Abstract
During late gestation, the fetal heart primarily relies on glucose and lactate to support rapid growth and development. Although numerous studies describe changes in heart metabolism to utilize fatty acids preferentially a few weeks after birth, little is known about metabolic changes of the heart within the first day following birth. Therefore, we used the ovine model of pregnancy to investigate metabolic differences between the near-term fetal and the newborn heart. Heart tissue was collected for metabolomic, lipidomic, and transcriptomic approaches from the left and right ventricles and intraventricular septum in 7 fetuses at gestational day 142 and 7 newborn lambs on the day of birth. Significant metabolites and lipids were identified using a Student’s t-test, whereas differentially expressed genes were identified using a moderated t-test with empirical Bayes method [false discovery rate (FDR)-corrected P < 0.10]. Single-sample gene set enrichment analysis (ssGSEA) was used to identify pathways enriched on a transcriptomic level (FDR-corrected P < 0.05), whereas overrepresentation enrichment analysis was used to identify pathways enriched on a metabolomic level (P < 0.05). We observed greater abundance of metabolites involved in butanoate and propanoate metabolism, and glycolysis in the term fetal heart and differential expression in these pathways were confirmed with ssGSEA. Immediately following birth, newborn hearts displayed enrichment in purine, fatty acid, and glycerophospholipid metabolic pathways as well as oxidative phosphorylation with significant alterations in both lipids and metabolites to support transcriptomic findings. A better understanding of metabolic alterations that occur in the heart following birth may improve treatment of neonates at risk for heart failure.
Keywords: birth, fetus, heart, metabolomics, newborn
INTRODUCTION
The last thirty days of gestation mark a time of rapid fetal cardiac growth and remodeling, as cardiomyocytes transition from a proliferative state to terminal maturation (29). To supply energy for this rapid growth, the fetal heart relies primarily on glucose and lactate supplied by the mother and placenta. However, mRNA transcripts for fatty acid oxidation increase in the fetal heart throughout gestation (27), resulting in primary utilization of fatty acids for energy in the postnatal heart, a switch that persists in the healthy adult (35, 36). The use of fatty acids as an energy source by the neonatal heart is an essential adaptation, as this metabolic process is energetically favorable, yielding ~2.4 times more ATP per gram of palmitic acid than per gram of glucose (31).
Previous studies have suggested that within the first day of life, the newborn heart retains the same glycolytic capacity as that of the fetal heart, with the switch to fatty acid utilization occurring 1–2 wk following birth (3, 5, 35, 36). Yet, a previous study in our laboratory revealed that chronic maternal stress leads to fetal stillbirth in the peripartal period, possibly because of alterations in fetal cardiac metabolism, showing the importance of cardiac metabolic shifts during labor (50). To study alterations in cardiac metabolism that occur at the time of birth further, we utilized a sheep model of pregnancy to investigate differences in metabolism present in newborn hearts as compared with near-term fetal hearts (gestational day 142; 3–6 days before term) of healthy, unstressed ewes. We hypothesized that normal labor results in a shift of cardiac metabolism that can be modeled by the patterns of change in gene expression of metabolic pathways, along with corresponding changes in cardiac metabolites and lipids. To test this hypothesis, we utilized a multiomics approach integrating transcriptomic, metabolomic, and lipidomic platforms to study global alterations in cardiac metabolism.
MATERIALS AND METHODS
Animals.
Dorset ewes and their lambs or fetuses were studied. All animal use was approved by the University of Florida Institutional Animal Care and Use Committee. To control for metabolic variation in the blood, obese or very lean ewes (judged on the basis of body condition score, the ovine equivalent of body mass index) were excluded from the study. All ewes and their newborns were housed in a temperature- and light-controlled animal facility before use and fed a ruminant diet based on the National Research Council calculations of energy requirements in the late-gestation ewe. Feed and animal husbandry were provided each morning. Ewes were assigned to one of two groups: 1) a fetal group (n = 7, 3 singleton and 2 multiple gestation pregnancies) or 2) a newborn group (n = 7, 2 singleton and 2 multiple gestation pregnancies). Newborn samples were collected on day of birth (within 24 h), whereas fetal samples were collected on gestational day 142. Tissues were collected identically for both groups. Ewes and their fetuses or newborn lambs were euthanized using intravenous administration of Euthasol (pentobarbital sodium and phenytoin sodium; Virbac AH, Inc). Immediately following euthanasia, the heart was rapidly excised, and ~100 mg of tissue were dissected from the right ventricular (RV) free wall, left ventricular (LV) free wall, and intraventricular septum and immediately frozen in liquid nitrogen. Samples were stored at −80°C until analysis.
Nuclear magnetic resonance metabolomics analysis.
High-resolution magic angle spinning (HR-MAS) proton nuclear magnetic resonance (1H-NMR) was used to determine metabolites present in RV and LV free walls and the intraventricular septum. Metabolites identified with this technique were not found to be significantly altered between different areas of the heart and were therefore treated as technical replicates (Supplemental Table S1; Supplemental Data for this article can be found on the AJP-Endocrinology and Metabolism website). Samples were prepared as previously described (6). Briefly, 30 μl of deuterium oxide (D2O) were added to 17–56 mg (mean 34.8 mg) of intact heart tissue before being placed into a 4-mm HR-MAS rotor (Bruker Biospin, Billerica, MA) and spun at 6 kHz with a spectral width of 10 parts per million. Data were acquired on an Avance III 600 MHz Bruker NMR spectrometer equipped with a 4-mm HR-MAS probe at the University of Florida Advanced Magnetic Resonance Imaging and Spectroscopy Facility using a one-dimensional nuclear Overhauser effect pulse sequence with presaturation of water resonance.
Two-dimensional 1H-NMR was used to confirm metabolites identified in HR-MAS. One hundred milligrams of pooled tissue samples were created for newborn (n = 4) and fetal (n = 3) specimens. Three 3.5-mm glass and 325 mg of 1.4-mm steel beads were added to 1.5-ml tubes containing tissue (BioSpec Products, Inc., Bartlesville, OK). One milliliter of 80:20 methanol/H2O (HPLC-grade) were added to each sample before homogenization 3 times at 18,000 revolutions/min for 30 s. Samples were incubated on dry ice for 5 min between homogenization rounds. Samples were then spun at 10,000 rcf for 10 min at 4°C. The supernatant was transferred to a new microcentrifuge tube and concentrated overnight using a CentriVap Benchtop Vacuum Concentrator (Labconco, Kanas City, MO). The resulting dried extracts were frozen at −80°C until further analysis, when they were thawed and prepared for NMR metabolomic analysis as previously described (60). Briefly, 600 μl of sodium phosphate buffer at pH 7.0 with 0.33 mM 4,4-dimethyl-4-silapentane-1-sulfonic acid (DSS) was added to each sample and vortexed until pellets dissolved. The samples were then centrifuged at 14,000 rcf for 15 min at 4°C, and 590 μl of the resulting supernatant was transferred into a 5-mm NMR tube (Bruker Biospin). Data were acquired on an Avance III HD 600 MHz Bruker NMR spectrometer at the University of Georgia using two-dimensional 1H-13C heteronuclear single quantum correlation and heteronuclear single quantum correlation-spectroscopy pulse sequences. A total of 25 metabolites were identified using Bruker AssureNMR software (Bruker Biospin) with BBiorefcode metabolite database and COLMARm (10). The metabolites were assigned a confidence level ranging from 1 to 5 as previously described (60). The metabolite confidence levels are reported in Supplemental Table S2. The spectra were processed using Bruker Topspin 3.6 software and in-house MATLAB scripts. All metabolites with relative means and standard error (SE), P values [raw and false discovery rate (FDR)-corrected] and log2 fold changes (newborn/fetal) following birth are listed in Supplemental Table S3. All raw and processed data are available on the Metabolomics Workbench (http://www.metabolomicsworkbench.org/), along with detailed experimental NMR and analysis methods.
Lipidomic analysis.
Homogenized RV tissue samples (100 mg from 7 fetuses and 7 newborns) were used for lipid extraction via the Folch method (24). Briefly, 2 μl of internal standard mixture was added to each sample before adding 1.2 ml of 2:1 chloroform/methanol solution (HPLC-grade). Internal and injection standard mixtures are listed in Supplemental Table S4. Samples were incubated at 4°C for 20 min with occasional vortexing. Following incubation, 200 μl of water (HPLC-grade) were added, and samples were incubated at 4°C for 10 min. Samples were centrifuged at 2,000 g for 5 min at 4°C, and the resulting chloroform layer was removed and transferred into a clean tube. The extraction process was repeated with 400 μl 2:1 chloroform/methanol (HPLC-grade), and the resulting chloroform layers were combined. Samples were dried under a nitrogen stream at 30°C and stored at −80°C until reconstitution. Samples were reconstituted with 200 μl of isopropanol and 2 μl injection standard mixture (Supplemental Table S4). Ammonium acetate and all analytical grade solvents (formic acid, chloroform, and methanol) were purchased from Fisher Scientific (Waltham, MA). All mobile phase solvents were Fisher Optima LC/MS-grade (acetonitrile, isopropanol, and water). Lipid analysis was performed on a Thermo Q-Exactive Orbitrap with Dionex Ultimate 3000 UHPLC and autosampler. The mass spectrometer was operated in the positive and negative ionization mode, using a heated electrospray ionization source. Spectra were collected from mass-to-charge ratio (m/z) 200–2200 at with a mass resolution setting of 35,000 (defined at m/z 200), and tandem mass spectra were collected using data-dependent scanning (top 10) and all-ion fragmentation. Feature finding and alignment was performed with MZmine 2.26 (46) as previously described (32). LipidMatch was used to identify lipids (33), which were normalized to their representative internal standard using an in-house R script, LipidMatch Quant, which can be accessed at (http://secim.ufl.edu/secim-tools/). A total of 292 lipids identified in positive and negative ion modes were used for statistical analyses. Supplemental Table S5 shows m/z, retention time, relative means following normalization (SE), raw and FDR-corrected P values, log2 fold change (newborn/fetal), LipidMatch results, lipid class, and adducts for all annotated lipids. All lipid classes with the number of lipids in each class, relative class means and standard error (SE), P values (raw and FDR-corrected) and log2 fold changes (newborn/fetal) following birth are listed in Supplemental Table S6. All raw and processed data are available on the Metabolomics Workbench (http://www.metabolomicsworkbench.org/), along with detailed experimental methods.
Transcriptomic analysis.
LV cardiac tissue samples (~200 mg from 7 fetuses and 7 newborns) were homogenized and mRNA was extracted with Trizol and purified with on-column DNase digestion (Qiagen RNeasy Plus Kits, Germantown, MD) as previously described (66). RNA integrity numbers ranged from 7.8 to 9.4. mRNA (200 ng) was labeled with Cy5 using Agilent QuickAmp Labeling kit (5190–0442, Santa Clara, CA) to generate cRNA. The labeled cRNA was analyzed on a NanoDrop-1000 spectrophotometer (ThermoFisher, Wilmington, DE), and the specific activities and yields ranged from 13.10 to 17.46 pmol Cy3/μg and from 4.83 to 6.64 μg. Microarray hybridization, washing, and scanning were performed as previously described (49). The Agilent-019921 Sheep Gene Expression Microarray 8x 15k, G4813A (GPL14112) platform was used for analysis of cRNA. One fetal specimen was removed before statistical analysis because of poor array quality as determined by spike in coefficient of variation. The microarray data were prepared for statistical analysis as previously described (48). A total of 4,991 unique genes were annotated and included for statistical analysis. All significant genes (FDR-corrected P < 0.10), raw and FDR-corrected P values, and log2 fold changes (newborn/fetal) are displayed in Supplemental Table S7. Microarray data have been submitted to the National Center for Biotechnology Information’s Gene Expression Omnibus (22) and can be found under accession no. GSE117343.
Statistics.
Metabolomic and lipidomic data were normalized using probabilistic quotient normalizationand all data (transcriptomic, metabolomic, and lipidomic) were range-scaled before multivariate statistical analysis (19, 23). Multivariate analyses of processed spectra were performed using in-house MATLAB scripts (https://github.com/artedison/Edison_Lab_Shared_Metabolomics_UGA) and Metaboanalyst 4.0, an online webserver for metabolomic data analysis (14). Univariate statistics were performed on metabolite and mean lipid classes after probabilistic quotient normalization. Mean lipid classes were defined as the average of all lipids identified in each class. All P values were subject to FDR using the Benjamini-Hochberg method (9). A Student’s t-test with an FDR correction was used to determine significant metabolites and lipid classes (FDR-corrected, P < 0.10). A one-way ANOVA was used to determine the metabolites that differed between areas of the heart for HR-MAS metabolomics data (raw P < 0.05). A Tukey-Kramer post hoc analysis was used to determine significant metabolite variations (raw P < 0.05) between different regions of the cardiac walls for metabolites identified by HR-MAS. Differentially expressed genes were identified using a moderated t-test with empirical Bayes method (54). Genes were considered differentially expressed with an FDR-corrected P < 0.10. Partial least squares discriminant analysis (PLS-DA) and heatmap correlations were conducted in Metaboanalyst 4.0 to identify metabolites and lipids that differed between fetal and newborn cardiac tissue (14). A 10-fold cross validation was used for all PLS-DA models generated. A heatmap of the top 50 significant lipids was generated using a Pearson correlation coefficients of autoscaled features, and clustering was determined using the Ward algorithm (62).
Pathway analysis for metabolomic, lipidomic, and transcriptomic data.
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways that were enriched in fetal or newborn tissues using significantly altered genes were determined using WebGestalt overrepresentation enrichment analysis (ORA) (61). Significance was defined as pathways with an FDR-corrected P < 0.05. Metscape 3.0 was utilized to determine pathways involved in correlations between significant metabolites and lipid classes (30). To identify pathways that showed enrichment for significantly altered compounds (metabolites and lipid classes) further, we utilized an ORA based on a hypergeometric test (67). Significant pathways were defined as pathways with raw P < 0.05. Finally, we applied single-sample gene set enrichment analyses (ssGSEA) (2, 56) to determine if pathways identified by metabolomics and lipidomics, as well as pathways important in the adult heart energetics (oxidative phosphorylation, fatty acid metabolism, TCA cycle) and lipid synthesis/metabolism (ether lipid metabolism and peroxisome), were also enriched in transcriptomic data. Significant pathways were defined with FDR-corrected P < 0.05. The Molecular Signatures Database (MSigDB) hallmark gene set pathways were used for oxidative phosphorylation, fatty acid metabolism, and peroxisome pathways, whereas KEGG was used for all remaining pathways (34). All pathways from ssGSEA are displayed in Supplemental Table S8, along with raw P values, FDR-corrected P values, mean enrichment scores for each group (fetal and newborn), and number of significant genes (FDR-corrected P < 0.10) in each group. It should be noted that our transcriptomic data set did not include transcripts for genes involved in taurine and hypotaurine metabolism (P < 0.05 following ORA), and therefore, this pathway is not shown in Supplemental Table S8.
RESULTS
Metabolites in intact cardiac tissue are altered following birth.
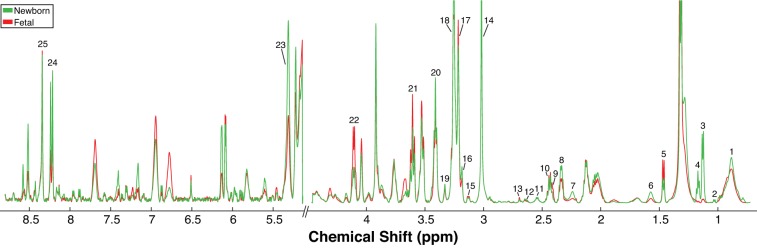
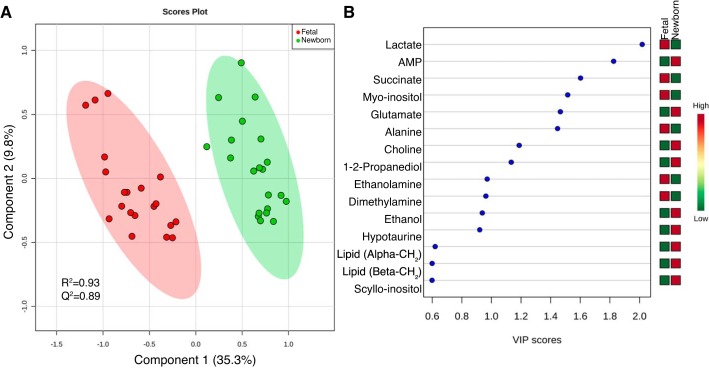
HR-MAS 1H-NMR revealed changes in the metabolite profile of cardiac tissue in the newborn heart compared with the term fetal heart (Fig. 1; exact spectral areas and confidence values for each metabolite are listed in Supplemental Table S2). A PLS-DA scores plot (R2 = 0.93; Q2 = 0.89) revealed clear separation of fetal and newborn cardiac tissue in principal component 1 based on metabolites (Fig. 2A). A variable importance projection (VIP) plot was used to identify the first 15 metabolites that contribute to the PLS-DA in component one. The VIP plot revealed that lactate, succinate, myo-inositol, and alanine were elevated in fetal cardiac samples with VIP scores greater than one. AMP, glutamate, choline, and 1,2-propanediol were elevated in newborn samples compared with fetal cardiac samples with VIP scores greater than one (Fig. 2B). Comparison of peaks by t-test also indicated higher levels of 1,2-propanediol, AMP, and glutamate in the newborn heart, as well as hypotaurine, choline, scyllo-inositol, and some lipid classes. Lactate, succinate, myo-inositol, alanine, dimethylamine, and ethanolamine were all higher in fetal heart (Table 1; see Supplemental Table S2 for all annotated metabolites).
Fig. 1.
High resolution magic angle spinning (HR-MAS) overlay of mean fetal (n = 20 from 7 animals, red) and mean newborn (n = 21 from 7 animals, green) spectra from ovine cardiac tissue. Metabolite areas that were used for quantification are highlighted with the following numbers: 1) lipid (−CH3), 2) valine, 3) 1,2-propanediol, 4) ethanol, 5) alanine, 6) lipid (βCH2), 7) lipid (αCH2), 8) glutamate, 9) succinate, 10) glutamine, 11) glutathione, 12) hypotaurine, 13) dimethylamine, 14) creatine, 15) ethanolamine, 16) choline, 17) glycerophosphocholine, 18) phosphocholinebetaine, 19) scyllo-inositol, 20) taurine, 21) myo-inositol, 22) lactate, 23) lipid (-CH=CH-), 24) adenosine 5′-monophosphate, 25) inosine.
Fig. 2.
Partial least squares discriminant analysis (PLS-DA) reveals separation of HR-MAS cardiac specimens from fetal sheep near term and newborn sheep immediately following birth. PLS-DA scores plot reveals separation of fetal (n = 20 from 7 animals, red) and newborn (n = 21 from 7 animals, green) cardiac tissue in the first principal component (A). Variable importance of projection (VIP) plot for the first 15 metabolites that contribute to separation in PLS-DA component 1 (B). A higher VIP score indicates a greater contribution of that metabolite to the separation of the groups.
Table 1.
Significantly altered metabolites in the newborn compared with fetal heart as identified by HR-MAS
| Relative Mean (SE) |
|||||
|---|---|---|---|---|---|
| Metabolite | Fetal | Newborn | P value | FDR-corrected P value | Log2(FC), N/F |
| 1,2-Propanediol | 0.163 (0.007) | 2.334 (0.336) | 1.08E−06 | 3.86E−06 | 3.84 |
| Ethanol | 0.087 (0.007) | 1.018 (0.220) | 5.27E−04 | 1.10E−03 | 3.55 |
| Lipid (β-CH2) | 0.328 (0.034) | 0.824 (0.171) | 0.01 | 0.03 | 1.33 |
| AMP | 0.067 (0.004) | 0.137 (0.005) | 1.09E−12 | 1.36E−11 | 1.02 |
| Lipid (α-CH2) | 0.454 (0.043) | 0.884 (0.146) | 0.02 | 0.03 | 0.96 |
| Lipid (-CH = CH-) | 0.358 (0.035) | 0.660 (0.124) | 0.04 | 0.07 | 0.88 |
| Glutamate | 1.309 (0.055) | 1.951 (0.073) | 1.94E−07 | 8.09E−07 | 0.58 |
| Hypotaurine | 0.143 (0.007) | 0.205 (0.010) | 5.47E−05 | 1.71E−04 | 0.52 |
| Choline | 0.718 (0.034) | 0.989 (0.043) | 6.82E−05 | 1.89E−04 | 0.46 |
| Scyllo-inositol | 0.434 (0.016) | 0.513 (0.024) | 0.02 | 0.03 | 0.24 |
| Lactate | 3.494 (0.131) | 1.692 (0.068) | 1.12E−13 | 2.79E−12 | −1.05 |
| Succinate | 0.558 (0.036) | 0.278 (0.015) | 6.88E−08 | 3.44E−07 | −1.00 |
| Alanine | 1.357 (0.051) | 0.853 (0.026) | 7.47E−10 | 6.22E−09 | −0.67 |
| Myo-inositol | 4.409 (0.166) | 2.971 (0.083) | 1.65E−08 | 1.03E−07 | −0.57 |
| Dimethylamine | 0.117 (0.007) | 0.079 (0.004) | 1.74E−04 | 4.35E−04 | −0.56 |
| Ethanolamine | 0.290 (0.011) | 0.226 (0.009) | 2.27E−04 | 5.16E−04 | −0.35 |
AMP, adenosine 5′-monophosphate; FDR, false discovery rate; HR-MAS, high-resolution magic angle spinning; FC, fold change; N/F, newborn/fetal.
Lipid profile of cardiac tissue is altered following birth.
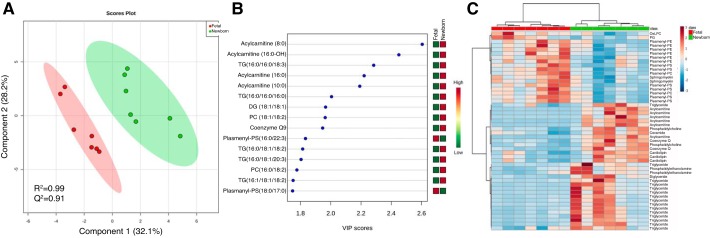
Ultra-high-performance liquid chromatography-mass spectrometry (UHPLC-MS) identified 292 lipids across 23 different lipid classes in fetal and newborn RV tissue (Supplemental Table S5). A PLS-DA scores plot (R2 = 0.99; Q2 = 0.91) of lipids reveals separation of fetal and newborn cardiac tissue in principal component 1 (Fig. 3A). A VIP plot of the first 15 lipids that contribute to this separation in the first component revealed that triglycerides (TGs), phosphatidylcholines (PCs), one diglyceride species (DG 18:1/18:1), coenzyme Q (CoQ)9, and acylcarnitines were more abundant in newborn cardiac tissue, whereas ether-linked phosphatidylserines [(PSs) plasmanyl- and plasmenyl-PSs] were more abundant in fetal cardiac tissue (Fig. 3B). Finally, a correlation heatmap was utilized to determine how lipid classes varied between fetal and newborn cardiac tissue (Fig. 3C). There were 8 lipid classes more highly elevated in newborn cardiac tissue as compared with the fetus: TGs were the main lipid class elevated, but acylcarnitines, PCs, CoQ9 and CoQ10, cardiolipins (CLs), and phosphatidylethanolamines (PEs) had more than one lipid species elevated in newborn cardiac tissue as compared with fetal cardiac tissue (Fig. 3C). There were 7 lipid classes more highly enriched in fetal cardiac tissue as compared with newborn tissue. These included sphingomyelin (SM), PC, PE, and PS species (Fig. 3C). Relative means of each lipid class following normalization were used for a Student’s t-test to reveal nine lipid classes that were significantly altered in neonatal compared with fetal cardiac tissue (Table 2).
Fig. 3.
Partial least squares discriminant analysis (PLS-DA) reveals separation of lipidomic cardiac specimens from fetal sheep near term and newborn sheep immediately following birth PLS-DA scores plot reveals separation of fetal (n = 7, red) and newborn (n = 7, green) cardiac tissue in the first principal component (A). Variable importance of projection (VIP) plot for the first 15 lipids that contribute to separation in PLS-DA component 1 (B). Heatmap of top 50 significant features (P < 0.05) reveals clustering of fetal and newborn samples based on lipid class (C). DG, diglyceride; PC, phosphatidylcholine; PS, phosphatidylserine; TG, triglyceride.
Table 2.
Significantly altered lipid classes in the newborn compared with fetal heart as identified by UHPLC-MS
| Relative Mean (SE) |
||||||
|---|---|---|---|---|---|---|
| Lipid Class | No. Lipids/Class | Fetal | Newborn | P value | FDR-corrected P value | Log2(FC), N/F |
| AcCa, long-chain | 4 | 0.003 (0.000) | 0.065 (0.008) | 1.13E−03 | 0.02 | 4.42 |
| AcCa, medium-chain | 3 | 0.009 (0.001) | 0.125 (0.017) | 2.13E−03 | 0.02 | 3.87 |
| TG | 45 | 0.027 (0.001) | 0.115 (0.018) | 0.02 | 0.06 | 2.10 |
| DG | 3 | 0.034 (0.003) | 0.112 (0.012) | 4.65E−03 | 0.02 | 1.73 |
| CoQ | 2 | 0.053 (0.004) | 0.142 (0.012) | 1.68E−03 | 0.02 | 1.42 |
| Ether-PC | 28 | 0.512 (0.020) | 0.356 (0.011) | 3.02E−03 | 0.02 | −0.52 |
| SM | 21 | 2.281 (0.102) | 1.548 (0.090) | 0.01 | 0.04 | −0.56 |
| OxLPC | 5 | 0.073 (0.004) | 0.044 (0.005) | 0.03 | 0.07 | −0.74 |
| Ether-PS | 12 | 0.042 (0.002) | 0.024 (0.002) | 5.67E−03 | 0.02 | −0.81 |
AcCa, acylcarnitine; CoQ, coenzyme Q; DG, diglyeride; FDR, false discovery rate; OxLPC, oxidized lysophosphatidylcholine; PC, phosphatidylcholine; PS, phosphatidylserine; SM, sphingomyelin; TG, triglyceride.
Transcriptomic profile in cardiac tissue is altered following birth.
A principal component analysis (PCA) scores plot revealed separation of genes from fetal cardiac tissue and newborn cardiac tissue following birth (data not shown). In 142-day fetal heart, 893 genes were significantly elevated compared with the newborn, whereas 940 genes were significantly elevated in the newborn heart as compared with the fetus. ORA of 893 genes significantly elevated in fetal compared with newborn heart revealed differential regulation of pathways associated with branched chain amino acid (BCAA) degradation, β-alanine, butanoate, and tryptophan metabolism, as well as fatty acid degradation (Table 3). In addition, genes in pathways related to cardiomyopathy, including dilated and arrhythmogenic RV cardiomyopathy were also highly expressed in the fetal heart, consistent with cardiac maturation and remodeling during this period. Whereas ORA revealed greater expression of genes in 31 pathways indicative of growth and metabolic development in the fetal heart, only 6 significant pathways were identified as more highly expressed in the newborn heart (Table 3). These six pathways included pathways involved in protein synthesis, including spliceosome, ribosome, protein processing in the endoplasmic reticulum, and glycosylphosphatidylinositol-anchor biosynthesis, a process that occurs in the endoplasmic reticulum.
Table 3.
KEGG pathways modeled as differently regulated in fetal and newborn hearts
| Description | Relative Abundance | No. Genes in Geneset/Total No. of Genes | P value | FDR-Corrected P Value |
|---|---|---|---|---|
| Proteasome | N > F | 21/44 | 1.35E−14 | 4.10E−12 |
| Protein processing in endoplasmic reticulum | N > F | 34/166 | 3.06E−10 | 4.64E−08 |
| Spliceosome | N > F | 22/134 | 2.05E−05 | 2.07E−03 |
| Ribosome | N > F | 22/154 | 1.77E−04 | 0.01 |
| Glycosylphosphatidylinositol-anchor biosynthesis | N > F | 7/25 | 5.92E−04 | 0.04 |
| p53 signaling pathway | N > F | 12/69 | 9.28E−04 | 0.05 |
| Valine, leucine, and isoleucine degradation | F > N | 17/48 | 1.18E−10 | 3.57E−08 |
| Adherens junction | F > N | 16/74 | 1.00E−06 | 1.52E−04 |
| Pathways in cancer | F > N | 43/397 | 3.62E−06 | 3.65E−04 |
| Proteoglycans in cancer | F > N | 27/205 | 8.05E−06 | 6.10E−04 |
| Endocrine resistance | F > N | 17/98 | 1.12E−05 | 6.79E−04 |
| Transcriptional misregulation in cancer | F > N | 24/180 | 2.10E−05 | 1.06E−03 |
| Arrhythmogenic right ventricular cardiomyopathy | F > N | 14/74 | 2.44E−05 | 1.06E−03 |
| Fatty acid degradation | F > N | 10/44 | 7.08E−05 | 2.68E−03 |
| HIF-1 signaling pathway | F > N | 16/103 | 8.23E−05 | 2.77E−03 |
| Longevity regulating pathway | F > N | 15/94 | 1.00E−04 | 2.80E−03 |
| Longevity regulating pathway, multiple species | F > N | 12/64 | 1.02E−04 | 2.80E−03 |
| β-alanine metabolism | F > N | 8/31 | 1.46E−04 | 3.69E−03 |
| FoxO signaling pathway | F > N | 18/134 | 2.07E−04 | 4.81E−03 |
| ECM-receptor interaction | F > N | 13/82 | 3.09E−04 | 6.68E−03 |
| Osteoclast differentiation | F > N | 17/132 | 5.10E−04 | 0.01 |
| AGE-RAGE signaling pathway in diabetic complications | F > N | 14/101 | 7.52E−04 | 0.01 |
| Dilated cardiomyopathy | F > N | 13/90 | 7.77E−04 | 0.01 |
| Focal adhesion | F > N | 22/203 | 9.33E−04 | 0.02 |
| Insulin signaling pathway | F > N | 17/140 | 1.01E−03 | 0.02 |
| Leukocyte transendothelial migration | F > N | 15/118 | 1.23E−03 | 0.02 |
| Thyroid hormone signaling pathway | F > N | 15/118 | 1.23E−03 | 0.02 |
| Prion diseases | F > N | 7/35 | 1.96E−03 | 0.03 |
| Prostate cancer | F > N | 12/89 | 2.25E−03 | 0.03 |
| Acute myeloid leukemia | F > N | 9/57 | 2.65E−03 | 0.03 |
| Butanoate metabolism | F > N | 6/28 | 2.84E−03 | 0.03 |
| Notch signaling pathway | F > N | 8/48 | 3.20E−03 | 0.04 |
| Lysine degradation | F > N | 9/59 | 3.37E−03 | 0.04 |
| Breast cancer | F > N | 16/146 | 4.04E−03 | 0.04 |
| Tryptophan metabolism | F > N | 7/40 | 4.33E−03 | 0.05 |
| Insulin resistance | F > N | 13/109 | 4.49E−03 | 0.05 |
| Platelet activation | F > N | 14/122 | 4.61E−03 | 0.05 |
F, fetal; N, newborn; ECM, extracellular matrix; HIF, hypoxia-inducible factor; KEGG, Kyoto Encyclopedia of Genes and Genomes; AGE, advanced glycation end product; RAGE, receptor for advanced glycation end product.
Integration of metabolomic, lipidomic, and transcriptomic data reveals altered cardiac metabolic pathways following birth.
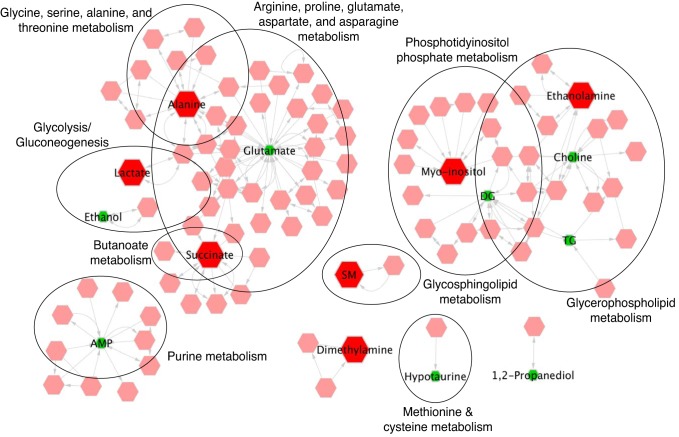
Metscape 3.0 was utilized to visualize pathways involved in significant metabolites and lipid classes (30). Using this method, we determined nine distinct pathway clusters (Fig. 4). Three of these included pathways involved in lipid metabolism: glycerophospholipid, phosphatidylinositol phosphate, and glycosphingolipid metabolism. In addition, pathways involved in amino acid metabolism, including arginine, proline, glutamate, and asparagine, as well as glycine, serine, and alanine and threonine metabolism revealed two clusters that were slightly overlapping. Lastly, we identified metabolites involved in glycolysis, butanoate metabolism, and purine metabolism. ORA with significantly altered metabolites and lipids revealed eight pathways that were significantly enriched (Table 4), including four that overlapped with pathways identified in Metscape (Fig. 4); these included alanine, aspartate, glutamate metabolism, glycolysis, glycerophospholipid, and butanoate metabolism.
Fig. 4.
Metabolite correlation network reveals 9 clusters that correspond to distinct metabolic pathways. Hexagons in red represent metabolites and lipid classes that are significantly upregulated in near-term fetal cardiac tissue whereas green hexagons represent metabolite and lipids that are upregulated in newborn cardiac tissue immediately following birth. All other correlated metabolites that were not identified or significantly altered are represented by pink hexagons. Pathways related to amino acid metabolism, glycolysis, and purine metabolism appear as clusters on the left, whereas pathways related to phospholipid and metabolism appear on the right.
Table 4.
Metabolic pathways significantly enriched in fetal and newborn cardiac tissue following ORA
| Metabolites |
||||
|---|---|---|---|---|
| Pathway | No. Metabolites/Total No. Metabolites | P value | F > N | N > F |
| Alanine, aspartate, and glutamate metabolism | 3/24 | 5.41E−04 | Alanine, Succinate | Glutamate |
| Taurine and hypotaurine metabolism | 2/20 | 8.28E−03 | Alanine | Hypotaurine |
| Glycolysis | 2/31 | 0.02 | Lactate | Ethanol |
| Pyruvate metabolism | 2/32 | 0.02 | Lactate | 1,2-Propanediol |
| Propanoate metabolism | 2/35 | 0.02 | Succinate, Lactate | — |
| Nitrogen metabolism | 2/39 | 0.03 | — | Glutamate, AMP |
| Glycerophospholipid metabolism | 2/39 | 0.03 | Ethanolamine | Choline |
| Butanoate metabolism | 2/40 | 0.03 | Succinate | Glutamate |
ORA, overrepresentation analysis.
To determine if pathways identified in metabolomic and lipidomic enrichment analyses were significantly altered on a transcriptomic level, ssGSEA was utilized. Gene expression was significantly elevated in the fetal heart as compared with the newborn heart in four metabolic pathways, including nitrogen metabolism, butanoate metabolism, glycolysis, and propanoate metabolism (Table 5). Expression of genes that utilize alcohol for production of acetyl CoA, including alcohol dehydrogenase 5 (ADH5) and multiple isoforms of aldehyde dehydrogenase (ALDH7A, ALDH9A1, ALDH2) were higher in fetal cardiac tissue (Supplemental Table S7). In contrast, gene expression was significantly increased in pathways involved in lipid metabolism, including fatty acid metabolism, peroxisome, and glycerophospholipid metabolism in the newborn heart (Table 5). In addition, the enrichment analysis indicated upregulation of oxidative phosphorylation and purine metabolism in the newborn heart.
Table 5.
Pathways significantly enriched in fetal and newborn cardiac following ssGSEA
| Number of Significant Genes (FDR-corrected P < 0.10) |
|||||
|---|---|---|---|---|---|
| Pathway | P value | FDR-corrected P value | Relative Abundance | F > N | N > F |
| Purine metabolism | 2.42E−05 | 4.35E−04 | N > F | 5 | 17 |
| Oxidative phosphorylation | 2.10E−04 | 1.26E−03 | N > F | 23 | 31 |
| Fatty acid metabolism | 6.64E−04 | 2.39E−03 | N > F | 18 | 21 |
| Peroxisome | 8.40E−04 | 2.52E−03 | N > F | 8 | 10 |
| Glycerophospholipid metabolism | 5.74E−03 | 0.01 | N > F | 3 | 8 |
| Nitrogen metabolism | 1.22E−04 | 1.10E−03 | F > N | 2 | 1 |
| Butanoate metabolism | 3.50E−04 | 1.58E−03 | F > N | 9 | 2 |
| Glycolysis | 5.82E−03 | 0.01 | F > N | 6 | 10 |
| Propanoate metabolism | 5.99E−03 | 0.01 | F > N | 8 | 2 |
FDR, false discovery rate; ssGEA, single-sample gene enrichment analysis.
DISCUSSION
Transcriptomic analysis reveals enrichment of BCAA degradation pathways in fetal heart.
Our transcriptomic analysis predicts that BCAA degradation is high in the fetal heart as compared with the newborn. BCAA degradation was the most enriched KEGG pathway in fetal heart tissue (Table 3). Although we were only able to identify valine utilizing HR-MAS, we did not find a significant difference in the cardiac valine concentration between newborn and fetal heart tissue (Table 1). This lack of difference between valine levels in the fetal and newborn heart may be because of the constant supply of fetal valine levels from the mother and placenta, causing valine to remain elevated despite increased breakdown in the fetal heart (16). However, 17 transcripts for genes associated with BCAA degradation were more highly expressed in fetal cardiac tissue compared with newborn (Table 3), whereas 2 transcripts for genes involved in acetyl-CoA synthesis (hydroxyacyl-CoA dehydrogenase; HADH) and acetyl-CoA utilization (3-hydroxy-3-methylglutaryl-CoA synthase 1; HMGCS1) were more highly expressed in newborn cardiac tissue (Supplemental Table S7). Therefore, future studies should aim at identifying metabolic intermediates of this pathway, as activation of BCAA degradation in the fetal heart could lead to increased BCAA breakdown products that stimulate protein synthesis and growth in the developing fetal heart (15).
Transcriptomic and metabolomic analyses reveal enrichment of carbohydrate metabolism in the fetal heart.
Transcripts for genes involved in butanoate and propanoate metabolism were more highly expressed in the fetal compared with the newborn heart (Table 5). These short-chain fatty acid degradation pathways are formed from nondigestible carbohydrates in the gut microbiome in adults (37). In both humans and sheep, propanoate and butanoate metabolism lead to the production of acetyl-CoA for entry into the TCA cycle, whereas in sheep, propionate metabolism has been shown to stimulate gluconeogenesis (20). Succinate, present in both butanoate and propanoate metabolism, was more abundant in fetal cardiac tissue (Table 1). Although TCA cycle metabolism does occur in the human placenta, as evident from placenta mitochondrial studies, it is unknown if fetal succinate is derived from maternal, placental, or fetal metabolism (40, 57). Our transcriptomic data revealed greater expression of succinate-semialdehyde dehydrogenase (ALDH5A1) in fetal cardiac tissue, a mitochondrial gene involved in butanoate metabolism (Supplemental Table S7). ALDH5A1 catalyzes the reaction of succinate semialdehyde to succinate and elevations of both ALDH5A1 transcripts and succinate in fetal cardiac tissue suggests formation of succinate in the fetal heart at term. This could lead to increased pools of succinate that can be utilized by the heart for TCA cycle metabolism and oxidative phosphorylation. In addition, lactate, which can contribute to propanoate production, was more abundant in the fetal heart, whereas glutamate, which can contribute to butanoate production, was more abundant in the newborn heart. However, no direct gene correlations were observed for either of these metabolites in butanoate or propanoate metabolic pathways.
Our analysis indicates a shift in the type of carbohydrates used by the heart at the time of birth. Glycolysis was identified by ORA of metabolites and lipids (Table 4). However, lactate was significantly elevated in fetal tissue, and ethanol was significantly elevated in newborn tissue (Table 1). This elevated lactate in the fetal heart can enter propanoate metabolism, creating substrates that can enter the TCA cycle via acetyl-CoA or succinate, contributing to carbohydrate metabolism in the term fetal heart. In the fetal heart, transcripts for genes identified in the glycolysis pathway were significantly elevated, including alcohol dehydrogenase 5 (ADH5) and multiple isoforms of aldehyde dehydrogenase (ALDH7A, ALDH9A1, ALDH2), all genes involved in alcohol degradation (Supplemental Table S7). Higher expression of transcripts for genes related to alcohol degradation in the fetal heart are consistent with the decreased cardiac alcohol levels observed in the fetal heart as compared with the newborn. In the newborn heart, transcripts for genes related to glycolysis of sugars, including pyruvate kinase (PKM2) and triosephosphate isomerase (TPI1) were more highly expressed compared with the fetal heart (Supplemental Table S7). In addition to glycolysis, TPI1 is also involved in TG formation, a class of lipids significantly elevated in the newborn compared with the fetal heart (Table 2). Furthermore, transcripts for lactate dehydrogenase (LDHA) were significantly elevated in newborn cardiac tissue, suggesting elevations in anaerobic glycolysis (Supplemental Table S7). LDHA catalyzes the reaction of pyruvate to lactate, a metabolite significantly elevated in the fetal heart (Table 1). Previous studies have shown that the newborn heart retains the same levels of lactate uptake and oxidation as the fetal heart up to 2 wk following delivery and that lactate uptake is linearly correlated to supply in the heart (4, 36). In addition, previous work in our laboratory has shown that blood lactate levels remain high in the newborn within 24 h following birth (data not published), suggesting that the decrease in cardiac lactate observed in this study is not because of a change in arterial levels. Plasma lactate levels are high beginning in late gestation because of limited return to the maternal circulation across the placenta as well as increased lactate production in the placenta and fetus, creating a plentiful supply for use by the fetal heart (7, 55). However, the decrease in cardiac lactate concentrations in the newborn may indicate higher utilization and lead to loss of feedback inhibition and activation of LDHA in the newborn heart to maintain lactate supply and oxidation. Our study supports previous findings that the fetal heart preferentially utilizes lactate and glucose in utero but that lactate utilization remains high in the newborn heart (4, 35, 36). In addition, our results suggest that the fetal heart has a higher activation of propanoate and butanoate metabolism as well as alcohol degradation. Ethanol readily crosses the near-term sheep placenta (18), and evidence presented here suggests that alcohols that cross the placenta may be important in maintaining energetics the fetal heart. However, more studies are needed to identify further the importance of alcohol degradation in the heart during this period, especially in humans and other nonruminant models.
Lipidomic analysis reveals elevations in sphingo- and ether lipids in the fetal heart.
Two species of ether lipids were significantly upregulated in fetal cardiac tissue (Table 2). These included both plasmanyl- and plasmenyl-phosphatidylserines (ether-PSs) and plasmanyl- and plasmenyl-PCs (ether-PCs). Although the presence of ether-PSs has not previously been previously reported in heart tissue, plasmenyl-PCs have been reported in the bovine heart (52). However, the function of these plasmalogens in the heart is less clear. Cellular studies suggest that plasmalogens are localized in lipid rafts in the cell membrane, which are important in cellular signaling and transduction (26, 45). In addition, plasmalogens are targets of oxidation and have been associated with free radical consumption and prevention of oxidative damage (12). In the porcine heart, oxidized plasmalogen products were elevated following cardiac infarction, adding to evidence of their protective role following stress in the heart (21). Furthermore, oxidative products of plasmalogens are unable to undergo lipid peroxidation in human brain tissue, which may prevent lipid oxidation (53). Although the function of ether-linked lipids in the heart remains unclear, their abundance in fetal cardiac tissue may be important in cell membrane signaling and oxidative protection, especially during birth as the fetus goes from a low- to high-oxygen environment.
SMs were another lipid class found to be significantly upregulated in the fetal heart (Table 2). SMs are found in the cellular membrane, particularly in lipid rafts and caveolae (11) and are important components of lung surfactant and fetal lung maturity (1, 8, 13, 41, 65). Although sphingolipid metabolic pathways were not found to be significantly enriched by ssGSEA (Supplemental Table S8), there was significantly greater expression of sphingomyelin synthase 1 (SGMS1) transcripts in fetal cardiac tissue (Supplemental Table S7). This enzyme catalyzes the conversion of ceramides to sphingomyelins. Upregulated levels of ceramides in cardiomyocytes have been linked to apoptosis (44, 58), suggesting that elevations in SMs may be cardio-protective to the developing fetus.
Multiomic analysis reveals importance of glycerophospholipid metabolism in the newborn heart.
Glycerophospholipid metabolism was significantly enriched following pathway analysis for identified metabolites and lipids in fetal and newborn hearts (Table 4), with ethanolamine significantly elevated in fetal cardiac tissue and choline significantly elevated in newborn cardiac tissue (Table 1). In addition, the glycerophospholipid metabolism pathway was significantly upregulated in newborn cardiac tissue following ssGSEA (Table 5). Transcripts for ethanolamine kinase 1 (ETNK1) were significantly upregulated in newborn cardiac tissue (Supplemental Table S7); ETNK1 catalyzes the conversion of ethanolamine to PEs. Therefore, the decrease in ethanolamine levels in newborn cardiac tissue suggests increased utilization of ethanolamine following birth. However, PEs were not found to be significantly altered between fetal and neonatal tissue as a class (Supplemental Table S6), suggesting that PE synthesis may occur from a separate mechanism in the fetal heart. Previous evidence suggests that ethanolamine uptake in the hamster heart is utilized for PE synthesis, a major phospholipid found in the cell membrane in mammalian hearts (39). PEs play a role in maintaining cellular integrity and preventing damage to sarcolemma during hypoxia in the heart (47). Therefore, ethanolamine degradation in the newborn heart may play an important role in maintaining cellular membrane composition following birth as the heart transitions from a low-oxygen environment in utero to a high-oxygen environment outside of the womb.
Transcripts for choline phosphotransferase 1 (CHPT1) were significantly elevated in newborn tissue (Supplemental Table S7). CHPT1 catalyzes the reaction of PCs to 1,2 diacyl-sn-glycerol, one of two forms of DGs. Although our methods were not able to distinguish between the two common forms of DGs (1,2 and 1,3-diacylglycerols), DGs as a class were more abundant in newborn cardiac tissue (Table 2) and play an important role in cellular signaling in the heart (43). In addition, TGs can be produced by DGs in cardiac tissue through diacylglycerol o-transferase 2 (DGAT2). DGs, TGs, and DGAT2 transcripts were all significantly upregulated in the newborn heart (Table 2 and Supplemental Table S7), suggesting increased activation of this pathway following birth. TG storages in the heart account for 10%–50% of fatty acids for energetics when circulating levels of fatty acids are low and are therefore, an important source of fatty acids during periods of low energy (51). In addition, plasma TG levels rise immediately following birth (25), which could lead to increased storage of TGs in the newborn heart as a fuel source during starvation. Our findings suggest that these storage forms of TGs are increased immediately following birth in the newborn heart, within the first day of life, and that these elevations in cardiac TGs may be from cardiac DG sources. This elevation in cardiac TGs may be an essential adaptation for the newborn heart to survive during periods of low circulating fatty acids for energy.
Transcriptomic and metabolomic analyses reveal purine metabolism is enriched in the newborn heart.
Purine metabolism was identified via Metscape correlations (Fig. 4) and was significantly enriched in newborn cardiac tissue following birth by ssGSEA with 17 genes being significantly upregulated in newborn cardiac tissue (Table 5). In addition, adenosine 5′-monophosphate (AMP) was significantly elevated in newborn cardiac tissue (Table 1). Two genes significantly upregulated in the newborn heart, adenine phosphoribosyltransferase (APRT) and adenosine kinase (ADK), produce AMP and are involved in the purine salvage pathway (Supplemental Table S7). In the heart, AMP is produced during stress when energy demand of the heart is high and exceeds energy supplies (28, 59). Therefore, it is not surprising that following the stress of birth, AMP is accumulated in the heart, as ATP is utilized for energy during this period.
Transcriptomic and lipidomic analyses reveal oxidative phosphorylation and fatty acid metabolism are enriched in the newborn heart.
Oxidative phosphorylation was significantly enriched in newborn cardiac tissue (Table 5). One gene in this pathway, NADH dehydrogenase flavoprotein 2 (NDUFV2), produces a mitochondrial enzyme involved in electron transport chain complex I, which utilizes CoQ10. CoQ10 was elevated in newborn cardiac tissue (Supplemental Table S5) as well as CoQ enzymes as a class (Table 2). In addition, transcripts for CoQ5 methyltransferase (COQ5) and CoQ3 methyltransferase (COQ3) were significantly upregulated in newborn cardiac tissue (Supplemental Table S7) and are involved in CoQ10 production, with COQ3 catalyzing the final formation of CoQ10. This adds to evidence that oxidative phosphorylation is elevated in the newborn heart following birth compared with the term fetus and that both substrates, particularly CoQ intermediates, and mitochondrial genes play a role in this adaptation. However, there is evidence that despite decreased mitochondrial structural organization and number, fetal cardiomyocytes have the same capacity for oxidative phosphorylation as cardiomyocytes from adult hearts (63). In our study, transcripts for 23 genes involved in oxidative phosphorylation were more highly expressed in cardiac tissue of term fetuses (Supplemental Table S8). This included succinate dehydrogenase complex subunit C (SDHC), one of four subunits of succinate dehydrogenase that utilizes succinate, a metabolite significantly elevated in fetal cardiac tissue (Table 1). Our data indicate that the term fetal heart has the capacity for oxidative phosphorylation (through succinate in the TCA cycle) but that this ability is enhanced following birth because of increased availability of CoQ enzymes and mitochondrial genes. Furthermore, consistent with an increase in mitochondria in the newborn heart, 3 cardiolipin species were increased in newborn cardiac tissue; however, these were not statistically significant following FDR correction (P < 0.20). In addition, transcripts for cardiolipin synthase 1 (CRLS1), an enzyme that produces CLs, were significantly enriched in newborn cardiac tissue, adding to evidence of increased CLs possibly because of increased mitochondrial biogenesis immediately following birth. However, future studies are needed to determine if birth alters the number and cellular integrity of mitochondria in the newborn heart.
In addition to oxidative phosphorylation, fatty acid metabolism was significantly elevated in newborn cardiac tissue (Table 5). Our lipidomic approach was able to identify two short-chain (AcCa 2:0 and AcCa 4:0), three medium-chain (AcCa 8:0, AcCa 10:0, and AcCa 12:0), and four long-chain acylcarnitines (AcCa 16:0, AcCa 16:0-OH/14:0, AcCa 18:0, and AcCa 18:1), with significant elevations in medium- and long-chain acylcarnitines in newborn cardiac tissue (Table 2). Medium- and long-chain acylcarnitines are produced from the breakdown of fatty acids in the mitochondria during β-oxidation and are abundant in cardiac tissue (38). A previous study in the bovine heart revealed that the fetal heart maintains the same ability to utilize short- and medium-chain fatty acids for oxidative phosphorylation as the neonatal heart but that long-chain fatty acid metabolism is only present in neonatal cardiac mitochondria (64). Our data support this previous finding, as no significant changes were observed in short-chain acylcarnitines between fetal and newborn cardiac tissue (Supplemental Table S6), whereas long-chain acylcarnitines were significantly upregulated in the newborn heart following birth (Table 2). In addition, transcripts for carnitine palmitoyltranfersferase 1A (CPT1A), which produces long-chain acylcarnitines during β-oxidation, were significantly upregulated in the newborn heart following birth as compared with the fetal heart (Supplemental Table S7). Thus, an elevation in medium- and long-chain acylcarnitines in the newborn heart may indicate increased β-oxidation within the first day of life, as opposed to 1–2 wk following birth (3, 5, 35, 36). However, as our model only looked at the newborn heart on the day of birth, future studies are needed to determine if β-oxidation is increased in the neonatal heart at subsequent times.
Conclusions.
This study is the first, to our knowledge, to utilize a multiomics approach to determine global metabolic changes that occur in the heart around the time of birth on both a transcriptional and small molecule level. Our study reveals that the fetal heart relies on substrates readily available from the mother and placenta and our model reveals enrichment in BCAA degradation and carbohydrate metabolism in the fetal heart. We observe increased abundance of both genes and metabolites involved in propanoate and butanoate metabolism in the fetal heart, a finding not previously reported. In addition, we found that although glycolysis is enriched in the fetal heart, it may be more efficiently utilizing alcohols for energy, whereas the newborn heart converts to more efficiently utilizing sugars. Further studies are needed to confirm altered flux in glycolysis in the fetal compared with newborn heart as well as the importance of alcohol degradation in nonruminant animals. Our study also indicates that the newborn heart has begun the shift toward an adult phenotype immediately following birth on the first day of life, as revealed by alterations in oxidative phosphorylation and fatty acid metabolism (35, 36). In addition, our data reveal that glycerophospholipid and purine metabolism may be important in maintaining energetics in the newborn heart, findings not previously reported.
Our data do show that numerous metabolic alterations occur in the heart immediately following delivery. Results presented here may serve as a basis for understanding normal cardiac metabolic development that occurs around the time of birth on a molecular and transcriptional level. Metabolic disruptions during pregnancy from the mother, as in the case of maternal diabetes, have been linked to increased risk of cardiac failure (42), whereas insufficient nutrient supplement in utero as seen during intrauterine growth restriction has been linked to increased risk of newborn and infant cardiac morbidities (17). Therefore, a better understanding of normal metabolic development of the heart in utero may shed light into mechanisms leading to heart failure in these populations and provide metabolic pathways that would be suitable for pharmacological intervention. In addition, by better understanding normal development of cardiac metabolism, we cannot only identify biomarkers for identifying fetuses and neonates at-risk for heart failure but also gain a better understanding of metabolic alterations that occur because of heart failure in the adult, ultimately leading to better diagnosis and treatment in both adults and newborns at-risk for cardiac metabolic failure.
GRANTS
This study was supported by NIH training Grant Nos. TL-1-TR-001428 and T32-HL-083810, as well as NIH Grant Nos. U-24-DK-097209 and HD-087306. A portion of this work was performed in the McKnight Brain Institute at the National High Magnetic Field Laboratory’s Advanced Magnetic Resonance Imaging and Spectroscopy Facility, which is supported by National Science Foundation Cooperative Agreement No. DMR-1157490 and the State of Florida. A. Edison was partially supported by the Georgia Research Alliance.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
J.M.W., A.S.E., and M.K.-W. conceived and designed research; J.M.W., J.P.K., and T.J.G. performed experiments; J.M.W. analyzed data; J.M.W., A.S.E., and M.K.-W. interpreted results of experiments; J.M.W. prepared figures; J.M.W. drafted manuscript; J.M.W., J.P.K., T.J.G., A.S.E., and M.K.-W. edited and revised manuscript; J.M.W., J.P.K., T.J.G., A.S.E., and M.K.-W. approved final version of manuscript.
ACKNOWLEDGMENTS
We thank the members of the Keller-Wood, Edison, and Garrett laboratories for their resources and expertise and Steven Robinette for the MATLAB Metabolomics Toolbox.
REFERENCES
- 1.Baig S, Lim JY, Fernandis AZ, Wenk MR, Kale A, Su LL, Biswas A, Vasoo S, Shui G, Choolani M. Lipidomic analysis of human placental syncytiotrophoblast microvesicles in adverse pregnancy outcomes. Placenta 34: 436–442, 2013. doi: 10.1016/j.placenta.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 2.Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF, Schinzel AC, Sandy P, Meylan E, Scholl C, Fröhling S, Chan EM, Sos ML, Michel K, Mermel C, Silver SJ, Weir BA, Reiling JH, Sheng Q, Gupta PB, Wadlow RC, Le H, Hoersch S, Wittner BS, Ramaswamy S, Livingston DM, Sabatini DM, Meyerson M, Thomas RK, Lander ES, Mesirov JP, Root DE, Gilliland DG, Jacks T, Hahn WC. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 462: 108–112, 2009. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartelds B, Gratama JW, Knoester H, Takens J, Smid GB, Aarnoudse JG, Heymans HS, Kuipers JR. Perinatal changes in myocardial supply and flux of fatty acids, carbohydrates, and ketone bodies in lambs. Am J Physiol Heart Circ Physiol 274: H1962–H1969, 1998. doi: 10.1152/ajpheart.1998.274.6.H1962. [DOI] [PubMed] [Google Scholar]
- 4.Bartelds B, Knoester H, Beaufort-Krol GC, Smid GB, Takens J, Zijlstra WG, Heymans HS, Kuipers JR. Myocardial lactate metabolism in fetal and newborn lambs. Circulation 99: 1892–1897, 1999. doi: 10.1161/01.CIR.99.14.1892. [DOI] [PubMed] [Google Scholar]
- 5.Bartelds B, Knoester H, Smid GB, Takens J, Visser GH, Penninga L, van der Leij FR, Beaufort-Krol GC, Zijlstra WG, Heymans HS, Kuipers JR. Perinatal changes in myocardial metabolism in lambs. Circulation 102: 926–931, 2000. doi: 10.1161/01.CIR.102.8.926. [DOI] [PubMed] [Google Scholar]
- 6.Beckonert O, Coen M, Keun HC, Wang Y, Ebbels TMD, Holmes E, Lindon JC, Nicholson JK. High-resolution magic-angle-spinning NMR spectroscopy for metabolic profiling of intact tissues. Nat Protoc 5: 1019–1032, 2010. doi: 10.1038/nprot.2010.45. [DOI] [PubMed] [Google Scholar]
- 7.Bell AW, Ferrell CL, Freetly HC. Pregnancy and fetal metabolism. In: Quantitative Aspects of Ruminant Digestion and Metabolism, edited by Dijkstra J, Forbes J, and France J. Cambridge, MA: CABI Publishing, 2005, p. 523–550. doi: 10.1079/9780851998145.0523. [DOI] [Google Scholar]
- 8.Bender TM, Stone LR, Amenta JS. Diagnostic power of lecithin/sphingomyelin ratio and fluorescence polarization assays for respiratory distress syndrome compared by relative operating characteristic curves. Clin Chem 40: 541–545, 1994. [PubMed] [Google Scholar]
- 9.Benjamini Y, Hochberg Y. Controlling the false discovery rate - a practical and powerful approach to multiple testing. J Roy Stat Soc B Met 57: 289–300, 1995. [Google Scholar]
- 10.Bingol K, Li DW, Zhang B, Brüschweiler R. Comprehensive metabolite identification strategy using multiple two-dimensional NMR spectra of a complex mixture implemented in the COLMARm web server. Anal Chem 88: 12411–12418, 2016. doi: 10.1021/acs.analchem.6b03724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Borodzicz S, Czarzasta K, Kuch M, Cudnoch-Jedrzejewska A. Sphingolipids in cardiovascular diseases and metabolic disorders. Lipids Health Dis 14: 55, 2015. doi: 10.1186/s12944-015-0053-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brosche T, Platt D. The biological significance of plasmalogens in defense against oxidative damage. Exp Gerontol 33: 363–369, 1998. doi: 10.1016/S0531-5565(98)00014-X. [DOI] [PubMed] [Google Scholar]
- 13.Chauvin S, Yinon Y, Xu J, Ermini L, Sallais J, Tagliaferro A, Todros T, Post M, Caniggia I. Aberrant TGFβ signalling contributes to dysregulation of sphingolipid metabolism in intrauterine growth restriction. J Clin Endocrinol Metab 100: E986–E996, 2015. doi: 10.1210/jc.2015-1288. [DOI] [PubMed] [Google Scholar]
- 14.Chong J, Soufan O, Li C, Caraus I, Li S, Bourque G, Wishart DS, Xia J. MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res 46: W486–W494, 2018. doi: 10.1093/nar/gky310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chua B, Siehl DL, Morgan HE. Effect of leucine and metabolites of branched chain amino acids on protein turnover in heart. J Biol Chem 254: 8358–8362, 1979. [PubMed] [Google Scholar]
- 16.Chung M, Teng C, Timmerman M, Meschia G, Battaglia FC. Production and utilization of amino acids by ovine placenta in vivo. Am J Physiol Endocrinol Metab 274: E13–E22, 1998. doi: 10.1152/ajpendo.1998.274.1.E13. [DOI] [PubMed] [Google Scholar]
- 17.Cohen E, Wong FY, Horne RS, Yiallourou SR. Intrauterine growth restriction: impact on cardiovascular development and function throughout infancy. Pediatr Res 79: 821–830, 2016. doi: 10.1038/pr.2016.24. [DOI] [PubMed] [Google Scholar]
- 18.Cumming ME, Ong BY, Wade JG, Sitar DS. Maternal and fetal ethanol pharmacokinetics and cardiovascular responses in near-term pregnant sheep. Can J Physiol Pharmacol 62: 1435–1439, 1984. doi: 10.1139/y84-238. [DOI] [PubMed] [Google Scholar]
- 19.Dieterle F, Ross A, Schlotterbeck G, Senn H. Probabilistic quotient normalization as robust method to account for dilution of complex biological mixtures. Application in 1H NMR metabonomics. Anal Chem 78: 4281–4290, 2006. doi: 10.1021/ac051632c. [DOI] [PubMed] [Google Scholar]
- 20.Dijkstra J, France J. Volatile Fatty Acid Production. In: Quantitative Aspects of Ruminant Digestion and Metabolism, edited by Dijkstra J, Forbes JM, and France J. Wallingford, UK: CABI Publishing, 2005, p. 157–175. doi: 10.1079/9780851998145.0000. [DOI] [Google Scholar]
- 21.Dudda A, Spiteller G, Kobelt F. Lipid oxidation products in ischemic porcine heart tissue. Chem Phys Lipids 82: 39–51, 1996. doi: 10.1016/0009-3084(96)02557-1. [DOI] [PubMed] [Google Scholar]
- 22.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30: 207–210, 2002. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eriksson L, Johansson E, Kettaneh-Wold N, Wold S. Multi- and Megavariate Data Analysis. Principles and Applications. Umea, Sweden: Umetrics AB, 2001. [Google Scholar]
- 24.Folch J, Lees M, Sloane Stanley GH. A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem 226: 497–509, 1957. [PubMed] [Google Scholar]
- 25.Hamosh M, Simon MR, Canter H JR, Hamosh P. Lipoprotein lipase activity and blood triglyceride levels in fetal and newborn rats. Pediatr Res 12: 1132–1136, 1978. doi: 10.1203/00006450-197812000-00006. [DOI] [PubMed] [Google Scholar]
- 26.Honsho M, Yagita Y, Kinoshita N, Fujiki Y. Isolation and characterization of mutant animal cell line defective in alkyl-dihydroxyacetonephosphate synthase: localization and transport of plasmalogens to post-Golgi compartments. Biochim Biophys Acta 1783: 1857–1865, 2008. doi: 10.1016/j.bbamcr.2008.05.018. [DOI] [PubMed] [Google Scholar]
- 27.Iruretagoyena JI, Davis W, Bird C, Olsen J, Radue R, Teo Broman A, Kendziorski C, Splinter BonDurant S, Golos T, Bird I, Shah D. Metabolic gene profile in early human fetal heart development. Mol Hum Reprod 20: 690–700, 2014. doi: 10.1093/molehr/gau026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jennings RB, Steenbergen C JR. Nucleotide metabolism and cellular damage in myocardial ischemia. Annu Rev Physiol 47: 727–749, 1985. doi: 10.1146/annurev.ph.47.030185.003455. [DOI] [PubMed] [Google Scholar]
- 29.Jonker SS, Zhang L, Louey S, Giraud GD, Thornburg KL, Faber JJ. Myocyte enlargement, differentiation, and proliferation kinetics in the fetal sheep heart. J Appl Physiol (1985) 102: 1130–1142, 2007. doi: 10.1152/japplphysiol.00937.2006. [DOI] [PubMed] [Google Scholar]
- 30.Karnovsky A, Weymouth T, Hull T, Tarcea VG, Scardoni G, Laudanna C, Sartor MA, Stringer KA, Jagadish HV, Burant C, Athey B, Omenn GS. Metscape 2 bioinformatics tool for the analysis and visualization of metabolomics and gene expression data. Bioinformatics 28: 373–380, 2012. doi: 10.1093/bioinformatics/btr661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Katz AM. Physiology of the Heart. Philadelphia, PA: Lippincott Williams & Wilkins, 2006. [Google Scholar]
- 32.Koelmel JP, Kroeger NM, Gill EL, Ulmer CZ, Bowden JA, Patterson RE, Yost RA, Garrett TJ. Expanding lipidome coverage using LC-MS/MS data-dependent acquisition with automated exclusion list generation. J Am Soc Mass Spectrom 28: 908–917, 2017. doi: 10.1007/s13361-017-1608-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koelmel JP, Kroeger NM, Ulmer CZ, Bowden J, Garland RP, Cochran JA, Beecher CW, Garrett TJ, Yost RA. LipidMatch: an automated workflow for rule-based lipid identification using untargeted high-resolution tandem mass spectrometry data. BMC Bioinformatics, 18: 331, 2017. doi: 10.1186/s12859-017-1744-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst 1: 417–425, 2015. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lopaschuk GD, Spafford MA. Energy substrate utilization by isolated working hearts from newborn rabbits. Am J Physiol Heart Circ Physiol 258: H1274–H1280, 1990. doi: 10.1152/ajpheart.1990.258.5.H1274. [DOI] [PubMed] [Google Scholar]
- 36.Lopaschuk GD, Spafford MA, Marsh DR. Glycolysis is predominant source of myocardial ATP production immediately after birth. Am J Physiol Heart Circ Physiol 261: H1698–H1705, 1991. doi: 10.1152/ajpheart.1991.261.6.H1698. [DOI] [PubMed] [Google Scholar]
- 37.Louis P, Flint HJ. Formation of propionate and butyrate by the human colonic microbiota. Environ Microbiol 19: 29–41, 2017. doi: 10.1111/1462-2920.13589. [DOI] [PubMed] [Google Scholar]
- 38.Makrecka-Kuka M, Sevostjanovs E, Vilks K, Volska K, Antone U, Kuka J, Makarova E, Pugovics O, Dambrova M, Liepinsh E. Plasma acylcarnitine concentrations reflect the acylcarnitine profile in cardiac tissues. Sci Rep 7: 17528, 2017. doi: 10.1038/s41598-017-17797-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McMaster CR, Tardi PG, Choy PC. Modulation of phosphatidylethanolamine biosynthesis by exogenous ethanolamine and analogues in the hamster heart. Mol Cell Biochem 116: 69–73, 1992. doi: 10.1007/BF01270571. [DOI] [PubMed] [Google Scholar]
- 40.Meigs RA, Sheean LA. Mitochondria from human term placenta. III. The role of respiration and energy generation in progesterone biosynthesis. Biochim Biophys Acta 489: 225–235, 1977. doi: 10.1016/0005-2760(77)90141-2. [DOI] [PubMed] [Google Scholar]
- 41.Melland-Smith M, Ermini L, Chauvin S, Craig-Barnes H, Tagliaferro A, Todros T, Post M, Caniggia I. Disruption of sphingolipid metabolism augments ceramide-induced autophagy in preeclampsia. Autophagy 11: 653–669, 2015. doi: 10.1080/15548627.2015.1034414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Narchi H, Kulaylat N. Heart disease in infants of diabetic mothers. Images Paediatr Cardiol 2: 17–23, 2000. [PMC free article] [PubMed] [Google Scholar]
- 43.Newton AC. Lipid activation of protein kinases. J Lipid Res 50, Suppl: S266–S271, 2009. doi: 10.1194/jlr.R800064-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parra V, Moraga F, Kuzmicic J, López-Crisosto C, Troncoso R, Torrealba N, Criollo A, Díaz-Elizondo J, Rothermel BA, Quest AFG, Lavandero S. Calcium and mitochondrial metabolism in ceramide-induced cardiomyocyte death. Biochim Biophys Acta 1832: 1334–1344, 2013. doi: 10.1016/j.bbadis.2013.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pike LJ, Han X, Chung KN, Gross RW. Lipid rafts are enriched in arachidonic acid and plasmenylethanolamine and their composition is independent of caveolin-1 expression: a quantitative electrospray ionization/mass spectrometric analysis. Biochemistry 41: 2075–2088, 2002. doi: 10.1021/bi0156557. [DOI] [PubMed] [Google Scholar]
- 46.Pluskal T, Castillo S, Villar-Briones A, Oresic M. MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinformatics 11: 395, 2010. doi: 10.1186/1471-2105-11-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Post JA, Bijvelt JJ, Verkleij AJ. Phosphatidylethanolamine and sarcolemmal damage during ischemia or metabolic inhibition of heart myocytes. Am J Physiol Heart Circ Physiol 268: H773–H780, 1995. doi: 10.1152/ajpheart.1995.268.2.H773. [DOI] [PubMed] [Google Scholar]
- 48.Rabaglino MB, Chang EI, Richards EM, James MO, Keller-Wood M, Wood CE. Genomic effect of triclosan on the fetal hypothalamus: evidence for altered neuropeptide regulation. Endocrinology 157: 2686–2697, 2016. doi: 10.1210/en.2016-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rabaglino MB, Richards E, Denslow N, Keller-Wood M, Wood CE. Genomics of estradiol-3-sulfate action in the ovine fetal hypothalamus. Physiol Genomics 44: 669–677, 2012. doi: 10.1152/physiolgenomics.00127.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Richards EM, Wood CE, Rabaglino MB, Antolic A, Keller-Wood M. Mechanisms for the adverse effects of late gestational increases in maternal cortisol on the heart revealed by transcriptomic analyses of the fetal septum. Physiol Genomics 46: 547–559, 2014. doi: 10.1152/physiolgenomics.00009.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saddik M, Lopaschuk GD. Myocardial triglyceride turnover and contribution to energy substrate utilization in isolated working rat hearts. J Biol Chem 266: 8162–8170, 1991. [PubMed] [Google Scholar]
- 52.Schmid HHO, Takahashi T. The alk-i-enyl ether and alkyl ether lipids of bovine heart muscle. Biochim Biophys Acta 164: 141–147, 1968. doi: 10.1016/0005-2760(68)90140-9. [DOI] [PubMed] [Google Scholar]
- 53.Sindelar PJ, Guan Z, Dallner G, Ernster L. The protective role of plasmalogens in iron-induced lipid peroxidation. Free Radic Biol Med 26: 318–324, 1999. doi: 10.1016/S0891-5849(98)00221-4. [DOI] [PubMed] [Google Scholar]
- 54.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: Article3, 2004. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 55.Sparks JW, Hay WW JR, Bonds D, Meschia G, Battaglia FC. Simultaneous measurements of lactate turnover rate and umbilical lactate uptake in the fetal lamb. J Clin Invest 70: 179–192, 1982. doi: 10.1172/JCI110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 102: 15545–15550, 2005. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Swierczyński J, Scislowski P, Aleksandrowicz Z, Zelewski L. Stimulation of citrate oxidation and transport in human placental mitochondria by L-malate. Acta Biochim Pol 23: 93–102, 1976. [PubMed] [Google Scholar]
- 58.Tepper CG, Jayadev S, Liu B, Bielawska A, Wolff R, Yonehara S, Hannun YA, Seldin MF. Role for ceramide as an endogenous mediator of Fas-induced cytotoxicity. Proc Natl Acad Sci USA 92: 8443–8447, 1995. doi: 10.1073/pnas.92.18.8443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ver Donck K. Purine metabolism in the heart. Strategies for protection against myocardial ischaemia. Pharm World Sci 16: 69–76, 1994. [DOI] [PubMed] [Google Scholar]
- 60.Walejko JM, Chelliah A, Keller-Wood M, Gregg A, Edison AS. Global Metabolomics of the Placenta Reveals Distinct Metabolic Profiles between Maternal and Fetal Placental Tissues Following Delivery in Non-Labored Women. Metabolites 8: 10, 2018. doi: 10.3390/metabo8010010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang J, Vasaikar S, Shi Z, Greer M, Zhang B. WebGestalt 2017: a more comprehensive, powerful, flexible and interactive gene set enrichment analysis toolkit. Nucleic Acids Res 45: W130–W137, 2017. doi: 10.1093/nar/gkx356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ward JH., JR Hierarchical grouping to optimize an objective function. J Am Stat Assoc 58: 236–244, 1963. doi: 10.1080/01621459.1963.10500845. [DOI] [Google Scholar]
- 63.Warshaw JB. Cellular energy metabolism during fetal development. I. Oxidative phosphorylation in the fetal heart. J Cell Biol 41: 651–657, 1969. doi: 10.1083/jcb.41.2.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Warshaw JB, Terry ML. Cellular energy metabolism during fetal development. II. Fatty acid oxidation by the developing heart. J Cell Biol 44: 354–360, 1970. doi: 10.1083/jcb.44.2.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Whitfield CR, Chan WH, Sproule WB, Stewart AD. Amniotic fluid lecithin: sphingomyelin ratio and fetal lung development. BMJ 2: 85–86, 1972. doi: 10.1136/bmj.2.5805.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wood CE, Rabaglino MB, Chang EI, Denslow N, Keller-Wood M, Richards E. Genomics of the fetal hypothalamic cellular response to transient hypoxia: endocrine, immune, and metabolic responses. Physiol Genomics 45: 521–527, 2013. doi: 10.1152/physiolgenomics.00005.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xia J, Wishart DS. MetPA: a web-based metabolomics tool for pathway analysis and visualization. Bioinformatics 26: 2342–2344, 2010. doi: 10.1093/bioinformatics/btq418. [DOI] [PubMed] [Google Scholar]