Abstract
Rice foot rot disease caused by the pathogen Dickeya zeae (formerly known as Erwinia chrysanthemi pv. zeae), is a newly emerging damaging bacterial disease in China and the southeast of Asia, resulting in the loss of yield and grain quality. However, the genetic resistance mechanisms mediated by miRNAs to D. zeae are unclear in rice. In the present study, 652 miRNAs including osa-miR396f predicted to be involved in multiple defense responses to D. zeae were identified with RNA sequencing. A total of 79 differentially expressed miRNAs were detected under the criterion of normalized reads ≥10, including 51 known and 28 novel miRNAs. Degradome sequencing identified 799 targets predicted to be cleaved by 168 identified miRNAs. Among them, 29 differentially expressed miRNA and target pairs including miRNA396f-OsGRFs were identified by co-expression analysis. Overexpression of the osa-miR396f precursor in a susceptible rice variety showed enhanced resistance to D. zeae, coupled with significant accumulation of transcripts of osa-miR396f and reduction of its target the Growth-Regulating Factors (OsGRFs). Taken together, these findings suggest that miRNA and targets including miR396f-OsGRFs have a role in resisting the infections by bacteria D. zeae.
Keywords: rice foot rot disease, Dickeya zeae, microRNA, RNA and degradome sequencing, disease resistance
1. Introduction
Plant microRNAs (miRNAs) are non-protein coding RNAs with 21–24 nucleotides (nt). Most recently, miRNAs were demonstrated to be a critical component of sophisticated plant defense system [1,2,3]. Plants are known to use a multifaceted defense system to prevent infections by pathogenic microbes through a co-evolutionary mechanism with pathogens. Plant immunity is involved in two tiers of defenses where the first tier is a pathogen-associated molecular pattern (PAMP) trigged immunity (PTI) [4,5]. The second tier of defense is often involved in pathogen effector-triggered immunity (ETI) mediated mostly by R proteins with leucine rich repeat and nucleotide binding site (NLR) [4,5]. Activation of ETI often reinforces PTI to achieve plant immunity to prevent damages by invaders [6,7,8]. It was predicted that targets of plant miRNAs can be NLR genes, or genes involved in PTI [2,3]. To name a few, Li et al showed that two miRNA, nta-miR6019 and nta-miR6020 co-regulated the transcripts of R genes to regulate resistance responses and productivity in tobacco [9]. In Arabidopsis, bacterial flagellin flg22-induced miR393 promoted resistance response to virulent Pseudomonas syringae pv. tomato Pto DC3000 by inhibiting predicted target-auxin receptor transcripts [10]. Three bacterial flagellin flg22-induced miRNAs, miR160, miR398b and miR773 were shown to be involved in resistance responses via regulating PAMP-induced callose deposition in Arabidopsis [11,12]. The stress-inducible miR163 and its target histone deacetylase were demonstrated to confer resistance to P. s. pv. Tomato [13].
In rice, recent studies also showed that miRNAs were involved in disease resistance [14,15,16]. Examples are 23 miRNAs and 59 targets were predicted to be involved in resistance against rice sheath blight disease [17]. A group of miRNAs and their targets functioned in an incompatible interaction between rice and fungal pathogen Magnaporthe oryzae (M. oryzae) [18]. In addition, the large differentially expressed miRNAs and their putative target genes were involved in hormone biosynthesis and signaling pathways in rice after infections by bacterial pathogens [19,20]. The osa-miR169, miR160a, miR398b, miR7695 and miR1861k repressed the expression of the predicted targets respectively, have a role in resistance to M. oryzae or Xanthomonas oryzae pv. oryzae (Xoo) [18,21,22,23].
Rice foot rot disease caused by the pathogenic bacteria D. zeae, is a newly emerged destructive bacterial disease threatening rice yield and quality [24]. The occurrence of rice foot rot disease was first reported in Japan in 70 s of the 20th century [25]. Recently, field incidence of rice foot rot disease shows an increasing trend in rice planting areas in Southeast Asia and South China [24]. Little is known about the pathogenicity of D. zeae and the genetic resistance mechanism to D. zeae [26,27,28,29], and it is unknown if miRNAs are involved in the defense responses to D. zeae in rice. Uncovering the resistance mechanism will lead to developing better control methods to this disease.
We previously isolated and identified an isolate of D. zeae JS2012 from diseased roots of a rice variety [30]. The objectives of the present study were to (1) determine temporal transcriptome profiles of miRNAs after D. zeae infection, (2) predict miRNA targets by degradome sequencing, (3) validate differentially expressed miRNAs and targets by qRT-PCR, and (4) overexpress the osa-miR396f precursor to determine if enhanced resistance to D. zeae occurs in rice. Through these efforts we demonstrated that a set of candidate miRNAs and relevant targets including miR396 and predicted target may play an important role in the defense responses to D. zeae.
2. Results
2.1. Overview of miRNA Sequencing
A total of 4 miRNA libraries were constructed from a resistant rice variety (an incompatible interaction) at 0 h (K0), 6 h (K1), 12 h (K2) and 48 h (K3) after D. zeae inoculation. The illumina sequencings revealed 1,053,550 (K0), 1,000,514 (K1), 1,023,748 (K2), 483,076 (K3) unique clean reads respectively after removing low resolution, length less than 18 nt or greater than 25 nt, junk reads, mRNA, rRNA, tRNA, snRNA and snoRNA (Table 1). A total number of 19–24 nt from K0 miRNA library accounted for 82.82%, in which 21, 24 nt unique miRNA accounted for 16.10% and 11.03% respectively (Table S1). After inoculation with D. zeae in a resistant rice variety, the length of unique miRNAs was found between the range of 20–24 nt. Among them, 21 and 24 nt unique miRNAs accounted for 15.25% and 18.61% (K1), 17.15% and 14.83% (K2), 16.13% and 9.64%, respectively (K3; Table S1). Differences in miRNA length and number from different time points after pathogen inoculation may reflect different biological roles during different stages of rice growth and development.
Table 1.
Overview of sequencing reads from raw data to cleaned sequences in four libraries.
| Sequence Type | K0 | K1 | K2 | K3 | ||||
|---|---|---|---|---|---|---|---|---|
| Total Counts | Unique Counts | Total Counts | Unique Counts | Total Counts | Unique Counts | Total Counts | Unique Counts | |
| Raw reads | 6,166,878 | 2,434,380 | 8,949,370 | 2,363,366 | 7,312,871 | 2,301,987 | 5,565,701 | 1,511,820 |
| 3ADT&length filter | 1,909,403 | 1,024,342 | 2,458,847 | 982,458 | 2,232,137 | 922,159 | 2,525,190 | 803,851 |
| Junk reads | 18,778 | 13,765 | 21,772 | 14,658 | 25,064 | 16,132 | 12,782 | 9,015 |
| Non-coding RNA | 559,796 | 169,225 | 600,364 | 129,071 | 625,544 | 132,521 | 359,131 | 91,856 |
| Rfam | 888,552 | 201,522 | 1,146,506 | 162,399 | 1,071,523 | 164,209 | 637,768 | 115,524 |
| mRNA | 1,279,886 | 170,257 | 2,684,763 | 234,424 | 1,778,770 | 204,532 | 1,146,100 | 122,153 |
| Repeats | 13,479 | 3,241 | 19,257 | 2,241 | 14,299 | 2,895 | 9,875 | 1,869 |
| rRNA | 641,748 | 127,477 | 857,965 | 102,851 | 749,376 | 98,155 | 491,401 | 73,325 |
| tRNA | 176,213 | 57,003 | 173,576 | 39,418 | 228,124 | 45,153 | 91,910 | 27,900 |
| snoRNA | 12,162 | 4,408 | 33,516 | 6,862 | 21,275 | 5,829 | 10,543 | 3,854 |
| snRNA | 9,586 | 2,845 | 24,758 | 3,638 | 14,329 | 3,441 | 7,713 | 2,389 |
| Other Rfam RNA | 48,843 | 9,789 | 56,691 | 9,630 | 58,419 | 11,631 | 36,201 | 8,056 |
| Clean reads | 2,385,536 | 1,053,550 | 3,164,367 | 1,000,514 | 2,637,057 | 1,023,748 | 1,512,623 | 483,076 |
2.2. Identification of Known and Novel miRNAs
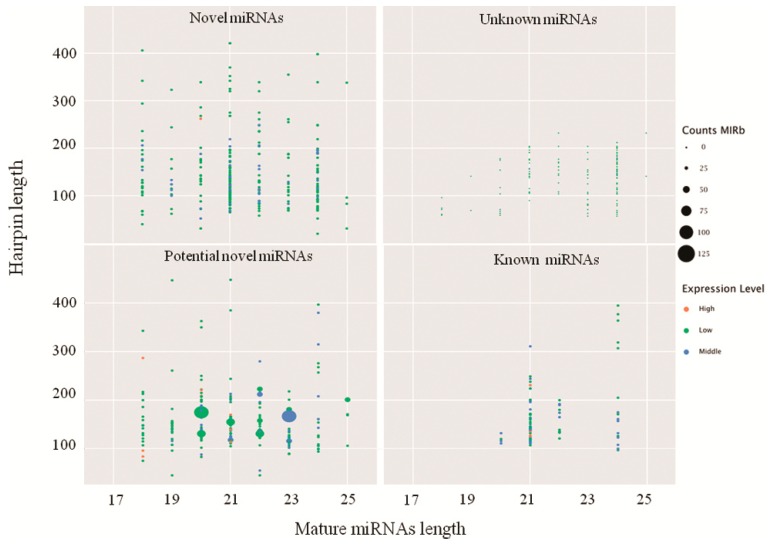
A total of 652 miRNAs including 250 novel, 78 known, and 185 predicted but not mapped at the plant genome (potential novel miRNAs), and 139 mapped on the plant genome not in the miRNA database were identified (unknown miRNAs; Figure 1; Table S2). A total of 10 known miRNAs, osa-miR1425-5p, osa-miR156b-3p, osa-miR156c-3p, osa-miR171b, osa-miR156a, osa-miR535-5p, osa-miR162a, osa-miR166a-5p, osa-miR166a-3p and osa-miR166b-5p with more than 100 reads was detected (Figure 1; Table S2). Among them, three miRNA families, osa-miR156, 166 and 167, with the highest frequencies were identified (Figure 1; Table S2). The miRNAs mapped to the rice genome but did not match to known pre-miRNAs as novel miRNAs. 250 novel miRNAs were predicted in four libraries (Figure 1; Table S2). The majority of novel miRNAs was found to be slowly accumulated after inoculation with D. zeae. However, osa-MIR5083-p5 was the most abundant one with reads of 135.04 (K0), 484.00 (K1), 713.11 (K2) and 233.71 (K3) in four libraries, respectively, suggesting its unique role for responding to D. zeae infection in a resistant rice variety.
Figure 1.
The four indicated groups of miRNAs identified from an incompatible interaction. CountsMIRb, the counts of miRNAs from miRBase. Expression level, low indicates <10, middle indicates >10 but less than average, high indicates over average.
2.3. D. zeae-Responsive miRNAs in Resistant Rice
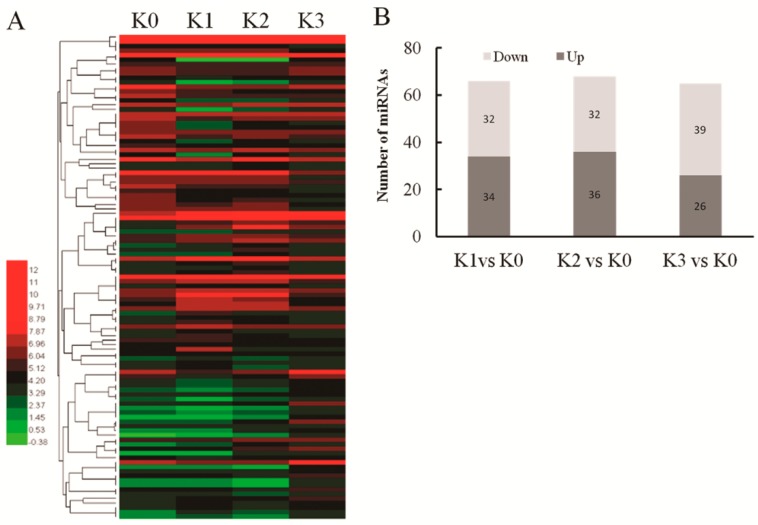
To identify miRNAs in resistant rice that respond to D. zeae, we analyzed the differential expressions of miRNAs in the four libraries with the normalized reads from deep sequencing. A total of 79 differentially expressed miRNAs was found in all four libraries including 51 known miRNAs and 28 novel miRNAs under the criterion of normalized reads ≥10 (Figure 2A; Table S3). Furthermore, 34 miRNAs were up-regulated and 32 miRNAs were down-regulated at six hours post inoculation (hpi). At 12 hpi, 36 miRNAs were up-regulated and 32 miRNAs were down-regulated. At 48 hpi, 26 miRNAs were up-regulated and 39 miRNAs were down-regulated (Figure 2B; Table S3). These results suggest that certain miRNAs were expressed at some time points after D. zeae infection in an incompatible interaction suggesting that they may play an important role during pathogen infection.
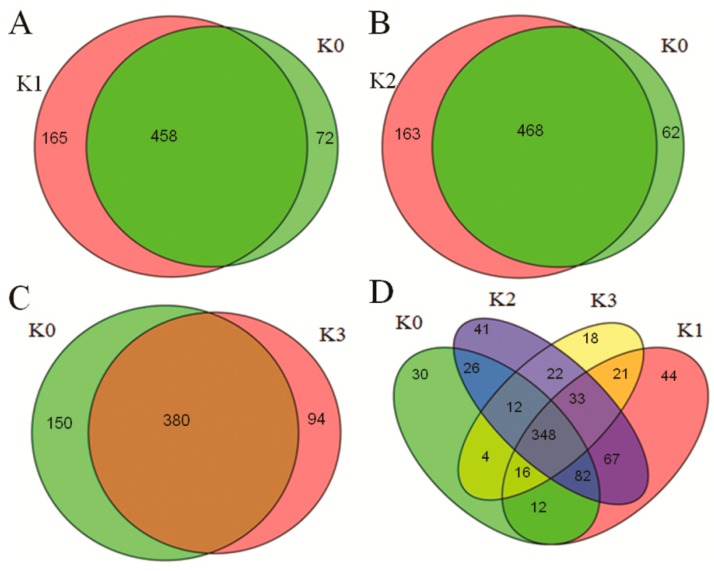
Figure 2.
A summary of unique miRNAs detected in four libraries. (A–C) The miRNAs responded specifically the D. zeae infection in an incompatible interaction at 6, 12 and 48 h compared with at 0 h respectively. (D) The specific miRNAs were detected among four libraries.
Sequence analysis revealed that there were 530 miRNAs in K0, 623 in K1, 631 in K2, and 474 in K3, respectively (Figure 3). When compared with K0, 165 unique miRNAs were identified at 6 hpi, (Figure 3A), 163 unique miRNAs were identified at 12 hpi, 94 unique miRNAs were detected at 48 hpi, respectively (Figure 3B,C). When compared among four libraries, 30 (K0), 44 (K1), 41 (K2) and 18 (K3) miRNAs were detected (Figure 3D). The presences of different miRNAs at different time points after inoculation suggest miRNAs have a role to D. zeae in an incompatible interaction.
Figure 3.
D. zeae-responsive miRNAs in resistant rice. (A) Heat plot showing D. zeae-responsive miRNAs identified in an incompatible interaction. (B) The number of differentially expressed miRNAs in four groups.
2.4. Target Prediction of miRNAs by Degradome Sequencing and Analysis
To identify the relevant targets of miRNAs, high-throughput degradome sequencing was used and the data were analyzed with the CleavLand 3 pipeline. Degradome sequencing analysis showed that 10931903 and 8679632 raw reads were detected from the control (T0 h) and infected (T6–48 h) rice samples, respectively (Table S4). After removing the adaptor reads and mapping the rice transcriptome, 35890 and 32146 covered transcripts were identified in the control (T0 h) and infected (T6–48) rice samples, respectively (Table S4). Further, a total of 799 targets were predicted to be cleaved by the 168 identified miRNAs in an incompatible interaction (Table S5). Many transcript targets mediated by miRNAs were closely related to the signal transduction involved in plant immunity, such as PR5 (LOC_Os03g14030.1), leucine-rich repeat (LRR) kinase (LOC_Os12g10740.1), calcium-dependent kinase (LOC_Os07g22640.), WRKY transcription factor (LOC_Os04g51560.1), AUX/IAA transcriptional regulator (LOC_Os12g40900.1), ethylene-responsive binding protein (LOC_Os07g42510.1), growth-regulating factor 5 (LOC_Os02g53690.1) and etc (Table 2). These findings demonstrate that miRNAs-target pairs may be involved in resistance responses through multiple defense pathways in rice.
Table 2.
The identified targets involved in rice defense responses by degradome sequencing.
| No. | Targets | Target Annotation | UP/Down | Cleavage Site |
|---|---|---|---|---|
| 1 | LOC_Os08g40900.1 | Auxin response factor 6 | up | 3466 |
| 2 | LOC_Os07g40290.1 | Auxin-responsive GH3 family protein | up | 2091 |
| 3 | LOC_Os06g39590.1 | AUX/IAA transcriptional regulator | up | 502 |
| 4 | LOC_Os03g51970.1 | Growth-regulating factor 1 | up | 430 |
| 5 | LOC_Os11g35030.1 | Growth-regulating factor 2 | up | 880 |
| 6 | LOC_Os02g53690.1 | Growth-regulating factor 5 | up | 581 |
| 7 | LOC_Os04g04330.1 | Leucine-rich repeat kinase | down | 3509 |
| 8 | LOC_Os10g33940.1 | Auxin response factor 16 | down | 2332 |
| 9 | LOC_Os12g10740.1 | Leucine-rich repeat protein kinase | down | 1219 |
| 10 | LOC_Os11g38440.1 | NB-ARC disease resistance protein | down | 3382 |
| 11 | LOC_Os04g59430.1 | Auxin response factor 16 | down | 1345 |
| 12 | LOC_Os04g38720.1 | NAC domain containing protein 80 | down | 811 |
| 13 | LOC_Os08g10080.1 | NAC domain containing protein 1 | down | 783 |
| 14 | LOC_Os12g40900.1 | AUX/IAA transcriptional regulator | down | 536 |
| 15 | LOC_Os07g42510.1 | Ethylene-responsive binding protein | down | 721 |
| 16 | LOC_Os03g08050.1 | GTP binding Elongation factor Tu protein | down | 969 |
| 17 | LOC_Os03g08020.1 | GTP binding Elongation factor Tu protein | down | 969 |
| 18 | LOC_Os03g08010.1 | GTP binding Elongation factor Tu protein | down | 969 |
| 19 | LOC_Os06g01620.1 | GRAS family transcription factor | down | 464 |
| 20 | LOC_Os02g44360.1 | GRAS family transcription factor | down | 1362 |
| 21 | LOC_Os02g09060.1 | Zinc finger family protein | down | 401 |
| 22 | LOC_Os08g41290.1 | Auxin-responsive family protein | down | 178 |
| 23 | LOC_Os04g51560.1 | WRKY DNA-binding protein 11 | down | 623 |
| 24 | LOC_Os10g01100.1 | Lectin protein kinase family protein | down | 2638 |
| 25 | LOC_Os02g50960.1 | Auxin efflux carrier family protein | down | 2109 |
| 26 | LOC_Os12g37760.1 | NB-ARC disease resistance protein | down | 3101 |
| 27 | LOC_Os07g33480.1 | Cytochrome P450 family 716 | down | 1606 |
| 28 | LOC_Os02g48080.1 | S-locus lectin protein kinase | down | 1135 |
| 29 | LOC_Os02g11980.1 | Leucine-rich repeat protein kinase | down | 3002 |
| 30 | LOC_Os12g43640.1 | Leucine-rich receptor-like protein kinase | down | 3203 |
| 31 | LOC_Os01g11340.1 | Cytochrome P450, family 710 | down | 1694 |
| 32 | LOC_Os02g14120.1 | Leucine-rich repeat protein kinase | down | 2050 |
| 33 | LOC_Os12g01510.1 | Leucine-rich repeat protein kinase | down | 2123 |
| 34 | LOC_Os12g37980.1 | Leucine-rich repeat transmembrane kinase | down | 2614 |
2.5. Functional Classification of Predicted Targets
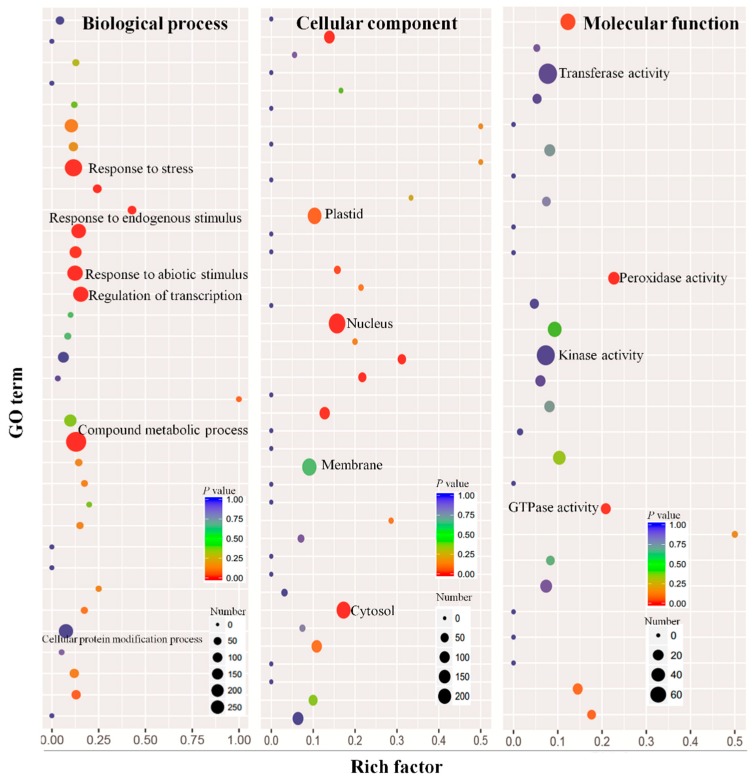
A gene ontology (GO) analysis was used to classify the biochemical functions of predicted targets (Figure 4). A total of 858 transcript targets were predicted as protein kinase, 852 as transferase, 171 as signal transducers, and 81 as protein receptors, suggesting their potential involvements in signal recognition and transduction in plant immunity (Figure 4). Similarly, a group of targets was predicted to be involved in multiple molecular function and all of them were predicted to be located in diverse cellular components of cells, suggesting that predicted target genes may also play a role in biological processes including hormone stimulus, signal transducer, sequence-specific DNA binding, secondary metabolic process, cell growth, and multicellular organismal development (Figure 4).
Figure 4.
GO functional classification of identified target genes.
2.6. Co-expression Analysis and Validation of miRNA-Targets by qRT-PCR
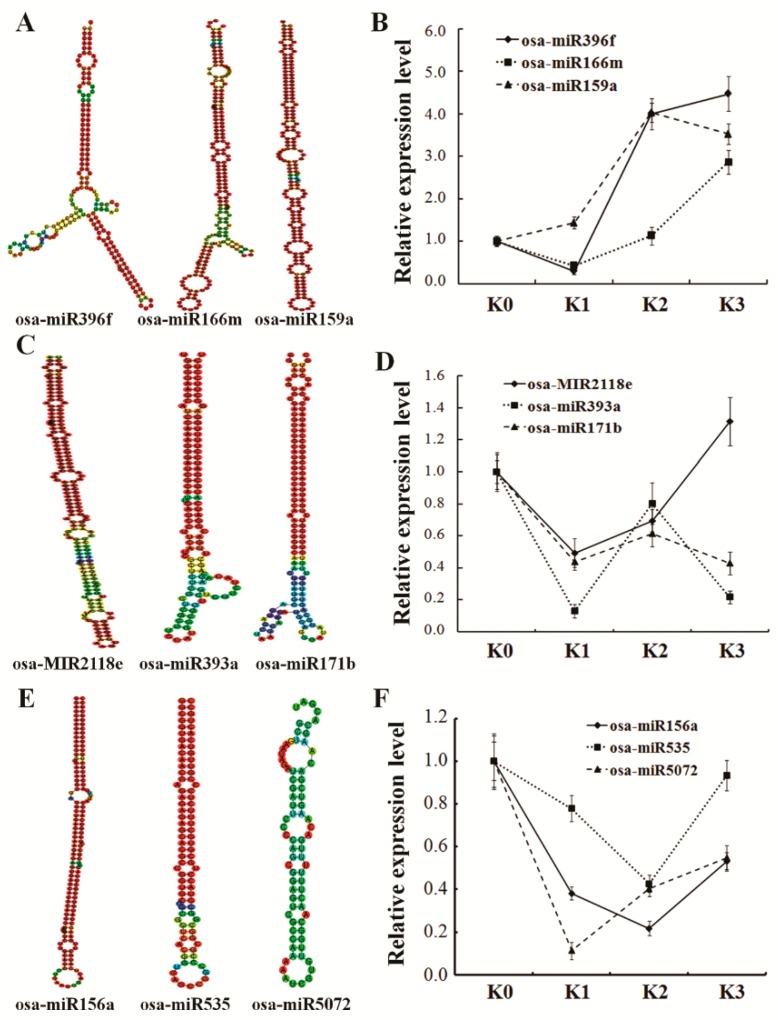
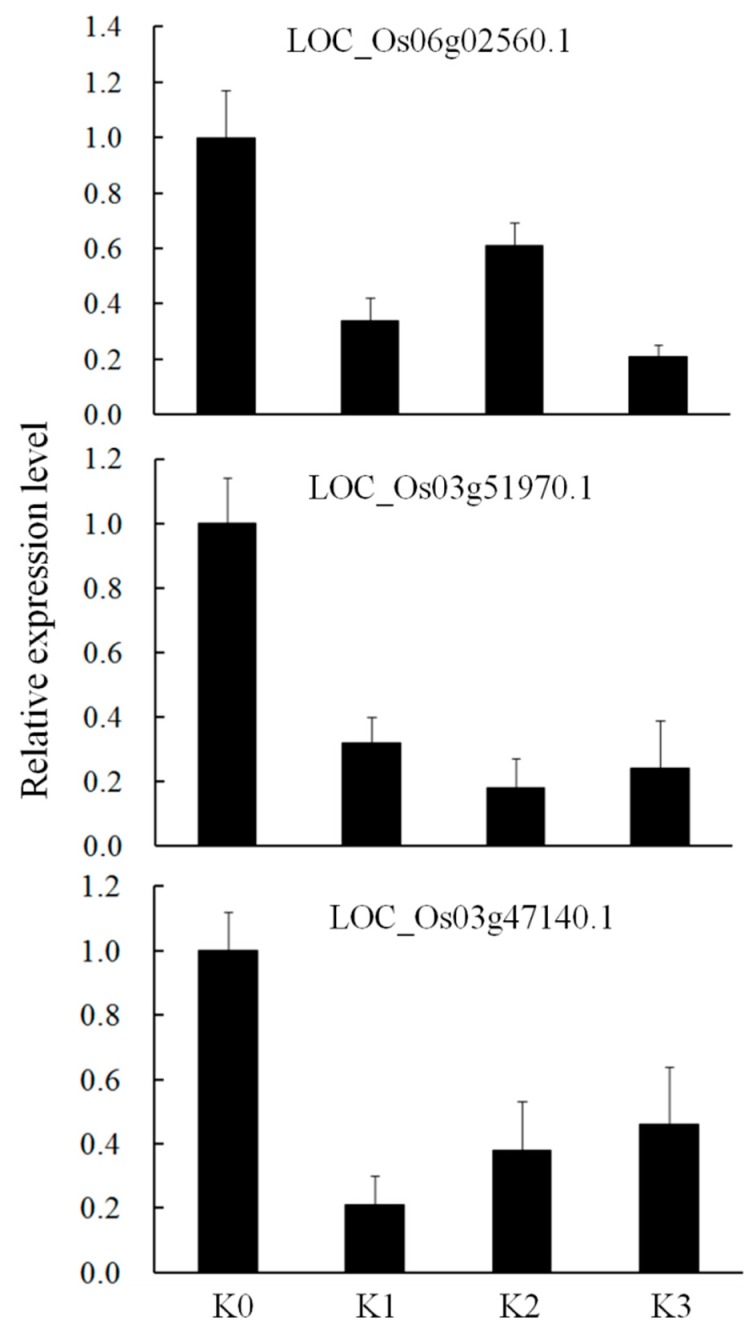
We analyzed the miRNAs obtained by miRNA sequencing, and verified the data with qRT-PCR and degradome sequencing. Firstly, 29 (36.71%) of 79 differentially expressed miRNAs detected by miRNA-seq were detected from the degradome sequencing (Table S6). A total of 82 genes resulting in 132 transcripts were predicted to be targets of these 29 miRNAs (Table S6). Moreover, 9 differentially expressed miRNAs based on the number of normalized reads were selected from 29 miRNAs (Table 3). Interestingly, we found that among 9 miRNAs, 3 miRNAs were induced and 6 were reduced (Table S2). Moreover, the secondary structures of 9 miRNAs precursor were predicted by RNAfold respectively (Figure 5A). Despite the difference in the fold changes, the expression patterns of 9 miRNAs analyzed by qRT-PCR were consistent with that of miRNA-seq (Figure 5B). Moreover, the degradome sequencing data revealed an opposite expression pattern of the cognate targets of these 9 miRNAs (Tables S2 and S6) suggesting that these miRNAs may regulate target gene expression at different time points post inoculation in an incompatible interaction. Reduced expressions of three target genes, LOC_Os06g02560.1, LOC_Os03g51970.1 and LOC_Os03g47140.1) predicted to be regulated by osa-miR396f were verified by qRT-PCR, suggesting that the osa-miR396f-targets pair may play a role for resistance to D. zeae in rice (Figure 6).
Table 3.
The miRNA-target pairs in multi-defense responses detected by miRNA and degradome sequencing.
| No. | miR_name | Targets | Target Annotation | T0 h | T6–48 h | Up/Down | Cleavage Site |
|---|---|---|---|---|---|---|---|
| 1 | Osa-miR2118e- | LOC_Os03g06680.1 | Protein of unknown function 506 | 91.48 | 0 | down | 1608 |
| p5_1ss13TA | LOC_Os02g50960.1 | Auxin efflux carrier protein | 91.48 | 0 | down | 2109 | |
| 2 | Osa-miR393a | LOC_Os03g36080.1 | – | 1829.51 | 0 | down | 741 |
| LOC_Os05g05800.1 | F-box/RNI-like protein | 182.95 | 460.85 | up | 1708 | ||
| LOC_Os04g32460.1 | Auxin signaling F-box 2 | 1234.92 | 172.82 | down | 2235 | ||
| 3 | Osa-miR396f | LOC_Os02g53690.1 | Growth-regulating factor 5 | 0 | 460.85 | up | 581 |
| LOC_Os03g51970.1 | Growth-regulating factor 1 | 0 | 115.21 | up | 430 | ||
| LOC_Os11g35030.1 | Growth-regulating factor 2 | 0 | 115.21 | up | 880 | ||
| 4 | Osa-miR166m_R-1 | LOC_Os12g41860.1 | Leucine zipper family protein | 640.33 | 76.81 | down | 888 |
| LOC_Os03g43930.1 | Leucine zipper family protein | 548.85 | 76.81 | down | 966 | ||
| LOC_Os10g33960.1 | Leucine zipper family protein | 922.38 | 249.63 | down | 935 | ||
| LOC_Os03g01890.1 | Leucine zipper family protein | 899.51 | 249.63 | down | 1100 | ||
| 5 | Osa-miR171b | LOC_Os02g44370.1 | GRAS family transcription factor | 457.38 | 0 | down | 1537 |
| LOC_Os02g44360.1 | GRAS family transcription factor | 274.43 | 0 | down | 1362 | ||
| LOC_Os10g40390.1 | GRAS family transcription factor | 182.95 | 0 | down | 179 | ||
| LOC_Os06g01620.1 | GRAS family transcription factor | 1189.18 | 460.85 | down | 467 | ||
| LOC_Os04g46860.1 | GRAS family transcription factor | 2378.36 | 691.27 | down | 1337 | ||
| 6 | Osa-miR156a | LOC_Os08g41940.1 | SBP transcription factor | 457.38 | 0 | down | 1064 |
| LOC_Os01g69830.1 | SBP transcription factor | 182.95 | 115.21 | down | 1163 | ||
| LOC_Os02g04680.1 | SBP transcription factor | 457.38 | 57.61 | down | 1975 | ||
| LOC_Os11g30370.1 | SBP transcription factor | 91.48 | 0 | down | 1101 | ||
| LOC_Os09g31438.1 | SBP transcription factor | 91.48 | 0 | down | 819 | ||
| LOC_Os02g07780.1 | SBP transcription factor | 45.74 | 0 | down | 997 | ||
| LOC_Os06g49010.1 | SBP transcription factor | 45.74 | 0 | down | 1696 | ||
| LOC_Os06g45310.1 | SBP transcription factor | 274.43 | 115.21 | down | 864 | ||
| 7 | Osa-miR535-5p | LOC_Os06g45310.1 | Squamosa promoter-like 11 | 91.48 | 0 | down | 863 |
| 8 | Osa-miR159a_1R-3 | LOC_Os01g59660.1 | Myb domain protein 33 | 411.64 | 288.03 | down | 135 |
| LOC_Os10g05230.1 | RING/U-box superfamily protein | 274.43 | 0 | down | 1271 | ||
| LOC_Os12g10740.1 | Leucine-rich repeat protein kinase | 182.95 | 0 | down | 346 | ||
| LOC_Os06g40330.1 | Myb domain protein 65 | 182.95 | 0 | down | 1219 | ||
| LOC_Os01g47530.1 | MAP kinase 20 | 68.61 | 28.80 | down | 2333 | ||
| 9 | Osa-miR5072 _L-4 | LOC_Os05g07050.1 | Pre-mRNA processing splicing factor | 45.74 | 0 | down | 5043 |
Figure 5.
Predicted secondary structures and verification of expression of miRNAs precursor. (A,C,E) Predicted secondary structures of nine miRNAs precursor by RNAfold (http://rna.tbi.univie.ac.at//cgi-bin/RNAWebSuite/RNAfold.cgi, 19 March 2018) respectively. (B,D,F) The expression of nine miRNAs precursor were tested by qRT-PCR. K0, K1, K2 and K3 represent a resistance rice variety at 0, 6, 12 and 48 hpi respectively. Data represent means of three replicates ± standard deviation for each miRNA in the four miRNAs libraries.
Figure 6.
The expression levels of three targets OsGRFs of osa-miR396f analyzed by qRT-PCR in an incompatible interaction. Data represent means of three replicates ± standard deviation for each target in the four miRNAs libraries from an incompatible interaction.
2.7. Overexpression of Osa-miR396f Precursor Enhanced Rice Resistance to D. zeae
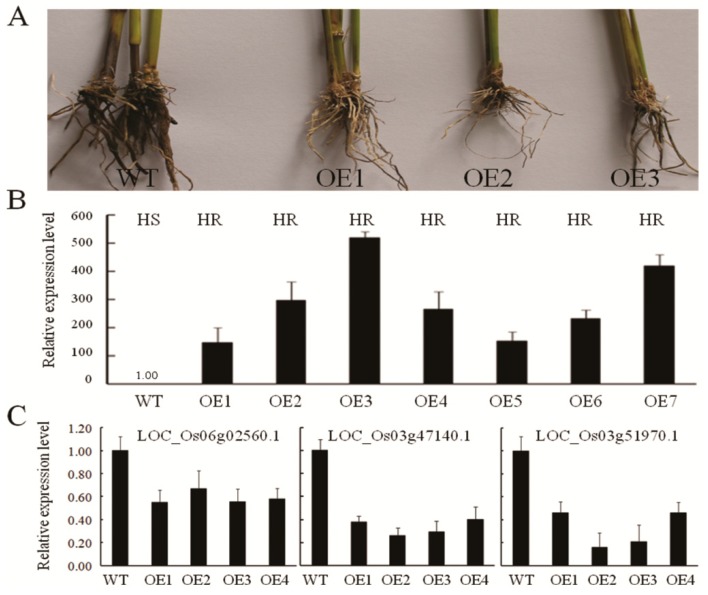
To validate if osa-miR396f was involved in defense response to D. zeae, the precursor sequences of osa-miR396f were isolated from the resistant cultivar Nanjing 40 and were expressed under the control of the constitutive CMV35S promoter and transformed into susceptible japonica rice cultivar Nipponbare. Seven positive osa-miR396f precursor-overexpressing rice plants were inoculated with D. zeae. The transgenic rice plants with the osa-miR396f precursor showed increased resistance to D. zeae in comparison with that of Nipponbare (Figure 7A). Consistently, the transcripts of osa-miR396f precursor in all seven independent transgenic lines were higher than that in wild-type plants (Figure 7B). As predicted, transcripts of all three targets OsGRFs of osa-miR396f were significantly reduced in these transgenics revealed by qRT-PCR (Figure 7C). These results suggest that miRNA396f is involved in resisting D. zeae infection in rice.
Figure 7.
Overexpression of the osa-miR396f precursor in rice enhanced resistance to D. zeae. (A) Resistant phenotype to D. zeae in overexpressing transgenic plants; (B) The relative expression level of osa-miR396f precursor and resistant reactions in overexpressing rice. HS, highly susceptibility. HR, highly resistance; (C) The expression levels of three targets of osa-miR396f were inhibited in overexpressing rice. WT, wild-type rice Nipponbare. OE1-7, the osa-miR396f precursor- overexpressing rice plants.
3. Discussion
Recently, rice bacterial foot rot disease caused by D. zeae has the potential to become one of the most important bacterial diseases, resulting in the loss of yield and grain quality in China, in the southeast of Asia and worldwide. In the present study we identified and characterized a number of miRNAs-target pairs involved in defense responses in rice by miRNA, degradome sequencing and qRT-PCR.
The fast evolved high-throughput sequencing technology allows rapid identification of miRNAs and targets in plants that are involved in plant immunity [3,31]. Accumulated studies suggest that miRNAs played an important role for regulating the biological process and stress responses in plants [32,33]. It is known that plant miRNAs expressed differently during the infection process of pathogens [7]. In the present study, we found 652 miRNAs including 79 significantly differentially expressed miRNAs at different time points post inoculation in an incompatible interaction (Figure 2A; Table S3). For instance, two miRNAs miR827n-5p and miR164 we identified were also identified in a resistant rice variety to M. grisea, as negative and positive regulator respectively [21]. Moreover, the expression levels of seven miRNAs, miR156a, 159b, 166e-3p, 394, 396c-3p, 812 and 827 were decreased in rice in resistance response to Xoo [20]. We showed a similar responsive mode of these miRNAs to fungal and bacterial pathogens infection demonstrating that miRNAs were indeed involved in resistance response to D. zeae in rice. We also validated the expression profiles of nine miRNAs (miR2118, 393, 396, 166, 171, 156, 535, 159 and 5072) related to defense responses by qRT-PCR (Figure 5). The relevant targets of these nine miRNAs were also identified and verified by degradome sequencing and qRT-PCR (Table S6; Figure 6). These 9 miRNAs were previously shown to respond to the pathogens infection in an incompatible interaction, probably resulting in the activation of multiple defense responses [34,35,36,37,38]. Our data suggest that these nine differentially expressed miRNAs may be involved in regulating the resistance to D. zeae in rice.
In the present study, we showed that overexpression of osa-miR396f precursor in rice enhanced resistance to D. zeae, correlated with significant reduction of transcripts of three predicted targets OsGRFs (Figure 7). Therefore, miR396 may modulate resistance to D. zeae by cleaving the transcripts of its targets OsGRFs. Rice miR396 family has eight members, osa-miR396a, 396b, 396c, 396d, 396e, 396f, 396g and 396h in the miRNA database (miRBase, http://www.mirbase.org/, 27 March 2018). Rice miR396-GRFs pair was previously shown to regulate plant growth and development by positively activating plant hormones indole-3-acetic acid (IAA), gibberellin (GA) and brassinolide biosynthesis pathways in rice [39,40,41,42]. The miR396 family members, osa-miR396e-3p and osa-miR396d/e-5p were induced by blast pathogen in a susceptible rice variety, and the expression level of osa-miR396c-5p was increased in a resistant rice variety [21]. Some members of miR396 were previously predicted to be a negative factor for the infection process of southern rice black-streaked dwarf virus (SRBSDV) in rice [38]. In contrast, overexpression of miR396a or miR396b significantly compromised the susceptibility to cyst Nematode in Arabidopsis [43]. The Growth-Regulating Factors (GRFs) were also the targets of miR396 family members and positively regulated the number and size of the rice grain [44]. The blocking of OsGRFs by miR396 generated a larger grain size, auxiliary branches and spikelets then enhanced grain yield through auxin in rice [41,45,46]. We speculate that miR396-OsGRFs pair may play a role for regulating the balance between yield and disease resistance via the auxin signaling pathway. However, the biological functions of miR396 on defense responses and rice grain development still need to be explored.
Plant miRNAs were predicted to be involved in the biological process and stress responses by inhibiting the transcripts of relevant targets [1,13,33]. In the present study, 132 miRNA responsive to the infection of D. zeae in an incompatible interaction were identified, and the predicted targets of 29 differentially expressed miRNAs were identified by degradome sequencing (Table S6). These identified targets might modulate rice resistance response to D. zeae. Most of the targets we identified were predicted to be involved in multiple defense response signal pathways such as OsGRFs, Leucine-rich repeat protein kinase, MYB domain protein, and auxin efflux carrier family protein (Table 2 and Table 3; Table S6). The genes, AtGRF1 and AtGRF3 were shown to be involved in multiple resistance-related signal pathways related with cell-wall modification, cytokinin biosynthesis and the accumulation of secondary metabolites in Arabidopsis [43,47]. Overexpression of miR396a or miR396b and miR396 targets grf1/grf2/grf3 triple mutants previously significantly compromised the susceptibility to cyst nematode in Arabidopsis [43]. The miR396-GRF6 interaction network was predicted to be involved in the development of inflorescence architecture by regulating the auxin biosynthesis and signaling pathways [41]. Moreover, the auxin-responsive GH3 family gene LOC_Os07g40290.1 as the target of osa-miR172d-5p_R-2 showed more cleavages in a resistant rice variety after inoculation with D. zeae, compared with that without inoculation (Table 2). Additionally, auxin as a negative regulator was shown to contribute to resistance to bacteria pathogen Xoo, which resulted in the change of the cell wall structure [45,46]. The Mybs1 gene encoding one MYB transcription factor was involved in a broad-spectrum resistance against M. oryzae in rice by increasing the accumulation of H2O2 through inhibition of the expression level of bsr-d1 [48]. Taken together, these findings suggest that miRNA and targets are involved in multiple defense responses, including auxin and active oxygen signal pathways.
In summary, knowledge of miRNAs and targets opens a new avenue to understand the complexity of plant immunity. In the present study, a group of differentially expressed miRNAs and targets in an incompatible interaction at early interphases of host-pathogen interaction was identified by miRNA and degradome sequencing and qRT-PCR. These miRNAs and targets were predicted to be involved in multiple defense signal and diverse cellular pathways in rice. Through a transgenic approach, we demonstrated that one of these miRNAs, osa-miR396f enhanced the defense responses against D. zeae by inhibiting the transcripts of the three growth related transcription factor genes, OsGRFs. These results not only pave the road for controlling the newly emerging damaging foot rot disease but also promise a better understanding of plant innate immunity.
4. Materials and Methods
4.1. Plant Materials and Measurement of Resistance Reactions to D. zeae
The foot rot disease resistant Japonica rice variety Nanjing 40 was used to analyze the candidate miRNAs involved in disease resistance and the relevant targets by high-through sequencing technologies. The individual positive rice lines overexpressing the osa-miR396f precursor were chosen for analyzing disease reactions to D. zeae. All the plants used in this study were grown in a greenhouse in Jiangsu Academy of Agricultural Sciences, Nanjing, Jiangsu Province, China. Disease reactions to D. zeae were measured by the basal stem and root inoculation methods, respectively [30,49]. Disease reaction was classified as five groups by measuring the percentage of diseased area at 7–10 days after inoculation. No visible disease symptom in the stem and foot indicates immunity. The disease index less than or equal to five indicates highly resistance (HR). The disease index 5.1 to 12.4 indicates moderate resistance (MR). The disease index, 12.5 to 19.9 indicates moderate susceptibility (MS). The disease index greater than or equal to 20 indicates highly susceptibility (HS) [30,49].
4.2. Construction of Small RNA Library, Sequencing and Data Analysis
Total RNA was extracted from four mixed rice roots at 0 (K0), 6 (K1), 12 (K2) and 48 (K3) hpi with D. zeae using Trizol reagent (Invitrogen, Shanghai, China) following the manufacturer’s procedure respectively. The rice roots sampled with two biological repeats were combined as K0, K1, K2 and K3 for total RNA extraction, small RNA library construction and sequencing. Approximately 1 mg of total RNAs was used to prepare the miRNA library according to the protocol of TruSeq Small RNA Sample Prep Kits (Illumina, San Diego, CA, USA). The libraries were sequenced with the single-end sequencing (36 bp) on an Illumina Hiseq2500 at the LC-Bio Co., Ltd (Hangzhou, China) following the vendor’s recommended protocol.
The raw reads were analyzed with the Illumina pipeline filter (Solexa 0.3), and the dataset was further processed with an in-house program, ACGT101-miR (LC Sciences, Houston, TX, USA) to remove adapter dimers, junk, low complexity, common RNA families (rRNA, tRNA, snRNA, snoRNA) and repeats. Subsequently, unique sequences with length in 18–25 nucleotides were mapped to specific species precursors in the miRBase database (http://www.mirbase.org/, 27 March 2018) using the BLAST algorithm to identify known miRNAs and novel 3p and 5p-derived miRNAs. The unique sequences mapping to specific species mature miRNAs in hairpin arms were identified as known miRNAs; the unique sequences mapping to the other arm of known specific species precursor hairpin opposite to the annotated mature miRNA-containing arm were considered to be novel 5p or 3p-derived miRNA candidates [50,51].
4.3. Identification and Function Analysis of Targets of miRNAs
The total RNA extracted from control T0h (0 hpi) and infected T6–48h (mixtures of 6, 12, 48 hpi) rice sample respectively were used for degradome sequencing on Illumina Hiseq 2500 at the LC-Bio Co., Ltd. (Hangzhou, China). The degradome reads were mapped to the rice transcriptome data. Publicly available software packages, CleaveL and 3.0 pipeline was used to predict and identify the potentially cleaved targets [50,52]. Then, the computational target prediction algorithms (Target Finder) were used to identify miRNA binding sites [53]. All of the predicted target genes were analyzed with NCBI BLASTX algorithm. Finally, the GO terms of these differentially expressed miRNA targets were also annotated by AgriGO program. The candidate miRNAs-related to defense responses and their targets with differential expression based on normalized deep-sequencing counts were analyzed by the Fisher exact test, Chi-squared test, Student’s t test, and ANOVA based on the experiments design [54]. The level of significance threshold was set at 0.01 and 0.05 for each test.
4.4. Validation of Differentially Expressed miRNAs and Targets by qRT-PCR
To validate miRNAs and their target expressions determined by high-through sequencing technology, the root fragments at different time points after inoculation were used to extract total RNA using the Trizol reagent (TaKaRa, Dalian, China). The qRT-PCR analysis was performed with the Applied Biosystems 7500 Real Time PCR System and SYBR Premix Ex TaqTM (TaKaRa, Dalian, China) according to the manufacturer’s instructions. The rice gene EF1-a and U6 was used as the internal reference gene to standardize RNA quantity for evaluating relative expression levels. For qRT-PCR assays, three independent biological samples were carried out with three technical replicates with a gene-specific primer (Table S7).
4.5. Vector Construction and Rice Transformation
The 176nt precursor sequence of osa-miR396f was isolated from Nanjing 40 by PCR amplification, and inserted into the pCAMBIA1301 binary vector driving by the constitutive CMV35s promoter for overexpression. The T-DNA recombinant plasmids were transformed into calli derived from the mature Nipponbare embryos by the Agrobacterium mediated transformation method [46].
Supplementary Materials
Supplementary materials can be found at http://www.mdpi.com/1422-0067/20/1/222/s1.
Author Contributions
Conceived and designed the experiments: W.L., Y.J., F.L., W.Z. and J.Y. Performed the experiments: W.L., F.W. and F.F. Analyzed the data: J.W. and J.Z. Contributed reagents/materials/analysis tools: Y.X. Wrote the paper: W.L., J.Y. and Y.J.
Funding
This work was supported by the National Key Research and Development Program (No. 2017YFD0100403), Genetically Modified Organisms Breeding Major Projects of P.R. China (2018ZX0801002B-003), the Jiangsu Key Laboratory of Agrobiology Program (No. 49114042015Z006), the Jiangsu Province Key Research and Development Program (Modern Agriculture) Project (No. BE2017368), the Exploratory Project of the Jiangsu Academy of Agricultural Sciences (No. ZX(17)2014), and the Jiangsu Province Natural Science Foundation (No. BK20171326). The United States Department of Agriculture (USDA) is an equal opportunity provider and employer
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Navarro L., Dunoyer P., Jay F., Arnold B., Dharmasiri N., Estelle M., Voinnet O., Jones J.D. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science. 2006;312:436–439. doi: 10.1126/science.1126088. [DOI] [PubMed] [Google Scholar]
- 2.Omidbakhshfard M.A., Proost S., Fujikura U., Mueller-Roeber B. Growth-Regulating Factors (GRFs): A small transcription factor family with important functions in plant biology. Mol. Plant. 2015;16:998–1010. doi: 10.1016/j.molp.2015.01.013. [DOI] [PubMed] [Google Scholar]
- 3.Baldrich P., San S.B. MicroRNAs in rice innate immunity. Rice. 2016;9:6. doi: 10.1186/s12284-016-0078-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jones J.D., Dangl J.L. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 5.Bellincampi D., Cervone F., Lionetti V. Plant cell wall dynamics and wall-related susceptibility in plant-pathogen interactions. Front Plant Sci. 2014;5:228. doi: 10.3389/fpls.2014.00228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo W., Wang C., Zuo Z.L., Qiu J.L. The roles of anion channels in Arabidopsis immunity. Plant Signal Behav. 2014;9:e29230. doi: 10.4161/psb.29230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu W.D., Liu J., Triplett L., Leach J.E., Wang G.L. Novel insights into rice innate immunity against bacterial and fungal pathogens. Annu. Rev. Phytopathol. 2014;52:213–241. doi: 10.1146/annurev-phyto-102313-045926. [DOI] [PubMed] [Google Scholar]
- 8.Gao Q.M., Zhu S., Kachroo P., Kachroo A. Signal regulators of systemic acquired resistance. Front Plant Sci. 2015;6:228. doi: 10.3389/fpls.2015.00228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li F., Pignatta D., Bendix C., Brunkard J.O., Cohn M.M., Tung J., Sun H., Kumar P., Baker B. MicroRNA regulation of plant innate immune receptors. Proc. Natl. Acad. Sci. USA. 2012;109:1790–1795. doi: 10.1073/pnas.1118282109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones-Rhoades M.W., Bartel D.P. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol. Cell. 2004;14:787–799. doi: 10.1016/j.molcel.2004.05.027. [DOI] [PubMed] [Google Scholar]
- 11.Li Y., Zhang Q.Q., Zhang J.G., Wu L., Qi Y.J., Zhou J.M. Identification of microRNAs involved in pathogen-associated molecular pattern-triggered plant innate immunity. Plant Physiol. 2010;152:2222–2231. doi: 10.1104/pp.109.151803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pumplin N., Voinnet O. RNA silencing suppression by plant pathogens: Defence, counter-defence and counter-counter-defence. Nat. Rev. Microbiol. 2013;11:745–760. doi: 10.1038/nrmicro3120. [DOI] [PubMed] [Google Scholar]
- 13.Chow H.T., Ng D.W. Regulation of miR163 and its targets in defense against Pseudomonas syringae in Arabidopsis thaliana. Sci. Rep. 2017;7:46433. doi: 10.1038/srep46433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eckardt N.A. A microRNA cascade in plant defense. Plant Cell. 2012;24:840. doi: 10.1105/tpc.112.240311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maudet C., Mano M., Eulalio A. MicroRNAs in the interaction between host and bacterial pathogens. FEBS Lett. 2014;588:4140–4147. doi: 10.1016/j.febslet.2014.08.002. [DOI] [PubMed] [Google Scholar]
- 16.Yang L., Huang H. Roles of small RNAs in plant disease resistance. J. Integr. Plant Biol. 2014;56:962–970. doi: 10.1111/jipb.12200. [DOI] [PubMed] [Google Scholar]
- 17.Lin R.M., He L.Y., He J.Y., Qin P.G., Wang Y.R., Deng Q.M., Yang X.T., Li S.C., Wang S.Q., Wang W.M., et al. Comprehensive analysis of microRNA-Seq and target mRNAs of rice sheath blight pathogen provides new insights into pathogenic regulatory mechanisms. DNA Res. 2016;23:415–425. doi: 10.1093/dnares/dsw024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Campo S., Peris-Peris C., Siré C., Moreno A.B., Donaire L., Zytnicki M., Notredame C., Llave C., San S.B. Identification of a novel microRNA (miRNA) from rice that targets an alternatively spliced transcript of the Nramp6 (Natural resistance-associated macrophage protein 6) gene involved in pathogen resistance. New Phytol. 2013;199:212–227. doi: 10.1111/nph.12292. [DOI] [PubMed] [Google Scholar]
- 19.Zhang W.X., Gao S., Zhou X., Chellappan P., Chen Z., Zhou X.F., Zhang X.M., Fromuth N., Coutino G., Coffey M., et al. Bacteria-responsive microRNAs regulate plant innate immunity by modulating plant hormone networks. Plant Mol. Biol. 2011;75:93–105. doi: 10.1007/s11103-010-9710-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hong H.M., Liu Y.Y., Zhang H.T., Xiao J.H., Li X.H., Wang S.P. Small RNAs and gene network in a durable disease resistance gene-mediated defense responses in rice. PLoS ONE. 2015;10:e0137360. doi: 10.1371/journal.pone.0137360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Y., Lu Y.G., Shi Y., Wu L., Xu Y.J., Huang F., Guo X.Y., Zhang Y., Fan J., Zhao J.Q., et al. Multiple rice microRNAs are involved in immunity against the blast fungus Magnaporthe oryzae. Plant Physiol. 2014;164:1077–1092. doi: 10.1104/pp.113.230052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang S., Mei J., Wang T., Wang C.C., Zhang W.L., Yang L. Identification and expression analysis of OsmiR1861k in rice leaves in response to Xanthomonas oryzae pv. oryzae. J. Gen. Plant Pathol. 2015;81:108–117. doi: 10.1007/s10327-015-0579-x. [DOI] [Google Scholar]
- 23.Li Y., Zhao S.L., Li J.L., Hu X.H., Wang H., Cao X.L., Xu Y.J., Zhao Z.X., Xiao Z.Y., Yang N., et al. Osa-miR169 negatively regulates rice immunity against the blast fungus Magnaporthe oryzae. Front. Plant Sci. 2017;17:2. doi: 10.3389/fpls.2017.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Q.G., Zhang Q., Wei C.D. Advances in research of rice bacterial foot rot. Sci. Agric. Sin. 2013;46:2923–2931. [Google Scholar]
- 25.Goto M. Bacterial foot rot of rice caused by a strain of Erwinia chrysanthemi. Phytopathology. 1979;69:213–216. doi: 10.1094/Phyto-69-213. [DOI] [Google Scholar]
- 26.Yang S., Peng Q., Francisco M.S., Wang Y., Zeng Q., Yang C.H. Type III secretion system genes of Dickeya dadantii 3937 are induced by plant phenolic acids. PLoS ONE. 2008;3:e2973. doi: 10.1371/annotation/91170966-226f-4678-999e-22f2c4a6bb8d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang S., Peng Q., Zhang Q., Yi X., JaeChoi C., Reedy R.M., Charkowski A.O., Yang C.H., et al. Dynamic regulation of GacA in type III secretion, pectinase gene expression, pellicle formation, and pathogenicity of Dickeya dadantii (Erwinia chrysanthemi 3937) Mol. Plant-Microbe Interact. 2008;21:133–142. doi: 10.1094/MPMI-21-1-0133. [DOI] [PubMed] [Google Scholar]
- 28.Zhou J.N., Zhang H.B., Wu J., Liu Q.G., Xi P.G., Lee J., Liao J.L., Jiang Z.D., Zhang L.H., et al. A novel multidomain polyketide synthase is essential for zeamine antibiotics production and the virulence of Dickeya zeae. Mol. Plant-Microbe Interact. 2011;24:1156–1164. doi: 10.1094/MPMI-04-11-0087. [DOI] [PubMed] [Google Scholar]
- 29.Chen X.F., Wei C.D., Zhang Q., Liu Q.G. Functional analysis of HrpX/HrpY in Dickeya zeae virulence. Sci. Agric. Sin. 2014;47:675–684. [Google Scholar]
- 30.Li W.Q., Wang J., Fan F.J., Zhu J.Y., Wang F.Q., Yang J., Zhong W.G. The isolation of rice bacterial foot rot disease pathogen in Jiangsu Province and identification of resistant rice varieties. Jiangsu Agric. Sci. 2014;42:139–142. [Google Scholar]
- 31.Padmanabhan C., Zhang X.M., Jin H.L. Host small RNAs are big contributors to plant innate immunity. Curr. Opin. Plant Biol. 2009;12:465–472. doi: 10.1016/j.pbi.2009.06.005. [DOI] [PubMed] [Google Scholar]
- 32.Sunkar R., Girke T., Jain P.K., Zhu J.K. Cloning and characterization of microRNAs from rice. Plant Cell. 2005;17:1397–1411. doi: 10.1105/tpc.105.031682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li S.J., Castillo-González C., Yu B., Zhang X.R. The functions of plant small RNAs in development and in stress responses. Plant J. 2017;90:654–670. doi: 10.1111/tpj.13444. [DOI] [PubMed] [Google Scholar]
- 34.Xin M.M., Wang Y., Yao Y.Y., Xie C.J., Peng H.R., Ni Z.F., Sun Q.X. Diverse set of microRNAs are responsive to powdery mildew infection and heat stress in wheat (Triticum aestivum L.) BMC Plant Biol. 2010;10:123. doi: 10.1186/1471-2229-10-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen L., Ren Y.Y., Zhang Y.Y., Xu J.C., Zhang Z.Y., Wang Y.W. Genome-wide profiling of novel and conserved Populus microRNAs involved in pathogen stress response by deep sequencing. Planta. 2012;235:873–883. doi: 10.1007/s00425-011-1548-z. [DOI] [PubMed] [Google Scholar]
- 36.Gupta O.P., Permar V., Koundal V., Singh U.D., Praveen S. MicroRNA regulated defense responses in Triticum aestivum L. during Puccinia graminis f.sp. tritici infection. Mol. Biol. Rep. 2012;39:817–824. doi: 10.1007/s11033-011-0803-5. [DOI] [PubMed] [Google Scholar]
- 37.Yin Z.J., Li Y., Han X.L., Shen F.F. Genome-wide profiling of miRNAs and other small non-coding RNAs in the Verticillium dahliae-inoculated cotton roots. PLoS ONE. 2012;7:e35765. doi: 10.1371/journal.pone.0035765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu D.G., Mou G.P., Wang K., Zhou G.H. MicroRNAs responding to southern rice black-streaked dwarf virus infection and their target genes associated with symptom development in rice. Virus Res. 2014;190:60–68. doi: 10.1016/j.virusres.2014.07.007. [DOI] [PubMed] [Google Scholar]
- 39.Choi D., Kim J.H., Kende H. Whole genome analysis of the Os-GRF gene family encoding plant-specific putative transcription activators in rice (Oryza sativa L.) Plant Cell Physiol. 2004;45:897–904. doi: 10.1093/pcp/pch098. [DOI] [PubMed] [Google Scholar]
- 40.Duan P.G., Ni S., Wang J.M., Zhang B.L., Xu R., Wang Y.X., Chen H.Q., Zhu X.D., Li Y.H. Regulation of OsGRF4 by OsmiR396 controls grain size and yield in rice. Nat. Plants. 2015;2:15203. doi: 10.1038/nplants.2015.203. [DOI] [PubMed] [Google Scholar]
- 41.Gao F., Wang K., Liu Y., Chen Y.P., Chen P., Shi Z.Y., Luo J., Jiang D.Q., Fan F.F., Zhu Y.G., et al. Blocking miR396 increases rice yield by shaping inflorescence architecture. Nat. Plants. 2015;2:15196. doi: 10.1038/nplants.2015.196. [DOI] [PubMed] [Google Scholar]
- 42.Hu J., Wang Y.X., Fang Y.X., Zeng L.J., Xu J., Yu H.P., Shi Z.Y., Pan J.J., Zhang D., Kang S.J., et al. A rare allele of GS2 enhances grain size and grain yield in rice. Mol. Plant. 2015;8:1455–1465. doi: 10.1016/j.molp.2015.07.002. [DOI] [PubMed] [Google Scholar]
- 43.Hewezi T., Maier T.R., Nettleton D., Baum T.J. The Arabidopsis microRNA396-GRF1/GRF3 regulatory module acts as a developmental regulator in the reprogramming of root cells during cyst nematode infection. Plant Physiol. 2012;159:321–335. doi: 10.1104/pp.112.193649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Debernardi J.M., Rodriguez R.E., Mecchia M.A., Palatnik J.F. Functional specialization of the plant miR396 regulatory network through distinct microRNA-target interactions. PLoS Genet. 2012;8:e1002419. doi: 10.1371/journal.pgen.1002419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ding X.H., Cao Y.L., Huang L.L., Zhao J., Xu C.G., Li X.H., Wang S.P. Activation of the indole-3-acetic acid-amido synthetase GH3-8 suppresses expansin expression and promotes salicylate- and jasmonate-independent basal immunity in rice. Plant Cell. 2008;20:228–240. doi: 10.1105/tpc.107.055657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li W.Q., Wang F.Q., Wang J., Fan F.J., Zhu J.Y., Yang J., Liu F.Q., Zhong W.G. Overexpressing CYP71Z2 enhances resistance to bacterial blight by suppressing auxin biosynthesis in rice. PLoS ONE. 2015;10:e0119867. doi: 10.1371/journal.pone.0119867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu J.Y., Rice J.H., Chen N.N., Baum T.J., Hewezi T. Synchronization of developmental processes and defense signaling by growth regulating transcription factors. PLoS ONE. 2014;9:e98477. doi: 10.1371/journal.pone.0098477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li W.T., Zhu Z.W., Chern M.S., Yin J.J., Yang C., Ran L., Cheng M.P., Wang K., Wang J., Zhou X.G., et al. A natural allele of a transcription factor in rice broad-spectrum blast resistance. Cell. 2017;170:114–126. doi: 10.1016/j.cell.2017.06.008. [DOI] [PubMed] [Google Scholar]
- 49.Pu X.M., Zhou J.N., Lin B.R., Shen H.F. First report of bacterial foot rot of rice caused by a Dickeya zeae in China. Plant Dis. 2012;96:1818. doi: 10.1094/PDIS-03-12-0315-PDN. [DOI] [PubMed] [Google Scholar]
- 50.Wu L., Zhang Q.Q., Zhou H.Y., Ni F.R., Wu X.Y., Qi Y.J. Rice microRNA effector complexes and targets. Plant Cell. 2009;213:3421–3435. doi: 10.1105/tpc.109.070938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Taylor R.S., Tarver J.E., Hiscock S.J., Donoghue P.C. Evolutionary history of plant microRNAs. Trends Plant Sci. 2014;19:175–182. doi: 10.1016/j.tplants.2013.11.008. [DOI] [PubMed] [Google Scholar]
- 52.Li Y.F., Zheng Y., Addo-Quaye C., Zhang L., Saini A., Jagadeeswaran G., Axtell M.J., Zhang W., Sunkar R. Transcriptome-wide identification of microRNA targets in rice. Plant J. 2010;62:742–759. doi: 10.1111/j.1365-313X.2010.04187.x. [DOI] [PubMed] [Google Scholar]
- 53.Zheng Y., Li Y.F., Sunkar R., Zhang W.X. SeqTar: An effective method for identifying microRNA guided cleavage sites from degradome of polyadenylated transcripts in plants. Nucleic Acids Res. 2012;40:e28. doi: 10.1093/nar/gkr1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou M., Gu L.F., Li P.C., Song X.W., Wei L.Y., Chen Z.Y., Cao X.F. Degradome sequencing reveals endogenous small RNA targets in rice (Oryza sativa L. ssp. indica) Front. Biol. 2010;5:67–90. doi: 10.1007/s11515-010-0007-8. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.