Abstract
This unit describes a protocol for Fluorescence Assisted Cell Sorting (FACS) of virus infected cerebral organoid cultures. The described method eliminates the need to grow individual cell populations and instead offers the possibility to isolate and study separate cell populations that where grown in circumstances that much closer resemble in vivo conditions. The protocol starts from stem cell derived mature brain organoids and includes steps on: preparing the culture for viral infection, production of the viral stocks, FACS sample preparation, and gating and sorting implementation. The protocol has been developed for ZIKA virus infection but can be extrapolated to other viruses or fluorescent marker expression as illustrated in an alternative protocol using a single cycle lentivirus expressing a fluorescent reporter protein.
Keywords: Cerebral organoids, FACS, virus, ZIKA, lentiviral vectors, HIV, human ESC
INTRODUCTION
In vitro generated, stem cell derived cerebral organoids closely mimic the characteristic of human fetal neurodevelopment in vivo while providing unlimited brain tissue comprising specific cell populations such as neuronal progenitors, neurons and astrocytes. This three- dimensional tissue allows for directly testing brain development and modeling complex human neurological disorders such as hereditary microcephaly, autism spectrum disorders and Alzheimer’s disease (Choi et al., 2014; Lancaster et al., 2013; Mariani et al., 2015). Brain organoids have also proven useful to dissect the general impact of viral infections on brain development. For example, Zika virus infection of cerebral organoids confirmed the neurotropic nature of the virus and provided rapid confirmation of the causal link between prenatal Zika virus infection and congenital microcephaly (Garcez et al., 2016).
However, with the creation of more complex, multilayered tissue models comes the challenge of developing assays that can translate complex experimental starting conditions into useful data outputs. Possibilities to reduce this complexity include the use of organoids confined to specific brain regions (Qian et al., 2016) or microdissection of certain cerebral organoid anatomy combined with single cell RNA sequencing (Camp et al., 2015). Nevertheless, a satisfying approach to study the effects of virus infection or other treatments in distinct cell populations of organoids has been lacking. We describe a protocol to isolate neural progenitors, astrocytes and neurons from virus infected cerebral organoids, validated for downstream epigenetic and gene expression assays (Janssens et al., 2018). This unit in detail outlines how to prepare Zika and single cycle lentiviral reporter viruses, prepare cerebral organoid cultures for viral infection, identify neuronal populations and subsequently isolate infected and non-infected neural progenitor cells, neurons and astrocytes suitable for downstream manipulations such as RNA and DNA extraction.
CAUTION: Manipulations of virus containing cultures have to be carried out in biological safety cabinets with appropriate PPE and infectious waste has to be collected and disposed according to the correct biosafety level (BSL) guidelines.
BASIC PROTOCOL 1. GENERATION OF 2D CEREBRAL ORGANOID CULTURES FOR OPTIMAL VIRAL INFECTION
This protocol describes the steps for the generation of a 2D culture that is optimal for ample viral infection while maintaining the original cell types and cell interactions typical of cerebral organoids (Fig. 1). The successful isolation of virus-infected cells from cerebral organoids strongly correlates with the initial infection rate. Fluorescence activated cell sorting requires for a big enough portion of cells to be separated from background noise, and downstream applications are limited by minimal material input. While developing this protocol, low infection rates were experienced when infecting mature cerebral organoids as a whole. Vibratome slicing and culturing the cerebral organoid as 500 μm thick slices did elevate infection rates to some extent by increasing surface exposure to the virus (Fig. 1C). Nevertheless, switching to two-dimensional culture conditions deemed necessary to reach satisfactory infection rates.
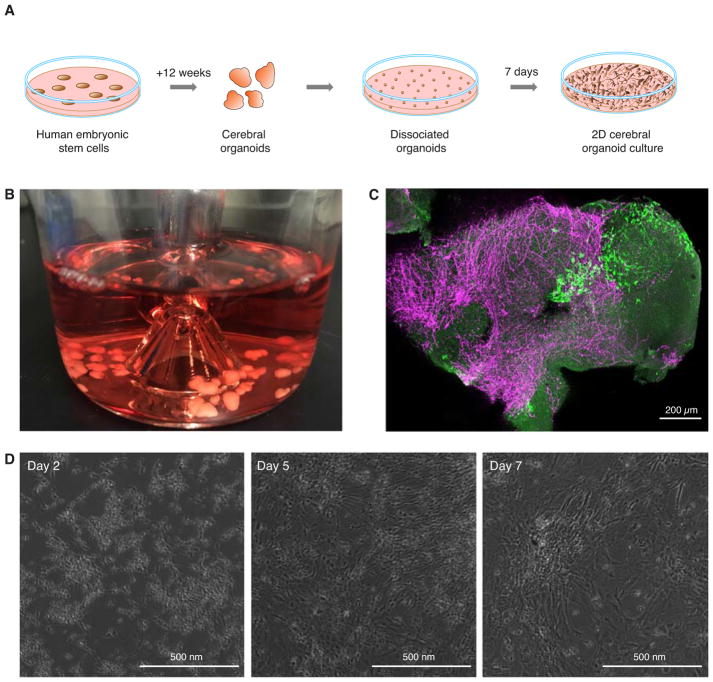
Figure 1.
Generation of 2D cerebral organoid cultures. (A) Schematic overview of the steps and duration to create 2D cerebral organoid cultures suitable for viral infection. (B) Example of 3-month old cerebral organoids (average size, 5 mm). (C) Confocal immunofluorescence maximum image projection of a 500 μm thick ZIKV infected cerebral organoid slide showing ZIKV-positive cells (E-antigen, green) and astrocytes (GFAP, purple). (D) Phase contrast images of dissociated cerebral organoids at different time points after dissociation.
Materials
-
hESC or hiPSC derived cerebral organoids.
NOTE: This protocol unit has been optimized for cerebral organoids generated according to Lancaster et al. (Lancaster et al., 2013). Growth factor reduced Matrigel (Corning, cat. no. 356230)
DMEM/F12 (Gibco, cat. no. 11330-032)
Cerebral organoid medium (see recipe)
Accutase (Innovative Cell Technologies, cat. no. AT104)
50 ml polypropylene centrifuge tubes
15 ml polypropylene centrifuge tubes
6-well flat bottom cell culture plates
25 ml serological pipets
Sterile low binding 1000 μl filter tips
Prepare Matrigel coated 6-well cell culture plates
-
1
Thaw Matrigel in ice water overnight at 4˚C and make 2 mg aliquots while keeping constantly on ice. Store at −80˚C.
-
2
Add one Matrigel aliquot to 12 ml ice cold DMEM/F12 in a 15 ml polypropylene centrifuge tube, dissolve and immediately add 1 ml per well into two 6-well plates.
-
3
Incubate the plates for one hour at room temperature and add 1 extra ml of DMEM/F12 per well.
-
4
Matrigel coated plates can be stored in a 37°C incubator for up to a week.
Collect and dissociate organoids
-
5
Pipet 2–3 mature organoids (Fig. 1B) per well into a 24 well plate using 25 ml serological pipets.
This protocol unit has been optimized for cerebral organoids generated according to Lancaster et al. (Lancaster et al., 2013), but is expected to be adaptable to alternative neural organoid growing methods. A step by step protocol for the generation of cerebral organoids has been published (Lancaster & Knoblich, 2014), and describes in detail the consecutive phases of making embryonic bodies, inducing neural fate and establishing a 3-dimensional culture. In our experiences, H9 (WAO9) hESC are most suitable for the generation of cerebral organoids containing high levels of brain tissues, while satisfactory results have also been obtained when using H1 (WA01) cells.
-
6
Remove excess medium that was transferred with the organoids, add 0.5 ml Accutase per well and place in a 37˚C (5% CO2) incubator for 3–5 min.
-
7
Initiate dissociation by pipetting the organoid/Accutase mixture up and down for a few times using low binding 1000 μl filter tips and incubate for another 3–5 min at 37˚C. Do not exceed 10 minutes of exposure to undiluted Accutase.
-
8
Further dissociate the organoids into little chunks by pipetting up and down a few more times and immediately add the solution to a 50 ml tube containing at least 5 ml Cerebral organoid medium per 0.5 ml used Accutase.
NOTE: It is important that dissociation is carried out in a gentle way to minimize damage to the cells. It is preferred to dissociate into clusters of cells rather than single cells to promote better cell survival. -
9
Centrifuge at 100 x g for 5 min at room temperature. Transfer supernatant to a new 50 ml tube while keeping the pellet.
-
10
Centrifuge the transferred supernatant at 300 x g for 3 min at room temperature. Transfer supernatant to a new 50 ml tube while keeping the pellet.
-
11
Centrifuge the transferred supernatant at 1000 x g for 3 min at room temperature. Discard supernatant and keep the pellet.
-
12
Combine pellets from step 11 – 13 and resuspend in cerebral organoid medium (1 ml per organoid transferred in step 5).
Plate cerebral organoids
-
13
Aspirate excess medium from the prepared Matrigel coated plates, and add 2 ml organoid medium to every well.
-
14
Using a 1 ml pipet slowly spread 0.5 ml of the organoid suspension into every well and place plates into a 37˚C incubator.
-
15
After 12–18 hours, replace medium by carefully removing half of the medium with a 1 ml pipet and slowly adding the same amount of fresh organoid medium dropwise into each well.
-
16
Culture for 6 more days changing half of the medium as described in step 15 every 2–3 days. During this time the cells will regrow their processes and reestablish cellular interactions (Fig. 1D).
BASIC PROTOCOL 2. VIRAL INFECTION AND SAMPLE PREPARATION FOR FACS
This protocol describes how to prepare viral stocks, infect cerebral organoid cultures and subsequently prepare the infected cultures for FACS sorting (Fig. 2). The main protocol describes infection with a replicating virus (ZIKA virus). Infection with a single cycle lentiviral reporter virus encoding GFP under the control of a CMV promoter in position of its envelope (env-defective HIV) and pseudotyped with the VSV-G envelope is included as an alternate protocol. The env-defective HIV virus was generated by cloning the CMV EGFP expression cassette into the ENV position of the virus, while preserving the remaining full length with intact gag/pol and accessory proteins. This insertion restricts the virus to a single round of infection, since HIV virus particles defective for the envelope gene are generated.
Figure 2.
Virus infection and Fluorescence Assisted Cell Sorting (FACS). (A) Schematic overview of virus infection and sorting of cerebral organoid cultures. (B) Example of a plaque assay readout; Vero cells were infected with 10-fold serial dilutions as indicated (C) Examples of cell sorting plots for isolating astrocytes, neurons and neural progenitor cells from human ESC derived cerebral organoid cultures. (D) Illustration of ZIKV- and ZIKV+ FACS gating for all cells and sorted neural progenitors, astrocytes and neurons.
Materials
VERO cells (ATCC® CCL-81™)
Vero cell culture medium (see recipe)
Vero cell infection medium (see recipe)
T175 culture flasks
ZIKA virus strain, for example Uganda 1947 (MR 766, ATCC® 1838™) or Puerto Rico 2015 (PRVABC59, ATCC® VR-1843™)
2 ml cryogenic vials
6-well flat bottom cell culture plates
2% Oxoid agar stock solution (see recipe)
2X Overlay medium (see recipe)
5% Sodium bicarbonate (Fisher Scientific, cat. no. L-14639) w/v in distilled water
1% DEAE Dextran (MP Biomedicals, cat. no. 195133) w/v in distilled water, sterilize by filtration
4% PFA in Phosphate-Buffered Saline (Boston BioProducts, cat. no. BM-155)
Dulbecco’s Phosphate-Buffered Saline (PBS) – no calcium or magnesium (Invitrogen, cat. no. 14190144)
1% Crystal violet (Fisher Scientific, cat. no. C581-100) w/v in distilled water
Methanol
2D cerebral organoid culture of 7 days (Step 16 of basic protocol 1)
Cerebral organoid medium (see recipe)
Penicillin/Streptomycin (Sigma, cat. no. P4333)
Accutase (Innovative Cell Technologies, cat. no. AT104)
DMEM/F12 (Gibco, cat. no. 11330-032)
Cell scrapers
EDTA solution (Invitrogen, cat. no. 15575-038)
Fetal bovine serum (HyClone, cat. no. SH30071.03)
PermWash buffer (BD Biosciences, cat. no. 554723)
Fluorochrome-conjugated antibodies (see Table 1)
Table 1.
Antibody conjugates used for flow cytometry and fluorescence-activated cell sorting
| Antibody | Conjugate | Catalog number | Dilution |
|---|---|---|---|
| anti-Flavivirus group E-antigen 4G2 | AF488 | Millipore MAB10216; conjugated with the Invitrogen antibody labeling kit A20181 | 1:200 |
| anti-human CD15 | V450 | BD Biosciences 561584 | 1:100 |
| anti-human CD24 | BUV395 | BD Biosciences 563818 | 1:100 |
| anti-human CD44 | PERCP-Cy5.5 | BD Biosciences 560531 | 1:100 |
| anti-human CD184 | APC | BD Biosciences 555976 | 1:50 |
| anti-human CD271 | PE | BD Biosciences 557196 | 1:50 |
ZIKA Virus production
-
1
Grow Vero cells in Vero cell culture medium to ~80% confluence, dissociate with EDTA/trypsin and seed 7.5 million cells in a T175 culture flask.
-
2
After 16–24 hours, remove the medium from the T175 flask and replace with 1 multiplicity of infection (MOI) of virus prepared in fresh Vero cell infection medium. Place back into a 37˚C, 5% CO2 incubator.
NOTE: Multiplicity of infection (MOI) = Plaque forming units (pfu) of virus used for infection / number of cells. The ZIKA virus titer is determined via a plaque assay on Vero cells as described below.NOTE: Vero cell infection medium contains lower FBS levels to prevent reduced infection efficiency from serum induced antiviral activity. -
3
After 72 hours collect the medium and centrifuge 10 minutes at 400 x g to remove cellular debris.
-
4
Keep the supernatant and store as ~1 ml aliquots in cryogenic vials at −80˚C.
NOTE: Cell supernatant from uninfected cells can be frozen as a mock infection control
Plaque assay to measure ZIKA virus titer
-
5
Seed Vero cells at 1 x 105 cells per well in 2 ml volume using a 6-well plate.
-
6
After 24 hours, verify if Vero cells have formed confluent monolayers without any gaps.
-
7
Prepare 10-fold serial dilutions of viral culture supernatant from step 4 in Vero cell infection medium.
-
8
Remove the culture medium from the Vero cells and add 500 μl of each dilution from step 7, moving from highest (106x) to lowest (101x). Place the 6-well plates back into the incubator. Cover the cells with the virus by tilting the 6-well plates in different directions every 15 minutes.
-
9
Prepare fresh agar overlay: microwave the 2% Oxoid agar stock solution until completely liquid. Mix 8.5 ml of distilled water, 25 ml of 2X Overlay medium, 0.5 ml 1% DEAE dextran, 1 ml 5% sodium bicarbonate, 15 ml of 2% Oxoid agar. Let the mixture cool until it can be handheld.
-
10
After 1 hour, remove the medium containing the virus and apply 2 ml agar overlay per well.
-
11
Let the agar overlay solidify for 5 minutes at room temperature before moving the 6-well plates to the incubator.
-
12
48 hours post-inoculation, count the viral foci using a phase contrast microscope. Calculate the viral titer as plaque forming units (pfu) per ml.
-
13
Fix the cells by applying 4 ml of 4% formaldehyde on the agar overlay for 15 minutes at room temperature.
-
14
Wash the cells/agar overlay once with 5 ml PBS.
-
15
Remove the agar overlay by flicking the plate upside down and stain the cells for 5 minutes with 2 ml per well of freshly prepared crystal violet staining solution (40 ml of 1% crystal violet solution, 80 ml of 100% methanol, 380 ml distilled water) for clear visualization of plaques.
NOTE: Figure 2B shows an example of a plaque assay after crystal violet staining.
Infection of cerebral organoid cultures with virus
-
16
Approximately 1 hour before infection, remove half of the medium by using a 1 ml pipet and replace with organoid medium supplemented with Penicillin/Streptomycin (1:50 dilution), place the culture back into a 37˚C incubator.
-
17
Thaw the appropriate amount of virus and control cell supernatant on ice. Using a 20 μl pipet, drip 5–20 μl (equaling the desired MOI) virus or control cell supernatant into each well in a cross pattern to enhance spread throughout the whole well.
NOTE: In order not to harm the axonal and dendritic cell connections, it is recommended not to shake the tissue culture plates to spread the virus. -
18
Place the culture back into a 37˚C incubator for 72 hours.
Cell collection and immunostaining for FACS
-
19
Prepare fresh FACS buffer: 1% FBS, and 2 mM EDTA in PBS.
-
20
Dilute 10X BD Perm/Wash buffer in distilled H2O to make a 1X solution.
-
21
Gently aspirate all medium from the culture with a pipet. Add 0.5 ml Accutase per well and incubate for 5 minutes at 37˚C.
-
22
Add 1 ml of DMEM/F12, pipet up and down using a 1 ml pipet to dislodge the cells and add the cells to 50 ml tubes containing 3 ml DMEM/F12 medium for every well that is collected. Wash each well one time with 1 ml DMEM/F12 to collect any remaining cells.
NOTE: When cells do not easily dislodge, using a cell scraper can facilitate collection of the cells. -
23
Centrifuge at 450 x g for 10 min at room temperature. Remove supernatant and gently resuspend the cells in 5 ml of FACS buffer.
NOTE: Washing volumes are anticipated on cell pellets derived from ~2–4 plates -
24
Centrifuge at 450 x g for 5 min at room temperature. Remove supernatant, gently resuspend the cells in 1 ml of FACS buffer and transfer into 1.5 ml microcentrifuge tubes.
-
25
Prepare 200 μl per sample of cell surface marker antibodies mixtures by adding the appropriate concentrations of fluorophore-conjugated antibodies for CD15, CD24, CD44, CD184 and CD271 to FACS buffer.
NOTE: See Table 1 for antibody conjugates and dilutions used for flow cytometry and fluorescence-activated cell sorting. -
26
Centrifuge the cells from step 24 at 450 x g for 5 min at room temperature. Remove supernatant and gently resuspend the cells in 200 μl of FACS buffer with fluorophore-conjugated antibodies. Incubate 45 minutes in the dark at room temperature.
NOTE: Since fluorophores are light sensitive it is recommended to avoid exposure of the samples to light from this step on. -
27
Wash the cells by adding 500 μl of FACS buffer, and centrifuge at 450 x g for 5 min at room temperature. Remove supernatant, gently resuspend the cells in 500μl of FACS buffer.
-
28
Centrifuge at 450 x g for 5 min at room temperature. Remove supernatant, gently resuspend the cells in 2% PFA in PBS (1:2 dilution of the 4% PFA solution in PBS). Incubate 5 minutes in the dark at room temperature.
-
29
Centrifuge at 450 x g for 5 min at room temperature. Remove supernatant, gently resuspend the cells in 500 μl PermWash buffer.
-
30
Repeat step 29.
-
31
Centrifuge at 450 x g for 5 min at room temperature. Remove supernatant, gently resuspend the cells in 200 μl PermWash buffer supplemented with antibody against ZIKV (fluorophore conjugated anti-E-antigen, 4G2). Incubate 1 hour in the dark at room temperature.
-
32
Centrifuge at 450 x g for 5 min at room temperature. Remove supernatant, gently wash the cells with 500 μl PermWash buffer. Repeat this step at least once.
-
33
Centrifuge at 450 x g for 5 min at room temperature. Remove supernatant, gently resuspend the cells in 1 ml FACS buffer. Place the samples on ice and shield from light for transportation to the flow cytometer.
BASIC PROTOCOL 3. FLUORESCENCE ASSISTED CELL SORTING
This protocol describes how to set the compensation parameters for multicolor FACS and set the appropriate gates to separate the cerebral organoid derived cells for cell type (neurons, neural progenitors and astrocytes) and infection based on their fluorescent labeling.
Materials
Falcon™ test tube with cell strainer snap cap (Corning, cat. no. 08-771-23)
Ultracomp eBeads® for compensation (Thermo Fisher Scientific cat. no. 01-2222-42)
Fluorophore-conjugated antibodies (Table 1)
Dulbecco’s Phosphate-Buffered Saline (PBS) – no calcium or magnesium (Invitrogen, cat. no. 14190144)
Fluorescence assisted cell sorter (FACSARIA III™, BD Biosciences)
Preparing compensation beads and sorter settings determination
-
1
Make single stain controls for every antibody used. Vortex compensation beads for 5 minutes before use. To 1 drop of compensation beads from the dispenser bottle, add the same amount of conjugated antibody or less, as used for staining one cell sample. Dilutions of conjugated antibodies are given in Table 1.
-
2
Incubate 15 minutes in the dark at 4˚C.
-
3
Wash the beads once: centrifuge beads with antibodies at 600 x g for 5 min, discard the supernatant and resuspend in 400 μl FACS buffer.
-
4
Use unstained cells and compensation beads to determine optimal settings for forward and side scatter, as well as for fluorescence detectors by adjusting the voltages for the photomultiplier tubes (PMT) until you see beads and cells.
-
5
Analyze all single stain controls to ensure that the positive peak for every antibody-fluorophore is on scale. Change PMT voltage if this is necessary. Try to have a maximum spread between the positive and negative peaks for every control.
-
6
Once optimal PMT voltages for all channels are determined, run all single stain controls again and record all data for compensation setup. Do not change PMT voltages anymore at this point. In case you need to change PMT voltages later, you will have to rerun all single stain controls again with the adjusted PMT voltages.
-
7
Calculate the compensation matrix, by using the software provided with the flow cytometer (FACSDiva™, BD Biosciences).
Prepare samples, set gates and sort samples
-
8
Filter the samples by pipetting the cell solution on top of the cell strainer caps of the test tubes. Add 0.5–1 ml PBS on top of the strainer to ensure all single cells went through the strainer.
-
9
Remove the cap and place the test tube into the tube holder of the sorter. Sort infected from non-infected cells based on detection of flavivirus E protein. Mock and infected cells are used to set the gates for positive and negative cells. In order to avoid false negative cells as much as possible, move the negative gate far to the left. Sorting efficiency should be 90% or higher. Collect virus-positive and negative cells in 15 ml Falcon tubes.
NOTE: Illustration of gates for infection and non-infection gates are shown in Figure 2D.NOTE: Cells can be first sorted for cell type or infection dependent on personal preference. If it is necessary to concentrate the cells after the first sorting round, centrifuge the samples at 450 x g and resuspend in FACS buffer. -
10
Sort the different cell types (astrocytes, neural progenitor cells, neurons) from the virus-positive and negative cells obtained in step 9 using the gating scheme given in Figure 2C. Gates are first applied for CD271 and CD44. CD271− CD44+ cells are then further gated for CD184+ to obtain cells with the astrocyte marker signature. CD271− CD44− cells are further gated for CD184, CD24 and CD15 to separate neural progenitor cells (CD271− CD44− CD184+ CD24+) and neurons (CD271− CD44− CD184− CD24+ CD15low).
-
11
Collect cells in three 5 ml FACS tubes with sorting efficiency of 90% or higher. Transfer cells to 15 ml Falcon tubes for further downstream applications.
Process samples for downstream applications
-
12
Centrifuge sorted cell samples at 450 x g for 10 min at 4˚C. Keep samples on ice and remove supernatant without disturbing the pellet
-
13
Directly add lysis buffer to the pellet for downstream DNA or RNA extraction or store the cells at −80˚C until further use.
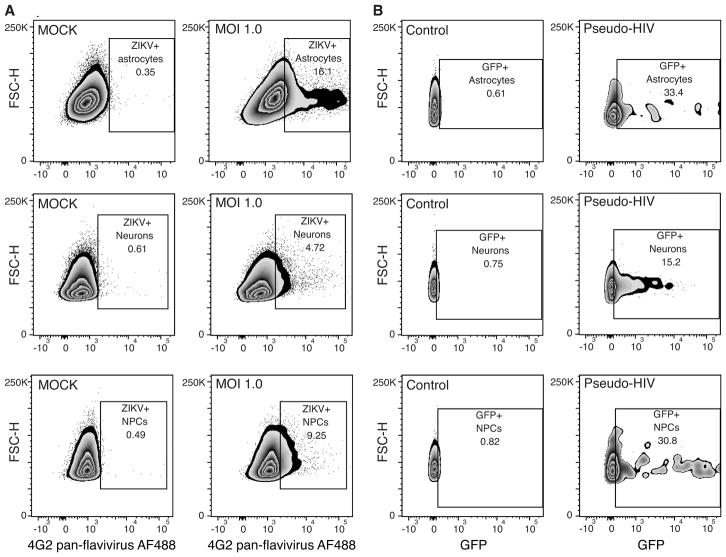
NOTE: Examples of Zika Virus infection rates per cell type are shown in Figure 3A.
Figure 3.
Examples of FACS gating and percentages of virus infected cells among isolated astrocytes, neurons, and neural progenitor cells (NPCs). (A) FACS gating and percentages from cerebral organoid cultures infected with ZIKV. (B) FACS gating and percentages from cerebral organoid cultures infected with pseudotyped HIV.
ALTERNATE PROTOCOL 1. SORTING OF PSEUDOTYPED HIV VIRUS INFECTED CEREBRAL ORGANOID CULTURES
To illustrate adaptation of the protocol to any virus that infects human neuronal cells, we included an alternative protocol for flow cytometry after infection with a single cycle HIV lentiviral reporter virus. HIV1 lacking a functional Env gene and expressing a GFP reporter gene under the control of a CMV promoter, was pseudotyped with the vesicular stomatitis virus (VSV-G) envelope glycoprotein to mediate cell entry but limit replication to a single round. The flow cytometry plots in Figure 3B show expression of the CMV promoter driven GFP reporter gene in neural progenitors, neurons and astrocytes, indicating that lentiviral integration is supported in these cell types.
Materials
293T cells (ATCC® CRL-3216™)
HEK293T cell culture medium (see recipe)
Plasmids: molecular clone HIV R7/3 Δenv CMV-EGFP, pHCMV-G coding for the G glycoprotein of VSV-G (Lloyd, Ng, Muesing, Simon, & Mulder, 2007).
T75 culture flask
Polyethylenimine – PEI 25K (Polysciences, cat.no. 23966)
Sterile Disposable Filter Units (0.45 μm)
2 ml cryogenic vials
Production of single cycle lentiviral reporter viruses
CAUTION: Appropriate biosafety precautions including standard operating procedures for work with lentiviruses need to be implemented prior to start work with lentiviruses. The described experiments were conducted in a BSL2+ facility by trained personnel.
-
1
Seed 5 x 106 HEK293T in 15 ml HEK293T cell culture medium into a T75 culture flask
-
2
The next day transfect the cells overnight with 20 μg of HIV R7/3 Δenv CMV-EGFP (replication defective) and 3 μg pHCMV-G using 3 μg/ml polyethylenimine.
-
3
Replace the medium in the morning
-
4
On day two and three after transfection, collect the culture supernatant and centrifuge at 450 x g for 5 minutes to remove cellular debris.
-
5
Filter the supernatant through a 0.45 μm filter and store as ~1 ml aliquots in cryogenic vials at −80˚C.
NOTE: Infectivity titers of the viral stocks can be determined by infecting TZM-bl reporter cells (RRID:CVCL_B478) with serial dilutions in triplicate.
Infection, sample preparation and sorting
-
6
Follow BASIC PROTOCOL 2, part “Infection of cerebral organoid cultures with virus” and “Cell collection and immunostaining for FACS” step 16 until step 28 using HIV R7/3 Δenv CMV-EGFP culture supernatant from step 5. Since the HIV reporter virus expresses GFP, which is detectable with the same settings as for AF488 or FITC fluorophores, there is no need to permeabilize the cells to allow for intracellular viral protein staining.
-
7
Centrifuge at 450 x g for 5 min at room temperature. Remove supernatant, gently resuspend the cells in 1 ml FACS buffer. Repeat this washing step twice. Place the samples on ice and shield from light for transportation to the flow cytometer.
-
8
Similar flow cytometer settings and gating strategy can be used as described above for ZIKV infected cells. Use uninfected cells to determine the background and setting the gate for virus-positive cells.
REAGENTS AND SOLUTIONS
Cerebral organoid medium (according to Lancaster & Knoblich, 2014)
125 ml DMEM/F12 (Gibco, cat. no. 11330-032)
125 ml Neurobasal medium (Gibco, cat. no. 21103049)
1.25 ml N-2 supplement (Gibco, cat. no. 17502001)
62.5 μl Insulin solution (Sigma, cat. no. I9278)
2.5 ml GlutaMAX supplement (Gibco, cat. no. 35050061)
1.25 ml MEM-NEAA (Gibco, cat. no. 11140050)
2.5 ml B27 supplement (Gibco, cat. no. 17504044)
Prepare a 1:100 dilution of 2-mercaptoethanol (Sigma, cat. no. M3148) in DMEM-F12 and add 87.5 μl of this to the medium.
Sterilize by filtration and store at 4˚C for up to two weeks.
Equilibrate to room temperature before use.
Vero cell culture medium
Dulbecco’s modified Eagle’s medium, DMEM (Corning, cat. no. 10-013-CV)
10% Fetal bovine serum (HyClone, cat. no. SH30071.03)
1:100 Penicillin/Streptomycin (Sigma, cat. no. P4333)
Sterilize by filtration and store at 4˚C for up to four weeks.
Equilibrate to room temperature before use.
Vero cell infection medium
Dulbecco’s modified Eagle’s medium, DMEM (Corning, cat. no. 10-013-CV)
2% fetal bovine serum (HyClone, cat. no. SH30071.03)
1:100 Penicillin/Streptomycin (Sigma, cat. no. P4333)
Sterilize by filtration and store at 4˚C for up to four weeks.
Equilibrate to room temperature before use.
2X overlay medium
200 ml of 10x MEM (Gibco, cat. no.21430020)
20 ml of 200 mM L-glutamine (Corning, cat. no. 25-005-CR)
48 ml of 5% sodium bicarbonate (Fisher Scientific, cat. no. L-14639) w/v in distilled water.
20 ml of 1 M HEPES (Fisher Scientific, cat. no. BP310-500)
20 ml Penicillin/Streptomycin (Sigma, cat. no. P4333)
20 ml Fetal bovine serum (HyClone, cat. no. SH30071.03)
672 ml of distilled H2O
Sterilize by filtration and store at 4˚C for up to four weeks.
2% Oxoid agar stock solution
2% Oxoid agar (Oxoid, cat. no. LP0028) w/v in distilled water.
-
Sterilize by autoclaving and store at room temperature for up to four weeks.
NOTE: The agar overlay will become solid when cooling down to room temperature and needs reheating before use.
HEK293T cell culture medium
Dulbecco’s modified Eagle’s medium, DMEM (Corning, cat. no. 10-013-CV)
10% fetal bovine serum
1:100 Penicillin/Streptomycin (Sigma, cat. no. P4333)
Sterilize by filtration and store at 4˚C for up to four weeks.
Equilibrate to room temperature before use.
COMMENTARY
Background Information
The human brain is unique in its structure and functionality compared to other mammals. To circumvent the limitations of animal models, ample effort has been made to establish human cell based models to study brain development, function and disorders. Preceded by many protocols describing the generation of neural rosettes, neural progenitors and different types of mature neuronal cells, the most comprehensive model resembling the human brain to date is the human stem cell-derived cerebral organoid model, which generates three dimensional brain-like tissues in vitro that not only mimic the diversity of neuronal cell types but also the structure and cellular interactions typical for human brain tissue. The fact that cells develop similar as in their in vivo environment, allowing the establishment of heterotypical neuronal interactions, distinguishes this model compared to the direct differentiation of human stem cells into one specific neural cell type. Challenges in studying these more complex multicellular tissues include potential higher variability and increased complexity of data analyses. To facilitate the analysis of defined cell types in complex brain organoids upon virus infection, we developed a FACS based protocol. FACS has been proven useful to sort neuronal cells from adult rat brains and to distinguish cell populations during neural induction of human embryonic stem cells (Yuan et al., 2011). Choosing FACS over microscopy based cell identification not only reduces output quantification errors, but also expands choices for downstream analyses of the identified cells. In this protocol we used 6-color cell sorting, which leaves the option to include more colors if the FACS/flow cytometer laser and filter setup allows for it. For example, inclusion of a viability dye to separate live from dead cells, could promote lower noise and better separation of cell populations. However, one should be cautious about interference of the dead cell reagents with further downstream processing. The protocol we describe includes cell fixation either to allow intracellular staining of viral proteins and/or as a safety measure. Although not tested by us, a modified version of this protocol without fixation, if staining of intracellular markers is not required, is anticipated to allow life cell sorting.
Critical Parameters and troubleshooting
The protocol includes three critical stages: preparing organoid culture for optimal infection, obtaining well prepared virus stocks, and fluorescence activated cell sorting of immunostained cell samples. Due to the advanced level sorting with at least 6 markers, for those users who do not yet have advanced FACS skills guidance or assistance of a FACS specialist might be required during this step. A guide to common problems with possible causes and solutions is summarized in Table 2.
Table 2.
Troubleshooting guide for common problems with FACS sorting of virus infected cerebral organoid cultures
| Problem | Possible cause | Solutions |
|---|---|---|
| Basic protocol I | ||
| Low reattachment and/or survival of cells after dissociation | Too long incubation with dissociation agent. | Restart the protocol and reduce incubation time dissociation agent. |
| Dissociation of organoids in single cells, cells are replated too sparse. | Reserve bigger cell clumps, increase seeding density. | |
| Cells got damaged during centrifugation. | Use lower centrifugation speed. | |
| Basis protocol II | ||
| Excessive cell death upon infection | Too high concentration of virus. Contamination of virus with toxic agents. | Check virus stock concentration. Harvest virus before massive cell death occurs or grow virus in insect cells to reduce toxins during virus production. |
| Low infection efficiency | Too low concentration of virus. Unhealthy cerebral organoid culture. | Check virus stock for concentration. Check cell culture for mycoplasma or other “hidden” contaminants. |
| Basic Protocol III | ||
| No events are displayed in the FSC-SSC scatter plot | Voltages of the photomultiplier tubes (PMT) need to be adjusted depending on cell size and granularity. | Adjust voltages for FSC and SSC PMT. |
| No or very low cell amounts after sorting | Too short centrifugation time of cells diluted in high volumes. | Increase centrifugation time appropriately when working with high volumes. |
| Clogging of the FACS system. | Carefully monitor during sorting for any anomalies of the system. | |
| Low sorting efficiency | Cells are too concentrated and/or flow settings are too high for efficient sorting. | Reduce flow speed and/or dilute cell sample more. |
Anticipated Results
In a typical experiment, starting from ~15 organoids seeded in four 6-well plates, with an overall infection efficiency of 15% one can expect to obtain 20.000–200.000 cells per group. For downstream applications, we have successfully extracted DNA suitable for bisulfite sequencing, or RNA suitable for whole transcriptome amplification and subsequent qRT-PCR from samples as low as ~4000 cells (Janssens et al., 2018).
Time Considerations
The protocol starts from mature organoids, which will take up to 12 weeks to develop. Growing a virus stock and performing plaque assays usually takes 5 days. Preparing the infected organoid culture will take 10 days. The last day of the protocol, which includes cell collection, immunostaining and FACS should be anticipated to be labor intensive and may take up to 10 hours. It is therefore recommended to split some of the tasks between two people to lower the work load but also to reduce the time spend between collection and final freezing of the cells.
Significance Statement.
Organoids – or pluripotent stem cell derived in vitro grown simplified mini organs – have become a tremendously important model to study human organ development and disease. While organoids ameliorate the study of organized tissues with multicellular interactions, their high level of cellular diversity and complexity also challenges experimental readouts. To restrict the noise inherent to the heterogeneous cell mixtures derived from organoid cultures, we developed a new technique of sorting specific cell populations from virus infected brain organoid cultures. This method still includes the advantage of growing cells in a more natural environment than traditional cell culture, but now renders samples suitable for downstream cell-type specific multi-omics analyses.
Acknowledgments
We acknowledge Christopher Bare, the flow cytometry core, and the microscopy core at the Icahn School of Medicine at Mount Sinai for assistance. We thank Dr. Ana Fernandez-Sesma, Dr. Rafael Fenutria, Dr. Vinod Balasubramaniam and Ms. Megan M. Schwarz for sharing conjugated anti-flavivirus group E antigen 4G2 antibody and virus. This work was supported by the Huffington foundation (T.P.Z.), NIH grants NIH/NIAID U19AI118610 (A.G-S.), R21AI129486 (A.G-S.), R21 AI125236 (VS), and the Belgian American Educational Foundation (fellowships to both S.J. and M.S.). The authors declare no competing financial interests.
LITERATURE CITED
- Camp JG, Badsha F, Florio M, Kanton S, Gerber T, Wilsch-Bräuninger M, … Treutlein B. Human cerebral organoids recapitulate gene expression programs of fetal neocortex development. Proceedings of the National Academy of Sciences of the United States of America. 2015;112(51):15672–15677. doi: 10.1073/pnas.1520760112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SH, Kim YH, Hebisch M, Sliwinski C, Lee S, D’Avanzo C, … Kim DY. A three-dimensional human neural cell culture model of Alzheimer’s disease. Nature. 2014;515(7526):274–278. doi: 10.1038/nature13800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcez PP, Loiola EC, Madeiro da Costa R, Higa LM, Trindade P, Delvecchio R, … Rehen SK. Zika virus impairs growth in human neurospheres and brain organoids. Science (New York, NY) 2016;352(6287):816–818. doi: 10.1126/science.aaf6116. [DOI] [PubMed] [Google Scholar]
- Janssens S, Schotsaert M, Karnik R, Balasubramaniam V, Dejosez M, Meissner A, … Zwaka TP. Zika Virus Alters DNA Methylation of Neural Genes in an Organoid Model of the Developing Human Brain. MSystems. 2018;3(1) doi: 10.1128/mSystems.00219-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster MA, Knoblich JA. Generation of cerebral organoids from human pluripotent stem cells. Nature Protocols. 2014;9(10):2329–2340. doi: 10.1038/nprot.2014.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster MA, Renner M, Martin CA, Wenzel D, Bicknell LS, Hurles ME, … Knoblich JA. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501(7467):373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd AG, Ng YS, Muesing MA, Simon V, Mulder LCF. Characterization of HIV-1 integrase N-terminal mutant viruses. Virology. 2007;360(1):129–135. doi: 10.1016/j.virol.2006.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariani J, Coppola G, Zhang P, Abyzov A, Provini L, Tomasini L, … Vaccarino FM. FOXG1-Dependent Dysregulation of GABA/Glutamate Neuron Differentiation in Autism Spectrum Disorders. Cell. 2015;162(2):375–390. doi: 10.1016/j.cell.2015.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X, Nguyen HN, Song MM, Hadiono C, Ogden SC, Hammack C, … Ming G-L. Brain-region-specific organoids using mini-bioreactors for modeling ZIKV exposure. Cell. 2016;165(5):1238–1254. doi: 10.1016/j.cell.2016.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan SH, Martin J, Elia J, Flippin J, Paramban RI, Hefferan MP, … Carson CT. Cell-surface marker signatures for the isolation of neural stem cells, glia and neurons derived from human pluripotent stem cells. PloS One. 2011;6(3):e17540. doi: 10.1371/journal.pone.0017540. [DOI] [PMC free article] [PubMed] [Google Scholar]