Abstract
Aiming at further systematically comparing the similarities and differences of the chemical components in ginseng of different ages, especially comparing the younger or the older and mountain-cultivated ginseng (MCG), 4, 5, 6-year-old cultivated ginseng (CG) and 12, 20-year-old MCG were chosen as the analytical samples in the present study. The combination of UPLC-QTOF-MSE, UNIFI platform and multivariate statistical analysis were developed to profile CGs and MCGs. By the screening analysis based on UNIFI, 126 chemical components with various structural types were characterized or tentatively identified from all the CG and MCG samples for the first time. The results showed that all the CG and MCG samples had the similar chemical composition, but there were significant differences in the contents of markers. By the metabolomic analysis based on multivariate statistical analysis, it was shown that CG4–6 years, MCG12 years and MCG20 years samples were obviously divided into three different groups, and a total of 17 potential age-dependent markers enabling differentiation among the three groups of samples were discovered. For differentiation from other two kinds of samples, there were four robust makers such as α-linolenic acid, 9-octadecenoic acid, linoleic acid and panaxydol for CG4–6 years, five robust makers including ginsenoside Re1, -Re2, -Rs1, malonylginsenoside Rb2 and isomer of malonylginsenoside Rb1 for MCG20 years, and two robust makers, 24-hydroxyoleanolic acid and palmitoleic acid, for MCG12 years were discovered, respectively. The proposed approach could be applied to directly distinguish MCG root ages, which is an important criterion for evaluating the quality of MCG. The results will provide the data for the further study on the chemical constituents of MCG.
Keywords: mountain-cultivated ginseng, identification, metabolomic analysis, UPLC-QTOF-MS
1. Introduction
Ginseng, the king of herbs in the Orient, has always received a lot of attention, not only as a therapeutic medicinal herb, but also as a health supplement. According to the different growing environments and diverse cultivation methods, there two kinds of ginseng are distinguished in the Chinese Pharmacopoeia: cultivated ginseng (CG) and mountain-cultivated ginseng (MCG). CG is cultivated artificially in gardens, while MCG is grown for at least 10 years [1,2]. MCG, also called “Lin-Xia-Shan-Shen”, can be regarded as a replacement of wild ginseng. MCG is of better quality than CG and offers more production than wild ginseng [3]. Actually, the adulteration or falsification of the cultivation age of MCG has always been a serious problem in the MCG commercial market. As we all know, the chemical components and biological activities of ginseng with different cultivation ages are distinct [4,5], and more aged ginseng is usually of higher economic value. In an investigation of the characteristic components for distinguishing CG (4–7-year of age) and MCG (with 15-years of growth), 12 compounds, including ginsenoside Ra3/isomer, gypenoside XVII, quinquenoside R1, ginsenoside Ra7, notoginsenoside Fe, ginsenoside Ra2, ginsenoside Rs6/Rs7, malonyl ginsenoside Rc, malonyl ginsenoside Rb1, malonyl ginsenoside Rb2, palmitoleic acid, and ethyl linoleate were regarded as the characteristic chemical markers for the discrimination [6]. Recently, a UPLC/QTOF- MS-based metabolomics approach was applied to the global metabolite profiling of MCG leaf samples aged from 6 to 18 years, and the authors claimed that the approach could also be applied to discriminate MCG root ages indirectly [7]. It is undoubted that the developed method can be used as a standard protocol for discriminating and predicting MCG leaf ages directly, but there might be some inaccuracy and uncertainty when discriminating MCG root ages indirectly.
In the past decades, some analytical methods focusing on ginsenosides had been used to distinguish MCG from CG, such as thin layer chromatography (TLC), or high performance liquid chromatography (HPLC) [8,9]. However, these technologies require lots of time and energy, and the results cannot provide a comprehensive or accurate discrimination between them. Currently, untargeted metabolomics, combined with multivariate statistical methods such as principal component analysis (PCA) and orthogonal partial least squares discriminant analysis (OPLS-DA), are widely used to profile diverse classes of metabolites and to better understand the chemical diversity and the multiple pharmacological effects of ginsenosides or ginseng [10,11]. Given the multi-component property, the combination of LC-MS-based metabolomic profiling with multivariate statistical analysis methods was used as a rapid means of characterization and was increasingly applied for analyzing ginseng from different herbs, cultivation environments/areas, cultivation ages or different parts [12,13]. As an example, for different herbs belonging to the same genus, specific biomarkers including chikusetsusaponin IVa, ginsenoside Rf and ginsenoside Rc were selected and verified for ginseng [14]. In another example of different parts analysis, the metabolic profiles of root, leaf, flower bud, berry and seed of ginseng were investigated [12,15]. In addition, the approach for the discrimination of different red ginseng root parts was reported. As a result, fine roots had the highest protopanaxadiol (PPD)/protopanaxatriol (PPT) ratio, which could clearly distinguish the main roots from the lateral roots and fine roots parts [16]. Such analysis was also applied to make metabolite profiling and age discrimination of 4- and 6-year- old red ginseng [17], or 1–6 years ginseng [18].
In addition, UNIFI, the automated data processing software, is an integrated informatics platform that possesses the ability to incorporate scientific library into a streamlined workflow, aiming at identifying chemical components from complex raw data [19]. The combination of UPLC separation, Q/TOF-MS detection and UNIFI platform has been frequently applied in the characterization of chemical constituents of herbs [20,21].
Normally, CG is harvested after a 4–6 years cultivation period, and MCG is collected at ages of 10–20 years. To develop a more direct and more efficient discrimination method for the cultivation ages and to explore potential age-dependent markers, we chose 4, 5, 6-year-old CG and 12, 20-year-old MCG as the analytical samples in the present study. UPLC-QTOF-MSE, UNIFI platform and multivariate statistical analysis were then used to profile these two kinds of ginseng. The aims were to systematically screen the chemical components and to perform the non-targeted metabolomic analysis, and in turn will lay the foundation for the establishment of CG and MCG quality criteria in the future. In one hand, this study will reveal the structural diversity of secondary metabolites and the different patterns in CG and MCG. In the other hand, the present study could provide a reference point for a reliable, accurate method for distinguishing among CG and MCG samples of different ages.
2. Materials and Methods
2.1. Materials and Reagents
A total of 40 batches of CG and MCG root products, including 24 batches of CGs and 16 batches of MCGs, were collected from different cultivation areas in Jilin Province, the main source of ginseng in China. A detailed sample list is given in Table 1. All samples were harvested and collected by Professor Li Ping-ya from Jilin University Institute of Frontier Medical Science, according to China Pharmacopoeia (2015 version) [22]. Voucher specimens have been deposited at the Research Center of Nature Drug, School of Pharmaceutical Sciences, Jilin University, Changchun, China.
Table 1.
Details of the MCG and CG samples.
| Sample No. | Source | Collection Time |
|---|---|---|
| CG3years-1, CG3years-2; CG4years-1, CG4years-2; CG5years-1, CG5years-2; MCG12years-1, MCG12years-2; MCG20years-1, MCG20years-2 | Ji′an City, Jilin Province, China | 2017.09–2017.10 |
| CG3years-3, CG3years-4; CG4years-3, CG4years-4; CG5years-3, CG5years-4; MCG12years-3, MCG12years-4; MCG20years-3, MCG20years-4 | Fusong County, Jilin Province, China | 2017.09–2017.10 |
| CG3years-5, CG3years-6; CG4years-5, CG4years-6; CG5years-5, CG5years-6; MCG12years-5, MCG12years-6; MCG20years-5, MCG20years-6 | Tonghua City, Jilin Province, China | 2017.09–2017.10 |
| CG3years-7, CG3years-8; CG4years-7, CG4yeasr-8; CG5years-7, CG5years-8; MCG12years-7, MCG12years-8; MCG20years-7, MCG20years-8 | Jingyu Country, Jilin Province, China | 2017.09–2017.10 |
Acetonitrile, methanol were all UPLC-MS pure grade (Fisher Scientific Inc., Geel, Belgium). Formic acid (MS grade) was purchased from Sigma-Aldrich (St. Louis, MO, USA). Leucine enkephaline was provided by Waters (Waters Technologies, Milford, MA., USA). Distilled water was prepared in-house via a Millipore water purification system (Millipore, Billerica, MA, USA). All other chemicals were analytical grade. For reference substances, ginsenoside F1 (R20151040), -F2 (R20151040), notoginsenoside R1 (R20170210), notoginseno- side R4 (R20170212) were provided by the Research Center of Natural Drugs, School of Pharmaceutical Sciences, Jilin University, China. Ginsenoside Rb1, -Rb2, -Rb3, -Rc, -Rd, -Re, -Rf, -F5, -Rg1, 20(R)-Rg2, 20(S)-Rh1, 20(R)-Rh1, 20(S)-Rg3, 20(R)-Rg3, -Ro, gypenoside XVII, ginsenoside Rs1, -Rs2 were isolated in our laboratory and identified by spectroscopic data. Adenine (101774299), tryptophane (73-22-2), palmitoleic acid (101491588) were purchased from Sigma-Aldrich. Notoginsenoside Fe (8105-29-5), D-adenosine (110879- 200502), histidine (624-200304) were purchased from the National Institutes for Food and Drug Control. Ginsenoside Rg5 (wkq16051002, Victory Biological Technology Co., Ltd., Sichuan, China), α-linoleic acid (B21469; Yuanye Biological Technology Co., Ltd., Shanghai, China), D-arginin (130701; Nuoye Biological Engineering Co., Ltd., Anhui, China) and phenylpropionic acid (A20160211), quillaic acid (A20171109) were purchased from Beijing Zhongke Quality Inspection Biotechnology Co., Ltd. (Beijing, China) with the Chinese National Standard Sieve No. 3 (R40/3 series).
2.2. Sample Preparation and Extraction
All the CG and MCG samples were air-dried, grinded (Baijie Stainless Steel Grinder, BJ-800A, Deqing Baijie Electric Apllicance Co. Ltd., Zhejiang, China) and sieved (Chinese National Standard Sieve No. 3, R40/3 series) to get the homogeneous powder respectively. Then, the powder of 40 samples (200 mg accurately weighed per sample) were refluxed respectively with 85% methanol (2 L) at 80 °C for three times (2 h, 2 h, 1 h each time, respectively). Then, the extracts of each sample were combined, concentrated and evaporated to dryness. Each powder was dissolved in 5.0 mL of 80% methonal. After being filtered, each methanolic solution was injected directly into UPLC system. Meanwhile, 20 μL aliquots of each CG and MCG sample were mixed to obtain a quality control (QC) sample, which contained all of the components in the analysis. The QC sample was run randomly to monitor the stability of the system. All of the above solutions were stored at 4 °C prior to LC-MS analysis and the injection volume was 2 μL.
2.3. UPLC/QTOF-MSE
The chromatographic separation and mass spectrometry detection were conducted on the Waters Acquity UPLC system coupled with a Xevo G2-S QTOF mass spectrometer equipped with an electrospray ionization source (ESI). Separation was performed on Waters ACQUITY UPLC BEH C18 column (100 mm × 2.1 mm, 1.7 μm) at 40 °C. The mobile phase consisted of eluent A (0.1% formic acid aqueous solution) and eluent B (0.1% formic acid in acetonitrile) at flow rate of 0.4 mL/min with the following gradient program: 0~2 min, 10% (B); 2~26 min, 10%~100% (B); 26~28 min, 100% (B); 28~28.1 min, 100%~10% (B); 28.1~30 min, 10% (B). Mixtures of 10/90 and 90/10 water/acetonitrile were the strong wash and the weak wash solvent, respectively. The optimized conditions were employed: source temperature was 120 °C, the desolvation temperature was 300 °C, capillary voltage was 2.6 kV(ESI+) or 2.2 kV (ESI−), cone voltage was 40 V, desolvation gas flow was 800.0 L/h, cone gas flow was 50 L/h. The energy of low energy function and the collision energy of high energy function were set at 6 V and 20 V~40 V respectively in MSE mode. The mass spectrometer was calibrated with sodium formate in the range of 200–1500 Da. The lockmass compound used was leucine- enkephaline (external reference to the ion m/z 556.2771 in positive mode and 554.2615 in negative mode). Data were collected with Masslynx™ V4.1 workstation in continuum mode.
2.4. Chemical Information Database for the Components of CG and MCG
In addition to the Waters Traditional Medicine Library in UNIFI software, a systematic investigation of chemical constituents from the target herbs based on the literature was conducted. A self-built database of compounds, such as saponins, flavonoids, volatile oil, amino acids and so on, isolated from CG and MCG was established by searching online databases such as China Journals of Full-Text Database (CNKI), PubMed, Medicine, Web of Science and ChemSpider. The name, molecular formula and structure of components from CG and MCG were obtained in the database.
2.5. The Screening Analysis Based on UNIFI Platform
UNIFI 1.7.0 software (Waters, Manchester, UK) was used to perform the screening analysis on the structural characteristics and MS fragmentation behaviors, especially for characteristic fragments. Main parameters were set as follows: peak intensity of high energy over 200 counts and the peak intensity of low energy over 1000 counts were the selected parameters in peak detection; mass error up to ±10 ppm for identified compound; retention time tolerance was set in the range of ±0.1 min; positive adducts containing +H, +Na or negative adducts containing −H, +HCOOH were all selected; the reference compound was leucine-enkephalin (556.2766 for positive ion, 554.2620 for negative ion). The MS raw data were processed using the streamlined workflow of UNIFI software to quickly identify the chemical components that met the match criteria with the in-house Traditional Medicine Library and the self-built database [20,21].
2.6. The Metabolomics Analysis Based on Multivariate Statistical Analysis
To differentiate MCG and CG, MarkerLynx XS V4.1 software (Waters, Milford, DE, USA) was used to process the raw data by deconvolution, alignment, data reduction and to perform the multivariate statistical analysis [20,21]. The following steps were performed: acquiring data, creating a MarkerLynx processing method, processing the acquired data and viewing results Extended Statistics (XS) Viewer. The main parameters in the method set to process the raw data were as follows: retention time range 5–28 min, mass range 200–1400 Da, mass tolerance 5 mDa, intensity threshold 2000 counts, mass window 0.05 Da, retention time window 0.20 min. In resulting database list, RT-m/z pairs represent an identifier of ion in the order of their elution time. The same value of RT and m/z in different batches of samples were regarded as the same compound. Multivariate statistical analysis was then performed to find the potential biomarkers that significantly contributed to the difference among the groups. During the analysis, principal component analysis (PCA) was firstly used to show the maximum variation and pattern recognition in order to get the overview and classification, and the orthogonal projections to latent structures discriminant analysis (OPLS-DA) was then performed aiming to get the maximum separation between two groups. S-plots was then available to provide visualization of the OPLS-DA predictive component loading to facilitate model interpretation. Variable importance for the projection (VIP) was also used to help screen the different components, and the metabolites with VIP value above 1.0 were considered as potential markers. Additionally, a permutation test was performed to provide reference distributions of the R2/Q2-values that could indicate the statistical significance. Simca 15.0 software (Umetrics, Malmö, Sweden) was used to show the analysis results.
3. Results and Discussion
3.1. Identification of Components from MCG and CG Based on UNIFI Platform
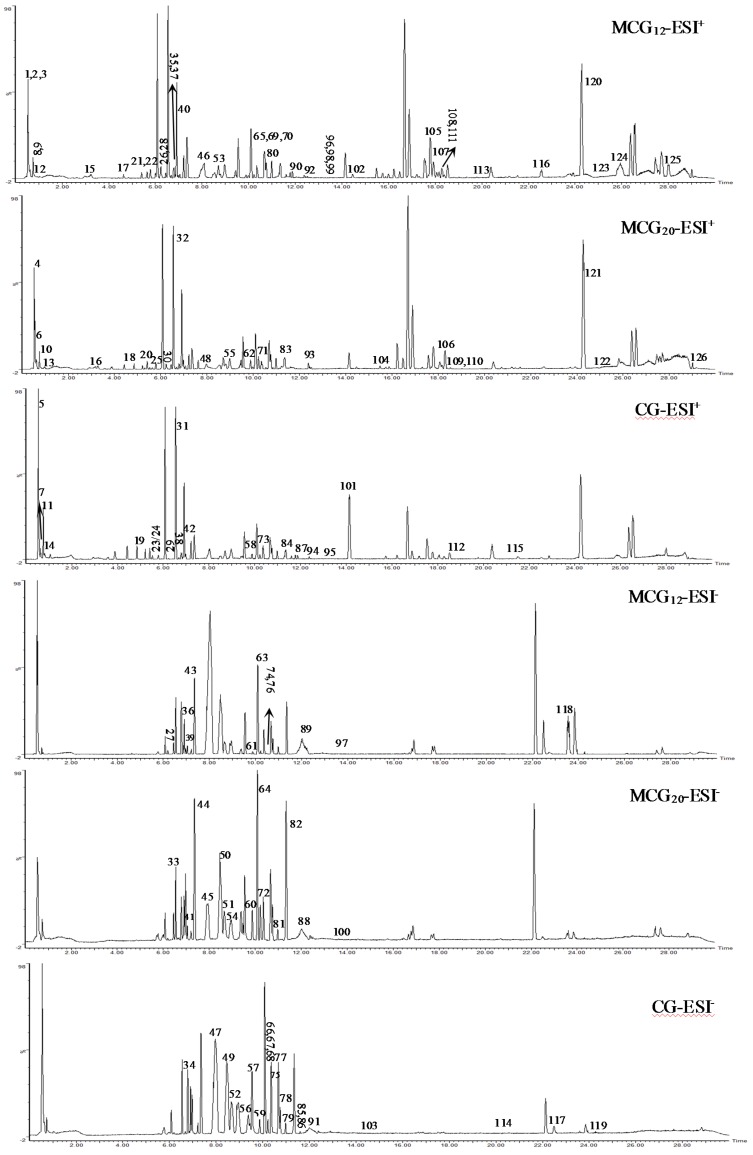
As a result of our analysis, a total of 126 compounds, including triterpenoids (the main ingredients), flavonoids, organic acids and organic acid esters, alcohol phenols, aldehyde ketones and amino acids, etc., were characterized or tentatively identified from the MCG and CG in both ESI+ and ESI− modes. 85 compounds were identified in ESI+ mode and 41 compounds were identified in ESI- mode. Base peak intensity (BPI) chromatograms are shown in Figure 1, the identification information is listed in Table 2, and the chemical structures are shown in Figure 2.
Figure 1.
The representative BPI chromatograms of CG and MCG in positive and negative modes.
Table 2.
Compounds identified from MCG and CG by UPLC-QTOF-MSE.
| No. | tR (min) | Formula | Calculated Mass (Da) | Theoretical Mass (Da) | Mass Error (ppm) | MSE Fragmentation | Identification | Sources | Ref. |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 0.49 | C5H8N2O5 | 176.0431 | 176.0433 | −1.5 | 177.0503[M + H]+; 130.0495[M – 2 × OH − NH2]+ | Dencichine | CG, MCG12, MCG20 | [23] |
| 2 | 0.54 | C6H14N4O2 | 174.1115 | 174.1117 | −1.1 | 175.1188[M + H]+;158.0912[M − NH2]+; 116.0704[M − NH2 − CN2H2]+;114.1015[M − NH2 − CHO2]+ | Adipodihydrazide | CG, MCG12, MCG20 | a |
| 3 | 0.55 | C30H22O10 | 542.1257 | 542.1213 | 8.1 | 543.1330[M + H]+; 273.0833[M − C15H10O5]+; 242.1025[M − OH − C15H10O6]+; 127.0388[M − C24H17O7]+; 116.0703[M − 2 × OH − C21H12O8]+; 109.0284[M − C24H15O8]+ | Chamaejasmine | CG, MCG12, MCG20 | a |
| 4 | 0.59 | C12H22O11 | 342.1156 | 342.1162 | −1.6 | 365.1055[M + Na]+; 203.0550[M − OH − C4H8O4]+; 185.0444[M − 2 × OH − C4H8O4]+ | α-Maltose | CG, MCG12, MCG20 | [24] |
| 5 | 0.69 | C6H14N4O2 | 174.1115 | 174.1117 | −0.9 | 175.1188[M + H]+ | d-Arginin | CG, MCG12, MCG20 | s |
| 6 | 0.74 | C19H18O11 | 422.0842 | 422.0849 | −1.7 | 423.0915[M + H]+; 268.1040[M + H − C6H6O5]+; 119.0349[M − C15H10O7]+ | Isomangiferin | CG, MCG12, MCG20 | [25] |
| 7 | 0.75 | C5H5N5 | 135.0546 | 135.0545 | 0.5 | 136.0618[M + H]+; 119.0352[M − NH2]+ | Adenine | CG, MCG12, MCG20 | s |
| 8 | 0.76 | C10H13N5O4 | 267.0974 | 267.0968 | 2.4 | 268.1050[M + H]+; 136.0618[M − C5H9O4]+; 119.0352[M − C5H9O4 − NH2]+ | d-Adenosine | CG, MCG12, MCG20 | s |
| 9 | 0.80 | C20H15NO6 | 365.0876 | 365.0899 | −6.3 | 366.0949[M + H]+ | Integriamide | CG, MCG12, MCG20 | a |
| 10 | 0.81 | C25H30O12 | 538.1677 | 538.1686 | −1.7 | 561.1569[M + Na]+; 393.1138[M − C6H8O4]+; 381.0788[M − CH3 − C7H10O3]+; 366.0930[M − OH − C8H10O3]+; 366.0930[M − C19H21O8]+ | Linearoside | CG, MCG12, MCG20 | [26] |
| 11 | 0.82 | C9H11NO2 | 165.0782 | 165.0790 | −0.5 | 166.0862[M + H]+; 120.0805[M − COOH]+; 103.0543[M − COOH − NH2]+ | Phenylpropionic acid | CG, MCG12, MCG20 | s |
| 12 | 0.91 | C6H9N3O2 | 155.0762 | 155.0695 | −3.5 | 156.0762[M + H]+ | Histidine | CG, MCG12, MCG20 | s |
| 13 | 1.05 | C11H12N2O2 | 204.0898 | 204.0899 | −0.5 | 205.0971[M + H]+; 188.0706[M − NH2]+; 143.0723[M − NH2 − COOH]+; 118.0649[M − NH2 − COOH − C2H3]+ | Tryptophan | CG, MCG12, MCG20 | s |
| 14 | 1.06 | C6H14N4O2 | 174.1117 | 174.1117 | 0.0 | 175.1190[M + H]+ | Argentine | CG, MCG12, MCG20 | [27] |
| 15 | 3.13 | C25H28O4 | 392.2009 | 392.1988 | 5.5 | 393.2082[M + H]+ | Glabrol | CG, MCG12, MCG20 | [28] |
| 16 | 3.27 | C25H24O5 | 404.1646 | 404.1624 | 5.1 | 405.1719[M + H]+ | Puerarol | CG, MCG12, MCG20 | [29] |
| 17 | 4.42 | C27H38O6 | 458.2716 | 458.2668 | 10.0 | 481.2609[M + Na]+; 436.2642[M − COOH]+ | Lucideric acid | CG, MCG12, MCG20 | a |
| 18 | 4.64 | C36H58O8 | 618.4107 | 618.4132 | −4.0 | 619.4180[M + H]+; 421.3446[M − Glc − OH]+ | β-d-Glcopyranosyl oleanolate | CG, MCG12, MCG20 | [30] |
| 19 | 4.89 | C48H82O19 | 962.5484 | 962.5450 | 3.3 | 985.5312[M + Na]+; 765.4795[M − Glc − OH]+; 541.2637[M − Glc − OH − C15H28O]+; 421.3463[M − Glc − Glc/Rha − 2 × OH]+ | Majoroside F6 | CG, MCG12, MCG20 | [31] |
| 20 | 5.21 | C31H46O8 | 546.3248 | 546.3193 | 9.7 | 569.3140[M + Na]+; 133.0859[M − C25H33O5]+ | Methyl ganoderate G | CG, MCG12, MCG20 | a |
| 21 | 5.53 | C35H48O9 | 612.3344 | 612.3298 | 7.4 | 613.3416[M + H]+; 582.3264[M − OCH3]+; 526.2986[M − C4H7O2]+ | Cinobufagin 3-hemisuberate methyl ester | CG, MCG12, MCG20 | a |
| 22 | 5.56 | C45H74O17 | 886.4877 | 886.4926 | −5.4 | 909.4769[M + Na]+; 745.4383[M − CH2OH − C6H10O2]+; 729.4136[M − OH − C8H14O2]+; 601.2768[M − 2 × OH − C16H26O4]+; 431.1870[M − Glc − C19H27O2]+ | Shatavarin IV | CG, MCG12, MCG20 | [32] |
| 23 | 5.64 | C25H32O13 | 540.1800 | 540.1843 | 3.0 | 541.1873[M + H]+; 347.0906[M − CH3O − C2H4 − C8H9O2]+; 195.1008[M − Glc − C2H4O2 − C6H5O2]+ | Oleuropein | CG, MCG12, MCG20 | [33] |
| 24 | 5.65 | C14H16O4 | 248.1025 | 248.1049 | −9.7 | 271.0917[M + Na]+; 195.1008[M − C2H4 − COH]+; 189.1348[M − OH − COOH]+ | Isohistiopterosin A | CG, MCG12, MCG20 | a |
| 25 | 5.79 | C45H74O17 | 886.4881 | 886.4926 | −5.0 | 909.4773[M + Na]+; 707.4360[M − Glc]+; 689.4262[M − Glc − OH]+; 657.3636[M − Glc − OH − 2 × CH3]+; 609.3646[M − Glc − C6H10O2]+; 523.3626[M − Glc/Glc − C3H6]+ | Malonylginsenoside Rf | CG, MCG12, ## MCG20 | [34] |
| 26 | 6.21 | C42H70O14 | 798.4778 | 798.4766 | 1.6 | 799.4851[M + H]+; 439.3563[M − Glc/Rha − 2 × OH]+; 421.3441[M − Glc/Rha − 3 × OH − H2O]+ | Ginsenoside Rg8 | CG, MCG12, MCG20 | [34] |
| 27 | 6.23 | C22H32O13 | 504.1840 | 504.1843 | −0.6 | 503.1767[M − H]−; 457.1715[M − OH − CH2OH]−; 293.0878[M − C2H4O − CH2OH − C8H9O2]− | Cistanoside H | CG, MCG12, MCG20 | [35] |
| 28 | 6.24 | C23H28O11 | 480.1594 | 480.1632 | −7.7 | 481.1667[M + H]+; 317.0803[M − C10H12O2]+ | Peoniflorin | CG, MCG12, MCG20 | a |
| 29 | 6.28 | C24H30O12 | 510.1702 | 510.1737 | −6.9 | 511.1775[M + H]+; 317.0803[M − C11H14O3]+ | Mudanpioside D | CG, MCG12, MCG20 | [36] |
| 30 | 6.39 | C54H92O24 | 1124.5943 | 1124.5978 | −3.1 | 1147.5835[M + Na]+; 585.2870[M − C25H47O12]+; 325.1130[M − C42H71O14]+ | Ginsenoside V | CG, MCG12, MCG20 | [31] |
| 31 | 6.49 | C48H82O19 | 962.5484 | 962.5450 | 3.3 | 985.5302[M + Na]+; 865.4789[M − C6H9O]+; 823.4787[M − C8H11O2]+; 805.4668[M − C8H13O3]+; 555.2763[M − C12H26O5]+; 423.3602[M − Glc − Glc/Glc − OH]+; 405.3507[M − Glc − Glc/Glc − 2 × OH]+ | Ginsenoside Re1 | CG, MCG12, ##,* MCG20 | [37] |
| 32 | 6.59 | C23H28O11 | 480.1587 | 480.1632 | −9.3 | 481.1660[M + H]+; 317.0810[M − C7H5O − C3H5O]+ | Mudanpioside I | CG, MCG12, MCG20 | [38] |
| 33 | 6.64 | C41H70O14 | 786.4762 | 786.4766 | −0.4 | 831.4744[M + HCOO]−; 653.4270[M − H − C5H8O4]−, 491.3710[M − H − C11H18O9]− | Notoginsenoside Rw2 | CG, MCG12, MCG20 | [39] |
| 34 | 6.67 | C47H80O18 | 932.5335 | 932.5345 | −1.0 | 977.5317[M + HCOO]−; 785.4693[M − Ara − CH3]−;653.4282[M − Glc − 2 × OH − C5H9]− | Quinquenoside F6 | CG, MCG12, MCG20 | [37] |
| 35 | 6.77 | C36H60O9 | 636.4217 | 636.4237 | −3.1 | 637.4290[M + H]+; 621.42740[M − OH]+; 423.3605[M − Glc − 2 × OH]+ | Ginsenoside Rh8 | CG, MCG12, MCG20 | [40] |
| 36 | 6.84 | C48H82O19 | 962.5469 | 962.5450 | 1.9 | 1007.5456[M + HCOO]−; 799.4848[M − Glc]−; 637.4317[M − Glc/Glc]−; 179.0545[Glc − H]− | 20-β-d-Glucopyranosyl-ginsenoside Rf | CG, MCG12, MCG20 | [41] |
| 37 | 6.80 | C42H70O13 | 782.4773 | 782.4816 | −5.4 | 805.4665[M + Na]+; 765.4734[M − OH]+; 677.4220[M − 2 × OH − C4H7O]+; 661.4265[M − 3 × OH − C4H7O]+; 439.3562[M − Glc − Man − OH]+ | Ginsenoside Rh14 | CG, MCG12, MCG20 | [40] |
| 38 | 6.82 | C17H24O8 | 356.1460 | 356.1472 | −3.1 | 379.1352[M + Na]+; 145.0495[M − OH − C11H13O3]+ | Erigeside II | CG, MCG12, MCG20 | [42] |
| 39 | 6.96 | C47H80O18 | 932.5410 | 932.5345 | 6.7 | 977.5392[M + HCOO]−; 799.4825[M − Xyl]−; 769.4724[M − H − Glc]−; 637.4291[M − (Glc/Xyl) ]−; 179.0539 [Glc − H]− | Notoginsenoside R1 | CG, MCG12, MCG20 | s |
| 40 | 6.99 | C28H44O12 | 572.2810 | 572.2833 | −3.9 | 573.2883[M + H]+; 555.2779[M − OH]+; 531.2860[M − C2H3O]+ | Picrasinoside G | CG, MCG12, MCG20 | a |
| 41 | 7.05 | C48H82O19 | 962.5425 | 962.5450 | −2.6 | 1007.5415[M + HCOO]−; 799.4822[M − Glc]−; 637.4333[M − (Glc/Glc) ]− | Notoginsenoside N | CG, MCG12, MCG20 | [43] |
| 42 | 7.20 | C48H82O19 | 962.5422 | 962.5450 | −2.9 | 985.5314[M + Na]+; 703.4371[M − Glc − 2 × OH − CH2OH]−; 439.3565[M − Glc − Glc/Glc − OH]− | Ginsenoside Re2 | CG, MCG12, ##,**MCG20 | [40] |
| 43 | 7.34 | C42H72O14 | 800.4934 | 800.4922 | 1.4 | 845.4916[M + HCOO]−; 637.4344[M − Glc]−; 475.3798[M − Glc − Glc]−; 179.0553[Glc − H]−; | Ginsenoside Rg1 | CG, MCG12, MCG20 | s |
| 44 | 7.36 | C48H82O18 | 946.5524 | 946.5501 | 2.3 | 991.5506[M + HCOO]−; 783.4912[M − Glc]−; 637.4344[M − (Glc/Rha)]−; 475.3798[M − Glc − (Glc/Rha)]− | Ginsenoside Re | CG, MCG12, MCG20 | s |
| 45 | 7.74 | C45H74O17 | 886.4925 | 886.4926 | −0.1 | 885.4853[M − H]−; 781.4740[M − HOCOCH2COOH]−; 619.4197[M − Glc(Mal)]−; 161.0438[Glc − H2O]− | Malonylginsenoside Rg1 | CG, MCG12, MCG20 | [39] |
| 46 | 7.93 | C48H76O19 | 956.4960 | 956.4981 | −2.2 | 979.4852[M + Na]+; 799.4161[M − CO2 − CH2OH − C6H12]+; 641.4008[M − Glc − C4H6O5]+;439.3562[M − Glc − Glc/Glc(mal)]+; 145.0493[Glc − OH]+ | Isomer of ginsenoside Ro | # CG, MCG12, MCG20 | [31] |
| 47 | 8.04 | C51H84O21 | 1032.5532 | 1032.5505 | 2.6 | 1031.5460[M − H]−; 987.5564[M − CO2]−; 927.5337[M − HOCOCH2COOH]−; 781.4759[M − Rha(Mal) ]−; 619.4222[M − (Rha(Mal)/Glc]− | Malonylginsenoside Re | CG, MCG12, MCG20 | [39] |
| 48 | 8.08 | C48H76O19 | 956.4950 | 956.4981 | −3.1 | 979.4842[M + Na]+; 817.4311[M − Glc]+; 439.3571[M − Glc/Glc − Glc − OH]+ | Isomer of ginsenoside Ro | CG, MCG12, MCG20 | [44] |
| 49 | 8.09 | C44H74O15 | 842.5032 | 842.5028 | 0.5 | 841.4959[M − H]−; 799.4861[M − CH2O]+; 781.4741[M − CH2O − OH]+; 637.4316[M − Xyl(mal)]+; 619.4228[M − Xyl(mal) − OH]+; 475.3798[M − Xyl(mal) − Glc]+; 179.0550[Glc − H]+; 161.0439[Glc − OH]+ | Yesanchinoside D | CG, MCG12, MCG20 | [45] |
| 50 | 8.10 | C45H74O17 | 886.4931 | 886.4926 | 0.6 | 885.4858[M − H]−; 781.4741[M − H − HOCOCH2COOH]−; 619.4228[M − H − Glc(Mal) ]−; 161.0439[Glc − H − H2O]− | Isomer of malonylginsenoside Rg1 | CG, MCG12, MCG20 | [39] |
| 51 | 8.49 | C41H70O13 | 770.4801 | 770.4816 | −1.5 | 815.4784[M + HCOO]−; 637.4321[M − Xyl]− | Notoginsenoside R2 | CG, MCG12, MCG20 | [39] |
| 52 | 8.50 | C56H94O24 | 1150.6124 | 1150.6135 | −1.1 | 1149.6051[M − H]−; 1119.5951[M − CH2OH − 2 × OH]−; 807.4861[M − Glc/Glc − OH]−; 605.4423[M − Glc/Glc − Glc(mal) ]−; 325.1119[Glc/Glc − OH]− | Quinquenoside R1 | CG, MCG12, ## MCG20 | [46] |
| 53 | 8.60 | C22H30O47 | 406.1957 | 406.1992 | −8.5 | 407.2030[M + H]+; 376.1859[M − OCH3]+ | Nigakilactone K | CG, MCG12, MCG20 | [47] |
| 54 | 8.87 | C48H82O19 | 962.5445 | 962.5450 | −0.5 | 1007.5427[M + HCOO]−; 797.4706[M − Glc]− | Ginsenoside Re3 | CG, MCG12, MCG20 | [37] |
| 55 | 8.96 | C56H92O25 | 1164.5929 | 1164.5928 | 0.1 | 1187.5821[M + Na]+; 1147.5803[M − OH]+; 805.4305[M − Ara/Glc − CH2OH − CH3]+; 443.3868[M − Ara/Glc − Glc/Glc(mal)]+ | Malonylginsenoside Rb2 | CG, MCG12, ##,* MCG20 | [44] |
| 56 | 9.41 | C59H100O27 | 1240.6488 | 1240.6452 | 2.8 | 1285.6740[M + HCOO]−; 945.5421[M − (Ara/Xyl) ]−; 913.5184[M − (Glc/Glc)]−; 783.4900[M − (Ara/Xyl) − Glc]− | Notoginsenoside R4 | CG, MCG12, MCG20 | s |
| 57 | 9.56 | C42H72O14 | 800.4921 | 800.4922 | −0.1 | 845.4903[M + HCOO]−; 637.4319[M − Glc]−; 475.3786[M − (Glc/Glc)]−; 1,3A2β221.0658; 161.0439[Glc – H − H2O]−;2,5A1β101.0235 | Ginsenoside Rf | CG, MCG12, MCG20 | s |
| 58 | 9.79 | C18H34O5 | 330.2398 | 330.2406 | −2.3 | 353.2290[M + Na]+; 213.1459[M + H – COOH – C5H11]+ | 12,13,15-Trihydroxy-9-octadecenoic acid | # CG, MCG12, MCG20 | [48] |
| 59 | 9.87 | C41H70O13 | 770.4809 | 770.4816 | −1.0 | 815.4791[M + HCOO]−; 475.3783[M − (Glc /Xyl)]−; 161.0437[Glc – H – H2O]− | Ginsenoside F5 | CG, MCG12, MCG20 | s |
| 60 | 9.89 | C60H102O28 | 1270.6635 | 1270.6558 | 5.9 | 1315.6617[M + HCOO]−; 841.4991[M − Glc/Glc – OH – C4H4]−; 769.4777[M − Glc/Glc/Glc – CH3]− | Ginsenoside Ra0 | CG, MCG12, MCG20 | [49] |
| 61 | 9.94 | C58H98O26 | 1210.6358 | 1210.6346 | 1.0 | 1255.6340[M + HCOO]−; 1077.5833[M – Xyl]−; 1047.5719[M – Glc]−; 955.4871[M – Glc – OH – C4H7]−; 783.4892[M – Glc/Xyl/Rha]− | Ginsenoside Ra2 | CG, MCG12, MCG20 | [50] |
| 62 | 10.00 | C22H22O10 | 446.1192 | 446.1213 | −4.5 | 469.1084[M + Na]+; 429.1154[M – OH]+; 385.0884[M – OH – CH3 – CH2OH]+; 341.0661[M – C4H8O3]+; 237.0746[M – C10H13O5]+; 193.0483[M – C12H16O6]+ | Glycitin | CG, MCG12, MCG20 | a |
| 63 | 10.01 | C59H100O27 | 1240.6462 | 1240.6452 | 0.8 | 1285.6444[M + HCOO]−; 1107.5964[M-Xyl]−; 945.5424[M – (Glc/Xyl)]−; 783.4912[M – Xyl – GlcGlc]− | Notoginsenoside Fa | CG, MCG12, MCG20 | [50] |
| 64 | 10.05 | C54H92O23 | 1108.6101 | 1108.6029 | 6.2 | 1153.6083[M + HCOO]−; 945.5437[M – Glc]−; 783.4888[M – (Glc/Glc)]−; 621.4382[M – (Glc/Glc) – Glc]−; 459.3835[M – (Glc/Glc) – (Glc/Glc)]−; 2,5A1β101.0235 | Ginsenoside Rb1 | CG, MCG12, MCG20 | s |
| 65 | 10.10 | C42H70O12 | 766.4863 | 766.4867 | −0.5 | 767.4936[M + H]+; 443.3866[M – Rha – Glc]+; 425.3762[M – Rha – Glc – OH]+ | Ginsenoside Rg4 | CG, MCG12, MCG20 | [34] |
| 66 | 10.20 | C57H94O26 | 1194.6087 | 1194.6033 | 4.5 | 1193.6015[M – H]−; 1149.6098[M – CO2]−; 783.4908[M – Glc/Glc)]−; 179.0545[Glc – H]− | Isomer of malonylginsenoside Rb1 | CG, MCG12, ##,** MCG20 | [39] |
| 67 | 10.22 | C42H72O13 | 784.4997 | 784.4973 | 2.9 | 829.4979[M + HCOO]−; 637.4336[M − Rha]−; 475.3809[M – (Glc/Rha)]−; 161.0449 [Rha – H]− | 20(R)-Ginsenoside Rg2 | CG, MCG12, MCG20 | s |
| 68 | 10.25 | C36H62O9 | 638.4407 | 638.4394 | 2.9 | 683.4389[M + HCOO]−; 161.0449[Glc − H − H2O]− | Ginsenoside Rh1 | CG, MCG12, MCG20 | s |
| 69 | 10.27 | C41H70O13 | 770.4779 | 770.4816 | −4.7 | 793.4672[M + Na]+; 587.4276[M − Ara(p) − 2 × OH]+; 423.3589[M − Ara(p)/Glc − 2 × OH]+ | Ginsenoside F3 | CG, ∆∆ MCG12, MCG20 | [34] |
| 70 | 10.29 | C36H60O8 | 620.4292 | 620.4288 | 0.7 | 621.4365[M + H]+; 390.2277[M − C17H26]+; 187.1473[M − OH − Glc − C16H24O]+ | Ginsenoside Rh4 | CG, MCG12, MCG20 | [40] |
| 71 | 10.32 | C53H90O22 | 1078.5939 | 1078.5924 | 1.3 | 1101.5805[M + Na]+; 939.5312[M − Glc]+; 929.5452[M − Ara(f)]+; 789.4784[M − Ara(f) − Glc]+ | Ginsenoside Rc | CG, MCG12, MCG20 | s |
| 72 | 10.34 | C58H98O26 | 1210.6356 | 1210.6346 | 0.7 | 1255.6338[M + HCOO]−; 1077.5851[M − Xly]−; 1047.5702[M − Glc]−; 945.5396[M − (Xly/ Ara(p))]−; 621.4323[M − (Xly/ Ara(p)/Glc − Glc]− | Ginsenoside Ra1 | CG, MCG12, MCG20 | [50] |
| 73 | 10.38 | C42H70O12 | 766.4872 | 766.4867 | 0.6 | 767.4945[M + H]+;605.4423[M − Glc]+;443.3870[M − Glc/Xyl]+;407.3660[M − Glc − 2 × OH]+; 163.0591[Glc − OH]+;145.04901[Glc − OH − H2O]+ | Ginsenoside Rg5 | CG, MCG12, MCG20 | s |
| 74 | 10.47 | C56H92O25 | 1164.5947 | 1164.5928 | 1.6 | 1163.5874[M − H]−; 1119.5961[M − CO2]−; 927.5320[M − Ara(f) − HOCOCH2COOH]− | Malonylginsenoside Rc | CG, MCG12, MCG20 | [44] |
| 75 | 10.51 | C48H76O19 | 956.5001 | 956.4981 | 2.1 | 955.4928[M − H]−; 793.4399[M − Glc]−; 613.3739[M − Glc − Glc − OH]− | Ginsenoside Ro | # CG, MCG12, MCG20 | s |
| 76 | 10.57 | C57H94O26 | 1194.6059 | 1194.6033 | 2.2 | 1193.5986[M − H]−; 1149.6062[M − CO2]−; 1089.5851[M − HOCOCH2COOH]−; 945.5428[M − Glc(Mal)]−; 783.4926[M − (Glc/Glc)]− | Malonylginsenoside Rb1 | CG, MCG12, MCG20 | [39] |
| 77 | 10.63 | C53H90O22 | 1078.5979 | 1078.5924 | 4.9 | 1123.5961[M + HCOO]−; 945.5448[M − Ara(p)]−; 783.4896[M − (Ara/Glc)]−; 149.0443[Ara(p) − H]− | Ginsenoside Rb2/Rb3 | CG, MCG12, MCG20 | s |
| 78 | 10.77 | C56H92O25 | 1164.5986 | 1164.5928 | 5.0 | 1163.5913[M − H]−; 1101.5822[M − CO2]−; 765.4782[M − H − Glc(Mal) − Ara(p) − OH]− | Malonylginsenoside Rb2 | CG, MCG12, MCG20 | [44] |
| 79 | 11.06 | C36H62O9 | 638.4391 | 638.4394 | −0.4 | 683.4373[M + HCOO]− | 20(R)-Ginsenoside Rh1 | CG, MCG12, MCG20 | s |
| 80 | 11.14 | C36H62O9 | 638.4399 | 638.4394 | 0.7 | 661.4291[M + Na]+; 376.2462[M − C17H24O2]+ | Ginsenoside F1 | CG, MCG12, MCG20 | s |
| 81 | 11.15 | C56H92O25 | 1164.5971 | 1164.5928 | 3.7 | 1163.5898[M − H]−; 1119.6000[M − CO2]−; 1059.5772[M − H − C3H4O4]−; | Malonylginsenoside Rb3 | CG, MCG12, MCG20 | [39] |
| 82 | 11.27 | C48H82O18 | 946.5482 | 946.5501 | −1.9 | 991.5464[M + HCOO]−; 783.4878[M − Glc]−; 621.4350[M − (Glc/Glc)]−; 161.0435[Glc − H]− | Ginsenoside Rd | CG, MCG12, MCG20 | s |
| 83 | 11.31 | C55H92O23 | 1120.6049 | 1120.6029 | 1.7 | 1143.5941[M + Na]+; 831.4874[M − Glc(mal)]− | Ginsenoside Rs1 | CG, MCG12, ##,* MCG20 | s |
| 84 | 11.36 | C42H70O12 | 766.4875 | 766.4867 | 1.0 | 767.4947[M + H]+; 605.4423[M − Rha]+; 587.4300[M − Rha − OH]+; 569.4211[M − Rha − 2 × OH]+; 443.3866[M − Rha/Glc]+; 425.3769[M − Rha/Glc − OH]+; 145.0491[Rha − H − H2O]+ | Ginsenoside Rg6 | CG, MCG12, MCG20 | [44] |
| 85 | 11.42 | C51H84O21 | 1032.5515 | 1032.5505 | 0.9 | 1131.5442[M − H]−; 765.4785[M − Glc(mal) − OH]−; 621.4372[M − (Glc/Glc(mal)]− | Malonylginsenoside Rd | CG, MCG12, MCG20 | [45] |
| 86 | 11.53 | C55H92O23 | 1120.6065 | 1120.6029 | 3.0 | 1165.6047[M + HCOO]−; 1077.5851[M − Ac]−; 1059.5745[M − Ac − OH]− | Ginsenoside Rs2 | CG, MCG12, MCG20 | s |
| 87 | 11.69 | C42H70O13 | 782.4738 | 782.4816 | −9.7 | 805.4631[M + Na]+; 621.4354[M − Glc]+; 311.0902[Glc/Glc − CH2OH]+ | Ginsenoside Rg10 | CG, MCG12, MCG20 | a |
| 88 | 11.79 | C48H82O18 | 946.5494 | 946.5501 | −0.7 | 991.5476[M + HCOO]−; 927.5308[M − OH]−; 783.4926[M − Glc]−; 621.4412[M − (Glc/Glc) ]− | Gypenoside XVII | CG, MCG12, MCG20 | s |
| 89 | 11.81 | C51H84O21 | 1032.5504 | 1032.5505 | −0.1 | 1031.5431[M − H]−; 987.5535[M − CO2]−; 621.4412[M − (Glc/Glc(mal))]−; 179.0546[Glc − H]− | Isomer of malonylginsenoside Rd | CG, MCG12, MCG20 | [49] |
| 90 | 11.88 | C48H82O18 | 946.5476 | 946.55021 | −2.6 | 969.5368[M + Na]+; 605.4394[M − Glc/Glc]+; 587.4312[M − Glc/Glc − OH]+; 425.3744[M − Glc/Glc − Glc]+; 407.3661[M − Glc/Glc − OH − Glc]+ | Chikusetsusaponin FK1 | CG, MCG12, MCG20 | [40] |
| 91 | 12.18 | C47H80O17 | 916.5398 | 916.5396 | 0.2 | 961.5380[M + HCOO]−; 783.4870[M − Xyl]−; 621.4388[M − (Xyl/glc)]− | Notoginsenoside Fe | CG, MCG12, MCG20 | s |
| 92 | 12.39 | C50H84O19 | 988.5565 | 988.5607 | −4.1 | 1011.5458[M + Na]+; 831.4819[M − Glc]+; 425.3763[M − Glc/Glc(ace) − Glc]+ | Quinquenoside III | CG, MCG12, MCG20 | [51] |
| 93 | 12.45 | C47H80O17 | 916.5376 | 916.5396 | −2.1 | 939.5268[M + Na]+; 789.4754[M − 2 × OH − CH6O3]+ | Vinaginsenoside R16 | CG, MCG12, MCG20 | [40] |
| 94 | 12.59 | C47H80O17 | 916.5361 | 916.5396 | −3.7 | 939.5253[M + Na]+; 407.3672[M − Glc − (Glc/Xyl) − OH]+ | Gypenoside IX | CG, MCG12, MCG20 | [52] |
| 95 | 12.91 | C50H84O19 | 988.5569 | 988.5607 | −3.8 | 1011.5461[M + Na]+; 789.4784[M − Glc − 2 × OH]+ | Quinquenoside III isomer | CG, MCG12, MCG20 | [51] |
| 96 | 13.29 | C52H86O19 | 1014.5753 | 1014.5763 | −1.0 | 1037.5645[M + Na]+; 857.5032[M − C4H8O4 − 2 × OH]+; 393.1376[Glc/Glc(ace) − OH]+ | Quinquenoside I | CG, MCG12, MCG20 | [53] |
| 97 | 13.34 | C42H72O13 | 784.4984 | 784.4973 | 1.4 | 829.4966[M + HCOO]−; 621.4373[M − Glc]−; 161.0437[Glc − H − H2O]− | Ginsenoside F2 | CG, MCG12, MCG20 | s |
| 98 | 13.55 | C42H72O13 | 784.4977 | 784.4973 | 0.5 | 807.4869[M + Na]+; 605.4402[M − Glc]+; 587.4286[M − Glc − OH]+; 425.3765[M − Glc/Glc − OH]+; 407.3659[M − Glc/Glc − 2 × OH]+ | 20(R)-Ginsenoside Rg3 | CG, MCG12, MCG20 | s |
| 99 | 13.57 | C17H24O2 | 260.1774 | 260.1776 | −0.8 | 261.1847[M + H]+ | Panaxydol | ∆∆,## CG, MCG12, MCG20 | [54] |
| 100 | 13.77 | C42H66O14 | 794.4464 | 794.4453 | 1.4 | 793.4391[M − H]−; 731.4375[M − CO2 − OH]−; 613.3746[M − Glc]− | Chikusetsusaponin Iva | CG, MCG12, MCG20 | [51] |
| 101 | 14.02 | C52H86O19 | 1014.5750 | 1014.5763 | −1.3 | 1037.5642[M + Na]+; 789.4732[M − Glc − C2H4O2]+ | Isomer of Quinquenoside I | CG, MCG12, MCG20 | [51] |
| 102 | 14.38 | C17H26O3 | 278.1879 | 278.1882 | −1.1 | 279.1952[M + H]+ | Panaxtriol | CG, MCG12, MCG20 | [55] |
| 103 | 14.46 | C42H72O13 | 784.4970 | 784.4973 | −0.3 | 829.4966[M + HCOO]−; 621.4373[M − Glc]−; 407.3672[M − Glc/Glc − 2 × OH]+ | 20(S)-Ginsenoside Rg3 | CG, MCG12, MCG20 | s |
| 104 | 15.05 | C18H34O4 | 314.2444 | 314.2457 | −3.8 | 337.2336[M + Na]+ | Dibutyl sebacate | CG, MCG12, MCG20 | a |
| 105 | 17.90 | C16H22O4 | 278.1516 | 278.1518 | −0.7 | 301.1408[M + Na]+; 149.0230[M − C4H9 − C4H9O]+ | n-Butyl isobutyl phthalate | CG, MCG12, MCG20 | a |
| 106 | 17.93 | C30H52O4 | 476.3856 | 476.3866 | −2.2 | 499.3747[M + Na]+; 441.3728[M − 2 × OH]+; 423.3590[M − 3 × OH]+; 317.2049[M − 2 × CH3 − C8H15O]+ | 20(R)-Protopanaxatriol | CG, MCG12, MCG20 | [56] |
| 107 | 17.95 | C16H30O2 | 254.2246 | 254.2268 | 8.2 | 277.2161[M + Na]+ | Palmitoleic acid | CG, ∆,** MCG12, MCG20 | s |
| 108 | 18.07 | C19H18O3 | 294.1258 | 294.1256 | 0.5 | 317.1150[M + Na]+ | Tashinone IIA | CG, MCG12, MCG20 | [57] |
| 109 | 18.08 | C30H48O4 | 472.3546 | 472.3553 | −1.4 | 495.3438[M + Na]+ | β-Amyrone | CG, MCG12, MCG20 | [58] |
| 110 | 18.08 | C6H6O3 | 126.0331 | 126.0317 | 9.4 | 149.0223[M + Na]+ | Pyrogallol | CG, MCG12, MCG20 | a |
| 111 | 18.09 | C30H48O4 | 472.3546 | 472.3553 | −1.8 | 495.3438[M + Na]+ | 24-Hydroxyoleanolic acid | CG, ∆∆,** MCG12, MCG20 | [59] |
| 112 | 18.09 | C24H38O5 | 406.2720 | 406.2719 | 0.3 | 429.2613[M + Na]+; 319.1950[M − CH3 − C4H7O]+; 261.2213[M − 2 × C2H4O2 − C2H3]+; | Vitetrifolin | CG, MCG12, MCG20 | a |
| 113 | 20.14 | C31H46O2 | 450.3535 | 450.3498 | 8.0 | 473.3428[M + Na]+; 430.2889[M − C3H7]+ | Vitamin K1 | CG, MCG12, MCG20 | [60] |
| 114 | 20.97 | C18H30O2 | 278.2224 | 278.2252 | −7.9 | 277.2151[M − H]−; 232.2172[M − COOH]− | α-Linolenic acid | ∆∆,## CG, MCG12, MCG20 | [61] |
| 115 | 21.18 | C21H38O4 | 354.2758 | 354.2770 | −3.1 | 377.2650[M + Na]+ | β-Monolinolein | CG, MCG12, MCG20 | [62] |
| 116 | 22.11 | C18H32O | 264.2452 | 264.2453 | −0.5 | 265.2525[M + H]+; 149.1320[M − CH2 − C6H12O]+; 135.1166[M − CH2 − C7H13O]+; 121.1008[M − CH2 − C8H15O]+; 109.1010[M − C8H15O − C2H3]+ | (Z)-9,17-Octadecadienal | CG, MCG12, MCG20 | [63] |
| 117 | 22.49 | C18H32O2 | 280.2386 | 280.2402 | −5.9 | 279.2313[M − H]−; 234.2325[M − COOH]− | Linoleic acid | ∆∆,## CG, MCG12, MCG20 | s |
| 118 | 23.85 | C14H20O2 | 220.1478 | 220.1463 | 5.6 | 265.1460[M + HCOO]− | Thymyl isobutyrate | CG, MCG12, MCG20 | [64] |
| 119 | 24.25 | C18H34O2 | 282.2541 | 282.2559 | −6.3 | 281.2468[M − H]−; 236.2481[M − COOH]− | 9-Octadecenoic acid | ∆∆,## CG, MCG12, MCG20 | a |
| 120 | 24.40 | C36H62O8 | 622.4454 | 622.4445 | 1.6 | 623.4527[M + H]+; 316.2842[M − OH − Glc − C8H14]+; | Compound K | CG, MCG12, MCG20 | [40] |
| 121 | 24.89 | C40H56O4 | 600.4219 | 600.4179 | 6.7 | 601.4292[M + H]+; 557.4021[M − C2H4]+ | Violaxanthin | CG, MCG12, MCG20 | [65] |
| 122 | 25.31 | C20H38O2 | 310.2862 | 310.2872 | −3.2 | 311.2935[M + H]+; 277.1995[M − C6H13]+ | Ethyloleate | CG, MCG12, MCG20 | a |
| 123 | 25.35 | C40H56O4 | 600.4212 | 600.4179 | 5.6 | 601.4285[M + H]+; 497.3800[M − OH − C4H8O2]+ | Neoxanthine | CG, MCG12, MCG20 | [65] |
| 124 | 26.38 | C24H38O4 | 390.2758 | 390.2770 | −2.8 | 413.2653[M + Na]+; 301.1406[M − 3 × C2H5]+; 189.0153[M − C2H5 − C4H9 − C8H17]+; 167.0327[M − 2 × C2H17]+ | Bis(2-ethylhexyl) phthalate | CG, MCG12, MCG20 | a |
| 125 | 28.01 | C30H46O5 | 486.3334 | 486.3345 | −2.2 | 509.3226[M + Na]+ | Quillaic acid | CG, MCG12, MCG20 | s |
| 126 | 29.04 | C5H8O2 | 100.0512 | 100.0524 | −10.0 | 123.0404[M + Na]+ | Pentanedial | CG, MCG12, MCG20 | [66] |
s Identified with standard. a Compared with spectral data obtained from Wiley Subscription Services, Inc. (USA). ∆, ∆∆: Represented the content either in CG4–6 years group or in MCG12 years group was significantly higher than the other one (∆ p < 0.05, ∆∆ p < 0.001) #,##: Represented the content either in CG4–6 years group or in MCG20 years group was significantly higher than the other one (# p < 0.05, ## p < 0.001) *, **: Represented the content either in MCG12 years group or in MCG20 years group was significantly higher than the other one (* p < 0.05, ** p < 0).
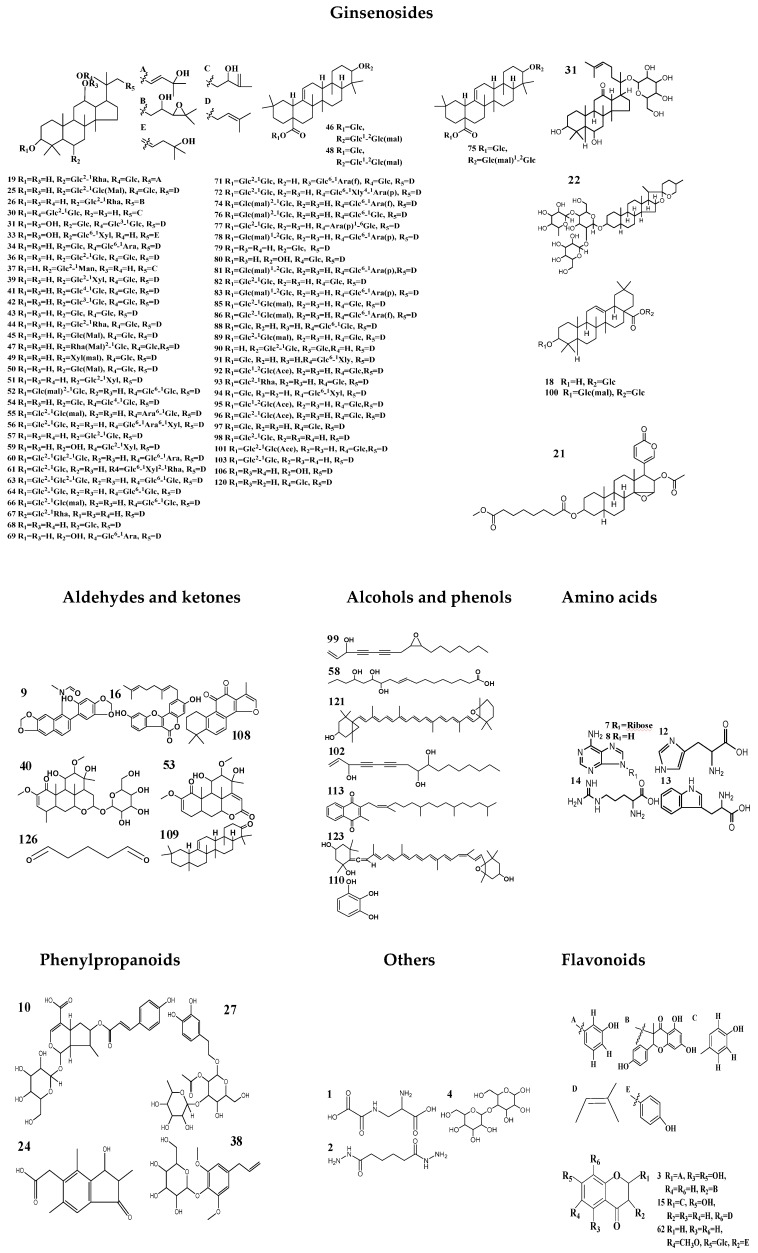
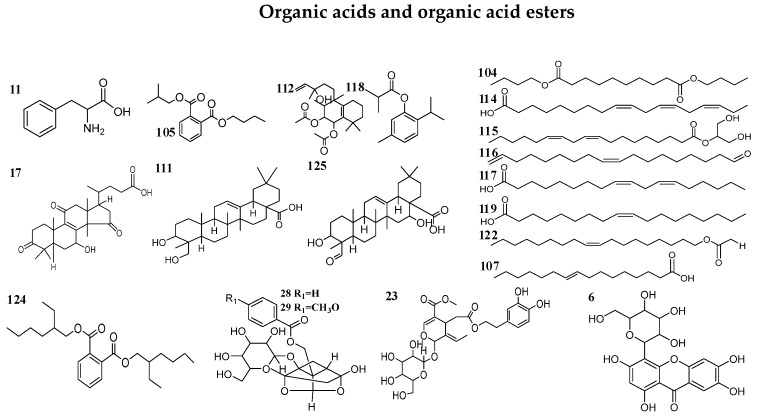
Figure 2.
Chemical structures of compounds identified in MCG and CG.
For the isomers, they could be compared with the retention time of the standards or distinguished by the characteristic MS fragmentation patterns reported in literature. Taking compounds 82 and 88 as example, both of them had the same protonated ion [M + HCOO]− at m/z 991.5464 and 991.5476. In a result, one of them was identified as ginsenoside Rd due to the same retention time, and the other one was tentatively identified as gypenoside XVII because it was matched with the characteristic MS fragmentation pattern of gypenoside XVII reported in the literature [31].
3.2. Biomarker Discovery for Distinguishing MCG and CG
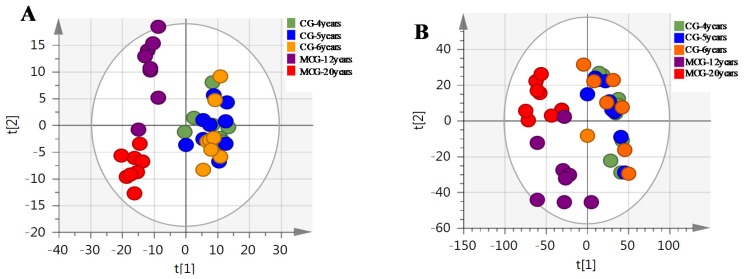
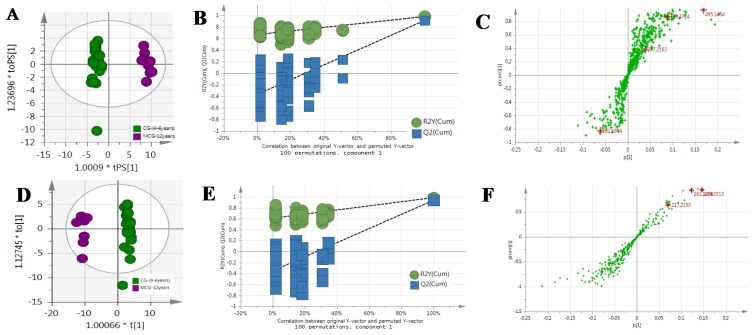
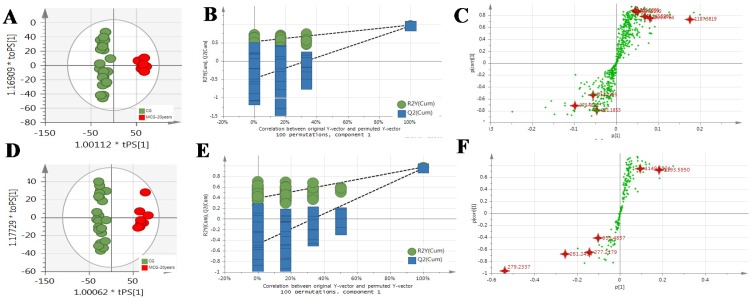
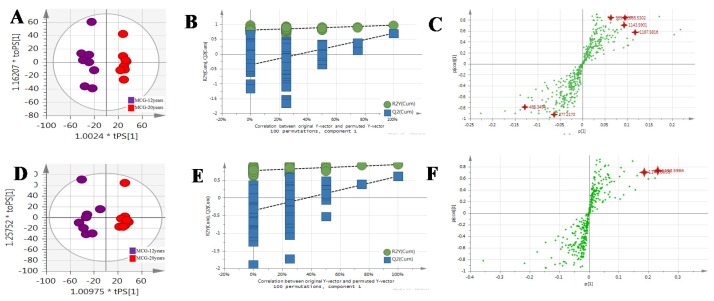
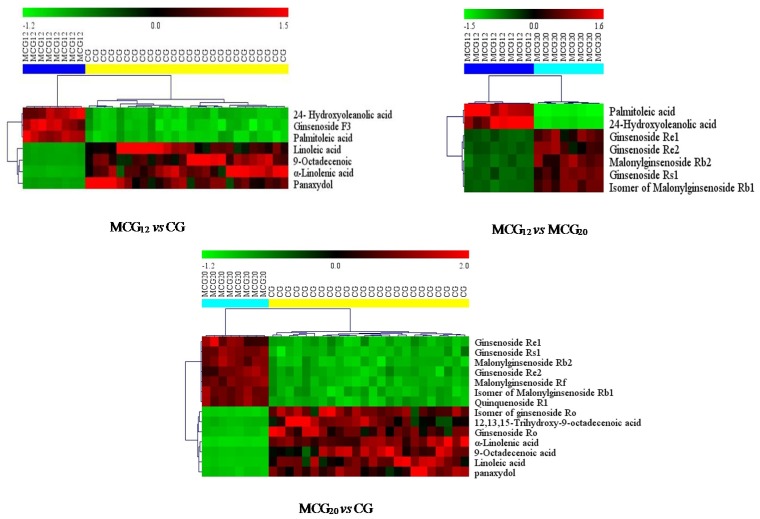
The MSE data of CG and MCG samples were statistically analyzed via PCA and OPLS-DA. As seen in PCA 2D plots (Figure 3), there was no obvious difference among of 4–6-year-old CG samples, but the MCG20 years, MCG12 years and CG4–6 years groups were obviously separated, indicating that these three groups could be differentiated. With the aim of distinguishing MCG from CG, or MCG20 years from MCG12 years, OPLS-DA plot, permutation test, and S-plot, VIP values were obtained to understand which variables were responsible for the separation (Figure 4, Figure 5 and Figure 6). The variables showing VIP > 1 and p < 0.05 (in t-test) were considered as potential biomarkers. The robust known biomarkers enabling the differentiation between CG and MCG were discovered and marked in S-plots. In order to systematically evaluate the biomarkers, heatmaps (Figure 7) were generated from these biomarkers. The hierarchical clustering heatmaps, intuitively visualizing the differential levels of potential biomarkers concentration in different ginseng groups, are shown in Figure 7. The larger contents were represented by red squares and smaller values by green squares.
Figure 3.
The PCA of CG and MCG in positive mode (A) and negative mode (B).
Figure 4.
The OPLS-DA/Permutation test/S-Plot of CG4–6 years and MCG12 years in positive mode (A/B/C) and negative mode (D/E/F).
Figure 5.
The OPLS-DA/Permutation test/S-Plot of CG4–6 years and MCG20 years. in positive mode (A/B/C) and negative mode (D/E/F).
Figure 6.
The OPLS-DA/Permutation test/S-Plot of MCG12 years and MCG20 years in positive mode (A/B/C) and negative mode (D/E/F).
Figure 7.
The heatmaps visualizing the intensities of potential biomarkers.
Between the CG4–6 years and MCG12 years groups, the contents of 24-hydroxyoleanolic acid, ginsenoside F3 and palmitoleic acid in MCG12 samples were significantly higher. While, the contents of α-linolenic acid, 9-octadecenoic acid, linoleic acid and panaxydol in all the CG samples were significantly higher.
Between the CG4–6 years and MCG20 years groups, the contents of ginsenoside Re1, -Re2, -Rs1, malonylginsenoside Rb2, -Rf, isomer of malonylginsenoside-Rb1 and quinquenoside R1 in the samples of MCG20 years were higher. On the contrary, the contents of ginsenoside Ro and the isomer of ginsenoside Ro, 12,13,15-trihydroxy-9-octadecenoic acid, linoleic acid, 9-octadecenoic acid, α-linolenic acid, panaxydol were rather higher in CG samples.
Between the MCG12 years and MCG20 years groups, the contents of palmitoleic acid and 24-hydroxyoleanolic acid in MCG12 years samples were significantly high, while the contents of ginsenoside Re1, -Rs1, malonylginsenoside Rb2, -Re2 and isomer of malonylginsenoside Rb1 were rather higher in MCG20 years samples.
Overall, on one hand, the contents of α-linolenic acid, linoleic acid, 9-octadecenoic acid and panaxydol in CG samples were significantly higher than those in all MCG samples. On the other hand, ginsenoside Re1, -Re2, -Rs1, malonylginsenoside Rb2 and isomer of malonylginsenoside Rb1 in MCG20 years samples were really higher than those both in MCG12 years and in all of CG samples, but there is no significant difference between MCG12 years and CG4–6 years samples. The summary with variable identity, VIP and p value were shown in Table 3.
Table 3.
The summary table with variable identity, VIP and p value.
| Groups for Comparison | Marker’ Name | VIP Value | p Value | |
|---|---|---|---|---|
| CG4–6 years vs. MCG12 years | CG4–6 years | α-linolenic acid | 1.23 | <0.001 |
| 9-octadecenoic acid | 2.17 | <0.001 | ||
| linoleic acid | 2.57 | <0.001 | ||
| panaxydol | 1.49 | <0.001 | ||
| MCG12 years | 24-hydroxyoleanolic acid | 4.13 | <0.001 | |
| ginsenoside F3 | 2.15 | <0.001 | ||
| palmitoleic acid | 1.54 | 0.037 | ||
| CG4–6 years vs. MCG20 years | MCG20 years | ginsenoside Re1 | 1.60 | <0.001 |
| ginsenoside Re2 | 1.75 | <0.001 | ||
| ginsenoside Rs1 | 1.59 | <0.001 | ||
| malonylginsenoside Rb2 | 4.10 | <0.001 | ||
| ginsenoside Rf | 1.83 | <0.001 | ||
| isomer of malonylginsenoside Rb1 | 2.30 | <0.001 | ||
| quinquenoside R1 | 1.21 | <0.001 | ||
| CG4-6 years | ginsenoside Ro | 1.39 | 0.017 | |
| isomer of ginsenoside Ro | 2.31 | 0.022 | ||
| 12,13,15-trihydroxy-9-octadecenoic acid | 1.25 | 0.003 | ||
| linoleic acid | 7.08 | <0.001 | ||
| 9-octadecenoic acid | 3.45 | <0.001 | ||
| α-linolenic acid | 1.86 | <0.001 | ||
| panaxydol | 1.12 | <0.001 | ||
| MCG12 years vs. MCG20 years | MCG12 years | palmitoleic acid | 2.07 | <0.001 |
| 24-hydroxyoleanolic acid | 3.26 | <0.001 | ||
| MCG20 years | ginsenoside Re1 | 1.16 | 0.002 | |
| ginsenoside Rs1 | 1.89 | 0.024 | ||
| malonylginsenoside Rb2 | 2.76 | 0.026 | ||
| ginsenoside Re2 | 1.60 | <0.001 | ||
| isomer of malonylginsenoside-Rb1 | 3.87 | <0.001 | ||
4. Discussion
Although MCG and CG both belong to Panax ginseng, their chemical ingredients and pharmacological activities are different due to their significantly different growth environment [3,67]. As we all know, MCG has been regarded as a replacement of wild ginseng. Recently, the UPLC-QTOF-MS/MS-based approach has been developed to distinguish MCG (grown for 15 years) and CG (grown for 4–7 years) [6]. As a result, 40 ginsenosides in both MCG and CG were unambiguously identified and tentatively assigned, and the potential chemical markers identifying different ginseng products were characterised [6]. Additionally, the study on 6–18-year-old Mountain Cultivated Ginseng Leaves (MGL) samples showed that the MGL were obviously divided into three main groups according to different age brackets (6~10, 11~13 and 14~18 years) [7]. Although the sample of the study was the leaf of MCG, it could be indirectly speculated that the MCG roots with different cultivation ages are also different. In order to further systematically compare the similarities and differences at the chemical level between different ages of ginseng, especially to compare the younger or the older MCG, 4, 5, 6-year-old CG and 12, 20-year-old MCG were chosen as the analytical samples in the present study.
Firstly, based on UNIFI platform, intelligent and automatic workflows, the screening analysis of metabolites in different cultivation ages of ginseng were rapidly performed. As a result, a total of 126 compounds were characterized from CG4–6 years, MCG12 years and MCG20 years samples. Among of them, ginsenosides were the main ingredients. Both CG and MCG had the similar chemical composition, but the components were variously distributed in CG and MCG samples at different contents. That means in CG and MCG, the secondary metabolites had the features of structural diversity and the different content patterns. As far as we know, this is the first time that the comprehensive screening analysis of MCG12 years and MCG20 years samples by using UPLC-QTOF-MSE combined with UNIFI platform. It could provide the scientific data for clarifying the chemical composition of MCG.
Secondly, the combination of LC-MS based metabolomic profiling with multivariate statistical analysis method was used to profile the CG, MCG12 years and MCG20 years samples. A total of 17 potential age-dependent markers enabling differentiation among the CG and MCG samples were discovered. (1) There were four robust markers including α-linolenic acid, 9-octadecenoic acid, linoleic acid and panaxydol being the characteristic components for CG samples, that distinguished them from both MCG12 years and MCG20 years samples. The results showed that CG samples contained more non-ginsenosides. Both linoleic acid and α-linolenic acid, the main products of the acetate-malonate pathway, are two essential fatty acids necessary for health. Linoleic acid is used in the biosynthesis of arachidonic acid and thus some prostaglandins, leukotrienes, and thromboxane [68,69]. Panaxydol, one of the C17 polyacetylenic compounds, originates from acetyl-CoA/malonyl-CoA via fatty acids with crepenynate as the intermediate [70]. It is considered a potential antitumor agent due to its significant anticancer activity [71]. (2) In CG samples, there were three other characteristic components such as ginsenoside Ro, the isomer of ginsenoside Ro, and 12,13,15-trihydroxy-9-octadecenoic acid, that could be used to differentiate them from MCG20 years samples. From this, we could draw a conclusion that pentacyclic triterpenoids decreased significantly in older MCG samples. (3) Five robust biomarkers including ginsenoside Re1, -Re2, -Rs1, malonylginsenoside Rb2 and isomer of malonylginsenoside Rb1 were found to enable differentiation of MCG20 years from CG and MCG12 years samples. These five compounds might be used for rapid identification of MCG20 years samples. A proposed biosynthetic pathway of ginsenosides is as follows: with the action of squalene epoxidase, squalene was converted to 2,3-oxidosqualene. Dammaranes can be synthesized by dammarenediol synthase, and oleananes by β-amyrin synthase [72]. Ginsenosides were found to have both antimicrobial and antifungal properties and the molecules are naturally bitter-tasting, discouraging insects and other animals from consuming the plant, so ginsenosides likely serve as mechanisms for plant defense [73,74]. (4) In MCG20 years samples, another two markers, ginsenoside Rf and quinquenoside R1, were discovered that distinguished them from all CG samples. (5) In MCG12 years samples, 24-hydroxyoleanolic acid and palmitoleic acid were the two robust markers for distinguished from both CG and MCG20 years samples. These two compounds might be used for rapid identification of MCG12 years samples. Palmitoleic acid is biosynthesized from palmitic acid by the action of the enzyme stearoyl-CoA desaturase-1, a key enzyme in fatty acid metabolism [75]. (6) Ginsenoside F3 was another marker for MCG12 years samples that differentiated them from CG samples. However, there are still some unresolved issues. For example, as shown in BPI chromatograms, though 126 compounds were identified, there are still some unidentified components. there are still some unidentified components. Further research should be carried out based on the formula of these unknown compounds.
5. Conclusions
By combining the UPLC-Q/TOF-MSE and UNIFI platform, 126 chemical components with various structural types, such as triterpenoids, flavonoids, organic acids and organic acid esters, etc., were characterized or tentatively identified from CG4–6 years, MCG12 years and MCG20 years samples for the first time. All the CG and MCG samples had the similar chemical composition, but there were significant differences in the content of each component. Further nontarget metabolomic analysis combined with multivariate statistical analysis showed that CG4–6 years, MCG12 years and MCG20 years samples were obviously divided into three different groups. A total of 17 potential age-dependent markers enabling differentiation among the CG and MCG samples were discovered. Among of these markers, four robust markers, including α-linolenic acid, 9-octadecenoic acid, linoleic acid and panaxydol, could be the characteristic components for differentiation of CG from all other MCG samples. Five robust markers including ginsenoside Re1, -Re2, -Rs1, malonylginsenoside Rb2 and isomer of malonylginsenoside Rb1 were found to enable differentiate MCG20 years samples from all other samples, while 24-hydroxyoleanolic acid and palmitoleic acid were the robust markers for distinguishing MCG12 years samples from all the CG samples and MCG20 years samples. The proposed approach could be applied to directly distinguish MCG root ages, which is an important criterion for evaluating the quality of MCG. The results will provide the data for the deficient study on the chemical constituents of MCG and provide reference for the quantitative determination in the quality control criterion of MCG.
Author Contributions
The individual contributions of authors are specified as following: Data curation, InvestigationWriting-original draft, H.Z.; Methodology, Software, H.L.; Formal analysis, Writing-original draft, J.T.; Components identification, Writing editing, C.W.; Conceptualization, Methodology, H.W.; Investigation, F.W.; Data curation, Q.D.; Writing-review and editing, Y.L.; Funding acquisition, P.L.; Supervision, J.L.
Funding
This research was supported by the Jilin Province Science and Technology Department for Science and Technology Development Project of Jilin Province [No. 20160307008YY].
Conflicts of Interest
The authors declare that they have no conflict of interest concerning this article.
Footnotes
Sample Availability: Samples of the compounds are available from the authors.
References
- 1.Liu D., Li Y.G., Xu H., Sun S.Q., Wang Z.T. Differentiation of the root of Cultivated Ginseng, Mountain Cultivated Ginseng and Mountain Wild Ginseng using FT-IR and two-dimensional corre- lation IR spectroscopy. J. Mol. Struct. 2008;883:228–235. doi: 10.1016/j.molstruc.2008.02.025. [DOI] [Google Scholar]
- 2.Jung C.H., Seog H.M., Choi I.W., Cho H.Y. Antioxidant activities of cultivated and wild Korean ginseng leaves. Food Chem. 2005;92:535–540. doi: 10.1016/j.foodchem.2004.08.021. [DOI] [Google Scholar]
- 3.Kim S.J., Shin S.S., Seo B.I., Jee S.Y. Effect of mountain grown ginseng radix, mountain cultivated ginseng radix, and cultivated ginseng radix on apoptosis of HL-60 cells. J. Herb. 2004;19:19–41. [Google Scholar]
- 4.Pan H.Y., Qu Y., Zhang J.K., Kang T.G., Dou D.Q. Antioxidant activity of ginseng cultivated under mountainous forest with different growing years. J. Ginseng Res. 2013;37:355–360. doi: 10.5142/jgr.2013.37.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xiao D., Yue H., Xiu Y., Sun X.L., Wang Y.B., Liu S.Y. Accumulation characteristics and correlation analysis of five ginsenosides with different cultivation ages from different regions. J. Ginseng Res. 2015;39:338–344. doi: 10.1016/j.jgr.2015.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu X.F., Cheng X.L., Lin Q.H., Li S.S., Jia Z., Han T., Lin R.C., Wang D., Wei F., Li X.R. Identification of mountain-cultivated ginseng and cultivated ginseng using UPLC/oa- TOF MSE with a multivariate statistical sample-profiling strategy. J. Ginseng Res. 2016;40:344–350. doi: 10.1016/j.jgr.2015.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang X.W., Zhang J.J., Li D.K., Zou D.Z., Zhang Y.L., Wang J.C., Hu B., Ju A.C., Ye Z.L. Nontargeted metabolomics approach for the differentiation of cultivation ages of mountain cultivated ginseng leaves using UHPLC/QTOF-MS. J. Pharm. Biomed. Anal. 2017;141:108–122. doi: 10.1016/j.jpba.2017.04.009. [DOI] [PubMed] [Google Scholar]
- 8.Corthout J., Naessens T., Apers S., Vlietinck A.J. Quantitative determination of ginsenosides from Panax ginseng roots and ginseng preparations by thin layer chromatography-densitometry. J. Pharm. Biomed. Anal. 1999;21:187–192. doi: 10.1016/S0731-7085(99)00109-0. [DOI] [PubMed] [Google Scholar]
- 9.Wei S., Wang Y.T., Li J., Zhang H.Q., Ding L. Investigation of ginsenosides in different parts and ages of Panax ginseng. Food Chem. 2007;102:664–668. [Google Scholar]
- 10.Yong E.C., Yong S.K., Yi M.J., Park W.G., Yi J.S., Chun S.R., Han S.S., Lee S.J. Physiological and chemical characteristics of field-and mountain-cultivated ginseng roots. J. Plant Biol. 2007;50:198–205. [Google Scholar]
- 11.Wu W., Jiao C.X., Li H., Ma Y., Jiao L.L., Liu S.Y. LC-MS based metabolic and metabonomic studies of Panax ginseng. Phytochem. Anal. 2018;29:331–340. doi: 10.1002/pca.2752. [DOI] [PubMed] [Google Scholar]
- 12.Qiu S., Yang W.Z., Yao C.L., Qiu Z.D., Shi X.J., Zhang J.X., Hou J.J., Wang Q.R., Wu W.Y., Guo D.A. Nontargeted metabolomic analysis and “commercial-homophyletic” comparison-induced biomarkers verification for the systematic chemical differentiation of five different parts of Panax ginseng. J. Chromatogr. A. 2016;33:78–87. doi: 10.1016/j.chroma.2016.05.051. [DOI] [PubMed] [Google Scholar]
- 13.Pace R., Martinelli E.M., Sardone N., Combarieu E.D.E. Metabolomic evaluation of ginsenosides distribution in Panax genus (Panax ginseng and Panax quinquefolius) using multivariate statistical analysis. Fitoterapia. 2015;101:80–91. doi: 10.1016/j.fitote.2014.12.013. [DOI] [PubMed] [Google Scholar]
- 14.Wang H.P., Liu Y., Chang C., Xiao H.B. Screening Specific Biomarkers of Herbs Using a Metabolomics Approach: A Case Study of Panax ginseng. Sci. Rep. 2017;7:1–9. doi: 10.1038/s41598-017-04712-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang X.W., Li D.K., Wang T., Wu Y.C., Zhao Y., Zhou D.Z., Zhang T., Ye Z.L. Application of metabolomics approach to study of different parts of Mountain Cultivated Ginseng using UHPLC-QTOF/MS. Acta Pharm. Sin. B. 2016;51:1609–1615. [PubMed] [Google Scholar]
- 16.In G., Seo H.K., Park H.W., Jang K.H. A Metabolomic Approach for the Discrimination of Red Ginseng Root Parts and Targeted Validation. Molecules. 2017;22:471. doi: 10.3390/molecules22030471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shin J.S., Park H.W., In G., Seo H.K., Won T.H., Jang K.H., Cho B.G., Han C.K., Shin J.H. Metabolomic approach for discrimination of four- and six-year-old red ginseng (Panax ginseng) using UPLC-QToF-MS. Chem. Pharm. Bull. 2016;64:1298–1303. doi: 10.1248/cpb.c16-00240. [DOI] [PubMed] [Google Scholar]
- 18.Kim N., Kim K., Choi B.Y., Lee D.H., Shin Y.S., Bang K.H., Cha S.W., Lee J.W., Choi H.K., Jang D.S., et al. Metabolomic approach for age discrimination of Panax ginseng using UPLC-Q-TOF MS. J. Agric. Food Chem. 2011;59:10435–10441. doi: 10.1021/jf201718r. [DOI] [PubMed] [Google Scholar]
- 19.Zhang F.X., Li M., Qiao L.R., Yao Z.H., Li C., Shen X.Y., Wang Y., Yu K., Yao X.S., Dai Y. Rapid characterization of Ziziphi Spinosae Semen by UPLC/Qtof MS with novel informatics platform and its application in evaluation of two seeds from Ziziphus species. J. Pharm. Biomed. Anal. 2016;122:59–80. doi: 10.1016/j.jpba.2016.01.047. [DOI] [PubMed] [Google Scholar]
- 20.Wang C.Z., Zhang N.Q., Wang Z.Z., Qi Z., Zhu H.L., Zheng B.Z., Li P.Y., Liu J.P. Nontargeted Metabolomic Analysis of Four Different Parts of Platycodon grandiflorum Grown in Northeast China. Molecules. 2017;22:1280. doi: 10.3390/molecules22081280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y.R., Wang C.Z., Lin H.Q. Discovery of the Potential Biomarkers for Discrimination between Hedyotis diffusa and Hedyotis corymbosa by UPLC-QTOF/MS Metabolome Analysis. Molecules. 2018;23:1525. doi: 10.3390/molecules23071525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.National Pharmacopoeia Commission . Pharmacopoeia of the People’s Republic of China, 2015 Version. China Medical Science and Technology Press; Beijing, China: 2015. [Google Scholar]
- 23.Koh L.H., Lau A.J., Chan C.Y. Hydrophilic interaction liquid chromatography with tandem mass spectrometry for the determination of underivatized dencichine (β-N-oxalyl-l-α,β-diaminopropionic acid) in Panax medicinal plant species. Rapid Commun. Mass Spectrom. 2005;19:1237–1244. doi: 10.1002/rcm.1928. [DOI] [PubMed] [Google Scholar]
- 24.Tüting W., Adden R., Mischnick P. Fragmentation pattern of regioselectively O-methylated maltooligosaccharides in electrospray ionisation-mass spectrometry/collision induced dissociation. Int. J. Mass Spectrom. 2004;232:107–115. doi: 10.1016/j.ijms.2003.12.004. [DOI] [Google Scholar]
- 25.Wang H., Sun H., Zhang A., Li Y., Wang L., Shi H., Li Dizou X., Wang X. Rapid identification and comparative analysis of the chemical constituents and metabolites of Phellodendri amurensis cortex and Zhibai dihuang pill by ultra-performance liquid chromatography with quadrupole TOF-MS. J. Sep. Sci. 2013;36:3874–3882. doi: 10.1002/jssc.201300794. [DOI] [PubMed] [Google Scholar]
- 26.Chen M.L., Chang W.Q., Zhou J.L., Yin Y.H., Xia W.R., Liu J.R., Liu L.F., Xin G.Z. Comparison of three officinal species of Callicarpa based on a biochemome profiling strategy with UHPLC-IT-MS and chemometrics analysis. J. Pharm. Biomed. Anal. 2017;145:666–674. doi: 10.1016/j.jpba.2017.07.054. [DOI] [PubMed] [Google Scholar]
- 27.Fuhrer T., Heer D., Begemann B., Zamboni N. High-throughput, accurate mass metabolome profiling of cellular extracts by flow injection-time-of-flight mass spectrometry. Anal. Chem. 2011;83:7074–7080. doi: 10.1021/ac201267k. [DOI] [PubMed] [Google Scholar]
- 28.Song W., Qiao X., Chen K., Wang Y., Ji S., Feng J., Li K., Lin Y., Ye M. Biosynthesis-Based Quantitative Analysis of 151 Secondary Metabolites of Licorice to Differentiate Medicinal Glycyrrhiza Species and Their Hybrids. Anal. Chem. 2017;89:3146–3153. doi: 10.1021/acs.analchem.6b04919. [DOI] [PubMed] [Google Scholar]
- 29.Li Y., Li C., Yu J., Gao Y., Zhao Y., Xue D., Zhang G.Q., Chai Y.F., Ke Y., Zhang H. Rapid separation and characterization of comprehensive ingredients in Yangxinshi tablet and rat plasma by ultrahigh-performance liquid chromatography–quadrupole time-of-flight mass spectrometry. J. Liq. Chromatogr. Relat. Technol. 2017;59:339–354. doi: 10.1080/10826076.2017.1335213. [DOI] [Google Scholar]
- 30.Wang H.Y., Hua H.Y., Liu X.Y., Liu J.H., Yu B.Y. In vitro biotransformation of red ginseng extract by human intestinal microflora: Metabolites identification and metabolic profile elucidation using LC-Q-TOF/MS. J. Pharm. Bio. Anal. 2014;98:296–306. doi: 10.1016/j.jpba.2014.06.006. [DOI] [PubMed] [Google Scholar]
- 31.Yang W.Z., Ye M., Qiao X., Liu C.F., Miao W.J., Bo T., Tao H.Y., Guo D.A. A strategy for efficient discovery of new natural compounds by integrating orthogonal column chromatography and liquid chromatography/mass spectrometry analysis: Its application in Panax ginseng, Panax quinquefolium and Panax notoginseng to characterize 437 potential new ginsenosides. Anal. Chim. Acta. 2012;739:56–66. doi: 10.1016/j.aca.2012.06.017. [DOI] [PubMed] [Google Scholar]
- 32.Jaiswal Y., Liang Z., Ho A., Chen H., Zhao Z. A Comparative Tissue-specific Metabolite Analysis and Determination of Protodioscin Content in Asparagus Species used in Traditional Chinese Medicine and Ayurveda by use of Laser Microdissection, UHPLC–QTOF/MS and LC–MS/MS. Phytochem. Anal. Pca. 2014;25:514–528. doi: 10.1002/pca.2522. [DOI] [PubMed] [Google Scholar]
- 33.Li H., Yao W., Liu Q., Xu J., Bao B., Shsn M.Q., Cao Y.D., Cheng F.F., Ding A., Zhang L. Application of UHPLC-ESI-Q-TOF-MS to Identify Multiple Constituents in Processed Products of the Herbal Medicine Ligustri Lucidi Fructus. Molecules. 2017;22:689. doi: 10.3390/molecules22050689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu W., Sun L., Zhang Z., Guo Y.Y., Liu S.Y. Profiling and multivariate statistical analysis of Panax ginseng based on ultra-high-performance liquid chromatography coupled with quadrupole-time-of-flight mass spectrometry. J. Pharm. Bio. Anal. 2015;107:141–150. doi: 10.1016/j.jpba.2014.12.030. [DOI] [PubMed] [Google Scholar]
- 35.Yang L., Ying P., Wang M.Y., Zhou G.S., Zhang Y.L., Li X. Rapid screening and identification of the differences between metabolites of Cistanche deserticola, and C. tubulosa, water extract in rats by UPLC-Q-TOF-MS combined pattern recognition analysis. J. Pharm. Bio. Anal. 2016;131:364–372. doi: 10.1016/j.jpba.2016.09.018. [DOI] [PubMed] [Google Scholar]
- 36.Zhang Y., Cheng Y., Liu Z., Ding L.Q., Qiu T.Y., Chai L.W., Qiu F., Wang Z.Z., Xiao W., Zhao L.S., Chen X.H. Systematic screening and characterization of multiple constituents in Guizhi Fuling capsule and metabolic profiling of bioactive components in rats using ultra-high-performance liquid chromatography/quadrupole-time-of-flight mass spectrometry. J. Chromatogr. B. 2017;1061:474–486. doi: 10.1016/j.jchromb.2017.07.021. [DOI] [PubMed] [Google Scholar]
- 37.Tang S.Y., Liu S., Liu Z.Q., Song F.R., Liu S.Y. Analysis and Identification of the Chemical Constituents of Ding-Zhi-Xiao-Wan Prescription by HPLC-IT-MSn and HPLC-Q-TOF-MS. Chinese J. Chem. 2015;33:451–462. doi: 10.1002/cjoc.201400789. [DOI] [Google Scholar]
- 38.Chen L.L., Qi J., Chang Y.X., Zhu D.N., Yu B.Y. Identification and determination of the major constituents in Traditional Chinese Medicinal formula Danggui-Shaoyao-San by HPLC-DAD-ESI-MS/MS. J. Pharm. Biomed. Anal. 2009;50:127–137. doi: 10.1016/j.jpba.2009.03.039. [DOI] [PubMed] [Google Scholar]
- 39.Chen Y.J., Xu L., Zhao Y.C., Zhao Z.Z., Chen H.B., Yi T., Qin M.J., Liang Z.T. Tissue-specific metabolite profiling and quantitative analysis of ginsenosides in Panax quinquefolium, using laser microdissection and liquid chromatography–quadrupole/time of flight-mass spectrometry. Chem. Cent. J. 2015;9:66–72. doi: 10.1186/s13065-015-0141-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qiu S., Yang W.Z., Shi X.J., Yao C.L., Yang M., Liu X., Jiang B.H., Wu W.Y., Guo D.A. A green protocol for efficient discovery of novel natural compounds: Characterization of new ginsenosides from the stems and leaves of Panax ginseng as a case study. Anal. Chim. Acta. 2015;893:65–76. doi: 10.1016/j.aca.2015.08.048. [DOI] [PubMed] [Google Scholar]
- 41.Dan M., Su M.M., Gao X.F., Zhao T., Zhao A.H., Xie G.X., Qiu Y.P., Zhou M.M., Liu Z., Jia W. Metabolite profiling of Panax notoginseng using UPLC-ESI-MS. Phytochemistry. 2008;69:2237–2244. doi: 10.1016/j.phytochem.2008.04.015. [DOI] [PubMed] [Google Scholar]
- 42.Wu Q.L., Wang M.F., Simon J.E., Yu S.C., Xiao P.G., Ho C.T. Studies on the Chemical Constituents of Loquat Leaves (Eriobotrya japonica) ACS Sym. 2003;28:292–306. [Google Scholar]
- 43.Wang L.L., Han L.F., Yu H.S., Sang M.M., Liu E.W., Zhang Y., Fang S.M., Wang T., Gao X.M. Analysis of the Constituents in “Zhu She Yong Xue Shuan Tong” by Ultra High Performance Liquid Chromatography with Quadrupole Time-of-Flight Mass Spectrometry Combined with Preparative High Performance Liquid Chromatography. Molecules. 2015;20:20518–20537. doi: 10.3390/molecules201119712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li S.L., Lai S.F., Song J.Z., Qiao C.F., Liu X., Zhou Y., Cai H., Cai B.C., Xu H.X. Decocting-induced chemical transformations and global quality of Du-Shen-Tang, the decoction of ginseng evaluated by UPLC-Q-TOF-MS/MS based chemical profiling approach. J. Pharm. Biomed. Anal. 2010;53:946–957. doi: 10.1016/j.jpba.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 45.Liang Z.T., Chen Y.J., Liang X., Qin M.J., Yi T., Chen H.B., Zhao Z.Z. Localization of ginsenosides in the rhizome and root of Panax ginseng, by laser microdissection and liquid chromatography–quadrupole/time of flight-mass spectrometry. J. Pharm. Biomed. Anal. 2015;105:121–133. doi: 10.1016/j.jpba.2014.12.005. [DOI] [PubMed] [Google Scholar]
- 46.Peng J., Dou S.S., Liu L., Zhang W.D., Chen Z.L., Xu R.L., Ding J.M. Identification of Multiple Constituents in the TCM-Formula Shexiang Baoxin Pill by LC Coupled with DAD-ESI-MS-MS. Chromatographia. 2009;70:133–142. [Google Scholar]
- 47.Murae T., Sugie A., Moriyama Y., Tsuyuki T., Takahashi T. Mass spectra of the bitter principles fromPicrasma ailanthoides Planchon. J. Mass Spectrom. 1974;8:291–301. doi: 10.1002/oms.1210080132. [DOI] [Google Scholar]
- 48.Castro O.N., Benites J.L., Rodilla J., Santiago J., Simirgiotis M., Sepulveda B., Areche C. Metabolomic Analysis of the Lichen Everniopsis trulla Using Ultra High Performance Liquid Chromatography-Quadrupole-Orbitrap Mass Spectrometry (UHPLC-Q-OT-MS) Chromatographia. 2017;80:1–7. doi: 10.1007/s10337-017-3304-4. [DOI] [Google Scholar]
- 49.Peng L., Yu H.S., Zhang L.J., Song X.B., Kang L.P., Liu J.Y., Zhang J., Cao M., Yu K., Kang T.J., Ma B.P. A rapid method for chemical fingerprint analysis of Pan Panax notoginseng powders by ultra performance liquid chromatography coupled with quadrupole time-of-flight mass spectrometry. Chin. J. Nat. Med. 2015;13:471–480. doi: 10.1016/S1875-5364(15)30042-X. [DOI] [PubMed] [Google Scholar]
- 50.Lai C.M., Li S.P., Yu H., Wan K.W., Wang Y.T. A rapid HPLC-ESI-MS/MS for qualitative and quantitative analysis of saponins in "XUESETONG" injection. J. Pharm. Biomed. Anal. 2006;40:669–678. doi: 10.1016/j.jpba.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 51.Sun J.H., Chen P. Differentiation of Panax quinquefolius, grown in the USA and China using LC/MS-based chromatographic fingerprinting and chemometric approaches. Anal. Bioanal. Chem. 2011;399:1877–1889. doi: 10.1007/s00216-010-4586-7. [DOI] [PubMed] [Google Scholar]
- 52.Wan J.Y., Wang C.Z., Liu Z., Zhang Q.H., Musch M., Bissonnette M., Chang E.B., Li P., Qi L.W., Yuan C.S. Determination of American ginseng saponins and their metabolites in human plasma, urine and feces samples by liquid chromatography coupled with quadrupole time-of-flight mass spectrometry. J. Chromatogr. B. 2016;62:1015–1016. doi: 10.1016/j.jchromb.2016.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Qi L.W., Wang H.Y., Zhang H., Wang C.Z., Li P., Yuan C.S. Diagnostic ion filtering to characterize ginseng saponins by rapid liquid chromatography with time-of-flight mass spectrometry. J. Chromatogr. A. 2012;1230:93–99. doi: 10.1016/j.chroma.2012.01.079. [DOI] [PubMed] [Google Scholar]
- 54.Xu G.H., Choo S.J., Ryoo I.J., Kim Y.H., Paek K.Y., Yoo I.D. Polyacetylenes from the tissue cultured adventitious roots of Panax ginseng C.A. meyer. Nat. Prod. Sci. 2008;14:177–181. [Google Scholar]
- 55.Yang H., Liu L., Gao W., Liu K., Qi L.W., Li P. Direct and comprehensive analysis of ginsenosides and diterpene alkaloids in Shenfu injection by combinatory liquid chromatography-mass spectrometric techniques. J. Pharm. Biomed. Anal. 2014;92:13–21. doi: 10.1016/j.jpba.2013.12.041. [DOI] [PubMed] [Google Scholar]
- 56.Yan L., Guan T.Y., Zhou Y.Y., Liu Y.N., Xing L., Zheng X., Dai C., Du P., Rao T., Zhou L.J., Yu X.Y., Hao K., Xie L., Wang G.J. Effect of mobile phase additives on qualitative and quantitative analysis of ginsenosides by liquid chromatography hybrid quadrupole-time of flight mass spectrometry. J. Chromatogr. A. 2013;1297:29–36. doi: 10.1016/j.chroma.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 57.Yang X.H., Cheng X.L., Qin B., Cai Z.Y., Cai X., Liu S., Wang Q., Qin Y. Ultra-high performance liquid chromatography coupled with quadrupole/time of flight mass spectrometry based chemical profiling approach for the holistic quality control of complex Kang-Jing formula preparations. J. Pharm. Biomed. Anal. 2016;124:319–336. doi: 10.1016/j.jpba.2016.03.012. [DOI] [PubMed] [Google Scholar]
- 58.Coqueiro A., Regasini L.O., Leme G.M., Polese L., Nogueira C.T., Cistia M.D., Graminha M.A.S. Leishmanicidal activity of Brosimum glaziovii (Moraceae) and chemical composition of the bioactive fractions by using high-resolution gas chromatography and GC-MS. J. Braz. Chem. Soc. 2014;25:1839–1847. [Google Scholar]
- 59.Wang C.Z., Zhang N.Q., Wang Z.Z., Qi Z., Zheng B.Z., Li P.Y., Liu J.P. Rapid characterization of chemical constituents of Platycodon grandiflorum and its adulterant Adenophora stricta by UPLC-QTOF-MS/MS. J. Mass Spectrom. 2017;52:643–657. doi: 10.1002/jms.3967. [DOI] [PubMed] [Google Scholar]
- 60.Huang B.F., Zheng F.F., Fu S.S., Yao J.H., Tao B.H., Ren Y.P. UPLC-ESI-MS/MS for determining trans-and cis-vitamin K1 in infant formulas: Method and applications. Eur. Food Res. Tech. 2012;235:873–879. doi: 10.1007/s00217-012-1823-7. [DOI] [Google Scholar]
- 61.Hurtadofernández E., Pacchiarotta T., Gómezromero M. Ultra high performance liquid chromatography-time of flight mass spectrometry for analysis of avocado fruit metabolites: Method evaluation and applicability to the analysis of ripening degrees. J. Chromatogr. A. 2011;1218:7723–7738. doi: 10.1016/j.chroma.2011.08.059. [DOI] [PubMed] [Google Scholar]
- 62.Doshi G.M., Nalawade V.V., Mukadam A.S. Structural elucidation of chemical constituents from Benincasa hispidaseeds andCarissa congestaroots by gas chromatography: Mass spectroscopy. Pharmacognosy Res. 2015;7:282–293. doi: 10.4103/0974-8490.157179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Montserrat R.A., Liliana V., Stefania V., Josep M.G., Elvira L.T., Susana B. Characterisation of volatile composition of white salsify (Tragopogon porrifolius L.) by headspace solid-phase microextraction (HS-SPME) and simultaneous distillation-extraction (SDE) coupled to GC-MS. Food Chem. 2011;129:557–564. doi: 10.1016/j.foodchem.2011.04.061. [DOI] [PubMed] [Google Scholar]
- 64.Mathela C.S., Singh K.K., Gupta V.K. Synthesis and in vitro antibacterial activity of thymol and carvacrol derivatives. Acta Pol. Pharm. 2010;67:375–380. [PubMed] [Google Scholar]
- 65.Wang N., Manabe Y.K., Sugawara T., Paul N.A., Zhao J. Identification and biological activities of carotenoids from the freshwater alga Oedogonium intermedium. Food Chem. 2017;20:247–255. doi: 10.1016/j.foodchem.2017.03.115. [DOI] [PubMed] [Google Scholar]
- 66.Lee M.J., Chung I.M., Kim H.J., Jung M.J. High resolution LC–ESI-TOF-mass spectrometry method for fast separation, identification, and quantification of 12 isoflavones in soybeans and soybean products. Food Chem. 2015;176:254–262. doi: 10.1016/j.foodchem.2014.12.073. [DOI] [PubMed] [Google Scholar]
- 67.Hwang J.W., Oh J.H., Yoo H.S., Lee Y.W., Cho C.K., Kwon K.R., Yoon J.H., Park J.S., Her S., Lee Z.W. Mountain ginseng extract exhibits anti-lung cancer activity by inhibiting the nuclear translocation of NF-κB. Am. J. Chin. Med. 2012;40:187–202. doi: 10.1142/S0192415X12500152. [DOI] [PubMed] [Google Scholar]
- 68.Carvalho E.B.T., Melo I.L.P., Mancini-Filho J. Chemical and physiological aspects of isomers of conjugated fatty acids. Food Sci. Technol. 2010;30:295–307. doi: 10.1590/S0101-20612010000200002. [DOI] [Google Scholar]
- 69.Harwood J.L. Recent advances in the biosynthesis of plant fatty acids. Biochim. Biophysica. Acta. 1996;1301:7–56. doi: 10.1016/0005-2760(95)00242-1. [DOI] [PubMed] [Google Scholar]
- 70.Kim H.S., Lim J.M., Kim J.Y., Park S., Sohn J. Panaxydol, a component of Panax ginseng, induces apoptosis in cancer cells through EGFR activation and ER stress and inhibits tumor growth in mouse models. Int. J. Cancer. 2016;138:1432–1441. doi: 10.1002/ijc.29879. [DOI] [PubMed] [Google Scholar]
- 71.Nihat K., Elena O., Nicholas S., Huber C., Luis M., Bonfill M. Biosynthesis of Panaxynol and Panaxydol in Panax ginseng. Molecules. 2013;18:7686–7698. doi: 10.3390/molecules18077686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liang Y., Zhao S. Progress in understanding of ginsenoside biosynthesis. Plant Biol. 2010;10:415–421. doi: 10.1111/j.1438-8677.2008.00064.x. [DOI] [PubMed] [Google Scholar]
- 73.Kim Y.J., Zhang D., Yang D.C. Biosynthesis and biotechnological production of ginsenosides. Biotechnol. Adv. 2015;33:717–735. doi: 10.1016/j.biotechadv.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 74.Leung K.W., Wong A.S. Pharmacology of ginsenosides: A literature review. Chin. Med. 2010;5:20–27. doi: 10.1186/1749-8546-5-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Velisek J., Cejpek K. Biosynthesis of food constituents: Lipids. 1. Fatty acids and derived compounds—A review. Czech J. Food Sci. 2006;24:193–216. doi: 10.17221/3317-CJFS. [DOI] [Google Scholar]