Abstract
We recently demonstrated that the circadian clock component CRY2 is an essential cofactor in the SCFFBXL3-mediated ubiquitination of c-MYC. Because our demonstration that CRY2 recruits phosphorylated substrates to SCFFBXL3 was unexpected, we investigated the scope of this role by searching for additional substrates of FBXL3 that require CRY1 or CRY2 as cofactors. Here, we describe an affinity purification mass spectrometry (APMS) screen through which we identified more than one hundred potential substrates of SCFFBXL3+CRY1/2, including the cell cycle regulated Tousled-like kinase, TLK2. Both CRY1 and CRY2 recruit TLK2 to SCFFBXL3, and TLK2 kinase activity is required for this interaction. Overexpression or genetic deletion of CRY1 and/or CRY2 decreases or enhances TLK2 protein abundance, respectively. These findings reinforce the idea that CRYs function as co-factors for SCFFBXL3, provide a resource of potential substrates, and establish a molecular connection between the circadian and cell cycle oscillators via CRY-modulated turnover of TLK2.
Introduction
Circadian clocks are present in virtually all cells in the body and maintain an intrinsic 24-hour period that allows organisms to predict recurring daily changes in their environment1. The core of the mammalian clock is composed of a transcriptional and translational negative feedback loop, in which the transcription factors CLOCK and BMAL1 heterodimerize and promote the expression of their own repressors cryptochrome (Cry1&2) and period (Per1-3)2,3. The clock is reset by several post-translational modifications, including ubiquitination and subsequent degradation of CRY1 and CRY2 in the nucleus by the SKP-CULLIN-FBOX (SCF) family E3 ligase containing the F-box and leucine rich repeat (FBXL3) substrate adaptor (SCFFBXL3)4–6. AMP-activated protein kinase (AMPK)-mediated phosphorylation of CRY1 promotes its association with SCFFBXL3, and subsequent degradation7.
In addition to the function of CRYs as transcriptional repressors in the circadian clock, we recently discovered a previously unappreciated role for CRY2 as an essential cofactor in the SCFFBXL3 mediated ubiquitination of the oncoprotein c-MYC, which may contribute to the mechanistic basis underlying enhanced tumorigenesis in the context of disrupted circadian rhythms8. Here, we describe an approach using affinity purification mass spectrometry (APMS) to identify additional substrates recruited to SCFFBXL3 by CRY1 or CRY2. We found that SCFFBXL3+CRY1/2 interacts with and destabilizes the Tousled-like kinase, TLK2.
TLK1 and TLK2 are highly homologous cell cycle regulated kinases with peak activity in S phase9. They were initially characterized after their plant homolog Tousled (TSL) was found to be necessary for proper flower and leaf formation, likely by regulating cell division during development9–12. TLK1 and TLK2 regulate chromatin assembly by phosphorylating ASF1 histone chaperone proteins13–15 and are catalytically activated and targeted for proteasomal degradation in response to DNA damage9,13. TLK2 is uniquely important for checkpoint recovery in response to DNA damage15. Additionally, TLK2 deregulation has been linked to cancer in multiple studies16–18. Our finding that SCFFBXL3+CRY1/2 enhances turnover of TLK2 suggests another mechanistic link by which deregulation of clock components could lead to a cancer-prone phenotype.
Results
Search for targets of SCFFBXL3+CRY1/2
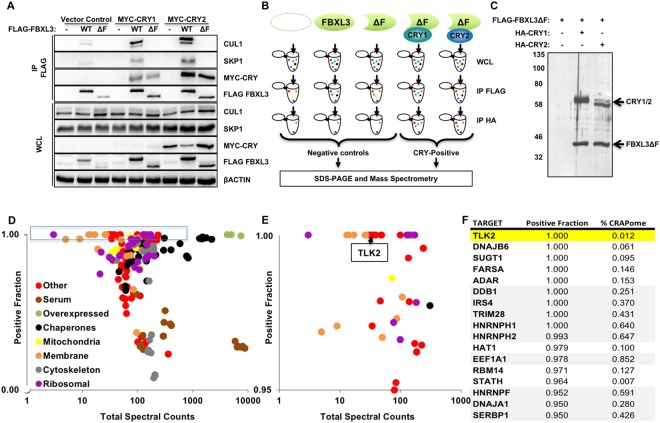
Following our identification of a CRY and SCFFBXL3 complex that recruits c-MYC for ubiquitination, we performed a proteomic screen to identify additional substrates, using truncated FBXL3 lacking the F-box (FBXL3ΔF) to prevent its association with the SKP-CULLIN E3 ligase complex and ubiquitination and degradation of targets8,19. To confirm the validity of this approach, we showed that FBXL3ΔF does not interact with SKP1 or CUL1, but maintains the ability to bind CRY1 and CRY2 (Fig. 1A). We used sequential immunoprecipitation (IP) to first purify FLAG-tagged FBXL3 (wildtype or ΔF), followed by purification of HA-tagged CRY1/2-containing complexes from the FLAG peptide eluate to focus only on proteins present in complexes containing both FBXL3 and either CRY1 or CRY2 (Fig. 1B). We included three control samples to exclude proteins associated non-specifically with the solid support or with FBXL3 independent of CRY1/2. This two-step, sequential IP greatly reduced the presence of non-specific partners in our final analysis, thus increasing confidence in identified targets (Fig. 1C).
Figure 1.
Unbiased screen for FBXL3 partners recruited by CRY1 and/or CRY2. (A) CUL1, SKP1, βACTIN, MYC-CRY1, MYC-CRY2, FLAG-FBXL3 detected by immunoblot (IB) in whole cell lysate (WCL) or following immunoprecipitation (IP) of FLAG tagged proteins from cells transiently expressing the indicated plasmids. (B) Screening conditions and workflow. (C) Proteins detected by silver stain in samples prepared as shown in (B). Molecular weight markers (kDa) at left. Uncropped gel in Supplementary Fig. 4. (D) Each symbol represents one of 141 proteins for which at least 30 peptides were detected in CRY-positive samples. Colors denote association with subcellular compartments or protein complexes as indicated. (E) Magnified view of the data in the blue rectangle in (D). (F) List of top candidates from the screen, with TLK2 highlighted in yellow. Gray shading indicates proteins frequently detected my mass spectrometry in Flag IPs from 293 T cells. In (D,E) Total spectral counts indicates the number of peptides detected in all samples in all three experiments combined. Positive fraction indicates the number of peptides detected in CRY-positive samples divided by the number detected in all samples.
We performed the experiments and analysis described above in triplicate (Supplementary Fig. 1) and used Integrated Proteomics Pipeline - IP2 (Integrated Proteomics Applications, Inc., San Diego, CA. http://www.integratedproteomics.com/) to identify >6000 proteins in all of our samples combined. For 141 of these proteins, we detected at least 30 spectral counts spread across at least five of the six experimental samples (those purified from cells expressing FLAG-FBXL3ΔF and either HA-CRY1 or HA-CRY2, Supplementary Table 1). We were surprised to find several mitochondrial proteins among those identified in our initial screen (Fig. 1D). We purified mitochondria from livers of Cry1−/− and Cry2−/− mice collected at zeitgeber time 16 (ZT16, sixteen hours after lights on) during the dark phase when CRY protein levels are high, but were unable to detect CRY1 or CRY2 in mitochondria (Supplementary Fig. 2). Therefore, we excluded mitochondrial and other membrane-associated proteins from further analysis. We focused on candidates for which at least 95% of the peptides detected in our experiments were found in the CRY-positive fraction (rather than negative control) samples (Fig. 1E). Furthermore, we used publicly available data in the so-called Contaminant Repository of Affinity Purification (CRAPome) to exclude from further consideration all proteins typically detected in more than 25% of negative control samples in APMS experiments using a similar setup20. These stringent filters left us with a short list of high confidence components of FBXL3- and CRY-containing complexes, including Tousled-like kinase TLK2 (Fig. 1F). We expect that the interactions of several of these proteins with FBXL3 and CRY1/2 are biologically meaningful. Here, we focus on TLK2 to demonstrate that CRY1 and CRY2 act as co-factors for SCFFBXL3 to target proteins involved in the DNA damage response and cell cycle, as an extension of our earlier work involving c-MYC8.
TLK2 interacts with FBXL3 and CRY
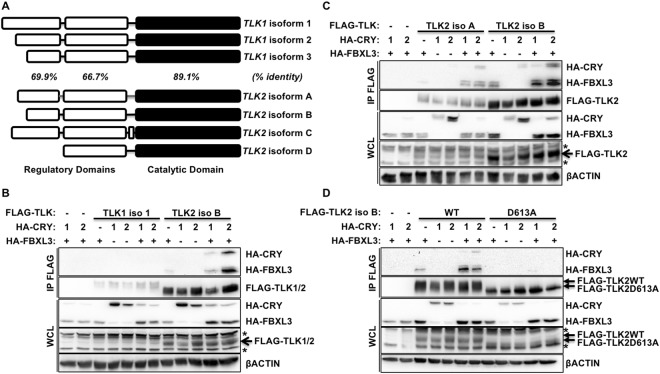
TLK1 and TLK2 are highly homologous proteins (approximate overall homology is 80%) with the highest conservation at the C-terminus, which includes the catalytically active domain (Fig. 2A)9. TLK1 and TLK2 are each expressed from several transcripts, resulting in three or four unique protein isoforms, respectively (Fig. 2A). To assess whether TLK1 and/or TLK2 interact with FBXL3 and CRY1/2, we ectopically expressed epitope-tagged proteins in 293 T cells. While we were unable to express comparable amounts of FLAG-TLK1 and FLAG-TLK2, we did not detect interaction of overexpressed TLK1 with FBXL3 and/or CRY1 or CRY2 (Fig. 2B and Supplementary Fig. 3). Both the A and B isoforms of TLK2 interact with FBXL3, and this interaction is enhanced in the presence of CRY1 or CRY2 (Fig. 2C).
Figure 2.
TLK2 interacts with FBXL3 and CRY. (A) TLK1 and TLK2 isoform homology in the indicated domains based on amino acid sequence. Percentages indicate amino acid identity of TLK1 and TLK2 in the indicated domains. (B–D) Detection of indicated proteins by IB in WCL or following FLAG IP from 293 T cells expressing the indicated plasmids. In (D) D613A is a catalytically inactive mutant of TLK2 iso B. * denotes non-specific signal.
Kinase activity of TLK2 is required for interaction with SCFFBXL3+CRY
Like many kinases, TLK2 is prone to autophosphorylation, in addition to its role in phosphorylating additional substrates9. We previously found that interaction of phosphorylated threonine 58 (pT58) in c-MYC with a phosphate binding loop (P loop) of CRY2 is critical for association of c-MYC with FBXL3 + CRY28. Therefore, we examined whether the interaction with CRY and FBXL3 was affected by TLK2 kinase activity. Indeed, a catalytically inactive mutant of TLK2 (D613A) fails to associate with FBXL3 and CRY1 or CRY2 (Fig. 2D). These data indicate that TLK2 kinase activity is required for its interaction with FBXL3 and CRY. This may reflect a requirement for autophosphorylation of TLK2 or phosphorylation of another component of the complex by TLK2. Mutation of several individual serine or threonine residues in TLK2 did not prevent interaction with FBXL3 and CRYs (not shown).
CRY and/or FBXL3 decrease TLK2 protein expression
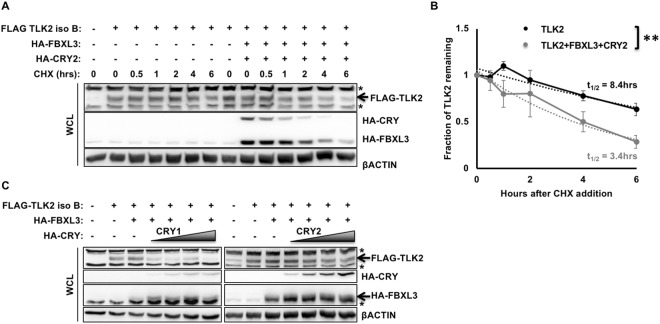
After determining that CRY and FBXL3 interact with TLK2, we wanted to determine whether they modulate TLK2 stability. First, we examined the effect of CRY and FBXL3 overexpression on turnover of overexpressed TLK2. In 293 T cells treated with cycloheximide, transient overexpression of CRY and FBXL3 increased TLK2 turnover (Fig. 3A,B). We also detected decreased TLK2 protein inversely correlated with the amount of CRY1 or CRY2 expressed in the presence of FBXL3 (Fig. 3C). Inhibition of the proteasome prevented the FBXL3 and CRY mediated decrease in FLAG-TLK2 protein levels (Supplementary Fig. 4).
Figure 3.
CRY1/2 and FBXL3 affect TLK2 stability. (A,C) Proteins detected by IB in WCL or following FLAG IP from 293 T cells expressing the indicated plasmids and following treatment with CHX for the indicated times. In (C) shaded triangles indicate increasing HA-CRY1 or HA-CRY2 expression. * denotes non-specific signal. (B) Quantitation of IB data from three experiments as shown in (A). Each data point represents the mean ± s.e.m. of the signal for FLAG-TLK2 overexpressed alone (black) or with overexpressed FBXL3 + CRY2 (gray) divided by the signal for βACTIN from the same sample, normalized to the sample collected before addition of CHX (t = 0). The dotted lines represent the best exponential fit to each data set, from which the half-lives were calculated according to the formula t1/2 = Tln(2), where T is derived from the exponential fit equation N(t) = N0exp(−t/T). **P < 0.01 by two-way ANOVA.
Endogenous CRYs influence TLK2 protein levels
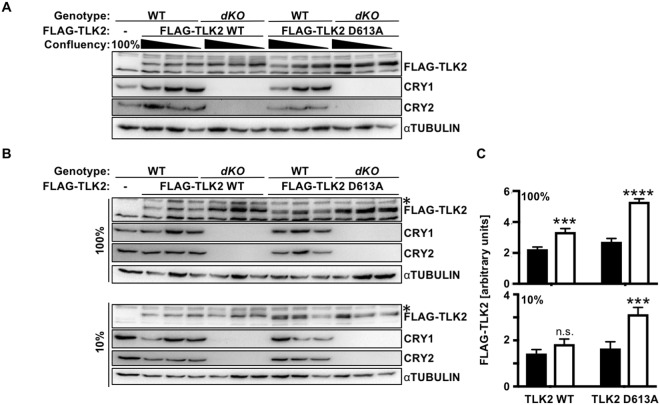
To investigate the impact of endogenous CRY1 and CRY2 on TLK2, we stably expressed tetracycline inducible FLAG-tagged human TLK2 in adult mouse skin fibroblasts. While studying TLK2 in fibroblasts, we found that cell density altered the amount of TLK2 detected (Fig. 4A). Regardless, TLK2 protein levels are consistently elevated in dKO fibroblasts compared to wildtype cells (Fig. 4A–C and Supplementary Figs 5 and 6A). Deletion of either Cry1 or Cry2 also seems to enhance TLK2 but the effects were less clear (Supplementary Fig. 5). While overexpressed catalytically inactive TLK2 did not interact with overexpressed FBXL3 and CRY1/2, its expression was enhanced in dKO fibroblasts (Fig. 4A–C, and Supplementary Figs 5 and 6A). Cry1/2 genotype had no effect on the expression of endogenous Tlk2 mRNA (Supplementary Fig. 6B), but to our surprise doxycycline-induced expression of FLAG-TLK2 mRNA was somewhat elevated in CRY-deficient cells compared to wildtype (Supplementary Fig. 6C). Together, these data support the conclusion that CRY1 or CRY2 can promote destabilization of TLK2 by recruiting it to SCFFBXL3, though enhanced transcription of the virally encoded exogenous TLK2 may contribute to the observed increase in FLAG-TLK2 protein detected in CRY-deficient cells as well.
Figure 4.
Endogenous CRY1 and CRY2 impact TLK2 protein expression. (A,B) Detection of indicated proteins in lysates prepared from mouse adult skin fibroblasts from WT and Cry1−/−;Cry2−/− (dKO) mice stably expressing doxycycline-inducible FLAG-TLK2 WT or FLAG-TLK2D613A (KD) and at the plating confluencies as indicated. In (A), triangles indicate decreasing plating density (100%, 20%, 10%) 48 hours prior to lysis. (C) Quantitation of data shown in (B). Data represent mean ± s.d. for biological triplicates FLAG-TLK2 normalized to non-specific band (*) in lysates from wildtype (black bars) or Cry1−/−;Cry2−/− (white bars) cells. P values were determined using two-way ANOVA followed by Sidak’s multiple comparison testing. ***,****P < 0.001, 0.0001 vs. WT cells.
Discussion
Here we report the identification of substrates recruited to SCFFBXL3 by CRY1 and/or CRY2 in addition to the CRY2-dependent targeting of phosphorylated c-MYC that we previously reported8. While the APMS approach yielded many non-specific partners as others have reported20, we used stringent filters to focus on the most likely meaningful candidates, including the Tousled-like kinase TLK2.
In addition to TLK2, our data suggest several additional proteins as likely targets of SCFFBXL3+CRY-mediated ubiquitination and degradation. We identified the RNA editing enzyme ADAR as a potential target of FBXL3 + CRY-mediated ubiquitination (Fig. 1F). ADAR expression and RNA editing activity exhibit circadian oscillation in mouse livers21. In addition, ADAR regulates CRY2 expression by modulating microRNA targeting of its 3′ untranslated region (3′UTR), such that Adarb1−/− mice express elevated levels of CRY2 protein21. Our data indicate that SCFFBXL3+CRY may target ADAR for proteasomal degradation, suggesting another connection between circadian clocks and RNA editing. We also detected several ribosomal proteins as potential substrates for SCFFBXL3+CRY ubiquitination (Fig. 1D,E). Recent findings have demonstrated that ribosome assembly and the translation of subsets of RNAs are regulated by circadian clocks22–24. SCFFBXL3+CRY-driven rhythms in turnover of ribosome components could contribute to such observations. The DNA Damage-Binding Protein 1, DDB1, is a component of CUL4-containing E3 ligase complexes and was recently shown to interact with and destabilize CRY125. Its appearance in our screen may reflect FBXL3-independent CRY1 interaction or it could be a false positive given its relatively frequent occurrence in APMS screens performed in 293 T cells20.
TLK1 and TLK2 are regulated by the cell cycle and function in chromatin assembly and DNA damage response pathways9,13,15,26. Unlike c-MYC, which is specifically targeted by CRY2, TLK2 seems to be recruited to SCFFBXL3 by either CRY1 or CRY2. Similar to c-MYC, TLK2 interacts with FBXL3 and that interaction is enhanced in the presence of CRY. Since a catalytically inactive mutant of TLK2 (D613A) does not interact with FBXL3 + CRY, their interaction seems to depend on phosphorylation of one or more of the components. However, the stable expression of TLK2 D613A was enhanced in Cry1−/−;Cry2−/− cells compared to wildtype cells, at least as much as that of wildtype TLK2 (Fig. 4). We suspect that this reflects phosphorylation of the relevant substrate by endogenous TLK2 or a related kinase but cannot exclude other possible explanations (Supplementary Fig. 7). Previous studies demonstrated that TLK2 kinase activity peaks when its protein level is at a minimum, during S phase9. Intriguingly, we observed an inverse correlation between cell density and exogenously expressed TLK2 protein levels. Interestingly, plating density had a stronger impact on the protein level of catalytically inactive TLK2 than on wildtype TLK2. We suspect that expression of exogenous wildtype TLK2 could mask the inactivation of endogenous TLK2 in confluent cells. The interactions between cell plating density, Cry1/2 genotype, and TLK2 kinase activity in determining TLK2 protein levels suggest a complex interplay between these factors in the regulation of TLK2 degradation.
While we did not detect a strong preference of TLK2 for CRY1 or CRY2, FBXL3 and CRY may preferentially interact with TLK2 compared to TLK1. TLK1 and TLK2 are highly homologous proteins and both function in DNA replication by regulating chromatin assembly through phosphorylation of the ASF1 histone chaperone proteins13. Both TLK1 and TLK2 regulate cell cycle progression in undamaged cells, function in the DNA damage response, and modulate DNA replication. However, two unique findings distinguish them. First, TLK2 uniquely regulates checkpoint recovery after DNA damage during the G2 phase of the cell cycle. Also, TLK2 is frequently amplified in breast cancers13,15–18,26–29, while the same has not been observed for TLK1. Since TLK1 expression was much lower than that of TLK2 in our experiments, TLK1 may associate with SCFFBXL3+CRY but at a level below our limit of detection. If SCFFBXL3 and CRY specifically regulate TLK2 and not TLK1, that could point to circadian regulation of checkpoint recovery in the cell cycle after DNA damage. Circadian rhythms are intimately intertwined with the cell cycle in diverse organisms30–32, and the mechanistic connection described here could contribute to their interrelationship.
The kinase activity and phosphorylation state of TLK2 change throughout the cell cycle9, though the mechanisms underlying this modulation are unknown. TLK2 activity is also suppressed in response to DNA damage by ATM-dependent signaling29. Intriguingly, FBXL3 is one of only four F-box proteins that exhibited significantly enhanced association with CUL1 after DNA damage in a recent study33, suggesting a critical role for SCFFBXL3 in response to DNA damage. Both c-MYC and TLK2 are degraded after DNA damage, and are targeted by SCFFBXL3+CRY1/2 complexes, which are expected to be more active in response to damage33,34. Further investigation will determine whether SCFFBXL3+CRY1/2 regulates TLK2, c-MYC, or other substrates during the cell cycle and/or in response to DNA damage. Also, the relationship between DNA damage signaling, TLK2 kinase activity, and SCFFBXL3+CRY-mediated ubiquitination is not yet clear. The SCFFBXL3+CRY-mediated post-translational regulation of TLK2 described here suggests another mechanistic link between clocks and cell cycle progression and timing.
Methods
Mass spectrometry
Reagents and chemicals
Deionized water (18.2 MΩ, Barnstead, Dubuque, IA) was used for all preparations. Buffer A consists of 5% acetonitrile 0.1% formic acid, buffer B consists of 80% acetonitrile 0.1% formic acid, and buffer C consists of 500 mM ammonium acetate and 5% acetonitrile.
Sample preparation
Proteins were precipitated in 23% TCA (Sigma-Aldrich, St. Louis, MO, Product number T-0699) at 4 °C O/N. After 30 min. centrifugation at 18000 × g, protein pellets were washed 2 times with 500 ul ice-cold acetone. Air-dried pellets were dissolved in 8 M urea/100 mM Tris pH 8.5. Proteins were reduced with 1 M Tris (2-carboxyethyl) phosphine hydrochloride (Sigma-Aldrich, St. Louis, MO, product C4706) and alkylated with 500 mM 2-Chloroacetamide (Sigma-Aldrich, St. Louis, MO, product 22790-250G-F). Proteins were digested for 18 hrs. at 37 °C in 2 M urea, 100 mM Tris pH 8.5, 1 mM CaCl2 with 2 ug trypsin (Promega, Madison, WI, product V5111). Digest was stopped with formic acid, 5% final concentration. Debris was removed by centrifugation, 30 min 18000 × g.
MudPIT microcolumn
A MudPIT microcolumn35 was prepared by first creating a Kasil frit at one end of an undeactivated 250 μm ID/360 μm OD capillary (Agilent Technologies, Inc., Santa Clara, CA). The Kasil frit was prepared by briefly dipping a 20–30 cm capillary in well-mixed 300 μL Kasil 1624 (PQ Corporation, Malvern, PA) and 100 μL formamide, curing at 100 °C for 4 hrs, and cutting the frit to ~2 mm in length. Strong cation exchange particles (SCX Partisphere, 5 μm dia., 125 Å pores, Phenomenex, Torrance, CA) was packed in-house from particle slurries in methanol 2.5 cm. An additional 2.5 cm reversed phase particles (C18 Aqua, 3 µm dia., 125 Å pores, Phenomenex) were then similarly packed into the capillary using the same method as SCX loading, to create a biphasic column. An analytical RPLC column was generated by pulling a 100 μm ID/360 μm OD capillary (Polymicro Technologies, Inc, Phoenix, AZ) to 5 μm ID tip. Reversed phase particles (Aqua C18, 3 μm dia., 125 Å pores, Phenomenex, Torrance, CA) were packed directly into the pulled column at 800 psi until 12 cm long. The MudPIT microcolumn was connected to an analytical column using a zero-dead volume union (Upchurch Scientific (IDEX Health & Science), P-720-01, Oak Harbor, WA). LC-MS/MS analysis was performed using an Agilent Technologies 1200 HPLC pump and a Thermo Orbitrap Velos Pro using an in-house built electrospray stage. MudPIT experiments were performed with steps of 0% buffer C, 30% buffer C, 70% buffer C, 100% C, and 90/10% buffer C/B, being run for 3 min at the beginning of each gradient of buffer B. Electrospray was performed directly from the analytical column by applying the ESI voltage at a tee (150 μm ID, Upchurch Scientific)35. Electrospray directly from the LC column was done at 2.5 kV with an inlet capillary temperature of 325 °C. Data-dependent acquisition of MS/MS spectra with the Orbitrap Velos Pro were performed with the following settings: MS/MS on the 10 most intense ions per precursor scan; 1 microscan; reject unassigned charge state and charge state 1; dynamic exclusion repeat count, 1; repeat duration, 30 second; exclusion list size 300; and exclusion duration, 20 second.
Data analysis
Protein and peptide identification and protein quantitation were done with Integrated Proteomics Pipeline - IP2 (Integrated Proteomics Applications, Inc., San Diego, CA. http://www.integratedproteomics.com/). Tandem mass spectra were extracted from raw files using RawConverter36 and were searched against a Uniprot human protein database with fusion proteins and reversed sequences using ProLuCID37,38. The search space included all fully-tryptic and half-tryptic peptide candidates with a fixed modification of 57.02146 on cysteine. Peptide candidates were filtered using DTASelect39.
Sequential immunoprecipitation
Samples were prepared for mass spectrometry analysis following the sequential immunoprecipitation protocol previously outlined8. After the anti-HA beads were then spun down and washed 3X with lysis buffer containing 1%TX-100, 50 mM HEPES (pH 7.4), 138 mM KCl, 4 mM NaCl, 50 mM sodium pyrophosphate, 100 mM sodium fluoride, 10 mM EDTA, 1 mM EGTA pH 8, 50 μM PMSF, 1 mM β-Glycerophosphate, 1 mM sodium orthovanadate, and protease inhibitors (Roche cat # 11697498001) similar to previously described40, they were incubated in 150uL HA peptide (Sigma cat# I2149), diluted 1:50 in PBS, for 1.5 hours at 4 °C while nutating. 90% of the eluates were flash frozen for subsequent mass spectrometry analysis, while 10% were boiled in SDS sample buffer, run on an SDS-PAGE gel and stained using a silver stain kit (Thermo Scientific Pierce Biotech cat# P124612).
Plasmids
pcDNA3-HA-CRY2, pcDNA3-HA-FBXL3, pcDNA3-MYC-CRY2, pcDNA3-2xFLAG-FBXL3, and pcDNA3-2xFLAG-FBXL3ΔF are as described previously8. Cry1 and Fbxl3ΔF coding sequences were transferred to pcDNA3.1-based HA-epitope or pcDNA3.1-based MYC-epitope vectors using standard protocols. cDNA encoding human Tlk2 isoform A was generated by RT-PCR from RNA extracted from 293 T cells. Plasmids expressing TLK2 isoform A were made by cloning the Tlk2 isoform A cDNA into a pcDNA3.1-based V5-epitope tagged vector using standard protocols. pcDNA3-2xFLAG-TLK2 isoform A and pcDNA3-2xFLAG-TLK2 isoform B were generated using Q5 Site-Directed Mutagenesis (New England Biolabs Inc. cat # E0554S). pDONR223-TLK1, originally generated by was a gift from William Hahn & David Root (Addgene plasmid # 23727)41 and was cloned into a pcDNA3.1-based V5-epitope tagged vector using the pcDNA Gateway Directional TOPO Expression Kit (Invitrogen #K2440-20). pcDNA3.1-2xFLAG-TLK1 was then similarly generated using Q5 Site-Directed Mutagenesis. psPAX plasmid (Addgene plasmid 12260) and pMD2.G plasmid (Addgene plasmid 12259) deposited by Dr. Didier Trono, and used for infection, were also purchased from Addgene. pCW-Cas9 was a gift from Eric Lander & David Sabatini (Addgene plasmid # 50661)42 and the Cas9 sequence was replaced with Tlk2 cDNA using the Gibson Assembly Ultra Kit (Synthetic Genomics Inc. cat # GA1200-10). pCW-2xFLAG-TLK2 was then generated by introducing a 2xFLAG sequence using Q5 Site-Directed Mutagenesis. Plasmids containing TLK2 KD mutant was generated by mutating TLK2 WT to TLK2 D613A using Q5 Site-Directed Mutagenesis.
Cell culture and transfection
All cells were grown in complete Dulbecco’s Modified Eagle Medium (DMEM) (cat #10569; Invitrogen) unless otherwise indicated. 293 T cell media was supplemented with 10% fetal bovine serum, and 1% penicillin and streptomycin. 293 T cells were grown in a 37 °C incubator maintained at 5% CO2 and 20% O2 (high oxygen). Adult skin fibroblasts were prepared from ear biopsies of adult mice of the indicated genotypes, as described previously34. Adult skin fibroblasts (ASFs) were used as primary (passaged no more than 10 times and grown in 3% oxygen). ASF media were supplemented with 15% fetal bovine serum, and 1% penicillin and streptomycin. ASFs were grown in a 37 °C incubator maintained at 5% CO2 and 3% O2 (low oxygen). Transfections for mass spectrometry analysis were carried out using calcium phosphate, while all other transfections were performed using polyethylenimine (cat #23966-2; PEI; Polysciences Inc, Warrington, PA) by standard protocols. For calcium phosphate transfections of a 10 cm plate of 293 T cells: 1–10 μg DNA was mixed with 450 μL sterile water, 150 μL 1 M CaCl2 was added, and the tube contents mixed by vortex. 600 μL 2xHBS pH 7.02 (140 mM NaCl, 1.5 mM Na2HPO4, 50 mM HEPES) was pipetted into a separate tube, and the DNA/CaCl2 mix was added dropwise to the 2xHBS and the contents of the tube were mixed by forcing air bubbles through the solution. For PEI transfections, 1–10 µg DNA was added to 1 mL serum free DMEM and mixed with 20 µl PEI. Solutions were mixed by vortex and incubated for 20 mins. For both transfection methods, the transfection solution was added dropwise to each plate after the 293 T cell media was changed. After 24 hours, the 293 T cell media was again replaced and protein extracts were isolated 48 hours post transfection. Cycloheximide (CHX) (Fisher cat # 50255724) was used at a concentration of 100 μg/ml as indicated.
Generation of viruses and stable cell lines
Lentiviral pCW-2xFLAG-TLK2 was produced by transient transfection into HEK 293 T cells using psPAX and pMD2.G packaging plasmids for virus generation. Lentiviral supernatants were harvested 48 hours after transfection, filtered through a 0.45 μm filter, supplemented with 6 μg/ml polybrene (Sigma) and added to parental cell lines. The medium was replaced 24 hours after the virus and polybrene were added to the parental cells. 48 hours after viral transduction, the infected cells were split into selection media containing 2 μg/ml of puromycin (Sigma cat # P9620-10ML). Selection media were replaced every 2–3 days until selection was complete (2 days–1 week).
Doxycycline induction of stable cell lines
ASF cells of the indicated genotypes stably expressing pCW-2xFLAG-TLK2 WT and pCW-2xFLAG-TLK2 KD were treated with 1 μM doxycycline (Sigma cat # D9891) for 48 hours to induce 2xFLAG-TLK2 expression.
Immunoprecipitation and western blotting
293 T whole cell extracts were prepared using lysis buffer containing 1%TX-100, 50 mM HEPES (pH 7.4), 138 mM KCl, 4 mM NaCl, 50 mM sodium pyrophosphate, 100 mM sodium fluoride, 10 mM EDTA, 1 mM EGTA pH 8, 50 μM PMSF, 1 mM β-Glycerophosphate, 1 mM sodium orthovanadate, and protease inhibitors (Roche cat # 11697498001 or Thermo Scientific cat # 88265) as previously described (Lamia et al.40). ASF cell extracts were prepared from RIPA buffer containing 1% TX-100, 147 mM NaCl, 12 mM sodium deoxycholate, 0.1% SDS, 50 mM Tris pH 8.0, 10 mM EDTA, 50 μM PMSF, 1 mM β-Glycerophosphate, 1 mM sodium orthovanadate, 1 mM sodium fluoride, and protease inhibitors (Thermo Scientific cat # 88265). Antibodies used for immunoprecipitation were anti-FLAG M2 agarose beads (Sigma cat # A2220) and monoclonal anti-HA agarose beads (Sigma cat # A2095). Antibodies for Western blot were anti-FLAG polyclonal (Sigma cat # F7425), anti-HA polyclonal (Sigma cat # H6908), anti-MYC polyclonal (Sigma cat # C3956), anti-SKP1 (BD Biosciences cat # BDB610530), anti-CUL1 (Life Technologies cat # 71–8700), anti-αTubulin (Sigma cat # T5168), anti-βactin (Sigma cat # A1978), anti-AFG3L2[N1N2] (GeneTex cat # GTX102036), and CRY1-CT and CRY2-CT as described (Lamia et al., 2011).
Mitochondrial fractionation
Mice were sacrificed after two weeks of entrainment to 12:12 LD cycles. The right lobe of each liver was extracted from the mice at the indicated zeitgeber times (ZT, hours after lights on), and washed 3X in cold PBS. All steps were done on ice or at 4 °C. 1 mL of mitochondrial lysis buffer containing 220 mM sorbitol, 70 mM sucrose, 50 mM MOPS pH 7.4, 5 mM EGTA and protease inhibitors (cat #11697498001; Roche, Switzerland), was added to each liver sample. The samples were then homogenized and samples centrifuged at 1000 × g for 10 minutes at 4 °C to pellet nuclei and debris. The supernatant was collected as the post-nuclear lysate. The sample was then centrifuged at 9500 × g for 10 minutes at 4 °C to pellet mitochondria. The supernatant (cytosolic fraction) was removed, and the mitochondrial pellet was resuspended in mitochondria wash buffer containing 220 mM sorbitol, 70 mM sucrose, 50 mM MOPS pH 7.4 and centrifuged again at 9500 × g for 10 minutes at 4 °C. This wash step was then repeated twice more. Finally, TX-100 was added to all samples at a final concentration of 1%, and samples were incubated on ice for 10 mins. Mitochondrial lysates were boiled in SDS sample buffer and 60 μg per sample were run on SDS-PAGE.
Measurement of gene expression
RNA was extracted from adult mouse ear fibroblasts with Qiazol reagent using standard protocols (Qiagen cat # 799306). cDNA was prepared using iScript RT Supermix for RT-qPCR (Bio-Rad cat # 1708841) and analyzed for gene expression using quantitative real-time PCR with iQ SYBR Green Supermix (Biorad cat # 1708885). Primers used for qPCR were:
| Forward (5′-3′) | Reverse (5′-3′) | |
| mTlk2 | ACTAGCGCAAAGGAACTCAATGTG | CCAGACCAGGCAAGACTCAGAC |
| hTLK2 | GACGATGACGATAAAATGATGG | CACTCAAGGATCCGACGC |
| mU36b4 | AGATGCAGCAGATCCGCA | GTTCTTGCCCATCAGCACC |
Mice
Cry1−/−;Cry2−/− mice were from Dr. Aziz Sancar43. All animal care and treatments were in accordance with The Scripps Research Institute guidelines and regulations for the care and use of animals. All procedures involving experimental animals were approved by The Scripps Research Institute Institutional Animal Care and Use Committee (IACUC) under protocol #10-0019.
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE [1] partner repository with the dataset identifier PXD009522 and 10.6019/PXD009522.
Electronic supplementary material
Acknowledgements
This work was supported by NIH grants DK097164, CA211187, and DK112927 (to K.A.L.) and 8 P41 GM103533 (to J.R.Y. III), a Searle scholars award from the Kinship Foundation (to K.A.L.), and by fellowships from the American Heart Association (16PRE3041001 to S.P.C.) and the Scripps’ Structure and Function Summer Institute (SFSI) funding for V.P. (NSF/DBI-1359160). We thank Drs Luke Wiseman and Justine Lebeau for help with mitochondrial fractionation and Drs. Eros Lazzerini-Denchi, David Sabatini, and Ben Turk for helpful discussions and sharing valuable reagents.
Author Contributions
K.A.L., S.P.C. and A.L.H. conceived the experiment(s); S.P.C., J.J.M., J.R.Y.III developed the methods; S.P.C., A.B.C., N.Z., M.V., V.P., A.L.H., A.K., J.J.M. conducted the experiment(s); S.J.P. and K.A.L. wrote the manuscript; S.P.C., K.A.L., J.J.Y.III acquired funding; J.J.Y.III provided critical resources and equipment; K.A.L. supervised the project; all authors reviewed and edited the manuscript.
Data Availability
All data generated or analyzed during this study are either included in this published article (and its Supplementary Information files) or are available from the corresponding author on reasonable request.
Competing Interests
Dr. Papp Correia became an employee at Celgene, Inc., after her role in this project was completed. Celgene, Inc., had no involvement with or influence on this study, financial or otherwise. The other authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Stephanie Papp Correia and Alanna B. Chan contributed equally.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-36618-3.
References
- 1.Mohawk JA, Green CB, Takahashi JS. Central and peripheral circadian clocks in mammals. Annual review of neuroscience. 2012;35:445–462. doi: 10.1146/annurev-neuro-060909-153128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Partch CL, Green CB, Takahashi JS. Molecular architecture of the mammalian circadian clock. Trends in cell biology. 2014;24:90–99. doi: 10.1016/j.tcb.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takahashi JS. Transcriptional architecture of the mammalian circadian clock. Nature reviews. Genetics. 2017;18:164–179. doi: 10.1038/nrg.2016.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Siepka SM, et al. Circadian mutant Overtime reveals F-box protein FBXL3 regulation of cryptochrome and period gene expression. Cell. 2007;129:1011–1023. doi: 10.1016/j.cell.2007.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Busino L, et al. SCFFbxl3 controls the oscillation of the circadian clock by directing the degradation of cryptochrome proteins. Science. 2007;316:900–904. doi: 10.1126/science.1141194. [DOI] [PubMed] [Google Scholar]
- 6.Godinho SI, et al. The after-hours mutant reveals a role for Fbxl3 in determining mammalian circadian period. Science. 2007;316:897–900. doi: 10.1126/science.1141138. [DOI] [PubMed] [Google Scholar]
- 7.Lamia KA, et al. AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science. 2009;326:437–440. doi: 10.1126/science.1172156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huber AL, et al. CRY2 and FBXL3 Cooperatively Degrade c-MYC. Molecular cell. 2016;64:774–789. doi: 10.1016/j.molcel.2016.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sillje HH, Takahashi K, Tanaka K, Van Houwe G, Nigg EA. Mammalian homologues of the plant Tousled gene code for cell-cycle-regulated kinases with maximal activities linked to ongoing DNA replication. The EMBO journal. 1999;18:5691–5702. doi: 10.1093/emboj/18.20.5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roe JL, Nemhauser JL, Zambryski PC. TOUSLED participates in apical tissue formation during gynoecium development in Arabidopsis. The Plant cell. 1997;9:335–353. doi: 10.1105/tpc.9.3.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roe JL, et al. TOUSLED is a nuclear serine/threonine protein kinase that requires a coiled-coil region for oligomerization and catalytic activity. The Journal of biological chemistry. 1997;272:5838–5845. doi: 10.1074/jbc.272.9.5838. [DOI] [PubMed] [Google Scholar]
- 12.Roe JL, Rivin CJ, Sessions RA, Feldmann KA, Zambryski PC. The Tousled gene in A. thaliana encodes a protein kinase homolog that is required for leaf and flower development. Cell. 1993;75:939–950. doi: 10.1016/0092-8674(93)90537-Z. [DOI] [PubMed] [Google Scholar]
- 13.Sillje HH, Nigg EA. Identification of human Asf1 chromatin assembly factors as substrates of Tousled-like kinases. Current biology: CB. 2001;11:1068–1073. doi: 10.1016/S0960-9822(01)00298-6. [DOI] [PubMed] [Google Scholar]
- 14.Klimovskaia IM, et al. Tousled-like kinases phosphorylate Asf1 to promote histone supply during DNA replication. Nature communications. 2014;5:3394. doi: 10.1038/ncomms4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bruinsma W, van den Berg J, Aprelia M, Medema RH. Tousled-like kinase 2 regulates recovery from a DNA damage-induced G2 arrest. EMBO reports. 2016;17:659–670. doi: 10.15252/embr.201540767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim JA, et al. Comprehensive functional analysis of the tousled-like kinase 2 frequently amplified in aggressive luminal breast cancers. Nature communications. 2016;7:12991. doi: 10.1038/ncomms12991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim JA, et al. Amplification of TLK2 Induces Genomic Instability via Impairing the G2-M Checkpoint. Molecular cancer research: MCR. 2016;14:920–927. doi: 10.1158/1541-7786.MCR-16-0161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mertins P, et al. Proteogenomics connects somatic mutations to signalling in breast cancer. Nature. 2016;534:55–62. doi: 10.1038/nature18003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yumimoto K, Matsumoto M, Oyamada K, Moroishi T, Nakayama KI. Comprehensive identification of substrates for F-box proteins by differential proteomics analysis. Journal of proteome research. 2012;11:3175–3185. doi: 10.1021/pr201216u. [DOI] [PubMed] [Google Scholar]
- 20.Mellacheruvu D, et al. The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nature methods. 2013;10:730–736. doi: 10.1038/nmeth.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Terajima H, et al. ADARB1 catalyzes circadian A-to-I editing and regulates RNA rhythm. Nature genetics. 2017;49:146–151. doi: 10.1038/ng.3731. [DOI] [PubMed] [Google Scholar]
- 22.Atger F, et al. Circadian and feeding rhythms differentially affect rhythmic mRNA transcription and translation in mouse liver. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:E6579–6588. doi: 10.1073/pnas.1515308112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Janich P, Arpat AB, Castelo-Szekely V, Lopes M, Gatfield D. Ribosome profiling reveals the rhythmic liver translatome and circadian clock regulation by upstream open reading frames. Genome research. 2015;25:1848–1859. doi: 10.1101/gr.195404.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sinturel F, et al. Diurnal Oscillations in Liver Mass and Cell Size Accompany Ribosome Assembly Cycles. Cell. 2017;169:651–663 e614. doi: 10.1016/j.cell.2017.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tong X, et al. DDB1-Mediated CRY1 Degradation Promotes FOXO1-Driven Gluconeogenesis in Liver. Diabetes. 2017;66:2571–2582. doi: 10.2337/db16-1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Groth A, et al. Human Tousled like kinases are targeted by an ATM- and Chk1-dependent DNA damage checkpoint. The EMBO journal. 2003;22:1676–1687. doi: 10.1093/emboj/cdg151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kelly R, Davey SK. Tousled-like kinase-dependent phosphorylation of Rad9 plays a role in cell cycle progression and G2/M checkpoint exit. PloS one. 2013;8:e85859. doi: 10.1371/journal.pone.0085859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Canfield C, Rains J, De Benedetti A. TLK1B promotes repair of DSBs via its interaction with Rad9 and Asf1. BMC molecular biology. 2009;10:110. doi: 10.1186/1471-2199-10-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krause DR, et al. Suppression of Tousled-like kinase activity after DNA damage or replication block requires ATM, NBS1 and Chk1. Oncogene. 2003;22:5927–5937. doi: 10.1038/sj.onc.1206691. [DOI] [PubMed] [Google Scholar]
- 30.Johnson CH. Circadian clocks and cell division: what’s the pacemaker? Cell cycle. 2010;9:3864–3873. doi: 10.4161/cc.9.19.13205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feillet C, et al. Phase locking and multiple oscillating attractors for the coupled mammalian clock and cell cycle. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:9828–9833. doi: 10.1073/pnas.1320474111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bieler J, et al. Robust synchronization of coupled circadian and cell cycle oscillators in single mammalian cells. Molecular systems biology. 2014;10:739. doi: 10.15252/msb.20145218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reitsma JM, et al. Composition and Regulation of the Cellular Repertoire of SCF Ubiquitin Ligases. Cell. 2017;171:1326–1339 e1314. doi: 10.1016/j.cell.2017.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Papp, S. J. et al. DNA damage shifts circadian clock time via Hausp-dependent Cry1 stabilization. eLife4, 10.7554/eLife.04883 (2015). [DOI] [PMC free article] [PubMed]
- 35.Wolters DA, Washburn MP, Yates JR., 3rd An automated multidimensional protein identification technology for shotgun proteomics. Analytical chemistry. 2001;73:5683–5690. doi: 10.1021/ac010617e. [DOI] [PubMed] [Google Scholar]
- 36.He L, Diedrich J, Chu YY, Yates JR., 3rd Extracting Accurate Precursor Information for Tandem Mass Spectra by RawConverter. Analytical chemistry. 2015;87:11361–11367. doi: 10.1021/acs.analchem.5b02721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peng J, Elias JE, Thoreen CC, Licklider LJ, Gygi SP. Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. Journal of proteome research. 2003;2:43–50. doi: 10.1021/pr025556v. [DOI] [PubMed] [Google Scholar]
- 38.Xu T, et al. ProLuCID: An improved SEQUEST-like algorithm with enhanced sensitivity and specificity. Journal of proteomics. 2015;129:16–24. doi: 10.1016/j.jprot.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tabb DL, McDonald WH, Yates JR., 3rd DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. Journal of proteome research. 2002;1:21–26. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lamia KA, et al. Increased insulin sensitivity and reduced adiposity in phosphatidylinositol 5-phosphate 4-kinase beta−/− mice. Molecular and cellular biology. 2004;24:5080–5087. doi: 10.1128/MCB.24.11.5080-5087.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johannessen CM, et al. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature. 2010;468:968–972. doi: 10.1038/nature09627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thresher RJ, et al. Role of mouse cryptochrome blue-light photoreceptor in circadian photoresponses. Science. 1998;282:1490–1494. doi: 10.1126/science.282.5393.1490. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are either included in this published article (and its Supplementary Information files) or are available from the corresponding author on reasonable request.