Abstract
In this Research Article, a novel inkjet printing technique, micro impact printing (MI printing), is applied for the first time to combinatorial peptide microarray synthesis on amine functionalized microdisc arrays through standard Fmoc chemistry. MI printing shows great advantages in combinatorial peptide microarray synthesis compared with other printing techniques, including (1) a disposable cartridge; (2) a small spot size (80 μm) increases array density; (3) minimal loading volume (0.6 μL) and dead volume (<0.1 μL), reduce chemical waste; and (4) multiplexibility of 5 channels/cartridge and capacity of multiple cartridges. Using this synthesis platform, a tetrapeptide library with 625 permutations was constructed and then applied for the screening of ligands targeting α4β1 integrin on Jurkat cells.
Keywords: peptide synthesis, microfluidic, peptide microarrays, printing, cancer targeting, integrin binding
Introduction
In the early 1960s, Merrifield developed solid phase peptide synthesis (SPPS) by growing peptides tethered on a solid support.1 This established a solid foundation for the field of multipeptide synthesis and combinatorial chemistry2−6 and became an essential research tool for various biomedical and material applications, such as drug discovery7 and ligand–receptor interaction study.8 Over the years, several advances have occurred in the development of peptide synthesis. The one-bead-one-compound (OBOC) method4 has been widely applied for peptide library synthesis, but suffers from a slow and relatively expensive decoding procedure that is required to obtain the peptide sequences of positive beads.9 In contrast, peptide arrays are spatially addressable.10 The diversity of peptide arrays, however, is more limited, and therefore this technology is more suitable for ligand optimization and SAR studies. Peptide arrays prepared by SPOT synthesis11,12 method uses standard solid phase peptide synthesis (SPPS) chemistry. It is robust and can be performed by automatic equipment, but the array density and throughput are limited.13 The lithographic method14,15 and particle-based techniques16−18 can greatly increase the peptide array density but can only be applied to amino acids with nonstandard protecting groups due to their dependence on special chemical reaction conditions, such as light irradiation or high temperatures, which may also result in low yield compared with traditional t-butyloxycarbonyl (Boc) or 9-fluorenylmethoxycarbonyl (Fmoc) protection chemistry. Inkjet printing19−22 shows great potential because of its ability to generate droplets at a picoliter level with a frequency at the kilohertz level, leading to high array density and throughput. Furthermore, it is compatible with standard chemistry, which guarantees high coupling yield. Current development in inkjet printing synthesis, such as piezoelectric dispensing19,20 and acoustic droplet ejection,21,22 are in slow progression mainly due to high costs, the complexity of the printing systems, and required cartridge replacement. Thus, far, only the feasibility of these two methods has been tested by synthesizing peptide arrays with identical sequences using a single-channel device; large scale combinatorial peptide synthesis has yet to be explored.15
Our group has been working on a modified inkjet printing technique, referred to as microfluidic printing, which includes microfluidic impact printing and microfluidic pneumatic printing. Recently we have demonstrated the use of a microfluidic pneumatic printing platform to generate peptide microarrays.23 The resulting repeatability was larger than 90% according to analysis for both intrachip and interchip replicates, which indicates good synthesis yield. As an alternative approach, this paper describes the application of our recently developed microfluidic impact printing (MI Printing) technique24 for combinatorial peptide microarray synthesis. Compared with the pneumatic printing platform, the MI printing platform can implement smaller footprint due to its faster response. More importantly, in this study, we verified the amino acid coupling efficiency by sequencing the resulting sample on a Procise protein sequencer. Results indicated good coupling efficiency, which further verified the potential of the MI printing platform in peptide array synthesis. Taking advantage of the fully detachable/disposable polydimethylsiloxane (PDMS) cartridge, the MI-printing system greatly reduces the cost of cartridge replacement.
Results and Discussion
Microfluidic Impact Printing Synthesis
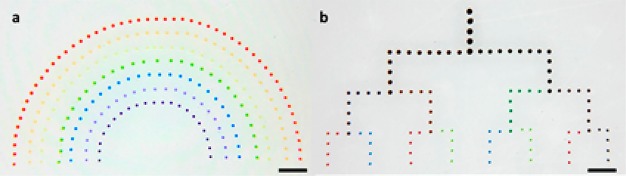
The microfluidic cartridge (Figure 1a) is mounted on a commercial dot matrix printer head with five pins. Upon printing, pins in the dot matrix printer head squeeze the cartridge membrane, thus generating a droplet (Figure 1b). In traditional inkjet printing techniques, a piezoelectric or thermal electric material is usually integrated with the cartridge, resulting in a high cartridge cost. However, for MI printing, the cartridge is purely composed of low-cost PDMS material, which can easily be detached from the dot matrix printer. This results in a disposable cartridge design and greatly reduces cost, while also avoiding contamination. Small size (80 μm), multiplexed (5 channels per cartridge) droplet arrays (Figure 2) can be achieved using the MI printer, demonstrating its capability of producing combinatorial peptide libraries. Moreover, due to the geometry of the cartridge, the loading volume in each channel is only 0.54 μL, while the dead volume is less than 0.05 μL.24 Such features are highly desirable for peptide synthesis research considering the high cost of some unusual amino acids. For large-scale array synthesis, the system can also be easily adjusted by expanding the reservoirs to contain large amounts of chemicals and using multiple cartridges.
Figure 1.
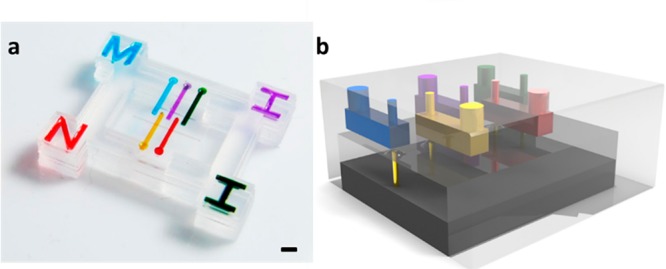
Microfluidic cartridge (a) and its working principle (b) (scale bar = 2 mm).
Figure 2.
Multiplexed droplet array in rainbow (a) and mixer (b) format (scale bar = 1 mm).
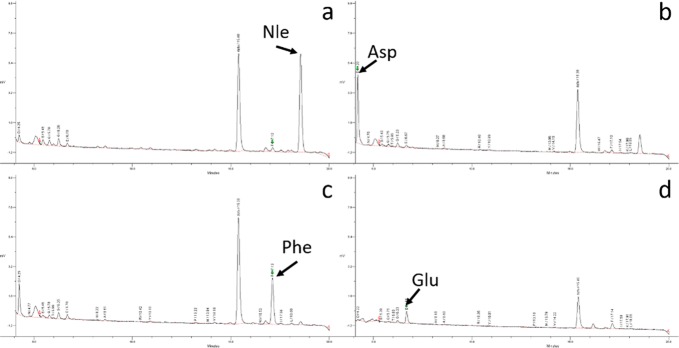
To demonstrate MI printing synthesis technique and its biomedical utility, we implemented the synthesis of a combinatorial tetrapeptide library with 625 permutations, which was then screened with Jurkat lymphoid malignant T-cells for α4β1 integrin targeting.25 To achieve high yield from peptide synthesis, an amino-functionalized polyethylene glycol (PEG) microdisc array was first fabricated on a glass slide support as the solid support for peptide synthesis. Compared with conventional peptide synthesis directly performed on glass slide, the PEG-based microdisc has several advantages: (i) it has very low nonspecific binding compared to glass surface, (ii) it has better visibility and more flexibility in patterning (i.e., we can control the pattern and thickness of the PEG microdisc), and (iii) it has higher peptide-loading than glass surface or glass microdisc, and is therefore suitable for solution-phase releasable assay.26 The fabricated micro discs were 200 μm in diameter, 5 μm in height, 1.2 mm in center-to-center distance, and had an amine loading of 0.24 mmol/g. A PDMS frame was then attached to prevent crosstalk. Amino acids were then dispensed into the PDMS wells by the MI printer and coupled to the microdisc array. Standard Fmoc-protected chemistry was used: after the protecting group was removed, the next Fmoc-amino acid was coupled. The coupling cycle was repeated until the last amino acid was coupled. The entire synthesis procedure is illustrated in Figure 3. Bromophenol blue was used to detect the presence of free amine groups before and after the coupling reaction and after the removal of Fmoc. The reaction time was found to be optimal at 1 h, thus total time consumption could be less than 2 h for each coupling cycle, which includes printing (20 min, may vary on library size) Fmoc deprotection (10 min) and washing (10 min). The coupling yield was verified by synthesizing a tetrapeptide, Nle-Asp-Phe-Glu, followed by sequencing the resulting microdiscs on a Procise protein sequencer. Sequencing results show very little impurities (Figure 4, with standard absorption included in Figure S1 and high-resolution absorption data in Figures S2–S5), thus demonstrating the coupling efficiency of the MI printing synthesis method. To target α4β1 integrin, which is highly expressed on malignant lymphoid cells, we designed and synthesized a tetrapeptide library with the N-terminus capped with 4-[(N′-2-methylphenyl)ureido]phenylacetic acid (UPA). The library is referred to as UPA-X1X2X3X4, where X1, X2, X3, and X4 represent the last, third, second, and first amino acids that were coupled to the substrate, respectively. Leu-Asp-Val-Pro is a known binding motif for α4β1 integrin.8 To keep the diversity of the peptide microarrays small, we picked three of the four amino acid in the motif: Leu, Asp, and Pro. Val was chosen not to be used because it is very similar to Leu. To add to the diversity, we also chose Ala and Arg. With a total of five building blocks (Leu, Asp, Pro, Ala, and Arg) in each coupling cycle, and a total of four cycles, the peptide microarray library had 54 = 625 permutations.
Figure 3.
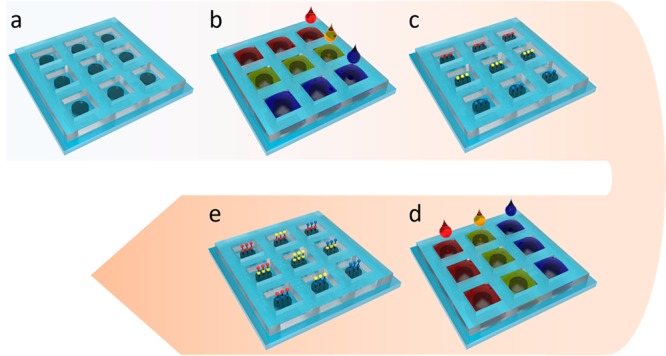
MI printing synthesis procedure: (a) amine functionalized micro disc array, (b, c), first coupling cycle, and (d, e) second coupling cycle.
Figure 4.
Sequencing result for Nle-Asp-Phe-Glu for verification of coupling effectiveness.
Cell Binding Results
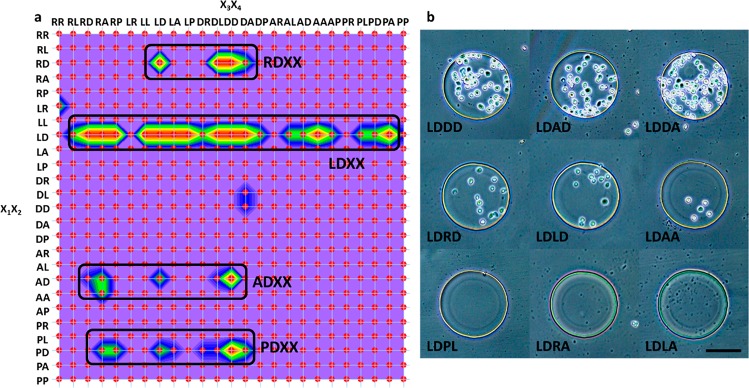
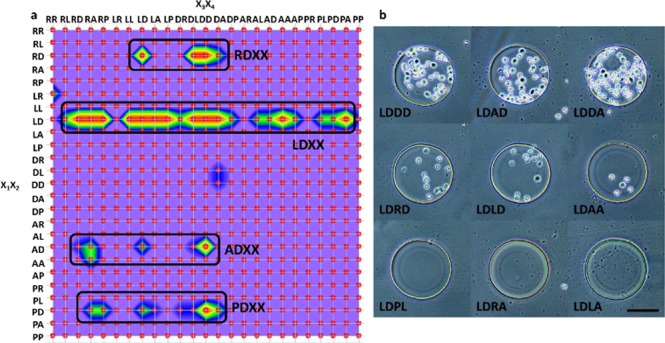
To identify positive sequences that can target α4β1 integrin within the library, the entire microarray was screened simultaneously, in situ, against live Jurkat cells, which is known to have a high expression level of activated α4β1 integrin.27 After incubation, selective cell attachment on microdisc was observed and summarized (Figures S6 and S7). The cell binding affinity was evaluated according to the area covered with cells on each microdisc, where strong, medium, weak, and no binding were represented by >50%, 10%–50%, <10%, and 0% of the disc area covered with cells, respectively. A color contour map (Figure 5a) was drawn (3Dfield Pro) to display the binding results. Four regions with strong binding can be observed, corresponding to preference of Arg-Asp, Leu-Asp, Ala-Asp, and Arg-Asp sequences at X1X2 position. Among them, Leu-Asp and Pro-Asp were already identified as positive motifs targeting α4β1 integrin by existing publications in the Leu-Asp-Val-Pro library,8,28 showing our results to be consistent with previously published results. To further verify that both sequences, Arg-Asp and Ala-Asp, were targeting α4β1 integrin instead of other surface receptors displayed on Jurkat cells, a competitive screening method with high stringency was carried out.8 In this competitive screening method, 100 pM of BIO-1211, a known high-affinity α4β1 antagonists,29 was added to the Jurkat cell suspension. Selected pictures of microdiscs, obtained under this higher stringent screening condition are shown in Figure 5b; only microdiscs with Leu-Asp-Arg-Leu, Leu-Asp-Arg-Asp, Leu-Asp-Arg-Ala, Leu-Asp-Leu-Leu, Leu-Asp-Leu-Asp, Leu-Asp-Asp-Leu, Leu-Asp-Asp-Asp, Leu-Asp-Asp-Ala, and Leu-Asp-Ala-Asp sequences still had cells attached, while no cell binding occurred with the other microdiscs. This can be explained by the presence of the high affinity BIO-1211, which targeted and blocked some of the α4β1 integrin displayed on the cell surface, leaving fewer binding sites for the peptide array to interact with the cells. Under such conditions, peptides with Arg-Asp and Ala-Asp motifs had no cells attach, indicating that both of these sequences target the α4β1 integrin specifically instead of other surface receptors. The reason why there were cells still attached to the aforementioned sequences is most likely due to the strong binding affinity of the Leu-Asp motif. The results obtained from the above feasibility study clearly demonstrates that the MI printer can generate peptide microarrays with high array density, high coupling efficiency, and low cost. Such peptide microarray platform can provide invaluable information on SAR of peptide-integrin interactions.
Figure 5.
(a) Color contour map of the binding results. (b) Selective stringent screening results with presence of BIO-1211 (scale bar = 100 μm).
Conclusion
We have reported the utilization of our previously developed microfluidic impact printing technique, in combination with the PEG based microdisc array substrate, for combinatorial peptide microarray synthesis and screening. The MI printing possesses several distinct favorable features including disposable cartridge, very low dead volume and multichannel integration. The PEG microdisc provides a preferable substrate for synthesis and screening due to its low nonspecific binding and compatibility with both aqueous and organic conditions. As a proof of concept, a tetrapeptide library with 625 permutation was synthesized targeting α4β1 integrin highly expressed on Jurkat T lymphoid cancer cell. Future work may include increasing the microarray density, expanding the library size for higher throughput screening, including multiple cartridges so that over 20 amino acids can be used, and increasing the printing speed with a better on-the-fly printing mode for the synthesis of larger libraries.
Experimental Procedures
Glass slide (2 in × 3 in) was plasma-treated for 15 min, and immersed in 1% (3-acryloxypropyl)trichlorosilane for 2 h for silane cross-linking. 200 μL of polyethylene glycol diacrylate (PEGDA) (MW = 700), 112 μL of cross-linker (trimethylolpropane ethoxylate triacrylate), 8 μL of photo initiator (2-hydroxyl-2-methylpropiophenone), 7.2 mg of 2-aminoethyl methacrylate·HCl, and 300 μL of deionized water was mixed as prepolymer for microdisc cross-linking. The solution mixture was then sandwiched between the glass substrate and a photomask. Cross-linked amino-PEG microdiscs were then formed upon UV polymerization through a photomask under 365 nm UV light, with a total exposure energy of 150 mJ/cm2. The amine loading was measured by reacting the PEG discs (scrapped off the glass slide with razor blade) with Fmoc-OSU followed by thorough washing and then removal of the Fmoc with 20% 4-methylpiperidine. The released Fmoc in solution was quantified by measuring its UV absorption at 276 nm (Nanodrop). Amine loading can then be calculated based on the weight of the dried PEG discs and the amount of Fmoc quantified by UV measurement. 0.5 mm thick PDMS membrane (40 mm × 40 mm) was laser cut into hole arrays and attached to the glass slide by self-stiction with each hole aligned to a microdisc to prevent crosstalk.
MI-Printing Synthesis
The MI-printer consists of a multichannel microfluidic cartridge, a commercial dot matrix printer head, and a three-dimensional traveling stage. A detailed assembly of the printing system, fabrication of the microfluidic cartridge, and its working principles can be found in our previous paper.24 Fmoc-protected amino acids (120 μM) were dissolved in a solution of 1-hydroxy-6-chloro-benzotriazole (6-Cl HOBt) (120 μM) and 1,3-diisopropylcarbodiimide (DIC) (120 μM) in anhydrous N,N′-dimethylformamide (DMF). 100 nL of selected amino acid solution was printed into specific sites on the chip, and incubated for 30 min at room temperature. The printing was then repeated immediately for double coupling to maximize the coupling efficiency. After another 30 min of incubation, the chip was submerged in a solution of 2% Acetic anhydride and 2% N, N-diisopropylethylamine (DIPEA) (v: (v) in DMF solution, in which the PDMS frame was removed by peeling it off from the side, and the chip was immersed for 15 min to cap the remaining unreacted amine groups. The Fmoc protection group was then removed with 20% (v: (v) 4-methyl peperidine in DMF solution for 10 min, during which the solution was refreshed after 5 min. The resulting chip was covered by a PDMS frame again and aligned manually according to the microdisc array stained by bromophenol blue. The chip was then used for the next round of peptide synthesis. The whole procedure was repeated until the last amino acid was coupled. Afterward, 4-[(N′-2-methylphenyl)ureido]phenylacetic acid (UPA) was coupled to the N-terminus of the peptide chain on every spot using the same protocol. As the final step, the side-chain protection group was removed with a mixture of 5% phenol, 5% water, 5% thioanisole, 2.5% triisopropylsilane, and 82.5% trifluoroacetic acid (TFA) (v/v). After each step, the chip was washed sequentially with DMF (3 times) and ethanol (3 times), and then dried in the fume hood.
Cell Binding Assay
Jurkat malignant T cell line was maintained in a 95% air/5% CO2 humidified incubator at 37 °C. Cells were harvested and resuspended to a concentration of 2 million cells per milliliter. The peptide chip was then incubated with suspended Jurkat cells in culture medium for 2 h at 37.5 °C. Extra cells were gently washed three times with culture medium. The chip was then kept in fresh culture medium and examined under an EVOS transmitted microscope (AMG). After the first screen, the chip was recycled by adding 6 M guanidine hydrochloride, and then thoroughly washed with ethanol and DI water. For screening under higher stringency, the recycled chip was screened again against the same concentration of Jurkat cell solution but in the presence of 100 pM BIO-1211 in the culture medium. The chip was then gentle washed examined again under an EVOS transmitted microscope (AMG).
Acknowledgments
We acknowledge the assistance from Dr. Tsung-Chieh Shih for cell culturing and binding study and Mr. Vladimir Galouchko for developing the online software 3Dfield Pro.
Glossary
Abbreviations
- Fmoc
9-fluorenylmethoxycarbonyl
- MI
microfluidic impact
- PEG
polyethylene glycol
- PDMS
polydimethylsiloxane
- UPA
4-[(N′-2-methylphenyl)ureido]phenylacetic acid
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acscombsci.8b00125.
Materials information, whole cell binding map, and selected cell binding pictures (PDF)
Author Present Address
∥ G.Y.: X-Therma, Inc., Richmond, CA, USA.
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
This research work has been supported in part by the National Science Foundation Awards ECCS-0846502 and DBI-1256193, National Institute of Health Awards NCI-R21CA173243 and NIEHS-P42ES004699, and the US NSF-and industry-funded I/UCRC Center for Biophotonic Sensors and Systems, NSF Grant 1650588.
The authors declare no competing financial interest.
Supplementary Material
References
- Merrifield R. B. Solid Phase Peptide Synthesis. 1. Synthesis of a Tetrapeptide. J. Am. Chem. Soc. 1963, 85 (14), 2149–2154. 10.1021/ja00897a025. [DOI] [Google Scholar]
- Lam K. S.; Renil M. From combinatorial chemistry to chemical microarray. Curr. Opin. Chem. Biol. 2002, 6 (3), 353–8. 10.1016/S1367-5931(02)00326-5. [DOI] [PubMed] [Google Scholar]
- Houghten R. A.; Pinilla C.; Blondelle S. E.; Appel J. R.; Dooley C. T.; Cuervo J. H. Generation and Use of Synthetic Peptide Combinatorial Libraries for Basic Research and Drug Discovery. Nature 1991, 354 (6348), 84–86. 10.1038/354084a0. [DOI] [PubMed] [Google Scholar]
- Lam K. S.; Salmon S. E.; Hersh E. M.; Hruby V. J.; Kazmierski W. M.; Knapp R. J. A New Type of Synthetic Peptide Library for Identifying Ligand-Binding Activity. Nature 1991, 354 (6348), 82–84. 10.1038/354082a0. [DOI] [PubMed] [Google Scholar]
- Furka A.; Sebestyen F.; Asgedom M.; Dibo G. General-Method for Rapid Synthesis of Multicomponent Peptide Mixtures. Int. J. Pept. Protein Res. 1991, 37 (6), 487–493. 10.1111/j.1399-3011.1991.tb00765.x. [DOI] [PubMed] [Google Scholar]
- Geysen H. M.; Meloen R. H.; Barteling S. J. Use of Peptide-Synthesis to Probe Viral-Antigens for Epitopes to a Resolution of a Single Amino-Acid. Proc. Natl. Acad. Sci. U. S. A. 1984, 81 (13), 3998–4002. 10.1073/pnas.81.13.3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R. W.; Li X. C.; Lam K. S. Combinatorial chemistry in drug discovery. Curr. Opin. Chem. Biol. 2017, 38, 117–126. 10.1016/j.cbpa.2017.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L.; Liu R. W.; Marik J.; Wang X. B.; Takada Y.; Lam K. S. Combinatorial chemistry identifies high-affinity peptidomimetics against alpha(4)beta(1) integrin for in vivo tumor imaging. Nat. Chem. Biol. 2006, 2 (7), 381–389. 10.1038/nchembio798. [DOI] [PubMed] [Google Scholar]
- Liu R. W.; Wang X. B.; Song A. M.; Bao T.; Lam K. S. Development and applications of topologically segregated bilayer beads in one-bead one-compound combinatorial libraries. QSAR Comb. Sci. 2005, 24 (10), 1127–1140. 10.1002/qsar.200540010. [DOI] [PubMed] [Google Scholar]
- Szymczak L. C.; Kuo H. Y.; Mrksich M. Peptide Arrays: Development and Application. Anal. Chem. 2018, 90 (1), 266–282. 10.1021/acs.analchem.7b04380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank R. Spot-Synthesis - an Easy Technique for the Positionally Addressable, Parallel Chemical Synthesis on a Membrane Support. Tetrahedron 1992, 48 (42), 9217–9232. 10.1016/S0040-4020(01)85612-X. [DOI] [Google Scholar]
- Hundsberger H.; Onder K.; Schuller-Gotzburg P.; Virok D. P.; Herzog J.; Rid R. Assembly and use of high-density recombinant peptide chips for large-scale ligand screening is a practical alternative to synthetic peptide libraries. BMC Genomics 2017, 18, 450. 10.1186/s12864-017-3814-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitling F.; Nesterov A.; Stadler V.; Felgenhauer T.; Bischoff F. R. High-density peptide arrays. Mol. BioSyst. 2009, 5 (3), 224–234. 10.1039/b819850k. [DOI] [PubMed] [Google Scholar]
- Fodor S. P.; Read J. L.; Pirrung M. C.; Stryer L.; Lu A. T.; Solas D. Light-directed, spatially addressable parallel chemical synthesis. Science 1991, 251 (4995), 767–73. 10.1126/science.1990438. [DOI] [PubMed] [Google Scholar]
- Pellois J. P.; Zhou X. C.; Srivannavit O.; Zhou T. C.; Gulari E.; Gao X. L. Individually addressable parallel peptide synthesis on microchips. Nat. Biotechnol. 2002, 20 (9), 922–926. 10.1038/nbt723. [DOI] [PubMed] [Google Scholar]
- Stadler V.; Felgenhauer T.; Beyer M.; Fernandez S.; Leibe K.; Guttler S.; Groning M.; Konig K.; Torralba G.; Hausmann M.; Lindenstruth V.; Nesterov A.; Block I.; Pipkorn R.; Poustka A.; Bischoff F. R.; Breitling F. Combinatorial synthesis of peptide arrays with a laser printer. Angew. Chem., Int. Ed. 2008, 47 (37), 7132–7135. 10.1002/anie.200801616. [DOI] [PubMed] [Google Scholar]
- Breitling F.; Felgenhauer T.; Nesterov A.; Lindenstruth V.; Stadler V.; Bischoff F. R. Particle-Based Synthesis of Peptide Arrays. ChemBioChem 2009, 10 (5), 803–808. 10.1002/cbic.200800735. [DOI] [PubMed] [Google Scholar]
- Loeffler F. F.; Foertsch T. C.; Popov R.; Mattes D. S.; Schlageter M.; Sedlmayr M.; Ridder B.; Dang F. X.; von Bojnicic-Kninski C.; Weber L. K.; Fischer A.; Greifenstein J.; Bykovskaya V.; Buliev I.; Bischoff F. R.; Hahn L.; Meier M. A.; Brase S.; Powell A. K.; Balaban T. S.; Breitling F.; Nesterov-Mueller A. High-flexibility combinatorial peptide synthesis with laser-based transfer of monomers in solid matrix material. Nat. Commun. 2016, 7, 11844. 10.1038/ncomms11844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antohe B. V.; Cooley P. W. In situ synthesis of peptide microarrays using ink-jet microdispensing. Methods Mol. Biol. 2007, 381, 299–312. 10.1007/978-1-59745-303-5_15. [DOI] [PubMed] [Google Scholar]
- Cooley P.; Wallace D.; Antohe B. Applications of ink-jet printing technology to BioMEMS and microfluidic systems. Proc. SPIE 2001, 4560, 177–188. 10.1117/12.443057. [DOI] [Google Scholar]
- Choe Y.; Chen S. J.; Kim E. S. Peptide Synthesis on Glass Substrate Using Acoustic Droplet Ejector. IEEE Trans. Biomed. Eng. 2014, 61 (3), 705–710. 10.1109/TBME.2013.2287218. [DOI] [PubMed] [Google Scholar]
- Lee C. Y.; Yu H.; Kim E. S. Acoustic ejector with novel lens employing air-reflectors. Proc. Ieee Micr Elect 2006, 170–173. 10.1109/MEMSYS.2006.1627763. [DOI] [Google Scholar]
- Li J.; Carney R. P.; Liu R.; Fan J.; Zhao S.; Chen Y.; Lam K. S.; Pan T. Microfluidic Print-to-Synthesis Platform for Efficient Preparation and Screening of Combinatorial Peptide Microarrays. Anal. Chem. 2018, 90 (9), 5833–5840. 10.1021/acs.analchem.8b00371. [DOI] [PubMed] [Google Scholar]
- Ding Y. Z.; Huang E.; Lam K. S.; Pan T. R. Microfluidic impact printer with interchangeable cartridges for versatile non-contact multiplexed micropatterning. Lab Chip 2013, 13 (10), 1902–1910. 10.1039/c3lc41372a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aina O. H.; Sroka T. C.; Chen M. L.; Lam K. S. Therapeutic cancer targeting peptides. Biopolymers 2002, 66 (3), 184–199. 10.1002/bip.10257. [DOI] [PubMed] [Google Scholar]
- Townsend J. B.; Shaheen F.; Liu R. W.; Lam K. S. Jeffamine Derivatized TentaGel Beads and Poly(dimethylsiloxane) Microbead Cassettes for Ultrahigh-Throughput in Situ Releasable Solution-Phase Cell-Based Screening of One-Bead-One-Compound Combinatorial Small Molecule Libraries. J. Comb. Chem. 2010, 12 (5), 700–712. 10.1021/cc100083f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyano J. V.; GarciaGila M.; DominguezJimenez C.; GarciaPardo A. Fibronectin type III5 repeat contains a novel cell adhesion sequence which binds activated alpha 4 beta 1 and alpha 4 beta 7 integrins. Mol. Biol. Cell 1997, 8, 399–399. [DOI] [PubMed] [Google Scholar]
- Lin K. C.; Ateeq H. S.; Hsiung S. H.; Chong L. T.; Zimmerman C. N.; Castro A.; Lee W. C.; Hammond C. E.; Kalkunte S.; Chen L. L.; Pepinsky R. B.; Leone D. R.; Sprague A. G.; Abraham W. M.; Gill A.; Lobb R. R.; Adams S. P. Selective, tight-binding inhibitors of integrin alpha 4 beta 1 that inhibit allergic airway responses. J. Med. Chem. 1999, 42 (5), 920–934. 10.1021/jm980673g. [DOI] [PubMed] [Google Scholar]
- Kim D. H.; Shin D. S.; Lee Y. S. Spot arrays on modified glass surfaces for efficient SPOT synthesis and on-chip bioassay of peptides. J. Pept. Sci. 2007, 13 (10), 625–633. 10.1002/psc.884. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.