Abstract
In flies, the olfactory information is carried from the first relay in the brain, the antennal lobe, to the mushroom body (MB) and the lateral horn (LH). Olfactory associations are formed in the MB. The LH was ascribed a role in innate responses based on the stereotyped connectivity with the antennal lobe, stereotyped physiological responses to odors, and MB silencing experiments. Direct evidence for the functional role of the LH is still missing. Here, we investigate the behavioral role of the LH neurons (LHNs) directly, using the CO2 response as a paradigm. Our results show the involvement of the LH in innate responses. Specifically, we demonstrate that activity in two sets of neurons is required for the full behavioral response to CO2. Tests of the behavioral response to other odors indicate the neurons are selective to CO2 response. Using calcium imaging, we observe that the two sets of neurons respond to CO2 in a different manner. Using independent manipulation and recording of the two sets of neurons, we find that the one that projects to the superior intermediate protocerebrum (SIP) also outputs to the local neurons within the LH. The design of simultaneous output at the LH and the SIP, an output of the MB, allows for coordination between innate and learned responses.
Author summary
Behavioral responses to a stimulus in the environment may be hardwired or acquired with experience. In the olfactory system of the fruit fly Drosophila melanogaster, associations of odors are formed in a brain structure called the mushroom body. A different brain region, the lateral horn, is thought to process innate responses, but this has not been shown directly. Here, we demonstrate that activity in neurons in the lateral horn does mediate innate responses. We used a behavioral experiment in which fruit flies avoid carbon dioxide to test the involvement in this response of different neurons in the lateral horn. We found that two sets of neurons, one local and one with projections to a separate region called the superior intermediate protocerebrum, mediate carbon dioxide response. These neurons do not mediate the avoidance response to the other aversive odors tested, indicating selectivity for carbon dioxide. By independent manipulation of and by recording from these two sets of neurons in the lateral horn, we revealed that the local neurons receive input from the output neurons. Thus, these output neurons of the lateral horn simultaneously output at the lateral horn and at the superior intermediate protocerebrum, which is also an area of mushroom body output. This connection between areas may allow coordination between innate and learned responses.
Introduction
Animals use the olfactory system to find partners or food and to avoid predators. To a certain extent, the ability to navigate the olfactory environment is hardwired. This ability is expanded with life experiences that result in olfactory associations. The architecture of the olfactory system is comprehensively characterized in the fruit fly, and it is remarkably similar to the mammalian olfactory system [1,2]. Typically, each olfactory sensory neuron expresses a single odorant receptor that confers to the neuron its response profile [3–5]. With a few exceptions, the response profile of an odorant receptor is broad, meaning that an odor activates a combination of odorant receptors [6–10]. Olfactory sensory neurons expressing the same receptor project to the same glomerulus in the first olfactory center in the brain called the antennal lobe in the fly [3,4]. Most projection neurons (PNs) innervate a single glomerulus and carry the information to higher brain centers: the mushroom body (MB) and the lateral horn (LH) [11,12]. The MB is critical for olfactory associations [13]. The LH was ascribed the role of innate response based on MB silencing experiments [14,15]. PNs from the antennal lobe connect to the MB without apparent spatial selection, whereas at the LH, axonal arbors from different glomeruli interdigitate in a stereotyped fashion [11,12,16–19]. The stereotypy is consistent with the proposed role for the LH as the center for innate olfactory processing. While connectivity at the LH is being scrutinized, direct evidence for the functional role of the LH is still missing.
One of the strongest innate olfactory responses on a T-maze is the response to CO2. Unlike most insects, Drosophila melanogaster avoids CO2 when tested in a T-maze [20]. The aversive response may correspond to an avoidance of hypoxic environments, avoidance of exhalation of a large animal, or the recognition of the stress odor released by other flies. The aversive response up to 2% CO2 is solely mediated by antennal neurons, expressing the CO2 gustatory receptor (Gr) complex Gr21a-Gr63a, which connect to the V-glomerulus in the antennal lobe [7,21]. Synaptic inhibition of Gr21a-Gr63a–expressing neurons abolishes the avoidance response to low concentrations of CO2 [20]. Conversely, artificial stimulation of CO2-sensing neurons with light elicits the avoidance behavior typically observed in response to CO2 [22]. Three PNs that innervate the V-glomerulus (VPNs) clearly project to the LH [17,18,23]. One VPN innervates only the LH, the second one innervates the LH and the MB calyx on both brain hemispheres (biVPN or PNv-1), and the third one connects to the LH and the superior intermediate protocerebrum (SIP) (PNv-3). PNv-3 dendrites innervate many glomeruli in the antennal lobe. It was shown that this PN gates the behavioral response of the biVPN, allowing a behavioral response at 0.5% CO2 but not at 2% CO2. A number of other PNs connect the V-glomerulus with different brain areas. Notably, PNv-2 has dendrites in the V-glomerulus alone and connects to the SIP, an area also innervated by the MB output neurons [24,25]. PNv-2 participates in the avoidance response to 2% CO2. The complexity of connectivity at higher brain levels is probably related with the double valence of CO2. CO2 generates an aversive response in a T-maze, but it is a product of fermentation, therefore indicating the presence of food. Starvation changes how the CO2 response is processed. Under starvation, the MB activity is required for the response, but not in fed flies [26].
Not all responses to CO2 are mediated by Gr21a-Gr63a neurons. Higher CO2 concentrations elicit an aversive response to acid that is processed in a separate glomerulus [27]. Also, a recent study shows that the response to CO2 is state dependent, with high-activity flies moving towards CO2 and low-activity flies avoiding it [28]. These results explain previous reports that CO2 can elicit an attractive response in flying individuals [29,30]. The attractive response does not require Gr21a-Gr63a receptors; instead, it is mediated by the ionotropic receptor (IR) 25a[28].
Here, we address directly the behavioral role of the LH neurons (LHNs) using the CO2 avoidance on a T-maze to low concentrations to probe the requirement of the LH for innate responses. We demonstrate that activity in two sets of neurons is required selectively in the behavioral response to CO2. Using calcium imaging, we observe the neurons responding to CO2 in a different concentration-dependent manner. Activity manipulation experiments combined with calcium imaging reveal that the two sets of neurons are connected in a circuit that receives input from the V-glomerulus.
Results
Neurons labeled by lines 21G11 and 23C09 process CO2 avoidance
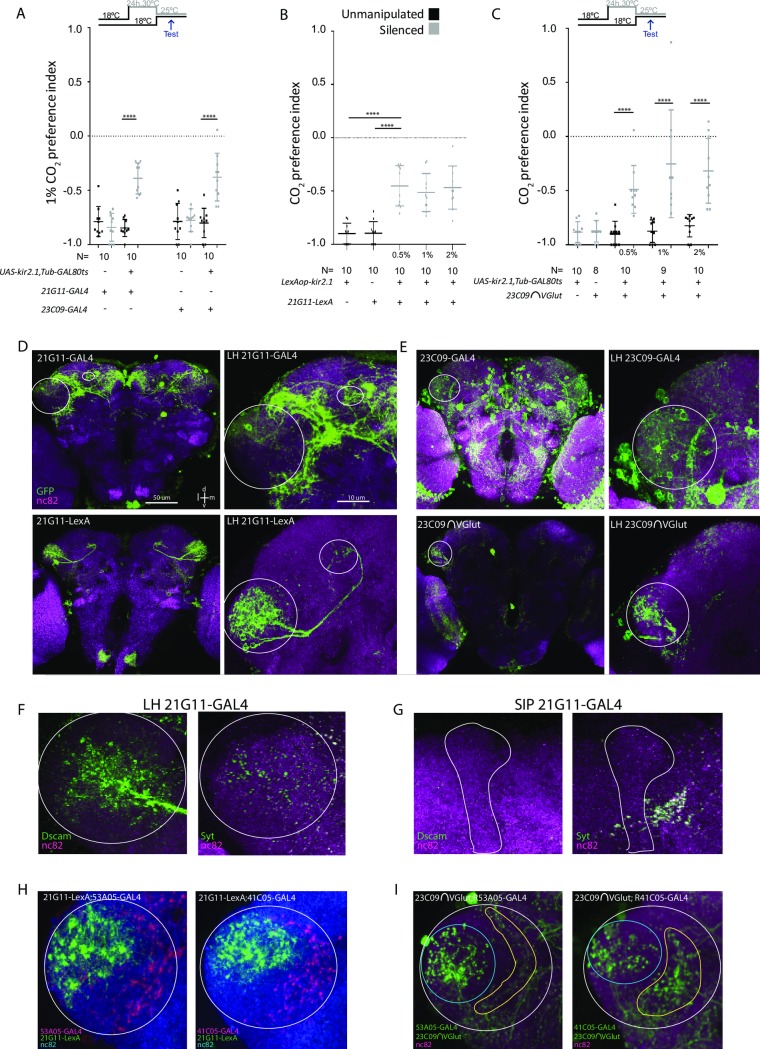
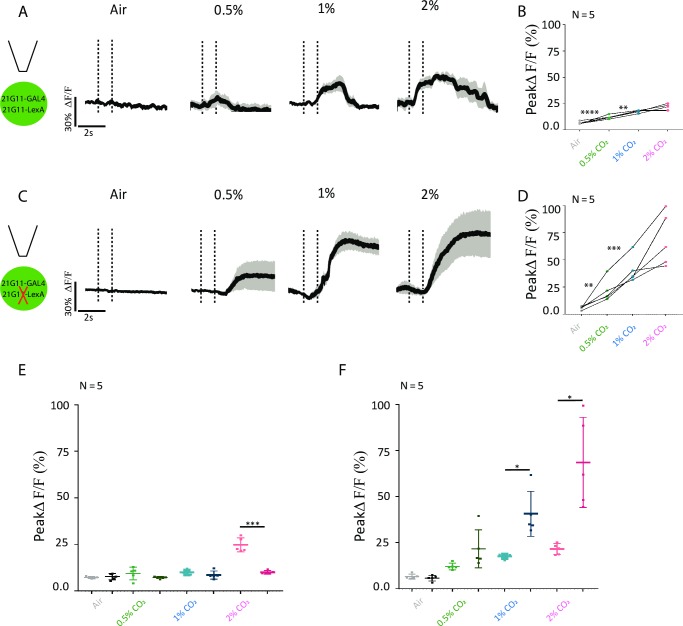
We chose to investigate the role of LHNs in the context of the response to CO2 because of the strength of the innate response on a T-maze. To identify neurons involved in CO2 avoidance, we performed an inhibitory screen of fly lines labeling LHNs (S1 Fig). Through visual inspection of the expression pattern of Janelia’s collection of GAL4 lines, we selected 32 lines with obvious LH innervation [31,32]. To silence the neurons, we expressed the inward-rectifier potassium channel Kir2.1 [33], which hyperpolarizes neurons and thus decreases the probability of firing an action potential. In the screen and in other behavioral experiments with GAL4 lines, we restricted Kir2.1 expression to the adult stage by using a temperature-sensitive GAL80 (GAL80TS, see Materials and Methods) [34]. The 32 lines were tested on a T-maze in which flies were allowed to choose between air and 0.5% CO2 (S1A Fig). Eight lines showed a significant reduction in avoidance (multiple t test corrected with Holm-Sidak method, p < 0.05,) and when retested, three of them exhibited a consistent reduction in avoidance to CO2 (S1B Fig). Line 65D12 was discarded because of innervation in the V-glomerulus (see laser scanning microscope file at http://flweb.janelia.org/cgi-bin/view_flew_imagery.cgi?line=R65D12). Neurons in lines 21G11 and 23CO9 (which we will henceforth call 21G11 and 23CO9 neurons) are necessary for the behavioral response to CO2 (Fig 1A, here tested to 1% CO2). We observe a similar reduction in the behavioral response to CO2 when we express Kir2.1 only in the brain neurons labeled by these lines (S2A and S2B Fig). Since the requirement of the biVPNs and the MB for CO2 avoidance is feeding-state dependent [18], we tested whether feeding state also affects the contribution of 21G11 and 23C09 neurons in the avoidance response of the fly. We observe that starvation does not alter the phenotype, indicating that the involvement of 21G11 and 23C09 neurons in CO2 response is independent of the feeding state of the fly (S2C Fig).
Fig 1. Activity in 21G11 and 23C09 neurons is required for behavioral response to CO2.
(A) T-maze response to 1% CO2 of 21G11-GAL4 and 23C09-GAL4 flies driving UAS-Kir2.1, TubGAL80TS expression. (A and C) Black dots, no heat induction of Kir2.1 expression (see Materials and Methods). Gray dots, heat induction of Kir2.1 expression before test. The top and bottom lines represent the first and the third quartiles. The line across the box is the median. (B and C) T-maze response to three CO2 concentrations—0.5%, 1%, and 2%—of the flies with 21G11-LexA driving LexAopKir2.1 expression (B) and 23C09∩VGlut driving UAS-Kir2.1,TubGAL80TS expression (C). For 21G11-LexA driving LexAopkir2.1 expression, black dots represent parental controls, and gray dots represent constitutive Kir2.1 expression. For 23C09∩VGlut driving UAS-Kir2.1, TubGAL80TS expression, black dots represent no heat induction of Kir2.1 expression, and gray dots represent both parental controls and heat induction of Kir2.1 expression before test. Post hoc two-way ANOVA showed no significance when comparing among expressions for both control and test samples. (D) Brain and LH UAS-mCD8-GFP expression of 21G11-GAL4 and 21G11-LexA. (E) Brain and LH UAS-mCD8-GFP expression of 23C09-GAL4 and 23C09∩VGlut. (D–E) Circle highlights the LH, and oval highlights the SIP. (F and G) Dscam17.1-GFP and syt-HA expression in the LH and SIP of 21G11-GAL4 (green). In (F), circle highlights the LH. In (G), the vertical lobe of the MB is drawn to facilitate visualization of the adjacent SIP. (H) LH showing expression of 21G11-LexA (green) and the VPN lines 53A05-GAL4 (red) and 41C05-GAL4 (red). Circle highlights the LH. (I) LH expression of 23C09∩VGlut (blue circles) and the VPN lines 53A05-GAL4 (yellow line) and 41C05-GAL4 (yellow line). White circle highlights the LH. For all images, the brain neuropil was stained with nc82 (magenta and blue). ±SEM ****p < 0.0001. All p values are calculated with one-way ANOVA. d, dorsal; Dscam17.1, Down syndrome cell adhesion molecule with isoform 1 of the transmembrane domain; l, lateral; LH, lateral horn; m, medial; MB, mushroom body; nc82, monoclonal antibody to Bruchpilot; PN, projection neuron; SEM, standard error of the mean; SIP, superior intermediate protocerebrum; v, ventral; VPN, V-glomerulus–innervating PN.
Given that CO2 avoidance is reduced but not abolished for either line, we tested flies with both sets of neurons silenced (S2D Fig). We observe no change in the phenotype, indicating that the two sets of neurons do not complement each other, i.e., the activity of these populations may not be independent to drive avoidance responses (post hoc two-way ANOVA comparing individual and combined expressions not significant both for control and test samples). It has been previously shown that different PNs of the V-glomerulus are required for the behavioral response to different CO2 concentrations [17]. Therefore, we tested whether the requirement of 21G11 and 23C09 neurons for avoidance to CO2 was concentration dependent. For this experiment, we used lines 21G11-LexA and 23C09∩VGlut, which have a restricted expression when compared to the 21G11-GAL4 and 23C09-GAL4 lines, respectively (Fig 1B and 1C, see below). We observe that silencing the activity of these neurons reduces the avoidance behavior of the flies in a comparable manner across odor concentrations (post hoc two-way ANOVA comparing across odor concentrations not significant, both for control and test samples). These results suggest that 21G11 and 23C09 neurons contribute to CO2 avoidance independently of concentration (within the range that does not engage the acid-sensing response).
Anatomical inspection reveals that line 21G11 labels one cluster of neurons that innervate the dorsoposterior area of the LH and project to the SIP (Fig 1D). The 21G11 cluster, with its posterior cell bodies and projections to the SIP, appears to correspond to the posterior ventral (PV) 5a tract described by Frechter and colleagues [35]. We generated a LexA version of the line to allow independent manipulation of the 21G11-LexA neurons and other neurons labeled with GAL4. The LexA version of 21G11 is very sparse. Additionally, the number of neurons labeled in the LH cluster is smaller. We counted 10 cell bodies in the LexA version and 16 to 18 cell bodies in the GAL4 version (n = 5). When we overlaid the expression of both lines, we found that seven to nine cells were specific to 21G11-GAL4, three cells are specific to 21G11-LexA, and seven cells are common to both lines (S3 Fig, n = 9). Nevertheless, activity in 21G11-LexA neurons is necessary to elicit full CO2 avoidance (Fig 1B). Line 23C09 labels more than one cluster of neurons at the LH (Fig 1E). To narrow down the expression of line 23C09, we generated a splitGAL4 version, and then we intersected the expression with that of different neurotransmitter–splitGAL4 lines [36,37]. We found that a glutamatergic cluster located posteriorly is involved in the response (Fig 1C–1E). This cluster, which we will call 23C09∩VGlut, has 8 to 10 cell bodies (n = 3) with processes only within the LH. It appears to correspond to the primary neurite tract PV4a described in Frechter and colleagues [35]. For expression of restricted lines in 10 μm sections across the brain and ventral nerve cord (VNC) expression in all lines, see S4 Fig. In order to determine the polarity of 21G11 and 23C09∩VGlut neurons, we used the neural compartment markers Down syndrome cell adhesion molecule with isoform 1 of the transmembrane domain (Dscam17.1)-green fluorescent protein (GFP) [38] for dendrites and synaptotagmin-hemagglutinin (HA) [39] for presynaptic areas. Dscam17.1-GFP signal localized exclusively to the LH in 21G11, which indicates that these neurons receive inputs there, presumably olfactory. The synaptotagmin-HA signal, on the other hand, is localized both to the LH and the SIP. To exclude the possibility that the GAL4 cluster holds a mixed population of neurons with different polarities, we marked the more restricted 21G11-LexA neurons and observed the same distribution of synaptotagmin-HA (S5A Fig). These results suggest that 21G11 neurons output both in the SIP and the LH. For 23C09∩VGlut neurons, the signal is localized in the LH for both Dscam17.1-GFP and synaptotagmin-HA (S5B Fig). Finally, we asked whether 21G11 or 23C09 contact projections from the V-glomerulus. Three distinct VPNs at the antennal lobe innervate the medial border of the LH [17,18]. We tested two VPNs for which there are lines available with a strong visible projection. We do not see clear overlap at the LH between the innervation of the VPNs and the innervation of the LHNs we identified (Fig 1H and 1I). This observation, together with the fact that the reduction in avoidance is not complete, indicates that additional LHNs are involved in the response.
21G11 and 23C09∩VGlut neurons respond to CO2 in different concentration-dependent manners
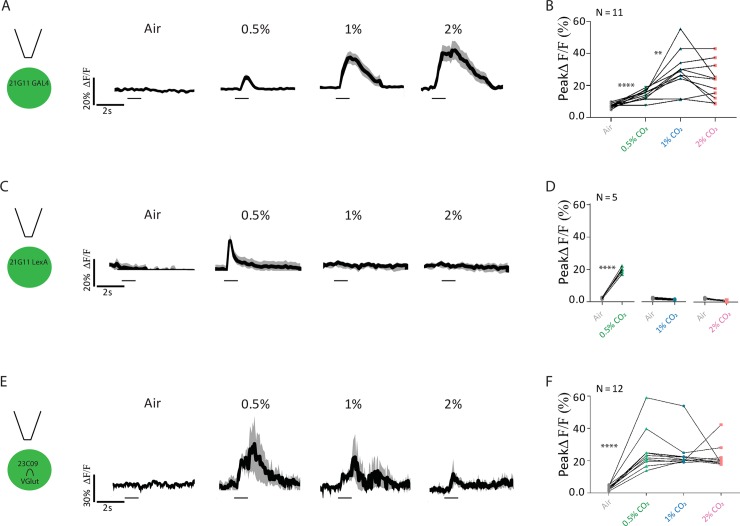
Having demonstrated that 21G11 and 23C09 neurons are required for the behavioral response to CO2, we next addressed the physiological response of these neurons. We measured the changes in internal free-calcium levels in 21G11 and 23C09∩VGlut neurons upon stimulation with CO2. For this, we expressed the genetically encoded calcium indicator GCaMP6m [40] in 21G11 and 23C09∩VGlut neurons and recorded the calcium dynamics in a live fly preparation at the two-photon microscope. Within each LH imaged, we manually delineated the region of interest as the area around the observable innervation to measure the changes in fluorescence. The neurons labeled by 21G11 respond to all concentrations of CO2, with the peak ΔF/F increasing from 0.5 to 1% CO2 (Fig 2A and 2B, Wilcoxon signed-rank test w = 66.00, p = 0.0011). The peak response to 1% and 2% are not significantly different, though the length of the response appears to be larger at 2% (Wilcoxon signed-rank test w = 38.00, p = 0.0727). We also tested responses to CO2 in the subset of 21G11 neurons labeled by 21G11-LexA (Fig 2C and 2D). To our surprise, this subset responds only to 0.5% CO2 (Wilcoxon signed-rank test w = 15.00, p < 0.0001). This observation suggests that within the 21G11 cluster, different neurons are sensitive to different concentrations of CO2. However, in Fig 1C, we observed that when these same neurons are silenced, the behavioral response to different CO2 concentrations does not change. We will address this discrepancy between physiology and behavioral requirement three sections below. Recordings of 23C09∩VGlut neurons show that they respond to all CO2 concentrations tested. Though the curve of the response appears larger for lower concentrations, there is no significant difference between peak amplitudes of ΔF/F of different concentrations (Fig 2E and 2F, 0.5%–1%: Wilcoxon signed-rank test w = 71.00, p = 0.1801; 1%–2%: Wilcoxon signed-rank test w = 68.00, p = 0.6377). In summary, 21G11-GAL4 and 23C09∩VGlut clusters respond to CO2 stimulation at different concentrations, with each set of neurons exhibiting a different pattern of free calcium response to CO2 stimulation.
Fig 2. Physiological response to CO2 of 21G11 and 23C09 neurons.
(A, C, and E) LH activity of 21G11-GAL4, 21G11-LexA and 23C09∩VGlut, respectively, to air, 0.5%, 1%, and 2% of CO2. Schematics of the experiment and average time course of GCaMP6m intensity change. The black bar indicates the time of the stimulus. The black trace represents the average, while the gray shows the range of individual traces. (B, D, and F) Peak GCaMP6m intensity change after stimulation with air, 0.5%, 1%, and 2% of CO2. **p < 0.01, ****p < 0.0001. Comparisons are made between tested concentrations. All p values are calculated with Wilcoxon signed-rank test. LH, lateral horn.
Physiological response of 23C09∩VGlut depends on output of 21G11-LexA
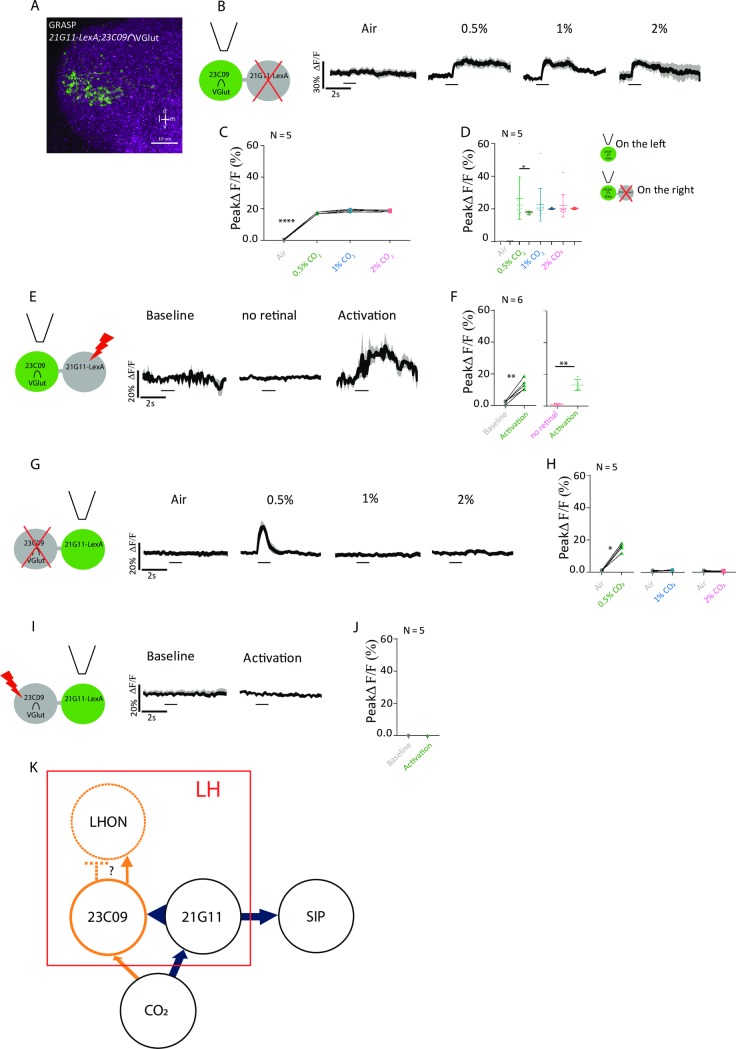
The two sets of neurons that we identified innervate a similar region of the LH and contribute to the same behavioral response. When we silenced both clusters simultaneously, we saw no additive effect, indicating they are not independent from each other to drive avoidance (S2D Fig, post hoc two-way ANOVA comparing individual and combined expressions not significant both for control and test samples). We therefore asked whether the two clusters are connected. To this end, we used GFP reconstitution across synaptic partners (GRASP), which reveals membrane contact between two sets of neurons [41]. We observe a strong signal in the LH, indicating that the membranes of the two clusters contact each other (Fig 3A). To assess functional connectivity, we manipulated the activity of one cluster while imaging activity on the second cluster. We silenced 21G11-LexA neurons with the expression of Kir2.1 using the LexA/LexAOp expression system and imaged 23C09∩VGlut neurons expressing GCaMP6m with the GAL4/UAS system (schematic in Fig 3B). To control for silencing, we coexpressed Kir2.1 and GCaMP6m in 21G11-LexA neurons and confirmed that no calcium signal is observed with CO2 stimulation (S6A and S6B Fig). Upon presentation of CO2 at the concentrations 0.5%, 1%, and 2%, we observed a very consistent response across trials and across concentrations in 23C09∩VGlut neurons when 21G11-LexA neurons are silenced (Fig 3B and 3C, Mann–Whitney, not significant). When we compared the peak responses of 23C09∩VGlut while 21G11-LexA is intact (Fig 2F) or silenced (Fig 3C), we found that there is pronounced reduction for 0.5% CO2 responses, which corresponds to the profile of 21G11-LexA responses (Fig 3D, Wilcoxon signed-rank test w = 17.00, p = 0.0337). The results indicate that the output of 21G11-LexA neurons contributes to 23C09∩VGlut activity. To test this directly, we expressed the red-shifted channelrhodopsin Chrimson [42] in 21G11-LexA neurons to allow activation of 21G11-LexA neurons with 720 nm light while recording 23C09∩VGlut calcium concentration with GCaMP6m. We observe that indeed, activation of 21G11-LexA neurons with light generates a strong calcium response in 23C09∩VGlut neurons (Fig 3E and 3F, Mann–Whitney, U = 94, p = 0.0012). No calcium response was observed in 23C09∩VGlut neurons when flies were not fed retinal, which is necessary for Chrimson function (Fig 3E and 3F, Mann–Whitney, not significant). To check that Chrimson indeed activates 21G11-LexA neurons, we expressed both Chrimson and GCaMP6m in these neurons and could see a response with light stimulation (S6C and S6D Fig, Mann–Whitney, U = 63, p = 0.0321).
Fig 3. 21G11-LexA neurons are presynaptic to 23C09∩VGlut neurons.
(A) LH showing GRASP, a measure of contact between the membranes of neurons, in 21G11-LexA and 23C09∩VGlut. (B) Schematics of the experiment and calcium response at the LH, using GCaMP6m, of 23C09∩VGlut neurons to air, 0.5%, 1%, and 2% of CO2 while 21G11-LexA neurons are silenced by expression of Kir2.1. (C) Peak GCaMP6m intensity change after stimulation with air, 0.5%, 1%, and 2% of CO2. (D) Peak GCaMP6m intensity change between Fig 2F and Fig 3C after stimulation with air, 0.5%, 1%, and 2% of CO2. (E) Schematics of the experiment and calcium response at the LH of 23C09∩VGlut during baseline, activity without retinal, and upon activation with 720 nm light of 21G11-LexA driving expression of Chrimson. (F) Peak GCaMP6m intensity change of 23C09∩VGlut during baseline, activity without retinal, and upon activation with 720 nm light of 21G11-LexA driving expression of Chrimson. (G) Schematics of the experiment and calcium response at the LH, using GCaMP6m, of 21G11-LexA neurons to air, 0.5%, 1%, and 2% of CO2 while 23C09∩VGlut neurons are silenced by expression of Kir2.1. (H) Peak GCaMP6m intensity change after stimulation with air, 0.5%, 1%, and 2% of CO2. (I) Schematics of the experiment and LH activity of 21G11-LexA upon activation of 23C09∩VGlut neurons, expressing Chrimson, with 720 nm light. (J) Peak GCaMP6m intensity change of 21G11-LexA during baseline and activation with 720 nm light of 23C09∩VGlut driving expression of Chrimson. (K) Proposed model of the LHNs processing CO2 information. Elements in dashed lines are hypothetical. For (B), (E), (G), and (I), the average time course of GCaMP6m intensity change is shown. The black bar indicates the time of the stimulus. Scale bar = 10 μm. *p < 0.05, **p < 0.01, ****p < 0.0001. All p values are calculated with Wilcoxon signed-rank test. d, dorsal; GFP, green fluorescent protein; GRASP, GFP reconstitution across synaptic partners; l, lateral; LH, lateral horn; LHN, LH neuron; LHON, LH output neuron; m, medial; SIP, superior intermediate protocerebrum; v, ventral.
We next did the converse experiments in which we manipulate activity in 23C09∩VGlut neurons and image the activity in 21G11-LexA neurons, using the same tools with the expression systems reversed. Silencing 23C09∩VGlut neurons does not change 21G11-LexA response to CO2 presentation (Fig 3G and 3H, Air–0.5%: Wilcoxon signed-rank test w = 15.00, p = 0.0002). These results indicate that 23C09∩VGlut neurons do not output into 21G11-LexA neurons. To control for silencing, we coexpressed Kir2.1 and GCaMP6m in 23C09∩VGlut neurons and confirmed that no calcium signal is observed with CO2 stimulation (S7A and S7B Fig, Wilcoxon signed-rank test not significant). We then performed optogenetic activation of 23C09∩VGlut neurons and recorded the calcium response of 21G11-LexA neurons. We did not observe a calcium response in 21G11-LexA neurons upon light stimulation of 23C09∩VGlut neurons (Fig 3I and 3J, Mann–Whitney not significant). To control for activation of 23C09∩VGlut neurons, we expressed both Chrimson and GCaMP6m in these neurons and could see a response with light stimulation (S7C and S7D Fig, Mann–Whitney, U = 67, p = 0.0361). The activation results further support the notion that 23C09∩VGlut does not output into 21G11-LexA neurons.
Taken together, these results indicate that 21G11-LexA neurons are presynaptic to 23C09∩VGlut neurons. The presence of a presynaptic marker at the LH processes of 21G11-LexA neurons is consistent with these observations (Fig 1F). Based on our findings, we propose a model in which there are two outputs to the CO2 response (Fig 3K). 21G11-LexA neurons outputs at the LH to activate 23C09∩VGlut local neurons. These, in turn, activate or inhibit another set of LH output neurons that will initiate a motor response. 21G11-LexA neurons also output at the SIP, where coordination with the MB signal is likely taking place.
21G11-LexA neurons receive inputs from the V-glomerulus
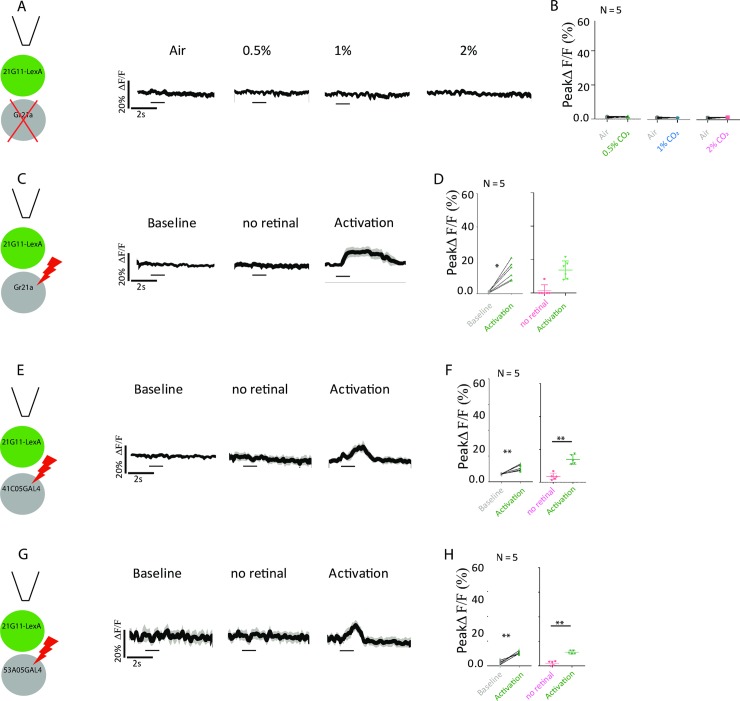
We wished to confirm that the CO2 responses we observe originate in the V-glomerulus because the CO2 response is flexible and may have different origins[28]. We silenced the neurons expressing the CO2 coreceptor Gr21a that converge in the V-glomerulus while we recorded the response to CO2 in 21G11-LexA neurons using similar methodology to that used in the previous section. We find that CO2 response is abolished in 21G11-LexA neurons (Fig 4A and 4B, Wilcoxon signed-rank test not significant). Conversely, optogenetic activation of Gr21a neurons induces activation of 21G11-LexA neurons (Fig 4C and 4D, Mann–Whitney, U = 86, p = 0.0172). We found that VPNs that innervate specifically the V-glomerulus and project to the LH do not directly synapse onto 21G11-LexA (Fig 1H). We asked whether, even so, they induce activation of 21G11-LexA. Stimulation of each VPN leads to activation of 21G11-LexA neurons (Fig 4F and 4H, 4F: Mann–Whitney, U = 78, p = 0.0188; 4H: Mann–Whitney, U = 82, p = 0.0124). Our results indicate that 21G11-LexA neurons are an integral part of a CO2 response that originates in the V-glomerulus.
Fig 4. 21G11-LexA neurons receive input from the V-glomerulus.
(A) Schematics of the experiment and calcium response at the LH, using GCaMP6m, of 21G11-LexA neurons to air, 0.5%, 1%, and 2% of CO2 while Gr21a-GAL4 neurons are silenced by expression of Kir2.1. (B) Peak GCaMP6m intensity change after stimulation with air, 0.5%, 1%, and 2% of CO2. (C, E, and G) Schematics of the experiment and calcium response at the LH of 21G11-LexA neurons during baseline, activity without retinal, and upon activation with 720 nm light of Gr21a-GAL4 (C), 41C05-GAL4 (E), and 53A05-GAL4 (G) driving expression of Chrimson. (D, F, and H) Peak GCaMP6m intensity change of 21G11-LexA during baseline, activity without retinal, and upon activation with 720 nm light of Gr21a-GAL4 (D), 41C05-GAL4 (F), and 53A05-GAL4 (H) driving expression of Chrimson. For (A), (C), (E), and (G), the average time course of GCaMP6m intensity change is shown. The black bar indicates the time of the stimulus. Gr, gustatory receptor; LH, lateral horn.
Output of 21G11-LexA neurons regulates activity of other 21G11-GAL4 neurons
Next, we explored how activity in 21G11-LexA neurons is required for the behavioral response to CO2 at three different concentrations when the neurons respond only to the lowest concentration (0.5% CO2). Since 21G11-LexA neurons are essentially a subset of a cluster defined by expression of 21G11-GAL4, we reasoned that perhaps subthreshold activity in 21G11-LexA neurons influences activity of 21G11-GAL4 neurons that respond to 1% and 2% CO2. We silenced 21G11-LexA neurons while recording calcium activity in 21G11-GAL4 neurons to CO2 stimulation (Fig 5). When 21G11-LexA neurons are silenced, the responses within the stimulation period are all rather low, though the difference to control only reaches significance at 2% CO2 (Fig 5E, 0.5%: Wilcoxon signed-rank test w = −13.00, p = 0.2231; 1%: Wilcoxon signed-rank test w = −13.00, p = 0.338; 2%: Wilcoxon signed-rank test w = 13.00, p = 0.0003). However, a few seconds after stimulation, we observe for all concentrations a large and persistent rise in free calcium (Fig 5F, 0.5%: Wilcoxon signed-rank test w = −15.00, p = 0.1272; 1%: Wilcoxon signed-rank test w = 17.00, p = 0.0188; 2%: Wilcoxon signed-rank test w = 15.00, p = 0.0124). The persistence of calcium is so extended that the stimulation protocol had to be readjusted from 20 to 80 s between stimulations. Thus, the response profile changes considerably when 21G11-LexA neurons are silenced, which could explain the change in the behavioral response. The exact nature of the interaction between subsets of 21G11 neurons still needs to be defined.
Fig 5. 21G11 neurons change their response profile when their 21G11-LexA subset is silenced.
(A) Schematics of the experiment and calcium response at the LH, using GCaMP6m, of 21G11 neurons to air, 0.5%, 1%, and 2% of CO2. (B) Peak GCaMP6m intensity change after stimulation with air, 0.5%, 1%, and 2% of CO2. (C) Schematics of the experiment and calcium response at the LH of 21G11 neurons to air, 0.5%, 1%, and 2% of CO2 while 21G11-LexA neurons are silenced by expression of Kir2.1. (D) Peak GCaMP6m intensity change after stimulation with air, 0.5%, 1%, and 2% of CO2. (E–F) Peak GCaMP6m intensity change during stimulation period (E) and poststimulation period (F), with air, 0.5%, 1%, and 2% of CO2 for both (B) values on the left and (D) values on the right. For (A) and (C), the average time course of GCaMP6m intensity change is shown. The vertical dashed lines indicate the time of the stimulus. *p < 0.05, **p < 0.01, ****p < 0.0001. All p values are calculated with Wilcoxon signed-rank test. LH, lateral horn.
21G11 and 23C09 neurons are selectively involved in processing CO2 avoidance
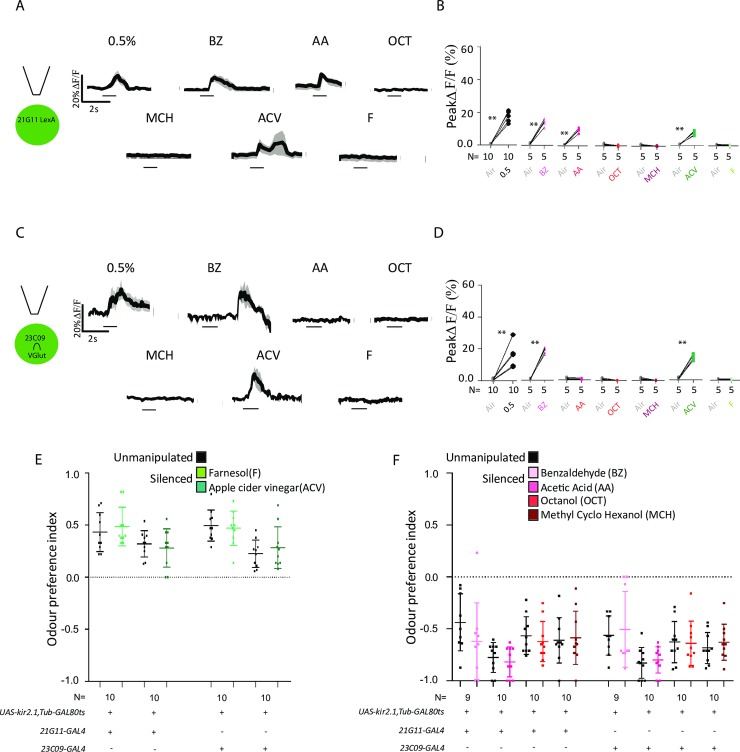
We then asked how specific the circuit we identified at the LH is. Is it generally involved in odor responses or is it involved specifically in avoidance responses? To answer this question, we first measured the calcium responses to different odors (Fig 6). To test attractive odor responses, we used farnesol (F), an attractant present in the rind of ripe citrus and processed through a single glomerulus [10], and apple cider vinegar (ACV), a complex attractive stimulus [43]. While ACV elicits a small response in both sets of neurons, F does not elicit a response in either set of neurons (Fig 6A–6D, F: Wilcoxon signed-rank test not significant; ACV: Wilcoxon signed-rank test w = 16.00, p = 0.0004). To test aversive responses, we used benzaldehyde (BZ), which smells of bitter almond, and acetic acid (AA), which elicits the acid-sensing response in the antennal lobe [27]. We also used octanol (OCT) and methylcyclohexanol (MCH), which are aversive at higher concentrations [44]. 21G11 neurons respond to both BZ and AA (Fig 6A and 6B, BZ: Wilcoxon signed-rank test w = 18.00, p = 0.0001; AA: Wilcoxon signed-rank test w = 19.00, p = 0.0001). 23C09∩VGlut neurons respond only to BZ in an atypical fashion (Fig 6C and 6D). The rise in calcium concentration happens a few seconds after stimulus presentation. It is not clear what the origin of the delayed response is. It could represent rebound excitation but also could stem from a delayed response to BZ at the PN level, as it has been reported in some instances [8]. 21G11 neurons and 23C09∩VGlut neurons do not respond to OCT and MCH (Wilcoxon signed-rank test not significant). Though the peak ΔF/F in all these responses is low, the physiological response is broad and includes responses to aversive and attractive odors. How do these physiological responses translate into a behavioral response? To address this question, we tested the requirement of activity in 23C09 or 21G11 neurons for the behavioral response to other odors. We used the original GAL4 lines because they have broader expression. Similarly to what we did in the screen, we tested the flies using a T-maze. Also following the screen conditions, we silenced the neurons with Kir2.1 only in the adult stage. We allowed the flies to choose between air and F, ACV, AA, or BZ at 1/1000 dilution and octanol and MCH at 1/500 dilution. It was reported that there is a nonolfactory component to BZ avoidance at 1/100 dilution. We confirmed that we are not including a nonolfactory component in our experiment by testing the response of flies without olfactory organs to our working dilution of BZ (S8 Fig, Mann–Whitney, U = 45, p = 0.0007). We observe that activity in 23C09 or 21G11 neurons is not required either for attraction to F or ACV, indicating that activity in these LHNs is not involved in general odor responses (Fig 6E, Mann–Whitney not significant). Silencing 23C09 or 21G11 neurons also does not affect avoidance to BZ, AA, OCT, and MCH (Fig 6F, Mann–Whitney not significant). Interestingly, though the arborization at the LH of the acid-sensing PNs is very similar to the arborization of the VPNs [23], it appears that different LHNs process these responses. The results indicate that activity in 21G11 and 23C09 neurons is selectively required for the response to CO2.
Fig 6. Physiological and behavioral response to attractive and repulsive compounds of 21G11 and 23C09 neurons.
(A and C) Schematics of the experiment and calcium response at the LH of 21G11-LexA and of 23C09∩VGlut to the repulsive compounds—BZ, AA, OCT, and MCH—and the attractive compounds—F and ACV. The image shows the average time course of GCaMP6m intensity change. The black bar indicates the time of the stimulus. (B and D) Peak GCaMP6m intensity change after stimulation with 0.5% CO2, BZ, AA, OCT, MCH, ACV, and F. (E) T-maze response to F and ACV of 21G11 and 23C09. (F) T-maze response to BZ, AA, OCT, and MCH of 21G11 and 23C09. (E–F) Black dots, no heat induction of Kir2.1 expression (see materials and methods). Colored dots, heat induction of Kir2.1 expression before test. The top and bottom lines represent the first and the third quartiles. The line across the box is the median. For (B) and (D), **p < 0.01. p values are calculated with Wilcoxon signed-rank test. For (E) and (F), comparisons calculated with one-way ANOVA are nonsignificant. AA, acetic acid; ACV, apple cider vinegar; BZ, benzaldehyde; F, farnesol; MCH, methylcyclohexanol; OCT, octanol.
Discussion
In early studies, LHNs that respond to the male pheromone 11-cis-vaccenyl acetate were identified based on the expression of the male-specific form of the transcription factor fruitless [45,46]. One cluster of male LHNs responds specifically to the pheromone. These results opened the possibility that each odor has a cognate LHN. However, activity and anatomy analysis on larger sets of LHNs suggests a mixed model of connectivity [35,47,48]. Two very recent studies on large sets of LHNs convey important insights into LH organization [35,48]. Both studies show that genetically labeled LHNs show stereotyped responses from animal to animal. Jeanne and colleagues have photostimulated PNs at the antennal lobe while recording from 110 LHNs of 39 morphological types [48]. They find that certain combinations of glomeruli are over-represented at LHN recordings. These combinations represent an odor scene, for instance, odors relevant for egg-laying site selection rather than a single odorant. Frechter and colleagues have characterized the cellular composition of LH using a combination of anatomical analysis of genetic driver lines and whole-brain electron microscopy data [35]. They find that the LH contains over 1,300 neurons, of which 580 are LH local neurons and 830 are LH output neurons. LHNs were clustered into 150 cell types, revealing unanticipated complexity in this brain region. LHN recordings indicate that LHNs are better odor categorizers than PNs.
In this study, we used a behavioral readout to directly address the role of the LH in olfactory responses. We showed that the activity of two clusters of LHNs is required for D. melanogaster innate avoidance of CO2. 23C09 is a cluster of LH local neurons and 21G11 is a cluster of output neurons. Both clusters show a physiological response to CO2. We were able to show that 21G11-LexA, a subset of 21G11 neurons, outputs to 23C09 within the LH. 21G11-LexA activity in response to CO2 originates in the activity of Gr21a neurons that converge to the V-glomerulus. The response of 21G11-LexA neurons is shaped by the activity of VPNs, although they are not directly connected. Our results define a circuit within the LH region specific for CO2 avoidance responses in the lower concentration range that starts at the V-glomerulus. However, the avoidance to CO2 is not completely abolished by silencing these neurons. This observation could have three origins: not all cells are labeled within the identified cluster; other unidentified clusters are involved; or the LH has intricate connectivity, and silencing of any LHN leads to disruption of the behavior. The latter is unlikely because we silenced and tested 32 lines labeling different LHNs, some of them innervating very large sections of the LH, and only the two lines described here consistently had an effect. Though we cannot eliminate any of the former possibilities, the lack of contact between the LHNs we identified and the VPNs that carry CO2 stimulus information argues for the involvement of additional neurons within the LH. In addition, a contribution from the MB cannot be excluded since MB output neuron (MBON) activity is required for naïve avoidance of odors, including CO2, and intrinsic MB neurons participate in the CO2 response when flies are starved [18,44,49].
The LH is required for the execution of learned aversion [50]. Hence, the roles of the LH and MB centers are not as segregated as previously thought. They must be connected to coordinate the innate, the context-dependent, and the learned responses. There are at least two LH outputs to the CO2 response. 21G11 neurons output within the LH and connect to the SIP. The SIP is an area highly innervated by MBON terminals, suggesting a location for integration of LH and MB output [25,51]. A preprint published while this manuscript was under revision suggests that presynaptic labeling (as well as postsynaptic labeling) in dendrites of LH output neurons, such as we observe in 21G11 neurons, is a common occurrence and estimates that one third of LH projections converge to sites of MB outputs [52].
21G11 and 23C09 show specificity to CO2 behavioral response. Activity in these neurons is not required for the T-maze response to other four aversive odors. The most logical organization for a center that processes innate behavioral responses is that inputs that elicit a particular behavioral outcome (such as avoidance) converge into a particular region. However, our work indicates that not all avoidance responses are processed equally. Still, it is possible that the CO2 response converges with the response to other aversive stimuli downstream of 23C09 neurons. A candidate region of the LH tuned to repulsive odors has been described [53].
Our findings revealed a disparity between activity of the LHNs we identified and the behavioral requirement for a few of the tested odors. The result indicates that not all activity in these LHNs will drive the behavior. This means that the LH is more complex than previously anticipated. We speculate that since innate responses can be flexible, in a different context, the activity here revealed will shape the behavioral response to other odors.
A vast search has identified compounds that either increase or decrease the activity of the CO2 receptors in mosquitoes [54]. A later study in the fly revealed that odors increasing activity of Gr21-Gr63a receptor neurons generate avoidance responses and odors decreasing receptor activity generate attraction responses, indicating that the CO2 receptor pathway has a strong weight in establishing odor valence [55]. Further work should elucidate how the LHNs identified here contribute to these responses.
In summary, we demonstrated a role of the LH in an innate behavioral response. A preprint posted while this manuscript was under revision confirms this conclusion, looking at egg-laying aversion [56]. Moving forward, it will be interesting to explore whether a similar organization at the LH is used to generate responses to other odors and how the response is coordinated with the MB.
Materials and methods
Contact for reagent and resource sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Maria Luísa Vasconcelos (maria.vasconcelos@neuro.fchampalimaud.org).
Experimental model and subject details
Flies were maintained on standard cornmeal–agar medium at 18°C or 25°C and 70% relative humidity under a 12 h light/dark cycle. Fly strains used were as follows: UAS-Kir2.1 [33]; UAS>stop>Kir2.1 [57]; UAS>stop>mCD8GFP [58]; tub-GAL80TS [34]; UAS-Chrimson [42]; UAS-GCaMP6m [40]; UAS-Dscam17.1-GFP [38]; UAS-syt-HA [39]; UAS-CD4-GFP1-10 [41]; UAS-CD8-GFP [59]; VGlut-DBD [37]; UAS-myr-tdTomato [60]; LexAop-Kir2.1 (provided by Barry Dickson) [61]; LexAop-Chrimson [62]; LexAop-GCaMP6m [40]; LexAop-syt-HA [63]; LexAop-CD4-GFP11 [41]; LexAop-mCD2-GFP [64]. We used the following GAL4 lines: 21G11, 23C09, 84A06, 65D12, 19B07, 13A11, 30A10, 37G11, 33E01, 93D02, 93D05, 41F11, 85C07, 64B02, 36E10, 30H02, 25B07, 36G09, 23F06, 54G12, 16C09, 13A07, 22B02, 29F04, 16C06, 82E01, 25A01, 84G12, 25G10, 26C12, 20C09, 20B0, 53A05, 41C05 from the Janelia farm collection [31,32], Gr21a-GAL4 [65], and otdFLP [66]. 21G11-LexA and 23C09-AD were generated for this work (see Method details).
Genotypes per figure
Fig 1. Panel A
w1118; ; 21G11-GAL4/+
w1118;UAS-Kir2.1,tub-Gal80TS/+;21G11-GAL4/+
w1118; ; 23C09-GAL4/+
w1118;UAS-Kir2.1,tub-Gal80TS/+;23C09-GAL4/+
Panel B
w1118;LexAop-Kir2.1/+;+
w118; 21G11-LexA/+;21G11-LexA/+
w1118; 21G11-LexA/LexAop-Kir2.1;21G11-LexA/+
Panel C
w1118;UAS-Kir2.1,tub-Gal80TS/+;+
w1118; 23C09-AD,VGlut-DBD/+;+
w1118;UAS-Kir2.1,tub-Gal80TS/23C09-AD,VGlut-DBD;+
Panel D
w1118; ;21G11-GAL4/UAS-mCD8-GFP
w1118;LexAop-mCD2-GFP/+;21G11-LexA/+
Panel E
w1118; ;23C09-GAL4/UAS-mCD8-GFP
w1118;23C09-AD/VGlut-DBD;UAS-mCD8-GFP/+
Panels F and G
w1118;UAS-Dscam-17.1-GFP/+;21G11-GAL4/+
w1118;UAS-syt-HA/+;21G11-GAL4/+
Panel H
w1118;21G11-LexA/LexAop-CD2-GFP;UAS-myr-tdTomato/53A05-GAL4
w1118; 21G11-LexA/ LexAop-CD2-GFP;UAS-myr-tdTomato /41C05-GAL4
Panel I
w1118; 23C09-AD/VGlut-DBD;UAS-mCD8-GFP/53A05-GAL4
w1118; 23C09-AD/VGlut-DBD;UAS-mCD8-GFP/41C05-GAL4
Fig 2. Panels A and B
w1118;UAS-GCaMP6m/+;21G11-GAL4/+
Panels C and D
w1118;21G11-LexA/+;21G11-LexA/LexAop-GCaMP6m
Panels E and F
w1118;23C09-AD/VGlut-DBD;UASGCaMP6m/+
Fig 3. Panel A
w1118;LexAop-CD4-GFP11/21G11-LexA;UAS-mCD4-GFP1-10/23C09-GAL4
Panels B, C, and D
w1118;21G11-LexA,LexAop-Kir2.1/23C09-AD,VGlut-DBD;21G11-LexA/UAS-GCaMP6m
Panels E and F
w1118;21G11-LexA,LexAop-Chrimson/23C09-AD,VGlut-DBD;21G11-LexA/UASGCaMP6m
Panels G and H
w1118;21G11-LexA/23C09-AD,VGlut-DBD;21G11-LexA,LexAop-GCaMP6m/UAS-Kir2.1
Panel I
w1118;21G11-LexA/23C09-AD,VGlut-DBD;21G11-LexA,LexAop-GCaMP6m/UAS-Chrimson
Fig 4. Panels A and B
w1118;21G11-LexA,LexAop-GCaMP6m/Gr21a-GAL4;21G11-LexA/UAS-Kir2.1
Panels C and D
w1118;21G11-LexA,LexAop-GCaMP6m/Gr21a-GAL4;21G11-LexA/UAS-Chrimson
Panels E and F
w1118;21G11-LexA/UAS-Chrimson;21G11-LexA,LexAop-GCaMP6m/41C05-GAL4
Panels G and H
w1118;21G11-LexA/UAS-Chrimson;21G11-LexA,LexAop-GCaMP6m/53A05-GAL4
Fig 5. Panels A and B
w1118;21G11-LexA/UAS-GCaMP6m;21G11-LexA/21G11-GAL4
Panels C and D
w1118;21G11-LexA,LexAop-Kir2.1/UAS-GCaMP6m;21G11-LexA/21G11-GAL4
Panel E
w1118;21G11-LexA/UAS-GCaMP6m;21G11-LexA/21G11-GAL4
w1118;21G11-LexA,LexAop-Kir2.1/UAS-GCaMP6m;21G11-LexA/21G11-GAL4
Fig 6. Panels A and B
w1118;21G11-LexA/+;21G11-LexA/LexAop-GCaMP6m
Panels C and D
w1118;23C09-AD/VGlut-DBD;UASGCaMP6m/+
Panels E and F
w1118;UAS-Kir2.1,tub-Gal80TS/+;21G11-GAL4/+
w1118;UAS-Kir2.1,tub-Gal80TS/+;23C09-GAL4/+
S1 Fig. Panel A
w1118;UAS-Kir2.1,tub-Gal80TS/+;84A06-GAL4/+
w1118;UAS-Kir2.1,tub-Gal80TS/+;65D12-GAL4/+
w1118;UAS-Kir2.1,tub-Gal80TS/+;21G11-GAL4/+
w1118;UAS-Kir2.1,tub-Gal80TS/+;23C09-GAL4/+
w1118;UAS-Kir2.1,tub-Gal80TS/+;19B07-GAL4/+
w1118;UAS-Kir2.1,tub-Gal80TS/+;13A11-GAL4/+
w1118;UAS-Kir2.1,tub-Gal80TS/+;30A10-GAL4/+
w1118;UAS-Kir2.1,tub-Gal80TS/+;37G11-GAL4/+
w1118;UAS-Kir2.1,tub-Gal80TS/+;33E01-GAL4/+
w1118;UAS-Kir2.1,tub-Gal80TS/+;93D02-GAL4/+
w1118;UAS-Kir2.1,tub-Gal80TS/+;93D05-GAL4/+
w1118;UAS-Kir2.1,tub-Gal80TS/+;41F11-GAL4/+
w1118;UAS-Kir2.1,tub-Gal80TS/+;85C07-GAL4/+
w1118;UAS-Kir2.1,tub-Gal80TS/+;64B02-GAL4/+
w1118;UAS-Kir2.1,tub-Gal80TS/+;36E10-GAL4/+
w1118;UAS-Kir2.1,tub-Gal80TS/+;30H02-GAL4/+
w1118;UAS-Kir2.1,tub-Gal80TS/+;25B07-GAL4/+
w1118;UAS-Kir2.1,tub-Gal80TS/+;36G09-GAL4/+
w1118;UAS-Kir2.1,tub-Gal80TS/+;23F06 GAL4/+
w1118;UAS-Kir2.1,tub-Gal80TS/+;54G12-GAL4/+
w1118;UAS-Kir2.1,tub-Gal80TS/+;16C09-GAL4/+
w1118;UAS-Kir2.1,tub-Gal80TS/+;13A07-GAL4/+
w1118;UAS-Kir2.1,tub-Gal80TS/+;22B02-GAL4/+
w1118;UAS-Kir2.1,tub-Gal80TS/+;29F04-GAL4/+
w1118;UAS-Kir2.1,tub-Gal80TS/+;16C06-GAL4/+
w1118;UAS-Kir2.1,tub-Gal80TS/+;82E01-GAL4/+
w1118;UAS-Kir2.1,tub-Gal80TS/+;25A01-GAL4/+
w1118;UAS-Kir2.1,tub-Gal80TS/+;84G12-GAL4/+
w1118;UAS-Kir2.1,tub-Gal80TS/+;25G10-GAL4/+
w1118;UAS-Kir2.1,tub-Gal80TS/+;26C12-GAL4/+
w1118;UAS-Kir2.1,tub-Gal80TS/+;20C09-GAL4/+
w1118;UAS-Kir2.1,tub-Gal80TS/+;20B07-GAL4/+
Panel B
w1118;UAS-Kir2.1,tub-Gal80TS/+;65D12-GAL4/+
w1118;UAS-Kir2.1,tub-Gal80TS/+;21G11-GAL4/+
w1118;UAS-Kir2.1,tub-Gal80TS/+;23C09-GAL4/+
w1118;UAS-Kir2.1,tub-Gal80TS/+;13A11-GAL4/+
w1118;UAS-Kir2.1,tub-Gal80TS/+;33E01-GAL4/+
w1118;UAS-Kir2.1,tub-Gal80TS/+;85C07-GAL4/+
w1118;UAS-Kir2.1,tub-Gal80TS/+;36G09-GAL4/+
w1118;UAS-Kir2.1,tub-Gal80TS/+;16C09-GAL4/+
S2 Fig. Panel A
UAS>stop>Kir2.1/ w118;otdFLP/+;+
w118; ;21G11-GAL4/+
w1118; ;23C09-GAL4/+
UAS>stop>Kir2.1/ w118;otdFLP/+;21G11-GAL4/+
UAS>stop>Kir2.1/ w118;otdFLP/+;23C09-GAL4/+
Panel B
w1118; otdFLP /+;21G11-GAL4/ UAS>stop>mCD8-GFP
w1118; otdFLP /+;23C09-GAL4/ UAS>stop>mCD8-GFP
Panel C
w1118;UAS-Kir2.1,tub-Gal80TS/+;21G11-GAL4/+
w1118;UAS-Kir2.1,tub-Gal80TS/+;23C09-GAL4/+
w1118;UAS-Kir2.1,tub-Gal80TS/+;21G11-GAL4/23C09-GAL4
Panel D
w118; ;21G11-GAL4/+
w1118;UAS-Kir2.1,tub-Gal80TS/+;21G11-GAL4/+w1118; ;23C09-GAL4/+
w1118;UAS-Kir2.1,tub-Gal80TS/+;23C09-GAL4/+
w1118; ;21G11-GAL4/23C09-GAL4
w1118;UAS-Kir2.1,tub-Gal80TS/+;21G11-GAL4/23C09-GAL4
w1118; 21G11-LexA /LexAop-mCD2-GFP;21G11-LexA/21G11-GAL4,UAS-myr-tdTomato
S4 Fig. Panel A
w1118;LexAop-mCD2-GFP/+;21G11-LexA/+
Panel B
w1118;23C09-AD/VGlut-DBD;UAS-mCD8-GFP/+
Panel C
w1118; ;21G11-GAL4/UAS-mCD8-GFP
w1118;LexAop-mCD2-GFP/+;21G11-LexA/+
Panel D
w1118; ;23C09-GAL4/UAS-mCD8-GFP
w1118;23C09-AD/VGlut-DBD;UAS-mCD8-GFP/+
S5 Fig. Panel A
w1118;LexAop-syt-HA/+;21G11-LexA/+
Panel B
w1118;UAS-Dscam-17.1-GFP/23C09-AD,VGlut-DBD;+
w1118;UAS-syt-HA/23C09-AD,VGlut-DBD;+
S6 Fig. Panels A and B
w1118;21G11-LexA,LexAop-Kir2.1/+;21G11-LexA/LexAop-GCaMP6m
Panels C and D
w1118;21G11-LexA,LexAop-Chrimson/+;21G11-LexA/LexAop-GCaMP6m
S7 Fig. Panels A and B
w1118;23C09-AD,VGlut-DBD/+;UASGCaMP6m/UAS-Kir2.1
Panels C and D
w1118;23C09-AD,VGlut-DBD/+;UASGCaMP6m/UAS-Chrimson
w1118;UAS-Kir2.1,tub-Gal80TS/+;+
Method details
Generating transgenic flies
For the establishment of the 21G11-LexA lines, we used the Gateway System. The LexA vector used was purchased from Addgene (plasmid #26230; Watertown, MA, USA). We carried out a genomic PCR with the primers (5′ to 3′) F-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCGCGCAGCACGTGAAGAACAAGGC and R-GGGGACCACTTTGTACAAGAAAGCTGGGTCATGGCAACGTACTTCCAGTCCTCT. The fragment was inserted in a pDONR221 vector and then recombined into the pBPnlsLexA::p65Uw vector (Addgene, plasmid #26230). To construct the 23C09AD line, we use the fragment in a pDON that was kindly provided by the Rubin Lab. We recombined it as described above. 21G11-LexA was injected into attp40 and attp2 flies. 23C09-AD was injected into attp40 flies.
Immunostaining
Adult fly brains were dissected, fixed, and stained using standard protocols. Briefly, tissue was dissected in phosphate-buffered saline (PBS), fixed in 4% PFA in PBL (PBS and 0.12M lysine) for 30 min at room temperature, washed 3× for 5 min in PBT (PBS and 0.5% Triton X-100), and blocked for 15 minutes in 10% normal goat serum in PBT (Sigma Aldrich, cat# G9023; St. Louis, MO, USA). Samples were then incubated with primary antibodies for 72 h at 4°C. After incubation, they were washed 3× for 10 min in PBT and incubated with secondary antibodies for 72 h at 4°C. Finally, the samples were washed 3× for 10 min in PBT and mounted in Vectashield (Vector Laboratories, cat# H-1000; Burlingame, CA, USA). As primary antibodies, we used: rabbit anti-GFP (1:2000, Molecular Probes, cat# 11122; Eugene, OR, USA), chicken anti-GFP (1:2000, Molecular Probes, cat# A10262), rabbit anti-DsRed (1:2000, Molecular Probes, cat# 710530), and mouse anti-nc82 (1:10, Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA, USA). The secondary antibodies used were anti-rabbit or anti-chicken IgG conjugated to Alexa 488, anti-mouse or anti-rabitt IgG conjugated to Alexa 594, and anti-mouse IgG conjugated to Alexa 405. All microscopy of immunostainings was performed with a Zeiss LSM 710 confocal microscope (Zeiss, Oberkochen, Germany). Images were processed with ImageJ.
Behavioral experiments
Neuronal silencing: Flies were kept at 18°C for 8 to 16 d. When using the UASKir2.1,TubGAL80ts, tester flies were placed at 30°C 24 h before the experiment, while control flies were always kept at 18°C. On the day of the experiment, both tester and control flies were transferred to 25°C where we quantified their response in a T-maze [67]. When using the LexAopKir2.1 flies, both tester and control flies were always kept at 25°C. We quantified flies’ response to air, three concentrations of CO2 (0.5%, 1%, and 2%), and two known attractive (ACV and F) and four known repulsive compounds (BZ, AA, OCT, and MCH). To obtain the CO2 concentrations, we mixed bottled synthetic air with bottled CO2 (Linde, Danbury, CT, USA). The flow rate used was of 0.12 l per min. All other compounds were diluted 1:1000 in paraffin oil (Sigma Aldrich) with the exception of OCT and MCH, for which the dilution was 1:500 in paraffin oil. To test for starvation, adult flies were removed from vials with food and placed in vials with humidified paper 24 h prior to the experiment. To test for the nonolfactory component to BZ avoidance, the olfactory organs were removed manually (antennae and maxillary palps) 24 h before the experiment. For all experiments, flies were tested in groups of 20 individuals. Flies were placed on the T-maze elevator and dropped to the choice area, where they were given 45 seconds to choose an arm. To control whether the T-maze was balanced, we tested flies to air on both sides. For control and tester flies, one arm of the T-maze released air, while the other arm released the testing compound. After the experiment, flies were counted, and the odor preference index was calculated by subtracting the number of flies on the air side from the number of flies on the other side and dividing it with the total number of flies.
Calcium imaging experiments
Preparation: For all calcium imaging experiments, flies expressed the calcium indicator GCaMP6m. The preparation was based on walking behavior preparation [68] but without the ball. To image the LH in an in vivo preparation, we glued the fly head to a microscope base (Scientifica, Uckfield, UK) with bee’s wax (Sigma Aldrich). We then opened a window that corresponded to half of the fly brain and removed all fat and trachea. We made sure that both antennae were untouched and healthy.
Microscopy: We used an Ultima two-photon laser-scanning microscope from Prairie Technologies (now Bruker; Billerica, MA, USA) and a Coherent Chameleon XR laser (Coherent, Santa Clara, CA, USA). All images were acquired every 0.2 ms with an Olympus BX61 microscope (Olympus, Tokyo, Japan) equipped with a 40 × 0.8 NA objective. The image was zoomed to allow the selection of a region of interest (ROI) in the LH where the stimulation was performed. To measure the fluorescent intensity at the LH, we used ImageJ. In the image recorded at the time of the experiment, we delineated by hand and around the observable innervation of the LH a second ROI. The resulting time trace from the second ROI was used for further analysis. To calculate the normalized change in the relative fluorescence intensity, we used ΔF/F = 100(Fn − F0)/F0, where Fn is the nth frame after stimulation and F0 is the average basal fluorescence before the stimulation. Images with visible rhythmic movements of the animal were discarded.
Olfactory stimulation: For olfactory stimulation, a custom-made delivery system consisting of a four-way solenoid valve (Parker Hannifin, Cleveland, OH, USA) connected to a peristaltic pump (Ismatec, Wertheim, Germany) creating a continuous airstream (1800 ml/min) that was delivered to the antennae with chemically inert tubing (Ismatec). The valve stimulation was commanded through the PrairieView software. For the CO2 stimulation, dilutions were placed in Tedlar gas sampling bags (#24634, Sigma Aldrich) that were then connected to the valve. For stimulation with other compounds dilutions were made in glass vials with rubber taps (Thermo Fisher Scientific, Waltham, MA, USA). At the rubber tap, we inserted two venofix needles (B. Braun, Melsungen, Germany): one to connect the vial to the valve, the other to connect the vial to the air in the room for airflow in the vial. We set up the system so that when a stimulus is triggered, the odor dilutions replace only 50% of the airflow in order to minimize the turbulence. For this reason, all dilutions were prepared to double of the desired concentrations. In all experiments, stimuli were delivered for one second. The interval between stimuli was of 20 s for all experiments except for Fig 5C and 5D, in which the recovery time proved to be longer. For this experiment, the interval was set at 80 s. To control for calcium changes with the airflow, all stimulations were done twice. In addition, we performed experiments both in and outside the LH and imaged the LH in neurons with both Kir2.1 and GCaMP6m. No interference from the airflow in the calcium response was ever observed.
Neuronal activation: For the neuronal activation, we used 8 LEDs of 720 nm wavelength in a custom-made ring placed beneath the stage that surrounded the fly. The delivery of light to the fly was commanded through the PrairieView software. The stimulus was delivered for one second at 5-Hz and 40-ms pulses.
Neuronal silencing: For the neuronal silencing experiments, all flies expressed Kir2.1.
Quantification and statistical analysis
All behavioral data was statistically analyzed by one-way analysis of variance and a Sidak’s multiple comparisons test. For all imaging data, a Wilcoxon signed-rank test comparison was performed. For all analysis and statistical tests, we used the GraphPad Prism Software version 6.0 (GraphPad Software).
The numerical data used in all figures are included in S1 Data.
Supporting information
(A) T-maze response to 0.5% CO2 of 32 fly lines from the Janelia farm collection chosen by their innervation at the LH. Avoidance responses to CO2 are significantly reduced in eight lines (84A06, 65D12, 21G11, 23C09, 33E01, 85C07, 36G09, and 16C09) when silenced with UAS-Kir2.1,Tub-GAL80ts. (A–B) The top and bottom lines represent the first and the third quartiles. The line across the box is the median. (B) Retest of T-maze response to 0.5% CO2 of the eight fly lines that showed reduced avoidance on (A). Avoidance responses to CO2 are significantly reduced in three lines (65D12-GAL4, 21G11-GAL4, and 23C09-GAL4) when silenced with UAS-Kir2.1,Tub-GAL80ts. *p < 0.05. All p values are calculated with multiple t test corrected with the Holm–Sidak method. LH, lateral horn; LHN, LH neuron.
(TIF)
(A) T-maze response to 0.5%, 1%, and 2% CO2 of 21G11 and 23C09 with Kir2.1 expression restricted to the brain using UAS>stop>Kir2.1;otdFLP. Post hoc two-way ANOVA comparing behavioral response to different concentrations reveals no significance. (B) Brain and VNC expression of 21G11-GAL4 and of 23C09-GAL4 intersected with otdFLP. (C) T-maze response to 1% CO2 of starved flies of 21G11 and 23C09. The box represents the first and the third quartiles, and the whiskers the 10th and 90th percentiles. The line across the box is the median. N = 10. All p values are calculated via one-way ANOVA. (D) Retest of T-maze response to 1% CO2 of same lines as Fig 1A plus flies with all genetic elements combined so that neurons of both line 21G11 and 23C09 are manipulated. Post hoc two-way ANOVA comparing behavioral response of individual and combined expressions both for control and test samples reveals no significance. otdFLP, orthodenticle-flipase; VNC, ventral nerve cord.
(TIF)
LH cells of 21G11-LexA (red) and 21G11-GAL4 (green) and the merge of both. Scale bar = 10 μm. LH, lateral horn.
(TIF)
(A and B) Brain expression every 10 μm of 21G11-LexA (A) and 23C09∩VGlut (B) lines. (C) VNC expression of 21G11 and 21G11-LexA. (D) VNC expression of 23C09 and 23C09∩VGlut. For all images, green = GFP, magenta = nc82. GFP, green fluorescent protein; nc82, monoclonal antibody to Bruchpilot; VNC, ventral nerve cord.
(TIF)
(A) Syt-HA expression in the LH and SIP of 21G11-LexA (green). The circle highlights the LH. The vertical lobe of the MB is drawn to facilitate visualization of the adjacent SIP. (B) Dscam17.1-GFP and syt-HA expression in the LH of 23C09∩VGlut (green). The circle highlights the LH. Scale bar = 10 μm. d, dorsal; Dscam17.1, Down syndrome cell adhesion molecule with isoform 1 of the transmembrane domain; GFP, green fluorescent protein; HA, hemagglutinin; l, lateral; LH, lateral horn; m, medial; MB, mushroom body; SIP, superior intermediate protocerebrum; syt, synaptotagmin; v, ventral.
(TIF)
(A and B) Schematics of the experiment and calcium response at the LH, using GCaMP6m, of 21G11-LexA neurons to air, 0.5%, 1%, and 2% of CO2 while 21G11-LexA neurons are silenced by expression of Kir2.1. (C) Schematics of the experiment and LH activity of 21G11-LexA upon activation of 21G11-LexA neurons, expressing Chrimson, with 720 nm light. (D) Peak GCaMP6m intensity change upon activation. For (C), the average time course of GCaMP6m intensity change is shown. The black bar indicates the time of the stimulus. *p < 0.05. All p values are calculated with Wilcoxon signed-rank test. LH, lateral horn.
(TIF)
(A and B) Schematics of the experiment and calcium response at the LH, using GCaMP6m, of 23C09∩VGlut neurons to air, 0.5%, 1% and 2% of CO2, while 23C09∩VGlut neurons are silenced by expression of Kir2.1. (C) Schematics of the experiment and LH activity of 23C09∩VGlut upon activation of 23C09∩VGlut neurons, expressing Chrimson, with 720 nm light. (D) Peak GCaMP6m intensity change upon activation. For (C), the average time course of GCaMP6m intensity change is shown. The black bar indicates the time of the stimulus. *p < 0.05. All p values are calculated with Wilcoxon signed-rank test. LH, lateral horn.
(TIF)
T-maze response to benzaldehyde at 1:1000 dilution (BZ). White box, flies with olfactory organs. Gray box, flies without olfactory organs. The box represents the first and the third quartiles, and the whiskers the 10th and 90th percentiles. The line across the box is the median. N = 10. Error bars indicate ±SEM ****p<0.0001. p values are calculated with Wilcoxon signed-rank test. BZ, benzaldehyde; SEM, standard error of the mean.
(TIF)
(XLSX)
Acknowledgments
We thank Eugenia Chiappe, Susana Lima, Marta Moita, and members of the Vasconcelos Laboratory for discussion and comments on the manuscript. Gerald M. Rubin provided a promoter plasmid. We would like to thank the Fly and Molecular platforms of Champalimaud Centre for the Unknown (CCU) for generating the transgenic flies and scientific hardware and software platforms of CCU for support with the optogenetic activation experiments.
Abbreviations
- AA
acetic acid
- ACV
apple cider vinegar
- BZ
benzaldehyde
- CCU
Champalimaud Centre for the Unknown
- Dscam17.1
Down syndrome cell adhesion molecule with isoform 1 of the transmembrane domain
- F
farnesol
- GFP
green fluorescent protein
- Gr
gustatory receptor
- GRASP
GFP reconstitution across synaptic partners
- HA
hemagglutinin
- IR
ionotropic receptor
- LH
lateral horn
- LHN
LH neuron
- LHON
LH output neuron
- MB
mushroom body
- MBON
MB output neuron
- MCH
methylcyclohexanol
- nc82
monoclonal antibody to Bruchpilot
- OCT
octanol
- PBS
phosphate-buffered saline
- PN
projection neuron
- PV
posterior ventral
- ROI
region of interest
- SIP
superior intermediate protocerebrum
- VNC
ventral nerve cord
- VPN
V-glomerulus–innervating PN
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
Fundação para Ciência e tecnologia (FCT) (grant number PTDC/BIA-BCM/104898/2008). The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Fundação para Ciência e tecnologia (FCT) (grant number SFRH/BDP/77360/2011). The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Fundação para Ciência e tecnologia (FCT), Lisboa2020, under the PORTUGAL2020 agreement (European Regional Development Fund) (grant number Congento LISBOA-01-0145-FEDER-022170). The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Vosshall LB, Stocker RF. Molecular architecture of smell and taste in Drosophila. Annu Rev Neurosci. 2007;30: 505–33. 10.1146/annurev.neuro.30.051606.094306 [DOI] [PubMed] [Google Scholar]
- 2.Masse NY, Turner GC, Jefferis GSXE. Olfactory Information Processing in Drosophila. Curr Biol. Elsevier Ltd; 2009;19: R700–R713. 10.1016/j.cub.2009.06.026 [DOI] [PubMed] [Google Scholar]
- 3.Gao Q, Yuan B, Chess A. Convergent projections of Drosophila olfactory neurons to specific glomeruli in the antennal lobe. Nat Neurosci. 2000;3: 780–785. 10.1038/77680 [DOI] [PubMed] [Google Scholar]
- 4.Vosshall LB, Wong AM, Axel R. An olfactory sensory map in the fly brain. Cell. 2000;102: 147–159. 10.1016/j.ydbio.2008.05.014 [DOI] [PubMed] [Google Scholar]
- 5.Hallem E a, Ho MG, Carlson JR. The molecular basis of odor coding in the Drosophila antenna. Cell. 2004;117: 965–79. 10.1016/j.cell.2004.05.012 [DOI] [PubMed] [Google Scholar]
- 6.Hallem EA, Carlson JR. Coding of Odors by a Receptor Repertoire. Cell. 2006;125: 143–160. 10.1016/j.cell.2006.01.050 [DOI] [PubMed] [Google Scholar]
- 7.Kwon JY, Dahanukar A, Weiss LA, Carlson JR. The molecular basis of CO2 reception in Drosophila. Proc Natl Acad Sci U S A. 2007;104: 3574–8. 10.1073/pnas.0700079104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schlief ML, Wilson RI, Orn D. Olfactory processing and behavior downstream from highly selective receptor neurons. Nat Neurosci. 2007;10: 623–630. 10.1038/nn1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stensmyr MC, Dweck HKM, Farhan A, Ibba I, Strutz A, Mukunda L, et al. A conserved dedicated olfactory circuit for detecting harmful microbes in drosophila. Cell. Elsevier Inc.; 2012;151: 1345–1357. 10.1016/j.cell.2012.09.046 [DOI] [PubMed] [Google Scholar]
- 10.Ronderos DS, Lin C-CC, Potter CJ, Smith DP. Farnesol-Detecting Olfactory Neurons in Drosophila. J Neurosci. 2014;34: 3959–3968. 10.1523/JNEUROSCI.4582-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wong AM, Wang JW, Axel R. Spatial representation of the glomerular map in the Drosophila protocerebrum. Cell. 2002;109: 229–41. 10.1016/S0092-8674(02)00707-9 [DOI] [PubMed] [Google Scholar]
- 12.Marin EC, Jefferis GSXE, Komiyama T, Zhu H, Luo L. Representation of the glomerular olfactory map in the Drosophila brain. Cell. 2002;109: 243–255. 10.1016/S0092-8674(02)00700-6 [DOI] [PubMed] [Google Scholar]
- 13.Keene AC, Waddell S. Drosophila olfactory memory: Single genes to complex neural circuits. Nat Rev Neurosci. 2007;8: 341–354. 10.1038/nrn2098 [DOI] [PubMed] [Google Scholar]
- 14.Heimbeck G, Bugnon V, Gendre N, Keller A, Stocker RF. A central neural circuit for experience-independent olfactory and courtship behavior in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2001;98: 15336–15341. 10.1073/pnas.011314898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parnas M, Lin AC, Huetteroth W, Miesenböck G. Odor Discrimination in Drosophila: From Neural Population Codes to Behavior. Neuron. 2013;79: 932–944. 10.1016/j.neuron.2013.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Datta SR, Vasconcelos ML, Ruta V, Luo S, Wong A, Demir E, et al. The Drosophila pheromone cVA activates a sexually dimorphic neural circuit. Nature. 2008;452: 473–477. 10.1038/nature06808 [DOI] [PubMed] [Google Scholar]
- 17.Lin H-H, Chu L-A, Fu T-F, Dickson BJ, Chiang A-S. Parallel neural pathways mediate CO2 avoidance responses in Drosophila. Science. 2013;340: 1338–41. 10.1126/science.1236693 [DOI] [PubMed] [Google Scholar]
- 18.Bräcker LB, Siju KP, Varela N, Aso Y, Zhang M, Hein I, et al. Essential role of the mushroom body in context-dependent CO2 avoidance in Drosophila. Curr Biol. 2013;23: 1228–34. 10.1016/j.cub.2013.05.029 [DOI] [PubMed] [Google Scholar]
- 19.Caron SJC, Ruta V, Abbott LF, Axel R. Random convergence of olfactory inputs in the Drosophila mushroom body. Nature. Nature Publishing Group; 2013;497: 113–117. 10.1038/nature12063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suh GSBB, Wong AM, Hergarden AC, Wang JW, Simon AF, Benzer S, et al. A single population of olfactory sensory neurons mediates an innate avoidance behaviour in Drosophila. Nature. 2004;431: 854–859. 10.1038/nature02980 [DOI] [PubMed] [Google Scholar]
- 21.Jones WD, Cayirlioglu P, Kadow IG, Vosshall LB. Two chemosensory receptors together mediate carbon dioxide detection in Drosophila. Nature. 2007;445: 86–90. 10.1038/nature05466 [DOI] [PubMed] [Google Scholar]
- 22.Suh GSB, Ben-Tabou de Leon S, Tanimoto H, Fiala A, Benzer S, Anderson DJ. Light Activation of an Innate Olfactory Avoidance Response in Drosophila. Curr Biol. 2007;17: 905–908. 10.1016/j.cub.2007.04.046 [DOI] [PubMed] [Google Scholar]
- 23.Min S, Ai M, Shin S a, Suh GSB. Dedicated olfactory neurons mediating attraction behavior to ammonia and amines in Drosophila. Proc Natl Acad Sci U S A. 2013;110: E1321–9. 10.1073/pnas.1215680110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin H-H, Chu L-A, Fu T-F, Dickson BJ, Chiang A-S. Parallel Neural Pathways Mediate CO2 Avoidance Responses in Drosophila. Science (80-). 2013;340: 1338–1341. 10.1126/science.1236693 [DOI] [PubMed] [Google Scholar]
- 25.Aso Y, Sitaraman D, Ichinose T, Kaun KR, Vogt K, Belliart-Guérin G, et al. Mushroom body output neurons encode valence and guide memory-based action selection in Drosophila. Elife. 2014;3: e04580 10.7554/eLife.04580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bräcker LBLB, Siju KPP, Arela N, So Y, Hang MM, Hein I, et al. Essential role of the mushroom body in context-dependent CO2 avoidance in drosophila. Curr Biol. 2013;23: 1228–1234. 10.1016/j.cub.2013.05.029 [DOI] [PubMed] [Google Scholar]
- 27.Ai M, Min S, Grosjean Y, Leblanc C, Bell R, Benton R, et al. Acid sensing by the Drosophila olfactory system. Nature. Nature Publishing Group; 2010;468: 691–695. 10.1038/nature09537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Breugel F, Huda A, Dickinson MH. Distinct activity-gated pathways mediate attraction and aversion to CO2 in Drosophila. Nature. 2018;564: 420–424. 10.1038/s41586-018-0732-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Faucher CP, Hilker M, de Bruyne M, Faucher P, Hilker M, Bruyne M De. Interactions of Carbon Dioxide and Food Odours in Drosophila: Olfactory Hedonics and Sensory Neuron Properties. PLoS One. 2013;8: e56361 10.1371/journal.pone.0056361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wasserman S, Salomon A, Frye M a. Drosophila tracks carbon dioxide in flight. Curr Biol. Elsevier Ltd; 2013;23: 301–6. 10.1016/j.cub.2012.12.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pfeiffer BD, Jenett A, Hammonds AS, Ngo T-TBT-TB, Misra S, Murphy C, et al. Tools for neuroanatomy and neurogenetics in Drosophila. Proc Natl Acad Sci. 2008;105: 9715–9720. 10.1073/pnas.0803697105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jenett A, Rubin GM, Ngo TTB, Shepherd D, Murphy C, Dionne H, et al. A GAL4-Driver Line Resource for Drosophila Neurobiology. Cell Rep. The Authors; 2012;2: 991–1001. 10.1016/j.celrep.2012.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baines R a, Uhler JP, Thompson a, Sweeney ST, Bate M. Altered electrical properties in Drosophila neurons developing without synaptic transmission. J Neurosci. 2001;21: 1523–1531. 21/5/1523 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McGuire SE, Mao Z, Davis RL. Spatiotemporal Gene Expression Targeting with the TARGET and Gene-Switch Systems in Drosophila. Sci Signal. 2004;2004: pl6–pl6. 10.1126/stke.2202004pl6 [DOI] [PubMed] [Google Scholar]
- 35.Frechter S, Bates AS, Tootoonian S, Dolan M-J, Manton JD, Jamasb A, et al. Functional and Anatomical Specificity in a Higher Olfactory Centre. bioRxiv. 2018; 336982 10.1101/336982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luan H, Peabody NC, Vinson CR, White BH. Refined Spatial Manipulation of Neuronal Function by Combinatorial Restriction of Transgene Expression. Neuron. 2006;52: 425–436. 10.1016/j.neuron.2006.08.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Diao FF, Ironfield H, Luan H, Diao FF, Shropshire WC, Ewer J, et al. Plug-and-Play Genetic Access to Drosophila Cell Types using Exchangeable Exon Cassettes. Cell Rep. The Authors; 2015;10: 1410–1421. 10.1016/j.celrep.2015.01.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang J, Ma X, Yang JS, Zheng X, Zugates CT, Lee CHJ, et al. Transmembrane/juxtamembrane domain-dependent Dscam distribution and function during mushroom body neuronal morphogenesis. Neuron. 2004;43: 663–672. 10.1016/j.neuron.2004.06.033 [DOI] [PubMed] [Google Scholar]
- 39.Robinson IM, Ranjan R, Schwarz TL. Synaptotagmins I and IV promote transmitter release independently of Ca(2+) binding in the C(2)A domain. Nature. 2002;418: 336–40. 10.1038/nature00915 [DOI] [PubMed] [Google Scholar]
- 40.Chen TW, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 2013;499: 295–300. 10.1038/nature12354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gordon MD, Scott K. Motor Control in a Drosophila Taste Circuit. Neuron. Elsevier Ltd; 2009;61: 373–384. 10.1016/j.neuron.2008.12.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klapoetke NC, Murata Y, Kim SS, Pulver SR, Birdsey-Benson A, Cho YK, et al. Independent optical excitation of distinct neural populations. Nat Methods. Nature Publishing Group; 2014;11: 338–346. 10.1038/nmeth.2836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Semmelhack JL, Wang JW. Select Drosophila glomeruli mediate innate olfactory attraction and aversion. Nature. 2009;459: 218–23. 10.1038/nature07983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Owald D, Felsenberg J, Talbot CB, Das G, Perisse E, Huetteroth W, et al. Activity of defined mushroom body output neurons underlies learned olfactory behavior in Drosophila. Neuron. The Authors; 2015;86: 417–427. 10.1016/j.neuron.2015.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ruta V, Datta SRSR, Vasconcelos MLML, Freeland J, Looger LLLL, Axel R. A dimorphic pheromone circuit in Drosophila from sensory input to descending output. Nature. Nature Publishing Group; 2010;468: 686–690. 10.1038/nature09554 [DOI] [PubMed] [Google Scholar]
- 46.Kohl J, Ostrovsky AD, Frechter S, Jefferis GSXE. A Bidirectional Circuit Switch Reroutes Pheromone Signals in Male and Female Brains. Cell. The Authors; 2013;155: 1610–1623. 10.1016/j.cell.2013.11.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fişek M, Wilson RI. Stereotyped connectivity and computations in higher-order olfactory neurons. Nat Neurosci. 2014;17: 280–288. 10.1038/nn.3613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jeanne JM, Fişek M, Wilson RI. The Organization of Projections from Olfactory Glomeruli onto Higher-Order Neurons. Neuron. 2018;98: 1198–1213.e6. 10.1016/j.neuron.2018.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lewis LPC, Siju KP, Aso Y, Friedrich AB, Bulteel AJB, Rubin GM, et al. A Higher Brain Circuit for Immediate Integration of Conflicting Sensory Information in Drosophila. Curr Biol. Elsevier Ltd; 2015;25: 2203–2214. 10.1016/j.cub.2015.07.015 [DOI] [PubMed] [Google Scholar]
- 50.Dolan M, Belliart-Guérin G, Bates AS, Frechter S, Lampin-Saint-Amaux A, Aso Y, et al. Communication from Learned to Innate Olfactory Processing Centers Is Required for Memory Retrieval in Drosophila. Neuron. 2018; 167312 10.1016/j.neuron.2018.08.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aso Y, Hattori D, Yu Y, Johnston RM, Iyer NA, Ngo TTB, et al. The neuronal architecture of the mushroom body provides a logic for associative learning. Elife. 2014;3: e04577 10.7554/eLife.04577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dolan M-J, Frechter S, Bates AS, Dan C, Huoviala P, Ruairi J.V. Roberts RJV, et al. Neurogenetic dissection of the Drosophila innate olfactory processing center. BioRxiv 404277 [Preprint]. 2018. [cited 2018 September 10]. Available from: https://www.biorxiv.org/content/early/2018/09/12/404277. [Google Scholar]
- 53.Strutz A, Soelter J, Baschwitz A, Farhan A, Grabe V, Rybak J, et al. Decoding odor quality and intensity in the Drosophila brain. Elife. 2014;3: e04147 10.7554/eLife.04147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tauxe GM, Macwilliam D, Boyle SM, Guda T, Ray A. Targeting a dual detector of skin and CO2 to modify mosquito host seeking. Cell. Elsevier; 2013;155: 1365–1379. 10.1016/j.cell.2013.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.MacWilliam D, Kowalewski J, Kumar A, Pontrello C, Ray A. Signaling Mode of the Broad-Spectrum Conserved CO2Receptor Is One of the Important Determinants of Odor Valence in Drosophila. Neuron. Elsevier Inc.; 2018;97: 1153–1167.e4. 10.1016/j.neuron.2018.01.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huoviala P, Dolan M-J, Love F, Frechter S, Roberts RJV, Mitrevica Z, et al. Neural circuit basis of aversive odour processing in Drosophila from sensory input to descending output. BioRxiv 394403 [Preprint]. 2018. [cited 2018 September 10]. Available from: https://www.biorxiv.org/content/early/2018/08/26/394403. [Google Scholar]
- 57.Yang C hui, Rumpf S, Xiang Y, Gordon MD, Song W, Jan LY, et al. Control of the Postmating Behavioral Switch in Drosophila Females by Internal Sensory Neurons. Neuron. Elsevier Ltd; 2009;61: 519–526. 10.1016/j.neuron.2008.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hong W, Zhu H, Potter CJ, Barsh G, Kurusu M, Zinn K, et al. Leucine-rich repeat transmembrane proteins instruct discrete dendrite targeting in an olfactory map. Nat Neurosci. 2009;12: 1542–50. 10.1038/nn.2442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee T, Luo L. Mosaic analysis with a repressible neurotechnique cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22: 451–461. 10.1016/S0896-6273(00)80701-1 [DOI] [PubMed] [Google Scholar]
- 60.Pfeiffer BD, Ngo TTB, Hibbard KL, Murphy C, Jenett A, Truman JW, et al. Refinement of tools for targeted gene expression in Drosophila. Genetics. 2010;186: 735–755. 10.1534/genetics.110.119917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Feng K, Palfreyman MT, Häsemeyer M, Talsma A, Dickson BJ. Ascending SAG neurons control sexual receptivity of Drosophila females. Neuron. 2014;83: 135–148. 10.1016/j.neuron.2014.05.017 [DOI] [PubMed] [Google Scholar]
- 62.Fosque BF, Sun Y, Dana H, Yang C, Ohyama T, Tadross MR, et al. Labeling of active neural circuits in vivo with designed calcium integrators. 2015;347. [DOI] [PubMed] [Google Scholar]
- 63.Nern A, Pfeiffer BD, Rubin GM. Optimized tools for multicolor stochastic labeling reveal diverse stereotyped cell arrangements in the fly visual system. Proc Natl Acad Sci. 2015;112: E2967–E2976. 10.1073/pnas.1506763112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lai S-L, Lee T. Genetic mosaic with dual binary transcriptional systems in Drosophila. Nat Neurosci. 2006;9: 703–9. 10.1038/nn1681 [DOI] [PubMed] [Google Scholar]
- 65.Cayirlioglu P, Kadow IG, Zhan X, Okamura K, Suh GSB, Gunning D, et al. Hybrid neurons in a microRNA mutant are putative evolutionary intermediates in insect CO2 sensory systems. Science (80-). 2008;319: 1256–1260. 10.1126/science.1149483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Asahina K, Watanabe K, Duistermars BJ, Hoopfer E, González CR, Eyjólfsdóttir EA, et al. Tachykinin-expressing neurons control male-specific aggressive arousal in drosophila. Cell. 2014;156: 221–235. 10.1016/j.cell.2013.11.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tully T, Quinn WG. Classical conditioning and retention in normal and mutant Drosophila melanogaster. J Comp Physiol a-Neuroethology Sens Neural Behav Physiol. 1985;157: 263–277. 10.1007/BF01350033 [DOI] [PubMed] [Google Scholar]
- 68.Chiappe ME, Jayaraman V. Genetically Encoded Functional Indicators. 2012;72: 83–101. 10.1007/978-1-62703-014-4 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) T-maze response to 0.5% CO2 of 32 fly lines from the Janelia farm collection chosen by their innervation at the LH. Avoidance responses to CO2 are significantly reduced in eight lines (84A06, 65D12, 21G11, 23C09, 33E01, 85C07, 36G09, and 16C09) when silenced with UAS-Kir2.1,Tub-GAL80ts. (A–B) The top and bottom lines represent the first and the third quartiles. The line across the box is the median. (B) Retest of T-maze response to 0.5% CO2 of the eight fly lines that showed reduced avoidance on (A). Avoidance responses to CO2 are significantly reduced in three lines (65D12-GAL4, 21G11-GAL4, and 23C09-GAL4) when silenced with UAS-Kir2.1,Tub-GAL80ts. *p < 0.05. All p values are calculated with multiple t test corrected with the Holm–Sidak method. LH, lateral horn; LHN, LH neuron.
(TIF)
(A) T-maze response to 0.5%, 1%, and 2% CO2 of 21G11 and 23C09 with Kir2.1 expression restricted to the brain using UAS>stop>Kir2.1;otdFLP. Post hoc two-way ANOVA comparing behavioral response to different concentrations reveals no significance. (B) Brain and VNC expression of 21G11-GAL4 and of 23C09-GAL4 intersected with otdFLP. (C) T-maze response to 1% CO2 of starved flies of 21G11 and 23C09. The box represents the first and the third quartiles, and the whiskers the 10th and 90th percentiles. The line across the box is the median. N = 10. All p values are calculated via one-way ANOVA. (D) Retest of T-maze response to 1% CO2 of same lines as Fig 1A plus flies with all genetic elements combined so that neurons of both line 21G11 and 23C09 are manipulated. Post hoc two-way ANOVA comparing behavioral response of individual and combined expressions both for control and test samples reveals no significance. otdFLP, orthodenticle-flipase; VNC, ventral nerve cord.
(TIF)
LH cells of 21G11-LexA (red) and 21G11-GAL4 (green) and the merge of both. Scale bar = 10 μm. LH, lateral horn.
(TIF)
(A and B) Brain expression every 10 μm of 21G11-LexA (A) and 23C09∩VGlut (B) lines. (C) VNC expression of 21G11 and 21G11-LexA. (D) VNC expression of 23C09 and 23C09∩VGlut. For all images, green = GFP, magenta = nc82. GFP, green fluorescent protein; nc82, monoclonal antibody to Bruchpilot; VNC, ventral nerve cord.
(TIF)
(A) Syt-HA expression in the LH and SIP of 21G11-LexA (green). The circle highlights the LH. The vertical lobe of the MB is drawn to facilitate visualization of the adjacent SIP. (B) Dscam17.1-GFP and syt-HA expression in the LH of 23C09∩VGlut (green). The circle highlights the LH. Scale bar = 10 μm. d, dorsal; Dscam17.1, Down syndrome cell adhesion molecule with isoform 1 of the transmembrane domain; GFP, green fluorescent protein; HA, hemagglutinin; l, lateral; LH, lateral horn; m, medial; MB, mushroom body; SIP, superior intermediate protocerebrum; syt, synaptotagmin; v, ventral.
(TIF)
(A and B) Schematics of the experiment and calcium response at the LH, using GCaMP6m, of 21G11-LexA neurons to air, 0.5%, 1%, and 2% of CO2 while 21G11-LexA neurons are silenced by expression of Kir2.1. (C) Schematics of the experiment and LH activity of 21G11-LexA upon activation of 21G11-LexA neurons, expressing Chrimson, with 720 nm light. (D) Peak GCaMP6m intensity change upon activation. For (C), the average time course of GCaMP6m intensity change is shown. The black bar indicates the time of the stimulus. *p < 0.05. All p values are calculated with Wilcoxon signed-rank test. LH, lateral horn.
(TIF)
(A and B) Schematics of the experiment and calcium response at the LH, using GCaMP6m, of 23C09∩VGlut neurons to air, 0.5%, 1% and 2% of CO2, while 23C09∩VGlut neurons are silenced by expression of Kir2.1. (C) Schematics of the experiment and LH activity of 23C09∩VGlut upon activation of 23C09∩VGlut neurons, expressing Chrimson, with 720 nm light. (D) Peak GCaMP6m intensity change upon activation. For (C), the average time course of GCaMP6m intensity change is shown. The black bar indicates the time of the stimulus. *p < 0.05. All p values are calculated with Wilcoxon signed-rank test. LH, lateral horn.
(TIF)
T-maze response to benzaldehyde at 1:1000 dilution (BZ). White box, flies with olfactory organs. Gray box, flies without olfactory organs. The box represents the first and the third quartiles, and the whiskers the 10th and 90th percentiles. The line across the box is the median. N = 10. Error bars indicate ±SEM ****p<0.0001. p values are calculated with Wilcoxon signed-rank test. BZ, benzaldehyde; SEM, standard error of the mean.
(TIF)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.