Abstract
It is widely accepted that different strains of Mycobacterium tuberculosis have variable degrees of pathogenicity and induce different immune responses in infected hosts. Similarly, different strains of Mycobacterium bovis have been identified but there is a lack of information regarding the degree of pathogenicity of these strains and their ability to provoke host immune responses. Therefore, in the current study, we used a mouse model to evaluate various factors involved in the severity of disease progression and the induction of immune responses by two strains of M. bovis isolated from cattle. Mice were infected with both strains of M. bovis at different colony-forming unit (CFU) via inhalation. Gross and histological findings revealed more severe lesions in the lung and spleen of mice infected with M. bovis N strain than those infected with M. bovis C68004 strain. In addition, high levels of interferon-γ (IFN-γ), interleukin-17 (IL-17), and IL-22 production were observed in the serum samples of mice infected with M. bovis N strain. Comparative genomic analysis showed the existence of 750 single nucleotide polymorphisms and 145 small insertions/deletions between the two strains. After matching with the Virulence Factors Database, mutations were found in 29 genes, which relate to 17 virulence factors. Moreover, we found an increased number of virulent factors in M. bovis N strain as compared to M. bovis C68004 strain. Taken together, our data reveal that variation in the level of pathogenicity is due to the mutation in the virulence factors of M. bovis N strain. Therefore, a better understanding of the mechanisms of mutation in the virulence factors will ultimately contribute to the development of new strategies for the control of M. bovis infection.
Keywords: M. bovis, immune response, pathogenicity, virulence factors, mutation
1. Introduction
Tuberculosis (TB) remains one of the most important infectious diseases of humans and animals worldwide. The disease is caused by Mycobacterium tuberculosis complex (MTBC), especially, Mycobacterium tuberculosis (M. tuberculosis) and Mycobacterium bovis (M. bovis). It is reported that 10.4 million new cases and 1.4 million deaths were caused by TB in 2015, and 2.8% of new cases of tuberculosis are attributed to M. bovis infection [1,2]. Like many other developing countries, control of bovine tuberculosis remains a challenge in China, which is mainly caused by M. bovis.
Tuberculosis consists of multiple stages that depend on both the pathogen and the host immune response to the infection [3,4]. Innate immune responses play an important role in bacterial control. There is increasing evidence that genetic variation among mycobacterial isolates contributes to differences in immune response induction and disease progression [5,6,7,8]. However, variability in the innate immune responses to genetic differences among M. bovis strains remains poorly understood. A better understanding of the genetic factors involved in immune response to infection will contribute to the control of bovine tuberculosis.
Since the first genomic study of M. tuberculosis in 1998, many additional studies have shown the genomic details of MTBC strains [9,10]. Though various strains of MTBC exhibit a high degree of genetic conservation, even then they exhibit a variable degree of pathogenicity [11,12,13]. This variation in the pathogenicity of MTBC strains is probably caused by the diversity in genetic and evolutionary background [14,15]. Therefore, it is necessary to understand the role of genetic diversity of different pathogenic strains of M. bovis in disease development.
In this study, we investigated the severity of disease in mice infected with M. bovis C68004 strain and M. bovis N strain. Mycobacterium bovis N strain was isolated from brain tissue of infected cattle with typical signs of systemic tuberculosis. Based on the clinical and histopathological observations in mice, we hypothesized that this strain of M. bovis may have distinct virulence factors and would show different pathogenesis in mice compared with the M. bovis C68004 strain.
As the natural route of infection of M. bovis is inhalation, we compared these two M. bovis strains with different genetic and evolutionary background using an intranasal infection mouse model [16,17]. We observed substantial variation in the disease progression and cytokine expression profiles in mice infected with two different strains of M. bovis, which showed altered degreesof pathogenesis. We further compared the whole-genome sequence (WGS) of both strains to identify the virulence factors that may contribute in the pathogenesis of these two strains.
We found that mice infected with M. bovis N strain showed acute death and severe inflammatory signs than those infected with M. bovis C68004 strain. The multiplex cytokine analysis assay for the serum samples of mice infected with M. bovis N strain also resulted in an increased production of IFN-γ, IL-17, and IL-22 in comparison to M. bovis C68004 strain. Comparative genomic analysis showed 750 single nucleotide polymorphisms (SNPs) and 145 small insertions/deletions (InDels) between the two M. bovis strains. After matching with the Virulence Factors Database (VFDB) database [18], we found mutations in 29 genes, which relate to 17 virulence factors. Expression of these genes was mainly related to the secretion system and cell surface transport. Out of 29 mutated genes, nine were present in C68004 while 20 existed in the N strain. Overall, our results demonstrate that mutations in the virulence factors contribute to the variation in the pathogenicity of M. bovis strains. The identification of mutated virulence factors in the pathogenic strains of M. bovis lead toward a better understanding of disease progression and further studies are suggested to develop new strategies for the control of M. bovis infection.
2. Materials and Methods
2.1. Ethics Statement
All animal experiments were performed according to the Chinese Regulations of Laboratory Animals − The Guidelines for the Care of Laboratory Animals (Ministry of Science and Technology of People’s Republic of China) and Laboratory Animal Requirements of Environment and Housing Facilities (GB 14925±2010, National Laboratory Animal Standardization Technical Committee).
The animal studies and research protocols were approved by The Laboratory Animal Ethical Committee of China Agricultural University under the license number cau20170108-2 (8 January, 2017).
2.2. Mice and M. bovis Strains
Female BALB/C mice aged between 6–8 weeks were obtained from specific-pathogen-free facilities at Charles River Laboratories and were maintained in BSL3 facilities.
In the present study, we used two virulent strains of M. bovis. Mycobacterium bovis C68004 strain was obtained from the China Institute of Veterinary Drug Control and this strain has been used for years with stable virulence. The M. bovis N strain was isolated recently from the brain tissue of cattle with generalized bovine tuberculosis (Figure S1). These strains were identified as M. bovis by multilocus sequence typing analysis as described before [19]. The M. bovis N strain was isolated in our laboratory and only cultured for few passages, while the other strain was obtained from cold storage and was passaged for three times after re-isolated from experimentally infected animals. Both strains were grown to mid-log phase in Middlebrook 7H9 broth (BD Biosciences) supplemented with 10% v/v OADC (Oleic acid, Albumin, Dextrose, Catalase) enrichment medium (BD Biosciences) and Tween 80. Before animals were infected with M. bovis strains, we calculated the colony-forming unit (CFU) by obtaining the optical density (OD) value for each isolate.
2.3. Genomic DNA Extraction, Sequencing, and Analysis
Genomic DNA from the two M. bovis strains were extracted using a TIANamp Bacterial Genomic DNA Kit (Tiangen Biotech Co. Ltd., Beijing, China), and were sequenced on an Illumina HiSeq 2000 platform. Raw sequencing reads were aligned to the reference genome Mycobacterium bovis AF2122/97 (LT708304.1) using the bwa mem alignment algorithm version 0.7.12 with default settings [20]. Single nucleotide polymorphisms (SNPs) and insertion/deletion (INDELs) events were identified using the Genome Analysis Tool Kit Haplotype Caller [21]. After combining variant call files for both strains, SNPs were filtered with the setting QD < 3.0 || FS > 60.0 || MQ < 40.0 || MQRankSum < −12.5 || ReadPosRankSum < −8.0 and INDELs were filtered with QD < 3.0 || FS > 200.0 || ReadPosRankSum < −20.0 as recommended in software documentation. Variants common to both isolates were then removed leaving only mutations variable among strains M. bovis C68004 and M. bovis N. Variants were annotated according to the reference genome gene annotations using snpEff version 4.3t [22]. Large deletions are difficult to assess using GATK, so we used the PILON genome analysis tool to look for additional larger deletion sequences [23].
2.4. Bacterial Infection
Mice (n = 6 per group) were anesthetized by Zoletil 50 (Virbac, France), which was first diluted 5-fold and 50-μL per mouse (0.5mg) was injected intraperitoneally. The BALB/c mice were divided into fifteen groups, and each group contain six mice. The animals were challenged via intranasal route with 200, 500, 1000, 2000, 5000, 10,000 and 20,000 CFU of M. bovis C68004 or M. bovis N diluted in phosphate buffer saline (PBS) according to the protocol described elsewhere [16]. The inoculum size was quantified after 24 h of infection by determining CFU in the lung tissue of 3 animals after infection with 200 CFU for both isolates and we found that the animals were successfully infected with M. bovis strains. We observed mean 159 ± 19 CFU in animals infected with M. bovis N strain while 172 ± 31 CFU in the lung tissue of animals infected with M. bovis C68004 strain. The control group of mice was challenged with PBS. All mice were maintained in the cages fitted with micro-isolators. Mice health was controlled by daily checking for smoothness of the fur, movement and activity, and reaction to the external stimuli. All of the mice inoculated with 10,000 and 20,000 CFU of both strains contracted acute death around the fourth week post inoculation. Therefore, we selected the animal groups infected with 5000 CFU for the long-term survival experiment. We observed a clear difference in the body weight of mice infected with 2000 CFU of M. bovis N showing a weight loss of 15% or more around four weeks post-inoculation as compared to the control group. In addition, previous studies showed that bacterial replication in the lung, spleen, and liver of mice challenged with the low dose of M. bovis reached a plateau at four weeks post infection [16,24,25]. Therefore, we sacrificed all the 2000 CFU and below infected mice by day 30 post-inoculation. Similar results were observed from two independent experiments.
2.5. Histopathology
All the mice infected with a CFU of 2000 or below were weighed and sacrificed on day 30 post-inoculation. We referred to a previous study and developed a clinical score method to calculate the severity of disease progression [26]. The mice were scored in terms of general behavior, feeding habits, and weight gain or loss. Animals reaching a clinical score of ≥4.0 were at the time point that followed by the time of death. The weight of each lung and spleen was measured, and organ coefficient was obtained by using the following formula: organ coefficient = organ weight (g)/mouse weight (g) [27]. Blood was obtained ante-mortem from eye route and serum was collected after clotting. The blood serum samples were stored at −80 °C until further experiment. The same lobe of the lung from each study group was fixed in 4% phosphate-buffered formaldehyde and embedded in paraffin. Sections of ~5 μm thickness were prepared and stained with haematoxylin and eosin (H&E), and with Ziehl-Neelsen (ZN) for histological examination and for the detection of acid-fast bacilli, respectively [28]. The digital images of the whole slide were acquired by Hamamatsu NanoZoomer S60 slide scanner and its viewing platform (NDP Viewer).
2.6. Multiplex Cytokine Assay
The concentrations of various cytokines and chemokines in blood serum were determined by ProcartaPlexTM Multiplex Immunoassay (eBioscience, San Diego, CA, USA) according to the manufacturer instructions, which contains 18 cytokines (GM-CSF, IFN-γ, IL-β, TNF-α, IL-1β, IL-2, IL-4, IL-5, IL-6, IL-9, IL-10, IL-12 P70, IL-13, IL-17, IL-18, IL-22, IL-23, and IL-27). We preferably used multiplex immunoassay for the determination of various cytokines and chemokines because of the limited quantity of serum samples from each animal [29,30].
2.7. Statistical Analysis
Results are presented as mean and SEM (standard error) within each group (n = 6). For analysis of organ coefficients, clinical score, and cytokine production, we used student’s unpaired t-test (two-tailed). Data were analyzed using the SPSS Statistics version 20.0 (IBM, Armonk, NY, USA), the ImageJ version 1.45 (National Institutes of Health, Bethesda, MA, USA) and the GraphPad Prism version 5.01 program (GraphPad Software Inc., San Diego, CA, USA). A p < 0.05 was considered as statistically significant. The p-values shown in the figures are presented with *** (p < 0.001), ** (p < 0.01), * (p < 0.05), and n.s. indicates statistically non-significant.
3. Results
3.1. Clinical Presentation and Necropsy Lesions
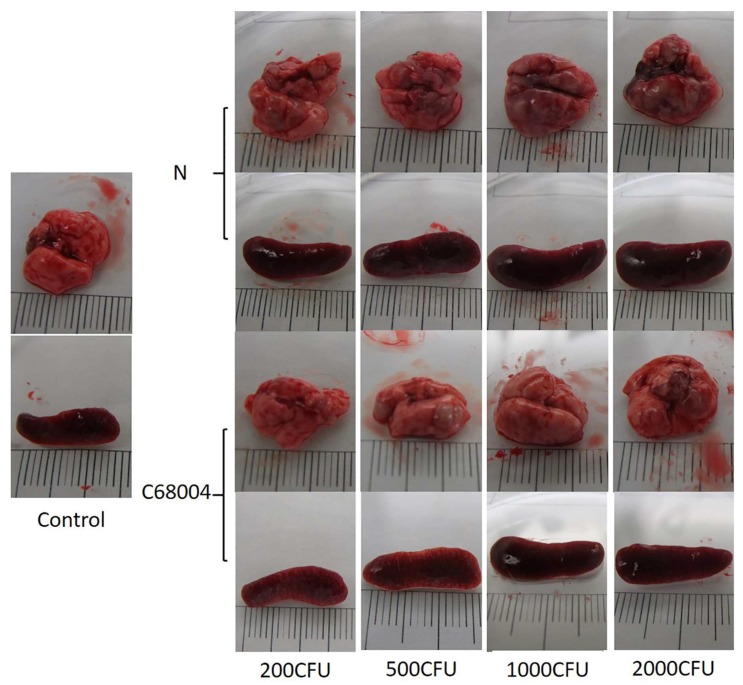
The infected animals showed clinical signs around 19 days post-inoculation. The animals infected with M. bovis N with 5000 CFU and above showed obvious weight loss, disordered hair, somnolence, inappetence, and conjunctivitis. The animals infected with 2000 CFU of M. bovis N showed similar signs when sacrificed on day 30 post-inoculation, while no visible signs were observed in animals infected with the same CFU of M. bovis C68004 strain (Figure S2). On gross examination of organs, we observed that the longitudinal size of the lung and spleen were 2–5 millimeters longer in the infected groups as compared with the control group. Normal tissue could barely be seen in the lung of M. bovis N strain infected animals while several nodular lesions were present in M. bovis C68004 infected animals (Figure 1). Overall, the mice infected with M. bovis N showed more severe organ lesions and symptoms of the disease than M. bovis C68004. Similar results were observed in two independent experiments.
Figure 1.
Representative photographs showing gross lesions of the lung and spleen of mice infected with different strains of Mycobacterium bovis. The lung and spleen were aseptically removed from the mice at the end of the clinical observation (30 days post-inoculation) before death. Pictures of the lung and spleen of mice infected with 200, 500, 1000, and 2000 colony-forming unit (CFU) of M. bovis N and M. bovis C68004 strains are shown individually. Control shows the lung of the mice inoculated with phosphate buffer saline (PBS).
3.2. Survival Time
To assess the difference of pathogenesis between the two M. bovis strains, we also observed the survival time of mice post infection [31]. We evaluated the survival time for mice infected with 5000 CFU of both strains. A dose of 5000 CFU of M. bovis N strain resulted in acute death (median survival time 20–22 days post challenge). The mice showed clinical symptoms around 19 days post-inoculation and acute death happened in three days. On the other hand, mice infected with 5000 CFU of M. bovis C68004 survived significantly longer than those infected with M. bovis N, only two mice died around 40 days post-inoculation (Figure 2). The animal survival experiment was also repeated twice and similar results were obtained.
Figure 2.
The survival rates of mice infected with 5000 CFU of M. bovis N and M. bovis C68004 strains. Each group had six mice. Observation times (day post-inoculation) are indicated in the x-axis. The experiment was repeated two times independently.
3.3. Organ Coefficient and Clinical Score of Mice Infected with Different Strains of M. bovis
In order to evaluate the effect of M. bovis infection on lung and spleen, we measured the weight of these organs. We observed that the lung and spleen of mice infected with either strain of M. bovis with a CFU of 2000 and below were heavier as compared with the uninfected control group. The organ coefficient of M. bovis N infected mice showed that the weight of lung and spleen was positively correlated with the dose of infection. Moreover, the organ coefficient also demonstrated that lung and spleen of mice infected with 500 and 2000 CFU of M. bovis N strain were heavier than of those infected with M. bovis C68004 (Figure 3A,B). On the other hand, no significant difference was observed in the weight of lung and spleen of mice infected with M. bovis N and M. bovis C68004 with 200 and 1000 CFU. The lung and spleen of mice challenged with 2000 CFU seemed lighter than the groups receiving lower doses but no significant difference was observed between 2000 CFU and below CFU infected mice with M. bovis C68004 strain. The mice were monitored for clinical signs of disease on a daily basis as described in Materials and Methods section. The clinical symptoms were more severe in mice infected with 2000 and 1000 CFU of M. bovis N than of those infected with M. bovis C68004 strain, as reflected by the clinical score (Figure 3C). By comparing with the uninfected mice, the significant body weight loss was only observed in mice infected with 2000 CFU of M. bovis N (Figure 3D). Similar results were observed in two independent experiments.
Figure 3.
Organ coefficient of the lung (A) and spleen (B), score of clinical symptoms (C) and body weight (D) from mice infected with different CFU of M. bovis N and M. bovis C68004 strains or PBS 30 days post-inoculation. The experiment was repeated two times independently. Statistical analysis was performed using student’s unpaired t-test (two-tailed). Bars represent means SEM (n = 6); * p < 0.05, ** p < 0.01; *** p < 0.001.
3.4. Histopathological Examination
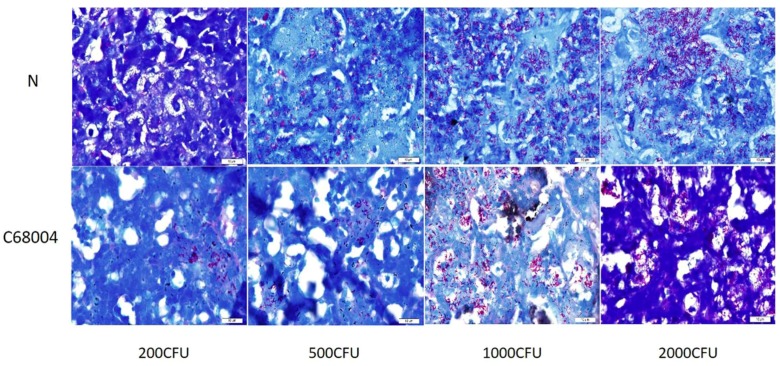
For histopathological examination, the lung tissues of infected and un-infected control mice were stained using H&E staining and were observed under light microscope. We found multiple dispersed inflammatory areas in the lung tissue of all animals infected with 2000 CFU and below. We observed clear difference of the lesions in the lung of mice infected with 1000 CFU and 2000 CFU of the two strains and larger inflammatory areas were seen in lung of mice infected with M. bovis N as compared with M. bovis C68004 (Figure 4A). The percentage of lung surface area inflammation had a statistically significant increase in M. bovis N treated mice as compared with M. bovis C68004 (Figure 4B). All the inflammatory areas were infiltrated with lymphocytes and macrophages in all infected animals compared with control animals (Figure 4C). In addition, we observed several inflammatory regions with multiple necrotic foci in the lung of animals infected with high CFU of M. bovis N strain than those infected with M. bovis C68004 strain (Figure 4C). In addition, we observed visible acid-fast bacilli in the lung sections of all infected mice showing that infection occurred in all group of mice at different administered doses for both strains of M. bovis (Figure 5). These experiments were repeated twice and found similar results for histological changes in the lung tissue.
Figure 4.
Development of lung lesions in mice infected with M. bovis N and M. bovis C68004 strains. The lung and spleen were aseptically removed from the mice at the end of the clinical observation (30 days post-inoculation) before death. (A) The whole-slide digital images of H&E-stained lung sections. Pictures of the lung of mice infected with 200, 500, 1000, and 2000 CFU of the both strains are shown individually. (B) Morphometric analysis demonstrated affected lungs in mice treated with the both M. bovis strains. Results are represented as percentage of lung surface area involved, calculated using ImageJ viewer software (obtained from six animals per group). (C) Granulomatous lesions in lung infected with 2000 CFU of the both strains. Pictures of the lung of mice infected with 2000 CFU of M. bovis N and M. bovis C68004 strains and uninfected control are shown individually. The arrows indicate multiple necrotic areas in the lung of mice infected with 2000 CFU of M. bovis N (arrows: necrosis). Scale bar: 100 μm. * p < 0.05, ** p < 0.01; *** p < 0.001.
Figure 5.
Acid fast bacilli in infected lung. High magnification of central necrotic areas showing M. bovis bacilli within cellular debris of infected lung. Scale bar: 10 μm.
3.5. Cytokine Profiles in Serum
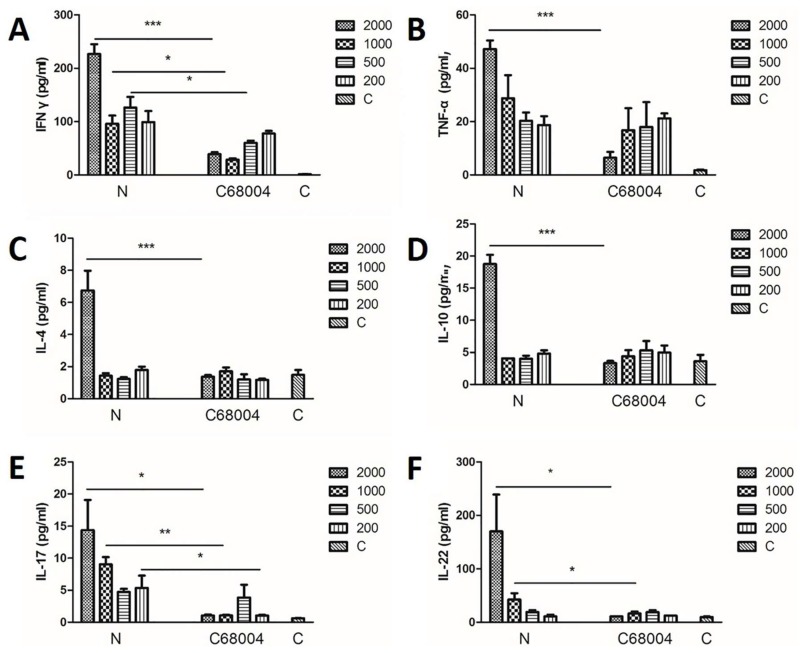
In order to investigate immune responses induced by the both M. bovis strains, we assessed the level of various cytokines in the serum samples. The cytokine analysis was done by multiplex cytokine assay. We found significant difference in cytokines (IFN-γ, IL-4, IL-10, IL-17, IL-22, and TNF-α) expression in all infected animals that have roles in the regulation of T cell responses including T helper1 (Th1), T helper2 (Th2), and T helper17 (Th17) (Figure 6). While no significant difference was observed for the remaining cytokines among various infected animal groups (Figure S3). The level of IFN-γ in the mice infected with M. bovis N was significantly increased when compared with the M. bovis C68004 infected mice, except the animals challenged with 200 CFU for both strains (Figure 6A). Mycobacterium bovis N strain also induced higher levels of IL-17 production in mice than M. bovis C68004 infected mice, except for the mice challenged with 500 CFU (Figure 6E). In addition, we observed an increased level of IL-22 only in mice infected with 2000 CFU and 1000 CFU of M. bovis N than M. bovis C68004 strain infected mice, while no significant difference was recorded in the animal groups infected with CFU of 200 and 500 of both strains of M. bovis (Figure 6F). Similarly, the production of TNF-α, IL-4, and IL-10 was only significantly high in the M. bovis N exposed group when the animals were infected with 2000 CFU (Figure 6B–D). These results show that an increased inflammatory response was observed in mice infected with M. bovis N strain as compared with M. bovis C68004 strain.
Figure 6.
Cytokines expression profiles in mice infected with different CFU of M. bovis N and M. bovis C68004 strains. The blood was harvested ante-mortem via eye route and serum was used for cytokines expression analysis. Th1 immune response cytokines shown are IFN-γ (A) and TNF-α (B). Th2 immune response cytokines shown are IL-4 (C) and IL-10 (D). Th17 immune response cytokines shown are IL-17 (E) and IL-22 (F). Statistical analysis was performed using student’s unpaired t-test (two-tailed). Bars represent means SEM (n = 6); * p < 0.05, ** p < 0.01; *** p < 0.001.
3.6. Genomic Sequence Analysis
Multilocus sequence typing (MLST) analysis confirmed that both strains were members of M. bovis, so we next performed whole-genome sequencing (WGS) to identify genetic differences that may explain the observed differences in pathology and immune responses induction between these isolates. Comparing both isolates with the reference M. bovis strain AF2122 (ATCC##), we identified 750 single nucleotide polymorphisms (SNPs) and 145 small insertions/deletions (InDels) that differentiated the two isolates (Table 1, Supplementary Materials Tables S1 and S2). Of these, 467 SNPs and 86 InDels happened in M. bovis N strain and the rest existed in M. bovis C68004 strain. In addition, 115 SNPs and 55 InDels belonged to the PPE/PE-PGRS gene families [32].
Table 1.
Mutation type of single nucleotide polymorphisms (SNPs) and insertions/deletions (InDels).
| SNP | InDel | ||
|---|---|---|---|
| Intergenic region | 86 | Conservative inframe deletion | 13 |
| Missense variant | 409 | Disruptive inframe insertion | 26 |
| Stop gained | 7 | Frameshift variant | 60 |
| Stop lost & splice region variant | 2 | Frameshift variant & stop gained | 4 |
| Synonymous variant | 246 | Intergenic region | 31 |
| Pilon deletion | 11 | ||
3.7. Virulence Factors Mutation in Both Strains
We then searched for virulence factors by comparing these changes against 257 virulence factors collected from the VFDB database [18]. We found that 20 genes were mutated in M. bovis N strain and nine genes were mutated in M. bovis C68004 strain in comparison to the reference strain. The results showed that 17 virulence factors were variable between the two strains of M. bovis (Table 2). These data unveiled that higher level of mutations was found in M. bovis N strain in comparison to C68004 strain and these mutations in the virulence factors seems to be responsible for the high degree of pathogenicity of the N strain in the mouse model of infection.
Table 2.
Related genes of M. bovis C68004 and N strains.
| Gene Mb# | VFclass | Virulence factors | Strain C68004 | Strain N |
|---|---|---|---|---|
| Mb0607 | Mammalian cell entry (mce) operons | Mce2 | - | mce2D |
| Mb1214 | Cell surface components | Glycopeptidolipid (GPL) locus | - | papA3 |
| Mb3856 | Cell surface components | Trehalose-recycling ATP-binding cassette (ABC) transporter | fad23 | - |
| Mb3906 | Secretion system | ESAT-6 secretion system (ESX)-1 (T7SS) | espI | - |
| Mb3916c-Mb3917c | Secretion system | ESX-2 (T7SS) | - | mycP2 |
| Secretion system | ESX-2 (T7SS) | - | eccD2 | |
| Mb3924C | Secretion system | ESX-2 (T7SS) | eccC2 | - |
| Mb0208C | Metal uptake | Heme uptake | - | mmpL11 |
| Mb0250C | Cell surface components | GPL locus | - | fadE5 |
| Mb0661C | Cell surface components | Methyltransferase | mmaA4 | - |
| Mb0994 | Metal exporters | Copper exporter | - | ctpV |
| Mb1193 | Anaerobic respiration | Nitrate reductase | - | narG |
| Mb1267 | Cell surface components | Trehalose-recycling ABC transporter | - | lpqY |
| Mb1553C | Cell surface components | GPL locus | - | gtf1 |
| Mb1766C | Anaerobic respiration | Nitrate/nitrite transporter | narK2 | - |
| Mb2139C | Proteases | Proteasome-associated proteins | mpa | - |
| Mb2404C | Metal uptake | Mycobactin | - | mbtB |
| Mb2439C | Secreted proteins | Enhanced intracellular survival protein | - | eis |
| Mb2958 | Cell surface components | Phthiocerol dimycocerosate (PDIM) and Phenolic glycolipid (PGL) biosynthesis and transport | - | ppsC |
| Mb2959 | Cell surface components | PDIM and PGL biosynthesis and transport | - | ppsD |
| Mb2960 | Cell surface components | PDIM and PGL biosynthesis and transport | - | ppsE |
| Mb2966 | Cell surface components | PDIM and PGL biosynthesis and transport | - | fadD28 |
| Mb2982C | Cell surface components | PDIM and PGL biosynthesis and transport | - | - |
| Mb3298 | Metal exporters | P(1)-type Mn2+ transporting ATPase | - | ctpC |
| Mb3631C | Lipid and fatty acid metabolism | Pantothenate synthesis | panD | - |
| Mb3851 | Cell surface components | Trehalose-recycling ABC transporter | sap | - |
| Mb3896 | Regulation | RegX3 | espG1 | - |
| Mb3912C | Secretion system | ESX-1 (T7SS) | - | eccE1 |
| Mb0606 | mce operons | Mce2 | - | mce2C |
| Mb0609 | mce operons | Mce2 | - | mce2E |
4. Discussion
Bovine tuberculosis represents a significant animal health problem having economic impact and also imposing zoonotic threat to human health [2,33]. It is estimated that the prevalence of bovine tuberculosis is 9% worldwide, especially in the developing countries [24]. Therefore, it is important to study the factors related to pathogenicity of different M. bovis strains, which could help in the early diagnostic procedure and adopting new strategies for the control of the disease. Although many studies have been reported on the difference in virulence of M. tuberculosis strains, only a few studies provided information on the variation in virulence among different strains of M. bovis [33,34,35,36]. In the current study, we analyzed the genetic differences of two M. bovis strains in mouse models that were isolated from cattle with different backgrounds.
To date, BALB/c and C57Bl/6 mice are the most classical tuberculosis resistant animal models and are widely used for M. tuberculosis or M. bovis studies, although both of them show limited features of tuberculosis pathophysiology when compared with the C3HeB/FeJ mouse model [24,37,38]. Based on previous studies and experience in our laboratory, the BALB/c mouse model is the best choice for M. bovis infection through the nostril [16,39]. In our current study, we infected the animals with different CFUs to determine the suitable dose of both M. bovis strains that cause clinical disease. Therefore, we infected the animals with various CFU starting from 200 to maximal 20000 CFU. We found that the animals exposed to 10000 CFU and above with both strains of M. bovis died acutely around four weeks post infection (data not shown). Based on these observations we focused on animal groups infected with 200 to 5000 CFU to analyze the pathogenicity difference between two M. bovis isolates in our study. We found that all animals had died after three weeks of infection with 5000 CFU of M. bovis N strain. This finding of early-mortality of mice is similar to a previous study in C3HeB/Fej mice infected with M. tuberculosis [24]. On the other hand, animals challenged with M. bovis C68004 strain survived for much longer time post-inoculation. It showed that both strains of M. bovis were not similar regarding their pathogenicity level in mice. In a previous study, BABL/C mice inoculated with more than 1000 CFU virulent M. bovis strain via lateral tail vein suffered from acute death with a median survival time of 22 days [16]. Previous studies on non-human models of mycobacterial infection suggest that the key period to determine whether the mice will show the progression of acute or chronic infection is between 3–6 weeks post infection [40,41]. Consistent with that, we found that the mice infected with the highest dose of M. bovis N showed an acute onset of infection while the animals infected with M. bovis C68004 developed progressive chronic infection. Animals infected with lower CFU than 5000 showed clinical symptoms after three weeks of infection. When the mice were sacrificed on day 30 post-inoculation, we found a variable number of small and large tubercles in the lung of all infected animals. In addition, the lung and the spleen were also enlarged in mice exposed with both strains of M. bovis compared with the control uninfected mice. Like the reference strain M. bovis AF2122 (ATCC##), both of the strains caused inflammatory response areas in BABL/c mice [24]. Considering the discrepancy of the organ coefficient, clinical scores, and the gross lesion examination, M. bovis N may cause a more severe inflammatory reaction in lung and spleen than M. bovis C68004. Based on the findings of acid-fast staining of lung sections for all infected mice, we found significantly more M. bovis bacilli in mice infected with 2000 CFU of M. bovis N strain than mice infected with M. bovis C68004. The above findings suggest that M. bovis N strain is more pathogenic compared to M. bovis C68004 strain.
This conclusion was supported by further analysis of cytokine levels in the serum samples of mice infected with both strain of M. bovis. Cytokines are critical for recruitment and activation of immune modulating cells to the site of infection and are essential for the control of bacterial growth and survival [42]. Both IFN-γ and TNF-α have been accepted as the most important Th1 cells cytokines playing a key role in the progress and outcome of tuberculosis infection [43,44]. IFN-γ has been considered as a potential biomarker for the identification of various stages of M. tuberculosis infection [45,46]. Recent studies revealed that IFN-γ and IL-17 play an important role in pro-inflammatory responses and chemokine regulation, and a stronger IFN-γ and IL-17 response correlates with the severity of the disease in humans [44,47]. In vitro and in vivo generation of Th17 cells can lead to stronger IL-17 and IL-22 expression [48]. Our finding showed that M. bovis N strain induced stronger IFN-γ, IL-17, and IL-22 responses than M. bovis C68004 strain. In line with the histopathological observation, we considered stronger IFN-γ, IL-17, and IL-22 responses in mice induced by different M. bovis strains may be related with the severity of the disease. TNF-α is a protective cytokine and most virulent mycobacterial strains elicit an increased expression of TNF-α than avirulent mycobacterial strains [49,50]. In our study, we only found significant difference of TNF-α response in the 2000 CFU level of infection between the two strains, same as other cytokines including IL-4, IL-10, GM-CSF, IL-1β, IL-2, IL-6, IL-13, and IL-18. It is generally accepted that the outcome of mycobacterial infection depends on the induction of host immune responses but unrestricted immune responses are deleterious and may lead to the disease exacerbation [51,52]. Therefore, this could be interpreted as chaotic immune responses when the mice infected with 2000 CFU of M. bovis N were close to death. In addition, we found no significant difference in the expression of TNF-α, IL-4, and IL-10 induced by two different strains of M. bovis. Unexpectedly, in our study, we found significantly low levels of IFN-γ in animals infected with high CFU (1000 to 2000) of M. bovis C68004 strain than animals infected with low CFU of the same strain. This may be due to faster activation of adaptive immunity in the higher doses of M. bovis C68004 compared with lower doses.
The increasing availability of WGS data for M. tuberculosis has shown that the evolution of M. tuberculosis strains contributes to lineage-specific adaptation of human populations, disease pathogenesis, and drug resistance [13,53]. That is why tuberculosis remains one of the leading causes of death worldwide. We hypothesized that the epidemic of M. bovis strains nowadays may have stronger pathogenesis and better bovine adaptation. Therefore, we investigated the genome difference between the two strains for further understanding of variation in the degree of pathogenicity. Upon WGS analysis for both strains of M. bovis, we found many SNPs and InDels between these two strains that may account for the observed differences in pathology and immune responses. After the comparison with the reference virulence factors collected from the VFDB, there were 17 virulence factors related to 29 genes in the two strains. Of these, nine existed in M. bovis C68004 strain and 20 existed in M. bovis Nstrain. The proteins expressed by these genes are mainly involved in secretion and cell surface transport, like trehalose-recycling ABC transporter and phthiocerol dimycocerosates (PDIMs) and phenolic glycolipids (PGLs). Acquisition of host sugars is quite important during some stages of M. tuberculosis infection, trehalose-recycling ABC transporter is highly specific for uptake of the disaccharide trehalose and essential for virulence of M. tuberculosis [54]. Phthiocerol dimycocerosates (PDIMs) and phenolic glycolipids (PGLs) are structurally-related complex lipids in the mycobacterial cell wall and present only in pathogenic mycobacteria, such as M. tuberculosis and M. bovis [55]. The loss of PDIMs and PGLs has been associated with decrease in virulence of M. bovis, although the contribution of individual PDIMs and PGLs to virulence in these organisms has not been evaluated [56]. In addition, some studies have focused on the mutations in mice of these genes and resulted virulence variation. For example, mutation in mice operons, espG1 gene, and ppsD gene may attenuate M. tuberculosis virulence [57,58,59]. However, in our study, the mutations of PDIMs, PGLs-related genes, and the above three genes happened in M. bovis N strain. We suggest that the mutations in these genes may not change the gene function. After all, most studies have focused on the relation between the loss of the virulence factors or related genes and the virulence change in M. tuberculosis. It is necessary for us to identify these virulence factors in M. bovis and find out what variants play key roles for the different clinical presentation in mice.
According to the limited reports, cattle maintain both M. bovis and M. tuberculosis in China and the infection with non-tuberculosis mycobacterium was also reported recently [60,61]. The M. bovis N strain has more genome differences compared with the reference M. bovis strain AF2122 (ATCC##) than the M. bovis C68004 strain. In addition, considering its strong pathogenicity, it may have well developed bovine adaptation, can aggravate the problems in bovine tuberculosis eradication, and might have a potential zoonotic risk to human in China.
Taken together, the data demonstrate that both strains of M. bovis are diverse in terms of intracellular adaptation and the progression of disease. In addition, the data elucidate that different strains of M. bovis induce a varied level of immune responses in the infected animals. Genomic-based analysis reveals several mutations in the virulence factors responsible for the degree of pathogenicity. The study widens our knowledge of M. bovis strains associated virulence factors, and the possible role of these factors in pathogenicity and determining the outcomes of the infection.
Acknowledgments
We thank the China Agricultural University for providing the Biosafety Level-3 Laboratories for experiments using M. bovis culture and infection.
Supplementary Materials
The supplementary materials are available online at http://www.mdpi.com/1422-0067/20/1/5/s1.
Author Contributions
G.C. wrote the manuscript. T.H. and N.S. rewrote and edited the manuscript. D.Z. and X.Z. designed experiments. G.C. and J.N. performed experiments. M.L. assisted to complete experiments.
Funding
This work was supported by China Agriculture Research System (No. CARS-36), the National Key Research and Development Program (Project No. 2017YFD0500901), National Natural Science Foundation of China (Project No. 31572487); and the MoSTRCUK international cooperation project (Project No. 2013DFG32500); and the High-end Foreign Experts Recruitment Program (Project No. GDW20151100036, GDW20161100071).
Conflicts of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- 1.WHO . Global Tuberculosis Report. Geneva. WHO; Geneva, Switzerland: 2016. [Google Scholar]
- 2.Muller B., Durr S., Alonso S., Hattendorf J., Laisse C.J., Parsons S.D., van Helden P.D., Zinsstag J. Zoonotic Mycobacterium bovis-induced tuberculosis in humans. Emerg. Infect. Dis. 2013;19:899–908. doi: 10.3201/eid1906.120543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O’Garra A., Redford P.S., McNab F.W., Bloom C.I., Wilkinson R.J., Berry M.P. The immune response in tuberculosis. Annu. Rev. Immunol. 2013;31:475–527. doi: 10.1146/annurev-immunol-032712-095939. [DOI] [PubMed] [Google Scholar]
- 4.Roy Chowdhury R., Vallania F., Yang Q., Lopez Angel C.J., Darboe F., Penn-Nicholson A., Rozot V., Nemes E., Malherbe S.T., Ronacher K., et al. A multi-cohort study of the immune factors associated with M. Tuberculosis infection outcomes. Nature. 2018;560:644–648. doi: 10.1038/s41586-018-0439-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barreiro L.B., Tailleux L., Pai A.A., Gicquel B., Marioni J.C., Gilad Y. Deciphering the genetic architecture of variation in the immune response to Mycobacterium tuberculosis infection. Proc. Natl. Acad. Sci. USA. 2012;109:1204–1209. doi: 10.1073/pnas.1115761109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang Y., Liu H., Wang X., Xiao S., Li M., Li G., Zhao L., Zhao X., Dou X., Wan K. Genetic diversity of immune-related antigens in region of difference 2 of Mycobacterium tuberculosis strains. Tuberculosis. 2017;104:1–7. doi: 10.1016/j.tube.2016.05.002. [DOI] [PubMed] [Google Scholar]
- 7.Portevin D., Gagneux S., Comas I., Young D. Human macrophage responses to clinical isolates from the Mycobacterium tuberculosis complex discriminate between ancient and modern lineages. PLoS Pathog. 2011;7:e1001307. doi: 10.1371/journal.ppat.1001307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dong H., Lv Y., Sreevatsan S., Zhao D., Zhou X. Differences in pathogenicity of three animal isolates of Mycobacterium species in a mouse model. PLoS ONE. 2017;12:e0183666. doi: 10.1371/journal.pone.0183666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cole S.T., Brosch R., Parkhill J., Garnier T., Churcher C., Harris D., Gordon S.V., Eiglmeier K., Gas S., Barry C.E., 3rd, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 10.Jia X., Yang L., Dong M., Chen S., Lv L., Cao D., Fu J., Yang T., Zhang J., Zhang X., et al. The bioinformatics analysis of comparative genomics of Mycobacterium tuberculosis complex (MTBC) provides insight into dissimilarities between intraspecific groups differing in host association, virulence, and epitope diversity. Front. Cell. Infect. Microbiol. 2017;7:88. doi: 10.3389/fcimb.2017.00088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garbaccio S., Macias A., Shimizu E., Paolicchi F., Pezzone N., Magnano G., Zapata L., Abdala A., Tarabla H., Peyru M., et al. Association between spoligotype-VNTR types and virulence of Mycobacterium bovis in cattle. Virulence. 2014;5:297–302. doi: 10.4161/viru.27193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peters J.S., Calder B., Gonnelli G., Degroeve S., Rajaonarifara E., Mulder N., Soares N.C., Martens L., Blackburn J.M. Identification of quantitative proteomic differences between Mycobacterium tuberculosis lineages with altered virulence. Front. Microbiol. 2016;7:813. doi: 10.3389/fmicb.2016.00813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coscolla M., Gagneux S. Does M. Tuberculosis genomic diversity explain disease diversity? Drug Discov. Today Dis. Mech. 2010;7:e43–e59. doi: 10.1016/j.ddmec.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gagneux S., Small P.M. Global phylogeography of Mycobacterium tuberculosis and implications for tuberculosis product development. Lancet Infect. Dis. 2007;7:328–337. doi: 10.1016/S1473-3099(07)70108-1. [DOI] [PubMed] [Google Scholar]
- 15.Coscolla M., Gagneux S. Consequences of genomic diversity in mycobacterium tuberculosis. Semin. Immunol. 2014;26:431–444. doi: 10.1016/j.smim.2014.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Logan K.E., Gavier-Widen D., Hewinson R.G., Hogarth P.J. Development of a Mycobacterium bovis intranasal challenge model in mice. Tuberculosis. 2008;88:437–443. doi: 10.1016/j.tube.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 17.Salguero F.J., Gibson S., Garcia-Jimenez W., Gough J., Strickland T.S., Vordermeier H.M., Villarreal-Ramos B. Differential cell composition and cytokine expression within lymph node granulomas from BCG-vaccinated and non-vaccinated cattle experimentally infected with Mycobacterium bovis. Transbound. Emerg. Dis. 2017;64:1734–1749. doi: 10.1111/tbed.12561. [DOI] [PubMed] [Google Scholar]
- 18.Chen L., Zheng D., Liu B., Yang J., Jin Q. VFDB 2016: Hierarchical and refined dataset for big data analysis—10 years on. Nucleic Acids Res. 2016;44:D694–697. doi: 10.1093/nar/gkv1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huard R.C., Lazzarini L.C., Butler W.R., van Soolingen D., Ho J.L. PCR-based method to differentiate the subspecies of the Mycobacterium tuberculosis complex on the basis of genomic deletions. J. Clin. Microbiol. 2003;41:1637–1650. doi: 10.1128/JCM.41.4.1637-1650.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H., Durbin R. Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van der Auwera G.A., Carneiro M.O., Hartl C., Poplin R., Del Angel G., Levy-Moonshine A., Jordan T., Shakir K., Roazen D., Thibault J., et al. From FASTQ data to high confidence variant calls: The genome analysis toolkit best practices pipeline. Curr. Protoc. Bioinform. 2013;43:11.10.1–11.10.33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cingolani P., Platts A., Wang le L., Coon M., Nguyen T., Wang L., Land S.J., Lu X., Ruden D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEFF: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly. 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walker B.J., Abeel T., Shea T., Priest M., Abouelliel A., Sakthikumar S., Cuomo C.A., Zeng Q., Wortman J., Young S.K., et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE. 2014;9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boute M., Carreras F., Rossignol C., Doz E., Winter N., Epardaud M. The C3HeB/FeJ mouse model recapitulates the hallmark of bovine tuberculosis lung lesions following Mycobacterium bovis aerogenous infection. Vet. Res. 2017;48:73. doi: 10.1186/s13567-017-0477-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kato-Maeda M., Shanley C.A., Ackart D., Jarlsberg L.G., Shang S., Obregon-Henao A., Harton M., Basaraba R.J., Henao-Tamayo M., Barrozo J.C., et al. Beijing sublineages of Mycobacterium tuberculosis differ in pathogenicity in the guinea pig. Clin. Vaccine Immunol. CVI. 2012;19:1227–1237. doi: 10.1128/CVI.00250-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dibbern J., Eggers L., Schneider B.E. Sex differences in the C57BL/6 model of Mycobacterium tuberculosis infection. Sci. Rep. 2017;7:10957. doi: 10.1038/s41598-017-11438-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y., Yang C., He Y., Zhan X., Xu L. IPR1 modified bcg as a novel vaccine induces stronger immunity than bcg against tuberculosis infection in mice. Mol. Med. Rep. 2016;14:1756–1764. doi: 10.3892/mmr.2016.5447. [DOI] [PubMed] [Google Scholar]
- 28.Wang J., Li B.X., Ge P.P., Li J., Wang Q., Gao G.F., Qiu X.B., Liu C.H. Mycobacterium tuberculosis suppresses innate immunity by coopting the host ubiquitin system. Nat. Immunol. 2015;16:237–245. doi: 10.1038/ni.3096. [DOI] [PubMed] [Google Scholar]
- 29.Yang T., Wu X., Hu J., Hu M., Xu H., Jiang H., Ding Q. Maternal high-fat diet promotes the development and progression of prostate cancer in transgenic adenocarcinoma mouse prostate offspring. Cell. Physiol. Biochem. 2018;47:1862–1870. doi: 10.1159/000491066. [DOI] [PubMed] [Google Scholar]
- 30.Sigdel K.R., Duan L., Wang Y., Hu W., Wang N., Sun Q., Liu Q., Liu X., Hou X., Cheng A., et al. Serum cytokines Th1, Th2, and Th17 expression profiling in active lupus nephritis-iv: From a southern chinese han population. Mediat. Inflamm. 2016;2016:4927530. doi: 10.1155/2016/4927530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.North R.J., Jung Y.J. Immunity to tuberculosis. Annu. Rev. Immunol. 2004;22:599–623. doi: 10.1146/annurev.immunol.22.012703.104635. [DOI] [PubMed] [Google Scholar]
- 32.Comas I., Chakravartti J., Small P.M., Galagan J., Niemann S., Kremer K., Ernst J.D., Gagneux S. Human T cell epitopes of Mycobacterium tuberculosis are evolutionarily hyperconserved. Nat. Genet. 2010;42:498–503. doi: 10.1038/ng.590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiong X., Wang R., Deng D., Chen Y., Liu H., Wang T., Wang J., Zhu X., Zhu X., Zhu Y., et al. Comparative genomics of a bovine Mycobacterium tuberculosis isolate and other strains reveals its potential mechanism of bovine adaptation. Front. Microbiol. 2017;8:2500. doi: 10.3389/fmicb.2017.02500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paolino M., Brindisi M., Vallone A., Butini S., Campiani G., Nannicini C., Giuliani G., Anzini M., Lamponi S., Giorgi G., et al. Development of potent inhibitors of the Mycobacterium tuberculosis virulence factor ZMP1 and evaluation of their effect on mycobacterial survival inside macrophages. ChemMedChem. 2018;13:422–430. doi: 10.1002/cmdc.201700759. [DOI] [PubMed] [Google Scholar]
- 35.Montoya-Rosales A., Provvedi R., Torres-Juarez F., Enciso-Moreno J.A., Hernandez-Pando R., Manganelli R., Rivas-Santiago B. LYSX gene is differentially expressed among Mycobacterium tuberculosis strains with different levels of virulence. Tuberculosis. 2017;106:106–117. doi: 10.1016/j.tube.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 36.Vargas-Romero F., Mendoza-Hernandez G., Suarez-Guemes F., Hernandez-Pando R., Castanon-Arreola M. Secretome profiling of highly virulent Mycobacterium bovis 04-303 strain reveals higher abundance of virulence-associated proteins. Microb. Pathog. 2016;100:305–311. doi: 10.1016/j.micpath.2016.10.014. [DOI] [PubMed] [Google Scholar]
- 37.Rhoades E.R., Frank A.A., Orme I.M. Progression of chronic pulmonary tuberculosis in mice aerogenically infected with virulent mycobacterium tuberculosis. Tuber. Lung Dis. 1997;78:57–66. doi: 10.1016/S0962-8479(97)90016-2. [DOI] [PubMed] [Google Scholar]
- 38.Hoff D.R., Ryan G.J., Driver E.R., Ssemakulu C.C., De Groote M.A., Basaraba R.J., Lenaerts A.J. Location of intra- and extracellular M. Tuberculosis populations in lungs of mice and guinea pigs during disease progression and after drug treatment. PLoS ONE. 2011;6:e17550. doi: 10.1371/journal.pone.0017550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Garcia-Pelayo M.C., Bachy V.S., Kaveh D.A., Hogarth P.J. BALB/c mice display more enhanced bcg vaccine induced Th1 and Th17 response than C57BL/6 mice but have equivalent protection. Tuberculosis. 2015;95:48–53. doi: 10.1016/j.tube.2014.10.012. [DOI] [PubMed] [Google Scholar]
- 40.Kimmey J.M., Campbell J.A., Weiss L.A., Monte K.J., Lenschow D.J., Stallings C.L. The impact of isgylation during Mycobacterium tuberculosis infection in mice. Microbes Infect. 2017;19:249–258. doi: 10.1016/j.micinf.2016.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Coleman M.T., Maiello P., Tomko J., Frye L.J., Fillmore D., Janssen C., Klein E., Lin P.L. Early changes by (18)fluorodeoxyglucose positron emission tomography coregistered with computed tomography predict outcome after Mycobacterium tuberculosis infection in cynomolgus macaques. Infect. Immun. 2014;82:2400–2404. doi: 10.1128/IAI.01599-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Domingo-Gonzalez R., Prince O., Cooper A., Khader S.A. Cytokines and chemokines in Mycobacterium tuberculosis infection. Microbiol. Spectr. 2016:4. doi: 10.1128/microbiolspe. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schwander S., Dheda K. Human lung immunity against mycobacterium tuberculosis: Insights into pathogenesis and protection. Am. J. Respir. Crit. Care Med. 2011;183:696–707. doi: 10.1164/rccm.201006-0963PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zuniga J., Torres-Garcia D., Santos-Mendoza T., Rodriguez-Reyna T.S., Granados J., Yunis E.J. Cellular and humoral mechanisms involved in the control of tuberculosis. Clin. Dev. Immunol. 2012:193923. doi: 10.1155/2012/193923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Greveson K., Goodhand J., Capocci S., Woodward S., Murray C., Cropley I., Hamilton M., Lipman M. Yield and cost effectiveness of mycobacterial infection detection using a simple IGRA-based protocol in uk subjects with inflammatory bowel disease suitable for anti-TNFalpha therapy. J. Crohns Colitis. 2013;7:412–418. doi: 10.1016/j.crohns.2012.08.010. [DOI] [PubMed] [Google Scholar]
- 46.Weiner J., 3rd, Kaufmann S.H. Recent advances towards tuberculosis control: Vaccines and biomarkers. J. Intern. Med. 2014;275:467–480. doi: 10.1111/joim.12212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jurado J.O., Pasquinelli V., Alvarez I.B., Pena D., Rovetta A.I., Tateosian N.L., Romeo H.E., Musella R.M., Palmero D., Chuluyan H.E., et al. IL-17 and ifn-gamma expression in lymphocytes from patients with active tuberculosis correlates with the severity of the disease. J. Leukoc. Biol. 2012;91:991–1002. doi: 10.1189/jlb.1211619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liang S.C., Tan X.Y., Luxenberg D.P., Karim R., Dunussi-Joannopoulos K., Collins M., Fouser L.A. Interleukin (IL)-22 and IL-17 are coexpressed by Th17 cells and cooperatively enhance expression of antimicrobial peptides. J. Exp. Med. 2006;203:2271–2279. doi: 10.1084/jem.20061308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu L., Fu R., Yuan X., Shi C., Wang S., Lu X., Ma Z., Zhang X., Qin W., Fan X. Differential immune responses and protective effects in avirulent mycobacterial strains vaccinated BALB/c mice. Curr. Microbiol. 2015;71:129–135. doi: 10.1007/s00284-015-0837-3. [DOI] [PubMed] [Google Scholar]
- 50.Silva D., Silva M.V.D., Barros C.C.O., Alexandre P.B.D., Timoteo R.P., Catarino J.S., Sales-Campos H., Machado J.R., Rodrigues D.B.R., Oliveira C.J., et al. TNF-alpha blockade impairs in vitro tuberculous granuloma formation and down modulate Th1, Th17 and Treg cytokines. PLoS ONE. 2018;13:e0194430. doi: 10.1371/journal.pone.0194430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dorhoi A., Reece S.T., Kaufmann S.H. For better or for worse: The immune response against Mycobacterium tuberculosis balances pathology and protection. Immunol. Rev. 2011;240:235–251. doi: 10.1111/j.1600-065X.2010.00994.x. [DOI] [PubMed] [Google Scholar]
- 52.Kaufmann S.H., Dorhoi A. Inflammation in tuberculosis: Interactions, imbalances and interventions. Curr. Opin. Immunol. 2013;25:441–449. doi: 10.1016/j.coi.2013.05.005. [DOI] [PubMed] [Google Scholar]
- 53.Warner D.F., Koch A., Mizrahi V. Diversity and disease pathogenesis in mycobacterium tuberculosis. Trends Microbiol. 2015;23:14–21. doi: 10.1016/j.tim.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 54.Kalscheuer R., Weinrick B., Veeraraghavan U., Besra G.S., Jacobs W.R., Jr. Trehalose-recycling abc transporter LpqY-SugA-SugB-SugC is essential for virulence of mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA. 2010;107:21761–21766. doi: 10.1073/pnas.1014642108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Onwueme K.C., Vos C.J., Zurita J., Ferreras J.A., Quadri L.E. The dimycocerosate ester polyketide virulence factors of mycobacteria. Prog. Lipid Res. 2005;44:259–302. doi: 10.1016/j.plipres.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 56.Hotter G.S., Wards B.J., Mouat P., Besra G.S., Gomes J., Singh M., Bassett S., Kawakami P., Wheeler P.R., de Lisle G.W., et al. Transposon mutagenesis of MB0100 at the ppe1-nrp locus in Mycobacterium bovis disrupts phthiocerol dimycocerosate (PDIM) and glycosylphenol-PDIM biosynthesis, producing an avirulent strain with vaccine properties at least equal to those of M. Bovis BCG. J. Bacteriol. 2005;187:2267–2277. doi: 10.1128/JB.187.7.2267-2277.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Marjanovic O., Miyata T., Goodridge A., Kendall L.V., Riley L.W. Mce2 operon mutant strain of Mycobacterium tuberculosis is attenuated in C57BL/6 mice. Tuberculosis. 2010;90:50–56. doi: 10.1016/j.tube.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kirksey M.A., Tischler A.D., Simeone R., Hisert K.B., Uplekar S., Guilhot C., McKinney J.D. Spontaneous phthiocerol dimycocerosate-deficient variants of Mycobacterium tuberculosis are susceptible to gamma interferon-mediated immunity. Infect. Immun. 2011;79:2829–2838. doi: 10.1128/IAI.00097-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bottai D., Majlessi L., Simeone R., Frigui W., Laurent C., Lenormand P., Chen J., Rosenkrands I., Huerre M., Leclerc C., et al. ESAT-6 secretion-independent impact of ESX-1 genes espf and espg1 on virulence of mycobacterium tuberculosis. J. Infect. Dis. 2011;203:1155–1164. doi: 10.1093/infdis/jiq089. [DOI] [PubMed] [Google Scholar]
- 60.Cheng G., Xu D., Wang J., Liu C., Zhou Y., Cui Y., Liu H., Wan K., Zhou X. Isolation and identification of multiple drug resistant nontuberculous mycobacteria from organs of cattle produced typical granuloma lesions. Microb. Pathog. 2017;107:313–316. doi: 10.1016/j.micpath.2017.03.047. [DOI] [PubMed] [Google Scholar]
- 61.Chen Y., Chao Y., Deng Q., Liu T., Xiang J., Chen J., Zhou J., Zhan Z., Kuang Y., Cai H., et al. Potential challenges to the stop tb plan for humans in china; cattle maintain M. Bovis and M. Tuberculosis. Tuberculosis. 2009;89:95–100. doi: 10.1016/j.tube.2008.07.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.