Abstract
Kinetoplastid parasites, included Trypanosoma cruzi, the causal agent of Chagas disease, present a unique genome organization and gene expression. Although they control gene expression mainly post-transcriptionally, chromatin accessibility plays a fundamental role in transcription initiation control. We have previously shown that High Mobility Group B protein from Trypanosoma cruzi (TcHMGB) can bind DNA in vitro. Here, we show that TcHMGB also acts as an architectural protein in vivo, since the overexpression of this protein induces changes in the nuclear structure, mainly the reduction of the nucleolus and a decrease in the heterochromatin:euchromatin ratio. Epimastigote replication rate was markedly reduced presumably due to a delayed cell cycle progression with accumulation of parasites in G2/M phase and impaired cytokinesis. Some functions involved in pathogenesis were also altered in TcHMGB-overexpressing parasites, like the decreased efficiency of trypomastigotes to infect cells in vitro, the reduction of intracellular amastigotes replication and the number of released trypomastigotes. Taken together, our results suggest that the TcHMGB protein is a pleiotropic player that controls cell phenotype and it is involved in key cellular processes.
Introduction
Trypanosoma cruzi is a hemoflagellate protozoan parasite and it is the causative agent of Chagas disease, a neglected disease endemic in Latin America. There are currently limited options for safe pharmacological treatment: the only available drugs for treatment, Benznidazole and Nifurtimox, have proven to be efficient during the acute phase while its use during the chronic phase is still controversial1. T. cruzi has a complex life cycle that alternates between replicative and non-replicative forms in insect and mammalian hosts. Intracellular amastigotes and bloodstream trypomastigotes are present in the mammalian host, whereas epimastigotes and the infective metacyclic trypomastigotes are present in insect vectors from the Reduviidae family2. Through its life cycle, the trypanosome’s nuclear structure undergoes several changes. The epimastigote form, which divides by binary fission in the triatomine insect gut, presents a rounded nucleus with a defined nucleolus and relatively small amounts of peripheral heterochromatin. A similar pattern is found in the intracellular amastigotes nuclei. On the other hand, the non-replicative trypomastigote forms, exhibit an elongated nucleus, no identifiable nucleolus and heterochromatin distributed quite homogeneously through the whole nucleoplasm. These changes are accompanied by a decrease in transcription rates when the replicative forms transform into trypomastigote forms3,4. It is not fully understood, however, how these differences in the nuclear structure are achieved during the differentiation process.
High Mobility Group B (HMGB) proteins are highly abundant ubiquitous non-histone chromatin proteins. They play fundamental roles both inside the nucleus, where they act as architectural factors and outside the cell, where they function as alarmins participating in cell signaling and inflammation5–7. These proteins possess one or two HMG-box domains capable of recognizing and binding altered DNA structures with high affinity. Upon binding, HMGBs bend the DNA helix thus being able to alter the chromatin structure. Thus, HMGBs are considered architectural factors and they are involved in key nuclear processes like transcriptional control, DNA replication, recombination and repair8,9. Mammalian HMGB1, as well as most higher eukaryotic HMGBs, bear two “HMG-box” domains in tandem named “A-box” and “B-box” followed by about 30 glutamic and aspartic amino acids known as the “C-terminal acidic tail”, which modulates the DNA-binding properties and functioning of these proteins10. Kinetoplastid parasites, including the Trypanosoma cruzi, also encode ortholog proteins from the HMGB family with two HMG box domains, in contrast to other HMGBs identified so-far in protozoan parasites like Plasmodium falciparum, Toxoplasma gondii, and Entamoeba histolytica that bear only one HMG-box11–14. The HMGBs from kinetoplastid protozoa lack the typical acidic tail in the C-terminus, and have, instead, a unique sequence of 110 amino acids in the N-terminus conserved among trypanosomatid HMGBs and absent in all other HMGB family members. According to Pfam (http://pfam.sanger.ac.uk/) and SUPERFAMILY (http://supfam.cs.bris.ac.uk/SUPERFAMILY/), the trypanosomatid HMGBs contain a “DEK-C terminal domain”, defined as a DNA binding structural domain found in the C-terminal region of the chromatin-associated oncoprotein DEK15. This N-terminal region also bears a predicted Nuclear Localization Signal (NLS), which differs, in sequence and in location, from human HMGB1’s NLSs16.
In our previous work, we demonstrated that TcHMGB is a nuclear protein expressed in all T. cruzi life cycle stages. Interestingly, replicative forms of the parasite showed higher levels of TcHMGB than the infective non replicative trypomastigote forms, what may be related to the chromatin structure changes in the parasite16. Using a recombinant TcHMGB protein, we showed that the T. cruzi HMGB, has architectural features like the ability to bend linear DNA and to bind non-canonical structures16. Finally, we also showed that TcHMGB can act as an inflammatory mediator like other HMGB family members17. Then, we decided to perform functional studies for TcHMGB in the parasite. The complete genome sequence of T. cruzi has been published in 2005 allowing genome-wide and in silico studies18. However, many biological aspects of this parasite remain unveiled due to its unusual characteristics and genome complexity and because the available tools for genetic manipulation of T. cruzi are relatively scarce, particularly compared to other members of the trypanosomatid family, such as Trypanosoma brucei19,20. The toolkit available to be used in T. cruzi research is limited to a low number or episomal and integrative constitutive expression vectors and the tetracycline (Tet)-inducible system based on plasmid pTcINDEX21. The RNA interference system is not active in T. cruzi and gene knock out by homologous recombination is very inefficient. Recently, CRISPR/Cas9 nuclease system has been used to disrupt several genes in T. cruzi, but it cannot be used to obtain stable null mutants of essential genes22–24. In fact, obtaining stable knock-outs may be almost impossible even if the gene of interest is not completely essential but its deletion confers a selective disadvantage or impaired growth preventing knock out selection.
Therefore, in the present work we used the pTcINDEX-GW inducible vector (a GATEWAY® Cloning Technology-adapted pTcINDEX version built in our laboratory) to continue studying TcHMGB in vivo25. Data showed that TcHMGB can alter the nuclear structure in T. cruzi epimastigotes and seems to be important for fundamental processes like replication, cell cycle progression, infection and metacyclogenesis. Overexpression of TcHMGB caused a marked decrease in epimastigotes growth and an accumulation of parasites in G2/M phase, probably associated to a failure in cytokinesis. Also, we observed a decrease in the infection rate in Vero cells, amastigotes proliferation and the number of trypomastigotes released from infected cells in vitro in TcHMGB-overexpressing parasites. These results suggest that, as well as other HMGB family members, the T. cruzi HMGB can be considered as a pleiotropic factor involved in key cellular processes that may play a role in Chagas disease pathogenesis.
Results
Nuclear ultrastructure and chromatin state are affected by TcHMGB protein levels
To gain insight into the TcHMGB protein function in vivo, we constructed parasite strains capable of overexpressing TcHMGB. We used the inducible vector pTcINDEX-GW21,25 to obtain transgenic parasites expressing the protein fused to a C-terminal double hemagglutinin tag (HA)2 under the control of a Tetracycline (Tet)-inducible promoter. Overexpression after Tet-induction was tested by Western blot and qRT-PCR, showing approximately 12-fold protein overexpression and ∼200 change fold in mRNA levels relative to non-induced control in epimastigotes (Fig. S1A,B). We also analyzed the plasmid-derived protein expression by western blot with antibodies directed to the HA-tag and confirmed that TcHMGB protein is overexpressed in all life cycle stages and there is almost no leaky expression in the absence of tetracycline (Fig. S1C). To rule out possible side-effects of the overexpression induction, we constructed a control strain that, under the same conditions, overexpresses a mutant TcHMGB that bears the two HMGB-box domains characteristic of the HMGB family but lacks the N-terminal domain necessary to direct the protein to the nucleus (Dm28c/pTcINDEX-GW-ΔN-TcHMGB(HA)2) (see Fig. S2).
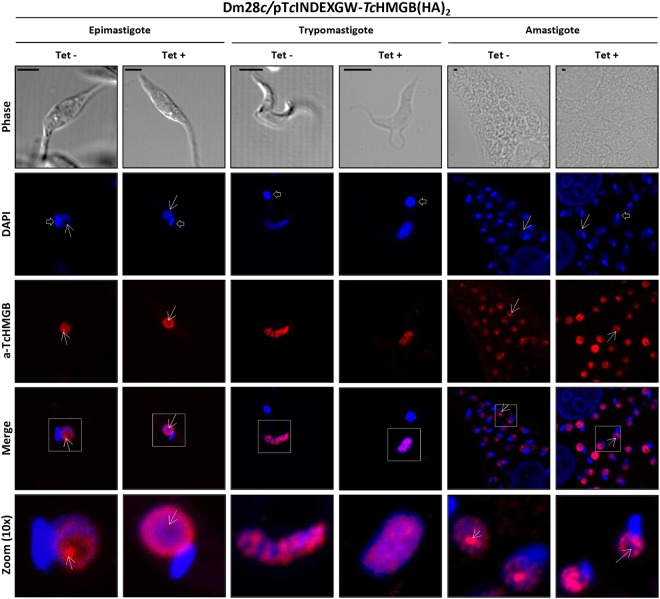
Like other HMGB proteins, included the endogenous TcHMGB16, the plasmid-derived TcHMGB(HA)2 was directed to the nucleus, giving a strong nuclear signal by immunofluorescence confocal microscopy both using specific antibodies and commercial anti-HA monoclonal antibodies in all the parasite life cycle stages (Fig. S1D). In contrast, the truncated protein localized in the cytoplasm as expected (Fig. S2D). Surprisingly, despite keeping the nuclear localization, TcHMGB showed a different labeling pattern after Tet-induction, being particularly evident in epimastigote and amastigote forms (Fig. 1). Under normal (non-induced) conditions, TcHMGB appears as a strong signal localized in the nucleolus accompanied by other spots of lower intensity distributed through the nucleoplasm in both replicative forms. In contrast, in Tet-induced TcHMGB-overexpressing epimastigotes, the protein concentrates adjacent to the nuclear envelope and appears to be excluded from the nucleolus. Similarly, in overexpressing amastigotes, TcHMGB label is seen as fluorescent spots of similar intensity regularly distributed along the whole nucleus except for the nucleolar region, from where the protein seems to be also excluded. In trypomastigotes, the non-replicative stage, there is no defined nucleolar structure and TcHMGB is more regularly distributed through the nucleus. However, the protein localization also changed after Tet-induction: while in non-induced trypomastigotes TcHMGB concentrates in discrete nuclear territories (regularly distributed in the nucleus), the fluorescent label appears more diffuse in overexpressing parasites.
Figure 1.
TcHMGB overexpression affects nuclear structure and the protein localization. Confocal Immunofluorescence microscopy of non-induced (Tet−) and induced (Tet+) (0.5 μg/ml tetracycline, 12 h) T. cruzi Dm28c/pTcINDEX-GW-TcHMGB(HA)2 epimastigotes, trypomastigotes and amastigotes. The nucleus and kinetoplast were labeled with DAPI (blue); rabbit anti-TcHMGB (a-TcHMGB) was revealed with Cy3-conjugated anti-rabbit IgG antibodies (red). Arrows indicate the nucleolar region and open arrows the kinetoplast. Scale bar: 1 μm.
It is worth noting the apparent relationship between TcHMGB and the nucleolus, whose primary function is rRNA transcription and ribosomes assembly. The nucleolus can be easily identified in DAPI (4′,6-diamidino-2-phenylindole)-stained cells as the nuclear region without labeling, just because this region is essentially composed by ribonucleoproteins. Interestingly, this nuclear subdomain appears to be reduced in TcHMGB-overexpressing epimastigotes (Fig. 1).
Transmission electron microscopy (TEM) images of Tet-induced parasites (Fig. 2A panels D to F), also showed a reduction of the nucleolus (nu) associated to TcHMGB overexpression, when compared to non-induced parasites (Fig. 2A panels A to C). This nucleolar reduction was confirmed by area measurements performed on TEM images (Table 1). In relation to non-induced (Tet−) parasites, the overexpressing epimastigotes (Tet+) presented a 28% increase in the total nuclear area. Considering nuclear domains, the euchromatin (eu) area augmented in 109%, whereas the nucleolus was reduced in a 55%. The heterochromatin (ht) area remained the same in both cell types, suggesting that the increase value obtained for euchromatin is not due to DNA unpacking, but results from nucleolar disorganization and the increase in the total nuclear area.
Figure 2.
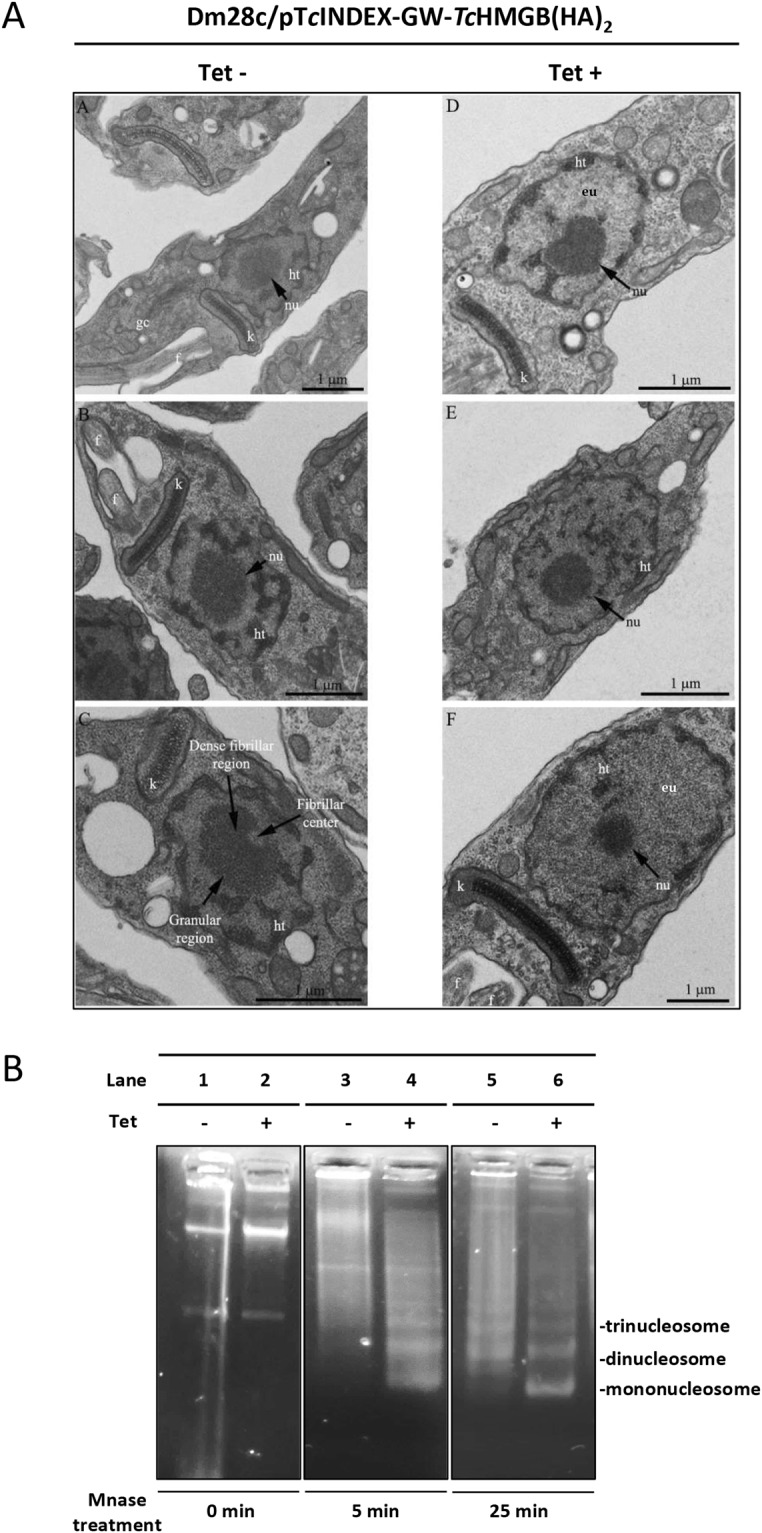
Nuclear ultrastructure and chromatin state are affected by TcHMGB protein levels. (A) Transmission Electron Microscopy (TEM) analysis of the ultrastructure of the T. cruzi Dm28c/pTcINDEX-GW-TcHMGB(HA)2 epimastigotes in the absence (Tet−, Panels A–C) or presence (Tet+, Panels D–F) of tetracycline. Note the augmented space occupied by euchromatin (eu) and the reduction of the nucleolus in induced (Tet+) (E,F) in relation to non-induced parasites (Tet−) (B,C). Arrows indicate the nucleolus and in panel C its distinct domains. Kinetoplast (k), flagellum (f), Golgi complex (gc) and the different nucleolus regions are also indicated. (B) Analysis of chromatin isolated from T. cruzi Dm28c/pTcINDEX-GW-TcHMGB(HA)2 epimastigotes non-induced (−, lanes 1, 3, 5) or induced (+, lanes 2, 4, 6) with tetracycline (Tet). Chromatin from an equal number of cells was isolated and digested with 1 unit of micrococcal nuclease (MNase) for 0 (lanes 1, 2), 5 (lanes 3, 4) and 25 minutes (lanes 5, 6). Equal amounts of DNA were loaded on an ethidium bromide-stained 1% agarose gel. Note that in Tet−induced parasites, the characteristic ladder pattern is observed at shorter times compared to non-induced (Tet−). A representative experiment is shown, digestion products corresponding to DNA that has been bound to mono- di- and tri-nucleosomes are indicated. The corresponding full-length gel is presented in Supplementary Fig. S4.
Table 1.
Measurements of the total nucleus area and nucleolar domains in µm2.
| Measured area (μm2) | Nucleus | Nucleolus | Heterochromatin | Euchromatin |
|---|---|---|---|---|
| Tet− | 1.753 ± 0.112 | 0.475 ± 0.299 | 0.590 ± 0.037 | 0.687 ± 0.102 |
| Tet+ | 2.245 ± 0.109 | 0.215 ± 0.073 | 0.590 ± 0.086 | 1.440 ± 0.182 |
| Percentage change | +28% | −55% | — | +109% |
| Statistically different? | Yes | Yes | No | Yes |
One hundred cells were measured in each one of three independent experiments. Statistics were calculated using the unpaired t test with Welch’s correction in GraphPad Prism 6 software. P values less than 0.05 were considered statistically significant.
Afterwards, we investigated the effect of TcHMGB overexpression on the chromatin structure using micrococcal nuclease enzyme (MNase). MNase preferentially cleaves the double stranded naked DNA located between nucleosomes, thus giving a typical ladder pattern corresponding to mono-, di-, tri- and so on -nucleosomal species in DNA gel electrophoresis (Fig. 2B). A more relaxed or open chromatin structure is more accessible to the enzyme and consequently more prone to be digested than highly condensed chromatin. Accordingly, we verified that overexpression of TcHMGB after Tet-induction results in a chromatin more sensitive to MNase treatment evidenced by the typical ladder pattern obtained at shorter times compared to non-induced control. Indeed, after a 5 minute-treatment, the smallest band corresponding to one nucleosome-DNA is already visible in the induced sample (Fig. 2B, lane 4) while, in the non-induced control, longer times are required to observe a similar pattern (Fig. 2B, lane 5). The enrichment in smaller fragments and disappearance of larger DNA molecules after 25 min treatment in the induced sample (Fig. 2B, lane 6) means that the MNase was able to reach more spacer DNA between nucleosomes. This data is in accordance with the increased area of euchromatin observed by TEM in TcHMGB overexpressing cells.
TcHMGB overexpression decreases cell proliferation and affects cell division and morphology of epimastigotes
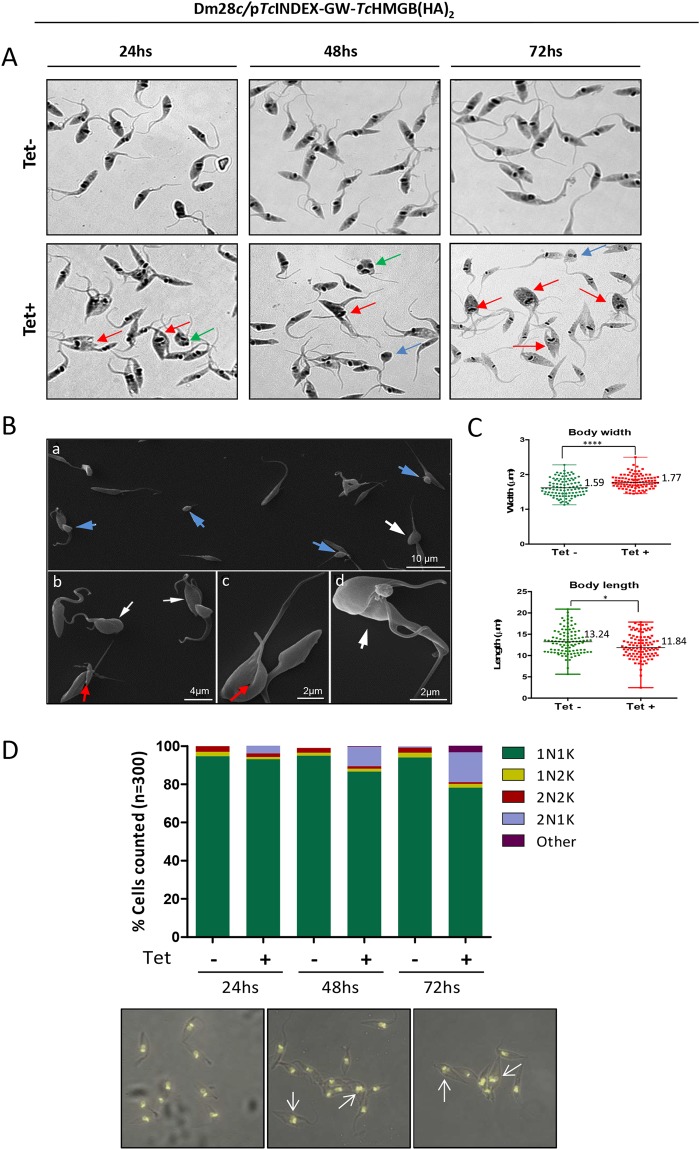
The TcHMGB overexpression clearly affected epimastigote growth, as can be seen in Fig. 3. Indeed, as soon as 24 hs post-induction, the parasite number appears reduced in induced cultures compared to the non-induced ones. Even though the difference seems minimal at this time point (2.12 × 106 vs 2.58 × 106 parasites/ml, that is only 0, 46 parasites/ml difference), it is already statistically significant. TcHMGB-overexpressing epimastigotes proliferation was slower in relation to control (non-induced) cells during the whole growth curve, increasing the difference in the parasite number with time. At day 2 post-induction (pi), the average difference in the number of parasites was 2.7 × 106 parasites/ml, reaching 43 × 106 parasites/ml at day 7 pi (comparing the mean values in each time point). This growing difference was not seen after Tet-treatment of wild type parasites or those transfected with pTcINDEX-GW-GFP (Fig. S3) or other constructions in the same pTcINDEX vector25–28. Regarding the overexpression of the mutant TcHMGB, although we observed a slight delay in the growth curve, the duplication time of the induced vs. non-induced parasites, doesn’t show a significant difference until the fifth day post-induction and the difference in parasite number in each time point is low during the whole period (0.24 × 106 parasites/ml at day 2 pi to 5 × 106 parasites/ml at day 7 pi) (Fig. S2). This data suggests that the higher levels of TcHMGB inside the nucleus and the associated changes in the nuclear and chromatin structure are responsible for the growing defect observed for TcHMGB-overexpressing epimastigotes. In contrast, the observed slight reduction in the ΔN-TcHMGB-overexpressing epimastigotes growth could be associated to non-nuclear functions of TcHMGB, similar to those described for other HMGB family proteins orthologs, not yet studied in T. cruzi29,30.
Figure 3.
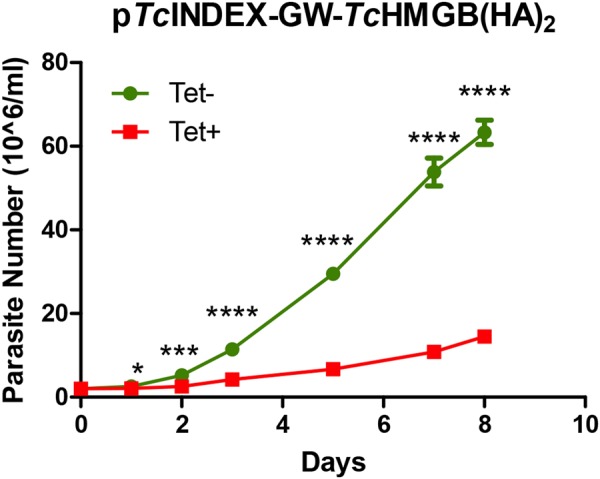
TcHMGB overexpression decreases cell proliferation. Growth curve of epimastigotes transfected with pTcINDEX-GW-TcHMGB(HA)2 in the absence (green circles, Tet−) or presence (red squares, Tet+) of 0.5 µg/ml tetracycline, counted daily for 8 days. Values represent mean ± SD of 3 independent replicates, *p < 0.05, ***p < 0.0005, ****p < 0.00005 (Student t test).
Interestingly, Tet-induced TcHMGB overexpressing parasites also showed morphological differences when compared to non-induced epimastigotes according to data obtained by optical and scanning electron microscopy (SEM). For example, we observed an increasing number of parasites where the kDNA has been duplicated, but the kinetoplast has not been segregated, thus impairing the cytokinesis (Fig. 4A, red arrows). Parasites with aberrant morphologies or reduced dimensions were also observed (Fig. 4A,B, blue arrows), as well as a population of epimastigotes with two nuclei and only one kinetoplast (Fig. 4A, green arrows; Fig. 4D, white arrows). By SEM, it was also possible to identify TcHMGB overexpressing cells in cytokinesis (Fig. 4B, panels b and d, white arrows) or undergoing asymmetric divisions (Fig. 4B, red arrows on panels b and c). Finally, a quantitative analysis of the parasite measures from SEM images showed that an increased proportion of TcHMGB overexpressing cells presented a slight decrease in length and a small increase in the width of the cell body, which may be related to division impairment (Fig. 4C).
Figure 4.
TcHMGB overexpression affects cell morphology and division. (A) May Grünwald Giemsa-stained T. cruzi Dm28c/pTcINDEX-GW-TcHMGB(HA)2 epimastigotes in the absence (−) or presence (+) of Tet at 24, 48 and 72 h post-induction. Red arrows indicate parasites with apparent impaired cytokinesis. Some of them present altered cell morphology for example, two nuclei and one kinetoplast (green arrows) and reduced sizes (blue arrows). (B) Images obtained by Scanning Electron Microscopy (SEM) showed that the induction of TcHMGB overexpression promoted the appearance of cells with reduced sizes (panel a, blue arrows) and a high number of parasites in cytokinesis, presenting an enlargement or widening of the cell body (white arrows). Some cells presented an asymmetric division (panels b and c, red arrows), which can generate parasites with reduced sizes (panel a, blue arrows). (C) Analysis of the parasite body dimensions. The body widths and lengths of control (Tet−) and induced (Tet+) epimastigotes were measured from the SEM images. Wilcoxon-Mann-Whitney Test confirmed that the values for width and length between the (Tet−) and (Tet+) groups are significantly different (p < 0.05), N = 101, Dot plot graphs represent individual values, median and range are indicated; *p < 0.05, ****p < 0.0001. (D) Nucleus/Kinetoplast content (N/K) of Dm28c/pTcINDEX-GW-TcHMGB(HA)2 epimastigote cultures in the absence (Tet−) or presence (Tet+) of tetracycline at different time points (24, 48 and 72 hours). Data from two independent experiments were considered in the analysis (n = 300 cells for each column). Below, representative pictures of the microscopy images analyzed to perform the graph. White arrows show examples of epimastigotes with 2 segregated nuclei and 1 kinetoplast (2N1K).
The ordered progression of the cell cycle, in which kinetoplast segregation precedes nuclear division, allows the identification of three normal states regarding nuclear/kinetoplast (N/K) content: 1N1K, 1N2K and 2N2K31. Under normal conditions, most epimastigotes in a non-synchronous exponentially growing culture contain one nucleus and one kinetoplast (1N1K, usually ∼80–95%), corresponding to parasites in G1 or S phase of the cell cycle32. A smaller proportion exhibits two kinetoplasts and one nucleus (1N2K ∼2.3%), these correspond to parasites in G2 phase or the beginning of mitosis. Finally, cells presenting two kinetoplasts and two nuclei (2N2K ∼3%) are those that have finished mitosis and are undergoing cytokinesis or ready to do so. Thus, the appearance of cells with abnormal N/K content (for example, 0N1K, 1N0K or 2N1K) is indicative of a possible cell cycle deffect31. In fact, after analyzing the N/K content of the TcHMGB-overexpressing epimastigotes, we found an increase of 5% of cells presenting the phenotype 2N1K, 24 hs post-induction. This percentage augmented with time, reaching approximately 18% of the total population after 72 hs of tetracycline induction. These data are in accordance to cytokinesis impairment and suggests a failure in the cell cycle progression in TcHMGB overexpressing parasites (Fig. 4D).
TcHMGB overexpression alters cell cycle progression
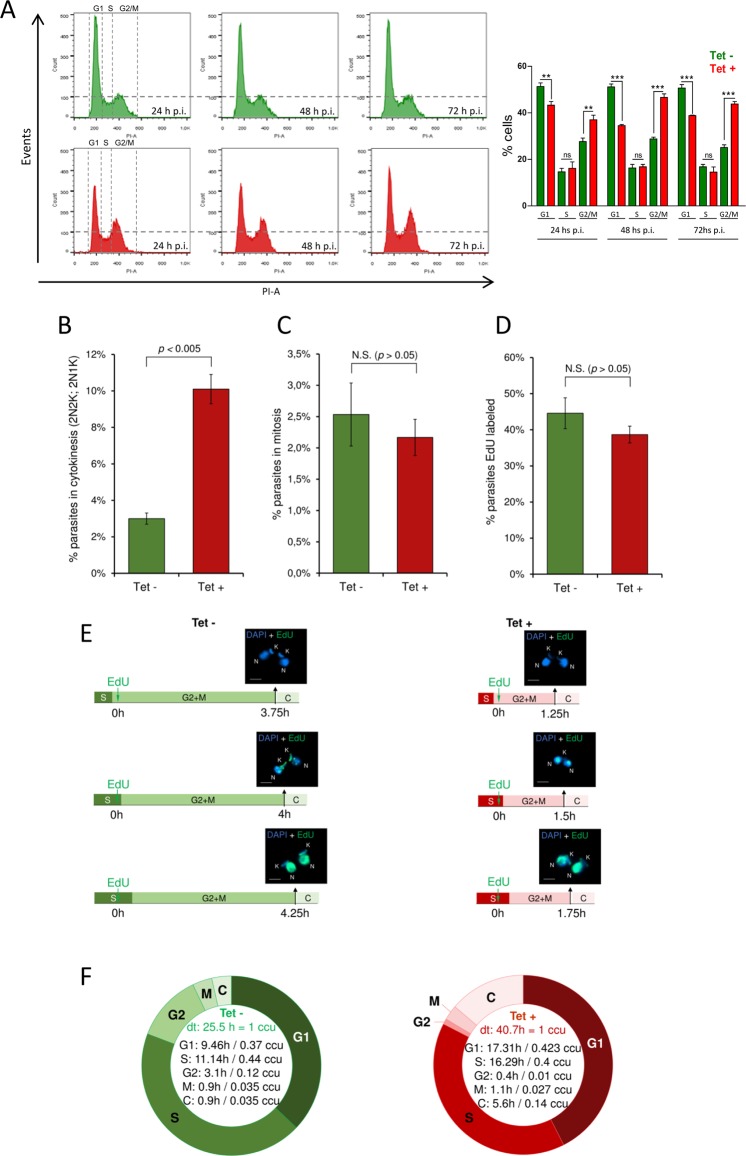
The cell cycle progression of TcHMGB overexpressing epimastigotes was analyzed by flow cytometry with Propidium iodide (PI) staining for 72 h post-induction and compared with non-induced control parasites (Fig. 5A). As expected, in the absence of Tet we did not observe major changes with time in the counts for each population. The main peak corresponds to the parasites on G1 phase of the cell cycle (∼50% of the total), that is, parasites with the DNA content corresponding to one nucleus (2n). A second minor peak represents the parasites in G2/M phase (∼25–28%), which corresponds to epimastigotes with the double of DNA content (4n), including those on cytokinesis. In the valley between the two peaks are the cells on S phase (∼15%). As can be seen in the lower (red) panel of Fig. 5A, the peak of cells in G2/M increased after Tet-induction ranging from 38% at 24 post-induction to 48% at 48–72 h (Fig. 5A), suggesting that the TcHMGB overexpression results in an arrest of cells containing duplicated DNA (4n). Our hypothesis, in accordance with the previous microscopy observations, is that cytokinesis is impaired in overexpressing parasites and causes the accumulation of parasites in the G2/M peak, which in fact includes parasites that have not finished the cell division.
Figure 5.
TcHMGB overexpression alters cell cycle progression. (A) Flow Cytometry analysis of the cell cycle progression. On the left, Flow Cytometry analysis of cultured T. cruzi Dm28c/pTcINDEX-GW-TcHMGB(HA)2 epimastigotes at different times post-tetracycline induction (p.i.). Histograms are plotted as number of events vs. propidium iodide absorbance (PI-A). On the right, bar graph with the percentages of cells in the different phases of the cell cycle. **p < 0.005, ***p < 0.0001 (Student t test). (B–F) Estimation of the duration of each cell cycle phase. T. cruzi Dm28c/pTcINDEX-GW-TcHMGB(HA)2 epimastigotes in exponential phase of culture induced (Tet+) or not (Tet−) with tetracycline, were used in these analysis. (B) DAPI-labeled parasites (2N2K and 2N1K) were used to measure the percentage of parasites in cytokinesis, which was estimated to be 3.0% ± 0.3 for Tet− group (green) and 10.1% ± 0.8 for Tet+ group (red). Error bars represent SD. The values shown represent the average of three independent assays. These values were used in Williams (1971) equation to estimate the duration of cytokinesis phase. (C) Parasites with nuclei in division but not yet segregated were used to estimate the proportion of parasites performing mitosis [2.5% ± 0.5 for Tet− (green), and 2.2% ± 0.2 for Tet + (red)]. Error bars represent SD. The values shown represent the average of three independent assays. These values were used in Williams (1971) equation to estimate the duration of mitosis phase. (D) Parasites EdU-labeled after 1 h of EdU pulse were used to estimate the percentage of parasites replicating DNA [44.6% ± 4.3 for Tet– (green), and 38.7% ± 2.3 for Tet+ (red)]. Error bars represent SD. The values shown represent the average of three independent assays. These values were used in Stanners and Till (1960) equation to estimate the duration of S phase. (E) To estimate the duration of G2 + M phases, the thymidine analog EdU was added to the culture and parasites were collected every 15 min until parasites containing two EdU-labeled nuclei were observed (2N2K or 2N1K). In Tet– (green), this pattern was observed after 4 h, and in Tet + (red) after 1.5 h. This assay was carried out in triplicate and in all of them, we found a parasite containing two EdU-labeled nuclei at the same time. The scale bar on the fluorescence images corresponds to 2 µm. (F) Schematic representation showing the duration of each cell cycle phase established using EdU. Of note, ccu means cell cycle unit, where one unit corresponds to the specific doubling time (dt) for each strain. The statistical analysis for Fig. 5B-D was made with Student t test using GraphPad Prism 6. As expected, the significant statistical difference (comparing Tet− and Tet + ) relative to parasites performing cytokinesis (Fig. 5B) is reflected in the cytokinesis phase lengths estimated for both Tet− and Tet + (Fig. 5F). Of note, there was no significant statistical difference between parasites EdU-labeled or parasites performing mitosis (p > 0.05).
Afterwards, we made use of DAPI and the thymidine analog EdU (5-Ethynyl-2´-deoxyuridine) to follow the cell cycle progression at the microscope and estimate both the duration and the percentage of parasites in each cell cycle phase33. In accordance to our previous results, we confirmed using DAPI staining the presence of an increased percentage of parasites in cytokinesis presenting typical (2N2K) or atypical (2N1K) cellular patterns (Fig. 5B). Parasites with nuclei in division but not yet segregated were used to estimate the proportion of parasites performing mitosis, which did not show major differences compared to non-induced parasites (Fig. 5C). Finally, the thymidine analog EdU was added to the culture and parasites labeled after 1 h were used to estimate the percentage of cells replicating DNA (Fig. 5D). The duration of cytokinesis and mitosis phases were estimated through Williams equation34, using the percentage of parasites in each respective phase. To estimate the duration of G2/M phases, we made an EdU 1h-pulse and then collected parasites every 15 min in order to observe parasites containing two EdU-labeled nuclei (2N2K or 2N1K) (Fig. 5E). In non-induced (Tet−) cultures, this N/K pattern was observed after 4 h, while in Tet-induced (Tet+) after 1.5 h. The difference between this value and the duration of mitosis previously calculated by Williams equation34 corresponds to G2 phase duration. The duration of S phase was estimated according to the Stanners and Till equation35. Figure 5F shows a schematic representation of the duration of each cell cycle phase. The whole cycle lasts 40.7 h in TcHMGB overexpressing parasites culture versus 25.5 h for the non-induced control culture. This difference in duplication times for the induced vs non-induced parasites is consistent with the delay in the growth curve described previously (Fig. 3). Also, as expected, the cytokinesis lasts longer for the Tet−induced culture, contributing to the whole cell cycle duration, together with the G1 phase, which is actually estimated by the difference between the duplication time and the estimated times for the other phases. The S phase lasts 16.29 h for Tet+ vs. 11.14 h for Tet−. The M phase showed no great differences, but it is noteworthy the shortening in G2 for Tet+. We must emphasize that the DNA content of cells in G2, M, or cytokinesis is exactly the same (4n). Thus, G2 + M + C takes 2.2 h more hours for the TcHMGB overexpressing parasites, supporting the accumulation of 4n-containing cells observed by flow cytometry in the TcHMGB overexpressing parasites. Together, our data support the hypothesis of an impaired or delayed cytokinesis caused by TcHMGB overexpression.
TcHMGB levels can affect cell infection, amastigote replication and metacyclogenesis
To investigate if TcHMGB can be important for Chagas disease pathogenesis, we needed to focus on the parasite life cycle stages present in the mammalian host. As a first approach, we analyzed in vitro the performance of transgenic parasites overexpressing TcHMGB to invade and replicate inside the host cells. We used different Tet-induction schemes to analyze the TcHMGB-overexpression effect over the different time points during the whole in vitro infection process (see Methods section).
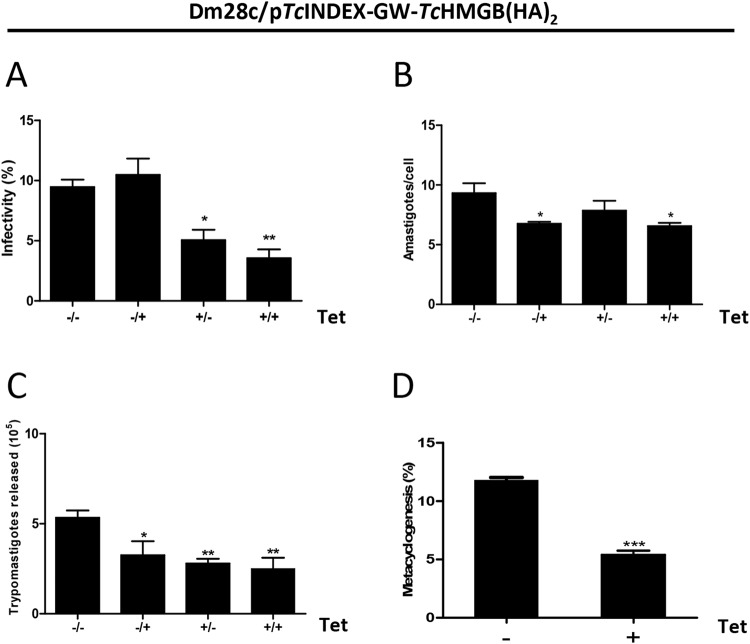
To study if trypomastigote ability to invade and infect cells on a monolayer was affected by TcHMGB overexpression, trypomastigotes were pre-incubated with Tet (Fig. 6, +/− and +/+ condition) or not (−/+ and −/− condition) and then allowed to infect a monolayer of Vero cells. After 6 hs infection, free trypomastigotes were washed out and fresh medium with (+/+ and −/+ condition) or without Tet (+/− and −/− condition) was added to the cells and incubated for 3 days. The −/− condition represents the non-induced control where Tet was never added and the +/+ condition shows the effect of protein overexpression during the whole infection assay (from prior to invasion “+/” and through the 3 days left for amastigote replication inside the host cell “/+”). The trypomastigotes infection performance was analyzed by the proportion of infected Vero cells (Fig. 6A) and intracellular amastigotes replication ability was measured as the average number of intracellular amastigotes per infected cell at 72 hs post-infection (Fig. 6B).
Figure 6.
TcHMGB levels affect cell infection, amastigote replication and metacyclogenesis. The infection performance of T. cruzi Dm28c/pTcINDEX-GW-TcHMGB(HA)2 was analyzed in the absence (Tet−) or presence (Tet+) of 0.5 μg/ml tetracycline. To test the effect over the different phases of infection, we designed different induction protocols as follows: (−/−), Tet was never added to the medium, non-induced control; (+/−), trypomastigotes were pre-treated with Tet for 2 hours prior to infection, and during the invasion incubation period but not after; (−/+), trypomastigotes were not induced, Tet was only added after 48 hours post-infection to see the effect on the amastigote stage; (+/+), trypomastigotes were pre-treated and Tet was present during the whole infection assay, to see the overexpression effect both in trypomastigotes and amastigotes. The percentage of infected cells (A), the number of amastigotes per cell (B) and the number of trypomastigotes released 6 days post-infection (C) were determined by counting Giemsa-stained slides using an optical microscope. Results are expressed as mean ± SD of triplicates. Statistical analysis of the data was carried out using one-way ANOVA, *p < 0.05, **p < 0.001 and ***p < 0.0005. (D) In vitro metacyclogenesis using TAU medium of the pTcINDEX-GW-TcHMGB(HA)2 parasites non-induced (−) or induced (+) with 0.5 µg/ml Tet for 96 h. The bar graph represents the mean ± SD from three independent experiments; ***p < 0.0005 (Student t test).
Overexpression of TcHMGB on trypomastigotes by pre-treatment with Tet before infection caused a dramatic decrease (∼52%) in the infection rate (Fig. 6A, compare +/− and +/+ conditions with −/−). As expected, the addition of Tet after the incubation period to allow trypomastigotes invasion of Vero cells (−/+), showed no significant difference in the proportion of infected cells compared to the negative control (−/−). In contrast, amastigote duplication was affected in this condition (−/+). As can be seen in Fig. 6B, the addition of Tet once the trypomastigotes have entered the host cells (−/+) or when Tet was always present (+/+), caused a reduction in the average number of intracellular amastigotes per cell (compared to −/−) showing an effect on this parasite stage too. Furthermore, the number of trypomastigotes released to the supernatant at day 5 post-infection due to the lysis of infected cells (Fig. 6C), was also lowered in TcHMGB-overexpression conditions, either when Tet was added only to pre-incubate trypomastigotes (+/−), only to intracellular amastigotes (−/+) or when it was always present (+/+). The number of released trypomastigotes is influenced by the infectivity of trypomastigotes and by the amastigotes replication rate, but also the efficiency in the transformation between the different life cycle forms (from trypomastigote to amastigote and back to the trypomastigote form finally released) may contribute to the final number of released parasites. It is difficult to assess how much each of these processes affects the final number of released trypomastigotes, in particular it is not easy to measure the transformation between the parasite life cycle stages contribution. However, we can easily evaluate in vitro the epimastigote to metacyclic trypomastigote transformation process to see if it is affected by TcHMGB overexpression. Indeed, in vitro metacyclogenesis was performed in the absence or presence of Tet, and TcHMGB overexpression resulted in a reduced number of metacyclic trypomastigotes (Fig. 6D).
Discussion
As in other eukaryotic cells, nuclear architecture is dynamic in trypanosomes. Chromatin shows differences through the parasites life cycles and also through cell cycle in proliferating stages regarding its position inside the nucleus, condensation state and protein composition3,36,37. These changes thus modify DNA accessibility to protein complexes involved in nuclear functions like transcription and replication. The histones suffer different post-translational modifications through the parasite life cycle, such as methylation or acetylation, that contribute to chromatin remodeling and epigenetic control of gene expression38. Several epigenetic control mechanisms have been already described in trypanosomatids39–44. Interestingly, it has recently been reported that chaetocin, a histone methyltransferases inhibitor, promoted irreversible inhibition of protozoa growth presumably as a consequence of the unpacking of nuclear heterochromatin and intense nucleolus fragmentation, which is associated with the parasite cell cycle arrest and RNA transcription blockage45. It is clear that chromatin structure must be well regulated for an accurate performance of the parasite functions, however, there are still many unsolved issues regarding how these parasites control their gene expression and how chromatin and nuclear structure changes are achieved during the parasite differentiation.
High mobility group B proteins are important players in chromatin structure. These proteins have shown to facilitate chromatin remodeling thus increasing accessibility of the nucleosomal DNA to transcription factors and protein complexes involved in transcription, replication, recombination, DNA repair and genomic stability9. TDP1, the ortholog HMGB from T. brucei, showed to be directly involved in epigenetic control of RNA Polymerase I (RNAP I)-transcription of rDNA and VSG genes43,46. In our previous study, we demonstrated that TcHMGB is an architectural protein capable of modifying DNA structure in vitro16. Here, we attempted to study the putative roles of the protein in the parasite. Protein overexpression is a valuable approach25,28,47, considering that silencing by RNAi is not possible in T. cruzi, and our failure to obtain knockout parasites, thus suggesting that the gene may be essential or, at least, that its deletion results in a growth disadvantage. Using a pTcINDEX-GW vector-derived construction, we were able to overexpress TcHMGB in T. cruzi. Tet−induced parasites presented modifications not only in the cell morphology but also in its nuclear structure, diminishing its replication rate and affecting several functions related to the parasite biology, as infectivity and differentiation. It is worth mentioning that the observed behavior of TcHMGB overexpressing parasites is due to the increased TcHMGB protein in the nucleus where it can interact with the DNA since none of these phenomena were observed after overexpression of a truncated version of the protein, which has the two functional HMG-Box domains but localizes in the cytoplasm of the parasites (Fig. S2).
Taking into account the architectural properties of HMGB protein family and our previous in vitro evidence, it was expected that TcHMGB interaction with genomic DNA would also occur in vivo16. Thus, an altered chromatin structure would be expected in parasites with an increased content of this protein in their nuclei. Not surprisingly, the chromatin in these overexpressing parasites showed to be in a more relaxed or open state, as determined by its MNase sensitivity, suggesting that TcHMGB can contribute to chromatin remodeling in the parasite. Accordingly, TEM analysis showed that higher TcHMGB content results in a reduction of the nucleolus and an increase of the euchromatin region considering the total nuclear area. The nucleolus is the most evident nuclear domain and can be easily distinguished by microscopy techniques in eukaryotic cells. It is characterized by its resident proteins that play essential roles, as ribosomal RNAs transcription, processing and assemblage into ribonucleoprotein (RNP) that results in ribosome biogenesis. More recently, additional functions have been proposed for the nucleolus such us regulation of mitosis, cell cycle progression and stress response48.
Changes in nucleolar shape and size have been described in T. cruzi under stress conditions, like the induction of the stationary phase in cultured epimastigotes49 or when replicative forms transform into the non-proliferative trypomastigotes3. It is worth mentioning that these changes in the nucleus correlate with the parasite replication and transcription rates50. In transcriptionally active epimastigotes and dividing amastigotes the rounded nucleus contains the heterochromatin organized around the central nucleolus and in the nuclear periphery, while in trypomastigotes the nucleus is elongated, lacks an evident nucleolus and presents a disperse heterochromatin3,4. In our previous report, we showed that TcHMGB is a nuclear protein expressed in all T. cruzi life cycle stages, although the protein content is higher in epimastigote and amastigote forms in comparison to the non-replicative stage16. The reduced TcHMGB content in trypomastigotes correlates with increased amounts of heterochromatin and may be associated to their lower transcriptional activity. In accordance to this idea, the overexpression TcHMGB would contribute to relaxation of chromatin, increasing the euchromatin in the nucleus and thus affecting different nuclear functions.
Regarding the observed reduction of the nucleolus, it is interesting to note the TcHMGB immunofluorescence microscopy labeling pattern changes in overexpressing parasites. In wild type and non-induced parasites, TcHMGB signal appears localized in the whole nucleus as several spots quite regularly distributed, except for a strong signal located in the nucleolus. Similarly, T. brucei TDP1, showed to be distributed throughout the nucleus in both bloodstream and procyclic forms, but enriched in either one or two discrete spots corresponding to the nucleolus and to specific expression site bodies (ESB) in bloodstream forms46. There, TDP1 presumably facilitates RNAP I dependent transcription of rRNA and VSG genes respectively46. Analogously, the strong signal of TcHMGB in control cells may correspond to the rRNA transcription site in the T. cruzi nucleolus, whereas in overexpressing cells the redistribution of this protein could result from nucleolar disassembly or disorganization, as a consequence of higher production of rRNA transcripts. It is also worth noting that histone H1 showed to be depleted from the nucleolus in T. brucei presumably as a consequence of the extremely high rates of transcription of the ribosomal DNA (rDNA)51. In other organisms, HMGB proteins have shown to facilitate nucleosome remodeling and accessibility of the nucleosomal DNA through the displacement of histone H1. Thus, taking into account our observations and the evidence from homologous proteins, it seems likely that TcHMGB can contribute to RNAP I-transcription of rDNA genes in T. cruzi. Finally, it cannot be ruled out that TcHMGB may also contribute to the RNAPII-dependent transcription. In fact, RNAPII was found to concentrate in a domain close to the parasite nucleolus containing the spliced leader genes and the remaining protein was diffusely distributed in the nucleoplasm52, showing an immunofluorescence pattern similar to the one observed for TcHMGB in wild type (or non-induced) parasites16.
When overexpression of TcHMGB is induced by Tet, the protein appears more regularly distributed showing a strong immunofluorescence signal along the whole nucleus, but reduced or even excluded from the nucleolar region. This relocalization of TcHMGB can be consequence of an ectopic aggregation of the protein as a consequence of the higher expression levels. This anomalous distribution of TcHMGB may be responsible for the reduction or disassembly of the nucleolus observed by TEM and DAPI staining in overexpressing parasites. Indeed, the nucleolus is a dynamic structure and it is intimately linked to the processes that take place there and to their associated proteins. It has been already described that nucleolar markers are seen dispersed in the nucleoplasm when the nucleolus is reorganized during the life cycle or when dividing epimastigotes reach the stationary phase49,53. However, it is not clear which process is cause and which is consequence, that is, does nucleolar proteins redistribution cause the nucleolus to disassemble or vice versa? In other organisms, it has been shown that introduction of rDNA genes on non-integrating plasmids led to the formation of mini-nucleoli and inhibition of the RNAPI results in disassembly of nucleoli54. Thus, nucleolar proteins association and functions are linked to nucleolar structure formation.
If we assume that TcHMGB concentrates in the nucleolus in replicating epimastigotes and amastigotes and facilitate rRNA transcription by maintaining an open chromatin state, it is possible that ectopically localized aggregates of the overexpressed protein results in the destabilization of nucleolar structure. Moreover, the overexpression of TcHMGB and its anomalous localization in T. cruzi may be also interfering with the normal RNAP II transcriptional activity. Thus, we cannot rule out that a failure in the control of specific genes transcription may influence the observed phenotypes, since indeed all HMGB family members have pleiotropic functions. Additional work and a detailed study of both global and individual genes transcription in overexpressing parasites would be necessary to better understand the exact role of TcHMGB in gene expression control. The overexpression of TcHMGB can also interfere with proper DNA replication, thus affecting the normal development of the parasite functions. In fact, the replication origins have been localized at the boundaries of polycistronic transcription units suggesting a functional interaction between DNA replication and transcription initiation in T. brucei genome55, thus it is likely that an overexpression of TcHMGB in the trypanosome nucleus causes an imbalance of both processes. This would explain the reduction in proliferation, the appearance of atypical phenotypes and cell cycle arrest in parasites where the TcHMGB is displaced.
In TcHMGB overexpressing cells, the destabilization of chromatin DNA structure might favor nuclear DNA replication, but this process would proceed uncoupled from kinetoplast DNA replication and cytokinesis. In accordance to this idea is the higher proportion of Tet-induced parasites presenting 2N1K and the delayed cytokinesis phenotype, which supports the hypothesis of a failure in DNA replication control. Somehow, this uncontrolled DNA replication may impair the proper progression of the cell cycle since when we analyzed the parasites by flow cytometry, we observed that TcHMGB overexpression results in an accumulation of cells in G2/M phase of the cell cycle, which also includes parasites in cytokinesis. A higher number of cells in cytokinesis were observed by optical microscopy and SEM, corroborating the idea that this process was impaired in Tet-induced parasites. Furthermore, a detailed analysis of the cell cycle revealed that epimastigotes overexpressing TcHMGB present a longer generation time (almost double) and the period spent on cytokinesis is five times higher in relation to control cells. The control of cytokinesis initiation in trypanosomatids is predominantly made by CIF1, Aurora B kinase, and Polo-like kinase56–58. In T. brucei, the disruption of CIF1 promotes an alternative cytokinesis pathway, which promotes cell division in an opposite direction of the typical cytokinesis56. The Aurora B kinase plays essential roles in mitosis and cytokinesis in model eukaryotes59,60, but in T. brucei the mutation in some key amino acids and its overexpression caused problems predominantly in mitosis61. Curiously, the Polo-like kinase in T. brucei was shown to be essential for kDNA segregation and also for cytokinesis62, two characteristics that have been impaired in our study after the induction of TcHMGB (Figs 4 and 5). Whether the TcHMGB overexpression disrupts the Polo-like kinase impairing cytokinesis and kDNA segregation in T. cruzi is an opened question that requires further investigation.
Finally, we also found that TcHMGB is important for proper functions during T. cruzi life cycle. TcHMGB overexpressing trypomastigotes infected Vero cells less efficiently than the control. Also, amastigotes duplication rates and the number of trypomastigotes released after infected-cells lysis were diminished. The whole performance of the parasite regarding host cell-infection seems to be negatively affected after TcHMGB overexpression. The reduced number of released trypomastigotes can be a consequence of impaired trypomastigote infectivity, lowered amastigote replication rate and/or the capacity of the parasite to go through its life cycle forms, that is, to transform from the infective trypomastigote to the replicative amastigote and back again to trypomastigote to be finally released upon cell lysis. It cannot be easily determined if all of these processes are responsible for the final number of trypomastigotes released or to what extent, but undoubtedly, TcHMGB overexpressing parasites are less efficient than the wild type to infect cells in vitro. Finally, metacyclogenesis, that is the transformation of epimastigotes to the infective metacyclic trypomastigotes, which occurs in the insect vector, was also reduced by the overexpression of TcHMGB, as measured by an in vitro artificial model.
In conclusion, the results presented here suggest that TcHMGB protein levels in the nucleus should be regulated for proper function of the T. cruzi cellular processes and let us propose a role for TcHMGB in DNA replication and cell cycle progression control.
Methods
Molecular cloning of TcHMGB(HA)2
TcHMGB gene (TCDM_04259) was amplified by PCR from T. cruzi Dm28c genomic DNA using the following oligonucleotides: TcHMGB-Fw (5′-AAGGATCCATGTCCACTGAACTAAAGTCAG-3′), TcHMGB-HA-Rv (5′-AACTCGAGTTAAGCGTAATCTGGAACATCGTATGGGTAAGCGTAATCTGGAACATCGTATGGGTAGCTCGCCCTTGCAG-3′), designed to add two hemagglutinin (HA) tags at the C-terminus of the protein. Truncated protein was constructed using the following oligonucleotides: ΔN-TcHMGB(HA)2-Fw (5′-AAGGATCCACCCAAAGGCGGCGCTCTCGCC-3′), ΔN-TcHMGB(HA)2-Rv (5′-AACTCGAGTTAAGCGTAATCTGGAACATCGTATGGGTAAGCGTAATCTGGAACATCGTATGGGTAGCTCGCCCTTGCAG-3′). The PCR products were first cloned into pCR2.1-TOPO vector (Invitrogen) and sequenced. The coding sequences were sub cloned into pENTR-3C vector (Invitrogen) using BamHI/XhoI restriction sites included in the oligonucleotides (underlined) and then transferred to the pTcINDEX-GW vector by recombination using LR clonase II enzyme mix (Invitrogen).
Real time PCR (qRT-PCR)
For qRT-PCR primers were designed to amplify a 58 bp fragment of TcHMGB (TcHMGB-Fw 5′-CGAGGTACCGCATGGAGTTC-3′ and TcHMGB-Rv 5′-CTTCGTAATGATGCCCTCTATGG-3′) and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (5′-TGGAGCTGCGGTTGTCATT-3′ and 5′-AGCGCGCGTC TAAGACTTACA-3′) as an endogenous control. TRIzol reagent (Invitrogen) was used to extract total RNA from epimastigotes (1 × 108 cells) and then RNA was treated with RQ1 RNase-free DNase I (Promega, Madison, WI, USA). First-strand cDNA was synthesized using the First Strand cDNA Synthesis kit (Life Technologies) according to the manufacturer’s instructions. The reactions were performed with 200 nM forward and reverse TcHMGB primers or 200 nM forward and reverse GAPDH primers, SYBR Green Master Mix (Applied Biosystems, Life Technologies, Argentina) and epimastigote cDNA in triplicates. An ABI PRISM 7000 (Applied Biosystems) thermocycler was used following standard cycling conditions. The data were analyzed by the 2−ΔΔCT method normalizing with GAPDH using the 7000 SDS software (Applied Biosystems).
Polyclonal antibodies
Rabbit polyclonal antibodies against TcHMGB were affinity-purified from antisera obtained as described in our previous work16.
Parasite cultures
T. cruzi Dm28c epimastigotes were cultured at 28 °C in LIT medium (5 g/L liver infusion, 5 g/L bacto-tryptose, 68 mM NaCl, 5.3 mM KCl, 22 mM Na2HPO4, 0.2% (w/v) glucose and 0.002% (w/v) hemin) supplemented with 10% (v/v) heat-inactivated, UV-irradiated Fetal Calf Serum (FCS) (Internegocios S.A, Argentina).
Transgenic parasites generation
Epimastigotes from T. cruzi Dm28c were transfected with the pLEW13 plasmid to generate parasites expressing T7 RNA polymerase and the Tet repressor using a standard electroporation method21. Briefly, epimastigotes were cultured in LIT medium at 28 °C to a final concentration of 3–5 × 107 parasites/ml. Then, parasites were harvested by centrifugation at 1500 g for 5 min at room temperature, washed twice with phosphate buffered saline (PBS) and resuspended in 0.35 ml transfection buffer (0.5 mM MgCl2, 0.1 mM CaCl2 in PBS, pH 7.5) to a density of 1 × 108 cells/ml for each transfection. Electroporation was performed in a 0.2 cm gap cuvette (Bio-Rad) with ~40 μg of plasmid DNA added to a final volume of 400 μl. The parasite-DNA mixture was kept on ice for 25 min and subjected to a 450 V, 500 μF pulse using GenePulser II (Bio-Rad, Hercules, USA). After electroporation, cells were transferred into 3 ml of LIT medium containing 10% FCS, maintained at room temperature for 15 minutes and then incubated at 28 °C. Geneticin (G418; Life Technologies) was added at a concentration of 200 μg/ml, and parasites were incubated at 28 °C. After selection, pLEW13 transfected epimastigotes were maintained in the presence of 200 μg/ml of G418. This parental cell line was then transfected with pTcINDEX-GW-TcHMGB construct following a similar protocol and transgenic parasites were obtained after 3 weeks of selection with 100 μg/ml G418 and 200 μg/ml Hygromycin B (Sigma).
In vitro metacyclogenesis
Metacyclic trypomastigotes were obtained in vitro using chemically defined conditions as described previously63. Briefly, exponential epimastigotes were washed with PBS and resuspended in TAU medium (190 mM NaCl, 17 mM KCl, 2 mM MgCl2, 2 mM CaCl2, 8 mM phosphate buffer pH 6.0) to a density of 5 × 108 parasites/ml with and without Tet (0.5 μg/ml) for 2 hs at 28 °C. Then, they were diluted 1:100 in TAU3AAG Medium (TAU medium plus 10 mM Glucose, 2 mM L-Aspartic Acid, 50 mM L-Glutamic Acid and 10 mM L-Proline) and incubated at 28 °C for 72 hours in the absence or presence of Tet. Parasites were collected, fixed in 4% paraformaldehyde solution and stained with Giemsa to be visualized with a Nikon Eclipse Ni-U microscope and counted using ImageJ software64. Only parasites with a fully elongated nucleus and rounded kinetoplast at the posterior end of the parasite were considered as metacyclic forms. Five hundred parasites from each triplicate were counted and the experiment was repeated three times.
T. cruzi infection of Vero cells
Vero cells (ATCC CCL-81) were cultured in Dulbecco’s Modified Eagle Medium (DMEM) (ThermoFisher), supplemented with 2 mM L-glutamine, 10% FCS, 100 U/ml penicillin and 100 μg/ml streptomycin. For the first round of infection, metacyclic trypomastigotes were obtained by spontaneous differentiation from late-stationary phase cultured epimastigotes at 28 °C. Cell-derived trypomastigotes were obtained by infection with metacyclic trypomastigotes in Vero cell monolayers. For the infection and amastigote proliferation experiments, we used cell-derived trypomastigotes released from the second round of infection. Trypomastigotes were collected from the supernatant of the infected cells culture, harvested by centrifugation at 5000 × g for 10 min at room temperature, resuspended in DMEM and counted in a Neubauer chamber. When indicated, the parasites were pre-incubated with Tet (0.5 μg/ml) for 2 hs in order to allow protein induction and a new monolayer of cultured Vero cells was infected with a MOI of 10:1. After a 6h-incubation at 37 °C, the free trypomastigotes were removed by washes with PBS. This pre-treatment of trypomastigotes with Tetracycline prior to infection and during the 6 h-incubation to allow invasion is indicated with a “+” before the bar “(+/)”. Then, infected Vero cell cultures were incubated in DMEM supplemented with 2% FCS with or without Tet (0.5 μg/ml) for two days. The presence of Tet during this incubation period when amastigotes replicate inside the infected cell is indicated by the “+” after the bar “(/+)”. The experiments were stopped by cell fixation with methanol and the percentage of infected cells and the mean number of amastigotes per infected cell was determined by direct slide counting (Nikon Eclipse Ni-U microscope). Giemsa staining was used for amastigotes visualization and approximately 800 cells were counted per slide.
Western blot
Protein extracts were fractioned in SDS-PAGE and transferred to a nitrocellulose membrane. Transferred proteins were visualized with Ponceau S staining. Membranes were treated with 10% non-fat milk in PBS for 2 hours and then incubated with specific antibodies diluted in 0.5% Tween20 in PBS (PBS-T) for 3 hours. Antibodies used were: rat monoclonal anti-HA (ROCHE), affinity-purified rabbit polyclonal anti-TcHMGB, mouse monoclonal anti-trypanosome α-tubulin clone TAT-1 (a gift from K. Gull, University of Oxford, UK). Bound antibodies were detected using peroxidase-labeled anti-mouse, anti-rabbit IgG (GE Healthcare), anti-mouse IgG (GE Healthcare) or anti-rat IgG (Thermo Scientific) and developed using ECL Prime kit (GE Healthcare) according to manufacturer’s protocols. Immunoreactive bands were visualized by autoradiography, photographed and images were processed with Adobe Photoshop 6.0.
Immunofluorescence
Mid-log epimastigotes were harvested by centrifugation at 1500 xg for 5 min at room temperature and washed twice prior to fixation in 4% paraformaldehyde solution. Fixed parasites were placed on a coverslip pre-coated with poly L-lysine for 20 min and then washed with PBS. Permeabilization was done with 0.2% Triton X-100 solution for 10 min. After washing with PBS, parasites were incubated with the appropriate primary antibody diluted in 1% BSA in PBS for 2 hs at room temperature. Non-bound antibodies were washed with PBS-T and then the slides were incubated with anti-rat IgG::FITC (Life Technologies) and anti-rabbit IgG::Cy3 (Life Technologies) conjugated antibodies and 2 μg/ml 4′,6-diamidino-2-fenilindol (DAPI) for 1 hour. The slides were washed again with PBS-T and mounted with VectaShield (Vector Laboratories). Images were acquired with a confocal microscope ZEISS LSM 880. Adobe Photoshop CS and ImageJ software were used to process all images.
Ultrastructural analysis
Transmission electron microscopy (TEM)
Parasites were washed twice in PBS and fixed in 2.5% (w/v) glutaraldehyde in 0.1 M cacodylate buffer (pH 7.2) for 1 h. Then, cells were washed in 0.1 M cacodylate buffer (pH 7.2) and post fixed for 1 h in 1% (w/v) osmium tetroxide, 0.8% potassium ferrocyanide and 5 mM CaCl2 in 0.1 M cacodylate buffer. After post fixation, cells were washed in the same buffer, dehydrated in a series of increasing acetone concentrations and embedded in Epon, first as a mixture of Epon and acetone (1:1) and then as pure Epon. Ultrathin sections were obtained using an Ultracut Reichert Ultramicrotome and mounted on 400-mesh copper grids. Samples were stained with uranyl acetate and lead citrate and then analyzed using a Zeiss 900 transmission electron microscope. Measurements of the total nucleus area, as well as nucleolar domains, such as nucleolus, heterochromatin and euchromatin, were made in images obtained by transmission electron microscopy using ImageJ software. First we delimited the nucleus, then the nucleolus and the heterochromatin region. The euchromatin region corresponds to the total nuclear area minus those occupied by the nucleolus and heterochromatin regions. Statistics were calculated using the unpaired t test with Welch’s correction in GraphPad Prism 6 software (GraphPad Software). P values less than 0.05 were considered statistically significant.
Scanning electron microscopy (SEM)
Parasite processing was carried out using glass coverslips pre-coated with 1 mg/ml poly-L-lysine. Cells were fixed for 1 h in 2.5% glutaraldehyde diluted in cacodylate buffer [0.1 M (pH 7.2)] and then were adhered to coverslips, post-fixed for 1 h with 1% osmium tetroxide diluted in cacodylate buffer. Samples were dehydrated in a graded alcohol series (50%, 70%, 90%, and two exchanges of 100% ethanol for 10 min each step) and then were critical-point dried in a Leica EM CPD030 apparatus (Leica, Wetzlar, Germany). Specimens were coated with platinum in a Leica EM SCD050 before visualization using a Zeiss EVO 40 VP scanning electron microscope. Measurements of cells lengths were made using the program AxioVision4 and were based upon the SEM images. Statistics were calculated using the Wilcoxon-Mann-Whitney test in GraphPad Prism 6 software (GraphPad Software). P values less than 0.05 were considered statistically significant.
Micrococcal nuclease assay
Micrococcal nuclease (MNase) assay was performed as described previously37 with slight modifications. Briefly, 108 parasites were washed in lysis buffer (1 mM potassium L-glutamate, 250 mM Sucrose, 2.5 mM CaCl2, 1 mM PMSF) and resuspended in the same buffer containing 0.1% Triton X-100. The supernatant was discarded and the nuclei-containing-pellet washed twice in lysis buffer without detergent. Samples were incubated with 1U of MNase (Thermo Scientific) for 0, 5, 25 minutes at 37 °C, and the reaction was stopped with 2 mM EDTA-EGTA. Then, 10 μl of proteinase K (20 mg/ml) was added and incubated at 56 °C for 3 hs. Subsequently, DNA was purified by ethanol precipitation after phenol/chloroform extraction and analyzed in a 1.5% agarose gel stained with ethidium bromide, visualized under UV light and photographed with a Nikon 3200 digital camera. Images were processed with Adobe Photoshop 6.0.
Analysis of the cell cycle
Cell cycle progression of parasites was analyzed by flow cytometry. One million cells were fixed with cold 70% ethanol and then washed with PBS and stained with 20 μg/ml Propidium Iodide (PI) in buffer K (0.1% sodium citrate, 0.02 mg/ml RNAse A (Sigma), and 0.3% NP-40. Ten thousand events per sample were acquired using BD Cell Sorter BD FACSAria II. Results were analyzed using WinMDi, Cylchred and FlowJo software.
Formaldehyde-fixed and DAPI-stained exponentially growing epimastigotes forms of T. cruzi Dm28c/pTcINDEX-GW-HMGB(HA)2 [induced with tetracycline (Tet+), and non-induced (Tet -)] were examined under an Olympus BX51 fluorescent microscope (Olympus, Japan) (100x oil objective) to observe the profile of organelles that contain DNA (nucleus and kinetoplast). To estimate the duration of mitosis (M) and cytokinesis (C), we used the Williams (1971) equation:
where x is the cumulative time within the cycle until the end of the stage in question, y is the cumulative % of cells up to and including the stage in question (expressed as a fraction of one unit), and α is the specific growth rate.
To estimate the G2 + M phases, we used an EdU pulse (1 h) and then collected parasites every 15 min until a parasite containing two EdU-labeled nuclei (2N2K or 2N1K) were observed. The difference between this value and the duration of mitosis previously calculated corresponds to G2 phase duration. The duration of S phase was estimated according to the Stanners and Till (1960) equation:
where L is the proportion of cells exhibiting EdU-labeled nuclei, α = ln 2/T (T = doubling time expressed in hours), Z = G2 + M + cytokinesis, and t is the duration of the EdU labeling period in hours. Finally, the duration of G1 phase was estimated by the difference between the doubling time and the sum of the remaining phases.
Statistical analysis
Statistical analysis of the data was carried out using GraphPad Prism version 6.0.
Electronic supplementary material
Acknowledgements
We would like to thank Rodrigo Vena, Dolores Campos, Romina Manarin and Mara Ojeda for their technical assistance in confocal microscopy, cells and parasites culture and flow cytometry and Dr. Javier De Gaudenzi for the pTcINDEX-eGFP strain. This work was supported by Agencia Nacional de Promoción Científica y Tecnológica, Ministerio de Ciencia, Tecnología e Innovación Productiva, Argentina [PICT 2008–1871]; Consejo Nacional de Investigaciones Científicas y Técnicas, [PIP 114-201101-00372]; São Paulo Research Foundation (FAPESP) and Center of Toxins, Immune Response and Cell Signaling (CeTICS), grants 2014/24170-5, 2013/07467-1.
Author Contributions
P.C., L.E.T., M.C.M.M., M.C.E. conceived and designed experiments; L.E.T., C.S.G., V.L.A., M.C.M.M., P.C., M.S.d.S. performed experiments; L.E.T. and P.C. wrote the manuscript; L.E.T., V.L.A., P.C., M.S.d.S., M.C.E. and M.C.M.M. prepared the figures; P.C., M.C.M.M., L.E.T., V.L.A., M.S.d.S., M.C.E., E.S. discussed the results; all authors reviewed the manuscript.
Data Availability
No datasets were generated or analyzed during the current study.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-36718-0.
References
- 1.Urbina JA, Docampo R. Specific chemotherapy of Chagas disease: Controversies and advances. Trends in Parasitology. 2003;19:495–501. doi: 10.1016/j.pt.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 2.De Souza W. Basic cell biology of Trypanosoma cruzi. Curr. Pharm. Des. 2002;8:269–85. doi: 10.2174/1381612023396276. [DOI] [PubMed] [Google Scholar]
- 3.Elias MCQB, Marques-Porto R, Freymüller E, Schenkman S. Transcription rate modulation through the Trypanosoma cruzi life cycle occurs in parallel with changes in nuclear organisation. Mol. Biochem. Parasitol. 2001;112:79–90. doi: 10.1016/S0166-6851(00)00349-2. [DOI] [PubMed] [Google Scholar]
- 4.Gonçalves CS, Ávila AR, de Souza W, Motta MCM, Cavalcanti DP. Revisiting the Trypanosoma cruzi metacyclogenesis: morphological and ultrastructural analyses during cell differentiation. Parasit. Vectors. 2018;11:83. doi: 10.1186/s13071-018-2664-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Czura CJ, Wang H, Tracey KJ. Dual roles for HMGB1: DNA binding and cytokine. J. Endotoxin Res. 2001;7:315–21. doi: 10.1177/09680519010070041401. [DOI] [PubMed] [Google Scholar]
- 6.Magna M, Pisetsky DS. The Alarmin Properties of DNA and DNA-associated Nuclear Proteins. Clin. Ther. 2016;38:1029–1041. doi: 10.1016/j.clinthera.2016.02.029. [DOI] [PubMed] [Google Scholar]
- 7.Klune J, Dhupar R, Cardinal J, Billiar TR, Tsung A. HMGB1: endogenous danger signaling. Mol. Med. 2008;14:1. doi: 10.2119/2008-00034.Klune. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bianchi ME, Agresti A. HMG proteins: dynamic players in gene regulation and differentiation. Curr. Opin. Genet. Dev. 2005;15:496–506. doi: 10.1016/j.gde.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 9.Stros M. HMGB proteins: interactions with DNA and chromatin. Biochim. Biophys. Acta. 2010;1799:101–13. doi: 10.1016/j.bbagrm.2009.09.008. [DOI] [PubMed] [Google Scholar]
- 10.Sessa L, Bianchi ME. The evolution of High Mobility Group Box (HMGB) chromatin proteins in multicellular animals. Gene. 2007;387:133–40. doi: 10.1016/j.gene.2006.08.034. [DOI] [PubMed] [Google Scholar]
- 11.Stillman DJ. Nhp6: a small but powerful effector of chromatin structure in Saccharomyces cerevisiae. Biochim. Biophys. Acta. 2010;1799:175–80. doi: 10.1016/j.bbagrm.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Briquet S, et al. High-Mobility-Group Box Nuclear Factors of Plasmodium falciparum †. Eukaryot Cell. 2006;5:672–682. doi: 10.1128/EC.5.4.672-682.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang H, et al. A nuclear factor of high mobility group box protein in Toxoplasma gondii. PLoS One. 2014;9:e111993. doi: 10.1371/journal.pone.0111993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abhyankar MM, et al. Characterization of an Entamoeba histolytica high-mobility-group box protein induced during intestinal infection. Eukaryot. Cell. 2008;7:1565–72. doi: 10.1128/EC.00123-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kappes F, Scholten I, Richter N, Gruss C, Waldmann T. Functional domains of the ubiquitous chromatin protein DEK. Mol. Cell. Biol. 2004;24:6000–10. doi: 10.1128/MCB.24.13.6000-6010.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cribb P, Perozzi M, Villanova GV, Trochine A, Serra E. Characterization of TcHMGB, a high mobility group B family member protein from Trypanosoma cruzi. Int. J. Parasitol. 2011;41:1149–56. doi: 10.1016/j.ijpara.2011.06.009. [DOI] [PubMed] [Google Scholar]
- 17.Cribb P, et al. Trypanosoma cruzi High Mobility Group B (TcHMGB) can act as an inflammatory mediator on mammalian cells. PLoS Negl. Trop. Dis. 2017;11:e0005350. doi: 10.1371/journal.pntd.0005350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.El-Sayed NM, et al. The genome sequence of Trypanosoma cruzi, etiologic agent of Chagas disease. Science. 2005;309:409–15. doi: 10.1126/science.1112631. [DOI] [PubMed] [Google Scholar]
- 19.Burle-Caldas G, et al. Expanding the tool box for genetic manipulation of Trypanosoma cruzi. Mol. Biochem. Parasitol. 2015;203:25–33. doi: 10.1016/j.molbiopara.2015.10.004. [DOI] [PubMed] [Google Scholar]
- 20.Taylor MC, Huang H, Kelly JM. Genetic Techniques in Trypanosoma cruzi. Adv. Parasitol. 2011;75:231–250. doi: 10.1016/B978-0-12-385863-4.00011-3. [DOI] [PubMed] [Google Scholar]
- 21.Taylor MC, Kelly JM. pTcINDEX: a stable tetracycline-regulated expression vector for Trypanosoma cruzi. BMC Biotechnol. 2006;6:32. doi: 10.1186/1472-6750-6-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peng D, Kurup SP, Yao PY, Minning TA, Tarleton RL. CRISPR-Cas9-Mediated Single-Gene and Gene Family Disruption in Trypanosoma cruzi. MBio. 2014;6:e02097–14. doi: 10.1128/mBio.02097-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lander, N., Chiurillo, M., Vercesi, A. & Docampo, R. Endogenous C-terminal Tagging by CRISPR/Cas9 in Trypanosoma cruzi. BIO-PROTOCOL7, (2017). [DOI] [PMC free article] [PubMed]
- 24.Romagnoli BAA, et al. Improvements in the CRISPR/Cas9 system for high efficiency gene disruption in Trypanosoma cruzi. Acta Trop. 2018;178:190–195. doi: 10.1016/j.actatropica.2017.11.013. [DOI] [PubMed] [Google Scholar]
- 25.Alonso VL, Ritagliati C, Cribb P, Serra EC. Construction of three new Gateway® expression plasmids for Trypanosoma cruzi. Mem. Inst. Oswaldo Cruz. 2014;109:1081–5. doi: 10.1590/0074-0276140238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laverrière M, Cazzulo JJ, Alvarez VE. Antagonic activities of Trypanosoma cruzi metacaspases affect the balance between cell proliferation, death and differentiation. Cell Death Differ. 2012;19:1358–69. doi: 10.1038/cdd.2012.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Romaniuk MA, Frasch AC, Cassola A. Translational repression by an RNA-binding protein promotes differentiation to infective forms in Trypanosoma cruzi. PLOS Pathog. 2018;14:e1007059. doi: 10.1371/journal.ppat.1007059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ritagliati C, Alonso VL, Manarin R, Cribb P, Serra EC. Overexpression of Cytoplasmic TcSIR2RP1 and Mitochondrial TcSIR2RP3 Impacts on Trypanosoma cruzi Growth and Cell Invasion. PLoS Negl. Trop. Dis. 2015;9:e0003725. doi: 10.1371/journal.pntd.0003725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kang R, Zhang Q, Zeh HJ, Lotze MT, Tang D. HMGB1 in cancer: good, bad, or both? Clin. Cancer Res. 2013;19:4046–57. doi: 10.1158/1078-0432.CCR-13-0495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vizoso-Vázquez A, et al. HMGB proteins involved in TOR signaling as general regulators of cell growth by controlling ribosome biogenesis. Curr. Genet. 2018;0:3. doi: 10.1007/s00294-018-0842-8. [DOI] [PubMed] [Google Scholar]
- 31.Morriswood B, Engstler M. Let’s get fISSical: fast in silico synchronization as a new tool for cell division cycle analysis. Parasitology. 2018;145:196–209. doi: 10.1017/S0031182017000038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Elias MC, et al. Morphological Events during the Trypanosoma cruzi Cell Cycle. Protist. 2007;158:147–157. doi: 10.1016/j.protis.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 33.da Silva MS, Muñoz PAM, Armelin HA, Elias MC. Differences in the Detection of BrdU/EdU Incorporation Assays Alter the Calculation for G1, S, and G2 Phases of the Cell Cycle in Trypanosomatids. J. Eukaryot. Microbiol. 2017;64:756–770. doi: 10.1111/jeu.12408. [DOI] [PubMed] [Google Scholar]
- 34.Williams, A. F. In System Analysis and Simulation Ecology (ed. Patten, B.) 247–262 (New York: Academic Press, 1971).
- 35.Stanners CP, Till JE. DNA synthesis in individual L-strain mouse cells. Biochim. Biophys. Acta. 1960;37:406–419. doi: 10.1016/0006-3002(60)90496-0. [DOI] [PubMed] [Google Scholar]
- 36.Elias MC, Nardelli SC, Schenkman S. Chromatin and nuclear organization in Trypanosoma cruzi. Future Microbiol. 2009;4:1065–1074. doi: 10.2217/fmb.09.74. [DOI] [PubMed] [Google Scholar]
- 37.Leandro de Jesus TC, et al. Quantitative Proteomic Analysis of Replicative and Nonreplicative Forms Reveals Important Insights into Chromatin Biology of Trypanosoma cruzi. Mol. Cell. Proteomics. 2017;16:23–38. doi: 10.1074/mcp.M116.061200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Figueiredo LM, Cross GAM, Janzen CJ. Epigenetic regulation in African trypanosomes: a new kid on the block. Nat. Rev. Microbiol. 2009;7:504–13. doi: 10.1038/nrmicro2149. [DOI] [PubMed] [Google Scholar]
- 39.Maree JP, Patterton H-G. The epigenome of Trypanosoma brucei: a regulatory interface to an unconventional transcriptional machine. Biochim. Biophys. Acta. 2014;1839:743–50. doi: 10.1016/j.bbagrm.2014.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Elias MC, Faria M. Are there epigenetic controls in Trypanosoma cruzi? Ann. N. Y. Acad. Sci. 2009;1178:285–90. doi: 10.1111/j.1749-6632.2009.05008.x. [DOI] [PubMed] [Google Scholar]
- 41.Ekanayake D, Sabatini R. Epigenetic regulation of polymerase II transcription initiation in Trypanosoma cruzi: modulation of nucleosome abundance, histone modification, and polymerase occupancy by O-linked thymine DNA glucosylation. Eukaryot. Cell. 2011;10:1465–72. doi: 10.1128/EC.05185-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ekanayake DK, et al. Epigenetic regulation of transcription and virulence in Trypanosoma cruzi by O-linked thymine glucosylation of DNA. Mol. Cell. Biol. 2011;31:1690–700. doi: 10.1128/MCB.01277-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aresta-Branco F, Pimenta S, Figueiredo LM. A transcription-independent epigenetic mechanism is associated with antigenic switching in Trypanosoma brucei. Nucleic Acids Res. 2016;44:3131–3146. doi: 10.1093/nar/gkv1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Respuela P, Ferella M, Rada-Iglesias A, Aslund L. Histone acetylation and methylation at sites initiating divergent polycistronic transcription in Trypanosoma cruzi. J. Biol. Chem. 2008;283:15884–92. doi: 10.1074/jbc.M802081200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zuma AA, et al. Chaetocin—A histone methyltransferase inhibitor—Impairs proliferation, arrests cell cycle and induces nucleolar disassembly in Trypanosoma cruzi. Acta Trop. 2017;170:149–160. doi: 10.1016/j.actatropica.2017.02.007. [DOI] [PubMed] [Google Scholar]
- 46.Narayanan MS, Rudenko G, Shankar Narayanan M, Rudenko G. TDP1 is an HMG chromatin protein facilitating RNA polymerase i transcription in African trypanosomes. Nucleic Acids Res. 2013;41:2981–92. doi: 10.1093/nar/gks1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alonso, V. L., Ritagliati, C., Cribb, P., Cricco, J. A. & Serra, E. C. Overexpression of bromodomain factor 3 in Trypanosoma cruzi (TcBDF3) affects differentiation of the parasite and protects it against bromodomain inhibitors. FEBS J. 283, (2016). [DOI] [PubMed]
- 48.Dubois, D. P. & Boisvert, F.-M. In The Functional Nucleus (eds Bazett-Jones, D. P. & Dellaire, G.) 29–49 (Springer 2016).
- 49.Nepomuceno-Mejía T, et al. The Trypanosoma cruzi nucleolus: a morphometrical analysis of cultured epimastigotes in the exponential and stationary phases. FEMS Microbiol. Lett. 2010;313:41–46. doi: 10.1111/j.1574-6968.2010.02117.x. [DOI] [PubMed] [Google Scholar]
- 50.dos Santos, C. M. B. et al. Trypanosoma cruzi transcriptome during axenic epimastigote growth curve. Mem. Inst. Oswaldo Cruz113, (2018). [DOI] [PMC free article] [PubMed]
- 51.Povelones ML, Gluenz E, Dembek M, Gull K, Rudenko G. Histone H1 Plays a Role in Heterochromatin Formation and VSG Expression Site Silencing in Trypanosoma brucei. PLoS Pathog. 2012;8:e1003010. doi: 10.1371/journal.ppat.1003010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dossin F, de M, Schenkman S. Actively Transcribing RNA Polymerase II Concentrates on Spliced Leader Genes in the Nucleus of Trypanosoma cruzi. Eukaryot. Cell. 2005;4:960–970. doi: 10.1128/EC.4.5.960-970.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gluenz E, Taylor MC, Kelly JM. The Trypanosoma cruzi metacyclic-specific protein Met-III associates with the nucleolus and contains independent amino and carboxyl terminal targeting elements. Int. J. Parasitol. 2007;37:617–25. doi: 10.1016/j.ijpara.2006.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Olson MO, Dundr M, Szebeni A. The nucleolus: an old factory with unexpected capabilities. Trends Cell Biol. 2000;10:189–96. doi: 10.1016/S0962-8924(00)01738-4. [DOI] [PubMed] [Google Scholar]
- 55.Tiengwe C, et al. Genome-wide Analysis Reveals Extensive Functional Interaction between DNA Replication Initiation and Transcription in the Genome of Trypanosoma brucei. CELREP. 2012;2:185–197. doi: 10.1016/j.celrep.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou Q, Li Z. A backup cytokinesis pathway in Trypanosoma brucei. Cell Cycle. 2016;15:2379–2380. doi: 10.1080/15384101.2016.1181882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tu X, Kumar P, Li Z, Wang CC. An aurora kinase homologue is involved in regulating both mitosis and cytokinesis in Trypanosoma brucei. J. Biol. Chem. 2006;281:9677–9687. doi: 10.1074/jbc.M511504200. [DOI] [PubMed] [Google Scholar]
- 58.Kumar P, Wang CC. Dissociation of cytokinesis initiation from mitotic control in a eukaryote. Eukaryot. Cell. 2006;5:92–102. doi: 10.1128/EC.5.1.92-102.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shimada, M. et al. Essential role of autoactivation circuitry on Aurora B-mediated H2AX-pS121 in mitosis. Nat. Commun. 7, (2016). [DOI] [PMC free article] [PubMed]
- 60.Basant A, et al. Aurora B Kinase Promotes Cytokinesis by Inducing Centralspindlin Oligomers that Associate with the Plasma Membrane. Dev. Cell. 2015;33:204–215. doi: 10.1016/j.devcel.2015.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hu H, et al. The Aurora B kinase in Trypanosoma brucei undergoes post-translational modifications and is targeted to various subcellular locations through binding to TbCPC1. Mol. Microbiol. 2014;91:256–274. doi: 10.1111/mmi.12458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hammarton TC, Kramer S, Tetley L, Boshart M, Mottram JC. Trypanosoma brucei Polo-like kinase is essential for basal body duplication, kDNA segregation and cytokinesis. Mol. Microbiol. 2007;65:1229–1248. doi: 10.1111/j.1365-2958.2007.05866.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Contreras VT, Salles JM, Thomas N, Morel CM, Goldenberg S. In vitro differentiation of Trypanosoma cruzi under chemically defined conditions. Mol. Biochem. Parasitol. 1985;16:315–27. doi: 10.1016/0166-6851(85)90073-8. [DOI] [PubMed] [Google Scholar]
- 64.Rasband, W. ImageJ [Software]. U. S. Natl. Institutes Heal. Bethesda, Maryland, USA //imagej.nih.gov/ij/ (2015).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No datasets were generated or analyzed during the current study.