Conspectus
The unique physiochemical properties and multiscale organization of layered materials draw the attention of researchers across a wide range of scientific disciplines. Layered structures are commonly found in diverse biological systems where they fulfill various functions. A prominent example of layered biological materials is the organization of proteins and polypeptides into the archetypal aggregated amyloidal structures. While the organization of proteins into amyloid structures was initially associated with various degenerative disorders, it was later revealed that proteins not related to any disease could also form identical layered assemblies. Thus, it appears that the ability of peptides and proteins to produce amyloid-like aggregates represents a generic property of polyamides to assemble into higher order fibrillar structures. In the aggregated state, the peptide backbone forms β-sheet structures which are further organized into layered arrangements.
We have recently extended the identified amyloidogenic building blocks to include not only peptides or proteins, but also single amino acids and other metabolites. High resolution spectroscopy and crystallography analyses confirm the clear potential of amino acids and other metabolites to form layered amyloid-like aggregates showing biophysical and biochemical properties similar to protein amyloids. Therefore, the generic propensity of peptides and proteins backbones to assemble into layered organizations may emanate from their basic building block, the amino acid. In this Account, we aim to introduce the concept of supramolecular β-sheet organization of single amino acids and to present an analysis of their layered-structure organization based on single crystal structures. We demonstrate that, despite the different side-chains that considerably vary in their chemical properties, all coded amino acids display a layer-like assembly stabilized by α-amine to α-carboxyl H-bonds, resembling supramolecular β-sheet structures, while the side-chains determine the higher order organization of the layers. Our work presents the first analysis of the β-sheet propensity of single amino acids in their unbound form, indicating an evolutionary predisposition. We classify the amino acids β-sheet propensity on the basis of the interlayer separation distance in the crystal packing, which correlates well with previously reported classifications based on various criteria, such as hydrophobicity, steric bulkiness, and folding. In addition, we demonstrate that the relative direction of α-amine to α-carboxyl H-bonding pattern provides critical insights regarding the stabilization of parallel versus antiparallel β-sheet structures by the various amino acids. Taken together, our analysis of amino acid crystals provides substantial information regarding protein folding and dynamics and could serve as basic rules set for the design of potential building blocks for molecular self-assembly to produce functional materials of tunable properties, an important objective of bottom-up nanotechnology.
1. Introduction
1.1. Synthetic and Biological Layered Structures
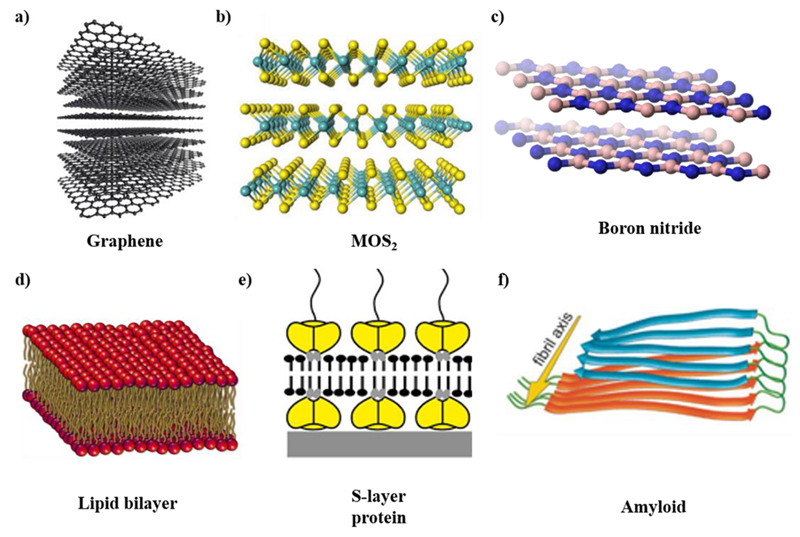
The formation of layered structures by different materials is one of the most important areas of advanced material research.1 Layered structures formed by two-dimensional (2D) materials offer a great opportunity for fundamental studies as well as practical applications. Graphene is the first material found to exist as truly two-dimensional layered structure (Figure 1a). Other important materials with 2D properties include MoS2 (Figure 1b), boron nitride (Figure 1c), and other dichalcogenides. Various functional materials, such as highly selective sensors and drug delivery systems, can be efficiently fabricated using nanomaterials arranged in ordered layered structures. Layered structures generated by controlled sequences and layer spacing lead to novel materials with different properties compared to the corresponding bulk materials.
Figure 1.
Layer structured materials. (a) Graphene. Reproduced with permission from ref 4. Copyright 2012 OpenStax. (b) MoS2. Reproduced with permission from ref 5. Copyright 2013 National Academy of Sciences, U.S.A. (c) Boron nitride. Reproduced with permission from ref 6. Copyright 2010 OpenStax-CNX. (d) Lipid bilayer. Reproduced with permission from ref 7. Copyright 2016 Nature Publishing Group. (e) S-layer protein. Reproduced with permission from ref 3. Copyright 2014 Oxford University Press. (f) Amyloid. Reproduced with permission from ref 8. Copyright 2002 National Academy of Sciences, U.S.A.
Beyond material science and engineering, layered structures are very common to evolutionarily developed biological systems. A most notable example is phospholipid membranes, which are composed of two basic parts, a polar head comprising a phosphate group, and two nonpolar fatty acid tails. In their energy minimum state, these cell membranes form a bilayer structure, with their polar heads pointing outward and bound with water molecules through hydrogen bonding, and their hydrophobic tails situated inside and bound via van der Waals interactions, thus further stabilizing the lipid bilayer structure (Figure 1d).2 This structure accounts for the selective permeability of hydrophilic molecules and ions.
Specifically, layered protein structures have been known for many years. The surface layer (S-layer) of almost all archaea and many bacteria is composed of regularly ordered isoporous lattices of protein layers completely covering the cell surface, thus constituting a component of the cell wall, an essential structural feature of prokaryotic organisms (Figure 1e).3 These layered structures provide microorganisms with diverse selective advantages, performing various functions, such as molecular sieves, ion traps, antifouling layers, and protective coats, as well as playing a role in cell adhesion and surface recognition.
Even under normal physiological conditions, many peptides and proteins fold into amyloid-like, rather than globular, structures (Figure 1f).8,9 Noncovalent interactions, which do not involve electrons sharing by molecules, but rather more dispersed variations of electromagnetic interactions, play crucial roles in the stabilization of amyloid structures. These non-covalent interactions mainly include hydrogen bonding, π−π stacking, electrostatic and hydrophobic interactions, as well as various other types. The assembly into ordered amyloid structures is associated with many degenerative disorders, such as Alzheimer’s disease, Parkinson’s disease, and Type II diabetes. Although amyloid aggregation was initially thought to be only associated with disease state, it was later shown that peptides and proteins not associated with any disease can also form amyloid-like assemblies with chemical and biophysical properties similar to the disease-associated structures.10 Applying a reductionist approach, it was identified that very short motifs (as short as dipeptides) are also able to self-assemble into ordered structures showing common features with protein amyloids, including amide bond conformation and binding to indicative dyes.11–14 Amyloids are also believed to have played an important role in the early evolution of proteins,15 suggesting that the amyloid state of polypeptides and proteins backbones is generic in nature.
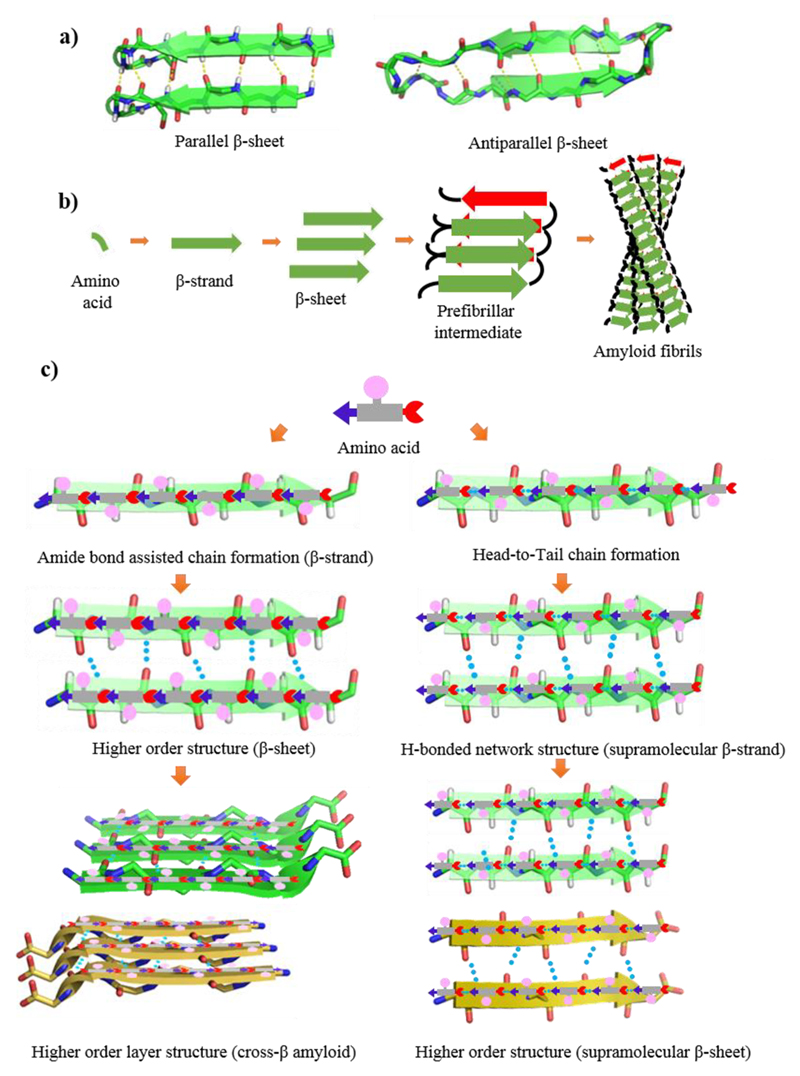
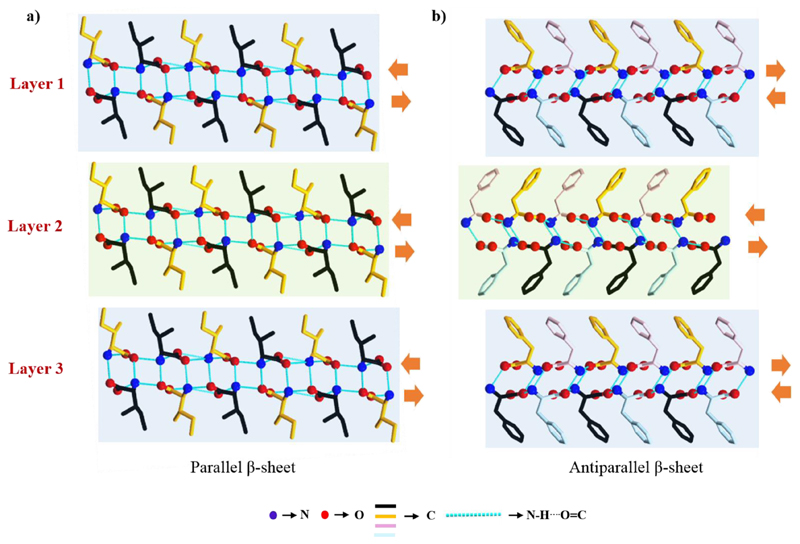
Amyloid structure has been investigated using various methods, including cryo-electron microscopy, solid-state NMR spectroscopy, and X-ray fiber diffraction.9 These studies have shown that in spite of differences in chain length and amino acid sequences, all amyloid fibrils share a common structure, comprised of β-sheet conformation in which the β-strands are perpendicular to the fiber axis and the hydrogen bonding direction is parallel. Therefore, the prevalence of β-sheet structures is attributed to the inherent stability of the peptide backbone. The β-sheet structure is the extended H-bonding network of peptide backbones in which the N−H groups of one backbone interact with the C = O groups of the neighbor. In this structure, H-bond forming groups are situated in a position that allows the formation of ideal H-bonds between peptide chains by placing the side-chain residues in optimal configuration for favorable interactions.16 Some of these interactions include hydrophobic forces between adjacent β-branched side chains, hydrophobic and π-stacking interactions among aromatic side chains and electrostatic attractions between charged amino acid pairs. Depending on the arrangement of N−H⋯O═C bonds of adjacent peptide backbones, β-strands are classified into parallel (same direction) and antiparallel (opposite direction) arrangements (Figure 2a). This layered cross-β-structure is stabilized by H-bonding and side-chain interactions, forming a higher order nanofibrillar assembly with excellent mechanical and physical robustness (Figure 2b).
Figure 2.
(a) Parallel and antiparallel β-sheet structure, PBD ID: 2MUS and 5GOC. (b) Generalized pathway for amyloid fibril formation from a single amino acid. (c) Amino acid−based amide bond mediated β-sheet (left) and H-bond mediated supramolecular β-sheet (right) structures.
Recently, our group has demonstrated that the study of amyloidogenic building blocks can be extended to amino acids and nucleobases related to inborn error of metabolism disorders.17–20 Amino acids like phenylalanine (Phe), cysteine (Cys), tryptophan (Trp), and tyrosine (Tyr) can form amyloid-like aggregates with the characteristic biophysical, biochemical, and cytotoxic properties. Single amino acids are thus able to aggregate into β-sheet-like layered structures. These studies suggested that systematic analysis of single crystal X-ray structures of amino acids may provide valuable information regarding their H-bonding patterns and layer formation. In pioneer work, Görbitz carefully investigated amino acid crystal structures and provided important insights.21 Several other review articles have also discussed various aspects of amino acids crystal structures.22,23 However, to date, no attempts have been made to define the crystal packing of amino acids as layered structured materials, reminiscent of cross-β architecture, and the effects of such organizations on protein folding. Moreover, all the reported supramolecular β-Sheet structures comprise peptides, where amino acids are connected through amide bonds. In this Account, we will introduce the concept of supramolecular secondary structure formed by amino acids without any covalent bonds between them and discuss their supramolecular assembly into layered structures based on this concept, providing a novel and robust perspective to analyzing the tendency of the amino acids to form layered structural materials. In the context of this publication, “layered” means propagating as a single layer. Furthermore, we attempt to draw a correlation between the supramolecular secondary structure and the propensity of the amino acids to adopt a β-sheet conformation when incorporated inside polypeptide chains. This analysis lays the basis for providing a prediction of parallel or antiparallel β-sheet formation based on peptide or protein sequence, which is fundamental to advance our understanding of protein folding. This Account also suggests answers for unresolved phenomena, such as the higher frequency of nonpolar, aliphatic hydrocarbon amino acids, such as valine (Val), isoleucine (Ile), and leucine (Leu), in parallel strands as compared to antiparallel strands.24 Finally, we outline some of the applications of layered amino acid structures in material science, nanotechnology and the design of peptide-based biomaterials.
1.2. Supramolecular Secondary Structure
Peptides and proteins are polymers of amino acids connected by amide bonds. This polymeric module can arrange in a linear chain known as β-strand and subsequently form H-bonds mediated interactions with the adjacent strand to form a β-sheet. Such structures may form a highly stable layered cross-β structure with optimized side chains packing (Figure 2c, left panel). It can be envisioned that individual amino acids also have the potential to form head-to-tail α-amine to α-carboxyl H-bonding in their zwitterionic form, as shown in Figure 2c (right panel), to generate H-bond assisted chain-like structure irrespective of the side-chain (hydrophobic or hydrophilic). These chain-like structures further assemble to H-bonded single layer comprising of two adjacent chains. We termed this assembly as supramolecular β-strand. The H-bonded β-strand network can further assemble with the adjacent strand to form supramolecular β-sheets. The crystals of amino acids, excluding glycine (Gly), are optically active, enantiomorphic, and exist in two possible molecular configurations, namely levorotated (l) and dextrorotated (d). However, the exclusive presence of l-amino acids in proteins of all living organisms still remains a mystery. Here, we consider only zwitterionic crystal structures of naturally occurring 20 amino acids, including 19 coded l-amino acids and Gly. The formation of supramolecular layered structures resulting from the assembly of head-to-tail H-bonding between α-amine and α-carboxyl groups is discussed. The solvent can impose a powerful and subtle effect on the molecular self-assembly of a nucleating system. Moreover, several other factors, including temperature, concentration, and additives, control the nucleation of crystallization, resulting in the generation of different polymorphs of the same amino acid. Here, we explore the packing pattern of all the polymorphs to gain insights regarding comparative β-sheet propensity of the different polymorphs of a particular amino acid. The supramolecular β-sheet concept introduced here comprises stacked single layers of H-bonded amino acids (denoted here as “supramolecular β-strands”) forming a network (Figure 2c, right). This organization is highly similar to the layered higher order organization of amyloid cross-β structure (Figure 2c, left). As the packing of the side chains is essential for the exceptional stability of the amyloid cross-β structure, the notion of supramolecular β-sheet is highly instrumental for understanding amino acids higher-order organization and determining their propensity in the context of protein secondary structures. Thus, we show that all coded amino acids have a tendency to organize into supramolecular β-sheet-like layered structures. The different parameters of layered geometry, such as interlayers separation and the relative directions of adjacent N−H⋯O═C supramolecular β-strands, have direct consequence on their tendency to form β-sheet-like secondary structures.
2. Layered Structure Formation by Amino Acids
2.1. Layered Structure Formation by Hydrophobic Amino Acids
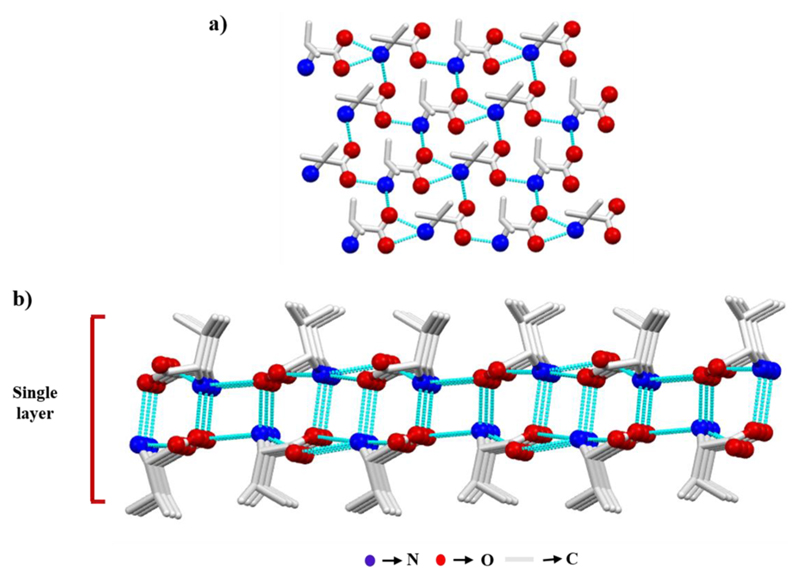
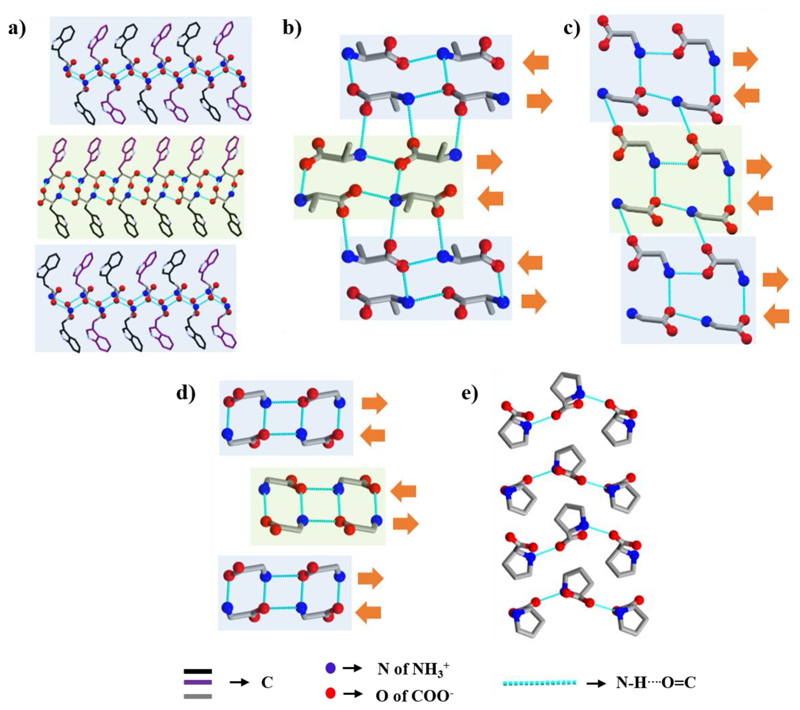
In aliphatic amino acids and Phe, the only functional groups available to form H-bonds are the α-amine and α-carboxyl groups. Indeed, in the crystal, individual amino acids interact with each other via head-to-tail intermolecular H-bonds between the polar head groups to form a two-dimensional molecular chain. Among the three amino hydrogens, one is involved in molecular chain formation, whereas the second connects the parallel molecular chains to fabricate a sheet (Figure 3a) in which all the side chains are positioned in the same direction. In an original work, Görbitz and colleagues showed four distinct H-bonding patterns giving rise to four different classes of hydrophilic sheets (L1, L2, L3, and Lx) observed in the crystal structure of hydrophobic amino acids.25 Notably, an acceptor for the third amino hydrogen is required which is compensated by intermolecular interactions with adjacent sheet, thereby producing a single layer (Figure 3b). The monolayers pack up to form a double layer assembly, in which side-chain to side-chain van der Waals forces connect the hydrophobic edges of the planes.
Figure 3.
l-Val crystal demonstrating the packing in the crystal structure of hydrophobic amino acids. (a) Sheet formation by a parallel set of head-to-tail H-bonded chains. (b) Single layer produced by two adjacent molecular sheets.
2.1.1. l-Isoleucine
The reported crystal structure of l-Ile has two molecules in the asymmetric unit.26 The α-amine to α-carboxyl H-bonds generate a chain and then two-dimensional sheet-like structure. Two adjacent sheets interact through H-bonds, generating single layer that resembles a single β-strand. Adjacent monolayers pack up to form a layer-by-layer structure where weak van der Waal interactions between hydrophobic side-chains keep the interlayer at a distance (Figure 4a). This layered crystal arrangement of l-Ile is the higher order organization of β-stands, similar to supramolecular β-sheet-like secondary structure. Accordingly, like peptide β-sheet classification, the β-sheet structure can be classified as parallel or antiparallel based on the N−H⋯O═C bonding direction inside each β-strand with respect to its neighbor β-strand. For l-Ile, the direction of H-bonds inside the top and bottom sheet of one β-strand is the same as that of the adjacent strand. Thus, the crystal packing of l-Ile represents a supramolecular parallel β-sheet-like secondary structure. The crystal packing pattern of l-Val,27 l-Leu, 28 and l-Met29 is highly similar.
Figure 4.
(a) Higher order layered structure of l-Ile in the b-direction resembles parallel β-sheet-like organization. (b) Antiparallel β-sheet-like structural arrangement of l-Phe (F−III) viewed along the b-axis. Orange arrows indicate the direction of N−H⋯O═C bonds within chains.
2.1.2. l-Phenylalanine
Görbitz and co-workers30 thoroughly analyzed the crystal structure of four different l-Phe polymorphs (F−I, F−II,31 F−III,32 and F−IV). F−I and F−III contain four molecules in the asymmetric unit and their packing is identical. Similar to other hydrophobic amino acids, individual β-strand is produced by two sheets, whereas the interstrand interactions between hydrophobic phenyl rings are thought to be related to edge-to-face π−π stacking interactions (Figure 4b). Two different β-strands consecutively repeat throughout the crystal packing, resulting in higher order stacking of β-strands. The direction of N−H⋯O═C bond in two sheets constituting a β-strand is opposite, whereas in the hydrophobic region it is parallel. Hence, the crystal packing represents a supramolecular antiparallel β-sheet-like secondary structure (Figure 4b), a very rare structure among the hydrophobic amino acids, previously observed only for α-Gly.
The F−II structure was elucidated using powder X-ray diffraction and found to be stable only under rigorously dry conditions. A large elongated wedges-shaped crystal of F−IV was solved with several disorders in the side chain, showing parallel organization of adjacent β-strands.
2.1.3. l-Tryptophan
Although l-Trp contains an extra N−H group in the pyrrole ring, this group does not take part in the N−H⋯O or N−H⋯N interactions.33 Considering α-amine to α-carboxyl H-bonding, a layer-by-layer arrangement (β-sheet) of Trp molecules was identified (Figure 5a). However, two nearby layers are perpendicular to each other, unlike Phe and other hydrophobic amino acids. The crystal packing is thus composed of two different kinds of repeating β-strands, comprising an antiparallel β-sheet-like structure.
Figure 5.
Supramolecular β-sheet-like secondary layered structure of (a) l-Trp viewed nearly along the ac-plane, (b) l-Ala in the a-direction, (c) β-Gly and (d) α-Gly in the ac-plane, and (e) l-Pro in the ab-plane.
2.1.4. l-Alanine and Glycine
Both l-Ala and Gly do not contain a H-bond forming side-chain. Yet, their packing pattern is different from that of other hydrophobic amino acids. In the crystal of l-Ala, a linear molecular chain is originated by head-to-tail H-bonding interactions along the c-axis (Figure 5b).34 Adjacent chains are connected through H-bonds generated by a 21 screw operation about the a-axis, producing a single layer (β-strand). Repetition of this β-strand along the b-direction fabricates a pleated β-sheet-like structure in which the direction of H-bonds inside each strand is opposite to the nearby strand.
In the β-Gly crystal, a single layer is produced by two branches of molecular chains fabricated by head-to-tail H-bonding interactions (Figure 5c).35 The direction of the N−H⋯O═C bond is opposite within each pair of chains forming a single β-strand and parallel to the neighbor β-strand. The crystal packing of β-Gly thus resembles a parallel β-sheet-like structure. A similar type of molecular chain and β-strand formation is also observed in the α-Gly (Figure 5d).35 Based on the direction of the N−H⋯O═C bond of adjacent β-strands, it represents an antiparallel β-sheet-like structure.
2.1.5. l-Proline
In the zwitterionic state, only two l-Pro amino H atoms are available for hydrogen bonding due to the side-chain ring.36 Thus, even though l-Pro forms wavy sheets by head-to-tail H-bonds (Figure 5e), two nearby sheets cannot interact via H-bonds. Thus, the crystal packing of l-Pro does not show a layered structure and does not resemble a supramolecular β-sheet-like structural arrangement.
2.2. Layered Structure Formation by Polar Amino Acids
Along with the amine and carboxylate head groups, polar amino acids generally contain functional groups capable of H-bond formation, resulting in diverse crystal packings, largely depending on the nature of H-bond donation or acceptance by the side-chain. Herein, we will consider the formation of a layer only through α-amine-to-α-carboxylate H-bond interactions. We will demonstrate that in spite of containing side-chains with different properties, all polar amino acids are also able to form a single layer (β-strand) only by head-to-tail arrangement. Also, by higher order self-assembly of β-strands, all polar amino acids form a β-sheet-like structural arrangement.
2.2.1. l-Cysteine
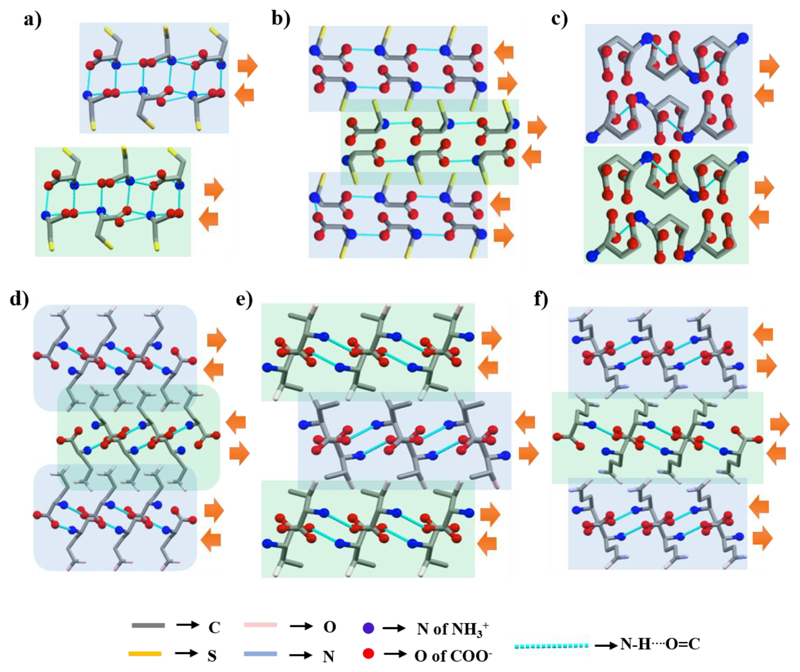
As the side-chain S−H group is not involved in H-bonding, the crystal packing of monoclinic l-Cys37 is similar to those of hydrophobic amino acids (Figure 6a) and represents a supramolecular parallel β-sheet-like structure.
Figure 6.
Supramolecular β-sheet-like layered secondary structure of (a) l-Cys (II) in the ac-plane, (b) l-Cys (I) in the bc-plane, (c) l-Glu α-form in the a-direction, (d) l-Glu β-form in the c-direction, and (e) l-Thr and (f) l-Gln in the ac-plane.
The crystal packing of orthorhombic l-Cys37 is completely different. A single layer (β-strand) is simultaneously formed by two branches of molecular chains produced by head-to-tail interactions (Figure 6b) and two adjacent layers are similarly stabilized by intermolecular H-bonds. The crystal packing resembles supramolecular antiparallel β-sheet-like secondary structural arrangement.
2.2.2. l-Glutamic Acid, l-Threonine, and l-Glutamine
Layered structure of l-Glu in the α-form is generated by two branches of molecular chains (Figure 6c).38 Two adjacent chains are connected by H-bonds involving both head groups and the side-chain, thereby comprising a single layer (β-strand). Two nearby layers are also connected through intermolecular H-bonds. The direction of the N−H⋯O═C bonds is identical in each β-strand, thus representing a parallel β-sheet-like structural arrangement.
In the β-form of l-Glu, the bonding pattern brings the head groups to the center of the layer and pushes the side-chain outward (Figure 6d).38 Two adjacent layers are connected by both O−H⋯O and O⋯H−N H-bonds formed by the side-chain carboxylic group. This crystal packing results in a zigzag pattern of the side-chain designated a herringbone-like structural arrangement. The direction of the N−H⋯O═C bonds in two adjacent β-strands is opposite, thus representing an antiparallel arrangement of β-strands. The crystal packing of l-Thr39 (Figure 6e) and l-Gln40 (Figure 6f) is highly similar to the β-form of l-Glu.
2.2.3. l-Aspartic Acid, l-Asparagine, and l-Histidine
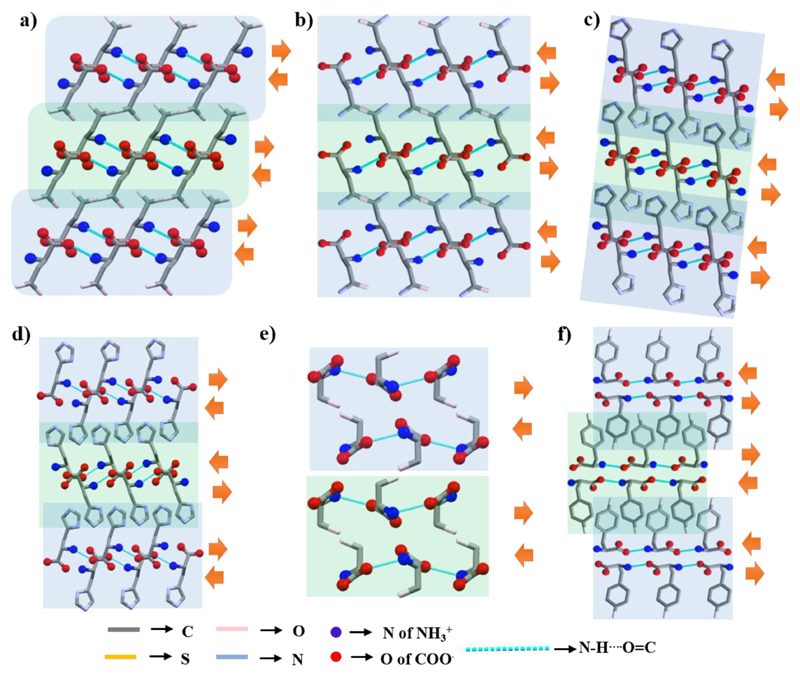
The molecular arrangement in the crystal packing of l-Asp,41 l-Asn,42 and both forms of l-His (monoclinic and orthorhombic)43 is very similar to the β-form of l-Glu (Figure 7a−d). Based on the direction of N−H⋯O═C bonds, this arrangement represents a parallel orientation of β-strands, except for orthorhombic l-His, which is antiparallel.
Figure 7.
Supramolecular β-sheet-like layered secondary structure of (a) l-Asp, (b) l-Asn, (c) l-His (monoclinic), and (d) l-His (orthorhombic) near the ac-plane, (e) l-Ser in the ab-plane, and (f) l-Tyr in the bc-plane.
2.2.4. l-Serine and l-Tyrosine
In addition to head-to-tail H-bonding, the higher order packing of the l-Ser crystal revealed that both the side-chain −OH group and the head groups are alternatively involved in bonding to construct a single β-strand as well as a β-sheet (Figure 7e).44 Based on the direction of the N−H⋯O═C H-bond within the single β-strand, l-Ser crystal structure represents a parallel β-sheet-like arrangement.
The well-ordered layered (β-strand) structure of l-Tyr is generated through the formation of a molecular chain (Figure 7f).45 Two consecutive layers are stabilized by intermolecular H-bonds involving the phenolic−OH group. According to the direction of N−H⋯O═C bonds within each strand, l-Tyr displays an antiparallel arrangement of β-strands.
The single crystal structure of l-Arg and l-Lys is yet to be reported. In the powder X-ray structures, no H-bond interaction between α-amine-to-α-carboxyl head groups was found. Therefore, we did not include these two amino acids in this analysis.
3. Amino Acid Predisposition to Parallel/Antiparallel β-Sheet Structure
Based on the direction of N−H⋯O═C bonds within each β-strand, our predictions of parallel and antiparallel amino acids β-sheet structure highly correlate with a previous report. Otaki and Tsutsumi constructed a parallel strand database (PS-DB) and an antiparallel strand database (AS-DB) using the structure of 1641 proteins.24 They found a P/A ratio (i.e., ratio between PS-DB occurrence and AS-PD occurrence) > 1 for Ile, Val, Met, Leu, His, and Ala, for which, with the exception of Ala, our analysis also predicts a parallel arrangement of β-strands in the crystal structure. The lowest P/A values, implying a preference for an antiparallel β-strand arrangement, were calculated for Gln, Trp, Glu, Ser, Tyr, and Thr, as also predicted by our analysis, with the exception of Ser. The P/A ratio of other amino acids was calculated to be very close to 1, indicating a similar abundance of parallel and antiparallel strands, with a slightly higher frequency of the antiparallel strands. Likewise, we predicted an antiparallel β-sheet conformation for Phe and Cys. This classification of amino acids based on their preferred organization could be potentially applied for prediction of the directionality of proteins secondary structure.
4. Analysis of β-Sheet Propensity Based on Interlayer Separations
Understanding the β-sheet propensity of single amino acids would be highly useful for the prediction of protein folding, as well as for the design of different secondary structural arrangements. Minor and co-workers showed this property to be comparatively high for Thr, Ile, Val, Met, Tyr, and Phe, and low for His, Asp, Asn, Gly, and Pro.46 Side chain hydrophobicity was initially suggested as the key determinant, but Fujiwara and co-workers later found this to be true only for buried residues, not exposed ones.47 Street and colleagues demonstrated a more local phenomenon, namely the steric interaction between the side-chain and its local backbone.48 While several factors are thus assumed to play a role in β-sheet formation, a general trend is common to all reports. For a well-ordered β-sheet structure, there should be a minimal separation of two consecutive β-strands. Amino acids with layered structures indicate that the β-sheet propensity will be high for those with well-separated β-strands, but low for those where strands overlap.
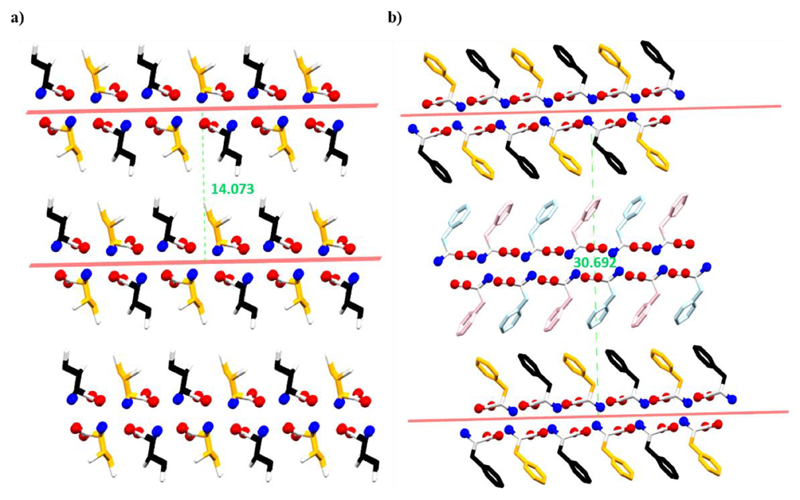
To measure the distance, we drew a plane through the center of each β-strand. For parallel β-sheet, the interlayer distance is defined as the distance between two adjacent planes (Figure 8a). For antiparallel β-sheet, half of the distance between two consecutive similar β-strands was considered as the average interlayer separation distance (Figure 8b). The interlayer distances of the natural amino acids are listed in Table 1.
Figure 8.
Interlayer distance in (a) parallel and (b) antiparallel β-sheet of l-Ile and l-Phe, respectively.
Table 1. Interlayer Separation Distance in Amino Acids Crystal Structure.
| amino acid | interlayer distance (Å) | β-sheet propensity |
|---|---|---|
| l-Trp | 17.695 | |
| l-Phe | ||
| F–I | 15.351 | |
| F–II | 12.881 | |
| F–III | 15.396 | high |
| F–IV | 15.393 | |
| l-Met | 15.009 | |
| l-Ile | 14.039 | |
| l-Leu | 14.073 | |
| l-Val | 12.059 | |
| l-Cys | ||
| monoclinic | 10.938 | |
| orthorhombic | 6.099 | |
| l-Tyr | 10.545 | |
| l-His | ||
| monoclinic | 9.424 | |
| orthorhombic | 9.427 | moderate |
| l-Ser | 9.323 | |
| l-Glu | ||
| α-form | 8.776 | |
| β-form | 8.670 | |
| l-Asn | 8.047 | |
| l-Gln | 8.002 | |
| l-Asp | 7.537 | |
| l-Thr | 6.803 | |
| l-Ala | 6.172 | low |
| Gly | ||
| α-form | 5.967 | |
| β-form | 4.923 |
The β-strands of two aromatic amino acids, Trp and Phe, are separated by largest distances, 17.695 and ~15 Å, respectively. Met, Ile, Leu, and Val show a high interlayer separation distance of 15−12 Å and are thus predicted to have a higher β-sheet propensity. l-Cys adopts a variable interlayer separation, indicating alternating roles in protein conformation. l-Tyr, l-His, l-Ser, l-Glu, l-Asn, and l-Gln have interlayer separation distances in the range of 10 Å − 8 Å and are expected to have a moderate β-sheet propensity. The β-strands of l-Asp, l-Thr, l-Ala, and Gly are separated by very low distances, below 8 Å, predicted to have a low β-sheet propensity. To conclude, our prediction of amino acids β-sheet propensity based on interlayer separation is well correlated with previous reports.46−48
5. Conclusions and Future Outlook
Natural evolution endows diverse functionalities to amino acids as reflected by their side-chains, ranging from hydrophobic to hydrophilic, from H-bond donor to acceptor, and so forth. Yet, all amino acids display α-amine to α-carboxyl H-bonded chain structures in the crystalline state, reminiscence of a β-strand. The chain structure is further arranged into a layer-like assembly and the interlayer separations and interactions are dictated by the side-chain properties. The specific layer geometry characteristic of individual amino acids may impose different constrains in the context of a peptide or protein chain, thereby enforcing different secondary structures, such as α-helix, β-sheet or random conformations. The different interlayer distances and their remarkable correlation with β-sheet propensity indicate that further analysis of amino acid crystals will greatly aid in understanding protein folding and dynamics.
Furthermore, amino acids and peptides are currently emerging as excellent building blocks for the design and exploration of various biomaterials and nanomaterials. Amino acids layered structure formation can be used to decorate a surface to achieve different functionalities or to produce desired solids via physical or chemical vapor deposition techniques. For example, hydrophobic amino acids can impart water repellant properties on a surface, as well as be used to design materials with low interlayer frictions owing to the presence of only hydrophobic interactions and high separations between two adjacent layers. Hydrophilic surfaces can be engineered using polar amino acids with very high interlayer distances. The extent of hydrophilicity or hydrophobicity can be controlled by selecting different amino acids. Such vapor deposition techniques could also alter the chemical nature of individual amino acids, for instance, through polymerization, thereby affording new unexplored functionalities.
The design of predictable self-assembling peptide motifs is crucial for advancement of their uses in biotechnology. To date, small peptide modules have been mainly designed by a trial and error approach. Ulijn and co-workers made a notable progress toward predicting the supramolecular behavior of small peptides based on sequence alone by employing computational tools or novel experimental methodology involving enzymatic reactions.49,50 The crystal structures analysis presented here reveals that different amino acids align with different interlayer distances. Based on this observation, it can be hypothesized that arrangement of amino acids with comparable interlayer distances will have a higher propensity to aggregate into a stable β-sheet structure, forming distinct supramolecular nanostructures. Furthermore, coassembly of two different amino acids with similar layer distances could afford unique self-assembled structures not obtainable via organization of a single amino acid. Several highly aggregating peptides identified by Ulijn et al. using molecular dynamics showed quite a good correlation with the hypothesis postulated here, which may further lead to the identification of additional novel sequences. Another very interesting direction in the design of new synthetic peptide materials is using noncoded and non-natural amino acids, such as phenylglycine and homophenylalanine, 2-amino-5-phenyl-pentanoic acid, 2-amino-6-phenylhexanoic acid, and so on.51,52 Crystallographic analysis of these amino acids at the molecular level could provide a new tool for the design of novel chemical entities based on modified peptides, with implications ranging from material engineering and supramolecular polymers to drug design and bionanotechnology. Finally, it is conceivable that the amino acid crystal structures present a wealth of information still to be explored.
Acknowledgments
S.B. thanks Tel Aviv University for fellowship. S.M. thanks the Planning and Budgeting Committee (PBC) Program for scholarship. This project received funding from ERC under the European Union Horizon 2020 Research (BISON project).
Biographies
Santu Bera joined Prof. Ehud Gazit’s research group as a postdoctoral fellow in 2016. His areas of research include exploration of the supramolecular structures of the simplest natural building blocks, namely, amino acids and related biomolecules.
Sudipta Mondal is a postdoctoral fellow at the lab of Prof. Ehud Gazit since 2013. His research explores small peptides self-assembly into ordered nanostructures and the organization of conformationally constrained minimal helical sequences into super helical structures.
Sigal Rencus-Lazar joined Prof. Ehud Gazit’s group in 2017 as a scientific writer and editor. She completed her B.Sc., M.Sc. (both summa cum laude), and Ph.D. studies at Tel Aviv University.
Prof. Ehud Gazit is the incumbent of the Chair for Biotechnology of Neurodegenerative Diseases at Tel Aviv University. Prof. Gazit completed his B.Sc. (summa cum laude) at the Special Program for Outstanding Students at Tel Aviv University and his Ph.D. (with highest distinction) at the Weizmann Institute of Science. In 2000, after completing his postdoctoral studies at The Massachusetts Institute of Technology (MIT), he established his research lab at Tel Aviv University. Gazit has received numerous honors and awards, including The Landau Research Award, Hesterin Award and Kadar Family Award. He is a member of the European Molecular Biology Organization (EMBO) and a fellow of the Royal Society of Chemistry (FRSC).
Footnotes
ORCID
Ehud Gazit: 0000-0001-5764-1720
Notes
The authors declare no competing financial interest.
References
- (1).Low T, Chaves A, Caldwell JD, Kumar A, Fang NX, Avouris P, Heinz TF, Guinea F, Martin-Moreno L, Koppens F. Polaritons in layered two-dimensional materials. Nat Mater. 2017;16:182–194. doi: 10.1038/nmat4792. [DOI] [PubMed] [Google Scholar]
- (2).Nagle JF, Tristram-Nagle S. Lipid bilayer structure. Curr Opin Struct Biol. 2000;10:474–480. doi: 10.1016/s0959-440x(00)00117-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Sleytr UB, Schuster B, Egelseer E-M, Pum D. S-layers: principles and applications. FEMS Microbiol Rev. 2014;38:823–864. doi: 10.1111/1574-6976.12063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).BC Open Textbooks, Chemistry. Chapter 10 Rice University, OpenStax; 2012. Liquids and Solids. [Google Scholar]
- (5).Wang H, Lu Z, Xu S, Kong D, Cha JJ, Zheng G, Hsu P-C, Yan K, Bradshaw D, Prinz FB, Cui Y. Electrochemical tuning of vertically aligned MoS2 nanofilms and its application in improving hydrogen evolution reaction. Proc Natl Acad Sci U S A. 2013;110:19701–19706. doi: 10.1073/pnas.1316792110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Barron A. Chemistry of the Main Group Elements. OpenStax-CNX; 2010. Boron Compounds with Nitrogen Donors. [Google Scholar]
- (7).Zhernenkov M, Bolmatov D, Soloviov D, Zhernenkov K, Toperverg BP, Cunsolo A, Bosak A, Cai YQ. Revealing the mechanism of passive transport in lipid bilayers via phonon-mediated nanometre-scale density fluctuations. Nat Commun. 2016;7 doi: 10.1038/ncomms11575. 11575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Petkova AT, Ishii Y, Balbach JJ, Antzutkin ON, Leapman RD, Delaglio F, Tycko R. A structural model for Alzheimer’s β-amyloid fibrils based on experimental constraints from solid state NMR. Proc Natl Acad Sci U S A. 2002;99:16742–16747. doi: 10.1073/pnas.262663499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Knowles TP, Vendruscolo M, Dobson CM. The amyloid state and its association with protein misfolding diseases. Nat Rev Mol Cell Biol. 2014;15:384–396. doi: 10.1038/nrm3810. [DOI] [PubMed] [Google Scholar]
- (10).Dobson CM. Protein folding and misfolding. Nature. 2003;426:884–890. doi: 10.1038/nature02261. [DOI] [PubMed] [Google Scholar]
- (11).Reches M, Porat Y, Gazit E. Amyloid fibril formation by pentapeptide and tetrapeptide fragments of human calcitonin. J Biol Chem. 2002;277:35475–35480. doi: 10.1074/jbc.M206039200. [DOI] [PubMed] [Google Scholar]
- (12).Reches M, Gazit E. Casting metal nanowires within discrete self-assembled peptide nanotubes. Science. 2003;300:625–627. doi: 10.1126/science.1082387. [DOI] [PubMed] [Google Scholar]
- (13).Reches M, Gazit E. Self-assembly of peptide nanotubes and amyloid-like structures by charged-termini-capped diphenylalanine peptide analogues. Isr J Chem. 2005;45:363–371. [Google Scholar]
- (14).Adler-Abramovich L, Aronov D, Beker P, Yevnin M, Stempler S, Buzhansky L, Rosenman G, Gazit E. Self-assembled arrays of peptide nanotubes by vapour deposition. Nat Nanotechnol. 2009;4:849–854. doi: 10.1038/nnano.2009.298. [DOI] [PubMed] [Google Scholar]
- (15).Carny O, Gazit E. A model for the role of short self-assembled peptides in the very early stages of the origin of life. FASEB J. 2005;19:1051–1055. doi: 10.1096/fj.04-3256hyp. [DOI] [PubMed] [Google Scholar]
- (16).Richardson JS. The anatomy and taxonomy of protein structure. Adv Protein Chem. 1981;34:167–339. doi: 10.1016/s0065-3233(08)60520-3. [DOI] [PubMed] [Google Scholar]
- (17).Adler-Abramovich L, Vaks L, Carny O, Trudler D, Magno A, Caflisch A, Frenkel D, Gazit E. Phenylalanine assembly into toxic fibrils suggests amyloid etiology in phenylketonuria. Nat Chem Biol. 2012;8:701. doi: 10.1038/nchembio.1002. [DOI] [PubMed] [Google Scholar]
- (18).Shaham-Niv S, Adler-Abramovich L, Schnaider L, Gazit E. Extension of the generic amyloid hypothesis to nonproteinaceous metabolite assemblies. Sci Adv. 2015;1 doi: 10.1126/sciadv.1500137. e1500137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Shaham-Niv S, Rehak P, Vuković L, Adler-Abramovich L, Kral P, Gazit E. Formation of apoptosis-inducing amyloid fibrils by tryptophan. Isr J Chem. 2017;57:729–737. [Google Scholar]
- (20).Berger O, Adler-Abramovich L, Levy-Sakin M, Grunwald A, Liebes-Peer Y, Bachar M, Buzhansky L, Mossou E, Forsyth VT, Schwartz T, Ebenstein Y, et al. Light-emitting self-assembled peptide nucleic acids exhibit both stacking interactions and Watson-Crick base pairing. Nat Nanotechnol. 2015;10:353–360. doi: 10.1038/nnano.2015.27. [DOI] [PubMed] [Google Scholar]
- (21).Görbitz CH. Crystal structures of amino acids: from bond lengths in glycine to metal complexes and high pressure polymorphs. Crystallogr Rev. 2015;21:160–212. [Google Scholar]
- (22).Boldyreva EV. Crystalline amino acids. In: Boeyens JCA, Ogilvie JF, editors. Models, Mysteries and magic of molecules. Springer; Dordrecht: 2008. pp. 167–192. [Google Scholar]
- (23).Schade B, Fuhrhop J-H. Amino acid networks. New J Chem. 1998;22:97–104. [Google Scholar]
- (24).Tsutsumi M, Otaki JM. Parallel and antiparallel β-strands differ in amino acid composition and availability of short constituent sequences. J Chem Inf Model. 2011;51:1457–1464. doi: 10.1021/ci200027d. [DOI] [PubMed] [Google Scholar]
- (25).Görbitz CH, Vestli K, Orlando R. A solution to the observed Ź = 2 preference in the crystal structures of hydrophobic amino acids. Acta Crystallogr, Sect B: Struct Sci. 2009;B65:393–400. doi: 10.1107/S0108768109014700. [DOI] [PubMed] [Google Scholar]
- (26).Torii K, Iitaka Y. The crystal structure of L-isoleucine. Acta Crystallogr, Sect B: Struct Crystallogr Cryst Chem. 1971;B27:2237–2246. [Google Scholar]
- (27).Torii K, Iitaka Y. The crystal structure of L-valine. Acta Crystallogr, Sect B: Struct Crystallogr Cryst Chem. 1970;B26:1317–1326. doi: 10.1107/s0567740870004065. [DOI] [PubMed] [Google Scholar]
- (28).Harding MM, Howieson RM. L-leucine. Acta Crystallogr, Sect B: Struct Crystallogr Cryst Chem. 1976;B32:633–634. [Google Scholar]
- (29).Torii K, Iitaka Y. Crystal structures and molecular conformations of L-methionine and L-norleucine. Acta Crystallogr, Sect B: Struct Crystallogr Cryst Chem. 1973;B29:2799–2807. [Google Scholar]
- (30).Ihlefeldt FS, Pettersen FB, von Bonin A, Zawadzka M, Gorbitz CH. The polymorphs of L-Phenylalanine. Angew Chem, Int Ed. 2014;53:13600–13604. doi: 10.1002/anie.201406886. [DOI] [PubMed] [Google Scholar]
- (31).Williams PA, Hughes CE, Buanz ABM, Gaisford S, Harris KDM. Expanding the solid-state landscape of L-phenylalanine: discovery of polymorphism and new hydrate phases, with rationalization of hydration/dehydration processes. J Phys Chem C. 2013;117:12136–12145. [Google Scholar]
- (32).Mossou E, Teixeira SCM, Mitchell EP, Mason SA, Adler-Abramovich L, Gazit E, Forsyth VT. The self-assembling zwitterionic form of L-phenylalanine at neutral pH. Acta Crystallogr, Sect C: Struct Chem. 2014;70:326–331. doi: 10.1107/S2053229614002563. [DOI] [PubMed] [Google Scholar]
- (33).Görbitz CH, Törnroos KW, Day GM. Single-crystal investigation of L-tryptophan with Ź=16. Acta Crystallogr, Sect B: Struct Sci. 2012;B68:549–557. doi: 10.1107/S0108768112033484. [DOI] [PubMed] [Google Scholar]
- (34).Funnell NP, Dawson A, Francis D, Lennie AR, Marshall WG, Moggach SA, Warren JE, Parsons S. The effect of pressure on the crystal structure of L-alanine. CrystEngComm. 2010;12:2573–2583. [Google Scholar]
- (35).Weissbuch I, Torbeev VY, Leiserowitz L, Lahav M. Solvent effect on crystal polymorphism: why addition of methanol or ethanol to aqueous solutions induces the precipitation of the least stable beta form of glycine. Angew Chem, Int Ed. 2005;44:3226–3229. doi: 10.1002/anie.200500164. [DOI] [PubMed] [Google Scholar]
- (36).Kayushina RL, Vainshtein BK. X-ray determination of the structure of L-proline. Kristallografiya. 1965;10:833–844. [Google Scholar]
- (37).Görbitz CH, Dalhus B. L-Cysteine, monoclinic form, redetermination at 120 K. Acta Crystallogr, Sect C: Cryst Struct Commun. 1996;C52:1756–1759. [Google Scholar]
- (38).Liang S, Duan X, Zhang X, Qian G, Zhou X. Insights into polymorphic transformation of L-glutamic acid: a combined experimental and simulation study. Cryst Growth Des. 2015;15:3602–3608. [Google Scholar]
- (39).Shoemaker DP, Donohue J, Schomaker V, Corey RB. The crystal structure of Ls-threonine. J Am Chem Soc. 1950;72:2328–2349. [Google Scholar]
- (40).Cochran W, Penfold BR. The crystal structure of L-glutamine. Acta Crystallogr. 1952;5:644–653. [Google Scholar]
- (41).Derissen JL, Endeman HJ, Peerdeman AF. The crystal and molecular structure of L-aspartic acid. Acta Crystallogr, Sect B: Struct Crystallogr Cryst Chem. 1968;B24:1349–1354. [Google Scholar]
- (42).Yamada K, Hashizume D, Shimizu T, Yokoyama S. L-asparagine. Acta Crystallogr, Sect E: Struct Rep Online. 2007;E63:o3802–o3803. [Google Scholar]
- (43).Wantha L, Flood AE. Growth and dissolution kinetics of A and B polymorphs of L-histidine. Chem Eng Technol. 2015;38:1022–1028. [Google Scholar]
- (44).Benedetti E, Pedone C, Sirigu A. The crystal structure of l-(−)-serine. Gazz Chim Ital. 1973;103:555–561. [Google Scholar]
- (45).Mostad A, Nissen HM, Romming C, Olli M, Pilotti A. Crystal structure of L-tyrosine. Acta Chem Scand. 1972;26:3819–3833. doi: 10.3891/acta.chem.scand.26-3819. [DOI] [PubMed] [Google Scholar]
- (46).Minor DL, Jr, Kim PS. Measurement of the β-sheet-forming propensities of amino acids. Nature. 1994;367:660–663. doi: 10.1038/367660a0. [DOI] [PubMed] [Google Scholar]
- (47).Fujiwara K, Toda H, Ikeguchi M. Dependence of α-helical and β-sheet amino acid propensities on the overall protein fold type. BMC Struct Biol. 2012;12:18. doi: 10.1186/1472-6807-12-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Street AG, Mayo SL. Intrinsic β-sheet propensities result from van der Waals interactions between side chains and the local backbone. Proc Natl Acad Sci U S A. 1999;96:9074–9076. doi: 10.1073/pnas.96.16.9074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Frederix PWJM, Scott GG, Abul-Haija YM, Kalafatovic D, Pappas CG, Javid N, Hunt NT, Ulijn RV, Tuttle T. Exploring the sequence space for (tri) peptide self-assembly to design and discover new hydrogels. Nat Chem. 2015;7:30–37. doi: 10.1038/nchem.2122. [DOI] [PubMed] [Google Scholar]
- (50).Pappas CG, Shafi R, Sasselli IR, Siccardi H, Wang T, Narang V, Abzalimov R, Wijerathne N, Ulijn RV. Dynamic peptide libraries for the discovery of supramolecular nanomaterials. Nat Nanotechnol. 2016;11:960–967. doi: 10.1038/nnano.2016.169. [DOI] [PubMed] [Google Scholar]
- (51).Reches M, Gazit E. Formation of closed-cage nanostructures by self-assembly of aromatic dipeptides. Nano Lett. 2004;4:581–585. [Google Scholar]
- (52).Pellach M, Mondal S, Shimon LJW, Adler-Abramovich L, Buzhansky L, Gazit E. Molecular engineering of self-assembling diphenylalanine analogues results in the formation of distinctive microstructures. Chem Mater. 2016;28:4341–4348. [Google Scholar]