Abstract
The pathological consequences of ischemic heart disease involve signaling through the autonomic nervous system. Although early activation may serve to maintain hemodynamic stability, persistent aberrant sympathoexcitation contributes to the development of lethal arrhythmias and heart failure. We hypothesized that as the myocardium reacts and remodels to ischemic injury over time, there is an analogous sequence of gene expression changes in the thoracic spinal cord dorsal horn, the processing center for incoming afferent fibers from the heart to the central nervous system. Acute and chronic myocardial ischemia (MI) was induced in a large animal model of Yorkshire pigs, and the thoracic dorsal horn of treated pigs, along with control nonischemic pigs, was harvested for transcriptome analysis. We identified 32 differentially expressed genes between healthy and acute ischemia cohorts and 46 differentially expressed genes between healthy and chronic ischemia cohorts. The canonical immediate-early gene c-fos was upregulated after acute MI, along with fosB, dual specificity phosphatase 1 and 2 (dusp1 and dusp2), and early growth response 2 (egr2). After chronic MI, there was a persistent yet unique activation of immediate-early genes, including fosB, nuclear receptor subfamily 4 group A members 1−3 (nr4a1, nr4a2, and nr4a3), egr3, and TNF-α-induced protein 3 (tnfaip3). In addition, differentially expressed genes from the chronic MI signature were enriched in pathways linked to apoptosis, immune regulation, and the stress response. These findings support a dynamic progression of gene expression changes in the dorsal horn with maturation of myocardial injury, and they may explain how early adaptive autonomic nervous system responses can maintain hemodynamic stability, whereas prolonged maladaptive signals can predispose patients to arrhythmias and heart failure.
NEW & NOTEWORTHY Activation of the autonomic nervous system after myocardial injury can provide early cardiovascular support or prolonged aberrant sympathoexcitation. The later response can lead to lethal arrhythmias and heart failure. This study provides evidence of ongoing changes in the gene expression signature of the spinal cord dorsal horn as myocardial injury progresses over time. These changes could help explain how an adaptive nervous system response can become maladaptive over time.
Keywords: autonomic nervous system, gene expression/regulation, myocardial ischemia, spinal cord
INTRODUCTION
The central nervous system is engaged after myocardial ischemia (MI) through indirect and direct chemical and electrical signals. A principal component of this signaling is afferent input from ischemia-sensitive cardiac neurons to the dorsal root ganglia (DRG) and upper thoracic dorsal horn of the spinal cord (4). Efferent pathways in the spinal cord then trigger sympathoexcitation, which is essential to maintain hemodynamic stability in the presence of injured myocardium during the acute period. Unfortunately, aberrant signaling through the same pathway can lead to deadly arrhythmias and heart failure (9, 34). Strategies to prevent these pathological consequences of sympathoexcitation have targeted the efferent limb of this neural network, but results have been equivocal perhaps because of redundant parallel processes (7, 17, 22, 28, 35, 38). As a result, upstream targeting of the afferent limb at the dorsal horn might be more successful. Such strategies have been limited by a lack of understanding of the chemical, molecular, and genetic changes that take place in the dorsal horn after MI.
In general, afferent signaling to the dorsal horn involves the propagation of action potentials from the periphery but can also involve activation of new gene expression networks (26). These latter responses contribute to the plasticity of neurons and can lead to phenotypic changes, such as variation in their landscape of ion channels (25), de novo nerve sprouting (23), memory (18), and even cell death (10, 24). These networks generally entail sequential activation of distinct groups of genes. The first group is known as immediate-early genes (IEGs), as they are usually activated rapidly and for a short period time in the absence of new protein synthesis. IEGs mostly comprise master regulators, such as transcription factors that control expression of late response genes that typically encode proteins with specific biological functions (6, 26, 29). This basic model of gene activation has been well studied in animal models of peripheral nerve injury, which have consistently shown rapid and transient upregulation of the well-known IEG c-Fos in the dorsal horn after noxious stimulation (11, 13, 36). The spinal cord response to MI is no exception to this pattern of neural activation. In rats undergoing intermittent coronary artery occlusion, c-Fos is upregulated in lamina I–V of the dorsal horn within 90 min (8, 12). Nevertheless, a full description of the changes in gene expression that take place in the dorsal horn after MI has yet to be described. In the present study, we performed genome-wide transcriptome analysis on thoracic dorsal horn tissue after an acute episode of MI and reperfusion injury in a large animal porcine model. Additionally, we conducted similar transcriptome profiling in the dorsal horn of a chronic ischemic model to determine how persistent myocardial injury impacts gene expression. These genetic signatures could be crucial to the activation and maintenance of sympathoexcitation and thus may serve as potential novel targets to curtail potential novel tarts to curtain progression to heart failure and/or sudden cardiac death caused by arrhythmias.
METHODS
Animal subjects.
Yorkshire pigs weighing on average 30 ± 15 kg were randomized to healthy control (n = 10), acute ischemia (n = 10), and chronic ischemia (n = 8) cohorts. All experiments were performed in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals and approved by the University of California-Los Angeles Animal Research Committee.
Chronic ischemia protocol.
Chronic ischemia was induced as previously described (20). Briefly, pigs were sedated with telazol (8 mg/kg im) and then induced with general endotracheal anesthesia and maintained on 1–2% end-tidal isoflurane with intermittent fentanyl boluses (total: 10–30 μg/kg). All subjects were monitored with standard 12-lead ECG and pulse oximetry. The right femoral vein was catheterized for venous access, and an 8-Fr sheath was placed in the right femoral artery for continuous arterial blood pressure monitoring and percutaneous intervention. An angioplasty balloon catheter (3 mm, FoxCross PTA Catheter, Abbott Vascular, Temecula, CA) was guided over a wire into the third diagonal of the left anterior descending coronary artery. The balloon was inflated for 30 s followed by injection of a 5 ml suspension of saline containing 1 ml of polystyrene microspheres (Polybead, 90-μm diameter, Polysciences, Warrington, PA) into the distal segment of the artery. Occlusion of the vessel was confirmed by contrast angiography and ST-segment elevations (0.1 mV above baseline) in lead II of the ECG. Close monitoring for arrhythmias was performed after extubation for up to 2 h, and arrhythmias were treated with esmolol (5 mg boluses) and lidocaine (10–20 mg boluses). Animals were then returned to their pen and subjected to daily monitoring for 8 wk. A mean percent infarction of 17.6 ± 4.5% of left ventricular volume has been previously described using this method (20).
Acute ischemia protocol.
Acute ischemia was induced as previously described (10). Briefly, after the induction of anesthesia as described above, animals were placed in the supine position followed by a median sternotomy. The second diagonal branch of the left anterior descending coronary artery was ligated with a 4-0 suture. Ischemia was confirmed by the presence of ST-segment elevations (0.1 mV above baseline) in greater than five contiguous limb and precordial leads, direct discoloration of the heart in the field, and regional hypokinesis of the left ventricular anteroapical walls distal to the ligation site. After 1 h of ischemia, the 4-0 prolene suture was removed and reperfusion was permitted for 3 h.
Tissue collection.
Chronic ischemia and healthy pigs were induced with general anesthesia followed by a median sternotomy as described above. Animals were then maintained under general anesthesia to replicate the surgical time of the acute ischemia cohort. All pigs were then placed in the prone position, and a dorsal spinal laminectomy was performed to expose the T1–T4 and L1–L4 spinal segments. Animals were then euthanized by terminal cardiac arrhythmia under general anesthesia. Each spinal cord segment was then quickly harvested and frozen in liquid nitrogen followed by storage at −80°C.
RNA sequencing.
Under a dissecting microscope, the thoracic and lumbar dorsal horn (T2 and L2) of 0.5-cm thick spinal segments were cut from frozen tissue and immediately placed in ice-cold TRIzol (ThermoFisher, Waltham, MA) for homogenization and RNA extraction according to the manufacturer’s protocol. RNA cleanup was conducted using the RNeasy Mini Kit (Qiagen, Germantown, MD), and quality was assessed using the Agilent Bioanalyzer 2200 (Agilent, Santa Clara, CA). Libraries were prepared with the KAPA Stranded RNA-Seq kit (KAPABiosystems, Wilmington, MA), which consists of mRNA enrichment, cDNA generation, end repair to generate blunt ends, A-tailing, adaptor ligation, and PCR amplification. Single-end sequencing proceeded with the Illumina HiSeq 3000 (Illumina, San Diego, CA). The read depth was on average 27.5 million reads/sample.
Read alignment and differential expression analysis.
Raw reads were analyzed for quality followed by trimming of low-quality bases and adaptor removal. The tuxedo pipeline was then used for downstream analysis as previously described (31). In brief, alignment was performed using TopHat version 2.1.0 under default settings with an average mapping rate of 92% (14). Differential expression proceeded from Cufflinks version 2.1.1 (32) under default settings using the most up-to-date annotation file for the pig genome from the Ensembl database (Sscrofa 11.1). Genes with P values adjusted for multiple comparison of <0.1 (Benjamini-Hochberg method) were considered differentially expressed. Only genes with mean fragments per kilobase per million mapped reads of >1 in at least one cohort were considered. Volcano plots were generated in RStudio version 0.99.896 (30).
Immunofluorescence staining.
Spinal cord segments fixed in paraformaldehyde and stored in 70% ethanol were embedded in paraffin and sectioned at 5µm. Slides were then deparaffinized in an oven for 2 h at 60°C followed by rehydration through a series of xylene and alcohol washes. Antigen retrieval was performed on the slides in hot (98°C) sodium citrate buffer (pH 6.9). After being cooled, slides were washed four times in PBS (pH 7.3) for 5 min. Blocking took place for 2 h in 1% BSA, 0.4% Triton X-100, and 5% normal donkey serum. Slides were then incubated overnight in primary antibodies in 1% BSA, 0.4% Triton X-100, and 5% normal donkey serum. After an additional wash in PBS, slides were incubated for 2 h in 1% BSA, 0.4% Triton X-100, 5% normal donkey serum, and secondary antibody. After another round of washes, slides were then coverslipped using mounting medium with DAPI and sealed with clear nail polish. Antibodies used include microtubule-associated protein 2 (MAP2; ab-5392, Abcam, 1:2,000), c-Fos (catalog no. 226003, Synaptic System, 1:5,000), donkey anti-rabbit secondary Alexa-fluor 488-conjugated antibody (ThermoFisher Scientific, 1:200), and donkey anti-chicken secondary Alexa-Fluor 594-conjugated antibody (Jackson ImmunoResearch, 1:200). Images were obtained with a confocal scanning microscope (Nikon). For each field, double-positive cytosolic MAP2/nuclear c-Fos-stained neurons were counted as a percentage of the total number of MAP2-stained cells in the dorsal horn. For each condition, cells were counted in a total of 33 fields over 10 slides. Significance was determined by a two-tailed Student’s t-test.
Quantitative real-time PCR.
RT-PCR was performed according to the manufacturer’s protocol using iScript Reverse Transcription Supermix (Bio-Rad, Hercules, CA). cDNA from these reactions was then used for quantitative real-time PCR using SYBR Green Mastermix (Bio-Rad) and cycled using the CFX96 Real-Time PCR Detection System (Bio-Rad). Custom primers were designed using primer3 and included c-fos (forward 5′-ACTACGAGGCGTCATCATCC-3′ and reverse 5′-GGCTGGTCGAGATAGCAGTC-3′), early growth response 3 (egr3; forward 5′-CTTCGACTCCCCTTCCAACT-3′ and reverse 5′-AAGACACAGGCTCCGAGTAG-3′), fosB (forward 5′-GAGAACGGAACAAACTGGCA-3′ and reverse 5′-GATCTTGCAACCCGGTTTGT-3′), and nuclear receptor subfamily 4 group A members 1 (nr4a1; forward 5′-CTGCCTGGCTAACAAGGACT-3′ and reverse 5′-CAGGGAGGTGAGGAGATTGG-3′). Hydroxymethylbilane synthase (hmbs) was used as an internal control (forward 5′-TGGAGGAGTCTGGAGTCTGA-3′ and reverse 5′-TCTTGGCACCTTTGTTCAGC-3′). High efficiency and specificity were established with standard curves and melting curves, respectively. Reactions were performed in duplicate, and threshold cycle (CT) values were averaged followed by transformation according to the ΔCT method (ΔCT = CT of the gene − CT of the internal control). Significance was assessed by a one-tailed Student’s t-test. Dot plots were generated using RStudio version 0.99.896 (30).
Gene Ontology enrichment and pathway analysis.
Gene Ontology (GO) analysis was conducted using the R package clusterProfiler (37) on differentially expressed genes between healthy versus acute cohorts and healthy versus chronic cohorts. P values were corrected for multiple comparison using the Benjamini-Hochberg method. Pathway association of differentially expressed genes between healthy versus chronic subjects was generated from Ingenuity Pathway Analysis (Ingenuity Systems, Redwood City, CA).
Western blot analysis.
Whole cell protein extracts from thoracic and lumbar dorsal horn (T2 and L2) tissue were prepared in 50 µM Tris (pH 7.4), 250 µM NaCl, 10% glycerol, 0.5% Triton X-100, and a cocktail of protease inhibitors (P-8340, Sigma-Aldrich, St. Louis, MO). Protein extract (75 µg) was run on acrylamide gels and transferred to a nitrocellulose membrane. Membranes were probed with antibodies against β-actin (no. 4970, Cell Signaling, 1:1,000) and cleaved caspase-3 (no. 9661, Cell Signaling, 1:1,000). Secondary antibodies were used at a concentration of 1:5,000 before development with LI-COR systems (LI-COR Biosciences, Lincoln, NE).
RESULTS
Transcriptome changes in the dorsal horn after MI.
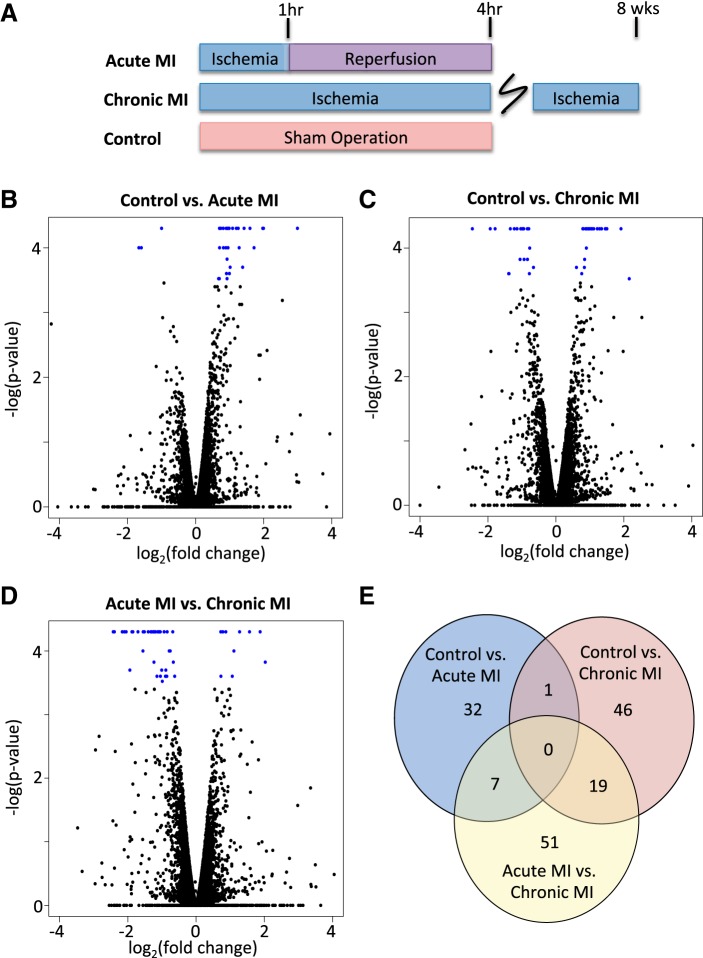
T2 thoracic dorsal horn tissue was extracted from 10 pigs subjected to ischemia-reperfusion injury, 8 pigs subjected to chronic MI, and 10 healthy pigs that underwent sham operations (Fig. 1A). There were no major adverse hemodynamic events noted in any protocol and no mortality before euthanasia. Tissue samples were used for whole genome transcriptome experiments. There were a total of 25,878 genes in the most up-to-date pig genome annotation file considered for differential expression analysis. Compared with healthy controls, acute ischemia resulted in 32 significant genes with an adjusted P value of <0.1 (Table 1). The absolute log2(fold change) ranged from 0.67 to 2.99. Of these genes, 3 genes were downregulated and 29 genes were upregulated after ischemia (Fig. 1B). In comparison, chronic ischemia compared with healthy control pigs resulted in 46 differentially expressed genes under the same adjusted P value cutoffs (Table 2). The absolute log2(fold change) ranged from 0.60 to 2.46; 21 of these genes were downregulated and 25 were upregulated (Fig. 1C). fosB was the only gene shared by both profiles, and it was slightly more upregulated in the acute MI group compared with the chronic MI group. When we directly compared the gene expression profile of acute MI pigs with chronic MI pigs, there were 51 differentially expressed genes (Fig. 1D). The majority of these genes (40 genes) were downregulated from acute to chronic MI. Twenty-six of these genes were shared with the previously identified differentially expressed genes from comparisons with the control cohort (Fig. 1E).
Fig. 1.
Acute and chronic myocardial ischemia (MI) leads to gene expression changes in the dorsal horn compared with healthy controls. A: Yorkshire pigs were randomized to healthy controls (n = 10), 1 h of acute MI and 3 h of reperfusion injury (n = 10), and 8 wk of chronic MI through catheter-mediated coronary artery occlusion (n = 8). B−D: volcano plots showing log2(fold change) and −log(P value) for all genes when control with acute MI cohorts (B), control with chronic MI cohort (C), and acute MI with chronic MI cohorts (D) were compared. Blue dots represent those differentially expressed genes that meet the threshold for multiple comparison testing (Benjamini-Hochberg < 0.1). E: Venn diagram illustrating the number of differentially expressed genes when control with acute MI cohorts (blue), control with chronic MI cohorts (pink), and acute MI with chronic MI (yellow) cohorts were compared. Overlapping sections display shared genes between comparisons.
Table 1.
Differentially expressed genes in acute myocardial ischemia versus control groups
| Gene Name | Mean FPKM Acute MI | Mean FPKM Control | Log2(Fold Change) | Adjusted P Value |
|---|---|---|---|---|
| ENSSSCG00000036224 | 3.26868 | 26.0231 | 2.99301 | 0.027325 |
| CXCL9 | 1.53946 | 6.13762 | 1.99526 | 0.027325 |
| ENSSSCG00000039758 | 0.411301 | 1.61426 | 1.97261 | 0.027325 |
| IL1R2 | 1.00432 | 3.04015 | 1.59792 | 0.027325 |
| LAMC2 | 1.01493 | 2.72421 | 1.42446 | 0.027325 |
| DUSP2 | 0.991545 | 2.36293 | 1.25283 | 0.027325 |
| SCN4A | 0.520871 | 1.19341 | 1.19609 | 0.027325 |
| CEMIP | 2.17406 | 4.61843 | 1.08701 | 0.027325 |
| FosB | 24.2775 | 48.0798 | 0.985814 | 0.027325 |
| COL1A1 | 25.8304 | 49.7963 | 0.94697 | 0.027325 |
| COL12A1 | 1.79667 | 3.42936 | 0.932616 | 0.027325 |
| EDN3 | 4.52509 | 8.43376 | 0.898229 | 0.027325 |
| cFos | 75.2914 | 132.081 | 0.810871 | 0.027325 |
| CTGF | 13.8012 | 23.028 | 0.738601 | 0.027325 |
| RELN | 3.15722 | 5.13255 | 0.701019 | 0.027325 |
| COL11A2 | 2.26798 | 1.13377 | −1.00028 | 0.027325 |
| MMP8 | 0.479562 | 1.57743 | 1.71779 | 0.0465106 |
| TGM3 | 7.46493 | 18.1301 | 1.28019 | 0.0465106 |
| PTGS2 | 1.86534 | 3.61183 | 0.953292 | 0.0465106 |
| OSGIN1 | 14.597 | 27.0637 | 0.890688 | 0.0465106 |
| DUSP1 | 47.0782 | 82.922 | 0.816698 | 0.0465106 |
| RGS1 | 10.0266 | 16.3331 | 0.703971 | 0.0465106 |
| IDO1 | 1.47227 | 0.487352 | −1.59501 | 0.0465106 |
| ENSSSCG00000006286 | 1.01102 | 0.318086 | −1.66831 | 0.0465106 |
| POSTN | 21.9545 | 41.6646 | 0.924306 | 0.0636699 |
| ENSSSCG00000001252 | 7.97156 | 20.786 | 1.38268 | 0.0787748 |
| DLL4 | 1.39979 | 2.83034 | 1.01576 | 0.0787748 |
| HSPA1L | 2.49817 | 4.97079 | 0.992604 | 0.086746 |
| CD163 | 2.40293 | 4.50363 | 0.906296 | 0.086746 |
| SLC16A9 | 2.44071 | 4.63081 | 0.923968 | 0.0993636 |
| COL1A2 | 32.3967 | 52.5392 | 0.697546 | 0.0993636 |
| EGR2 | 8.5098 | 13.5815 | 0.674445 | 0.0993636 |
Differentially expressed genes between acute myocardial ischemia (MI; n = 10) and control pigs (n = 10) with adjusted P values of <0.1 are shown. Adjusted P value, adjusted P value by Benjamini-Hochberg; FPKM, fragments per kilobase per million mapped reads.
Table 2.
Differentially expressed genes in chronic myocardial ischemia versus control groups
| Gene Name | Mean FPKM Control | Mean FPKM Chronic MI | Log2(Fold Change) | Adjusted P Value |
|---|---|---|---|---|
| ENSSSCG00000036836 | 193.397 | 729.194 | 1.91474 | 0.027325 |
| EGR3 | 7.29248 | 20.6288 | 1.50018 | 0.027325 |
| CXCL2 | 8.08219 | 22.2903 | 1.4636 | 0.027325 |
| COL6A5 | 0.758159 | 2.07294 | 1.45111 | 0.027325 |
| NR4A3 | 1.96634 | 4.98727 | 1.34274 | 0.027325 |
| CENPE | 0.620469 | 1.51267 | 1.28567 | 0.027325 |
| SFRP4 | 6.32537 | 14.7165 | 1.21822 | 0.027325 |
| MPZ | 9.85831 | 22.9047 | 1.21623 | 0.027325 |
| NR4A1 | 27.2766 | 58.999 | 1.11302 | 0.027325 |
| NR4A2 | 5.45997 | 11.4228 | 1.06495 | 0.027325 |
| MGP | 15.6685 | 32.3172 | 1.04443 | 0.027325 |
| PMP2 | 303.429 | 605.926 | 0.997784 | 0.027325 |
| ENSSSCG00000007527 | 4.52509 | 8.88267 | 0.973046 | 0.027325 |
| CHRNA4 | 3.15805 | 6.07507 | 0.943868 | 0.027325 |
| FosB | 24.2775 | 45.4682 | 0.90524 | 0.027325 |
| BTG2 | 18.6393 | 34.1701 | 0.874389 | 0.027325 |
| TLCD2 | 25.5482 | 46.8275 | 0.874137 | 0.027325 |
| ADAMTS1 | 6.78085 | 11.8755 | 0.808449 | 0.027325 |
| COX7B | 49.1576 | 85.0035 | 0.790108 | 0.027325 |
| MX2 | 14.1256 | 8.11949 | −0.798851 | 0.027325 |
| OAS2 | 24.9147 | 13.8876 | −0.843199 | 0.027325 |
| FAM84A | 27.784 | 14.4033 | −0.947857 | 0.027325 |
| SAMD9 | 4.79184 | 2.44723 | −0.969431 | 0.027325 |
| RGS20 | 1.99631 | 1.00276 | −0.993358 | 0.027325 |
| PANX1 | 4.00116 | 1.95053 | −1.03655 | 0.027325 |
| ISG15 | 22.6618 | 10.2576 | −1.14357 | 0.027325 |
| ACTC1 | 8.42141 | 3.80225 | −1.14721 | 0.027325 |
| HTR2A | 2.14839 | 0.917864 | −1.2269 | 0.027325 |
| TEAD4 | 6.03033 | 2.37845 | −1.34221 | 0.027325 |
| GLDN | 5.86291 | 1.69466 | −1.79063 | 0.027325 |
| RSAD2 | 5.45731 | 1.42844 | −1.93375 | 0.027325 |
| TNFSF18 | 9.39156 | 1.7066 | −2.46024 | 0.027325 |
| TNFAIP3 | 2.88778 | 5.36972 | 0.894886 | 0.0465106 |
| PARP12 | 4.87329 | 2.85286 | −0.772488 | 0.0465106 |
| KLF4 | 5.44863 | 9.78261 | 0.844327 | 0.0636699 |
| ENSSSCG00000033089 | 2.82248 | 1.57891 | −0.838032 | 0.0636699 |
| CMPK2 | 4.91795 | 2.56769 | −0.937588 | 0.0636699 |
| RASL11B | 2.89599 | 1.39235 | −1.05654 | 0.0636699 |
| IGFBP6 | 25.4766 | 45.5083 | 0.836957 | 0.0787748 |
| PDK4 | 37.893 | 57.5803 | 0.603646 | 0.0787748 |
| MX1 | 46.1784 | 29.198 | −0.66135 | 0.0787748 |
| ENSSSCG00000022246 | 4.19374 | 7.09691 | 0.758953 | 0.086746 |
| ARID5A | 12.0509 | 7.00777 | −0.782113 | 0.086746 |
| ZBP1 | 4.03983 | 1.55017 | −1.38186 | 0.086746 |
| C1QTNF7 | 2.39948 | 0.914228 | −1.3921 | 0.086746 |
| CSF3 | 0.493867 | 2.20436 | 2.15816 | 0.0993636 |
Differentially expressed genes between chronic myocardial ischemia (MI; n = 8) and control pigs (n = 10) with adjusted P values of <0.1 are shown. Adjusted P value, adjusted P value by Benjamini-Hochberg; FPKM, fragments per kilobase per million mapped reads.
Acute MI triggers IEG activation.
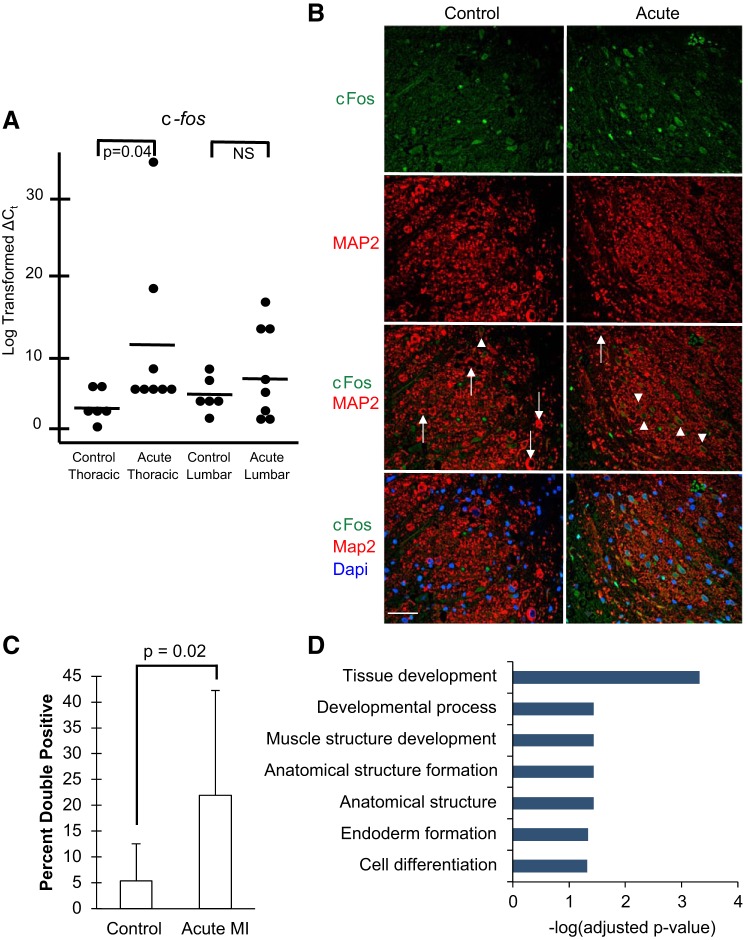
The IEG c-fos was among the most significant differentially expressed genes in the acute MI group compared with the control group [log2(fold change): 0.81, adjusted P value: 0.027]. We validated this finding with quanitative PCR and also showed that these changes were specific to the thoracic dorsal horn, as a similar upregulation of c-fos expression was not present in lumbar dorsal horn tissue from the same animals (Fig. 2A). Similarly, we used immunofluorescence to determine if there was a similar increase in c-Fos protein among neurons in the dorsal horn. To this end, we stained T2 thoracic dorsal horn segments from healthy and acute MI pigs for c-Fos and the neuron-specific marker microtubule-associated protein 2 (MAP2) (Fig. 2B). The percentage of c-Fos-labeled neurons was significantly higher in the acute MI cohort compared with the healthy cohort [22.0% (SD 20.3) vs. 5.3% (SD 7.2), P = 0.02; Fig. 2C). This is consistent with previous studies in rats undergoing coronary artery occlusion, which demonstrated increased c-Fos protein in the thoracic dorsal horn compared with nonischemic controls (8, 12). Ontology analysis of all dysregulated genes showed enrichment for categories associated with development and cell differentiation (Fig. 2D). Genes in these categories included IEGs known to be activated during stress responses in the spinal cord, including c-fos, fosB, dual specificity phosphatase 1 and 2 (dusp1 and dusp2), and egr2 (33).
Fig. 2.
Acute myocardial ischemia (MI) increases c-Fos in the dorsal horn. A: quantitative PCR of c-fos expression in thoracic and lumbar dorsal horn tissue from control (n = 6) and acute (n = 8) MI pigs. Each dot represents the log-transformed ∆CT value (where CT is threshold cycle) for each biological replicate. Solid lines represent the mean for each group. B: immunofluorescence staining of c-Fos (green), microtubule-associated protein 2 (MAP2; red), and DAPI (blue) in dorsal horn tissue from a representative control pig and a pig exposed to acute MI. Arrows denote c-Fos-negative neurons; arrowheads denote c-Fos-positive neurons. Scale bar = 50 μm. C: quantification of double-positive MAP2- and c-Fos-positive cells as a percentage of MAP2-positive cells in control (n = 3) and acute MI (n = 4) cohorts. Error bars depict SDs. D: biological process Gene Ontology enrichment terms for all differentially expressed genes in the dorsal horn after acute MI compared with control pigs. Terms are ranked by significance using −log(adjusted P value). NS, not significant.
Chronic MI results in prolonged upregulation of IEGs.
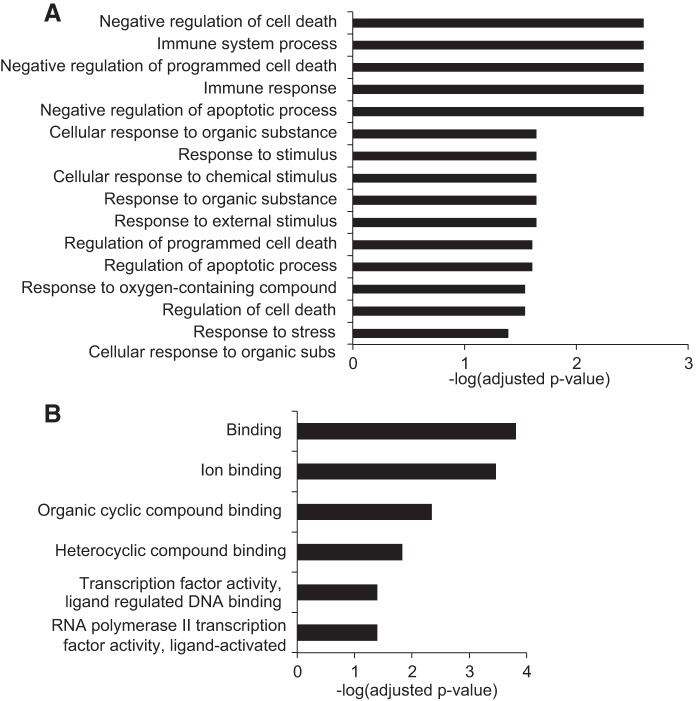
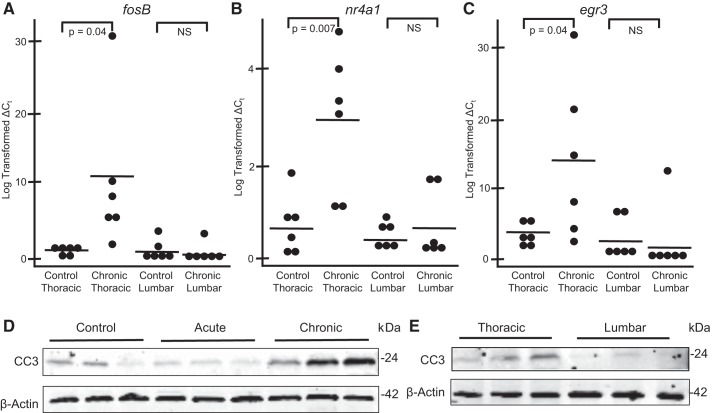
Since spinal cord signaling continues to influence cardiovascular hemostasis weeks to years after myocardial infarction, we wanted to closely examine the gene expression signature of the dorsal horn after chronic MI. Surprisingly, the expression profile in this model demonstrated persistent activation of genes linked to an early stress response. This included upregulation of many IEGs, such as fosB, nr4a1, nr4a2, nr4a3, egr3, and TNF-α-induced protein 3 (tnfaip3). This was supported by molecular function GO enrichment analysis, as many of these genes were included in significant categories linked to transcription factors (Fig. 3B). We validated these findings with quantitative PCR and demonstrated that these changes were specific to the thoracic dorsal horn, as a similar upregulation was not shown in lumbar segments (Fig. 4, A–C).
Fig. 3.
Chronic myocardial ischemia (MI) triggers changes in gene expression linked to the stress response, immunity, and apoptosis. A and B: biological process (A) and molecular function Gene Ontology enrichment terms (B) for all differentially expressed genes in the dorsal horn after chronic MI compared with control pigs. Terms are ranked by significance using −log(adjusted P value).
Fig. 4.
Validation of chronic myocardial ischemia (MI)-induced activation of immediate-early genes and apoptosis in the dorsal horn. A−C: quantitative PCR of fosB (A), nuclear receptor subfamily 4 group A member 1 (nr4a1; B), and early growth response 3 (egr3; C) expression in thoracic and lumbar dorsal horn tissue from control (n = 6) and chronic (n = 6) MI pigs. Each dot represents the log-transformed ∆CT value (where CT is threshold cycle) for each biological replicate. Solid lines represent the mean for each group. D: Western blots for cleaved caspase-3 (CC3) in thoracic dorsal horn tissue from control, acute MI, and chronic MI pigs. E: Western blots for cleaved caspase-3 in thoracic and matched lumbar dorsal horn tissue from chronic MI pigs. β-Actin and cleaved caspase-3 blots were derived from the same gel to ensure accurate representation from the β-actin loading control. Three biological replicates from distinct pigs are shown for each condition. NS, not significant.
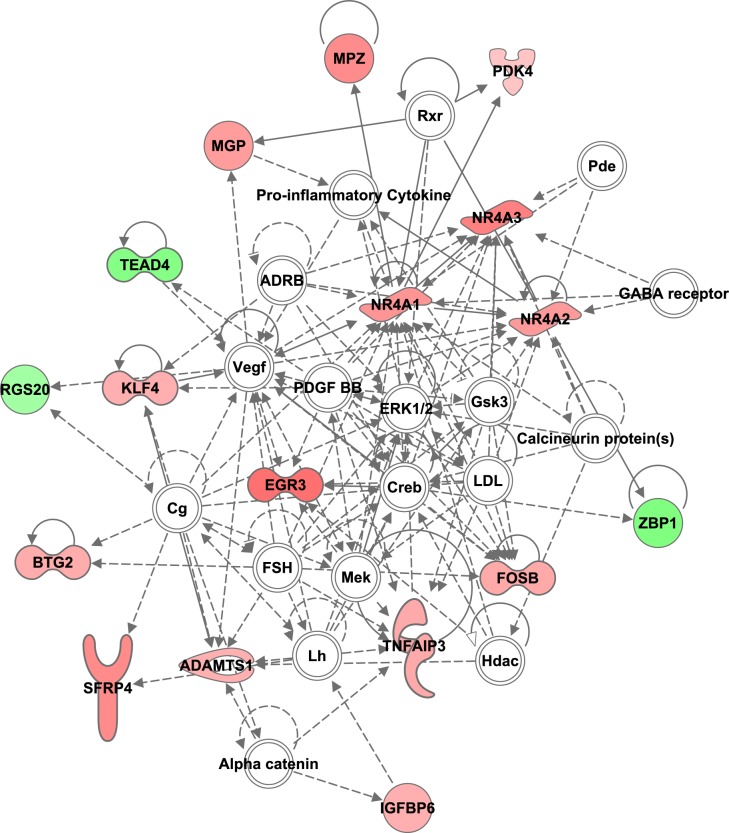
Biological process GO identified additional enrichment categories for the set of differentially expressed genes including those linked to activation of the immune response and cell death (Fig. 3A). Protein extracts from thoracic dorsal horn tissue revealed increased expression of cleaved caspase-3 in animals that were subjected to chronic MI compared with healthy and acute MI animals (Fig. 4D). This apoptosis seems to be isolated to the thoracic segments, as a similar increase in cleaved caspase-3 was not observed in lumbar segments from the same pigs exposed to chronic MI (Fig. 4E). Furthermore, ingenuity pathway analysis of all 46 dysregulated genes from our chronic model grouped many genes into a network that integrates the following cellular processes: cell cycle, cellular development, cell death, and survival (Fig. 5).
Fig. 5.
Dysregulated genes in the dorsal horn after chronic myocardial ischemia are joined in a network that regulates cell cycle, cellular development, cell death, and survival. Those genes that are upregulated compared with control samples are labeled in pink, and those downregulated are labeled in green.
We also examined the enrichment processes for the genes that were differentially expressed when comparing the acute ischemia MI group directly with the chronic MI group without using healthy animals. These genes were enriched in cellular differentiation and immune response (Benjamini-Hochberg-corrected P value of 0.04 for both), which were already categories uncovered from the previous enrichment analysis.
DISCUSSION
In the present study, we identified gene signatures in the thoracic spinal cord dorsal horn after acute and chronic MI. The ability of neurons to undergo transient and long-lasting phenotypic changes in the setting of stimulation is the hallmark of neural plasticity (26). After MI, such phenotypic changes have been documented throughout the peripheral nervous system including the stellate ganglion (SG) (2, 3, 10, 16, 20, 21, 35), DRG (20), and intrinsic neurons of the heart (5, 20, 27). We recently described changes in global gene transcription in the SG and DRG after myocardial injury that likely contribute to the generation of these pathological and molecular phenotypes (10). Within the central nervous system, similar profiles are lacking, and their identification is important given the prominent role of dorsal horn processing of afferent neural signaling from the heart. To our knowledge, this study is the first to describe the global changes in gene transcription that occur within the thoracic spinal cord dorsal horn after acute and chronic MI. These profiles are unique but contain some overlapping enrichment themes including activation of IEGs.
For many years, it has been known that the plasticity of spinal neurons can be tracked, as their initial stimulation leads to the rapid activation of IEGs including Fos and Jun family members (13, 26). Our acute MI model is no exception. Four hours after the initiation of injury, we identified an increasing number of c-Fos-positive neurons in the thoracic dorsal horn. Whole genome transcriptome analysis of the same tissue also identified c-fos as one of the top upregulated differentially expressed genes. Not surprisingly, of the 31 remaining dysregulated genes, 4 genes were also upregulated IEGs, including fosB, dusp1, dusp2, and egr2. Thus, early activation of gene expression changes in the thoracic dorsal horn after MI appears similar to gene activation in the central nervous system after other types of peripheral stimulation and injury. Of course, there may be differences in the degree to which these genes are upregulated, the precise location and identity of the cells that undergo these changes, and the exact identity of IEGs induced. Further studies are required to shed light on these unknowns.
Within the thoracic dorsal horn in animals exposed to chronic MI, there appears to be a persistent activation of stress response genes, as 6 of the 46 dysregulated genes are well-known IEGs. Other models of chronic pain and inflammation have shown similar activation of IEGs days to weeks after the initiation of injury. Time-course studies of IEG expression in animal models of subacute and chronic inflammation have also shown that the duration of IEG expression is consistent with the duration of inflammation (1, 15, 19). This suggests that injury likely continued throughout the entire 8-wk duration of ischemia in our chronic model. Even more, these same studies demonstrated that some IEGs peak early and have transient expression (i.e., c-Fos), whereas others peak late and have longer, more persistent expression (i.e., FosB) (15). This is consistent with our results, as fosB was significantly activated in acute and chronic ischemic models, whereas c-fos was significantly upregulated in the acute model alone. Nevertheless, further studies examining the level of expression of these IEGs at various time points throughout chronic ischemia are needed to fully trend their expression pattern.
In addition to stress response, immune system activation and apoptosis were also enrichment categories for the set of differentially expressed genes from the chronic MI model. This provides additional evidence to suggest that there is ongoing injury throughout the entire 8-wk incubation period after coronary artery occlusion. Previous work on the same model noted similar enrichment in the SG and DRG (10), and thus these findings could be a generalized nervous system response to chronic MI. These results could be affected by exposure to ischemia during tissue harvesting or episodes of poor spinal cord perfusion in the setting of MI, although healthy tissues were harvested in a similar fashion serving as controls, and no major hemodynamic events were noted in the animals during their 8-wk incubation. Furthermore, we also cannot eliminate the potential influence of transsynaptic-independent processes on changes in gene transcription. For example, the release of cytokines and growth factors at the site of ischemic injury could travel systemically to the spinal cord or a gradual progression to heart failure in the setting of left ventricular dysfunction could lead to total body phenotypic shifts that secondarily stress the central nervous system. Nevertheless, quantitative PCR showing no significant activation of select differentially expressed genes in lumbar dorsal horn tissue would suggest that there exists some local specific mechanism to gene transcription changes, such as transsynaptic regulation.
In conclusion, we describe the global transcriptome changes in the dorsal horn after acute and chronic MI. These signatures highlight the importance of a specific profile of IEGs that are activated as MI progresses over time from acute to chronic injury. In the future, it will be essential to determine the functional significance of why some IEGs have early transient upregulation (c-fos and dusp1) and others peak late (nr4a1 and egr3), whereas fosB uniquely has sustained activation. These patterns may provide insight into the mechanisms behind the plasticity of the central nervous system after organ injury. They may also act as potential targets for limiting the negative consequences of MI that act through the nervous system, such as fatal arrhythmias and heart failure.
GRANTS
This work was funded by intramural department funds. A. Mahajan is supported by National Heart, Lung, and Blood Institute (NHLBI) Research Project Grant R01-HL-084261. K. Howard-Quijano is supported by a Foundation for Anesthesia Education and Research Mentored Research Training Grant and NHLBI Research Project Grant K08-HL-135418. L. A. Saddic is supported by a Society of Cardiovascular Anesthesiologists-International Anesthesia Research Society Starter Grant.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
L.A.S., K.H.-Q., K.S., M.E., and A.M. conceived and designed research; L.A.S., K.H.-Q., J.K., Y.K., E.A.D., D.H., and K.S. performed experiments; L.A.S., K.H.-Q., J.K., Y.K., E.A.D., D.H., K.S., M.E., and A.M. analyzed data; L.A.S., K.H.-Q., D.H., K.S., M.E., and A.M. interpreted results of experiments; L.A.S. and A.M. prepared figures; L.A.S. drafted manuscript; L.A.S., K.H.-Q., J.K., Y.K., E.A.D., D.H., K.S., M.E., and A.M. edited and revised manuscript; L.A.S., K.H.-Q., J.K., Y.K., E.A.D., D.H., K.S., M.E., and A.M. approved final version of manuscript.
ACKNOWLEDGMENTS
We thank Christian Wooten and Andyshea Afyouni for the technical assistance with this project.
REFERENCES
- 1.Abbadie C, Besson JM. c-fos expression in rat lumbar spinal cord during the development of adjuvant-induced arthritis. Neuroscience 48: 985–993, 1992. doi: 10.1016/0306-4522(92)90287-C. [DOI] [PubMed] [Google Scholar]
- 2.Ajijola OA, Yagishita D, Patel KJ, Vaseghi M, Zhou W, Yamakawa K, So E, Lux RL, Mahajan A, Shivkumar K. Focal myocardial infarction induces global remodeling of cardiac sympathetic innervation: neural remodeling in a spatial context. Am J Physiol Heart Circ Physiol 305: H1031–H1040, 2013. doi: 10.1152/ajpheart.00434.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ajijola OA, Yagishita D, Reddy NK, Yamakawa K, Vaseghi M, Downs AM, Hoover DB, Ardell JL, Shivkumar K. Remodeling of stellate ganglion neurons after spatially targeted myocardial infarction: neuropeptide and morphologic changes. Heart Rhythm 12: 1027–1035, 2015. doi: 10.1016/j.hrthm.2015.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Armour JA. Myocardial ischaemia and the cardiac nervous system. Cardiovasc Res 41: 41–54, 1999. doi: 10.1016/S0008-6363(98)00252-1. [DOI] [PubMed] [Google Scholar]
- 5.Armour JA, Linderoth B, Arora RC, DeJongste MJ, Ardell JL, Kingma JG Jr, Hill M, Foreman RD. Long-term modulation of the intrinsic cardiac nervous system by spinal cord neurons in normal and ischaemic hearts. Auton Neurosci 95: 71–79, 2002. doi: 10.1016/S1566-0702(01)00377-0. [DOI] [PubMed] [Google Scholar]
- 6.Bahrami S, Drabløs F. Gene regulation in the immediate-early response process. Adv Biol Regul 62: 37–49, 2016. doi: 10.1016/j.jbior.2016.05.001. [DOI] [PubMed] [Google Scholar]
- 7.Deer TR, Mekhail N, Provenzano D, Pope J, Krames E, Leong M, Levy RM, Abejon D, Buchser E, Burton A, Buvanendran A, Candido K, Caraway D, Cousins M, DeJongste M, Diwan S, Eldabe S, Gatzinsky K, Foreman RD, Hayek S, Kim P, Kinfe T, Kloth D, Kumar K, Rizvi S, Lad SP, Liem L, Linderoth B, Mackey S, McDowell G, McRoberts P, Poree L, Prager J, Raso L, Rauck R, Russo M, Simpson B, Slavin K, Staats P, Stanton-Hicks M, Verrills P, Wellington J, Williams K, North R; Neuromodulation Appropriateness Consensus Committee . The appropriate use of neurostimulation of the spinal cord and peripheral nervous system for the treatment of chronic pain and ischemic diseases: the Neuromodulation Appropriateness Consensus Committee. Neuromodulation 17: 515–550, 2014. doi: 10.1111/ner.12208. [DOI] [PubMed] [Google Scholar]
- 8.Ding X, Ardell JL, Hua F, McAuley RJ, Sutherly K, Daniel JJ, Williams CA. Modulation of cardiac ischemia-sensitive afferent neuron signaling by preemptive C2 spinal cord stimulation: effect on substance P release from rat spinal cord. Am J Physiol Regul Integr Comp Physiol 294: R93–R101, 2008. doi: 10.1152/ajpregu.00544.2007. [DOI] [PubMed] [Google Scholar]
- 9.Fukuda K, Kanazawa H, Aizawa Y, Ardell JL, Shivkumar K. Cardiac innervation and sudden cardiac death. Circ Res 116: 2005–2019, 2015. doi: 10.1161/CIRCRESAHA.116.304679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao C, Howard-Quijano K, Rau C, Takamiya T, Song Y, Shivkumar K, Wang Y, Mahajan A. Inflammatory and apoptotic remodeling in autonomic nervous system following myocardial infarction. PLoS One 12: e0177750, 2017. doi: 10.1371/journal.pone.0177750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herdegen T, Kovary K, Leah J, Bravo R. Specific temporal and spatial distribution of JUN, FOS, and KROX-24 proteins in spinal neurons following noxious transsynaptic stimulation. J Comp Neurol 313: 178–191, 1991. doi: 10.1002/cne.903130113. [DOI] [PubMed] [Google Scholar]
- 12.Hua F, Harrison T, Qin C, Reifsteck A, Ricketts B, Carnel C, Williams CA. c-Fos expression in rat brain stem and spinal cord in response to activation of cardiac ischemia-sensitive afferent neurons and electrostimulatory modulation. Am J Physiol Heart Circ Physiol 287: H2728–H2738, 2004. doi: 10.1152/ajpheart.00180.2004. [DOI] [PubMed] [Google Scholar]
- 13.Hunt SP, Pini A, Evan G. Induction of c-fos-like protein in spinal cord neurons following sensory stimulation. Nature 328: 632–634, 1987. doi: 10.1038/328632a0. [DOI] [PubMed] [Google Scholar]
- 14.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36, 2013. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lantéri-Minet M, de Pommery J, Herdegen T, Weil-Fugazza J, Bravo R, Menétrey D. Differential time course and spatial expression of Fos, Jun, and Krox-24 proteins in spinal cord of rats undergoing subacute or chronic somatic inflammation. J Comp Neurol 333: 223–235, 1993. doi: 10.1002/cne.903330208. [DOI] [PubMed] [Google Scholar]
- 16.Li Z, Wang M, Zhang Y, Zheng S, Wang X, Hou Y. The effect of the left stellate ganglion on sympathetic neural remodeling of the left atrium in rats following myocardial infarction. Pacing Clin Electrophysiol 38: 107–114, 2015. doi: 10.1111/pace.12513. [DOI] [PubMed] [Google Scholar]
- 17.Liao S-Y, Liu Y, Zuo M, Zhang Y, Yue W, Au K-W, Lai W-H, Wu Y, Shuto C, Chen P, Siu C-W, Schwartz PJ, Tse H-F. Remodelling of cardiac sympathetic re-innervation with thoracic spinal cord stimulation improves left ventricular function in a porcine model of heart failure. Europace 17: 1875–1883, 2015. doi: 10.1093/europace/euu409. [DOI] [PubMed] [Google Scholar]
- 18.Matthies H. In search of cellular mechanisms of memory. Prog Neurobiol 32: 277–349, 1989. doi: 10.1016/0301-0082(89)90024-5. [DOI] [PubMed] [Google Scholar]
- 19.Menétrey D, Gannon A, Levine JD, Basbaum AI. Expression of c-fos protein in interneurons and projection neurons of the rat spinal cord in response to noxious somatic, articular, and visceral stimulation. J Comp Neurol 285: 177–195, 1989. doi: 10.1002/cne.902850203. [DOI] [PubMed] [Google Scholar]
- 20.Nakamura K, Ajijola OA, Aliotta E, Armour JA, Ardell JL, Shivkumar K. Pathological effects of chronic myocardial infarction on peripheral neurons mediating cardiac neurotransmission. Auton Neurosci 197: 34–40, 2016. doi: 10.1016/j.autneu.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nguyen BL, Li H, Fishbein MC, Lin SF, Gaudio C, Chen PS, Chen LS. Acute myocardial infarction induces bilateral stellate ganglia neural remodeling in rabbits. Cardiovasc Pathol 21: 143–148, 2012. doi: 10.1016/j.carpath.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Odenstedt J, Linderoth B, Bergfeldt L, Ekre O, Grip L, Mannheimer C, Andréll P. Spinal cord stimulation effects on myocardial ischemia, infarct size, ventricular arrhythmia, and noninvasive electrophysiology in a porcine ischemia-reperfusion model. Heart Rhythm 8: 892–898, 2011. doi: 10.1016/j.hrthm.2011.01.029. [DOI] [PubMed] [Google Scholar]
- 23.Oh YS, Jong AY, Kim DT, Li H, Wang C, Zemljic-Harpf A, Ross RS, Fishbein MC, Chen PS, Chen LS. Spatial distribution of nerve sprouting after myocardial infarction in mice. Heart Rhythm 3: 728–736, 2006. doi: 10.1016/j.hrthm.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 24.Oo TF, Henchcliffe C, James D, Burke RE. Expression of c-fos, c-jun, and c-jun N-terminal kinase (JNK) in a developmental model of induced apoptotic death in neurons of the substantia nigra. J Neurochem 72: 557–564, 1999. doi: 10.1046/j.1471-4159.1999.0720557.x. [DOI] [PubMed] [Google Scholar]
- 25.Pellegrini-Giampietro DE, Zukin RS, Bennett MV, Cho S, Pulsinelli WA. Switch in glutamate receptor subunit gene expression in CA1 subfield of hippocampus following global ischemia in rats. Proc Natl Acad Sci USA 89: 10499–10503, 1992. doi: 10.1073/pnas.89.21.10499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pérez-Cadahía B, Drobic B, Davie JR. Activation and function of immediate-early genes in the nervous system. Biochem Cell Biol 89: 61–73, 2011. [DOI] [PubMed] [Google Scholar]
- 27.Rajendran PS, Nakamura K, Ajijola OA, Vaseghi M, Armour JA, Ardell JL, Shivkumar K. Myocardial infarction induces structural and functional remodelling of the intrinsic cardiac nervous system. J Physiol 594: 321–341, 2016. doi: 10.1113/JP271165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schwartz PJ, Motolese M, Pollavini G, Lotto A, Ruberti U, Trazzi R, Bartorelli C, Zanchetti A; Italian Sudden Death Prevention Group . Prevention of sudden cardiac death after a first myocardial infarction by pharmacologic or surgical antiadrenergic interventions. J Cardiovasc Electrophysiol 3: 2–16, 1992. doi: 10.1111/j.1540-8167.1992.tb01090.x. [DOI] [Google Scholar]
- 29.Sheng M, Greenberg ME. The regulation and function of c-fos and other immediate early genes in the nervous system. Neuron 4: 477–485, 1990. doi: 10.1016/0896-6273(90)90106-P. [DOI] [PubMed] [Google Scholar]
- 30.Team RC. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing, 2015. [Google Scholar]
- 31.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7: 562–578, 2012. doi: 10.1038/nprot.2012.016. [Erratum in Nat Protoc 9: 2513, 2014.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28: 511–515, 2010. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tullai JW, Schaffer ME, Mullenbrock S, Sholder G, Kasif S, Cooper GM. Immediate-early and delayed primary response genes are distinct in function and genomic architecture. J Biol Chem 282: 23981–23995, 2007. doi: 10.1074/jbc.M702044200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vaseghi M, Shivkumar K. The role of the autonomic nervous system in sudden cardiac death. Prog Cardiovasc Dis 50: 404–419, 2008. doi: 10.1016/j.pcad.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang S, Zhou X, Huang B, Wang Z, Liao K, Saren G, Lu Z, Chen M, Yu L, Jiang H. Spinal cord stimulation protects against ventricular arrhythmias by suppressing left stellate ganglion neural activity in an acute myocardial infarction canine model. Heart Rhythm 12: 1628–1635, 2015. doi: 10.1016/j.hrthm.2015.03.023. [DOI] [PubMed] [Google Scholar]
- 36.Williams S, Evan G, Hunt SP. C-fos induction in the spinal cord after peripheral nerve lesion. Eur J Neurosci 3: 887–894, 1991. doi: 10.1111/j.1460-9568.1991.tb00100.x. [DOI] [PubMed] [Google Scholar]
- 37.Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16: 284–287, 2012. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang L, Lu Y, Zhou X, Tang B. Spinal cord stimulation: a potential therapeutic approach for post-myocardial infarction patients. Int J Cardiol 203: 1129–1130, 2016. doi: 10.1016/j.ijcard.2015.09.060. [DOI] [PubMed] [Google Scholar]