Abstract
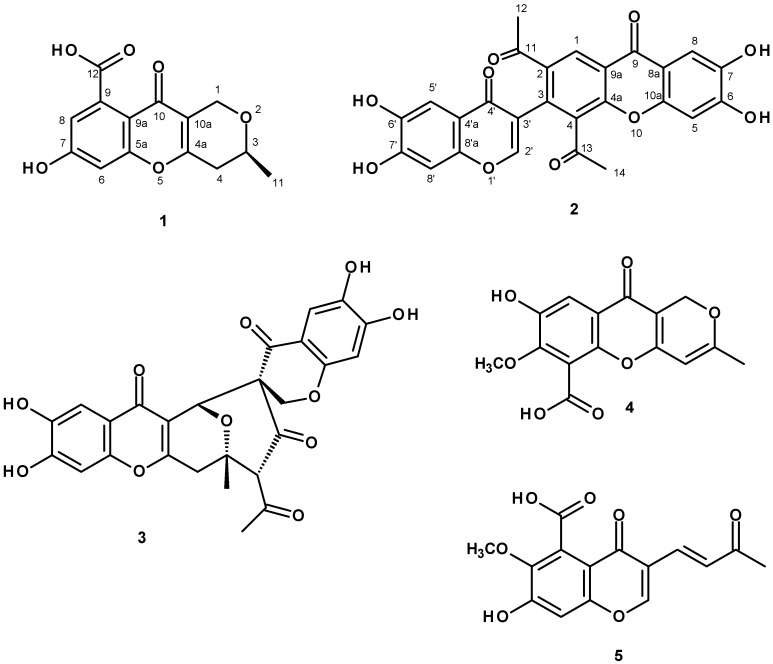
A new polyketide erubescensoic acid (1), and the previously reported xanthonopyrone, SPF-3059-26 (2), were isolated from the uninvestigated fractions of the ethyl acetate crude extract of the marine sponge-associated fungus Penicillium erubescens KUFA0220. The structures of the new compound, erubescensoic acid (1), and the previously reported SPF-3059-26 (2), were elucidated by extensive analysis of 1D and 2D-NMR spectra as well as HRMS. The absolute configuration of the stereogenic carbon of erubescensoic acid (1) was determined by X-ray analysis. Erubescensoic acid (1) and SPF-3059-26 (2), together with erubescenschromone B (3), penialidin D (4), and 7-hydroxy-6-methoxy-4-oxo-3-[(1E)-3-oxobut-1-en-1-yl]-4H-chromen-5-carboxylic acid (5), recently isolated from this fungus, were assayed for their antibacterial activity against gram-positive and gram-negative reference strains and the multidrug-resistant (MDR) strains from the environment. The capacity of these compounds to interfere with the bacterial biofilm formation and their potential synergism with clinically relevant antibiotics for the MDR strains were also investigated.
Keywords: Penicillium erubescens, marine sponge-associated fungus, polyketides, erubescensoic acid, SPF-3059-267, antibacterial activity, antibiofilm activity, antibiotic synergy
1. Introduction
Penicillium (Family Aspergillaceae) is a diverse genus with more than 300 known species today, which are widely present throughout the world. Its species play important roles as decomposers of organic materials and cause destructive rots in the food industry where they produce a wide range of mycotoxins. Other species are considered enzyme factories or are common indoor allergens [1]. The biggest impact and claim to fame is the production of penicillin, which revolutionized the pharmaceutical industry and saved millions of lives around the world. Moreover, compactin, the first member of the anticholestolemic drug “statins”, was first isolated from P. citrinum [2]. Species of Penicillium are found in both terrestrial and marine environments. The marine-derived Penicillium species, normally associated with a variety of marine invertebrates, mangroves, and sediments, are a source of structurally diverse classes of secondary metabolites such as polyketides, sterols, terpenoids, and alkaloids, most of which exhibit a myriad of biological activities [3]. Although members of the genus Penicillium from terrestrial environments have been extensively investigated for their secondary metabolites, their marine counterparts are still underexplored.
During our search for antibiotics from marine-derived fungi from the Gulf of Thailand and the Andaman Sea, we have reported isolation of several previously undescribed chromone and chromene derivatives as well as a chromone dimer, from the culture of Penicillium erubescens strain KUFA 0220, isolated from the marine sponge Neopetrosia sp., which was collected from the coral reef at Samaesan Island in the Gulf of Thailand [4]. Reexamination of the column fractions of P. erubescens, which have not been investigated in the previous study led us to further isolate one previously unreported polyketide which we have named erubescensoic acid (1) and the xanthonopyrone, SPF-3059-26 (2), which was previously reported from the culture of Penicillium sp. SPF-3059 [5] (Figure 1). Compounds 1 and 2, were tested for their antibacterial activity against different strains of gram-positive and gram-negative bacteria, including reference strains and environmental multidrug-resistant isolates, together with erubescenschromone B (3), penialidin D (4), and 7-hydroxy-6-methoxy-4-oxo-3-[(1E)-3-oxobut-1-en-1-yl]-4H-chromen-5-carboxylic acid (5), which were isolated in our previous study [4] but were not tested for antibacterial activity. Compounds 1–5 were also evaluated for their capacity to prevent biofilm formation of the four reference strains as well as for their potential synergy between the compounds and clinically relevant drugs against the multidrug-resistant isolates.
Figure 1.
Structures of erubescensoic acid (1), SPF-3059-26 (2), erubescenschromone B (3), penialidin D (4), and 7-hydroxy-6-methoxy-4-oxo-3-[(1E)-3-oxobut-1-en-1-yl]-4H-chromen-5-carboxylic acid (5).
2. Results and Discussion
Compound 1 was isolated as a white crystal (mp. 218–220 °C), and displayed its (+)-HRESIMS m/z at 277.0719 [M + H]+, (calculated 277.0712 for C14H13O6). Therefore, its molecular formula was established as C14H12O6, indicating nine degrees of unsaturation. However, the 13C-NMR spectrum (Table 1, see Supplementary materials, Figure S2) displayed only thirteen carbon signals which, according to DEPTs and HSQC (Supplementary materials, Figure S2), can be classified as one conjugated ketone carbonyl (δC 173.0), one conjugated carboxyl (δC 161.8), three oxyquaternary sp2 (δC 160.0, 157.4, 138.2), two quaternary sp2 (δC 119.7, 115.3), two methine sp2 (δC 111.8, 102.0), one oxymethylene sp3 (δC 61.6), one methylene sp3 (δC 33.5), one oxymethine sp3 (δC 69.4), and one methyl (δC 20.8) carbons. That means one quaternary sp2 carbon signal was not observed, and this is characteristic of the carboxyl-bearing aromatic carbon. The 1H- and 13C-NMR data of 1 resembled those of anhydrofulvic acid [6]; however the benzene ring of the chromone moiety of 1 has only one hydroxyl group, as evidenced by the presence of two broad singlets of the meta-coupled protons at δH 6.78 (H-6/δC 102.0) and 6.27 (H-8/δC 111.8), instead of two hydroxyl groups. Moreover, the double bond between C-2 and C-3 of the 3-methyl-2H-pyran ring was saturated as corroborated by the presence of the methylene group (δC 33.5/δH 2.66, d, J = 17.3 Hz/2.56, dd, J = 17.3, 9.8 Hz). Therefore, the planar structure of 1 was elucidated as 7,8-dihydroxy-3-methyl-10-oxo-4,10-dihydro-1H,3H-pyrano[4,3-b]chromene-9-carboxylic acid. This was confirmed by HMBC correlations (Table 1, Supplementary materials, Figure S4) from the methyl protons at δH 1.28, d (J = 6.2 Hz, Me-11) to C-3 (δC 69.4) and C-4 (δC 33.5), H-3 (δH 3.83, m) to C-1 (δC 61.6), H2-1 (δH 4.56/4.33) to C-3, C-4a (δC 160.0), C-10a (δC 115.3) as well as H2-4 (δH 2.56/2.66) to C-3, C-4a and C-10a. The saturation of the double bond between C-2 and C-3 makes C-3 stereogenic, whose absolute configuration needs to be determined.
Table 1.
The 1H- and 13C-NMR (DMSO-d6, 500 and 125 MHz) and HMBC assignment for 1.
| Position | δC, Type | δC (J in Hz) | HMBC |
|---|---|---|---|
| 1 | 61.6, CH2 | 4.56, d (14.9) | C-3, 4a, 10a |
| 4.33, d (14.9) | C-10a | ||
| 3 | 69.4, CH | 3.83, m | C-1 |
| 4 | 33.5, CH2 | 2.66, d (17.3) | C-3, 4a, 11 |
| 2.56, dd (17.3, 9.8) | C-3, 4a, 10a | ||
| 4a | 160.0, C | - | |
| 5a | 157.4, C | - | |
| 6 | 102.0, CH | 6.78, s | 4 |
| 7 | 138.2, C | - | |
| 8 | 111.8, CH | 6.27, s | |
| 9 | - | - | |
| 9a | 119.7, C | - | |
| 10 | 173.0, CO | - | |
| 10a | 115.3, C | - | |
| 11 | 20.8, CH3 | 1.28, d (6.2) | C-3, 4 |
| 12 | 161.8, CO | - |
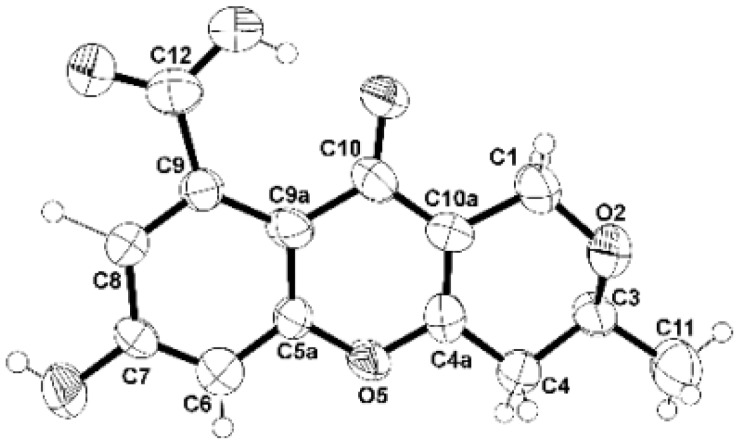
Since 1 was obtained as a suitable crystal, the X-ray analysis was carried out. The Ortep diagram of 1 (Figure 2) not only confirms its structure but establishes the absolute configuration of C-3 as 3S. Since 1 has never been previously reported, it was named erubescensoic acid.
Figure 2.
Ortep view of 1.
Compound 2 was isolated as a pale yellow viscous oil, and its molecular formula C26H16O10 was established based on its (+)-HRESIMS m/z 489.0818 [M + H]+, (calculated 489.0822 for C26H17O10), indicating nineteen degrees of unsaturation. The infrared (IR) spectrum showed absorption bands for the hydroxyl (3445 cm−1), conjugated ketone (1650 cm−1), olefin (1625 cm−1), aromatic (1605, 1542 cm−1), and ether (1262 cm−1). The 13C-NMR spectrum of 2 (Table 2, Supplementary materials, Figure S7) displayed twenty six carbon signals which, in combination with DEPTs and HSQC spectra (Supplementary materials, Figures S8–S10), can be categorized as four conjugated ketone carbonyls (δC 201.3, 199.2, 173.7 and 173.4), seven oxyquaternary sp2 (δC 154.5,152.8, 152.5, 151.1, 150.7, 145.0, 144.6), seven quaternary sp2 (δC 135.9, 133.5, 132.7, 120.8, 119.8, 115.7, 113.4), six methine sp2 (δC 152.9, 126.4, 108.6, 107.9, 103.1, 102.9), and two methyl (δC 32.3 and 29.2) carbons. The 1H- and 13C-NMR data of 2 resemble those of SPF-3059-30, also isolated from this fungus [4], except for the absence of the oxymethylene sp3 carbon at δC 66.2 and the appearance of the oxymethine sp2 carbon at δC 152.9 in 2. The presence of the 3-substituted 6,7-dihydroxy-4H-chromen-4-one was substantiated by HMBC correlations (Supplementary materials, Figures S11 and S12) from H-5′ (δH 7.28, brs/δC 107.9) to C-4′ (δC 173.7), C-6′ (δC 152.8), C-7′ (δC 145.0) and C-8′a (δC 151.1), H-8′ (δH 6.94, s/δC 103.1) to C-4′a (δC 113.4), C-6′, and from H-2′ (δH 8.13, s/ δC 152.9) to C-3′ (δC 120.7), C-4′ and C-8′a. That another part of the molecule was a 2,3,4-trisubstituted 6,7-dihydroxyxanthone, resembles that of SPF-3059-30 [5] was supported by HMBC correlations (Supplementary materials, Figures S11 and S12) from H-5 (δH 6.93, s/δC 102.9) to C-7 (δC 144.6), C-8a (δC 115.7), and from H-8 (δH 7.48, s/δC 108.6) to C-6 (150.5), C-9 (δC 173.4) and C-10a (δC 154.5). That the substituents on C-2 and C-4 of the benzene ring of the xanthone moiety were acetyl groups was corroborated by HMBC correlations (Supplementary materials, Figures S11 and S12) from H-1 (δH 8.58, s/δC 126.4) to C-3 (δC 132.7), C-4a (δC 152.5), C-9 (δC 173.4), C-11 (δC 199.2), from Me-12 (δH 2.55,s/δC 29.2) to C-11) and Me-14 (δH 2.53,s/δC 32.3) to C-13 (δC 201.3). Finally, the 6,7-dihydroxy-4H-chromen-4-one and the 2,4-diacetyl-6,7-dihydroxyxanthone are linked through C-3′ of the former and C-3 of the latter was confirmed by HMBC correlation from H-2′ to C-3. iterature search revealed that the planar structure of 2 is the same as that of SPF-3059-26, another polyketide isolated from the acetone extract of the mycelium of Penicilium sp. SPF-3050 (FERM BB-7663), cultured in the liquid medium [5]. However, there were no assignments of 1H and 13C chemical shift values for any protons and carbons of the structure of SPF-3059-26. Analysis of the structure of 2 revealed that the existence of the acetyl groups on C-2 and C-4 of the benzene ring of the xanthone moiety can impose a restriction of the rotation of the C-3 and C-3’ bond, thus creating a phenomenon of atropoisomerism. Optical rotation measurement revealed that 2 is dextrorotatory, presenting [α]25D +266 in MeOH. Due to the interesting activity of this class of compounds, SPF-3059-26 was later obtained, together with vinaxanthone and its derivatives, by ynone coupling reaction by Chin et al. [7]. Examination of the HRMS (ESI) data, 1H- and 13C-NMR spectra of SPF-3059-26 (compound 29 in Ref. 7) from the supporting information of the article by Chin et al. [7] revealed that they are compatible with those of 2. However, neither optical rotation nor electronic circular dichroism (ECD) spectrum was mentioned in the discussion or provided in this supporting information. SPF-3059-26 (2) can be perceived as a decarboxylated derivative of vinaxanthone, which was previously isolated from the culture of P. vinaceum NR6815, isolated from soil [8], P. glabrum (Wehmer) Westling [9] and Penicillium sp. strain SPF-3059 [10]. It is noteworthy to mention that the structure elucidation of vinaxanthone in all these articles was based on analyses of the 1D-and 2D-NMR data, nothing was mentioned about its optical rotation or ECD spectrum.
Table 2.
The 1H- and 13C-NMR (DMSO-d6, 500 and 125 MHz) and HMBC assignment for 2.
| Position | δC, Type | δC (J in Hz) | HMBC |
|---|---|---|---|
| 1 | 126.4, CH | 8.58, s | C-3, 4a, 9, 11 |
| 2 | 135.9, C | - | |
| 3 | 132.7, C | - | |
| 4 | 133.5, C | - | |
| 4a | 152.5, C | - | |
| 5 | 102.9, CH | 6.93, s | C-7, 8a, 9, 10a |
| 6 | 150.7, C | - | |
| 7 | 144.6, C | - | |
| 8 | 108.6, CH | 7.48, s | C-7, 8a, 9, 10a |
| 8a | 115.7, C | - | |
| 9 | 173.4, CO | - | |
| 9a | 119.8, C | ||
| 10a | 154.5, C | ||
| 11 | 199.2, CO | ||
| 12 | 29.2, CH3 | 2.55, s | C-2, 11 |
| 13 | 201.3, CO | - | |
| 14 | 32.3, CH3 | 2.53, s | C-4, 13 |
| 2′ | 152.9, CH | 8.13, s | C-3′, 4′, 8′a, 9′ |
| 3′ | 120.8, C | - | |
| 4′ | 173.7, CO | - | |
| 4′a | 113.4, C | - | |
| 5′ | 107.9, CH | 7.28, brs | C-4’, 6′, 7′, 8′a |
| 6′ | 152.8, C | - | |
| 7′ | 145.0, C | - | |
| 8′ | 103.1, CH | 6.94, s | C-4′, 6′, 7′ |
| 8′a | 151.1, C |
Compounds 1 and 2, were evaluated, together with erubescenschromone B (3), penialidin D (4), and 7-hydroxy-6-methoxy-4-oxo-3-[(1E)-3-oxobut-1-en-1-yl]-4H-chromen-5-carboxylic acid (5) (Figure 1), for their antibacterial activity against different strains of gram-positive and gram-negative bacteria, including reference strains and multidrug-resistant environmental isolates. However, in the range of concentrations tested, none of the compounds were active. The ability of 1–5 to prevent biofilm formation was also evaluated on four reference strains by measuring the total biomass. Since it was not possible to determine MIC (minimal inhibitory concentration) values of these compounds, the highest concentration tested in previous assays was used (64 mg/L or 32 mg/L for 3). The results were interpreted using a comparative classification that divides adherence capabilities of tested strains into four categories: Non-adherent, weakly adherent, moderately adherent and strongly adherent [11]. The use of this classification, which uses the negative control as a starting point, instead of using the positive control as a reference, reduces the risk of inconsistencies due to external factors that influence biofilm production [12]. None of the compounds inhibited biofilm formation of Pseudomonas aeruginosa ATCC 27853, Staphyllococus aureus ATCC 29213, or Enterococcus faecalis ATCC 29212. Nonetheless, all the compounds tested were capable of impairing the biofilm forming ability of Escherichia coli ATCC 25922, which was classified as a strong biofilm producer (Table 3). These results suggest that the mechanism for impairing biofilm formation might be other than bactericidal activity. Other mechanisms for anti-biofilm activity have been described, such as inhibition of bacterial surface attachment, interference with quorum sensing signaling or even inhibition of biosynthesis of matrix components [13,14].
Table 3.
Classification of the ability of E. coli ATCC 25922 to adhere to and form a biofilm after an exposure to 1–5.
| Compound | Concentration (mg/L) | OD ± SD | Classification |
|---|---|---|---|
| None | 0 | 0.361 ± 0.159 | strong |
| 1 | 64 | 0.188 ± 0.012 | moderate |
| 2 | 64 | 0.195 ± 0.012 | moderate |
| 3 | 32 | 0.246 ± 0.038 | moderate |
| 4 | 64 | 0.172 ± 0.024 | weak |
| 5 | 64 | 0.194 ± 0.013 | moderate |
OD, optical density; SD, standard deviation; ODc, optical density cut-off value. Average OD value for negative control was found to be 0.065 ± 0.007, therefore ODc equals 0.065 + (3 × 0.007) = 0.086; 2 × ODc = 0.172; 4 × ODc = 0.344.
Potential synergy between the tested compounds and clinically relevant antimicrobial drugs were also screened using different methodologies. No associations were found with the disc diffusion assay. These results were obtained by determination of the MIC for each antibiotic in the presence of a fixed concentration of each compound, as it was not possible to determine MIC values for the test compounds. The concentration of each compound used was the highest concentration tested in previous assays (64 mg/L or 32 mg/L for 3), which did not inhibit the growth of the three multidrug-resistant strains under study. This method allows to determine that 2 causes a four-fold reduction in the cefotaxime (CTX) MIC of this strain (Table 4). However, this compound increased the oxacillin (OXA) MIC of methicillin-resistant Staphylococcus aureus (MRSA) S. aureus 66/1 by two-fold.
Table 4.
Combined effect of clinically used antibiotics with 1–5 against multidrug-resistant strains. Minimal inhibitory concentration (MICs) are expressed in mg/L.
| E. coli SA/2 (ESBL) | E. faecalis B3/101 (VRE) | S. aureus 66/1 (MRSA) | ||||
|---|---|---|---|---|---|---|
| CTX | VAN | OXA | ||||
| Compound | Distribution | MIC | Distribution | MIC | Distribution | MIC |
| Antibiotic | - | 512 | - | 1024 | - | 64 |
| Antibiotic +1 | - | 512 | - | 1024 | - | 64 |
| Antibiotic +2 | - | 128 | - | 1024 | - | 128 |
| Antibiotic +3 | - | 512 | - | 1024 | - | 64 |
| Antibiotic +4 | - | 512 | - | 1024 | - | 64 |
| Antibiotic +5 | - | 512 | - | 1024 | - | 64 |
MIC, minimal inhibitory concentration; (-), no inhibition halo or no increase in the inhibition halo; CTX, cefotaxime; VAN, vancomycin; OXA, oxacillin; ESBL, extended-spectrum β-lactamase producer; VRE, vancomycin-resistant Enterococcus; MRSA, methicillin-resistant Staphylococcus aureus.
3. Experimental Section
3.1. General Experimental Procedures
Melting points were determined on a Stuart Melting Point Apparatus SMP3 (Bibby Sterilin, Stone, Staffordshire, UK) and are uncorrected. Optical rotations were measured on an ADP410 Polarimeter (Bellingham + Stanley Ltd., Tunbridge Wells, Kent, UK). 1H- and 13C-NMR spectra were recorded at ambient temperature on a Bruker AMC instrument (Bruker Biosciences Corporation, Billerica, MA, USA), operating at 300 or 500 and 75 or 125 MHz, respectively. High resolution mass spectra were measured with a Waters Xevo QToF mass spectrometer (Waters Corporations, Milford, MA, USA) coupled to a Waters Aquity UPLC system. A Merck (Darmstadt, Germany) silica gel GF254 was used for preparative TLC, and a Merck Si gel 60 (0.2–0.5 mm) was used for column chromatography.
3.2. Fungal Material
Isolation, identification and cultivation of the fungus as well as preparation of the crude fungal extract were previously described by us [4].
3.3. Extraction and Isolation
Chromatographic isolation of the compounds from the crude EtOAc extract of P. erubescens KUFA 0220 was recently described by us [4]. For isolation of 1 and 2, sub-fractions 185–251 from the silica gel column frs 445–529 were combine (658 mg) and applied on a Sephadex LH-20 column (20 g) and eluted with MeOH, wherein 2 mL fractions were collected. Frs 25–30 were combined (40.0 mg) and purified by TLC (Silica gel G254, CHCl3:MeOH:HCO2H, 9:1:0.01) to give 1 (7 mg). Sfrs 295–344 were combined (3.0 g) and applied on a Sephadex LH-20 column (20 g) and eluted with a 1:1 mixture of CHCl3:MeOH, wherein 20 mL fractions were collected. Frs 1–30 were combined (217 mg) and re-applied on another Sephadex LH-20 column (20 g) and eluted with MeOH, wherein 30 sfrs of 2 mL were collected. Sfrs 8–26 were combined to give 2 (7.2 mg).
3.3.1. Erubescensoic Acid (1)
White crystal. Mp 218–220 °C; [α]25D: −100.0 (MeOH, c 0.04 g/mL); IR (KBr) νmax 3445, 2921, 1733, 1716, 1698, 1683, 1652, 1635, 1558, 1540, 1506, 1472 cm−1.For 1H- and 13C-NMR data, see Table 1; (+)-HRESIMS m/z 277.0719 [M + H]+ (calculated for C14H13O6, 277.0712).
3.3.2. SPF-3059-26 (2)
Pale yellow viscous oil; [α]25D +266 (MeOH, c = 0.03 g/mL), IR (KBr) νmax 3445, 2958, 2922 1650, 1605, 1262 cm−1; For 1H- and 13C-NMR data, see Table 2; (+)-HRESIMS m/z 489.0818 [M + H]+ (calculated for C26H17O10, 489.0822).
3.4. X-Ray Crystal Structure of 1
A single crystal was mounted on a cryoloop using paratone. X-ray diffraction data was collected at 288 K with a Gemini PX Ultra equipped with CuKα radiation (λ = 1.54184 Å). The crystal was orthorhombic, space group P212121, cell volume 1413.65(12) Å3 and unit cell dimensions a = 6.7568(4) Å, b = 13.0791(5) Å and c = 15.9964(6) Å (uncertainties in parentheses). The structure was solved by direct methods using SHELXS-97 and refined with SHELXL-97 [15]. One molecule of the compound and two water molecules were found in the asymmetric unit. Carbon and oxygen atoms were refined anisotropically. Hydrogen atoms either directly found from difference Fourier maps and were refined freely with isotropic displacement parameters or placed at their idealized positions using appropriate HFIX instructions in SHELXL and included in subsequent refinement cycles. Hydrogens of one of the water molecules were not observed in the difference Fourier maps. The refinement converged to R (all data) = 10.43% and wR2 (all data) = 16.95%. Full details of the data collection and refinement and tables of atomic coordinates, bond lengths and angles, and torsion angles have been deposited with the Cambridge Crystallographic Data Centre (CCDC 1870933).
3.5. Antibacterial Activity Bioassays
3.5.1. Bacterial Strains and Testing Conditions
Four reference strains and three multidrug-resistant (MDR) strains were used in this study. Gram-negative strains included Escherichia coli ATCC 25922, Pseudomonas aeruginosa ATCC 27853 and the clinical isolate SA/2, an extended-spectrum β-lactamase producer (ESBL). Gram-positive bacteria comprised Staphylococcus aureus ATCC 29213, Enterococcus faecalis ATCC 29212, methicillin-resistant Staphylococcus aureus (MRSA) 66/1, isolated from public buses [16] and vancomycin-resistant Enterococcus faecalis (VRE) B3/101, isolated from river water [17]. All strains were kept in Trypto-Casein Soy agar (TSA, Biokar Diagnostics, Allone, Beauvais, France) slants, at room temperature, in the dark. Before each assay, all strains were cultured in Mueller-Hinton agar (MH, Biokar Diagnostics, Allone, Beauvais, France) and incubated overnight at 37 °C. Stock solutions of the compounds were prepared in DMSO (Alfa Aesar, Kandel, Germany) and kept at −20 °C. With the exception of compound 3, 10 mg/mL stock solutions were prepared. Compound 3 was less soluble in DMSO than other compounds, so a 2 mg/mL stock solution was prepared. In all experiments, the final in-test concentration of DMSO was maintained below 1%, as recommended by the Clinical and Laboratory Standards Institute [18].
3.5.2. Antimicrobial Susceptibility Testing
The antimicrobial activity of the compounds was screened using the Kirby-Bauer method, as recommended by the CLSI [19]. Briefly, 6 mm blank paper discs (Liofilchem, Roseto degli Abruzzi, Teramo, Italy) were impregnated with 15 µg of each compound, and blank paper discs impregnated with DMSO were used as negative control. MH inoculated plates were incubated for 18–20 h at 37 °C. The results were evaluated by measuring the inhibition halos. Minimal inhibitory concentrations (MIC) for each compound were accessed in accordance with the CLSI standard [20]. Two-fold serial dilutions of the compounds were prepared in cation-adjusted Mueller-Hinton broth (CAMHB, Sigma-Aldrich, St. Louis, MO, USA) within the concentration range 64–2 mg/L, except for 3, for which the highest concentration tested was 32 mg/L. The initial inoculum size (which should be approximately 5 × 105 CFU/mL) was determined by colony forming unit counts. The 96-well U-shaped untreated polystyrene plates were incubated for 16–20 h at 37 °C and the MIC was defined as the lowest concentration of compound that prevented visible growth. These assays were conducted for reference and MDR strains.
3.5.3. Biofilm Formation Inhibition Assay
The effect of 1–5 on biofilm formation was evaluated using the crystal violet method, as previously described [4]. Briefly, the highest concentration of compound tested in the MIC assay was added to bacterial suspensions of 1 × 106 CFU/mL prepared in unsupplemented Tryptone Soy broth (TSB, Biokar Diagnostics, Allone, Beauvais, France) or TSB supplemented with 1% (p/v) glucose [D(+)-Glucose anhydrous for molecular biology PanReac AppliChem, Barcelona, Spain] for Gram-positive strains. A control with appropriate concentration of DMSO, as well as a negative control (TSB alone) were included. Sterile 96-well flat-bottomed untreated polystyrene plates were used. After a 24 h incubation at 37 °C, the biofilms were stained and their biomass was quantified by measuring the absorbance of each sample at 570 nm in a microplate reader (Thermo Scientific Multiskan® EX, Thermo Fisher Scientific, Waltham, MA, USA). This assay was performed for reference strains.
3.5.4. Antibiotic Synergy Testing
In order to screen for potential synergy between the compounds and clinically relevant antimicrobial drugs, the Kirby-Bauer method was used, as previously described [21]. A set of antibiotic discs (Oxoid, Basingstoke, England) to which the isolates were resistant was selected: cefotaxime (CTX, 30 µg) for E. coli SA/2, vancomycin (VAN, 30 µg) for E. faecalis B3/101, and oxacillin (OXA, 1 µg) for S. aureus 66/1. Antibiotic discs impregnated with 15 µg of each compound were placed on seeded MH plates. The controls used included antibiotic discs alone, blank paper discs impregnated with 15 µg of each compound alone and blank discs impregnated with DMSO. Plates with CTX were incubated for 18–20 h and plates with VAN and OXA were incubated for 24 h at 37 °C [18]. Potential synergy was considered when the inhibition halo of an antibiotic disc impregnated with compound was greater than the inhibition halo of the antibiotic or compound-impregnated blank disc alone. The combined effect of the compounds and clinical relevant antimicrobial drugs was also evaluated by determining the antibiotic MIC in the presence of each compound. Briefly, when it was not possible to determine a MIC value for the test compound, the MIC of CTX (Duchefa Biochemie, Haarlem, The Netherlands), VAN (Oxoid, Basingstoke, England), and OXA (Sigma-Aldrich, St. Louis, MO, USA) for the respective multidrug-resistant strain was determined in the presence of the highest concentration of each compound tested in previous assays. For 3 the concentration used was 32 mg/L, while it was 64 mg/L for the other compounds. The antibiotic tested was serially diluted whereas the concentration of each compound was kept fixed. Antibiotic MICs where determined as described above. Potential synergy was considered when the antibiotic MIC was lower in the presence of compound.
4. Conclusions
We have recently described the first chemical investigation and antibacterial activity assay of the constituents isolated from the culture on the solid medium (cooked rice) of the marine-derived fungus Penicillium erubescens strain KUFA 0220, isolated from the marine sponge Neopetrosia sp., which was collected from the coral reef at Samaesan Island in the Gulf of Thailand. Although nineteen compounds (five of which were reported for the first time) have been isolated, some column fractions were very complex and difficult to purify and were left over for further study. Repetition of chromatographic fractionations by silica gel and Sephadex LH-20 columns, in combination with preparative TLC of silica gel, allowed us to retrieve a previously unreported metabolite which was named erubescenoic acid (1) and another polyketide called SPF-3059-26 (2), previously reported in a European patent of the nerve regeneration promotors containing semaphoring inhibitors as active ingredient, from Penicillium sp. SPF-3050 (FERM BB-7663). Since we have not yet evaluated the antibacterial activity of erubescenschromone B (3), penialidin D (4) and 7-hydroxy-6-methoxy-4-oxo-3-[(1E)-3-oxobut-1-en-1-yl]-4H-chromen-5-carboxylic acid (5), isolated from the same extract in our previous study, we evaluated these compounds, together with the newly isolated rubescenoic acid (1) and SPF-3059-26 (2), against Gram-positive and Gram-negative reference strains and environmental multidrug-resistant (MDR) strains, as well as their capacity to interfere with the bacterial biofilm formation and their potential synergism with clinically relevant antibiotics for the MDR strains. Although all the tested compounds were neither active against the reference and multidrug-resistant strains nor able to inhibit a biofilm formation of Pseudomonas aeruginosa ATCC 27853, Staphyllococus aureus ATCC 29213 or Enterococcus faecalis ATCC 29212, they were capable of impairing the biofilm forming ability of a strong biofilm producer, Escherichia coli ATCC 25922. Interestingly, screening of potential synergy with antibiotics revealed that SPF-3059-26 (2) was able to reduce the CTX MIC of E. coli SA/2 (ESBL) for four-fold while it increased the OXA MIC of MRSA S. aureus 66/1 by two-fold. Given the capacity of the neuronal regenerative effects of some of these compounds isolated from this fungus, it is desirable to test the extract of this fungus and its constituents for this effect.
Acknowledgments
We thank the Foundation for Science and Technology of the Ministry of Science, Technology and Higher Education (PIDDAC) and European Regional Development Fund (ERDF) through the COMPETE-Programa Operacional Factores de Competitividade (POFC) programme, under the project PTDC/MAR-BIO/4694/2014 (reference POCI-01-0145-FEDER-016790; Project 3599-Promover a Produção Científica e Desenvolvimento Tecnológico e a Constituição de Redes Temáticas (3599-PPCDT)) in the framework of the programme PT2020. Decha Kumla thanks the Alfabet Project of the Erasmus Mundus for a PhD’s scholarship. We thank Michael Lee of the Department of Chemistry, University of Leicester, UK for providing HRMS spectra.
Supplementary Materials
The following are available online, Figures S1–S8: 1D- and 2D-NMR spectra of 1 and 2.
Author Contributions
A.K. and M.M.M.P. conceived, designed the experiment and elaborated the manuscript. D.K. performed isolation, purification and structure elucidation of the compounds. T.D. isolated, identified and cultured the fungus and also prepared a crude fungal extract. J.F.-S. and P.M.C. performed antibacterial activity assays. L.G. obtained crystal structure. A.M.S.S provided 1D- and 2D-NMR spectra. J.A.P. assisted in structure elucidation of the compounds.
Funding
This research was partially funded the project PTDC/MAR-BIO/4694/2014 (reference POCI-01-0145-FEDER-016790); Project 3599-Promover a Produção Científica e Desenvolvimento Tecnológico e a Constituição de Redes Temáticas (3599-PPCDT).
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Sample Availability: Not available.
References
- 1.Visagle C.M., Houbraken J., Frisvad J.C., Hong S.B., Klaassen C.H.W., Perrone G., Seifert K.A., Vatga J., Yaguchi T., Samson R.A. Identification and nomenclature of the genus Penicillium. Sud. Mycol. 2014;78:343–371. doi: 10.1016/j.simyco.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Endo A. A historical perspective on the discovery of statins. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2010;86:484–493. doi: 10.2183/pjab.86.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ma H.G., Liu Q., Zhu G.L., Liu H.S., Zhu W.M. Marine natural products sources from marine-derived Penicillium fungi. J. Asian Nat. Prod. Res. 2016;18:92–115. doi: 10.1080/10286020.2015.1127230. [DOI] [PubMed] [Google Scholar]
- 4.Kumla D., Pereira J.A., Dethoup T., Gales L., Freitas-Silva J., Costa P.M., Lee M., Silva A.M.S., Sekeroglu N., Pinto M.M.M., et al. Chromone derivatives and other constituents from cultures of the marine sponge-associated fungus Penicilium erubescens KUFA 0220 and their antibacterial activity. Mar. Drugs. 2018;16:289. doi: 10.3390/md16080289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nerve Regeneration Promoters Containing Semaphorin Inhibitor as the Active Ingredient. [(accessed on 7 July 2018)]; Available online: https://data.epo.org/publication-server/rest/v1.0/publication-dates/20071003/patents/EP1306093NWB1/document.html.
- 6.Fujita K.I., Nagamine Y., Ping X., Taniguchi M. Mode of action of anhydrofulvic acid against Candida utilis ATCC 42402 under acidic condition. J. Antibiot. 1999;52:628–634. doi: 10.7164/antibiotics.52.628. [DOI] [PubMed] [Google Scholar]
- 7.Chin M.R., Zlotkowski. C., Han M., Patel S., Eliasen A.M., Axelrod A., Siegel D. Expedited access to vinaxanthone and chemically edited derivatives possessing neuronal tegenerative effects through ynone coupling reactions. ACS Chem. Neurosci. 2015;6:542–550. doi: 10.1021/cn500237z. [DOI] [PubMed] [Google Scholar]
- 8.Aoki M., Itezono Y., Shirai H., Nakayama N., Sakai A., Tanaka Y., Yamaguchi A., Shimma N., Yokose K. Structure of a novel phospholipase C inhibitor, vinaxanthone (Ro 09-1450), produced by Penicillium vinaceum. Tetrahedron Lett. 1991;32:4737–4740. doi: 10.1016/S0040-4039(00)92295-0. [DOI] [Google Scholar]
- 9.Wrigley S.K., Latif M.A., Gibson T.M., Chicarelli-Robinson M.I., Williams D.H. Structure elucidation of xanthone derivatives with CD4-binding activity from Penicillium glabrum (Wehmer) Westling. Pure Appl. Chem. 1994;66:2383–2386. doi: 10.1351/pac199466102383. [DOI] [Google Scholar]
- 10.Kumagai K., Hosotani N., Kikuchi K., Kimura T., Saji I. Xanthofulvin, a novel semaphoring inhibitor produced by a strain of Penicillium. J. Antibiot. 2003;56:610–616. doi: 10.7164/antibiotics.56.610. [DOI] [PubMed] [Google Scholar]
- 11.Stepanović S., Vuković D., Dakic I., Savić B., Švabic-Vlahović M. A modified -plate test for quantification of staphylococcal biofilm formation. J. Microbiol. Methods. 2000;40:175–179. doi: 10.1016/S0167-7012(00)00122-6. [DOI] [PubMed] [Google Scholar]
- 12.Stepanović S., Vuković D., Hola V., Di Bonaventura G., Djukić S., Ćirković I., Ruzicka F. Quantification of biofilm in microtiter plates: Overview of testing conditions and practical recommendations for assessment of biofilm production by staphylococci. Apmis. 2007;115:891–899. doi: 10.1111/j.1600-0463.2007.apm_630.x. [DOI] [PubMed] [Google Scholar]
- 13.Cegelski L., Marshall G.R., Eldridge G.R., Hultgren S.J. The biology and future prospects of of antivirulence therapies. Nat. Rev. Microbiol. 2008;6:17–27. doi: 10.1038/nrmicro1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miquel S., Lagrafeuille R., Souweine B., Forestier C. Anti-biofilm activity as a health issue. Front. Microbiol. 2016;7:1–14. doi: 10.3389/fmicb.2016.00592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sheldrick G.M. A short story of SHELX. Acta Cryst. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 16.Simões R.R., Aires-de-Sousa M., Conceicao T., Antunes F., da Costa P.M., de Lencastre H. High prevalence of EMRSA-15 in Portuguese public buses: A worrisome finding. PLoS ONE. 2011;6:e17630. doi: 10.1371/journal.pone.0017630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bessa L.J., Barbosa-Vasconcelos A., Mendes A., Vaz-Pires P., Martins da Costa P. High prevalence of multidrug-resistant Escherichia coli and Enterococcus spp. in river water, upstream and downstream of a wastewater treatment plant. J. Water Health. 2014;12:426–435. doi: 10.2166/wh.2014.160. [DOI] [PubMed] [Google Scholar]
- 18.Clinical and Laboratory Standards Institute (CLSI) Performance Standards for Antimicrobial Susceptibility Testing. 27th ed. Wayne; Wayne, PA, USA: 2017. [Google Scholar]
- 19.Clinical and Laboratory Standards Institute (CLSI) Performance Standards for Antimicrobial Disk Susceptibility Tests. 11th ed. Wayne; Wayne, PA, USA: 2012. [Google Scholar]
- 20.Clinical and Laboratory Standards Institute (CLSI) Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically. 10th ed. Wayne; Wayne, PA, USA: 2015. [Google Scholar]
- 21.Buttachon S., Ramos A.A., Inácio Â., Dethoup T., Gales L., Lee M., Costa P.M., Silva A.M.S., Sekeroglu N., Rocha E., et al. Bis-indolyl benzenoids, hydroxypyrrolidine derivatives and other constituents from cultures of the marine sponge-associated fungus Aspergillus candidus KUFA0062. Mar. Drugs. 2018;16:119. doi: 10.3390/md16040119. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.