Abstract
DNA repair processes are involved in both the onset and treatment efficacy of colorectal cancer (CRC). A change of a single nucleotide causing an amino acid substitution in the corresponding protein may alter the efficiency of DNA repair, thus modifying the CRC susceptibility and clinical outcome. We performed a candidate gene approach in order to analyze the association of non-synonymous single nucleotide polymorphisms (nsSNPs) in the genes covering the main DNA repair pathways with CRC risk and clinical outcome modifications. Our candidate polymorphisms were selected according to the foremost genomic and functional prediction databases. Sixteen nsSNPs in 12 DNA repair genes were evaluated in cohorts from the Czech Republic and Austria. Apart from the tumor-node-metastasis (TNM) stage, which occurred as the main prognostic factor in all of the performed analyses, we observed several significant associations of different nsSNPs with survival and clinical outcomes in both cohorts. However, only some of the genes (REV3L, POLQ, and NEIL3) were prominently defined as prediction factors in the classification and regression tree analysis; therefore, the study suggests their association for patient survival. In summary, we provide observational and bioinformatics evidence that even subtle alterations in specific proteins of the DNA repair pathways may contribute to CRC susceptibility and clinical outcome.
Keywords: DNA repair genes, functional single nucleotide polymorphism, colorectal cancer susceptibility, survival analysis
1. Introduction
Colorectal cancer (CRC) is the third most common malignancy and the second leading cause of cancer death worldwide [1]. In Europe, the highest incidence rates are reported in Eastern and Central European countries, such as the Czech Republic and Austria [2]. CRC represents a multifactorial disease associated with several genetic and environmental factors [3].
The prognosis for patients with CRC is heavily dependent on stage at diagnosis; the five-year survival rate is up to 90% for stage I, but only <15% for stage IV [4]. Over half of the cases are diagnosed at an advanced stage of disease (III and IV), with treatment usually involving complete primary tumor resection and appropriate chemotherapy. While the treatment can reduce the risk of relapse and increase patients’ survival, it can also cause severe side effects and impair quality of life [5]. The differences in medication response are considerably affected by individual inherited genetic susceptibility. Current approaches to choose and implement chemotherapy regimens for CRC patients are primarily determined by tumor staging and histopathological examination. Developing prognostic and predictive biomarkers based on a personal genetic background would greatly aid the selection of an optimal treatment by oncologists, so as to improve clinical outcome for each patient.
Genetics plays a key role in predisposition to CRC, its initiation, and progression [6]. Several studies provided evidence that single nucleotide polymorphisms (SNPs) in DNA repair genes could alter DNA repair function, modulate its capacity, and thus induce genetic instability or unregulated cell growth and cancer [7,8,9]. In the last decade, while association studies (including genome-wide) have identified multiple SNPs involved in CRC susceptibility, none have been validated as biomarkers for clinical use [10,11,12,13,14]. Furthermore, most of the anticancer agents are targeted to induce DNA damage, which overwhelms the cellular DNA repair capacity and thus leads to apoptosis. The most affected are the rapidly dividing cells, such as cancer cells. Treatment efficacy is therefore influenced by the DNA repair capacity of cancer cells, and the differences in treatment response might be affected by the inherited variations of genes encoding DNA repair enzymes [15].
In this study, we hypothesized that SNPs causing amino acid substitution (non-synonymous SNPs—nsSNPs) in DNA repair genes that are known to be involved in maintaining genome stability (cancer prevention) and in chemotherapy response (cancer treatment), may influence CRC susceptibility and modulate the clinical outcome after cancer diagnosis. We evaluated the association of 16 nsSNPs in 12 DNA repair genes with CRC risk, post-diagnosis survival, and therapy outcomes in a discovery set of 1832 patients and 1172 controls from the Czech Republic and in an independent replication set comprising 950 patients and 820 controls from Austria.
2. Results
2.1. SNP Selection
In total, sixteen nsSNPs in 12 genes passed the selection criteria and were successfully genotyped and analyzed in the Czech cohort (Table 1). The same nsSNPs were analyzed in the replication Austrian cohort, except for two nsSNPs (FAAP24 rs3816032 and MUS81 rs545500), where the genome-wide association study (GWAS) data were not available.
Table 1.
Selected single nucleotide polymorphisms (SNPs) in DNA repair genes, with a minor allele frequency of a > 0.10 in the European population.
| Genomic Annotation | Functional Genomics | Comparative Genomics | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene ID | DNA Repair Pathway | UniProtKB | SNP ID | Base Change | Amino Acid Change | MAF in EUR a | LD with Other SNPs Associated with CRC | LD within the Same Gene | F-SNP Prediction Result (on Protein Coding) | ELASPIC (∆∆G) | DUET (∆∆G) | Element GERP RS Score >800 | SIPHYs |
| EME1 | DSB | Q96AY2 | rs12450550 | T > C | Ile350Thr | 0.24 | no | no | deleterious | Destabilizing (Core 1.646) | Destabilizing (−3.002 Kcal/mol) | ||
| FAAP24 | DSB | Q9BTP7 | rs3816032 b | T > C | Ile192Thr | 0.11 | no | no | deleterious | Destabilizing (Core 1.133) | Destabilizing (−1.653 Kcal/mol) | X | |
| FANCI | DSB | Q9NVI1 | rs2283432 | C > G | Cys742Ser | 0.38 | no | no | deleterious | NA | NA | X | X |
| MUS81 | DSB | Q96NY9 | rs545500 b | C > G | Arg180Pro | 0.33 | no | no | deleterious | NA | NA | X | |
| NEIL3 | BER | Q8TAT5 | rs7689099 | C > G | Pro117Arg | 0.12 | no | no | deleterious | NA | NA | X | X |
| POLE | BER, DSB, NER | Q07864 | rs5744934 | A > G | Asn1396Ser | 0.13 | no | no | deleterious | NA | NA | X | |
| POLN | DSB | Q7Z5Q5 | rs2353552 | C > A | Gln121His | 0.13 | no | no | deleterious | NA | NA | ||
| rs9328764 | A > G | Arg425Cys | 0.12 | no | no | deleterious | NA | Destabilizing (−1.765 Kcal/mol) | X | ||||
| POLQ | DSB | O75417 | rs1381057 | C > T | Gln2513Arg | 0.33 | no | no | deleterious | Destabilizing (Core 1.648) | NA | X | |
| rs3218649 | C > G | Thr982Arg | 0.39 | no | no | deleterious | NA | NA | X | ||||
| rs3218651 | T > C | His1201Arg | 0.15 | no | no | damaging | NA | NA | X | ||||
| RAD51D | DSB | O75771 | rs4796033 | C > T | Arg165Gln | 0.13 | no | no | deleterious | Destabilizing (Core 1.843) | NA | X | |
| REV1 | DSB | Q9UBZ9 | rs3087386 | G > A | Phe257Ser | 0.43 | no | no | deleterious | NA | NA | X | |
| rs3087399 | A > G | Asn373Ser | 0.12 | no | no | deleterious | NA | Destabilizing (−0.596 Kcal/mol) | X | X | |||
| REV3L | DSB | O60673 | rs3204953 | G > A | Val2986Ile | 0.17 | no | no | deleterious | Destabilizing (Core 1.965) | NA | X | X |
| RPA1 | BER, DSB, NER | P27694 | rs5030755 | A > G | Thr351Ala | 0.10 | no | no | deleterious | NA | Destabilizing (−1.037 Kcal/mol) | X | |
SNP—single nucleotide polymorphism; MAF—minor allele frequency; EUR—European population; LD—linkage disequilibrium; CRC—colorectal cancer; GERP—Genomic Evolutionary Rate Profiling; DSB—double strand break repair pathway; BER—base excision repair pathway; NER—nucleotide excision repair pathway; NA—not applicable; X—evolutionary conserved position. a https://www.ensembl.org/index.html. b Data for SNPs were not available in the Austrian cohort.
2.2. Case-Control Study
The characteristics of the study participants are shown in Table 2. Compared with controls, CRC cases in the Czech cohort had a slightly higher prevalence of male individuals, and were more likely to be older, to smoke, to have diabetes mellitus, and a positive family history of CRC. In the Austrian cohort, CRC cases were more often males and smokers.
Table 2.
Characteristics of the study populations.
| Czech Republic | Austria | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Variables | Controls | Cases | OR | 95% CI | p-Value | Controls | Cases | OR | 95% CI | p-Value | |
| No. (%) | No. (%) | No. (%) | No. (%) | ||||||||
| Sex | Female | 478 (40.8) | 696 (38.1) | Ref | 353 (43.1) | 389 (40.9) | Ref | ||||
| Male | 694 (59.2) | 1133 (61.9) | 1.20 | 1.02–1.40 | 0.03 | 467 (56.9) | 561 (59.1) | 1.39 | 1.09–1.78 | 0.01 | |
| Age (years) | <50 | 269 (22.9) | 140 (9.2) | Ref | 92 (11.2) | 129 (13.5) | Ref | ||||
| (50, 60] | 546 (46.6) | 433 (28.4) | 1.52 | 1.20–1.94 | 0.0006 | 147 (17.9) | 208 (21.9) | 2.18 | 0.72–1.43 | 0.92 | |
| (60, 70] | 183 (15.6) | 639 (42.0) | 6.71 | 5.16–8.72 | <0.0001 | 282 (34.4) | 323 (34.0) | 0.82 | 0.60–1.13 | 0.22 | |
| >70 | 174 (14.9) | 310 (20.4) | 3.42 | 2.60–4.51 | <0.0001 | 299 (36.5) | 291 (30.6) | 0.70 | 0.51–0.96 | 0.03 | |
| BMI | (18.5, 25] | 93 (8.0) | 358 (23.5) | Ref | 189 (23.7) | 296 (35.7) | Ref | ||||
| <18.5 | 334 (28.5) | 370 (24.3) | 3.22 | 2.45–4.24 | <0.0001 | 2 (0.3) | 17 (2.0) | 5.43 | 1.24–23.76 | 0.02 | |
| (25, 30] | 529 (45.1) | 508 (33.4) | 0.84 | 0.69–1.02 | 0.08 | 364 (45.6) | 336 (40.5) | 0.59 | 0.47–0.75 | <0.0001 | |
| >30 | 213 (18.4) | 286 (18.8) | 1.13 | 0.89–1.43 | 0.31 | 243 (30.4) | 181 (21.8) | 0.48 | 0.37–0.62 | <0.0001 | |
| Smoking habit | No | 638 (57.6) | 769 (53.1) | Ref | 447 (55.7) | 251 (48.8) | Ref | ||||
| Yes a | 470 (42.4) | 679 (46.9) | 1.33 | 1.13–1.56 | <0.001 | 356 (44.3) | 263 (51.2) | 1.30 | 0.97–1.73 | 0.08 | |
| DM | No | 555 (85.5) | 1076 (80.4) | Ref | 370 (82.8) | 817 (86.0) | Ref | ||||
| Yes | 94 (14.5) | 263 (19.6) | 1.41 | 1.09–1.84 | 0.01 | 77 (17.2) | 133 (14.0) | 0.62 | 0.42–0.92 | 0.02 | |
| Family history of CRC | No | 942 (89.3) | 1103 (84.1) | Ref | NDA | NDA | |||||
| Yes | 113 (10.7) | 209 (15.9) | 1.65 | 1.28–2.12 | <0.001 | NDA | NDA | ||||
| Diagnosis | Colon | 1192 (65.8) | 586 (62.6) | ||||||||
| Rektum | 621 (34.2) | 350 (37.4) | |||||||||
| tnm stage | I | 277 (16.8) | 188 (21.2) | ||||||||
| II | 498 (30.2) | 227 (25.5) | |||||||||
| III | 491 (29.8) | 354 (39.8) | |||||||||
| IV | 384 (23.3) | 120 (13.5) | |||||||||
| Chemotherapy | None | 795 (49.9) | 389 (43.0) | ||||||||
| 5-FU | 494 (31.0) | 253 (28.0) | |||||||||
| 5-FU combined with oxaliplatin | 303 (19.1) | 262 (29.0) | |||||||||
OR—odds ratio; CI—confidence interval; BMI—body mass index; DM—diabetes mellitus; CRC—colorectal cancer; TNM—tumor-node-metastasis; 5-FU—5-fluorouracil; NDA—no data available. Significant results are in bold. Numbers may not add up to 100% of available subjects because of missing data. a Ex-smokers included in smokers.
For all of the SNPs, the distribution of the genotypes within the studied genes in controls was in agreement with the Hardy–Weinberg equilibrium. The SNPs significantly associated with CRC risk are presented in Table 3.
Table 3.
Associations of SNPs in DNA repair genes with the risk of CRC and its major sub-sites (colon/rectum).
| All CRC Patients | Colon Cancer Patients | Rectal Cancer Patients | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Genotype | Controls a | Cases a | OR b | 95% CI | p-Value | Cases a | OR b | 95% CI | p-Value | Cases a | OR b | 95% CI | p-Value | HWE c |
| SNP | Χ2, p-Value | ||||||||||||||
| Czech Republic | |||||||||||||||
|
EME1 rs12450550 |
TT | 678 | 815 | Ref | 526 | Ref | 284 | Ref | 1.07, 0.58 | ||||||
| TC | 410 | 570 | 1.19 | 1.00–1.40 | 0.05 | 363 | 1.17 | 0.97–1.40 | 0.11 | 198 | 1.20 | 0.96–1.50 | 0.11 | ||
| CC | 73 | 108 | 1.24 | 0.90–1.70 | 0.20 | 64 | 1.15 | 0.80–1.65 | 0.46 | 41 | 1.38 | 0.91–2.09 | 0.13 | ||
| TC+CC | 483 | 678 | 1.19 | 1.02–1.40 | 0.03 | 427 | 1.16 | 0.97–1.39 | 0.10 | 239 | 1.23 | 0.99–1.52 | 0.06 | ||
| TT+TC | 1088 | 1385 | Ref | 889 | Ref | 482 | Ref | ||||||||
| CC | 73 | 108 | 1.16 | 0.84–1.59 | 0.36 | 64 | 1.08 | 0.76–1.55 | 0.66 | 41 | 1.28 | 0.86–1.93 | 0.23 | ||
|
REV3L rs3204953 |
GG | 839 | 1049 | Ref | 666 | Ref | 371 | Ref | 4.68, 0.10 | ||||||
| GA | 304 | 405 | 1.09 | 0.91–1.30 | 0.37 | 261 | 1.12 | 0.91–1.37 | 0.27 | 139 | 1.06 | 0.83–1.34 | 0.66 | ||
| AA | 15 | 43 | 2.32 | 1.27–4.25 | 0.006 * | 30 | 2.59 | 1.36–4.91 | 0.004 * | 13 | 1.97 | 0.92–4.22 | 0.08 | ||
| GA+AA | 319 | 448 | 1.14 | 0.96–1.36 | 0.13 | 291 | 1.19 | 0.98–1.44 | 0.08 | 152 | 1.10 | 0.87–1.39 | 0.42 | ||
| GG+GA | 1143 | 1454 | Ref | 927 | Ref | 510 | Ref | ||||||||
| AA | 15 | 43 | 2.28 | 1.24–4.17 | 0.008 * | 30 | 2.52 | 1.33–4.77 | 0.005 * | 13 | 1.95 | 0.91–4.18 | 0.09 | ||
| Austria | |||||||||||||||
|
POLQ rs1381057 |
CC | 372 | 413 | Ref | 267 | Ref | 134 | Ref | 1.49, 0.47 | ||||||
| CT | 349 | 423 | 1.09 | 0.90–1.34 | 0.38 | 250 | 1.00 | 0.80–1.25 | 1.00 | 166 | 1.32 | 1.01–1.74 | 0.04 | ||
| TT | 99 | 114 | 1.05 | 0.77–1.42 | 0.76 | 65 | 0.93 | 0.65–1.32 | 0.68 | 49 | 1.40 | 0.94–2.08 | 0.10 | ||
| CT+TT | 448 | 537 | 1.08 | 0.90–1.31 | 0.40 | 315 | 0.98 | 0.80–1.22 | 0.89 | 215 | 1.34 | 1.04–1.74 | 0.03 | ||
| CC+CT | 721 | 836 | Ref | 517 | Ref | 300 | Ref | ||||||||
| TT | 99 | 114 | 1.00 | 0.75–1.34 | 0.98 | 65 | 0.93 | 0.66–1.29 | 0.66 | 49 | 1.21 | 0.84–1.75 | 0.32 | ||
|
REV1 rs3087399 |
AA | 593 | 673 | Ref | 414 | Ref | 243 | Ref | 0.02, 0.99 | ||||||
| AG | 208 | 259 | 1.10 | 0.89–1.36 | 0.39 | 151 | 1.04 | 0.81–1.32 | 0.78 | 105 | 1.25 | 0.95–1.66 | 0.11 | ||
| GG | 19 | 18 | 0.83 | 0.43–1.60 | 0.58 | 17 | 1.27 | 0.65–2.48 | 0.48 | 1 | 0.13 | 0.02–0.96 | 0.05 | ||
| AG+GG | 227 | 277 | 1.08 | 0.87–1.32 | 0.50 | 168 | 1.06 | 0.83–1.34 | 0.65 | 106 | 1.16 | 0.88–1.53 | 0.30 | ||
| AA+AG | 801 | 932 | Ref | 565 | Ref | 348 | Ref | ||||||||
| GG | 19 | 18 | 0.81 | 0.42–1.56 | 0.53 | 17 | 1.26 | 0.65–2.44 | 0.50 | 1 | 0.12 | 0.02–090 | 0.04 | ||
OR—odds ratio; CI—confidence interval. Nominally significant results are in bold. Results that passed the Benjamini–Hochberg test for multiple comparisons are marked with an asterisk. a Numbers may not add up to 100% of subjects due to genotyping failure. All of the samples that did not give a reliable result in the first round of genotyping were retested in up to two additional rounds. Samples failing these procedures were omitted from the analysis. b Logistic regression analysis values are adjusted for age. c X2 and p-values for the deviation of the observed and of the numbers expected from the Hardy–Weinberg equilibrium (HWE) in the controls.
Czech cohort. The carriers of the TC genotype in EME1 rs12450550 had an associated increased CRC risk (TC vs. TT; odds ratio (OR) 1.19; 95% confidence interval (CI) 1.00–1.40; p = 0.05), with the same tendency observed for the presence of the variant C allele in the dominant model (TC+CC vs. TT; OR 1.19; 95% CI 1.02–1.40; p = 0.03). However, this association should be considered cautiously due to the low frequency of the variant CC genotype in this study group. The variant AA genotype in REV3L rs3204953 was associated with an increased risk of CRC in the codominant and recessive model (AA vs. GG; OR 2.32; 95% CI 1.27–4.25; p = 0.006; and AA vs. GG+GA; OR 2.28; 95% CI 1.24–4.17; p = 0.008). After stratification according to the tumor site, the association was similar in colon cancer patients (AA vs. GG; OR 2.59; 95% CI 1.36–4.91; p = 0.004; and AA vs. GG+GA; OR 2.52; 95% CI 1.33–4.77; p = 0.005). By considering multiple testing correction using the Benjamini–Hochberg false discovery rate (FDR) measure, only the association for rs3204953 in REV3L remained significant (q = 0.01).
Austrian cohort. The association of the SNP genotypes with CRC risk was observed only for the rectal cancer patients. Carriers of the CT genotype in POLQ rs1381057 were at an increased risk of the disease (CT vs. CC; OR 1.32; 95% CI 1.01–1.74; p = 0.04), with the same tendency observed in the presence of the variant T allele in the dominant model (CT+TT vs. CC; OR 1.34; 95% CI 1.04–1.74; p = 0.03). The variant GG genotype in REV1 rs3087399 was associated with a decreased risk of rectal cancer in the codominant and recessive model (GG vs. AA; OR 0.13; 95% CI 0.02–0.96; p = 0.05; and GG vs. AA+AG; OR 0.12; 95% CI 0.02–0.90; p = 0.04). None of the associations remained significant after the Benjamini–Hochberg correction.
2.3. Survival Analyses
In total, 1832 Czech and 950 Austrian CRC cases were included in the survival analyses. In the univariate assessment, several covariates were associated with survival, including established prognostic factors such as male sex, higher age, smoking habit, and cancer stage, which were associated with decreased patients’ survival and increased risk of recurrence (Table 4).
Table 4.
Clinical characteristics significantly affecting overall survival (OS) and event free survival (EFS) in CRC patients with complete follow up.
| Czech Republic | Austria | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Variables | N a | OS | EFS | N a | OS | EFS | |||||
| HR (95% CI) | p-Value | HR (95% CI) | p-Value | HR (95% CI) | p-Value | HR (95% CI) | p-Value | ||||
| Sex | Female | 696 | Ref | Ref | 389 | Ref | Ref | ||||
| Male | 1133 | 1.47 (1.20–1.80) | 0.0002 | 1.29 (1.09–1.52) | 0.003 | 561 | 1.37 (1.03–1.83) | 0.03 | 1.43 (1.11–1.83) | 0.005 | |
| Age (years) | <50 | 149 | Ref | Ref | 129 | Ref | Ref | ||||
| (50, 60] | 433 | 0.96 (0.62–1.50) | 0.87 | 1.06 (0.76–1.49) | 0.72 | 208 | 1.44 (0.77–2.69) | 0.26 | 1.62 (0.99–2.65) | 0.05 | |
| (60, 70] | 639 | 1.08 (0.71–1.65) | 0.72 | 0.90 (0.65–1.25) | 0.54 | 323 | 2.14 (1.21–3.79) | 0.01 | 1.68 (1.05–2.68) | 0.03 | |
| >70 | 610 | 1.47 (0.97–2.24) | 0.07 | 1.05 (0.76–1.46) | 0.77 | 291 | 3.11 (1.77–5.47) | <0.0001 | 2.53 (1.60–4.00) | <0.0001 | |
| BMI | (18.5, 25] | 434 | Ref | Ref | 296 | Ref | Ref | ||||
| <18.5 | 456 | 0.99 (0.77–1.27) | 0.92 | 1.06 (0.86–1.32) | 0.58 | 17 | 1.12 (0.41–3.07) | 0.83 | 1.31 (0.57–3.00) | 0.52 | |
| (25, 30] | 626 | 0.83 (066–1.06) | 0.13 | 0.94 (0.77–1.15) | 0.54 | 336 | 0.79 (0.55–1.12) | 0.18 | 0.85 (0.63–1.15) | 0.29 | |
| >30 | 315 | 0.58 (0.43–0.80) | 0.0008 | 0.83 (0.65–1.06) | 0.13 | 181 | 1.13 (0.77–1.65) | 0.54 | 1.04 (0.74–1.46) | 0.83 | |
| Smoking habit | No | 967 | Ref | Ref | 251 | Ref | Ref | ||||
| Yes b | 777 | 1.266 (1.049–1.529) | 0.01 | 1.27 (1.08–1.48) | 0.003 | 263 | 0.93 (0.65–1.32) | 0.67 | 1.02 (0.75–1.39) | 0.91 | |
| Stage | I | 277 | Ref | Ref | 188 | Ref | Ref | ||||
| II | 498 | 1.75 (1.10–2.80) | 0.02 | 1.99 (1.41–2.82) | 0.0001 | 227 | 1.00 (0.57–1.76) | 1.00 | 0.90 (0.56–1.45) | 0.67 | |
| III | 491 | 3.46 (2.22–5.39) | <0.0001 | 3.45 (2.47–4.83) | <0.0001 | 354 | 1.51 (0.92–2.45) | 0.10 | 1.55 (1.03–2.32) | 0.03 | |
| IV | 384 | 8.91 (5.78–13.74) | <0.0001 | 6.00 (4.30–8.38) | <0.0001 | 120 | 7.98 (4.95–12.88) | <0.0001 | 9.33 (6.21–14.02) | <0.0001 | |
| 5FU-based chemotherapy | No | 765 | Ref | Ref | 389 | Ref | Ref | ||||
| Yes | 797 | 1.022 (0.84–1.24) | 0.82 | 1.387 (1.18–1.63) | <0.0001 | 515 | 1.33 (0.99–1.78) | 0.06 | 1.79 (1.38–2.32) | <0.0001 | |
HR—hazard ratio; CI—confidence interval; BMI—body mass index. Significant results are in bold. a Numbers may not add up to 100% of the available subjects because of missing information. b Ex-smokers included in smokers.
Czech cohort. Overall, no SNPs were associated with either the overall survival (OS) or event free survival (EFS). However, after stratification according to tumor localization, nominally significant associations were detected for six SNPs in the univariate assessment (Table S1). In colon cancer patients, two SNPs were associated with increased EFS (rs3816032 and rs2283432; p = 0.02 for either variant genotype). In rectal cancer patients, one SNP was associated with an increased EFS (rs7689099; p = 0.02), and three with decreased OS or EFS (rs545500, rs3218649, and rs3087386; p = 0.02, 0.02, and 0.03, respectively).
Austrian cohort. Four SNPs were associated with either OS or EFS (Table S1). Three SNPs were observed in association with increased OS or EFS in CRC patients (rs12450550, rs2283432, and rs3204953; p = 0.03, p = 0.02, and p = 0.02, respectively). Rs3087386 was found to be significantly associated with decreased OS and EFS in colon cancer patients (p = 0.02).
2.4. Survival and Therapy
To examine the association of SNPs with the therapy outcome, we further stratified patients according to the treatment received into the following three separate groups: (1) CRC patients receiving no treatment or (2) patients receiving a 5-Fluorouracil (5-FU) regimen without or (3) in combination with oxaliplatin. The group of patients treated with a combination of 5-FU and oxaliplatin was investigated separately, because the latter drug induces a different type of DNA damage compared to 5-FU alone, and thus different DNA repair pathways and genes may be involved [16,17]. The univariate model for survival and therapy showed several genotypes nominally significantly associated with OS or EFS (detailed description in supplementary text and Tables S2, S3, and S4).
2.5. Classification and Regression Tree Survival Analysis
In order to assess the prognostic utility of the investigated DNA repair gene polymorphisms, the interactive effects of genotypes and clinico-pathological parameters in association with five-year OS and EFS were explored using a classification and regression tree (CART) analysis. Only patients with complete data for all of the parameters described in the Material and Methods were included in the analysis (n = 1105 (60%) for the Czech CRCs, and n = 841 (88%) for the Austrian CRCs). The results indicated that the tumor–node–metastasis (TNM) stage was chosen as the initial optimal split factor for predicting both OS and EFS in both of the cohorts (Figure 1, Figure 2, Figure 3 and Figure 4).
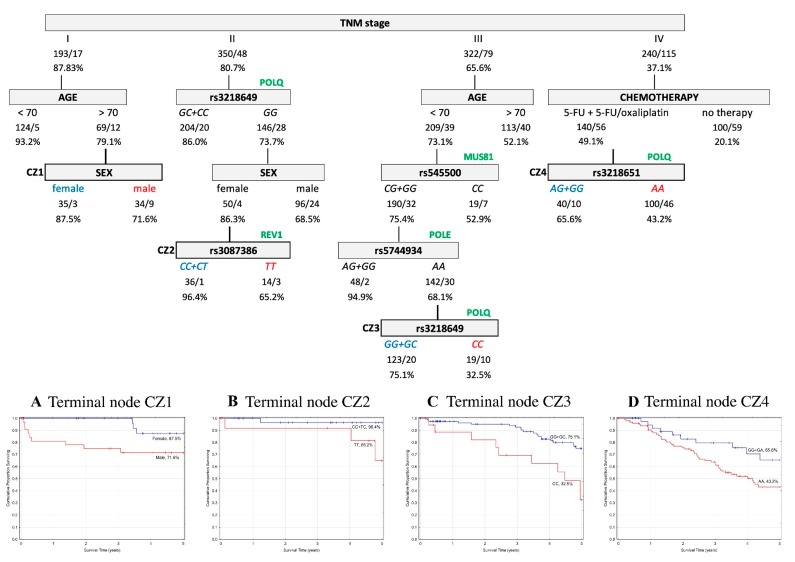
Figure 1.
Overall survival (OS) classification and regression tree analysis of colorectal cancer patients from the Czech Republic. Numbers under each node indicate the total number of cases in the subcategory/number of events and percentage of patients with five-year OS. Terminal nodes are bordered in bold, and the corresponding Kaplan–Meier curves are shown underneath. (A) Terminal node CZ1; (B) Terminal node CZ2; (C) Terminal node CZ3; (D) Terminal node CZ4.
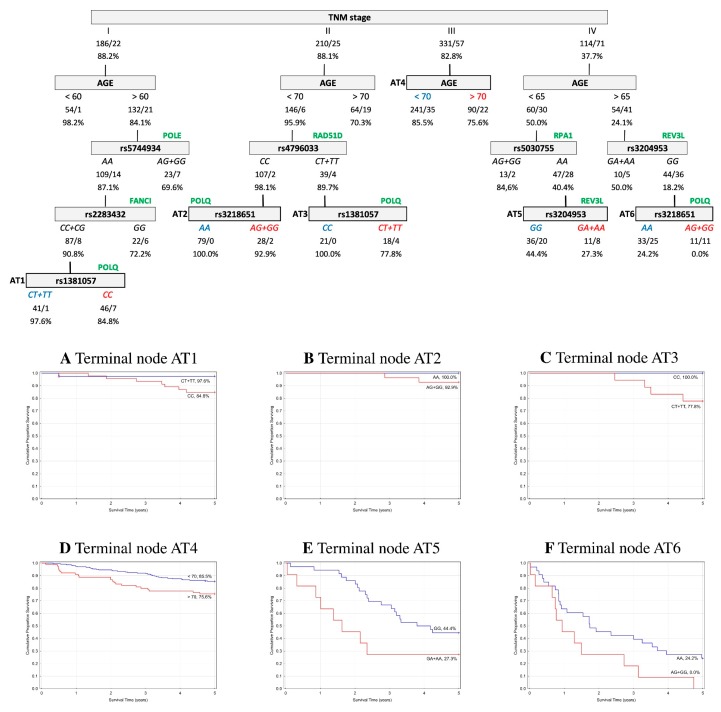
Figure 2.
Overall survival (OS) classification and regression tree analysis of colorectal cancer patients from Austria. Numbers under each node indicate the total number of cases in the subcategory/number of events, and the percentage of patients with five-year OS. Terminal nodes are bordered in bold, and the corresponding Kaplan–Meier curves are shown underneath. (A) Terminal node AT1; (B) Terminal node AT2; (C) Terminal node AT3; (D) Terminal node AT4; (E) Terminal node AT5; (F) Terminal node AT6.
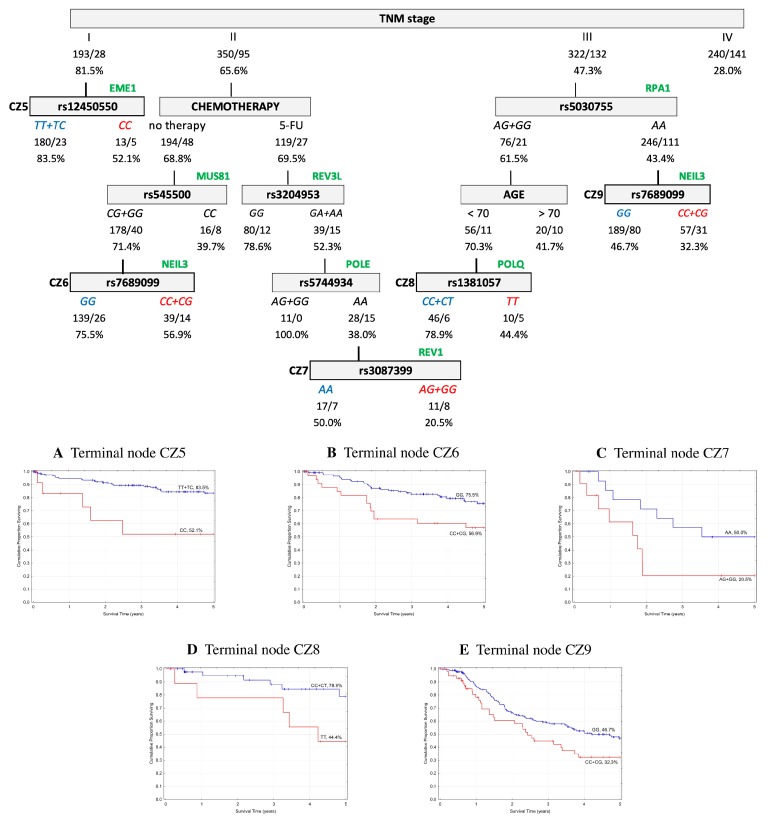
Figure 3.
Event free survival (EFS) classification and regression tree of colorectal cancer patients from the Czech Republic. Numbers under each node indicate the total number of cases in the subcategory/number of events, and the percentage of patients with five-year EFS. Terminal nodes are bordered in bold, and the corresponding Kaplan–Meier curves are shown underneath. (A) Terminal node CZ5; (B) Terminal node CZ6; (C) Terminal node CZ7; (D) Terminal node CZ8; (E) Terminal node CZ9.
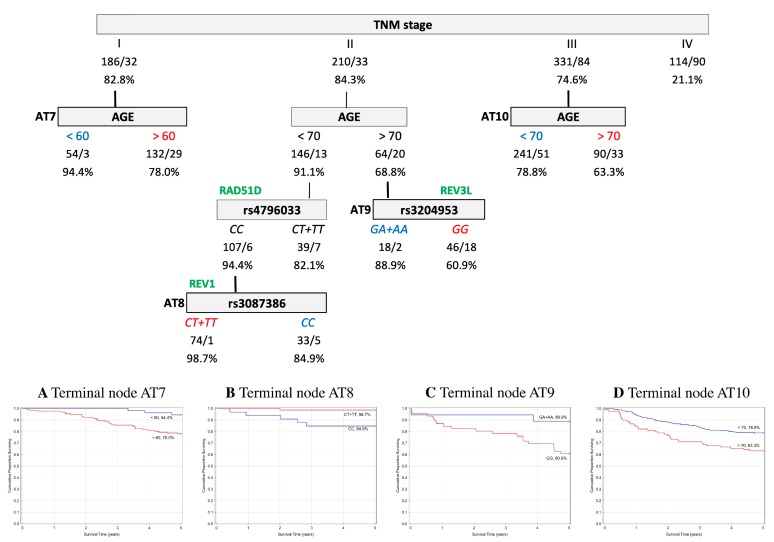
Figure 4.
Event free survival (EFS) classification and regression tree of colorectal cancer patients from Austria. Numbers under each node indicate the total number of cases in the subcategory/number of events and the percentage of patients with five-year EFS. Terminal nodes are bordered in bold, and the corresponding Kaplan–Meier curves are shown underneath. (A) Terminal node AT7; (B) Terminal node AT8; (C) Terminal node AT9; (D) Terminal node AT10.
2.5.1. Overall Survival
Czech cohort. The five-year OS analysis resulted in four terminal nodes. Variables determining the structure of the tree included TNM stage, age, sex, chemotherapy, and five SNPs—rs3087386, rs3218649, rs3218651, rs545500, and rs5744934. Among the stage I CRC patients, the subsequent split showed interactions between age and sex. In stage II, the carriers of GC+CC genotypes in POLQ rs3218649 were associated with a better prognosis. However, the GG genotype in females showed almost similar OS prognosis and an even better prognosis when in combination with CC+CT genotypes in REV1 rs3087386 (CC+CT 96.4% vs. TT 65.2%). In stage III, the subsequent split was age, which was seen to interact with GG+GC genotypes in MUS81 rs545500 and AG+GG genotypes in POLE rs5744934 (AG+GG 94.9% vs. AA 68.1%). The AA genotype of POLE gene further interacted with POLQ rs3218649 (GG+GC genotypes 75.1% vs. CC genotype 32.5%). In stage IV, chemotherapy was the next most significant factor and the level of OS increased when in combination with the rs3218651 variant in POLQ gene (AG+GG 65.6% vs. AA 43.2%). The structure of the tree and corresponding survival curves from terminal nodes are presented in Figure 1.
Austrian cohort. The final tree structure contained six terminal nodes and included nine variables (age, TNM stage, and seven SNPs—rs1381057, rs2283432, rs3204953, rs3218651, rs4796033, rs5030755, and rs5744934). Among CRC patients at stage I, age as the subsequent split showed interactions with the AA genotype in POLE rs5744934 and CC+CG genotypes in FANCI rs2283432. The interaction was concluded by CT+TT genotypes in POLQ rs1381057 (CT+TT 97.6% vs. CC 84.8%). In stage II, age was the next most significant factor, and the level of OS increased when in combination with the CC genotype in RAD51D rs4796033 and the AA genotype in POLQ rs3218649 (AA 100% vs. AG+GG 92.9%). Carriers of the CT+TT genotype in RAD51D rs4796033 showed a better prognosis in combination with the CC genotype in POLQ rs1381057 (CC 100% vs. CT+TT 77.8%). In stage III, age was the most significant factor for OS. In stage IV, age was further associated with three SNPs (RPA1 rs5030755 combined with REV3L rs3204953 as a terminal node: GG 44.4% vs. GA+AA 27.3%; and REV3L rs3204953 combined with POLQ rs3218651 as a terminal node: AA 24.2% vs. AG+GG 0%). The structure of the tree and corresponding survival curves from terminal nodes are presented in Figure 2.
2.5.2. Event-Free Survival
Czech cohort. Regarding the five-year EFS, the final tree structure contained five terminal nodes and included 11 variables (age, TNM stage, chemotherapy, and eight SNPs—rs12450550, rs1381057, rs3087399, rs3204953, rs5030755, rs545500, rs5744934, and rs7689099). Among CRC patients at stage I, the subsequent split was for EME1 rs12450550 (TT+TC 83.5% vs. CC 52.1%). In stage II, chemotherapy was the first split, when patients with no treatment and those with 5-FU-based therapy without oxaliplatin showed almost the same prognosis level. Patients without treatment had a better prognosis when associated with CG+GG genotypes in MUS81 rs545500 in combination with the GG genotype in NEIL3 rs7689099 (GG 75.5% vs. CC+CG 56.9%). Patients treated only with 5-FU had a better prognosis when in association with the GG genotype in REV3L rs3204953 (GG 78.6% vs. GA+AA 52.3%). On the other hand, the negative association for rs3204953 with the prognosis level was further worsened by the AA genotype in POLE rs5744934 and AG+GG genotype in REV1 rs3087399 (AA 50.0% vs. AG+GG 20.5%). In stage III, the subsequent split AG+GG genotype in RPA1 rs5030755 was seen to interact with patients under 70 years of age and the CC+CT genotype in POLQ rs1381057 (CC+CT 78.9% vs. TT 44.4%). Patients with the AA genotype for rs5030755 were further associated with a worse prognosis level in combination with the wild type allele C in NEIL3 (GG 46.7% vs. CC+CG 32.3%). The structure of the tree and corresponding survival curves from terminal nodes are presented in Figure 3.
Austrian cohort. The final tree structure contained four terminal nodes determined by five variables (age, TNM stage, and three SNPs—rs3087386, rs3204953, and rs4796033). In stage I, a better EFS prognosis was shown within patients under 60 years of age. In stage II, the subsequent split was age, which was seen to interact with the CC genotype in RAD51D rs4796033 and CT+TT genotypes in REV1 rs3087386 (CT+TT 98.7% vs. CC 84.9%). Furthermore, GA+AA genotypes in REV3L rs3204953 showed a better EFS prognosis in stage II patients over the age of 70 (GA+AA 88.9% vs. GG 60.9%). In stage III, the subsequent split showed an interaction with age only. The structure of the tree and corresponding survival curves from terminal nodes are presented in Figure 4.
3. Discussion
DNA repair has an essential role in maintaining genome integrity and preventing carcinogenesis. Amino acid alterations by nsSNPs in DNA repair genes can cause changes to the function or level of the coded proteins, resulting in abrogated DNA repair, which in combination with continuous endogenous DNA damage over time could lead to genomic damage and carcinogenesis [7,18]. In the present study, we sought to identify associations between 16 potentially functional genetic polymorphisms in 12 DNA repair genes with CRC risk, patients’ survival, and response to chemotherapy in Czech and Austrian cohorts. To our knowledge, no similar studies have previously examined these selected SNPs in relation to CRC susceptibility and clinical outcomes after diagnosis.
In the discovery set from the Czech Republic, the results showed an association between the variant AA genotype in REV3L rs3204953 and an increased risk of CRC. REV3L encodes a catalytic subunit of an error-prone DNA polymerase ζ, whose involvement in both double strand break (DSB) repair and translesion synthesis (TLS) pathways may explain why it is the only known specialized DNA polymerase reducing spontaneous tumor development [19,20]. DSBs, i.e. breaks in both DNA strands, are one of the most cytotoxic lesions for genetic integrity, and if not adequately repaired, DSB can result in mutagenic events or cell death [21]. TLS is a DNA damage tolerance process that allows cells to continue replication past DNA templates containing bulky lesions without resulting in stalled replication forks and therefore preventing DNA strand breaks.
Disrupted REV3L in cancer cell lines showed the importance of accurately regulated REV3L expression, when its inhibition induced DNA damage and growth arrest in cancer cells, whereas overexpression led to increased spontaneous mutation rates [22]. Expression levels of this polymerase have also been linked to sensitivity to chemotherapy. While defects in the protein resulted in an increased sensitivity to therapy in multiple tumor cell lines, its overexpression induced increased therapy resistance [23,24,25]. Furthermore, a decreased expression of REV3L has also been reported in tumor compared with the adjacent non-malignant tissue in colon cancer [26,27].
An association of rs3204953 was observed with a higher risk of breast cancer in a Swedish cohort, however, the results were not replicated in a Polish cohort [28]. Other genetic variants in REV3L have been found to be associated with both disease development risk and patients’ survival for different tumor types, such as breast cancer, stomach cancer, and CRC [28,29,30]. None of the other associated SNPs were found in linkage disequilibrium with rs3204953 examined here.
In addition to the in silico predictions of the F-SNP database of the deleterious nature of the rs3204953 SNP for REV3L protein function, we also used web-servers ELASPIC and DUET to assess the energetic impact of the amino acid change. In ELASPIC, the valine to isoleucine substitution was predicted to decrease the protein stability, resulting in a protein favoring an unfolded state (as the Gibbs free energy of folding for the domain affected by the SNP is changed by ∆∆G = 1.97).
Regarding the clinical outcome, results of the Czech five-year EFS CART analysis showed that rs3204953 in REV3L was chosen as the optimal split for the CRC stage II patients receiving 5-FU-based chemotherapy. This finding indicates its possible use in personalized treatment strategies by identifying CRC stage II patients who are likely to benefit from adjuvant therapy.
Despite the promising results in the Czech population, an association of REV3L SNP with CRC risk could not be confirmed in the Austrian replication set. However, REV3L emerged several times as the optimal split in the Austrian CART analyses as well. Thus, according to all of the available data, we suggest that the REV3L gene may impact CRC susceptibility, survival, and therapy outcomes and warrants further investigation.
In survival CART analyses, TNM stage and age were shown as the most significant prognostic factors in both of the study cohorts. Apart from these clinico-pathological factors, we observed significant associations of several nsSNPs with patients’ survival and clinical outcomes. However, a few of these were shown as significant more than once in the CART analysis, suggesting their potentially greater relevance on patients’ survival. For example, POLQ gene polymorphisms appeared four times as the optimal split factor in the Czech CART analyses (rs1381057, rs3218649 twice, and rs3218651) and four times in the Austrian CART analyses (rs1381057 twice and rs3218651 twice). Polymerase θ encoded by POLQ is an error-prone polymerase with a similar role to polymerase ζ, and is involved in the base excision repair (BER) and DSB repair [31]. In addition to DNA repair, this polymerase also plays a crucial role in TLS [32].
The expression of polymerase θ is tightly regulated. A complementary body of literature reported an upregulation of POLQ in different tumor tissues (breast cancer, non-small cell lung cancer, oral squamous cell carcinoma, stomach cancer, and CRC), and this overexpression acted as a strong prognostic factor [33,34,35,36].
Strikingly, at least nine polymorphisms out of 23 known SNPs in the human POLQ gene are predicted to alter protein function [32]. Several POLQ SNPs have also been associated with a risk of different tumors, such as breast cancer, esophageal cancer, and Non-Hodgkin’s Lymphoma [28,37,38,39,40]. While only some of the breast cancer studies included rs3218649, no significant association was detected [28,38,40] and none of the other associated SNPs were found in the linkage disequilibrium with our selected SNPs.
The abovementioned studies highlighted the significance of adequate POLQ functioning and regulation for tumor suppression. Furthermore, the protein stability prediction for rs1381057 by ELASPIC estimated a change of the Gibbs free energy to ∆∆G = 1.65, suggesting that the substitution of glutamine to arginine decreases the altered protein stability. Unfortunately, we could not perform a protein stability prediction of rs3218649 and rs3218651 by ELASPIC, as these SNPs do not fall within the domain boundaries required by the software.
Another SNP, rs7689099 in NEIL3 gene, emerged twice in the Czech five-year EFS CART analysis as the optimal split factor after the TNM stratification, suggesting its significance in patients´ survival. The NEIL3 encodes a DNA glycosylase, playing an important role in the first step of the BER pathway [41]. The process of eliminating damaged nucleotides by BER is crucial to evade mutations at these sites, which is likely to aid tumor suppression [42].
The upregulation of NEIL3 appears to be involved in the maintenance of cancer cell growth or the progression of malignancy. Significantly elevated expression levels in tumors, compared to corresponding non-malignant tissues, were reported in 20 cancer sites, including CRC [43,44]. The overexpression was further observed in association with the progression of primary melanoma to distant metastasis [45].
Sequence variability in different DNA glycosylases have been proposed as susceptibility factors for different malignancies [46]. Specifically, NEIL3 SNPs were associated with the risk of glioma, prostate, and thyroid cancer [47,48,49], with rs7689099 being associated with a reduced risk of differentiated thyroid carcinoma and prostate cancer [47,49]. None of the other associated SNPs were found in linkage disequilibrium with rs7689099.
As rs7689099 in NEIL3 gene does not fall within the domain boundaries, we could not use ELASPIC protein stability prediction. Again, the association of NEIL3 SNP with the survival of CRC patients was not replicated in the Austrian sample set. However, considering the available data, we suggest that the variation of the NEIL3 gene also has relevance for CRC susceptibility, survival, and therapy outcome.
In agreement with the in silico predictions about the functionality of the SNPs, we observed several significant associations of different genetic variants with survival and clinical outcomes of CRC patients both from the Czech Republic and Austria. However, we were not able to confirm the particular associations of individual SNPs between the discovery and the replication set. One might argue that the failure to replicate the association results might be due to differential gene–environmental interactions, and the differences in the clinical composition between the case-control populations of the discovery and the replication set (Table 2). Furthermore, it is also possible that other factors might have biased the results, for example earlier CRC detection in Austrian patients thanks to a better general awareness of the disease and a high standard medical care. This assumption is supported by the results in five-year OS CART analyses for stage III patients, where we observed a substantial difference between survival in the Czech and Austrian patients (65.6% vs. 82.8%). Our conclusion was based on the fact that CRC stage III is further divided into three more separate categories (IIIA, IIIB, and IIIC) according to the extent to which cancer has spread (i.e., number of lymph nodes affected). The survival rate then significantly decreases with the disease advancement. For example, in colon cancer patients, the survival rate for stage IIIA is about 90%, for stage IIIB 72%, and for stage IIIC only about 53% [50]. The strengths of the present work include the recruitment of a considerable number of cases and controls at the same centers, homogeneous for their ancestry (all Caucasian from the Czech Republic and Austria), and clinically well-defined (follow-up data collected by the same physicians), thus minimizing any possible population bias.
In conclusion, this is the first study to evaluate the association of genetic variants in DNA repair genes, selected by likely functional relevance with CRC. We identified several nsSNPs potentially affecting either CRC susceptibility or patients’ survival. Our data provide observational evidence of the potential role of nsSNPs in CRC pathogenesis, and suggest that even subtle alterations in the specific proteins that function in DNA repair pathways may lead to inaccurate DNA repair, and thus contribute to carcinogenesis.
Due to the lack of replication of significant associations, further studies on independent populations are warranted. This is underlined by the involvement of the same DNA repair genes in both Czech and Austrian CRC populations. Moreover, it is important to functionally characterize these candidate genetic variants, and to find biological mechanisms underlying the associations in order to assess these nsSNPs as prognostic and/or predictive biomarkers in CRC. Potential clinical uses are to help define individual CRC risk and tailor disease management based on the unique molecular profile of each patient.
4. Material and Methods
4.1. SNP Selection and In Silico Analysis of Functional Relevance and Conservation
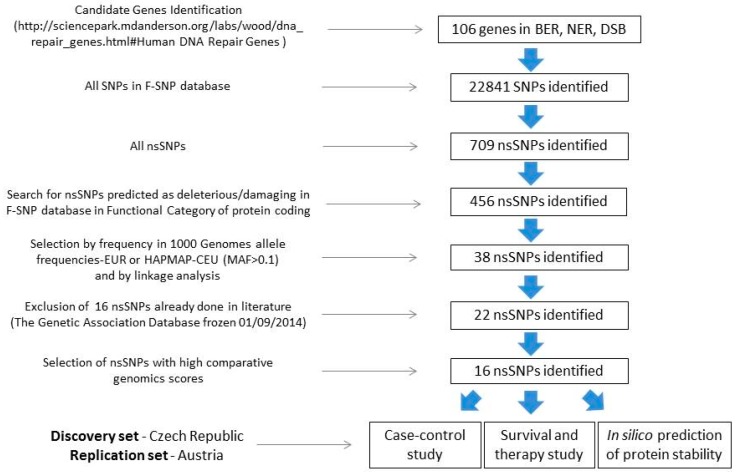
From the complete list of DNA repair genes available online (http://sciencepark.mdanderson.org/labs/ wood/DNA_Repair_Genes.html, March 2014 version), all of the genes involved in repairing DNA damage caused by 5-FU or oxaliplatin were retrieved, as these are common chemotherapeutic treatment regimens for CRC.
In total, 106 genes of BER, nucleotide excision repair (NER), and DSB (including interstrand cross-links repair (ICL), fanconi anemia (FA), and TLS pathways) were searched for nsSNPs in the freely available F-SNP database [51]. The database also provides integrated information about possible effects of the base change on the coded protein, and thus helps to identify nsSNPs with a potential pathological effect on human health. The F-SNP data are obtained from several genomic databases, like SIFT, PolyPhen2, SNPeffect, and SNPs3D. The variants predicted as deleterious or damaging were further studied.
Selected relevant nsSNPs were then filtered for a MAF >10% in European populations to provide sufficient study power with the size of our case-control study, in order to uncover moderate genetic effects. The information was primarily derived from the Ensembl 2015 database—1000 Genomes Project Phase 3, EUR population (https://www.ensembl.org/index.html). Whenever this was not possible, other reference populations were considered (i.e., HAPMAP CEU population).
The SNPs with the required MAF were tested for linkage disequilibrium (LD) with the data from HapMap (v. 3, release R2 in the CEU population, ftp://ftp.ncbi.nlm.nih.gov/hapmap/). The 38 identified nsSNPs were further searched within the Genetic Association Database (http://geneticassociationdb.nih.gov, accessed on 9 January 2014). From these, sixteen nsSNPs were already investigated elsewhere in relation to CRC, and therefore were excluded from this study.
The 22 remaining nsSNPs were tested by comparative genomics to evaluate the probability that the nucleotide is located in an evolutionary conserved position or within a constrained element, using the Genomic Evolutionary Rate Profiling GERP++ RS (Rejected Substitutions) score. An element with a GERP++ RS score >800 defines ultra-conserved regions among mammals. SiPhy evaluates the conservation of the motif around the SNPs.
After this selection, sixteen nsSNPs in 12 DNA repair genes complied with the required selection criteria. The workflow for the selection is depicted in Figure 5.
Figure 5.
Workflow strategy for the selection and analyses of functional non-synonymous polymorphisms in DNA repair genes. BER—base excision repair; NER—nucleotide excision repair; DSB—double strand break repair; SNP—single nucleotide polymorphism; nsSNP—non-synonymous SNP; MAF—minor allele frequency.
To evaluate the stability of the final protein affected by nsSNP, we further utilized web-server tools ELASPIC and DUET to assess the energetic impact of the amino acid change (http://elaspic.kimlab.org/ and http://biosig.unimelb.edu.au/duet/stability). The main output is the predicted variation in the Gibbs free energy (ΔΔG) of folding and/or binding for every domain affected by the SNP.
4.2. Study Populations and Data Collection
Patients included in the study were newly diagnosed histologically confirmed individuals with sporadic CRC. The exclusion criteria were as follows: (1) hereditary CRC forms (Lynch syndrome and familial adenomatous polyposis) and (2) a personal history of previous malignant disease. Personal data, such as date of birth, sex, lifestyle habits, body mass index (BMI), diabetes mellitus, and family/personal history of cancer, were obtained using a structured questionnaire in order to determine potential risk factors for CRC. For all subjects, clinical data including tumor-related parameters, such as the location of the tumor, International Union Against Cancer (UICC) TNM stage system, degree of tumor differentiation, and adjuvant chemotherapy treatment details, were collected, along with information about distant metastasis, relapse, and date of death.
Patients were divided into three subgroups according to the therapy received. The first group of patients did not receive any adjuvant chemotherapy after surgery. The second group of patients received a 5-FU-based adjuvant regimen as a postoperative therapy (based either on a Mayo, a simplified DeGramont, or a Xeloda regimen). The third group of subjects received adjuvant 5-FU treatment combined with oxaliplatin (based either on a FOLFOX or a XELOX regimen).
The study was approved by the local ethics committee of each participating hospital, and written informed consent to participate in the study and to approve the use of their biological samples for genetic analyses was obtained from all patients, according to the 1964 Helsinki declaration.
4.2.1. Discovery Set—Czech Republic
Patients (n = 1832) were recruited at several oncological and gastroenterological departments of different hospitals all over the Czech Republic from September 2003 to January 2014. The last update of the patients’ follow-up for this study was in December 2015. Characteristics of the study participants are shown in Table 2 (partially described in the literature [52,53].
The control group consisted of 659 healthy blood donors and 513 colonoscopy-negative controls, which were collected during the same time period as the cases. Healthy blood donor volunteers were recruited at the Faculty Hospital Kralovske Vinohrady in Prague and the Vojkov hospital. The group of colonoscopy-negative controls consisted of subjects admitted to the hospital gastroenterology departments who had negative colonoscopy results for malignancy or idiopathic bowel diseases. The reasons for undergoing the colonoscopy were as follows: (i) positive fecal occult blood test, (ii) hemorrhoids, (iii) abdominal pain of unknown origin, and (iv) macroscopic bleeding. All individuals were subjected to standard examinations so as to verify the health status for blood donation, and were cancer-free at the time of the sampling.
DNA was extracted from the peripheral blood lymphocytes using standard procedures. When blood was not available (for 690 cases), healthy colon/rectal tissue was used to obtain DNA by using the DNeasy Blood and Tissue Kit (Qiagen, Courtaboeuf, France). Genotyping was performed at LGC Genomics (Hoddesdon, Herts, UK), using the KASP™, a competitive allele-specific PCR genotyping system. For quality control purposes, duplicate samples (5% of the total numbers of samples) were repeated for each SNP. Two no-template controls were included in each plate. The genotype correlation between the duplicate samples was >98%. Two CRC cases were eliminated due to low genotyping rates.
4.2.2. Replication Set—Austria
In the ongoing Colorectal Cancer Study of Austria (CORSA), over 13,000 participants comprising CRC cases (stages I–IV); adenomas; and population-based, colonoscopy-negative controls have been recruited since 2003, in cooperation with the province-wide screening program “Burgenland Prevention Trial of Colorectal Disease with Immunological Testing” (B-PREDICT). All inhabitants of the Austrian province Burgenland aged between 40 and 80 years are invited annually to participate in fecal immunochemical testing (FIT). FIT-positive tested individuals are offered a complete colonoscopy and are asked to participate in CORSA at the time of colonoscopy. Only the individuals with histologically confirmed sporadic CRC were included in this study.
Further CRC cases were recruited at multiple centers in Vienna, including the Medical University of Vienna (Department of Surgery), the Sozialmedizinisches Zentrum Süd, the Hospital Rudolfstiftung, and the Medical University of Graz (Department of Internal Medicine). The replication set comprised 950 CRC patients and 820 colonoscopy-negative controls from CORSA. The last update of the patients’ follow-up was performed in August 2018. Baseline characteristics of this cohort are presented in Table 2 , and the study has previously been described [54].
The genomic DNA isolation from peripheral blood was performed using the QIAamp DNA Blood Midi Kit, according to the manufacturer’s recommendations (Qiagen, Valencia, CA, USA), and was stored at −80 °C. Genotyping was performed using the population-optimized Axiom Genome-Wide CEU 1 Array (Affymetrix, Santa Clara, CA, USA). The arrays were processed at the Institute of Human Genetics, Helmholtz Center Munich, Germany, and genotype assignment was performed as described in Hofer et al. [54]. Data for two SNPs (FAAP24 rs3816032 and MUS81 rs545500) were not covered on the array, and therefore could not be included in further analyses.
4.3. Statistical Analysis
In controls, the genotype frequencies for each polymorphism were tested for deviation from the Hardy–Weinberg equilibrium, using a Pearson χ2-test (1 degree of freedom) with a type-I error threshold set at α = 0.05.
The association between nsSNPs and CRC risk was determined by logistic regression, and was calculated by estimating the ORs, and their 95% CIs were adjusted for age. The ancestral allele (evolutionary primal) was used as a reference. For all nsSNPs, co-dominant, dominant, and recessive models were calculated.
In this study, the outcome variables measured were OS and EFS. OS was defined as the time from the surgery to the date of death, or the date of last follow up (for the Czech cohort it was December 2015, for the Austrian cohort it was August 2018). EFS was defined as the time from surgery to the occurrence of distant metastasis, local recurrence, or death, whichever came first. The survival curves for OS and EFS were derived by the Kaplan–Meier log-rank test. The relative risk of death and recurrence was estimated as a hazard ratio (HR) with 95% CIs, using Cox regression (no covariates adjustment was applied).
A multivariate analysis, referred to as a CART [55], was used to assess the prognostic value of interactions between the standard clinico-pathological variables and the genetic variants in relation to their impact on five-year survival in CRC patients. The analysis constructs a set of decision rules that stratify the homogenous risk groups of the responsive variable. Splits for each variable were examined, and the variable (predictor) that provides the best or “optimal” split was selected. Each subgroup was further divided in the same manner. In the Czech sample set, CART was implemented using nine common clinical and pathological variables, including age, sex, smoking habit (non-smokers vs. smokers vs. ex-smokers), diabetes mellitus, positive family history of CRC, diagnosis (colon vs. rectal cancer), TNM stage, grade, and therapy (no therapy vs. 5-FU-based without oxaliplatin vs. 5-FU in combination oxaliplatin), and all examined nsSNPs. In the Austrian sample set, because the information for five of the variables (smoking habit, positive family history of CRC, grade, and nsSNPs rs3816032 and rs545500) were only available for a small number of patients, only six common clinico-pathological variables and 14 nsSNPs were implemented for the CART analysis of this cohort.
Statistical analyses were performed using SAS software (SAS Institute, Cary, NC, USA). Graphs were performed using SW STATISTICA (StatSoft, Inc., Tulsa, OK, USA). Multiple testing corrections were performed using the Benjamini–Hochberg FDR [56].
Acknowledgments
This article is based upon work from COST Action CA17118, supported by COST (European Cooperation in Science and Technology, www.cost.eu).
Abbreviations
| 5-FU | 5-Fluorouracil |
| BER | Base excision repair |
| BMI | Body mass index |
| CART | Classification and regression tree analysis |
| CI | Confidence intervals |
| CRC | Colorectal cancer |
| DSB | Double strand break repair |
| EFS | Event-free survival |
| FA | Fanconi anemia |
| FDR | False discovery rate |
| GERP | Genomic evolutionary rate profiling |
| GWAS | Genome-wide association study |
| HRs | Hazard ratios |
| ICL | Interstrand cross-links repair |
| LD | Linkage disequilibrium |
| MAF | Minor allele frequency |
| NER | Nucleotide excision repair |
| nsSNP | Non-synonymous single nucleotide polymorphism |
| ORs | Odds ratios |
| OS | Overall survival |
| RS | Rejected substitutions |
| TLS | Translesion synthesis |
| TNM | Tumor–node–metastasis stage system |
| UICC | International union against cancer |
Supplementary Materials
Supplementary materials can be found at http://www.mdpi.com/1422-0067/20/1/97/s1.
Author Contributions
K.J., V.V. (V. Vymetalkova), and P.V. conceived the study. V.V. (V. Veskrnova), T.B., M.S., M.L., and V.L. performed hospital-based sample and data collection for the discovery set. K.J. performed data curation for the discovery set. S.B. and T.G. performed data curation for the replication set. K.J. and C.D.G. performed a computational prediction of the functional SNPs. K.J. and S.V. performed data analyses and interpreted the results with assistance from V.V. (V. Vymetalkova), A.N., and B.P. K.J. wrote the manuscript. P.V., D.J.H., and A.G. revised the manuscript. All of the authors read and approved the final manuscript for publication.
Funding
This study was supported by the Grant Agency of Charles University (GAUK112515) (KJ), the Health Research Board of Ireland project grant HRA_PHS/2015/1142 (DJH), the Centrum of Clinical and Experimental liver surgery (UNCE/MED/006), the National Sustainability Program I (NPU I) Nr. LO1503 provided by the Ministry of Education Youth and Sports of the Czech Republic (VL), the Grant Agency of the Czech Republic (GACR 17-16857S) (AN), the Fondazione Umberto Veronesi “Post-doctoral fellowship Year 2014–2017” (BP), the Internal Grant Agency of the Ministry of Health of the Czech Republic (AZV MZ 15-26535A and AZV 17-30920A) (VV), the FFG BRIDGE (grant 829675), and the Herzfelder´sche Familienstiftung (AG) and the Grant Agency of the Czech Republic (GACR 15-14789S) (PV).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018 doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J., Steliarova-Foucher E., Lortet-Tieulent J., Rosso S., Coebergh J.W., Comber H., Forman D., Bray F. Cancer incidence and mortality patterns in Europe: Estimates for 40 countries in 2012. Eur. J. Cancer. 2013;49:1374–1403. doi: 10.1016/j.ejca.2012.12.027. [DOI] [PubMed] [Google Scholar]
- 3.Aran V., Victorino A.P., Thuler L.C., Ferreira C.G. Colorectal Cancer: Epidemiology, Disease Mechanisms and Interventions to Reduce Onset and Mortality. Clin. Colorectal Cancer. 2016;15:195–203. doi: 10.1016/j.clcc.2016.02.008. [DOI] [PubMed] [Google Scholar]
- 4.Binefa G., Rodriguez-Moranta F., Teule A., Medina-Hayas M. Colorectal cancer: From prevention to personalized medicine. World J. Gastroenterol. 2014;20:6786–6808. doi: 10.3748/wjg.v20.i22.6786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gustavsson B., Carlsson G., Machover D., Petrelli N., Roth A., Schmoll H.J., Tveit K.M., Gibson F. A review of the evolution of systemic chemotherapy in the management of colorectal cancer. Clin. Colorectal Cancer. 2015;14:1–10. doi: 10.1016/j.clcc.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 6.Brenner H., Kloor M., Pox C.P. Colorectal cancer. Lancet. 2014;383:1490–1502. doi: 10.1016/S0140-6736(13)61649-9. [DOI] [PubMed] [Google Scholar]
- 7.Vodicka P., Stetina R., Polakova V., Tulupova E., Naccarati A., Vodickova L., Kumar R., Hanova M., Pardini B., Slyskova J., et al. Association of DNA repair polymorphisms with DNA repair functional outcomes in healthy human subjects. Carcinogenesis. 2007;28:657–664. doi: 10.1093/carcin/bgl187. [DOI] [PubMed] [Google Scholar]
- 8.He J., Shi T.Y., Zhu M.L., Wang M.Y., Li Q.X., Wei Q.Y. Associations of Lys939Gln and Ala499Val polymorphisms of the XPC gene with cancer susceptibility: A meta-analysis. Int. J. Cancer. 2013;133:1765–1775. doi: 10.1002/ijc.28089. [DOI] [PubMed] [Google Scholar]
- 9.Slyskova J., Naccarati A., Pardini B., Polakova V., Vodickova L., Smerhovsky Z., Levy M., Lipska L., Liska V., Vodicka P. Differences in nucleotide excision repair capacity between newly diagnosed colorectal cancer patients and healthy controls. Mutagenesis. 2012;27:225–232. doi: 10.1093/mutage/ger088. [DOI] [PubMed] [Google Scholar]
- 10.Peters U., Jiao S., Schumacher F.R., Hutter C.M., Aragaki A.K., Baron J.A., Berndt S.I., Bezieau S., Brenner H., Butterbach K., et al. Identification of Genetic Susceptibility Loci for Colorectal Tumors in a Genome-Wide Meta-analysis. Gastroenterology. 2013;144:799–807.e24. doi: 10.1053/j.gastro.2012.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Whiffin N., Hosking F.J., Farrington S.M., Palles C., Dobbins S.E., Zgaga L., Lloyd A., Kinnersley B., Gorman M., Tenesa A., et al. Identification of susceptibility loci for colorectal cancer in a genome-wide meta-analysis. Hum. Mol. Genet. 2014;23:4729–4737. doi: 10.1093/hmg/ddu177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shen C., Yan T., Wang Z., Su H.C., Zhu X., Tian X., Fang J.Y., Chen H., Hong J. Variant of SNP rs1317082 at CCSlnc362 (RP11-362K14.5) creates a binding site for miR-4658 and diminishes the susceptibility to CRC. Cell Death Dis. 2018;9:1177. doi: 10.1038/s41419-018-1222-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tanskanen T., van den Berg L., Valimaki N., Aavikko M., Ness-Jensen E., Hveem K., Wettergren Y., Bexe Lindskog E., Tonisson N., Metspalu A., et al. Genome-wide association study and meta-analysis in Northern European populations replicate multiple colorectal cancer risk loci. Int. J. Cancer. 2018;142:540–546. doi: 10.1002/ijc.31076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huyghe J.R., Bien S.A., Harrison T.A., Kang H.M., Chen S., Schmit S.L., Conti D.V., Qu C., Jeon J., Edlund C.K., et al. Discovery of common and rare genetic risk variants for colorectal cancer. Nat. Genet. 2018 doi: 10.1038/s41588-018-0286-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Slyskova J., Cordero F., Pardini B., Korenkova V., Vymetalkova V., Bielik L., Vodickova L., Pitule P., Liska V., Matejka V.M., et al. Post-treatment recovery of suboptimal DNA repair capacity and gene expression levels in colorectal cancer patients. Mol. Carcinog. 2015;54:769–778. doi: 10.1002/mc.22141. [DOI] [PubMed] [Google Scholar]
- 16.Wyatt M.D., Wilson D.M., 3rd Participation of DNA repair in the response to 5-fluorouracil. Cell. Mol. Life Sci. 2009;66:788–799. doi: 10.1007/s00018-008-8557-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin L.P., Hamilton T.C., Schilder R.J. Platinum resistance: The role of DNA repair pathways. Clin. Cancer Res. 2008;14:1291–1295. doi: 10.1158/1078-0432.CCR-07-2238. [DOI] [PubMed] [Google Scholar]
- 18.De Mattia E., Cecchin E., Toffoli G. Pharmacogenomics of intrinsic and acquired pharmacoresistance in colorectal cancer: Toward targeted personalized therapy. Drug Resist. Updat. 2015;20:39–70. doi: 10.1016/j.drup.2015.05.003. [DOI] [PubMed] [Google Scholar]
- 19.Vaisman A., Woodgate R. Translesion DNA polymerases in eukaryotes: What makes them tick? Crit. Rev. Biochem. Mol. Biol. 2017;52:274–303. doi: 10.1080/10409238.2017.1291576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wittschieben J.P., Patil V., Glushets V., Robinson L.J., Kusewitt D.F., Wood R.D. Loss of DNA polymerase zeta enhances spontaneous tumorigenesis. Cancer Res. 2010;70:2770–2778. doi: 10.1158/0008-5472.CAN-09-4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aparicio T., Baer R., Gautier J. DNA double-strand break repair pathway choice and cancer. DNA Repair (Amst.) 2014;19:169–175. doi: 10.1016/j.dnarep.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knobel P.A., Marti T.M. Translesion DNA synthesis in the context of cancer research. Cancer Cell Int. 2011;11:39. doi: 10.1186/1475-2867-11-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Galluzzi L., Senovilla L., Vitale I., Michels J., Martins I., Kepp O., Castedo M., Kroemer G. Molecular mechanisms of cisplatin resistance. Oncogene. 2012;31:1869–1883. doi: 10.1038/onc.2011.384. [DOI] [PubMed] [Google Scholar]
- 24.Wang H., Zhang S.Y., Wang S., Lu J., Wu W., Weng L., Chen D., Zhang Y., Lu Z., Yang J., et al. REV3L confers chemoresistance to cisplatin in human gliomas: The potential of its RNAi for synergistic therapy. Neuro Oncol. 2009;11:790–802. doi: 10.1215/15228517-2009-015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roos W.P., Tsaalbi-Shtylik A., Tsaryk R., Guvercin F., de Wind N., Kaina B. The translesion polymerase Rev3L in the tolerance of alkylating anticancer drugs. Mol. Pharmacol. 2009;76:927–934. doi: 10.1124/mol.109.058131. [DOI] [PubMed] [Google Scholar]
- 26.Stallons L.J., McGregor W.G. Translesion synthesis polymerases in the prevention and promotion of carcinogenesis. J. Nucleic Acids. 2010;2010:643857. doi: 10.4061/2010/643857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brondello J.M., Pillaire M.J., Rodriguez C., Gourraud P.A., Selves J., Cazaux C., Piette J. Novel evidences for a tumor suppressor role of Rev3, the catalytic subunit of Pol zeta. Oncogene. 2008;27:6093–6101. doi: 10.1038/onc.2008.212. [DOI] [PubMed] [Google Scholar]
- 28.Varadi V., Bevier M., Grzybowska E., Johansson R., Enquist K., Henriksson R., Butkiewicz D., Pamula-Pilat J., Tecza K., Hemminki K., et al. Genetic variation in genes encoding for polymerase zeta subunits associates with breast cancer risk, tumour characteristics and survival. Breast Cancer Res. Treat. 2011;129:235–245. doi: 10.1007/s10549-011-1460-z. [DOI] [PubMed] [Google Scholar]
- 29.Pan J., Chi P., Lu X., Xu Z. Genetic polymorphisms in translesion synthesis genes are associated with colorectal cancer risk and metastasis in Han Chinese. Gene. 2012;504:151–155. doi: 10.1016/j.gene.2012.05.042. [DOI] [PubMed] [Google Scholar]
- 30.Hussain S.K., Mu L.N., Cai L., Chang S.C., Park S.L., Oh S.S., Wang Y., Goldstein B.Y., Ding B.G., Jiang Q., et al. Genetic variation in immune regulation and DNA repair pathways and stomach cancer in China. Cancer Epidemiol. Biomark. Prev. 2009;18:2304–2309. doi: 10.1158/1055-9965.EPI-09-0233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yousefzadeh M.J., Wood R.D. DNA polymerase POLQ and cellular defense against DNA damage. DNA Repair (Amst.) 2013;12:1–9. doi: 10.1016/j.dnarep.2012.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beagan K., McVey M. Linking DNA polymerase theta structure and function in health and disease. Cell. Mol. Life Sci. 2016;73:603–615. doi: 10.1007/s00018-015-2078-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kawamura K., Bahar R., Seimiya M., Chiyo M., Wada A., Okada S., Hatano M., Tokuhisa T., Kimura H., Watanabe S., et al. DNA polymerase theta is preferentially expressed in lymphoid tissues and upregulated in human cancers. Int. J. Cancer. 2004;109:9–16. doi: 10.1002/ijc.11666. [DOI] [PubMed] [Google Scholar]
- 34.Lemee F., Bergoglio V., Fernandez-Vidal A., Machado-Silva A., Pillaire M.J., Bieth A., Gentil C., Baker L., Martin A.L., Leduc C., et al. DNA polymerase theta up-regulation is associated with poor survival in breast cancer, perturbs DNA replication, and promotes genetic instability. Proc. Natl. Acad. Sci. USA. 2010;107:13390–13395. doi: 10.1073/pnas.0910759107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Allera-Moreau C., Rouquette I., Lepage B., Oumouhou N., Walschaerts M., Leconte E., Schilling V., Gordien K., Brouchet L., Delisle M.B., et al. DNA replication stress response involving PLK1, CDC6, POLQ, RAD51 and CLASPIN upregulation prognoses the outcome of early/mid-stage non-small cell lung cancer patients. Oncogenesis. 2012;1:e30. doi: 10.1038/oncsis.2012.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pillaire M.J., Selves J., Gordien K., Gourraud P.A., Gentil C., Danjoux M., Do C., Negre V., Bieth A., Guimbaud R., et al. A ‘DNA replication’ signature of progression and negative outcome in colorectal cancer. Oncogene. 2010;29:876–887. doi: 10.1038/onc.2009.378. [DOI] [PubMed] [Google Scholar]
- 37.Li W.Q., Hu N., Hyland P.L., Gao Y., Wang Z.M., Yu K., Su H., Wang C.Y., Wang L.M., Chanock S.J., et al. Genetic variants in DNA repair pathway genes and risk of esophageal squamous cell carcinoma and gastric adenocarcinoma in a Chinese population. Carcinogenesis. 2013;34:1536–1542. doi: 10.1093/carcin/bgt094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brandalize A.P., Schuler-Faccini L., Hoffmann J.S., Caleffi M., Cazaux C., Ashton-Prolla P. A DNA repair variant in POLQ (c.-1060A > G) is associated to hereditary breast cancer patients: A case-control study. BMC Cancer. 2014;14:850. doi: 10.1186/1471-2407-14-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rendleman J., Antipin Y., Reva B., Adaniel C., Przybylo J.A., Dutra-Clarke A., Hansen N., Heguy A., Huberman K., Borsu L., et al. Genetic variation in DNA repair pathways and risk of non-Hodgkin’s lymphoma. PLoS ONE. 2014;9:e101685. doi: 10.1371/journal.pone.0101685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Family L., Bensen J.T., Troester M.A., Wu M.C., Anders C.K., Olshan A.F. Single-nucleotide polymorphisms in DNA bypass polymerase genes and association with breast cancer and breast cancer subtypes among African Americans and Whites. Breast Cancer Res. Treat. 2015;149:181–190. doi: 10.1007/s10549-014-3203-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krokeide S.Z., Laerdahl J.K., Salah M., Luna L., Cederkvist F.H., Fleming A.M., Burrows C.J., Dalhus B., Bjoras M. Human NEIL3 is mainly a monofunctional DNA glycosylase removing spiroimindiohydantoin and guanidinohydantoin. DNA Repair (Amst.) 2013;12:1159–1164. doi: 10.1016/j.dnarep.2013.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wallace S.S., Murphy D.L., Sweasy J.B. Base excision repair and cancer. Cancer Lett. 2012;327:73–89. doi: 10.1016/j.canlet.2011.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hildrestrand G.A., Neurauter C.G., Diep D.B., Castellanos C.G., Krauss S., Bjoras M., Luna L. Expression patterns of Neil3 during embryonic brain development and neoplasia. BMC Neurosci. 2009;10:45. doi: 10.1186/1471-2202-10-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shinmura K., Kato H., Kawanishi Y., Igarashi H., Goto M., Tao H., Inoue Y., Nakamura S., Misawa K., Mineta H., et al. Abnormal Expressions of DNA Glycosylase Genes NEIL1, NEIL2, and NEIL3 Are Associated with Somatic Mutation Loads in Human Cancer. Oxid. Med. Cell. Longev. 2016;2016:1546392. doi: 10.1155/2016/1546392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kauffmann A., Rosselli F., Lazar V., Winnepenninckx V., Mansuet-Lupo A., Dessen P., van den Oord J.J., Spatz A., Sarasin A. High expression of DNA repair pathways is associated with metastasis in melanoma patients. Oncogene. 2008;27:565–573. doi: 10.1038/sj.onc.1210700. [DOI] [PubMed] [Google Scholar]
- 46.D’Errico M., Parlanti E., Pascucci B., Fortini P., Baccarini S., Simonelli V., Dogliotti E. Single nucleotide polymorphisms in DNA glycosylases: From function to disease. Free Radic. Biol. Med. 2017;107:278–291. doi: 10.1016/j.freeradbiomed.2016.12.002. [DOI] [PubMed] [Google Scholar]
- 47.Barry K.H., Koutros S., Berndt S.I., Andreotti G., Hoppin J.A., Sandler D.P., Burdette L.A., Yeager M., Freeman L.E., Lubin J.H., et al. Genetic variation in base excision repair pathway genes, pesticide exposure, and prostate cancer risk. Environ. Health Perspect. 2011;119:1726–1732. doi: 10.1289/ehp.1103454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bethke L., Webb E., Murray A., Schoemaker M., Johansen C., Christensen H.C., Muir K., McKinney P., Hepworth S., Dimitropoulou P., et al. Comprehensive analysis of the role of DNA repair gene polymorphisms on risk of glioma. Hum. Mol. Genet. 2008;17:800–805. doi: 10.1093/hmg/ddm351. [DOI] [PubMed] [Google Scholar]
- 49.Cipollini M., Figlioli G., Maccari G., Garritano S., De Santi C., Melaiu O., Barone E., Bambi F., Ermini S., Pellegrini G., et al. Polymorphisms within base and nucleotide excision repair pathways and risk of differentiated thyroid carcinoma. DNA Repair (Amst.) 2016;41:27–31. doi: 10.1016/j.dnarep.2016.03.011. [DOI] [PubMed] [Google Scholar]
- 50.Allione A., Pardini B., Viberti C., Oderda M., Allasia M., Gontero P., Vineis P., Sacerdote C., Matullo G. The prognostic value of basal DNA damage level in peripheral blood lymphocytes of patients affected by bladder cancer. Urol. Oncol. 2018;36:241.e15–241.e23. doi: 10.1016/j.urolonc.2018.01.006. [DOI] [PubMed] [Google Scholar]
- 51.Lee P.H., Shatkay H. F-SNP: Computationally predicted functional SNPs for disease association studies. Nucleic Acids Res. 2008;36:D820–D824. doi: 10.1093/nar/gkm904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vymetalkova V., Pardini B., Rosa F., Jiraskova K., Di Gaetano C., Bendova P., Levy M., Veskrnova V., Buchler T., Vodickova L., et al. Polymorphisms in microRNA binding sites of mucin genes as predictors of clinical outcome in colorectal cancer patients. Carcinogenesis. 2017;38:28–39. doi: 10.1093/carcin/bgw114. [DOI] [PubMed] [Google Scholar]
- 53.Schneiderova M., Naccarati A., Pardini B., Rosa F., Gaetano C.D., Jiraskova K., Opattova A., Levy M., Veskrna K., Veskrnova V., et al. MicroRNA-binding site polymorphisms in genes involved in colorectal cancer etiopathogenesis and their impact on disease prognosis. Mutagenesis. 2017;32:533–542. doi: 10.1093/mutage/gex026. [DOI] [PubMed] [Google Scholar]
- 54.Hofer P., Hagmann M., Brezina S., Dolejsi E., Mach K., Leeb G., Baierl A., Buch S., Sutterluty-Fall H., Karner-Hanusch J., et al. Bayesian and frequentist analysis of an Austrian genome-wide association study of colorectal cancer and advanced adenomas. Oncotarget. 2017;8:98623–98634. doi: 10.18632/oncotarget.21697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lemon S.C., Roy J., Clark M.A., Friedmann P.D., Rakowski W. Classification and regression tree analysis in public health: Methodological review and comparison with logistic regression. Ann. Behav. Med. 2003;26:172–181. doi: 10.1207/S15324796ABM2603_02. [DOI] [PubMed] [Google Scholar]
- 56.Benjamini Y., Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Statist. Soc. B. 1995;57:289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.