APEC causes a range of infections in poultry, collectively called colibacillosis, and is the leading cause of mortality and is associated with major economic significance in the poultry industry. A growing number of studies have suggested APEC as an external reservoir of human ExPEC, including UPEC, which is a reservoir. ExPEC belonging to ST95 is considered one of the most important pathogens in both poultry and humans. This study is the first in-depth whole-genome-based comparison of ST95 E. coli which investigates both the core genomes as well as the accessory genomes of avian and human ExPEC. We demonstrated that multiple lineages of ExPEC belonging to ST95 exist, of which the majority may cause infection in humans, while only part of the ST95 cluster seem to be avian pathogenic. These findings further support the idea that urinary tract infections may be a zoonotic infection.
KEYWORDS: E. coli, genomics, pathogenicity, sequencing, zoonosis
ABSTRACT
Avian-pathogenic Escherichia coli (APEC) is a subgroup of extraintestinal pathogenic E. coli (ExPEC) presumed to be zoonotic and to represent an external reservoir for extraintestinal infections in humans, including uropathogenic E. coli (UPEC) causing urinary tract infections. Comparative genomics has previously been applied to investigate whether APEC and human ExPEC are distinct entities. Even so, whole-genome-based studies are limited, and large-scale comparisons focused on single sequence types (STs) are not available yet. In this study, comparative genomic analysis was performed on 323 APEC and human ExPEC genomes belonging to sequence type 95 (ST95) to investigate whether APEC and human ExPEC are distinct entities. Our study showed that APEC of ST95 did not constitute a unique ExPEC branch and was genetically diverse. A large genetic overlap between APEC and certain human ExPEC was observed, with APEC located on multiple branches together with closely related human ExPEC, including nearly identical APEC and human ExPEC. These results illustrate that certain ExPEC clones may indeed have the potential to cause infection in both poultry and humans. Previously described ExPEC-associated genes were found to be encoded on ColV plasmids. These virulence-associated plasmids seem to be crucial for ExPEC strains to cause avian colibacillosis and are strongly associated with strains of the mixed APEC/human ExPEC clusters. The phylogenetic analysis revealed two distinct branches consisting of exclusively closely related human ExPEC which did not carry the virulence-associated plasmids, emphasizing a lower avian virulence potential of human ExPEC in relation to an avian host.
IMPORTANCE APEC causes a range of infections in poultry, collectively called colibacillosis, and is the leading cause of mortality and is associated with major economic significance in the poultry industry. A growing number of studies have suggested APEC as an external reservoir of human ExPEC, including UPEC, which is a reservoir. ExPEC belonging to ST95 is considered one of the most important pathogens in both poultry and humans. This study is the first in-depth whole-genome-based comparison of ST95 E. coli which investigates both the core genomes as well as the accessory genomes of avian and human ExPEC. We demonstrated that multiple lineages of ExPEC belonging to ST95 exist, of which the majority may cause infection in humans, while only part of the ST95 cluster seem to be avian pathogenic. These findings further support the idea that urinary tract infections may be a zoonotic infection.
INTRODUCTION
Escherichia coli is an important pathogen of both poultry and humans and may cause both intestinal and extraintestinal infections (1, 2). E. coli bacteria that are causing extraintestinal infections are known as extraintestinal pathogenic E. coli (ExPEC). ExPEC include uropathogenic E. coli (UPEC), which is the primary cause of urinary tract infections in humans,and neonatal meningitis E. coli (NMEC), which is an important cause of bacterial meningitis in human neonates (1–3). Avian-pathogenic E. coli (APEC) is an ExPEC that causes a range of infections in poultry collectively known as avian colibacillosis, which is one of the leading causes of infection in the poultry industry throughout the world (4–6). Furthermore, a growing number of infections caused by antimicrobial-resistant human ExPEC strains has complicated treatment of extraintestinal infections (2, 3).
In humans, the main reservoir of ExPEC is the host’s own intestinal tract, but a number of recent studies have suggested that the external reservoir of ExPEC may be of animal origin, including broilers, where the ExPEC is transmitted via the animal food chain (2, 7, 8). These speculations have spawned a large number of comparative studies that have demonstrated an overlap in serogroups, sequence types (STs) and virulence-associated genes, which have previously been suggested to be used for classification or even as a diagnostic tool of APEC and human ExPEC (5, 9, 10). Studies have demonstrated that some human ExPEC strains cause disease in chicken infection models, and some APEC strains cause disease in mammalian models of human disease (7, 11–13). Furthermore, identical or nearly identical pulsed-field gel electrophoresis (PFGE) profiles have been identified in specific serotype-phylogroup-sequence type groups from human extraintestinal infections and avian colibacillosis (14).
ExPEC strains belonging to ST95 have been prevalent causes of human disease (15, 16). However, strains of ST95 have also been found to be among the predominant STs causing avian colibacillosis (8, 17–19). Studies have demonstrated a close relationship between ST95 APEC and human ExPEC isolates belonging to the same sero-phylo groups, including O1/O2/O18/O45:H7-B2. Certain APEC and human ExPEC have also been found with highly similar PFGE profiles (11, 14, 20). Additionally, comparative whole-genome sequencing-based studies have demonstrated a close relationship between the ST95 APEC O1 type strain and human ExPEC strains (21–23). However, it is evident that to accurately investigate the genetic properties of APEC and human ExPEC, comparative genomic analyses of a large and diverse collection from both humans and birds are required. A better understanding of ExPEC isolates belonging to ST95 may reveal whether certain sublineages have adapted to constitute unique host-specific “subpathotypes.”
The present study aimed to compare the genetic composition of 32 APEC and 291 human ExPEC genome sequenced isolates belonging to ST95. Using these data, we investigated whether distinct genomic features of ST95 strains from different hosts exist and how these may provide insight into host adaptation as it pertains to ExPEC.
RESULTS
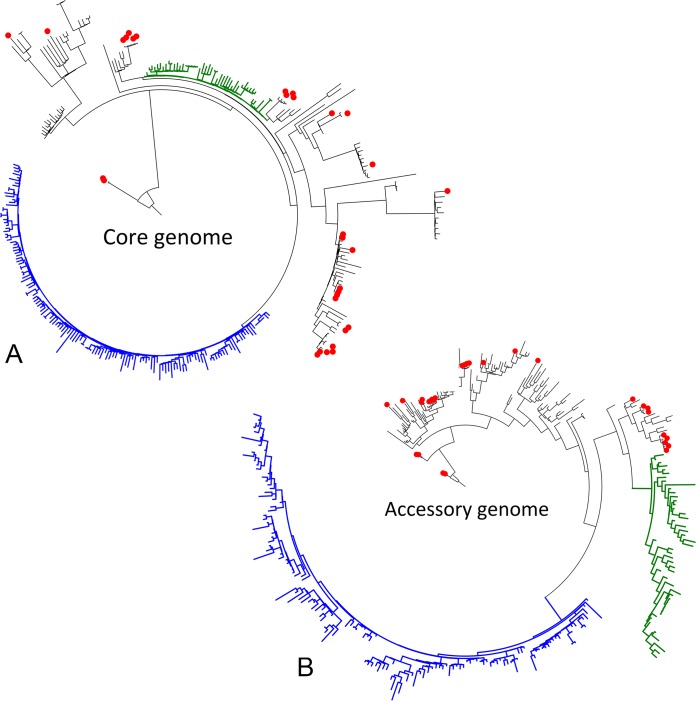
The initial pan-genomic analysis identified 16,786 genes, of which 3,789 belonged to the core genome. Diversity within the core genome of the APEC and human ExPEC strains were characterized by 17,468 single nucleotide polymorphisms (SNPs), while differences in the accessory genome were based on the presence or absence of the genes from the analysis of the pan-genome. A purged SNP phylogeny was constructed based on the identified SNPs with recombination regions removed (Fig. 1A). Meanwhile, presence or absence of the genes in the accessory genome was used to generate an accessory-genome-derived tree (Fig. 1B). Both trees demonstrated similar clustering, with APEC strains located on several different branches mixed with human ExPEC strains. Analysis of the SNPs by univariate and multivariate analysis could not identify single SNPs or a combination of SNPs which explicitly defined either APEC or human ExPEC. No single gene or combination of genes could separate and describe either the APEC or human ExPEC genome.
FIG 1.
Rooted phylogenetic trees of the core genome (A) and accessory genome (B) of 323 genomes in the study. APEC genomes are marked with red dots on the branch ends. Subclades consisting exclusively of human ExPEC were identified in the core and accessory phylogenies and indicated by colored branches. The human ExPEC/HExPEC-1 subclade (blue) was distinct in both the core and accessory genome phylogenies. The HExPEC-2 subclade (green) was identified in the accessory-genome-derived tree, while a less distinct clustering was identified in the core genome phylogeny. Branches with mixes of both APEC and human ExPEC were designated A/E-PEC branches.
Thus, APEC belonging to ST95 did not constitute a unique branch, and it was not associated with any SNP(s) or accessory genome features. APEC was located on multiple branches of mixed APEC and human ExPEC strains in both the SNP phylogeny and the accessory-gene-derived tree (Fig. 1A and B), demonstrating a great overall genetic overlap with certain human ExPEC strains, including closely related APEC and human ExPEC strains. These branches of mixed APEC and human ExPEC strains were designated A/E-PEC. Three minor A/E-PEC clades of closely related strains, which included both APEC and human ExPEC, were identified in both the SNP phylogeny and the accessory-gene-derived tree (clades 1 to 3) (see Fig. S1A and B in the supplemental material). SNP analysis indicated that genomes within the clades differed by no more than 32 SNPs (clade 1), 57 SNPs (clade 2), or 30 SNPs (clade 3), which further underlined the close relationship of certain APEC and human ExPEC strains. The associated metadata regarding the year of sampling and country of isolation presented no indication of the clinical relationship between the isolates but rather indicated highly dispersed clones.
Supplementary phylogenetic trees of the core genome (A) and accessory genome (B) of 323 genomes in the study. APEC genomes are marked with red dots on the branch ends. HExPEC subclades are indicated by colored branches as follows: HExPEC-1 subclade is shown in blue, and HExPEC-2 subclade is shown in green. Surrounding symbols indicate FimH type (squares), isolation country (triangles), and isolation year (circles). Download FIG S1, PDF file, 1.4 MB (1.5MB, pdf) .
Copyright © 2019 Jørgensen et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Two clusters exclusively containing human ExPEC strains were identified in the two phylogenies. A large cluster consisting solely of closely related human ExPEC strains (n = 145) was observed in both the SNP phylogeny and accessory-genome-derived tree and designated human ExPEC 1 (HExPEC-1) and represented strains over a period of 67 years from multiple countrie sbut with an overrepresentation of strains collected in the United States (USA) in 2012 (62%) (Fig. 1A and B). An additional human ExPEC cluster (HExPEC-2) was identified in the accessory-genome-derived tree (n = 48) (Fig. 1A). Strains of the HExPEC-2 cluster showed a distinct separate branch in the accessory-genome-derived tree; however, the cluster did not demonstrate the same distinct separation as in the SNP-based phylogeny (Fig. 1B). Almost all strains in the HExPEC-2 cluster (94%) were collected in North America over a period of 38 years and also demonstrated an overrepresentation of strains from the USA in 2012 (46%).
Multivariate analysis was performed to distinguish the A/E-PEC, HExPEC-1, and HExPEC-2. The analysis identified two canonical but nonsynonymous SNPs both found in the nadR gene, which separated the large HExPEC-1 cluster from the remaining strains (Fig. 1). Furthermore, two presence/absence gene combinations were identified that could characterize and separate the HExPEC-1 cluster from the remaining genomes. The first gene combination included the copresence of yjjW and an identified gene labeled gene3477 (see Table S1 in the supplemental material), but an absence of yaeP (Table 1, gene profile A/B). Gene3477 encoded a putative transposase (GenBank accession no. CAV01995.1). The second gene combination reported the absence of the six genes belonging to the fec protein operon (fecA2, fecB, fecC, fecD, fecE, and fecI) and presence of putative gene8134, which could not be further characterized by either Prokka, NCBI BLAST, or UniProt (Table 1, gene profile C) (Table S1). The HExPEC-2 was described by the absence of both the fec operon and gene8134 (Table 1, gene profile E). Furthermore, the HExPEC-2 could be identified by the copresence of cutC and cutD (Table 1, gene profile F). The mixed A/E-PEC clusters could be separated from the two HExPEC clusters by the presence of the fec gene operon (Table 1, gene profile D).
TABLE 1.
Gene presence/absence profile for the discriminating the human ExPEC clusters HExPEC-1 and HExPEC-2 apart from the mixed A/E-PEC cluster
| Prevalence (%) |
Gene presence/absence profilea |
Gene profile |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A/E-PEC | HExPEC-1 | HExPEC-2 | yjjW | gene3477 | yaeP | gene8134 | fecA2 | fecB | fecC | fecD | fecE | fecI | cutC | cutD | |
| 98 | 0 | 98 | − | − | + | A | |||||||||
| 0 | 98 | 0 | + | + | − | B | |||||||||
| 0 | 96 | 6 | + | − | − | − | − | − | − | C | |||||
| 92 | 2 | 2 | − | + | + | + | + | + | + | D | |||||
| 0 | 0 | 94 | − | − | − | − | − | − | − | E | |||||
| 0 | 0 | 98 | + | + | F | ||||||||||
Open or blank gene cells were not relevant for the gene profile.
Putative gene sequences. Download Table S1, DOCX file, 0.01 MB (12.6KB, docx) .
Copyright © 2019 Jørgensen et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
FimH typing.
Subdivision by FimH typing revealed a correlation between the FimH variants and the individual branches in both the SNP phylogeny and accessory-genome-derived tree (Fig. S1A and B). The mixed branches of APEC and ExPEC belonging to A/E-PEC carried genes encoding multiple FimH variants, including FimH15, FimH27, FimH30, and FimH54, and APEC strains were represented among all these FimH variants. Three genomes encoded two novel FimH variants, FimH526 and FimH525, each differing only by one nucleotide from FimH15 and FimH30, respectively, and grouped together with these. All strains encoding the FimH41 variant belonged to the HExPEC-1 cluster. Furthermore, all but one strain encoding the FimH18 variant belonged to the HExPEC-2 cluster.
ExPEC-associated genes.
All strains were investigated for 55 genes previously described to be distinct for ExPEC (Table 2). Interestingly, 10 genes of these were present in all or >95% of all investigated genomes. Six additional genes were also highly frequent and were present in >75% of all genomes. Conversely, 14 genes were not identified or found in <5% of the genomes.
TABLE 2.
Prevalence of the investigated ExPEC-associated genes
| Category and genea | Description | Prevalence (%) |
GenBank accession no. | ||||
|---|---|---|---|---|---|---|---|
| APEC (n = 32) |
ExPEC (n = 291) |
A/E-PEC (n = 127) |
HExPEC-1 (n = 147) |
HExPEC-2 (n = 49) |
|||
| A/E-PEC-associated genes | |||||||
| iucC1 | Aerobactin synthesis | 96.9 | 35.4 | 85.4 | 14.5 | 4.2 | AAS66995.1 |
| iucD1 | Aerobactin synthesis | 96.9 | 35.4 | 86.2 | 13.8 | 4.2 | AAA23196.1 |
| iutA1 | Iron transport | 96.9 | 35.1 | 84.6 | 14.5 | 4.2 | AAS66997.1 |
| cvaA1 | Colicin V | 96.9 | 33.0 | 83.1 | 13.1 | 0.0 | CAA11514.1 |
| etsA1 | ABC transport system | 96.9 | 30.6 | 77.7 | 13.1 | 0.0 | YP_444079.1 |
| hlyF1 | Hemolysin | 93.8 | 34.4 | 85.4 | 13.1 | 0.0 | AAO49613.1 |
| ompT1/2 | Outer membrane protease | 93.8 | 34.0 | 84.6 | 13.1 | 0.0 | ADK70174.1 |
| cvaB1 | Colicin V | 87.5 | 33.3 | 81.5 | 13.1 | 0.0 | CAA11515.1 |
| cvaC1 | Colicin V | 84.4 | 32.3 | 78.5 | 13.1 | 0.0 | CAA40746.1 |
| cvi1 | Colicin V immunity | 84.4 | 33.3 | 80.8 | 13.1 | 0.0 | CAA11513.1 |
| HExPEC-1- and A/E-PEC- associated genes |
|||||||
| tia2 | Invasion determinant protein | 71.9 | 75.6 | 84.6 | 91.7 | 0.0 | ABJ02397.1 |
| papE2 | P fimbria | 71.9 | 75.3 | 83.8 | 91.7 | 0.0 | CAA43568.1 |
| ireA2 | Siderphore receptor | 71.9 | 74.9 | 83.1 | 91.7 | 0.0 | AMR36194.1 |
| papG-II2 | P fimbria adhesin variant | 71.9 | 74.9 | 83.8 | 91.0 | 0.0 | AAA24293.1 |
| papA2 | P fimbria shaft | 68.8 | 74.6 | 81.5 | 91.7 | 0.0 | AAZ04426.1 |
| HExPEC-2-associated genes | |||||||
| ibeA2 | Brain epithelium invasion | 40.6 | 21.3 | 20.0 | 0.7 | 100 | AAA92443.1 |
| sfaS2 | S fimbria adhesin | 28.1 | 19.6 | 13.8 | 0.7 | 97.9 | AAB25046.1 |
| sfaG2 | S fimbria | 28.1 | 18.6 | 11.5 | 0.7 | 97.9 | AAB25045.1 |
| cnf12 | Cytotoxic necrotizing factor | 0.0 | 17.2 | 3.1 | 0.7 | 93.8 | CAA50007.1 |
| papG-III2 | P fimbria adhesin variant | 0.0 | 15.8 | 0.8 | 0.0 | 93.8 | WP_001468556.1 |
| HExPEC-2- and A/E-PEC- associated gene |
|||||||
| iroN1 | Siderophore | 96.9 | 49.1 | 83.1 | 13.1 | 97.9 | AAN76093.1 |
| Highly frequent (genes present in >75% of all strains) |
|||||||
| csgA1/2 | Curli | 100 | 100 | 100 | 100 | 100 | AAA23616.1 |
| fyuA2 | Siderophore | 100 | 100 | 100 | 100 | 100 | CAA86211.1 |
| ompA2 | Serum resistance | 100 | 100 | 100 | 100 | 100 | AAN79561.1 |
| iss1 | Serum survival | 100 | 100 | 100 | 100 | 100 | X52665 |
| feoB3 | Iron uptake | 100 | 99.7 | 100 | 99.3 | 100 | NP_417868.1 |
| fimH2 | Type 1 fimbria adhesin | 100 | 99.7 | 100 | 99.3 | 100 | CAA12423.1 |
| usp2 | Uropathogenic-specific protein (bacteriocin) | 100 | 99.3 | 99.2 | 99.3 | 100 | BAA93674.1 |
| irp22 | Yersiniabactin | 100 | 98.6 | 98.5 | 98.6 | 100 | OAC27446.1 |
| kpsMT-K12 | Group 2 capsule kpsM variant | 100 | 99.7 | 99.2 | 100 | 100 | AAA24046.1 |
| malX (PAI)2 | Pathogenicity island | 81.3 | 98.3 | 91.5 | 100 | 100 | AF003742.1 |
| papC2 | P fimbria usher | 71.9 | 91.1 | 84.6 | 91.0 | 95.8 | CAA43564.1 |
| papF2 | P fimbria subunit | 71.9 | 91.4 | 85.4 | 91.7 | 93.8 | AAZ04419.1 |
| vat2 | Vacuolating autotransporter toxin | 59.4 | 88.0 | 80.0 | 96.6 | 64.6 | AAO21903.1 |
| bor3 | Serum resistance | 96.9 | 77.3 | 74.6 | 88.3 | 64.6 | AGD80583.1 |
| sitA2 | Iron transport | 53.1 | 78.0 | 54.6 | 88.3 | 93.8 | CAJ21596.1 |
| Genes of variable presence | |||||||
| traT1 | Serum resistance | 43.8 | 57.4 | 50.0 | 66.9 | 39.6 | AAA26075.1 |
| eitA1 | Iron ABC transport system | 31.3 | 11.7 | 10.8 | 9.7 | 33.3 | ABD51672.1 |
| cma1 | Colicin M | 34.4 | 2.4 | 13.1 | 0.7 | 0.0 | AFK30933.1 |
| cbi1 | Colicin B immunity | 31.3 | 2.7 | 13.1 | 0.7 | 0.0 | YP_003937726.1 |
| cdtB2 | Cytolethal distending toxin B | 0.0 | 5.8 | 12.3 | 0.0 | 2.1 | BAL47227.1 |
| Low frequency (genes identified in <5% of all investigated genomes) | |||||||
| astA2 | EAST1 toxin | 12.5 | 1.0 | 4.6 | 0.7 | 0.0 | AAG18472.1 |
| tsh1 | Hemagglutinin | 12.5 | 0.0 | 3.1 | 0.0 | 0.0 | AAA24698.1 |
| iha1 | Nonfimbrial Iha adhesin | 0.0 | 0.3 | 0.0 | 0.7 | 0.0 | ABB17254.1 |
| focG2 | F1C fimbria | 0.0 | 0.3 | 0.8 | 0.0 | 0.0 | AAB20438.1 |
| afa2 | Nonfimbrial adhesin | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | CAA54113.1 |
| fliC-H72 | Flagellin | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | AGA03821.1 |
| hlyD2 | α-Hemolysin | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | YP_002756568.1 |
| papG-I2 | P fimbria adhesin variant | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | CAA43570.1 |
| bfp1 | Bundle-forming pilus | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | AID54475.1 |
| bma | Blood group M adhesin | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | AAA23523.1 |
| gafD | G-fimbriae | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | AAA69514.1 |
| rfc2 | Lipopolysaccharide synthesis | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | AAC43898.1 |
| stgA2 | Stg fimbria | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | AAS99229.1 |
| stx2 | Shiga toxin | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | AB012101.1 |
The category is the gene location or gene frequency class. Genes localized on plasmid are indicated by superscript 1, genes localized on chromosome are indicated by superscript 2, and genes carried on phage are indicated by superscript 3.
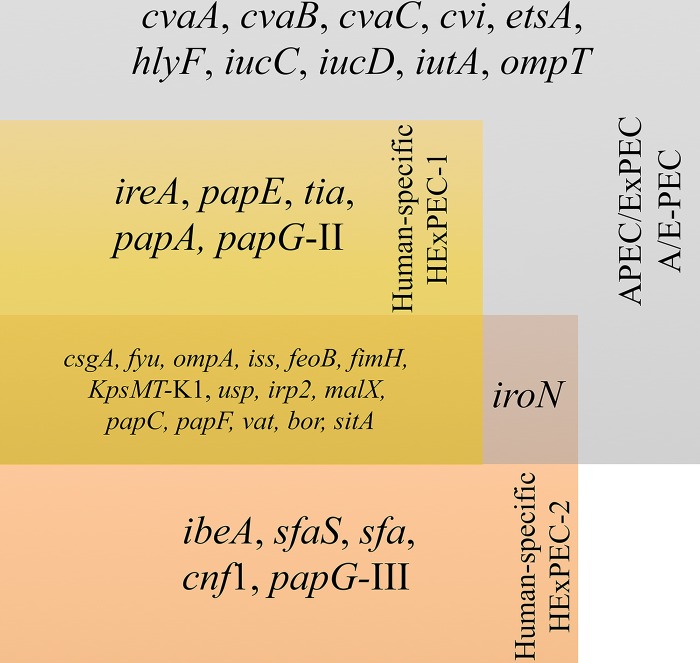
A correlation was observed between the virulence-associated genes and the A/E-PEC group and the two HExPEC groups (Fig. 2). Ten genes (cvaA, cvaB, cvaC, cvi, etsA, hlyF, iucC, iucD, iutA, and ompT) were significantly more prevalent (P < 0.01) in the A/E-PEC compared to the strains from either of the two HExPEC clusters. Five genes (ireA, papA, papE, papG-II, and tia) were found in almost all and significantly more HExPEC-1 and A/E-PEC, while were absent in the strains belonging to HExPEC-2 (P < 0.001). Meanwhile, five genes (ibeA, sfaS, sfa, cnf1, and papG-III) were significantly more prevalent in HExPEC-2 (P < 0.001). The iroN gene was detected in significantly fewer HExPEC-1 strains compared to A/E-PEC and HExPEC-2 (P < 0.0001).
FIG 2.
Diagram of ExPEC-associated gene distribution in the three groups; the two human ExPEC clusters HExPEC-1 and HExPEC-2 and the mixed avian/human ExPEC A/E-PEC cluster. Genes associated with the A/E-PEC are located in the gray box. HExPEC-1-associated genes are located in the yellow box. HExPEC-2- associated genes are located in the orange box.
Plasmids.
All genomes were investigated for virulence-associated plasmids using PlasmidSeeker (24). Multiple closely related variants of the pAPEC-O2-ColV, pAPEC-O1, p1ColV5155, and pAPEC-O78-ColV plasmid types were identified. All APEC strains harbored ColV plasmids, while only 25% of the human ExPEC strains carried plasmids. None of HExPEC-2 strains harbored ColV plasmids, while 11.7% of the HExPEC-1 strains contained the pAPEC-O78-ColV plasmid. Meanwhile, 70% of the human ExPEC strains belonging to the A/E-PEC clusters harbored ColV plasmids, corresponding to 78% of all A/E-PEC strains that contained plasmids.
Antibiotic resistance.
The levels of antibiotic resistance were investigated in silico (Table 3). There was no statistical difference in the number of APEC and human ExPEC resistant strains for any of the classes of antibiotics. HExPEC-2 demonstrated the highest prevalence of beta-lactam, sulfonamide, trimethoprim, tetracycline, and macrolide antibiotic resistance genes. However, only the prevalence of beta-lactam and tetracycline genes were significantly higher in HExPEC-2 than in A/E-PEC (P < 0.05). Furthermore, significantly more A/E-PEC carried sulfonamide and aminoglycoside resistance genes than HExPEC-1 (P < 0.05). Strains with genes coding for ≥3 classes of antibiotic resistance genes were considered multiantibiotic resistant (Table 3). While there was no difference in the number of putative multidrug-resistant strains between the APEC and human ExPEC, there were a significantly higher number of multiantibiotic-resistant HExPEC-2 strains compared to both the HExPEC-1 and A/E-PEC (P < 0.01 and P < 0.05, respectively).
TABLE 3.
Prevalence antibiotic classes of the different ExPEC subtypes
| Antibiotic resistance class |
Prevalence (%) |
|||||
|---|---|---|---|---|---|---|
| Total (n = 323) |
APEC (n = 32) |
Human ExPEC (n = 291) |
A/E-PEC (n = 127) |
HExPEC-1 (n = 147) |
HExPEC-2 (n = 49) |
|
| Beta-lactams | 18.3 | 6.3 | 19.6 | 14.6 | 17.9 | 29.2 |
| Sulfonamides | 16.1 | 12.5 | 16.5 | 20.0 | 9.0 | 27.1 |
| Trimethoprims | 8.7 | 3.1 | 9.3 | 9.2 | 4.8 | 18.8 |
| Aminoglycosides | 7.4 | 9.4 | 7.2 | 11.5 | 3.4 | 8.3 |
| Tetracyclines | 4.6 | 3.1 | 4.8 | 3.1 | 3.4 | 12.5 |
| Macrolides | 1.2 | 0.0 | 1.4 | 0.8 | 1.4 | 2.1 |
| Quinolones | 0.3 | 0.0 | 0.3 | 0.8 | 0.0 | 0.0 |
| Multiresistance | 11.0 | 6.3 | 11.0 | 10.0 | 6.9 | 22.9 |
DISCUSSION
Previous studies have reported a genetic overlap in APEC and human ExPEC strains belonging to ST95 and suggested a zoonotic potential of APEC (21–23). However, these studies were solely based on comparing the APEC-O1 type strain to human ExPEC strains. Here, using whole-genome sequencing (WGS) data on a large collection of ST95 strains, APEC belonging to ST95 did not constitute a unique branch and were not associated with single SNPs or specific accessory genome features. The APEC genomes were generally diverse and located on multiple branches together with closely related human ExPEC, including nearly identical APEC and human ExPEC strains in both the SNP phylogeny and accessory-genome-derived trees. These results clearly show that APEC and human ExPEC belonging to ST95 could not be distinguished. The large overall genetic overlap and identification of dispersed clones of closely related APEC and human ExPEC support the previous findings and demonstrate that certain ST95 ExPECs do not exhibit host specificity or have host specificity against multiple hosts, including humans and birds. Furthermore, the genetic diversity between the investigated APEC genomes also reveal that no single ST95 APEC strain can be used as a ST95 APEC type strain to represent the entire ST95 APEC pathotype.
Two clusters consisted entirely of human ExPEC strains, the HExPEC clusters. There was a strong overrepresentation of strains collected in the USA in 2012 in the two HExPEC clusters compared to the mixed human ExPEC and APEC cluster, A/E-PEC (62% of HExPEC-1, 46% of HExPEC-2, and 24% of A/E-PEC) (P < 0.001), Thus, the clustering could be the result of skewed overrepresentation of clonally related strains. However, both HExPEC-1 and HExPEC-2 represented strains from multiple countries over a 67-year and 38-year period, HExPEC-1 and HExPEC-2, respectively, and resulted in closely related clusters in the SNP-based phylogenies. Though human ExPEC and APEC A/E-PEC genomes demonstrated a great genetic overlap suggesting low host association, these findings suggest that certain subclades of genetically stable ExPEC belonging to ST95 could have adapted a degree of host predilection.
The HExPEC-1 cluster was distinctly separated from the remaining genomes in both the SNP phylogeny and accessory-genome-derived tree, while the HExPEC-2 cluster demonstrated a clear separation only in the accessory-genome-derived tree. The HExPEC-1 was distinguishable from other strains by two canonical SNPs as well as several accessory genome features, while the HExPEC-2 cluster constituted a distinct subclade in the accessory genome, defined by the presence of two cut genes. The two canonical SNPs that differentiated the HExPEC-1 were both in the nicotinamide ribose kinase domain of the nadR gene and were both nonsynonymous. NadR is a trifunctional protein involved in regulation of NAD and associated fermentation pathways and anaerobic growth (25, 26). Two combinations of genes were found to be unique to the HExPEC-1. The first consisted of yjjW, yaeP, and gene3477 (Table 1, gene profile A/B), and the second consisted of gene8134 and the fec operon (Table 1, gene profile C/D/E). The YjjW protein has been associated with the activation of YjjI, a protein hypothesized to be involved in nitrogen metabolism. The yaeO gene has been suggested to be expressed through translational coupling with yaeP (27). YaeO has been shown to be a Rho-specific inhibitor of transcription termination, an essential process for the regulation of bacterial gene expression (28). The gene3477 gene encodes a putative transposase. The fec genes are signaling and transport genes for the binding and uptake of diferric dicitrate (29). Evidence indicates that fec genes are acquired by horizontal gene transfer and have been reported to be located in pathogenicity islands in Shigella flexneri. Meanwhile, gene8134 could not be annotated by Prokka, NCBI BLAST, or UniProt. For HExPEC-2, the presence of cutC and cutD genes was unique. The cut gene cluster has been described and associated with disease-associated microbial metabolic activity in humans and has been suggested to be horizontally transferred between bacteria in the human gut (30).
The prevalence of 55 genes, which previously have been suggested to demonstrate APEC or human ExPEC association were investigated (Table 2) (5, 9, 10). Interestingly, in this study, nine genes were found to be present in all investigated genomes (csgA, fyu, ompA, iss, feoB, fimH, usp, kpsMT-K1, and irp2). Additionally, six genes (malX, papC, papF, vat, bor, and sitA) were identified in >75% of all genomes and did not show any host or cluster association (Fig. 2). All investigated genomes originated from isolates from diseased birds or were clinical human isolates; thus, a degree of virulence potential in the strains was expected. However, several of the 15 frequently identified genes have previously been suggested to be host associated. The iss, papC, vat, and sitA genes have been suggested to be APEC associated, while csgA and fimH have been suggested to have stronger association with human ExPEC (5, 9, 10, 31). Additionally, fyu and irp2 have been suggested to be associated with both host types. These results demonstrate that these frequently identified genes possibly have lower host association in ST95 ExPECs than previously predicted. Meanwhile, 14 of the 55 genes were not identified or found in very low numbers in the investigated strains, including the astA, tsh, iha, and afa genes which have been suggested to be host associated (9, 31) and generic for ExPEC; however, these genes appear not to be significant for ExPEC belonging to ST95.
The 10 A/E-PEC-associated genes (cvaA, cvaB, cvaC, cvi, etsA, hlyF, iucC, iucD, iutA, and ompT) were identified in the majority of included APEC isolates (84.4 to 96.6%) and seemed essential for the avian virulence potential (Table 2). These 10 genes were also identified in the majority (77.7 to 86.2%) of the human ExPEC strains belonging to the A/E-PEC clusters, supporting their close relatedness to APEC. Previous studies have described the function and pathogenic significance of the 10 A/E-PEC genes (5, 9, 10). Furthermore, the four ColV plasmids, pAPEC-O2-ColV, pAPEC-O1, p1ColV5155, and pAPEC-O78-ColV, and closely related variants have been described to be closely related, with large strongly conserved core genes, including the 10 A/E-PEC-associated genes (32–34). ColV plasmids were identified in all APEC and 70% of the human ExPEC strains belonging to the A/E-PEC clusters, corresponding to 78% of all A/E-PEC strains containing ColV plasmids. These results further underline the significance of these 10 genes and the ColV plasmids which have previously been described to be associated with pathogenicity (5, 9, 10, 32–34). Meanwhile, 11.7% of the HExPEC-1 strains were sporadically identified to harbor the pAPEC-O78-ColV plasmid. It is generally accepted that transconjugation of virulence-associated plasmids can induce the recipient’s virulence potential, and transmission of pAPEC-O2-ColV into commensal E. coli strains has been demonstrated to significantly increase the virulence potential in in ovo experiments (12). This suggests that the avian virulence potential can possibly be introduced into human clones by the uptake of such plasmids.
The identified ireA, papE, tia, papA, and papG-II virulence genes encoded by HExPEC-1 could be host species related or associated. However, the majority of the A/E-PEC also carried these genes, which emphasizes A/E-PEC’s potential to infect humans as well as birds. While few HExPEC-2 carried the A/E-PEC- and HExPEC-1-associated genes, five genes (ibeA, sfaS, sfa, cnf1, and papG-III) were significantly (P < 0.001) and almost exclusively found in the strains of the HExPEC-2 cluster compared to the A/E-PEC and HExPEC-1. These findings could indicate a lower virulence potential of HExPEC-2 strains toward birds, but an increase in the clinical virulence potential in a putative human subclade (Fig. 2). Interestingly, the papG-II variant was exclusively identified in the HExPEC-1 and A/E-PEC, while papG-III was almost only identified in the HExPEC-2. One may speculate that the P-fimbria adhesin variant PapG-II might have a significant role in the pathogenicity in both birds and humans, while one might suggest that the PapG-III variant had less avian pathogenicity potential.
Antimicrobial resistance genes toward the beta-lactam, sulfonamide, and trimethoprim classes were the most prevalent. Beta-lactams are among the most frequently used antimicrobial agents in both poultry and humans, which consequently can be reflected by high levels of beta-lactam resistance (35). Trimethoprim-sulfonamide combinations are broad-spectrum antimicrobial agents frequently used in poultry production as well as for treatment against urinary tract infection (UTI) (35). The prevalence of beta-lactam- and tetracycline-resistant HExPEC-2 strains was significant compared to that of A/E-PEC (P < 0.05), while A/E-PEC demonstrated significantly more sulfonamide- and aminoglycoside-resistant strains compared to HExPEC-1 (P < 0.05). The overall prevalence of antimicrobial resistance was observed in similar or lower levels as currently observed in poultry and clinical cases and did not show any strong correlation with any of the observed clusters (35). The aminoglycoside resistance gene aac3Via was the only gene found in significantly higher numbers of APEC compared to human ExPEC (P < 0.01) (see Fig. S1 in the supplemental material). Furthermore, the prevalence of strA, strB, tetA, tetB, sul2, dfrA5, and aac3VI genes were identified in significant higher numbers in A/E-PEC than in the HExPEC clusters. Tetracyclines are commonly used antimicrobials in poultry as well as in other animal species, which could explain the high prevalence of the tetA and tetB genes (35).
Typing of E. coli using concatenated FimH and MLST sequences has been suggested to have a greater discriminatory power than MLST alone (36). FimH typing correlated highly with the clustering of the accessory-derived tree, the presence of virulence-associated plasmids and additional virulence genes. APEC and human ExPEC strains belonging to A/E-PEC were found to have genes encoding FimH15/FimH526, FimH27, FimH30/FimH525, and FimH54 variants which correlated highly with the subclades of the A/E-PEC. While the strains encoding the FimH18 and FimH41 variants belonged to the HExPEC-2 and HExPEC-1, respectively, possibly this was due to the result of the strong overrepresentation of North American strains in the two clusters; the A/E-PEC-associated FimH groups were represented by temporally and spatially diverse strains (Table S2 and Fig. S1A and B).
List of strains used in this study. Download Table S2, DOCX file, 0.1 MB (58.3KB, docx) .
Copyright © 2019 Jørgensen et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
While limited information is available on FimH15 and FimH54, previous studies have reported the FimH18, FimH27, FimH30, and FimH41 variants belonging to ST95 that are able to cause extraintestinal infections in humans (37, 38). One might suggest that combined MLST and FimH typing could give a greater overview of the subclades of ST95 E. coli strains capable of infecting both birds and humans.
In summary, APEC belonging to ST95 did not constitute a unique cluster, and these strains exhibited high genetic diversity. A large genetic overlap was observed between APEC and certain human ExPEC, including closely related clones. These results suggest a genotype that is not host specific and may be pathogenic in both birds and humans. Ten ExPEC genes were highly associated with avian pathogenicity. The genes were encoded on transferable plasmids, which were closely related to APEC-associated pAPEC-O2-ColV, pAPEC-O1, p1ColV5155, and pAPEC-O78-ColV. Additionally, aminoglycoside and sulfonamide resistance, including specific antimicrobial resistance-associated genes, tended to be more prevalent in A/E-PEC than in HExPEC.
Meanwhile, the large human exclusive ExPEC cluster, HExPEC-1, was identified by both SNP phylogeny and accessory-genome-derived tree, which represented a genetically stable human-specific clone. The additional HExPEC-2 cluster was identified in the accessory-genome-derived tree. Neither of the HExPEC-2 genomes harbored any of the virulence-associated plasmids related to avian hosts and carried fewer ExPEC-associated virulence genes in general. However, the geotemporal relationship of the strains of the HExPEC-1 and HExPEC-2 clusters suggests that these human ExPEC clusters do not necessarily reflect specific large human host-adapted genotypes but could merely be clonally related isolates from historical outbreak investigations. Though HExPEC might denote a lower virulence potential toward birds, transmission of virulence-associated genes carried on plasmids, including A/E-PEC-associated genes, are capable of introducing avian virulence potential as well as increasing the human virulence potential. The high genomic plasticity and rapid transmission of genes in E. coli further underline the challenges when defining host associations.
These findings suggest that multiple lineages of ExPEC belonging to ST95 exists, of which the majority may cause infection in humans, while only part of the ST95 cluster seem to be avian pathogenic. These results add s to a growing body of evidence that suggests that urinary tract infection may be a foodborne disease.
MATERIALS AND METHODS
Bacterial strains and genomes.
The genomes from 323 E. coli ExPEC strains belonging to the ST95 lineage were selected in accordance with the method of Johnson et al. (39) by carrying ≥2 ExPEC-associated genes. In total, 291 human ExPEC genomes and 32 APEC genomes were included in this study (see Table S2 in the supplemental material). A total of 146 genomes that fulfilled the criteria were obtained from EnteroBase (http://enterobase.warwick.ac.uk/species/index/ecoli), whereas 17 genomes were from GenBank. A total of 141 genomes were obtained from a 1-year prospective study in Flagstaff, Arizona. Seven genomes were obtained from the National Veterinary Institute, Technical University of Denmark. These seven isolates were previously included in another study, and the raw reads have been made available in the NCBI SRA under the study accession number SRP092633 (isolation IDs: E25, E28, E33, E37, E41, E42, and E43) (40). An additional 13 bacterial isolates were sequenced in-house, five E. coli isolates were from the University of Georgia, and eight isolates were from the strain collection at the Department of Veterinary and Animal Sciences, University of Copenhagen. The assemblies have been made available on NCBI under the BioProject accession number PRJNA431453. The 13 bacterial isolates were sequenced by 2x250-bp paired-end sequencing on a MiSeq instrument (Illumina) and assembled using SPAdes v3.5.0 (41). Initial analysis on all isolates was performed with MLST, PlasmidFinder, SerotypeFinder, FimTyper, and ResFinder (42–45).
Analysis of the core genome.
Single nucleotide polymorphisms (SNPs) of the core genome were identified using NASP (46). All raw reads were aligned, and the sequence data of each genome were analyzed against the E. coli reference APEC O1 genome (GenBank accession no. CP000468). All positions with coverage less than 10× or if the variant was present in <90% of the base calls were excluded. For the univariate and multivariate analyses, all bases identical to the reference were set at “0,” while bases differing from the reference base were set at “1.” Dense areas of polymorphisms due to recombination or misalignment were removed using CleanRecomb by identification of stretches of consecutive sites with identical SNP profiles (47). An approximately maximum likelihood phylogeny was inferred using the FastTree 2.1.5 implementation in Geneious 9.1.8 (Biomatters, Auckland, New Zealand) on the purged SNPs. The tree was rooted using E. coli ST140 IMT5155 (GenBank accession ID CP005930) strain as an outgroup. All trees were visualized using iTol (48). Univariate analysis of the identified SNPs was performed by Fisher’s exact test. Significant P values (<0.05) were corrected for multiple testing using the false-discovery rate (FDR) method. Multivariate analysis of SNPs was performed by a discriminant analysis of principal components (DAPC) in R (version), using the adegenet package (49).
Analysis of the accessory genome.
Open reading frames were identified and annotated with Prokka 1.12-beta with default settings, using barrnap 0.7 for rRNA prediction (50). Unannotated CDS were designated “geneXXXX.”
The pan-genome of annotated bacterial genomes was investigated using Roary 3.7.0 (51). The binary presence/absence data of accessory genes produced in Roary was used for the construction of an accessory binary tree as well as for univariate and multivariate analyses.
ExPEC-associated genes and plasmid analysis.
The content of 55 previously described ExPEC-associated genes was investigated using BLASTn as implemented in Geneious 9.1.8 (10, 14, 21, 52). Reference genes were obtained from GenBank (Table 2). Genes were considered present with ≥85% coverage and ≥90% identity. Significant associations were analyzed using the Fisher exact test.
The genomes were investigated for large virulence-associated plasmids using PlasmidSeeker, with APEC-O1 as the reference (GenBank accession no. CP000468) (24). Identified plasmids with at least 80% Kmers found were considered a closely related variant of the respective plasmid.
Percent abundance of antibiotic resistance genes for each of the groups; APEC, ExPEC, A/E-PEC, HExPEC-1, and HExPEC-2. Significant P values are indicated by asterisks as follows; *, P > 0.05; **, P > 0.01; ***, P > 0.001. Download FIG S2, TIF file, 0.1 MB (147.1KB, tif) .
Copyright © 2019 Jørgensen et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
REFERENCES
- 1.Smith JL, Fratamico PM, Gunther NW. 2007. Extraintestinal pathogenic Escherichia coli. Foodborne Pathog Dis 4:134–163. doi: 10.1089/fpd.2007.0087. [DOI] [PubMed] [Google Scholar]
- 2.Singer RS. 2015. Urinary tract infections attributed to diverse ExPEC strains in food animals: evidence and data gaps. Front Microbiol 6:28. doi: 10.3389/fmicb.2015.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Terlizzi ME, Gribaudo G, Maffei ME. 2017. UroPathogenic Escherichia coli (UPEC) infections: virulence factors, bladder responses, antibiotic, and non-antibiotic antimicrobial strategies. Front Microbiol 8:1566. doi: 10.3389/fmicb.2017.01566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dho-Moulin M, Fairbrother JM. 1999. Avian pathogenic Escherichia coli. Vet Res 30:299–316. [PubMed] [Google Scholar]
- 5.Rodriguez-Siek KE, Giddings CW, Doetkott C, Johnson TJ, Nolan LK. 2005. Characterizing the APEC pathotype. Vet Res 33:239–250. doi: 10.1051/vetres:2004057. [DOI] [PubMed] [Google Scholar]
- 6.Nolan LK, Barnes HJ, Vaillancourt J-P, Abdul-Aziz T, Louge CM. 2013. Colibacillosis, p 751–805. In Swayne DE, Glisson JR, McDougald LR, Nolan LK, Suarez DL, Nair V (ed), Diseases of poultry, 13th ed. Wiley-Blackwell, Ames, Iowa. [Google Scholar]
- 7.Jakobsen L, Garneau P, Bruant G, Harel J, Olsen SS, Porsbo LJ, Hammerum AM, Frimodt-Møller N. 2012. Is Escherichia coli urinary tract infection a zoonosis? Proof of direct link with production animals and meat. Eur J Clin Microbiol Infect Dis 31:1121–1129. doi: 10.1007/s10096-011-1417-5. [DOI] [PubMed] [Google Scholar]
- 8.Manges AR. 2016. Escherichia coli and urinary tract infections: the role of poultry-meat. Clin Microbiol Infect 22:122–129. doi: 10.1016/j.cmi.2015.11.010. [DOI] [PubMed] [Google Scholar]
- 9.Ewers C, Janssen T, Kiessling S, Philipp H-C, Wieler LH. 2005. Rapid detection of virulence-associated genes in avian pathogenic Escherichia coli by multiplex polymerase chain reaction. Avian Dis 49:269–273. doi: 10.1637/7293-102604R. [DOI] [PubMed] [Google Scholar]
- 10.Johnson TJ, Wannemuehler Y, Doetkott C, Johnson SJ, Rosenberger SC, Nolan LK. 2008. Identification of minimal predictors of avian pathogenic Escherichia coli virulence for use as a rapid diagnostic tool. J Clin Microbiol 46:3987–3996. doi: 10.1128/JCM.00816-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moulin-Schouleur M, Reperant M, Laurent S, Bree A, Mignon-Grasteau S, Germon P, Rasschaert D, Schouler C. 2007. Extraintestinal pathogenic Escherichia coli strains of avian and human origin: link between phylogenetic relationships and common virulence patterns. J Clin Microbiol 45:3366–3376. doi: 10.1128/JCM.00037-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Skyberg JA, Johnson TJ, Johnson JR, Clabots C, Logue CM, Nolan LK. 2006. Acquisition of avian pathogenic Escherichia coli plasmids by a commensal E. coli isolate enhances its abilities to kill chicken embryos, grow in human urine, and colonize the murine kidney. Infect Immun Immun 74:6287–6292. doi: 10.1128/IAI.00363-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tivendale KA, Logue CM, Kariyawasam S, Jordan D, Hussein A, Li G, Wannemuehler Y, Nolan LK. 2010. Avian-pathogenic Escherichia coli strains are similar to neonatal meningitis E. coli strains and are able to cause meningitis in the rat model of human disease. Infect Immun 78:3412–3419. doi: 10.1128/IAI.00347-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mora A, López C, Dabhi G, Blanco M, Blanco JE, Alonso MP, Herrera A, Mamani R, Bonacorsi S, Moulin-Schouleur M, Blanco J. 2009. Extraintestinal pathogenic Escherichia coli O1:K1:H7/NM from human and avian origin: detection of clonal groups B2 ST95 and D ST59 with different host distribution. BMC Microbiol 9:132. doi: 10.1186/1471-2180-9-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mahjoub-Messai F, Bidet P, Caro V, Diancourt L, Biran V, Aujard Y, Bingen E, Bonacorsi S. 2011. Escherichia coli isolates causing bacteremia via gut translocation and urinary tract infection in young infants exhibit different virulence genotypes. J Infect Dis 203:1844–1849. doi: 10.1093/infdis/jir189. [DOI] [PubMed] [Google Scholar]
- 16.Yun KW, Kim DS, Kim W, Lim IS. 2015. Molecular typing of uropathogenic Escherichia coli isolated from Korean children with urinary tract infection. Korean J Pediatr 58:20–27. doi: 10.3345/kjp.2015.58.1.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poulsen LL, Thøfner I, Bisgaard M, Christensen JP, Olsen RH, Christensen H. 2017. Longitudinal study of transmission of Escherichia coli from broiler breeders to broilers. Vet Microbiol 207:13–18. doi: 10.1016/j.vetmic.2017.05.029. [DOI] [PubMed] [Google Scholar]
- 18.Pires-dos-Santos T, Bisgaard M, Christensen H. 2013. Genetic diversity and virulence profiles of Escherichia coli causing salpingitis and peritonitis in broiler breeders. Vet Microbiol 162:873–880. doi: 10.1016/j.vetmic.2012.11.008. [DOI] [PubMed] [Google Scholar]
- 19.Pires dos Santos T. 2014. Population genetics, evolution and molecular epidemiology of Escherichia coli isolated from chickens. PhD thesis. University of Copenhagen, Copenhagen, Denmark. [Google Scholar]
- 20.Mora A, Viso S, López C, Alonso MP, García-Garrote F, Dabhi G, Mamani R, Herrera A, Marzoa J, Blanco M, Blanco JE, Moulin-Schouleur M, Schouler C, Blanco J. 2013. Poultry as reservoir for extraintestinal pathogenic Escherichia coli O45:K1:H7-B2-ST95 in humans. Vet Microbiol 167:506–512. doi: 10.1016/j.vetmic.2013.08.007. [DOI] [PubMed] [Google Scholar]
- 21.Johnson TJ, Kariyawasam S, Wannemuehler Y, Mangiamele P, Johnson SJ, Doetkott C, Skyberg JA, Lynne AM, Johnson JR, Nolan LK. 2007. The genome sequence of avian pathogenic Escherichia coli strain O1:K1:H7 shares strong similarities with human extraintestinal pathogenic E. coli genomes. J Bacteriol 189:3228–3236. doi: 10.1128/JB.01726-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rasko DA, Rosovitz MJ, Myers GSA, Mongodin EF, Fricke WF, Gajer P, Crabtree J, Sebaihia M, Thomson NR, Chaudhuri R, Henderson IR, Sperandio V, Ravel J. 2008. The pangenome structure of Escherichia coli: comparative genomic analysis of E. coli commensal and pathogenic isolates. J Bacteriol 190:6881–6893. doi: 10.1128/JB.00619-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu Ge X, Jiang J, Pan Z, Hu L, Wang S, Wang H, Leung FC, Dai J, Fan H. 2014. Comparative genomic analysis shows that avian pathogenic Escherichia coli isolate IMT5155 (O2:K1:H5; ST complex 95, ST140) shares close relationship with ST95 APEC O1:K1 and human ExPEC O18:K1 strains. PLoS One 9:e112048. doi: 10.1371/journal.pone.0112048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roosaare M, Puustusmaa M, Möls M, Vaher M, Remm M. 2018. PlasmidSeeker: identification of known plasmids from bacterial whole genome sequencing reads. PeerJ 6:e4588. doi: 10.7717/peerj.4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grose JH, Bergthorsson U, Roth JR. 2005. Regulation of NAD synthesis by the trifunctional NadR protein of Salmonella enterica. J Bacteriol 187:2774–2782. doi: 10.1128/JB.187.8.2774-2782.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Finn TJ, Shewaramani S, Leahy SC, Janssen PH, Moon CD. 2017. Dynamics and genetic diversification of Escherichia coli during experimental adaptation to an anaerobic environment. PeerJ 5:e3244. doi: 10.7717/peerj.3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pichoff S, Alibaud L, Guédant A, Castanié MP, Bouché JP. 1998. An Escherichia coli gene (yaeO) suppresses temperature-sensitive mutations in essential genes by modulating Rho-dependent transcription termination. Mol Microbiol 29:859–869. doi: 10.1046/j.1365-2958.1998.00981.x. [DOI] [PubMed] [Google Scholar]
- 28.Gutiérrez P, Kozlov G, Gabrielli L, Elias D, Osborne MJ, Gallouzi IE, Gehring K. 2007. Solution structure of YaeO, a rho-specific inhibitor of transcription termination. J Biol Chem 282:23348–23353. doi: 10.1074/jbc.M702010200. [DOI] [PubMed] [Google Scholar]
- 29.Mahren S, Schnell H, Braun V. 2005. Occurrence and regulation of the ferric citrate transport system in Escherichia coli B, Klebsiella pneumoniae, Enterobacter aerogenes, and Photorhabdus luminescens. Arch Microbiol 184:175–186. doi: 10.1007/s00203-005-0035-y. [DOI] [PubMed] [Google Scholar]
- 30.Martínez-del Campo A, Bodea S, Hamer HA, Marks JA, Haiser HJ, Turnbaugh PJ, Balskusa EP. 2015. Characterization and detection of a widely distributed gene cluster that predicts anaerobic choline utilization by human gut bacteria. mBio 6:e00042-15. doi: 10.1128/mBio.00042-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maluta RP, Logue CM, Casas MRT, Meng T, Guastalli EAL, Rojas TCG, Montelli AC, Sadatsune T, de Carvalho Ramos M, Nolan LK, da Silveira WD. 2014. Overlapped sequence types (STs) and serogroups of avian pathogenic (APEC) and human extra-intestinal pathogenic (ExPEC) Escherichia coli isolated in Brazil. PLoS One 9:e105016. doi: 10.1371/journal.pone.0105016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nicholson BA, West AC, Mangiamele P, Barbieri N, Wannemuehler Y, Nolan LK, Logue CM, Li G. 2016. Genetic characterization of ExPEC-like virulence plasmids among a subset of NMEC. PLoS One 11:e0147757. doi: 10.1371/journal.pone.0147757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mellata M, Touchman JW, Curtis R. 2009. Full sequence and comparative analysis of the plasmid pAPEC-1 of avian pathogenic E. coli 7122 (O78:k80:H9). PLoS One 4:e4232. doi: 10.1371/journal.pone.0004232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Olsen RH, Christensen H, Bisgaard M. 2012. Comparative genomics of multiple plasmids from APEC associated with clonal outbreaks demonstrates major similarities and identifies several potential vaccine-targets. Vet Microbiol 158:384–393. doi: 10.1016/j.vetmic.2012.03.008. [DOI] [PubMed] [Google Scholar]
- 35.The Danish Integrated Antimicrobial Resistance Monitoring and Research Programme (DANMAP). 2017. Danmap 2016 - use of antimicrobial agents and occurrence of antimicrobial resistance in bacteria from food animals, food and humans in Denmark. Statens Serum Institut, Copenhagen, Denmark. [Google Scholar]
- 36.Dias RCS, Moreira BM, Riley LW. 2010. Use of fimH single-nucleotide polymorphisms for strain typing of clinical isolates of Escherichia coli for epidemiologic investigation. J Clin Microbiol 48:483–488. doi: 10.1128/JCM.01858-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gomi R, Matsuda T, Matsumura Y, Yamamoto M, Tanaka M, Ichiyama S, Yoneda M. 2017. Whole-genome analysis of antimicrobial-resistant and extraintestinal pathogenic Escherichia coli in river water. Appl Environ Microbiol 83:e02703-16. doi: 10.1128/AEM.02703-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kallonen T, Brodrick HJ, Harris SR, Corander J, Brown NM, Martin V, Peacock SJ, Parkhill J. 2017. Systematic longitudinal survey of invasive Escherichia coli in England demonstrates a stable population structure only transiently disturbed by the emergence of ST131. Genome Res 27:1437–1449. doi: 10.1101/gr.216606.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnson JR, Murray AC, Gajewski A, Sullivan M, Snippes P, Kuskowski MA, Smith KE. 2003. Isolation and molecular characterization of nalidixic acid-resistant extraintestinal pathogenic Escherichia coli from retail chicken products. Antimicrob Agents Chemother 47:2161–2168. doi: 10.1128/AAC.47.7.2161-2168.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ronco T, Stegger M, Olsen RH, Sekse C, Nordstoga AB, Pohjanvirta T, Lilje B, Lyhs U, Andersen PS, Pedersen K. 2017. Spread of avian pathogenic Escherichia coli ST117 O78:H4 in Nordic broiler production. BMC Genomics 18:13. doi: 10.1186/s12864-016-3415-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carattoli A, Zankari E, García-Fernández A, Larsen MV, Lund O, Villa L, Aarestrup FM, Hasman H. 2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Joensen KG, Tetzschner AMM, Iguchi A, Aarestrup FM, Scheutz F. 2015. Rapid and easy in silico serotyping of Escherichia coli isolates by use of whole-genome sequencing data. J Clin Microbiol 53:2410–2426. doi: 10.1128/JCM.00008-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roer L, Tchesnokova V, Allesøe R, Muradova M, Chattopadhyay S, Ahrenfeldt J, Thomsen MCF, Lund O, Frank H, Hammerum AM, Sokurenko EV, Hasman H. 2017. Development of a web tool for Escherichia coli subtyping based on fimH alleles. J Clin Microbiol 55:2538–2543. doi: 10.1128/JCM.00737-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sahl JW, Roe C, Smith DE, Williamson CHD, Aziz M, Keim P, Hepp CM, Driebe EM, Lemmer D, Travis J, Hicks ND, Schupp JM, Wagner DM, Engelthaler DM, Gillece JD, Sahl JW, Drees KP, Lemmer D, Travis J, Schupp JM, Gillece JD, Aziz M, Driebe EM, Drees KP, Hicks ND, Williamson CHD, Hepp CM, Smith DE, Engelthaler DM, Wagner DM, Keim P. 2016. NASP: an accurate, rapid method for the identification of SNPs in WGS datasets that supports flexible input and output formats. Microb Genomics 2:e000074. doi: 10.1099/mgen.0.000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Østerlund M, Kiil K. 2018. CleanRecomb, a quick tool for recombination detection in SNP based cluster analysis. bioRxiv https://www.biorxiv.org/content/early/2018/05/09/317131.
- 48.Letunic I, Bork P. 2016. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jombart T. 2012. An introduction to adegenet 1.3-4. R vignette, R-Forge, The R Project for Statistical Computing.
- 50.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 51.Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MTG, Fookes M, Falush D, Keane JA, Parkhill J. 2015. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maturana VG, de Pace F, Carlos C, Mistretta Pires M, Amabile de Campos T, Nakazato G, Guedes Stheling E, Logue CM, Nolan LK, Dias da Silveira W. 2011. Subpathotypes of avian pathogenic Escherichia coli (APEC) exist as defined by their syndromes and virulence traits. Open Microbiol J 5:55–64. doi: 10.2174/1874285801105010055. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary phylogenetic trees of the core genome (A) and accessory genome (B) of 323 genomes in the study. APEC genomes are marked with red dots on the branch ends. HExPEC subclades are indicated by colored branches as follows: HExPEC-1 subclade is shown in blue, and HExPEC-2 subclade is shown in green. Surrounding symbols indicate FimH type (squares), isolation country (triangles), and isolation year (circles). Download FIG S1, PDF file, 1.4 MB (1.5MB, pdf) .
Copyright © 2019 Jørgensen et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Putative gene sequences. Download Table S1, DOCX file, 0.01 MB (12.6KB, docx) .
Copyright © 2019 Jørgensen et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
List of strains used in this study. Download Table S2, DOCX file, 0.1 MB (58.3KB, docx) .
Copyright © 2019 Jørgensen et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Percent abundance of antibiotic resistance genes for each of the groups; APEC, ExPEC, A/E-PEC, HExPEC-1, and HExPEC-2. Significant P values are indicated by asterisks as follows; *, P > 0.05; **, P > 0.01; ***, P > 0.001. Download FIG S2, TIF file, 0.1 MB (147.1KB, tif) .
Copyright © 2019 Jørgensen et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.