Abstract
Short sleep duration has been linked to negative health effects, but is a complex phenotype with many contributing factors, including genetic. We evaluated 27 common single nucleotide polymorphisms (SNPs) in candidate genes previously reported to be associated with other sleep variables for association with self-reported habitual sleep duration in the UK Biobank in 111 975 individuals of European ancestry. Genetic variation in DAT1 (rs464049) was significantly associated with sleep duration after correction for multiple testing (p = 4.00 × 10−5), whereas SNPs correlated to a previously studied variable number tandem repeat (VNTR) in DAT1 were not significant in this population. We also replicated a previously reported association in DRD2. Independent replication of these associations and a second signal in DRD2 (rs11214607) was observed in an additional 261 870 participants of European ancestry from the UK Biobank. Meta-analysis confirmed genome-wide significant association of DAT1 rs464049 (G, beta [standard error, SE] = −0.96 [0.18] minutes/allele, p = 5.71 × 10−10) and study-wide significant association of DRD2 (rs17601612, C, beta [SE] = −0.66 [0.18] minutes/allele, p = 1.77 × 10−5; rs11214607, C, beta [SE] = 1.08 (0.24) minutes/allele, p = 1.39 × 10−6). Overall, SNPs in two dopamine-related genes were significantly associated with sleep duration, highlighting the important link of the dopamine system with adult sleep duration in humans.
Keywords: single nucleotide polymorphism, dopamine, dopamine receptors, sleep, genetic association study
Statement of Significance
Short sleep duration is known to negatively affect human health. Inter-individual variation in sleep duration has a genetic component. Genetic variation in the dopamine transporter Dopamine Transporter 1 (Solute Carrier Family 6 Member 3) (DAT1 (SLC6A3)) has been shown to be associated with sleep irregularity in humans and with sleep duration in Drosophila. This study demonstrates a significant link between DAT1 polymorphisms and self-reported sleep duration in humans. Additionally, although variation in DRD2, a dopamine receptor, has been shown to be associated with sleep duration in humans in a moderately sized dataset and confirmed in this study, this result requires further validation. This study’s results reinforce the role of dopamine in influencing sleep and arousal. These data serve as a basis for further studies on the functional mechanisms of dopamine in regulating sleep.
Introduction
Chronic sleep deprivation, which is increasingly prevalent in the United States [1], has been linked to negative effects on memory, attention, and executive function and causes changes in brain architecture [2, 3]. Therefore the factors that influence short sleep are important to investigate.
Sleep duration is a complex behavioral phenotype influenced by numerous exogenous factors, such as work schedule, social demands, personality, and biological factors. Sleep duration is heritable, with heritability estimates of 15%–40% [4–6]. The goal of this study was to test if there is an association between common polymorphisms in candidate genes previously implicated in sleep systems and sleep duration in the UK Biobank. The outcomes of this study will inform future research on sleep and health.
In this study, a set of 27 common single nucleotide polymorphisms (SNPs; >20% minor allele frequency) from 20 candidate genes were surveyed for association with sleep duration. They were chosen from a broad range of sources in an attempt to cover the diversity of genes that may contribute to sleep. Common variants in CLOCK, Neuronal PAS Domain Protein 2 (NPAS2), and Cryptochrome Circadian Regulator 1 (CRY1) were chosen because they are molecular components of the mammalian circadian clock [7]. Adenosine A1 receptor (ADORA1) and Adenosine A2a receptor (ADORA2a) were chosen because of their role in maintaining the homeostatic sleep drive, as they are adenosine receptors, and the activation of ADORA1 and ADORA2a leads to an increase in somnolence [8]. Brain derived neurotrophic factor (BDNF) is an important factor in determining neural development and genetic variation in BDNF has been previously linked to inter-individual differences in EEG patterns during sleep [9].
Additionally, dopamine-related genes were included (tyrosine hydroxylase [TH], catechol-O-methyltransferase [COMT], dopamine receptor D1 [DRD1], dopamine receptor D2 [DRD2], Vesicle Monoamine Transporter Type 2 [VMAT2], and DAT1), given that a subset of dopaminergic neurons in humans are integral in sleep/wake, and dopaminergic drugs affect wakefulness in a dose-dependent manner [10]. Genetic variation in the dopamine receptor DRD2 has strong associations with sleep duration [11]. Also, a recent study reported that variation in the dopamine transporter DAT1 is associated with reported daytime sleepiness in humans [12]. TH is involved in the synthesis of dopamine and COMT in its degradation, and variation in COMT has been linked to sleep irregularity as well as inter-individual differences in EEG patterns [9, 12]. However, these previous studies were performed in smaller datasets, and need further validation in independent samples.
In this study, the association between sleep duration and common genetic variation in candidate genes from pathways previously implicated in sleep was investigated in a large dataset of 111 975 individuals. A follow-up study of DAT1 and DRD2 variants was performed in an additional cohort of 261 840 individuals. We found an association between sleep duration and SNPs located in DAT1 and DRD2.
Methods
UK Biobank subject selection
In the United Kingdom, ~500 000 people between the ages of 40–69 were recruited by UK Biobank researchers to provide phenotypic and genotypic data in order to create a large database that will be a resource for health researchers [13]. Sample size in the interim genotype release UK Biobank dataset (UKB1), after excluding relatives, individuals of non-European genetic ancestry, and those individuals with no or poor genetic data or those who reported use of sleep medication or shift work, was 111 975 subjects (Table 1) [14].
Table 1.
UK Biobank dataset statistics
| Characteristic | UKB1 N = 111 975 | UKB2 N = 261 840 |
|---|---|---|
| Sex (M), N (%) | 52 817 (47%) | 136 939 (52%) |
| Age, years, Mean (SD) | 57.69 (7.90) | 56.71 (8.14) |
| Sleep duration, hours, Mean (SD) | 7.19 (1.08) | 7.14 (1.09) |
Descriptive statistics on sleep and demography in the two subsets of the UK Biobank sample used in the current study.
Recently, genotype data from the full UK Biobank population (~500 000 individuals) has been released, and a follow-up genetic association study was also performed using an additional 261 840 individuals of self-reported British white ancestry (termed here UKB2). This group excluded anyone who was part of the first study or related to those persons (pi-hat or proportion identity by descent, calculated as P(identity by descent [IBD] = 2) + 0.5*P(IBD = 1)) > 0.125). Related people within this group were also removed. A pi-hat cutoff of 0.125 was chosen because this excludes third-degree or closer relatives from the study.
Sleep duration was self-reported and was assessed with the question: “About how many hours sleep do you get in every 24 hours (including naps)?” with responses in 1-hour increments. The average length reported was 7.19 hours with a standard error of 1.08 hours (Table 1).
UK Biobank genetic association analysis
Linear regression analysis using an additive genetic model adjusting for age, sex, 10 principal components of ancestry, and genotyping array was used to test for association of SNPs with hours of sleep duration treated as a continuous outcome variable. Analysis was done using the software PLINK [15]. Initial analysis was performed in the interim genetic data release for a population of 111 975 individuals of European ancestry (UKB1), and was repeated in the secondary group of 261 840 unrelated subjects of European ancestry (UKB2) to confirm the findings. A fixed-effects, inverse-variance meta-analysis was conducted with METAL using summary statistics of both association analyses to estimate the effect size and significance of DRD2 and DAT1 variants in the total population [16]. For this study, we focused on 27 SNPs from candidate genes selected on the basis of a literature review (Supplementary Table S1). The 27 SNPs were abundant throughout all ethnic groups and were located in genes that represented a broad swathe of biological systems implicated in sleep or were loci identified in recent Genome-Wide Association Study (GWAS) sleep studies. To ensure that the variants were common in the population, we chose SNPs with a minor allele frequency of 20% or above. We used a Bonferroni correction to set the threshold for statistical significance (p < 0.00185), since this would be the most rigorous cutoff considering that we tested 27 SNPs.
Results
DAT1 gene region association testing
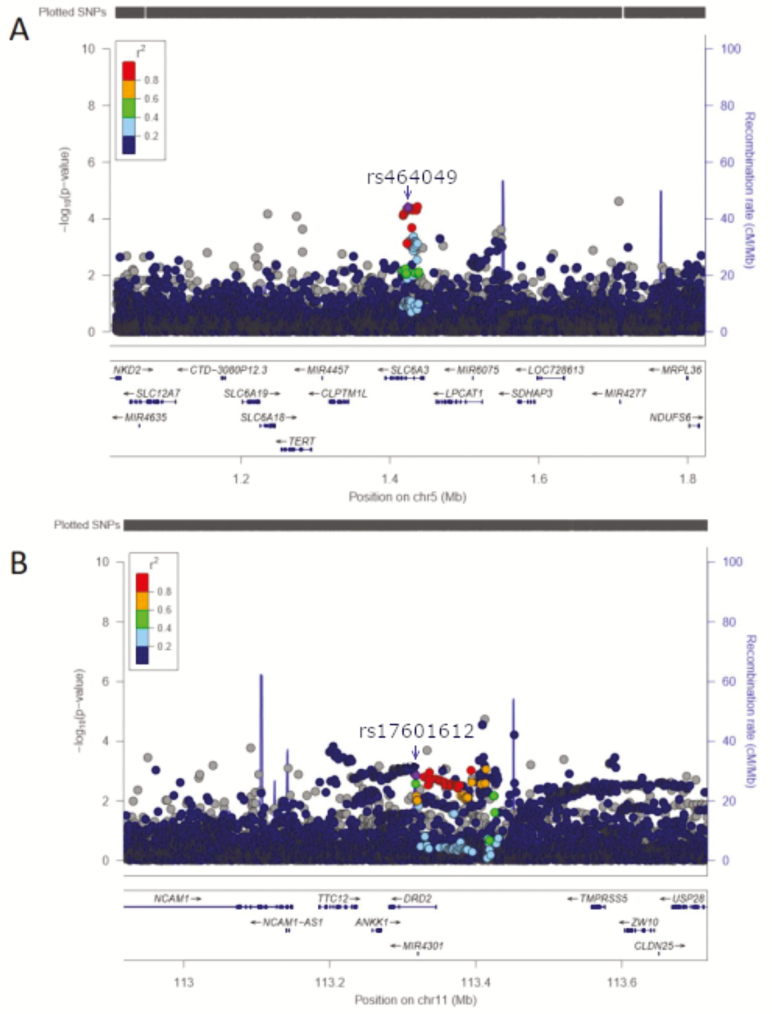
A significant association (rs464049, G, effect allele frequency [EAF] = 0.430, beta [standard error, SE] = −1.20 [0.30] minutes, p = 1.72 × 10−5; rs460000, T, EAF = 0.204, p = 5.44 × 10−4) was found between sleep duration and two correlated SNPs found in the 5′ region of DAT1—rs464049 and rs460000 (r2 = 0.44 in the 1000 Genomes Project, the European population [CEU] [17], Table 2, Figure 1). The remaining SNPs were not significantly associated with sleep duration (Supplementary Table S1). We replicated this association in an additional 261 840 subjects from the UK Biobank (rs464049, G, beta [SE] = −0.84 [0.18] minutes, p = 4.24 × 10−7) [18]. We then performed a meta-analysis (rs464049, G, beta [SE] = −0.96 [0.18] minutes, p = 5.71 × 10−10) (Tables 2 and 3). This result meets the genome-wide association threshold of significance (5 × 10−8), decreasing the likelihood of our having observed a false positive association [19].
Table 2.
Results of the association study in the initial UK Biobank sample (UKB1), the UKB2 sample, and a meta-analysis of the two studies
| Gene | SNP | CHR:BP | EFF/ALT | UKB1 (N ~ 110 000) | UKB2 (N ~ 255 000) | META | |||
|---|---|---|---|---|---|---|---|---|---|
| Beta (SE) in minutes | P-value | Beta (SE) in minutes | P-value | Beta (SE) in minutes | P-value | ||||
| DAT1 | rs40184 | 5:1,395,077 | T/C | 0.12 (0.30) | 0.603 | 0.24 (0.18) | 0.137 | 0.24 (0.18) | 0.126 |
| rs11133767 | 5:1,401,580 | T/C | 0.36 (0.30) | 0.239 | 0.48 (0.18) | 0.015 | 0.42 (0.18) | 7.30 × 10−3 | |
| rs464049 | 5:1,423,905 | G/A | −1.20 (0.30) | 1.72 × 10−5 | −0.84 (0.18) | 4.24 × 10−7 | −0.96 (0.18) | 5.71 × 10−10 | |
| rs460000 | 5:1,432,825 | T/G | −1.20 (0.36) | 5.44 × 10−4 | −0.72 (0.24) | 1.59 × 10−3 | −0.84 (0.18) | 5.77 × 10−6 | |
| DRD2 | rs11214607 | 11:113,312,139 | G/T | 0.84 (0.36) | 0.025 | 1.08 (0.24) | 1.73 × 10−5 | 1.02 (0.18) | 1.39 × 10−6 |
| rs17601612 | 11:113,317,745 | C/G | −0.90 (0.30) | 1.23 × 10−3 | −0.54 (0.18) | 2.56 × 10−3 | −0.66 (0.18) | 1.77 × 10−5 | |
CHR = chromosome; BP = base pair position in hg19; EFF = effect allele; ALT = alternate allele. Linear regression analysis in PLINK in the primary population of 111 975 and the secondary population of 261 870 people, and a meta-analysis of both the initial and follow-up studies done in METAL. Beta and standard error are in minutes.
Figure 1.
Locus Zoom Plots for DAT1 and DRD2. (A) Regional association plot for rs464049 in DAT1, based on data presented in “Genome-wide association analysis identifies novel loci chronotype in 100 420 individuals from the UK Biobank” [14]. The x-axis is the physical position of the SNP on the chromosome, while the y-axis is the negative log of the p-value for the association between sleep duration and SNP. Rs464049 and highly correlated SNPs on the 5′ end of the gene are highly associated with sleep duration (p = 4.00 × 10−5). (B) Regional association plot for DRD2. The association is significant after Bonferroni correction (p = 1.41 × 10−3).
Table 3.
Sleep duration statistics by genotype
| Gene | SNP | CHR:BP | EFF/ALT | Genotype | N | Mean (hour) | SD (hour) | SE |
|---|---|---|---|---|---|---|---|---|
| DAT1 | rs40184 | 5:1,395,077 | T/C | CC | 92 805 | 7.141 | 1.094 | 0.004 |
| CT | 143 725 | 7.153 | 1.100 | 0.003 | ||||
| TT | 55 937 | 7.136 | 1.093 | 0.005 | ||||
| rs11133767 | 5:1,401,580 | T/C | CC | 136 382 | 7.146 | 1.089 | 0.003 | |
| CT | 119 615 | 7.154 | 1.094 | 0.003 | ||||
| TT | 26 944 | 7.146 | 1.099 | 0.007 | ||||
| rs464049 | 5:1,423,905 | G/A | AA | 88 573 | 7.168 | 1.085 | 0.004 | |
| AG | 141 373 | 7.150 | 1.092 | 0.003 | ||||
| GG | 58 995 | 7.111 | 1.115 | 0.005 | ||||
| rs460000 | 5:1,432,825 | T/G | GG | 177 244 | 7.161 | 1.088 | 0.003 | |
| GT | 98 297 | 7.131 | 1.101 | 0.004 | ||||
| TT | 15 435 | 7.084 | 1.134 | 0.009 | ||||
| DRD2 | rs11214607 | 11:113,312,139 | G/T | TT | 113 558 | 7.137 | 1.111 | 0.003 |
| GT | 127 995 | 7.152 | 1.090 | 0.003 | ||||
| GG | 39 063 | 7.149 | 1.073 | 0.005 | ||||
| rs17601612 | 11:113,317,745 | C/G | GG | 204 092 | 7.139 | 1.098 | 0.002 | |
| CG | 74 031 | 7.162 | 1.093 | 0.004 | ||||
| CC | 7113 | 7.180 | 1.104 | 0.013 |
CHR = chromosome; BP = base pair position in hg19; EFF = effect allele; ALT = alternate allele. The mean, SD, and SE of self-reported sleep duration in individuals in the UK Biobank, divided by genotype in SNPs in DAT1 and DRD2.
Previously, a variable number tandem repeat (VNTR) polymorphism in the 3′ region of DAT1 has been associated with daytime sleepiness but not with sleep duration [12]. Previous literature confirms that rs464049, located in the 5′ region of DAT1 is independent of the VNTR (r2 = 0.16) [20]. Consistently, there was no association with a SNP correlated to the previously studied VNTR (proxy SNP rs11133767, r2 = 0.75, p = 0.168).
Replication of DRD2 association
This study also focused on replicating two previously reported independent association signals for sleep in DRD2, represented by rs17601612 and rs11214607 [11]. There is a suggestive association between rs17601612 and sleep duration in our initial sample (rs17601612, C, EAF = 0.383, beta [SE] = −0.90 (0.30) minutes, p = 1.23 × 10−3) (Table 2, Figure 1) in the same direction as previously reported (rs17601612, C, beta (SE) = −3.07 (0.63) minutes, p = 9.75 × 10−7) [11]. Analysis in the remaining UK Biobank subset showed a significant association with the other DRD2 variant (rs11214607, G, beta [SE] = 1.08 [0.24] minutes, p = 1.73 × 10−5), again with a direction of effect concordant with the published Cade et al. study (rs11214607, G, beta [SE] = 4.04 [0.90] minutes, p = 7.76 × 10−6) [11].
Meta-analysis in UK Biobank confirms the significant association between variation in both SNPs and sleep duration (rs17601612, C, beta [SE] = −0.66 [0.18] minutes, p = 1.77 × 10−5; rs11214607, G, beta [SE] = 1.08 [0.24] minutes, p = 1.39 × 10−6) (Tables 2 and 3).
The associations in DAT1 and DRD2 did not substantially change in magnitude of effect or in significance in sensitivity analyses of healthy participants free of chronic disease or psychiatric conditions (Supplementary Table 2). Although genetic variation in both DAT1 and DRD2 was significantly associated with sleep duration, there was no significant pair-wise epistatic interaction between the associated SNPs in DAT1 and DRD2 (p > 0.114).
Discussion
We identified a genome-wide significant association (below the genome-wide statistical threshold of significance p = 5 × 10−8) [19] between genetic variation in DAT1 and sleep duration [20]. Another dopamine-related gene has been linked to sleep duration before, namely DRD2, and variants in this gene were also found to be significantly associated with sleep duration in this study in the same direction, replicating the previous association.
Each variant confers a small effect on sleep duration, with approximately 1-minute change in sleep duration per allele, and this is consistent with sleep duration being a complex behavioral trait with small genetic contributions from multiple genes and a strong environmental, behavioral and social component. In agreement, the first well-replicated genome-wide significant association (at Paired Box Gene 8 [PAX8]) influenced sleep duration by 3.1 minutes per allele in the discovery sample [4]. Regardless of effect size, variants identified from GWAS point to new or suspected biological candidates that influence inter-individual trait variability, because they suggest that modulation of the genes at association peaks can modulate the phenotype. Therefore, biological follow-up of genetic associations can provide novel insights into molecular processes underlying sleep regulation, valuable both for enhancing scientific knowledge of sleep and for informing new therapeutic approaches. Variants of small effect size are not individually useful for prediction of sleep patterns in people, but polygenic risk profiles are beginning to be useful in predicting individual risk for some disease traits and guiding therapy [21].
It is likely that the association signals at DAT1 and DRD2 reflect inter-individual differences in dopamine signaling that influence sleep duration. However, further genetic and experimental studies will be necessary to find the causal variant and causal genes at both associated regions of the genome. DAT1 has been linked to shorter sleep in Drosophila, as flies with a mutation in their orthologue of DAT1, called DAT, were found to have fragmented and short sleep, as well as a lack of sleep rebound after sleep restriction [22]. The neurological dopaminergic pathway that controls wakefulness has been charted out in Drosophila, but considering the complexity of the human brain, it is not surprising that similar discoveries have not yet been made in humans [23]. While it is obvious that dopamine plays a role in wakefulness, the specific mechanisms of its neurological function in regard to sleep are not entirely clear. The bulk of the previous research done on DAT1 genetic variation in human populations focused on a series of 40-bp repeats in its 3′ untranslated region. It was thought that having nine or 10 repeats could influence the expression of this transporter and a variety of phenotypic traits have been assessed for association with this region of repeats, including sleep architecture [24]. The results of studies that focus on this repeating sequence are often contradictory. Interestingly, the SNPs most associated with sleep duration are located at the 5′ end of the gene, and are not close to the repeating region in the 3′ untranslated region (UTR). When an SNP in linkage disequilibrium with the 3′ VNTR was tested for association with sleep duration, there was no significant association. In this study, the data suggests that although genetic variation in DAT1 is associated with sleep duration, the cause of this association is unlikely to be the string of repeats in the gene, whatever their impact on its expression [25–27]. Further studies that directly investigate the 3′ VNTR of DAT1 are required to confirm or deny whether this polymorphism modulates subjective sleep duration.
Furthermore, two independent SNPs in DRD2 have been previously found to be associated with sleep duration and this study adds support to those findings. The effect sizes reported previously are in the same direction as the effect sizes reported in this study [11]. Dopaminergic neurons are involved in the wake/sleep switch, which is the neurological mechanism that enforces the binary state of wake and sleep and mediates switching between the two states [10]. Thus, genetic variation in transporter genes that work to transport dopamine out of the synapse, like DAT1, or in dopamine receptors, like DRD2, could be affecting this wake/sleep switch, leading to a tendency for longer or shorter sleep.
This study has important strengths. First, the large sample size of the UK Biobank allows high statistical power to detect a genetic association. Second, based on genome-wide genotyping, the population of European genetic ancestry could be carefully defined to exclude population outliers and we could adequately control for population stratification. Third, habitual sleep duration was assessed using the same question across the large sample, limiting differences in the quality of phenotyping across individuals.
However, our study also has several limitations that need to be addressed in future studies. One limitation in this study is that the sleep duration responses were collected in hour increments—so someone could not answer that they typically slept 6.5 hours a night, only that they slept 6 or 7 hours on the average night. In a smaller study, such a drastic rounding of the phenotypic data could severely affect the statistical outcome. However, in this large population, the amount that the sleep duration is rounded is about equal to the standard deviation of the population (the standard deviation is 1.09 hours). A.R. Tricker defines the rounding variable r as ϖ/σ, where ϖ is the interval between rounded extremes, in this case, 1 hour, and σ is the population standard deviation—in this case, 1.09 hours [28]. Thus, r is 0.92, a little less than 1. Tricker discovered that when r is under 1, the power of t-tests and chi-square tests in populations of 10 individuals is not significantly decreased. It can be extrapolated that in a population of about 450 000 individuals that the same would hold true. Still, considering the effect size of the significant SNPs is around a minute, the rounding of the sleep duration data should be noted. Despite this limitation, a previously reported genetic association with PAX8 was replicated by Lane et al. using the UKB1 sample and validates the phenotype and analytic approach used here [4, 29].
Another limitation with this study is the reliance on self-reported sleep duration. In a 2008 study, researchers found that subjective sleep duration is moderately correlated to objective sleep duration (r = 0.47), and that people who sleep less tend to overestimate their own sleep [30]. Thus, although variation in these genes is associated with subjective sleep duration, that does not necessarily mean that we have found a connection to objective sleep duration—we could be seeing association with an altered perception of sleep, for example. Depression, chronic pain, and insomnia symptoms are often comorbid, and dopamine systems have been implicated in these conditions as well [31]. It is possible that variation in dopaminergic genes could be associated with conditions that influence the perceptions of sleep. It will not be possible to know if this is the case until these results are compared to analyses done in a group with objective sleep measures, however we note that the associations in DAT1 and DRD2 did not substantially change in magnitude of effect or in significance in sensitivity analyses of healthy participants free of chronic disease or psychiatric conditions, suggesting that for the most part, these variants are in fact modulating sleep duration, and not disease processes that secondarily influence sleep.
Moving forward, it will be fruitful to investigate DAT1 and DRD2 variation in datasets with objective sleep duration, as well as datasets that contain more physiological data and data collected in controlled environments to attempt to understand how DRD2 and DAT1 influence sleep duration. These data were obtained from people in an uncontrolled environment with many additional factors, like work, social demands, and light availability that could have affected their sleep duration and our study cannot assess how environmental context alters the impact of these genetic variants on sleep duration. Variation in DAT1 and DRD2 could be associated with an intrinsic need for less sleep, or it could be modulating the effects of sleep deprivation. A study focusing on DRD2 and the 3′ VNTR of DAT1 suggests that variation in these regions is associated with differing functional consequences of sleep deprivation [32]. Thus, more detailed and rigorously controlled datasets will be necessary to separate the variables that could have contributed to the association between DAT1 and DRD2 and sleep duration. Additionally, since the region of the DAT1 gene that is associated with sleep duration is in a less studied part of the gene, more detailed functional studies on the 5′ end of DAT1 and its relation to sleep and sleep duration would assist in determining how these dopaminergic genes are involved in modulating sleep duration. Overall, the significant association between DAT1 and DRD2 genetic variation and sleep duration demonstrates that variation in dopamine-related genes is significantly associated with self-selected sleep in humans.
Supplementary Material
Acknowledgments
This research has been conducted using the UK Biobank Resource under application number 6818. We would like to thank the participants and researchers from the UK Biobank who contributed or collected data. Results from GWAS data for sleep duration from UKB1 are publicly available and were used for this study (Lane et al., Nat. Genet. 2016; https://sleepgenetics.org/downloads/). We thank Hassan Dashti for sharing results on the three DAT1 and DRD2 variants from GWAS of sleep duration in healthy participants.
Funding
R.S. was funded by National Institutes of Health (NIH) grants R01DK107859, R01DK102696, R01DK105072, R21HL121728, and R01HL113338.
Conflict of interest statement. Financial and Non-financial Disclosure: none for J.A.R., J.M.L., I.M.V., M.K.R., and R.S. C.A.C. is/was a paid consultant to Bose, Boston Celtics, Boston Red Sox, Columbia River Bar Pilots, Institute of Digital Media and Child Development, Jazz Pharma, Merck, Purdue Pharma, Quest Diagnostics, Samsung, Teva, Vanda Pharmaceuticals, Inc. and V-Watch/PPRS; has received lecture fees from Global Council on Brain Health/AARP, Integritas Communications Group, Maryland Sleep Society, National Sleep Foundation and Zurich Insurance Company, Ltd.; holds equity in Vanda Pharmaceuticals, Inc.; receives research/education support from Cephalon, Mary Ann & Stanley Snider via Combined Jewish Philanthropies, Jazz Pharma, Optum, ResMed, San Francisco Bar Pilots, Schneider, Simmons, Sysco, Koninklijke Philips Electronics, Vanda Pharmaceuticals, Inc.; is/was an expert witness in legal cases, including those involving Bombardier, Columbia River Bar Pilots, Continental Airlines, Fedex, Greyhound, Purdue Pharma, UPS; serves as the incumbent of a professorship endowed by Cephalon; and receives royalties from McGraw Hill, Houghton Miflin Harcourt, and Philips Respironics for the Actiwatch-2 & Actiwatch Spectrum devices. C.A.C.’s interests were reviewed and are managed by Brigham and Women’s Hospital and Partners HealthCare in accordance with their conflict of interest policies.
References
- 1. Ford ES, et al. . Trends in self-reported sleep duration among US adults from 1985 to 2012. Sleep. 2015;38(5):829–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Liu C, et al. . Long-term total sleep deprivation reduces thalamic gray matter volume in healthy men. Neuroreport. 2014;25(5):320–323. [DOI] [PubMed] [Google Scholar]
- 3. Killgore WD. Effects of sleep deprivation on cognition. Prog Brain Res. 2010;185:105–129. [DOI] [PubMed] [Google Scholar]
- 4. Lane JM, et al.. Genome-wide association analyses of sleep disturbance traits identify new loci and highlight shared genetics with neuropsychiatric and metabolic traits. Nat Genet. 2017;49(2):274–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Watson NF, et al.. A twin study of sleep duration and body mass index. J Clin Sleep Med. 2010;6(1):11–17. [PMC free article] [PubMed] [Google Scholar]
- 6. Gottlieb DJ, et al.. Genome-wide association of sleep and circadian phenotypes. BMC Med Genet. 2007;8 (Suppl 1):S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Buhr ED, et al. . Molecular components of the mammalian circadian clock. Handb Exp Pharmacol. 2013;217:3–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Huang ZL, et al.. The role of adenosine in the regulation of sleep. Curr Top Med Chem. 2011;11(8):1047–1057. [DOI] [PubMed] [Google Scholar]
- 9. Landolt HP. Genetic determination of sleep EEG profiles in healthy humans. Prog Brain Res. 2011;193:51–61. [DOI] [PubMed] [Google Scholar]
- 10. Fuller PM, et al. . Dopamine. In: The Neuroscience of Sleep. Philadelphia, PA: Elsevier; 2009:125–130. [Google Scholar]
- 11. Cade BE, et al.. Common variants in DRD2 are associated with sleep duration: the CARe consortium. Hum Mol Genet. 2016;25(1):167–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Valomon A, et al.. Genetic polymorphisms of DAT1 and COMT differentially associate with actigraphy-derived sleep-wake cycles in young adults. Chronobiol Int. 2014;31(5):705–714. [DOI] [PubMed] [Google Scholar]
- 13. About UK Biobank. UK Biobank 2010. http://www.ukbiobank.ac.uk/about-biobank-uk/ Accessed February 13, 2017.
- 14. Lane JM, et al. . Genome-wide association analysis identifies novel loci chronotype in 100,420 individuals from the UK Biobank. Nat Commun. 2016;7:10889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Purcell S, et al.. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Willer CJ, et al. . METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26(17):2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Auton A, et al. ; 1000 Genomes Consortium A global reference for human genetic variation. Nature. 2015;526(7571): 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bycroft C, et al. . Genome-wide genetic data on ~500,000 UK Biobank participants. 2017. bioRxiv doi:10.1101/166298 [Google Scholar]
- 19. Pe’er I, et al.. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet Epidemiol. 2008;32(4):381–385. [DOI] [PubMed] [Google Scholar]
- 20. Le Strat Y, et al.. The 3’ part of the dopamine transporter gene DAT1/SLC6A3 is associated with withdrawal seizures in patients with alcohol dependence. Alcohol Clin Exp Res. 2008;32(1):27–35. [DOI] [PubMed] [Google Scholar]
- 21. Natarajan P, et al.. Polygenic risk score identifies subgroup with higher burden of atherosclerosis and greater relative benefit from statin therapy in the primary prevention setting. Circulation. 2017;135(22):2091–2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kume K. A Drosophila dopamine transporter mutant, fumin (fmn), is defective in arousal regulation. Sleep Biol Rhythms. 2006;4(3):263–273. [Google Scholar]
- 23. Ueno T, et al.. Identification of a dopamine pathway that regulates sleep and arousal in Drosophila. Nat Neurosci. 2012;15(11):1516–1523. [DOI] [PubMed] [Google Scholar]
- 24. Guindalini C, et al.. Influence of genotype on dopamine transporter availability in human striatum and sleep architecture. Psychiatry Res. 2010;179(2):238–240. [DOI] [PubMed] [Google Scholar]
- 25. Costa A, et al.. Relationship between SLC6A3 genotype and striatal dopamine transporter availability: a meta-analysis of human single photon emission computed tomography studies. Synapse. 2011;65(10):998–1005. [DOI] [PubMed] [Google Scholar]
- 26. Fuke S, et al.. The VNTR polymorphism of the human dopamine transporter (DAT1) gene affects gene expression. Pharmacogenomics J. 2001;1(2):152–156. [DOI] [PubMed] [Google Scholar]
- 27. Van de Giessen EM, et al. . Striatal dopamine transporter availability associated with polymorphisms in the dopamine transporter gene SLC6A3. J Nucl Med. 2009;50(1):45–52. [DOI] [PubMed] [Google Scholar]
- 28. Tricker AR. The effect of rounding on the power level of certain normal test statistics. J Appl Stat. 1990;17(2):219–228. [Google Scholar]
- 29. Gottlieb DJ, et al.. Novel loci associated with usual sleep duration: the CHARGE Consortium Genome-Wide Association Study. Mol Psychiatry. 2015;20(10):1232–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lauderdale DS, et al.. Self-reported and measured sleep duration: how similar are they? Epidemiology. 2008;19(6):838–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Finan PH, et al.. The comorbidity of insomnia, chronic pain, and depression: dopamine as a putative mechanism. Sleep Med Rev. 2013;17(3):173–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Holst SC, et al.. Functional polymorphisms in dopaminergic genes modulate neurobehavioral and neurophysiological consequences of sleep deprivation. Sci Rep. 2017;7:45982. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.