Abstract
A new polyketide containing the benzoisoquinoline-9-one moiety, peyronetide A (1), and three other new derivatives peyronetides B–D (2–4), as well as one known compound (5) were purified from the cultured broth of the endophytic fungus Peyronellaea sp. FT431, which was isolated from the Hawaiian indigenous plant, Verbena sp. The structures of the new compounds were determined through the analysis of HRMS and NMR spectroscopic data. Compounds 1, 2, and 5 showed cytotoxic activities against TK-10 (human kidney adenocarcinoma cells), cisplatin sensitive A2780S (human ovarian carcinoma cells), and cisplatin resistant A2780CisR cell lines, with IC50 values between 6.7 to 29.2 μM.
Keywords: endophytic fungi, Peyronellaea, benzoisoquinoline-9-one, antiproliferative, Hawaii
1. Introduction
Endophytic fungi are wonderful producers of various secondary metabolites, which have attracted great interest in the past decades to identify structurally unique and biologically active small molecules [1,2,3,4,5,6]. Our previous investigation of Hawaiian endophytic fungi had led to the identification of many new and/or bioactive compounds [7,8,9,10,11,12,13,14,15,16,17,18], including verbenanone from Peyronellaea sp. FT431 [13]. The crude extract of FT431 showed antiproliferative activity at 20 μg/mL against the A2780 cancer cell line, but verbenanone was inactive, so we decided to study FT431 further to identify the antiproliferative compounds.
The fermented whole broth (4.5 L) was filtered through filter paper to separate the supernatant from the mycelia. The latter was extracted with 80% acetone/H2O (×3), and the extract was concentrated under reduced pressure to afford an aqueous solution. The aqueous solution was passed through HP-20 eluted with MeOH-H2O (10%, 50%, 90%, 100%) to afford four fractions (Fr. A–D). The active fraction (Fr. C) was further separated by preparative HPLC and semi-preparative HPLC to get compounds 1–5 (Figure 1). Three of them (1, 2, and 5) showed antiproliferative activity against different cancer cell lines. Herein, we report the isolation, structure elucidation, and bioactivities of these isolated compounds.
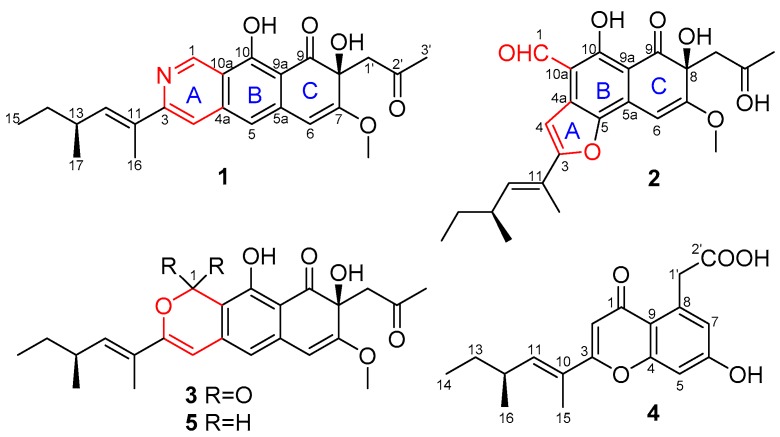
Figure 1.
Structures of compounds 1–5.
2. Results and Discussions
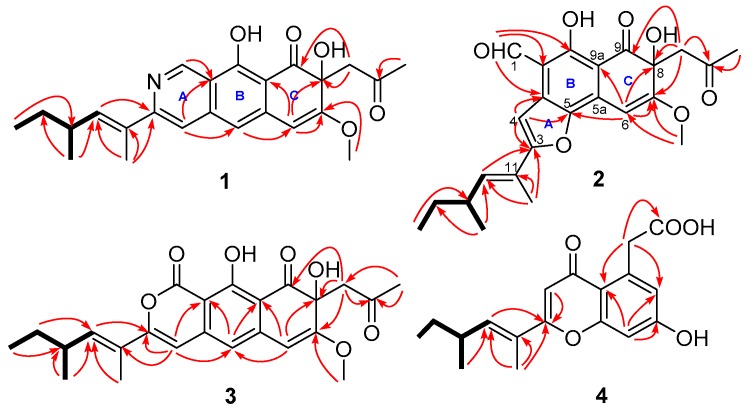
Compound 1 was isolated as a brown solid. Its molecular formula, C24H27NO5, was determined by HRESIMS (High-Resolution Electrospray Ionization Mass Spectrometry) at m/z 410.1964 [M + H]+ (calcd 410.1968), requiring 12 degrees of unsaturation. A comprehensive analysis of the 1D and 2D NMR spectra indicated the presence of four methyls, one methoxy group, two methylenes, six methines (five olefinic or aromatic), and eleven carbons with no hydrogen connected, including two ketones (δC 205.7, 202.8) (Table 1). The spin systems, C-12‒C-13(C-17)‒C-14‒C-15, were established by the 1H-1H COSY spectrum as shown in Figure 2, which was also verified by the corresponding HMBC correlations from H3-17 to C-12 and C-14, and from H3-15 to C-13. Meanwhile, HMBC correlations from the singlet methyl H3-16 (δH 2.16) to C-11 and C-12, C-3, as well as from H-12 to C-3 implied that the side-chain CH3-CH2-CH(CH3)-CH=C(CH3)- was connected to the ring system at C-3. HMBC correlations from the methyl group H3-3′ to the ketone C-2′ (δC 205.7) and methylene C-1′ (δC 50.6), and from H2-1′ to the oxygenated aromatic carbon C-7 (δC 161.6), an oxygenated quaternary C-8 (δC 72.8), and a ketone C-9 (δC 202.8) indicated the presence of another side-chain C-1′−C-3′, which was connected to C-8. The only methoxy group was assigned at 7-position by an HMBC correlation from the methoxy group to C-7. In addition, the specific de-shielded aromatic methine resonating at δH 9.43/δC 148.4 implied that it should be a nitrogenated atom. HMBC correlations from H-1 to C-4a, C-10, and C-10a, and from the aromatic proton H-4 to C-3, C-5, and C-10a suggested the presence of an isoquinoline ring system (rings A and B). Moreover, the HMBC correlations from H-5 to C-6, and from H-6 to the oxygenated olefinic carbon C-7, C-8, and C-9a established the naphthalen-1-(2H)-one rings B and C, and rings A–C were linearly aligned to form a benzoisoquinoline-9-one moiety (rings A–C) as shown. Literature research indicated that compound 1 had a similar ring system to the compound O-dihydroquinone (5) that was obtained as an epimer mixture from a marine ascomycete strain, which was the only report of this type of compound [19]. In spite of this, the presence of the nitrogen atom at 2-position in compound 1 instead of the oxygen in that of the reported compound was unusual. The configuration of the double bond C11(12) on the side-chain was determined to be E by the NOE correlations between H3-16 and H3-17. Hence, the planar structure of 1 was determined as shown.
Table 1.
NMR spectroscopic data for 1 and 2 in acetone-d6.
| No. | 1 | 2 | ||
|---|---|---|---|---|
| δH, J (Hz) a | δC b | δH, J (Hz) a | δC b | |
| 1 | 9.43, s | 148.4 | 10.52, s | 187.6 |
| 3 | 158.5 | 166.87 | ||
| 4 | 7.61, s | 115.0 | 7.42, s | 104.2 |
| 4a | 143.3 | 112.2 | ||
| 5 | 7.05, s | 114.7 | 144.2 | |
| 5a | 139.0 | 136.3 | ||
| 6 | 6.00, s | 98.9 | 6.33, s | 90.7 |
| 7 | 161.6 | 166.92 | ||
| 8 | 72.8 | 73.2 | ||
| 9 | 202.8 | 202.6 | ||
| 9a | 107.7 | 106.4 | ||
| 10 | 164.7 | |||
| 10a | 117.9 | 130.0 | ||
| 11 | 133.3 | 124.6 | ||
| 12 | 6.70, d, 10 | 140.1 | 141.0 | |
| 13 | 2.60, m | 35.7 | 2.64, m | 35.5 |
| 14 | 1.49, m;1.41, m | 31.0 | 1.49, m;1.42, m | 30.8 |
| 15 | 0.92, t, 7.4 | 12.4 | 0.92, t, 7.4 | 12.4 |
| 16 | 2.16, d, 1.3 | 14.5 | 1.45, d, 1.4 | 13.6 |
| 17 | 1.07, d, 6.7 | 20.8 | 1.10, d, 6.6 | 20.5 |
| 1′ | 3.52, s | 50.6 | 3.59, d, 5.5 | 51.3 |
| 2′ | 205.7 | 206.5 | ||
| 3′ | 2.09, s | 29.6 | 2.13, s | 29.8 |
| 7-MeO | 3.79, s | 56.2 | 3.91, s | 56.9 |
a Spectra recorded at 400 MHz. b Spectra recorded at 100 MHz. Data based on 1H, 13C, HSQC, and HMBC experiments.
Figure 2.
dqfCOSY (bolds) and selected HMBC (single head arrows, red) correlations of compounds 1–4.
Compound 2 was isolated as a brown solid. Its molecular formula was determined to be C24H26O7 by HRESIMS at m/z 427.1751 [M + H]+ (calcd 427.1757), with 12 degrees of unsaturation. A comprehensive analysis of the 1D and 2D NMR spectra indicated the presence of four methyls, one methoxy group, two methylenes, five methines (including one aldehyde), and twelve carbons with no hydrogen connected, including two ketones (δC 206.5, 202.6) and four oxygenated aromatic carbons (Table 1). The 1H-1H COSY implied that 2 had the same spin system as 1, which was verified by HMBC correlations as shown in Figure 2. The similarity of the NMR data of 2 to those of 1 implied that both had similar moieties. The key HMBC correlations from H3-3′ and H2-1′ to the ketone at δC 206.5, from H2-1′ to C-7, C-8, and C-9, and from H-6 to C-8, C-9a, and C-5, as well as from the methoxy group to C-7 implied the presence of ring C and the same substituents at 7- and 8-positions as those of 1. Moreover, the HMBC correlations from the proton of the aldehyde proton H-1 (δH 10.52 ppm) to C-10, C-10a, and C-4a placed the aldehyde group at C-10a, which implied that ring A in the molecule of 1 was opened in 2. A combined analysis of the observed HMBC correlations from H-4 to C-4a, and to the two oxygenated carbons C-3 and C-5 suggested the formation of a furan ring (ring A) as shown. The side-chain at 3-position was the same as that of 1. The configuration of the double bond was assigned as E at 11(12)-position by NOESY spectrum. Hence, the planar structure of 2 was determined as shown.
Compound 3 was isolated as a brown solid. The positive HRESIMS quasi-molecular ion peak at m/z 427.1760 [M + H]+ (calcd 427.1757) suggested the molecular formula of 3 as C24H26O7, which was same as that of compound 2. A comprehensive comparison of the NMR data (Table 2) of 3 with those of 1 indicated that the main difference was the absence of the nitrogenated methine (-N=CH-) and the presence of a lactone carbonyl group (-O-CO-) at δC 164.8 in 3. The configuration of the double bond at C-11 was assigned as E by NOESY spectrum. Hence, the planar structure of 3 was determined as shown.
Table 2.
NMR spectroscopic data for compounds 3 (acetone-d6) and 4 (MeOH-d4).
| No. | 3 a | 4 b | ||
|---|---|---|---|---|
| δH, J (Hz) | δC | δH, J (Hz) | δC | |
| 1 | 164.8 | 181.6 | ||
| 2 | 6.16, s | 107.4 | ||
| 3 | 158.0 | 164.3 | ||
| 4 | 6.56 | 101.6 | 160.3 | |
| 4a | ||||
| 5 | 6.77, s | 114.1 | 6.78, d, 2.0 | 102.1 |
| 5a | ||||
| 6 | 5.89, s | 97.1 | 163.1 | |
| 7 | 163.1 | 6.69, d, 2.6 | 118.5 | |
| 8 | 72.1 | 140.2 | ||
| 9 | 199.9 | 115.4 | ||
| 9a | 109.1 | |||
| 10 | 159.1 | 126.8 | ||
| 10a | 104.4 | |||
| 11 | 125.6 | 142.8 | ||
| 12 | 6.34, d, 9.7 | 140.3 | 2.59, m | 35.8 |
| 13 | 2.58, m | 34.7 | 1.52, m; 1.42, m | 30.6 |
| 14 | 1.48, m; 1.41, m | 29.8 | 0.92, t, 7.6 | 12.0 |
| 15 | 0.89, t, 7.4 | 11.4 | 1.97, s | 12.7 |
| 16 | 1.98, s | 11.9 | 1.08, d, 6.7 | 20.3 |
| 17 | 1.05, d, 6.6 | 19.6 | ||
| 1′ | 3.46, br.d, 5.9 | 50.6 | 4.11, s | 42.8 |
| 2′ | 205.7 | 176.9 | ||
| 3′ | 2.10, s | 29.0 | ||
| 7-MeO | 3.79, s | 56.2 | 107.4 | |
a Spectra recorded at 400 MHz. b Spectra recorded at 100 MHz. Data based on 1H, 13C, HSQC, and HMBC experiments.
Compound 4, a brown solid, was determined to have a molecular formula of C18H20O5 by HRESIMS at m/z 317.1390 [M + H]+ (calcd 317.1389), which was six carbons less than those of compounds 1–3. 1H-1H COSY and HMBC spectra suggested that 4 had the same side-chain (C-10–C-16) as compounds 1–3 (C-11–C-17). A comprehensive analysis of the NMR data (Table 2), and especially the HMBC spectrum, implied that 4 was a chromone derivative, which has the same ring system as that of 2-methyl-5-carboxymethyl-7-hydroxychromone [20]. The main difference between 4 and 2-methyl-5-carboxymethyl-7-hydroxychromone was the long side-chain instead of a methyl group in the molecule of 4. HMBC correlations from H-2 to C-3 and C-10, and from the olefinic H-10 and single methyl H3-15 to C-3 confirmed the position of the side-chain. The E configuration of the double bond was determined by NOESY spectrum. Hence, the planar structure of 4 was determined as shown.
The known compound 5 (O-dihydroquinone) was identified by comparison of its physical data with reported values in the literature [19].
We tried to determine the configuration of the new compounds including chemical reactions and crystallization, but it was unsuccessful. Then we purchased both (S)-(+)- and (R)-(−)- 2-methylbutanoic acids, and compared the optical rotation of 4 with those of (S)-(+)- and (R)-(−)- 2-methylbutanoic acids. Compound 4 showed a positive sign of optical rotation, indicating that 4 should also have an S configuration at 13-position. Hence, assuming a 13-S configuration in compounds 1–3, the relative configuration of the 8-position remained unknown. To solve this task, we relied on GIAO 13C-NMR calculations, a strategy that has been extensively employed in recent publications to settle structural and stereochemical issues of complex organic molecules [21,22,23,24,25]. Several strategies have been developed to determine the most likely stereostructure among several candidates, including DP4, [22] and DP4+, an updated version of DP4 including scaled and unscaled NMR shifts computed at higher levels of theory [23]. The capacity of these methodologies to discriminate among candidates featuring rigid structures and contiguous or near-by stereocenters is often excellent [24], but when two or more steroclusters are separated the determination of the relative configuration becomes much more challenging [25]. In any case, we decided to explore this approach to suggest a sound stereochemical assignment of the new natural products herein reported and to validate our assignment of the planar structure of 1 discussed above. Initially, we carried out preliminary DP4 calculations of the two possible diastereoisomers of 1 (1a = 1–8S,13S and 1b = 1–8S,13R [equivalent to 1–8R,13S], see structures of 1a and 1b in Supplementary Materials) at the affordable B3LYP/6-31G**//MMFF level of theory [21b]. As shown in the Supporting Information, compound 1a displayed a slightly better fit between experimental and calculated NMR data, and was identified by DP4 as the most probable candidate (55% for 1a and 45% for 1b). Most of the calculated shifts agreed well with our experimental values, providing further evidence of our proposed connectivity analysis. However, we noticed alarmingly high errors (defined as Δδ = abs[δexp − δcalc]) in the signals assigned to C-8 (Δδ = 8.2 ppm), C-1′ (Δδ = 9.9 ppm), and C-2′ (Δδ = 9.1 ppm). After a detailed examination of the computational data, we noticed that such discrepancies arose from the conformations bearing intramolecular H-bonding between the OH group at C-8 with the ketone oxygen at C-2′, which in turn represented > 93% of the corresponding Boltzmann distributions according to the B3LYP/6-31G** energies. However, since the experimental NMR data were collected in acetone-d6, the real conformational landscape of the system might be shifted toward more flexible structures. Hence, following a similar approach recently employed in a related situation [14], we recomputed the NMR shifts by neglecting all conformations featuring intramolecular H-bonding between C8-OH and C2′=O. In excellent agreement with our hypothesis, a much better fit was computed for C-8 (Δδ = 2.5 ppm), C-1′ (Δδ = 1.9 ppm), C-2′ (Δδ = 2.2 ppm), and H-1′ (Δδ = 0.3 ppm). Nevertheless, the slight preference to 1a (52%) remained almost constant. We next refined the computational results by performing full geometry optimizations at the B3LYP/6-31G* level of theory followed by NMR calculations at the PCM/mPW1PW91/6-31+G** level, the recommended method for DP4+ calculations [21c–e]. Here again, the conformational preferences of 1a and 1b were considerably shifted toward intramolecular H-bonded structures. As expected, strong deviations from the experimental values were computed for the 13C-NMR resonances of C-8, C-1′, and C-2′ (Δδ = 5.1 − 11.8 ppm). Since the Boltzmann distributions hardly changed upon performing full geometry optimizations in water, we decided to recompute the NMR data by removing all the conformations showing H-bonding. In this reduced system, a much better agreement between experimental and calculated NMR data was observed, with a slight preference toward 1a (CMAE = 1.5 ppm for 1a and 1.6 ppm for 1b). As a result, the DP4+ values identified 1a as the most probable candidate (69%), in line with the previous DP4 results. From a biogenetic point of view, 2 and 3 should have the same configuration as 1 at the corresponding chiral centers. However, given the separation of the two stereocenters, the other relative configurations cannot be completely ruled out.
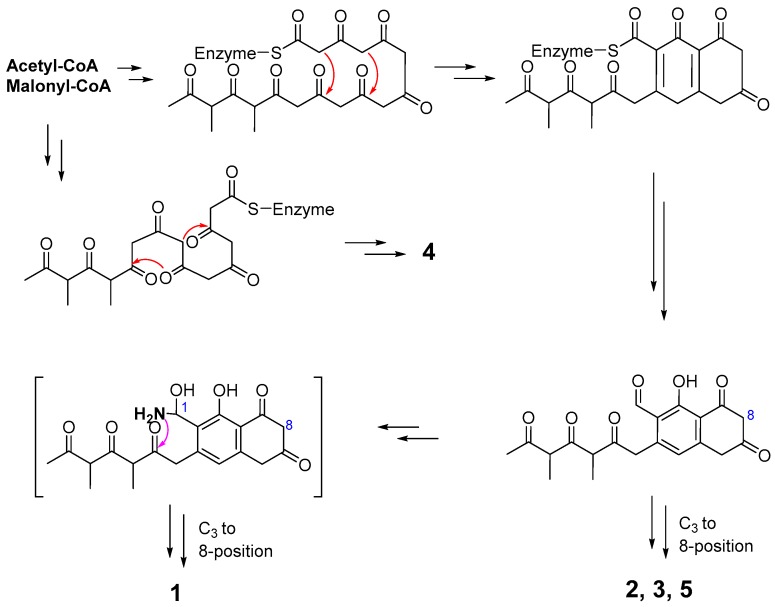
Biogenetically, all the new compounds could be derived from acetyl CoA and malonyl CoA. However, it is worthy to investigate how the nitrogen atom was introduced [26] into 1-position of compound 1, and the C3 side-chain to 8-position of compounds 1–3 and 5 (Figure 3).
Figure 3.
Proposed biosynthesis of compounds 1–5.
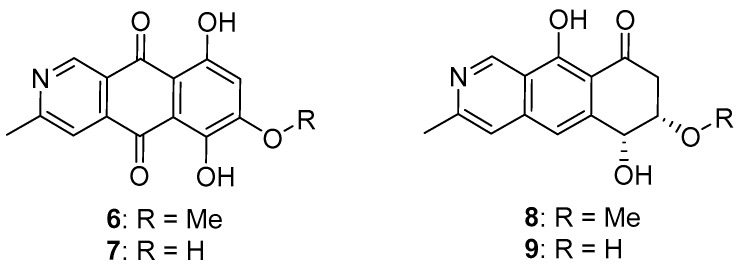
Natural azaanthraquinone derivatives were not rare, for example, bostrycoidin (6) and tolypocladin (7) [27,28]. However, natural products containing a benzoisoquinoline-9-one moiety are very uncommon. To the best of our knowledge, pyrenolines A (8) [29,30] and B (9) [30] were the only two known benzoisoquinoline-9-one derivatives (Figure 4) that were isolated from the culture fluid of Pyrenophora teres, a pathogen of barley.
Figure 4.
Some known natural azaanthraquinones and benzo-isoquinoline-9-one derivatives.
All the compounds were tested against ovarian cancer cell lines A2780S and A2780CisR, and renal cancer cell TK-10. Compounds 1, 2, and 5 were active, and compound 5 showed moderate activities against those cell lines (Table 3, see anti-proliferative data in Supplementary Materials).
Table 3.
Antiproliferative activities of compounds 1, 2, and 5 against different cell lines.
| Compounds | IC50 (μM) | ||
|---|---|---|---|
| A2780S | A2780CisR | TK-10 | |
| 1 | 24.1 ± 0.8 | 28.3 ± 7.2 | 29.2 ± 2.9 |
| 2 | 21.5 ± 0.3 | 27.2 ± 1.3 | 22.7 ± 1.3 |
| 5 | 7.1 ± 0.8 | 6.7 ± 1.2 | 8.5 ± 0.9 |
Cisplatin had IC50 values of 0.36, 1.1, and 13.2 μM against A2780S, A2780CisR, and TK-10, respectively.
3. Materials and Methods
3.1. General Experimental Procedures
Optical rotation was measured with a Rudolph Research Analytical AutoPol IV Automatic Polarimeter (Hackettstown, NJ, USA). UV and IR spectra were obtained with a Shimadzu UV-1800 spectrophotometer (Kyoto, Japan) and a Thermo Fisher Scientific Nicolet iS50 FTIR spectrometer (Madison, WI, USA), respectively. NMR spectra including 1D and 2D experiments were recorded in acetone-d6 or MeOH-d4 on a Bruker 400 MHz NMR (Fällanden, Switzerland). High resolution mass spectra were obtained on a Waters Micromass Q-Tof Ultima ESI-TOF mass spectrometer (Milford, MA, USA), or an Agilent Technologies 6530 Accurate-Mass Q-TOF LC/MS (Santa Clara, CA, USA). HPLC was carried out on a Thermo Fisher Scientific Ultimate 3000 LC system (Germering, Germany), and all solvents were HPLC grade. Column chromatography used a Diaion HP-20 (Alfa Aesar, Ward Hill, MA, USA).
3.2. Isolation and Identification of Fungal Strain
The fungal strain was isolated on PDA medium from a healthy leaf of a Hawaiian indigenous plant, Verbena sp., which was collected in the Lyon Botanical Garden in 2014. The strain FT431 was identified as Peyronellaea sp. based on the analysis of the DNA sequence of the nuclear ribosomal internal transcribed spacer, which has been deposited in GenBank with the accession no. KY971272. A voucher specimen was deposited at the Daniel K. Inouye College of Pharmacy, University of Hawaii at Hilo, USA (accession no. FT431).
3.3. Cultivation
The fungus was grown under static conditions at room temperature for 30 days in a 1 L conical flask containing a liquid medium (300 mL/flask) composed of mannitol (20 g/L), sucrose (20 g/L), monosodium glutamate (5 g/L), KH2PO4 (0.5 g/L), and MgSO4.
3.4. Isolation of Compounds 1–5
The whole fermented broth (4.5 L) was filtered through filter paper to separate the supernatant from the mycelia. The mycelia were extracted by 80% acetone/H2O three times, and the extracts were condensed under vacuum to get an aqueous solution. The solution was passed through a Diaion HP-20 column (Alfa Aesar, Ward Hill, MA, USA), eluted with MeOH-H2O (10%, 50%, 90%, and 100% methanol in H2O) to afford four fractions (Fr. A‒D). Fraction C (517.8 mg) was separated with a preparative HPLC column (C18 column, 5 µ, 100.0 × 21.2 mm; 10 mL/min; 10–100% methanol in H2O in 40 min) to generate 40 sub-fractions (C1‒40). C35 (27.4 mg) was subjected to the semi-preparative HPLC (C18 column, 5 µ, 250.0 × 10.0 mm; 4 mL/min; with 0.1% formic acid in 75% methanol in H2O) to obtain compounds 4 (7.12 mg, tR 31.5 min) and 5 (1.56 mg, tR 33.5 min). Fraction D (347.2 mg) was separated with a preparative HPLC column (C18 column, 5 µ, 100.0 × 21.2 mm; 10 mL/min; 30–100% methanol in H2O in 30 min) to generate 30 sub-fractions (D1‒30). D20 (8.47 mg) was subjected to the semi-preparative HPLC (C18 column, 5 µ, 250.0 × 10.0 mm; 3 mL/min; with 0.1% formic acid in 58% methanol in H2O) to afford compound 1 (1.34 mg, tR 35.0 min). D26 (18.28 mg) was subjected to the semi-preparative HPLC (C18 column, 5 µ, 250.0 × 10.0 mm; 3 mL/min; with 0.1% formic acid in 75% methanol in H2O) to afford compounds 2 (8.51 mg, tR 20.8 min) and 3 (1.38 mg, tR 25.6 min).
3.5. Charaterization of Compounds 1–4
Peyronetide A (1): Brown solid; + 73.3 (c = 0.06, MeOH); UV (MeOH) λmax (log e) 298 (4.21), 417 (3.56) nm; IR νmax3388, 2959, 2927, 2871, 1710, 1586, 1478, 1461, 1452, 1383, 1354, 1316, 1280, 1232, 1166, 1200, 1067, 873 cm−1; 1H(acetone-d6, 400 MHz) and 13C-NMR (acetone-d6, 100 MHz) data, see Table 1; positive HRESIMS m/z 410.1964 [M + H]+ (calcd for C24H28NO5, 410.1968).
Peyronetide B (2): Brown solid; + 68.8 (c = 0.08, MeOH); UV (MeOH) λmax (log e) 246 (4.13), 254 (4.11), 296 (4.26), 399 (4.22) nm; IR νmax3393, 2960, 2926, 2871, 1711, 1646, 1626, 1560, 1529, 1455, 1404, 1377, 1323, 1261, 1212, 1182, 1149, 1097, 1027, 991, 831 cm−1; 1H(acetone-d6, 400 MHz) and 13C-NMR (acetone-d6, 100 MHz) data, see Table 1; positive HRESIMS m/z 427.1751 [M + H]+ (calcd for C24H27O7, 427.1757).
Peyronetide C (3): Brown solid; + 79.1 (c = 0.09, MeOH); UV (MeOH) λmax (log e) 285 (4.57), 394 (4.20) nm; IR νmax3400, 2960, 2927, 2872, 1709, 1651, 1611, 1538, 1489, 1455, 1403, 1363, 1335, 1278, 1215, 1170, 1073, 1004, 869, 821, 780 cm−1; 1H(acetone-d6, 400 MHz) and 13C-NMR (acetone-d6, 100 MHz) data, see Table 1; positive HRESIMS m/z 427.1760 [M + H]+ (calcd for C24H27O7, 427.1757).
Peyronetide D (4): Brown solid; + 45.0 (c = 0.02, MeOH); UV (MeOH) λmax (log e) 215 (3.99), 238 (3.83), 257 (3.77), 304 (3.74) nm; IR νmax3382, 2959, 2928, 2872, 2360, 2342, 1617, 1578, 1506, 1452, 1384, 1340, 1315, 1280, 1163, 1110 cm−1; 1H(acetone-d6, 400 MHz) and 13C-NMR (acetone-d6, 100 MHz) data, see Table 1; positive HRESIMS m/z 317.1390 [M + H]+ (calcd for C18H21O5, 317.1389).
3.6. Anti-Proliferative Activity
The viability of A2780 and TK-10 (from the NCI) and the cisplatin-resistant, A2780CisR [31], was determined using the CyQuant cell proliferation assay kit, according to the manufacturer’s instructions (Life Technologies, Eugene, OR, USA). Briefly, cells in 96-well plates, seeded 24 h prior, were treated with or without compounds for 72 h, and subjected to CyQuant cell viability assay (Life Technologies, Eugene, OR, USA) [32,33,34]. Each cell line was cultured in 96-well plates at 6000 cells per well with the following conditions: 0 (no treatment, vehicle (DMSO)) and increasing concentrations of compounds for 72 h. Cisplatin was used as a positvie control. Viable cells were analyzed by subjecting the plates to the CyQuant, as previously reported [32,33,34]. Relative viability of the treated cells was normalized to the DMSO-treated control cells. All experiments were performed in triplicate.
3.7. DP4+ Calculations
All of the quantum mechanical calculations were performed using Gaussian 09 [35]. The conformational search was done in the gas phase using the MMFF (Merck Molecular Force Field) force field (implemented in Macromodel) [36]. All conformers within 10 kcal/mol from the global minima (more than 900 different structures) were kept for further calculations. After an exhaustive exploration of the conformational space of the two possible diastereoisomers of 1, namely, 1a = 1–8S,13S and 1b = 1–8S,13R (equivalent to 1–8R,13S), we were able to locate more than 900 unique conformations for both compounds. In order to narrow down the number of geometries for B3LYP/6-31G* optimizations, a previous HF/3-21G geometry optimization stage was carried out, and all confomers within 6 kcal/mol from the global minima were submitted to full geometry optimizations at the B3LYP/6-31G* level. The isotropic magnetic shielding constants (σ) were computed using the gauge including the atomic orbitals (GIAO) method [37,38,39,40], at the B3LYP/6-31G**//MMFF level (for DP4 calculations) [22] and PCM/mPW1PW91/6-31+G**//B3LYP/6-31G* level (for DP4+ calculations) [23] using methanol as the solvent. The unscaled chemical shifts (δu) were computed using TMS (Tetramethylsilane) as a reference standard according to δu = σ0 − σx, where σx is the Boltzmann averaged shielding tensor (over all significantly populated conformations) and σ0 is the shielding tensor of the TMS computed at the same level of theory employed for σx. The scaled chemical shifts (δs) were calculated as δs = (δu − b)/m, where m and b are the slope and intercept, respectively, deduced from a linear regression calculation on a plot of δu against δexp. The DP4+ calculations were run by the Excel spreadsheet available for free at sarotti-nmr.weebly.com or as part of the Supporting Information of the original paper [23], and the DP4 calculations were done according to the original reference [22].
4. Conclusions
In conclusion, five compounds (1–5) including four new ones (1–4) were isolated from a Hawaiian plant-asssociated endophytic fungus Peyronellaea sp. FT431. Compound 1 is a unique benzoisoquinoline-9-one derivative with two side-chains, 1,3-dimethyl-1-pentene and 2-propanone at 3- and 8-positions, respectively, which were diagonal to each other in the benzoisoquinoline-9-one. Compounds 1–5 were evaluated for their antiproliferative activity, and compound 5 was the most active one with IC50 values of 7.1, 6.7, and 8.5 μM against A2780S, A2780CisR, and TK-10, respectively. The results indicated that Hawaiian fungi are a good resource of new and bioactive compounds.
Acknowledgments
The authors would like to thank Marcus Tius for the ozonolysis as part of their attempt to determine the configuration of 13-position of compound 1 using chemical reaction. They are also grateful to Justin Reinicke at the Daniel K. Inouye College of Pharmacy, University of Hawaii at Hilo for help collecting the NMR data.
Supplementary Materials
The supplementary materials (NMR [including 1H, 13C, COSY, HSQC, HMBC, and NOESY], HRESIMS, IR spectra, anti-proliferative, and NMR calculation data) are available online.
Author Contributions
C.L. performed most of the experiments; A.M.S. performed the NMR calculations; X.W., B.Y., and J.T. carried out the bioassays; Y.C. and Q.L. carried out part of the configuration determination; C.L. and S.C. analyzed the data and wrote the paper. All authors read and approved the final manuscript.
Funding
This work was financially supported by start-up funding from the University of Hawaii Cancer Center (SC), funding from the University of Hawai’i at Hilo (SC), and grants from the Victoria S. and Bradley L. Geist Foundation (15ADVC-74420 and 17CON-86295) (SC) and the National Institutes of Health (NIH)/National Cancer Institute (NCI), Grant CA128865 (JT).
Conflicts of Interest
The authors declare no conflict of interest. The funding sponsors had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; and in the decision to publish the results.
Footnotes
Sample Availability: Samples of the compounds are available from the authors.
References
- 1.Khaarwar R.N., Mishra A., Gond S.K., Stierle A., Stiele D. Anticancer compounds derived from fungal endophytes: Their importance and future challenges. Nat. Prod. Rep. 2011;28:1208–1228. doi: 10.1039/c1np00008j. [DOI] [PubMed] [Google Scholar]
- 2.Zhang H.W., Song Y.C., Tan R.X. Biology and chemistry of endophytes. Nat. Prod. Rep. 2006;23:753–771. doi: 10.1039/b609472b. [DOI] [PubMed] [Google Scholar]
- 3.Cao S., McMillin D.W., Tamayo G., Delmore J., Mitsiades C.S., Clardy J. Inhibition of tumor cells interacting with stromal cells by xanthones isolated from a Costa Rican Penicillium sp. J. Nat. Prod. 2012;75:793–797. doi: 10.1021/np2009863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cao S., Cryan L., Habeshian K.A., Murillo C., Tamayo-Castillo G., Rogers M.S., Clardy J. Phenolic compounds as antiangiogenic CMG2 inhibitors from Costa Rican endophytic fungi. Bioorg. Med. Chem. Lett. 2012;22:5885–5888. doi: 10.1016/j.bmcl.2012.07.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cao S., Clardy J. New naphthoquinones and a new δ-lactone produced by endophytic fungi from Costa Rica. Tetrahedron Lett. 2011;52:2206–2208. doi: 10.1016/j.tetlet.2010.11.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao S., Ross L., Tamayo G., Clardy J. Asterogynins: Secondary metabolites from a Costa Rican endophytic fungus. Org. Lett. 2010;12:4661–4663. doi: 10.1021/ol101972g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li C.S., Yang B.J., Fenstemacher R., Turkson J., Cao S. Lycopodiellactone, an unusual δ-lactone-isochromanone from a Hawaiian plant-associated fungus Paraphaeosphaeria neglecta FT462. Tetrahedron Lett. 2015;56:1724–1727. doi: 10.1016/j.tetlet.2015.02.076. [DOI] [Google Scholar]
- 8.Li C.S., Ding Y., Yang B.J., Miklossy G., Yin H.Q., Walker L.A., Turkson J., Cao S. A New Metabolite with a Unique 4-Pyranone-γ-Lactam-1,4-Thiazine Moiety from a Hawaiian-Plant Associated Fungus. Org. Lett. 2015;17:3556–3559. doi: 10.1021/acs.orglett.5b01650. [DOI] [PubMed] [Google Scholar]
- 9.Li C.S., Ding Y., Yang B.J., Hoffman N., Yin H.Q., Mahmud T., Turkson J., Cao S. Eremophilane sesquiterpenes from Hawaiian endophytic fungus Chaetoconis sp. FT087. Phytochemistry. 2016;126:41–46. doi: 10.1016/j.phytochem.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 10.Li C.S., Ren G., Yang B.J., Miklossy G., Turkson J., Fei P., Ding Y., Walker L.A., Cao S. Meroterpenoids with Antiproliferative Activity from a Hawaiian-Plant Associated Fungus Peyronellaea coffeae-arabicae FT238. Org. Lett. 2016;18:2335–2338. doi: 10.1021/acs.orglett.6b00685. [DOI] [PubMed] [Google Scholar]
- 11.Fei-Zhang D.J., Li C.S., Cao S. Hawaii natural compounds are promising to reduce ovarian cancer deaths. Cancer Biol. Ther. 2016;17:709–712. doi: 10.1080/15384047.2016.1178428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li C.S., Yang B.J., Turkson J., Cao S. Anti-proliferative ambuic acid derivatives from Hawaiian endophytic fungus Pestalotiopsis sp. FT172. Phytochemistry. 2017;140:77–82. doi: 10.1016/j.phytochem.2017.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li C.S., Sarotti A.M., Turkson J., Cao S. Verbenanone, an octahydro-5H-chromen-5-one from a Hawaiian-plant associated fungus FT431. Tetrahedron Lett. 2017;58:2290–2293. doi: 10.1016/j.tetlet.2017.04.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang P., Li C.S., Sarotti A.M., Turkson J., Cao S. Sphaerialactonam, a γ-lactam–isochromanone from the Hawaiian endophytic fungus Paraphaeosphaeria sp. FT462. Tetrahedron Lett. 2017;58:1330–1333. doi: 10.1016/j.tetlet.2017.02.052. [DOI] [Google Scholar]
- 15.Li C.S., Sarotti A.M., Yang B.J., Turkson J., Cao S. A New N-methoxypyridone from the Co-Cultivation of Hawaiian Endophytic Fungi Camporesia sambuci FT1061 and Epicoccum sorghinum FT1062. Molecules. 2017;22:1166. doi: 10.3390/molecules22071166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li C.S., Sarotti A.M., Huang P., Dang U.T., Hurdle J.G., Kondratyuk T.P., Pezzuto J.M., Turkson J., Cao S. NF-κB inhibitors, unique γ-pyranol-γ-lactams with sulfide and sulfoxide moieties from Hawaiian plant Lycopodiella cernua derived fungus Paraphaeosphaeria neglecta FT462. Sci. Rep. 2017;7:10424. doi: 10.1038/s41598-017-10537-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li C.S., Sarotti A.M., Yoshida W., Cao S. Two new polyketides from Hawaiian endophytic fungus Pestalotiopsis sp. FT172. Tetrehedron Lett. 2018;58:42–45. doi: 10.1016/j.tetlet.2017.11.045. [DOI] [Google Scholar]
- 18.Li C.S., Hu Z., Liu Q., Wu X., Cao S. Two new tricycloalternarenes from Hawaiian endophytic fungus Didymella sp. FT433. Tetrahedron Lett. 2018;59:3381–3383. doi: 10.1016/j.tetlet.2018.07.061. [DOI] [Google Scholar]
- 19.Guerriero A., Amrosio M., Cuomo V., Pietra F. A Novel, Degraded Polyketidic Lactone, Leptosphaerolide, and Its Likely Diketone Precursor, Leptosphaerodione. Isolation from Cultures of the Marine Ascomycete Leptosphaeria oraemaris (LINDER) Helv. Chim. Acta. 1991;74:1445–1450. doi: 10.1002/hlca.19910740707. [DOI] [Google Scholar]
- 20.Kashiwada Y., Nonaka G.I., Nishioka I. Studies on Rhubarb (Rhei Rhizoma). V. Isolation and Characterization of Chromone and Chromanone Derivatives. Chem. Pharm. Bull. 1984;32:3493–3500. doi: 10.1248/cpb.32.3493. [DOI] [Google Scholar]
- 21.Grimblat N., Sarotti A.M. Computational Chemistry to the Rescue: Modern Toolboxes for the Assignment of Complex Molecules by GIAO NMR Calculations. Chem. Eur. J. 2016;22:12246–12261. doi: 10.1002/chem.201601150. [DOI] [PubMed] [Google Scholar]
- 22.Smith S.G., Goodman J.M. Assigning stereochemistry to single diastereoisomers by GIAO NMR calculation: The DP4 probability. J. Am. Chem. Soc. 2010;132:12946–12959. doi: 10.1021/ja105035r. [DOI] [PubMed] [Google Scholar]
- 23.Grimblat N., Zanardi M.M., Sarotti A.M. Beyond DP4: An Improved Probability for the Stereochemical Assignment of Isomeric Compounds using Quantum Chemical Calculations of NMR Shifts. J. Org. Chem. 2015;80:12526–12534. doi: 10.1021/acs.joc.5b02396. [DOI] [PubMed] [Google Scholar]
- 24.Zanardi M.M., Suárez A.G., Sarotti A.M. Determination of the Relative Configuration of Terminal and Spiroepoxides by Computational Methods. Advantages of the Inclusion of Unscaled Data. J. Org. Chem. 2017;82:1873–1879. doi: 10.1021/acs.joc.6b02129. [DOI] [PubMed] [Google Scholar]
- 25.Zanardi M.M., Biglione F.A., Sortino M.A., Sarotti A.M. General Quantum-Based NMR Method for the Assignment of Absolute Configuration by Single or Double Derivatization: Scope and Limitations. J. Org. Chem. 2018;83:11839–11849. doi: 10.1021/acs.joc.8b01749. [DOI] [PubMed] [Google Scholar]
- 26.Parisot D., Devys M., Barbier M. 5-Deoxybostrycoidin, a New Metabolite Produced by the Fungus Nectria haematococca (Berk, and Br.) Wr. Z. Naturforsch. 1989;44b:1473–1474. doi: 10.1515/znb-1989-1125. [DOI] [Google Scholar]
- 27.Arsenault G.P. The structure of bostrycoidin, a β-aza-anthraquinone from Fusariumsolani D2 purple. Tetrahedron Lett. 1965;45:4033–4037. doi: 10.1016/S0040-4039(01)99610-8. [DOI] [Google Scholar]
- 28.Graefe U., Ihn W., Tresselt D., Miosga N., Kaden U., Schlegel B., Bormann E.J., Sedmera P., Novak J. Tolypocladin—A new metal-chelating 2-aza-anthraquinone from Tolypocladium inflatum. Biol. Met. 1990;3:39–44. doi: 10.1007/BF01141176. [DOI] [Google Scholar]
- 29.Albinati A., Arnone A., Assante G., Meille S.V., Nasini G. Isoflavans from Millettia racemosa. Phytochemistry. 1989;28:923–927. doi: 10.1016/0031-9422(89)80144-X. [DOI] [Google Scholar]
- 30.Coval S.J., Hradil C.M., Lu H.S.M., Clardy J., Satouri S., Strobel G.A. Pyrenoline-A and -B, two new phytotoxins from Pyrenophora teres. Tetrahedron Lett. 1990;31:2117–2120. doi: 10.1016/0040-4039(90)80086-2. [DOI] [Google Scholar]
- 31.Yue P., Zhang X., Paladino D., Sengupta B., Ahmad S., Holloway R.W., Ingersoll S.B., Turkson J. Hyperactive EGF receptor, Jaks and Stat3 signaling promote enhanced colony-forming ability, motility and migration of cisplatin-resistant ovarian cancer cells. Oncogene. 2012;31:2309–2322. doi: 10.1038/onc.2011.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lopez-Tapia F., Brotherton-Pleiss C., Yue P., Murakami H., Costa Araujo A.C., Reis Dos Santos B., Ichinotsubo E., Rabkin A., Shah R., Lantz M., et al. Linker Variation and Structure-Activity Relationship Analyses of Carboxylic Acid-based Small Molecule STAT3 Inhibitors. ACS Med. Chem. Lett. 2018;9:250–255. doi: 10.1021/acsmedchemlett.7b00544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yue P., Lopez-Tapia F., Paladino D., Li Y., Chen C.-H., Namanja A.T., Hilliard T., Chen Y., Tius M., Turkson J. Hydroxamic acid and benzoic acid-based Stat3 inhibitors suppress human glioma and breast cancer phenotypes in vitro and in vivo. Cancer Res. 2016;76:652–663. doi: 10.1158/0008-5472.CAN-14-3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang X., Yue P., Page B.D., Li T., Zhao W., Namanja A.T., Paladino D., Zhao J., Chen Y., Gunning P.T., et al. Orally bioavailable small-molecule inhibitor of transcription factor Stat3 regresses human breast and lung cancer xenografts. Proc. Natl. Acad. Sci. USA. 2012;109:9623–9628. doi: 10.1073/pnas.1121606109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frisch M.J., Trucks G.W., Schlegel H.B., Scuseria G.E., Robb M.A., Cheeseman J.R., Scalmani G., Barone V., Petersson G.A., Nakatsuji H., et al. Gaussian 09. C.01 ed. Gaussian, Inc.; Wallingford, CT, USA: 2009. [Google Scholar]
- 36.MacroModel Schrodinger Release 2018-3. Schrodinger LLC; New York, NY, USA: 2018. [Google Scholar]
- 37.Ditchfield R. Molecular Orbital Theory of Magnetic Shielding and Magnetic Susceptibility. J. Chem. Phys. 1972;56:5688–5691. doi: 10.1063/1.1677088. [DOI] [Google Scholar]
- 38.Ditchfield R. Self-consistent perturbation theory of diamagnetism I. A gauge-invariant LCAO method for NMR chemical shifts. Mol. Phys. 1974;27:789–807. doi: 10.1080/00268977400100711. [DOI] [Google Scholar]
- 39.McMichael Rohlfing C., Allen L.C., Ditchfield R. Proton and carbon-13 chemical shifts: Comparison between theory and experiment. Chem. Phys. 1984;87:9–15. doi: 10.1016/0301-0104(84)85133-2. [DOI] [Google Scholar]
- 40.Wolinski K., Hinton J.F., Pulay P. Efficient implementation of the gauge-independent atomic orbital method for NMR chemical shift calculations. J. Am. Chem. Soc. 1990;112:8251–8260. doi: 10.1021/ja00179a005. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.