Abstract
A simple, in vitro intestinal model recapitulating key aspects of crypt architecture and physiology would facilitate our understanding the impact of drugs, foods and microbial metabolites on the intestine. To address the limitations of previously reported intestinal in vitro platforms, we developed a planar crypt array that replicated the spatial segregation and physiologic responses of primary mouse intestinal epithelial cells in the large intestine. Collagen was coated across an impermeable film possessing an array of microholes creating two regions of distinct stiffness and porosity (above and outside the microholes). Primary mouse colon epithelial cells formed a continuous monolayer across the array with a proliferative cell zone above the microholes and a nonproliferative or differentiated cell region distant from the microholes. Formation of a chemical gradient of growth factors across the array yielded a more complete or in vivo-like cell segregation of proliferative and differentiated cells with cell migration outward from the proliferative cell zone into the differentiated zone to replace apoptotic dying cells much as occurs in vivo. Short chain fatty acids (microbial metabolites) applied to the luminal surface of the crypt array significantly impacted the proliferation and differentiation of the cells replicating the known in vivo effects of these fatty acids. Importantly this planar crypt array was readily fabricated and maintained, easily imaged with properties quantified by microscopy, and compatible with reagent addition to either the luminal or basal fluid reservoirs. The ability to observe simultaneously stem/proliferative and differentiated cell behavior and movement between these two compartments in response to drugs, toxins, inflammatory mediators or microbial metabolites will be of widespread utility.
Introduction
The mammalian colon is lined with a single layer of epithelial cells which invaginate into the underlying mesenchyme to form tubular structures known as crypts. The proliferative compartment of the colon is located at the crypt base where the stem cells and transit-amplifying cells reside. These cells fuel the rapid renewal (5 days in mice1) of intestinal epithelial cells on the luminal intestinal surface where most of the non-proliferative cells reside.2 This polarity of cellular organization is thought to be maintained by a balance of biochemical and biophysical microenvironments in vivo.3–5 Growth factor concentrations are high or low at the crypt base or luminal surface, respectively, while differentiation factors possess an inverse gradient concentration.3–5 Gradients of extracellular matrix (ECM) proteins in the underlying lamina propria along the crypt’s long axis are also believed to be factors in stem cell self-renewal, proliferation, and differentiation.6, 7
Despite intense research, much remains to be understood about cellular patterning in the intestine due to the challenges in the study of this tissue in vitro. 8, 9 Stem cells dervived from the colonic epithelium can be cultured as organoids embedded in Matrigel with addition of soluble growth factors (Wnt-3A, R-spondin, Noggin and epidermal growth factor [EGF]).10–15 While this transformative method enables long-term colonic cell culture with all cell lineages found in vivo, the organoids lack the biochemical and biophysical gradients requited to form full colonic crypts identical to that in vivo. Proliferative and differentiated cells are not fully segregated as they are in vivo, although microdomains within the spherical structure form buds that have some structural similarities to crypts.10 Murine colonoids (organoids formed from the large intestine) cultured in specially designed microfluidic systems placing the organoids under a linear gradient of Wnt3A and R-spondin1 polarize forming distinct proliferative and differentiated cell zones. Thus a simple linear gradients of two growth factors is sufficient to recreate many aspects of intestinal cell compartmentalization observed in vivo.16 Varying biomechanical properties such as stiffness using synthetic scaffolds have been shown to be necessary for modulating stem cell renewal and differentiation,17 however, mimicking in vivo ECM properties for cell culture in embedded systems such as organoids is very challenging. An additional limitation is the complex and enclosed, budding structure of organoids that makes it difficult to image this tissue especially in a high-throughput manner. Furthermore, by being buried within the hydrogel with the cells’ luminal brush border facing the interior of the structure, molecular transport studies and exposure of the luminal cell surface to compounds of interest such as drugs or microbial metabolites, cannot be readily performed.
A traditional monolayer culture system with a stem/proliferative compartment and a differentiated cell zone would enable ready access to the luminal colonic epithelial surface of the cells as well as enable facile imaging and molecular transport measurements. Several groups have dissociated intestinal organoids and then cultured them on a thin ECM coating over a porous membrane.18, 19 These systems replicate either the stem cell or differentiated cell compartment depending on the media composition. Recently, our group developed a three-dimensional platform to recreate the crypt architecture and cell zones. The hydrogel scaffolds with the same spacing, size and shape of mouse20 and human21 colonic crypts were also constructed on a porous membrane so that the luminal and basal surfaces were accessible and in contact with different fluid compartments. Mouse and human colonic epithelial cells readily grew across the scaffold covering its surface and lining the microwells with a monolayer of cells. The growth factor gradient induced by placing the growth factors only in basal and not luminal reservoirs drove polarization of the in vitro crypts with stem/proliferative cells restricted to the crypt base where high growth factor concentrations were present. These basal proliferative cells migrated towards the lumen, differentiating and ceasing to proliferate as they moved towards the luminal tissue surface (with its low growth factor concentration) and formed a cell region populated exclusively with differentiated cells. An advantage of this platform, in addition to recreating an in vitro crypt, was the accessibility of both the luminal and basal reservoirs for either reagent addition or sampling for subsequent assay. Despite this advance, challenges existed in using these arrays for high-throughput screening. The in vitro crypts were best viewed with high resolution confocal microscopy due to their three-dimensional nature followed by image reconstruction to fully interrogate the cells within the crypts. This approach significantly limited the throughput of assays used in screening compounds such as drugs and microbial products.21
To address these challenges, we report an in vitro, intestinal cell culture platform that replicates the cell compartmentalization of crypts but as a monolayer i.e. a flattened or two-dimensional crypt with controllable dimensions. A microdevice with an array of microholes in an impermeable film was created and then overlaid with a collagen layer so that a hydrogel covered the film and microholes. The stiffness of the surface overlaying the microholes vs. that over the impermeable film was distinct owing to the very thin depth of the collagen. Primary murine colonic epithelial cells were cultured on the collagen-covered microhole array and the impact of the change in stiffness on the cell phenotype was measured. In addition, the microarray was constructed so that both the luminal and basal surfaces were in contact with different fluid reservoirs. When the basal but not luminal reservoir possessed growth factors, the microholes acted as a growth factor source to support nearby intestinal stem cells. The location of stem/proliferative and differentiated cells within the flattened crypt structures was assessed as well as the distance of the various cell types from the microholes when Wnt, Noggin and R-spondin were supplied via the microholes to the overlying tissue. The compatibility of the flattened crypts for facile, high-quality imaging was assessed by assaying cellular changes induced by short chain fatty acids in cells adjacent to and distant from a microhole. Cell migration outward from the proliferating cells above the microholes was quantified and compared to that of in vivo crypts. We envision this platform to be an efficient system for screening the impact on both stem/proliferative and differentiated cells in the rapidly growing area of gastrointestinal pharmaceuticals and nutriceuticals.
Materials and Methods
Microdevice fabrication
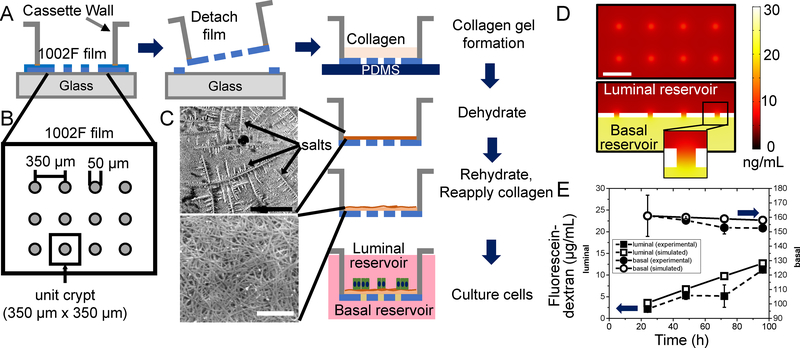
An array of 10X10 microholes (50 μm in diameter) in a thin photoresist film was fabricated by patterning through holes in a thin photoresist film. The patterned photoresist film was mounted onto a modified Transwell cassette and overlaid with a contiguous but thin collagen layer for cell culture. To create the patterned photoresist film, a layer (20 μm thick) of 1002F50 photoresist22 was spincoated onto a glass slide at 1500 rpm for 30 s and then solidified by baking for 30 min at 95°C. The photoresist was exposed to ultraviolet light (500 mJ) through a photomask with an array of open circles (50 μm diameter). The exposed photoresist was then developed in propylene glycol methyl ether acetate (PGMEA) and baked at 95°C for 12 h to create an array of microwells of very shallow depth (20 μm). Then the film on the glass slide was soaked for greater than 15 h to weaken the adhesion of the film to the glass so that the film could be readily transferred to a Transwell frame. The membrane on the base of a standard 12-well Transwell (Corning) was completely removed using a tweezer and then the Transwell frame was attached to the photoresist side of the patterned microwell array using 3M double-sided medical tape. At this point, the Transwell was rigidly affixed to the 1002F film overlying the glass slide. The slide was detached from the Transwell-1002F film by slowly lifting the Transwell with attached film from the glass slide and then trimming any film extending outside of the Transwell.
To prepare the final microdevice for mouse colonic cell culture, a thin layer of collagen was formed across the surface of the on the microwell array. First, the 1002F film within the Transwell was plasma treated for 5 min to improve its hydrophilicity and aqueous wetting properties. Then the plasma-treated Transwells with patterned 1002F film were placed onto a PDMS (Dow corning Sylgard 184 kit)-coated petri dish. Neutralized collagen (200 μL of rat tail collagen I in 0.02N acetic acid solution, Corning, neutralized with sodium hydroxide, sodium bicarbonate, HEPES and PBS23) was dispensed onto the 1002F film surface within the Transwell. The collagen was gelled by incubation for 1 h at 37°C. The collagen gel was then dried in a 40°C oven for 16 h resulting a thin layer of collagen covered with salt crystals and overlaying the photopatterned 1002F film. The thin collagen membrane spanned the gap across the holes on the 1002F film. The collagen-coated 1002F surface in the Transwell was gently rinsed with water to remove the salt, sterilized with 70% ethanol and washed with sterile PBS. Just prior to cell culture, 50 μg/mL of rat tail collagen I (Corning) in PBS was added and incubated for at least 1 h at 37°C with a goal of further enhancing cell adhesion to the collagen film.
COMSOL simulation and fluorescein-dextran diffusion experiments
COMSOL Multiphysics (COMSOL Inc., Burlington, MA) was used to model the diffusion of the growth factors through the collagen covered microholes. The geometry of the microhole array and the luminal/basal reservoirs was incorporated into the model. The diffusion coefficient in the thin collagen layer was assumed to be equivalent to that in the aqueous medium. This assumption is reasonable since the collagen layer in this platform was only 5 μm thick (and the average time through the membrane for even a large growth factor would be under 1 s). To simulate the diffusion of the growth factors (39.7 kDa, Wnt-3A; 40 kDa, R-spondin) through the microhole array, diffusion of fluorescein-dextran (40 kDa) was used. Fluorescein-dextran (Sigma, St. Louis, MO) was dissolved in PBS and placed in the basal reservoir (1.5 mL, 200 μg/mL) of the microdevice while the luminal reservoir contained PBS (0.5 mL) without the fluorescent dextran. Samples (50 μL) from the basal and luminal reservoirs were collected every 24 h. The fluorescence intensities of the samples were measured to estimate the concentration of the fluorescent dextran in each reservoir. The measured concentrations were compared to the simulated values in the microdevice using the previously reported diffusivity of 40 kDa fluorescein-dextran, 7.4×10−11 m2/s.24 Since the experimental data and the simulated value of the fluorescent dextran diffused through the collagen in the microdevice were similar, the same diffusion coefficient was used to simulate the diffusion of the growth factors in the microdevice.
Cell culture and EdU/alkaline phosphatase/immunofluorescence staining
Crypts from male and female murine intestines were isolated in buffer (2.0 mM EDTA and 0.5 mM DTT) as described previously.23 Mice were housed at the University of North Carolina, Chapel Hill. All cells used in these experiments had undergone fewer than 5 sub-cultures to insure chromosomal integrity.23 Mouse intestinal cells were cultured as monolayer on a flat surface of neutralized collagen gel (1 mL placed into a 6-well plate)23 under expansion medium (EM, Table S1). EM possesses Wnt3A, R-spondin3 and Noggin to support stem cells within the culture. EM was replaced with fresh EM every 2 days. The cells were subcultured every 3–5 days by degrading collagen with collagenase and dissociating the cells with 0.5 mM EDTA into clumps of cells.23 The monolayers were not fully dissociated into single cells since this results in a high stem cell mortality rate. When placing cells onto the surface of the microdevice, the cell suspension was diluted 1 to 2 in EM and overlaid onto the microdevice surface. The cells were cultured for 2 days with EM placed into the luminal and basal reservoirs of the Transwell cassette. On day 2, the luminal and basal media were changed as described in the text and Table 1, and the cells incubated for a day. On day 3, the luminal and basal media were replaced with fresh media (identical to that from day 2 replacement) and the cells incubated for another day. In some experiments, the medium in the luminal compartment was replaced differentiation medium (DM, Table S1) which is similar to EM but lacking in key growth factors required to support intestinal stem cells. For experiments with short chain fatty acids, each short chain fatty acid was added in the luminal medium at the concentration of 24 mM for acetate, 6 mM for propionate and 1 mM for butyrate.23
Table 1.
Media Conditions
| Name | First 2 days | Last 2 days |
|---|---|---|
| EM/EM | EM in the luminal / EM in the basal | EM in the luminal / EM in the basal |
| DM/EM | EM in the luminal / EM in the basal | DM in the luminal / EM in the basal |
| DM/DM | EM in the luminal / EM in the basal | DM in the luminal / DM in the basal |
EM: expansion medium with the growth factors (Wnt-3A, R-spondin, Noggin)
DM: differentiation medium without the growth factors
Cells in S phase of the cell cycle were incubated with a pulse of 5-ethynyl-2´-deoxyuridine (EdU) pulse following the manufacturers’ protocols (Click-iT EdU Alexa Fluor 647 Imaging kit, C10340, Thermo Fisher Scientific). EdU is a nucleotide analog incorporated into DNA as it is replicated in S phase. In brief, EdU (10 μM) was added to the luminal and basal media and the cells were incubated for 3 h at 37°C. The cells then were washed with PBS and fixed with 4% paraformaldehyde in PBS for 15 min and permeabilized with 0.5% Triton-X for 20 min at 25°C. EdU incorporated into cellular DNA was visualized by reaction with Alexa 647 via click chemistry following the manufacturer’s protocol (Click-iT EdU Alexa Fluor 647 Imaging kit, C10340, Thermo Fisher Scientific).
For pulse-chase experiments with EdU, cells were grown in EM for 2 days. On day 2, the luminal and basal media were removed and DM was then added to the luminal compartment while EM was added to the basal compartment, and the cells were incubated for a day. The media was replaced with fresh media (identical to that from day 2 replacement) and the cells incubated for another day. On day 4, EdU was added to the luminal and basal media and the cells incubated for 3 h as described above. The luminal and basal media then were replaced with fresh DM and EM, respectively and the cells incubated for 2 more days with media exchanges every day.
Cell monolayers were assayed for alkaline phosphatase (ALP) activity on day 4 of culture by incubation with ALP substrate mixture (Vector Red AP substrate kit, SK-5100, Vector Laboratories) in Tris buffer (pH 8.4) for 30 min at 37°C. Then cells were washed with PBS, fixed and permeabilized as described above. To label cell DNA, Hoechst 33342 (2 μg/mL, Sigma Aldrich, B2261) was incubated with cells (1 h, 25°C).
For immunofluorescence staining, primary antibodies against E-cadherin (1:200, 20874–1-AP, Proteintech), β-catenin (1:200, sc-7963, Santa Cruz Biotech), mucin 2 (1:200, sc-15334, Santa Cruz Biotechnology), and chromogrannin A (1:1500, ab15160, Abcam) and Alexa 488-conjugated goat anti-rabbit antibody (1:500, A11008, Life Technologies, used for E-cadherin, chromogrannin A and mucin 2), or Alexa 647-conjugated donkey anti-mouse antibody (1:500, 715–605-150, Jackson ImmunoResearch for β-catenin) were used. The cells were grown on the microdevices in EM for 2 days and then further cultured as described in the manuscript. On day 4, the cells were fixed with 4% paraformaldehyde in PBS for 15 min, and permeabilized with 0.5% Triton-X for 20 min at 25°C. To minimize nonspecific binding of antibodies, the cells were blocked with 3% BSA in PBS at °C for 1 h. Then each primary antibody that was diluted in 3% BSA in PBS with the dilution ratio recommended by the manufacturers as indicated above was added to the cells and incubated for at least 16 h at 4°C. The cells were then washed with 3% BSA in PBS 3 times, incubated with secondary antibody and Hoechst 33342 diluted both at 1:500 ratio with 3% BSA in PBS for 1 h at 25°C and finally washed with 3% BSA in PBS twice and then in PBS alone.
Cell imaging
The Transwell with cell layer from the microdevice was placed into a 12-well plate under PBS and imaged using an Olympus Fluoview FV3000 confocal microscope (10X, objective, numerical aperture of 0.4). Alexa 647, Vector-Red-ALP, and Hoechst 33342 were excited with a 640, 561, and 405 nm laser, respectively and the fluorescence emission was collected at 650–750 nm, 570–590 nm, and 430–470 nm, respectively. The images were analyzed with the Fiji software package.25 For all experiments, the numbers of sample sizes were estimated using a statistical power analysis26 based on the data from Wang et al 23 measuring EdU+ and ALP activities under varying conditions (α=0.05, β=0.85) using G*Power.27 A one-way ANOVA analysis was used for all statistical analyses of data obtained from the fluorescence microscopy images. Scanning electron microscopy (SEM) was performed using a FEI Quanta 200 ESEM microscope. For the SEM imaging, the samples were first fixed with 4% paraformaldehyde, dried with a critical point dryer (PVT-3, Tousimis Semidri, Rockville, MD) and then coated with 10 nm metal by a sputter coater (Cressington 108, Cresington Scientific Instruments, Watford, UK).
Stiffness measurements
The stiffness of the collagen above and in regions adjacent to the microholes in the 1002F film was measured in fluid (1X PBS) using an atomic force microscope (MFP3D, Asylum Research) to collect force vs indentation curves (2 samples, 10 points above and adjacent to the microholes for each sample). The silicon cantilever (0.03 N/m nominal spring constant) with an attached polystyrene bead (4.5 μm in diameter) was purchased from Novascan Technologies, Inc. All calibrations, data collection, and data analysis were performed using the Asylum software. The spring constant of the cantilever was more accurately determined by recording its thermal motion (0.03175 N/m). Force vs indentation curves were recorded by applying 10 nN to the sample, and the sample stiffness (kPa) obtained by fitting 90% of each curve (140 nm average indent) into the Hertz model.
Results and Discussion
Design, fabrication, and characterization of a microdevice to support planar intestinal crypts
We sought to develop a microdevice that could support a planar monolayer of primary murine intestinal cells with distinct stem/proliferative and differentiated cell zones. Stem cells require provision of growth factors such as Wnt3A, R-spondin and Noggin to maintain their ability to self-renew and produce proliferative progeny. In the absence of these growth factors, the cells rapidly differentiate to form the various nondividing cell types, for example, absorptive enterocytes. Thus, the planar-crypt microdevice must spatially control the location of these growth factors to provide growth factors such as Wnt3A, R-spondin and Noggin to support the stem cells while removing this factors from the differentiated cell zones. Even in the presence of these factors, stem cells rapidly differentiate upon loss of extracellular matrix (ECM) contacts or upon placement on matrices with inappropriate stiffness. For this reason, the microdevice must also supply the stem cells with a suitable ECM-containing surface of the appropriate stiffness (100 to 1.3 kPa for organoids, for example17). Finally, an optical clear device would support high quality optical imaging for assay readout and a simple operation protocol would insure wide-spread adoption.
To meet these demanding requirements, a layer of collagen was coated onto an impermeable film patterned with microholes. Growth factors might then be supplied selectively to the cells growing on the permeable collagen layer above the microholes by placement of factor-containing media below the impermeable film in the basal reservoir (Fig 1A). To fabricate the device, a layer (40 μm thick by SEM) of 1002F was coated onto a glass slide, and photopatterned using a mask with an array of 10 X 10 opaque circles (50 μm diameter, edge to edge distance of 300 μm, Fig 1B). The 1002F was removed from the glass slide by incubation in water and the film was transferred to the base of a Transwell insert (from which the commercially attached membrane was removed) (Fig. 1A). The insert with attached 1002F film was placed onto a PDMS surface and collagen loaded into the reservoir to form a 1 mm thick collagen gel above the micropatterned 1002F. The collagen was then dehydrated leaving a compacted collagen layer spanning the surface and bridging across the microholes, and covered with salt crystals (the arrow in Fig. 1C upper panel). Rehydration of the collagen provided a dense yet porous collagen membrane (4.7 μm thick by confocal fluorescence microscopy) across the entire upper surface of the device (Fig. 1C lower panel). The stiffness of the collagen layer in regions above the microhole and the 1002F surface was significantly different at 55.83±19.21 kPa and 146.38±42.27 kPa (n=3, 10 points per sample, p < 0.0001), respectively, when measured by AFM with a 4.5 μm-diameter probe tip. The dehydration and rehydration process increased the collagen density across the 1002F surface since this step decreased the collagen thickness from 1 mm to 4.7 μm. In turn, this increased density likely increased the collagen stiffness which when not compacted possesses a reported stiffness of 10–1000 Pa depending on the concentration.23 The higher stiffness of the collagen-coated 1002F surface suggested that the collagen coating did not entirely masked the stiffness of the 1002F film (typical stiffness of EPON resin is a few GPa28, 29). It should be noted that the indentation depth by AFM (~150 nm in this study) is typically shallower than that sensed by cells; therefore, it is possible that the cells may “feel” the underlying stiff 1002F film more than can be sensed by the AFM tip.30, 31
Figure 1.
Fabrication and characterization of the planar-crypt array microdevice. A) Fabrication of the microdevice. Shown is a side view of the 1002F film (blue) patterned with an array of microholes on the surface of a glass slide. After film removal from the glass surface, a layer of collagen (tan) is overlaid onto the 1002F film and dehydrated leaving a compacted collagen layer (brown). After rehydration cells (green) and media (pink) are placed onto the collagen-coated 1002F film. B) Top-view schematic of the film providing the pattern dimensions. A unit crypt is defined as 350 μm x 350 μm square with a microhole in the center. C) Scanning electron micrograph of the top surface of the collagen matrix on the 1002F film before (upper) and after (lower) rehydration. The arrows indicate salt crystals. The black bar (upper panel) represents 200 μm and the white bar (lower panel) 2 μm. D) COMSOL model of the diffusion of a 40-kDa molecule from the basal to the luminal compartment through the microholes. Top panel: Concentration profile of the 40-kDa molecules in the plane just above (10 μm) the luminal surface of the 1002F film. Shown is the concentration profile near 8 microholes in the center of the array at 24 h after addition of the molecule (30 ng/mL) to the basal reservoir. The white scale bar indicates 300 μm. Lower panel: Cross-sectional view through 4 microholes in the 1002F film. Inset shows a cross-section through a single microhole. E) Experimental measurement (solid shapes, dashed lines) and simulation results (open shapes, solid line) of the diffusion of fluorescein-dextran (40 kDa) between the luminal (squares) and basal (circles) compartments over time. At time 0, fluorescein-dextran was added to the basal but not luminal reservoir. The data points represent the average and the standard deviation of the measured data points (n=2) while the lines indicate the simulated concentrations.
The ability of growth factors to transit through the 1002F microholes and collagen film was simulated with COMSOL (Fig. 1D, 1E). The diffusion coefficient of a 40-kDa fluorescein-dextran (similar to the molecular weight of Wnt-3A and R-spondin) in a hydrogel was previously measured as 7.4×10−11 m2/s.24 As a control, the movement of a fluorescein-dextran (40 kDa) over time from the basal to the luminal compartment was measured by sampling these reservoirs and compared to that predicted by the simulation. The results were in excellent agreement (Fig. 1E). The simulation predicts that 24 h after loading fluorescein-dextran (30 ng/mL) into only the basal compartment, the fluorescein-dextran concentration in the luminal compartment was at its greatest level (7.3–8.3 ng/mL) over the collagen-coated microhole (10 μm above the surface) and declining rapidly in all directions to 4.5–5.5 ng/mL at a point equidistant between the microholes. The steepest gradient of growth-factor concentration drop was 18 pg/mL/μm suggesting that this value is sufficient to induce colonic epithelial cells to form two cell compartments, a stem/proliferative and differentiated cell compartment. Prior measurements reported that a gradient of 75 pg/mL/μm was sufficient to polarize a 200 μm-long murine colonoid24; however, this previously published work did not incorporate a stiffness or porosity gradient which may explain the requirement for a steeper gradient than that measured in this work. Since the above simulation did not incorporate turbulent or convective mixing, the gradient predicted by the simulation likely represents the minimal possible concentration difference along the 1002F surface. The simulation does, however, suggest that the collagen-coated, arrayed microholes could act as a localized source of growth-factors establishing a gradient of growth factor from the center of the microhole outward.
Growth of primary mouse intestinal epithelial cells on the collagen-coated, patterned film
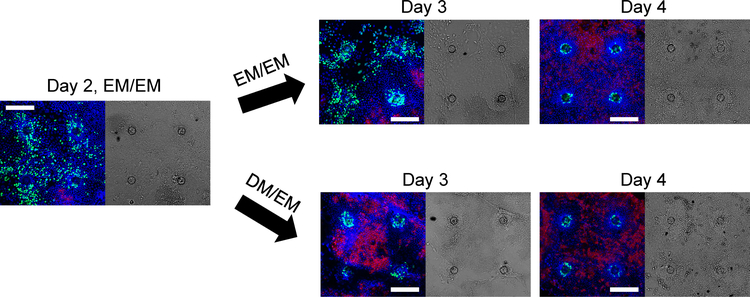
Cells were cultured on the collagen-coated 1002F films with patterned microholes for 4 days with EM or stem-cell supporting media in the luminal (upper) and basal (lower) reservoirs (EM/EM). Four days of culture time was chosen to mimic the typical life time (3–5 days) of murine colonic epithelial cells.32 Cell proliferation and differentiation as indicated by EdU incorporation and alkaline phosphatase (ALP) activity, respectively, was tracked over time (Fig. 2). At day 2, the cells exhibited robust proliferation as demonstrated by incorporation of EdU in cells across the film surface. In contrast, very little ALP activity was detected in the epithelial cells suggesting that under these conditions and at this time point, very few differentiated cells were present. At day 3 of culture, EdU+ cells remained present throughout the array accompanied by low levels of ALP activity. However, by day 4 of culture, distinctive cell patterning was present with circular regions of EdU+ cells extending 50–60 μm out from the microhole centers (Fig. 2, 3A, 3J). No EdU+ cells were present at distances greater than 70 μm from the through microcenter (Fig. 2, 3A, 3J). In contrast, low levels of ALP activity in cells at a distance greater than 90 μm from a microhole center remained present (Fig. 2, 3A, 3K). No cells within 70 μm of a microhole center expressed measurable ALP activity. Cells in the transition zone between 60–70 μm from a microhole center possessed little EdU incorporation or ALP activity suggesting that in these regions proliferative and differentiation signals were equally balanced.
Figure 2.
The time course of intestinal epithelial cell growth across the collagen-coated 1002F film of four microholes in an array. Confocal microscopy images along a single XY plane were taken on day 2, 3 and 4 with the media conditions indicated in the images (see also Table 1). Left panels are overlaid fluorescence images with green, red, blue representing EdU-incorporation, ALP activity and DNA-Hoechst 33342, respectively. Right panels are differential interference contrast images. After 2 days of growth in EM/EM, the media was switched to either fresh EM/EM or DM/EM. The white scale bar represents 200 μm with all images at the same magnification.
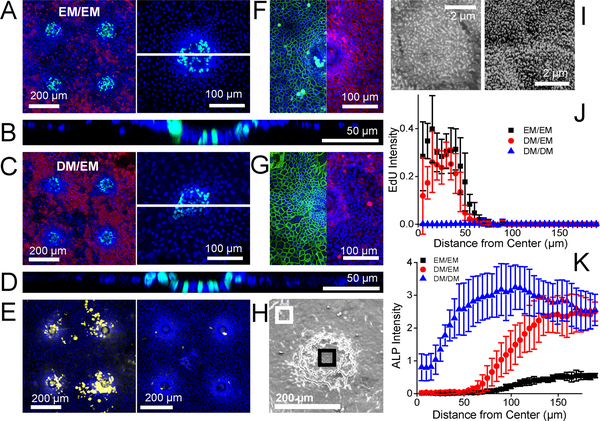
Figure 3.
Primary murine intestinal epithelial cell growth on the microdevice. A-D) Representative confocal fluorescence microscopy images of the cells on the array under EM/EM (A,B) or DM/EM (C,D). Left panels in A, C: Projected image of four planar crypts. Green, red, and blue represent EdU-incorporation, ALP activity and Hoechst 33342, respectively. Right panels in A, C: Projected images of a single planar crypt at higher magnification. The dashed white lines mark the locations of the cross-sectional confocal views through the monolayers shown in B and D. Green and blue represent EdU and Hoechst 33342, respectively. E) Fluorescent images of four planar crypts under DM/EM with yellow and blue representing mucin-2 (left panel) or chromogranin A (right panel) and Hoechst 33342, respectively. F-G) Projected split images of E-cadherin and β-catenin immunofluorescence staining of a single planar crypt under EM/EM (F) or DM/EM (G). Green, red and blue represent E-cadherin, β-catenin and Hoechst 33342. Each image was taken individually and then stitched at the same scale. H) Representative scanning electron microscopy image of a single planar crypt under DM/EM. Black and white boxes indicate the center region and the edge regions of the crypt shown at higher magnification in panel I. I) Representative high magnification SEM images of different regions in the planar crypt: cells over the center (left panel) or at the edge (right panel) of the crypt. J-K) Normalized fluorescence intensity of incorporated EdU (J) and ALP activity (K) in the radial direction outward from the center of the hole located at X = 0. The data points reflect the average and error bars the standard deviation (n=5). The fluorescence of a fluorescence image was measured in concentric rings of width 5 μm from the microhole center.
To understand this distinctive patterning despite the provision of EM with growth factors to all cells, cells were cultured on a collagen-coated 1002F film without microholes under EM/EM so that all cells experienced the stiff and impermeable underlying 1002F surface. At day 4 of culture, the cells demonstrated no EdU incorporation suggesting that the under these conditions the impact of the 1002F surface dominated and forced the cells to cease dividing (Fig. S1). Cells were also cultured on a collagen layer without the underlying 1002F film for 4 days under EM/EM (Fig. S1). Nearly all of these cells remained EdU+ during this time period. In the presence of a lower stiffness and higher porosity substrate, the growth factors were competent to drive cell proliferation. Thus, the changes in the material properties of the underlying matrix alone were sufficient to create two cellular zones on the array: a highly proliferative region and a nonproliferative compartment with evidence of cell differentiation. Porosity has a significant impact on stem cell behavior, though often in hydrogel systems its effect is coupled with stiffness, since crosslinking frequently is used to increase stiffness (also reducing pore size). As an example, the migration of mesenchymal stem cells was shown using PEG hydrogels to depend on scaffold porosity as well as stiffness and adhesivity.33 Additionally the porosity of a rigid polystyrene alters the growth of neurites derived from human stem cells.34 Porosity also impacts cell morphology of and cytoskeletal organization within epithelial cells. MDCK II cells, a model epithelial cell line, adopt a more flattened, spread out morphology and develop thicker actin stress fibers on nonporous substrate compared to that on a porous substrate.35, 36 The stiffness change between the collagen-covered microholes and the collagen-coated 1002F while small may also impact cell behavior. Stiffer surfaces have been shown to decrease protein levels of the Oct3/4 or Nanog pluripotency stem-cell markers in embryonic stem cells relative to softer surfaces suggesting that the harder surfaces can increase differentiation.37, 38 Finally, work by Wang and colleagues demonstrated that mouse and human intestinal stem cells ceased to proliferate and appeared to adopt a more differentiated phenotype on very stiff surfaces such as polystyrene or poly(dimethyl siloxane) relative to softer surfaces.23 In summary, these experiments are in good agreement with previous work by others showing that changes in porosity and stiffness can combine to alter cell fate. It should be noted that the EM contains ROCK inhibitor Y-27632, which is commonly used to inhibit anoikis in primary intestinal cell culture. Since ROCK signaling is known to modulate mechanosensing and mechanotransduction,39, 40 it is possible that the presence of Y-27632 alters the response to substrate stiffness.
The properties of the cells in the different regions of the arrays were further investigated to gain insight into the surface impact on cell behavior. A cross-section through the cells along the Z-axis confirmed that all regions on the surface grew as a monolayer (Fig. 3B). The cell outlines visualized by E-cadherin staining in Fig. 3F show that the cell density over the microholes (2.2±0.1 cells/100 μm2) was significantly different (n=3 crypts, p < 0.0005) from that in regions between the microholes (0.8±0.1 cells/100 μm2) suggesting that the cells over the microholes were more columnar in shape (as in the in vivo intestinal epithelium) while those in the intermediate regions adopted a wider more flattened shape. These results are consistent with the known responses of epithelial cells to porous and nonporous substrates noted above. However, cells in all regions of the surface expressed the adherens junction marker E-cadherin suggesting that cell-cell adhesion was maintained throughout the surface (Fig. 3F, green).41 β-catenin, a downstream effector of Wnt signaling that binds to cadherins in the adherens junction complex42 was also prominent on cells in all regions (Fig. 3F, red) indicating that all cells continue to experience Wnt-3A binding and signaling even though the cells far from the microholes fail to undergo Wnt-3-driven proliferation. These data demonstrate that stiffness and/or porosity were sufficient to initiate cell segregation into two regions: i) a proliferative region with few enterocytes and a high density of cells and ii) a nonproliferatve region higher in the enterocyte marker ALP and adopting a more flattened shape.
Creation of stem/proliferative cell compartment and differentiated cell zone
The absence of high ALP activity in the differentiated cells was possibly due to the continued presence of the growth factors, Wnt-3, Noggin and R-Spondin. Cells were cultured on the arrays in EM/EM for 2 days to enable the cells to attach and spread across the collagen-coated surface and then switched to DM/EM (luminal/basal) for an additional 2 days. DM or differentiation medium does not contain Wnt3A, R-spondin or Noggin and force-differentiates intestinal epithelial cells towards the enterocyte or absorptive cell lineage.20 To determine the fate of the cells exposed to DM/EM, cell proliferation and differentiation were tracked over time (Fig. 2). At day 3 of culture, proliferative cells were largely localized to the surface regions above the microholes while cells with ALP activity were restricted to other regions. By day 4, two distinct cells compartments were readily visualized. The proliferative cell compartment located only above the microholes was not statistically different in size from that created after 4 days of EM/EM (Fig. 3C, 3J). This suggests that cells over the microholes had ready access to the growth factors in EM via diffusion through the microhole. In contrast, the ALP activity in cells began to increase at 60 μm from the microhole center reaching a maximum activity at 130 μm. In these outer regions, the ALP activity was significantly increased (4.5-fold, n=5 crypts, p < 0.0005) from that produced by EM/EM (Fig. 3K). The DM in the upper reservoir along with the underlying surface directed the intestinal epithelial cells to a nonproliferative phenotype with greatly increased ALP activity creating differentiated cell zones (>130 μm) surrounding the proliferative cell zones (0–50 μm) above the microholes. This compartmentalized culture lasted at least up to day 6.
To confirm that the central proliferative zones were created by diffusion of growth factors through the microholes from the underlying EM, several controls were performed. Each control experiment cultured the cells on EM/EM for 2 days on the surface indicated below and then for a further 2 days in media as indicated below. In the first experiment, intestinal cells were cultured on a flexible collagen membrane without an underlying 1002F film and then placed into DM/EM for days 3–4 (Fig. S3A). Proliferative cells were present throughout the array surface at day 4 again suggesting that the growth factors readily diffused through the collagen layer. In the second control experiment, cells were cultured on a collagen-coated 1002F film without microholes and on day 3–4 placed into DM/EM. As expected, no EdU+ cells were present but ALP activity was apparent across the surface (Fig. S3B). In the third experiment, cells were cultured on a collagen-coated array of microholes and then on day 3–4 placed into DM/DM. These devices possessed no EdU+ cells at day 4 indicating that the collagen film above the microhole was not sufficient to support cell proliferation in the absence of growth factors (Fig. S2). The highest ALP activity was observed on these arrays with the ALP activity at 100–110 μm from the microhole center significantly greater (2.2X, n=5 crypts, p < 0.005) than that for the DM/EM culture (Fig. 3K). The wide difference in ALP activity of the DM/EM and DM/DM suggests that small amounts of growth factors likely diffused into the differentiated cell zones of the DM/EM cultures.
The planar crypt with proliferative and differentiation zones was further characterized by immunofluorescence and SEM. As shown in Fig. 3G, the cells in the planar crypt under DM/EM maintained cell-cell adhesions throughout the area as indicated by the presence of E-cadherin (Fig. 3G, green) while Wnt signaling, visualized by β-catenin (Fig. 3G, red), was localized predominantly near the microholes. Under DM/EM, cells in the differentiated zones lost access to growth factors enabling these cells to progress farther down the enterocyte differentiation pathway. SEM images revealed prominent microvilli, a characteristic of absorptive colonocyte, on cells away from the microhole in the differentiated zone and less dense and shorter microvilli on cells over the center of the microholes suggesting a less differentiated cell type (Fig. 3H,I). The presence of goblet cells in the planar crypt was also confirmed by immunofluorescence staining of mucin-2 (Fig. 3E, left). In contrast to the enterocytes the goblet cells were predominantly located near the microholes consistent with the placement of large numbers of goblet cells deep within the crypts in vivo.43 Lastly, the planar crypt possessed a small number (about 2 per planar crypt) of chromogrannin-A expressing enteroendocrine cells (Fig. 3E, right) which is consistent with the sparsity of this cell type in vivo.44 The planar crypt array reproduces the cell segregation found in vivo but in two-dimensions and in an easy to fabricate and maintain format.
Migration and death of intestinal epithelial cells on the planar crypt arrays
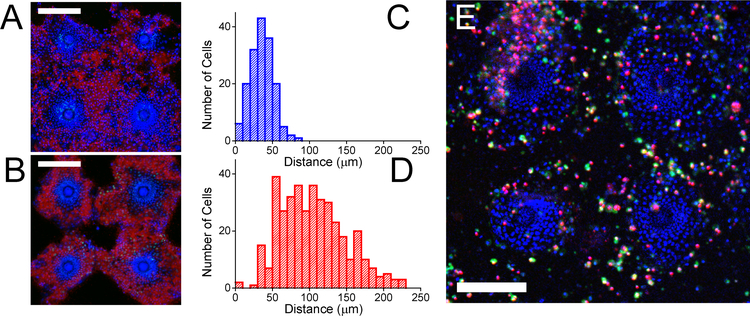
In the intestine in vivo, stem cells at the base of the crypt proliferate giving rise to transit amplifying cells that continue to divide and migrate up the long axis of the crypt toward the luminal surface of the intestine. As the cells approach the lumen, the cells differentiate into the nondividing lineages (enterocytes, goblet cells and enteroendocrine cells).45 To determine whether cells on the planar crypt arrays might recapitulate this orderly cell migrate from the proliferative cell compartment to the differentiated cell regions, cells on the collagen-coated microhole arrays were cultured in EM/EM for 2 days (days 1–2) and then polarized in DM/EM for a further 2 days (days 3–4). The cells were then incubated with EdU for three hours after which time the EdU was washed away and the cells cultured for an additional 2 days in DM/EM (days 5–6). The location of the EdU+ cells was measured on day 4 immediately after EdU incubation and again on day 6 (after the 2-day chase time). As expected on day 4, all EdU+ cells were located above the microholes in the proliferative cell zone (mean cell location of 36±15 μm from the microhole center, n=165 cells, Fig. 4A,C). However at day 6, the majority of EdU+ cells were located in the differentiated cell zones (mean cell location of 105±44 μm from the microhole center, n=386 cells, Fig. 4B,D) indicating that these cells migrated from the proliferative zone (pulse location) into the differentiated cell zone during the intervening 2 days. The locations of the EdU+ cells at day 6 were statistically different from their position at day 4 (p < 0.0001). Many of the EdU+ cells were also co-localized with the ALP activity measured at day 6 indicating that these day-4, proliferative cells differentiated during their outward migration. The average speed of these cells was 1.6±1.1 μm/h (the difference in the mean EdU+ cell location at 0 and 44 h after EdU washout divided by 44 h) which is similar to the cell migration velocity in the lower crypt in vivo (1.3 μm/h).32 The flow of cells from the stem/proliferative niche at the crypt base upward to the luminal surface of the intestine was recreated in the planar crypt array.
Figure 4.
Migration and viability of cells within the planar crypts. A) Cells were cultured under DM/EM for 4 days and then incubated with EdU for 3 h followed by cell fixation and staining. B) Cells were cultured and pulsed with EdU as in panel A. The cells were then cultured for an additional 2 days in EdU-free medium prior to fixation and staining. In panels A and B, EdU incorporation is shown in green, ALP activity in red, and Hoechst 33342 in blue while the white scale bar represents 200 μm. C,D) Histograms of the number of EdU+ cells at different distances from the center of the microholes immediately after the 3 h pulse (C) and after washout for 2 days (D) (n=5). E) Cell viability on the planar crypt arrays at day 4 of culture under DM/EM. Green, red, and blue represent fluorescein-annexin V, propidium iodide, and Hoechst 33342 fluorescence, respectively. The white scale bar depicts 200 μm.
In vivo epithelial cells on the luminal surface of the murine large intestine live an average of 5 days.32 After these luminal cells die, they are sloughed into the intestinal lumen making room for newly arriving cells.46 To understand the fate of the differentiated cells on the planar crypt arrays, cells were cultured in EM/EM for 2 days and then DM/EM for an additional 2 days. Necrotic and apoptotic cells were visualized by staining the arrays with propidium iodide (marking all dead cells) and fluorescein-annexin V (marking apoptotic cells) (Fig. 4E). Very few cells in the proliferative zones exhibited propidium iodide or annexin V fluorescence. In contrast, dead and dying cells were readily observed in the differentiated cell regions. In regions distant from the through holes, dying cells sometimes detached in a group leaving exposed regions of collagen-coated photoresist (Fig. 4). Taken together these data suggest that the progeny of the dividing cells in the proliferative zone migrate outward into the differentiated cell regions maturing into enterocytes as they move outward. At the end of their lifetime, these nondividing cells die by apoptosis and are replaced by newly arriving cells from the proliferative zone. These features replicate the life cycle of cells as they move from the crypt base to the luminal epithelium in vivo.
Effect of short chain fatty acids on the proliferation and differentiation of mouse intestinal epithelial cells
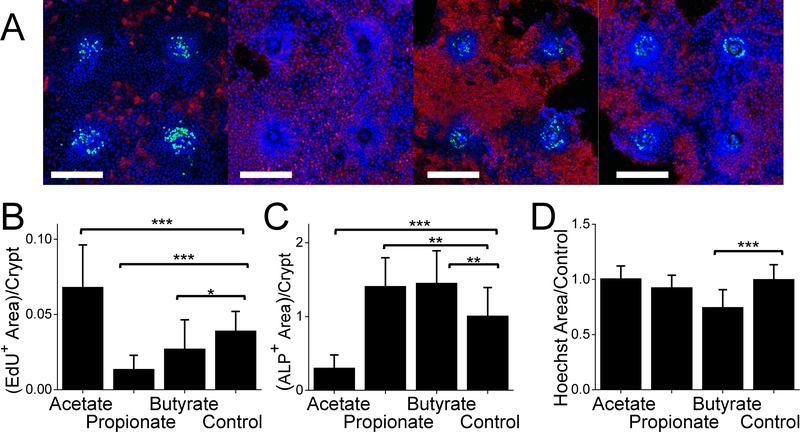
Colonic epithelial proliferation and differentiation is impacted by microbial products such as short chain fatty acids which are produced during bacterial fermentation of fibrous materials.47 For instance, propionate and butyrate suppress proliferation in intestinal tumor cell lines.48, 49 Butyrate also decreases proliferation in primary murine intestinal organoids.50 To evaluate the effect of short chain fatty acids on proliferation and differentiation of intestinal epithelial cells on the planar crypt arrays, short chain fatty acids (24 mM of acetate, 6 mM of propionate, or 1 mM of butyrate)23 was added in the DM luminal medium during the 2-day polarization time i.e. day 3–4 of cell culture on the arrays. The EdU+ and ALP+ areas were measured and normalized to the area positive for Hoechst 33342 (total cell nuclei area as a proxy for cell number). Acetate significantly increased the normalized area of EdU+ cells. Propionate and butyrate significantly decreased the normalized area of EdU+ cells compared to that of the control without the short chain fatty acid (Fig. 5B). This is consistent with the behavior of mouse intestinal cells cultured as a monolayer on a collagen gel23, mouse intestinal organoids within a Matrigel patty50 and tissue-cultured HT29 human colon cancer cells.48 In contrast, acetate significantly decreased ALP activity while propionate and butyrate significantly increased ALP activity relative to the control (Fig. 5C). Butyrate and propionate significantly increased ALP activity, similar to the impact of these two short chain fatty acids when applied as a gradient to three-dimensional crypts formed from primary human colonic cells.21 Butyrate increased ALP activity significantly more than did Butyrate was the only short chain fatty acid to decrease the number of Hoechst 33342+ cells per crypt by about 17% (p < 0.001) (Fig. 5D) which is consistent with the previous observation that butyrate induces apoptosis in human colon cancer cell lines.51–53 These data demonstrate that the planar crypt arrays reproduce key intestinal epithelial cell responses to microbial metabolites.
Figure 5.
Impact of short chain fatty acids on the planar crypts. A) Representative fluorescence images of 4 planar crypts under DM/EM, DM+acetate/EM, DM+propionate/EM, or DM+butyrate/EM culture conditions. EdU-incorporation, ALP activity and DNA stained with Hoechst 33342 represented in green, red, and blue, respectively. The white scale bar represents 200 μm. B,C) The EdU+ area (B) and ALP+ area (C) per crypt was normalized against the Hoechst 33342+ area per unit crypt. D) Ratio of Hoechst 33342+ area for the different fatty acids per unit crypt to that of control. Ten crypts each from 2 different mice (total 20 crypts) were analyzed. *, ** and *** indicate P ≤ 0.05, P ≤ 0.01 and P ≤ 0.001, respectively in panels B-D.
Discussion
In the current work, we describe a platform for the patterned culture of proliferative and differentiated primary murine colonic intestinal epithelial cells in a monolayer format simulating the intestinal crypt. The approach combines a collagen substrate of varying stiffness and porosity with the localized delivery of soluble growth factors through microholes in an impermeable membrane. The collagen substrate enables the adhesion, growth and migration of a cell monolayer by supplying the appropriate stiffness and ECM contacts. A suitably scaled linear gradient of growth factors delivered via the microholes supports the adjacent cells to establish and maintain stem/proliferative cell zones yet permits cells at a distance to undergo differentiation. Under these conditions, the cells nearest the microholes are exposed to high concentrations of growth factors while cells at a distance see progressively lower concentrations in a manner similar to the gradients believed to exist within the intestinal crypt.7 As the cells proliferate, they migrate radially from the microhole region, become differentiated and then undergo apoptosis. This process of proliferation, migration, differentiation and apoptosis occurs on a timescale similar to that seen in vivo. The platform was used to assay the impact of short chain fatty acids on the proliferation and differentiation of primary cells in vitro. Application of these microbiota-derived fermentation products drove alterations in the size of the proliferative and differentiated regions as predicted to occur in vivo.50 Screens using this simple bench-top platform can be undertaken to estimate in vivo concentrations and gradient profiles of soluble growth and morphogenic factors. The two-dimensional nature of cell patterning on the platform will enable high-throughput and high-content screens to study the impact of drugs, dietary factors, nutrients, prebiotics, and microbiota on primary intestinal tissue while maintaining precise control of numerous variables.
Supplementary Material
Acknowledgement
Research reported in this publication was supported by the National Institutes of Health under Award R01DK109559 to N.L.A. and S.T.M. We thank Ian Williamson and Scott Magness for kindly providing murine intestines. This work was performed in part at the Chapel Hill Analytical and Nanofabrication Laboratory, CHANL, a member of the North Carolina Research Triangle Nanotechnology Network, RTNN, which is supported by the National Science Foundation, Grant ECCS-1542015, as part of the National Nanotechnology Coordinated Infrastructure, NNCI.
Footnotes
Conflict of Interest
The authors declare the following competing financial interest(s): N.L.A., Y.W., C.E.S. have a financial interest in Altis Biosystems LLC.
References
- 1.Barker N, van de Wetering M and Clevers H, Genes & development, 2008, 22, 1856–1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fuchs E and Chen T, EMBO reports, 2013, 14, 39–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brittan M and Wright N, Gut, 2004, 53, 899–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kosinski C, Li VS, Chan AS, Zhang J, Ho C, Tsui WY, Chan TL, Mifflin RC, Powell DW and Yuen ST, Proceedings of the National Academy of Sciences, 2007, 104, 15418–15423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yen T-H and Wright NA, Stem cell reviews, 2006, 2, 203–212. [DOI] [PubMed] [Google Scholar]
- 6.Benoit YD, Groulx J-F, Gagné D and Beaulieu J-F, Journal of signal transduction, 2012, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Y, Kim R, Hinman SS, Zwarycz B, Magness ST and Allbritton NL, Cellular and Molecular Gastroenterology and Hepatology, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eastwood GL and Trier JS, Gastroenterology, 1973, 64, 375–382. [PubMed] [Google Scholar]
- 9.Autrup H, Barrett LA, Jackson FE, Jesudason ML, Stoner G, Phelps P, Trump B and Harris CC, Gastroenterology, 1978, 74, 1248–1257. [PubMed] [Google Scholar]
- 10.Sato T, Vries RG, Snippert HJ, Van De Wetering M, Barker N, Stange DE, Van Es JH, Abo A, Kujala P and Peters PJ, Nature, 2009, 459, 262. [DOI] [PubMed] [Google Scholar]
- 11.Sato T, Van Es JH, Snippert HJ, Stange DE, Vries RG, Van Den Born M, Barker N, Shroyer NF, Van De Wetering M and Clevers H, Nature, 2011, 469, 415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sato T, Stange DE, Ferrante M, Vries RG, Van Es JH, Van Den Brink S, Van Houdt WJ, Pronk A, Van Gorp J and Siersema PD, Gastroenterology, 2011, 141, 1762–1772. [DOI] [PubMed] [Google Scholar]
- 13.Jung P, Sato T, Merlos-Suárez A, Barriga FM, Iglesias M, Rossell D, Auer H, Gallardo M, Blasco MA and Sancho E, Nature medicine, 2011, 17, 1225. [DOI] [PubMed] [Google Scholar]
- 14.Yui S, Nakamura T, Sato T, Nemoto Y, Mizutani T, Zheng X, Ichinose S, Nagaishi T, Okamoto R and Tsuchiya K, Nature medicine, 2012, 18, 618. [DOI] [PubMed] [Google Scholar]
- 15.Stelzner M, Helmrath M, Dunn JC, Henning SJ, Houchen CW, Kuo C, Lynch J, Li L, Magness ST and Martin MG, American Journal of Physiology-Gastrointestinal and Liver Physiology, 2012, 302, G1359–G1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Attayek PJ, Ahmad AA, Wang Y, Williamson I, Sims CE, Magness ST and Allbritton NL, PLoS One, 2016, 11, e0153795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gjorevski N, Sachs N, Manfrin A, Giger S, Bragina ME, Ordóñez-Morán P, Clevers H and Lutolf MP, Nature, 2016, 539, 560. [DOI] [PubMed] [Google Scholar]
- 18.Moon C, VanDussen KL, Miyoshi H and Stappenbeck TS, Mucosal immunology, 2014, 7, 818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.VanDussen KL, Marinshaw JM, Shaikh N, Miyoshi H, Moon C, Tarr PI, Ciorba MA and Stappenbeck TS, Gut, 2015, 64, 911–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y, Gunasekara DB, Attayek PJ, Reed MI, DiSalvo M, Nguyen DL, Dutton JS, Lebhar MS, Bultman SJ and Sims CE, ACS Biomaterials Science & Engineering, 2017, 3, 2502–2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y, Kim R, Gunasekara DB, Reed MI, DiSalvo M, Nguyen DL, Bultman SJ, Sims CE, Magness ST and Allbritton NL, Cellular and Molecular Gastroenterology and Hepatology, 2018, 5, 113–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pai J-H, Wang Y, Salazar GTA, Sims CE, Bachman M, Li G and Allbritton NL, Analytical chemistry, 2007, 79, 8774–8780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y, DiSalvo M, Gunasekara DB, Dutton J, Proctor A, Lebhar MS, Williamson IA, Speer J, Howard RL and Smiddy NM, Cellular and molecular gastroenterology and hepatology, 2017, 4, 165–182. e167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ahmad AA, Wang Y, Sims CE, Magness ST and Allbritton NL, RSC Advances, 2015, 5, 74881–74891. [Google Scholar]
- 25.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S and Schmid B, Nature methods, 2012, 9, 676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cohen J, Journal, 1988. [Google Scholar]
- 27.Faul F, Erdfelder E, Lang A-G and Buchner A, Behavior research methods, 2007, 39, 175–191. [DOI] [PubMed] [Google Scholar]
- 28.Engelberg P and Tesoro G, Polymer Engineering & Science, 1990, 30, 303–307. [Google Scholar]
- 29.Vallo CI, Frontini PM and Williams RJ, Polymer Gels and Networks, 1993, 1, 257–266. [Google Scholar]
- 30.Qu F, Li Q, Wang X, Cao X, Zgonis MH, Esterhai JL, Shenoy VB, Han L and Mauck RL, Scientific reports, 2018, 8, 3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Franck C, Maskarinec SA, Tirrell DA and Ravichandran G, PloS one, 2011, 6, e17833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsubouchi S, Developmental Dynamics, 1981, 161, 239–246. [Google Scholar]
- 33.Peyton SR, Kalcioglu ZI, Cohen JC, Runkle AP, Van Vliet KJ, Lauffenburger DA and Griffith LG, Biotechnology and bioengineering, 2011, 108, 1181–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hayman M, Smith K, Cameron N and Przyborski S, Journal of biochemical and biophysical methods, 2005, 62, 231–240. [DOI] [PubMed] [Google Scholar]
- 35.Rother J, Büchsenschütz-Göbeler M, Nöding H, Steltenkamp S, Samwer K and Janshoff A, Journal of The Royal Society Interface, 2015, 12, 20141057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Janshoff A, Lorenz B, Pietuch A, Fine T, Tarantola M, Steinem C and Wegener J, Journal of Adhesion Science and Technology, 2010, 24, 2287–2300. [Google Scholar]
- 37.Chowdhury F, Li Y, Poh Y-C, Yokohama-Tamaki T, Wang N and Tanaka TS, PloS one, 2010, 5, e15655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lü D, Luo C, Zhang C, Li Z and Long M, Biomaterials, 2014, 35, 3945–3955. [DOI] [PubMed] [Google Scholar]
- 39.Yim EK and Sheetz MP, Stem cell research & therapy, 2012, 3, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Provenzano PP and Keely PJ, J Cell Sci, 2011, 124, 1195–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schneider MR, Dahlhoff M, Horst D, Hirschi B, Trülzsch K, Müller-Höcker J, Vogelmann R, Allgäuer M, Gerhard M and Steininger S, PloS one, 2010, 5, e14325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Valenta T, Hausmann G and Basler K, The EMBO journal, 2012, 31, 2714–2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Birchenough GM, Nyström EE, Johansson ME and Hansson GC, Science, 2016, 352, 1535–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sternini C, Anselmi L and Rozengurt E, Current opinion in endocrinology, diabetes, and obesity, 2008, 15, 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barker N, Nature reviews Molecular cell biology, 2014, 15, 19. [DOI] [PubMed] [Google Scholar]
- 46.Barkla DH and Gibson PR, Pathology, 1999, 31, 230–238. [DOI] [PubMed] [Google Scholar]
- 47.Koh A, De Vadder F, Kovatcheva-Datchary P and Bäckhed F, Cell, 2016, 165, 1332–1345. [DOI] [PubMed] [Google Scholar]
- 48.Gamet L, Daviaud D, Denis‐Pouxviel C, Remesy C and Murat JC, International Journal of Cancer, 1992, 52, 286–289. [DOI] [PubMed] [Google Scholar]
- 49.Whitehead R, Young G and Bhathal P, Gut, 1986, 27, 1457–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kaiko GE, Ryu SH, Koues OI, Collins PL, Solnica-Krezel L, Pearce EJ, Pearce EL, Oltz EM and Stappenbeck TS, Cell, 2016, 165, 1708–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ruemmele F, Schwartz S, Seidman E, Dionne S, Levy E and Lentze M, Gut, 2003, 52, 94–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fung KY, Brierley GV, Henderson S, Hoffmann P, McColl SR, Lockett T, Head R and Cosgrove L, Journal of proteome research, 2011, 10, 1860–1869. [DOI] [PubMed] [Google Scholar]
- 53.Xu S, Liu C-X, Xu W, Huang L, Zhao J-Y and Zhao S-M, Signal transduction and targeted therapy, 2017, 2, 16035. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.