Abstract
Resistance to 5-Fluorouracil chemotherapy is a major cause of therapeutic failure in colon cancer cure. Development of combined therapies constitutes an effective strategy to inhibit cancer cells and prevent the emergence of drug resistance. For this purpose, we investigated the anti-tumoral effect of thirteen phenolic compounds, from the Tunisian quince Cydonia oblonga Miller, alone or combined to 5-FU, on the human 5-FU-resistant LS174-R colon cancer cells in comparison to parental cells. Our results showed that only Kaempferol was able to chemo-sensitize 5-FU-resistant LS174-R cells. This phenolic compound combined with 5-FU exerted synergistic inhibitory effect on cell viability. This combination enhanced the apoptosis and induced cell cycle arrest of both chemo-resistant and sensitive cells through impacting the expression levels of different cellular effectors. Kaempferol also blocked the production of reactive oxygen species (ROS) and modulated the expression of JAK/STAT3, MAPK, PI3K/AKT and NF-κB. In silico docking analysis suggested that the potent anti-tumoral effect of Kaempferol, compared to its two analogs (Kaempferol 3-O-glucoside and Kampferol 3-O-rutinoside), can be explained by the absence of glucosyl groups. Overall, our data propose Kaempferol as a potential chemotherapeutic agent to be used alone or in combination with 5-FU to overcome colon cancer drug resistance.
Introduction
Colorectal cancer (CRC) is one of the most frequently occurring malignancies worldwide1. According to GLOBOCAN data, there were over 1.8 million new colorectal cancer cases and 881,000 deaths in 2018, accounting for about 1 in 10 cancer cases and deaths2. Globally, colorectal cancer ranks third in terms of incidence but second in terms of mortality since 40–50% of patients develop metastatic disease (mCRC)2,3. Although several chemotherapeutic agents have been identified to improve survival and quality of life of CRC patients4, 5-Fluorouracil (5-FU) remains recommended as the drug of a first choice after more than 30 years of clinical research5. The antimetabolite drug elicits its cytotoxic effect mainly through inhibition of Thymidylate Synthase (TS), a key enzyme for catalyzing the novo synthesis of thymine6.
In CRC, 5-FU was used in monotherapy or in combination with oxaliplatin (Folfox), irinotecan (Folfiri), or irinotecan and bevacizumab (Folfiri-bevacizumab). Unfortunately, the adjuvant chemotherapeutic regimens rarely cure cancer and disease relapses from the drug-resistant cells7.
Thus, resistance, either intrinsic or acquired during the course of treatment, is a major challenge for cancer therapy8. The development of chemoresistance can be attributed to a wide variety of mechanisms including drug influx and efflux, enhancement of drug inactivation and mutation of the drug target9. Acquired 5-FU resistance is generally caused by alteration in its metabolism. Overexpression of Thymidylate Synthase, for example, was mainly associated with 5-FU resistance in colorectal cancer10. Microarray analyses have shown that non-coding microRNAs (miRNAs) may enhance 5-FU resistance by regulating 5-FU-metabolizing enzymes11. The miR-433, miR-203, miR-192 and miR-215 regulate post-transcriptional expression of TS and modulate 5-FU chemosensitivity in colon cancer cells. Dihydropyrimidine dehydrogenase (DPD), the initial enzyme of 5-FU catabolism, can also be regulated by some miRNAs, including miR-27a, miR-27b, miR-582-5p, and miR-13411. Moreover, other mechanisms were implicated in conferring drug resistance to colorectal cancer cells such as the protection from apoptosis through the inhibition of pro-apoptotic and/or overexpression of survival proteins. Perturbation of cell cycle, preventing incorporation of 5-FU metabolites, and adaptive response to Reactive oxygen species (ROS) production have been also reported to cause 5-FU resistance6,12. Overexpression of ATP-binding cassette (ABC) transporters proteins including ATP-binding cassette sub-family G member 2 (ABCG2) and multidrug resistance-associated protein 1 (MDR1), known to mediate cellular efflux of the cytotoxic metabolite of 5-FU on cell membrane, is one of the key molecular mechanisms resulting in chemotherapeutic resistance13.
In colon cancer cells, the acquisition of invasive behavior was also related to Epithelial-mesenchymal transition (EMT) as a mechanism for 5-FU chemotherapy resistance14. Recent studies highlighted that overexpression of ABC transporters may be caused by the EMT as an important biological process that promotes drug resistance and tumor dissemination through deregulated expression of EMT mediators15.
Consequently, development of alternate strategies to improve the effectiveness of 5-FU chemotherapy and to overcome drug resistance are critically required16. Several studies have clearly shown that dietary polyphenols are among the naturally occurring substances that have shown promising anti-cancer properties and low toxicity in comparison to standard chemotherapeutic agents. Phenolic compounds exhibited anti-tumorigenic activities in multiple carcinogenesis pathways including the inhibition of cell proliferation, induction of apoptosis, modulation of oxidative stress, blockade of pro-inflammatory cascades and pathological angiogenesis and stimulation of anti-tumoral immune responses, which finally resulted in the arrest of cancer progression and metastasis17,18. An increase in the efficacy of chemotherapy and prevention of multidrug resistance are among other important effects of dietary polyphenols19. These compounds can not only kill cancer cells but also restore drug sensitivity20. Therefore, patients with colorectal cancer often adopt natural antioxidants or dietary supplements in their regimen as adjuncts to the conventional chemotherapy based on the belief that they would exhibit beneficial effects21. In fact, it has been shown that a combination of selected natural compounds improves the treatment efficacy of chemotherapy and increases the drug sensitivity in cancer cells22.
We have previously reported that peel polyphenolic extract (Peph) from the Tunisian quince (Cydonia oblonga Miller) displays a potent anti-tumoral effect in human colon adenocarcinoma LS174 cells. In the present study, we extend this work to investigate the anti-proliferative potentiality of each phenolic compound from total Peph extract on an in-house generated 5-FU-resistant cell line (LS174-R), in comparison with the parental 5-FU-sensitive LS174 cells. Interestingly, we found that Kaempferol, one of the phenolic compounds, can be proposed as a potential chemotherapeutic agent by its own and/or in combination to improve the sensitivity of 5-FU-resistant colon cancer cells.
Results
The 5-FU-resistant LS174-R cells display different characteristics from the parental one
To mimic development of 5-Fluorouracil-resistant colorectal tumors, a 5-FU-resistant LS174-R cells were generated in-house by continuous exposure of parental sensitive cells to increasing concentrations of 5-FU (10–100 μM). Refractive cells to 60 μM of 5-Fluorouracil were obtained after 8 months of treatment and were characterized morphologically by the acquisition of an appearance different from the parental LS174 cells (Fig. 1a). They became elongated and asteroid shaped compared to the parental cell type.
Figure 1.
Characteristics of 5-FU-resistant LS174-R cells in comparison to the parental ones. (a) Representative microscopic images of both sensitive and 5-FU resistant cancer cells photographed under phase contrast microscope. (b) Proliferation rate of parental and 5-FU resistant cells for 24 h, 48 h and 72 h was determined by MTT assay. (c) Colony-forming capacity of parental and 5-FU-resistant cells was measured using clonogenic survival assay. 2000 viable cells from each group were cultured in six-well plates for additional 10 days. Colonies were stained with crystal violet and each assay was photographed. (d) Representative microscopy images of colon cancer cells in 3D cultures (x10). Both colon cancer cells were analyzed for spheroid formation capacity in ultra-low attachment (ULA) round bottom 96-well plates coated with agarose. (e) Cell cycle analysis of 5-FU-resistant cells compared to parental cells by flow cytometry using propidium iodide assay. (f) Protein expression of different cellular effectors was analyzed in both parental LS174 and 5-FU resistant LS174-R cells by western blot using specific antibodies. β-actin was used as a reference protein for equal loading. (g) Sensitive LS174 and resistant LS174-R cells were treated with different concentrations of 5-FU. The IC50 (50% inhibitory concentration) values were calculated at 72 h time post treatment with MTT assay. Results were normalized to each control in percentage and represented as mean ± SE of three independent experiments, each performed at least in triplicate. *p < 0.05, **p < 0.01, ***p < 0.005 and ns: non significant.
For comparative study, the proliferation rate of parental LS174 cells and chemo-resistant cancer LS174-R cells was measured, over 3 days, using MTT cell viability assay. Our results showed that 5-FU-resistant cells grow slowly compared to the parental LS174 cells (Fig. 1b). Accordingly, the sensitive cells were able to produce higher number of colonies (463 colonies vs 318 for 5-FU-resistant LS174-R cells) (Fig. 1c) and larger spheroids in comparison to resistant cells which produced grape-like spheroids with small, round, single cells at the border of the compact spheres (Fig. 1d).
On the basis of the morphologic changes observed in LS174-R cells, we explored the characteristics and the molecular mechanisms driving the changes in resistant cells by analyzing the cell cycle distribution and investigating the levels of some proteins linked to 5-FU resistance such as ABC transporters, EMT protein markers and apoptosis effectors in both parental and resistant cells. As shown in Fig. 1e, the proportion of LS174-R cells (26.5%) in the S phase was higher than that of the parental LS174 cells (7.5%) and decreased in the G2/M phase (20.7% vs 34.7% in parental cells).
Western blot analysis indicated that chemo-resistant LS174-R cells expressed high levels of MDR1, ABCG2, c-Src and FAK proteins along with reduced expression of Bad and E-cadherin proteins relative to parental LS174 cells. Bcl2 expression was similar in both cell lines (Fig. 1f). However, we could not detect vimentin and Bax proteins in both sensitive and resistant cells, which is in accordance with previous work reporting that LS174 cells lack expression of these two proteins23,24.
To highlight the adaptation and acquired resistance of refractive cells to high concentrations of 5-Fluorouracil, the viability of parental and resistant cells at different doses of the drug was investigated using MTT assay. As shown in Fig. 1g, the IC50 value of 5-FU in resistant LS174-R cells (IC50 = 706 µM) was 26-fold higher than that of sensitive LS174 cells in which 26.9 µM were needed to kill 50% of the parental cells (Fig. 1g).
Kaempferol inhibits the viability of 5-FU-resistant LS174-R cancer cells
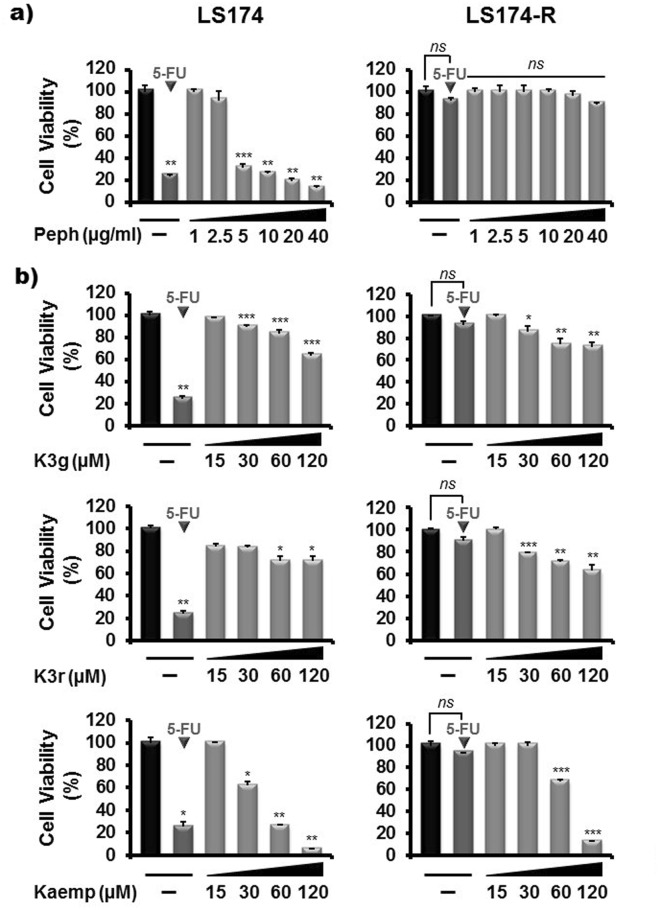
To assess the effect of quince peel polyphenolic extract on growth of 5-FU-resistant LS174-R cancer cells, MTT assay was employed. For comparative study, both parental and chemo-resistant cancer cells were treated for 72 h with increasing concentrations (0–40 µg/ml) of Peph polyphenolic extract. Consistent with our previous findings25, Peph significantly reduced the viability of parental LS174 cells in a dose-dependent manner. However, no significant inhibitory effect was observed in the growth of 5-FU-resistant LS174-R cells (Fig. 2a). Thereby, we proposed to evaluate the effect of each phenolic compound identified in the total peel polyphenolic extract on the viability of chemo-resistant LS174-R cells compared to parental LS174 cells. As summarized in Tables 1 and 2, phenolic compounds tested at equivalent concentrations to that present in 5 µg/ml, 10 µg/ml and 20 µg/ml of the total peel polyphenolic extract failed to induce any significant effect on cell viability in both parental and resistant cells.
Figure 2.
Kaempferol overcomes 5-FU resistance in LS174-R cancer cells. Parental LS174 cells and resistant LS174-R cells were cultured in 96-well plates and treated with increasing concentrations of (a) Cydonia oblonga Miller peel polyphenolic extract (Peph) (1–40 µg/ml) and (b) Kaempferol and its analogs, Kaempferol 3-O-glucoside (K3g) and Kaempferol 3-O-rutinoside (K3r), (15–120 µM) for 72 h. Cell viability was measured by MTT assay. The absorbance was measured at 540 nm. Results were normalized to each control in percentage and represented as mean ± SE of three independent experiments. *p < 0.05, **p < 0.01, ***p < 0.005 when compared to their respective CN and ns: non significant.
Table 1.
Effect of quince peel compounds on the viability of sensitive LS174 cells.
| Treatment (µM) | Cell Viability (%) | ||||||
|---|---|---|---|---|---|---|---|
| Mock | 100 ± 0.3 | ||||||
| 5-FU (60 µM) | 25 ± 0.9 | ||||||
| Compounds | % 5 µg/ml | % 10 µg/ml | % 20 µg/ml | 15 µM | 30 µM | 60 µM | 120 µM |
| Q | 100 ± 1.2 | 97 ± 0.2 | 96 ± 0.6 | 63 ± 1.3 | 41 ± 0.3 | 33 ± 0.7 | 20 ± 0.2 |
| R | 90 ± 0.5 | 50 ± 0.9 | 44 ± 0.9 | 41 ± 1.1 | 42 ± 0.23 | 38 ± 0.3 | 31 ± 1.3 |
| +C | 100 ± 0.43 | 100 ± 0.32 | 97 ± 0.81 | 98 ± 0.54 | 41 ± 1.63 | 40 ± 0.52 | 37 ± 0.5 |
| −C | 100 ± 0.63 | 91 ± 0.3 | 72 ± 0.63 | 56 ± 0.44 | 48 ± 0.61 | 47 ± 0.42 | 38 ± 0.21 |
| H | 100 ± 0.51 | 100 ± 0.91 | 99 ± 0.56 | 84 ± 1.4 | 38 ± 1.3 | 37 ± 1.63 | 35 ± 1.7 |
| I | 100 ± 0.3 | 100 ± 0.53 | 91 ± 0.57 | 50 ± 0.64 | 48 ± 0.81 | 47 ± 0.29 | 39 ± 0.77 |
| ChA | 93 ± 0.48 | 91 ± 0.6 | 90 ± 0.72 | 84 ± 0.7 | 85 ± 0.66 | 84 ± 0.35 | 41 ± 0.58 |
| CrA | 100 ± 1.6 | 100 ± 0.33 | 100 ± 0.9 | 89 ± 1.6 | 51 ± 1.6 | 30 ± 1.4 | 25 ± 0.93 |
| NeA | 100 ± 0.77 | 100 ± 0.63 | 90 ± 0.42 | 80 ± 0.3 | 46 ± 0.23 | 38 ± 0.53 | 34 ± 0.74 |
| PcA | 100 ± 0.73 | 98 ± 1.3 | 94 ± 1.2 | 64 ± 0.99 | 58 ± 0.92 | 48 ± 0.71 | 32 ± 0.81 |
| K3g | 100 ± 0.33 | 100 ± 0.51 | 100 ± 0.14 | 97 ± 0.26 | 89 ± 0.4 | 83 ± 0.21 | 63 ± 1.1 |
| K3r | 100 ± 0.66 | 100 ± 0.3 | 100 ± 0.42 | 84 ± 0.73 | 83 ± 0.3 | 79 ± 1.1 | 71 ± 1.3 |
| Kaemp | 100 ± 0.13 | 100 ± 0.23 | 100 ± 0.76 | 100 ± 0.16 | 62 ± 0.83 | 26 ± 0.91 | 5 ± 0.13 |
Each phenolic compounds [Quercetin (Q), Rutin (R), (+)-Catechin (+C), (−)-Catechin (−C), Hyperin (H), Isoquercitrin (I), Chlorogenic acid (ChA), Cryptochlorogenic acid (CrA), Neochlorogenic acid (NeA), p-coumaric acid (PcA), Kaempferol-3-O-glucoside (K3g), Kaempferol-3-O-rutinoside (K3r) and Kaempferol (Kaemp)] was tested at equivalent concentrations to that present in 5 µg/ml, 10 µg/ml and 20 µg/ml of the total peel polyphenolic extract and at increasing concentrations (15–120 µM). Cell viability was determined by MTT assay after 72 h of treatment. The absorbance was measured at 540 nm. Results were normalized to each control in percentage and represented as mean ± SE of three independent experiments.
Table 2.
Effect of quince peel compounds on the viability of resistant LS174-R cells.
| Treatment (µM) | Cell Viability (%) | ||||||
|---|---|---|---|---|---|---|---|
| Mock | 100 ± 0.61 | ||||||
| 5-FU (60 µM) | 93 ± 0.17 | ||||||
| Compounds | % 5 µg/ml | % 10 µg/ml | % 20 µg/ml | 15 µM | 30 µM | 60 µM | 120 µM |
| Q | 100 ± 0.12 | 100 ± 0.71 | 100 ± 0.3 | 100 ± 0.9 | 100 ± 1 | 100 ± 0.7 | 100 ± 0.4 |
| R | 100 ± 0.21 | 100 ± 0.12 | 100 ± 0.22 | 100 ± 0.29 | 98 ± 0.38 | 87 ± 0.49 | 86 ± 0.6 |
| +C | 100 ± 0.63 | 100 ± 0.24 | 100 ± 0.34 | 100 ± 0.5 | 100 ± 1 | 100 ± 1.3 | 98 ± 0.39 |
| −C | 100 ± 1.4 | 100 ± 0.7 | 100 ± 0.98 | 100 ± 0.13 | 98 ± 0.77 | 91 ± 0.52 | 84 ± 0.31 |
| H | 100 ± 0.43 | 100 ± 0.18 | 100 ± 0.22 | 100 ± 0.43 | 100 ± 0.48 | 94 ± 0.18 | 91 ± 0.7 |
| I | 100 ± 0.41 | 100 ± 0.45 | 100 ± 0.22 | 100 ± 0.7 | 100 ± 0.59 | 93 ± 0.19 | 90 ± 0.46 |
| ChA | 100 ± 0.26 | 100 ± 0.48 | 100 ± 0.49 | 100 ± 0.8 | 100 ± 0.32 | 100 ± 0.22 | 100 ± 0.63 |
| CrA | 100 ± 0.48 | 100 ± 0.46 | 100 ± 0.84 | 100 ± 0.7 | 100 ± 0.53 | 100 ± 0.3 | 82 ± 0.23 |
| NeA | 100 ± 0.7 | 100 ± 0.12 | 100 ± 0.3 | 100 ± 0.26 | 91 ± 0.14 | 92 ± 0.37 | 82 ± 0.12 |
| PcA | 100 ± 1 | 100 ± 0.45 | 100 ± 1.01 | 100 ± 0.7 | 99 ± 1.06 | 98 ± 0.38 | 94 ± 0.7 |
| K3g | 100 ± 0.2 | 100 ± 0.46 | 100 ± 0.2 | 100 ± 0.54 | 86 ± 0.6 | 74 ± 0.13 | 70 ± 0.32 |
| K3r | 100 ± 0.53 | 100 ± 0.32 | 100 ± 0.59 | 100 ± 0.4 | 79 ± 0.6 | 71 ± 0.59 | 63 ± 0.13 |
| Kaemp | 100 ± 0.21 | 100 ± 0.49 | 100 ± 0.56 | 100 ± 0.12 | 100 ± 0.24 | 67 ± 0.17 | 13 ± 0.56 |
Each phenolic compounds [Quercetin (Q), Rutin (R), (+)-Catechin (+C), (−)-Catechin (−C), Hyperin (H), Isoquercitrin (I), Chlorogenic acid (ChA), Cryptochlorogenic acid (CrA), Neochlorogenic acid (NeA), p-coumaric acid (PcA), Kaempferol-3-O-glucoside (K3g), Kaempferol-3-O-rutinoside (K3r) and Kaempferol (Kaemp)] was tested at equivalent concentrations to that present in 5 µg/ml, 10 µg/ml and 20 µg/ml of the total peel polyphenolic extract and at increasing concentrations (15–120 µM) after 72 h of treatment. Cell viability was determined by MTT assay. The absorbance was measured at 540 nm. Results were normalized to each control in percentage and represented as mean ± SE of three independent experiments.
Interestingly, we found that increasing concentrations (0–120 µM) of each phenolic compound inhibit the growth of sensitive LS174 cells in a dose-dependent manner after 72 h of treatment. However, only Kaempferol and its analogs (Kaempferol 3-O-glucoside and Kaempferol 3-O-rutinoside) were able to reduce the viability of 5-FU-resistant cells (Table 2 and Fig. 2b). As shown in Fig. 2b, at a concentration of 120 µM, Kaempferol induced the most effective inhibitory effect with more than 80% of growth inhibition while this effect did not exceed 30% and 37% for Keampferol 3-O-glucoside (K3g) and Kaempferol 3-O-rutinoside (K3r), respectively, after 72 h of treatment.
Kaempferol chemo-sensitizes resistant cancer cells to 5-FU chemotherapy
For comparative chemosensitivity study, the parental and 5-FU-resistant cells were analyzed by MTT and colony formation assays for cellular growth following treatment with 5-FU alone or combined with increasing concentrations (0–120 µM) of Kaempferol. Both colon cancer cells were pretreated for different time (8 h, 12 h and 24 h) with Kaempferol (1–75 µM) and then exposed to 60 µM of 5-FU after removing or preserving the phenolic compound in the culture medium for additional 72 h. We found that Kaempferol pretreatment decreased the growth of parental and 5-FU-resistant cancer cells in dose and time-dependent manners (see Supplementary Fig. S1). Kaempferol, at 75 µM concentration, was able to induce 40% of growth inhibition in 5-FU-resistant LS174-R cells after 24 h of pretreatment. This effect was improved to about 70% when Kaempferol was not removed from the culture medium (see Supplementary Fig. S1).
Interestingly, when cells were treated with Kaempferol at concentrations exceeding 75 µM (15–120 µM) for 72 h, this phenolic compound significantly decreased cell proliferation of both parental and chemo-resistant cells in a concentration-dependent manner. The IC50 of Kaempferol was approximately 75 µM on LS174-R cells after 72 h of exposure. This inhibitory effect was enhanced to about 65% after the combined treatment with 60 µM of 5-FU (Fig. 3a). It should be noted that 5-FU alone did not affect the viability of LS174-R cells at 60 µM concentration (Fig. 3a).
Figure 3.
Kaempferol chemo-sensitizes resistant colon cancer cells to 5-FU chemotherapy. (a) Parental LS174 cells and resistant LS174-R cells were cultured in 96-well plates and treated with a concentration range from 15 µM to 120 µM of Kaempferol alone, 5-FU (60 µM) alone and the combination of both for 72 h. Cell viability was measured by MTT assay. The absorbance was measured at 540 nm. Assays were performed in triplicate. (b) Kaempferol effect on parental LS174 cells and resistant LS174-R cells colony-forming capacity was measured using clonogenic survival assay. Colon cancer cells were treated with vehicle (control) or Kaempferol (15 and 75 µM) combined or not to 60 µM of 5-FU for 72 h. After removal of the medium, 2000 viable cells from each group were cultured in six-well plates for additional 10 days. Colonies were stained with crystal violet and each assay was photographed. (c) The number of colonies was analyzed and scored by CFU scope quantification software. Results are expressed as the number of colony forming cells per well in percentage and normalized to control (vehicle, considered to represent 100%). Graphs are represented as mean ± SD of three independent experiments. *p < 0.05, **p < 0.01 and ***p < 0.005 when compared to their respective controls. #p < 0.05, ##p < 0.01 (Kaempferol + 5-FU groups vs Keampferol groups).
Thus, on the basis of this result, the Kaempferol’s concentration of 75 µM was selected for further investigations. Our data showed that Kaempferol notably sensitized the chemo-resistant LS174-R cells to 5-FU treatment starting from low dose (15 µM) (Fig. 3), which suggests a synergistic or additive anti-tumor effect between 5-FU and Kaempferol in both 5-FU-sensitive and resistant cells.
Pharmacological interaction between 5-Fluorouracil and Kaempferol
Because certain drug interactions can lead to a loss of therapeutic efficacy26, a combination studies between Kaempferol and 5-Fluorouracil were performed on the viability of 5-FU-resistant LS174-R and parental LS174 cells. Our result showed that combined treatment of 5-FU with Kaempferol reduced the 5-FU-IC50 values in parental LS174 cells (from 29.9 to 20.9 µM) and 5-FU-resistant LS174-R cells (from 706 to 274.2 µM). Interestingly, the analysis of drug interaction after calculating the Combination Index (CI) revealed an additive effect (CI = 1) in sensitive cells and confirmed a synergism (CI < 1) in 5-FU-resistant cells (Table 3).
Table 3.
Drug and compound interaction in parental and 5-FU-resistant LS174 colon cancer cells.
| Cell line | Treatment | IC50 (µM) | Combination Index (CI) |
|---|---|---|---|
| LS174 | Kaempferol | 44.76 ± 1.06 | 1 |
| 5-FU | 26.9 ± 0.36 | ||
| LS174-R | Kaempferol | 72.98 ± 0.19 | 0.6 |
| 5-FU | 706 ± 0.28 |
Combination index values for Kaempferol and 5-Fluorouracil was calculated after 72 h of treatment of sensitive LS174 and 5-FU-resistant LS174-R cells at 50% growth inhibition.
CI <1 indicates synergism, CI = 1 additive effect and CI >1 antagonism.
Kaempferol induces cell cycle arrest of colon cancer cells
Cell cycle perturbation can be involved in acquired 5-FU resistance27. In accordance with this, we analyzed the cell cycle phase distribution of both resistant and parental cancer cells after 72 h of treatment with Kaempferol alone or combined with 60 µM of 5-Fluorouracil. As shown in Fig. 4a, 5-FU treatment induced accumulation of sensitive LS174 cells in the S phase of the cell cycle (from 7.5 to 20.6%) along with a decrease of cells in the G2/M phase (from 34.7 to 4.6%), compared to the mock-treated cells. However, no significant effect was observed on refractive LS174-R cells to 60 µM of 5-FU. Kaempferol at 75 µM alone or in combination with 5-FU increased parental cells proportion in the S phase (from 7.5 to 9.2% and to 10.7% respectively) and induced a decrease in the G2/M phase (from 34.7 to 18.5% and to 12.9% respectively). However, the resistant LS174-R cells were accumulated in G2/M phase only after the concomitant use of Kaempferol with 60 µM of 5-FU (from 20.7 to 28.7%). This effect was associated with an increase in sub-G0 population (more than 30%) in both sensitive and 5-FU resistant cancer cells (Fig. 4a).
Figure 4.
Kaempferol induces cell cycle arrest of sensitive and 5-FU-resistant cancer cells. (a) Cell cycle phase distribution of cancer cells cultured in the absence (mock) and the presence of Kaempferol (75 µM) alone or combined to 60 µM of 5-FU for 72 h were analyzed by flow cytometry using propidium iodide assay. Results were represented as mean ± SE of three independent experiments. *p < 0.05, **p < 0.01, ***p < 0.005 when compared to their respective CN, #p < 0.05, ##p < 0.01 (Kaempferol + 5-FU groups vs Keampferol groups) and ns: non significant. Cell cycle-related proteins were analyzed by western blotting in (b) 5-FU resistant cells versus parental cells and (c) after cell treatments with Kaempferol, 5-FU, and the combination of both for 72 h. β-actin was used as a loading control. The data shown are representative of three independent experiments.
To further characterize the mechanism by which Kaempferol induced cell growth arrest, we investigated the levels of some proteins involved in cell cycle arrest in both parental and 5-FU-resistant cancer cells. As shown in Fig. 4b, p21, p27 and cdc2 proteins were expressed at the same levels in both sensitive and chemo-resistant cells. However, chemo-resistant LS174-R cells expressed high level of the phosphorylated form of retinoblastoma (Rb), compared to parental LS174 cells where the level of phospho-p53 protein was increased. Kaempferol treatment of sensitive LS174 cells decreased the expression of p27 and the phospho-Rb, cdc2 and enhanced p53 and phospho-p53 (at Ser15) expression. The combined treatment (Kaempferol and 5-Fluorouracil) induced an increase in the expression level of p21, p53 and its phosphorylated form (at Ser15) along with a decrease in phospho-cdc2 and p27 expression levels (Fig. 4c). No effect was observed on the expression of phospho-Rb protein. Interestingly, while Kaempferol slightly decreased phospho-cdc2 and phospho-Rb proteins along with induced expression of phospho-p53 (at Ser15) and p21 in LS174-R cells, the combined treatment-mediated arrest of resistant cells in the G2-M phase was associated with the loss of the phosphorylated forms of Rb and cdc2 without any effect on the levels of p21 and p27 proteins (Fig. 4c).
Kaempferol induces a caspase-dependent apoptosis of resistant cancer cells
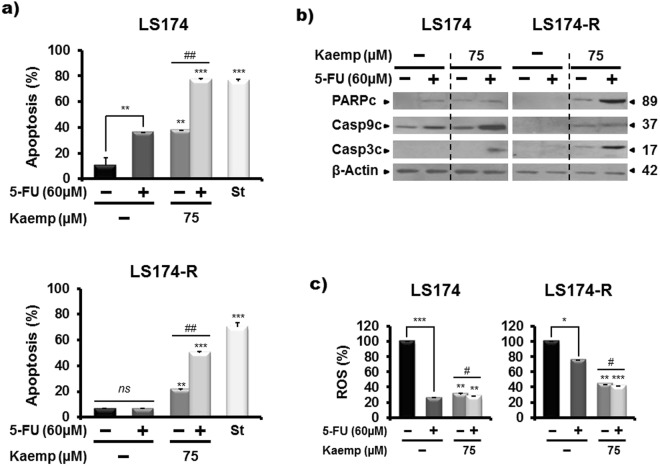
Resistance to apoptosis is one of the most important features of cancer28. The increase of sub-G0 population after Kaempferol treatment suggests that growth inhibition of colon cancer cells may be explained by cell apoptosis. Flow cytometric analysis (Fig. 5a) showed that, as expected, 5-FU treatment had no apoptotic effect on chemo-resistant LS174-R cells while 39% of the sensitive LS174-treated cells underwent apoptosis. Interestingly, the combined treatment with 5-FU and 75 µM Kaempferol increased the percentage of apoptotic cells to 76.8% in sensitive LS174 cells and to 50% in chemo-resistant LS174-R cells (Fig. 5a). The pro-apoptotic effect was associated with the activation of caspase 3 and caspase 9 and cleavage of PARP (Fig. 5b), suggesting that Kaempferol treatment chemo-sensitized resistant cancer cells to apoptosis through caspases-dependent mechanisms.
Figure 5.
Kaempferol chemosensitizes 5-FU-resistant cancer cells to apoptosis and reduces the ROS production. (a) Apoptosis detection in mock and treated-cells with Kaempferol alone or combined to 5-FU for 72 h by flow cytometry analysis using annexin-V/7-AAD staining. Staurosporin (2 µM, St) was used as a positive control of apoptosis. Results were represented as mean ± SE of three independent experiments. *p < 0.05, **p < 0.01, ***p < 0.005 when compared to their respective CN, #p < 0.05, ##p < 0.01 (Kaempferol + 5-FU groups vs Keampferol groups) and ns: non significant. (b) Western blot analysis of some apoptosis-related proteins in both parental LS174 and 5-FU resistant LS174-R cells after 72 h of treatment with Kaempferol, 5-FU and the combination of both. β-actin was used as a reference protein for equal loading. The data shown are representative of three independent experiments. (c) Histograms analysis of ROS production measured with CMH2DCFDA staining after 72 h of treatment with Kaempefrol, 5-FU and the combination of both in parental LS174 and 5-FU-resistant LS174-R cells. Detection of ROS was related to the quantity of subsequent oxidation leading emitting fluorescence. Data are reported as the means ± S.E.M of three independent experiments. *p < 0.05, **p < 0.01, *** p < 0.005 with respect to mock-treated controls; #p < 0.05, ##p < 0.01 (Kaempferol + 5-FU groups vs Keampferol groups).
Antioxidant activity of Kaempferol in colon cancer cells
It has been reported that Kaempferol reduces the cancer risk by increasing the body’s antioxidant defense against free radicals29. We thus assessed the effect of Kaempferol alone or combined with 5-FU on the intracellular redox status of parental LS174 cells and 5-FU-resistant cells. Our finding showed that Kaempferol at 75 μM reduced the reactive oxygen species (ROS) production by 69% and 56% after 72 h of treatment, in the sensitive and resistant cells, respectively (Fig. 5c). Combined treatment increased the inhibitory effect of Kaempferol on ROS production in both sensitive (to 73%) and chemo-resistant cells (to 60%).
Kaempferol blocks survival signaling pathways in colon cancer cells
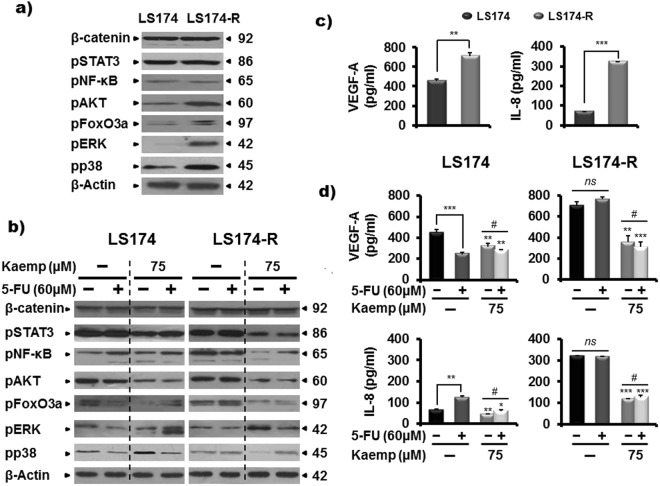
To further characterize the mechanisms by which Kaempferol chemosensitizes the resistant colon cancer cells to 5-FU treatment, we analyzed the expression level of some proteins involved in JAK/STAT3, Wnt/β-catenin, mitogen activated protein kinases (MAPK) cascade, phosphatidyl-3-phosphate kinase (PI3K)/AKT and NF-κB signaling pathways that play critical role in the development and progression of colorectal cancer30. Western blot analysis showed increased phosphorylation of extracellular-regulated kinases 1/2 (ERK1/2), p38 MAPK and AKT in chemo-resistant LS174-R cells compared to sensitive LS174 cells (Fig. 6a), suggesting the acquisition of more aggressive cell features related to the activation of MAPK and PI3K/AKT signaling pathways.
Figure 6.
Kaempferol modulates survival signaling pathways in both sensitive and resistant colon cancer cells and reduces the production of the two angiogenic factors, VEGF-A and IL-8 in 5-FU-refractive LS174-R cells. (a) Different cellular effectors were monitored in chemo-resistant LS174-R cells and sensitive LS174 cells by western blot using specific antibodies. β-actin was used as a loading control. (b) 5-FU-sensitive and resistant cancer cells were treated with Kaempferol, 5-FU, and the combination of both for 72 h. Protein extracts (30 µg) from whole cell lysates were analysed by western blotting using specific antibodies. β-actin was used as a reference protein for equal loading. One representative experiment of three independent ones was shown. (c) VEGF and IL-8 secretion were determined by human VEGF and IL-8 ELISA Kit in both control sensitive and 5-FU-resistant LS174-R cells. (d) Supernatants from parental LS174 and 5-FU resistant LS174-R cells cultured in the absence (vehicle) or presence of Kaempferol (75 µM) combined or not to 60 µM of 5-FU were collected and analyzed by Human VEGF and IL-8 specific ELISA. Results are reported as the mean ± SE of three independent experiments each run in triplicate (*p < 0.05, **p < 0.01, *** p < 0.005; #p < 0.05, ##p < 0.01 (Kaempferol + 5-FU groups vs Keampferol groups and ns: non significant). The data were corrected to the cell number.
After 72 h of treatment, 5-FU reduced the activation of ERK1/2 and p38 in sensitive tumor cells without any noticeable effect in chemo-resistant LS174-R cells. Interestingly, Kaempefrol alone or combined with 5-FU modulated the phosphorylation levels of the tested proteins in both parental and chemo-resistant cancer cells. As shown in Fig. 6b, Kaempferol treatment increased the expression of phospho-ERK1/2 associated to the inhibition of phospho-p38 kinase in resistant LS174-R cell line. The same effect was detected in sensitive LS174 cells with the combined treatment. This data suggests the existence of a cross-talk between ERK and p38 activation31.
Moreover, Kaempferol alone or combined with 5-FU reduced the phosphorylated form of STAT3 and the one of the pro-survival kinase AKT and its target tumor suppressor FOXO3a transcription factor, in both sensitive and resistant tumor cells (Fig. 6b). Our result also supports the inhibition of NF-κB in the refractive cancer cell line model, whereas it was slightly activated in treated-parental LS174 cells with the Kaempferol combined to 5-FU. However, no effect was detected on β-catenin activity for both parental and resistant cancer cells (Fig. 6b).
Kaempferol inhibits the production of VEGF-A and IL-8 angiogenic factors
Tumor growth depends in angiogenesis32. Therefore, we assessed the production of vascular endothelial growth factor (VEGF) and interleukin-8 (IL-8) known as the major positive regulators of angiogenesis. ELISA analysis highlighted that the secretion of IL-8 by chemo-resistant LS174-R cells was five times higher than that of LS174 sensitive cells. Similarly, LS174-R cells produced VEGF-A (707 pg/ml) to a greater extent than the parental LS174 cells (450 pg/ml) (Fig. 6c). This finding supports that LS174-R cells acquired other malignant characteristics than parental cells through expressing high amounts of IL-8 and VEGF-A proteins.
The treatment of sensitive cells with 5-FU modulated the VEGF-A and IL-8 cytokines secretion without affecting their levels in chemo-resistant tumor cells. Interestingly, Kaempferol alone or combined with 5-FU inhibited the production of the two angiogenic regulators in both colon cancer cells. ELISA analysis showed a 50% decrease of VEGF-A (from 707 to 355 pg/ml) and IL-8 (from 319 to 116 pg/ml) secretion after 72 h of treatment with Kaempefrol in chemo-resistant LS174-R cells. Interestingly, the combined treatment reduced also the production of VEGF-A (from 707 to 308 pg/ml) and IL-8 (from 319 to 127 pg/ml) (Fig. 6d) in 5-FU-resistant cells (Fig. 6d).
Kaempferol modulates the expression levels of enzymes involved in 5-FU metabolism
To further identify the molecular mechanism of 5-FU resistance in the generated chemo-resistant cancer cells, we investigated the expression level of five genes involved in drug metabolism such as, thymidylate synthase (TS), thymidine kinase (TK), dihydrofolate reductase (DHFR) and the folylpolyglutamate synthetase (FPGS) implicated in 5-FU anabolism and dihydropyrimidine dehydrogenase (DPD), the key enzyme of 5-FU catabolism33. Quantitative PCR analysis indicated an increase in mRNA levels of TS, TK, DHFR and FPGS in chemo-resistant cancer cells with a slight decrease in DPD expression compared to parental LS174 cells (Fig. 7a). Treatment of the cells with Kaempferol alone or combined with 5-FU significantly reduced the mRNA levels of the studied genes associated to 5-FU metabolism in 5-FU-resistant cells (Fig. 7b). These results were confirmed by western blotting analysis that showed a high protein levels of TS and TK in the 5-FU-resistant cells, compared to sensitive LS174 cells (Fig. 7c), while the Kaempferol alone or in combination with 60 µM of 5-FU decreased the expression of the two proteins in the resistant cells (Fig. 7d).
Figure 7.
Kaempferol modulates the expression levels of enzymes involved in 5-FU metabolism. (a) The amounts of mRNA transcripts of five genes, Thymidylate synthase (TS), Thymidine kinase (TK), Dihydrofolate reductase (DHFR), Folylpolyglutamate synthetase (FPGS) and Dihydropyrimidine dehydrogenase (DPD), involved in 5-FU metabolism were quantified by real time PCR in control sensitive and 5-FU-resitant cells and (b) in LS174 and LS174-R cells cultured in the absence (mock) and the presence of Kaempferol (75 µM) and/or 5-FU (60 µM) for 72 h. The values are normalized to GAPDH gene and the control value was taken as 1. Results are reported as the mean ± SE of three independent experiments each run in duplicate (*p < 0.05, **p < 0.01, *** p < 0.005; #p < 0.05, ##p < 0.01 (Kaempferol + 5-FU treatments vs Keampferol groups), ns: non significant) (c) TS and TK protein levels were assessed by western blotting in both control 5-FU-sensitive LS174 and resistant LS174-R cells and in (d) colon cancer cells after treatment with vehicle (mock), Kaempferol (75 µM), 5-FU (60 µM), and the combination of both for 72 h. β-actin was used as a reference protein for equal loading.
MAPK14 (p38) and PIM1 are two putative targets for Kaempferol
Considering the contribution of the Thymidylate synthase in acquiring the resistance to the treatment with 5-FU, we explored the putative interaction of the enzyme with Kaempferol in the cell signaling context. We established the protein-protein interaction network of 10 different signaling members, which expression levels were confirmed by western blot. The network consisted of 61 edges connecting 20 nodes, corresponding to interacting proteins. The highest betweenness scores were those for TP53, UBC and MAPK14 (p38) evaluated at 29.44, 16.3 and 16.03, respectively. Thymidylate synthase (TYMS) was situated on the edge of the network establishing connectivities with MAPK14, UBC and SP1 only with low betweenness score of 0.14 (Fig. 8a).
Figure 8.
Computational interaction study of Kaempferol and its analogs with putative human cell proteins. (a) Interaction network of the different proteins from which expression was evaluated by the western blot assay (represented by yellow nodes). Other proteins (blue nodes) emerged from the analysis as connectors of the input proteins. The degree of centrality is proportional to the node radius. (b) Docking of Kaempferol (yellow sticks) with PIM1. The first panel shows the good superposition of Kaempferol with Fisetin structure (Green sticks). The second and the third panel show the docking complexes of Kaempferol 3-O-rutinoside (K3r) and Kaempferol 3-O-glucoside (K3g) with PIM1 respectively, compared to the Kaempferol. (c) Docking solutions of Kaempferol (Yellow sticks) with the ATP binding site in MAPK14 (p38). The two other panels show the position of Kaempferol in the interaction site in comparison to the retained complexes for K3g and K3r. Unlike Kaempferol, these two ligands cannot attain the depth of the binding pocket.
We then verified if any of the network nodes were previously identified as a target for Kaempferol or other members of the flavonoid family by exploring the BindingDB34. From the 61 entries described for the Kaempferol, MAPK14 (p38α) was identified as putative target for Kaempferol along with two other proteins, CDK6 and PIM1 which are related to RBI/MAPK14 and FOXO3/STAT3, respectively members of the INPUT protein list.
The PharMApper server also was used to predict the potential cellular targets of Kaempferol starting from its 3D structure. The PharMApper report showed that uridine cytidine kinase 2 is a potential target with the highest fit score. CDK6 was situated at the 4th rank of the list, while MAPK14 (p38α) was ranked at the 30th position. MAPK14 occurred six other times in the list, probably because of the redundancy in the PDB data bank. However, the Thymidylate synthase and PIM1 had no appearances in the returned list even though their structure were previously solved35.
Molecular docking of Kaempferol and its analogs with their putative targets
We further investigated the putative interaction of Kaempferol with two of the identified protein targets, MAPK14 (p38) and PMI1, by proceeding with molecular docking. We also studied the interaction of the two other flavonoid components, Kaempferol-3-O-glucoside (K3g) and Kaempferol-3-O-rutinoside (K3r) (Figs 8b and S2).
Interestingly, we found that the co-crystals of four flavonoids, Myrecitin, Pentahydroxy-flavone, Quercetin and Quercetagetin with PIM1 were previously solved (PDB codes 2O3P, 2O63, 2O64, 2O65, respectively). To conduct the docking with Kaempferol, K3g and K3r, we used the bound structure of PIM1 with Myricetin (PDB code 2O63).
The spherical docking domain on PIM1 structure was centered on the binding sites of Myricetin. For MAPK14 (p38α), we proceeded according to Goettert et al.36.
The best docking scores among the retained solutions for Kaempferol against PIM1 reproduced the same interaction mode of Myricetin (Fig. 8b). For K3r, its orientation preserved the geometry of the phenyl group while the hetero-cyclic moiety was flipped in comparison to Kaempferol. For K3g, both the phenyl group and the hetero-cyclic moiety were disposed in opposing directions relative to Kaempferol. For both K3r and K3g, it seems that the glycosyl groups were crowding, disabling the phenyl and the heterocyclic groups from optimizing the steric interactions of the amino acid residues of the PIM1.
The molecular docking study on MAPK14 (p38α) revealed that the three ligands can adapt their conformation in the interaction site. However, only Kaempferol was able to reach the depth of the pocket while the steric void space for the other ligands was more important (Fig. 8c).
Discussion
After 30 years of clinical use, 5-Fluorouracil remains the cornerstone therapy for colorectal cancer. However, development of 5-FU resistant tumor cells is a limiting factor in CCR successful chemotherapy. Indeed chemo-resistant cells preserve unlimited proliferative potential, are protected from apoptosis and stimulate pathological angiogenesis, promoting then the progression of metastatic disease. Thus, novel therapeutic strategies to improve the effectiveness of 5-FU chemotherapy and to overcome drug resistance are critically needed.
One possible way to overcome or delay the emergence of resistant cancer cells is to co-administer drugs with different molecular mechanisms37. Combination treatments of natural polyphenols with anticancer drugs showed promising results greatly accepted by cancer researchers38. In the present report, we relied on our previous work in which we have shown a potent anti-tumoral effect of quince peel polyphenolic extract (Peph) through induction of apoptosis and cell cycle arrest, inhibition of angiogenic factors and improvement of therapeutic efficiency of 5-FU on human colon adenocarcinoma LS174 cells25. Here, we proposed to evaluate the effect of Peph extract in 5-FU-resistant colon cancer cells. For this, we established 5-FU-resistant cells (LS174-R) generated from parental sensitive colon cancer LS174 cells after continuous exposure to 60 µM of 5-FU. The 5-FU-resistant cells displayed the typical morphological changes associated to growth decrease relative to parental cells. Usually, drug-resistant cancer cells present slow growth39 which is in accordance with the study of Dallas et al.40 highlighting reduced proliferative rate in both 5-FU- and oxaliplatin-resistant colon cancer HT29 cells in comparison to parental one. We found that both colon cancer cells were able to form multicellular three-dimensional spheres when grown in low-adherent conditions. However, 5-FU-resistant cells produced less-condensed spheroids with a small amount of cancer cells non-incorporated into the compact cell aggregates. These cells known as non-spheroid forming (NSF) cells present loss of cell-cell adhesion associated to the inhibition of E-cadherin expression41. Interestingly, our result supports the reduced level of E-cadherin protein in our 5-FU-resistant cells in comparison to parental LS174 cells. The loss of E-cadherin expression promotes the epithelial-mesenchymal transition (EMT) process in epithelial tumor progression42. The EMT is a complex process that involves several signaling pathways, including FAK and Src activation43. Interestingly, our finding indicates the up-regulation of these proto-oncogenes suggesting the acquisition of more aggressive cell features in the 5-FU-resistant cancer cells.
ATP-binding cassette (ABC) transporters play a crucial role in the development of resistance by the efflux of anticancer agents outside the cancer cells13. Our result highlights the up-regulation of ABC sub-family G member 2 (ABCG2) and multidrug resistance-associated protein 1 (MDR1) in 5-FU-resistant LS174-R cells, which suggests that these two proteins affect the intracellular concentration of 5-FU conferring resistant phenotype to LS174-R cells.
The protection from apoptosis also constitutes an effective mechanism implicated in colorectal cancer resistance12. Our data showed the reduced expression of the pro-apoptotic mediator Bad in the refractive cells relative to parental one. This effect may be in part regulated by Src activation, which could reduce pro-apoptotic stimuli through the inhibition of death accelerators, such as Bad, Bax and caspase-9 proteins44.
Although 5-FU exerts its cytotoxic effect during the S phase of the cell cycle, prolonged G1 and S phases may provide cancer cells more time to repair the damage induced by this drug45,46. This correlates with the prolonged S phase and reduced proliferative rate in refractive cancer LS174-R cells in comparison to parental LS174 cells.
As comparative studies highlighted the resistant phenotype of 5-FU-resistant LS174-R cells, we studied their sensitivity to combination treatment of quince extract or its phenolic compounds with 5-Fluorouracil. Our data demonstrated that Peph extract did not affect the viability of resistant cells contrary to parental cells. Thus, we proposed to assess the effect of different phenolic compounds from total Peph extract on the viability of both sensitive LS174 cells and 5-FU-resistant LS174-R cells. We found that the thirteen phenolic compounds exhibited a dose-dependent inhibition effect on the viability of parental LS174 cells. However, Quercetin, Rutin, (+)- Catechin, (−)- Catechin, Hyperin, Isoquercitrin, Chlorogenic acid, Cryptochlorogenic acid, Neochlorogenic acid and p-coumaric acid compounds failed to induce significant inhibitory effect on the viability of 5-FU-resistant LS174-R cells. Interestingly, only Kaempferol and its analogs, Kaempferol 3-O-glucoside and Kaempferol 3-O-rutinoside were able to reduce the viability of chemo-resistant cells. Kaempferol presented the highest anti-proliferative activity with about 50% of inhibition at a concentration of 75 µM compared to its analogs that induced a slight inhibitory effect at concentration of 120 µM.
Interestingly, Kaempferol in combination with 5-FU improved the effectiveness of individual treatment and caused more decrease in colony formation of both sensitive and 5-FU-resistant cells even at low concentration (15 µM) of the compound. This chemosensitizing effect of Kaempferol to 5-FU chemotherapy highlighted an additive effect between 5-fluorouracil and Kaempferol in parental cells and synergistic interaction in 5-FU-resistant cells.
To investigate the putative cell targets of Kaempferol, we conducted a series of computational analysis. Our results suggest three intracellular protein targets CDK6, PIM1 and MAPK/p38 that have been reported to play an important role in colorectal carcinogenesis31,47,48. MAPK/p38 is positively correlated with typical features of cancer aggressiveness, such as migration and invasion. The p38 is highly expressed in CRC biopsies and inflammatory bowel disease-associated human CRC specimens31. MAPK14/p38 was shown to be inhibited by Kaempferol with an IC50 of 18 µM36.
It has been reported that the CDK6 expression increases from non-neoplastic mucosa through adenoma to submucosal invasive carcinoma to regulate the progression and metastasis of CRC47. CDK6 was found to interact with RB149, and its expression is also controlled by the MAPK14/p38 signaling pathway50. CDK6 is inhibited by Kaempferol with an IC50 of 22 µM51. However, we did not detect the expression of CDK6 in vitro, excluding any role played by this factor in our cellular model.
Kaempferol was also identified as an inhibitor of the proto-oncogene PIM1 with an IC50 of 1.288 µM52. This Serine/threonine kinase is implicated in the cell signaling process of FOXO3 and STAT353 which are members of the interaction network. PIM1 promoted the proliferation, differentiation and cell survival of colon cancer and its inhibition has been suggested as a possible target for therapeutic intervention48.
PIM1 could be considered as a highly potential target since Kaempferol reproduced the interaction model of Myricetin in a near exact manner. Pharmapper analysis also suggested a putative interaction of Kaempferol with MAPK14/p38 while no appearance was noticed for PIM1.
On the other hand, our docking study suggests that the glucosyl groups carried by K3g and K3r analogs of Kaempferol are too bulky to allow the fitting of the ligands inside the interaction pockets of the two putative targets. However, Kaempferol structure allows it to optimize the interactions with several amino acids including the most inner residues of the interaction site. Thus, this in silico study could explain why Kaempferol was endowed with the best inhibitory effect on the viability of resistant colon cancer LS174 R cells in vitro compared to its analogs.
Kaempferol, a natural flavonoid, is known for its antioxidant property29. Interestingly, this compound alone or combined with 5-FU significantly reduced ROS production in both sensitive and 5-FU-resistant cells. Over the past several years, researchers have associated ROS to malignant transformation. The generation of ROS mediated the expression of proteins involved in proliferation, tumor cell death or survival, invasion, angiogenesis, and metastasis54. We then tried to uncover the mechanisms of resistance to 5-FU in LS174-R cells and to shed light on the cellular targets through which Kaempferol restored 5-FU sensitivity in unresponsive LS174-R cells to drug.
The retinoblastoma protein (pRb) and p53 are known as tumor suppressor proteins that inhibit the transcription of different oncogenes involved in tumor growth and metastasis55. Our results suggest the acquisition of other malignant properties at least through increased phosphorylation of the protein Rb and dephosphorylation of p53 in chemo-resistant LS174-R cells. It has been reported that the hypophosphorylated form of pRb binds to the transactivation domain of E2F to inhibit cell cycle progression through the inhibition of CDK activity56. Interestingly, the combined treatment was effective in blocking 5-FU-resistant cells in G2/M phase which involves dephosphorylation of Rb and cdc2 (CDK1) proteins. According to our results, previous reports demonstrated that the arrest of G2/M checkpoint transition is controlled by down-regulation of cdc-family proteins57. In sensitive cells, our data suggest that the cooperative mechanism exploited by Kaempferol and 5-FU caused cell cycle arrest in S phase, which depends on p53, and p21 activation. Accordingly, this effect is in line with the work of Wiegering et al.58 describing the major role of several small molecules (identified from the compound library of the National Cancer Institute) in inducing p53 activation and tumor apoptosis (RITA) which in turn enhance the antiproliferative response to 5-FU and increase the expression levels of p53 protein in colorectal cancer cells.
Cell cycle arrest and apoptosis constitute the primary mechanisms that inhibit tumor formation by preventing inappropriate expansion of cells with malignant potential59. Acquired resistance to apoptosis represents an essential feature related to 5-FU resistance. Our finding highlights a pro-apoptotic effect of 5-FU treatment in sensitive cells without any considered activity in chemo-resistant cells. Interestingly, Kaempferol treatment increased the percentages of apoptotic cells approximately to 40% and 25% in sensitive and 5-FU resistant cells respectively. The combination of Kaempferol with 5-FU was crucial for effective sensitization of LS174-R cells to apoptosis showing that they act synergistically to trigger cell death. The increased level of PARP expression associated to the activation of caspase-3 and caspase-9 further confirmed the effectiveness of combined treatment in both parental and resistant cells in inducing apoptosis. Several therapeutics tend to eliminate transformed cells by the induction of apoptosis through activation of the effector caspases60. Our result suggests the therapeutic role of concomitant use of Kaempferol and 5-Fluorouracil for resistant colorectal cancer to 5-fluorouracil.
A number of signaling molecules and transcription factors such as the JAK/STAT3, Wnt/β-catenin, PI3K/Akt, MAPK and NF-κB signaling pathways are involved in the dysregulation of death machinery and progression of colorectal cancer30. Our results showed that MAPK (ERK1/2, p38) and PI3K/AKT (AKT) signaling pathways were activated in 5-FU-resistant LS174-R cells in comparison to the parental LS174 cells. This data suggests that acquisition of 5-FU resistance in generated 5-FU-resistant cancer cells is associated with more aggressive cell features related to the activation of AKT and ERK1/2/p38 MAPK signaling pathways. Various studies have revealed the upregulation of survival signaling pathways, including PI3K and ERK to prevent anti-cancer agents-induced cell apoptosis in multidrug resistant cancer cells61. The p38/MAPK pathway was also identified as a mediator of drug resistance in colorectal cancer31. Our finding indicated that the anti-apoptotic status of the chemo-resistant LS174-R cells to 5-FU treatment may be mediated by the intense activation of AKT and ERK/p38 MAPK pathways.
We observed that the 5-Fluorouracil treatment inhibited ERK1/2 and p38 activations in parental cells but did not affect their activity in chemo-resistant LS174-R cells. Interestingly, we found that Kaempferol alone or combined to 5-FU was able to modulate the expression of such protein kinases in both sensitive and 5-FU-resistant cells. We observed an up-regulation of the expression level of ERK1/2 associated to the inhibition of the one of p38 kinase. Generally, ERK activation has been associated to cell proliferation, differentiation and survival, but activated ERK has also been involved in growth arrest and induction of apoptotic cell death by various cytotoxic agents 62. Our result is in accordance with several studies that reported the activation of the MEK/ERK pathway upon the inhibition of p38/MAPK to lead to cell cycle arrest and apoptosis in colorectal cancer cells. A study by Zhang et al.63 demonstrated that, in combination with cisplatin treatment, the pharmacological inhibitor of ERK increased the phosphorylation of p38 and that p38 inhibition promoted the activation of ERK in Hela cells. The crosstalk between p38α and ERK pathway could represent a useful tool in CRC therapy31. Our data suggest that Kaempferol exerts its sensitizing effect through the regulation of ERK and p38 kinases. Our result also supports in silico analysis that explained the putative interaction of p38 and Kaempferol, which can adapt its conformation in the p38 binding site and thereby causes its inhibition.
We found that Kaempferol, by its own or combined with 5-FU, induced the inhibition of AKT along with the dephosphorylation of its target, the transcription factor FOXO3a. The hypophosphorylated active form of FOXO3a is known to bind to promoters of target genes involved in apoptosis and cell cycle arrest, inducing their transcription and thus contributed to tumor suppression64. The first negative control mechanism of FOXO3a is mediated by Phosphatidylinositol 3-kinase (PI3k)-activated AKT that stimulates the phosphorylation of FOXO3a promoting its interaction with nuclear export protein and its proteasomal degradation. Moreover, the study of Zhang et al.63 supported that inhibition of p38 pathway enhanced the expression of FOXO3a. The blocking of p38α-induced chemosensitization of tumor cells passes through nuclear accumulation of FOXO3a and activation of its pro-apoptotic gene targets31. Thus, Kaempferol inhibited the AKT and p38 kinases, activating then the tumor suppressor FOXO3a. It is well documented that compounds that reactivated FOXO3 based on its tumor suppressor property are considered as very attractive anti-cancer therapy65. Therefore, our finding suggests that the therapeutic potential of Kaempferol passes at least in part through the activation of FOXO3a.
Since AKT activation promotes the expression of pro-survival transcription factors and inhibits the pro-apoptotic FOXO3a66, we investigated the involvement of two other transcription factors, nuclear factor-κB (NF-κB) and STAT3 known to be activated in a wide variety of cancers, including colorectal cancer67. These regulatory proteins provide a survival mechanism by regulating the expression of target genes involved in inflammation, cell proliferation, survival, angiogenesis, invasion and metastasis, thereby representing a major causative factors for drug resistance68,69. Interestingly, our result indicated that Kaempferol alone or its concomitant use with 5-Fluorouracil reduced the protein level of phospho-STAT3 in both parental and chemo-resistant cells. Surprisingly, Kaempferol, 5-FU or their combined treatment induced the activation of NF-κB in sensitive LS174 cells whereas this compound alone or in combination with the drug decreased the level of its phosphorylated form in resistant LS174-R cells. In this context, Samuel et al.70 have reported that NF-κB activation varies with the cellular make up. The activation of NF-κB by several chemotherapeutic agents, including 5-Fluorouracil may be functionally uncoupled with anti-apoptotic outcomes70,71. In the other hand, several studies suggest the involvement of NF-κB activation in the resistance of colon cancer cells to 5-FU72,73. Targeting NF-κB inhibition may be used as a novel preventive and therapeutic strategy against chemoresistant human cancers73.
Given that cytokines and growth factors production is tightly intertwined to NF-κB and STAT3 activation67, we have consequently investigated the protein expression of VEGF-A and IL-8, which promote angiogenesis and pathological neovascularization in colorectal cancer74. Importantly, we found that the two angiogenic mediators were highly overproduced in the generated 5-FU-resistant cells in comparison to the parental LS174 cells. The VEGF is known to confer resistance to chemotherapy by inducing the expression of anti-apoptotic proteins75. It has been demonstrated that high levels of IL-8 are also linked to tumor development, angiogenesis, metastasis and anti-cancer drug resistance8. Consequently, our result suggests that acquisition of resistance to 5-FU could be related in part to up-regulation of the angiogenic factors. Interestingly, Kaempferol alone or its concomitant use with 5-FU significantly reduced the VEGF-A and IL-8 production in 5-FU-resistant LS174-R cells. However, we observed only a slight decrease in VEGF-A production without any effect on the secretion of IL-8 in parental cells. This effect could be explained by the fact that VEGF is the direct target gene of STAT369. Furthermore, NF-κB activation regulates the expression of downstream target genes including cytokines (IL-6, IL-8) and growth factors (VEGF)76. Our data suggest that the potent chemosensitizing effect of Kaempferol in our chemo-resistant cells passes through the inhibition of NF-κB and STAT3 transcription factors, leading then to VEGF-A and IL-8 downregulation.
The conversion of 5-Fluorouracil to active metabolites passes through several enzymatic steps77. Previous studies have reported the closely relationship between 5-FU metabolism and drug resistance78. Then, to better identify the mechanism by which Kaempferol sensitizes chemo-resistant LS174-R cells to 5-FU, we analyzed the expression of five genes involved in the metabolic pathways of 5-FU such as TS, TK, FPGS, DHFR and DPD. Our result showed that acquired resistance in the 5-FU-resistant cells was associated with significant increase in the mRNA levels of these enzymes, especially, for TS (with 91.77 fold induction). Only the DPD, implicated in 5-FU catabolism, was slightly decreased in comparison with parental cells. This result was confirmed by western blotting analysis for TS and TK proteins levels. The overexpression of TS is one of the most well-known mechanisms of acquired resistance after 5-FU exposure79. Previous study indicated that low expression of TS in CRC patients was associated with improved median survival compared with those with higher levels of TS enzyme77. The overexpression of TK also represents a potential mechanism of 5-FU resistance80. Interestingly, Kaempferol alone or combined with 5-FU significantly decreased the mRNA expression of the five genes implicated in 5-FU metabolism only in chemo-resistant LS174-R cells. This effect was associated with the inhibition of the expression of TS and TK proteins, which play a key role in dTMP (deoxythymidine monophosphate) formation, essential for DNA synthesis and cellular proliferation. Our result is in line with the study of De la Cueva et al.81 that reported that down modulation of TS and TK mRNA and protein levels affects the mechanisms of resistance in 5-FU-resistant colon cancer cells through dephosphorylation of Rb protein associated to E2F1 inhibition. Further investigation also revealed that PI3K/AKT activation induced the overexpression of TS in CRC cells resistant to 5-FU chemotherapy82. In addition, the low expressions of TS, TK, DHFR and DPD were linked to tumor progression in CRC patients.
The present study provides evidence for the chemosensitizing effect of Kaempferol to 5-FU chemotherapy of LS174-R cells by exerting synergistic inhibitory effect through blocking the production of ROS and modulating JAK/STAT3, MAPK, PI3K/AKT and NF-κB signaling pathway, involved in the progression and development of colorectal cancer. Kaempferol alone or in combination with 5-FU decreased the production of IL-8 and VEGF-A and reduced the expression of five genes involved in 5-FU metabolism in chemo-resistant cells. Taken together, our results suggest a potential chemotherapeutic role of Kaempferol, which could represent a novel concept for overcoming 5-FU resistance in patients diagnosed with colon cancer. Moreover, pharmacokinetic and pharmacodynamics studies demonstrated that Kaempferol is a non-toxic natural agent, which is safe in vivo and bioavailable83.
Although our results are encouraging, our resistant cell model did not take into account the presence in vivo of multiple cell types in addition to the complexity of the tumor environment where new interactions could be involved in the resistance of colorectal cancers. Further investigation is warranted to better verify if the in vitro chemosensitizing effect of Kampferol in 5-FU-resistant LS174-R colon cancer cells can be extended to the in vivo setting.
Materials and Methods
Quince peel polyphenolic compounds
Quince peel polyphenolic compounds identified in the total polyphenolic extract (Peph) [Quercetin (Q), Rutin (R), (+)-Catechin (+C), (−)-Catechin (−C), Hyperin (H), Isoquercitrin (I), Chlorogenic acid (ChA), Cryptochlorogenic acid (CrA), Neochlorogenic acid (NeA), p-coumaric acid (PcA), Kaempferol (Kaemp), Kaempferol-3-O-glucoside (K3g) and Kaempferol-3-O-rutinoside (K3r)] were purchased from Sigma (Sigma-Aldrich, St. Louis, MO).
Cell culture and generation of 5-FU-Resistant Colon Cancer Cells
Human colon adenocarcinoma LS174 cell line (CL-188), were obtained from American Type Culture Collection (ATCC, Manassas, VA). The cells were cultured in DMEM (Dulbecco’s Modified Eagle’s Medium) supplemented with 10% fetal bovine serum (FBS; GIBCO) and 50 U/ml penicillin and 50 µg/ml streptomycin. The 5-FU-Resistant cells maintained in 100 mm2 petri dishes were generated by continuous exposure of LS174 cells to increasing doses of 5-FU (10–100 μM). Medium was changed three times a week and adherent cells were passaged using trypsin/EDTA. Afterwards, the surviving 5-FU-resistant cells were maintained in complete culture medium containing 60 μM of 5-Fluorouracil.
Cell viability
The growth of parental LS174 cells and 5-FU-resistant cells was assessed by 3-(4,5-dimethylthiazol-2yl)-2,5-diphenyltetrazolium bromide (MTT) assay. Cells were seeded in 96-well plates (1000 cells/well) for 24 h and then incubated for 72 h in the presence of vehicle (control) and testing agents. At the end of the treatments, 50 µl of MTT solution (1 mg/ml final) were added into each 96-well plate and the cells were incubated for a further 3 h at 37 °C. Thereafter, the medium was removed and 100 µl of dimethyl sulfoxide (DMSO) was added to each well to dissolve the formazan crystals. The optical density (OD) at 540 nm was measured with a microplate reader (MULTISKAN, Labsystems). The cell viability was expressed as percentage of the viable cell number in treated cells relative to mock-treated cells (control). Cell viability was calculated using the following formula:
Colony formation assay
Parental and resistant cells were seeded in six-well plates (2 × 105 cells/well). After 24 h, cells were treated with 15 µM or 75 µM of Kaempferol alone or combined to 60 μM of 5-Fluorouracil. After 72 h of treatment, the medium was removed and cells were trypsinized and plated at low density of 2000 cells per six-well plate. Cells were then cultivated for 10 days. Colonies were stained with crystal violet and clones for each condition were photographed. The number of colonies was scored by CFU scope quantification software. Results are expressed as the number of colony forming cells per well in percentage and normalized to control (vehicle, considered to represent 100%).
Spheroid generation
The colon cancer cells LS174 and LS174-R were analyzed for spheroid formation capacity in ultra-low attachment (ULA) round bottom 96-well plates. Each well was coated with 100 µl of agarose. After 30 min at 37 °C, cells were seeded (1000 cells/well) for 5 days under standard conditions (37 °C, 5% CO2). After the incubation period, cells were photographed under light microscopy.
Sensitization assay
Both colon cancer cells were seeded in 96-well culture plates (1000 cells/well). After 24 h, the cells were treated with serial concentrations of Kaempferol (1, 5, 10, 15, 30, 60, 75 µM) for various periods (8 h, 12 h and 24 h). Cells were then exposed to 60 µM of 5-FU after removing and/or maintaining Kaempferol in cell culture medium. After 72 h of treatment, the viability of parental and 5-FU-resistant cells was determined by MTT assay as described above. The drug concentration that resulted in a 50% growth inhibition (GI50) was determined graphically from sigmoidal dose-response curves.
Drug and compound interaction: Determination of the Combination Index (CI) value
Parental and resistant cancer cells were seeded at 1000 cells/100 µl in 96-well plates and allowed to attach for 24 h. Cells were treated for 72 h with increasing concentrations of Kaempferol alone, 5-FU alone and the combination of both. Concentrations for drug combination studies were based on IC50 values (IC50/4, IC50/2, IC50, IC50*2, IC50*4). Following indicated treatments, the viability of both colon cancer cells was determined by MTT assay as described above. The interaction between 5-FU and Kaempferol was determined after calculating the Combination Index values, where a CI < 1 indicates synergism, CI = 1 additive effect and CI > 1 antagonism84.
The Combination index (CI) values were calculated based on the following formula:
CA,X: the concentrations of drug A to produce the given effect (e.g., IC50).
ICX,A: the concentrations of drug A and B in combination to provide the same effect (e.g., IC50).
Measurement of reactive oxygen species (ROS)
The intracellular ROS assay employs a cell-permeable fluorogenic probe, CMH2DCF-DA (life technologies, Oregon, USA). This molecule passively diffuses into cells and is deacetylated by cellular esterases to non-fluorescent 2′,7′-dichlorodihydrofluorescin (DCF), which is rapidly oxidized to highly fluorescent adduct in the presence of ROS. Colon cancer cells were cultured in 96-well plates (2000 cells/well) and treated with the listed agents for 72 h. Cells were washed with PBS (1X), resuspended in HBSS (GIBCO) and then incubated with 10 µM of CMH2DCFDA at 37 °C for 30 min in dark and in a CO2 incubator. Fluorescence was measured by the Varioskan Flash microplate reader (Thermo Scientific) with an excitation and emission wavelengths of 492 and 517 nm, respectively.
Cell cycle analysis
Sensitive and chemo-resistant colon cancer cells were seeded into six-well plates and then exposed for 72 h to testing agents. Thereafter, cells were harvested by trypsinization and fixed with 70% ice-cold ethanol. After washing twice with 1X ice-cold PBS, cells were resuspended in propidium iodide (PI)/RNase staining solution (Cell Signaling Technology; Danvers, MA) for 30 min in the dark at 37 °C. Cell cycle progression was analyzed on a Becton–Dickinson FACScanto II flow cytometer and further analyzed with BD FACSDiva 6 software (Becton–Dickinson). The PI fluorescence signal at FL2-A peak versus counts was used to determine cell cycle distribution and the data were analyzed using the Modfit software.
Assessment of Apoptosis
Apoptosis was quantitatively assessed using the annexin V/PE apoptosis detection kit (BD-Pharmingen) according to the manufacturer’s protocol. Briefly, the cells treated with the molecules of interest or vehicle as a negative control were collected by centrifugation, washed with PBS (1X) and resuspended in 100 μl of binding buffer (1X). Thereafter, cells were treated with 4 μl of Annexin V conjugated to phycoerythrin (PE) and 4 µl of 7-AAD and then rapidly incubated in the dark for 15 min. Stained cells were analyzed on a Becton–Dickinson FACScanto II flow cytometer and further analyzed with BD FACSDiva 6 software (Becton–Dickinson). Cell death was quantitatively evaluated by measuring the proportion of annexin V-positive cells, regardless of their staining for 7-AAD in order to include both early apoptotic and dead cells. Values are given in percent of total cell number. Percentage of apoptotic cells (%) was calculated as follows: early apoptotic cells (%) + late apoptotic cells (%).
Western blotting analysis
After 72 h of treatment with the molecules of interest, cells were solubilized in 100 µl of Laemmli buffer (1X) at room temperature. Protein content of the cell lysates was quantified using the BCA method (Bicinchoninic Acid Protein Assay kit, Sigma). Equal amounts of protein (30 µg/sample) were resolved on sodium dodecyl sulfate polyacrylamide gels (SDS-PAGE). After electrophoresis, proteins were transferred onto polyvinylidene difluoride (PVDF) membrane (Immobilon-Millipore) and incubated for 1 h at room temperature with blocking buffer (5% non fat dry milk). The membranes were probed overnight with primary antibodies (1:1,000 dilutions): [ABCG2 (MAB4146) was obtained from Millipore. β-actin, MDR1, cleaved PARP, cleaved caspase 3, cleaved caspase 9, Bcl-2, Bad, E-cadherin, vimentin, β-catenin, c-Src, FAK, p53, phospho p53 (Ser 15), phospho-FOXO3a, phospho-Stat3, phospho-ERK1/2, phospho-p38, phospho-AKT, phospho-NF-κB, Thymidylate Synthase and Thymidine Kinase were obtained from Cell Signaling Technology (Danvers, MA)] and incubated with a horseradish peroxidase-conjugated anti-IgG (Promega, Madison, WI) in a blocking buffer for 1 h. After washing, the blots were developed with enhanced chemiluminescence (ECL) (Millipore) and exposed to X-ray film.
Real time quantitative RT-PCR
To assess the expression of several genes involved in 5-FU metabolism, total RNA was isolated from both sensitive and 5-FU-resistant colon cancer cells after 72 h of treatment with molecules of interest. Reverse transcription was realized with 1 µg of RNA from each sample. After cDNA preparation, real-time PCR was carried out using the LightCycler System (Roche Applied Science, Mannheim, Germany). PCR was set up at 2.5 mM MgCl2, 10 μM of each primer (Eurofins Genomics, USA), 4 μl of recover DNA and 5 µl of Master Mix (KAPA Biosystems) in a final volume of 10 μl. Data analysis was essentially performed using “Fit Point Method” in the LightCycler software version 3.5.3. GAPDH gene was used as the internal reference.
To calculate the relative expression of transcripts, the 2[−ΔΔC (T)] method was used85. Each calculated fold was determined from the average of 3 independent experiments run in duplicate. The sequences of PCR primers are available upon request.
Determination of cytokines concentration
Seventy-two hours post-treatment with the testing agents, cell culture supernatants from parental LS174 cells and 5-FU-resistant cells were centrifuged at 1000 rpm for 5 min and the cells were counted. The VEGF-A and IL-8 concentrations were quantified using an ELISA kit Quantikine human Immunoassay (Thermo SCIENTIFIC) following the manufacturer’s guidelines and normalized to cell number.
In silico analysis of Kaempferol interaction with the cellular molecular targets
To get insight about the interaction network within which the Thymidylate synthase exerts its function, we retrieved the accession numbers of all the proteins whose expression was evaluated by western blot (FOXO3a: O43524, NF-κb1: P19838, STAT3: P40763, P53: P04637, phospho-rb: P06400, AKT: P31749, ERK (MAK1): P28482, Thymidylate synthase: P04818, MAPK14 (p38 14): Q16539, MAPK10 (p38 10): P53779). The list of the proteins was submitted to the NetworkAnalyst server86. The analysis was performed based on the protein-protein interaction data of the InnateDB87, which compiles more than 136000 experimentally validated interactions representing 3000 pathways from human, mouse and bovine genes.
Ligands structure
The 3D coordinates of Kaempferol (ChEMBL150) and Kaempferol-3-O-glucoside (ChEMBL453290) were obtained from the ChEMBL database. Those of Kaempferol-3-O-rutinoside were obtained from the ChEBI database using the identifier ChEBI69657. All the structures were converted to the SYBYL mol2 file format.
Prediction of cellular target by pharmacophore mapping
Given a ligand structure, the PharmMapper server88 assigns its pharmacophore representation to identify a list of potential protein targets by using the reverse pharmacophore mapping approach. The Kaempferol coordinates were first submitted to the PharMapper server which generates multiple conformations for the ligand structure. The pharmacophore models of each conformation of the ligand and those of the protein targets (considering only the human protein targets: 2241 proteins) were then aligned against each other. A score was then calculated describing the pairwise fit level between the pharmacophore models of the ligand and each target protein in the database. A report was finally generated in which all the potential targets are listed according to their fit score.
Molecular docking
The molecular docking of Kaempferol, Kaempferol-3-O-rutinoside and Kaempferol-3-O-glucoside were processed with the program PLANTS (Protein-Ligand ANT System)89. The ligands and the receptors were first treated with SPORES90,91 to generate the topologies of the interacting partners affecting the hydrogen atoms and assigning the hybridization states of the atoms. PLANTS uses the so called colony optimization algorithms to sample the rotational and translational degrees of freedom of the ligand and search for a low energy configuration. The internal ligand flexibility was also modeled by sampling the rotational bonds degrees of freedom. The docking domain was set to a sphere of 10 Å radius centered at the catalytic binding site of the target proteins. The software returned an ensemble of putative complexes, which were then clustered to account for the geometrical similarity based on a cutoff difference of 2 Å. The putative docking solutions were ranked according to the chemPLP scoring function, which was dimensionless. The lowest values correspond to the most favorable docking solutions.
Statistical analysis
Data from individual experiments are expressed as means ± S.E. Differences between means were evaluated using Student’s t-test. Differences were considered to be statistically significant at P < 0.05.
Electronic supplementary material
Acknowledgements
We thank Dr. Gilles Pagès and Dr. Sandy Giuliano for kindly providing ABCG2 and MDR1 antibodies. This work was supported by the Tunisian Ministry of Higher Education and Scientific Research (LR11IPT04/LR16IPT04) and the Institut Pasteur de Tunis (Collaborative Internal Project PCI_04_2012). The funders had no role in study design, data analysis, decision to publish, or preparation of the manuscript.
Author Contributions
Khadija Essafi-Benkhadir conceived and designed the experiments; Ichrak Riahi-Chebbi, Soumaya Souid and Houcemeddine Othman performed the experiments; Khadija Essafi-Benkhadir, Ichrak Riahi-Chebbi, Najet Srairi-Abid, Houcemeddine Othman, Meriam Haoues and Makram Essafi analyzed the data; Khadija Essafi-Benkhadir, Najet Srairi-Abid, Alain Morel, Makram Essafi and Habib Karoui contributed reagents/materials/analysis tools; Khadija Essafi-Benkhadir and Ichrak Riahi-Chebbi wrote the paper; Khadija Essafi-Benkhadir edited the manuscript. All authors read and approved the final manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Soumaya Souid and Houcemeddine Othman contributed equally.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-36808-z.
References
- 1.Murphy, N. et al. Heterogeneity of Colorectal Cancer Risk Factors by Anatomical Subsite in 10 European Countries: A Multinational Cohort Study. Clin Gastroenterol Hepatol, 10.1016/j.cgh.2018.07.030 (2018). [DOI] [PMC free article] [PubMed]
- 2.Bray, F. et al. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin, 10.3322/caac.21492 (2018). [DOI] [PubMed]
- 3.Landre T, et al. Doublet chemotherapy vs. single-agent therapy with 5FU in elderly patients with metastatic colorectal cancer. a meta-analysis. Int J Colorectal Dis. 2015;30:1305–1310. doi: 10.1007/s00384-015-2296-5. [DOI] [PubMed] [Google Scholar]
- 4.Mármol I, Sánchez-de-Diego C, Pradilla Dieste A, Cerrada E, Rodriguez Yoldi M. Colorectal Carcinoma: A General Overview and Future Perspectives in Colorectal Cancer. Int J Mol Sci. 2017;18:197. doi: 10.3390/ijms18010197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Webber EM, Kauffman TL, O’Connor E, Goddard KA. Systematic review of the predictive effect of MSI status in colorectal cancer patients undergoing 5FU-based chemotherapy. BMC Cancer. 2015;15:156. doi: 10.1186/s12885-015-1093-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xie T, et al. MicroRNAs as Regulators, Biomarkers and Therapeutic Targets in the Drug Resistance of Colorectal Cancer. Cell Physiol Biochem. 2016;40:62–76. doi: 10.1159/000452525. [DOI] [PubMed] [Google Scholar]
- 7.Touil Y, et al. Colon Cancer Cells Escape 5FU Chemotherapy-Induced Cell Death by Entering Stemness and Quiescence Associated with the c-Yes/YAP Axis. Clin Cancer Res. 2014;20:837–846. doi: 10.1158/1078-0432.CCR-13-1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jones VS, et al. Cytokines in cancer drug resistance: Cues to new therapeutic strategies. Biochim Biophys Acta - Rev Cancer. 2016;1865:255–265. doi: 10.1016/j.bbcan.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 9.Tang J-C, Feng Y-L, Liang X, Cai X-J. Autophagy in 5-Fluorouracil Therapy in Gastrointestinal Cancer: Trends and Challenges. Chin Med J (Engl) 2016;129:456–63. doi: 10.4103/0366-6999.176069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng Y, Zhou J, Tong Y. Gene signatures of drug resistance predict patient survival in colorectal cancer. Pharmacogenomics J. 2015;15:135–43. doi: 10.1038/tpj.2014.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deng J, Wang Y, Lei J, Lei W, Xiong JP. Insights into the involvement of noncoding RNAs in 5-fluorouracil drug resistance. Tumor Biol. 2017;39:101042831769755. doi: 10.1177/1010428317697553. [DOI] [PubMed] [Google Scholar]
- 12.Zhang N, Yin Y, Xu S-J, Chen W-S. 5-Fluorouracil: mechanisms of resistance and reversal strategies. Molecules. 2008;13:1551–69. doi: 10.3390/molecules13081551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.El Khoury F, Corcos L, Durand S, Simon B, Le Jossic-Corcos C. Acquisition of anticancer drug resistance is partially associated with cancer stemness in human colon cancer cells. Int J Oncol. 2016;49:2558–2568. doi: 10.3892/ijo.2016.3725. [DOI] [PubMed] [Google Scholar]
- 14.Kim A-Y, Kwak J-H, Je NK, Lee Y-H, Jung Y-S. Epithelial-mesenchymal Transition is Associated with Acquired Resistance to 5-Fluorocuracil in HT-29 Colon Cancer Cells. Toxicol Res. 2015;31:151–6. doi: 10.5487/TR.2015.31.2.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang Z-S, Sun Y-Z, Wang S-M, Ruan J-S. Epithelial-mesenchymal transition: potential regulator of ABC transporters in tumor progression. J Cancer. 2017;8:2319–2327. doi: 10.7150/jca.19079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.González-Sarrías A, Tomé-Carneiro J, Bellesia A, Tomás-Barberán FA, Espín JC. The ellagic acid-derived gut microbiota metabolite, urolithin A, potentiates the anticancer effects of 5-fluorouracil chemotherapy on human colon cancer cells. Food Funct. 2015;6:1460–9. doi: 10.1039/C5FO00120J. [DOI] [PubMed] [Google Scholar]
- 17.Abdal Dayem A, et al. The Anti-Cancer Effect of Polyphenols against Breast Cancer and Cancer Stem Cells: Molecular Mechanisms. Nutrients. 2016;8:581. doi: 10.3390/nu8090581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abaza MSI, Orabi KY, Al-Quattan E, Al-Attiyah RJ. Growth inhibitory and chemo-sensitization effects of naringenin, a natural flavanone purified from Thymus vulgaris, on human breast and colorectal cancer. Cancer Cell Int. 2015;15:46. doi: 10.1186/s12935-015-0194-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hussain SA, et al. Natural Polyphenols in Cancer Chemoresistance. Nutr Cancer. 2016;68:879–891. doi: 10.1080/01635581.2016.1192201. [DOI] [PubMed] [Google Scholar]
- 20.Aung T, Qu Z, Kortschak R, Adelson D. Understanding the Effectiveness of Natural Compound Mixtures in Cancer through Their Molecular Mode of Action. Int J Mol Sci. 2017;18:656. doi: 10.3390/ijms18030656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee J, Kang J-S, Choi B-Y, Keum Y-S. Sensitization of 5-Fluorouracil-Resistant SNUC5 Colon Cancer Cells to Apoptosis by α-Mangostin. Biomol Ther (Seoul) 2016;24:604–609. doi: 10.4062/biomolther.2016.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taylor WF, Jabbarzadeh E. The use of natural products to target cancer stem cells. Am J Cancer Res. 2017;7:1588–1605. [PMC free article] [PubMed] [Google Scholar]
- 23.Findlay VJ, et al. SNAI2 modulates colorectal cancer 5-fluorouracil sensitivity through miR145 repression. Mol Cancer Ther. 2014;13:2713–26. doi: 10.1158/1535-7163.MCT-14-0207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Violette S, et al. Resistance of colon cancer cells to long-term 5-fluorouracil exposure is correlated to the relative level of Bcl-2 and Bcl-X(L) in addition to Bax and p53 status. Int J cancer. 2002;98:498–504. doi: 10.1002/ijc.10146. [DOI] [PubMed] [Google Scholar]
- 25.Riahi-Chebbi I, et al. Quince peel polyphenolic extract blocks human colon adenocarcinoma LS174 cell growth and potentiates 5-fluorouracil efficacy. Cancer Cell Int. 2015;16:1. doi: 10.1186/s12935-016-0276-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yaheya, M. & Ismail, M. Review Article DRUG-FOOD INTERACTIONS AND ROLE OF PHARMACIST. Asian Journal of Pharmaceutical and Clinical Research2, 1–10 (2009).
- 27.Ijichi K, Adachi M, Ogawa T, Hasegawa Y, Murakami S. Cell-cycle distribution and Thymidilate Synthatase (TS) expression correlate with 5-FU resistance in head and neck carcinoma cells. Anticancer Res. 2014;34:2907–11. [PubMed] [Google Scholar]
- 28.Hassan M, Watari H, AbuAlmaaty A, Ohba Y, Sakuragi N. Apoptosis and Molecular Targeting Therapy in Cancer. Biomed Res Int. 2014;2014:1–23. doi: 10.1155/2014/150845. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29.Chen AY, Chen YC. A review of the dietary flavonoid, kaempferol on human health and cancer chemoprevention. Food Chem. 2013;138:2099–2107. doi: 10.1016/j.foodchem.2012.11.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang C-Y, Yu LC-H. Pathophysiological mechanisms of death resistance in colorectal carcinoma. World J Gastroenterol. 2015;21:11777–92. doi: 10.3748/wjg.v21.i41.11777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grossi V, Peserico A, Tezil T, Simone C. p38α MAPK pathway: A key factor in colorectal cancer therapy and chemoresistance. World J Gastroenterol. 2014;20:9744. doi: 10.3748/wjg.v20.i29.9744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Z, et al. Broad targeting of angiogenesis for cancer prevention and therapy. Semin Cancer Biol. 2015;35:S224–S243. doi: 10.1016/j.semcancer.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aubry K, Labourey JL, Bessède JP, Tubiana-Mathieu N, Rigaud M. Expression levels of thymidylate synthase, thymidylate phosphorylase and dihydropyrimidine dehydrogenase in head and neck squamous cell carcinoma: preliminary study. Clin Med Oncol. 2008;2:27–35. [PMC free article] [PubMed] [Google Scholar]
- 34.Liu T, Lin Y, Wen X, Jorissen RN, Gilson MK. BindingDB: a web-accessible database of experimentally determined protein-ligand binding affinities. Nucleic Acids Res. 2007;35:D198–D201. doi: 10.1093/nar/gkl999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Phan J, et al. Structure of human thymidylate synthase suggests advantages of chemotherapy with noncompetitive inhibitors. J Biol Chem. 2001;276:14170–7. doi: 10.1074/jbc.M009493200. [DOI] [PubMed] [Google Scholar]
- 36.Goettert M, Schattel V, Koch P, Merfort I, Laufer S. Biological Evaluation and Structural Determinants of p38α Mitogen-Activated-Protein Kinase and c-Jun-N-Terminal Kinase 3 Inhibition by Flavonoids. ChemBioChem. 2010;11:2579–2588. doi: 10.1002/cbic.201000487. [DOI] [PubMed] [Google Scholar]
- 37.Anreddy N, et al. Tyrosine Kinase Inhibitors as Reversal Agents for ABC Transporter Mediated Drug Resistance. Molecules. 2014;19:13848–13877. doi: 10.3390/molecules190913848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cao, J., Han, J., Xiao, H., Qiao, J. & Han, M. Effect of Tea Polyphenol Compounds on Anticancer Drugs in Terms of Anti-Tumor Activity, Toxicology, and Pharmacokinetics. Nutrients8 (2016). [DOI] [PMC free article] [PubMed]
- 39.van Dijk D, et al. Slow-growing cells within isogenic populations have increased RNA polymerase error rates and DNA damage. Nat Commun. 2015;6:7972. doi: 10.1038/ncomms8972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dallas NA, et al. Chemoresistant colorectal cancer cells, the cancer stem cell phenotype, and increased sensitivity to insulin-like growth factor-I receptor inhibition. Cancer Res. 2009;69:1951–7. doi: 10.1158/0008-5472.CAN-08-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stadler M, et al. Exclusion from spheroid formation identifies loss of essential cell-cell adhesion molecules in colon cancer cells. Sci Rep. 2018;8:1151. doi: 10.1038/s41598-018-19384-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178–96. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Canel, M., Serrels, A., Frame, M. C. & Brunton, V. G. E-cadherin-integrin crosstalk in cancer invasion and metastasis. 10.1242/jcs.100115. [DOI] [PubMed]
- 44.Fu Y, et al. Antioxidants decrease the apoptotic effect of 5-Fu in colon cancer by regulating Src-dependent caspase-7 phosphorylation. Cell Death Dis. 2014;5:e983. doi: 10.1038/cddis.2013.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Focaccetti C, et al. Effects of 5-Fluorouracil on Morphology, Cell Cycle, Proliferation, Apoptosis, Autophagy and ROS Production in Endothelial Cells and Cardiomyocytes. PLoS One. 2015;10:e0115686. doi: 10.1371/journal.pone.0115686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guo, X. et al. Cell cycle perturbation and acquired 5-fluorouracil chemoresistance. Anticancer Res28, 9–14 (2008). [PubMed]
- 47.Tadano T, et al. MicroRNA-320 family is downregulated in colorectal adenoma and affects tumor proliferation by targeting CDK6. World J Gastrointest Oncol. 2016;8:532–42. doi: 10.4251/wjgo.v8.i7.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peng Y, et al. Expression of pim-1 in Tumors, Tumor Stroma and Tumor-Adjacent Mucosa Co-Determines the Prognosis of Colon Cancer Patients. PLoS One. 2013;8:e76693. doi: 10.1371/journal.pone.0076693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Anders L, et al. A Systematic Screen for CDK4/6 Substrates Links FOXM1 Phosphorylation to Senescence Suppression in Cancer Cells. Cancer Cell. 2011;20:620–634. doi: 10.1016/j.ccr.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moro T, et al. Inhibition of Cdk6 expression through p38 MAP kinase is involved in differentiation of mouse prechondrocyte ATDC5. J Cell Physiol. 2005;204:927–933. doi: 10.1002/jcp.20350. [DOI] [PubMed] [Google Scholar]
- 51.Lu H, Chang DJ, Baratte B, Meijer L, Schulze-Gahmen U. Crystal Structure of a Human Cyclin-Dependent Kinase 6 Complex with a Flavonol Inhibitor, Fisetin. J Med Chem. 2005;48:737–743. doi: 10.1021/jm049353p. [DOI] [PubMed] [Google Scholar]
- 52.Holder, S. et al. Characterization of a potent and selective small-molecule inhibitor of the PIM1 kinase. 10.1158/1535-7163.MCT-06-0397. [DOI] [PubMed]
- 53.Magnuson, N. S., Wang, Z., Ding, G. & Reeves, R. Why target PIM1 for cancer diagnosis and treatment? 10.2217/fon.10.106. [DOI] [PMC free article] [PubMed]
- 54.Gupta SC, et al. Upsides and downsides of reactive oxygen species for cancer: the roles of reactive oxygen species in tumorigenesis, prevention, and therapy. Antioxid Redox Signal. 2012;16:1295–322. doi: 10.1089/ars.2011.4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Du M-D, He K-Y, Qin G, Chen J, Li J-Y. Adriamycin resistance-associated prohibitin gene inhibits proliferation of human osteosarcoma MG63 cells by interacting with oncogenes and tumor suppressor genes. Oncol Lett. 2016;12:1994–2000. doi: 10.3892/ol.2016.4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dick FA, Rubin SM. Molecular mechanisms underlying RB protein function. Nat Rev Mol Cell Biol. 2013;14:297–306. doi: 10.1038/nrm3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Singh SK, Banerjee S, Acosta EP, Lillard JW, Singh R. Resveratrol induces cell cycle arrest and apoptosis with docetaxel in prostate cancer cells via a p53/p21<sup>WAF1/CIP1</sup> and p27<sup>KIP1</sup> pathway. Oncotarget. 2017;8:17216–17228. doi: 10.18632/oncotarget.15303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wiegering A, et al. Reactivating p53 and Inducing Tumor Apoptosis (RITA) Enhances the Response of RITA-Sensitive Colorectal Cancer Cells to Chemotherapeutic Agents 5-Fluorouracil and Oxaliplatin. Neoplasia. 2017;19:301–309. doi: 10.1016/j.neo.2017.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li T, et al. Tumor suppression in the absence of p53-mediated cell-cycle arrest, apoptosis, and senescence. Cell. 2012;149:1269–83. doi: 10.1016/j.cell.2012.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kiedrowski M, Mroz A. The effects of selected drugs and dietary compounds on proliferation and apoptosis in colorectal carcinoma. Contemp Oncol (Poznan, Poland) 2014;18:222–6. doi: 10.5114/wo.2014.44296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xie CQ, et al. Triptolide exerts pro-apoptotic and cell cycle arrest activity on drug-resistant human lung cancer A549/Taxol cells via modulation of MAPK and PI3K/Akt signaling pathways. Oncol Lett. 2016;12:3586–3590. doi: 10.3892/ol.2016.5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yadav V, Varshney P, Sultana S, Yadav J, Saini N. Moxifloxacin and ciprofloxacin induces S-phase arrest and augments apoptotic effects of cisplatin in human pancreatic cancer cells via ERK activation. BMC Cancer. 2015;15:581. doi: 10.1186/s12885-015-1560-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang L, et al. Butein sensitizes HeLa cells to cisplatin through the AKT and ERK/p38 MAPK pathways by targeting FoxO3a. Int J Mol Med. 2015;36:957–66. doi: 10.3892/ijmm.2015.2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nho RS, Hergert P. FoxO3a and disease progression. World J Biol Chem. 2014;5:346. doi: 10.4331/wjbc.v5.i3.346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cautain B, et al. Discovery of a Novel, Isothiazolonaphthoquinone-Based Small Molecule Activator of FOXO Nuclear-Cytoplasmic Shuttling. PLoS One. 2016;11:e0167491. doi: 10.1371/journal.pone.0167491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Das TP, Suman S, Alatassi H, Ankem MK, Damodaran C. Inhibition of AKT promotes FOXO3a-dependent apoptosis in prostate cancer. Cell Death Dis. 2016;7:e2111–e2111. doi: 10.1038/cddis.2015.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rinkenbaugh, A. L. & Baldwin, A. S. The NF-κB Pathway and Cancer Stem Cells. Cells5 (2016). [DOI] [PMC free article] [PubMed]
- 68.Ruiz de Porras V, et al. Curcumin mediates oxaliplatin-acquired resistance reversion in colorectal cancer cell lines through modulation of CXC-Chemokine/NF-κB signalling pathway. Sci Rep. 2016;6:24675. doi: 10.1038/srep24675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li W, Chen C, Liu Z, Hua B. Targeting Signal Transducer and Activator ofTranscription 3 for Colorectal Cancer Prevention and Treatment with Natural Products. Cancer Transl Med. 2016;2:48. doi: 10.4103/2395-3977.184320. [DOI] [Google Scholar]
- 70.Samuel T, Fadlalla K, Gales DN, Putcha BD, Manne U. Variable NF-κB pathway responses in colon cancer cells treated with chemotherapeutic drugs. BMC Cancer. 2014;14:599. doi: 10.1186/1471-2407-14-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shin J-Y, Kim J-O, Lee SK, Chae H-S, Kang J-H. LY294002 may overcome 5-FU resistance via down-regulation of activated p-AKT in Epstein-Barr virus-positive gastric cancer cells. BMC Cancer. 2010;10:425. doi: 10.1186/1471-2407-10-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Körber MI, Staribacher A, Ratzenböck I, Steger G, Mader RM. NFκB-Associated Pathways in Progression of Chemoresistance to 5-Fluorouracil in an In Vitro Model of Colonic Carcinoma. Anticancer Res. 2016;36:1631–9. [PubMed] [Google Scholar]
- 73.Kang Y, et al. Curcumin sensitizes human gastric cancer cells to 5-fluorouracil through inhibition of the NFκB survival-signaling pathway. Onco Targets Ther. 2016;9:7373–7384. doi: 10.2147/OTT.S118272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shi J, Wei P-K. Interleukin-8: A potent promoter of angiogenesis in gastric cancer. Oncol Lett. 2016;11:1043–1050. doi: 10.3892/ol.2015.4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gao H, Lan X, Li S, Xue Y. Relationships of MMP-9, E-cadherin, and VEGF expression with clinicopathological features and response to chemosensitivity in gastric cancer. Tumor Biol. 2017;39:101042831769836. doi: 10.1177/1010428317698368. [DOI] [PubMed] [Google Scholar]
- 76.Deng Y, Verron E, Rohanizadeh R. Molecular Mechanisms of Anti-metastatic Activity of Curcumin. Anticancer Res. 2016;36:5639–5648. doi: 10.21873/anticanres.11147. [DOI] [PubMed] [Google Scholar]
- 77.Hammond WA, Swaika A, Mody K. Pharmacologic resistance in colorectal cancer: a review. Ther Adv Med Oncol. 2016;8:57–84. doi: 10.1177/1758834015614530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Valenzuela, M. M. A., Neidigh, J. W. & Wall, N. R. Antimetabolite Treatment for Pancreatic Cancer. Chemotherapy3 (2014). [DOI] [PMC free article] [PubMed]
- 79.Housman G, et al. Drug Resistance in Cancer: An Overview. Cancers (Basel) 2014;6:1769–1792. doi: 10.3390/cancers6031769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.González-Sarrías A, Ma H, Edmonds ME, Seeram NP. Maple polyphenols, ginnalins A–C, induce S- and G2/M-cell cycle arrest in colon and breast cancer cells mediated by decreasing cyclins A and D1 levels. Food Chem. 2013;136:636–642. doi: 10.1016/j.foodchem.2012.08.023. [DOI] [PubMed] [Google Scholar]
- 81.de la Cueva A, et al. Combined 5-FU and ChoKα Inhibitors as a New Alternative Therapy of Colorectal Cancer: Evidence in Human Tumor-Derived Cell Lines and Mouse Xenografts. PLoS One. 2013;8:e64961. doi: 10.1371/journal.pone.0064961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jiang B, Liu F, Liu Z, Zhang T, Hua D. B7-H3 increases thymidylate synthase expression via the PI3k-Akt pathway. Tumor Biol. 2016;37:9465–9472. doi: 10.1007/s13277-015-4740-0. [DOI] [PubMed] [Google Scholar]
- 83.Barve A, et al. Metabolism, oral bioavailability and pharmacokinetics of chemopreventive kaempferol in rats. Biopharm Drug Dispos. 2009;30:356–65. doi: 10.1002/bdd.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhao L, Au JL-S, Wientjes MG. Comparison of methods for evaluating drug-drug interaction. Front Biosci (Elite Ed) 2010;2:241–9. doi: 10.2741/e86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Livak KJ, Schmittgen TD. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 86.Xia J, Benner MJ, Hancock REW. NetworkAnalyst - integrative approaches for protein–protein interaction network analysis and visual exploration. Nucleic Acids Res. 2014;42:W167–W174. doi: 10.1093/nar/gku443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Breuer K, et al. InnateDB: systems biology of innate immunity and beyond—recent updates and continuing curation. Nucleic Acids Res. 2013;41:D1228–D1233. doi: 10.1093/nar/gks1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang, X. et al. PharmMapper 2017 update: a web server for potential drug target identification with a comprehensive target pharmacophore database. Nucleic Acids Res45 (2017). [DOI] [PMC free article] [PubMed]
- 89.Korb, O., Stützle, T. & Exner, T. E. PLANTS: Application of Ant Colony Optimization to Structure-Based Drug Design. In 247–258 10.1007/11839088_22 (Springer, Berlin, Heidelberg, 2006).
- 90.ten Brink T, Exner TE. Influence of Protonation, Tautomeric, and Stereoisomeric States on Protein−Ligand Docking Results. J Chem Inf Model. 2009;49:1535–1546. doi: 10.1021/ci800420z. [DOI] [PubMed] [Google Scholar]
- 91.ten Brink T, Exner TE. A combined combinatorial and pKa-based approach to ligand protonation states. J Cheminform. 2010;2:P43. doi: 10.1186/1758-2946-2-S1-P43. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.