Supplemental Digital Content is available in the text
Keywords: colorectal cancer, epidermal growth factor (EGF), meta-analysis, rs4444903
Abstract
Background:
Colorectal cancer was a complex disease with multiple causative factors including genetic and environmental factors, as well as the interaction of the 2 factors. Relationship between epidermal growth factor (EGF) A61G polymorphism and colorectal cancer risk has been widely investigated previously, whereas results derived from these studies were inconclusive and controversial. The aim of this study was to investigate the association between the EGF A61G polymorphism and colorectal cancer using a meta-analysis of existing literature.
Methods:
Literature search was conducted from PubMed, EMBASE, China National Knowledge Infrastructure, Wanfang, and Cochrane library databases before July 2017. Pooled odds ratios (ORs) with 95% confidence intervals (CIs) were used to evaluate the strength of the association between EGF A61G and colorectal cancer.
Results:
A total of 9 studies that involved 1448 cases and 1928 healthy controls and found allelic (OR = 1.18, P = .04) and recessive models (OR = 1.36, P = .03) of EGF A61G were significantly associated with the risk of colorectal cancer. Stratification analyses by ethnicity indicated that the EGF 61G significantly increased the risk of colorectal cancer in the Caucasian subgroup (OR = 1.24, P = .02), but not in Asian subgroup (OR = 1.12, P = .08). And the frequency of GG genotype of EGF A61G significantly increased in cases than that in healthy controls in both Caucasian (OR = 1.40, P = .04) and Asian subgroups (OR = 1.27, P = .01). Furthermore, the sample sources and genotyping methods seem to have no influence on the correction of EGF A61G and colorectal cancer susceptibility (P > .05).
Conclusion:
The results indicate that EGF A61G might increase the risk of colorectal cancers.
1. Introduction
Colorectal cancer (CRC) is one of the major cause of morbidity, mortality, and death worldwide especially in Caucasian and Asian populations.[1–3] The information on mechanism of this cancer is not clear yet. Previous studies have indicated that lifestyle, personal exposures including dietary, and genetic factors might increase the susceptibility to CRC.[4] Hypothesis supported that genetic factors may be an important contributor to the pathology of colorectal cancer. Alterations in certain key genes have already been linked to increase the risk of CRC.
Recently, large amount of genes have been reported to be associated with the susceptibility of CRC in elevated studies.[5,6] One of the most important cancer-related gene should be the epidermal growth factor (EGF).[7] The EGF gene encodes epidermal growth factor, which performs as a key role in promoting survival, proliferation, and differentiation of epithelial cells by binding to its receptor (EGFR).[8] Shahbazi et al[9] firstl reported a functional variation involving an A-to-G mutation at position 61 (rs4444903 -61A > G) of 5’-untranslated region of the EGF gene was associated with increased EGF expression and risk of malignant melanoma of skin. Subsequently, genetic association between EGF A61G and susceptibility of various cancers such as breast cancer,[10] cervical cancer,[11] lung cancer,[12] hepatocellular carcinoma,[13] gastric cancer,[14] esophageal cancer,[15] and colorectal cancer[16] were identified. Spindler et al firstly investigated association of EGF A61G and CRC in Denmark population and found no significant association.[17] Significant association between this polymorphism and risk of CRC was firstl identified by Wu et al in the German population,[18] while the positive result could not be replicated in most of other populations.[19,20] These discrepancies may be due to insufficient calculated power, different ethnicity, and limited sample sizes in individual studies.
In light of the inconclusive results of the previous studies and the insufficient statistical power of an individual study, we performed a meta-analysis by including the most recent and relevant articles to further evaluate the precise association of EGF +61A > G polymorphism and CRC risk.
2. Methods
2.1. Patient and public involvement
No patient and public involvement and ethical approval is necessary for the present meta-analysis.
2.2. Literature search strategy
This study was performed and reported according to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses guidelines.[21] The PubMed, EMBASE, China National Knowledge Infrastructure, Wanfang, and Cochrane library databases were searched with no language restrictions, using the following terms: “Epidermal growth factor” or “EGF” and “rs4444903” or “61A>G” or “A61G” and “polymorphism” or “variant” or “gene mutation” or “single nucleotide polymorphism (SNP)” or “gene variation” and “colorectal cancer” or “colorectal neoplasms” or “colorectal carcinoma”. In addition, the time period for literature searching was from the first available article until January 01, 2018. And all hits from the databases were screened by title first. Then, the abstracts of articles with titles fulfilling the study criteria were reviewed. Only full-text available articles were included; meeting or conference abstracts were excluded. Additional potentially relevant literature was identified by assessing the reference lists of eligible studies.
2.3. Inclusion/exclusion criteria
Inclusion criteria: studies were included if they fulfilled the following criteria:
-
(1)
case–control and/or cohort studies;
-
(2)
contained SNP genotype data; and
-
(3)
adequate data for the calculation of odds ratios (ORs) and 95% confidence intervals (CIs).
Exclusion criteria: studies were excluded if they:
-
(1)
not regarding the genetic association of EGF polymorphisms and risk of colorectal cancer;
-
(2)
were duplicate publications;
-
(3)
were case reports, letters, commentaries, meeting records, or review articles;
-
(4)
insufficient published data for calculating an OR with 95% CI.
2.4. Data extraction and quality assessment
Data extraction from eligible articles was independently performed by ZH Chen and HG Jiang. The extracted data included first author, year of publication, ethnicity of the study population, number of cases and controls; gender ratio in case and control, ages in case and control, genotyping methods, sample source, Hardy–Weinberg equilibrium in control group, quality scores. Any disagreements were resolved by a consensus achieved by the third author Y Zhu.
BH Lu and HG Jiang accessed the quality of included studies independently as proposed by the Newcastle–Ottawa Scale (NOS).[22] A quality score was calculated from group selection, comparability, and assessment of outcome or exposure. The quality scores ranged from 0 to 10 (0 being the least and 10 being the highest). The methodological quality assessment of the included studies was assessed using the risk-of-bias tool of Review Manager software (version 5.3, Nordic Cochrane Centre, Denmark).[23] Any discrepancies in the assessment were resolved by the third author (Y Zhu).
2.5. Statistical analyses
The Stata software (version 12.0; Stata Corp LP, College Station, TX) and RevMan (version 5.1 Copenhagen: The Nordic Cochrane Centre, The Cochrane Collaboration, 2014) were used in this meta-analysis. Crude OR and 95% CIs were calculated to test the strength of associations between the allelic, dominant, and recessive models of studied polymorphism and CRC susceptibility. The significance of the pooled OR was determined by Z test (P < .05 suggests a significant OR). A test of heterogeneity was conducted using Cochran's Q test and Higgins I-squared statistic. I2 values of >50% indicate heterogeneity among studies. A random effect model was applied if heterogeneity was observed (I2 > 50%, P < .05). Otherwise the fixed effect model was used. Subgroup analysis by ethnicity, genotyping methods, and sample source was also carried out. Sensitivity analysis was performed to assess the effects of each individual study on pooled results. Publication bias was examined visually by the funnel plot, and statistically using the Begg and Egger tests. Power analysis was performed by STATA (https://www.stat.ubc.ca/). And, a trial sequential analysis (TSA) was carried out to estimate the sample size in meta-analysis with the TSA software (www.ctu.dk/tsa/). P < .05 was considered statistically significant.
3. Results
3.1. Characteristics of the eligible studies
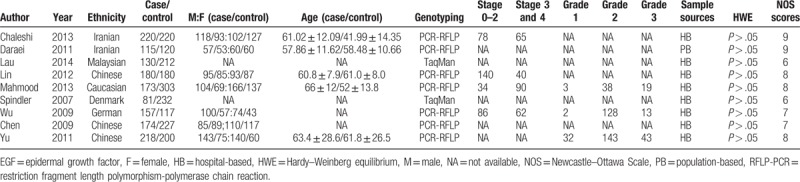
The search process and study selection are presented in Figure 1. A total of 352 articles were identified through the literature search. Three hundred thirty four were excluded as duplicate, irrelevant studies or not original articles, and 18 selected. After full-text scanning, 9 studies were removed for not case–control design, cohort design, or not regarding to the association between EGF A61G and CRC risk. Finally, 9 case–control studies with 1448 cases and 1928 controls were included in the meta-analysis.[16–27] The information for the selected studies was presented in Table 1. And the NOS quality assessment of these included studies is provided in Table 1 and Table s1. Only studies with NOS scores larger than 6 were selected in this meta-analysis.
Figure 1.
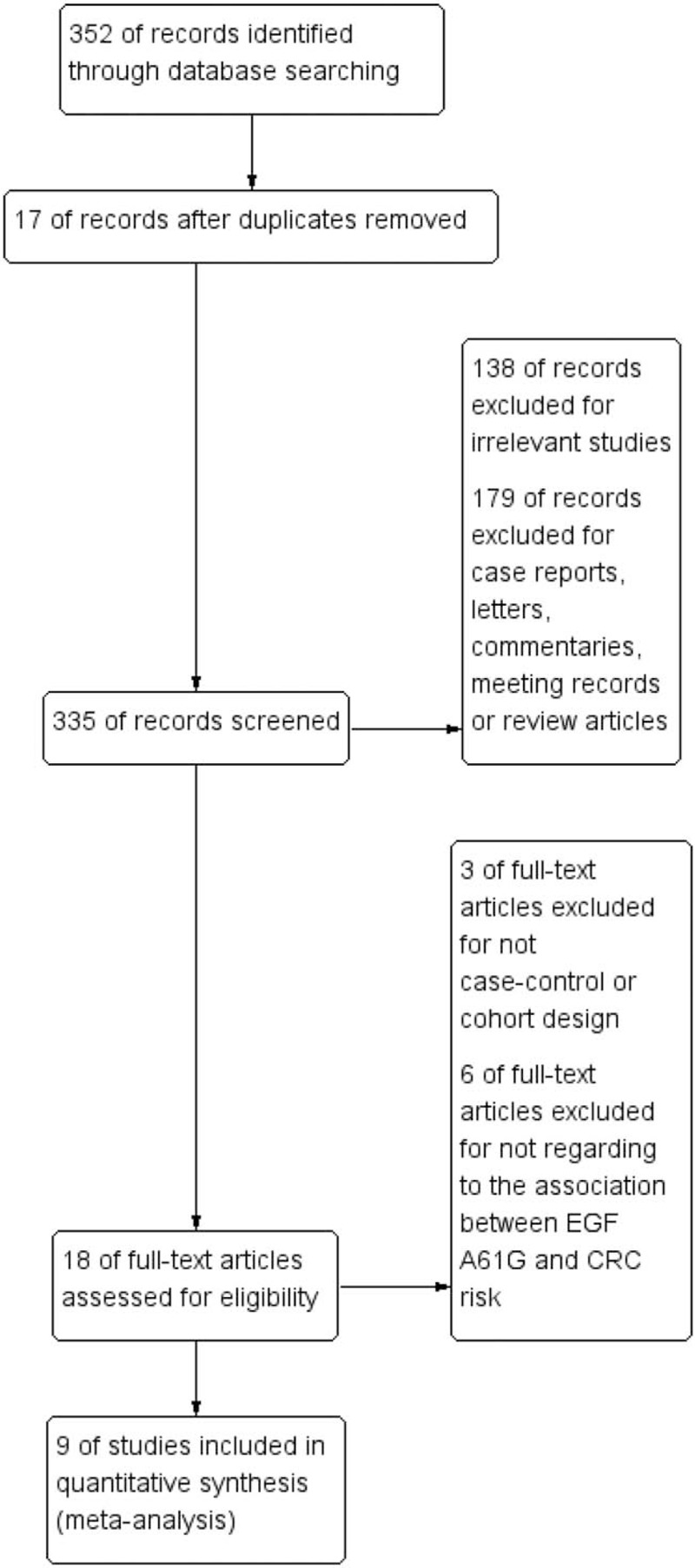
PRISMA flow chart of studies inclusion and exclusion. PRISMA = preferred reporting items for systematic reviews and meta-analyses.
Table 1.
The characters of individual studies.
3.2. Results of meta-analysis
Significant association was detected between allelic and recessive models of EGF A61G and the risk of CRC (allelic model: OR = 1.18, 95% CI [1.01, 1.37], P = .04; recessive model: OR = 1.36, 95% CI [1.04, 1.79], P = .03). And no significant association was observed between EGF A61G and CRC in dominant model (P > .05). Stratification analysis by ethnicity showed that the frequency of EGF 61G allele significantly increased in cases than that in healthy controls in Caucasian (OR = 1.24, 95% CI [1.03, 1.49], P = .02), but not in Asian (OR = 1.12, 95% CI [0.99, 1.27], P = .08). However, the recessive model of EGF A61G was significantly increased in cases than that in healthy control in both Caucasian (OR = 1.40, 95% CI [1.01, 1.93], P = .04) and Asian (OR = 1.27, 95% CI [1.05, 1.53], P = .01). Furthermore, subgroup analysis stratified by sample sources (population-based and hospital-based [HB]) and genotyping methods (restriction fragment length polymorphism-polymerase chain reaction [RFLP-PCR] and TaqMan) showed no significant association between the EGF A61G and CRC susceptibility in allelic, dominant, and recessive models (P > .05) (Table 2 and Fig. 2).
Table 2.
The main results of the association between EGF A61G and colorectal cancer.
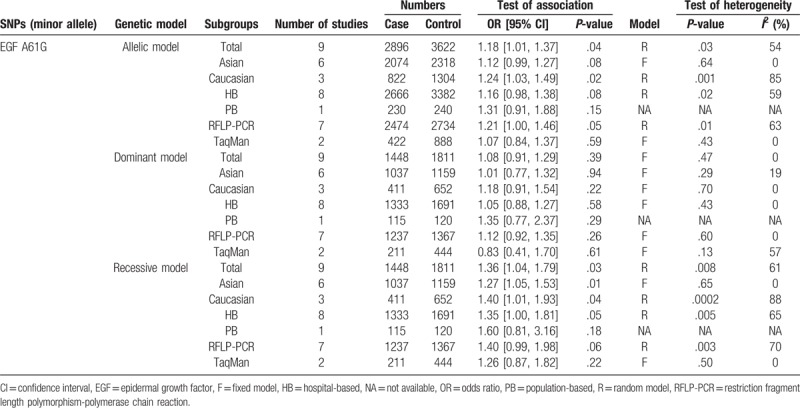
Figure 2.
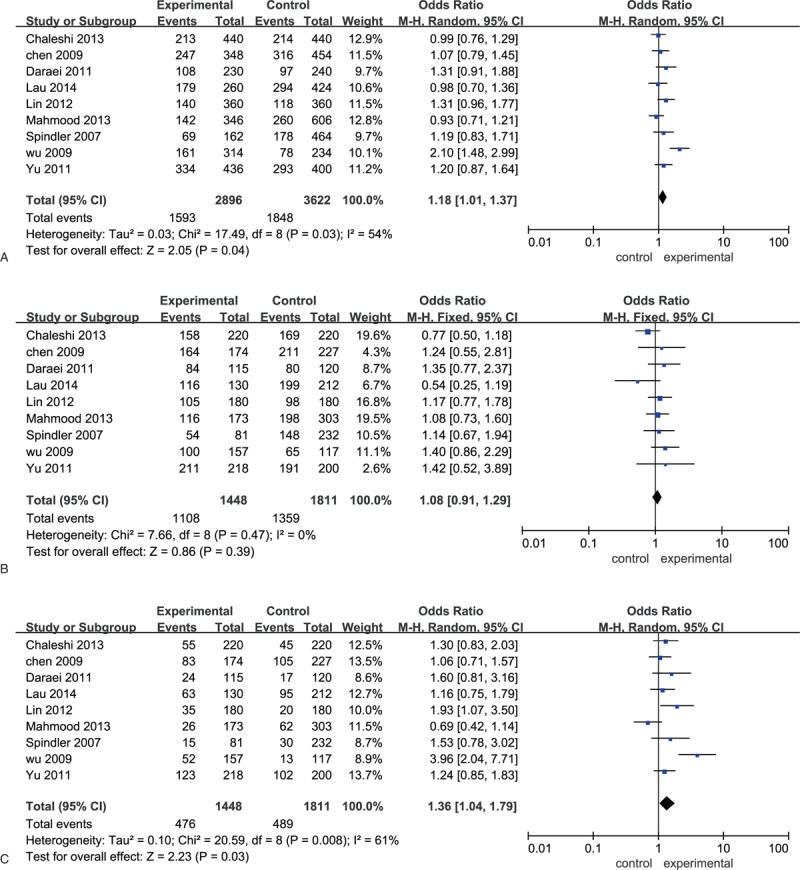
Forest plots of odds ratios for the association between EGF A61G and colorectal cancer A: allelic model; B: dominant model; C: recessive model. EGF = epidermal growth factor.
3.3. Test of heterogeneity
Significant heterogeneity was observed in allelic and recessive models of EGF A61G both in total group (allelic model: I2 = 54, P = .03; recessive model: I2 = 61, P = .008), Caucasian subgroup (allelic model: I2 = 85, P = .001; recessive model: I2 = 88, P = .0002), HB subgroup (allelic model: I2 = 59, P = .02; recessive model: I2 = 65, P = .005), and RFLP-PCR subgroup (allelic model: I2 = 63, P = .01; recessive model: I2 = 70, P = .003). The heterogeneity in all these comparisons was contributed mainly by Wu et al[18]. The removal of this study from the meta-analysis gave 0% to 40% (P > .05) heterogeneity and showed that it had the highest effect on EGF A61G in allelic, dominant, and recessive models in CRC (Table 2).
3.4. Power analysis
It is expected that the limited sample size causing serious power-loss. Before making a conclusion on the heterogeneity, power calculations about the meta-analysis should be performed. The power analysis suggested that power of 91% was determined for rs4444903. As shown in Figure s1, our meta-analysis collected 9 studies with 1448 cases. The required information size for this meta-analysis was 4312 with the indexes: type I error (α = 0.05), type II error (β = .2). However, the results of TSA showed the EGF A61G contribute to the risk of CRC was reliable.
3.5. Sensitivity analysis and publication bias
Sensitivity analysis which excluded the influence of a single study on the overall risk estimate by excluding 1 study at a time was confirmed. The ORs were significantly altered by excluding Wu et al (Fig. 3). After omitted this study, significant association was only detected between the recessive model of EGF A61G and CRC in Asian population (OR = 1.27, 95% CI [1.05, 1.53], P = .01) (Table s2). Sensitivity analysis based on these 8 studies showed that The ORs were not significantly altered in each genetic models (Fig. s2).
Figure 3.
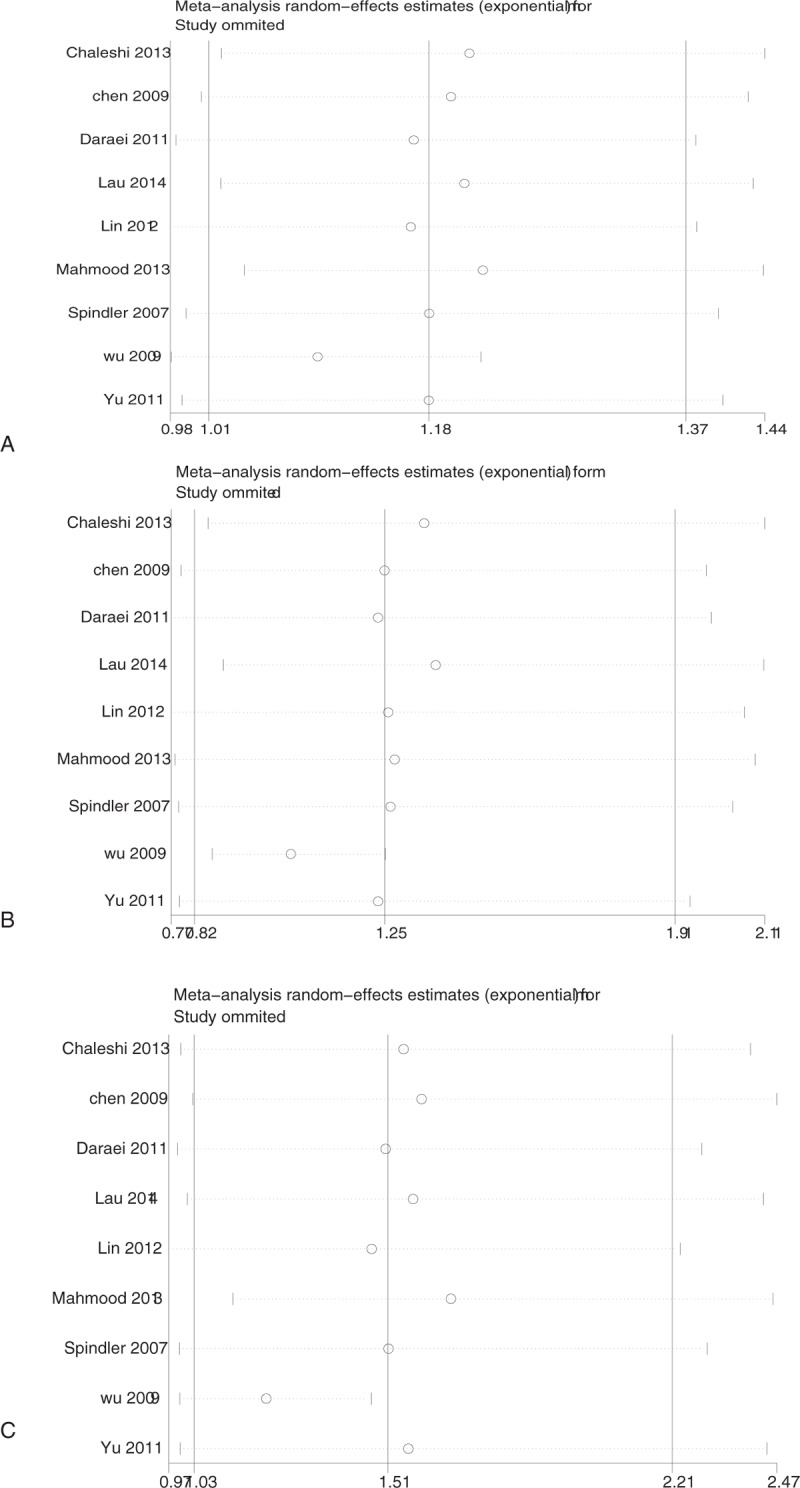
Sensitivity analyses between EGF A61G and colorectal cancer. A: allelic model; B: dominant model; C: recessive model. EGF = epidermal growth factor.
Funnel plots and Egger test were performed to assess publication bias. The results suggested that there was no publication bias for the comparison of EGF A61G in allelic, dominant, and recessive models (Fig. 4).
Figure 4.
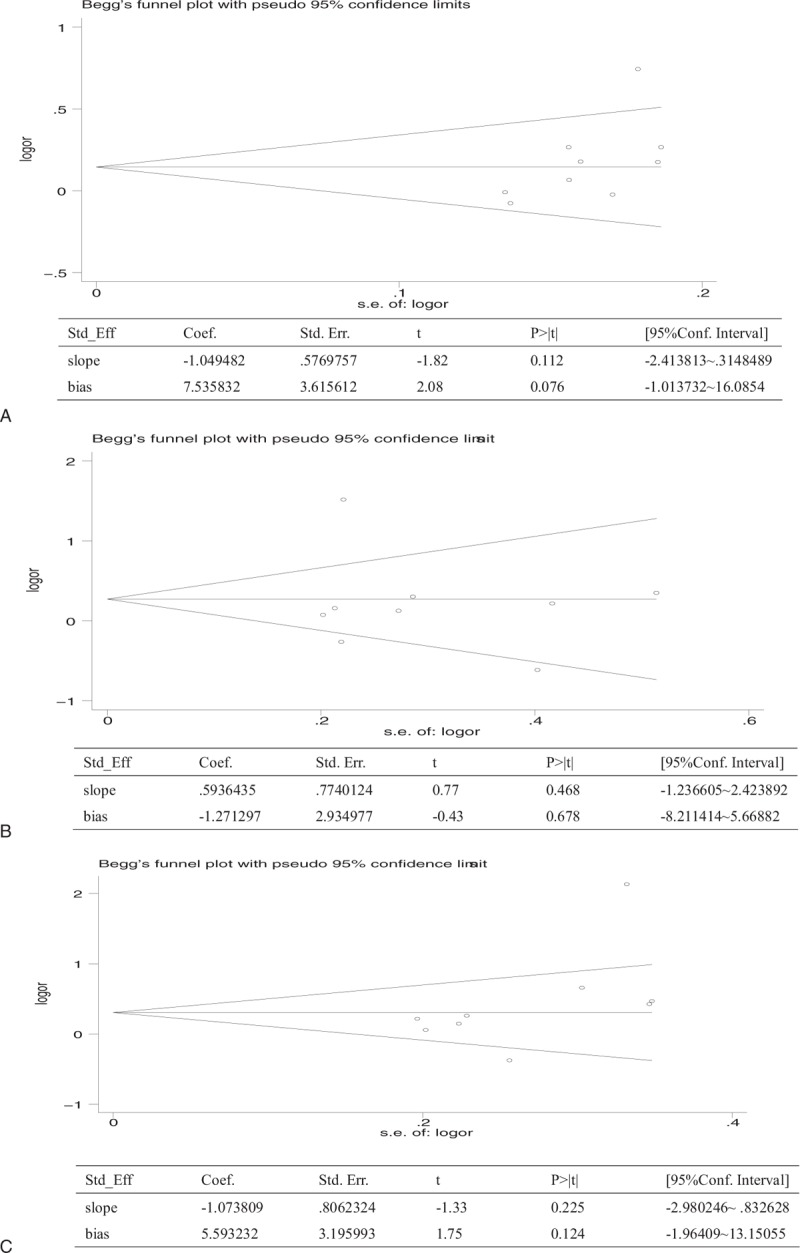
Publication bias of literatures for allelic, dominant and recessive models of EGF A61G were tested by Begg funnel plot and Egger test. A: allelic model; B: dominant model; C: recessive model. EGF = epidermal growth factor.
4. Discussion
This meta-analysis demonstrated that the EGF 61G allele was significantly associated with CRC in the Caucasian subgroup. And the GG genotype of EGF A61G was significantly associated with CRC in both Caucasian and Asian subgroups. The sample source and genotyping methods didn’t affected the significant association between EGF A61G and CRC in allelic, dominant and recessive models. Thus, the EGF A61G might be a risk factor for CRC susceptibility.
CRC is a multistep process, where genetic changes occur with mutations of several genes that accumulate with the progression from a normal epithelium to an adenoma to invasive cancer. The EGFR system is an important mediator in the tumor microenvironment that results in enhanced tumor growth.[28] EGF exerts effects on cell proliferation and differentiation by binding to the tyrosine kinase EGF receptor.[29] Both EGF and EGFR expression have been described to be significantly increased in patients with CRC compared with that in the normal individuals.[30] And the impact of EGF polymorphisms on cancers has been described. Recently, it has been shown that EGF A61G has a functional influence on the expression of EGF gene in CRC patients.[31] Subsequently, the genetic association between EGF A61G and CRC was also investigated by several studies with conflicting results.[16–27]
Notable, 8 of 9 included studies reported no significant association between EGF A61G and the risk of colorectal cancer.[16,17,19,20,24–27] While, positive result was detected between the allelic model of EGF A61G and CRC in our combined analysis, which was similar with the results of meta-analysis that conducted by Li et al[32] and Piao et al.[33] Notable, relatively small number of studies were included in Li et al[32] and Piao et al.[33] Our meta-analysis enrolled in 5 more studies with 1448 cases and 1928 controls and found that the significant association between the allelic model of EGF A61G and CRC can be only identified in the Caucasian subgroup when stratified by ethnicity, which may supply a new information about the influence of this polymorphism in the development of CRC in populations with different genetic background.
In addition, significant association was also observed between the recessive model (GG/AG + AA) of EGF A61G and CRC in both total group and subgroups stratified by ethnicity, which was partly similar with the results reported by Piao et al.[33] The G/G genotype was reported to lead to a higher production of EGF, and thus increase risk of colorectal cancer.[9] The mechanism that the GG genotype of EGF A61G increases the EGF production may due to the following reasons. First, G to A substitution might affect the DNA folding or processing of the mRNA transcript.[9] Second, this polymorphism might be closely linked to a functional polymorphism elsewhere in the gene.[34]
However, we failed to detect association between the dominant model (GG + AG/AA) of EGF A61G and CRC both in total group and subgroups analysis stratified by ethnicity, which was similar with the results reported by Li et al, but contrast to the results conducted by Piao et al. The inconsistent results in the previous meta-analysis may due to the following reasons. Firstl, the number of included studies in previous meta-analysis studies was relatively small. Although we included 6 more studies to investigate the association between the dominant model of EGF A61G and colorectal cancer, negative results were still obtained. Thus, more studies with larger number of cohorts and multiple ethnicity are still necessary. Second, differences in genetic and environmental background exist among different ethnicity. Third, different populations usually have different linkage disequilibrium patterns.
Furthermore, no significant association between the EGF A61G and the susceptibility of CRC was found in subgroup analysis stratified by both sample sources and genotyping methods, which was different from the results in Li et al.[32] This may indicate the sample sources and genotyping methods have no effect on the association between EGF A61G and colorectal cancer. And the small sample size included in study conducted by Li et al[32] might explain this difference.
Despite including case–control studies, the results of the present meta-analysis should be interpreted carefully because of the following limitations. Firstl, limited number of studies and subjects included in this meta-analysis were relatively small. Only 9 eligible studies with 1448 case and 1928 control were included, especially for eligible studies included in subgroups were relatively insufficient. It was necessary to confirm these results by including larger number of case–control studies regarding with the correction of EGF A61G and the risk of colorectal cancer. Second, we included studies only in Caucasian and Asian populations. The results may be need further accessed in multiple ethnicity groups such as African. Third, CRC is a multi-factorial disease. Gene–gene/gene–environment interactions may play important roles in the pathology of colorectal cancer, but most studies lack information about gene–gene/gene–environment interactions.
5. Conclusion
The current meta-analysis suggests an increased risk of CRC for EGF G allele in the Caucasian subgroup and EGF GG genotype in both Caucasian and Asian subgroups. To confirm these results, further study with larger sample size and multiple ethnicities is necessary.
Author contributions
Conceptualization: Yi Zhu.
Data curation: HongGang Jiang.
Funding acquisition: Yi Zhu.
Methodology: ZhiHeng Chen.
Software: HongGang Jiang.
Writing – original draft: Yi Zhu, BoHao Lu.
Writing – review and editing: Yi Zhu, ZhiHeng Chen, HongGang Jiang, BoHao Lu.
Supplementary Material
Footnotes
Abbreviations: CIs = confidence intervals, CRC = colorectal cancer, EGF = epidermal growth factor, HB = hospital-based, NOS = Newcastle–Ottawa Scale, ORs = odds ratios, RFLP-PCR = restriction fragment length polymorphism-polymerase chain reaction, SNP = single nucleotide polymorphism, TSA = trial sequential analysis.
YZ and ZHC contributed equally to this work.
The work was funded by the Foundation of Jiaxing department of Science and Technology (2016BY28010), the Basic Public Welfare Research Program of Zhejiang Province (LGF18H160033), the Medical and Health Science and Technology Project of Zhejiang Province (2019KY214), the No. 1 project of the first hospital of Jiaxing (2017-YA-52), and the innovation fund of Zhejiang colorectal cancer (2016).
The author(s) of this work have nothing to disclose.
Supplemental Digital Content is available for this article.
References
- [1].Siegel R, Naishadham D, Jemal A. Cancer statistics. CA Cancer J Clin 2012;62:10–29. [DOI] [PubMed] [Google Scholar]
- [2].Ko KP, Yeo Y, Yoon JH, et al. Plasma phytoestrogens concentration and risk of colorectal cancer in two different Asian populations. Clin Nutr 2017;96:1675–82. [DOI] [PubMed] [Google Scholar]
- [3].Choi BJ, Jeong WJ, Kim SJ, et al. Impact of obesity on the short-term outcomes of single-port laparoscopic colectomyfor colorectal cancer in the Asian population: a retrospective cohort study. Medicine 2017;96: e6649 (1–5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Haggar FA, Boushey RP. Colorectal cancer epidemiology: incidence,mortality, survival, and risk factors. Clin Colon Rectal Surg 2009;22:191–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Djansugurova L, Zhunussova G, Khussainova E, et al. Association of DCC, MLH1, GSTT1, GSTM1, and TP53 gene polymorphisms with colorectal cancer in Kazakhstan. Tumour Biol J Int Soc Oncodev Biol Med 2015;36:279–89. [DOI] [PubMed] [Google Scholar]
- [6].Wang ML, Gu DY, Du ML, et al. Common genetic variation in ETV6 is associated with colorectal cancer susceptibility. Nat Commun 2016;7:11478 (1–9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Kovar FM, Thallinger C, Marsik CL, et al. The EGF 61A/G polymorphism – a predictive marker for recurrence of liver metastases from colorectal cancer. Wien Klin Wochenschr 2009;121:638–43. [DOI] [PubMed] [Google Scholar]
- [8].Peng Q, Li S, Qin X, et al. EGF +61A/G polymorphism contributes to increased gastric cancer risk: evidence from a meta-analysis. Cancer Cell Int 2014;14:134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Shahbazi M, Pravica V, Nasreen N, et al. Association between functional polymorphism in EGF gene and malignant melanoma. Lancet 2002;359:397–401. [DOI] [PubMed] [Google Scholar]
- [10].Matsuda N, Lim B, Wang X, et al. Early clinical development of epidermal growth factor receptor targeted therapy in breast cancer. Expert Opin Investig Drugs 2017;26:463–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Araújo AP, Catarino R, Ribeiro R, et al. Epidermal growth factor genetic variation associated with advanced cervical cancer in younger women. Am J Clin Oncol 2012;35:247–50. [DOI] [PubMed] [Google Scholar]
- [12].Liu TC, Hsieh MJ, Wu WJ, et al. Association between survivin genetic polymorphisms and epidermal growth factor receptor mutation in non-small-cell lung cancer. Int J Med Sci 2016;13:929–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Jiang GP, Yu K, Shao LF, et al. Association between epidermal growth factor gene +61A/G polymorphism and the risk of hepatocellular carcinoma: a meta-analysis based on 16 studies. BMC Cancer 2015;15:1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Araújo AP, Costa BM, Pinto-Correia AL, et al. Association between EGF +61A/G polymorphism and gastric cancer in Caucasians. World J Gastroenterol 2011;17:488–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Cui L, Pan XM, Ma CF, et al. Association between epidermal growth factor polymorphism and esophageal squamous cell carcinoma susceptibility. Dig Dis Sci 2009;55:40–5. [DOI] [PubMed] [Google Scholar]
- [16].Daraei A, Salehi R, Salehi M, et al. Effect of rs6983267 polymorphism in the 8q24 region and rs4444903 polymorphism in EGF gene on the risk of sporadic colorectal cancer in Iranian population. Med Oncol 2012;29:1044–9. [DOI] [PubMed] [Google Scholar]
- [17].Spindler KLG, Nielsen JN, Ornskov D, et al. Epidermal growth factor (EGF) A61G polymorphism and EGF gene expression in normal colon tissue from patients with colorectal cancer. Acta Oncol 2007;46:1113–7. [DOI] [PubMed] [Google Scholar]
- [18].Wu GY, Hasenberg T, Magdeburg R, et al. Association between EGF, TGF-beta1, VEGF gene polymorphism and colorectal cancer. World J Surg 2009;33:124–9. [DOI] [PubMed] [Google Scholar]
- [19].Chen YF, Qi P, Ruan CP, et al. Relationship between polymorphism of epidermal growth factor 5’UTR variant G61A gene and colorectal in patients in China. Pract J cancer 2009;24:551–4. [Google Scholar]
- [20].Lin L, Li G, Zhang Z, et al. Association of epidermal growth factor +61 A/G polymorphism in Chinese patients with colon cancer. Genet Test Mol Biomarkers 2012;16:1142–5. [DOI] [PubMed] [Google Scholar]
- [21].Knobloch K, Yoon U, Vogt PM. Preferred reporting items for systematic reviews and meta-analyses (PRISMA) statement and publication bias. J Craniomaxillofac Surg 2011;39:91–2. [DOI] [PubMed] [Google Scholar]
- [22].Stang A. Critical evaluation of the Newcastle–Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol 2010;25:603–5. [DOI] [PubMed] [Google Scholar]
- [23].Robertson C, Ramsay C, Gurung T, et al. Practicalities of using a modified version of the Cochrane Collaboration risk of bias tool for randomised and non-randomised study designs applied in a health technology assessment setting. Res Synth Methods 2014;5:200–11. [DOI] [PubMed] [Google Scholar]
- [24].Mahmood S, Sivoňová MK, Matáková T, et al. Association of EGF and p53 gene polymorphisms and colorectal cancer risk in the Slovak population. Cent Eur J Med 2014;9:405–16. [Google Scholar]
- [25].Yu XF, Weng MW. Association between epidermal growth factor (EGF) G61A polymorphism and the susceptibility as well as clinicopathological characteristics of colorectal cancer. J Med Res 2011;40:92–4. [Google Scholar]
- [26].Lau TP, Roslani AC, Lian LH, et al. Association between EGF and VEGF functional polymorphisms and sporadic colorectal cancer in the Malaysian population. Genet Mol Res 2014;13:5555–61. [DOI] [PubMed] [Google Scholar]
- [27].Chaleshi V, Haghighi MM, Javadi GR, et al. The effect of 5′untranslated region polymorphism in EGF gene, rs4444903, on colorectal cancer. Gastroenterol Hepatol Bed Bench 2013;6:129–35. [PMC free article] [PubMed] [Google Scholar]
- [28].De LA, Carotenuto A, Rachiglio A, et al. The role of the EGFR signaling in tumor microenvironment. J Cell Physiol 2008;214:559–67. [DOI] [PubMed] [Google Scholar]
- [29].Boonstra J, Rijken P, Humbel B, et al. The epidermal growth factor. Cell Biol Int 2013;19:413–30. [DOI] [PubMed] [Google Scholar]
- [30].Pryczynicz A, Guzinskaustymowicz K, Kemona A, et al. Expression of EGF and EGFR strongly correlates with metastasis of pancreatic ductal carcinoma. Anticancer Res 2008;28:1399–404. [PubMed] [Google Scholar]
- [31].Spindler KL, Andersen RF, Jensen LH, et al. EGF61A>G polymorphism as predictive marker of clinical outcome to first-line capecitabine and oxaliplatin in metastatic colorectal cancer. Ann Oncol 2010;21:535–9. [DOI] [PubMed] [Google Scholar]
- [32].Li TF, Ren KW, Liu PF. Meta-analysis of epidermal growth factor polymorphisms and cancer risk: involving 9,779 cases and 15,932 controls. DNA Cell Biol 2012;31:568–74. [DOI] [PubMed] [Google Scholar]
- [33].Piao Y, Liu Z, Ding Z, et al. EGF +61A>G polymorphism and gastrointestinal cancer risk: a HuGE review and meta-analysis. Gene 2013;519:26–33. [DOI] [PubMed] [Google Scholar]
- [34].Jin G, Miao R, Deng Y, et al. Variant genotypes and haplotypes of the epidermal growth factor gene promoter are associated with a decreased risk of gastric cancer in a high-risk Chinese population. Cancer Sci 2010;98:864–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


