Abstract
Background
Despite their importance as vectors of zoonotic parasites that can impact human and animal health, Culicoides species distribution across different habitat types is largely unknown. Here we document the community composition of Culicoides found in an urban environment including developed and natural sites in east central Texas, a region of high vector diversity due to subtropical climates, and report their infection status with haemoparasites.
Results
A total of 251 individual Culicoides were collected from May to June 2016 representing ten Culicoides species, dominated by C. neopulicaris followed by C. crepuscularis. We deposited 63 sequences to GenBank among which 25 were the first deposition representative for six Culicoides species: C. arboricola (n = 1); C. nanus (n = 4); C. debilipalpis (n = 2); C. haematopotus (n = 14); C. edeni (n = 3); and C. hinmani (n = 1). We also record for the first time the presence of C. edeni in Texas, a species previously known to occur in the Bahamas, Florida and South Carolina. The urban environments with natural area (sites 2 and 4) had higher species richness than sites more densely populated or in a parking lot (sites 1 and 3) although a rarefaction analysis suggested at least two of these sites were not sampled sufficiently to characterize species richness. We detected a single C. crepuscularis positive for Onchocercidae gen. sp. DNA and another individual of the same species positive for Haemoproteus sacharovi DNA, yielding a 2.08% prevalence (n = 251) for both parasites in this species.
Conclusions
We extend the knowledge of the Culicoides spp. community in an urban environment of Texas, USA, and contribute to novel sequence data for these species. Additionally, the presence of parasite DNA (Onchocercidae gen. sp. and H. sacharovi) from C. crepuscularis suggests the potential for this species to be a vector of these parasites.
Electronic supplementary material
The online version of this article (10.1186/s13071-018-3283-9) contains supplementary material, which is available to authorized users.
Keywords: Biting midges, Avian hosts, Onchocercidae, Haemosporida, Vector-parasite association, Integrative taxonomy
Background
The family Ceratopogonidae (Diptera) includes the genus Culicoides, commonly known as biting midges or “no-see-ums”. Culicoides spp. are hematophagous pests and vectors of viruses, protozoans and filarial worms [1–3]. A major focus of research on Culicoides spp. is related to their roles as vectors of epizootic hemorrhagic disease virus (EHDV), bluetongue disease virus (BTV) and African horse sickness virus (AHSV) [3]. Beside their impact on livestock, Culicoides also transmit parasites to wildlife such as the avian trypanosome Trypanosoma bennetti [4] and Haemoproteus parasites [5, 6]; however, distributional patterns and infection prevalences of these parasites remain largely unknown.
One neglected area in the study of Culicoides spp. is their ecology in urban environments, where there is an interface with human populations. Prior studies have focused on specific species, such as pestiferous salt-marsh species [7] or species from specialized habitats such as zoos [8], and the vector-host-pathogen interactions in urban environments, could be of potential interest highlighting their potential as vectors of pathogens. Several species of Culicoides are known vectors of Haemosporida [9, 10], parasitic protozoans of amphibians, reptiles, birds and mammals [10]. These parasites can cause acute epizootic outbreaks that have severely altered avian communities by affecting long-term demographic processes such as reproductive rate and survivorship [10–12]. Culicoides are also involved in the transmission of filarial nematodes in the family Onchocercidae with infection report in the California quail, the American crow and the great-tailed grackle [13–15].
Recently, a high prevalence of haematozoan parasites was documented in a population of great-tailed grackles (Quiscalus mexicanus) in College Station, Texas, including Heamoproteus, Plasmodium, trypanosomes and filarial worms (Golnar et al., unpublished data). Previous work suggests that mosquitoes are not the vectors for these parasites and instead Culicoides are the more likely vectors [16]. Here, we document the Culicoides spp. inhabiting the urban environment of College Station, Texas, USA, and screen individuals for blood-borne parasites known to occur in local birds. The characterization of Culicoides species in this area where a high burden of avian parasites circulates provides an ideal opportunity to evaluate their potential role as vectors of haematozoan parasites.
Methods
Culicoides collection and identification
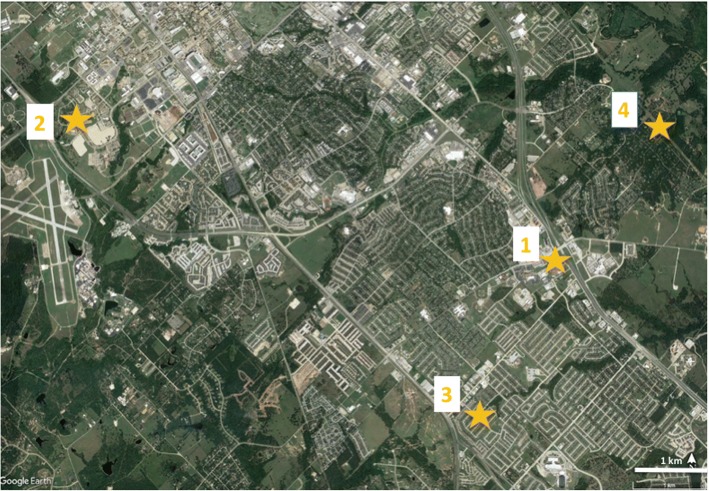
We collected Culicoides from College Station, Texas (30°37'40.7"N, 96°20'3.8"W) during the months of May and June, 2016. Centers for Disease Control and Prevention miniature light traps (CDC-LT, BioQuip model 2836BQ, with a 6 volt battery, USA) baited with 1.5 kg of dry ice were run from approximately 18:00 h to 8:00 h. Traps were placed in 4 localities: a grocery store parking lot (site 1), research park natural area (site 2), and two private homes in residential neighborhoods (site 3 and site 4) (Fig. 1). While site 1 and 3 are located in urban developed areas, sites 2 and 4 are located in urban natural environments with more vegetation in the surrounding landscape. In site 4, Culicoides traps were set in three locations, including one location next to a chicken coop. After collection, trap contents were chilled on ice for transport to the laboratory where specimens were identified to the species level based on wing pattern [17, 18] and stored at -20 °C until further processing.
Fig. 1.
Trapping sites across College Station, Texas, USA. Blue boxes and site numbers are locations where traps were set
We used an integrative taxonomic approach to identify Culicoides. Thirteen specimens were mounted on permanent slides to observe anatomic characteristics useful for species identification and were in parallel processed by a non-destructive DNA extraction method modified from the Gentra Puregene Kit (#D-5500A) (Gentra Systems, Inc., Minneapolis, USA). Whole specimens were added to individual Eppendorf tubes containing 100 μl of Cell Lysis Solution and 1 μl of Proteinase K and incubated overnight at 55 °C. Specimens were not crushed to preserve morphological features. The specimens were chilled at 0 °C for 20–30 min, after which 35 μl of 8.0 M ammonium acetate were added to each tube. The samples spun at 2200× rpm for 7 min in an Eppendorf 5424 Centrifuge. The supernatant was pipetted out of the tube and used to complete the DNA extraction protocol. Approximately 100 μl of 95% ethanol were added to the tube containing the exoskeleton to prevent further breakdown. The exoskeletons were separated into body regions (head, thorax, abdomen and wings) and transferred to 15.0% acetic acid for 10 min, 2-propanol for 10 min, and then 100.0% clove oil. Each body region of a single specimen was slide mounted in Canada balsam under its own coverslip. Slides were kept at 40 °C for 8–10 h and air dried for 72 h [19]. Slides were deposited in the Texas A&M University Entomology Museum under accession number 735. This method allowed for identification of specimens both molecularly and morphologically. Sequences of the cytochrome c subunit 1 (cox1) gene were obtained using the protocol described below.
All the remaining specimens (n = 238) were homogenized in a Mini-Beadbeater-96 (Bio Spec Products Inc., Bartlesville, OK, USA) with three 4 mm PYREX Solid Beads (Sigma-Aldrich, Inc., St. Louis, MO, USA) in 200 μl of Hank’s Buffer Salt Solution (ThermoFisher Scientific, Waltham, MA). DNA was extracted from 100 μl of the homogenized material (MagMAX CORE Nucleic Acid Purification Kit from Applied Biosystems, Foster City, CA). A PCR assay targeting a 710 bp region of the cox1 gene of invertebrates [20] was used for molecular identification of Culicoides species (Additional file 1: Table S1). The PCR consisted of 1.5 μl of genomic DNA, 0.5 μM of each primer, 12.5 μl of 1× Premix from the Epicentre Failsafe PCR purification kit (Epicentre Biotechnologies, Madison, WI, USA), and 1 unit of enzyme mix (total volume 25 μl). The thermal cycling profile consisted of denaturation at 95 °C for 3 min, followed by 35 cycles of 94 °C for 1 min, 45 °C for 1.5 min, 72 °C for 2 min, and a final extension step at 72 °C for 5 min. Amplicons were visualized on a 2% agarose gel. PCR products were purified using ExoSAP-IT PCR Product Cleanup (Affymetrix, Santa Clara, CA, USA) and bi-directional Sanger sequencing was performed (Eton Biosciences, San Diego, CA, USA). Forward and reverse sequence chromatographs were assessed for quality and sequences were aligned with sequences of Culicoides downloaded from NCBI’s GenBank database and analyzed using Geneious version 9.1.8 [21] using the Clustal-W method. Results from J-model test [22] indicated the best-fit selection model to run our analysis was GTR+G+I and therefore, maximum likelihood tree was constructed using Randomized Axelerated Maximum Likelihood (RAxML) with 1000 bootstrap replications. A phylogenetic tree was finalized using FigTree version 1.4.3 using a sequence of the cox1 gene of Atrichopogon levis (GenBank: KT092130.1) as the outgroup. In addition, fifteen previously published sequences of the cox1 gene of Culicoides species previously collected in Texas were included in this analysis (GenBank: KT794137.1; KT794138.1; KT794141.1-KT794144.1, KT794154.1; KT794155.1; KT794159.1; KT794161.1; KT794162.1; KT794164.1; KT794165.1; KT794167.1; KT794171.1) as well as 17 previously unpublished sequences (GenBank: MH751220, MH751222, MH751223, MH751224, MH751226, MH751227, MH751228, MH751235, MH751243, MH751244, MH751246, MH751248, MH751252, MH751258, MH751267, MH751273, MH751280) from specimens collected in Wisconsin, Wyoming and South Carolina and identified to species at the USDA-ARS Arthropod-Borne Animal Disease Research Unit in Manhattan, Kansas.
Parasite testing
Filarial nematodes PCR assays
DNA extracted from each specimen was screened for the presence of filarial nematodes using a PCR assay amplifying a 580 bp region of the filarial nematode 18S ribosomal gene (18S rRNA) ([23]; Additional file 1: Table S1). PCR cycling was performed at 94 °C for 2 min, 39 cycles of 94°C for 30 s, 57 °C for 30 s, 72 °C for 2 min, and a final extension step at 72 °C for 7 min. Samples that resulted in the successful amplification of the target pathogen were confirmed using a nested PCR targeting a 340 bp region of the cox1 gene ([16, 24, 25]; Additional file 1: Table S1). A touchdown cycling protocol was used and consisted of denaturation at 94 °C for 2 min; 8 cycles of 94 °C for 45 s, 51 °C for 45 s (reduced by 0.5 °C each cycle), and 72 °C for 1.5 min; followed by 25 cycles of 94 °C for 45 s, 45 °C for 45 s, and 72 °C for 1.5 min; and a final extension step at 72°C for 7 min. Amplicons were visualized and submitted for Sanger sequencing following the same protocol as above. Only those samples that resulted in amplification with the two PCR assays were considered positive in the determination of infection prevalence. When samples that screened positive using the 18S rRNA gene PCR assay could not be confirmed using the nested cox1 gene PCR, samples where re-extracted from the original homogenate and subjected a second time to the 18S rRNA gene PCR for confirmation.
Haemosporida PCR assays
Samples were screened for the presence of Haemosporida using a PCR assay targeting a 154 bp region of the 16S ribosomal gene (16S rRNA) ([26, 27]; Additional file 1: Table S1). Reactions contained 1.0 μl of genomic DNA,0.4 μM of each primer, 1× Premix from the Epicentre Failsafe PCR purification kit, and 1 unit of enzyme mix (10 μl total volume). Cycling conditions included 2 min at 94 °C; 35 cycles of 94 °C for 50 s, 55 °C for 50 s, and 72 °C for 25 s; and a final extension step at 72 °C for 2 min. Samples resulting in amplification were confirmed by a nested PCR assay targeting a 552 bp fragment of the cytochrome b gene [26, 28, 29, 38] (Additional file 1: Table S1). In this nested PCR assay, 1 μl of genomic DNA, 0.2 μM of each primer, 0.1 μg/μl BSA, 1 × Premix from the Epicentre Failsafe PCR purification kit, and 1 unit of enzyme mix was used. Cycling conditions were 4 min at 94 °C; 35 cycles of 94 °C for 20 s, 49 °C for 10 s, and 72 °C for 45 s; and a final extension step at 72 °C for 3 min for the outer reaction and 94 °C for 1 min; 28 cycles of 94 °C for 20 s, 52 °C for 10 s, and 68 °C for 50 s; and a final extension step at 72 °C for 7 min for the inner reaction. Amplicons were visualized and submitted for Sanger sequencing following the same protocol as above. Only those samples with sequences from both the screening and confirmatory assay were considered positive in the determination of infection prevalence.
Culicoides community composition
The community composition was compared for each site by reporting the species richness [30]. Rarefaction curves were created in R studio version 1.0.143 using the iNEXT package version 2.0.15 [31] to understand the weight of sampling size in the apparent species richness at each site.
Results
Culicoides species identification
A total of 251 Culicoides were collected among the four sites. Culicoides communities were characterized using three different approaches: integration of morphological and molecular characteristics, molecular characteristics only, and phylogenetic analysis (Fig. 2). Thirteen individuals were set aside for morphological identification coupled with molecular analysis. Using morphological identification, eight species were identified from the 13 samples: C. crepuscularis Malloch, 1915; C. neopulicaris Wirth, 1955; C. stellifer (Coquillett, 1901); C. haematopotus Malloch, 1915; C. sonorensis Wirth & Jones, 1957; C. paraensis (Goeldi, 1905); C edeni Wirth & Blanton, 1974; and C. arboricola Root & Hoffman, 1937. Characteristics wing markings can be seen in Additional file 2: Figure S1. Because we use a non-destructive dissection and extraction technique, we were able to recover DNA from these individuals that were then subject to our PCR assay targeting the cox1 gene. DNA sequences from specimens representative of these Culicoides species (Additional file 3: Figure S2) were used later in the phylogenetic analysis with the exception of C. arboricola from which no sequences were recovered.
Fig. 2.
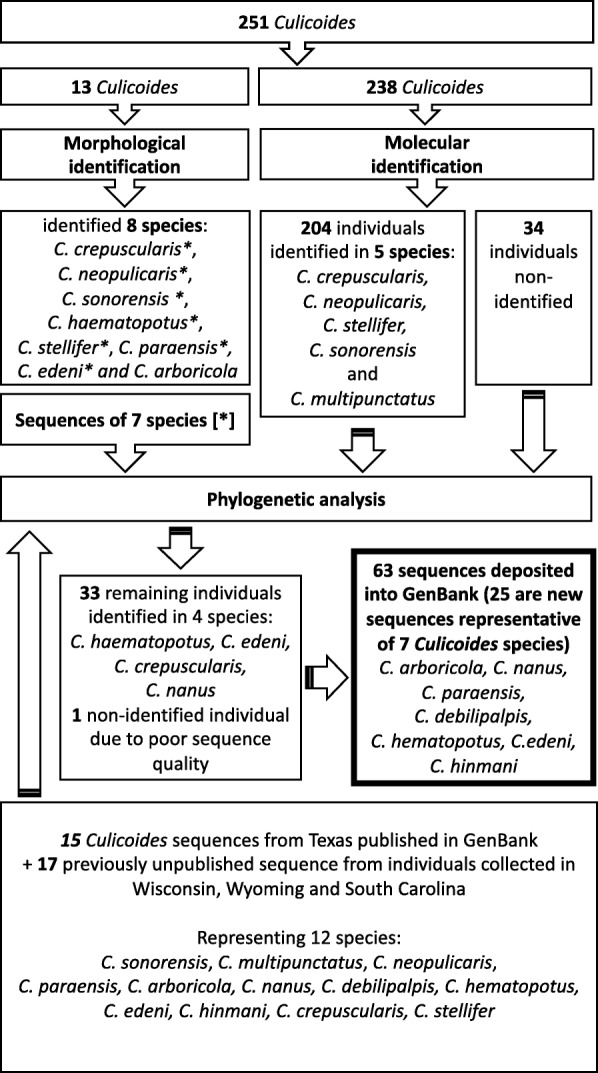
Flowchart of Culicoides morphological and molecular identification process
The 238 remaining samples were molecularly assessed, and we identified five species based on the cox1 gene. A BLAST search of NCBI GenBank database inferred the identity of the species as C. crepuscularis (n = 46, 99.2–100% identity), C. neopulicaris (n = 148, 99.2–100% identity), C. stellifer (n = 7, 95.6% identity), C. sonorensis (n = 1, 99.4% identity) and C. multipunctatus Malloch, 1915 (n = 1, 99.7% identity). Thirty-four sequences lacked sufficient sequence identity to infer species identification and one of the 34 sequences was removed from the analysis because of poor quality.
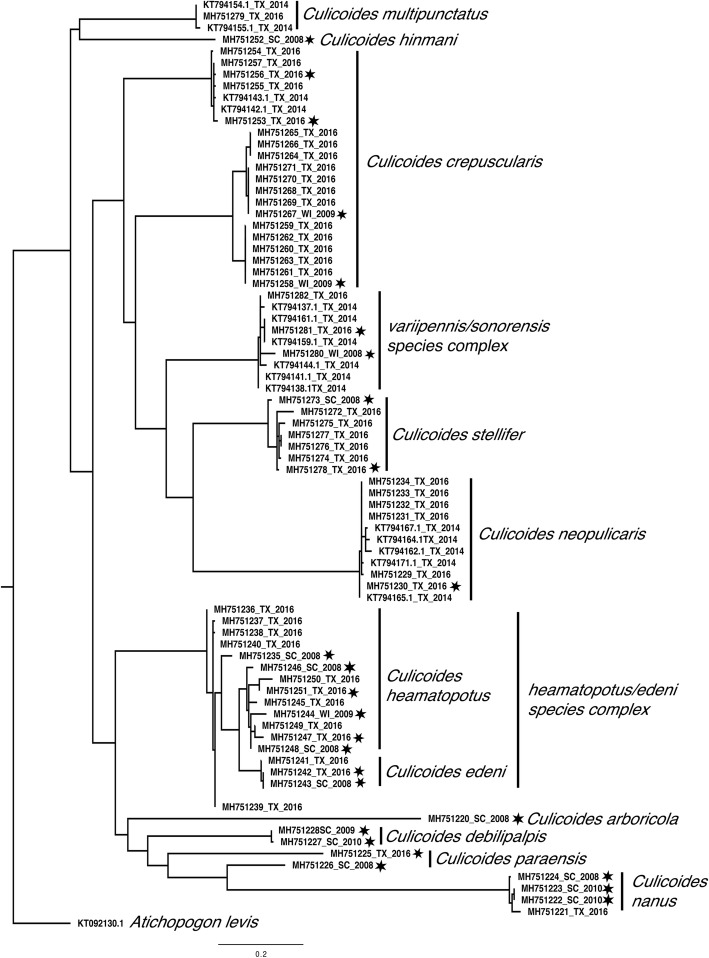
In order to identify the 33 remaining individuals, we performed a maximum likelihood analysis (Fig. 3) that included all the sequences obtained in this study 15 published sequences previously collected in Texas in 2015, and 17 otherwise unpublished North American Culicoides sequences from individuals collected between 2008 and 2010 in Wisconsin, Wyoming and South Carolina (Fig. 2) (GenBank: MH751220, MH751222-MH751224, MH751226-MH751228, MH751235, MH751243, MH751244, MH751246, MH751248, MH751252, MH751258, MH751267, MH751273, MH751280). All of our samples grouped in clades with known sequences and good bootstrap support, affording confidence of the species identity (Fig. 2). Two well-supported clades of C. crepuscularis (98% and 99% support) were observed, with one of them divided into two groups (82% and 95% support). Our sequences of C. sonorensis grouped together with C. variipennis and C. sonorensis with 99% support, but no further resolution between these species could be obtained using the cox1 gene. The sequences of C. stellifer grouped together in a well-supported clade (99% support). All C. neopulicaris grouped together with 100% support. Culicoides haematopotus and C. edeni were placed into a well-supported clade (96% support). Three groups were observed in this clade, with C. edeni (99% support), a group of eight C. haematopotus (90% support), and an unresolved second group of C. haematopotus (76% support). All C. nanus Root & Hoffman, 1937 grouped together in a single clade with 100% support, as did all C. multipunctatus (100% support). The C. paraensis collected from South Carolina did not group with the sequence of C. paraensis from this study. The 33 unknown sequences grouped within the well supported groups described above with one sequence attributed to C. edeni, 19 sequences to C. hematopotus and 13 sequences to C. crepuscularis. To simplify the phylogenetic tree, only a few representative of each species was mapped on the tree (Fig. 3).
Fig. 3.
Phylogenetic relationship between Culicoides species based on a 405 bp region of the cytochrome c oxidase subunit 1 gene. Culicoides sequences from individuals captured in Texas (GenBank: MH751225, MH751229-34, MH751236-42, MH751245, MH751247, MH751249-51, MH751253-57, MH751259-66, MH751268-72, MH751274-79, MH751281-82), Winsconsin, Wyoming and South Carolina (GenBank: MH751220, MH751222-24, MH751226-28, MH751235, MH751243, MH751244, MH751246, MH751248, MH751252, MH751258, MH751267, MH751273, MH751280) are listed and individually marked with a star to indicate sample for which morphological characterization and molecular sequence were performed. The bootstrap support values above < 90% are shown at the corresponding nodes. Additionally, publically available cox1 sequences from Texas were added to the phylogenetic tree (GenBank: KT794137.1; KT794138.1; KT794141.1; KT794142.1; KT794143.1; KT794144.1, KT794154.1; KT794155.1; KT794159.1; KT794161.1; KT794162.1; KT794164.1; KT794165.1; KT794167.1; KT7941371.1). The tree was rooted to the cox1 sequence of Atrichopogon levis (GenBank: KT092130.1)
Culicoides species community composition
Ten different species were present in the four trapping sites. The cumulative proportions of Culicoides collected from the four sites were principally C. neopulicaris (72.92%), C. crepuscularis (19.12%), C. haematopotus (3.98%) and C. stellifer (3.19%). On rare occasions (< 0.79%), C. multipunctatus, C. sonorensis, C. arboricola, C. paraensis, C. nanus and C. edeni were collected. Among individual sites (Table 1), species richness was the highest at site 2 with eight species. Culicoides haematopotus was the most abundant (28%), followed by C. neopulicaris (21%), C. crepuscularis (14%), C. stellifer (7%), C. sonorensis (7%), C. paraensis (7%), C. edeni (7%) and C. nanus (7%). Site 1 collections included C. crepuscularis (33.3%), C. neopulicaris (16.6%), C. haematopotus (16.6%), C. arboricola (16.6%) and C. sonorensis (16.6%). At site 4, the highest number of specimens were collected (n = 189) and represented five species (S = 5). Culicoides neopulicaris was the dominant species (73%) followed by C. crepuscularis (14%), C. haematopotus (8%), C. stellifer (4%), C. multipunctatus (1%) and C. edeni (1%). Site 3 had the lowest species richness (S = 3). The rarefaction analysis showed that the sampling effort in sites 3 and 4 was sufficient in explaining the richness of Culicoides present in these sites, whereas the sampling effort in sites 1 and 2 most likely resulted in an underestimation of the species richness (Fig. 4).
Table 1.
Culicoides species composition and richness in four sites in College Station, Texas, 2016
| Species | Site 1 n (%) |
Site 2 n (%) |
Site 3 n (%) |
Site 4 n (%) |
|---|---|---|---|---|
| C. paraensis b | 0 (0) | 1 (7) | 0 (0) | 0 (0) |
| C. edeni b,c | 0 (0) | 1 (7) | 0 (0) | 1 (1) |
| C. crepuscularis b,c | 2 (33) | 2 (14) | 33 (77) | 26 (14) |
| C. multipunctatus a | 0 (0) | 0 (0) | 0 (0) | 1 (1) |
| C. nanus b,c | 0 (0) | 1 (7) | 0 (0) | 0 (0) |
| C. neopulicaris b,c | 1 (16.6) | 3 (21) | 9 (21) | 138 (73) |
| C. sonorensis b | 1 (16.6) | 1 (7) | 0 (0) | 0 (0) |
| C. stellifer b | 0 (0) | 1 (7) | 0 (0) | 7 (4) |
| C. arboricola d | 1 (16.6) | 0 (0) | 0 (0) | 0 (0) |
| C. haematopotus b,c | 1 (16.6) | 4 (28) | 1 (2) | 16 (8) |
| No. of specimens trapped (N) | 6 | 14 | 43 | 189 |
| Species richness (S) | 5 | 8 | 3 | 5 |
aMolecular only
bMolecular and morphological
cUsing phylogenetic analysis
dMorphological only
Fig. 4.
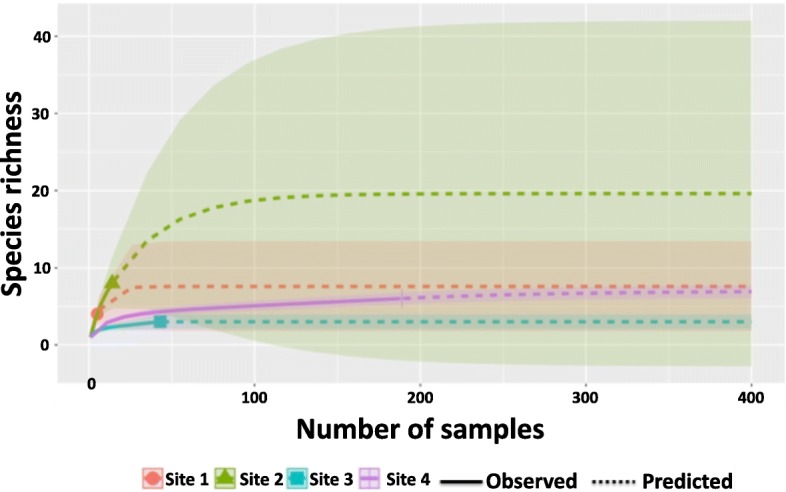
Rarefaction curves showing the influence of sample size on Culicoides species richness for each sampling site: Site 1 in red, Site 2 in green, Site 3 in blue and Site 4 in purple. Solid lines indicate the observed sample size and species richness whereas dashed lines represent the predicted sample size and species richness
Haemoparasite testing
One C. crepuscularis individual from site 3 tested positive for filarial nematodes with a sequence 100% similar to Onchocercidae gen. sp. isolated from common grackles (Q. quiscala) in Chicago (GenBank: JQ867040) [32]. The prevalence of this parasite in C. crepuscularis was 2.1%. One C. crepuscularis individual from site 3 tested positive for Haemosporida with a sequence 100% similar to Haemoproteus sacharovi isolated from a mourning dove in Arizona, USA (GenBank: KY653811, lineage hMODO1). Similarly, its prevalence in C. crepuscularis was 2.1%.
Discussion
Among the 151 Culicoides spp. described from the Nearctic region, north of Mexico, 40 are reported in Texas, USA [33–35]. This study documents the presence of ten Culicoides spp. in an urban region of east central Texas during summer 2016. The dominant species in our urban study sites were C. neopulicaris, C. crepuscularis, C. stellifer and C. haematopotus. This study illustrates the benefit of integrated morphological and molecular systematics for accurate characterization of Culicoides communities [36]. All Culicoides spp. reported in this study, other than C. edeni, are known to occur in Texas (Shuts PT, unpublished data; [17]) and many species were likely undetected due to different factors including the short sampling period, small sample area, limited habitat coverage and the collection method.
Culicoides neopulicaris has been recorded previously in Texas and Louisiana [34], but very little is known about the larval habitats, feeding behavior and vector status of this species. Culicoides crepuscularis, the second most abundant species in this study, is one of the most abundant species in North America [34, 37]. Here we document C. crepuscularis infected with Haemoproteus DNA and filarial nematode DNA. Culicoides crepuscularis is a known vector of H. danilewski and H. fringillae found in Passeriformes [38, 39]. The cycle of transmission of H. sacharovi involves the hippoboscid fly Pseudolynchia canariensis as a vector [40]. However, Culicoides have also been suggested as potential vectors [41], and our positive test of a single individual shows that there is vector-parasite contact. We also found evidence of infection with a filarial nematode from the family Onchocercidae in C. crepuscularis. Our sequence shares 100% homology with a sequence of Chandlerella quiscali previously isolated from a common grackle (Q. quiscala) in Chicago. The common grackle was reported infected by Ch. quiscali with members of the family Ceratopogonidae suggested as vectors [14]. Additionally, C. crepuscularis is a known vector of filarial nematodes including Ch. quiscali [14], Eufilaria and Splendidofilaria picacardina longicaudata (Hibler C, unpublished data). Based on abundance and a clear capacity to transmit a number of parasites, future studies should focus on the role of C. crepuscularis in disease transmission in urban systems. Our results also report several C. stellifer, a widespread species throughout most of the USA, previously recorded infected with BTV [42], vesicular stomatitis virus [43] and West Nile virus [44]. Additionally, C. haematopotus was present at certain sites in College Station, Texas. This species is primarily ornithophilic but has been reported to also feed on some mammals [17, 37, 44–46] and can transmit multiple parasites including Ch. quiscala [14] and Ch. striatospicula as well as E. longicaudata (Hibler C, unpublished data) and H. meleagridis [47].
Our study contributes 63 sequences for both under-represented species in GenBank including C. neopulicaris, C. crepuscularis, C. stellifer, C. multipunctatus, C. sonorensis and seven species unrepresented until our study in GenBank: C. arboricola, C. nanus, C. debilipalpis Lutz, 1913, C. paraensis, C. haematopotus, C.edeni and C. hinmani Khalaf, 1952. Our phylogenetic analysis based on the cox1 gene revealed well-supported clusters which were in agreement with the morphological determination with the exception of C. multipunctatus for which no morphological identification was performed. While most of the species characterized fall into distinct clades (e.g. C. stelliffer, C. neopulicaris, C. nanus, C. multipunctatus and C. paraensis), C. crepuscularis fell into two distinct clades, one of which was divided into two distinct groups. This observation suggests the presence of cryptic species within C. crepuscularis. Like many studies, our molecular analysis grouped C. variipennis and C. sonorensis into a single clade. Morphologically these two species are distinguishable [48], but the molecular analysis using the cox1 marker groups them in a single clade (Shults PT, unpublished data; [49]). Culicoides sonorensis is a known vector of BTV, EHDV and Main Drain virus [50–54]. Using molecular and morphological characteristics, we were able to separate C. edeni (n = 2) from C. haematopotus (n = 14) [37] (Fig. 3). Culicoides edeni is a species of interest due to its involvement in the transmission of H. danilewskyi in blue jays [55]. To our knowledge, this study represents the first record of C. edeni in Texas. Prior to this study, C. edeni was known from The Bahamas, Florida, and South Carolina (Swanson DA, unpublished data; [37]). Due to the morphological similarities, multiple genetic clades, and disjointed distribution, there is potential for undescribed cryptic species to exist within a C. edeni-C. haematopotus complex; species for which we know little from an epidemiological standpoint.
Among the other species found in our study, two are known vectors of pathogens. Culicoides paraensis is a species that preferentially breeds in tree holes [56] and feeds on a variety of birds and mammals, including humans [45, 56–58], and are known vector for Mansonella ozzardi [59] and Oropouche orthobunyavirus [60] in South America and the Caribbean. This is only the second record of C. paraensis in Texas. Our sequence of C. paraensis did not group with C. paraensis from South Carolina (82.28% nucleotide identity) highlighting possible cryptic species diversity, variation in cox1 sequences due to geographical location or a potential misidentification that will require further investigation. However, this situation exemplifies the need for integrated taxonomic approaches to species identification. The sequences of both of these specimens were obtained with non-destructive methods, and therefore reexamination of these two individuals is possible.
Culicoides multipunctatus is common and abundant throughout central and south Texas (Shults PT, unpublished data; Schoenthal C, unpublished data; [17]). Its rarity in this study could offer insight into the seasonal distribution of this species as well as the unsuitability of the habitat. Finally, we detected C. nanus, a species that was previously recorded in Texas [17, 61] but nothing is known of the biting records or vector capacity of this species [37].
Conclusions
We document the abundance of ten Culicoides spp. in an urban environment in Texas, USA. The identification of these species was enhanced by an integrative taxonomic approach, without which some species could have been misidentified when using morphological or molecular techniques only. Molecular barcoding sequences were submitted to GenBank for all species, including seven of which did not have previous sequence data in the GenBank database. One species, C. crepuscularis, was found to be positive for Onchocercidae gen. sp. and Haemoproteus sacharovi, supporting that they may be involved in the transmission of these avian parasites. Further investigations on vector competence of C. crepuscularis with H. sacharovi and filarial nematodes, is needed to support for this hypothesis.
Additional files
Table S1. Primer sequences for PCR. (DOCX 29 kb)
Figure S1. Wing markings of the eight morphologically identified Culicoides species collected in College Station. A, C. arboricola; B, C. crepuscularis; C, C. edeni; D, C. haematopotus; E, C. neopulicaris; F, C. paraensis; G, C. sonorensis; H, C. stellifer. (TIF 15482 kb)
Figure S2. Chromatogram presenting the sequence of cox1 gene for each of the seven Culicoides species collected in College Station and morphologically identified: A, C. crepuscularis; B, C. edeni; C, C. haematopotus; D, C. neopulicaris; E, C. paraensis; F, C. sonorensis; G, C. stellifer. On the left panel are the chromatographs representing sequence generated with the forward primer and on the right panel the chromatographs associated with the sequence generated with the reverse sequence. (TIFF 36312 kb)
Acknowledgements
We thank Ed Vargo for the use of laboratory space and equipment.
Funding
This study was funded by Texas A&M University teaching enhancement funds in the Department of Veterinary Integrative Biosciences, the Department of Entomology, Urban Entomology Endowment Fund, and the Montague-Center for Teaching Excellence award to SAH. The open access publishing fees for the article were covered by the Texas A&M University Open Access to Knowledge Fund (OAKFund), supported by the University Libraries and the Office of the Vice President for Research.
Availability of data and materials
Not applicable.
Abbreviations
- BTV
Bluetongue virus
- cox1
cytochrome c oxidase subunit 1
- EHD
Epizootic hemorrhagic disease
Authors’ contributions
MCIM, SCH and GLH design the research, EC, AG, JB, DK, PS, SP and CC performed field work, EC, AG, JB, DK, PS, SP, CC and EM processed the Culicoides for molecular identification. PS, DAS and EC processed Culicoides for the paired morphological and molecular identification. EM processed Culicoides for parasite identification. EM analyzed the data. EM, PS and AG wrote the manuscript. All authors revised the manuscript and read and approved the final draft.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Estelle Martin, Email: estellemartin@tamu.edu.
Elaine Chu, Email: elaine94@tamu.edu.
Phillip Shults, Email: ptshults@tamu.edu.
Andrew Golnar, Email: agolnar@tamu.edu.
Dustin A. Swanson, Email: Dustin.Swanson@ars.usda.gov
Jamie Benn, Email: jbenn@cvm.tamu.edu.
Dongmin Kim, Email: dongminkimkorea@gmail.com.
Peter Schneider, Email: pjs1991@tamu.edu.
Samantha Pena, Email: spena15@tamu.edu.
Cassie Culver, Email: cassiec2013@tamu.edu.
Matthew C. I. Medeiros, Email: mcmedeir@hawaii.edu
Sarah A. Hamer, Email: shamer@cvm.tamu.edu
Gabriel L. Hamer, Email: ghamer@tamu.edu
References
- 1.Mellor PS, Boorman J, Baylis M. Culicoides biting midges: their role as arbovirus vectors. Annu Rev Entomol. 2000;45:307–340. doi: 10.1146/annurev.ento.45.1.307. [DOI] [PubMed] [Google Scholar]
- 2.Carpenter S, Groschup MH, Garros C, Felippe-Bauer ML, Purse BV. Culicoides biting midges, arboviruses and public health in Europe. Antiviral Res. 2013;100:102–113. doi: 10.1016/j.antiviral.2013.07.020. [DOI] [PubMed] [Google Scholar]
- 3.Borkent A. The biting midges, the Ceratopogonidae (Diptera). In: Marquardt W, editor. Biology of Disease Vectors, 2nd edn. Cambridge: Academic Press; 2004. p. 113–26.
- 4.Svobodová M, Dolnik OV, Čepička I, Rádrová J. Biting midges (Ceratopogonidae) as vectors of avian trypanosomes. Parasit Vectors. 2017;10:224. doi: 10.1186/s13071-017-2158-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bukauskaitė D, Žiegytė R, Palinauskas V, Iezhova TA, Dimitrov D, Ilgūnas M, et al. Biting midges (Culicoides, Diptera) transmit Haemoproteus parasites of owls: evidence from sporogony and molecular phylogeny. Parasit Vectors. 2015;8:303. doi: 10.1186/s13071-015-0910-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Atkinson CT, Forrester DJ, Greiner EC. Epizootiology of Haemoproteus meleagridis (Protozoa: Haemosporina) in Florida: seasonal transmission and vector abundance. J Med Entomol. 1988;25:45–51. doi: 10.1093/jmedent/25.1.45. [DOI] [PubMed] [Google Scholar]
- 7.Kline DL. Seasonal abundance of adult Culicoides spp. (Diptera: Ceratopogonidae) in a salt marsh in Florida, USA. J Med Entomol. 1986;23:16–22. doi: 10.1093/jmedent/23.1.16. [DOI] [PubMed] [Google Scholar]
- 8.Nelder MP, Swanson DA, Adler PH, Grogan WL., Jr Biting midges of the genus Culicoides in South Carolina zoos. J Insect Sci. 2010;10:55. doi: 10.1673/031.010.5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Atkinson CT, Thomas NJ, Hunter DB. Parasitic Diseases of Wild Birds. Ames: Wiley-Blackwell; 2009. [Google Scholar]
- 10.Valkiunas G. Avian Malaria Parasites and Other Haemosporidia. Florida: CRC Press; 2004. [Google Scholar]
- 11.Bennett GF, Peirce MA, Ashford RW. Avian Haematozoa: mortality and pathogenicity. J Nat Hist. 1993;27:993–1001. doi: 10.1080/00222939300770621. [DOI] [Google Scholar]
- 12.van Riper C, van Riper SG, Goff ML, Laird M. The epizootiology and ecological significance of malaria in Hawaiian land bird. Ecol Monogr. 1986;56:327–344. doi: 10.2307/1942550. [DOI] [Google Scholar]
- 13.Atchley WR, Wirth WW. Two new western Culicoides (Diptera: Ceratopogonidae) which are vectors of filaria in the California valley quail. Can J Zool. 1975;53:1421–1423. doi: 10.1139/z75-171. [DOI] [PubMed] [Google Scholar]
- 14.Robinson EJ., Jr Culicoides crepuscularis (Malloch) (Diptera: Ceratopogonidae) as a host for Chandlerella quiscali (von Linstow, 1904) comb. n. (Filarioidea: Onchocercidae) J Parasitol. 1971;57:772–776. doi: 10.2307/3277795. [DOI] [PubMed] [Google Scholar]
- 15.Bartlett C, Anderson R. Filaroid nematodes (Filarioidea: Onchocercidae) of Corvus brachynrhynchos Brehm in southern Ontario, Canada and a consideration of the epizootiology of avian filariasis. Syst Parasitol. 1980;2:77–102. doi: 10.1007/BF00015096. [DOI] [Google Scholar]
- 16.Boothe E, Medeiros MC, Kitron UD, Brawn JD, Ruiz MO, Goldberg TL, et al. Identification of avian and hemoparasite DNA in blood-engorged abdomens of Culex pipiens (Diptera; Culicidae) from a West Nile virus epidemic region in suburban Chicago, Illinois. J Med Entomol. 2015;52:461–468. doi: 10.1093/jme/tjv029. [DOI] [PubMed] [Google Scholar]
- 17.Wirth WW, Bottimer LJ. A population study of the Culicoides midges of the Edwards plateau region of Texas. Mosq News. 1956;16:256–266. [Google Scholar]
- 18.Wirth WW, Dyce AL, Peterson BV. An atlas of wing photographs, with a summary of the numerical characters of the Nearctic species of Culicoides (Diptera: Ceratopogonidae) Contrib Am Entomol Inst. 1985;22:1–46. [Google Scholar]
- 19.Borkent A, Spinelli GR. Neotropical Ceratopogonidae (Diptera: Insecta) Rev Soc Entomol Argent. 2007;4:198. [Google Scholar]
- 20.Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. 1994;3:294–299. [PubMed] [Google Scholar]
- 21.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, et al. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Posada D. jModelTest: phylogenetic model averaging. Mol Biol Evol. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- 23.Hamer GL, Anderson TK, Berry GE, Makohon-Moore AP, Crafton JC, Brawn JD, et al. Prevalence of filarioid nematodes and trypanosomes in American robins and house sparrows, Chicago USA. Int Parasitol Parasites Wildl. 2013:42–9. [DOI] [PMC free article] [PubMed]
- 24.Bataille A, Fournie G, Cruz M, Cedeno V, Parker PG, Cunningham AA, Goodman SJ. Host selection and parasite infection in Aedes taeniorhynchus, endemic disease vector in the Galapagos Islands. Infect Genet Evol. 2012;12:1831–1841. doi: 10.1016/j.meegid.2012.07.019. [DOI] [PubMed] [Google Scholar]
- 25.Casiraghi M, Anderson TJ, Bandi C, Bazzocchi C, Genchi C. A phylogenetic analysis of filarial nematodes: comparison with the phylogeny of Wolbachia endosymbionts. Parasitology. 2001;122:93–103. doi: 10.1017/S0031182000007149. [DOI] [PubMed] [Google Scholar]
- 26.Fecchio A, Lima MR, Svensson-Coelho M, Marini MA, Ricklefs RE. Structure and organization of an avian haemosporidian assemblage in a Neotropical savanna in Brazil. Parasitology. 2013;140:181–192. doi: 10.1017/S0031182012001412. [DOI] [PubMed] [Google Scholar]
- 27.Fallon SM, Ricklefs RE, Swanson BL, Bermingham E. Detecting avian malaria: an improved polymerase chain reaction diagnostic. J Parasitol. 2003;89:1044–1047. doi: 10.1645/GE-3157. [DOI] [PubMed] [Google Scholar]
- 28.Perkins SL, Schall JJ. A molecular phylogeny of malarial parasites recovered from cytochrome b gene sequences. J Parasitol. 2002;88:972–978. doi: 10.1645/0022-3395(2002)088[0972:AMPOMP]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 29.Ricklefs RE, Swanson BL, Fallon SM, Martínez-Abraín A, Scheuerlein A, Gray J, Latta SC. Community relationships of avian malaria parasites in southern Missouri. 2005;75:543–59.
- 30.Magurran A. Ecological diversity and its measurement. Princeton: Princeton University Press; 1988. [Google Scholar]
- 31.Hsieh TC, Ma KH, Chao A. iNEXT: iNterpolation and EXTrapolation for species diversity. R package version 2.0.12 edn 2016. http://chao.stat.nthu.edu.tw/blog/software-download
- 32.Greiner EC, Bennett GF, White EM, Coombs RF. Distribution of the avian hematozoa of North America. Can J Zool. 1975;53:1762–1787. doi: 10.1139/z75-211. [DOI] [PubMed] [Google Scholar]
- 33.Fox I. A catalogue of the bloodsucking midges of the Americas (Culicoides, Leptoconops and Lasiohelea) with keys to the subgenera and Nearctic species, a geographic index and bibliography. J Agric Univ Puerto Rico. 1955;39:214–285. [Google Scholar]
- 34.Borkent A, Grogan WL., Jr Catalog of the New World biting midges north of Mexico (Diptera: Ceratopogonidae) Zootaxa. 2009;2273:515861. [Google Scholar]
- 35.Grogan WL, Jr, Lysyk T. A revision of the biting midges in the Culicoides (Monoculicoides) nubeculosus-stigma complex in North America with the description of a new species (Diptera: Ceratopogonidae) Insecta Mundi. 2015;441:1–24. [Google Scholar]
- 36.Valkiūnas G. Haemosporidian vector research: marriage of molecular and microscopical approaches is essential. Mol Ecol. 2011;20:3084–3086. doi: 10.1111/j.1365-294X.2011.05187.x. [DOI] [PubMed] [Google Scholar]
- 37.Blanton FS, Wirth WW. Arthropods of Florida and neighbouring land areas. The sand flies (Culicoides) of Florida (Diptera: Ceratopogonidae), vol. 10. Gainesville: Florida Department of Agriculture and Consumer Services, Division of Plant Industry; 1979. [Google Scholar]
- 38.Fallis AM, Bennett GF. Sporogony of Leucocytozoon and Haemoproteus in simuliids and ceratopogonids and a revised classification of the haemosporidiida. Can J Zool. 1961;39:215–228. doi: 10.1139/z61-026. [DOI] [Google Scholar]
- 39.Fallis AM, Bennett GF. Description of Haemoproteus canachites n. sp. (Sporozoa: Haemoproteidae) and sporogony in Culicoides (Diptera: Ceratopogonidae) Can J Zool. 1960;38:455–464. doi: 10.1139/z60-049. [DOI] [Google Scholar]
- 40.Huff CG. Studies on Haemoproteus of mourning doves. Am J Epidemiol. 1932;16:618–623. doi: 10.1093/oxfordjournals.aje.a117879. [DOI] [Google Scholar]
- 41.Bennett GF, Peirce MA. The haemoproteid parasites of the pigeons and doves (family Columbidae) J Nat Hist. 1990;24:311–325. doi: 10.1080/00222939000770231. [DOI] [Google Scholar]
- 42.Mullen GR. Biting midges (Ceratopogonidae). Med Vet Entomol. 2002:163–83.
- 43.Walton TE, Webb PA, Kramer WL, Smith GC, Davis T, Holbrook FR, et al. Epizootic vesicular stomatitis in Colorado, 1982: epidemiologic and entomologic studies. Am J Trop Med Hyg. 1987;36:166–176. doi: 10.4269/ajtmh.1987.36.166. [DOI] [PubMed] [Google Scholar]
- 44.Sabio IJ, Mackay AJ, Roy A, Foil LD. Detection of West Nile virus RNA in pools of three species of ceratopogonids (Diptera: Ceratopogonidae) collected in Louisiana. J Med Entomol. 2006;43:1020–1022. doi: 10.1603/0022-2585(2006)43[1020:downvr]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 45.Hair JA, Turner EC., Jr Preliminary host preference studies on Virginia Culicoides (Diptera: Ceratopogonidae) Mosq News. 1968;28:103–107. [Google Scholar]
- 46.Vigil S, Grogan WL Jr, Wlodkowski JC, Parris J, Edwards de Vargas S, Shaw D, et al. New records of biting midges of the genus Culicoides Latreille from the southeastern United States (Diptera: Ceratopogonidae). Gainesville: FL Center for Systematic Entomology Inc.; 2014;0394.
- 47.Atkinson CT. Epizootiology of Haemoproteus meleagridis (Protozoa: Haemosporina) in Florida: potential vectors and prevalence in naturally infected Culicoides (Diptera: Ceratopogonidae) J Med Entomol. 1988;25:39–44. doi: 10.1093/jmedent/25.1.39. [DOI] [PubMed] [Google Scholar]
- 48.Holbrook FR, Tabachnick WJ, Schmidtmann ET, McKinnon CN, Bobian RJ, Grogan WL., Jr Sympatry in the Culicoides variipennis complex (Diptera: Ceratopogonidae): a taxonomic reassessment. J Med Entomol. 2000;37:65–76. doi: 10.1603/0022-2585-37.1.65. [DOI] [PubMed] [Google Scholar]
- 49.Jewiss-Gaines A, Barelli L, Hunter FF. First records of Culicoides sonorensis (Diptera: Ceratopogonidae), a known vector of bluetongue virus, in southern Ontario. J Med Entomol. 2017;54:757–762. doi: 10.1093/jme/tjw215. [DOI] [PubMed] [Google Scholar]
- 50.Ruder MG, Lysyk TJ, Stallknecht DE, Foil LD, Johnson DJ, Chase CC, et al. Transmission and epidemiology of bluetongue and epizootic hemorrhagic disease in North America: current perspectives, research gaps, and future directions. Vector Borne Zoonotic Dis. 2015;15:348–363. doi: 10.1089/vbz.2014.1703. [DOI] [PubMed] [Google Scholar]
- 51.Tabachnick WJ. Culicoides variipennis and bluetongue-virus epidemiology in the United States. Annu Rev Entomol. 1996;41:23–43. doi: 10.1146/annurev.en.41.010196.000323. [DOI] [PubMed] [Google Scholar]
- 52.Foster NM, Breckon RD, Luedke AJ, Jones RH, Metcalf HE. Transmission of two strains of epizootic hemorrhagic disease virus in deer by Culicoides variipennis. J Wildl Dis. 1977;13:9–16. doi: 10.7589/0090-3558-13.1.9. [DOI] [PubMed] [Google Scholar]
- 53.Jones RH, Roughton RD, Foster NM, Bando BM. Culicoides, the vector of epizootic hemorrhagic disease in white-tailed deer in Kentucky in 1971. J Wildl Dis. 1977;13:2–8. doi: 10.7589/0090-3558-13.1.2. [DOI] [PubMed] [Google Scholar]
- 54.Mellor PS, Boorman J, Loke R. The multiplication of Main Drain Virus in two species of Culicoides (Diptera, Ceratopogonidae) Arch Gesamte Virusforsch. 1974;46:105–110. doi: 10.1007/BF01240210. [DOI] [PubMed] [Google Scholar]
- 55.Garvin MC, Greiner EC. Ecology of Culicoides (Diptera: Ceratopogonidae) in southcentral Florida and experimental Culicoides vectors of the avian hematozoan Haemoproteus danilewskyi Kruse. J Wildl Dis. 2003;39:170–178. doi: 10.7589/0090-3558-39.1.170. [DOI] [PubMed] [Google Scholar]
- 56.Snow WE, Pickard E, Moore JB. The Heleidae of the Tennessee River Basin. J Tenn Acad Sci. 1957;32:18–36. [Google Scholar]
- 57.Humphreys JG, Turner EC., Jr Blood-feeding activity of female Culicoides (Diptera: Ceratopogonidae) J Med Entomol. 1973;10:79–83. doi: 10.1093/jmedent/10.1.79. [DOI] [PubMed] [Google Scholar]
- 58.Snow WE. Feeding activities of some blood-sucking Diptera with reference to vertical distribution in bottomland forest. Ann Entomol Soc Am. 1955;48:512–521. doi: 10.1093/aesa/48.6.512. [DOI] [Google Scholar]
- 59.Veggiani Aybar CA, Dantur Jury MJ, Lizarralde De Grosso MS, Spinelli GR. Spatial and temporal distribution of Culicoides insignis and Culicoides paraensis in the subtropical mountain forest of Tucuman, northwestern Argentina. Fla Entomol. 2011;94:1018–1025. doi: 10.1653/024.094.0440. [DOI] [Google Scholar]
- 60.Pinheiro FP, Travassos da Rosa AP, Travassos da Rosa JF, Ishak R, Freitas RB, Gomes ML, et al. Oropouche virus. I. A review of clinical, epidemiological, and ecological findings. Am J Trop Med Hyg. 1981;30:149–160. doi: 10.4269/ajtmh.1981.30.149. [DOI] [PubMed] [Google Scholar]
- 61.Jones RH. Observations on the larval habitats of some North American species of Culicoides (Diptera: Ceratopogonidae) Ann Entomol Soc Am. 1961;54:702–710. doi: 10.1093/aesa/54.5.702. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Primer sequences for PCR. (DOCX 29 kb)
Figure S1. Wing markings of the eight morphologically identified Culicoides species collected in College Station. A, C. arboricola; B, C. crepuscularis; C, C. edeni; D, C. haematopotus; E, C. neopulicaris; F, C. paraensis; G, C. sonorensis; H, C. stellifer. (TIF 15482 kb)
Figure S2. Chromatogram presenting the sequence of cox1 gene for each of the seven Culicoides species collected in College Station and morphologically identified: A, C. crepuscularis; B, C. edeni; C, C. haematopotus; D, C. neopulicaris; E, C. paraensis; F, C. sonorensis; G, C. stellifer. On the left panel are the chromatographs representing sequence generated with the forward primer and on the right panel the chromatographs associated with the sequence generated with the reverse sequence. (TIFF 36312 kb)
Data Availability Statement
Not applicable.