Abstract
Facioscapulohumeral muscular dystrophy (FSHD) is a common, dominantly inherited disease caused by the epigenetic de-repression of the DUX4 gene, a transcription factor normally repressed in skeletal muscle. As targeted therapies are now possible in FSHD, a better understanding of the relationship between DUX4 activity, muscle pathology and muscle magnetic resonance imaging (MRI) changes is crucial both to understand disease mechanisms and for the design of future clinical trials. Here, we performed MRIs of the lower extremities in 36 individuals with FSHD, followed by needle muscle biopsies in safely accessible muscles. We examined the correlation between MRI characteristics, muscle pathology and expression of DUX4 target genes. Results show that the presence of elevated MRI short tau inversion recovery signal has substantial predictive value in identifying muscles with active disease as determined by histopathology and DUX4 target gene expression. In addition, DUX4 target gene expression was detected only in FSHD-affected muscles and not in control muscles. These results support the use of MRI to identify FSHD muscles most likely to have active disease and higher levels of DUX4 target gene expression and might be useful in early phase therapeutic trials to demonstrate target engagement in therapies aiming to suppress DUX4 expression.
Introduction
Facioscapulohumeral muscular dystrophy (FSHD) is the second most common form of adult muscular dystrophy with an estimated prevalence range of 2–7 per 100 000 people (1,2). The clinical spectrum of disease severity is wide, while the regional distribution of muscle weakness and the pattern of progression are unique. Symptomatic muscle weakness typically starts in the face and shoulders with a descending progression to involve the truncal muscles and then the lower extremity muscles. There is currently no effective treatment for FSHD (3). A unifying model has now emerged implicating epigenetic de-repression, caused by a critical contraction in the number of D4Z4 macrosatellite repeats on chromosome 4q35 (FSHD1), resulting in the aberrant expression of DUX4, a gene normally silenced in skeletal muscle (4,5). The reactivation of DUX4 causes disease by a toxic gain of function. The accuracy of this disease model is reinforced by the discovery of a phenotypically similar but genetically distinct form of FSHD (FSHD2) (6–8). In FSHD1, epigenetic de-repression and DUX4 expression occurs in the setting of contraction in the D4Z4 repeat number. In FSHD2, mutations in one of two chromatin modifier genes on different chromosomes, SMCHD1 and the DNMT3B, induce epigenetic de-repression of the D4Z4 repeats without a contraction in the number of D4Z4 repeats (6,9). The DUX4 protein is a transcription factor and its expression interferes with myogenic differentiation, which leads to apoptotic cell death and increases susceptibility to oxidative stress (10–13). This unified FSHD model has provided, for the first time, therapeutic targets for FSHD. As FSHD research enters a therapeutic-trial phase, the identification of markers of disease activity becomes critical for clinical trial design (14,15). Histopathologic analysis can grade the degree of disease activity based on specified criteria (16); however, within an individual, some muscle biopsies appear almost normal and others show active disease, and some muscle biopsies show evidence of DUX4 target expression and others do not. Our inability to predict muscles with active disease or DUX4 expression is a current limitation for clinical trial design.
Recently, several studies showing magnetic resonance imaging (MRI) changes in FSHD muscle suggest that MRI might be a measure of disease progression and a method to identify muscles with active disease based on histopathology and/or DUX4 expression. These studies suggest that MRI normal muscles transition to an early abnormality in the short tau inversion recovery (STIR) signal and that this leads to higher longitudinal relaxation (T1) signal as the disease progresses (17,18). Because STIR is sensitive to increased free water and T1 to fat content, this has led to the suggestion that FSHD muscle involvement begins with an edematous or inflammatory phase that progress to fatty infiltration and fibrosis. Just as this progression sequence requires further data for confirmation, it is critical to understand the underlying histopathology and the association with DUX4 mRNA and DUX4-regulated gene expression (detected in a subset of FSHD muscle biopsies (19)). Toward this end, we performed an MRI of the lower extremity muscles on 36 FSHD individuals and obtained a needle biopsy of a safely accessible muscle to understand the relationship between MRI characteristics, histopathologic score and the expression of DUX4 candidate biomarker genes.
Results
Study cohort
Recruitment for the study included 36 subjects (34% female) with genetically confirmed FSHD (34 FSHD1, 2 FSHD2). The mean subject age was 54 years (range, 20–75 years) and the mean Clinical Severity Score (CSS) score was 5 (range, 0–9). Supplementary Material, Table S1 presents subject demographics, genotype, disease severity, muscle biopsied, pathologic and MRI scores. All 36 subjects underwent MRIs of the lower extremities and needle muscle biopsies without complications. In three subjects, the histopathologic samples were inadequate for histopathologic scoring, and in two subjects, a muscle sample for RNA extraction and sequencing was not successfully obtained.
Quadriceps muscle biopsy control samples collected from six females and three males with a mean age of 35 years (range, 19–56 years) came from unaffected spouses, parents or siblings of FSHD individuals evaluated at the University of Rochester. All unaffected, at-risk individuals were genetically confirmed as such.
MRI characteristics of biopsied muscles
Of the 36 biopsied muscles, 2 were restricted to only T1 fatty/fibrous changes without STIR signal, 10 demonstrated regions of elevated STIR signal (STIR+, rating > 0) without T1-evidence for fatty/fibrous involvement, 14 had abnormalities in both STIR and T1 and 10 were rated as ‘normal-appearing’ muscle. A stepped progression was observed comparing fat replacement ratings versus DIXON fraction, with the caveat that the ratings reflect not simply percentage but also confluence of fatty infiltration [fat = 0 (N = 20): mean = 0.08, range 0.01–0.21; fat = 1 (N = 9): mean = 0.14, range 0.06–0.23; fat = 2 (N = 1): mean = 0.15; fat = 3 (N = 3): mean = 0.43, range = 0.42–0.45; fat = 4 (N = 2): mean = 0.43, 0.32–0.54; fat = 5 (N = 1): mean = 0.89]. This concordance was also observed in T1 rating and DIXON correlation (rs = 0.65, P < 0.001).
FSHD muscle histopathology
Histologic samples were collected from 33 subjects. Muscles biopsied included 6 vastus lateralis, 1 vastus medialis, 14 gastrocnemii, 14 tibialis anterior and 1 hamstring (biceps femoris). The range for pathologic score was 0–10 with a mean in the moderate range of severity at 5.18 (SD, 2.62). There was no significant difference in mean histopathologic severity between the three major muscle groups targeted for biopsy: quadriceps [mean = 4.33 ± 2.63 (SD)], tibialis anterior (mean = 5.08 ± 2.47) and gastrocnemii (mean = 5.31 ± 2.63).
Histopathologic changes included increased variability in fiber size observed in 97% (32/33), fibrosis observed in 67% (22/33) of samples and central nucleation in 82% (27/33) of muscle biopsies. Active myopathic changes, as defined by presence of inflammation or muscle fiber necrosis and regeneration, were seen in 55% (18/33) of samples. Inflammatory infiltrates, ranging from mild to severe, were observed in 33% (11/33). A total of 21% (7/33) of biopsies had evidence of muscle fiber necrosis and regeneration but did not have inflammatory infiltrates.
Correlation of MRI characteristics to histopathology
In the 10 FSHD biopsied muscles with normal whole muscle MRI, the pathologic changes were mild, with a mean pathology score of 3.5 (range 0–7) with only one (10%) biopsy showing mild active myopathic changes as defined by the presence of inflammation or the presence of muscle fiber necrosis or regeneration. Despite the lack of active myopathic changes in all but one muscle, 50% of the biopsies had mild to moderate fibrosis, suggestive of a chronic process.
In contrast, in the 22 STIR+ muscles, the mean pathology scores were moderate in severity (mean score of 5.9, range 1–10) and 16 (73%) of the 22 STIR+ muscles had active myopathic changes, including 10 (45%) with inflammation. Of the STIR+ muscles, the 12 with T1 changes had moderately severe pathologic changes (mean 7.0, range 2–10) compared to mild pathologic changes in muscles without T1 changes (mean 4.6, range 1–10). Of the muscles with both increased T1 signal and STIR positivity, all had fibrotic changes. Examples of MRI normal and STIR+ muscles and their corresponding biopsies are shown in Supplementary Material, Figure S1.
Pathology correlated with both fat and STIR measures, with whole muscle burden mostly associated with total pathology scores (T1 score: mean ± SD = 0.75 ± 1.19, rs = 0.61, P < 0.001; T1 fat fraction: mean ± SD = 0.14 ± 0.13, rs = 0.58, P = 0.001) (Supplementary Material, Fig. S2). Evaluating the subgroup of subjects with T1 scores of zero and correlating their corresponding DIXON fat fraction with pathology demonstrated a trend association (rs = 0.35, P = 0.14), supporting the increased discrimination possible with quantitative measurement; STIR muscle ratings were also associated with pathology, though the association was less robust (STIR score: mean ± SD = 1.38 ± 1.16, rs = 0.42, P = 0.017).
DUX4 candidate biomarkers elevated in FSHD samples
RNA-Seq was performed on muscle biopsy samples from 34 of the 36 FSHD subjects and on nine control quadriceps muscle biopsies from unaffected individuals. The initial analysis focused on the expression of candidate biomarkers regulated by DUX4 that we identified in a prior study, either a) an extended set of 54 genes or b) a restricted subset of four-candidate biomarkers (TRIM43, LEUTX, PRAMEF2, KHDC1L) that were previously shown to be as informative as the extended set at distinguishing FSHD from control biopsies (19). The control muscle biopsies had zero reads mapping to most of these candidate biomarkers with only a few genes showing a small number of reads (all less than 0.1 RPKM) for each sample, whereas most of the FSHD biopsies had higher numbers of reads mapping to each of these genes (Supplementary Material, Table S2 shows raw read counts, normalized counts and reads per kilobase per million (RPKM) for each gene).
Hierarchical clustering of the 9 controls and 34 FSHD samples, based either on the expression levels of the a) 54 DUX4-regulated genes or b) the subset of 4 candidate biomarkers, revealed that either 11 or 10 FSHD samples clustered with the controls (Supplementary Material, Fig. S3A and S3B). Further analysis of normalized read counts for the four-candidate biomarkers separated the control and FSHD samples into four groups (Fig. 1 and Supplementary Material, Fig. S4): group 1, 10 FSHD samples grouped with the controls and had few or no reads mapping to the biomarkers; group 2, five FSHD samples had minor but significantly increased biomarker read number; group 3, 13 FSHD samples had moderate amounts of biomarker RNA expression; and group 4, six FSHD samples with high levels of biomarker RNA expression. Although the biopsies were taken from the quadriceps in all the controls and from the quadriceps (6), gastrocnemius (14), anterior tibialis (13) or hamstring muscles (1) in the FSHD subjects, the group 1 FSHD muscles included quadriceps (1), gastrocnemius (4) and anterior tibialis (5), indicating that lower levels of DUX4 target gene expression in the controls were not determined by muscle type; however, it should be noted that these comparisons do not have matched controls for each type of muscle for the FSHD biopsies. In sum, these analyses indicate that the previously identified candidate FSHD biomarkers were either not detected or expressed at extremely low levels, in the nine new control samples, but were elevated in 24/34 (71%) of FSHD samples.
Figure 1.
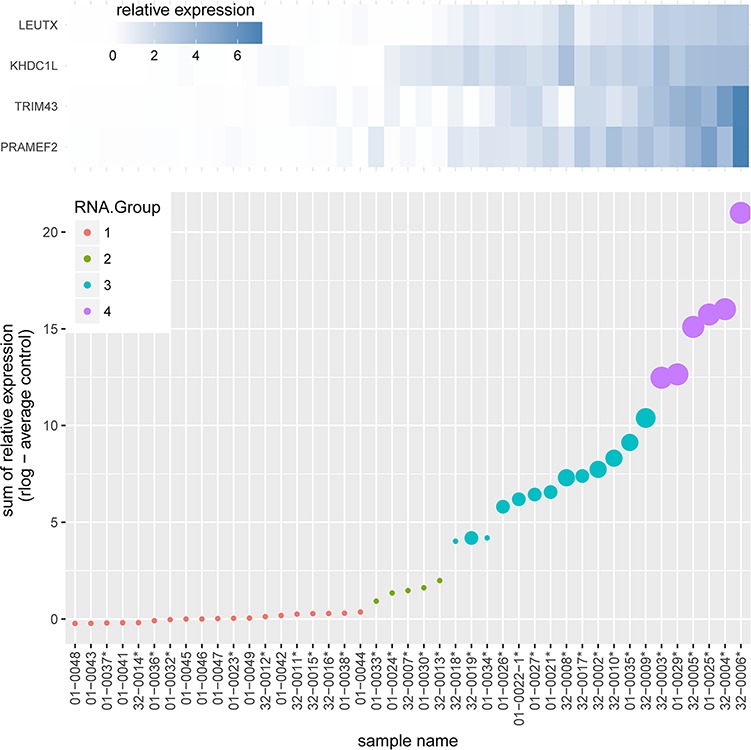
Relative expression of four candidate biomarkers in muscle biopsies from control and FSHD muscles. The sum of relative RNA-Seq expression for the genes LEUTX, KHDC1L, TRIM43 and PRAMEF2 was plotted for each RNA-Seq sample. Biopsies from FSHD individuals are indicated by an asterisk (*), whereas control muscle biopsies do not have an asterisk. A heat map of the relative expression of each of the four genes is shown at the top of the graph, and the graph plots the sum of reads for all four genes. The size of the spot reflects the number of candidate biomarker genes above a threshold level, therefore the smallest spot indicates no biomarkers above threshold, whereas the largest spot indicates all four biomarkers above threshold. The color coding divides the samples into four groups based on the relative expression for all four biomarkers.
Correlation of RNA-Seq to MRI and histopathologic changes
For the eight FSHD samples that did not have evidence of elevated DUX4 target genes on RNA-Seq (in group 1), the average pathology score was in the mild range at 3.4 (range 0–7), and none showed active disease as defined by the presence of necrosis, regeneration or inflammation. For the five samples in group 2 with a low but significant increase in the DUX4-target genes, the average pathology score was 5.2 (range 4–7), and three showed signs of active disease. For the 13 samples in group 3, the average pathology score was of moderate severity at 5.0 (range 1–10), with eight showing active disease, three of which showed inflammation. For the six samples in group 4, the average pathology score was 7.3 (range 4–10), with all samples showing active pathology in the form of muscle fiber necrosis/regeneration or inflammation (Supplementary Material, Fig. S5). Spearman rank analysis of the association between total pathology scores and RNA target expression shows a rs = 0.63 and P < 0.001 (Supplementary Material, Fig. S6).
For the 23 samples with abnormal whole muscle STIR+ signal, 20 (87%) showed expression of DUX4-target genes (groups 2–4). One that was STIR+ without DUX4-target gene expression (Sample 32-0016 in Fig. 1) had a high T1 signal and the RNASeq indicated that this sample had a higher representation of genes expressed in adipocytes. If this sample with a higher fat content is removed, then 91% of STIR+ muscles showed DUX4-target gene expression when biopsied. Moreover, there is a statistically significant difference (P < 0.001) between the mean (SD) RNA-Seq values in MRI normal muscles 1.4 (SD 2.4) compared to STIR+ muscles 7.7 (SD 5.7) (Fig. 2).
Figure 2.
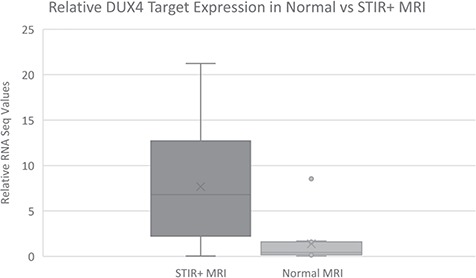
Biomarkers’ RNA-Seq expression in biopsy samples from MRI normal versus STIR+ samples: mean (SD) value in muscle biopsies from MRI normal muscle is 1.4 (SD 2.4) and in muscle biopsies from T2 STIR+ muscles is 7.7 (SD 5.7); the difference is statistically significant with a P < 0.001.
The overlap between STIR and RNA is also borne out in Spearman rank analyses. While fat ratings were associated with RNA measures at the whole muscle level (T1 whole rating-RNA score: rs = 0.49, P = 0.005; T1 whole rating-RNA rank: rs = 0.47, P = 0.008), there was only trend significance for quantitative fat fraction (fat fraction whole-RNA score: rs = 0.34, P = 0.06; fat fraction whole-RNA rank: rs = 0.32, P = 0.08). By contrast, STIR ratings were more associated with RNA (STIR rating whole-RNA score: rs = 0.61, P < 0.001; STIR rating whole-RNA rank: rs = 0.63, P < 0.001). The full set of Spearman rank analyses between MRI changes, pathology and RNA score is summarized in Supplementary Material, Figure 7.
Genes associated with extracellular matrix, immune response and inflammation distinguish FSHD from control samples
We assessed whether the FSHD groups differed from the control groups, or each other, based on global gene expression rather than limiting our analysis to genes previously identified as DUX4-induced candidate biomarkers. Principal component analysis showed the control samples clustered together, with FSHD groups 1–4 progressively further separated (Fig. 3). The exceptions were samples 01-0037 and 32-0016 in group 1, which were separated from the other group 1 samples. Samples 01-0037 and 32-0016 had very low or undetectable expression of DUX4-regulated candidate biomarker genes; however, gene set enrichment analysis (GSEA) showed that these samples were enriched in genes associated with extracellular matrix, inflammatory response, immune response and development and differentiation when compared to the control samples (Supplementary Material, Table S3). Notably, Myf5, MyoD and many muscle structural genes were decreased, whereas extracellular matrix, immune and inflammatory response genes were increased (Supplementary Material, Table S4). In the comparison of the control biopsies to the entire set of group 1 FSHD biopsies, although only a subset of these genes met the criteria to be called elevated (Supplementary Material, Table S5), many of these genes showed elevation in multiple group 1 samples as well as progressive elevation in groups 2–4 (examples are shown in Figs 4 and 5). It should be noted that the controls were biopsies from the quadriceps muscles and the FSHD biopsies were from the quadriceps (6), gastrocnemius (14), tibialis anterior (13) and hamstring (1) and some of the differential gene expression in Supplementary Material, Tables S4 and S5 might reflect differences in the type of muscle. Genes differentially expressed in FSHD group 2 compared to controls are shown in Supplementary Material, Table S6.
Figure 3.
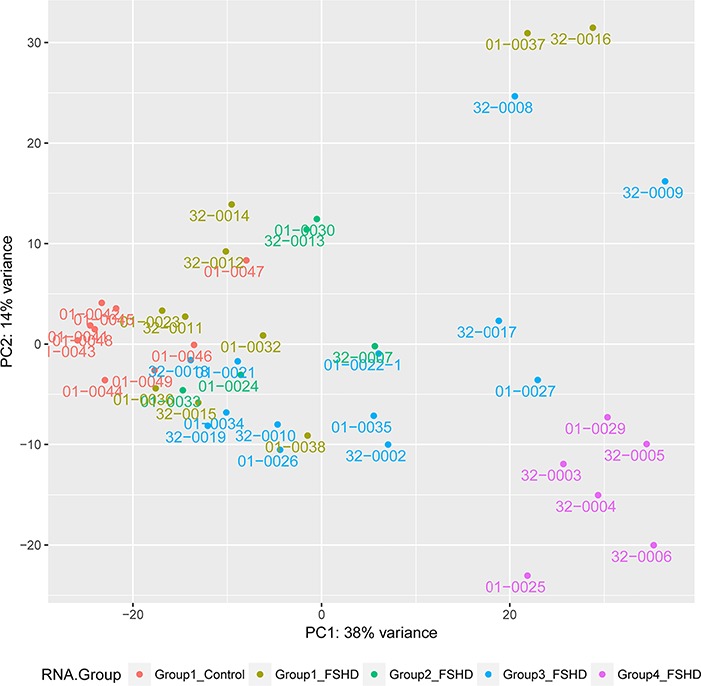
Principle component analysis based on total RNA sequencing data. Colors identify groups defined in Figure 1 based on the key at the bottom of this figure. Although FSHD groups 1, 2, 3 and 4 generally show progressively distinction from the group 1 controls, samples 01-0037 and 32-0016 represent outliers in the FSHD group 1 samples.
Figure 4.
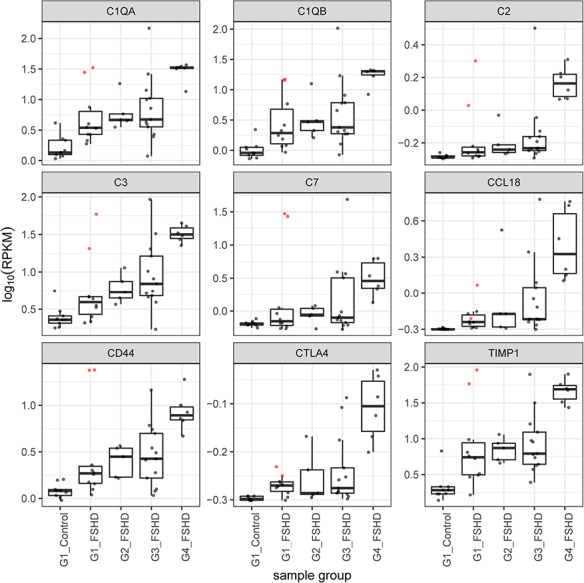
Boxplots showing the read counts scaled by log10 of RPKM for a subset of the immune/inflammation genes in the GSEA that distinguished the outlier group 1 FSHD samples (01-0037 and 32-0016) from control samples. Note that these same genes tend to be progressively elevated in groups 2, 3 and 4. The boxplots show the line for the median value, the box shows the interquartile range and the whiskers show the lowest and highest data point within the 1.5 interquartile range. The colored points represent expression in the two outlier samples.
Figure 5.
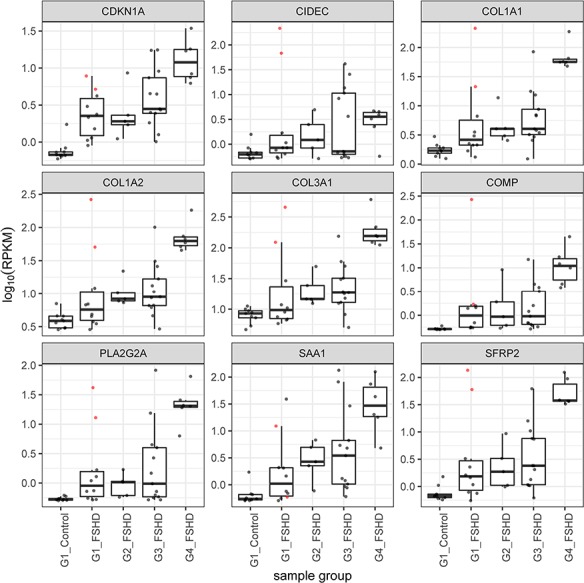
Boxplots showing the read counts scaled by log10 RPKM for a subset of the extracellular matrix and other genes in the GSEA that distinguished the outlier group 1 FSHD samples (01-0037 and 32-0016) from control samples. Note that these same genes tend to be progressively elevated in groups 2, 3 and 4. The boxplots show the line for the median value, the box shows the interquartile range and the whiskers show the lowest and highest data point within the 1.5 interquartile range. The colored points represent expression in the two outlier samples.
It was notable that elevated expression of many genes in the complement pathway was elevated in the group 1 outliers and also in many of the FSHD samples of groups 1 and 2 compared to the control samples (see Fig. 4). We proceeded to immunostain muscle sections from biopsies of four MRI normal muscles: three from group 1 samples (32-0012, 01-0037, 01-0038) and one from a single group 3 with a normal MRI and low pathology score (32-0010) with an antibody to the membrane attack complex (MAC) comprised of C5b-C9. Of the four biopsies, three showed distinct endomysial capillary staining for MAC (Fig. 6C, D and E). Sample 32-0010 also showed sarcolemmal staining of a single necrotic fiber (Fig. 6E and F). This indicates an early activation of the complement system in muscles with normal MRI, and in some muscles with low pathology scores and low or undetectable levels of DUX4 target gene expression.
Figure 6.
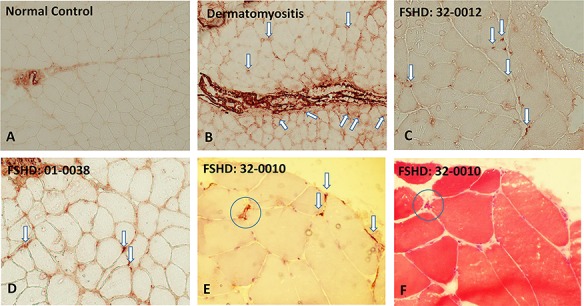
Immunohistochemical staining of muscle sections using C5b-9 (MAC) antibody (Agilent/Dako Clone aE11). (A) Muscle section from a normal control showing staining in larger perimysial vessels but no staining of endomysial capillaries. (B) A positive control section from an individual with dermatomyositis showing extensive, predominantly perifasicular, deposition of MAC in endomysial capillaries. (C, D, E) Show sections from FSHD subjects with positive MAC capillary staining (arrows); also visible in E is a small necrotic fiber (circle) with sarcolemmal MAC staining. (F) An adjacent section stained with hematoxylin and eosin showing the same atrophic necrotic fiber (circle).
In group 3 compared to the controls, GSEA identified biological processes related to negative regulation of cell death (Supplementary Material, Tables S7 and S8), but it should be noted that DUX4 induces some of the genes in this category, and that 47 of the 54 DUX4 candidate genes were differentially expressed in this group. GSEA for group 4 identified the inflammatory response, immune process, extracellular structure matrix and negative regulation of cell death as enriched pathways (Supplementary Material, Table S9 and S10). In addition, all 54 of the 54 DUX4-induced candidate biomarkers were differentially expressed.
Together, these data show that RNA expression changes associated with extracellular matrix, immune response and inflammatory response were elevated in FSHD muscle with only mildly abnormal pathology and very low or undetectable expression of DUX4-regulated genes. These changes likely represent the earliest global response that can be measured because many of these genes become progressively elevated with increased detection of DUX4-regulated genes.
Discussion
This is the first study to systematically examine the correlation between MRI changes, corresponding pathologic changes and DUX4-target gene expression in FSHD. The initial cross-sectional data show that a) expression of the set of DUX4-target genes is restricted to FSHD muscle, although not all FSHD muscles have expression of these target genes at significantly higher levels than controls; b) DUX4-target expression is generally higher in FSHD muscle with more advanced pathology; and c) STIR+ MRI measures might have substantial predictive value for identifying muscles with DUX4 expression and active disease. Indeed, in the analysis of this initial data, using an elevated STIR rating to select muscles with increased DUX4 target expression would yield positive results in ∼90% of the samples. The ability to follow these muscles over time will be valuable to create a full temporal model of disease progression.
Our data demonstrate that MRI T1 and STIR signals correlate with the presence of more advanced disease as determined by pathology. The presence of STIR signal in particular seems more associated with an active myopathic process with ongoing muscle fiber injury and repair with or without associated inflammatory changes, as well as with abnormal DUX4 target expression. One surprising result is that 9/10 of the normal-appearing MRI samples had mild-to-moderate pathologic changes. Consequently, early changes in FSHD muscle may not be visible on MRI until a threshold of injury is reached as evidenced in our fat fraction analyses of muscles rated as zero, a reason to use quantitative DIXON and possibly relaxometry measures as early as possible to create the most sensitive index possible for examining disease presence. In a prior paper, we observed that small non-confluent fat changes may precede confluent collections, something that will be instructive to evaluate in the longitudinal follow-up of this data.
Although 10 of the FSHD muscle biopsies did not show DUX4-target gene expression, these muscles did show elevated expression of extracellular matrix, inflammation and immune response genes (see Supplementary Material, Tables S3–5 and Figs 4 and 5), as well as showing mild-to-moderate histological changes. Many of these genes showed progressive elevation in FSHD groups showing higher levels of DUX4 target expression. It is also interesting that the decreased expression of MYF5, MYOD and muscle differentiation genes in some of these FSHD samples (see Supplementary Material, Table S4) is similar to the suppression of these genes and muscle differentiation caused by DUX4 expression. Together, this suggests a model where prior to disease progression in an individual muscle, the expression of DUX4 and DUX4-target genes might be below the threshold of detection in a muscle biopsy, e.g. expressed in a very small number of nuclei not evenly distributed through the muscle, or due to transient expression. This might be consistent with the tissue culture observation that only a minor fraction of nuclei in a culture of FSHD muscle express DUX4, likely reflecting occasional de-repression of DUX4 with an occasional burst of mRNA expression. In this model, the abnormal histological findings and gene expression in muscles without DUX4 target gene expression in the muscle biopsy might reflect the accumulated history of damage from occasional expression of DUX4 in a proportion of nuclei too small or transient to detect by RNA-Seq that is associated with either a primary or secondary immune response.
The increased DUX4 and DUX4-target gene expression in progressively affected muscle, based on pathological measures of active disease, inflammation and MRI abnormalities, further suggests that higher levels of DUX4 expression, possibly representing increased numbers of DUX4 expressing muscle nuclei, might cause disease progression. It remains possible that the higher levels of DUX4 might also occur in response to increased muscle damage. Nonetheless, the transition from a low basal level of DUX4 to higher levels of expression associated with disease progression suggests that treatments do not need to completely eliminate DUX4 expression, but rather it might be sufficient to prevent the progression to higher levels of expression. In the minimally affected FSHD muscles, e.g. the group 1 FSHD muscles (see Fig. 2), the abundance of genes associated with inflammation, including a large number of genes in the complement pathways (see Fig. 4), raises the possibility that complement inhibition or suppression of inflammation might suppress some of the early signs of FSHD and, possibly, slow the progression to the more active phases of the disease. In this regard, it is interesting to remember that components of the complement-mediated MAC were reported to be present in FSHD muscle and to be associated with structurally intact FSHD muscle fibers, suggesting a possibly early role for complement activation in FSHD pathology (20). Moreover, expression profiling showed differential expression of four complement pathway genes in affected versus unaffected FSHD muscles and, in a separate study, serum C3b was found to be elevated in individuals with FSHD (21,22). The capillary MAC deposition observed on immunostaining in a subset of our biopsies further reinforces the potential role of the complement pathway in FSHD. MAC deposition in capillaries is typically seen in dermatomyositis, but also in anti-SRP and HMGCR autoimmune necrotizing myopathies (23–25). This finding in FSHD suggests that an immune-mediated process may be an early contributing factor in FSHD muscle pathology.
A further point to consider is the relative concordance/discordance of MRI and pathological/RNA features. While it might seem surprising that T1 fat signal is more associated with pathology, and STIR signal with RNA-Seq, there was a selection bias to biopsy location of muscle tissue that appeared normal on T1 imaging but abnormal on STIR imaging. Finally, needle biopsies can only provide small samples, which are difficult to localize precisely on MRI images post hoc and may not be representative of the overall muscle pathology. Despite this target limitation, we have shown that whole muscle MRI fat and STIR elevations are a reliable predictor of the degree of muscle pathology and the presence of DUX4-target expression.
In considering rating scales versus quantification, rating scales have simplicity and ease of use compared to detailed measurement of quantitative parameters, especially in a priori site identification. However, early fatty changes are better characterized by quantitative DIXON measures than qualitative ratings, and this may allow for quantification of 0–20% of what was qualitatively rated as zero. As some pattern or degree of early fatty change is consistently associated with RNA-Seq expression, non-confluent fatty changes may represent more than normal aging or the consequence of inactivity, and may be important as a biomarker for early treatment efforts. Similarly, T2 data has been used in a range of studies to date to evaluate the range of changes in STIR signal (26–29). How quantified changes in T2 signal related to discrete categories of RNA-Seq will be useful to examine in future work with more samples.
In this study, some muscles had low or moderate levels of DUX4-target gene expression but did not show active disease on pathology, nor T1 or STIR signal on MRI; however, some of these did show capillary MAC deposition suggesting a low-level immune response. We will need to determine whether DUX4-target gene expression in relatively unaffected muscle is a transient event or the first step in a progressive process, a distinction that will help inform the use of DUX4-target genes as biomarkers in clinical trials. Although not all FSHD muscles express appreciable amounts of DUX4-regulated candidate biomarkers, the data presented suggests that, with MRI guidance, muscles most likely to have measurable DUX4 target expression can be more reliably identified. Consequently, DUX4 target expression measured by RNA-Seq can potentially be used as a pharmacodynamic biomarker for early phase FSHD therapeutic trials targeting DUX4 expression. However, further validation studies are needed to determine the test–retest variability of RNA-Seq DUX4 target expression and its sensitivity to change over time. The planned 1-year follow-up studies on these individuals, as well as other planned studies, will help address these questions.
Materials and Methods
The University of Washington and the University of Rochester jointly conducted the study through the Seattle Paul D. Wellstone Muscular Dystrophy Cooperative Research Center. Respective institutional IRBs approved the study protocol. A total of 36 subjects (18 per site) were recruited. All subjects underwent an MRI of the lower extremities, clinical evaluations of strength and function and needle biopsy of a muscle selected based on MRI characteristics. All procedures, including muscle biopsies, handling and processing of samples and muscle MRI protocols, were identical at both sites. Histologic muscle samples were processed and scored in Dr Tawil’s lab, RNA extraction for sequencing was performed in Dr Tapscott’s lab and all MRI interpretation and scoring were performed by Dr Shaw. Drs Tawil and Shaw were blinded to the corresponding imaging and histopathologic scoring, respectively.
Study subjects
As one of the primary objectives of the proposal was to develop biomarkers for future clinical trials, subject selection criteria reflected the type of subjects likely to be selected in early phase studies in FSHD. Such patients are typically mildly to moderately affected and able to ambulate independently.
Inclusion criteria. 1) Male and female subjects with the genetically confirmed FSHD1 or 2; 2) age: 18–75 years; 3) symptomatic: the subject has to have symptomatic lower limb weakness; 4) independently ambulatory: able to walk 30 feet without the support of another person.
Exclusion criteria. 1) Severe cardiac, respiratory or orthopedic conditions that preclude safe testing of muscle function; 2) use of warfarin anticoagulation; 3) malignancy with ongoing treatment with chemotherapeutic agents, use of immunosuppressants or anabolic agents; 4) pregnancy; 5) recent or ongoing infection; 6) presence of contraindication to performance of MRI: pacemaker, metallic foreign body in eye, brain aneurysm clip (unless documented as MRI compatible). All genetic testing was performed in the laboratory of Dr van der Maarel.
Clinical assessments
Clinical assessments included determination of the CSS (a validated, 10-grade overall clinical severity scale), and quantitative strength measurement of muscles selected for biopsy using a hand-held dynamometer (30,31).
MRI
Identical MRI muscle imaging protocols were performed at both sites on 3 T-T machines (Siemens VB17) utilizing phased array coils placed over the lower extremities (thighs, calves). First, multi-plane localizers (6-mm slice thickness) were acquired followed by T1 (TE = 8.9 ms, TR = 510 ms), STIR (TE = 38 ms, TR = 4790 ms, TI = 220), DIXON (TE = 1.3, 2.48, 3.73, 4.98, 7.38, 9.84 ms, TR = 11.3) and fat-saturated T2 (TE = 13.2, 26.4, 39.6 ms, TR = 1450) acquisitions at identical dimensions (5-mm slices, 40 image per location, 1.25-mm in-plane resolution). The tibial spine was used as a landmark for placing the slabs, a straightforward anatomical location used for slice registration on follow-up examination to be described in a subsequent report. MRI visible fiducials (gel stickers—SCH, vitamin E capsules—Rochester) were placed on the skin for the major muscles of interest (medial gastrocnemius, tibialis anterior, vastus lateralis, rectus femoris) to localize biopsy coordinates.
MRI measures
ITK-SNAP was used to label the full extent of imaged thigh and calf muscles as performed previously (32,33).
Qualitative. T1- and STIR severity score: assessments of fat infiltration in the T1 images used an established rating scale for fat: 0, normal appearance; 1, scattered small areas of abnormality; 2, numerous discrete areas of increased signal intensity, less than 30% of the muscle volume; 3, numerous discrete areas with early confluence, 30–60% volume; 4, > 60% with patchy loss of fascial structure; or 5, pronounced fatty replacement throughout with complete fascial structure loss. This imaging severity index was recently used in Duchenne dystrophy and correlated with histopathologic changes (34). It has also been used in oculopharyngeal muscular dystrophy where severity index correlated with clinical scores (35). This scale correlates well with the CSS in FSHD (18,36). STIR intensity was rated on a four-point scale as stage 0, normal appearance; stage 1, very mild diffuse elevation; stage 2, mild diffuse elevation with areas of moderate signal elevation, less than 30% of the muscle volume; stage 3, moderate areas of increased signal intensity, 30–60% volume; stage 4, moderate/severe involvement of entire muscle. The addition of stage 1 represents expansion on the scale described (29).
Quantitative. DIXON fat/water maps were processed in Matlab (Mathworks, Natick MA).
Muscle biopsy for pathologic grading and biomarker studies
Each subject underwent a lower extremity muscle biopsy in a muscle safely accessible by needle biopsy and included tibialis anterior, gastrocnemius muscles, vastus lateralis and one hamstring (biceps femoris) muscle. T2-STIR hyperintense muscles were preferentially biopsied when possible. Biopsies were obtained under sterile condition using either UCH modified Bergstrom needles (Rochester) or regular Bergstrom needles (Seattle). Biopsy specimens (75 to 150 mg) were divided into three aliquots. Two samples were flash-frozen in liquid nitrogen within 30 s of removal and stored in −80°C freezers for RNA expression studies, and the third was oriented under a dissecting microscope and frozen in isopentane cooled in liquid nitrogen for histologic evaluation. Muscle biopsies from unaffected control individuals obtained under identical conditions and banked at the University of Rochester were the source of the normal control samples for RNA expression studies.
Grading of muscle biopsy pathology
The histopathologic samples were graded for the severity of the pathologic changes based on 10-μm sections stained with hematoxylin & eosin and trichrome. A pathologic severity score is determined for each biopsy based on a 12-point scale giving a 0–3 score to each of the four major histologic features: variability in fiber size, percent of centrally located nuclei, interstitial fibrosis and muscle fiber necrosis/regeneration/inflammation. Pathologic scores of 1–4 are considered mild, 5–8 moderate and 9–12 severe (16).
RNA-Seq analysis
The preprocessing of RNA-Seq data started with filtering unqualified reads, followed by trimming Illumina universal adapter using Trimmomatic-0.32. Subsequently, the preprocessed reads were aligned to genome build hg38 using Tophat-21.0/Bowtie2-2.2.6. To profile the gene expression, the gene features and annotation were first obtained from GENCODE, version 24. Gene counts were then estimated by using Bioconductor function GenomicAlignment::summarizeOverlap with IntersectionStrict mode, which restrict read counts to those that fall completely within the range of exons.
The FSHD clustering and score of the RNA-Seq samples were based upon four FSHD-positive biomarkers that were previously found in Yao, 2014 [18, 28]: LEUTX, KHDC1L, PRAMEF2 and TRIM43. Using DESeq2’s regularized log2 scaling [30] on the biomarkers, the hierarchical clustering classified the RNA-seq samples into four categories, labeled group 1 to 4. Group 1 exhibited the lowest expression, whereas group 4 the highest. The FSHD score is evaluated by taking the sum of the four biomarkers’ log2 difference from the control average. Further downstream analysis for RNA-seq samples included differential and GSEA using DESeq2 and GOSeq Bioconductor packages, respectively.
The bioinformatics analysis and visualization were mostly performed using R/3.4 and Bioconductor 3.5 packages. The RNA-seq data and gene counts have been deposited to GEO series GSExxxxx. Codes for preprocessing pipeline and reproducible analysis can be found in TapscottLab’s GitHub repository, https://github.com/TapscottLab/FSHD-Biopsy-RNA-seq.
Biostatistical analysis
Initial analyses employed counts of MRI features by RNA-Seq groupings. Secondary analyses computed correlations between MRI and pathology and RNA-Seq features using Spearman correlations as the qualitative measures were ordered and the pathology and RNA scores were skewed.
Supplementary Material
Acknowledgement
This work is supported by National Institute of Health, National Institute of Arthritis and Musculoskeletal and Skin diseases [U54 AR065139] and a Friends of FSH Research grant.
Conflict of Interest statement. Some of the authors consult for companies performing or preparing clinical trials in FSHD: Fulcrum Therapeutics (R.T., S.M.vdM., S.J.T.), Acceleron Pharma (R.T.) and Genea Biocells (R.T.). R.T., S.M.vdM. and S.J.T. have filed provisional patents targeting DUX4 as a therapeutic and measuring DUX4-regulated genes as candidate biomarkers.
References
- 1. Flanigan K.M., Coffeen C.M., Sexton L., Stauffer D., Brunner S. and Leppert M.F. (2001) Genetic characterization of a large, historically significant Utah kindred with facioscapulohumeral dystrophy. Neuromuscul. Disord., 11, 525–529. [DOI] [PubMed] [Google Scholar]
- 2. Deenen J.C., Arnts H., Maarel S.M., Padberg G.W., Verschuuren J.J., Bakker E., Weinreich S.S., Verbeek A.L. and Engelen B.G. (2014) Population-based incidence and prevalence of facioscapulohumeral dystrophy. Neurology, 83, 1056–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tawil R., Kissel J.T., Heatwole C., Pandya S., Gronseth G. and Benatar M. (2015) Evidence-based guideline summary: evaluation, diagnosis, and management of facioscapulohumeral muscular dystrophy: Report of the Guideline Development, Dissemination, and Implementation Subcommittee of the American Academy of Neurology and the Practice Issues Review Panel of the American Association of Neuromuscular & Electrodiagnostic Medicine. Neurology, 85, 357–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lemmers R.J., Vliet P.J., Klooster R., Sacconi S., Camano P., Dauwerse J.G., Snider L., Straasheijm K.R., Ommen G.J., Padberg G.W. et al. (2010) A unifying genetic model for facioscapulohumeral muscular dystrophy. Science, 329, 1650–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Snider L., Geng L.N., Lemmers R.J., Kyba M., Ware C.B., Nelson A.M., Tawil R., Filippova G.N., Maarel S.M., Tapscott S.J. et al. (2010) Facioscapulohumeral dystrophy: incomplete suppression of a retrotransposed gene. PLoS Genet., 6, e1001181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lemmers R.J., Tawil R., Petek L.M., Balog J., Block G.J., Santen G.W., Amell A.M., Vliet P.J., Almomani R., Straasheijm K.R. et al. (2012) Digenic inheritance of an SMCHD1 mutation and an FSHD-permissive D4Z4 allele causes facioscapulohumeral muscular dystrophy type 2. Nat. Genet., 44, 1370–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Greef J.C., Lemmers R.J., Camano P., Day J.W., Sacconi S., Dunand M., Engelen B.G., Kiuru-Enari S., Padberg G.W., Rosa A.L. et al. (2010) Clinical features of facioscapulohumeral muscular dystrophy 2. Neurology, 75, 1548–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Greef J.C., Lemmers R.J., Engelen B.G., Sacconi S., Venance S.L., Frants R.R., Tawil R. and Maarel S.M. (2009) Common epigenetic changes of D4Z4 in contraction-dependent and contraction-independent FSHD. Hum. Mutat., 30, 1449–1459. [DOI] [PubMed] [Google Scholar]
- 9. Boogaard M.L., Lemmers R.J., Balog J., Wohlgemuth M., Auranen M., Mitsuhashi S., Vliet P.J., Straasheijm K.R., Akker R.F., Kriek M. et al. (2016) Mutations in DNMT3B modify epigenetic repression of the D4Z4 repeat and the penetrance of facioscapulohumeral dystrophy. Am. J. Hum. Genet., 98, 1020–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kowaljow V., Marcowycz A., Ansseau E., Conde C.B., Sauvage S., Matteotti C., Arias C., Corona E.D., Nunez N.G., Leo O. et al. (2007) The DUX4 gene at the FSHD1A locus encodes a pro-apoptotic protein. Neuromuscul. Disord., 17, 611–623. [DOI] [PubMed] [Google Scholar]
- 11. Snider L., Asawachaicharn A., Tyler A.E., Geng L.N., Petek L.M., Maves L., Miller D.G., Lemmers R.J., Winokur S.T., Tawil R. et al. (2009) RNA transcripts, miRNA-sized fragments and proteins produced from D4Z4 units: new candidates for the pathophysiology of facioscapulohumeral dystrophy. Hum. Mol. Genet., 18, 2414–2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wuebbles R.D., Long S.W., Hanel M.L. and Jones P.L. (2010) Testing the effects of FSHD candidate gene expression in vertebrate muscle development. Int. J. Clin. Exp. Pathol., 3, 386–400. [PMC free article] [PubMed] [Google Scholar]
- 13. Bosnakovski D., Xu Z., Gang E.J., Galindo C.L., Liu M., Simsek T., Garner H.R., Agha-Mohammadi S., Tassin A., Coppee F. et al. (2008) An isogenetic myoblast expression screen identifies DUX4-mediated FSHD-associated molecular pathologies. EMBO J., 27, 2766–2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tawil R., Padberg G.W., Shaw D.W., Maarel S.M. and Tapscott S.J. (2016) Clinical trial preparedness in facioscapulohumeral muscular dystrophy: clinical, tissue, and imaging outcome measures 29–30 May 2015, Rochester, New York. Neuromuscul. Disord., 26, 181–186. [DOI] [PubMed] [Google Scholar]
- 15. Tawil R., Shaw D.W., Maarel S.M. and Tapscott S.J. (2014) Clinical trial preparedness in facioscapulohumeral dystrophy: outcome measures and patient access: 8–9 April 2013, Leiden, The Netherlands. Neuromuscul. Disord., 24, 79–85. [DOI] [PubMed] [Google Scholar]
- 16. Statland J.M., Shah B., Henderson D., Van Der Maarel S., Tapscott S.J. and Tawil R. (2015) Muscle pathology grade for facioscapulohumeral muscular dystrophy biopsies. Muscle Nerve, 52, 521–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tasca G., Monforte M., Corbi M., Granata G., Lucchetti D., Sgambato A. and Ricci E. (2018) Muscle microdialysis to investigate inflammatory biomarkers in facioscapulohumeral muscular dystrophy. Mol. Neurobiol., 55, 2959–2966. [DOI] [PubMed] [Google Scholar]
- 18. Frisullo G., Frusciante R., Nociti V., Tasca G., Renna R., Iorio R., Patanella A.K., Iannaccone E., Marti A., Rossi M. et al. (2011) CD8(+) T cells in facioscapulohumeral muscular dystrophy patients with inflammatory features at muscle MRI. J. Clin. Immunol., 31, 155–166. [DOI] [PubMed] [Google Scholar]
- 19. Yao Z., Snider L., Balog J., Lemmers R.J., Van Der Maarel S.M., Tawil R. and Tapscott S.J. (2014) DUX4-induced gene expression is the major molecular signature in FSHD skeletal muscle. Hum. Mol. Genet., 23, 5342–5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Spuler S. and Engel A.G. (1998) Unexpected sarcolemmal complement membrane attack complex deposits on nonnecrotic muscle fibers in muscular dystrophies. Neurology, 50, 41–46. [DOI] [PubMed] [Google Scholar]
- 21. Rahimov F., King O.D., Leung D.G., Bibat G.M., Emerson C.P. Jr., Kunkel L.M. and Wagner K.R. (2012) Transcriptional profiling in facioscapulohumeral muscular dystrophy to identify candidate biomarkers. Proc. Natl. Acad. Sci. U. S. A., 109, 16234–16239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Petek L.M., Rickard A.M., Budech C., Poliachik S.L., Shaw D., Ferguson M.R., Tawil R., Friedman S.D. and Miller D.G. (2016) A cross sectional study of two independent cohorts identifies serum biomarkers for facioscapulohumeral muscular dystrophy (FSHD). Neuromuscul. Disord., 26, 405–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kissel J.T., Mendell J.R. and Rammohan K.W. (1986) Microvascular deposition of complement membrane attack complex in dermatomyositis. N. Engl. J. Med., 314, 329–334. [DOI] [PubMed] [Google Scholar]
- 24. Troyanov Y., Landon-Cardinal O., Fritzler M.J., Ferreira J., Targoff I.N., Rich E., Goulet M., Goulet J.R., Bourre-Tessier J., Robitaille Y. et al. (2017) Atorvastatin-induced necrotizing autoimmune myositis: an emerging dominant entity in patients with autoimmune myositis presenting with a pure polymyositis phenotype. Medicine, 96, e5694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dimitri D., Andre C., Roucoules J., Hosseini H., Humbel R.L. and Authier F.J. (2007) Myopathy associated with anti-signal recognition peptide antibodies: clinical heterogeneity contrasts with stereotyped histopathology. Muscle Nerve, 35, 389–395. [DOI] [PubMed] [Google Scholar]
- 26. Arpan I., Forbes S.C., Lott D.J., Senesac C.R., Daniels M.J., Triplett W.T., Deol J.K., Sweeney H.L., Walter G.A. and Vandenborne K. (2013) T(2) mapping provides multiple approaches for the characterization of muscle involvement in neuromuscular diseases: a cross-sectional study of lower leg muscles in 5-15-year-old boys with Duchenne muscular dystrophy. NMR Biomed., 26, 320–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Burakiewicz J., Sinclair C.D.J., Fischer D., Walter G.A., Kan H.E. and Hollingsworth K.G. (2017) Quantifying fat replacement of muscle by quantitative MRI in muscular dystrophy. J. Neurol., 264, 2053–2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Willcocks R.J., Arpan I.A., Forbes S.C., Lott D.J., Senesac C.R., Senesac E., Deol J., Triplett W.T., Baligand C., Daniels M.J. et al. (2014) Longitudinal measurements of MRI-T2 in boys with Duchenne muscular dystrophy: effects of age and disease progression. Neuromuscul. Disord., 24, 393–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yao L. and Gai N. (2012) Fat-corrected T2 measurement as a marker of active muscle disease in inflammatory myopathy. AJR Am. J. Roentgenol., 198, W475–W481. [DOI] [PubMed] [Google Scholar]
- 30. Ricci E., Galluzzi G., Deidda G., Cacurri S., Colantoni L., Merico B., Piazzo N., Servidei S., Vigneti E., Pasceri V. et al. (1999) Progress in the molecular diagnosis of facioscapulohumeral muscular dystrophy and correlation between the number of KpnI repeats at the 4q35 locus and clinical phenotype. Ann. Neurol., 45, 751–757. [DOI] [PubMed] [Google Scholar]
- 31. Tawil R., McDermott M.P., Mendell J.R., Kissel J. and Griggs R.C. (1994) Facioscapulohumeral muscular dystrophy (FSHD): design of natural history study and results of baseline testing. FSH-DY Group. Neurology, 44, 442–446. [DOI] [PubMed] [Google Scholar]
- 32. Yushkevich P.A., Piven J., Hazlett H.C., Smith R.G., Ho S., Gee J.C. and Gerig G. (2006) User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage, 31, 1116–1128. [DOI] [PubMed] [Google Scholar]
- 33. Friedman S.D., Poliachik S.L., Carter G.T., Budech C.B., Bird T.D. and Shaw D.W. (2012) The magnetic resonance imaging spectrum of facioscapulohumeral muscular dystrophy. Muscle Nerve, 45, 500–506. [DOI] [PubMed] [Google Scholar]
- 34. Kinali M., Arechavala-Gomeza V., Cirak S., Glover A., Guglieri M., Feng L., Hollingsworth K.G., Hunt D., Jungbluth H., Roper H.P. et al. (2011) Muscle histology vs MRI in Duchenne muscular dystrophy. Neurology, 76, 346–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fischmann A., Gloor M., Fasler S., Haas T., Rodoni Wetzel R., Bieri O., Wetzel S., Heinimann K., Scheffler K. and Fischer D. (2011) Muscular involvement assessed by MRI correlates to motor function measurement values in oculopharyngeal muscular dystrophy. J. Neurol., 258, 1333–1340. [DOI] [PubMed] [Google Scholar]
- 36. Kan H.E., Scheenen T.W., Wohlgemuth M., Klomp D.W., van Loosbroek-Wagenmans I., Padberg G.W. and Heerschap A. (2009) Quantitative MR imaging of individual muscle involvement in facioscapulohumeral muscular dystrophy. Neuromuscul. Disord., 19, 357–362. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


