Abstract
Genetics and epigenetics are tightly linked heritable information classes. Question arises if epigenetics provides just a set of environment dependent instructions, or whether it is integral part of an inheritance system. We argued that in the latter case the epigenetic code should share the universality quality of the genetic code. We focused on DNA methylation. Since availability of DNA methylation data is biased towards model organisms we developed a method that uses kernel density estimations of CpG observed/expected ratios to infer DNA methylation types in any genome. We show here that our method allows for robust prediction of mosaic and full gene body methylation with a PPV of 1 and 0.87, respectively. We used this prediction to complement experimental data, and applied hierarchical clustering to identify methylation types in ~150 eucaryotic species covering different body plans, reproduction types and living conditions. Our analysis indicates that there are only four gene body methylation types. These types do not follow phylogeny (i.e. phylogenetically distant clades can have identical methylation types) but they are consistent within clades. We conclude that the gene body DNA methylation codes have universality similar to the universality of the genetic code and should consequently be considered as part of the inheritance system.
Introduction
Living organisms are biological systems in which the complex interaction between different elements such as the nuclear genotype and epigenotype factors and the environment brings about a phenotype that develops and evolves over time1,2. For a complete understanding and potential control of biological processes such as development and evolution, it is therefore necessary to understand as many elements of biological systems as possible. In the present work, we focus on the epigenotype unit that we operationally define as any modification of the chromatin-DNA complex that has an impact on the expression and function of genes3. Epigenetic information can be stored in a multitude of bearers such as histone modifications, non-coding RNA, the topology of the nucleus, and methylation of DNA. DNA methylation has been one of the most studied epigenetic marks since its discovery in 19484. Methylation occurs at positions 4 and 5 of the pyrimidine ring of cytosine forming either 4-methyl-cytosine (4mC) or 5-methyl-cytosine (5mC), or at position 6 of the purine ring in 6-methyl-adenine (6mA). 6mA and 4mC were believed to occur only in bacteria but recent advances in sequencing technology made it possible to detect them also in eukaryotic species. A specific database (MethSMRT) was dedicated to these modifications5, and the available experimental data were used to train an algorithm to predict the occurrence of 4mC6 in DNA based on sequence features. We will focus here on 5mC and to facilitate the readability use the term DNA methylation for this purpose.
In most eukaryotes, 5mC is overrepresented or even restricted to the dinucleotide CpG context, where ‘p’ stands for the phosphodiester linkage between the cytosine (C) and the guanine (G). In plants, the 5mC can occur in other contexts such as CpHpG or CpHpH, where ‘H’ stands for A, C or T (reviewed in Vanyushin7). In contrast, in certain molds, methylation occurs preferentially (>60%) in CpAs8. DNA methylation is catalyzed by a family of enzymes called DNA methyltransferase (DNMT) composed of 3 canonicals members (DNMT 1, 2 and 3)9. After replication, 5mC will be maintained by the activity of DNMT1, which has a high affinity to hemi-methylated DNA, and that methylates immediately after replication the newly synthesized strand, reproducing methylation patterns in CpG dinucleotide with a fidelity of roughly 99.9%10 thus allowing for mitotic heritability of DNA methylation patterns. The role of DNMT2 is controversial because it has little DNA methylation activity11 but is able to methylate cytosine 38 in the anticodon loop of aspartic acid transfer RNA12 and some authors propose therefore to replace DNMT2 by tRNA (Cytosine(38)-C(5))-Methyltransferase TRDMT113. There are species, such as the model organism Drosophila melanogaster, that have only DNMT2 and do not possess 5-methyl-cytosine in their genome, or DNA methylations is so low that it is very difficult to detect11,14,15. These enzymes have distinct roles due to the presence of different domain structures. DNA methylation is established by DNMT3 that can methylate the two strands of the DNA i.e. has a de novo methylation function16. Various DNA methylation contexts are found across the plant and animal kingdoms. There are species where 5mC is present all over the genome (global methylation) while others can be entirely devoid of methylation. In species with global DNA methylation, only small regions, among them promoters and other regulatory elements, are methylation-free17. If 5mC occurs in the promoters of vertebrates, it has a repressive action on the gene transcription18. Invertebrates often have a mosaic-type methylation pattern with high methylation in almost all CpG in large blocks of genomic DNA interspersed with almost entirely unmethylated blocks. Changes in DNA methylation occur during organ regeneration19, aging20,21, in response to bacterial infection22, as well as flowering time and root length in Arabidopsis thaliana23, just to name a few examples. DNA methylation was therefore proposed as a language in which environment and genome talk to each other. Other authors have seen DNA methylation primarily as a genomic defense system against parasitic genomic elements17.
Given the apparent heterogeneity of DNA methylation patterns and the multitude of biological processes involved it was suggested that DNA methylation evolved in every phylogenetic clade towards a specific role in controlling gene expression. Essentially, the question is whether or not there is universality in the DNA methylation epigenetic code, conceptually similar to the universality of the genetic code, or if DNA methylation is non-universal and specific to every evolutionary unit. We wished to address this question through the analysis of the evolution of DNA methylation. Here, an obstacle is that a comprehensive analysis of DNA methylation patterns in a wide range of different species is missing. There are currently methods available that allow, in principle, for determining genome-wide DNA methylation patterns (“methylomes”) at a single base resolution. Since DNA methylation patterns can be different in different organisms of a species or even tissues of an individual, for a given species several methylomes can exist. A review of the available data in different databases and in the literature showed that there is a strong bias towards model organisms: there are at least 300 methylomes available for human, mouse and the model plant A. thaliana, but only 63 for a total of 16 other species (http://smithlabresearch.org/software/methbase/ and Céline Cosseau, pers. communication). As a consequence, global conclusions about the function and importance of DNA methylation are actually based on a very limited and biased amount of data. For this reason, it remains challenging to derive the general rules (if any) that govern DNA methylation in the different branches of the “tree of life”. A potential solution to the caveat that experimental “wet bench” data is missing is to infer DNA methylation indirectly with computational method24,25. The basis for this is that methylated CpG sites mutate relatively frequently compared to the other dinucleotides over evolutionary time26. If a cytosine is deamined, a deoxy-uracil will form, which is not stable in DNA: it will rapidly be excised by uracil glycosylase and replaced by cytosine. In contrast, 5mC deamination generates thymine, which is less efficiently processed by the DNA repair machinery. Despite the existence of a specific repair mechanism that restores G/C mismatch, the mutation rate from 5mC to T is therefore 10-fold to 50-fold higher than other transitions, depending on local GC content27. For Humans, it was estimated that within 20 years, 0.17% of 5mC in the genome were converted into thymine28. If 5mC occurs predominantly in CpG pairs, the above-mentioned mechanism will increase the mutation rate from CpG to TpG or CpA and induce an underrepresentation of CpG29. Therefore, CpG observed/expected ratio (CpG o/e) in gene bodies can be used to predict if a species’ DNA is methylated in gene bodies or not26,30,31. In other words, gene bodies of a species are not methylated when the CpG o/e ratio is in average close to 1, and methylated for an average ratio far below 1. Low CpG o/e is not a condition for methylation but a consequence of it. It is important to note that this is a species-level prediction that uses methylation signatures that pass through the germ-line and need several generations of mutation accumulation to be detectable. It cannot be used to predict methylation changes of individual genes within shorter periods.
These predictions were tested in at least 13 studies comparing CpGo/e to methylation levels obtained with various methods (Table 1 and Supplementary File 1). All came to the conclusion that CpGo/e ratios correlate well (inversely) with methylation levels when species were compared. Nevertheless, there remain technical challenges. For instant, in the past, prediction of in silico DNA methylation based on Gaussian distributions, that are relatively straightforward to implement, were used to describe the frequency distribution of CpGo/e ratios32–35. But in many species, frequency distributions of CpGo/e ratios are complex or skewed and Gaussian distribution is not suitable. In our hands, only for 58 out of 83 cases (65%) Gaussian mixtures allowed for description of the distribution36. These values are comparable to what was found by Bewick and colleagues who used CpG o/e ratios in transcriptomes of 124 species of which only 50 (40.32%) were described correctly with Gaussian mixtures37. We have also tested non-Gaussian distributions and the results were even less conclusive than Gaussian distributions: out of 83 only 41% delivered an exploitable result. Therefore, we have developed a new tool, called Notos, to identify DNA methylation signatures within CpGo/e ratios based on kernel density estimations36. This novel algorithm delivers robust descriptions of frequency distributions of CpGo/e ratios for up to 172,000 input sequences.
Table 1.
List of publications in which the authors investigated DNA methylation by a wet bench method and compared the results to CpGo/e ratios.
| Species | Formula | Sequences | Validation | References |
|---|---|---|---|---|
| Acropora millepora | Unknown | CDS | MBD-eq | 57 |
| Apis melifera | Unknown | CDS | BS-seq | 78 |
| Biomphalaria glabrata | Matsuo72 | RNAseq | Restriction enzyme, BS-seq (Nimbus retrotransposon), LC-MS | 79 |
| Crassostrea gigas | Matsuo72 | EST | Methylation sensitive PCR, BS-Seq | 32 |
| Solenopsis invicta | Unknown | Genome | MeDIP, BS-Seq (9 genes) | 80 |
| Gardiner-Garden and Frommer52 | Promoteur and Genes | BS-Seq | 81 | |
| Nasonia vitripennis | Matsuo72 | Refseq | Cloning and sequencing 18 genes at selected CpG sites, BS-seq | 33 |
| Unknown | Genome and coding sequences | Whole genome bisulfite sequencing | 82 | |
| Locusta migratoria | Unknown | cDNA, Unigene | Methylation-specific restriction enzyme assays | 83 |
| Acyrthosiphon pisum | Unknown | CDS and predicted genes | MeDIP, BS-seq, restriction enzyme | 35 |
| Bombyx mori | Unknown | Genes | MethylC-seq | 84 |
| Nicrophorus vespilloides | Unknown | Genes | Whole genome bisulfite sequencing | 85 |
| Ciona intestinalis | Unknown | Genes | BS-Seq | 86 |
| Unknown | EST | BS-Seq, Methylation-sensitive PCR | 31 | |
| Arabidopsis thaliana | Gardiner-Garden and Frommer52 | CDS | BS-seq | 87 |
Here, we have applied this software to predict DNA methylation with CpGo/e ratios in a total of 634 species and to use the results in combination with publicly available experimental data to infer evolution of DNA methylation over the eukaryotic tree of life. We applied Notos on coding sequences coming from three databases (dbEST, CleanEST, and CDS/cDNA). Our results show clearly (i) that DNA methylation prediction by CpGo/e ratio is robust, (ii) that only four types of DNA methylation can be identified in all species despite their wide range of genome sizes, environments, body plans, reproduction types etc., and (iii) that DNA methylation types does not follow phylogeny but is consistent within clades suggesting evolutionary constraints. Taken together our analysis delivers arguments for the idea of the universality of the role of DNA methylation that is preserved through evolution.
Results
CDS and cDNAs are the less biased data and thus the best choice for a pan-species study
We focused in this study on gene body DNA methylation. Annotated genomes are now available for many species, but messenger RNA sequence data is even more abundant and mRNA is representative of gene body DNA sequences. They could therefore be used instead of DNA sequence, but mRNA data is for historical reasons stored in different forms and in different databases. We reasoned that data quality will be critical for providing unbiased estimation of DNA methylation in gene bodies and therefore conducted a comparative pilot study to identify the best possible data source for the subsequent pan-species study. We used coding/transcript sequences from full genome annotations (CDS), dbEST, and cleanEST (details in Supplementary File 2). A total of 127 species are in common between CDS and dbEST, and 92 species were in common between dbEST and cleanEST. Only 29 species were common to all three databases (Supplementary File 3). We produced Notos CpGo/e profiles for all intersecting datasets and proceeded to visual inspection. In 11 out of 29 cases (38%) we identified discrepancies in at least one out of the three profiles and decided to clarify their origins by a detailed analysis of the sequences under the differential peaks. An in-depth analysis revealed that these discrepancies were either due to contaminations during the sequencing process, reflect co-occurrence of other species, or are due to bias in data acquisition. For instance, for Trichoplax adhaerens, Anolis carolinensis (green anole lizard) and Cordyceps militaris one or two additional shoulder peaks in dbEST and CleanEST datasets. We isolated the sequences contained in these peaks (dbEST and Clean EST) and performed a Blast2GO analysis with the aim to know their origins and functions. For the anole lizard (Supplementary Fig. 1), two peaks were isolated (peak 1: 0.92–1.08 and peak 2: 1.14–1.22), representing 7,030 and 4,922 sequences, respectively. The majority of sequences under peak 1 in the dbEST profile correspond to chloramphenicol O-acetytransferase used in bacterial cloning vectors. It is therefore likely that these sequences represent contaminations from the EST library generation procedure. Sequences under peak 2 present homologies with sequences from apicomplexans (plasmodium), and platyhelminths suggesting presence of such parasites in the initial biological sample. For T. adhaerens, a peak was isolated (1.22–1.35), which represents 1,609 ESTs. Most of the sequences under the dbEST peak were identified by Blast2GO as ‘other’. Since T. adherens is known to contain intracellular bacteria38 we believe that these sequences originate from them (Supplementary Fig. 2). For the mold C. militaris, two peaks were isolated (1.14–1.22, and 1.26–1.32). For the sequences under these peaks, homologies with other fungi sequences were found. We conclude that, in all three species, the additional modes occurred due to presence of sequences from other species, either through contamination during RNA extraction and library preparation, or as co-purification from naturally occurring symbionts or parasites. In other species, we identified other sources of bias in the transcript data. For instance, in Bombyx mori an ovarian library cleanEST showed an additional weak shoulder peak. We isolated the 769 sequences under this peak (0.40–0.60). The gene ontology showed that all sequences coded for the ribosomal protein SA (RPSA). In human, RPSA genes are indeed highly expressed in the ovary but no data are available for other species. Nevertheless, it seems unlikely that the high abundance of RPSA ESTs reflects an expression bias. We speculate that the research interest of the submitters focused on this particular gene and that therefore many individual EST were submitted (Supplementary Fig. 3). Also in the duck (Anas platyrhynchos), we found a shoulder peak at 0.57 and 0.59 in data from dbEST and Clean EST. Sequences under this peak corresponded to an EST library exclusively composed of immunoglobulins (146 sequences), reflecting probably a bias introduced by specific research interests (Supplementary Fig. 4). Interestingly, these sequences had a CpGo/e ratio between 0.5 and 0.8 suggesting hypomethylation, and in human, immunoglobulin genes in lymphoid cells are indeed undermethylated during differentiation39.
Finally, when we compared profiles with two peaks (bimodality), where we had noticed differences between CDS (derived from genomes) and dbEST/CleanEST (mRNA) for three invertebrate and one plant species (Crassostrea gigas, Nasonia vitripennis, Nematostella vectensis and Oryza sativa) (Supplementary Figs 5–8): mRNA derived profiles showed a higher peak in genes predicted to be methylated. Gene body methylation is suspected to increase transcription40,41. The principal differences between CDS and EST data is that for the former only one FASTA sequence per gene is considered while for the later potentially several FASTA sequences for a gene could be present. We therefore hypothesized that RNA abundance induced the bias in EST data. To test this hypothesis, we performed a RNA-seq analysis in these four species. We found that genes under the low CpGo/e peak (presumably hypermethylated) show higher median RNA amounts than genes under the high CpGo/e peak (this presumably hypomethylated). mRNA FPKM medians are 1.95 to 5.45 higher in presumably hypermethylated gene bodies (Supplementary Figs 5–8). We conclude that this expression difference is probably the origin of the bias in EST datasets.
In summary, dbEST and cleanEST have the advantage of being large repositories with data for many species, but for the purpose of our study we considered them too noisy. A complete list of species for each dataset is in Supplementary File 4.
CpGo/e clustering identifies four types of gene body DNA methylation
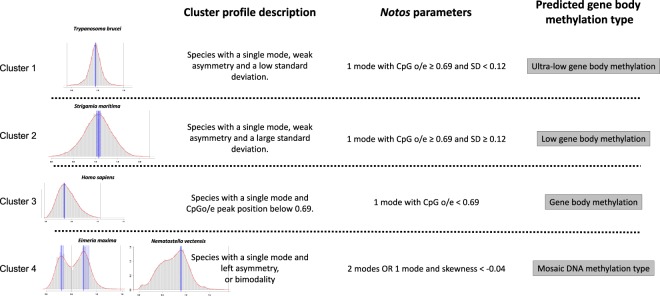
After having firmly established that cDNA provides an unbiased data basis, CpG o/e clustering was carried out on the 142 species for which CDS or cDNA were available. Parameters for mode number (n), mode positions (Mo), skewness (sk), and standard deviation (SD) of CpGo/e values were iteratively changed using species with known gene body DNA methylation. For further analysis, we used the following features that produced four clusters of CpGo/e: (cluster 1) species with one mode Mo ≥ 0.69 and SD an0.12, (cluster 2) species with one mode Mo ≥ 0.69 and SD ≥ 0.12, (cluster 3) species with one mode Mo s0.69, and (cluster 4) species with (a) two modes or (b) one mode and a skewness smaller than −0.04. Results are in Supplementary File 5 and Supplementary Fig. 9. We then associated the four clusters with known methylation types.
Fourteen species (9.72%), from different phylogenetic groups (e.g. Ascomycota, Apicomplexa, Basidiomycota, Plathyhelminthes and Arthropoda) constitute the cluster 1. All the species have a CpGo/e mode position mode above 0.69 (the mean CpGo/e peak position is 1.00), a weak negative skewness (meanabsolute Q50 skewness = −0.0019) and a narrow standard deviation (meanSD = Da0.11). For 4 species (29%), experimental data on DNA methylation was available in the literature. All these species showed either absence of DNA methylation in the gene bodies or extremely low levels (Supplementary File 5). The latter was found in only one species (Chlamydomonas reinhardtii) where WGBS revealed methylation in exons but still it was 20–30 times weaker compared to other plant species in the same study. We qualify cluster 1 as “ultra-low gene body methylation” (type 1).
It could be argued that absence of gene body methylation is simply a consequence of absence of enzymatic methylation activity. We therefore performed a metanalysis of existing literature data concerning DNMT presence. In cluster 1, 6 out of 14 species have DNMT2 or TRDMT1, and one specie has DNMT1 and DNMT2. Only the de-novo methylase DNMT3 is absent in this cluster. Absence of methylation does therefore not indicate necessarily absence of DNMT genes (Supplementary File 5).
Cluster 2 is constituted by 60 species (41.67%), also from different phylogenetic groups (apicomplexa, oobionta, rhodonbionta, ascomycota, basidiomycota, nematoda, platyhelminthes, annelida, arthropoda, ctenophora, chordata, embryophyta). As in the cluster 1, species present in the second cluster have a mode position > 0.69 with a mean mode position very close to the first cluster (mean CpGo/e position is 0.92) and a mean absolute Q50 skewness of 0.0012. However, in contrast to cluster 1, a wide standard deviation (meanSD = Da0.18) has been observed. Literature data were available for 18 species (30%). For 6 species (Schizosaccharomyces pombe, Aspergillus flavus, Brugia malayi, Meloidogyne incognita, Tribolium castaneum, Drosophila melanogaster) no methylation was reported. Methylation in 6 species (Schistosoma mansoni, Schistosoma japonicum, Fasciola hepatica, Petromyzon marinus, Caenorhabditis elegans and Saccharomyces cerevisiae) is controversial since different authors come to different conclusions. Nevertheless, methylation seems to be very low. Only three species (Trichinella spiralis, Solenopsis invicta, Physcomitrella patens) showed DNA methylation in gene bodies. DNMTs were studied in 37 species. In this cluster, 16 species have just DNMT2. One species has DNMT1 or TRDMT1 only, and 6 species have all the DNMT (DNMT1, 2 and 3). Interestingly, two species (Selaginella moellendorffii, Physcomitrella patens) has just DNMT1 and 3, and two just DNMT1 and 2 (Tribolium castaneum, Gasterosteus aculeatus). Also, for Strigamia martima DNMT1 and DNMT3 were found, but DNMT2 was not searched for (Supplementary File 5). We consider cluster 2 as “low gene body methylation” (type 2).
Species with a mode position ≤ 0.69 form the cluster 3 (mean CpGo/e position is 0.45). The skewness is positive and larger than in the two first clusters (meanabsolute Q50 skewness = 0.0483). The standard deviation (meanSD = Da0.19) is wider than the cluster 1 and 2. Forty-three species (29.86%) are present in this cluster, belonging to various phylogenetic groups (apicomplexa, sponges and nematodes, arthropoda, a large panel of chordata, and embryophytes). Many of them are important model organisms. For 26 (60%) literature data on DNA methylation was found. Gene bodies are methylated. In 19 species DNMTs were analyzed. Twelve species have all three DNMTs. One species (Naegleria gruberi) has DNMT1 and 2, but methylation of DNA was not yet studied. Four species have only DNMT2. Two species has DNMT1 and 3, and one only DNMT1 (Supplementary File 5). We qualify this cluster as with “gene body methylation” (type 3).
Finally, cluster 4 contains species that show bimodality or are strongly negatively skewed CpGo/e distributions (meanabsolute Q50 skewness = −0.0424, mean CpGo/e position of mode 1 is 0.54, of mode 2 0.85). Twenty-seven species (18.75%) from different phylogenetic groups compose this cluster (apicomplexa, cnidaria, nematoda, arthropoda, mollusca, tunicata, embryophyta). Five species are strongly negatively skewed and 15 species are bimodal. We found information on DNA methylation for 10 species (50%). All species show a mosaic type of methylation with DNA regions of ultra-low methylation interspersed with regions of strong methylation. Eleven species out of 15 that were studied have the three DNMT (1, 2 and 3), two had DNMT1 and 2, and two DNMT1 and 3 with uncertainty about DNMT2 (Supplementary File 5). Species in this cluster were considered as “mosaic type DNA methylation” (type 4).
Decision criteria are summarized in Fig. 1.
Figure 1.
Summary of decision grid for clustering of CpG o/e ratio distributions on species level.
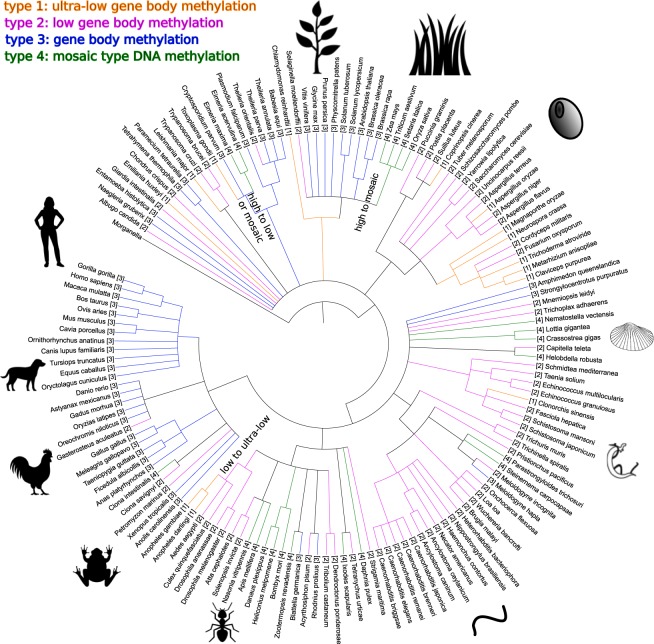
DNA methylation types do not follow the tree of life but are consistent within major clades
Among unicellular organisms, kinetoplastids are unmethylated, while alveolate protists are generally methylated (Fig. 2) with secondary loss of methylation that can lead to weaker methylation level or mosaic methylation. In flowering plant species, we differentiate either high (probably global) methylation in dicotyledons, and mosaic methylation in poaceae and potentially all monocotyledons. Fungi process in general ultra-low to weak methylation in the gene bodies. Also, platyhelminthes are characterized by low methylation in the coding regions. Gene bodies of deuterostomes are in general strongly methylated. There are, however, peculiar cases, e.g. two tunicate species (Ciona intestinalis and C. savignyi) that diverged from each other 184 (±15) Mya are in two different clusters (types 4 and 2, respectively)42 with mosaic and weak methylation, probably due to secondary loss of methylation. Within lophotrochozoa, annelids show low gene body methylation, and all studied mollusks are of the mosaic type. Nematodes have in general weak gene body methylation. A particular interesting and heterogenic clade concerning methylation types are arthropods. All tested diptera (and potentially all antliophora) belong to the low methylation clusters 1 and 2, certainly due to secondary loss of methylation after splitting from its insect sister clades. All other orders show weak to high methylation with occasionally mosaic type, probably through secondary gain of local methylation.
Figure 2.
Schematic representation of the “Tree of Life” for 147 species, associated with the four different types of DNA methylation that were identified in this work. Numbers in brackets indicate DNA methylation types (“clusters”) for each species. Line colors correspond to methylation types.
Discussion
Evolution is based on the selection of phenotypic variants that must (i) confer a reproductive advantage to the individual, and (ii) are heritable, i.e. information how to generate the phenotypic variants in response to an environment are passed from parents to offspring. Heritability has traditionally been thought to be exclusively genetic, i.e. based on variations in the DNA sequence. In this view, genetic information is then expressed under influence of environmental cues to bring about the phenotype, a process known as G × E→ P43. During the last 30 years it became however clear, that a substantial amount of heritable phenotypic variance can be coded by non-genetic means44. We had earlier conceptualized this view as a systems approach to inheritance, that includes genetic, epigenetic, cytoplasmic and microbial elements that are interrelated by forward and reverse interaction2. These elements interact mutually, and with the environment, to give raise to the phenotype. In this concept, genetic information (the genotype) is only one of many elements that as part of an inheritance system providing heritable information, that the environment will shape into a phenotype. We define here ‘inheritance system’ as a system that is able to write and store, transmit, and receive hereditary information45. The concept implies also that genotype and epigenotype cannot exist independent of each other, and are interrelated by forward action and feed-back. This is different from the idea that sees the genome as hard-wired information that is controlled by the epigenome46. In the latter, the epigenome is conceptually closer to the (molecular) phenotype (i.e. product of the genotype) than to an element of the inheritance system itself.
The introduction of the epigenotype notion did not really solve the question, since theoretically each phenotype could just be the visible expression of its underlying epigenotype. Given the multiple facets phenotypes can acquire in living organisms, it is remarkable that, with very few exceptions, the genetic material and the genetic code remains extremely constant and thus universal47. In other words, there exists a single ‘type’ of genome. The origin of this universality of the genetic code remains enigmatic and controversial but whatever the origin is, it allows to transmit coded information from one generation to the next. These generations can understand the code since it uses a universal and constant key.
Given the presumably close relation between genotype and epigenotype we and others reasoned that the epigenotype and the epigenetic code should equally possess universality. The high conservation of histones and histone marks, and the conservation of methylation of cytosines suggests indeed this. Nevertheless, one could argue that the epigenetic code is simply entirely genetically determined. If this were true, we would expect that different DNA methylation types would correspond to the clades in taxonomical tree that are based on DNA sequence similarity. Our results do not support this view. Alternatively, DNA methylation types could entirely be determined by environmental conditions. In this case, similar environments should impose similar DNA methylation types. Neither our results, nor recent analyses of DNA methylation in invertebrates provide evidence of this. E.g. a very comprehensive study of DNA methylation in insects37 did not find relations of methylation types to social behavior and the authors concluded that DNA methylation must have “more ubiquitous function”. However, compared to the tremendous amount of genomic data that is available, epigenomic data is relatively sparse and biased, which is an obstacle to answer the question conclusively. In the present study, we coped with this caveat by using a hybrid approach in which we combined available experimental data on DNA methylation with results coming from a newly developed software that predicts gene body DNA methylation types with CpG o/e ratios. Our algorithm (based on the number of species positive predicted and true positives based on the literature) allowed for including species for which no experimental DNA methylation data existed. The PPV of the algorithm is excellent for mosaic methylation (PPV = 1), and methylated gene bodies (PPV = 0.875), but decreases then to 0.75 (low methylated) and 0.5 for ultra-low gene body methylation. This is due to the fact that our algorithm does not differentiate well between low and ultra-low methylation. If we consider the dataset as a whole, out of the 54 species with known DNA methylation types, 41 were predicted correctly (total PPV = 0.76).
There are some particularly interesting cases of “wrong” prediction. Cryptosporium parvum is a monoxenic unicellular parasite of vertebrates. It belongs to the apicomplexan its exact phylogenetic position is controversial. Exysted oocysts are the only stage that can be used to produce host DNA free genomic DNA preparations. Notos predicts clearly high gene body methylation but LC-ESI-MS did not detect 5mC in exysted oocysts purified from infected cattle48. Genome analysis of C. hominis to which C. parvum has only 3–5% sequence divergence49, showed that the number of genes is reduced (3,952 genes) compared to other apicomplexan, relying heavily on host gene activity. The genome shows also traces of integration of genes by lateral transfer. We hypothesize that either the progenitor DNA was methylated, or that cryptosporidium methylates DNA in the intracellular stages using the vertebrate host DNMTs.
Another peculiar case is the ciliate Tetrahymena thermophile. Also for this species Notos predicted high methylation while radioisotope labeling showed that Tetrahymena contains only N6-methyl-adenine but not 5mC50. T. thermophila and other ciliates use DNA elimination to remove approximately one-third of the genome, when the somatic macronucleus differentiates from the germline micronucleus. Histone 3 lysine 9 trimethylation (H3K9me3) is deposited on DNA destined for this elimination (reviewed in Bracht51). Interestingly, in other ciliates, DNA methylation is used for the tagging of DNA to be eliminated. It might therefore be that Tetrahymena had used DNA methylation in the past and has lost this capacity relatively recently, so that we still see traces in the CpG o/e ratio.
In summary, Notos predicts very reliable mosaic and high gene body methylation without being entirely error free. We had earlier36 used only mode number (1 or 2, i.e. non-mosaic and mosaic methylation) and peak position of 0.75 to differentiate species with presumably methylated (<0.75) and non-methylated (≥0.75) gene bodies. For the present work we added skewness −0.04, and SD 0.12, and changed peak position threshold to 0.69 for better prediction.
Conceptually, our approach is based on the classical observation that CpN dinucleotides are observed in statistically expected frequency in low methylated regions or genomes. It was initially used to identify unmethylated CpG island in vertebrate promoters, and two major algorithms exist (Gardiner-Garden and Frommer52 and Takai and Jones53). These two algorithms use the CpG o/e ratios with a score above 0.60 and 0.65, respectively. ‘Score’ (here ‘mode position’ Mo) is one parameter of our clustering algorithm. We used a decision tree to iteratively adjust this score and reached 0.69. This value is close to what was used in previous studies (e.g. for C. intestinalis: 0.7054 and 0.8031, and Nematostella vectensis, 0.7054, or Apis mellifera, 1.054). It is conceivable that Mo could be slightly different for each major phylogenetical clade, and using more sophisticated clustering algorithms such as support vector machine clustering that can use multiple thresholds could still improve the PPV of our method. In addition, more experimental data on a wide range of organisms is urgently needed.
We find that there are four types of gene body methylation. Despite a much wider data basis in terms of phylogenetic clades, our results confirm earlier findings that concluded on three to four DNA methylation types17,24. This could be the result of a “frozen accident” situation in which methylation (e.g. type 1 and type 3) occurred randomly in early ancestors (since 5mC is coding neutral that would not have had an impact on translation), but with the establishment of a chromatin structure 5mC was recruited as epigenetic information carrier, and any change in DNA methylation type would have had a strong impact on genome function and thus fitness and was therefore maintained. Nevertheless, switching of methylation type has occurred in evolutionary time scales. Our findings indicate that there were at least three large events of secondary loss of DNA methylation: in archaeplastida (the “true” plants) where we find one branch with high methylation and another with mosaic methylation (in monocotyledons), ultra-low or mosaic methylation in the apicomplexa branch of “protists”, and one transition to ultra-low gene body methylation in Diptera (Fig. 2). For D. melanogaster in the dipteran branch it was shown experimentally that only the ‘writing’ capacity of the epigenetic inheritance element was lost, not the receiving (‘reading’) capacity55. The reason for evolutionary switching between methylation types is not clear and arguments are controversial.
It has been proposed that secondary loss of DNA methylation occurs because its mutational costs outweighed its adaptive value56. Indeed, in mosaic type methylation it is the evolutionary stable “old” genes that are in the methylated compartments meaning that there must be stabilizing mechanisms that prevent mutations there. Therefore, it might not be the mutational costs but the costs of maintaining such mechanisms that becomes an evolutionary burden. It was an early observation that CpG containing codons are used much less in coding sequences of vertebrates, and mutations due to CpG methylation was considered a major cause for such codon bias57 and therein. Codon bias was observed also recently in the reef-building coral Acropora millepora57, and linked to mosaic methylation in this species. Again, phylogenetically old genes which are constitutively expressed are methylated and CpG depleted. The authors conclude that CpG methylation leads to mutations that establish a set of preferred codons in constitutively expressed genes. Once such codon bias is fixed, then alleles that control the abundance of appropriate tRNAs could have stronger effects more amenable to natural selection. The authors hypothesize that an advantage of mutation-driven codon bias that it would be beneficial for organisms with small population size or otherwise inefficient selection. Still another explanation for mosaic methylation was advanced by Gavery and Roberts32 who speculated that hypo-methylated regions (here in the pacific oyster Crassostrea gigas) might have greater epigenetic flexibility and higher regulatory control than hyper-methylated ones. Mosaic methylation could also be the result of whole genome duplication (WGD) events as suggested for Oryza sativa56. In addition, we have shown that environmental conditions can influence on germ-line methylation in C. gigas that possess mosaic methylation, and that blocks of CpG methylation are added or removed preferentially in or around genes58. One should keep in mind that DNA methylation is only one of many bearers of epigenetic information. Another one, and probably the most difficult to capture is the topology of the interphase nucleus. Using Hi-C data, Lieberman-Aiden et al.59. established that the human genome is divided into two compartments (A-B) with pairs of loci in compartment B showing higher interaction frequency at a given genomic distance than pairs of loci in compartment A. They concluded that compartment B is more densely packed (heterochromatic) than compartment A. Higher average DNA methylation was later found to be a good predictor for the open compartment A in human cell lines60 but that link could be broken in cancer cells. This cannot be interpreted as DNA methylation being decisive for topologically associated domains (TAD) establishment since DNA methylation free organisms such as D. melanogaster also presents canonical A-B domains61. But in drosophila, such TAD organization is not driven by long-lived interactions but rather relies on the formation of transient, low-frequency contacts62. We hypothesize therefore that DNA methylation actually impacts on the relative dynamics of formation of contacts in A and B compartments, possibly stabilizing them. It is tempting to speculate that one consequence of compartmentation of genomes dynamics by methylation is that this might create additional units of selection. Results from tunicates support this idea: Ciona CpGo/e ratios have different profiles (bimodal for C. intestinalis and unimodal for C. savignyi). The C. intestinalis methylome is predicted to be mosaic that corresponds to experimental observations63. Our prediction for C. savignyi is low methylation (cluster 2). Both species diverged from each other 184 (±15) Mya42 and their genomes are very different64,65. For instance, analysis of 18S rRNA sequences shows that the pairwise divergence of the two ciona species is slightly greater than that between human and e.g. birds66. This is puzzling since developmental features, body plan, effective population size and environment are very similar, and even hybrids can be generated to the tadpole stage67. However, C. savignyi shows a genome wide average Single Nucleotide Polymorphism (SNP) heterozygosity of 4.5% while C. intestinalis, that has mosaic methylation, is genetically less polymorphic (1.5%) (reviewed in Veeman et al.68). It is conceivable that the methylated C. intestinalis genome can generate sufficiently stable TADs so that genome x epigenome interactions can serve as heritable unit of selection, while in C. savignyi TADs are more dynamic because the relative weight of DNA methylation in the generation of stable heritable phenotypic variants is less important. Our prediction concords with very recent results showing that stress-induced DNA methylation changes in C. savignyi can occur but are highly ephemeral (<48–120h), and thus not maintained through germline69.
In conclusion, our findings indicate that initially there were three types of gene body DNA methylation: ‘primary no methylation’, ‘primary whole genome methylation’, and ‘primary mosaic methylation’ that produced by secondary loss ‘weak methylation’, or ‘secondary no methylation’. These findings are in concordance with the idea that DNA methylation in gene bodies (i) uses three types of universal codes (low, high and mosaic)), and (ii) that it is an element of the inheritance system and not a molecular phenotype that results from genotype × environment interaction. This has immediate practical consequences: e.g. since there are three types of methylation codes, pan-species conclusions about the potential function of DNA methylation can only be drawn within the type (e.g. functional tests in vertebrates with high gene body methylation cannot be used to conclude on methylation function in mosaic type mollusks). In addition, if DNA methylation is part of the inheritance system then heritable phenotypic diversity can be produced by DNA methylation changes without changes in the DNA sequence. The notion that everything that is heritable is necessarily genetic should be abandoned.
Methods
Origin of sequences, data cleaning and Notos parameters
In this study, coding sequences (CDS) or cDNA sequences of 147 species were downloaded from Ensembl and VEGA databases. Expressed sequence tags (ESTs) were downloaded from two different databases: dbEST70 (605 species) and CleanEST71 (110 species) (Supplementary File 3). We used Notos36 for the calculation and modelling of CpGo/e distribution with the three datasets, with a minimal length L = mi200 bp and formula 172:
| 1 |
All the values outside the interval, and all the values with a score of 0 were removed. For each species, the number of mode, the position of mode(s), the Q50 skewness coefficient and the standard deviation (SD) were calculated.
Blast searches and gene ontology analysis
Database searches were done by Blastx searches against a local instance (ncbi-blast-2.2.30+) of non-redundant ‘nr’ with 20 maximum hits, an E-value of 0.001, and other parameters as default values. Gene ontology searches were performed with blast2go73.
RNA seq analysis
RNAseq datasets for Nematostella vectensis, Nasonia vitripennis, Crassostrea gigas and Oryza sativa japonica were downloaded as fastq files from the European Nucleotide Archive and NCBI (details in Supplementary File 3). For each dataset, the reads were filtered with a Fred quality score ≥26. Filtered reads were mapped on their reference genomes (downloaded from Ensembl, details in Supplementary File 3) with RNA STAR74 on a local Galaxy instance (v2.4.0d-2). Resulting BAM files and the gff files (downloaded from Ensembl, details in Supplementary File 3) with the coding sequences were used for FPKM estimations with Cufflinks75. Annotation gff-files were used to extract CDS in fasta format from their genomes and we calculated the CpGo/e ratios with Notos36 and detected modes (peaks). To compare FPKM for the genes under the peaks, a bandwidth of 0.2 (±0.1 around mode maximum) was arbitrarily chosen for the CpG o/e ratio. FPKMs were extracted and used for statistical analysis of expression level in gene bodies with low and high predicted methylation. Mood’s median test was used with R76.
Meta-analysis of DNA methylation using literature data
For each species for which data was available in the above-mentioned databases, we searched the literature on Google scholar (as of April–June 2016) with the following keywords: DNA methylation, 5-methyl-cytosine, gene body, mosaic methylation, global methylation, DNA methylation pattern. Articles were obtained from Bib CNRS (https://bib.cnrs.fr/) and manually curated to obtain gene body methylation, and presence of DNMTs.
Clustering
To identify distinct subgroups within the 147 analyzed species, we generated descriptive analyses, considering both the KDE of the CpGo/e ratios and aggregating statistics based on it. The statistics we used were (1) the number of modes of the KDE, (2) the position of the modes, (3) the standard deviation SD of the CpGo/e ratios, (4) absolute Q50 mode skewness of the CpGo/e ratios, i.e.,
| 2 |
with Q1 and Q3 the 25% and 75% quantile of the CpGo/e ratios, respectively, and Mo the global mode of the KDE. We investigated several formulas for the skewness, deeming the absolute Q50 mode skewness the most informative for our analysis36. For the sake of readability, we refer to the absolute Q50 mode skewness as “skewness” in what follows.
The four clusters into which we classify the species are specified in the result section. The values of the three thresholds used in the definition of the clusters were determined by evaluating the prediction performance of our approach depending on these three values, using 54 species for which the true methylation type had been determined experimentally (given in Supplementary File 5). Hereby, the clusters correspond to the patterns used in that file like follows: Cluster 1 - not methylated; Cluster 2 - low methylated; Cluster 3 - (high) methylated/global methylation; Cluster 4 - mosaic methylation.
To determine the optimal threshold values, we employed a two-step approach. First, we searched the whole parameter space (i.e. the space of all possible values the thresholds can assume) using a Metropolis-Hastings algorithm to ensure that strongly deviating from the threshold values we chose manually always leads to a poor prediction performance. Second, we systematically searched the parameter space around the manually chosen values. That is, we evaluated the prediction performance on a grid of size 21³ on9261, covering the following threshold values: skewness from −0.08 to −0.04 in steps of 0.002; peak position from 0.69 to 0.79 in steps of 0.005; SD from 0.11 to 0.21 in steps of 0.005. For the present work we used: skewness −0.04, peak position 0.69, and SD 0.12.
Due to the scarcity of the data, the optimal prediction (76%, 41 out of 54 true) is achieved for a rather large set of threshold values. To judge the performance of our algorithm, it should be noted that for 7 out the 13 misclassified species the true and the predicted classifications are “not methylated” and “low methylated”, or vice versa respectively. That is, the mistake made by the algorithm is rather small in these cases. The remaining four wrong predictions are actually peculiar cases that were discussed above.
The clustering was implemented using R version 3.4.0 (Supplementary Files 6 and 7). For visualizing the clustering, the R package dendextend has been used. Parameters were Rscript cluster.r −0.04 0.69 0.12 input_file_notos input_file_notos_bootstrap with first parameter [skewness], second [Mo], and third [SD], and Notos outputfiles as input. Further details on our method can be found in25.
Tree of life
We recovered the taxonomic IDs of all investigated species from the NCBI taxonomy database (https://www.ncbi.nlm.nih.gov/taxonomy) and created a tabular file (.txt). We used this file to generate a Phylip tree file based on the classification in the NCBI taxonomy database with the NCBI common taxonomy tree online tool (https://www.ncbi.nlm.nih.gov/Taxonomy/CommonTree/wwwcmt.cgi) and designed a tree with the interactive Tree of life (version 4.0.2) (https://itol.embl.de)76.
Supplementary information
Acknowledgements
The work on this tool was initiated during a meeting that had received funding of the French-Norwegian travel program AURORA. This work has been supported by Campus France and the Norges forskningsrad (program AURORA, nr. 34040YK) to C. Grunau and J. Bulla, the grant Felleslegat til fordel for biologisk forskning ved Universitetet i Bergen to J. Bulla, the ANR grants ANR-10-BLAN-1720 (EpiGEvol) and ANR‐17‐CE12‐0005‐01 (CHRONOGET), a PhD grant for disabled students by the French Ministry of Education and Research to B. Aliaga, and a DFG return grant to I. Bulla (BU 2685/4-1). The authors are grateful to Céline Cosseau and Cristian Chaparro for helpful discussion.
Author Contributions
C.G. and D.D. designed the study. B.A. performed the experiments. I.B. developed the mathematical models, wrote the associated code, and did the clustering. G.M. and B.A. worked on the phylogeny. All authors wrote and reviewed the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-37407-8.
References
- 1.Levine AJ. The Future of Systems Biology. Curr. Opin. Syst. Biol. 2017;1:v–vii. doi: 10.1016/j.coisb.2017.02.007. [DOI] [Google Scholar]
- 2.Cosseau C, et al. Epi)genetic Inheritance in Schistosoma mansoni: A Systems Approach. Trends Parasitol. 2017;33:285–294. doi: 10.1016/j.pt.2016.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nicoglou, A. & Merlin, F. Epigenetics: A way to bridge the gap between biological fields. Stud. Hist. Philos. Sci. Part C Stud. Hist. Philos. Biol. Biomed. Sci. 1–10 10.1016/j.shpsc.2017.10.002 (2017). [DOI] [PubMed]
- 4.Hotchkiss RD. The quantitative separation of purines, pyrimidines and nucleosides by paper chromatography. J. Biol. Chem. 1948;175:315–332. [PubMed] [Google Scholar]
- 5.Ye P, et al. MethSMRT: an integrative database for DNA N6-methyladenine and N4-methylcytosine generated by single-molecular real-time sequencing. Nucleic Acids Res. 2017;45:D85–D89. doi: 10.1093/nar/gkw950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen W, Yang H, Feng P, Ding H, Lin H. iDNA4mC: identifying DNA N4-methylcytosine sites based on nucleotide chemical properties. Bioinformatics. 2017;33:3518–3523. doi: 10.1093/bioinformatics/btx479. [DOI] [PubMed] [Google Scholar]
- 7.Vanyushin, B. F. In DNA Methylation: Basic Mechanisms 67–122, 10.1007/3-540-31390-7_4 (Springer-Verlag, 2006).
- 8.Cambareri E, Jensen B, Schabtach E, Selker E. Repeat-induced G-C to A-T mutations in Neurospora. Science (80-.). 1989;244:1571–1575. doi: 10.1126/science.2544994. [DOI] [PubMed] [Google Scholar]
- 9.Lyko, F. The DNA methyltransferase family: a versatile toolkit for epigenetic regulation. Nat. Rev. Genet. 10.1038/nrg.2017.80 (2017). [DOI] [PubMed]
- 10.Riggs AD, Xiong Z, Wang L, LeBon JM. Methylation dynamics, epigenetic fidelity and X chromosome structure. Epigenetics. 2008;793:214. doi: 10.1002/9780470515501.ch13. [DOI] [PubMed] [Google Scholar]
- 11.Hermann A, Schmitt S, Jeltsch A. The human Dnmt2 has residual DNA-(Cytosine-C5) methyltransferase activity. J. Biol. Chem. 2003;278:31717–31721. doi: 10.1074/jbc.M305448200. [DOI] [PubMed] [Google Scholar]
- 12.Goll MG. Methylation of tRNAAsp by the DNA Methyltransferase Homolog Dnmt2. Science (80−.). 2006;311:395–398. doi: 10.1126/science.1120976. [DOI] [PubMed] [Google Scholar]
- 13.Albalat R. Evolution of DNA-methylation machinery: DNA methyltransferases and methyl-DNA binding proteins in the amphioxus Branchiostoma floridae. Dev. Genes Evol. 2008;218:691–701. doi: 10.1007/s00427-008-0247-7. [DOI] [PubMed] [Google Scholar]
- 14.Schaefer M, Lyko F. Solving the Dnmt2 enigma. Chromosoma. 2010;119:35–40. doi: 10.1007/s00412-009-0240-6. [DOI] [PubMed] [Google Scholar]
- 15.Raddatz G, et al. Dnmt2-dependent methylomes lack defined DNA methylation patterns. Proc. Natl. Acad. Sci. 2013;110:8627–8631. doi: 10.1073/pnas.1306723110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Okano M, Xie S, Li E. Cloning and characterization of a family of novel mammalianDNA (cytosine-5) methyltransferases. Nat. Am. Inc. 1998;19:219–220. doi: 10.1038/890. [DOI] [PubMed] [Google Scholar]
- 17.Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat. Rev. Genet. 2008;9:465–76. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 18.Rivenbark AG, et al. Epigenetic reprogramming of cancer cells via targeted DNA methylation. Epigenetics. 2012;7:350–360. doi: 10.4161/epi.19507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Casimir CM, Gates PB, Patient RK, Brockes JP. Evidence for dedifferentiation and metaplasia in amphibian limb regeneration from inheritance of DNA methylation. Development. 1988;104:657 LP–668. doi: 10.1242/dev.104.4.657. [DOI] [PubMed] [Google Scholar]
- 20.Mugatroyd C, Wu Y, Bockmühl Y, Spengler D. The janus face of DNA methylation in aging. Aging (Albany. NY). 2010;2:107–110. doi: 10.18632/aging.100124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zampieri M, et al. Reconfiguration of DNA methylation in aging. Mech. Ageing Dev. 2015;151:60–70. doi: 10.1016/j.mad.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 22.Dowen RH, et al. Widespread dynamic DNA methylation in response to biotic stress. Proc. Natl. Acad. Sci. USA. 2012;109:E2183–91. doi: 10.1073/pnas.1209329109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cortijo S, et al. Mapping the Epigenetic Basis of Complex Traits. Science (80-.). 2014;343:1145 LP–1148. doi: 10.1126/science.1248127. [DOI] [PubMed] [Google Scholar]
- 24.Yi SV, Goodisman MAD. Computational approaches for understanding the evolution of DNA methylation in animals. Epigenetics. 2009;4:551–556. doi: 10.4161/epi.4.8.10345. [DOI] [PubMed] [Google Scholar]
- 25.Bulla I, et al. Notos - a galaxy tool to analyze CpN observed expected ratios for inferring DNA methylation types. BMC Bioinformatics. 2018;19:105. doi: 10.1186/s12859-018-2115-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bird AP. DNA methylation and the frequency of CpG in animal DNA. Nucleic Acids Res. 1980;8:1499–1504. doi: 10.1093/nar/8.7.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fryxell KJ, Moon WJ. CpG mutation rates in the human genome are highly dependent on local GC content. Mol. Biol. Evol. 2005;22:650–658. doi: 10.1093/molbev/msi043. [DOI] [PubMed] [Google Scholar]
- 28.Cooper DN, Krawczak M. Cytosine methylation and the fate of CpG dinucleotides in vertebrate genomes. Hum. Genet. 1989;83:181–188. doi: 10.1007/BF00286715. [DOI] [PubMed] [Google Scholar]
- 29.Jabbari K, Bernardi G. Cytosine methylation and CpG, TpG (CpA) and TpA frequencies. Gene. 2004;333:143–149. doi: 10.1016/j.gene.2004.02.043. [DOI] [PubMed] [Google Scholar]
- 30.Razin A, Cedar H. Distribution of 5-methylcytosine in chromatin. Proc. Natl. Acad. Sci. USA. 1977;74:2725–2728. doi: 10.1073/pnas.74.7.2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suzuki MM, Kerr ARW, De Sousa D, Bird A. CpG methylation is targeted to transcription units in an invertebrate genome. Genome Res. 2007;17:625–31. doi: 10.1101/gr.6163007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gavery MR, Roberts SB. DNA methylation patterns provide insight into epigenetic regulation in the Pacific oyster (Crassostrea gigas) BMC Genomics. 2010;11:483. doi: 10.1186/1471-2164-11-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park J, et al. Comparative analyses of DNA methylation and sequence evolution using Nasonia genomes. Mol. Biol. Evol. 2011;28:3345–3354. doi: 10.1093/molbev/msr168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dixon GB, Bay LK, Matz MV. Bimodal signatures of germline methylation are linked with gene expression plasticity in the coral Acropora millepora. BMC Genomics. 2014;15:1109. doi: 10.1186/1471-2164-15-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Walsh TK, et al. A functional DNA methylation system in the pea aphid, Acyrthosiphon pisum. Insect Mol. Biol. 2010;19:215–228. doi: 10.1111/j.1365-2583.2009.00974.x. [DOI] [PubMed] [Google Scholar]
- 36.Bulla, I. et al. Notos - a Galaxy tool to analyze CpN observed expected ratios for inferring DNA methylation types. bioRxiv 10.1101/180463 (2017). [DOI] [PMC free article] [PubMed]
- 37.Bewick AJ, Vogel KJ, Moore AJ, Schmitz RJ. Evolution of DNA Methylation across Insects. Mol. Biol. Evol. 2016;34:msw264. doi: 10.1093/molbev/msw264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Driscoll T, Gillespie JJ, Nordberg EK, Azad AF, Sobral BW. Bacterial DNA sifted from the Trichoplax adhaerens (Animalia: Placozoa) genome project reveals a putative rickettsial endosymbiont. Genome Biol. Evol. 2013;5:621–645. doi: 10.1093/gbe/evt036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Storb U, Arp B. Methylation patterns of immunoglobulin genes in lymphoid cells: correlation of expression and differentiation with undermethylation. Proc. Natl. Acad. Sci. USA. 1983;80:6642–6646. doi: 10.1073/pnas.80.21.6642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bewick AJ, Schmitz RJ. Gene body DNA methylation in plants. Curr. Opin. Plant Biol. 2017;36:103–110. doi: 10.1016/j.pbi.2016.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.He X-J, Chen T, Zhu J-K. Regulation and function of DNA methylation in plants and animals. Cell Res. 2011;21:442–465. doi: 10.1038/cr.2011.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.D’Onofrio, G., Berná, L. & Alvarez-Valin, F. How fast is the sessile Ciona? Comp. Funct. Genomics2009 (2009). [DOI] [PMC free article] [PubMed]
- 43.Bowman J. Genotype×environment interactions. Genet. Sel. Evol. 1972;4:117. doi: 10.1186/1297-9686-4-1-117. [DOI] [Google Scholar]
- 44.Danchin E, et al. Beyond DNA: integrating inclusive inheritance into an extended theory of evolution. Nat. Rev. Genet. 2011;12:475–486. doi: 10.1038/nrg3028. [DOI] [PubMed] [Google Scholar]
- 45.Lamm, E. In The Standford Encyclopedia of Philosophy (ed. Zalta, E. N.) (Metaphysics Research Lab, Stanford University). at https://plato.stanford.edu/archives/win2014/entries/inheritance-systems (2014).
- 46.Laland K, et al. Does evolutionary theory need a rethink? Nature. 2014;514:161–164. doi: 10.1038/514161a. [DOI] [PubMed] [Google Scholar]
- 47.Koonin EV, Novozhilov AS. Origin and evolution of the genetic code: The universal enigma. IUBMB Life. 2009;61:99–111. doi: 10.1002/iub.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gissot M, Choi SW, Thompson RF, Greally JM, Kim K. Toxoplasma gondii and Cryptosporidium parvum lack detectable DNA cytosine methylation. Eukaryot. Cell. 2008;7:537–540. doi: 10.1128/EC.00448-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu P, et al. The genome of Cryptosporidium hominis. Nature. 2004 doi: 10.1038/nature02977. [DOI] [PubMed] [Google Scholar]
- 50.Hattman S, Kenny C, Berger L, Pratt K. Comparative study of DNA methylation in three unicellular eucaryotes. J. Bacteriol. 1978;135:1156–1157. doi: 10.1128/jb.135.3.1156-1157.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bracht JR. Beyond transcriptional silencing: Is methylcytosine a widely conserved eukaryotic DNA elimination mechanism? BioEssays. 2014 doi: 10.1002/bies.201300123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gardiner-Garden M, Frommer M. CpG Islands in vertebrate genomes. J. Mol. Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 53.Takai D, Jones PA. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc. Natl. Acad. Sci. USA. 2002;99:3740–5. doi: 10.1073/pnas.052410099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nanty L, et al. Comparative methylomics reveals gene-body H3K36me3 in Drosophila predicts DNA methylation and CpG landscapes in other invertebrates. Genome Res. 2011;21:1841–1850. doi: 10.1101/gr.121640.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lyko F, et al. Mammalian (cytosine-5) methyltransferases cause genomic DNA methylation and lethality in Drosophila. Nat. Genet. 1999;23:363–366. doi: 10.1038/15551. [DOI] [PubMed] [Google Scholar]
- 56.Zemach, A., Mcdaniel, I., Silva, P. & Zilberman, D. Genome-Wide Evolutionary Analysis of Eukaryotic DNA Methylation. Sci. (New York, NY)11928, science.1186366v1 (2010). [DOI] [PubMed]
- 57.Dixon GB, Bay LK, Matz MV. Evolutionary Consequences of DNA Methylation in a Basal Metazoan. Mol. Biol. Evol. 2016;33:2285–2293. doi: 10.1093/molbev/msw100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rondon R, et al. Effects of a parental exposure to diuron on Pacific oyster spat methylome. Environ. Epigenetics. 2017;3:1–13. doi: 10.1093/eep/dvx004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lieberman-Aiden E, et al. Comprehensive Mapping of Long-Range Interactions Reveals Folding Principles of the Human Genome. Science (80-.). 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fortin J-P, Hansen KD. Reconstructing A/B compartments as revealed by Hi-C using long-range correlations in epigenetic data. Genome Biol. 2015;16:180. doi: 10.1186/s13059-015-0741-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sexton T, et al. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell. 2012;148:458–472. doi: 10.1016/j.cell.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 62.Cattoni, D. I. I. et al. Single-cell absolute contact probability detection reveals that chromosomes are organized by multiple, low-frequency yet specific interactions. Doi.Org 159814 10.1101/159814 (2017). [DOI] [PMC free article] [PubMed]
- 63.Suzuki MM, et al. Identical sets of methylated and nonmethylated genes in Ciona intestinalis sperm and muscle cells. Epigenetics Chromatin. 2013;6:38. doi: 10.1186/1756-8935-6-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Small KS, Brudno M, Hill MM, Sidow A. Extreme genomic variation in a natural population. Proc. Natl. Acad. Sci. USA. 2007;104:5698–703. doi: 10.1073/pnas.0700890104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kourakis, M. J. & Smith, W. C. An organismal perspective on C. intestinalis development, origins and diversification. Elife317 (2015). [DOI] [PMC free article] [PubMed]
- 66.Johnson DS, Davidson B, Brown CD, Smith WC, Sidow A. Noncoding regulatory sequences of Ciona exhibit strong correspondence between evolutionary constraint and functional importance. Genome Res. 2004;14:2448–2456. doi: 10.1101/gr.2964504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Byrd J, Lambert CC. Mechanism of the block to hybridization and selfing between the sympatric ascidiansCiona intestinalis andCiona savignyi. Mol. Reprod. Dev. 2000;55:109–116. doi: 10.1002/(SICI)1098-2795(200001)55:1<109::AID-MRD15>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 68.Veeman, M. T., Chiba, S. & Smith, W. C. In Vertebrate Embryogenesis: Embryological, Cellular, and Genetic Methods (ed. Pelegri, F. J.) 401–422 (Humana Press,). 10.1007/978-1-61779-210-6_15 (2011).
- 69.Huang X, et al. Rapid response to changing environments during biological invasions: DNA methylation perspectives. Mol. Ecol. 2017;12:3218–3221. doi: 10.1111/mec.14382. [DOI] [PubMed] [Google Scholar]
- 70.Boguski MS, Tolstoshev TMJLCM. dbEST-database for ‘expressed sequence tags’. Nat. Genet. 1993;4:332–333. doi: 10.1038/ng0893-332. [DOI] [PubMed] [Google Scholar]
- 71.Lee B, Shin G. CleanEST: A database of cleansed EST libraries. Nucleic Acids Res. 2009;37:686–689. doi: 10.1093/nar/gkn648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Matsuo K, Clay O, Takahashi T, Silke J, Schaffner W. Evidence for erosion of mouse CpG islands during mammalian evolution. Somat. Cell Mol. Genet. 1993;19:543–555. doi: 10.1007/BF01233381. [DOI] [PubMed] [Google Scholar]
- 73.Conesa A, et al. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 74.Dobin A, et al. Bioinformatics. 2013. STAR: Ultrafast universal RNA-seq aligner; pp. 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.R Development Core Team. R: A Language and Environment for Statistical Computing. At http://www.r-project.org (2008).
- 77.Letunic I, Bork P. Interactive tree of life (iTOL)v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lyko, F. et al. The honey bee epigenomes: Differential methylation of brain DNA in queens and workers. PLoS Biol. 8 (2010). [DOI] [PMC free article] [PubMed]
- 79.Fneich S, et al. 5-methyl-cytosine and 5-hydroxy-methyl-cytosine in the genome of Biomphalaria glabrata, a snail intermediate host of Schistosoma mansoni. Parasit. Vectors. 2013;6:167. doi: 10.1186/1756-3305-6-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wurm Y, et al. The genome of the fire ant Solenopsis invicta. Proc Natl Acad Sci USA. 2011;108:5679–5684. doi: 10.1073/pnas.1009690108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Simola DF, et al. Social insect genomes exhibit dramatic evolution in gene composition and regulation while preserving regulatory features linked to sociality. Genome Res. 2013;23:1235–1247. doi: 10.1101/gr.155408.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang, X. et al. Function and Evolution of DNA Methylation in Nasonia vitripennis. PLoS Genet. 9 (2013). [DOI] [PMC free article] [PubMed]
- 83.Robinson, K. L., Tohidi-Esfahani, D., Lo, N., Simpson, S. J. & Sword, G. A. Evidence for widespread genomic methylation in the migratory locust, Locusta migratoria (orthoptera: Acrididae). PLoS One6 (2011). [DOI] [PMC free article] [PubMed]
- 84.Xiang H, et al. Single base-resolution methylome of the silkworm reveals a sparse epigenomic map. Nat. Biotechnol. 2010;28:516–U181. doi: 10.1038/nbt.1626. [DOI] [PubMed] [Google Scholar]
- 85.Cunningham CB, et al. The Genome and Methylome of a Beetle with Complex Social Behavior, Nicrophorus vespilloides (Coleoptera: Silphidae) Genome Biol. Evol. 2015;7:3383–96. doi: 10.1093/gbe/evv194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Simmen MW, et al. Nonmethylated transposable elements and methylated genes in a chordate genome. Science. 1999;283:1164–1167. doi: 10.1126/science.283.5405.1164. [DOI] [PubMed] [Google Scholar]
- 87.Chen F-C, Chuang T-J, Lin H-Y, Hsu M-K. The evolution of the coding exome of the Arabidopsis species - the influences of DNA methylation, relative exon position, and exon length. BMC Evol. Biol. 2014;14:145. doi: 10.1186/1471-2148-14-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.