Abstract
Recent symbioses, particularly facultative ones, are well suited for unravelling the evolutionary give and take between partners. Here we look at variation in natural isolates of the social amoeba Dictyostelium discoideum and their relationships with bacterial symbionts, Burkholderia hayleyella and Burkholderia agricolaris. Only about a third of field-collected amoebae carry a symbiont. We cured and cross-infected amoebae hosts with different symbiont association histories and then compared host responses to each symbiont type. Before curing, field-collected clones did not vary significantly in overall fitness, but infected hosts produced morphologically different multicellular structures. After curing and reinfecting, host fitness declined. However, natural B. hayleyella hosts suffered fewer fitness costs when reinfected with B. hayleyella, indicating that they have evolved mechanisms to tolerate their symbiont. Our work suggests that amoebae hosts have evolved mechanisms to tolerate specific acquired symbionts; exploring host-symbiont relationships that vary within species may provide further insights into disease dynamics.
Research organism: Dictyostelium
eLife digest
A species can benefit or be hurt by other species. For example, honeybees and flowering plants help each other to flourish, while lions and gazelles behave in ways that decrease each other’s populations. Understanding these relationships is important for controlling pests and diseases.
Sometimes it is easiest to study such interactions by looking at simple ones that happen on a small scale. Amoebas are common soil organisms that have the same basic organization as human cells. They are much larger and more complex than the bacteria that also live in the soil. How exactly the amoebas and bacteria interact in the soil is an important question, particularly as some of the bacteria can also live inside amoebas. Does this intimate relationship help or harm the amoeba?
Shu, Brock, Geist et al. studied the relationship between a widely studied species of social amoeba and two species of bacteria that can live inside it. Some of the amoebas naturally contained one of the bacteria species, and others were infected with the bacteria in experiments.
Throughout the entire life cycle of the amoebas, the bacteria lived inside them. During one part of the life cycle, amoebas form so-called fruiting bodies, which release spores that can develop into new amoebas. Shu et al. found that both types of bacteria alter the structure of the fruiting bodies in ways that reduce how well the spores disperse.
One of the bacteria species, called Burkholderia hayleyella, harmed the amoebas a lot. It caused most harm to amoebas that do not naturally host the bacteria. This indicates that the amoebas that do host this species may have evolved to avoid its worst effects.
The amoebas have many similarities to the white blood cells that clear bacteria from the human body. Certain bacteria can get inside white blood cells, causing diseases such as tuberculosis. Understanding how bacteria harm amoebas might be useful for understanding such diseases, and developing treatments for them. Though the bacteria Shu et al. studied are not toxic to humans, they are closely related to bacteria that are harmful. It is therefore possible that some bacteria that infect humans first evolve to infect amoebas.
Introduction
Relationships are complicated because each party has evolved to maximize its own interests. Mutualisms arise when different parties can contribute unique abilities or resources to one another, under conditions where exploitation is controlled (Bronstein, 2015). Mutualisms where one or both parties are microbial fall under the general category of symbiosis. Symbioses are rife with potential conflict (Dale and Moran, 2006; Estrela et al., 2016; Garcia and Gerardo, 2014; Moran, 2007; Oliver et al., 2005). Despite this, symbiotic relationships are pervasive and persistent (Douglas, 2008; Moran et al., 1993; Wernegreen, 2017; Werner et al., 2015). The stability and ubiquity of these interactions implies that conflict can be managed or minimized by partners over multiple generations. However, stability does not imply stagnation. Recent studies suggest that examining extant symbiotic associations may not reveal the underlying mutualism to parasitism continuum that drives evolutionary change (Moran, 2007; Queller and Strassmann, 2018). Indeed, the view that symbiotic associations fluctuate along a mutualism-to-parasitism continuum is increasingly appreciated (McFall-Ngai et al., 2013).
Intracellular endosymbiosis involves a particularly intricate dance between partners. Intracellular endosymbionts must invade, survive, and replicate within host cells and move to new hosts. To exploit their host niche, mutualistic endosymbionts evolve specialized lifestyles that parallel those of intracellular pathogens (Casadevall, 2008; McCutcheon and Moran, 2011; Soto et al., 2009). Despite initial exploitative strategies, some endosymbionts become beneficial or even obligate for host survival, a situation common for many insect nutritional endosymbioses (Douglas, 2009). Strict vertical transmission of endosymbionts typically promotes a more tranquil relationship as the evolutionary fates of the two parties become increasingly interdependent (Ferdy and Godelle, 2005; Hosokawa et al., 2016). Host dependency also often leads to the reduction in size of symbiont genomes as they become more streamlined for life within their host, something true for both beneficial and pathogenic endosymbionts (Dale and Moran, 2006). Horizontally acquired facultative symbionts are more likely to retain active conflicts. This has been experimentally demonstrated by manipulating the jellyfish symbiont Symbiodinium microadriaticum towards solely vertical or horizontal transmission modes, leading to the evolution of more mutualistic or parasitic lineages respectively (Sachs and Wilcox, 2006). Though horizontal transmission can favor symbionts that behave more parasitically, selection can favor hosts that employ more severe countermeasures to limit symbiont entry or growth (Nyholm and McFall-Ngai, 2004; Ratzka et al., 2012; Reynolds and Rolff, 2008). Indeed, host-driven control can be extreme, as demonstrated by the ability of Paramecium bursaria to manipulate its facultative nutritional symbiont (Chlorella sp.) in a relationship that provides no apparent benefit for the imprisoned symbiont (Lowe et al., 2016).
Symbiotic relationships may be positive or negative under different environmental conditions (Leung and Poulin, 2008; Pérez-Brocal et al., 2011). An interesting example of context dependency occurs in the Acyrthosiphon pisum-Hamiltonella defensa symbiosis (Oliver et al., 2003; Oliver et al., 2005). H. defensa infection confers host resistance to parasitoid attack, with resistance being greater for hosts co-infected with Serratia symbiotica (Oliver et al., 2006). However, co-infection comes at a high fecundity cost, such that in parasitoid free environments, host fitness is reduced compared to uninfected counterparts (Oliver et al., 2006). This interplay between reproductive strategy, context, and evolutionary history illustrates the complexity and fluidity of symbiosis.
The facultative endosymbiosis between the social amoeba Dictyostelium discoideum and Burkholderia bacteria provides a promising system for revealing insights into symbiosis dynamics (Brock et al., 2011; DiSalvo et al., 2015). D. discoideum is a soil dwelling amoeba with an interesting life cycle involving unicellular and multicellular stages. During the unicellular stage, amoebae consume bacteria by phagocytosis and divide. When prey are scarce, amoebae aggregate by tens of thousands to form multicellular slugs that move towards heat and light, seeking out a location to form fruiting bodies. These fruiting bodies consist of a stalk of sacrificial dead cells which support a globular sorus containing hardy spore cells (Kessin, 2001). When spores are dispersed, they germinate into vegetative amoebae and the cycle continues. Processes employed by specialized immune-like cells (sentinel cells) during slug migration and fruiting body formation remove any remaining bacteria, typically producing bacteria-free sori (Brock et al., 2011; Chen et al., 2007; Cosson and Lima, 2014).
However, some D. discoideum isolates harvested from the wild are infected with Burkholderia symbionts (Brock et al., 2011). Infection persists in the lab throughout the social cycle, where intracellular bacteria can be visualized within spores and sori (DiSalvo et al., 2015). Infection can be terminated by treating hosts with antibiotics and induced by exposing naïve hosts to Burkholderia, thereby allowing us to easily mix and match partners and study subsequent fitness consequences (Brock et al., 2016a; DiSalvo et al., 2015).
D. discoideum also has a meiotic sexual process, that occurs much less frequently than the asexual proliferation process of binary fission that amoebae go through every few hours (Bloomfield et al., 2010). The asexual binary fission process results in lineages that can be quite different, though all of the same species. Thus, a lineage that has acquired a bacterial endosymbiont can evolve to tolerate it independent of uninfected lineages at least for the thousands of generations that might pass before sexual recombination occurs. Comparing such lineages can illuminate how natural selection operates in early stages of symbiosis.
The Dictyostelium-Burkholderia association is particularly compelling for studying the parasitism to mutualism continuum in endosymbiosis because the fitness consequences of infection are context dependent. Under standard laboratory growth conditions, Burkholderia infection is detrimental for hosts because it decreases slug migration and spore production (Brock et al., 2016a; DiSalvo et al., 2015). However, infection can be beneficial. Infection induces secondary carriage of edible bacteria (Burkholderia is typically inedible), which can increase host fitness in food scarce conditions when the carried food bacteria reseed new environments with a food source (Brock et al., 2011; DiSalvo et al., 2015). For this reason, field-infected host amoebae are also called ‘farmers’ and uninfected host amoebae are called ‘non-farmers.’ Additionally, infected hosts are less harmed by ethidium bromide exposure, possibly mediated by bacterial degradation of the toxin (Brock et al., 2016a).
We have identified three Burkholderia species associated with Dictyostelium: B. agricolaris, B. hayleyella, and B. bonniea (Brock et al., 2018). Most of our work has been conducted with the first two species, which differentially impact host fitness. Here we probe the interaction between host and symbiont genotypes (with regards to their association history) with infection outcomes using a set of previously field-collected and characterized amoebae isolates and their associated Burkholderia symbionts (Brock et al., 2011; DiSalvo et al., 2015). We use standard laboratory conditions in which symbiont infection is shifted towards host detrimental outcomes. Since all three Burkholderia species can be cultured on Petri plates with standard media, none of them are entirely dependent on Dictyostelium for survival.
We find that B. hayleyella is most detrimental for D. discoideum hosts in general, but is most costly to those first exposed to it in the lab. We also document symbiont localization and morphological symptoms in hosts throughout development. Although both species of bacteria can be observed within phagocytic vacuoles, B. hayleyella infects more cells and damages fruiting body structures. These morphological aberrations are also less severe in native-hayleyella hosts, suggesting that native hosts have evolved mechanisms to withstand symbiont colonization.
Results
Impact of B. agricola and B. hayleyella on D. discoideum spore production
Our first goal was to clarify how infection of Dictyostelium by Burkholderia symbionts differentially influences host fitness. We used 12 natural isolates of Dictyostelium, each from three original conditions, uninfected with Burkholderia, infected with B. agricolaris, or infected with B. hayleyella (Table 1). We refer to these three types respectively as naïve hosts, native-agricolaris hosts, and native-hayleyella hosts. The word ‘host’ always means D. discoideum, and sometimes refers to potential hosts not actually infected with Burkholderia. Prior to each treatment, all host types were developed on nutrient media supplemented with bacterial food (Klebsiella pneumoniae). Amoebae, spores, cells, slugs, fruiting bodies, stalks, and sori refer only to D. discoideum.
Table 1. Dictyostelium discoideum clones used for this study.
Clones are divided into specific sets each with naive, native-ag, and native-ha field-collected counterparts. They were collected from Virginia, North Carolina, and Texas as indicated.
| Set | Clone | Status | Burkholderia | Location collected | Date collected | GPS coordinates |
|---|---|---|---|---|---|---|
| 1 | QS9 | Naïve | None | Virginia-Mt Lake Biological Station | Oct. 15 2000 | N 37° 21’, W 80° 31’ |
| QS70 | Native | B. agricolaris | Texas- Houston Arboretum | Jul. 15 2004 | N 29° 46’, W 95° 27’ | |
| QS11 | Native | B. hayleyella | Virginia-Mt Lake Biological Station | Oct. 15 2000 | N 37° 21’, W 80° 31’ | |
| 2 | QS18 | Naïve | None | Virginia-Mt Lake Biological Station | Oct. 15 2000 | N 37° 21’, W 80° 31’ |
| QS159 | Native | B. agricolaris | Virginia-Mt Lake Biological Station | May. 2008 | N 37° 21’, W 80° 31’ | |
| QS23 | Native | B. hayleyella | Virginia-Mt Lake Biological Station | Sep. 25 2000 | N 37° 21’, W 80° 31’ | |
| 3 | QS17 | Naïve | None | Virginia-Mt Lake Biological Station | Oct. 15 2000 | N 37° 21’, W 80° 31’ |
| QS161 | Native | B. agricolaris | Virginia-Mt Lake Biological Station | May. 2008 | N 37° 21’, W 80° 31’ | |
| QS22 | Native | B. hayleyella | Virginia-Mt Lake Biological Station | Sep. 25 2000 | N 37° 21’, W 80° 31’ | |
| 4 | QS6 | Naïve | None | Virginia-Mt Lake Biological Station | Sep. 25 2000 | N 37° 21’, W 80° 31’ |
| NC21 | Native | B. agricolaris | North Carolina-Little Butts Gap | Oct. 1988 | N 35° 46’, W 82° 20’ | |
| QS21 | Native | B. hayleyella | Virginia-Mt Lake Biological Station | Oct. 15 2000 | N 37° 21’, W 80° 31’ |
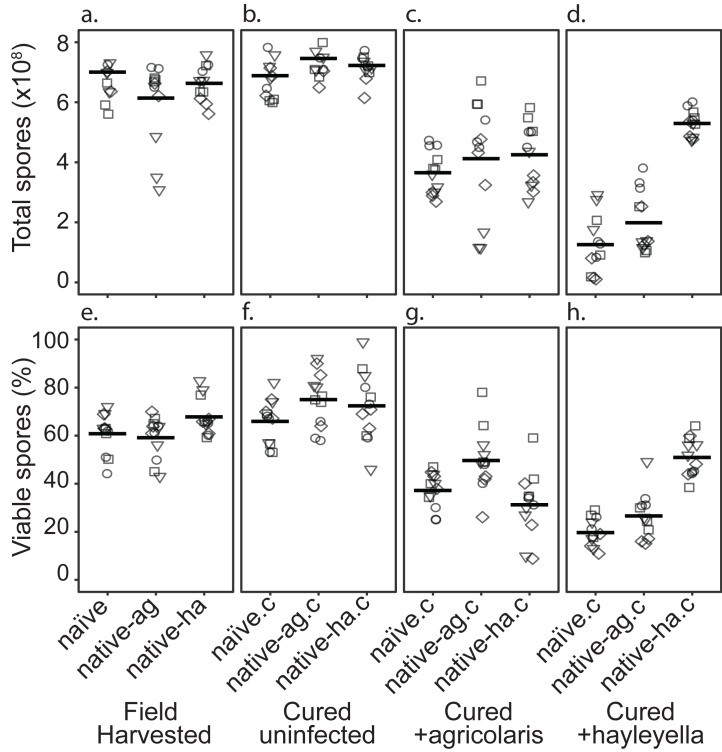
We looked at D. discoideum spore viability and other measures under the following conditions: a) natural field state (naïve, native-agricolaris, native-hayleyella), b) those same hosts cured of Burkholderia (antibiotic treated), and c) condition after curing and reinfecting (with either B. agicolaris or B. hayleyella) (Figure 1). We quantified percent spore viability and number of spores produced (Figure 2, and Supplementary files 1 and 2). We multiplied these two measures to get a single main measure of fitness, viable spores produced.
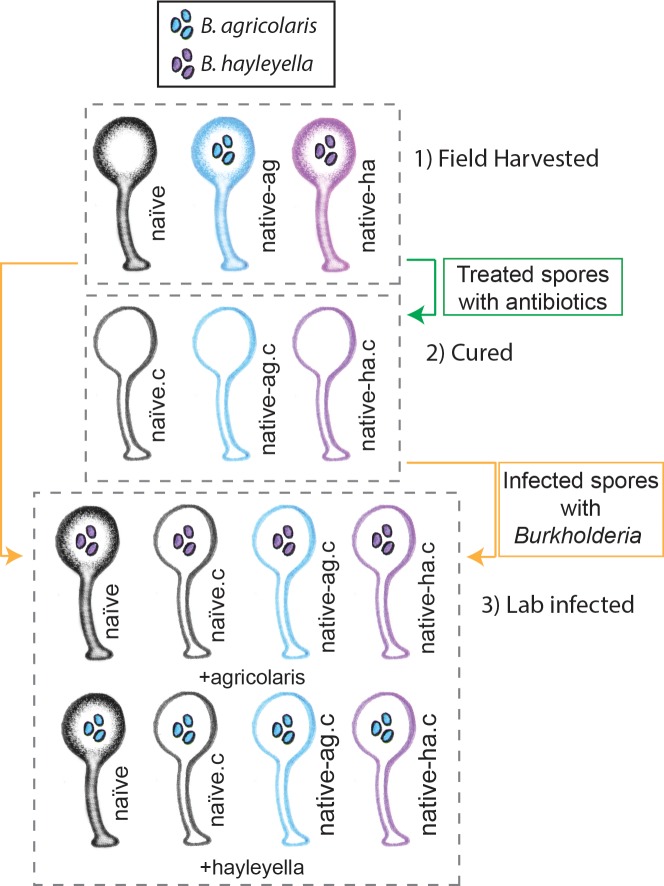
Figure 1. Illustration of host-symbiont pairs used throughout the study.
D. discoideum clones were originally harvested from the wild in three different states: uninfected (indicated as naïve), or naturally infected with B. agricolaris or B. hayleyalla (indicated as native-ag, and native-ha respectively). Clones were treated with antibiotics to eliminate symbionts and are indicated with a ‘.c’. Clones were subsequently exposed to Burkholderia to initiate new infections. Thus, experimental types include 1) Field harvested, 2) cured, and 3) lab infected hosts.
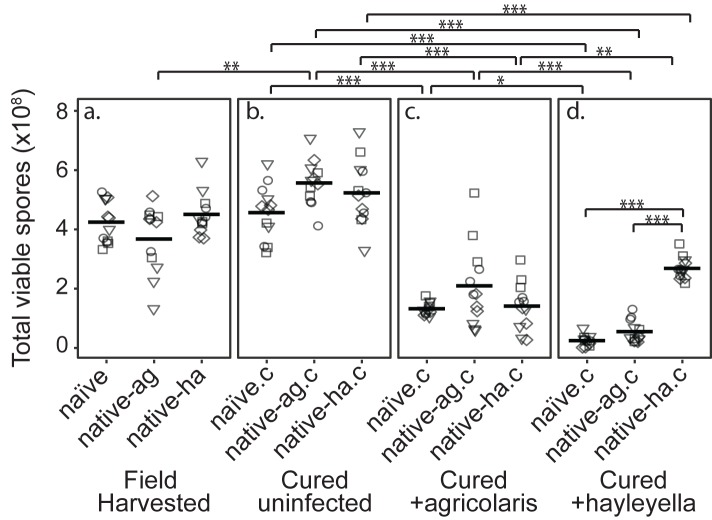
Figure 2. Burkholderia Infections Differentially Alter Spore Viability According to Burkholderia Species and Host Background.
Total viable spores were determined for naïve and native hosts in their field harvested (a), cured (b), B. agricolaris lab-infected (c), and B. hayleyella lab-infected state (d). Four clones were measured for each type with three replicates for each (squares, triangles, circles, and diamonds represent set 1–4 clones respectively). Spore viability for wild harvested B. agricolaris and B. hayleyella host clones is higher than their cured-re-infected counterparts. Notably, spores from infected B. agricolaris and B. hayleyella native hosts (either naturally infected or cured and re-infected with their original Burkholderia) have a higher fitness than Burkholderia infected non-native counterparts. Bars represent significant differences (p < 0.05, and as indicated in supplemental tables).
Figure 2—figure supplement 1. Total Spore Number and Percent of Viable Spores for Burkholderia Infections in Diverse Host Backgrounds.
D.discoideum fitness does not differ by wild burkholderia infection status
We found that infection status in the field did not affect total viable spore counts for naïve, native-agricolaris, or native-hayleyella hosts (Figure 2a and Supplementary file 1a) (linear mixed model (LMM), ΔAIC = −1.57, χ2 = 5.57, DF = 2, p = 0.062). Thus, field-infected native hosts do not seem to suffer any net fitness costs from infection by this measure.
D.discoideum fitness does not decrease with antibiotic treatment
To make parallel comparisons when we newly infected D. discoideum with either of the two Burkholderia species, we first had to cure all hosts and be sure that curing in itself did not decrease fitness. We found that wild-collected hosts of our three categories did not experience lowered fitness after being cured with antibiotics (Figure 2a and b) (viable spore production: LMM, ΔAIC = 1.74, χ2 = 2.26, DF = 2, p = 0.32, Supplementary file 1b). Antibiotic treatment actually increased viable spore production of native-agricolaris hosts compared to uncured native-agricolaris hosts (LMM, ΔAIC = −5.68, χ2 = 9.68, DF = 2, p = 0.008; Figure 2a and b).
D.discoideum fitness decreases with exposure to burkholderia
To test the effects of Burkholderia on all types of field-collected hosts, we grouped host types into four sets, with each set representing a unique naïve, native-agricolaris, and native-hayleyella host (Table 1). We then infected each cured host type within the set with each Burkholderia from the native hosts within the same set. We compared viable spore production of cured D. discoideum hosts versus those same hosts artificially infected with B. agricolaris or B. hayleyella (Figure 2b, c and d). We found an overall effect on total spore viability with the addition of Burkholderia to all antibiotic-cured hosts (LMM, ΔAIC = −198.81, χ2 = 210.81, DF = 6, P ≪ 0.001). Addition of either B. agricolaris (Wald t = −10.19, DF = 96, P ≪ 0.001) or B. hayleyella (t = −13.58, DF = 96, P ≪ 0.001) to any of the cured hosts decreased their fitness (Figure 2; Supplementary file 2).
We then asked whether D. discoideum hosts are adapted to the Burkholderia species they carried in the field (using the same set strategy described above). Overall, the addition of B. hayleyella to D. discoideum led to significantly lower viable spore production than did the addition of B. agricolaris (Wald t = −4.48, DF = 96, P ≪ 0.001) (Figure 2). We also tested for an interaction between native host status and which Burkholderia species was added. There was an interaction effect on total viable spore production (LMM, ΔAIC = −199.93, χ2 = 215.92, DF = 8, P ≪ 0.001) (Supplementary file 2e). To address specific adaptation, we performed separate tests for each Burkholderia species added. When B. hayleyella was added to the three cured hosts, native-hayleyella hosts had higher fitness than did either native-agricolaris or naïve hosts (both P ≪ 0.001, Figure 2d; Supplementary file 2e). In contrast, native-agricolaris hosts did not have significantly higher fitness with the addition of B. agricolaris than either native-hayleyella or naïve hosts (both p > 0.05, (Figure 2c; Supplementary 2e). However, there was a trend in the direction of native-agricolaris doing best (Figure 2c). These results indicate that native-hayleyella hosts are adapted to colonization by their field-acquired symbionts.
D.discoideum morphology and burkholderia infected state
We next examined host morphology and symbiont localization at several stages of the D. discoideum life cycle. Using transmission electronic and confocal microscopy, we examined one D. discoideum clone for each host type outlined above and in Figure 1 (QS9 for the naïve, QS70 for the native-agricolaris, and QS11 for the native-hayleyella host). These were either in an uninfected state or infected with one representative of B. agricolaris (Ba70 from QS70) or of B. hayleyella (Bh11 from QS11).
Food bacteria location inside D. discoideum uninfected with burkholderia
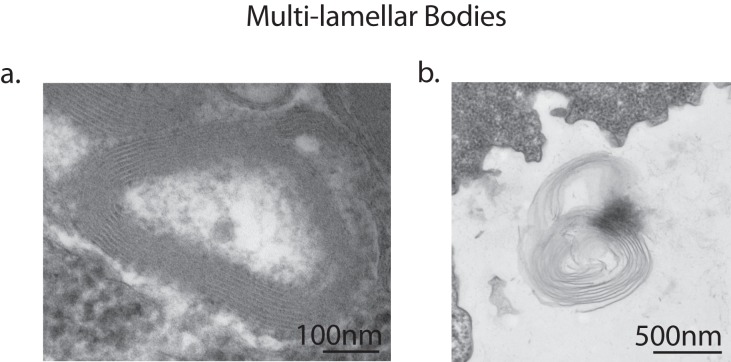
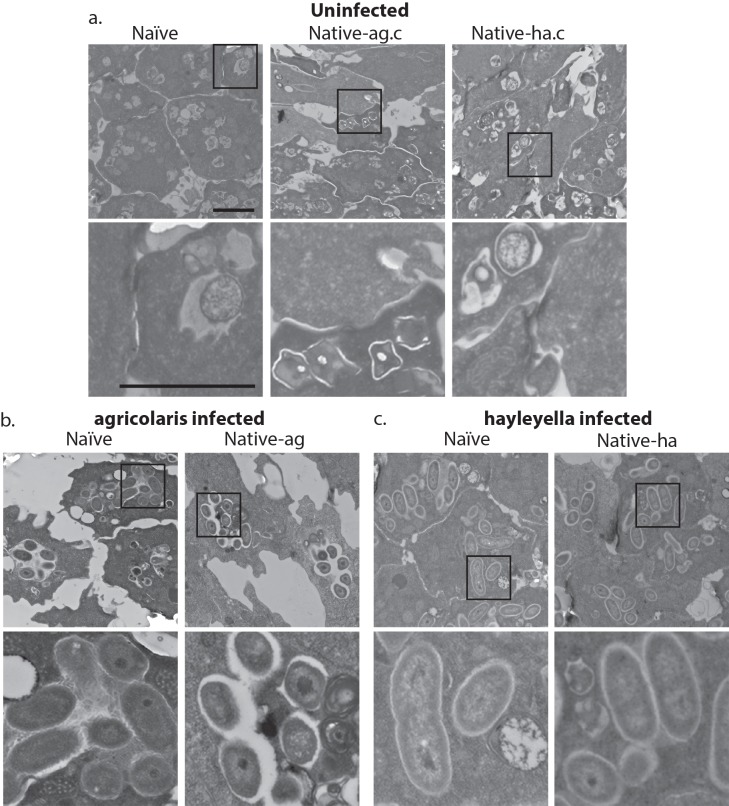
D.D. discoideum morphology without Burkholderia in both naïve and cured native hosts, has vegetative cells that harbor no intracellular bacteria but contain many empty multilamellar bodies inside vacuoles (Figure 3a and Figure 3–figure supplement 1). Multilamellar bodies are predominantly composed of amoebae membranes and are thought to be a byproduct of bacterial digestion as they are produced when amoebae are fed bacterial food (Denoncourt et al., 2016; Paquet et al., 2013). Confocal microscopy of vegetative amoebae from this same host set grown with GFP-labeled food bacteria (K. pneumoniae) contain little to no intracellular GFP, suggesting that they have digested food bacteria by the time of fixation (Figure 4a).
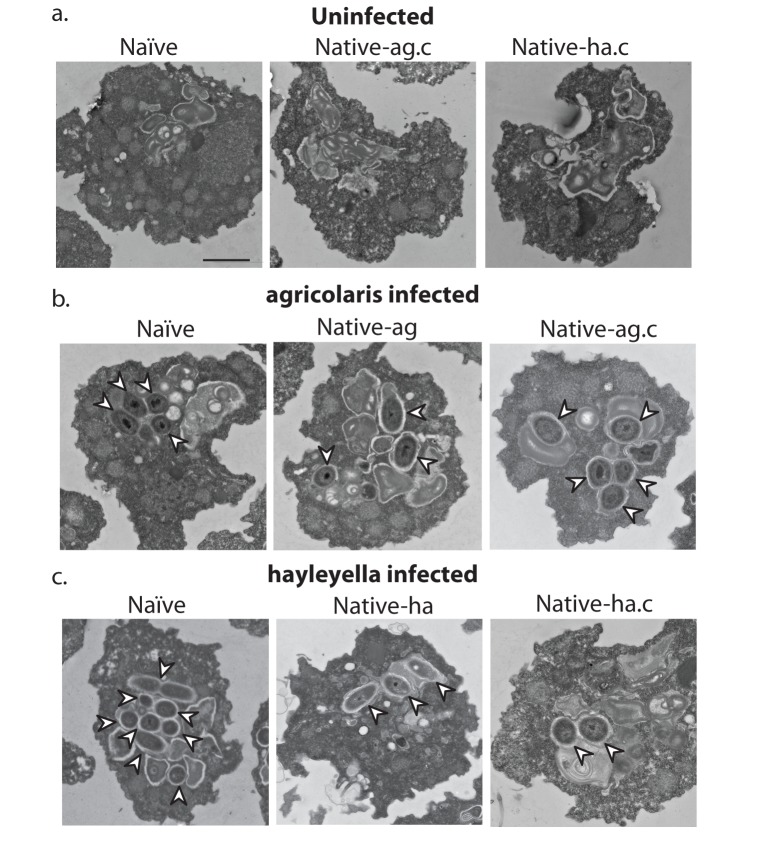
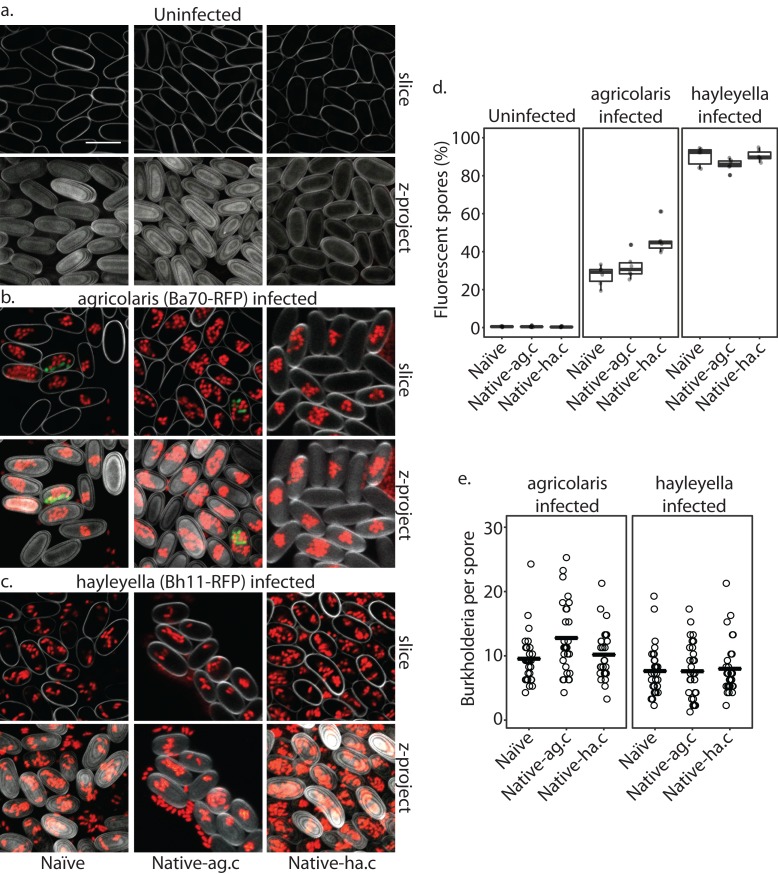
Figure 3. Bacterial cells are found within Burkholderia exposed vegetative amoebae.
Transmission electron micrographs of vegetative amoebae show naïve and cured native amoebae with intracellular morphologies suggestive of active bacterial digestion with no evidence of intact intracellular bacteria (a). In contrast, bacterial cells can be found within B. agricolaris (b) and B. hayleyella (c) infected hosts. Arrows point to bacterial cells. More bacteria are observed in the B. hayleyella infected naïve host than in field harvested native-hayleyella and cured and re-infected native-hayleyella hosts (c). Bacterial cells appear to be within vacuole-like compartments. Scale bar (applicable to all): 2 um.
Figure 3—figure supplement 1. Multi-lamellar bodies excreted by vegetative amoebae.
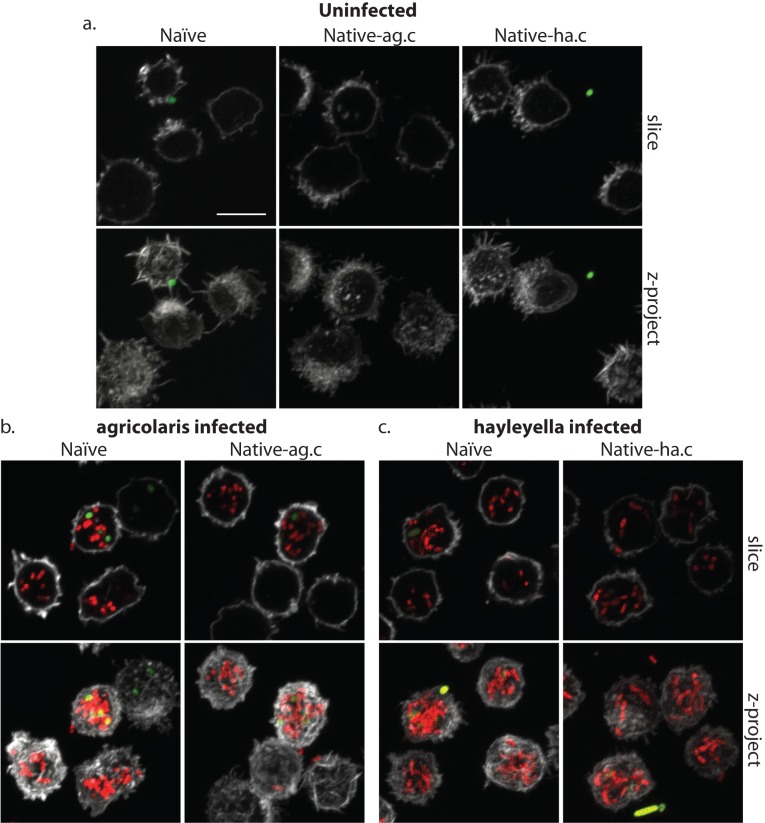
Figure 4. Burkholderia is found abundantly in colonized vegetative amoebae.
Confocal imaging of fixed and stained vegetative amoebas show little to no intracellular bacteria in uninfected clones (a). However, abundant Burkholderia (Burkholderia-RFP shown in red) is found in B. agricolaris (b) and B. hayleyella (c) infected hosts. Occasional intracellular food bacteria (Klebsiella-GFP shown in green) is seen in B. agricolaris hosts (c). Spore coats are stained with phalloidin shown in grey. Scale bar 10 um.
After bacterial food has been depleted, vegetative cells aggregate to form multicellular migratory slugs. In accordance with our observations that all bacteria were killed and digested by vegetative cells, we found no intact bacteria in naïve or cured native host slugs (Figure 5a). In addition, slug cells were in general compacted with electron dense materials and contained no vacuoles or multi-lamellar bodies (Figure 5a).
Figure 5. Intracellular bacteria are retained in naïve migrating slugs exposed to Burkholderia and in native Burkholderia hosts.
Transmission electron micrographs of uninfected (a) show closely packed amoebae with internal structures reminiscent of previous bacterial digestion but without evidence of intact internal bacteria. In contrast, B. agricolaris (b) and B. hayleyella (c) infected slugs retain intracellular bacteria. Bottom panels represent magnified versions (see box) of upper panels. Scale bar (applicable to all panels in row) 2 um.
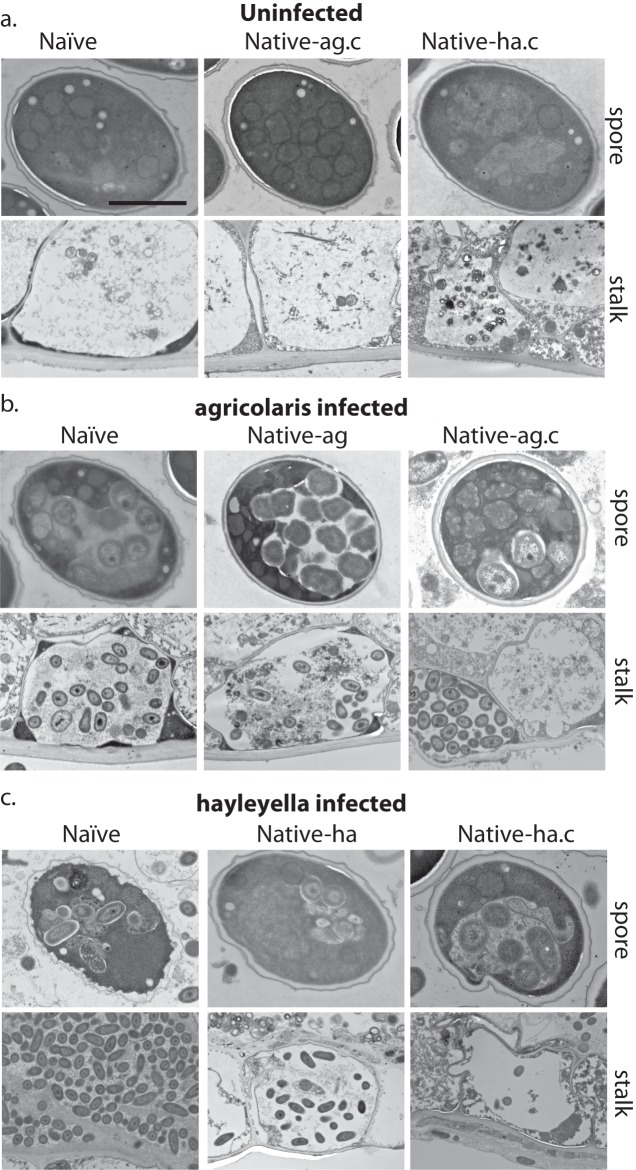
Ultimately, slug cells differentiate into fruiting bodies consisting of dead stalk cells that support a sorus containing reproductive spore cells. In uninfected fruiting bodies, we did not detect bacteria in stalk cells or spores (Figures 6a and 7a). Instead spores were packed with electron dense materials with no food vacuoles or multi-lamellar bodies (Figure 6a). Stalk cells showed plant cell-like characteristics, having a cellulosic cell wall and containing a single large vacuole (Figure 6a). Inside the large vacuole, there were some mitochondria and other cellular materials but no bacteria (Figure 6a). Taken together, these results suggest that the food bacterium we used, Klebsiella pneumoniae, was efficiently cleared during the social cycle from amoebae uninfected by Burkholderia and the amoebae then aggregate and produce bacteria-free fruiting bodies.
Figure 6. Bacterial cells are retained in spore and stalk cells from Burkholderia-exposed hosts.
As visualized through transmission electron microscopy, (a) uninfected hosts form sturdy spores and stalk cells with no detectable bacteria. Spores and stalk cells retain intracellular bacteria in B. agricolaris (b) and B. hayleyella (c) hosts. Naïve B. agricolaris hosts appear structurally similar to uninfected cells while naïve B. hayleyella hosts have compromised spore coats and collapsed stalk structures filled with bacteria. Scale bar: 2 um.
Figure 7. Burkholderia is retained in the sori of developed D. discoideum hosts and the percent of Burkholderia positive spores differs according to Burkholderia species.
Confocal images show no intra- or extracellular bacteria in uninfected spores (a) Abundant Burkholderia is seen in B. agricolaris (b) and B. hayleyella (c) hosts, with more infected spores seen for B. hayleyella (d) but more Burkholderia-RFP cells detected per infected spore for B. agricolaris hosts (e). Co-infection by food bacteria is occasionally observed in B. agricolaris infected spores (b). For a-c: Klebsiella-GFP shown in green, Burkholderia-RFP shown in red, and calcofluor stain shown in grey. Top panels are image slices; bottom panels are max intensity projections of z stacks. Scale bar: 10 um.
Burkholderia location inside D. discoideum
When D. discoideum is infected with Burkholderia, we find it in amoebae, slug cells, spores, and stalk cells. Using confocal microscopy and RFP labeled Burkholderia strains, we were able to specifically identify high levels of Burkholderia inside host amoebae (Figure 4b,c). B. hayleyella was present in more of the amoebae than B. agricolaris was (Figure 4b,c). The higher number of B. hayleyella bacteria may account for the more detrimental fitness consequences it imposes (Figure 2).
The food bacterium K. pneumoniae labelled with GFP was occasionally observed in Burkholderia infected amoebae, particularly those infected by B. agricolaris (Figures 4b and 7b). This suggests that Burkholderia colonization may partially impede food digestion, thereby allowing co-colonization of secondary bacteria and contributing to the proto-farming phenotype (Brock et al., 2011; DiSalvo et al., 2015). However, confocal images demonstrate that Burkholderia are much more abundant in amoebae than are K. pneumoniae, indicating that the majority of intact intracellular bacteria observed via transmission electron microscopy (TEM) are most likely Burkholderia cells (Figures 3, 5 and 6).
TEM of vegetative amoebae shows intact bacteria which we infer to be Burkholderia surrounded by multi-lamellar bodies and located inside what appear to be food vacuoles (Figure 3b,c). Their undamaged appearance suggests that they are resistant to phagocytic digestion. In addition to the presence of intact intracellular bacteria, we also observe empty multi-lamellar bodies inside food vacuoles of infected amoebae. This suggests that digestion is not completely arrested during colonization, an unsurprising finding given that hosts continue to grow and multiply. Interestingly, we did not find multi-lamellar bodies containing intact bacteria secreted into the extracellular environment, indicating that Burkholderia may not be expelled from host cells via multi-lamellar body excretion.
In Burkholderia infected hosts, intact bacteria are retained in food vacuoles throughout the transition to multicellular slugs, suggesting that bacteria stay within phagosomes throughout the aggregation stage (Figure 5b,c). Through TEM, we did not detect obvious morphological defects in infected slugs or differences between slugs infected with different Burkholderia species.
After fruiting body development, we find intracellular bacteria in both stalk and spore cells of Burkholderia infected hosts (Figure 6b,c). In infected stalk cells, intact bacteria reside in single large vacuoles inside the cellulosic cell wall (Figure 6b,c). In spore cells, bacteria remain within vacuoles (Figure 6b,c). We also observed bacterial cells outside spores but within the sorus, suggesting that bacteria may be carried interstitially into fruiting body structures, or they may be escaping pre-spore or spore cells within the sorus (Figure 7b,c).
We did not observe strikingly altered morphologies for B. agricolaris infected spore and stalk cells (Figure 6b). However, the spore and stalk cells of naïve hosts infected with B. hayleyella appeared to be morphologically aberrant (Figure 6c). We found numerous broken spores and signs that bacterial cells were escaping from damaged spores (Figure 6c). In addition, the whole stalk structure was often collapsed and filled with bacteria (Figure 6c). No clear cellulosic cell wall was observed in stalk cells, suggesting that B. hayleyella colonization inhibits the normal development of stalk cells or results in their disruption in naïve hosts and native-agricolaris hosts.
In line with the visualized differences between the abundance of the two species of Burkholderia in amoebae, we find significantly more spores infected with RFP labeled B. hayleyella (mean = 88.8%) than similarly labeled B. agricolaris (mean = 35.3%) (F2,4 = 191.33, p < 0.001) (Figure 7d). These results indicate that the degree of fitness detriment imposed by B. hayleyella could be a result of bacterial infection prevalence within the population. However, B. agricolaris displays a significantly higher intracellular density (mean = 10.55) within individual infected spores than B. hayleyella (mean = 7.45) (F1,177 = 23.845, p < 0.001), while host background does not play a significant role in Burkholderia intracellular density (F2,177 = 2.723, p = 0.068) (Figure 7e).
Burkholderia impact on fruiting body morphology
Since Burkholderia are found inside D. discoideum, it is no surprise they impact the morphology of fruiting bodies. In their field-collected state, the three clones carrying no Burkholderia, or B. agricolaris or B. hayleyella differed in both stalk height and stalk volume (F2,27 = 42.6, P ≪ 0.001, F2,27 = 50.8, P ≪ 0.001, Figure 8b, Supplementary file 3a). The main pattern is that both height and volume were significantly lower in native-hayleyella hosts (Figure 8; Supplementary file 3a).
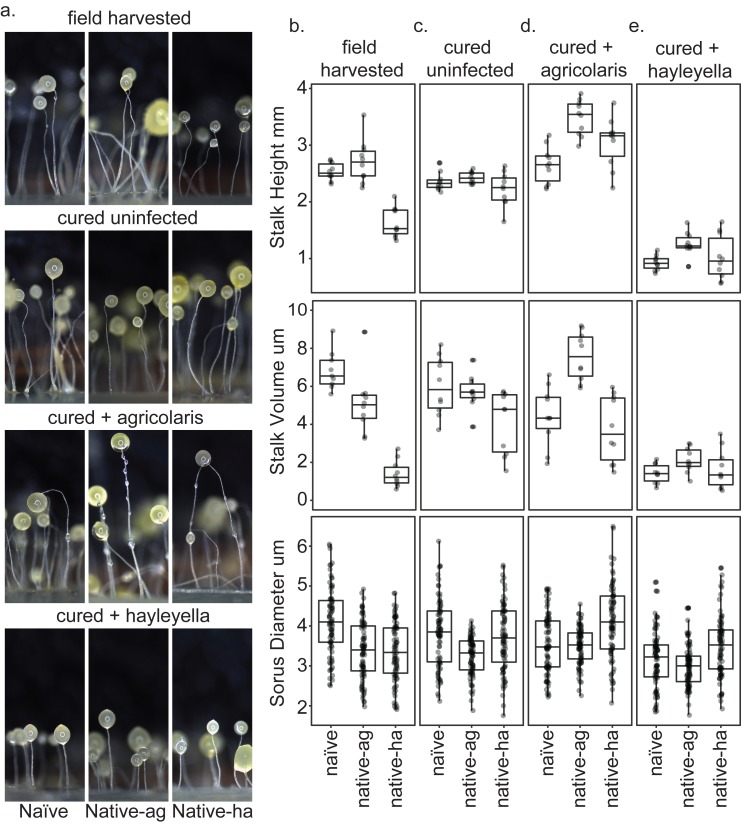
Figure 8. Fruiting body morphology is differentially altered by Burkholderia colonization.
Macro photographs of fruiting bodies (a) show slightly different morphologies according to Burkholderia infection status. Sori measurements demonstrate that field collected native-hayleyella hosts produce shorter stalks and less voluminous sori (b). Cured hosts produce similar fruiting body measurements across host background (c). Cured hosts subsequently infected with B. agricolaris produce slightly taller stalks, which is most noticeable in cured and re-infected native-agricolaris hosts (d). Cured hosts subsequently infected with B. hayleyella all produce significantly shorter stalks with overall smaller fruiting body dimensions (e).
Native-agricolaris fruiting bodies were generally similar to the naïve host but taller than the native-hayleyella host (p < 0.001). In the native-agricolaris host, the spore masses often slid down their stalks or the fruiting bodies fell over, though the stalks were not significantly taller than in the naïve host. If the spores fall off their stalks, they will not have the advantage of facilitated transport by a vector that they would have at the top of the stalk (Smith et al., 2014).
There was also an overall difference in sorus diameter and sorus volume among the three hosts in their field state (Figure 8; F2,236 = 25.4, P ≪ 0.001, F2,236 = 22.9, P ≪ 0.001, Supplementary file 4a). Compared to the naïve hosts, both the native-agricolaris and native-hayleyella hosts had smaller sorus sizes (both p < 0.001) but were not different from each other.
Curing with antibiotics caused no significant change in any stalk or spore measurements (Figure 8a,b,c; Supplementary file 3b, Supplementary file 4b). However, when Burkholderia bacteria were added to the cured hosts, we saw species-specific effects on morphology (Figure 8; Supplementary file 3c-d, Supplementary file 4c-d). Overall, the addition of B. agricolaris changed stalk height (F1,58 = 54.9, P ≪ 0.001) significantly increasing it in two of the hosts (Supplementary file 3c). Stalk volume was not affected. Addition of B. agricolaris also significantly changed both sorus diameter and sorus volume (F2,240 = 25.4, P ≪ 0.001, F2,240 = 21.2, P ≪ 0.001). Native-hayleyella hosts had larger sori when infected with B. agricolaris (Figure 8; Supplementary file 4c).
The addition of B. hayleyella decreased both stalk height (Figure 8; F1,58 = 366.4, P ≪ 0.001) and stalk volume (F1,58 = 120.2, P ≪ 0.001, Supplementary file 3d) significantly in all three host types. Addition of B. hayleyella also affected sorus diameter (F2,240 = 10.1, P ≪ 0.001) and volume (F2,240 = 11.3, P ≪ 0.001) with a significant specific effect of smaller sori in the naïve host (Figure 8; Supplementary file 4d).
There were interaction effects between the amoebae hosts’ native infection status and the species of Burkholderia added for all traits: stalk height (F4,81 = 4.9, p = 0.0015), stalk volume (F4,81 = 7.4, P ≪ 0.001; Figure 8; Supplementary file 3e), sorus diameter (F4,719 = 5.3, p = 0.0004) and sorus volume (F4,719 = 4.5, p = 0.0014; Figure 8; Supplementary file 4e). However, these interactions were matters of degree of change and did not involve sign changes: for all four measurements, fruiting bodies with B. agricolaris were taller than those with B. hayleyella (Figure 8; Supplementary file 3e, Supplementary file 4e).
Discussion
Here, we characterized D. discoideum infection by two symbiotic Burkholderia species, B. hayleyella and B. agricolaris (Brock et al., 2018). We looked at their impact on D. discoideum by comparing wild type, cured, and re-infected hosts. We assessed fitness measured as production of viable spores, and also evaluated morphological changes in amoebae, slugs, and fruiting bodies with numerical and microscopic data. We found that both Burkholderia species are a burden to D. discoideum under our experimental conditions. However, wild collected hosts did not differ in viable spore production according to whether or not they carried either species of Burkholderia. Even so, D. discoideum with their field-collected state of infection did differ in fruiting body dimensions, with uninfected hosts generally having taller, larger stalks, and larger sori. What explains the differences from experimental infection is unclear. Infection in the wild may be at a lower level than we used experimentally or may have initiated at a lower level that slowly amplified over time, allowing host acclimation to the metabolic costs of infection.
Once D. discoideum hosts are cured with antibiotics, so all comparisons can start from the same baseline, we found that there were few within treatment differences according to host type. The only exception to this is that native-hayleyella hosts produced more viable spores than did naïve or native-agricolaris hosts. This fitness difference is an indication of co-adaptation.
When we map naïve, native-agricolaris, and native-hayleyella host types from Haselkorn et al. (2018) on a previously generated phylogeny, we see no phylogenetic differentiation whatsoever between host types (Douglas et al., 2011). It is most likely that diverse amoebae isolates get infected with Burkholderia in nature, and then adaptations that provide these hosts with increased resilience are selected for over time. Perhaps the general lack of difference among host types with infection is due to sex, which not only recombines genes, but also exposes new clones horizontally to endosymbionts like Burkholderia. Dictyostelium recombination rates are high in natural populations (Flowers et al., 2010). The differences reported here among clones of D. discoideum indicate that specific co-adaptation in clones infected with B. hayleyella can occur during the long periods between sexual reproduction. Thus, reproduction by binary fission and vertical transmission of B. hayleyella is likely to occur much more often than sexual reproduction which could result in horizontal transmission.
Our previous work demonstrated that Burkholderia infections have contextually dependent costs and benefits for their hosts. In food abundant conditions (which we used here) Burkholderia infections are generally detrimental to host fitness (Brock et al., 2011). However, when dispersed to food scarce conditions, Burkholderia infected hosts are able to transport food bacteria with them which restocks their food source and results in higher host fitness (‘farming’). These fitness outcomes may result from the ongoing dynamics underlying long-term symbiosis.
A compelling question is what mediates this tolerance to B. hayleyella infection in its native host? It is possible that native hosts better inhibit infection events or better control intracellular replication of symbiont cells. The percent of spore cells infected in the population post symbiont exposure are not significantly different between a native-hayleyella host and naïve hosts, which does not support the idea that infection events are inhibited by hosts conditioned to that species. Further, we did not see a significantly different intracellular density of B. hayleyella-RFP across hosts from confocal micrographs, suggesting that intact spores from different hosts can accommodate similar numbers of bacterial cells. However, our TEM analysis qualitatively points to the idea that after infection, intracellular replication rates may differ between naïve and native-hayleyella hosts. TEM images of native and naïve B. hayleyella hosts consistently suggest a higher load of intracellular bacterial cells at each stage. For instance, naïve B. hayleyella hosts produce paltry stalks that appear to be overwhelmed by bacterial cells and infected spore cells that look on the verge of deteriorating. Neither of these extreme states were observed in the native host. This could be mediated by host countermeasures that control intracellular symbiont growth or disarm potential toxic symbiont byproducts.
Compared to B. hayleyella, B. agricolaris infections result in more modest (and statistically insignificant) drops in D. discoideum viable spore production for naïve and cured re-infected native-agricolaris hosts. B. agricolaris appears to be less invasive to the host population as it is found in only about a quarter of spore population after exposure. Despite this, it is maintained in infected populations throughout the social stage and over multiple social cycles (DiSalvo et al., 2015). However, B. agricolaris establishes a higher intracellular density within infected spores than B. hayleyella. These observations may be explained by reduced intracellular entrance efficiency coupled with higher intracellular replication rates. Alternatively, B. agricolaris could be more efficiently cleared by host mechanisms and requires a higher multiplicity of infection to overcome clearance, something that might help explain its more modest detrimental effect on host populations.
Earlier studies have identified other differences between naïve D. discoideum and those carrying B. hayleyella (Brock et al., 2013; Brock et al., 2016b; Stallforth et al., 2013). D. discoideum hosts carrying B. hayleyella harmed symbiont-free D. discoideum clones, causing them to lose in social competition (Brock et al., 2013). In another study we compared cured and uncured clones of B. hayleyella and found that slugs from uncured clones move less far across a Petri plate, a cost of infection (Bates et al., 2015). Interestingly, in the current study we detected no visible differences between cured and uncured slug cells. The recent discovery of sentinel cells as innate immune cells (Chen et al., 2007) made us wonder about how they fare with Burkholderia infected hosts. We found that D. discoideum hosts with Burkholderia do not produce as many sentinel cells but even so seem as resistant to toxins as uninfected lines with normal levels of sentinel cells (Brock et al., 2013; Brock et al., 2016b; Stallforth et al., 2013).
Another interesting recent result on the interaction between D. discoideum and bacteria involves the role of the lectin discoidin 1 (Dinh et al., 2018). Clones infected with Burkholderia produced much greater quantities of lectins early in the social stage compared to uninfected clones. These lectins coated the food bacterium K. pneumoniae allowing it to avoid digestion. This is undoubtedly just a beginning, though a fascinating one, in our understanding of how Burkholderia take over D. discoideum cellular machinery to change relationships with bacteria.
D.D. discoideum is already a popular system for examining the molecular mechanisms of bacterial pathogenesis for a variety of important pathogens (Cosson and Soldati, 2008). However, this work demonstrates several properties of this interaction that are distinct from other Dictyostelium-bacterial associations, possibly owing to its natural prevalence. For instance, Bordetella bronchiseptica can intracellularly infect vegetative D. discoideum amoebae and persist in sorus contents. However, in contrast to Burkholderia, B. bronchiseptica is localized extracellularly in sori rather than inside spore cells (Taylor-Mulneix et al., 2017). Infections of D. discoideum with other intracellular pathogens such as Legionella pneumophila often produce secreted multilamellar bodies which harbor the bacterial pathogen (Denoncourt et al., 2014; Paquet and Charette, 2016), while in this study we found no evidence of Burkholderia excreted in multilamellar bodies. In addition, many bacteria resistant to amoebae are found packaged in multilamellar bodies, a process speculated to enhance their resistance to environmental stress (Denoncourt et al., 2014). Thus, Burkholderia symbionts are most likely employing alternative tactics to not only evade digestion, but to also evade expulsion.
Burkholderia is important in another model symbiosis system, that with the bean bug, Riptortus pedestris (Takeshita and Kikuchi, 2017). These bugs acquire Burkholderia insecticola horizontally. They reside in the bean bug gut where they are presumably active in nutrition. They are a good model because it is a facultative symbiosis and both partners can be cultured independently.
We are in a new age of symbiosis studies where we can apply Koch’s principles of curing, re-infecting, and looking for evidence of disease. We can use genomics, experimental evolution and many other methods to ever more systems. The D. discoideum-Burkholderia system holds unique potential for studying eukaryote-bacterial associations. Given its natural occurrence, we can perform long-term ecological surveys, easily isolate new host-symbiont pairs, investigate naturally derived vs newly induced associations using a variety of partner pairing, and we can do experiments in evolution allowing each partner to evolve together or separately. In time, B. hayleyella and B. agricolaris in D. discoideum may be added to the classic symbioses of squid-vibrio, aphid-Buchnera, tsetse fly-Wiggelsworthia, legume-Rhizobia and more (Bennett and Moran, 2015; Bing et al., 2017; Koehler et al., 2018; Werner et al., 2015).
Materials and methods
Key resources table.
| Reagent type (species) or resource |
Designation | Source or reference |
Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strainbackground(Dictyostelium discoideum) | QS6 | Douglas et al., 2011, Brock et al., 2011 | Virginia-MtLake BiologicalStation | |
| Strain, strainbackground (D.discoideum) | QS9 | Douglas et al., 2011, Brock et al., 2011 | Virginia-Mt Lake Biological Station | |
| Strain, strainbackground (D. discoideum) | QS17 | Douglas et al., 2011, Brock et al., 2011 | Virginia-MtLake Biological Station | |
| Strain, strainbackground (D. discoideum) | QS18 | Douglas et al., 2011, Brock et al., 2011 | Virginia-Mt Lake BiologicalStation | |
| Strain, strainbackground (D. discoideum) | QS11 | Douglas et al., 2011, Brock et al., 2011 | Virginia-MtLake BiologicalStation | |
| Strain, strainbackground (D. discoideum) | QS21 | Douglas et al., 2011, Brock et al., 2011 | Virginia-MtLake BiologicalStation | |
| Strain, strainbackground (D. discoideum) | QS22 | Douglas et al., 2011, Brock et al., 2011 | Virginia-MtLake BiologicalStation | |
| Strain, strainbackground (D. discoideum) | QS23 | Douglas et al., 2011, Brock et al., 2011 | Virginia-MtLake BiologicalStation | |
| Strain, strainbackground (D. discoideum) | QS70 | Douglas et al., 2011 | Texas- HoustonArboretum | |
| Strain, strainbackground (D. discoideum) | QS159 | Brock et al., 2011 | Virginia-MtLake BiologicalStation | |
| Strain, strainbackground (D. discoideum) | QS161 | Brock et al., 2011 | Virginia-MtLake BiologicalStation | |
| Strain, strainbackground (D. discoideum) | NC21 | Francis and Eisenberg, 1993 | NC-Little ButtsGap | |
| Strain, strainbackground (Burkholderia hayleyella) | BhQS11 | Haselkorn et al., 2018 | isolated fromQS11 | |
| Strain, strainbackground (B. hayleyella) | BhQS21 | Haselkorn et al., 2018 | isolated fromQS21 | |
| Strain, strainbackground (B. hayleyella) | BhQS22 | Haselkorn et al., 2018 | isolated from QS22 | |
| Strain, strainbackground (B. hayleyella) | BhQS23 | Haselkorn et al., 2018 | isolated fromQS23 | |
| Strain, strainbackground (Burkholderia agricolaris) | BaQS70 | Haselkorn et al., 2018 | isolated fromQS70 | |
| Strain, strainbackground (B. agricolaris) | BaQS159 | Haselkorn et al., 2018 | isolated fromQS159 | |
| Strain, strainbackground (B. agricolaris) | BaQS161 | Haselkorn et al., 2018 | isolated fromQS161 | |
| Strain, strain background (B. agricolaris) | BaNC21 | Haselkorn et al., 2018 | isolated fromNC21 | |
| Strain, strain background (B. agricolaris) | BaQS70-RFP.1 | DiSalvo et al., 2015 | modified fromBaQS70 | |
| Strain, strain background (B. hayleyella) | BhQS11-RFP.2 | This paper | modified fromBaQS11 | |
| Strain, strainbackground (Klebsiella pneumoniae) | KpQS | Dictybase (http://dictybase.org/) | ||
| Strain, strainbackground (K. pneumoniae) | KpQS-GFP.1 | This paper | ||
| Recombinant DNA reagent | pmini-Tn7-KS-GFP | Teal et al., 2006 | ||
| Recombinant DNA reagent | pmini-Tn7-gat-P1-RFP | Su et al., 2014 |
D. discoideum strains and culture conditions
We collected D. discoideum isolates from the field that were uninfected, or infected with either B. agricolaris, or B. hayleyella. Table 1 describes host clone sets, location collected, and infection status. We used host sets 1–4 for the spore fitness assays and set one for all other experiments. We used Klebsiella pneumoniae obtained from the Dicty Stock Center (http://dictybase.org/StockCenter/StockCenter.html) as our food bacterium for D. discoideum. We grew all D. discoideum from spores on SM/5 agar plates (2 g glucose, 2 g BactoPeptone (Oxoid), 2 g yeast extract (Oxoid), 0.2 g MgCl2, 1.9 g KH2PO4, 1 g K2HPO4 and 15.5 g agar per liter) supplemented with K. pneumoniae at room temperature (21°C).
Symbiotic bacterial strains
We used D. discoideum-associated Burkholderia previously isolated and sequenced to verify closest 16S identity (Brock et al., 2011; DiSalvo et al., 2015). B. agricolaris and B. hayleyella strains were isolated from QS70, QS159, QS161, and NC21, and QS11, QS23, QS22, and QS21 D. discoideum hosts respectively. According to multi-locus sequence typing all B. hayleyella strains belong to the same haplotype, B. agricolaris from QS70 and QS161 are the same haplotype and B. agricolaris from NC21 and QS159 are unique haplotypes (Haselkorn et al., 2018).
Removal of symbiont from native D. discoideum hosts
We generated symbiont-free native host clones by tetracycline, or by ampicillin-streptomycin, treatment as previously described (Brock et al., 2011; DiSalvo et al., 2015). We confirmed loss of infection status using the spot test assay and PCR analysis of Burkholderia, and K. pneumoniae in D. discoideum sori as previously described.
Lab infections
We collected stationary phase bacteria in starvation buffer (2.2 g KH2PO4 monobasic and 0.7 g K2HPO4 dibasic/liter) from bacteria grown on SM/5 plates. We determined the bacterial absorbance (OD600) using a BioPhotometer (Eppendorf, NY) and set all suspensions to optical density (OD600 1.5). For experiments using lab-infected lines, we mixed the specified Burkholderia species at 5% and K. pneumoniae at 95% vol and plated D. discoideum spores (as indicated) with 200 µl of the bacterial mixture on SM/5 plates.
Spot test assay
We verified infection status by spot test assay as previously described (Brock et al., 2011). Briefly, we transferred sorus contents from individual D. discoideum fruiting bodies to SM/5 agar plates using a 10 μl filter pipet tip. We incubated at 21°C for one week and checked for bacterial growth as an indication of infection.
Fitness assay
We analyzed spore production and viability as a proxy for amoeba fitness using four sets of D. discoideum clones (Table 1). We tested three conditions: uninfected (naïve, cured naïve, cured native-agricolaris, and cured native-hayleyella), B. agricolaris infected (native-agricolaris, and naïve, native-agricolaris, and native-hayleyella first cured then infected with B. agricolaris) and B. hayleyella infected (native-hayleyella, and naïve, native-agricolaris, and native-hayleyella first cured then infected with B. hayleyella) across three temporal replicates.
To set up each assay, we plated 2 × 105 spores of each clone in each condition (with lab infected lines being plated on Burkholderia-Klebsiella mixtures as described) onto SM/5 agar plates in duplicate. All clones formed fruiting bodies by 3 days, so we performed data collection five days after fruiting. We used the first plate to ascertain total spore production as previously described (Brock et al., 2011). Briefly, spores were collected by washing plates with starvation buffer supplemented with 0.01% NP-40 alternative (Calbiochem). We counted spore dilutions on a hemocytometer using a light microscope and determined total spores according to total volume collected and dilution factor. To determine the proportion of viable spores we collected spores into starvation buffer only and determined spore density as above. We diluted suspensions to 104 and spread 100 spores over ten 100 × 15 mm2 SM/5 agar plates supplemented with 200 μl K. pneumoniae in starvation buffer (OD600 1.5). After 2 days the percentage of viable spores was determined by counting plaques formed on bacterial lawns.
Transmission electron microscopy
We prepared amoebae by plating 2 × 105 spores (with Klebsiella for uninfected or native-infected and for the indicated Burkholderia mixture for lab-infected). We harvested log-phase vegetative cells approximately 36 hr after plating and fruiting bodies 4 days after plating. To prepare migrating slugs, we mixed 200 µL of centrifuge-concentrated K. pneumoniae (absorbance, OD600 75) with 5 × 106 spores and plated the mixture in a straight line across a starving agar plate, which was then wrapped in aluminum foil with a small hole opposite the spore line. We incubated plates under a direct light and allowed slugs to migrate for about 80 hr before processing. We processed all stages by first adding fix solution (2% paraformaldehyde/2.5% glutaraldehyde (Polysciences Inc., Warrington, PA) in 100 mM cacodylate buffer, pH 7.2), followed by low melting agarose, over the plates to keep structures intact.
We fixed samples for 1–3 hr at room temperature then washed with cacodylate buffer and postfixed in 1% osmium tetroxide (Polysciences Inc.) for 1 hr. We then rinsed samples extensively in dH2O prior to en bloc staining with 1% aqueous uranyl acetate (Ted Pella Inc., Redding, CA) for 1 hr. Following several rinses in dH2O, we dehydrated samples in a graded series of ethanol and embedded them in Eponate 12 resin (Ted Pella Inc.). Sections of 95 nm were cut with a Leica Ultracut UCT ultramicrotome (Leica Microsystems Inc., Bannockburn, IL), stained with uranyl acetate and lead citrate, and viewed on a JEOL 1200 EX transmission electron microscope (JEOL USA Inc., Peabody, MA) equipped with an AMT eight megapixel digital camera (Advanced Microscopy Techniques, Woburn, MA).
Confocal microscopy
We constructed RFP labeled versions of B. agricolaris (from QS70) and B. hayleyella (from QS11) by performing triparental mating procedures with the E. coli helper strain E1354 (pTNS3-asdEc) and the E coli donor strain E2072 with pmini-Tn7-gat-P1-RFP and confirmed glyphosate resistant RFP positive Burkholderia conjugants using Burkholderia specific PCR as previously described (DiSalvo et al., 2015; Norris et al., 2009; Su et al., 2014). We constructed a GFP labeled version of K. pneumoniae using a triparental mating strategy with the donor E. coli donor strain WM3064 containing pmini-Tn7-KS-GFP and the E. coli helper strain E1354 helper pUXBF13 as previously described (Kikuchi and Fukatsu, 2014; Teal et al., 2006). We confirmed kanamycin resistant GFP positive recipient cells by 16S rRNA gene sequencing.
Using Burkholderia-RFP and Klebsiella-GFP we infected set one hosts (using cured native hosts) as previously described. Control samples were plated with K. pneumoniae-GFP only. We harvested log-phase amoebae approximately 36 hr after plating by flooding plates with 5 ml SorMC and washing 3x in PBS to remove residual bacteria. We set amoebae to 1 × 106 cells/ml and placed 200 μl onto #1.5 glass coverslips for 15 min to allow them to adhere before fixing in 4% formaldehyde for 10 min. We then washed with PBS, permeabilized with 0.5% triton-X, and stained with Alexa Fluor 680 phalloidin (lifetechnologies) for 30 min before mounting in Prolong Diamond antifade mountant (lifetechnologies). We prepared spores four days after plating by collecting sori into starving buffer with 1% calcofluor white and spreading the solution on a glass bottom culture dish under a 2% agarose overlay.
We collected images using a Nikon A1Si Laser Scanning confocal microscope with a CFI Plan Apo VC Oil 1.4 NA 100X objective and Nikon Elements software or an Olympus Fluoview FV1000 confocal microscope using Plan Apo Oil 1.4NA 60X objective and Olympus software. Z-sections were taken every 0.5 microns with an average of 2 at 1024 × 1024 resolution pixels or 600 × 600 pixels. We excited RFP using the 561 laser, GFP with the 488 laser, Calcoflour-white with the 408 laser, and Alexa Fluor 680 Phalloidin with the 640 laser. We created composite images in FIJI.
Infectivity quantification
We quantified bacterial density within individual spores by counting the number of visible Burkholderia-RFP cells within infected spores from confocal micrographs. We counted 15 infected spores for each condition from two individual replicates (30 spores total). We quantified the population of Burkholderia-RFP infected spores for the set one clones using the BD accuri C6 flow cytometer. We plated spores in duplicate as described in the confocal microscopy section. Four days after plating, we resuspended three sori from each plate into 500 µl of starving buffer with 0.01% NP-40 alternative. We ran 100 µl of each vortexed sample through the flow cytometer. We used non-fluorescent controls to establish an accurate gating between fluorescent and non-fluorescent boundaries. We measured and averaged duplicates for a total of 6 temporal replicates.
Morphometrics
We quantified fruiting body size and shape for each clone in each condition using set one clones. We plated the clones as described under fitness assays. Five days after fruiting, we carefully cut and removed a thin strip of agar approximately 5 mm wide from the central area of an experimental plate and laid it on its side in a Petri plate. We placed dampened Kimwipes around the agar slice to prevent desiccation. We used a Leica EC3 scope with the LASD core package LAS V4.1) to collect data. Fruiting body images were taken randomly along with graticule images for calibration. We took six measurements of each fruiting body: sorus width, sorus length, stalk height and the width of the stalk at its base, midpoint and at the top just below the sorus (Buttery et al., 2009). Stalk height was measured from the base of stalk to the tip of the sorus. We calculated sorus volume applying the formula for the volume of a sphere using diameter, V = 1/6 π d3. We calculated stalk volume using the formula of a cylinder, V= π r2 h, where height (h) is the stalk height and radius (r) is half the mean of the three stalk width measurements. We measured about 80 sori and 20 stalks for each clone for each condition.
Statistical analyses
All analyses were done in R. For fitness assays, we tested the effect of antibiotic treatment using random-slope linear mixed models (LMM) on those D. discoideum hosts not reinfected with Burkholderia. Our models included either spore numbers or proportion of viability as the response variable, host as random effect, and antibiotic treatment as a fixed effect. We similarly tested the effects of Burkholderia infection in the field and in the lab using random-slope LMMs on data from hosts cured with antibiotics. Our models included spore numbers or proportion viability as response variable, host as random effect, lab or field-infection status and Burkholderia type as fixed effects, as well as an interaction between field-infection status and Burkholderia infection. For all LMMs, we fitted models and assessed model fit with likelihood ratio tests executed with the lme4 package (Bates et al., 2015) in the R environment (v. 3.3.3, R Core Team 2017). We tested the significance of fixed effects with Wald tests using the t distribution, which we executed with the packager lmerTest (Kuznetsova et al., 2017). These tests use (Satterthwaite, 1946) approximation for denominator degrees of freedom to calculate p-values. Finally, for all post hoc multiple comparisons, we performed pairwise contrasts of least-square means with a multivariate t distribution adjustment as implemented with the package lsmeans (Lenth, 2016).
For morphometric analyses, we also tested the effect of antibiotic treatment on those hosts not re-infected with Burkholderia. We used a 2-way analysis of variance (ANOVA) with one of our four morphological measurements as the response variable and both Burkholderia colonization and antibiotic treatment as fixed effects. Similarly, we tested the effects of Burkholderia colonization from the field and Burkholderia infection in the lab on the amoeba hosts with 2-way ANOVAs on amoebae cured with antibiotics. Again, one of the four morphological measurements was the response variable with field-colonization status, Burkholderia infection, and an interaction between them as fixed effects. For all ANOVAs, when appropriate, we performed post hoc Tukey HSD tests for multiple comparisons. Sorus width data were square-root transformed as and sorus volume data were loge-transformed to meet test assumptions of normally distributed residuals.
We analyzed Burkholderia-RFP infectivity (population prevalence and intracellular density) with 2-way ANOVAs, followed by a Tukey HSD for multiple comparisons. We treated Burkholderia species and D. discoideum identity as fixed effects. All source data is available at https://doi.org/10.7936/wgnk-2c37.
Acknowledgements
Many thanks to the Strassmann/Queller laboratory group for useful discussion and Mountain Lake Biological Station where we collected the samples. Thanks to Kyle Skottke, Hassan Salem, and Tyler Larsen for helpful comments on the manuscript. We also thank the reviewers of this manuscript for their thorough analysis and insightful suggestions. All authors read and approved the final manuscript.
This material is based upon work supported by the National Science Foundation under grant nos. NSF DEB1146375, NSF IOS 1256416, NSF IOS 1656756, the Life Sciences Research Foundation and Simons Foundation (to LS), and the John Templeton Foundation grant no. 43667.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Contributor Information
Joan E Strassmann, Email: strassmann@wustl.edu.
Susanne DiSalvo, Email: sdisalv@siue.edu.
Tony Ruzzini, University of Saskatchewan, Canada.
Ian T Baldwin, Max Planck Institute for Chemical Ecology, Germany.
Funding Information
This paper was supported by the following grants:
National Science Foundation DEB1146375 to David C Queller, Joan E Strassmann.
Life Sciences Research Foundation to Longfei Shu.
John Templeton Foundation 43667 to David C Queller, Joan E Strassmann.
National Science Foundation IOS1256416 to David C Queller.
National Science Foundation IOS1656756 to David C Queller.
Simons Foundation to Longfei Shu.
Additional information
Competing interests
No competing interests declared.
Author contributions
Conceptualization, Investigation, Methodology.
Conceptualization, Validation, Investigation, Methodology.
Data curation, Formal analysis, Validation.
Investigation.
Conceptualization, Formal analysis, Supervision, Funding acquisition, Project administration, Writing—review and editing.
Conceptualization, Resources, Data curation, Supervision, Funding acquisition, Methodology, Project administration, Writing—review and editing.
Conceptualization, Data curation, Investigation, Visualization, Methodology, Writing—original draft, Writing—review and editing.
Additional files
The three fitness measures were percent of spores that were viable, the total number of spores produced by a clone, and total viable spores. Total viable spores is the product of the other two measures. For each pairwise contrast, the essential difference in treatments is in boldface, and a treatment that is significantly higher than the other is marked with an asterisk and printed in red. Each of the fitness measures was analyzed with a set of Generalized Linear Mixed Models (GLMMs). This table gives the p-values for each question asked about main or interaction effects and the post hoc pairwise comparisons made, as relevant. Details about the statistical tests used can be found in the main text.
he three fitness measures were again percent of spores that were viable, the total number of spores produced by a clone, and total viable spores. Total viable spores is the product of the other two measures. For each pairwise contrast, the essential difference in treatments is in boldface, and a treatment that is significantly higher than the other is marked with an asterisk and printed in red. Each of the fitness measures was analyzed with a set of Generalized Linear Mixed Models (GLMMs). This table gives the p-values for each question asked about main or interaction effects and the post hoc pairwise comparisons made, as relevant. Details about the statistical tests used can be found in the main text.
Each of the stalk measures was analyzed with a set of Generalized Linear Mixed Models (GLMMs). This table gives the p-values for each question asked about main or interaction effects and the post hoc pairwise comparisons made, as relevant. For each pairwise contrast, the essential difference in treatments is in boldface, and a treatment that is significantly higher than the other is marked with an asterisk and printed in red. Details about the statistical tests used can be found in the main text. One clone of each native type was tested: QS9 naïve; QS70 ag-infected; QS11 ha-infected.
Each of the spore measures was analyzed with a set of Generalized Linear Mixed Models (GLMMs). This table gives the p-values for each question asked about main or interaction effects and the post hoc pairwise comparisons made, as relevant. For each pairwise contrast, the essential difference in treatments is in boldface, and a treatment that is significantly higher than the other is marked with an asterisk and printed in red. Details about the statistical tests used can be found in the main text. One clone of each native type was tested: QS9 naïve; QS70 ag-infected; QS11 ha-infected.
Data availability
All raw data has been archived in the Washington University Library: https://doi.org/10.7936/wgnk-2c37
The following dataset was generated:
Strassmann J, DiSalvo S, Queller DC, Brock DA. 2018. Raw data from Symbiont location, host fitness, and possible coadaptation in a symbiosis between social amoebae and bacteria. Washington University Open Scholarship. 10.7936/wgnk-2c37
References
- Bates D, Mächler M, Bolker B, Walker S. Fitting linear mixed-effects models using lme4. Journal of Statistical Software. 2015;67 [Google Scholar]
- Bennett GM, Moran NA. Heritable symbiosis: the advantages and perils of an evolutionary rabbit hole. PNAS. 2015;112:10169–10176. doi: 10.1073/pnas.1421388112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bing X, Attardo GM, Vigneron A, Aksoy E, Scolari F, Malacrida A, Weiss BL, Aksoy S. Unravelling the relationship between the tsetse fly and its obligate symbiont Wigglesworthia : transcriptomic and metabolomic landscapes reveal highly integrated physiological networks. Proceedings of the Royal Society B: Biological Sciences. 2017;284:20170360. doi: 10.1098/rspb.2017.0360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloomfield G, Skelton J, Ivens A, Tanaka Y, Kay RR. Sex determination in the social amoeba Dictyostelium discoideum. Science. 2010;330:1533–1536. doi: 10.1126/science.1197423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock DA, Douglas TE, Queller DC, Strassmann JE. Primitive agriculture in a social amoeba. Nature. 2011;469:393–396. doi: 10.1038/nature09668. [DOI] [PubMed] [Google Scholar]
- Brock DA, Read S, Bozhchenko A, Queller DC, Strassmann JE. Social amoeba farmers carry defensive symbionts to protect and privatize their crops. Nature Communications. 2013;4:2385. doi: 10.1038/ncomms3385. [DOI] [PubMed] [Google Scholar]
- Brock DA, Jones K, Queller DC, Strassmann JE. Which phenotypic traits of Dictyostelium discoideum farmers are conferred by their bacterial symbionts? Symbiosis. 2016a;68:39–48. doi: 10.1007/s13199-015-0352-0. [DOI] [Google Scholar]
- Brock DA, Callison W. Éamon, Strassmann JE, Queller DC. Sentinel cells, symbiotic bacteria and toxin resistance in the social amoeba Dictyostelium discoideum. Proceedings of the Royal Society B: Biological Sciences. 2016b;283:20152727. doi: 10.1098/rspb.2015.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock DA, Hubert AM, Noh S, DiSalvo S, Geist KS, Haselkorn TS, Queller D, Strassmann JE. Endosymbiotic adaptations in three new bacterial species associated with Dictyostelium discoideum: Burkholderia agricolaris sp. nov., Burkholderia hayleyella sp. nov., and Burkholderia bonniea sp. nov. BioRxiv. 2018 doi: 10.1101/304352. [DOI] [PMC free article] [PubMed]
- Bronstein JL. Mutualism. Oxford University Press; 2015. [DOI] [Google Scholar]
- Buttery NJ, Rozen DE, Wolf JB, Thompson CR. Quantification of social behavior in D. discoideum reveals complex fixed and facultative strategies. Current Biology. 2009;19:1373–1377. doi: 10.1016/j.cub.2009.06.058. [DOI] [PubMed] [Google Scholar]
- Casadevall A. Evolution of intracellular pathogens. Annual Review of Microbiology. 2008;62:19–33. doi: 10.1146/annurev.micro.61.080706.093305. [DOI] [PubMed] [Google Scholar]
- Chen G, Zhuchenko O, Kuspa A. Immune-like phagocyte activity in the social amoeba. Science. 2007;317:678–681. doi: 10.1126/science.1143991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosson P, Lima WC. Intracellular killing of bacteria: is Dictyostelium a model macrophage or an alien? Cellular Microbiology. 2014;16:816–823. doi: 10.1111/cmi.12291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosson P, Soldati T. Eat, kill or die: when amoeba meets bacteria. Current Opinion in Microbiology. 2008;11:271–276. doi: 10.1016/j.mib.2008.05.005. [DOI] [PubMed] [Google Scholar]
- Dale C, Moran NA. Molecular interactions between bacterial symbionts and their hosts. Cell. 2006;126:453–465. doi: 10.1016/j.cell.2006.07.014. [DOI] [PubMed] [Google Scholar]
- Denoncourt AM, Paquet VE, Charette SJ. Potential role of bacteria packaging by protozoa in the persistence and transmission of pathogenic bacteria. Frontiers in Microbiology. 2014;5:240. doi: 10.3389/fmicb.2014.00240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denoncourt AM, Paquet VE, Sedighi A, Charette SJ. Identification of proteins associated with multilamellar bodies produced by Dictyostelium discoideum. PLOS ONE. 2016;11:e0158270. doi: 10.1371/journal.pone.0158270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinh C, Farinholt T, Hirose S, Zhuchenko O, Kuspa A. Lectins modulate the microbiota of social amoebae. Science. 2018;361:402–406. doi: 10.1126/science.aat2058. [DOI] [PubMed] [Google Scholar]
- DiSalvo S, Haselkorn TS, Bashir U, Jimenez D, Brock DA, Queller DC, Strassmann JE. Burkholderia bacteria infectiously induce the proto-farming symbiosis of Dictyostelium amoebae and food bacteria. PNAS. 2015;112:E5029–E5037. doi: 10.1073/pnas.1511878112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas AE. Conflict, cheats and the persistence of symbioses. New Phytologist. 2008;177:849–858. doi: 10.1111/j.1469-8137.2007.02326.x. [DOI] [PubMed] [Google Scholar]
- Douglas AE. The microbial dimension in insect nutritional ecology. Functional Ecology. 2009;23:38–47. doi: 10.1111/j.1365-2435.2008.01442.x. [DOI] [Google Scholar]
- Douglas TE, Kronforst MR, Queller DC, Strassmann JE. Genetic diversity in the social amoeba Dictyostelium discoideum: population differentiation and cryptic species. Molecular Phylogenetics and Evolution. 2011;60:455–462. doi: 10.1016/j.ympev.2011.05.007. [DOI] [PubMed] [Google Scholar]
- Estrela S, Kerr B, Morris JJ. Transitions in individuality through symbiosis. Current Opinion in Microbiology. 2016;31:191–198. doi: 10.1016/j.mib.2016.04.007. [DOI] [PubMed] [Google Scholar]
- Ferdy JB, Godelle B. Diversification of transmission modes and the evolution of mutualism. The American Naturalist. 2005;166:613–627. doi: 10.1086/491799. [DOI] [PubMed] [Google Scholar]
- Flowers JM, Li SI, Stathos A, Saxer G, Ostrowski EA, Queller DC, Strassmann JE, Purugganan MD. Variation, sex, and social cooperation: molecular population genetics of the social amoeba Dictyostelium discoideum. PLS Genetics. 2010;6:e1001013. doi: 10.1371/journal.pgen.1001013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis D, Eisenberg R. Genetic structure of a natural population of Dictyostelium discoideum, a cellular slime mould. Molecular Ecology. 1993;2:385–391. doi: 10.1111/j.1365-294X.1993.tb00031.x. [DOI] [PubMed] [Google Scholar]
- Garcia JR, Gerardo NM. The symbiont side of symbiosis: do microbes really benefit? Frontiers in Microbiology. 2014;5:e510. doi: 10.3389/fmicb.2014.00510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haselkorn TS, DiSalvo S, Miller JW, Bashir U, Brock DA, Queller DC, Strassmann JE. The specificity of Burkholderia symbionts in the social amoeba farming symbiosis: prevalence, species, genetic and phenotypic diversity. Molecular Ecology. 2018 doi: 10.1111/mec.14982. [DOI] [PubMed] [Google Scholar]
- Hosokawa T, Ishii Y, Nikoh N, Fujie M, Satoh N, Fukatsu T. Obligate bacterial mutualists evolving from environmental bacteria in natural insect populations. Nature Microbiology. 2016;1:15011. doi: 10.1038/nmicrobiol.2015.11. [DOI] [PubMed] [Google Scholar]
- Kessin RH. Dictyostelium: Evolution, Cell Biology, and the Development of Multicellularity. Cambridge, UK: Cambridge University Press; 2001. [DOI] [Google Scholar]
- Kikuchi Y, Fukatsu T. Live imaging of symbiosis: spatiotemporal infection dynamics of a GFP-labelled Burkholderia symbiont in the bean bug Riptortus pedestris. Molecular Ecology. 2014;23:1445–1456. doi: 10.1111/mec.12479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler S, Gaedeke R, Thompson C, Bongrand C, Visick KL, Ruby E, McFall-Ngai M. The model squid-vibrio symbiosis provides a window into the impact of strain- and species-level differences during the initial stages of symbiont engagement. Environmental Microbiology. 2018 doi: 10.1111/1462-2920.14392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsova A, Brockhoff PB, Christensen RHB. lmerTest Package: Tests in Linear Mixed Effects Models. Journal of Statistical Software. 2017;82 doi: 10.18637/jss.v082.i13. [DOI] [Google Scholar]
- Lenth RV. Least-Squares Means: The R Package lsmeans. Journal of Statistical Software. 2016;69 doi: 10.18637/jss.v069.i01. [DOI] [Google Scholar]
- Leung TLF, Poulin R. Parasitism, commensalism, and mutualism: exploring the many shades of symbiosis. Life Environ. 2008;2:107–115. [Google Scholar]
- Lowe CD, Minter EJ, Cameron DD, Brockhurst MA. Shining a Light on Exploitative Host Control in a Photosynthetic Endosymbiosis. Current Biology. 2016;26:207–211. doi: 10.1016/j.cub.2015.11.052. [DOI] [PubMed] [Google Scholar]
- McCutcheon JP, Moran NA. Extreme genome reduction in symbiotic bacteria. Nature Reviews Microbiology. 2011;10:13–26. doi: 10.1038/nrmicro2670. [DOI] [PubMed] [Google Scholar]
- McFall-Ngai M, Hadfield MG, Bosch TC, Carey HV, Domazet-Lošo T, Douglas AE, Dubilier N, Eberl G, Fukami T, Gilbert SF, Hentschel U, King N, Kjelleberg S, Knoll AH, Kremer N, Mazmanian SK, Metcalf JL, Nealson K, Pierce NE, Rawls JF, Reid A, Ruby EG, Rumpho M, Sanders JG, Tautz D, Wernegreen JJ. Animals in a bacterial world, a new imperative for the life sciences. PNAS. 2013;110:3229–3236. doi: 10.1073/pnas.1218525110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NA, Munson MA, Baumann P, Ishikawa H. A molecular clock in endosymbiotic bacteria is calibrated using the insect hosts. Proc. R. Soc. B Biol. Sci. 1993;253:167–171. doi: 10.1098/rspb.1993.0098. [DOI] [Google Scholar]
- Moran NA. Symbiosis as an adaptive process and source of phenotypic complexity. PNAS. 2007;104:8627–8633. doi: 10.1073/pnas.0611659104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris MH, Kang Y, Lu D, Wilcox BA, Hoang TT. Glyphosate resistance as a novel select-agent-compliant, non-antibiotic-selectable marker in chromosomal mutagenesis of the essential genes asd and dapB of Burkholderia pseudomallei. Applied and Environmental Microbiology. 2009;75:6062–6075. doi: 10.1128/AEM.00820-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyholm SV, McFall-Ngai MJ. The winnowing: establishing the squid-vibrio symbiosis. Nature Reviews Microbiology. 2004;2:632–642. doi: 10.1038/nrmicro957. [DOI] [PubMed] [Google Scholar]
- Oliver KM, Russell JA, Moran NA, Hunter MS. Facultative bacterial symbionts in aphids confer resistance to parasitic wasps. PNAS. 2003;100:1803–1807. doi: 10.1073/pnas.0335320100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver KM, Moran NA, Hunter MS. Variation in resistance to parasitism in aphids is due to symbionts not host genotype. PNAS. 2005;102:12795–12800. doi: 10.1073/pnas.0506131102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver KM, Moran NA, Hunter MS. Costs and benefits of a superinfection of facultative symbionts in aphids. Proceedings of the Royal Society B: Biological Sciences. 2006;273:1273–1280. doi: 10.1098/rspb.2005.3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquet VE, Lessire R, Domergue F, Fouillen L, Filion G, Sedighi A, Charette SJ. Lipid composition of multilamellar bodies secreted by Dictyostelium discoideum reveals their amoebal origin. Eukaryotic Cell. 2013;12:1326–1334. doi: 10.1128/EC.00107-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquet VE, Charette SJ. Amoeba-resisting bacteria found in multilamellar bodies secreted by Dictyostelium discoideum: social amoebae can also package bacteria. FEMS Microbiology Ecology. 2016;92:fiw025. doi: 10.1093/femsec/fiw025. [DOI] [PubMed] [Google Scholar]
- Pérez-Brocal V, Latorre A, Moya A. Symbionts and Pathogens: What isthe Difference? In: Dobrindt U, Hacker J. H, Svanborg C, editors. Between Pathogenicity and Commensalism. Berlin, Heidelberg: Springer Berlin Heidelberg; 2011. pp. 215–243. [DOI] [Google Scholar]
- Queller DC, Strassmann JE. Evolutionary Conflict. Annual Review of Ecology, Evolution, and Systematics. 2018;49:73–93. doi: 10.1146/annurev-ecolsys-110617-062527. [DOI] [Google Scholar]
- Ratzka C, Gross R, Feldhaar H. Endosymbiont Tolerance and Control within Insect Hosts. Insects. 2012;3:553–572. doi: 10.3390/insects3020553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds S, Rolff J. Immune function keeps endosymbionts under control. Journal of Biology. 2008;7:28. doi: 10.1186/jbiol88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs JL, Wilcox TP. A shift to parasitism in the jellyfish symbiont Symbiodinium microadriaticum. Proceedings of the Royal Society B: Biological Sciences. 2006;273:425–429. doi: 10.1098/rspb.2005.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterthwaite FE. An approximate distribution of estimates of variance components. Biometrics Bulletin. 1946;2:110–114. doi: 10.2307/3002019. [DOI] [PubMed] [Google Scholar]
- Smith J, Queller DC, Strassmann JE. Fruiting bodies of the social amoeba Dictyostelium discoideum increase spore transport by Drosophila. BMC Evolutionary Biology. 2014;14:105. doi: 10.1186/1471-2148-14-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto MJ, Domínguez-Ferreras A, Pérez-Mendoza D, Sanjuán J, Olivares J. Mutualism versus pathogenesis: the give-and-take in plant-bacteria interactions. Cellular Microbiology. 2009;11:381–388. doi: 10.1111/j.1462-5822.2009.01282.x. [DOI] [PubMed] [Google Scholar]
- Stallforth P, Brock DA, Cantley AM, Tian X, Queller DC, Strassmann JE, Clardy J. A bacterial symbiont is converted from an inedible producer of beneficial molecules into food by a single mutation in the gacA gene. PNAS. 2013;110:14528–14533. doi: 10.1073/pnas.1308199110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su S, Bangar H, Saldanha R, Pemberton A, Aronow B, Dean GE, Lamkin TJ, Hassett DJ. Construction and characterization of stable, constitutively expressed, chromosomal green and red fluorescent transcriptional fusions in the select agents, Bacillus anthracis, Yersinia pestis, Burkholderia mallei, and Burkholderia pseudomallei. MicrobiologyOpen. 2014;3:610–629. doi: 10.1002/mbo3.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshita K, Kikuchi Y. Riptortus pedestris and Burkholderia symbiont: an ideal model system for insect-microbe symbiotic associations. Research in Microbiology. 2017;168:175–187. doi: 10.1016/j.resmic.2016.11.005. [DOI] [PubMed] [Google Scholar]
- Taylor-Mulneix DL, Bendor L, Linz B, Rivera I, Ryman VE, Dewan KK, Wagner SM, Wilson EF, Hilburger LJ, Cuff LE, West CM, Harvill ET. Bordetella bronchiseptica exploits the complex life cycle of Dictyostelium discoideum as an amplifying transmission vector. PLOS Biology. 2017;15:e2000420. doi: 10.1371/journal.pbio.2000420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teal TK, Lies DP, Wold BJ, Newman DK. Spatiometabolic stratification of Shewanella oneidensis biofilms. Applied and Environmental Microbiology. 2006;72:7324–7330. doi: 10.1128/AEM.01163-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernegreen JJ. Ancient bacterial endosymbionts of insects: Genomes as sources of insight and springboards for inquiry. Experimental Cell Research. 2017;358:427–432. doi: 10.1016/j.yexcr.2017.04.028. [DOI] [PubMed] [Google Scholar]
- Werner GD, Cornwell WK, Cornelissen JH, Kiers ET. Evolutionary signals of symbiotic persistence in the legume-rhizobia mutualism. PNAS. 2015;112:10262–10269. doi: 10.1073/pnas.1424030112. [DOI] [PMC free article] [PubMed] [Google Scholar]