Abstract
In response to salinity and various other environmental stresses, plants accumulate reactive oxygen species (ROS). The ROS produced at very early stages of the stress response act as signaling molecules activating defense mechanisms, whereas those produced at later stages in an uncontrolled way are detrimental to plant cells by damaging lipids, DNA, and proteins. Multiple systems are involved in ROS generation and also in ROS scavenging. Their level and activity are tightly controlled to ensure ROS homeostasis and protect the plant against the negative effects of the environment. The signaling pathways responsible for maintaining ROS homeostasis in abiotic stress conditions remain largely unknown. Here, we show that in Arabidopsis thaliana, two abscisic acid- (ABA)-non-activated SNF1-releted protein kinases 2 (SnRK2) kinases, SnRK2.4 and SnRK2.10, are involved in the regulation of ROS homeostasis in response to salinity. They regulate the expression of several genes responsible for ROS generation at early stages of the stress response as well as those responsible for their removal. Moreover, the SnRK2.4 regulate catalase levels and its activity and the level of ascorbate in seedlings exposed to salt stress.
Keywords: antioxidant enzymes, Arabidopsis thaliana, ascorbate cycle, hydrogen peroxide, reactive oxygen species, salinity, SnRK2
1. Introduction
Plants growing in nature are exposed to ever changing environmental conditions. They experience various abiotic stresses, such as drought, temperature extremes, and salinity. Salinity and drought are among the most detrimental factors limiting plant growth and development. Salinity causes ion-related stress, limitations in nutrient uptake as well as osmotic stress.
A secondary effect of salt stress and several other stresses is the accumulation of reactive oxygen species (ROS) in plant cells. Various ROS, such as singlet oxygen (1O2), superoxide radical (O2−), hydroxyl radical (−OH), and hydrogen peroxide (H2O2), are produced at low levels in chloroplasts, mitochondria, peroxisomes, and the apoplast during plant growth in optimal conditions [1,2]. They are involved in the regulation of plant growth and development, acting as signaling molecules. Upon stress, however, ROS play a double role [1,3,4,5]. ROS production at a low level is needed at the first stages of the stress response for induction of the plant defense, e.g., activation of signaling cascades, expression of stress response genes encoding enzymes involved in the synthesis of osmoprotectants, and some enzymes responsible for ROS scavenging [6,7,8]. At the later stages, ROS that accumulate in a non-controlled way have a widespread toxic effect, causing peroxidation of lipids, and damaging proteins and DNA, eventually leading to cell death. In response to stress, ROS are produced by diverse enzymes, e.g., NADPH oxidases, glycolate oxidases, oxalate oxidase, xanthine oxidase, and some peroxidases [9,10]. In Arabidopsis thaliana subjected to salinity stress, mainly two NADPH oxidases, respiratory burst oxidases, AtRbohD and AtRbohF, are involved in ROS production. They generate O2− free radicals in the apoplastic space by transferring electrons from NADPH to O2. Then, the O2− is dismutated to H2O2 by superoxide dismutase (SOD) and the H2O2 molecules diffuse to adjacent cells, where they can play a role of signaling molecules, inducing plant defense, or they cause oxidative stress and cell damage. To achieve ROS homeostasis, which is required for efficient defense against the negative effects of harmful environmental conditions, plants have evolved several systems for ROS removal, both enzymatic, such as catalases (CATs), SODs, and various peroxidases (PRXs), and non-enzymatic, i.e., the ascorbate-glutathione cycle [9,11,12,13,14,15,16]. The enzymes involved in the ROS production and removal are encoded by multiple genes and are strictly regulated in response to stress, at the transcriptional, protein, and activity levels.
There are several reports showing that kinases from the SNF1-releted protein kinases 2 (SnRK2s) family are major regulators of the plant response to osmotic stress (drought, salinity). SnRK2s are plant-specific kinases activated in response to osmotic stress and some of them additionally in response to abscisic acid (ABA). They have been found in every plant species analyzed [17]. The SnRK2s are classified into three groups based on their phylogeny. The classification correlates well with their response to ABA: Group 1 comprises ABA-non-activated kinases; in group 2, are kinases weakly activated or non-activated by ABA (depending on plant species); and group 3 contains kinases strongly activated by ABA [18,19]. SnRK2s play a crucial role in the induction of defense mechanisms against drought [20,21] and salinity [22,23,24] via ABA-dependent and ABA-independent pathways. So far, the role of ABA-dependent SnRK2s (SnRK2.2, SnRK2.3, and SnRK2.6) has been mostly studied and found to be crucial for ABA signaling [25,26]. In response to drought, they regulate stomatal closure in an ABA-dependent manner by phosphorylating several ion channels [27,28,29], expression of stress-response genes by phosphorylating transcription factors activated in response to ABA [30,31,32], and the activity of aquaporins [33]. SnRK2.6 is involved in the ABA-dependent ROS production indispensable for stomatal closure [34], possibly by phosphorylating the NADPH oxidase, RbohF [35,36].
Much less is known about the involvement of SnRK2s in response to salinity, even though these kinases are known to be strongly activated in response to this stress. It has been shown that ABA-non-responsive kinases (belonging to group 1) are activated rapidly and transiently in response to salt stress [23,37,38]. McLoughlin et al. [23] showed that two kinases from group 1, SnRK2.4 and SnRK2.10, are fully active within seconds in roots of Arabidopsis plants after treatment with NaCl. Both kinases were found to be required for plant tolerance to salinity stress by regulating root growth and architecture [23]. Moreover, it has been shown that SnRK2s from group 1 influence the plant tolerance to salt stress via regulation of mRNA decay. They phosphorylate VARICOSE (VCS), a protein regulating mRNA decapping [24]. Very recently, using the phosphoproteomic approach, several potential ABA-non-activated SnRK2s’ targets that phosphorylated in response to salinity have been found [39]. Among them there were several proteins, e.g., RNA- and DNA- binding proteins, protein kinases, phosphatases, and dehydrins, Early Responsive to Dehydration 10 (ERD10) and ERD14, whose phosphorylation likely affects the plant tolerance to salt stress. It has also been suggested that kinases from this group could be involved in the regulation of tolerance to salt stress via regulation of oxidative stress generated in response to salinity. Diédhiou et al. [22] showed that transgenic rice overexpressing Stress-Activated Protein Kinase 4 (SAPK4), the rice ABA-non-activated SnRK2, exhibited improved tolerance to salt stress. Their results indicated that SAPK4 regulates Na+ and Cl− accumulation and ROS homeostasis; the enhanced level of the kinase caused up-regulation of the CatA gene encoding catalase. Additionally, it has been shown that SnRK2.4 positively regulates the accumulation of ROS in response to stress induced by cadmium ions [40].
These data suggest that at least some members of the group 1 of the SnRK2 family are likely to be involved in the regulation of plant tolerance to osmotic stress by controlling the ROS level. The aim of the present study was to establish the role of SnRK2.4 and SnRK2.10 in the regulation of the ROS homeostasis in response to salt stress in Arabidopsis thaliana.
2. Results
2.1. SnRK2.4 and SnRK2.10 Kinases Are Involved in H2O2 Accumulation in Response to Salt Stress
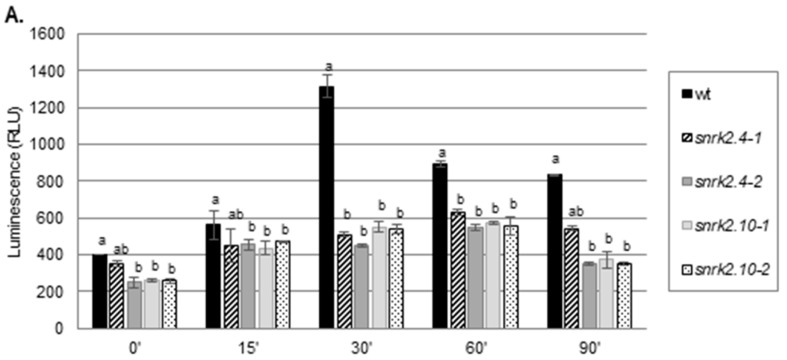
To check whether SnRK2.4 and/or SnRK2.10 are involved in ROS accumulation in the early response to salt stress, we compared the accumulation of H2O2 in leaves of four-week-old plants of the snrk2.4 and snrk2.10 knockout mutants and wild type Col-0 plants exposed to 150 mM NaCl for various time using a luminol-based assay. The mutant lines accumulated significantly less H2O2 than the wild type (wt) plants did in response to salinity (Figure 1A). The maximal level of H2O2 in Col-0 leaves was observed 30 min after the stressor application (over a three-fold increase in respect to the control level), whereas in the snrk2.4 and snrk2.10 mutants, the maximal H2O2 accumulation was only two-fold and occurred at 60 min. Notably, the level of H2O2 in control conditions was by ca. 30% lower in three out of four mutants tested relative to wt. The lower initial H2O2 content combined with the smaller increase resulted in the mutant plants having less than half of the H2O2 level found in the wt at the peak of the response to salt stress. Since the two independent lines of both mutants showed similar behavior for further studies, we decided to use only one line of each mutant, snrk2.4-1 and snrk2.10-1 (later referred to as snrk2.4 and snrk2.10 mutants), previously well characterized [23,39,40]. To verify the observed differences between the wt plants and the snrk2 mutants in ROS accumulation in response to salt stress, we monitored their level in Arabidopsis roots using the fluorescent dye, dichlorofluorescin diacetate (H2DCFDA). We analyzed the accumulation of ROS in roots of five-day-old seedlings of wt plants, and the snrk2.4 and snrk2.10 mutants (Figure 1B,C) exposed to 250 mM NaCl for 15 min. Similarly to what was observed for Arabidopsis leaves exposed to the salt treatment, in roots of the snrk2 mutant lines, the basal level of ROS was lower than in the roots of the wt plants and the ROS accumulation after the stress application was lower in those mutants in comparison with the one observed for wt seedlings. Thus, the obtained results strongly suggested the role of SnRK2.4 and SnRK2.10 in the regulation of ROS accumulation at the early stages of plant response to salinity.
Figure 1.
SnRK2.4 and SnRK2.10 affect ROS level in plants subjected to salt stress. (A). Leaves of wt plants and snrk2.4 and snrk2.10 mutant lines were subjected to 150 mM NaCl for the indicated time and H2O2 was determined using a luminol-based assay. Letters represent statistical differences in respect to the wt plants where a means no significant difference, and b means a significant difference [one way analysis of variance (ANOVA). Error bars represent standard deviation (SD). Three independent biological replicates, each with four samples per data point were performed. Results of all combined experiments are shown. B and C. Roots of five-day-old Arabidopsis seedlings (wt plants and snrk2.4 and snrk2.10 mutant lines) were stained with propidium iodide (PI; 20 µg/mL) and 2′,7′-Dichlorofluorescin diacetate (H2DCFDA; 30 µg/mL) and then treated for 15 min with 250 mM NaCl in ½ MS (+) or ½ MS only (−). (B) The production of ROS was monitored by imaging of H2DCFDA fluorescence in the roots using confocal microscopy; BF – bright field image, scale bars =50 µm (C) Fluorescence intensity of H2DCFDA was calculated from well-defined region of interest (4000 µm2) in the root meristematic zone on each single confocal section; stars represent statistically significant differences in respect to the wt plants (Mann-Whitney U test) where *** p < 0.0001; results represent data collected from at least 30 seedlings/line/conditions where each dot represents the sample value and a dash represents the median of measurements.
2.2. SnRK2.4 and SnRK2.10 Regulate Expression of Genes Involved in ROS Generation in Response to Salinity
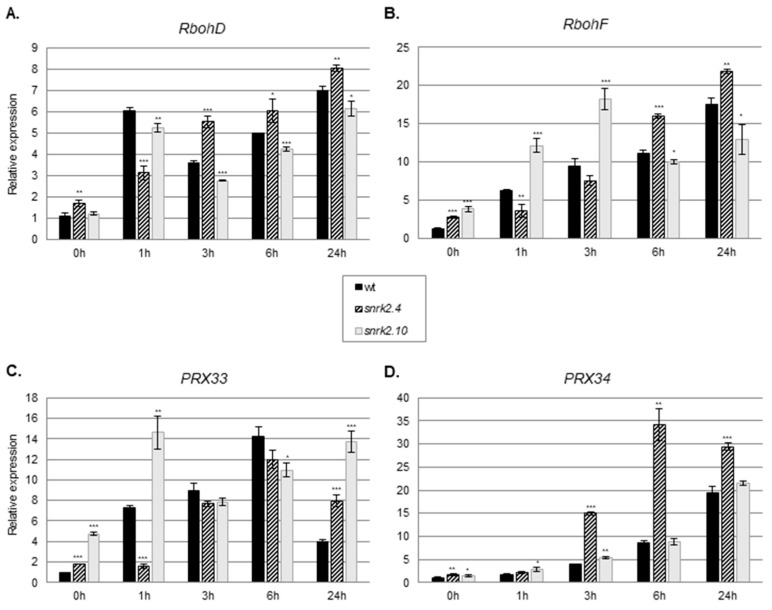
To determine the mechanism by which SnRK2.4 and SnRK2.10 affect the H2O2 homeostasis in salt-stressed plants, we investigated their impact on the enzymes involved in ROS production and scavenging. Since mitogen-activated protein kinase (MAPK, MPK) cascades regulate the ROS homeostasis by controlling the expression of genes encoding enzymes involved in ROS production [41,42], scavenging, as well as genes involved in ROS signaling [43], we studied the expression of several genes playing a role in the regulation of ROS levels. As the first approach, we analyzed the expression of genes whose products are responsible for ROS generation. Several cellular ROS generating enzymes, especially NADPH oxidases and apoplastic peroxidases, are involved in the plant response to environmental stresses [12]. Since, in the Arabidopsis, in response to salinity ROS, are generated mainly by the RbohD and RbohF oxidases [8,44,45], we analyzed transcript levels of RbohD and RbohF. For the studies, we used two-week-old Arabidopsis seedlings of the snrk2.4 and snrk2.10 mutant lines and wt plants treated with 150 mM NaCl up to 24 h. At the first stage of the response to salt stress (1 h), the transcript levels of RbohD and RbohF were significantly lower in the snrk2.4 mutant than in the wt plants (Figure 2A,B) in agreement with the lower level of H2O2 found in this mutant compared to the wt. At the later time points, this difference was reversed and the expression of RbohD and RbohF became higher in the mutant than in the wt plants. Notably, the two genes assayed showed markedly different kinetics and extents of activation during the salt treatment. The expression pattern of RbohD and RbohF in the snrk2.10 mutant in response to the stress differed significantly from the one observed for the snrk2.4 mutant. RbohD expression was hardly affected by the mutation (it was lower by only 10–15% than in the wt plants throughout the experiment), while the expression of RbohF was enhanced markedly at the early stages (up to 3 h) of the response and then fell to slightly below that in the wt. This behavior was opposite to that shown by the snrk2.4 mutant.
Figure 2.
SnRK2.4 and SnRK2.10 affect the expression of genes involved in ROS homeostasis during response to salt stress. Expression (mRNA level) of (A). RbohD—respiratory burst oxidase homolog protein D; (B) RbohF—respiratory burst oxidase homolog protein F; (C) PRX33—peroxidase 33; and (D) PRX34—peroxidase 34 was determined by RT-qPCR in wt plants and snrk2 mutant lines subjected to treatment with 150 mM NaCl at times indicated (h); error bars represent SD; stars represent statistically significant differences in comparison with the wt plants (Student t-test) where * p < 0.05; ** p < 0.001; *** p < 0.0001. At least two independent biological replicates of the experiment were performed. Results of one representative experiment are shown.
Additionally, we analyzed the expression of genes encoding two apoplastic peroxidases (PRX), PRX33 and PRX34, known to play an important role in the oxidative burst in response to biotic stresses [46,47]. The expression of both PRXs was induced in the wt plants in response to salinity stress, indicating their involvement in an abiotic stress response as well (Figure 2C,D). Notably, their expression patterns were affected in a complex manner in both snrk2 mutants indicating an involvement of SnRK2.4 and SnRK2.10 in their regulation upon salt stress. As for the two Rboh genes described above, also here, the two mutant lines showed contrasting responses. In the snrk2.4 mutant, the PRX33 gene showed a delayed induction compared to the wt, and at 24 h, its transcript level was double that in the wt. The induction of the PRX34 gene was enhanced several fold at 3 h and 6 h and was only slightly higher at 24 h relative to the wt. In the snrk2.10 mutant, PRX33 showed about a two-fold higher expression level than in the wt at 1 h, nearly identical ones at 3 and 6 h, and again much higher (over three-fold) at 24 h. One should note that also in control conditions, the expression of PRX33 was markedly up-regulated in the snrk2.10 mutant. The SnRK2.10 mutation had a negligible effect on the expression of the other PRX gene studied, PRX34.
These results indicate that both kinases have an impact on the expression of RbohD/F and PRX33/34 upon salt stress and that their roles are markedly different—they cannot substitute each other in this respect.
2.3. SnRK2.4 and SnRK2.10 Are Involved in Regulation of ROS Scavenging in Response to Salt Stress
Plants have evolved several ROS scavenging pathways, both enzymatic and non-enzymatic. We analyzed the involvement of SnRK2.4 and SnRK2.10 in the regulation of some of them upon salinity stress by comparing the responses of two-week-old snrk2.4 and snrk2.10 mutants and wt seedlings exposed to 150 mM NaCl.
2.3.1. SnRK2s Affect CAT1 Gene Expression, Catalase Level, and Activity
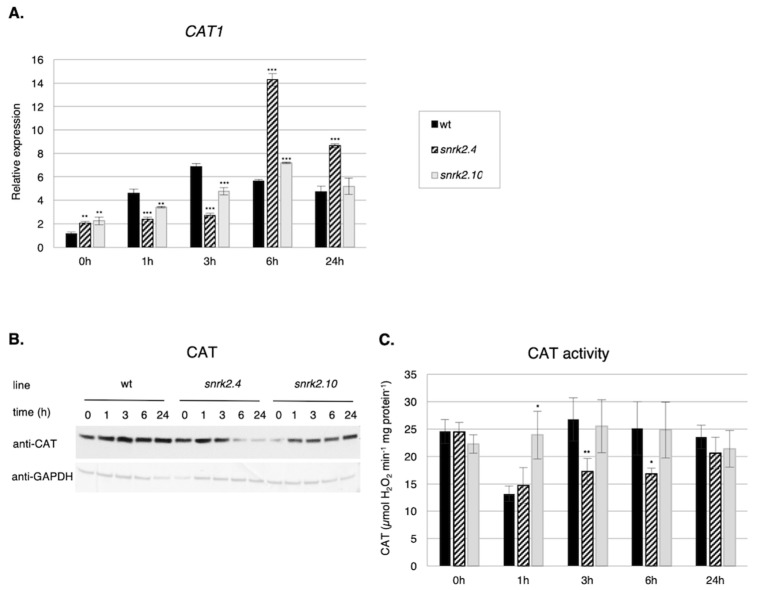
Among the most prominent ROS scavenging enzymes are catalases. The Arabidopsis genome encodes three catalases—CAT1, CAT2, and CAT3. Expression of all of them is induced by salinity, however, the strongest changes were observed for the CAT1 gene [48], therefore, we focused our studies on this gene. Our analysis revealed significant differences in the pattern of CAT1 expression in response to salt stress between the snrk2.4 and snrk2.10 seedlings and the wt ones (Figure 3A).
Figure 3.
SnRK2.4 and SnRK2.10 modulate catalase (CAT) on multiple levels during response to salt stress. Wild type and snrk2 mutants’ seedlings were subjected to treatment with 150 mM NaCl for times indicated. CAT1 expression was determined by RT-qPCR (A), total catalase protein was determined by immunoblot analysis (B), and total catalase activity assay was performed (C); error bars represent SD; stars represent statistically significant differences in comparison with the wt plants (Student t-test) where * p < 0.05; ** p < 0.001; *** p < 0.0001. After exposure, membranes were stripped and reused for glyceraldehyde 3-phosphate dehydrogenase (GAPDH) detection as a loading control. At least two independent biological replicates of the experiment were performed, each with four samples per data point. Results of one representative experiment are shown.
At the first stages of the response (up to 3 h), the CAT1 transcript level was significantly lower in both mutants than in the wt, whereas at the later stages, the reverse was true, especially in snrk2.4. This suggests that at first, the CAT1 expression is positively regulated by SnRK2.4 and SnRK2.10, whereas at the later stages of the stress response, the SnRK2s, especially SnRK2.4, exert an inhibitory action. Notably, the CAT1 transcript level was up-regulated ca. two-fold in the both mutants in control conditions. In that, the effect of a lack of either SnRK2s resembled the situation observed earlier for PRX33 and RbohD/RbohF genes. Unexpectedly, the differences in CAT1 transcript levels were not reflected by the amount of catalase protein (Figure 3B) or activity (Figure 3C). The catalase protein level was in fact lower in the snrk2 mutants exposed to salinity stress than in wt plants (Figure 3B); the lowest level was observed in the snrk2.4 mutant after salt treatment. These data apparently indicate discrepancies between the transcript and protein levels. However, the immunoblot analysis was performed using antibodies recognizing all three isoforms of catalase—CAT1, CAT2, and CAT3. It should be mentioned that in Arabidopsis rosettes, CAT2 and CAT3 transcripts are much more abundant than that for CAT1 [49,50]. Possibly the same is true for the corresponding proteins, therefore, the changes in the amount of the least abundant CAT1 were likely obscured by changes in the other two catalase isoenzymes. The latter, however, suggests that CAT2 and CAT3 could be also under the control of SnRK2s.
The catalase activity, reflecting the combined activity of the three isoenzymes, showed a different pattern for each line studied (Figure 3C); two mutants differed substantially from each other and also from the wt. Strikingly, at 1 h of salt treatment, there was a substantial drop of the catalase activity in the snrk2.4 and the wt, whereas in the snrk2.10 mutant, the activity basically did not change.
Taken together, our results show that during the plant response to salinity, SnRK2.4 and SnRK2.10 (especially SnRK2.4) regulate catalase at various levels, including gene expression, catalase protein level, and probably also its enzymatic activity.
2.3.2. SnRK2.4 and SnRK2.10 Regulate the Ascorbate Cycle
Ascorbate is a major antioxidant in plants. To check whether SnRK2.4 and SnRK2.10 play a role in the regulation of the ascorbate cycle in plants subjected to salinity, we compared the expression of genes and their protein products involved in the ascorbate cycle (ascorbate peroxidases, APXs, and dehydroascorbate reductase 1, DHAR1) as well as the APX activity in the snrk2.4 and snrk2.10 mutants and wt seedlings exposed to salt stress (Figure 4A–D). Expression of all the genes encoding cytoplasmic APXs (APX1, APX2, and APX6) and DHAR1 was highly induced in response to salinity and that response was markedly and in a complex manner affected in both snrk2 mutants. Notably, in control conditions, their expression was significantly higher in both mutants than in the wt.
Figure 4.
SnRK2.4 and SnRK2.10 regulate enzymes of the ascorbate cycle during response to salt stress. Wild type and snrk2 mutant seedlings were subjected to treatment with 150 mM NaCl for times indicated. Expression of (A) APX1—Ascorbate Peroxidase 1, (B) APX2—Ascorbate Peroxidase 2, (C) APX6—Ascorbate Peroxidase 6, and (D) DHAR1—Dehydroascorbate Reductase 1 was monitored by RT-qPCR; error bars represent SD; stars represent statistically significant differences in comparison with the wt plants (Student t-test) where * p < 0.05; ** p < 0.001; *** p < 0.0001. Total protein level of (E) APX and (F) DHAR1 was monitored with immunoblot analysis; after exposure, membranes were stripped and reused for GAPDH detection as a loading control. At least two independent biological replicates of the experiment were performed. Results of one representative experiment are shown.
APX1 expression was induced rapidly in response to salinity in wt seedlings, reached a maximum at 1 h and declined below control level at 6 h and 24 h, whereas in the snrk2.4 mutant, it increased gradually between 3 h and 24 h; in snrk2.10, the pattern was similar except for the presence of an early (1 h) peak of expression.
A different situation was observed for the APX2 gene. It underwent progressive very strong induction in wt plants (1000-fold increase at 24 h of salt treatment), and a similarly progressive, but much less pronounced, induction in the mutants.
APX6 expression was rapidly induced up to six-fold in response to salinity in the wt to reach the maximum at 3 h followed by a slight drop to four-times value at time 0. In contrast, in the snrk2.4 and snrk2.10 mutants, where the APX6 expression was only slightly (snrk2.10) or not at all (snrk2.4) induced upon salt application, and dropped below the initial level at 24 h in both mutants.
Immunoblot analysis (with antibodies recognizing all three APXs) showed a slight decrease of the APX amount during salt stress in the wt and substantial accumulation at the later stages (6 h and 24 h) in the snrk2 mutants, especially in the snrk2.4 (Figure 4E). This accumulation roughly parallels the transcript pattern of APX1 and APX2 and suggests that the SnRK2.4 and SnRK2.10 kinases are negative regulators of APX accumulation at the later, but not the early stages of the plant response to salinity.
DHAR1 expression was highly and progressively induced in wt seedlings exposed to salt stress, and to a lesser extent also in the mutants. Notably, in the latter, its level fell between 6 h and 24 h of salt stress. DHAR1 protein showed strong and rapid accumulation in the wt line in response to the stress, slightly lower in the snrk2.10 mutant, whereas in the snrk2.4 mutant, it was barely detectable in control conditions and then grew gradually until the end of the treatment (Figure 4F), but it was significantly lower than in the wt and the snrk2.10 mutant plants. These results suggest that the two SnRK2s, especially SnRK2.4, positively regulate DHAR1 accumulation.
The above results indicate that SnRK2.4, and to lesser extent also SnRK2.10, regulate the level of the enzymes of the ascorbate cycle during the plant response to salinity.
Next, we compared the total ascorbate (Asc) content, APX activity, and the ascorbate/dehydroascorbate (Asc/DHAsc) ratio in the snrk2.4 and snrk2.10 mutants and wt seedlings subjected to salinity (Figure 5).
Figure 5.
SnRK2.4 and SnRK2.10 regulate ascorbate cycle during response to salt stress. Wild type and snrk2 mutant seedlings were subjected to treatment with 150 mM NaCl for times indicated and (A) ascorbate content, (B) ascorbate peroxidase (APX) activity, and (C) ascorbate redox status were monitored. Asc—ascorbate, DHAsc—dehydroascorbate; error bars represent SD; stars represent statistically significant differences from wt plants (Student t-test for Asc and APX activity, Chi-square test for Asc/DHAsc ratio) where * p < 0.05; ** p < 0.001; *** p < 0.0001. At least two independent biological replicates of the experiment were performed, each with four samples per data point. Results of one representative experiment are shown.
The total Asc content in wt plants was slightly increased in response to salt stress (from 0.9 µmol g−1 FW in control conditions to 1.3 µmol g−1 FW after 6 h of the treatment) (Figure 5A). A similar pattern was observed for the snrk2.10 mutant, but with stronger Asc accumulation after 1 h of treatment (from 1 µmol g−1 FW in control conditions to 1.55 µmol g−1 FW after 1 h and 1.2 µmol g−1 FW after 3 h and 6 h of salt stress), while in the snrk2.4 mutant, the Asc content only slightly increased in response to salt stress (0.83–1.0 µmol g−1 FW).
The ratio between oxidized (DHAsc) and the reduced form of Asc was virtually identical in the wt and the snrk2.10 mutant and did not change upon salt stress (Figure 5C). In the snrk2.4 mutant line, the fraction of the reduced form of Asc was lower than that in the other lines at all time points of the treatment and additionally showed a significant decrease after 6 h and 24 h of the stress (from 69% in control to 53% after 24 h), suggesting an increased APX activity. Indeed, measurements of the APX activity confirmed this conjecture, albeit the activity pattern did not exactly match the DHAsc/Asc pattern (Figure 5B). Generally, the APX activity reflected the APX protein level (compare Figure 4E).
3. Discussion
Salinity imposes ion and osmotic stresses on plant cells and leads to accumulation of ROS. The understanding of the signaling pathways controlling redox homeostasis during salt stress remains limited. A majority of the data concerning this subject pertains to the plant responses to biotic stresses. Several kinases regulating ROS production in response to pathogen infection have been identified. Some of them act as positive regulators of ROS generation via direct or indirect regulation of RbohD and/or RbohF activity or of their transcript accumulation (e.g., MPK3/6, Flagellin-sensitive 2, EF-Tu receptor, Brassinosteroid insensitive 1 associated receptor kinase 1, Botrytis-induced kinase 1, CPK5), some positively regulate PRX activity (like ZmMPK7), whereas others inhibit ROS accumulation (e.g., MPK4, AtCPK28) [51,52,53,54,55,56,57,58,59,60]. Much less is known regarding the protein kinases involved in ROS production or scavenging in response to abiotic stresses. In response to salinity, ROS are generated in different cellular compartments: Chloroplasts (by the photosynthetic electron transport), mitochondria (by the respiratory electron transport), peroxisomes, and, in the apoplast, by the action of oxidases present in the plasma membrane. The ROS generated in the various cellular compartments cross talk with each other. Recently it has been shown that, similarly to animals, the ROS-induced ROS release (RIRR) process (e.g., ROS produced in one cellular organelle or compartment induce ROS production in another one) takes place also in plants [61]. An RIRR-generated ROS wave leads to ROS amplification and signal transduction to neighboring compartments and cells. Therefore, enzymes localized to the cytoplasm or nucleus (as is in the case of the SnRK2s studied here) can indirectly affect the ROS level also in other subcellular compartments.
3.1. Role of SnRK2.4/SnRK2.10 in ROS Accumulation in Response to Salinity Stress
Previously published results have indicated that ABA-non-responsive SnRK2s (from group 1) regulate ROS levels in response to abiotic stresses. Diédhiou et al. [22] showed that an ABA-non-activated SnRK2, SAPK4, regulates ROS homeostasis in rice in response to salt stress, and Kulik et al. [40] that SnRK2.4 and SnRK2.10 are involved in positive regulation of H2O2 accumulation in Arabidopsis roots in the early response to heavy metal stress.
Surprisingly, studies performed on Arabidopsis, the snrk2.2/3/6 triple, the septuple, and the decuple mutants defective in several SnRK2s did not show any correlation between SnRK2s level and H2O2 accumulation in response to osmotic stress (polyethylene glycol (PEG) treatment of Arabidopsis seedlings) [62]. However, first, the measurement was done 12 h after PEG addition. Moreover, it seems likely that individual SnRK2s might differently affect ROS production and scavenging and their defects in the multiple snrk2 mutants could effectively cancel out, resulting in no net change in H2O2 accumulation. Finally, the role of various SnRK2s might be different in response to ABA, PEG, and salinity.
Our present studies focused on the role of two ABA-non-responsive SnRK2 kinases, SnRK2.4 and SnRK2.10, in the regulation of ROS homeostasis in Arabidopsis exposed to salinity stress. Since these kinases localize to the cytoplasm (SnRK2.10) and the cytoplasm and nucleus (SnRK2.4), we studied those enzymes involved in ROS homeostasis that in principle could be regulated by cytoplasmic and nuclear kinases: Directly by phosphorylation or indirectly at the transcriptional level. In respect to ROS production, we studied two plasma membrane NADPH oxidases, RbohD and RbohF, and two apoplastic peroxidases, PRX33 and PRX34. It has been established that RbohD and RbohF are regulated at the activity and expression levels. Their activity is tightly controlled by phosphorylation and by Ca2+ binding. Several kinases capable of phosphorylating RbohD/F have been identified, but the role of these phosphorylations is not fully clear. Drerup et al. [63] showed that Calcineurin B-like protein 1/9 (CBL1/9)-CIPK26 (from CBL-interacting protein kinase 26) complexes phosphorylate and activate RbohF. Similarly, Han et al. [36] presented that CIPK11 and CIPK26 phosphorylate RbohF, constituting alternative paths for RbohF activation, whereas Kimura et al. [64] suggest that the binding of CIPK26 to RbohF decreases ROS production. It has also been shown that RbohD/F are regulated also by Ca2+-independent kinases, like MPK8, which inhibits RbohD activity in response to wounding [65]. Some data indicate an involvement of SnRK2s in the regulation of NADPH oxidase activity. OST1/SnRK2.6, an ABA-dependent kinase, regulates the ROS level required for the stomatal closure [34]. Sirichandra et al. [35] showed that OST1 phosphorylates RbohF in vitro and suggested that this phosphorylation plays a role in its activation and possibly in the regulation of stomatal movement in response to ABA. Recently, Han et al. [36] showed that OST1 together with CIPKs is involved in RbohF activation.
Our present results revealed that SnRK2.4 and SnRK2.10 positively regulate the ROS production at the early stages of the response to salinity; we observed significantly lower H2O2 levels in the snrk2.4 and snrk2.10 mutants than in wt plants salt-treated for up to 90 min. These results suggest that SnRK2.4 and SnRK2.10 might phosphorylate RbohD and/or RbohF in response to salinity and thereby regulate the ROS level. The phosphorylation of RbohD/F by SnRK2.4/10 is highly plausible since the substrate specificities of SnRK2s, CIPKs, and calcium-dependent protein kinases (CDPKs) are quite similar; all of them belong to the CDPK-SnRK superfamily [66]. The SnRK2.4 and SnRK210 kinases are activated rapidly in response to salinity, within seconds after the stressor application. Therefore, it is likely that they are involved in the earliest events of the response to salt stress, i.e., activation of the Rbohs and production of ROS responsible for triggering the defense mechanisms. However, this hypothesis needs further studies.
Our results pointed out to a role of SnRK2.4 and SnRK2.10 in the regulation of RbohD and RbohF expression. In Arabidopsis thaliana seedlings, the expression of RbohD and RbohF is induced in response to salinity [14,44,45,67]. At the first stages of the response (in our case, the treatment with NaCl for 1 h), the transcript levels of RbohD and RbohF were significantly lower in the snrk2.4 mutant than in wt plants, which is in agreement with the lower level of H2O2 found in the mutants. Later, during the salt treatment, this correlation was no longer sustained and the RbohD/F expression in the mutant became elevated above the level observed for the wt plants. The effect of disruption of the SnRK2.10 gene was more complex, as it has little effect on induction of RbohD expression, but actually enhanced that of RbohF.
Additionally, we analyzed the expression of genes encoding apoplastic peroxidases, PRX33 and PRX34, whose involvement in the response to salt stress has not been considered so far. Expression of both PRXs was induced in response to salinity stress, which indicates their role in the abiotic stress response, and this induction was apparently regulated by SnRK2s. Expression of PRX33 was significantly lower in the snrk2.4 mutant early in the response to salinity, but became highly elevated relative to the wt plants after prolonged salt treatment. For PRX34, the effect of the snrk2.4 mutation was visible only at later stages of the response and manifested as a several fold enhancement of the induction. This suggests an inhibitory role of SnRK2.4 on the salt-induced PRX34 expression.
As for the Rboh genes, also here, SnRK2.10 turned out to act differently to SnRK2.4. The snrk2.10 mutation had virtually no effect on PRX34 expression, but greatly stimulated the expression of PRX33 in control conditions and also at early and late response to salt stress.
These data indicate that even though in both the snrk2.4 and snrk2.10 mutants, the level of H2O2 produced early in response to salt stress is significantly lower than in wt plants, the mechanisms of the regulation of ROS accumulation by the two kinases seem to be different. It should be stressed here, that unlike SnRK2.4, SnRK2.10 does not localize to the nucleus, therefore, the different modes of regulation of gene expression by these kinases are not surprising (Figure 6).
Figure 6.
Possible roles of SnRK2.4 and SnRK2.10 in the regulation of the ROS homeostasis in Arabidopsis seedlings exposed to the salt stress. Proposed role of the SnRK2s in (A) early response and (B) late response to the salt stress. In response to salinity, SnRK2.4 along with SnRK2.10 regulate the ROS production/accumulation as well as ROS scavenging at the transcription as well as protein and/or activity levels. Detailed description in the text; dash lines—SnRK2.4 impact; dotted lines—SnRK2.10 impact; green question mark—probably indirect regulation; red question mark—plausible direct regulation by phosphorylation.
The expression of genes encoding ROS producing enzymes in the snrk2.4 and snrk2.10 (especially snrk2.4) is very different at early and late stages of the response. We propose that at the later stages the expression of genes studied might be regulated by other signaling pathways, for example, MAPK cascade(s), which are involved in controlling ROS homeostasis. Those pathways might be triggered to compensate for the low ROS level in the snrk2s mutants. In response to several stimuli MAPK cascade(s) control RbohD, RbohF, PRX33, and PRX34 expression [52,68]. MPK3/MPK6 phosphorylate and thus activate the ERF6 transcription factor, whose targets are RbohD and PRX33 in response to fungal pathogen [69,70,71]. Moreover, in Nicotiana benthamiana during the ETI (from effector-triggered immunity) and Elicitin-(INF1)-triggered PTI (from pattern-triggered immunity), salicylic acid induced protein kinase (SIPK, orthologue of Arabidopsis MPK6) phosphorylates four W-box binding transcription factors (WRKYs), which are responsible for the expression of RBOHB (ortholog of Arabidopsis RbohD), and positively regulates the RBOHB transcript level [72]. Since MPK6 and SIPK are activated in response to salinity and water deficits [37,73] and ERF6 is involved in response to the water limitation [74], it seems likely that the MPK6 pathway, and presumably some others, could overcompensate for the low expression of RbohD and PRX33 at early stages of the response in the snrk2.4 mutant.
3.2. Involvement of SnRK2.4/SnRK2.10 in ROS Removal under Salinity Stress Conditions
Data on the signaling pathways and protein kinases involved in the regulation of the antioxidant systems engaged in ROS scavenging in response to salinity or osmotic stress are scarce. It has been shown that GSK3 kinase (ASKα) regulates salt stress tolerance of Arabidopsis by phosphorylation and activation of glucose-6-phosphate dehydrogenase, an enzyme important for maintaining the cellular redox balance [75]. Zong et al. [76] have shown that ectopic expression of ZmMPK7 in Nicotiana tabaccum enhances peroxidase activity, which results in lower accumulation of H2O2 in response to osmotic stress. It has been reported that several protein kinases of MAPK cascades as well as CIPK are involved in the regulation of expression and/or activity of CAT1 [43,48,77].
We analyzed here the impact of SnRK2.4 and SnRK2.10 on several enzymes involved in ROS scavenging (CATs, APXs, and DHAR1). A comparison of the changes in the CAT1 transcript level in the snrk2.4, snrk2.10, and wt plants exposed to salinity indicated that SnRK2.4, and to a lesser extent also SnRK2.10, positively regulate the expression of CAT1 during the first stages of the stress response. However, similar to what was observed for RbohD, at the later stages of the response, the impact of the snrk2 mutations became just the opposite, which results in a higher CAT1 expression than in the wt plants. We conjecture that this effect was not due to a direct regulation of CAT1 expression by SnRK2.4/SnRK2.10, but rather because of reflected activation of some other signaling pathway(s) in order to compensate for the low CAT1 expression. One such pathway might be again the MPK6 cascade, known to mediate CAT1 expression and H2O2 production [77].
Besides the regulation of the CAT1 expression, SnRK2.4 and, less markedly, also SnRK2.10 positively regulated catalase protein accumulation and activity during salt stress, as they were both significantly lower in the snrk2 mutants exposed to the stress than in the wt. The discrepancy between the enhanced expression of CAT1 later in the response and the lower catalase protein level and activity indicates that in response to salinity, the SnRK2s affect not only CAT1, but most likely also the CAT2 and CAT3 levels, in opposing directions. Since CAT2 and CAT3 are more abundant than CAT1 in Arabidopsis seedlings, it is likely that the catalase activity, which we measured, represented mainly the activity of CAT2 and CAT3. It is plausible that SnRK2s regulate also the expression of CAT2 and/or CAT3 genes. We observed lower catalase activity in the snrk2.4 mutant exposed for up to 6 h to the salt stress. It is not clear whether SnRK2s modulate the enzyme(s) specific activity or only the catalases’ protein level or through the phosphorylation they impact the targeting of catalases into the peroxisomes (their final destination). Phosphorylation of catalases by SnRK2s is quite feasible, since it has been shown that salt overly sensitive 2 (SOS2), a kinase belonging to the SnRK3 subfamily, interacts with CAT2 and CAT3 and possibly influences H2O2 accumulation in response to salinity [78]. SOS2 localizes to the plasma membrane and cytoplasm and its substrate specificity is nearly the same as that of the SnRK2’s.
The available information on the regulation of the ascorbate cycle in response to abiotic stresses and the protein kinases is very limited. It has been suggested that in response to strong light, the SnRK2.6/OST1 kinase activates APX2 expression [79]. Pitzschke and Hirt [80] have speculated that MAPK cascades could be involved in ascorbate cycle regulation based on transcriptomic analysis performed on mapk mutants and wt plants. They revealed that several genes encoding enzymes involved in ascorbate biosynthesis and metabolism were differentially expressed compared to wt plants [43], but those results have not been confirmed so far.
Our data regarding the expression of genes encoding selected enzymes of the ascorbate cycle, APX (APX1, 2, 6) and DHAR1, indicate that SnRK2.4 and SnRK2.10 also modulate the ascorbate cycle. The two kinases had a significant impact on APXs’ expression upon salt stress, but the different APX genes were regulated differently. The effect of the snrk2 mutations on the APX1 expression profile was similar to those observed for RbohD or PRX33. At present, we do not know how the APX2 and APX6 up-regulation by SnRK2s affects the overall APX activity. The combined APX protein level in the mutants and wt plants exposed to the stress correlated well with the level of expression of APX1, but not of APX2 or APX6. These data indicate that most likely APX1 has the largest share in the overall cytoplasmic APX pool, and importantly, SnRK2.4 and SnRK2.10 play a role in its regulation. Furthermore, another enzyme of the ascorbate cycle that regenerates DHAsc to Asc, DHAR1, was strongly up-regulated at both the transcript and protein levels by SnRK2.4 and slightly less by SnRK2.10 in plants exposed to salinity.
The total Asc level and the Asc/DHAsc ratio were significantly lower in the snrk2.4 mutant in comparison with the wt plants in response to salinity, which suggests that SnRK2.4 kinase positively regulates Asc accumulation. In agreement with these data, the APX activity was higher in snrk2.4 than in the two other lines studied. Taken together, these data indicate that SnRK2.4 plays a substantial role in the regulation of the ascorbate cycle in response to salt stress, by direct or indirect regulation of APX and DHAR1 (Figure 6).
Discussing our results, one issue should be pointed out—the role of the circadian clock in the regulation of the redox homeostasis. Circadian clocks regulate the plant growth and development as well as responses to multiple environmental cues, both biotic as well as abiotic [81,82]. Numerous genes involved in the response to osmotic stress and ABA signaling have been identified as circadian clock-dependent, including SnRK2.6 and several genes encoding stress-responsive transcription factors (for review see [83]). It has been shown that the redox homeostasis (ROS production, scavenging, and expression of ROS-responsive genes) is tuned with diurnal and circadian rhythms, for example, the expression of CAT1 and CAT3 is highest at noon, whereas CAT2 is highest at dawn [84]. On the other hand, in the feedback response, ROS signals affect clock responses [84,85]. Since our results show that SnRK2.4 and SnRK2.10 regulate the ROS level, it is highly likely that the kinases have some impact on the circadian clock. We also conjecture that expression of SnRK2.4 and/or SnRK2.10 might be regulated by a circadian rhythm. It should be stressed at this point that the expression of some genes studied by us might be affected not only by the salt, but also, to some extent, by the diurnal/circadian rhythms.
It has been reported that the transcriptomic analysis using a circadian-guided network approach might be used for identification of the genes involved in the early sensing of mild drought [86], indicating again a close relation between the stress responses and the circadian clock. Salt stress and dehydration signaling pathways have several common elements, including SnRK2s. Importantly, dehydration accompanies salt stress and it has been shown that not only ABA-activated SnRK2s, but also SnRK2, which are not activated in response to ABA, e.g., SnRK2.10 are involved in the plant response to a water deficit [39]. Therefore, when analyzing the plant response to salinity stress, as well as all other stresses, one should be aware of circadian/diurnal rhythms, which play a role in tuning those responses [84,85,86].
Regulation of the plant response to salt stress by SnRK2s is complex, and our knowledge on this subject is very limited. To provide the full picture, presenting the role of SnRK2s in the salt stress response, additional extensive work is required, e.g., the elucidation of the interplay between SnRK2s, ROS, circadian clock, and various signaling pathways.
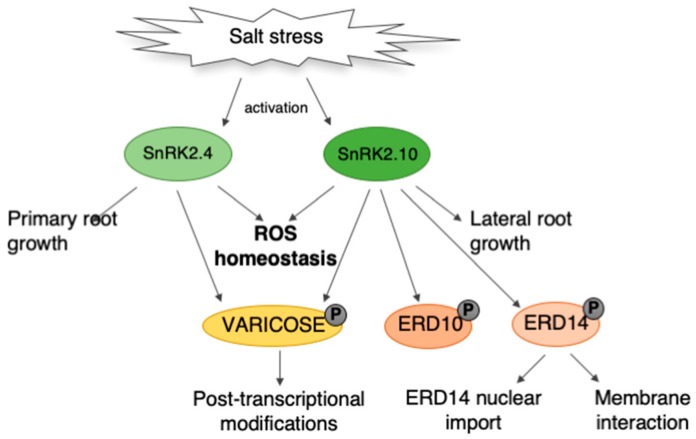
A model summarizing our knowledge on the involvement of SnRK2.4 and SnRK2.10 in the plant response to salt stress is presented in Figure 7. In response to salinity stress, the kinases regulate root growth and architecture [23], mRNA decay (by phosphorylation of VCS) [24], have an impact on dehydrin ERD14 localization and likely interactions with plant membranes [39], and on ROS homeostasis (the results described here).
Figure 7.
Schematic model illustrating the role of SnRK2.4 and SnRK2.10 in Arabidopsis’ response to salt stress. SnRK2.4 and SnRK2.10 modulate root growth under the salinity conditions. Moreover, in response to salt stress, the ABA-non-activated SnRK2s phosphorylate VARICOSE (VCS), a protein participating in mRNA decay, and two dehydrins, Early Responsive to Dehydration 10 (ERD10) and ERD14. Our results presented here revealed that SnRK2.4 and SnRK2.10 regulate the ROS homeostasis in the response to salinity.
In conclusion, our data described here show that SnRK2.4 along with SnRK2.10 positively regulate the first ROS wave that transduces the salt stress signal. The kinases regulate ROS accumulation as well as ROS scavenging, by modulating the catalase level and the ascorbate cycle (Figure 6). These results suggest that the two studied SnRK2s are involved in the fine tuning of the ROS level and thus contribute to the regulation of ROS homeostasis required for the plant acclimation to unfavorable environmental conditions.
4. Materials and Methods
4.1. Plant Material, Growth, and Treatment Conditions
The following Arabidopsis thaliana lines were used: Arabidopsis Col-0 ecotype (“wild type”; wt); homozygous T-DNA insertion lines snrk2.4-1 (SALK_080588), snrk2.4-2 (SALK_146522), and snrk2.10-1 (WiscDsLox233E9) kindly provided by Prof. C. Testerink (University of Amsterdam, The Netherlands), and snrk2.10-2 (SAIL_698_C05) from the Nottingham Arabidopsis Stock Center (NASC). Seedlings were grown in a sterile hydroponic culture as described Kulik et al. [40] for two weeks.
For luminol-based H2O2 determination, plants were grown for four weeks on Jiffy pods (Jiffy-7, Jiffy Group) in a growth chamber under 8 h of light /16 h dark conditions at 21 °C/18 °C.
For ROS production measurements with H2DCFDA, Arabidopsis seedling were grown on ½ MS plates supplemented with 0.8% agar for five days in a growth chamber under 8 h of light/16 h dark conditions at 21 °C/18 °C.
Two- or four-week-old plants were treated with 150 mM NaCl for the indicated time (as described in the results section; stress was applied 2 h after the light was turn on), harvested by sieving, and immediately frozen in liquid nitrogen. Plant material was kept at −80 °C until further analysis.
4.2. Determination of H2O2
Luminol-based assay for H2O2 was performed according to Rasul et al. [87]. Discs of 2-mm diameter were excised from leaves of four-week-old plants using a cork borer, from 5 leaves per sample per condition, and placed into assay vials with 200 μL of MQ water, sealed with parafilm, and incubated at RT overnight. Next, stress conditions were applied (either 150 mM NaCl or MQ water as a control) and 4 μL of luminol solution [3 mM luminol dissolved in dimethyl sulfoxide (DMSO); final concentration 60 μM] was added at appropriate time points. Vials were gently mixed and luminescence was measured using a luminometer for a total time of 120 s. The measurements were performed at selected time points up to 90 min post treatment. Statistical analysis was performed using one way analysis of variance (ANOVA).
ROS detection with H2DCFDA was performed as described previously by Kulik et al. [40] and Srivastava et al. [88] with minor modifications. Staining of five-day-old Arabidopsis seedlings roots with PI and H2DCFDA was performed before treatment with 250 mM NaCl in ½ MS or ½ MS only. Single confocal sections were collected with a 20× (NA 0.75) Plan Fluor multiimmersion objective mounted on an inverted epifluorescence TE 2000E microscope (Nikon, Tokyo, Japan) coupled with an EZ-C1 confocal laser-scanning head (Nikon). H2DCFDA fluorescence was excited with blue light at 488 nm emitted by a Sapphire 488 nm laser (Coherent, Santa Clara, CA, USA) and detected with a 515/30-nm band-pass-filter and rendered in false green, PI fluorescence was excited with green light at 543 nm emitted by a 1 mW He-Ne laser (Melles Griot, Carlsbad, CA, USA) and detected with a 610 nm long-pass filter and rendered in false magenta. All confocal parameters (laser power, gain, etc.) and conditions were the same during the experiment. EZ-C1 FreeViewer software was used to quantify the fluorescence intensity from the 4000 µm2 area of the root meristematic zone in each Arabidopsis seedling. Each experimental variant was repeated at least twice with a total of 30 single images collected. Statistical analysis was performed using the Mann-Whitney U test.
4.3. RNA Extraction and RT-qPCR Analysis
Total RNA was extracted with TRI Reagent® according to the manufacturer’s protocol (MRC). Approximately 150–200 mg of frozen ground plant material was used. DNA contamination was removed from the obtained RNA using a RapidOut DNA Removal kit (Thermo Scientific, Waltham, MA, USA). cDNA was synthesized from 4 μg of purified RNA using an Enhanced Avian HS RT-PCR Kit (Sigma-Aldrich, St. Louis, MO, USA) following the manufacturer’s protocol. RT-qPCR was performed on 50 ng of the cDNA using LightCycler® 480 SYBR Green I Master Mix (Roche, Basel, Switzerland) and a Roche LightCycler® 480 machine. Relative transcript levels were calculated according to Livak and Schmittgen [89] with UBQ10 (AT4G05320) and UBC (AT5G25760) as reference genes [90,91]. Statistical analysis was performed using Student t-test. All primers used in this study are listed in Table S1.
4.4. Protein Extraction and Immunoblot Analysis
Total proteins were extracted from frozen plant samples in two volumes of extraction buffer: 100 mM HEPES, pH7.5; 5 mM EDTA; 5 mM EGTA; 10 mM DTT; 1 mM Na3VO4; 10 mM NaF; 50 mM β-glycerophosphate; 10 mM pyridoxal 5-phosphate; 10% glycerol; and 1 × Complete protease inhibitors (EDTA-free, Roche) on a rotator for 30 min at 4 °C and then centrifuged at 12,000 rpm for 30 min at 4 °C. Protein concentration in the supernatant was measured using a Bradford Protein Assay. The extracts were used immediately or flash-frozen and kept at −80 °C for further analysis. The immunoblot blot analysis was based on a standard procedure described by Sambrook [92]. Protein samples (7–15 μg) were separated on 12% SDS-polyacrylamide gels and transferred to Immobilon®® P membrane by electroblotting in transfer buffer, TB (25 mM Tris base, 192 mM glycine), overnight at 18 V. Transferred proteins were visualized by staining the membranes with Ponceau S (2% Ponceau S in 3% trichloroacetic acid). Immunodetection with anti-APX rabbit IgG (AS08 368, Agrisera, Vännäs, Sweden), anti-CAT rabbit IgG (AS09 501, Agrisera), and anti-DHAR1 rabbit IgG (AS11 1746, Agrisera) was performed as described in the manufacturer’s protocols. Anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) rabbit IgG (raised against the CYDDIKAAIKEESEG peptide of GAPDH; BioGenes, Berlin, Germany) was used as described previously in Wawer et al. [93]. Secondary anti-rabbit antibodies (alkaline phosphatase (AP) conjugated—AS09 607, Agrisera; horseradish peroxidase (HRP) conjugated—AS09 602, Agrisera) were visualized using appropriate substrates—5-bromo-4-chloro-3-indolyl-phosphate/nitroblue tetrazolium (BCIP/NBT, Roche) for AP, and—ECL detection reagent (PierceTM ECL Western Blotting Substrate, Thermo Scientific) for HRP according to the manufacturer’s protocol. Membranes were reused for GAPDH protein detection used as a loading control for Western blots. Stripping of the membranes was performed according to Abcam online protocols.
For APX and CAT activity assays (see further), proteins were extracted from frozen plant samples (0.5 g FW) with 1 mL of ice-cold 50 mM sodium phosphate buffer, pH 7.5 or 100 mM potassium phosphate buffer, pH 7.0, respectively, containing 1 mM polyethylene glycol, 1 mM phenylmethylsulfonyl fluoride, 8% (w/v) polyvinylpolypyrolydone, and 0.01% (v/v) Triton X-100, according to Venisse et al. [94].
4.5. Determination of Ascorbate and Ascorbate/Dehydroascorbate Ratio
Ascorbate (Asc) and dehydroascorbate (DHAsc) was determined using a modified bipyridyl method described in detail by Polkowska-Kowalczyk et al. [95]. Statistical analysis was performed using the Student t-test and Chi-square test.
4.6. Determination of APX and CAT Activity
Ascorbate peroxidase (APX, EC 1.11.1.11) activity was assayed as described previously in Polkowska-Kowalczyk et al. [95]. Enzyme activity was expressed as µmol of oxidized ascorbate per min per mg of protein.
Catalase (CAT, EC 1.11.1.6) activity was assayed at 25 °C following the decomposition of H2O2 at 240 nm (extinction coefficient 0.036 mM−1 cm−1) according to a modified method of Aebi [96]. The reaction mixture contained 50 µL of plant extract in 1 mL 50 mM potassium phosphate buffer (pH 7.0) and 9.8 mM H2O2. Enzyme activity was expressed as µmol H2O2 decomposed per min per mg of protein.
Statistical analysis was performed using the Student t-test.
Acknowledgments
We kindly thank Christa Testerink for providing the snrk2 knockout mutants seeds.
Abbreviations
| APX | Ascorbate peroxidase |
| Asc | Ascorbate |
| CAT | Catalase |
| CDPK | Calcium-dependent protein kinase |
| CIPK | CBL-interacting protein kinase |
| DAsc | Dehydroascorbate |
| DHAR | Dehydroascorbate reductase |
| H2O2 | Hydrogen peroxide |
| MPK | Mitogen activated protein kinase |
| O2− | Superoxide radical |
| PRX | Peroxidase |
| Rboh | Respiratory burst oxidase homologue |
| ROS | Reactive oxygen species |
| SnRK2 | SNF-1 Related Protein Kinases type 2 |
Supplementary Materials
The following are available online at http://www.mdpi.com/1422-0067/20/1/143/s1, Table S1: List of primers used in this study.
Author Contributions
Conceptualization, K.P.S. and G.D.; Methodology, K.P.S. and L.P.-K.; Formal Analysis, K.P.S.; Investigation, K.P.S., L.P.-K. and M.L.; Resources, K.P.S. and J.M.; Visualization K.P.S.; Writing—Original Draft Preparation, G.D. and K.P.S.; Writing—Review & Editing K.P.S., L.P.-K., J.M. and G.D.; Funding Acquisition K.P.S.
Funding
This research was funded by the National Science Centre, grant number 2013/11/N/NZ1/02417 and 2015/16/T/NZ1/00164 to K.S. and Foundation for Polish Science (Program No. MPD/2009-3/2).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Mittler R. ROS Are Good. Trends Plant Sci. 2017;22:11–19. doi: 10.1016/j.tplants.2016.08.002. [DOI] [PubMed] [Google Scholar]
- 2.Mhamdi A., Van Breusegem F. Reactive Oxygen Species in Plant Development. Development. 2018;145:dev164376. doi: 10.1242/dev.164376. [DOI] [PubMed] [Google Scholar]
- 3.Foyer C.H., Ruban A.V., Noctor G. Viewing Oxidative Stress through the Lens of Oxidative Signalling Rather than Damage. Biochem. J. 2017;474:877–883. doi: 10.1042/BCJ20160814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Waszczak C., Carmody M., Kangasjärvi J. Reactive Oxygen Species in Plant Signaling. Annu. Rev. Plant Biol. 2018;69:209–236. doi: 10.1146/annurev-arplant-042817-040322. [DOI] [PubMed] [Google Scholar]
- 5.Noctor G., Reichheld J.P., Foyer C.H. ROS-Related Redox Regulation and Signaling in Plants. Semin. Cell Dev. Biol. 2018;80:3–12. doi: 10.1016/j.semcdb.2017.07.013. [DOI] [PubMed] [Google Scholar]
- 6.Foyer C.H., Noctor G. Redox Homeostasis and Antioxidant Signaling: A Metabolic Interface between Stress Perception and Physiological Responses. Plant Cell Online. 2005;17:1866–1875. doi: 10.1105/tpc.105.033589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sofo A., Scopa A., Nuzzaci M., Vitti A. Ascorbate Peroxidase and Catalase Activities and Their Genetic Regulation in Plants Subjected to Drought and Salinity Stresses. Int. J. Mol. Sci. 2015;16:13561–13578. doi: 10.3390/ijms160613561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ben Rejeb K., Lefebvre-De Vos D., Le Disquet I., Leprince A.S., Bordenave M., Maldiney R., Jdey A., Abdelly C., Savouré A. Hydrogen Peroxide Produced by NADPH Oxidases Increases Proline Accumulation during Salt or Mannitol Stress in Arabidopsis thaliana. New Phytol. 2015;208:1138–1148. doi: 10.1111/nph.13550. [DOI] [PubMed] [Google Scholar]
- 9.Mittler R. Oxidative Stress, Antioxidants and Stress Tolerance. Trends Plant Sci. 2002;7:405–410. doi: 10.1016/S1360-1385(02)02312-9. [DOI] [PubMed] [Google Scholar]
- 10.Baxter A., Mittler R., Suzuki N. ROS as Key Players in Plant Stress Signalling. J. Exp. Bot. 2014;65:1229–1240. doi: 10.1093/jxb/ert375. [DOI] [PubMed] [Google Scholar]
- 11.Mittler R., Vanderauwera S., Gollery M., Van Breusegem F. Reactive Oxygen Gene Network of Plants. Trends Plant Sci. 2004;9:490–498. doi: 10.1016/j.tplants.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 12.Miller G., Suzuki N., Ciftci-Yilmaz S., Mittler R. Reactive Oxygen Species Homeostasis and Signalling during Drought and Salinity Stresses. Plant Cell Environ. 2010;33:453–467. doi: 10.1111/j.1365-3040.2009.02041.x. [DOI] [PubMed] [Google Scholar]
- 13.Choudhury S., Panda P., Sahoo L., Panda S.K. Reactive Oxygen Species Signaling in Plants under Abiotic Stress. Plant Signal. Behav. 2013;8:e23681. doi: 10.4161/psb.23681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hossain M.S., Dietz K.-J. Tuning of Redox Regulatory Mechanisms, Reactive Oxygen Species and Redox Homeostasis under Salinity Stress. Front. Plant Sci. 2016;7:548. doi: 10.3389/fpls.2016.00548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Inupakutika M.A., Sengupta S., Devireddy A.R., Azad R.K., Mittler R. The Evolution of Reactive Oxygen Species Metabolism. J. Exp. Bot. 2016;67:5933–5943. doi: 10.1093/jxb/erw382. [DOI] [PubMed] [Google Scholar]
- 16.Czarnocka W., Karpiński S. Friend or Foe? Reactive Oxygen Species Production, Scavenging and Signaling in Plant Response to Environmental Stresses. Free Radic. Biol. Med. 2018;122:4–20. doi: 10.1016/j.freeradbiomed.2018.01.011. [DOI] [PubMed] [Google Scholar]
- 17.Kulik A., Wawer I., Krzywińska E., Bucholc M., Dobrowolska G. SnRK2 Protein Kinases-Key Regulators of Plant Response to Abiotic Stresses. OMICS. 2011;15:859–872. doi: 10.1089/omi.2011.0091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boudsocq M., Barbier-Brygoo H., Laurière C. Identification of Nine Sucrose Nonfermenting 1-Related Protein Kinases 2 Activated by Hyperosmotic and Saline Stresses in Arabidopsis thaliana. J. Biol. Chem. 2004;279:41758–41766. doi: 10.1074/jbc.M405259200. [DOI] [PubMed] [Google Scholar]
- 19.Kobayashi Y., Yamamoto S., Minami H., Kagaya Y., Hattori T. Differential Activation of the Rice Sucrose Nonfermenting1-Related Protein Kinase2 Family by Hyperosmotic Stress and Abscisic Acid. Plant Cell. 2004;16:1163–1177. doi: 10.1105/tpc.019943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fujii H., Zhu J.K. Arabidopsis Mutant Deficient in 3 Abscisic Acid-Activated Protein Kinases Reveals Critical Roles in Growth, Reproduction, and Stress. Proc. Natl. Acad. Sci. USA. 2009;106:8380–8385. doi: 10.1073/pnas.0903144106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fujita Y., Nakashima K., Yoshida T., Katagiri T., Kidokoro S., Kanamori N., Umezawa T., Fujita M., Maruyama K., Ishiyama K., et al. Three SnRK2 Protein Kinases Are the Main Positive Regulators of Abscisic Acid Signaling in Response to Water Stress in Arabidopsis. Plant Cell Physiol. 2009;50:2123–2132. doi: 10.1093/pcp/pcp147. [DOI] [PubMed] [Google Scholar]
- 22.Diédhiou C.J., Popova O.V., Dietz K.J., Golldack D. The SNF1-Type Serine-Threonine Protein Kinase SAPK4 Regulates Stress-Responsive Gene Expression in Rice. BMC Plant Biol. 2008;8:49. doi: 10.1186/1471-2229-8-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McLoughlin F., Galvan-Ampudia C.S., Julkowska M.M., Caarls L., Van Der Does D., Laurière C., Munnik T., Haring M.A., Testerink C. The Snf1-Related Protein Kinases SnRK2.4 and SnRK2.10 Are Involved in Maintenance of Root System Architecture during Salt Stress. Plant J. 2012;72:436–449. doi: 10.1111/j.1365-313X.2012.05089.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Soma F., Mogami J., Yoshida T., Abekura M., Takahashi F., Kidokoro S., Mizoi J., Shinozaki K., Yamaguchi-Shinozaki K. ABA-Unresponsive SnRK2 Protein Kinases Regulate MRNA Decay under Osmotic Stress in Plants. Nat. Plants. 2017;3:16204. doi: 10.1038/nplants.2016.204. [DOI] [PubMed] [Google Scholar]
- 25.Hubbard K.E., Nishimura N., Hitomi K., Getzoff E.D., Schroeder J.I. Early Abscisic Acid Signal Transduction Mechanisms: Newly Discovered Components and Newly Emerging Questions. Genes Dev. 2010;24:1695–1708. doi: 10.1101/gad.1953910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Umezawa T., Nakashima K., Miyakawa T., Kuromori T., Tanokura M., Shinozaki K., Yamaguchi-Shinozaki K. Molecular Basis of the Core Regulatory Network in ABA Responses: Sensing, Signaling and Transport. Plant Cell Physiol. 2010;51:1821–1839. doi: 10.1093/pcp/pcq156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Geiger D., Scherzer S., Mumm P., Stange A., Marten I., Bauer H., Ache P., Matschi S., Liese A., Al-Rasheid K.A.S., et al. Activity of Guard Cell Anion Channel SLAC1 Is Controlled by Drought-Stress Signaling Kinase-Phosphatase Pair. Proc. Natl. Acad. Sci. USA. 2009;106:21425–21430. doi: 10.1073/pnas.0912021106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee S.C., Lan W., Buchanan B.B., Luan S. A Protein Kinase-Phosphatase Pair Interacts with an Ion Channel to Regulate ABA Signaling in Plant Guard Cells. Proc. Natl. Acad. Sci. USA. 2009;106:21419–21424. doi: 10.1073/pnas.0910601106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sato A., Sato Y., Fukao Y., Fujiwara M., Umezawa T., Shinozaki K., Hibi T., Taniguchi M., Miyake H., Goto D.B., et al. Threonine at Position 306 of the KAT1 Potassium Channel Is Essential for Channel Activity and Is a Target Site for ABA-Activated SnRK2/OST1/SnRK2.6 Protein Kinase. Biochem. J. 2009;424:439–448. doi: 10.1042/BJ20091221. [DOI] [PubMed] [Google Scholar]
- 30.Furihata T., Maruyama K., Fujita Y., Umezawa T., Yoshida R., Shinozaki K., Yamaguchi-Shinozaki K. Abscisic Acid-Dependent Multisite Phosphorylation Regulates the Activity of a Transcription Activator AREB1. Proc. Natl. Acad. Sci. USA. 2006;103:1988–1993. doi: 10.1073/pnas.0505667103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang P., Xue L., Batelli G., Lee S., Hou Y.-J., Van Oosten M.J., Zhang H., Tao W.A., Zhu J.-K. Quantitative Phosphoproteomics Identifies SnRK2 Protein Kinase Substrates and Reveals the Effectors of Abscisic Acid Action. Proc. Natl. Acad. Sci. USA. 2013;110:11205–11210. doi: 10.1073/pnas.1308974110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Umezawa T., Sugiyama N., Takahashi F., Anderson J.C., Ishihama Y., Peck S.C., Shinozaki K. Genetics and Phosphoproteomics Reveal a Protein Phosphorylation Network in the Abscisic Acid Signaling Pathway in Arabidopsis thaliana. Sci. Signal. 2013;6:rs8. doi: 10.1126/scisignal.2003509. [DOI] [PubMed] [Google Scholar]
- 33.Grondin A., Rodrigues O., Verdoucq L., Merlot S., Leonhardt N., Maurel C. Aquaporins Contribute to ABA-Triggered Stomatal Closure through OST1-Mediated Phosphorylation. Plant Cell. 2015;27:1945–1954. doi: 10.1105/tpc.15.00421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mustilli A.-C., Merlot S., Vavasseur A., Fenzi F., Giraudat J. Arabidopsis OST1 Protein Kinase Mediates the Regulation of Stomatal Aperture by Abscisic Acid and Acts Upstream of Reactive Oxygen Species Production. Plant Cell. 2002;14:3089–3099. doi: 10.1105/tpc.007906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sirichandra C., Gu D., Hu H.C., Davanture M., Lee S., Djaoui M., Valot B., Zivy M., Leung J., Merlot S., et al. Phosphorylation of the Arabidopsis AtrbohF NADPH Oxidase by OST1 Protein Kinase. FEBS Lett. 2009;583:2982–2986. doi: 10.1016/j.febslet.2009.08.033. [DOI] [PubMed] [Google Scholar]
- 36.Han J.-P., Köster P., Drerup M.M., Scholz M., Li S., Edel K.H., Hashimoto K., Kuchitsu K., Hippler M., Kudla J. Fine Tuning of RBOHF Activity Is Achieved by Differential Phosphorylation and Ca2+ Binding. New Phytol. 2018 doi: 10.1111/nph.15543. [DOI] [PubMed] [Google Scholar]
- 37.Mikołajczyk M., Awotunde O.S., Muszyńska G., Klessig D.F., Dobrowolska G. Osmotic Stress Induces Rapid Activation of a Salicylic Acid-Induced Protein Kinase and a Homolog of Protein Kinase ASK1 in Tobacco Cells. Plant Cell. 2000;12:165–178. doi: 10.1105/tpc.12.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Burza A.M., Pekala I., Sikora J., Siedlecki P., Małagocki P., Bucholc M., Koper L., Zielenkiewicz P., Dadlez M., Dobrowolska G. Nicotiana tabacum Osmotic Stress-Activated Kinase Is Regulated by Phosphorylation on Ser-154 and Ser-158 in the Kinase Activation Loop. J. Biol. Chem. 2006;281:34299–34311. doi: 10.1074/jbc.M601977200. [DOI] [PubMed] [Google Scholar]
- 39.Maszkowska J., Dębski J., Kulik A., Kistowski M., Bucholc M., Lichocka M., Klimecka M., Sztatelman O., Szymańska K.P., Dadlez M., et al. Phosphoproteomic Analysis Reveals That Dehydrins ERD10 and ERD14 Are Phosphorylated by SNF1-Related Protein Kinase 2.10 in Response to Osmotic Stress. Plant Cell Environ. 2018 doi: 10.1111/pce.13465. [DOI] [PubMed] [Google Scholar]
- 40.Kulik A., Anielska-Mazur A., Bucholc M., Koen E., Szymanska K., Zmienko A., Krzywinska E., Wawer I., McLoughlin F., Ruszkowski D., et al. SNF1-Related Protein Kinases Type 2 Are Involved in Plant Responses to Cadmium Stress. Plant Physiol. 2012;160:868–883. doi: 10.1104/pp.112.194472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakagami H., Soukupová H., Schikora A., Zárský V., Hirt H. A Mitogen-Activated Protein Kinase Kinase Kinase Mediates Reactive Oxygen Species Homeostasis in Arabidopsis. J. Biol. Chem. 2006;281:38697–38704. doi: 10.1074/jbc.M605293200. [DOI] [PubMed] [Google Scholar]
- 42.Yang L., Ye C., Zhao Y., Cheng X., Wang Y., Jiang Y.Q., Yang B. An Oilseed Rape WRKY-Type Transcription Factor Regulates ROS Accumulation and Leaf Senescence in Nicotiana Benthamiana and Arabidopsis through Modulating Transcription of RbohD and RbohF. Planta. 2018;247:1323–1338. doi: 10.1007/s00425-018-2868-z. [DOI] [PubMed] [Google Scholar]
- 43.Pitzschke A., Djamei A., Bitton F., Hirt H. A Major Role of the MEKK1-MKK1/2-MPK4 Pathway in ROS Signalling. Mol. Plant. 2009;2:120–137. doi: 10.1093/mp/ssn079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ben Rejeb K., Benzarti M., Debez A., Bailly C., Savouré A., Abdelly C. NADPH Oxidase-Dependent H2O2 Production Is Required for Salt-Induced Antioxidant Defense in Arabidopsis thaliana. J. Plant Physiol. 2015;174:5–15. doi: 10.1016/j.jplph.2014.08.022. [DOI] [PubMed] [Google Scholar]
- 45.Ma L., Zhang H., Sun L., Jiao Y., Zhang G., Miao C., Hao F. NADPH Oxidase AtrbohD and AtrbohF Function in ROS-Dependent Regulation of Na+/K+ Homeostasis in Arabidopsis under Salt Stress. J. Exp. Bot. 2012;63:305–317. doi: 10.1093/jxb/err280. [DOI] [PubMed] [Google Scholar]
- 46.O’Brien J.A., Daudi A., Finch P., Butt V.S., Whitelegge J.P., Souda P., Ausubel F.M., Bolwell G.P. A Peroxidase-Dependent Apoplastic Oxidative Burst in Cultured Arabidopsis Cells Functions in MAMP-Elicited Defense. Plant Physiol. 2012;158:2013–2027. doi: 10.1104/pp.111.190140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Daudi A., Cheng Z., O’Brien J.A., Mammarella N., Khan S., Ausubel F.M., Bolwell G.P. The Apoplastic Oxidative Burst Peroxidase in Arabidopsis Is a Major Component of Pattern-Triggered Immunity. Plant Cell. 2012;24:275–287. doi: 10.1105/tpc.111.093039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xing Y., Jia W., Zhang J. AtMEK1 Mediates Stress-Induced Gene Expression of CAT1 Catalase by Triggering H2O2 Production in Arabidopsis. J. Exp. Bot. 2007;58:2969–2981. doi: 10.1093/jxb/erm144. [DOI] [PubMed] [Google Scholar]
- 49.Frugoli J.A., Zhong H.H., Nuccio M.L., McCourt P., McPeek M.A., Thomas T.L., McClung C.R. Catalase Is Encoded by a Multigene Family in Arabidopsis thaliana (L.) Heynh. Plant Physiol. 1996;112:327–336. doi: 10.1104/pp.112.1.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mhamdi A., Queval G., Chaouch S., Vanderauwera S., Van Breusegem F., Noctor G. Catalase Function in Plants: A Focus on Arabidopsis Mutants as Stress-Mimic Models. J. Exp. Bot. 2010;61:4197–4220. doi: 10.1093/jxb/erq282. [DOI] [PubMed] [Google Scholar]
- 51.Gao M., Liu J., Bi D., Zhang Z., Cheng F., Chen S., Zhang Y. MEKK1, MKK1/MKK2 and MPK4 Function Together in a Mitogen-Activated Protein Kinase Cascade to Regulate Innate Immunity in Plants. Cell Res. 2008;18:1190–1198. doi: 10.1038/cr.2008.300. [DOI] [PubMed] [Google Scholar]
- 52.Asai S., Ohta K., Yoshioka H. MAPK Signaling Regulates Nitric Oxide and NADPH Oxidase-Dependent Oxidative Bursts in Nicotiana Benthamiana. Plant Cell Online. 2008;20:1390–1406. doi: 10.1105/tpc.107.055855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dubiella U., Seybold H., Durian G., Komander E., Lassig R., Witte C.-P., Schulze W.X., Romeis T. Calcium-Dependent Protein Kinase/NADPH Oxidase Activation Circuit Is Required for Rapid Defense Signal Propagation. Proc. Natl. Acad. Sci. USA. 2013;110:8744–8749. doi: 10.1073/pnas.1221294110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kadota Y., Sklenar J., Derbyshire P., Stransfeld L., Asai S., Ntoukakis V., Jones J.D., Shirasu K., Menke F., Jones A., et al. Direct Regulation of the NADPH Oxidase RBOHD by the PRR-Associated Kinase BIK1 during Plant Immunity. Mol. Cell. 2014;54:43–55. doi: 10.1016/j.molcel.2014.02.021. [DOI] [PubMed] [Google Scholar]
- 55.Li L., Li M., Yu L., Zhou Z., Liang X., Liu Z., Cai G., Gao L., Zhang X., Wang Y., et al. The FLS2-Associated Kinase BIK1 Directly Phosphorylates the NADPH Oxidase RbohD to Control Plant Immunity. Cell Host Microbe. 2014;15:329–338. doi: 10.1016/j.chom.2014.02.009. [DOI] [PubMed] [Google Scholar]
- 56.Monaghan J., Matschi S., Shorinola O., Rovenich H., Matei A., Segonzac C., Malinovsky F.G.G., Rathjen J.P.P., Maclean D., Romeis T., et al. The Calcium-Dependent Protein Kinase CPK28 Buffers Plant Immunity and Regulates BIK1 Turnover. Cell Host Microbe. 2014;16:605–615. doi: 10.1016/j.chom.2014.10.007. [DOI] [PubMed] [Google Scholar]
- 57.Monaghan J., Matschi S., Romeis T., Zipfel C. The Calcium-Dependent Protein Kinase CPK28 Negatively Regulates the BIK1-Mediated PAMP-Induced Calcium Burst. Plant Signal. Behav. 2015;10:e1018497. doi: 10.1080/15592324.2015.1018497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu Y., He C. A Review of Redox Signaling and the Control of MAP Kinase Pathway in Plants. Redox Biol. 2017;11:192–204. doi: 10.1016/j.redox.2016.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kawasaki T., Yamada K., Yoshimura S., Yamaguchi K. Chitin Receptor-Mediated Activation of MAP Kinases and ROS Production in Rice and Arabidopsis. Plant Signal. Behav. 2017;12:e1361076. doi: 10.1080/15592324.2017.1361076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang M., Chiang Y.H., Toruño T.Y., Lee D.H., Ma M., Liang X., Lal N.K., Lemos M., Lu Y.J., Ma S., et al. The MAP4 Kinase SIK1 Ensures Robust Extracellular ROS Burst and Antibacterial Immunity in Plants. Cell Host Microbe. 2018;24:379–391.e5. doi: 10.1016/j.chom.2018.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zandalinas S.I., Mittler R. ROS-Induced ROS Release in Plant and Animal Cells. Free Radic. Biol. Med. 2018;122:21–27. doi: 10.1016/j.freeradbiomed.2017.11.028. [DOI] [PubMed] [Google Scholar]
- 62.Fujii H., Verslues P.E., Zhu J.-K. Arabidopsis Decuple Mutant Reveals the Importance of SnRK2 Kinases in Osmotic Stress Responses in Vivo. Proc. Natl. Acad. Sci. USA. 2011;108:1717–1722. doi: 10.1073/pnas.1018367108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Drerup M.M., Schlücking K., Hashimoto K., Manishankar P., Steinhorst L., Kuchitsu K., Kudla J. The Calcineurin B-like Calcium Sensors CBL1 and CBL9 Together with Their Interacting Protein Kinase CIPK26 Regulate the Arabidopsis NADPH Oxidase RBOHF. Mol. Plant. 2013;6:559–569. doi: 10.1093/mp/sst009. [DOI] [PubMed] [Google Scholar]
- 64.Kimura S., Kawarazaki T., Nibori H., Michikawa M., Imai A., Kaya H., Kuchitsu K. The CBL-Interacting Protein Kinase CIPK26 Is a Novel Interactor of Arabidopsis NADPH Oxidase AtRbohF That Negatively Modulates Its ROS-Producing Activity in a Heterologous Expression System. J. Biochem. 2013;153:191–195. doi: 10.1093/jb/mvs132. [DOI] [PubMed] [Google Scholar]
- 65.Takahashi F., Mizoguchi T., Yoshida R., Ichimura K., Shinozaki K. Calmodulin-Dependent Activation of MAP Kinase for ROS Homeostasis in Arabidopsis. Mol. Cell. 2011;41:649–660. doi: 10.1016/j.molcel.2011.02.029. [DOI] [PubMed] [Google Scholar]
- 66.Hrabak E.M., Chan C.W., Gribskov M., Harper J., Choi J., Halford N., Kudla J., Luan S., Nimmo H., Sussman M., et al. The Arabidopsis CDPK-SnRK Superfamily of Protein Kinases. Plant Physiol. 2003;132:666–680. doi: 10.1104/pp.102.011999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xie Y.J., Xu S., Han B., Wu M.Z., Yuan X.X., Han Y., Gu Q., Xu D.K., Yang Q., Shen W.B. Evidence of Arabidopsis Salt Acclimation Induced by Up-Regulation of HY1 and the Regulatory Role of RbohD-Derived Reactive Oxygen Species Synthesis. Plant J. 2011;66:280–292. doi: 10.1111/j.1365-313X.2011.04488.x. [DOI] [PubMed] [Google Scholar]
- 68.Arnaud D., Lee S., Takebayashi Y., Choi D., Choi J., Sakakibara H., Hwang I. Cytokinin-Mediated Regulation of Reactive Oxygen Species Homeostasis Modulates Stomatal Immunity in Arabidopsis. Plant Cell. 2017;29:543–559. doi: 10.1105/tpc.16.00583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Meng X., Xu J., He Y., Yang K.-Y., Mordorski B., Liu Y., Zhang S. Phosphorylation of an ERF Transcription Factor by Arabidopsis MPK3/MPK6 Regulates Plant Defense Gene Induction and Fungal Resistance. Plant Cell. 2013;25:1126–1142. doi: 10.1105/tpc.112.109074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sewelam N., Kazan K., Thomas-Hall S.R., Kidd B.N., Manners J.M., Schenk P.M. Ethylene Response Factor 6 Is a Regulator of Reactive Oxygen Species Signaling in Arabidopsis. PLoS ONE. 2013;8:e70289. doi: 10.1371/journal.pone.0070289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang P., Du Y., Zhao X., Miao Y., Song C.-P. The MPK6-ERF6-ROS-Responsive Cis-Acting Element7/GCC Box Complex Modulates Oxidative Gene Transcription and the Oxidative Response in Arabidopsis. Plant Physiol. 2013;161:1392–1408. doi: 10.1104/pp.112.210724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Adachi H., Nakano T., Miyagawa N., Ishihama N., Yoshioka M., Katou Y., Yaeno T., Shirasu K., Yoshioka H. WRKY Transcription Factors Phosphorylated by MAPK Regulate a Plant Immune NADPH Oxidase in Nicotiana Benthamiana. Plant Cell. 2015;27:2645–2663. doi: 10.1105/tpc.15.00213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ichimura K., Mizoguchi T., Yoshida R., Yuasa T., Shinozaki K. Various Abiotic Stresses Rapidly Activate Arabidopsis MAP Kinases ATMPK4 and ATMPK6. Plant J. 2000;24:655–665. doi: 10.1046/j.1365-313x.2000.00913.x. [DOI] [PubMed] [Google Scholar]
- 74.Dubois M., Skirycz A., Claeys H., Maleux K., Dhondt S., De Bodt S., Vanden Bossche R., De Milde L., Yoshizumi T., Matsui M., et al. Ethylene Response FACTOR6 Acts as a Central Regulator of Leaf Growth under Water-Limiting Conditions in Arabidopsis. Plant Physiol. 2013;162:319–332. doi: 10.1104/pp.113.216341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dal Santo S., Stampfl H., Krasensky J., Kempa S., Gibon Y., Petutschnig E., Rozhon W., Heuck A., Clausen T., Jonak C. Stress-Induced GSK3 Regulates the Redox Stress Response by Phosphorylating Glucose-6-Phosphate Dehydrogenase in Arabidopsis. Plant Cell. 2012;24:3380–3392. doi: 10.1105/tpc.112.101279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zong X.J., Li D.P.D.Q., Gu L.K., Li D.P.D.Q., Liu L.X., Hu X.L. Abscisic Acid and Hydrogen Peroxide Induce a Novel Maize Group C MAP Kinase Gene, ZmMPK7, Which Is Responsible for the Removal of Reactive Oxygen Species. Planta. 2009;229:485–495. doi: 10.1007/s00425-008-0848-4. [DOI] [PubMed] [Google Scholar]
- 77.Xing Y., Jia W., Zhang J. AtMKK1 Mediates ABA-Induced CAT1 Expression and H2O2 production via AtMPK6-Coupled Signaling in Arabidopsis. Plant J. 2008;54:440–451. doi: 10.1111/j.1365-313X.2008.03433.x. [DOI] [PubMed] [Google Scholar]
- 78.Verslues P.E., Batelli G., Grillo S., Agius F., Kim Y.-S., Zhu J.-K., Agarwal M., Katiyar-Agarwal S., Zhu J.-K. Interaction of SOS2 with Nucleoside Diphosphate Kinase 2 and Catalases Reveals a Point of Connection between Salt Stress and H2O2 Signaling in Arabidopsis thaliana. Mol. Cell. Biol. 2007;27:7771–7780. doi: 10.1128/MCB.00429-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Galvez-Valdivieso G., Fryer M.J., Lawson T., Slattery K., Truman W., Smirnoff N., Asami T., Davies W.J., Jones A.M., Baker N.R., et al. The High Light Response in Arabidopsis Involves ABA Signaling between Vascular and Bundle Sheath Cells. Plant Cell Online. 2009;21:2143–2162. doi: 10.1105/tpc.108.061507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pitzschke A., Hirt H. Disentangling the Complexity of Mitogen-Activated Protein Kinases and Reactive Oxygen Species Signaling. Plant Physiol. 2009;149:606–615. doi: 10.1104/pp.108.131557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hotta C.T., Gardner M.J., Hubbard K.E., Baek S.J., Dalchau N., Suhita D., Dodd A.N., Webb A.A.R. Modulation of Environmental Responses of Plants by Circadian Clocks. Plant Cell Environ. 2007;33:333–349. doi: 10.1111/j.1365-3040.2006.01627.x. [DOI] [PubMed] [Google Scholar]
- 82.Bhardwaj V., Meier S., Petersen L.N., Ingle R.A., Roden L.C. Defence Responses of Arabidopsis thaliana to Infection by Pseudomonas Syringae Are Regulated by the Circadian Clock. PLoS ONE. 2011;6:e26968. doi: 10.1371/journal.pone.0026968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Seung D., Risopatron J.P.M., Jones B.J., Marc J. Circadian Clock-Dependent Gating in ABA Signalling Networks. Protoplasma. 2012;249:445–457. doi: 10.1007/s00709-011-0304-3. [DOI] [PubMed] [Google Scholar]
- 84.Lai A.G., Doherty C.J., Mueller-Roeber B., Kay S.A., Schippers J.H.M., Dijkwel P.P. Circadian Clock-Associated 1 Regulates ROS Homeostasis and Oxidative Stress Responses. Proc. Natl. Acad. Sci. USA. 2012;109:17129–17134. doi: 10.1073/pnas.1209148109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li Z., Bonaldi K., Uribe F., Pruneda-Paz J.L. A Localized Pseudomonas Syringae Infection Triggers Systemic Clock Responses in Arabidopsis. Curr. Biol. 2018;28:630–639.e4. doi: 10.1016/j.cub.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Greenham K., Guadagno C.R., Gehan M.A., Mockler T.C., Weinig C., Ewers B.E., McClung C.R. Temporal Network Analysis Identifies Early Physiological and Transcriptomic Indicators of Mild Drought in Brassica Rapa. Elife. 2017;6:e29655. doi: 10.7554/eLife.29655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rasul S., Dubreuil-Maurizi C., Lamotte O., Koen E., Poinssot B., Alcaraz G., Wendehenne D., Jeandroz S. Nitric Oxide Production Mediates Oligogalacturonide-Triggered Immunity and Resistance to Botrytis Cinerea in Arabidopsis thaliana. Plant Cell Environ. 2012;35:1483–1499. doi: 10.1111/j.1365-3040.2012.02505.x. [DOI] [PubMed] [Google Scholar]
- 88.Srivastava A.K., Sablok G., Hackenberg M., Deshpande U., Suprasanna P. Thiourea Priming Enhances Salt Tolerance through Co-Ordinated Regulation of MicroRNAs and Hormones in Brassica Juncea. Sci. Rep. 2017;7:1–15. doi: 10.1038/srep45490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Livak K.J., Schmittgen T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2-ΔΔCT Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 90.Czechowski T. Genome-Wide Identification and Testing of Superior Reference Genes for Transcript Normalization in Arabidopsis. Plant Physiol. 2005;139:5–17. doi: 10.1104/pp.105.063743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Remans T., Smeets K., Opdenakker K., Mathijsen D., Vangronsveld J., Cuypers A. Normalisation of Real-Time RT-PCR Gene Expression Measurements in Arabidopsis thaliana Exposed to Increased Metal Concentrations. Planta. 2008;227:1343–1349. doi: 10.1007/s00425-008-0706-4. [DOI] [PubMed] [Google Scholar]
- 92.Sambrook J., Fritsch E.F., Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY, USA: 1989. [Google Scholar]
- 93.Wawer I., Bucholc M., Astier J., Anielska-Mazur A., Dahan J., Kulik A., Wysłouch-Cieszynska A., Zaręba-Kozioł M., Krzywinska E., Dadlez M., et al. Regulation of Nicotiana tabacum Osmotic Stress-Activated Protein Kinase and Its Cellular Partner GAPDH by Nitric Oxide in Response to Salinity. Biochem. J. 2010;429:73–83. doi: 10.1042/BJ20100492. [DOI] [PubMed] [Google Scholar]
- 94.Venisse J.S., Gullner G., Brisset M.N. Evidence for the Involvement of an Oxidative Stress in the Initiation of Infection of Pear by Erwinia Amylovora. Plant Physiol. 2001;125:2164–2172. doi: 10.1104/pp.125.4.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Polkowska-Kowalczyk L., Wielgat B., Maciejewska U. Changes in the Antioxidant Status in Leaves of Solanum Species in Response to Elicitor from Phytophthora Infestans. J. Plant Physiol. 2007;164:1268–1277. doi: 10.1016/j.jplph.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 96.Aebi H. Catalase in Vitro. Methods Enzymol. 1984;105:121–126. doi: 10.1016/s0076-6879(84)05016-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.