Abstract
Interferon-gamma (IFN-γ) plays an important role in innate and adaptive immunity against intracellular infections and is used clinically for the prevention and control of infections in chronic granulomatous disease (CGD) and inborn defects in the IFN-γ/interleukin (IL) −12 axis. Using transcriptome profiling (RNA-seq), we sought to identify differentially expressed genes, transcripts and exons in Epstein-Barr virus-transformed B lymphocytes (B-EBV cells) from CGD patients, IFN-γ receptor deficiency (IFNGRD) patients, and normal controls, treated in vitro with IFN-γ for 48 hours. Our results show that IFN-γ increased the expression of a diverse array of genes related to different cellular programs. In cells from normal controls and CGD patients, IFN-γ induced expression of genes relevant to oxidative killing, nitric oxide synthase pathway, proteasome-mediated degradation, antigen presentation, chemoattraction, and cell adhesion. IFN-γ also up-regulated genes involved in diverse stages of mRNA processing including pre-mRNA splicing, as well as others implicated in the folding, transport, and assembly of proteins. In particular, differential exon expression of WARS (encoding tryptophanyl-tRNA synthetase, which has an essential function in protein synthesis) induced by IFN-γ in normal and CGD cells suggests that this gene may have an important contribution to the benefits of IFN-γ treatment for CGD. Upregulation of mRNA and protein processing related genes in CGD and IFNRD cells could mediate some of the effects of IFN-γ treatment. These data support the concept that IFN-γ treatment may contribute to increased immune responses against pathogens through regulation of genes important for mRNA and protein processing.
Keywords: Transcriptome, Chronic granulomatous disease, Phagocyte, Interferon-gamma, Interferon-gamma receptor deficiency
INTRODUCTION
Interferon-γ (IFN-γ) is a proinflammatory cytokine with antiviral, antimicrobial, and anti-tumor properties. It is produced mainly by activated T-lymphocytes and natural killer cells [Liu and Cao, 2016]. T-lymphocytes release IFN-γ in response to antigen presentation and interleukin (IL)-12, while natural killer cells secrete IFN-γ in response to the recognition of microbial antigens and IL-12 [Liu and Cao, 2016]. IFN-γ treatment of neutrophils elicits a variety of responses including increased oxidative burst, differential gene expression, and induction of antigen presentation, depending on stimuli and environmental conditions, and thereby mediates the ability of these cells to mediate the transition between the innate and acquired immune responses [Ellis and Beaman, 2004]. In macrophages, IFN-γ activates a diverse array of functions, including antimicrobial activity [Nathan et al., 1983], increased killing of intracellular pathogens [Torrico et al., 1991], antigen processing and presentation to lymphocytes through induction of MHC antigens [Keller et al., 1988] and regulation of dendritic cell differentiation and maturation [Delneste et al., 2003; Ito et al., 2001; Pan et al., 2004]]. In addition, IFN-γ acts on B-lymphocytes to promote switching of IgG subclasses that bind to Fcγ receptors on phagocytes, activate complement, and contribute to phagocyte-mediated elimination of microbes [Ellis and Beaman, 2004].
IFN-γ finds clinical application in the prevention and control of infections in chronic granulomatous disease (CGD) [Bemiller et al., 1995; Ezekowitz et al., 1988; Filiz et al., 2015; Newburger and Ezekowitz, 1988] IFN-γ therapy is also used for hereditary disorders of the IFN-γ/IL12 axis, including defects in the IL-12 receptor and IL-12/p40 [Ramirez-Alejo and Santos-Argumedo, 2013]. Treatment with IFN-γ, along with prophylactic antibiotics and prolonged, aggressive treatment of infections, has significantly improved the prognosis of CGD [Holland, 2010; Johnston, 2001; Winkelstein et al., 2000]; however its mechanism of action remains unclear. The most striking responses to IFN-γ occur in patients with mutations in the CYBB gene (responsible for X-linked CGD) near splice sites, leading to to partial restoration of normal splicing upon IFN-γ treatment in vivo and in vitro [Condino-Neto and Newburger, 2000].
The present study investigated the effect of IFN-γ on gene expression in Epstein-Barr virus-transformed B-lymphocytes (B-EBV cells), which provide an in vitro model for CGD phagocytes [Condino-Neto and Newburger, 1998; Dusi et al., 1998; Volkman et al., 1984]. We studied cells lines derived from patients with X-linked CGD, patients with interferon-gamma receptor 1 or 2 deficiency (IFNGR1D/IFNGR2D) as negative controls for IFN-γ responses, and normal individuals. We also evaluated possible splicing modifications by detecting changes in transcript and exon levels upon IFN-γ stimulation. Our findings provide new insights into the molecular mechanisms of IFN-γ efficacy in CGD and provide a basis for further investigation of IFN-γ treatment for primary immunodeficiencies and acquired immune defects.
MATERIALS & METHODS
Patients -
All patients included in this study were diagnosed according to standards established by the International Consensus Document (“ICON”) [Routes et al., 2014], with confirmed molecular diagnoses. We evaluated B-EBV cell lines derived from peripheral blood of four X-linked CGD patients (two with splicing defects, one with a frameshift mutation, and one with a missense mutation; disease group D1–D4, respectively), three IFNGR1D patients with complete defects of the IFN-γ receptor α chain (one with a splicing defect, one with a missense mutation, and one with a small frameshift deletion; negative control group N1–N3, respectively), three IFNGR2D patients with complete defects of the IFN-γ receptor β subunit (one with a missense mutation and two with a small frameshift deletion; negative control group N4–N6, respectively), and five healthy individuals (normal control group, C1–C5). The IFNGRD patient cell lines were kindly provided by Prof. Dr. Jean-Laurent Casanova. Tables 1 and 2 present the genetic characteristics of the patients from whom the cell lines were derived.
Table 1:
Mutations in CGD patients.
| CGD Patients | Type of Mutation | Position | Nucleotide | Product |
|---|---|---|---|---|
| D1 | Splice region | Intron 1 | c 56+6 T>C | Mis-splicing |
| D2 | 5’ splice site | Intron 5 | gt>tt | Exon skipping |
| D3 | Frameshift | Exon 3 | G251del | Stop codon in exon 4 |
| D4 | Nonsense | Exon 11 | G1341A | Trp 443 stop |
Table 2:
Homozygous mutations in IFN-γ receptor deficiency patients.
| IFNGR1D Patients | Type of Mutation | Nucleotide | Reference |
| N1 | Missense | C77F/C77F | [Jouanguy et al., 2000] |
| N2 | Splice site | 202–1G>T/202–1G>T | |
| N3 | Small frameshift deletion | 523delT/523delT | [Dorman et al., 2004] |
| IFNGR2D Patients | Type of Mutation | Nucleotide | Reference |
| N4 | Small frameshift deletion | 212delA/212delA | |
| N5 | Missense | T168N/T168N | [Vogt et al., 2005] |
| N6 | Small frameshift deletion | 663del27/663del27 | [Vogt et al., 2005] |
Cell Culture -
B-EBV cell lines were seeded at 1–2 × 105/ml and cultured in RPMI 1640 complete medium supplemented with 10% heat inactivated fetal bovine serum, 2 mM L-glutamine, 100 units/ml penicillin and 100 mg/ml streptomycin, with or without IFN-γ (100 U/ml, R&D Systems, Minneapolis, MN, USA) in triplicate for 48 h in sterile polystyrene 25 cm3 flasks (BD-Becton Dickinscon, Sunnyvale, CA, USA). The manufacturers certified all reagents – serum, buffers, media – and containers as nonpyrogenic.
RNA Isolation -
Samples were taken in three biological triplicates. Cell culture samples (5–10 × 106) were immediately cooled on ice, centrifuged at 4 °C, washed with cold phosphate-buffered saline and total RNA extracted using TRIzol® Reagent (Life Technologies, Grand Island, NY, USA) in accordance to the manufacturer’s protocol. RNA quality was assayed by using BioAnalyzer (Agilent Technologies, Palo Alto, CA, USA).
RNA-Seq -
RNA-Seq libraries were prepared from isolated total RNA using Illumina TruSeq Stranded Total RNA Library Prep Kit with Ribo-Zero Gold TM (Illumina, California, USA) following the manufacturer’s high-throughput protocol. The synthesized libraries were strand-specific with cytoplasmatic and mitochondrial rRNA depletion, had paired-end reads with 100 bp size and were sequenced on the HISEQ2000 Illumina platform. Six bar-coded samples were multiplexed in each paired-end lane of the flowcell, yielding approximately 30 million reads per sequenced sample.
Bioinformatics -
A customized pipeline was developed for our analysis. Briefly, RNA high-throughput sequencing reads were first evaluated for quality metrics with FastQC, version 0.11.2, and mapped to the Ensembl Homo sapiens GRCh38/hg38 reference genome by Spliced Transcripts Alignment to a Reference (STAR), version 2.4.0g, [Dobin et al., 2013; Dobin and Gingeras, 2015]. Next, Samtools, version 1.2, [Li et al., 2009] was used to order mapped reads by position and used as input files for HTSeq, version 0.6.1, [Anders et al., 2015] to preprocess the RNA-Seq data for differential expression analysis by counting the overlay of mapped reads in the gene position. DESeq2, version 1.22.0, [Love et al., 2014] was used for differential gene expression analysis and DEXSeq, version 1.16.6, [Anders et al., 2012] for differential exons usage analysis. For identification of differentially expressed transcripts/isoforms, additional integrative analysis used STAR to align reads to Ensembl Homo sapiens GRCh38/hg38 reference genome and Kallisto, version 0.42.4, to quantify transcript abundance in combination with Sleuth, development version, for differential analysis that averaged the estimates produced by Kallisto. Significantly affected pathways were detected using the SetRank method [Simillion et al., 2017].
RESULTS
RNA-seq data collected from B-EBV cells derived from normal controls, X-linked CGD patients, and IFNGRD deficient patients, with or without treatment with IFN-γ in vitro for 48 hours, were analyzed as 15 different datasets (each one a set of triplicate determinations for untreated versus IFN-γ-treated B-EBV cells) for differential gene expression, differential exon usage, transcript abundance and gene set enrichment analysis of biological pathways.
Differential gene expression analysis
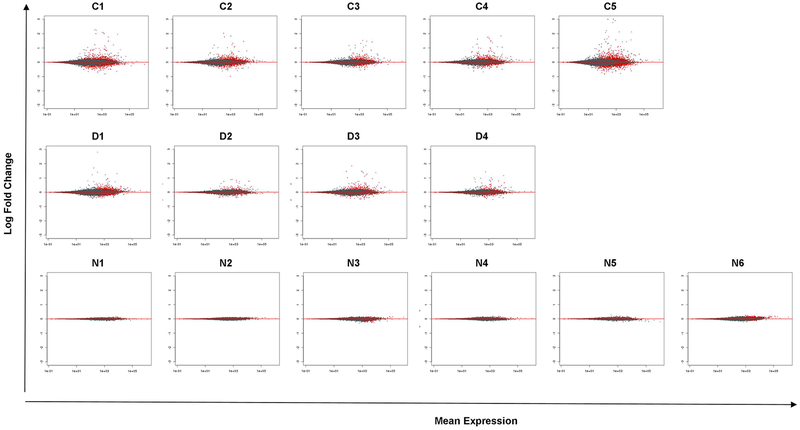
Using DESeq2 and applying regularized log transformation, a distinct separation of IFN-γ-treated versus untreated B-EBV cell lines was observed only for normal controls, X-linked CGD patients and two IFNGRD patients (N3 and N6) demonstrating a global change in gene expression on these cells caused by the IFN-γ in vitro treatment, represented in the principal component analysis in Supplemental Figure 1 and by hierarchical clustering of sample-to-sample distances represented on the heatmaps of Supplemental Figure 2. The MA plot (the distribution of the intensity ratio ‘M’ plotted by the average intensity ‘A’) of the log2-fold change over the mean of normalized counts, represented in Figure 1, demonstrates fewer differentially expressed genes in B-EBV cells of IFNGRD-patients compared to normal controls and X-linked-CGD-patients (red dots in the figure indicate genes with adjusted p-value < 0.1). There were no up- or down-regulated genes detected in our analysis of B-EBV cells of patient N2, confirming impairment of the IFN-γ signaling pathway, a characteristic feature of the patient’s genetic defect.
Figure 1: MA plot from each subject of the normal control group (C1–5 from left to right, top row), CGD patients (D1–4 from left to right, middle row) and of IFNGRD patients (N1–6 from left to right, bottom row).
These plots show the log2 fold changes from the treatment over the mean of normalized counts (the average of counts normalized by size factors). The plot shows the shrinkage of log2 fold changes resulting from the incorporation of zero-centered normal prior. The shrinkage is greater for the log2 fold change estimates from genes with low counts and high dispersion, as can be seen by the narrowing of spread of leftmost points in the right plot. Points colored red represents genes with adjusted p value less than 0.1 and points which fall out of the window are plotted as open triangles pointing either up or down. Less number of significant genes is detected in the IFNGRD group when compared to the healthy and CGD groups reflecting the impairment of the activation of the IFN-γ signaling pathway.
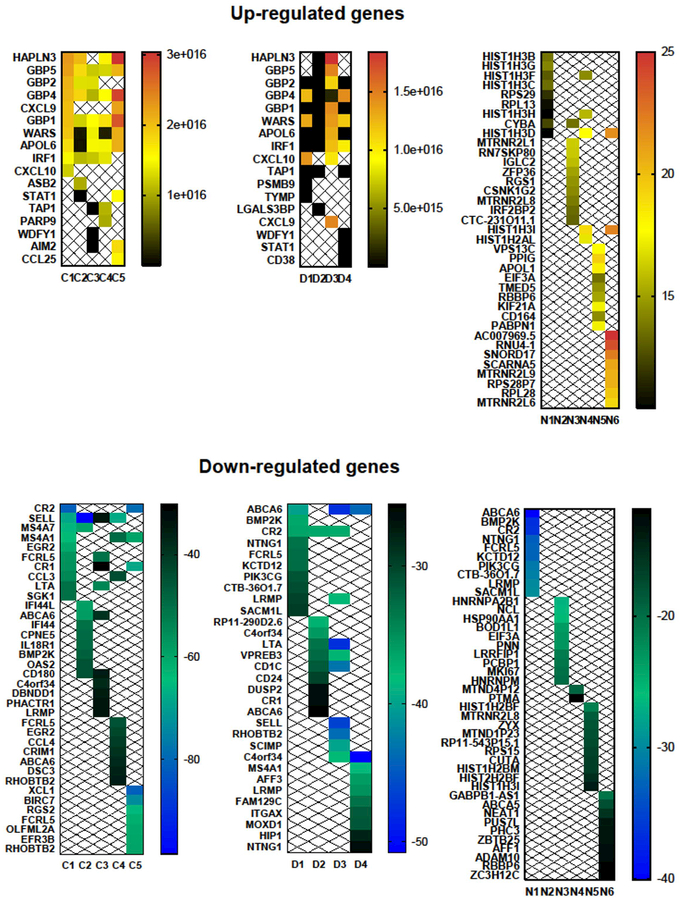
We next ranked up-regulated and down-regulated genes by highest fold change and lowest p-value (less than 0.05). The top ten up-regulated genes in response to in vitro IFN-γ treatment (Figure 2, Supplemental Table I) in B-EBV cells of normal controls and CGD patients (but not in IFNGRD patients) included APOL6; CXCL10; GBP family members (GBP−1, −2, −4 and −5); HAPLN3; IRF1; LGALS3BP; STAT1; TAP1; WARS and WDFY1. We also detected up-regulation of STAT1 and IRF1 (markers of IFN-γ stimulation) which supports the inference that the other detected changes represent an IFN-γ effect on gene expression. Among the top ten up-regulated genes, WARS was most consistently detected as up-regulated in B-EBV cells of both healthy subjects and CGD patients. The induction of these genes by IFN-γ has been reported previously, except for HAPLN3, which has only been reported to be induced by IFN-β [Khsheibun et al., 2014].
Figure 2: Heatmaps of the top 10 genes up- and down-regulated from each group (normal controls, CGD patients and IFNGRD patients).
These heatmaps show the log2 fold changes in expression.
Contrary to our initial hypothesis that IFNGRD patients would show little or no differential gene expression due to impairment of the IFN-γ signaling pathway, our findings reveal residual function of the IFN-γ receptor upon IFN-γ in vitro treatment. The IFNGR1D/IFNGR2D deficient group showed up-regulation of genes (Figure 2, Supplemental Table I) such as IRF2BP2, CSNK1G2 and genes encoding histone H3 core proteins such as HIST1H3B, HIST1H3C, HIST1H3D, HIST1H3F, HIST1H3G, HIST1H3H and HIST1H3I. These findings indicate residual transcriptional control of IFN-γ signaling in these receptor-deficient cells.
Other up-regulated genes, such as PABPN1, RN7SKP80, RNU4, SNORD17, RPL13, RPL28, RPS28P7, RPS29, SCARNA5, EIF3A and ZFP36, suggest that IFN-γ could have effects on poly(A) RNA binding, mRNA nonsense-mediated decay, structural assembly of ribosomes and Cajal body function to modify RNA, therefore acting in the regulation of pre-mRNA splicing. This finding corroborates previous results from our group that demonstrated partial correction of aberrant splicing in a form of variant CGD upon IFN-γ treatment [Condino-Neto and Newburger, 2000]. In addition, we observed up-regulation of genes with putative neuroprotective function, such as MTRNR2L1, MTRNR2L6, MTRNR2L8 and MTRNR2L9 [Bodzioch et al., 2009], as well as a gene, KIF21A, involved in neuronal axonal transport of essential cellular components along axonal and dendritic microtubules [Marszalek et al., 1999]. The latter finding may help explain how IFN-γ improves functional deficits in a mouse model of Friedreich ataxia [Tomassini et al., 2012] as well as in human patients [Seyer et al., 2015], in addition to its ability to improve frataxin levels.
Among the top ten genes down-regulated in response to in vitro IFN-γ (Fig. 2, Supplemental Table 2) with highest fold change and lowest p-value (less than 0.05), B-EBV cells of healthy individuals and X-linked CGD showed decreased expression of C4orf34, LTA, MS4A1 and SELL, which are known to have roles in endoplasmic reticulum functions and in the development, differentiation and adhesion of leucocytes on endothelial cells; these cellular functions underly inflammatory and antiviral responses. In B-EBV cells of IFNGR1D/IFNGR2D patients, IFN-γ treatment caused a wider number of genes to be down-regulated, including, among those most relevant to inflammation, ABCA5, ADAM10, LRRFIP1, MTND1P23, MTND4P12, MTRNR2L8, PIK3CG, PTMA, RBBP6, and ZYX.
Differential exon usage
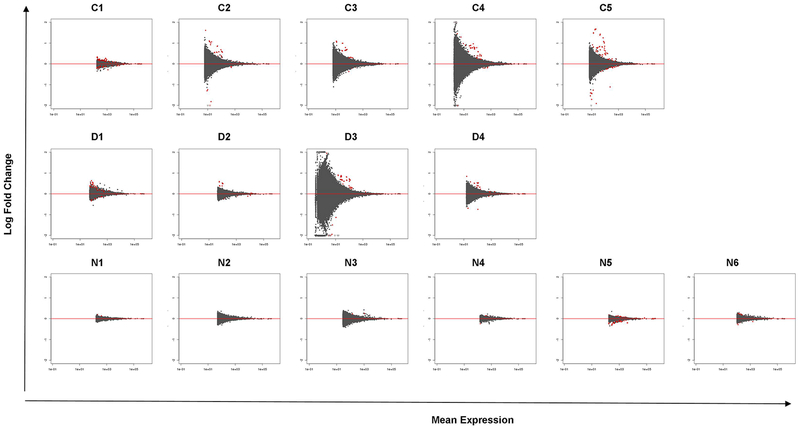
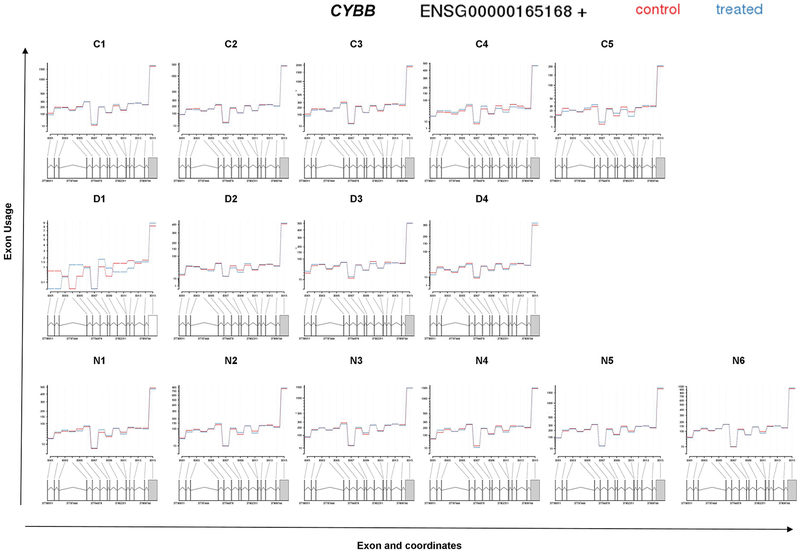
Analysis of differential exon usage (DEU) in our RNA-Seq data revealed a higher number of exons being differentially expressed (visualized by MA plot of the logarithm of fold change versus average normalized counts per exon) in B-EBV cells of normal controls and X-linked CGD patients compared to cells of IFNGRD patients (Figure 3). Supplemental Table 3 presents the top ten genes with highest log2 fold change in DEU due to in vitro treatment with IFN-γ in cells from each analyzed subject. As at the whole transcript level, no significant DEU was detected for any of the NADPH oxidase components; however, a trend towards increased DEU of exon 3 of CYBB in cells from patient D1, with CGD due to a splicing defect, was observed when cells were treated with IFN-γ (Figure 4).
Figure 3: MA plot of each analyzed subject in the normal control group (C1–5 from left to right, top row), CGD patients (D1–4 from left to right, middle row) and of IFNGRD patients (N1–6 from left to right, bottom row).
Mean expression versus log2 fold change plot. Significant hits (at padj < 0.1) are colored in red. Less number of significant exons is detected in the IFNGRD group when compared to the healthy and CGD groups reflecting the impairment of the activation of the IFN-γ signaling pathway.
Figure 4: Fitted expression of CYBB of each analyzed subject in the normal control group (C1–5 from left to right, top row), CGD patients (D1–4 from left to right, middle row) and of IFNGRD patients (N1–6 from left to right, bottom row).
The plot represents the expression estimates from a call to test for Differential Exon Usage (DEU). No significant differential exon usage was detected for CYBB, an interesting change of profile of DEU was detected in CGD patient D1.
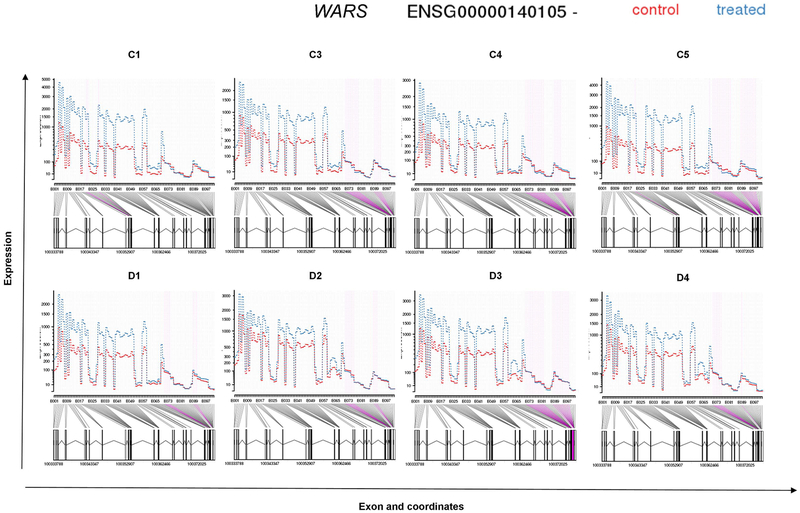
DEU analysis also demonstrated that WARS, the gene consistently up-regulated in B-EBV cells of healthy subjects and CGD patients in the gene level analysis, was also among the top genes with highest increment of DEU in those cells. Figure 5 presents a plot of DEU expression estimates for WARS; purple color indicates exons with significant DEU. No exons were detected differentially expressed in four of the six B-EBV cell lines of IFNGRD patients.
Figure 5: Fitted expression of WARS of each analyzed subject in the normal control group (C1–5 from left to right, top row) and CGD patients (D1–4 from left to right, middle row).
The plot represents the expression estimates from the Differential Exon Usage (DEU) test. In purple are shown the exons that presented significant DEU in normal controls and CGD patients. No significant differential exon usage was detected for WARS in the IFNGRD patients.
Differentially spliced transcripts and isoforms
Quantitation of transcript abundance confirmed the changes in gene expression that were detected as up-regulated on the gene level analysis, which included all known transcript isoforms for each gene. The most abundantly expressed transcripts (APOL6-001, GBP1-001, GBP2-002, GBP4-001, TAP1-001 and UBD-001) indicate induction of these genes at the individual transcript level. Supplemental Table 4 lists the most abundant expressed transcripts upon IFN-γ in vitro treatment for each B-EBV cell line.
Gene set enrichment analysis of biological pathways
Pathway analysis of gene expression data showed that in vitro IFN-γ treatment of B-EBV cells of CGD patients and normal controls activates pathways related to cell adhesion, protein modification processes (e.g. DTX3L targets), cell response to hypoxia conditions (e.g. HIF-1-alpha), immunoproteasome formation (which antigen-presenting cells express in response to oxidative stress and proinflammatory cytokines), and lymphocyte development (e.g. targets of transcription factor IKZF).
Identification of pathways for type II interferon signaling and mRNA processing, even in cells from IFNGRD deficient patients, suggests residual function of the defective IFN-γ receptors in B-EBV cells of IFNGRD deficient patients analysed in our study. In the gene sets of B-EBV cells of IFNGRD1/2 deficient patients, IFN-γ activated a broad array of cellular functions, including cytoskeleton organization, chromatin organization, histone modification, diverse signaling pathways, cell cycle control, nucleosome organization, transcription from RNA polymerase II promoter, transport, post-translational protein modification, ribosomal small subunit assembly, RNA processing, RNA splicing, and RNA transport. These changes indicate an IFN-γ-responsive, transcriptionally active state in cells derived from IFNGRD patients.
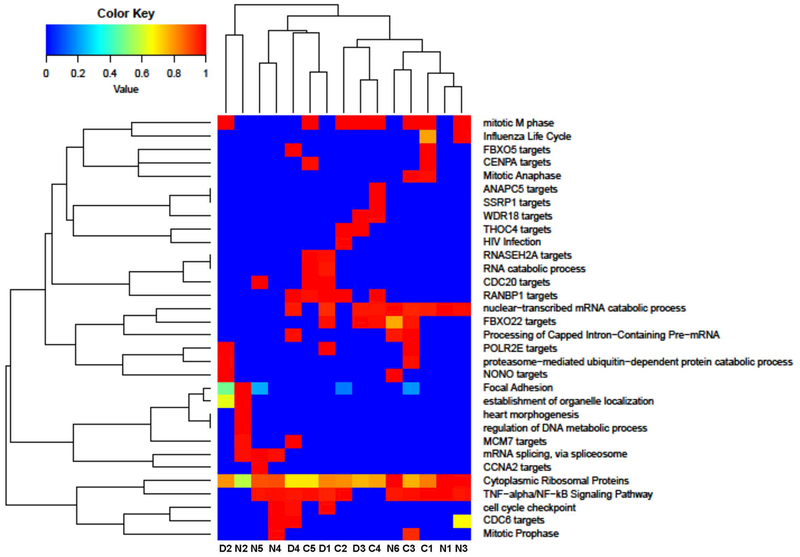
Supplemental Table 5 presents a list of pathways activated upon IFN-γ in vitro treatment. Comparisons of similarity of activated pathways between all samples of the three groups reveal that IFN-γ, EGFR1, TGF-beta receptor, and insulin signaling pathways are active at different intensities in all analyzed samples, as are pathways affecting ribosomal proteins and mRNA processing (Figure 6). These data demonstrate that IFN-γ coordinates a diverse array of cellular programs and is associated with an increased transcriptional active state of immunologically relevant genes through transcriptional regulation, and that IFNGRD1/2 patients may retain residual activity of the IFN-γ receptor.
Figure 6: Comparisons of similarity of activated pathways among samples in normal controls (C1–5), CGD patients (D1–4) and of IFNGRD patients (N1–6).
It was possible to detect that Cytoplasmatic Ribossomal proteins, mRNA processing, Type II Interferon signaling (IFNG), EGFR1 Signaling pathway, TGF-beta Receptor Signaling pathway and Insulin Signaling are present in different intensities in all samples.
DISCUSSION
IFN-γ coordinates a diverse array of cellular programs through transcriptional regulation of immunologically relevant genes [Schroder et al., 2004]; its receptor is expressed ubiquitously on almost all cell types. The striking effects of IFN-γ on neutrophils include increased oxidative burst, differential gene expression, and induction of antigen presentation [Ellis and Beaman, 2004], resulting in increased phagocytosis and cytotoxicity [Perussia et al., 1987; Petroni et al., 1988]. Previous studies from our group demonstrated that expression of the genes encoding phagocyte oxidase components gp91-phox (CYBB) and p47-phox (NCF1) in B-EBV lymphocytes correlated with their NADPH oxidase activity and cytochrome b558 content [Condino-Neto and Newburger, 1998]. Additional studies of our group using B-EBV cell lines as convenient models [Volkman et al., 1984] demonstrated that IFN-γ partially corrects the phenotype of a variant form of X-linked CGD by improving the fidelity of splicing of the first intron of the CYBB gene [Condino-Neto and Newburger, 2000], a finding confirmed by subsequent studies [Ishibashi et al., 2001; Nunoi et al., 2004]. However, the molecular processes responsible for these effects are not known. In the present study, we evaluated differential gene, transcript, and exon expression in B-EBV lymphocytes from normal controls, CGD and IFNGRD patients treated in vitro with IFN-γ, using RNA-Seq
Our current findings demonstrate up-regulation of genes related to lipid trafficking, chemotasis, cell adhesion, regulation of TLR-mediated signaling pathways, transport of antigens, and cellular responses including the regulation of IFN and IFN-inducible genes as well as genes related to antiviral activity, oxidative killing and delivery of antimicrobial peptides to autophagolysosomes. Among the up-regulated genes in B-EBV cell lines of normal controls and X-CGD patients are ones contributing to immune response against infection, such as APOL6, CXCL10, HAPLN3, IRF1, LGALS3BP, STAT1, TAP1, WDFY1 and genes of the GBP family members GBP−1, −2, −4 and −5. Most of the up-regulated genes were also detected as abundant transcripts, thus confirming the induction of these genes. Against our expectations, we also identified differential gene expression and exon usage in IFNGRD B-EBV cell lines despite their genetic defects, therefore suggesting residual function of these receptors in the patient-derived cells.
These results demonstrate that IFN-γ upregulates genes responsible for important cellular mechanisms and immunological functions and thus support the hypothesis that IFN-γ treatment contributes to increased immune responses against pathogens by inducing expression of a broad array of genes with functional relevance to both innate and acquired immune responses. A particularly prominent gene in our analysis, WARS, was up-regulated in B-EBV cells of healthy subjects and CGD patients; additionally it showed differentially expressed exons and the transcript WARS-004 was one of the most abundant expressed transcripts in normal controls and CGD patients. WARS encodes the cytoplasmic enzyme tryptophanyl-tRNA synthetase (TrpRS), which catalyzes the aminoacylation of tRNA(trp) with tryptophan and is known to be induced by IFN-γ [Buwitt et al., 1992; Fleckner et al., 1991]. WARS regulates the ERK, Akt, and eNOS activation pathways that are associated with angiogenesis, cytoskeletal reorganization and shear stress-responsive gene expression [Otani et al., 2002; Tzima et al., 2003; Wakasugi et al., 2002]. In humans, there are two forms of human TrpRS; the truncated isoform (mini TrpRS) induced by in vitro IFN-γ treatment in B-EBV cells is produced by alternative splicing [Tolstrup et al., 1995]. Both mini TrpRS and the full-length enzyme have been reported to be strongly up-regulated by IFN-γ [Tolstrup et al., 1995] and are catalytically active, but are distinguished by the anti-proliferative and anti-angiogenic activity specific to mini TrpRS. In our study, the detection of a lower level of induction by IFN-γ of the exons at the 3’ end of the WARS transcript suggests a possible change of function in the downstream pathway of activities dependent on the type of WARS transcript expressed. Additional studies are required to better assess the functional consequences of IFN-γ modulation of TrpRS expression in immune cells.
B-EBV cells of normal controls and X-linked CGD patients treated with IFN-γ showed decreased expression of genes with relevance to cytokine-mediated functions in apoptosis, regulation of B-cell activation and proliferation, and promotion of initial tethering and rolling of leukocytes over the endothelium. In IFNGRD cell lines, we detected decreased expression of genes with function related to cell growth, survival, proliferation, and motility, as well as a small number with possible neuroprotective functions. Additional functions of down-regulated genes related to cellular responses to DNA damage, bacterial lipopolysaccharide and double-stranded RNA – all involved in modulating immune function by conferring resistance to opportunistic infections. In B-EBV cells derived from IFNGRD patients we also found that some of the down-regulated genes may have additional consequences in transcriptional and post-transcriptional events, such as chromatin organization and histone modification, tRNA processing, mRNA splicing, protein synthesis and protein modification. Other genes – including EIF3A, GABPB1-AS1, HNRNPA2B1 and PCBP1 – down-regulated in B-EBV cells of IFNGR1D/IFNGR2D patients are involved in replication, transport, translation or expression of viral genes, which suggests at least some effect of Epstein-Barr virus on gene expression in this cell line model.
Among the common activated pathways, B-EBV cells from normal controls and from CGD and IFNGRD patients all displayed induction of the “Cytoplasmic Ribosomal Proteins” pathway, suggesting that IFN-γ stimulation induced expression of these proteins in combination with the activation of mRNA processing. Although the events that control IFN-γ-induced transcriptional activation are well characterized, the signals generated by the type II interferon receptor to regulate mRNA translation are still not well elucidated.
A limitation of the current study is its use of RNAseq technology, which may not always provide biologically meaningful gene expression data, especially in cells with regards to genes that present next to baseline expression and in cells with complex genomes. The identification of genes in RNAseq analyses may lead to misinterpretation of results due to over- and/or under-estimated expression levels, requiring a certain level of caution for the interpretation of results for certain genes. In addition, the B-EBV cell line model has limitations compared to primary cells, but major advantages in cell homogeneity and convenience to both patients and investigators. Furthermore, studies of RNA levels do not always correlate well with protein expression, although there is a close association during myeloid development [Lian et al., 2002; Lian et al., 2001], and functional effects are further inferred from those levels.
In summary, the present study reports a comprehensive whole transcriptome analysis of the effects of IFN-γ in vitro treatment of B-EBV cells of X-CGD and IFNGRD patient cell lines. Our data reveal that IFN-γ induced mRNA levels of a vast number of different gene sets related to distinct different cellular programs, including genes with antimicrobial functions of phagocytes (oxidative killing, nitric oxide synthase pathway activation, proteasome-mediated degradation, antigen presentation, chemoattraction, and cell adhesion) that may underlie some of the beneficial effects of IFN-γ therapy in CGD. Important genes relating to mRNA and protein processing were also upregulated in CGD and IFNRD cells suggesting that the regulation of pre-mRNA splicing, folding, transport, and assembly of proteins could also contribute to the effects of IFN-γ treatment, including the partial restoration of normal splicing of splice-mutant CYBB genes [Condino-Neto and Newburger, 2000; Ishibashi et al., 2001; Nunoi et al., 2004]. In addition, changes in expression of WARS induced by IFN-γ suggest that this gene may also contribute to immunomodulatory functions of IFN-γ treatment and merits further investigation.
Supplementary Material
ACKNOWLEDGMENTS
We thank Prof. Dr. Jean-Laurent Casanova for providing cell lines of IFNGR patients; Dr. Maria Zapp and the Deep Sequencing Core Facility of University of Massachusetts Medical School for advice and assistance in RNA-Seq library preparation and sequencing; Dr. Walid Gharib, Dr. Samuel Neuenschwander and Kurt Wyler for scientific advice and support; and the Swiss Institute of Bioinformatics for computational infrastructure.
Grant support: U.S. National Institutes of Health: R01GM115911, FAPESP (Fundação de Amparo à Pesquisa do Estado de São Paulo): 2014/15920–0, 2013/50460–8, 2012/51094–2 and 2008/58840–6.
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicts of interest to report.
REFERENCES
- Anders S, Pyl PT, Huber W. 2015. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31:166–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Reyes A, Huber W. 2012. Detecting differential usage of exons from RNA-seq data. Genome Res 22:2008–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bemiller LS, Roberts DH, Starko KM, Curnutte JT. 1995. Safety and effectiveness of long-term interferon gamma therapy in patients with chronic granulomatous disease. Blood Cells Mol Dis 21:239–47. [DOI] [PubMed] [Google Scholar]
- Bodzioch M, Lapicka-Bodzioch K, Zapala B, Kamysz W, Kiec-Wilk B, Dembinska-Kiec A. 2009. Evidence for potential functionality of nuclearly-encoded humanin isoforms. Genomics 94:247–56. [DOI] [PubMed] [Google Scholar]
- Buwitt U, Flohr T, Bottger EC. 1992. Molecular cloning and characterization of an interferon induced human cDNA with sequence homology to a mammalian peptide chain release factor. EMBO J 11:489–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condino-Neto A, Newburger PE. 1998. NADPH oxidase activity and cytochrome b558 content of human Epstein-Barr-virus-transformed B lymphocytes correlate with expression of genes encoding components of the oxidase system. Arch Biochem Biophys 360:158–64. [DOI] [PubMed] [Google Scholar]
- Condino-Neto A, Newburger PE. 2000. Interferon-gamma improves splicing efficiency of CYBB gene transcripts in an interferon-responsive variant of chronic granulomatous disease due to a splice site consensus region mutation. Blood 95:3548–54. [PubMed] [Google Scholar]
- Delneste Y, Charbonnier P, Herbault N, Magistrelli G, Caron G, Bonnefoy JY, Jeannin P. 2003. Interferon-gamma switches monocyte differentiation from dendritic cells to macrophages. Blood 101:143–50. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Gingeras TR. 2015. Mapping RNA-seq Reads with STAR. Curr Protoc Bioinformatics 51:11 14 1–11 14 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dusi S, Nadalini KA, Donini M, Zentilin L, Wientjes FB, Roos D, Giacca M, Rossi F. 1998. Nicotinamide-adenine dinucleotide phosphate oxidase assembly and activation in EBV-transformed B lymphoblastoid cell lines of normal and chronic granulomatous disease patients. J Immunol 161:4968–74. [PubMed] [Google Scholar]
- Ellis TN, Beaman BL. 2004. Interferon-gamma activation of polymorphonuclear neutrophil function. Immunology 112:2–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezekowitz RA, Dinauer MC, Jaffe HS, Orkin SH, Newburger PE. 1988. Partial correction of the phagocyte defect in patients with X-linked chronic granulomatous disease by subcutaneous interferon gamma. N Engl J Med 319:146–51. [DOI] [PubMed] [Google Scholar]
- Filiz S, Uygun DF, Koksoy S, Sahin E, Yegin O. 2015. In vitro interferon gamma improves the oxidative burst activity of neutrophils in patients with chronic granulomatous disease with a subtype of gp91phox deficiency. Cent Eur J Immunol 40:54–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleckner J, Rasmussen HH, Justesen J. 1991. Human interferon gamma potently induces the synthesis of a 55-kDa protein (gamma 2) highly homologous to rabbit peptide chain release factor and bovine tryptophanyl-tRNA synthetase. Proc Natl Acad Sci U S A 88:11520–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland SM. 2010. Chronic granulomatous disease. Clin Rev Allergy Immunol 38:3–10. [DOI] [PubMed] [Google Scholar]
- Ishibashi F, Mizukami T, Kanegasaki S, Motoda L, Kakinuma R, Endo F, Nunoi H. 2001. Improved superoxide-generating ability by interferon gamma due to splicing pattern change of transcripts in neutrophils from patients with a splice site mutation in CYBB gene. Blood 98:436–41. [DOI] [PubMed] [Google Scholar]
- Ito T, Amakawa R, Inaba M, Ikehara S, Inaba K, Fukuhara S. 2001. Differential regulation of human blood dendritic cell subsets by IFNs. J Immunol 166:2961–9. [DOI] [PubMed] [Google Scholar]
- Johnston RB Jr. 2001. Clinical aspects of chronic granulomatous disease. Curr Opin Hematol 8:17–22. [DOI] [PubMed] [Google Scholar]
- Keller R, Joller P, Keist R, Binz H, van der Meide PH. 1988. Modulation of major histocompatibility complex (MHC) expression by interferons and microbial agents. Independent regulation of MHC class II expression and induction of tumoricidal activity in bone marrow-derived mononuclear phagocytes. Scand J Immunol 28:113–21. [DOI] [PubMed] [Google Scholar]
- Khsheibun R, Paperna T, Volkowich A, Lejbkowicz I, Avidan N, Miller A. 2014. Gene expression profiling of the response to interferon beta in Epstein-Barr-transformed and primary B cells of patients with multiple sclerosis. PLoS One 9:e102331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian Z, Kluger Y, Greenbaum DS, Tuck D, Gerstein M, Berliner N, Weissman SM, Newburger PE. 2002. Genomic and proteomic analysis of the myeloid differentiation program: global analysis of gene expression during induced differentiation in the MPRO cell line. Blood 100:3209–20. [DOI] [PubMed] [Google Scholar]
- Lian Z, Wang L, Yamaga S, Bonds W, Beazer-Barclay Y, Kluger Y, Gerstein M, Newburger PE, Berliner N, Weissman SM. 2001. Genomic and proteomic analysis of the myeloid differentiation program. Blood 98:513–24. [DOI] [PubMed] [Google Scholar]
- Liu J, Cao X. 2016. Cellular and molecular regulation of innate inflammatory responses. Cell Mol Immunol 13:711–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marszalek JR, Weiner JA, Farlow SJ, Chun J, Goldstein LS. 1999. Novel dendritic kinesin sorting identified by different process targeting of two related kinesins: KIF21A and KIF21B. J Cell Biol 145:469–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan CF, Murray HW, Wiebe ME, Rubin BY. 1983. Identification of interferon-gamma as the lymphokine that activates human macrophage oxidative metabolism and antimicrobial activity. J Exp Med 158:670–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newburger PE, Ezekowitz RA. 1988. Cellular and molecular effects of recombinant interferon gamma in chronic granulomatous disease. Hematol Oncol Clin North Am 2:267–76. [PubMed] [Google Scholar]
- Nunoi H, Ishibashi F, Mizukami T, Hidaka F. 2004. Clinical evaluation of interferon-gamma treatment to chronic granulomatous disease patients with splice site mutations. Jpn J Infect Dis 57:S25–6. [PubMed] [Google Scholar]
- Otani A, Slike BM, Dorrell MI, Hood J, Kinder K, Ewalt KL, Cheresh D, Schimmel P, Friedlander M. 2002. A fragment of human TrpRS as a potent antagonist of ocular angiogenesis. Proc Natl Acad Sci U S A 99:178–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J, Zhang M, Wang J, Wang Q, Xia D, Sun W, Zhang L, Yu H, Liu Y, Cao X. 2004. Interferon-gamma is an autocrine mediator for dendritic cell maturation. Immunol Lett 94:141–51. [DOI] [PubMed] [Google Scholar]
- Perussia B, Kobayashi M, Rossi ME, Anegon I, Trinchieri G. 1987. Immune interferon enhances functional properties of human granulocytes: role of Fc receptors and effect of lymphotoxin, tumor necrosis factor, and granulocyte-macrophage colony-stimulating factor. J Immunol 138:765–74. [PubMed] [Google Scholar]
- Petroni KC, Shen L, Guyre PM. 1988. Modulation of human polymorphonuclear leukocyte IgG Fc receptors and Fc receptor-mediated functions by IFN-gamma and glucocorticoids. J Immunol 140:3467–72. [PubMed] [Google Scholar]
- Ramirez-Alejo N, Santos-Argumedo L. 2013. Innate defects of the IL-12/IFN-gamma axis in susceptibility to infections by mycobacteria and salmonella. J Interferon Cytokine Res 34:307–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Routes J, Abinun M, Al-Herz W, Bustamante J, Condino-Neto A, De La Morena MT, Etzioni A, Gambineri E, Haddad E, Kobrynski L, Le Deist F, Nonoyama S, Oliveira JB, Perez E, Picard C, Rezaei N, Sleasman J, Sullivan KE, Torgerson T. 2014. ICON: the early diagnosis of congenital immunodeficiencies. J Clin Immunol 34:398–424. [DOI] [PubMed] [Google Scholar]
- Schroder K, Hertzog PJ, Ravasi T, Hume DA. 2004. Interferon-gamma: an overview of signals, mechanisms and functions. J Leukoc Biol 75:163–89. [DOI] [PubMed] [Google Scholar]
- Seyer L, Greeley N, Foerster D, Strawser C, Gelbard S, Dong Y, Schadt K, Cotticelli MG, Brocht A, Farmer J, Wilson RB, Lynch DR. 2015. Open-label pilot study of interferon gamma-1b in Friedreich ataxia. Acta Neurol Scand 132:7–15. [DOI] [PubMed] [Google Scholar]
- Simillion C, Liechti R, Lischer HE, Ioannidis V, Bruggmann R. 2017. Avoiding the pitfalls of gene set enrichment analysis with SetRank. BMC Bioinformatics 18:151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolstrup AB, Bejder A, Fleckner J, Justesen J. 1995. Transcriptional regulation of the interferon-gamma-inducible tryptophanyl-tRNA synthetase includes alternative splicing. J Biol Chem 270:397–403. [DOI] [PubMed] [Google Scholar]
- Tomassini B, Arcuri G, Fortuni S, Sandi C, Ezzatizadeh V, Casali C, Condo I, Malisan F, Al-Mahdawi S, Pook M, Testi R. 2012. Interferon gamma upregulates frataxin and corrects the functional deficits in a Friedreich ataxia model. Hum Mol Genet 21:2855–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrico F, Heremans H, Rivera MT, Van Marck E, Billiau A, Carlier Y. 1991. Endogenous IFN-gamma is required for resistance to acute Trypanosoma cruzi infection in mice. J Immunol 146:3626–32. [PubMed] [Google Scholar]
- Tzima E, Reader JS, Irani-Tehrani M, Ewalt KL, Schwartz MA, Schimmel P. 2003. Biologically active fragment of a human tRNA synthetase inhibits fluid shear stress-activated responses of endothelial cells. Proc Natl Acad Sci U S A 100:14903–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkman DJ, Buescher ES, Gallin JI, Fauci AS. 1984. B cell lines as models for inherited phagocytic diseases: abnormal superoxide generation in chronic granulomatous disease and giant granules in Chediak-Higashi syndrome. J Immunol 133:3006–9. [PubMed] [Google Scholar]
- Wakasugi K, Slike BM, Hood J, Otani A, Ewalt KL, Friedlander M, Cheresh DA, Schimmel P. 2002. A human aminoacyl-tRNA synthetase as a regulator of angiogenesis. Proc Natl Acad Sci U S A 99:173–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkelstein JA, Marino MC, Johnston RB Jr., Boyle J, Curnutte J, Gallin JI, Malech HL, Holland SM, Ochs H, Quie P, Buckley RH, Foster CB, Chanock SJ, Dickler H. 2000. Chronic granulomatous disease. Report on a national registry of 368 patients. Medicine (Baltimore) 79:155–69. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.