated or upregulated genes in H9-derived GATA6Δ4/Δ4 mutant cells compared with H9∗ cells by RNA-seq. Key bound genes up- or downregulated are indicated in the table.
(D) Enriched gene ontology showing developmental pathways from direct target genes differentially expressed between H9∗ and H9-derived GATA6Δ4/Δ4 mutant cells derived from BETA analysis.
(E) Density heatmaps of GATA6-binding peak intensity at DE indicating direct overlap with known endodermal regulators, including SMAD2/3 and EOMES, within a 5-kb window centered at the transcription start site.
We also performed GATA6 chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-seq) on H9∗ and GATA64ins/+ cells at the DE stage. This analysis yielded 12,098 peaks (irreproducible discovery rate ≤0.05; median peak length = 417 bp) that are associated with 10,669 genes, 4,790 of which are protein coding (Table S1). Interestingly, we observe that GATA6 binding is enriched at the GATA4 locus in H9 cells, suggesting that GATA6 directly regulates GATA4 during DE specification (Figure 3B). Both qRT-PCR and RNA-seq show dose-dependent effects of GATA6 on GATA4 expression levels in GATA64ins/+ and GATA6Δ4/Δ4 mutant cells (Figures 2A, 3A, and 3B).
We next compared our RNA-seq and ChIP-seq datasets to identify those genes bound and regulated by GATA6. This analysis revealed 1,120 protein-coding genes that are bound by GATA6 in wild-type H9∗ but are downregulated in GATA6Δ4/Δ4 mutant cells, including pancreatic progenitor genes such as HNF1B and HNF4A (Figure 3C). In contrast, 745 genes are bound by GATA6 in H9∗ and upregulated in GATA6Δ4/Δ4. Similar overlaps were performed for GATA64ins/+ and patient A day 3 RNA-seq samples, yielding 337 and 607 GATA6-bound and downregulated genes, and 254 and 616 GATA6-bound and upregulated genes, respectively (Figures S3B and S3C). At the intersection of these comparisons are 143 commonly downregulated and 104 upregulated genes among GATA6Δ4/Δ4, GATA64ins/+, and patient A samples (Figures S3D and S3E). Key endoderm markers CXCR4, SOX17, GATA4, HNF1B, and HNF4A were among the 143 genes commonly downregulated (Figure S3D).
To infer genes that are directly targeted and regulated by GATA6, we performed binding and expression target analysis (BETA) to integrate our H9 ChIP-seq dataset with differential gene expression data from GATA6Δ4/Δ4, GATA64ins/+ and patient A (Wang et al., 2013). Targets predicted by BETA were then subjected to gene ontology analyses using the DAVID tool (Huang da et al., 2009a, Huang da et al., 2009b). We found that endoderm development is consistently upregulated in H9∗ compared with GATA6Δ4/Δ4 (Figure 3D), GATA64ins/+ (Figure S3F), and patient A (Figure S3G) mutant cells. In addition, mesoderm development is consistently upregulated in GATA6Δ4/Δ4 (Figure 3D), GATA64ins/+ (Figure S3F), and patient A (Figure S3G) mutant cells compared with H9∗. Motif analyses generated by BETA confirm that the GATA recognition motif is highly enriched in both “up” and “down” target genes (Figure S3H). We were unable to conclude from the BETA whether GATA6 has activating or repressive functions, or both, as the data were not significant. Thus, we propose that the most parsimonious explanation for the upregulation of mesodermal markers is aberrant differentiation. In the absence of GATA6, differentiation into the DE lineage is blocked, but differentiating cells remain bathed in high levels of two potent mesoderm inducers (Activin and BMP4) (Cho et al., 2012). Taken together, these results show that GATA6 is indispensable in driving the development of the human DE.
We previously established that the T-box transcription factor EOMES interacts with the Activin/Nodal effector proteins SMAD2/3 to deploy the GRN that directs DE formation. We thus sought to establish how GATA6 integrates into the SMAD2/3/EOMES signaling network by comparing our GATA6 day 3 ChIP-seq data with previously published SMAD2/3 and EOMES ChIP-seq (Brown et al., 2011, Teo et al., 2011). Remapping of the data resulted in 16,303, 20,089, and 2,613 peaks for GATA6, EOMES, and SMAD2/3, respectively. Of the 16,303 GATA6 ChIP-seq peaks, 950/2,613 (36.4%) overlap with SMAD2/3, and 8,126/20,089 (40.5%) overlap with EOMES in DE cells, with 858 common to all three datasets (Figure 3E, Table S1). In the EOMES/GATA6/SMAD2/3 intersection, we find almost all of the telltale endodermal regulator genes, including SOX17, EOMES, LHX1, MIXL1, FOXA2, HNF1B, and CXCR4. These data therefore place GATA6 centrally within the core nuclear transcriptional machinery that governs the acquisition of DE fate.
GATA6 Deficiency Impairs Pancreatic Lineage Commitment
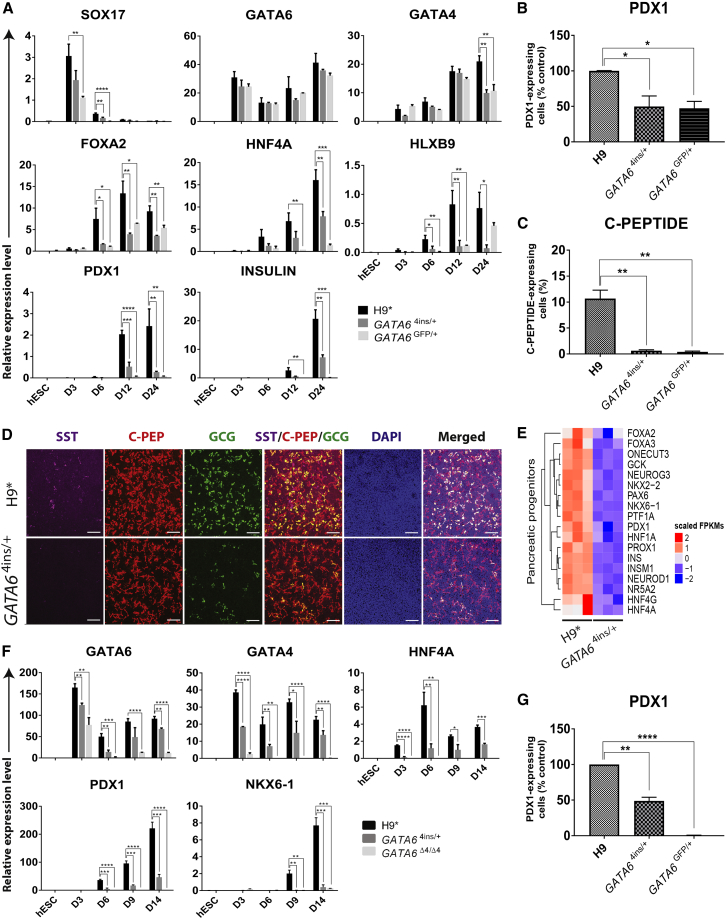
We further analyzed the effects of GATA6 heterozygous mutations on pancreatic lineage commitment at the PE (day 12) and endocrine progenitor (EP) (day 24) stages (Figure S1A). Key pancreatic markers such as HNF4A, HLXB9, PDX1, and INSULIN are significantly downregulated in GATA64ins/+ and GATA6GFP/+ mutant cells at both stages, with one exception: HLXB9 levels in GATA6GFP/+ are no different from those in H9∗ on day 24 (Figure 4A). HNF4A, PDX1, and INSULIN were also significantly decreased in 13.B-GATA6Δ14/+ and 13.B-GATA6GFP/+ and in patient A and B mutant cells on days 12 and 24 (Figures S4A and S4B). FACS analysis for PDX1 on day 12 reveals an approximately 50% decrease in the number of PDX1-positive GATA64ins/+ and GATA6GFP/+ cells and 13.B-GATA6GFP/+ cells (Figures 4B and S4C). 13.B-GATA6Δ14/+, patient A, and patient B cell lines exhibit an approximately 80%–90% decrease in PDX1 (Figures S4C and S4D). At the EP stage, all GATA6 heterozygous mutant cell lines share a common phenotype, with a strong decrease in the number of C-PEPTIDE+ cells (Figure 4C, S4C, and S4D). Immunostaining on H9∗ and GATA64ins/+ cells confirms the diminished number of SOMATOSTATIN-, C-PEPTIDE-, and GLUCAGON-positive cells in GATA64ins/+ cells (Figure 4D).
Figure 4.
Decreased Levels of GATA6 at the DE Stage Influence Downstream Pancreatic Differentiation
(A) Expression of DE (SOX17, GATA6, GATA4, and FOXA2), pancreatic (HNF4A, HLXB9, and PDX1), and endocrine (INSULIN) marker genes in H9∗ and H9-derived GATA64ins/+ and GATA6Δ4/Δ4 mutant cells at the four key stages of the 24-day pancreatic differentiation protocol (Figure S1A).
(B) Percentage of PDX1-positive cells in H9-derived GATA64ins/+ and GATA6GFP/+ lines on day 12 shown relative to H9∗ (100%) as measured by FACS.
(C) Percentage of C-PEPTIDE-positive cells in H9∗ and H9-derived GATA64ins/+ and GATA6GFP/+ mutant lines at the EP stage (day 24) as measured by FACS.
(D) Immunofluorescence analyses for the key PE markers SOMATOSTATIN (SST), C-PEPTIDE (C-PEP), and GLUCAGON (GCG) in H9∗ and H9-derived GATA64ins/+ cells at the EP stage (day 24). DAPI, 4′,6-diamidino-2-phenylindole dihydrochloride. Scale bars, 100 μm.
(E) Heatmap illustrating differential gene expression of key pancreatic progenitor markers via RNA-seq between H9∗cells and H9-derived GATA64ins/+ mutant cells at the PE stage. n = 3 biological replicates for each cell line.
(F) Expression of DE (GATA6 and GATA4) and pancreatic (HNF4A, PDX1, and NKX6-1) marker genes in H9∗ and H9-derived GATA64ins/+ and GATA6Δ4/Δ4 mutant cells at key stages of the differentiation protocol using the STEMdiff pancreatic progenitor kit.
(G) Percentage of PDX1-positive cells in GATA64ins/+ and GATA6GFP/+ lines on day 12 shown relative to H9∗ (100%) as measured by FACS in cells differentiated using the STEMdiff pancreatic progenitor kit.
(A–C, F, and G) Error bars represent the SE of three independent experiments. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
We also performed RNA-seq at the PE stage (day 12) for H9∗, GATA64ins/+, and patient A cells. H9∗ RNA-seq largely reproduced a previous dataset generated using both H9 and the same differentiation protocol (Spearman's rank correlation coefficient, ρ = 0.77 for in vitro multipotent pancreatic progenitor cells [MPCs] and ρ = 0.59 for in vivo MPCs isolated from Carnegie stage 16–18 human embryos, p < 2.2 × 10−16) (Cebola et al., 2015). Between H9∗ and GATA64ins/+ (Table S1), 1,423 genes were differentially expressed, and between H9∗ and patient A, 6,093 were differentially expressed (Table S1). We observed that, consistent with qRT-PCR and FACS results, GATA64ins/+ and patient A gene expression quantified by RNA-seq in mutant cells displays a decreased pancreatic signature (Figures 4E and S4E).
Finally, the above results were independently confirmed with H9∗, GATA64ins/+, and GATA6Δ4/Δ4 cells using the STEMdiff pancreatic progenitor kit: GATA64ins/+ cells show decreased PDX1 and NKX6-1 expression beginning on day 9, yielding ∼50% fewer PDX1+ pancreatic progenitors on day 12 (Figures 4F and 4G). Collectively, these in vitro findings strongly argue that decreased levels of GATA6 first influence the formation of DE, and predict that with fewer DE cells at the time of allocation to the pancreatic lineage in vivo, hypoplasia emerges.
Discussion
Deriving iPSC lines and the ability to rapidly engineer mutations in hPSCs have made human disease modeling in vitro commonplace. In the case of the well-characterized set of genes known to control mammalian pancreatic development, it is the expectation based on strong evolutionary conservation that phenotypes observed in knockout mouse models will be reproduced in human and in vitro. Indeed, the matching of human and mouse pancreatic and extrapancreatic phenotypes is seen for many recessive loss-of-function mutations in key pancreatic developmental transcription factors, e.g., PDX1, PTF1A, RFX6, NEUROD1, NGN3, and NKX2-2 (Ahlgren et al., 1996, Flanagan et al., 2014, Jonsson et al., 1994, Offield et al., 1996, Rubio-Cabezas et al., 2010, Rubio-Cabezas et al., 2011, Schwitzgebel et al., 2003, Sellick et al., 2004, Smith et al., 2010, Stoffers et al., 1997). Such consistency in phenotype is, however, not observed with Gata6. In mice, only the simultaneous inactivation of Gata6 and Gata4 results in pancreatic agenesis (Carrasco et al., 2012, Xuan et al., 2012), whereas in humans de novo heterozygous mutations in GATA6 underlie a similar pathology (De Franco et al., 2013, Lango Allen et al., 2011). However, GATA6 heterozygous phenotypes range from total pancreatic agenesis to isolated diabetes in young adulthood. This phenotypic diversity partly explains the difficulties in precisely modeling GATA6 haploinsufficiency in vitro across laboratories and across differentiation platforms, as evidenced by comparing our present findings with two recent reports from Shi et al. (2017) and Tiyaboonchai et al. (2017).
Here, we find a modest reduction (∼25%) in the production of DE after 3 days of directed differentiation irrespective of whether the GATA6 heterozygous line was patient derived or generated by gene editing in hPSCs. These findings are consistent with GATA6 expression in the DE, but contrast with the results of Shi et al. (2017) and Tiyaboonchai et al. (2017). These authors did not observe decreased DE formation using assorted GATA6 heterozygous hPSC lines. One potential explanation for these discrepant results is that GATA6 partial protein products, generated, for example, from the GATA64ins/+ allele in H9 cells, act in our hands in a dominant negative manner, further suppressing GATA6 levels and compromising normal DE formation (Figure 1I). The partial protein products encoded by the GATA64ins/+ locus are predicted to retain a long stretch of the N-terminal GATA6 transactivation domain but lack the zinc-finger DNA-binding domain and nuclear localization signal. As they are able neither to bind DNA nor to heterodimerize with GATA4 (Charron et al., 1999, Maeda et al., 2005), the biochemical mechanism by which such partial protein products interfere with GATA6 transcription or function is entirely unclear. Moreover, Tiyaboonchai et al. (2017) and Shi et al. (2017) also observe partial protein products in their GATA6 heterozygous hPSC lines, but do not observe effects during DE differentiation. The most significant evidence against dominant interference and in favor of a simple dosage effect comes from the fact that patient A and B iPSCs, whose mutations do not result in partial protein products (Figure 1I), also show decreased DE formation on day 3. Alternatively, because each group employed different hESC and iPSC lines, the specter of well-known line-to-line variations in differentiation efficiency could explain the results from the different laboratories (Cahan and Daley, 2013, Ortmann and Vallier, 2017).
Despite these differences among the GATA6 heterozygous phenotypes at the DE stage, complete loss of GATA6 was found by Tiyaboonchai et al., 2017, Shi et al., 2017, and us (Figure 2), as well as more recently by Liao et al. (2018) with short hairpin RNA targeting GATA6 in H1 cells, to unequivocally impair DE formation, a result highlighting not only the requirement for wild-type GATA6 gene dosage for robust DE specification in humans but also the dramatic species-specific differences between mice and humans. Importantly, our genome-wide studies place GATA6 among the core transcriptional machinery that controls DE formation. We previously reported that the pluripotency factors OCT4, SOX2, and NANOG bind cooperatively and control the expression of the T-box transcription factor gene EOMES (Teo et al., 2011). Upon activation, EOMES, jointly with SMAD2/3, the intracellular effectors of ACTIVIN/NODAL signaling, deploys a large part of the transcriptional network governing DE formation. Here, we find 858 genes that are bound within 5 kb of the transcription start site by EOMES, SMAD2/3, and GATA6. Importantly, such cooperation has not been evidenced in mouse development, suggesting major divergences between species in the molecular mechanisms controlling germ-layer specification. Considering the importance of GATA6 in specification of extraembryonic endoderm, this divergence in signaling pathways could result in the rewiring of downstream transcriptional networks with major consequences on the subsequent specification of DE.
With extended differentiation to the PE stage (day 12), we observe significantly decreased numbers of PDX1+ cells—between 50% and 90% fewer compared with wild-type depending on the GATA6 heterozygous line. This result is consistent with GATA6 expression in human pancreatic progenitors (Figure S1C) and with GATA6 directly regulating PDX1 transcription (Carrasco et al., 2012, Xuan et al., 2012) and also suggests that GATA6 plays a dual role in both early DE formation and allocation to the pancreatic lineage. The diminished numbers of GATA6 heterozygous PDX1+ progenitors that emerge at the PE stage ultimately differentiate into ≤10% C-PEPTIDE+ cells by the EP stage (day 24), across all cell types and across all alleles.
It is tempting to consider that the variation in clinical phenotype and the early phenotype in DE formation in vitro might be predominantly attributable to individual genetic backgrounds (Chen et al., 2016, Lek et al., 2016). GATA4 is an obvious choice for a genetic modifier, given its expression in the DE, genetic interaction with Gata6 in mice, and the identification of rare GATA4 heterozygous patients with pancreatic agenesis, as well as our finding that GATA4 is bound and regulated by GATA6 in vitro (Figure 3) (D'Amato et al., 2010, Freyer et al., 2015, Morrisey et al., 1996, Shaw-Smith et al., 2014). Indeed, Shi et al. (2017) elegantly show dosage-dependent effects of GATA4 alleles on phenotypes associated with GATA6 heterozygosity during in vitro differentiation. Despite reports of considerable phenotypic variation between family members with the same GATA6 mutation (Bonnefond et al., 2012, De Franco et al., 2013, Yau et al., 2017), in some cases a parent is a mosaic for the phenotype, so the variation between parental and offspring phenotypes can be explained by different mutation load in target tissues (Yau et al., 2017). If the variation in the human phenotype altered significantly with the genetic background, then most cases with severe pancreatic agenesis would likely be born to parents with the same mutation, but a 50% different (protective) genetic background would have a milder phenotype. However, this is not the case, as the vast majority of severe pancreatic agenesis is from de novo mutations (De Franco et al., 2013, Lango Allen et al., 2011). This means it is possible, but not likely, that genetic background explains why Shi et al. (2017) engineered, using CRISPR/Cas9, the common GATA6 agenesis mutation c.1366C>T (p.Arg456Cys) in HUES8 cells—the same allele present in our patient A-derived iPSC line (GATA6R456C/+)—and observed no heterozygous phenotype at the DE or pancreatic progenitor (PDX1+) stages, whereas we do, at both the DE stage and beyond.
In addition to line-to-line differentiation efficiencies in vitro (Cahan and Daley, 2013, Ortmann and Vallier, 2017), fundamental differences in the differentiation protocols themselves may underlie (or contribute to) the results we report here and those published by Shi et al. (2017) and Tiyaboonchai et al. (2017). For example, the growth factor and small-molecule components as well as medium formulations differ substantially for the first 3 days of DE differentiation among the three studies. Furthermore, our differentiation protocol relies on culture media devoid of serum or complex extracellular matrices such as Matrigel. Thus, the minimalist approach of our system could exacerbate the GATA6 phenotype, revealing a function for this gene that is otherwise masked. This possibility highlights the importance of culture conditions to study gene function in hPSCs and during their differentiation. Tiyaboonchai et al. (2017) additionally show that a GATA6 heterozygous iPSC line derived from an agenesis patient unexpectedly produced β-like cells in vitro. Simply reducing the concentration of retinoic acid 80-fold led to statistically significantly fewer PDX1+ cells compared with a wild-type iPSC line that showed negligible sensitivity to the same culture regime. Indeed, current hPSC pancreatic differentiation protocols have been highly tailored and refined, providing redundant and reinforcing signals that perhaps reconfigure underlying GRNs and bypass certain in vivo genetic requirements. Moreover, it must be acknowledged that adherent differentiation fails to achieve the 3D complexity of human endoderm formation in vivo. Thus, studies of early pancreatic lineage commitment would greatly benefit from universal protocols standardized intra- and inter-laboratory in an effort to minimize line-to-line and protocol-to-protocol differences.
Experimental Procedures
Human Pluripotent Stem Cell Culture and Pancreatic Differentiation
hESCs (H9 [WA09 from www.wicell.org]), hiPSCs (FSPS13.B derived in-house from human fibroblasts [http://www.hipsci.org/lines/#/lines/HPSI0813i-fpdm_2]), and GATA6 patient-derived iPSCs, from patients A and B, were routinely cultured under feeder-free conditions on vitronectin-coated (STEMCELL Technologies #07180) tissue culture plates (Corning) with Essential 8 Medium (Life Technologies #A1517001). All tissue culture was carried out in 5% CO2 at 37°C. Pancreatic differentiation was carried out as previously described (Cho et al., 2012), with modifications described in Supplemental Experimental Procedures.
GATA6 Patient Samples
The generation of GATA6 patient-derived hiPSCs was approved by the Great Ormond Street Hospital and Institute of Child Health Research Ethics Committee (ethics reference: 08/H0713/82), and informed consent was obtained from all patients. Skin punch biopsy samples were collected from patients and all hiPSC lines used were derived and validated by the Cambridge Biomedical Research Center hiPSC Core Facility. Reprogramming of the GATA6 patient fibroblasts to derive GATA6 patient iPSCs was done by the hiPSC core facility at the Anne McLaren Laboratory for Regenerative Medicine using Sendai virus reprogramming.
GATA6-Mutant and GATA6-emGFP Reporter hPSC Derivation
The construction of TALEN vectors, their introduction into H9 or 13.B cells via electroporation, and the screening of drug-resistant clones are described in detail in the Supplemental Experimental Procedures. Two TALEN pairs were generated, each targeting a different site within GATA6 exon 2. The first TALEN pair targets a site that is 6 bp downstream of the first GATA6 start codon. The second targets a site that is 149 bp downstream of the second GATA6 start codon. Primers used for TALEN construction and screening of genomic DNA are listed in Table S2.
RNA- and ChIP-Sequencing Analysis of Gene Expression
Library preparation and deep sequencing were performed at the Wellcome Trust Sanger Institute (Hinxton, UK). RNA-seq and ChIP-seq were run on Illumina Hiseq v.3 and v.4, respectively, with read length 75 bp and paired ends, and a library fragment size of 100–1,000 bp using a multiplex strategy. RNA-seq and ChIP-seq samples were run in biological triplicates and duplicates, respectively. Additional details of how RNA-seq and ChIP-seq reads were aligned and analyzed can be found in the Supplemental Experimental Procedures.
Author Contributions
L.V., N.R.D., C.Y.C., and M.C. designed the methods and experiments. C.Y.C. performed the experiments. C.Y.C., P.M., and S.L.I.J.D performed bioinformatics analyses. I.M. and J.G.-B. performed additional differentiation experiments for the revisions. A.T.H. and M.H.S identified the patients and designed the patient sample collection. M.H.S. and R.E.K. collected the patient samples. C.Y.C. and N.R.D. wrote the first draft. All authors approved the final draft and made modifications to the text.
Acknowledgments
We thank the NIHR Cambridge BRC hiPSC core facility for the derivation of GATA6 hiPSC lines, Dr. Norihiro Tsuneyoshi for advice and guidance on gene editing using TALENs, and Dr. Bruno Reversade for insightful discussions. This work was funded by an EDB Singapore Childhood Undiagnosed Diseases Program grant and an A∗STAR Strategic Positioning Fund (SPF) Genetic Orphan Diseases Adopted: Fostering Innovation Therapy (GODAFIT) grant and by an ERC starting grant Relieve IMDs and core grant funding from the Wellcome Trust and Medical Research Council (PSAG028) (L.V. and P.M.). A.T.H. is a Wellcome Trust Senior Investigator.
Published: January 8, 2019
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, four figures, and four tables and can be found with this article online at https://doi.org/10.1016/j.stemcr.2018.12.003.
Contributor Information
N. Ray Dunn, Email: ray.dunn@imb.a-star.edu.sg.
Ludovic Vallier, Email: lv225@cam.ac.uk.
Accession Numbers
Primary RNA-seq and ChIP-seq datasets generated here are available at ArrayExpress under accession numbers ArrayExpress: E-MTAB-5958 and E-MTAB-5959, respectively.
Supplemental Information
References
- Ahlgren U., Jonsson J., Edlund H. The morphogenesis of the pancreatic mesenchyme is uncoupled from that of the pancreatic epithelium in IPF1/PDX1-deficient mice. Development. 1996;122:1409–1416. doi: 10.1242/dev.122.5.1409. [DOI] [PubMed] [Google Scholar]
- Bonnefond A., Sand O., Guerin B., Durand E., De Graeve F., Huyvaert M., Rachdi L., Kerr-Conte J., Pattou F., Vaxillaire M. GATA6 inactivating mutations are associated with heart defects and, inconsistently, with pancreatic agenesis and diabetes. Diabetologia. 2012;55:2845–2847. doi: 10.1007/s00125-012-2645-7. [DOI] [PubMed] [Google Scholar]
- Brewer A., Gove C., Davies A., McNulty C., Barrow D., Koutsourakis M., Farzaneh F., Pizzey J., Bomford A., Patient R. The human and mouse GATA-6 genes utilize two promoters and two initiation codons. J. Biol. Chem. 1999;274:38004–38016. doi: 10.1074/jbc.274.53.38004. [DOI] [PubMed] [Google Scholar]
- Brown S., Teo A., Pauklin S., Hannan N., Cho C.H., Lim B., Vardy L., Dunn N.R., Trotter M., Pedersen R. Activin/Nodal signaling controls divergent transcriptional networks in human embryonic stem cells and in endoderm progenitors. Stem Cells. 2011;29:1176–1185. doi: 10.1002/stem.666. [DOI] [PubMed] [Google Scholar]
- Cahan P., Daley G.Q. Origins and implications of pluripotent stem cell variability and heterogeneity. Nat. Rev. Mol. Cell Biol. 2013;14:357–368. doi: 10.1038/nrm3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrasco M., Delgado I., Soria B., Martin F., Rojas A. GATA4 and GATA6 control mouse pancreas organogenesis. J. Clin. Invest. 2012;122:3504–3515. doi: 10.1172/JCI63240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebola I., Rodriguez-Segui S.A., Cho C.H., Bessa J., Rovira M., Luengo M., Chhatriwala M., Berry A., Ponsa-Cobas J., Maestro M.A. TEAD and YAP regulate the enhancer network of human embryonic pancreatic progenitors. Nat. Cell Biol. 2015;17:615–626. doi: 10.1038/ncb3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao C.S., McKnight K.D., Cox K.L., Chang A.L., Kim S.K., Feldman B.J. Novel GATA6 mutations in patients with pancreatic agenesis and congenital heart malformations. PLoS One. 2015;10:e0118449. doi: 10.1371/journal.pone.0118449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charron F., Paradis P., Bronchain O., Nemer G., Nemer M. Cooperative interaction between GATA-4 and GATA-6 regulates myocardial gene expression. Mol. Cell. Biol. 1999;19:4355–4365. doi: 10.1128/mcb.19.6.4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R., Shi L., Hakenberg J., Naughton B., Sklar P., Zhang J., Zhou H., Tian L., Prakash O., Lemire M. Analysis of 589,306 genomes identifies individuals resilient to severe Mendelian childhood diseases. Nat. Biotechnol. 2016;34:531–538. doi: 10.1038/nbt.3514. [DOI] [PubMed] [Google Scholar]
- Cho C.H., Hannan N.R., Docherty F.M., Docherty H.M., Joao Lima M., Trotter M.W., Docherty K., Vallier L. Inhibition of activin/nodal signalling is necessary for pancreatic differentiation of human pluripotent stem cells. Diabetologia. 2012;55:3284–3295. doi: 10.1007/s00125-012-2687-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Amato E., Giacopelli F., Giannattasio A., D'Annunzio G., Bocciardi R., Musso M., Lorini R., Ravazzolo R. Genetic investigation in an Italian child with an unusual association of atrial septal defect, attributable to a new familial GATA4 gene mutation, and neonatal diabetes due to pancreatic agenesis. Diabet. Med. 2010;27:1195–1200. doi: 10.1111/j.1464-5491.2010.03046.x. [DOI] [PubMed] [Google Scholar]
- Decker K., Goldman D.C., Grasch C.L., Sussel L. Gata6 is an important regulator of mouse pancreas development. Dev. Biol. 2006;298:415–429. doi: 10.1016/j.ydbio.2006.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan S.E., De Franco E., Lango Allen H., Zerah M., Abdul-Rasoul M.M., Edge J.A., Stewart H., Alamiri E., Hussain K., Wallis S. Analysis of transcription factors key for mouse pancreatic development establishes NKX2-2 and MNX1 mutations as causes of neonatal diabetes in man. Cell Metab. 2014;19:146–154. doi: 10.1016/j.cmet.2013.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Franco E., Shaw-Smith C., Flanagan S.E., Shepherd M.H., International N.D.M.C., Hattersley A.T., Ellard S. GATA6 mutations cause a broad phenotypic spectrum of diabetes from pancreatic agenesis to adult-onset diabetes without exocrine insufficiency. Diabetes. 2013;62:993–997. doi: 10.2337/db12-0885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freyer L., Schroter C., Saiz N., Schrode N., Nowotschin S., Martinez-Arias A., Hadjantonakis A.K. A loss-of-function and H2B-Venus transcriptional reporter allele for Gata6 in mice. BMC Dev. Biol. 2015;15:38. doi: 10.1186/s12861-015-0086-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W., Sherman B.T., Lempicki R.A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Jennings R.E., Berry A.A., Kirkwood-Wilson R., Roberts N.A., Hearn T., Salisbury R.J., Blaylock J., Piper Hanley K., Hanley N.A. Development of the human pancreas from foregut to endocrine commitment. Diabetes. 2013;62:3514–3522. doi: 10.2337/db12-1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson J., Carlsson L., Edlund T., Edlund H. Insulin-promoter-factor 1 is required for pancreas development in mice. Nature. 1994;371:606–609. doi: 10.1038/371606a0. [DOI] [PubMed] [Google Scholar]
- Jørgensen M.C., Ahnfelt-Rønne J., Hald J., Madsen O.D., Serup P., Hecksher-Sørensen J. An illustrated review of early pancreas development in the mouse. Endocr. Rev. 2007;28:685–705. doi: 10.1210/er.2007-0016. [DOI] [PubMed] [Google Scholar]
- Ketola I., Otonkoski T., Pulkkinen M.A., Niemi H., Palgi J., Jacobsen C.M., Wilson D.B., Heikinheimo M. Transcription factor GATA-6 is expressed in the endocrine and GATA-4 in the exocrine pancreas. Mol. Cell. Endocrinol. 2004;226:51–57. doi: 10.1016/j.mce.2004.06.007. [DOI] [PubMed] [Google Scholar]
- Koutsourakis M., Langeveld A., Patient R., Beddington R., Grosveld F. The transcription factor GATA6 is essential for early extraembryonic development. Development. 1999;126:723–732. [PubMed] [Google Scholar]
- Lango Allen H., Flanagan S.E., Shaw-Smith C., De Franco E., Akerman I., Caswell R., International Pancreatic Agenesis C., Ferrer J., Hattersley A.T., Ellard S. GATA6 haploinsufficiency causes pancreatic agenesis in humans. Nat. Genet. 2011;44:20–22. doi: 10.1038/ng.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O'Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lentjes M.H., Niessen H.E., Akiyama Y., de Bruine A.P., Melotte V., van Engeland M. The emerging role of GATA transcription factors in development and disease. Expert Rev. Mol. Med. 2016;18:e3. doi: 10.1017/erm.2016.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao C.M., Mukherjee S., Tiyaboonchai A., Maguire J.A., Cardenas-Diaz F.L., French D.L., Gadue P. GATA6 suppression enhances lung specification from human pluripotent stem cells. J. Clin. Invest. 2018;128:2944–2950. doi: 10.1172/JCI96539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda M., Ohashi K., Ohashi-Kobayashi A. Further extension of mammalian GATA-6. Dev. Growth Differ. 2005;47:591–600. doi: 10.1111/j.1440-169X.2005.00837.x. [DOI] [PubMed] [Google Scholar]
- Morrisey E.E., Ip H.S., Lu M.M., Parmacek M.S. GATA-6: a zinc finger transcription factor that is expressed in multiple cell lineages derived from lateral mesoderm. Dev. Biol. 1996;177:309–322. doi: 10.1006/dbio.1996.0165. [DOI] [PubMed] [Google Scholar]
- Morrisey E.E., Tang Z., Sigrist K., Lu M.M., Jiang F., Ip H.S., Parmacek M.S. GATA6 regulates HNF4 and is required for differentiation of visceral endoderm in the mouse embryo. Genes Dev. 1998;12:3579–3590. doi: 10.1101/gad.12.22.3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Offield M.F., Jetton T.L., Labosky P.A., Ray M., Stein R.W., Magnuson M.A., Hogan B.L., Wright C.V. PDX-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development. 1996;122:983–995. doi: 10.1242/dev.122.3.983. [DOI] [PubMed] [Google Scholar]
- Ortmann D., Vallier L. Variability of human pluripotent stem cell lines. Curr. Opin. Genet. Dev. 2017;46:179–185. doi: 10.1016/j.gde.2017.07.004. [DOI] [PubMed] [Google Scholar]
- Pan F.C., Wright C. Pancreas organogenesis: from bud to plexus to gland. Dev. Dyn. 2011;240:530–565. doi: 10.1002/dvdy.22584. [DOI] [PubMed] [Google Scholar]
- Patient R.K., McGhee J.D. The GATA family (vertebrates and invertebrates) Curr. Opin. Genet. Dev. 2002;12:416–422. doi: 10.1016/s0959-437x(02)00319-2. [DOI] [PubMed] [Google Scholar]
- Rubio-Cabezas O., Minton J.A., Kantor I., Williams D., Ellard S., Hattersley A.T. Homozygous mutations in NEUROD1 are responsible for a novel syndrome of permanent neonatal diabetes and neurological abnormalities. Diabetes. 2010;59:2326–2331. doi: 10.2337/db10-0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio-Cabezas O., Jensen J.N., Hodgson M.I., Codner E., Ellard S., Serup P., Hattersley A.T. Permanent neonatal diabetes and enteric anendocrinosis associated with biallelic mutations in NEUROG3. Diabetes. 2011;60:1349–1353. doi: 10.2337/db10-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrode N., Saiz N., Di Talia S., Hadjantonakis A.K. GATA6 levels modulate primitive endoderm cell fate choice and timing in the mouse blastocyst. Dev. Cell. 2014;29:454–467. doi: 10.1016/j.devcel.2014.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwitzgebel V.M. Many faces of monogenic diabetes. J. Diabetes Investig. 2014;5:121–133. doi: 10.1111/jdi.12197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwitzgebel V.M., Mamin A., Brun T., Ritz-Laser B., Zaiko M., Maret A., Jornayvaz F.R., Theintz G.E., Michielin O., Melloul D. Agenesis of human pancreas due to decreased half-life of insulin promoter factor 1. J. Clin. Endocrinol. Metab. 2003;88:4398–4406. doi: 10.1210/jc.2003-030046. [DOI] [PubMed] [Google Scholar]
- Sellick G.S., Barker K.T., Stolte-Dijkstra I., Fleischmann C., Coleman R.J., Garrett C., Gloyn A.L., Edghill E.L., Hattersley A.T., Wellauer P.K. Mutations in PTF1A cause pancreatic and cerebellar agenesis. Nat. Genet. 2004;36:1301–1305. doi: 10.1038/ng1475. [DOI] [PubMed] [Google Scholar]
- Shaw-Smith C., De Franco E., Lango Allen H., Batlle M., Flanagan S.E., Borowiec M., Taplin C.E., van Alfen-van der Velden J., Cruz-Rojo J., Perez de Nanclares G. GATA4 mutations are a cause of neonatal and childhood-onset diabetes. Diabetes. 2014;63:2888–2894. doi: 10.2337/db14-0061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Z.D., Lee K., Yang D., Amin S., Verma N., Li Q.V., Zhu Z., Soh C.L., Kumar R., Evans T. Genome editing in hPSCs reveals GATA6 haploinsufficiency and a genetic interaction with GATA4 in human pancreatic development. Cell Stem Cell. 2017;20:675–688.e6. doi: 10.1016/j.stem.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S.B., Qu H.Q., Taleb N., Kishimoto N.Y., Scheel D.W., Lu Y., Patch A.M., Grabs R., Wang J., Lynn F.C. Rfx6 directs islet formation and insulin production in mice and humans. Nature. 2010;463:775–780. doi: 10.1038/nature08748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanescu D.E., Hughes N., Patel P., De Leon D.D. A novel mutation in GATA6 causes pancreatic agenesis. Pediatr. Diabetes. 2015;16:67–70. doi: 10.1111/pedi.12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoffers D.A., Zinkin N.T., Stanojevic V., Clarke W.L., Habener J.F. Pancreatic agenesis attributable to a single nucleotide deletion in the human IPF1 gene coding sequence. Nat. Genet. 1997;15:106–110. doi: 10.1038/ng0197-106. [DOI] [PubMed] [Google Scholar]
- Suzuki S., Nakao A., Sarhat A.R., Furuya A., Matsuo K., Tanahashi Y., Kajino H., Azuma H. A case of pancreatic agenesis and congenital heart defects with a novel GATA6 nonsense mutation: evidence of haploinsufficiency due to nonsense-mediated mRNA decay. Am. J. Med. Genet. A. 2014;164A:476–479. doi: 10.1002/ajmg.a.36275. [DOI] [PubMed] [Google Scholar]
- Teo A.K., Arnold S.J., Trotter M.W., Brown S., Ang L.T., Chng Z., Robertson E.J., Dunn N.R., Vallier L. Pluripotency factors regulate definitive endoderm specification through eomesodermin. Genes Dev. 2011;25:238–250. doi: 10.1101/gad.607311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo A.K., Tsuneyoshi N., Hoon S., Tan E.K., Stanton L.W., Wright C.V., Dunn N.R. PDX1 binds and represses hepatic genes to ensure robust pancreatic commitment in differentiating human embryonic stem cells. Stem Cell Reports. 2015;4:578–590. doi: 10.1016/j.stemcr.2015.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiyaboonchai A., Cardenas-Diaz F.L., Ying L., Maguire J.A., Sim X., Jobaliya C., Gagne A.L., Kishore S., Stanescu D.E., Hughes N. GATA6 plays an important role in the induction of human definitive endoderm, development of the pancreas, and functionality of pancreatic beta cells. Stem Cell Reports. 2017;8:589–604. doi: 10.1016/j.stemcr.2016.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallier L., Touboul T., Chng Z., Brimpari M., Hannan N., Millan E., Smithers L.E., Trotter M., Rugg-Gunn P., Weber A. Early cell fate decisions of human embryonic stem cells and mouse epiblast stem cells are controlled by the same signalling pathways. PLoS One. 2009;4:e6082. doi: 10.1371/journal.pone.0006082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viger R.S., Guittot S.M., Anttonen M., Wilson D.B., Heikinheimo M. Role of the GATA family of transcription factors in endocrine development, function, and disease. Mol. Endocrinol. 2008;22:781–798. doi: 10.1210/me.2007-0513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Sun H., Ma J., Zang C., Wang C., Wang J., Tang Q., Meyer C.A., Zhang Y., Liu X.S. Target analysis by integration of transcriptome and ChIP-seq data with BETA. Nat. Protoc. 2013;8:2502–2515. doi: 10.1038/nprot.2013.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xuan S., Borok M.J., Decker K.J., Battle M.A., Duncan S.A., Hale M.A., Macdonald R.J., Sussel L. Pancreas-specific deletion of mouse Gata4 and Gata6 causes pancreatic agenesis. J. Clin. Invest. 2012;122:3516–3528. doi: 10.1172/JCI63352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yau D., De Franco E., Flanagan S.E., Ellard S., Blumenkrantz M., Mitchell J.J. Case report: maternal mosaicism resulting in inheritance of a novel GATA6 mutation causing pancreatic agenesis and neonatal diabetes mellitus. Diagn. Pathol. 2017;12:1. doi: 10.1186/s13000-016-0592-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorifuji T., Kawakita R., Hosokawa Y., Fujimaru R., Yamaguchi E., Tamagawa N. Dominantly inherited diabetes mellitus caused by GATA6 haploinsufficiency: variable intrafamilial presentation. J. Med. Genet. 2012;49:642–643. doi: 10.1136/jmedgenet-2012-101161. [DOI] [PubMed] [Google Scholar]
- Yu L., Bennett J.T., Wynn J., Carvill G.L., Cheung Y.H., Shen Y., Mychaliska G.B., Azarow K.S., Crombleholme T.M., Chung D.H. Whole exome sequencing identifies de novo mutations in GATA6 associated with congenital diaphragmatic hernia. J. Med. Genet. 2014;51:197–202. doi: 10.1136/jmedgenet-2013-101989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R., Watt A.J., Li J., Luebke-Wheeler J., Morrisey E.E., Duncan S.A. GATA6 is essential for embryonic development of the liver but dispensable for early heart formation. Mol. Cell. Biol. 2005;25:2622–2631. doi: 10.1128/MCB.25.7.2622-2631.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.