Abstract
In the ovary, the luteinizing hormone (LH) surge suppresses the proliferation and induces the luteinization of preovulatory granulosa cells (GCs), which is crucial for the survival of terminally-differentiated GCs. Krüppel-like factor 4 (Klf4) has been shown to play a role in regulating the cell cycle and apoptosis in various cell types. The rapid induction of Klf4 expressions by LH was observed in preovulatory GCs. To evaluate whether Klf4 affects GC proliferation and survival, primary rat GCs were isolated from pregnant mare serum gonadotropin-primed Sprague–Dawley rat ovaries and transfected with a Klf4 expression vector or Klf4-specific siRNA, followed by determination of the transcript levels of apoptosis-related and cell cycle-related genes. Cell proliferation, viability, and apoptosis were analyzed by BrdU incorporation, a Cell Counting Kit-8 assay, a bioluminescence caspase 3/7 assay, and flow cytometry. LH treatment increased Klf4 mRNA expression in preovulatory GCs. Transcripts of B-cell lymphoma 2 (Bcl-2) and cell cycle promoters (Cyclin D1 and Cyclin D2) decreased, whereas those of the cell cycle inhibitor, p21, increased. Altering the expression of Klf4 by overexpression or knockdown consistently affected the expression of Bcl-2 and Cyclin D1. In agreement with this, Klf4 overexpression reduced cell viability, increased the fraction of apoptotic cells, and arrested cell cycle progression in G1 phase. We conclude that Klf4 increases the susceptibility of preovulatory GCs to apoptosis by down-regulating Bcl-2, and promotes LH-induced cell cycle exit. It appears to be a key regulator induced by the LH surge that determines the fate of GCs in preovulatory follicles during the luteal transition.
Keywords: Klf4, granulosa cells, LH surge, apoptosis, cell cycle
1. Introduction
Whether the follicles are growing or atretic depends on the balance between anti-apoptotic and pro-apoptotic factors that determine the rate of granulosa cell (GC) apoptosis. Although apoptosis can occur at any stage of follicular development, follicles that are more differentiated are more prone to apoptosis than early-stage follicles [1]. In addition, the factors regulating follicular cell survival before and after the luteinizing hormone (LH) surge appear to differ. In response to the LH surge, the expression of diverse genes is rapidly and dramatically changed in preovulatory follicles [2]. For instance, highly expressed genes in growing preovulatory follicles are turned off by the LH surge, whereas various different genes that are relevant to luteinization increase dramatically during the periovulatory period [3].
The Krüppel-like factor family of transcription factors plays important roles in many biological processes, including proliferation, apoptosis, and differentiation [4,5]. Krüppel-like factor 4 (Klf4) expression was first reported in the epithelial lining of the gut and skin, and its effect on cell cycle arrest and apoptosis is different depending on the cellular context [6,7,8]. An LH-induced up-regulation of Klf4 expression in porcine GCs in vitro has been reported [9], and it is involved in apoptosis in several mammalian cell types [10]. Since luteinizing GCs cease to multiply, it is conceivable that Klf4 is induced by LH and influences the cell cycle transition and apoptosis of periovulatory GCs during this critical period. However, there is no direct evidence for anti-apoptotic or pro-apoptotic actions of Klf4, or its effects on cell cycle arrest, and its role in the ovary is largely unknown.
In this study, we investigated whether Klf4 is regulated by LH and whether it is involved in cell cycle arrest and the loss of cell viability during the luteal transition period in rat preovulatory GCs in vitro. Cell cycle arrest was analyzed by measuring the induction of cell cycle-related genes such as Cyclin D1, D2, p21, and p27 along with an analysis of 5-bromo-2′deoxyuridine (BrdU) incorporation and flow cytometry. Cell viability was analyzed by examining the expression of apoptosis-regulating genes, B-cell lymphoma 2 (Bcl-2) and Bcl-2-associated X (Bax) [11], accompanied by a cell counting kit-8 assay (CCK-8), a sensitive bioluminescence caspase 3/7 assay, and flow cytometry analysis. Here, we found that Klf4 increases the susceptibility of preovulatory GCs to apoptosis by down-regulating Bcl-2, and promotes LH-induced cell cycle exit.
2. Results
2.1. LH Regulates the Expression of Klf4, Bcl-2, and Cell Cycle Genes in Preovulatory GCs
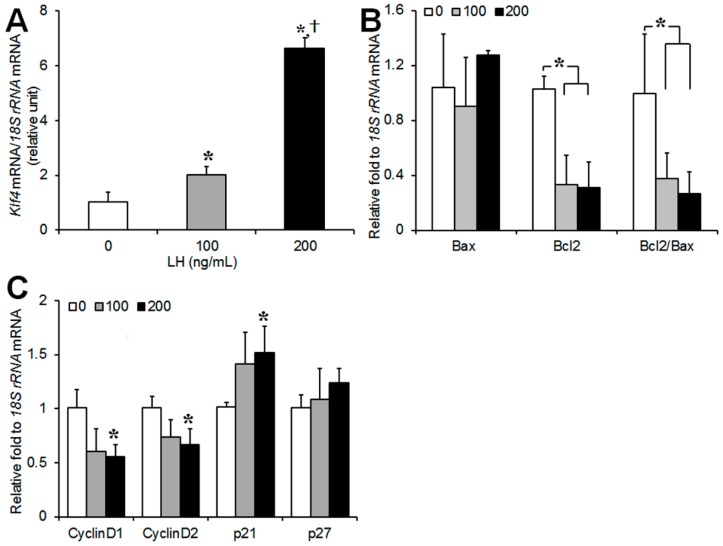
To investigate changes in the expression of Klf4 and the genes regulating apoptosis and the cell cycle in preovulatory GCs in the early phase of LH exposure, cells were cultured in the presence of LH (0, 100, 200 ng/mL) for 45 min, and real-time PCR analysis was performed (Figure 1). The culture time of 45 min was based on preliminary measurements of Klf4 expression after LH exposure for 15 min to 24 h. As shown in Figure 1A, LH treatment led to dose-dependent increases in Klf4 mRNA. For example an ovulatory dose of LH of 200 ng/mL led to a 6.6-fold increase in the Klf4 mRNA level compared to untreated control cells. LH treatment reduced the expression of Bcl-2 and the Bcl-2/Bax ratio in a dose-dependent manner, whereas no clear changes were found in the Bax transcript levels compared to the control (Figure 1B). LH treatment decreased the expression of cell cycle promoters (cyclin D1 and D2), and increased that of p21 at 200 ng/mL (Figure 1C).
Figure 1.
Effect of luteinizing hormone (LH) on Krüppel-like factor 4 (Klf4), transcripts of apoptosis- and cell cycle-related genes in cultured preovulatory granulosa cells (GCs). Real-time RT-PCR analysis of transcripts of (A) Klf4; (B) Bax, and Bcl-2; and (C) Cyclins D1, D2, p21, and p27 in preovulatory GCs in response to LH (0 ng/mL, 100 ng/mL, and 200 ng/mL). All of the expression levels were normalized to 18S rRNA levels. Values were calculated as fold changes relative to untreated cells and are expressed as means ± standard deviations (SDs) of three separate experiments. LH, luteinizing hormone. * p < 0.05, vs. untreated cells; † p < 0.05, vs. cells treated with 100 ng/mL of LH.
2.2. Effect of Klf4 on Expression of Apoptosis-Related and Cell Cycle-Related Genes in Preovulatory GCs
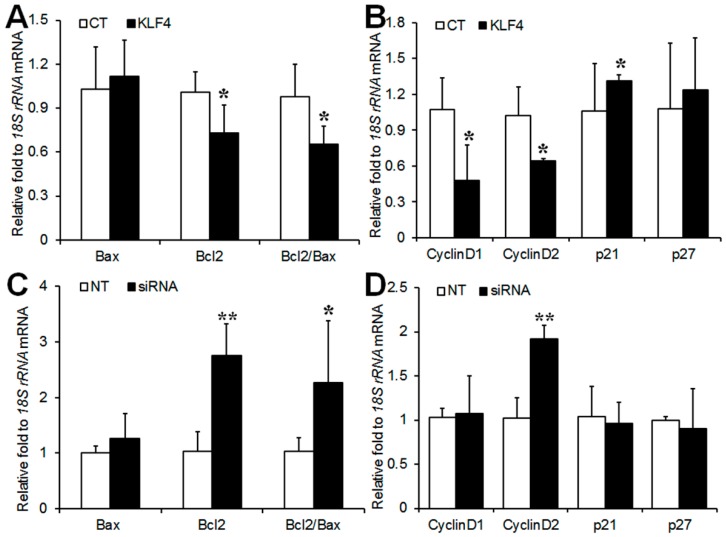
GCs from pregnant mare serum gonadotropin (PMSG)-primed rat ovaries were transfected with a Klf4 expression vector or Klf4-specific small interfering RNA (siRNA). The overexpression or knockdown of Klf4 in GCs was confirmed by real-time reverse transcription polymerase chain reaction (RT-PCR) and Western blotting analysis (Supplementary Figure S1). Klf4 overexpression caused significant decreases in Bcl-2 transcripts, which were accompanied by reduced Bcl-2/Bax ratios, with no effect on Bax expression itself (Figure 2A). Conversely, Klf4 knockdown increased Bcl-2 mRNA expression and the Bcl-2/Bax ratio (Figure 2C). Klf4 overexpression down-regulated cyclins D1 and D2, and up-regulated the cell cycle inhibitor p21 (Figure 2B), whereas only cyclin D2 was significantly up-regulated in response to Klf4 inhibition (Figure 2D), suggesting that Klf4 blocked cell cycle progression primarily by down-regulating cyclin D2.
Figure 2.
Regulation of apoptosis-related and cell cycle-related genes expression by Klf4 in preovulatory GCs. (A,C) Real-time RT-PCR analysis of Bax and Bcl-2 mRNA levels in GCs transfected with Klf4 expression vector (300 ng) and Klf4-specific siRNA (200 nM); (B,D) Real-time RT-PCR analysis of cyclins D1, D2, p21 and p27 mRNA levels in GCs transfected with Klf4 expression vector and Klf4-specific siRNA. 18S rRNA was used to normalize each reaction. Values are calculated as fold changes relative to empty vector (CT) or non-target siRNA (NT), and are expressed as the means ± SDs of three separate experiments. CT, cells transfected with empty vector; NT, cells transfected with non-target siRNA. * p < 0.05, ** p < 0.01 vs. CT or NT.
2.3. Klf4 Overexpression Reduces Cell Viability and Proliferation of GCs
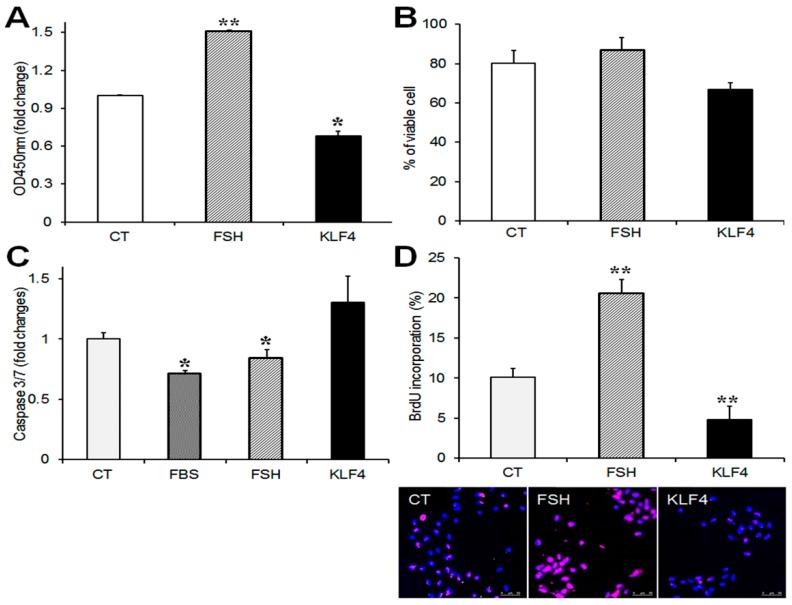
To determine the effects of Klf4 on GC viability and proliferation, cells were transfected with Flag-Klf4 or empty vector (CT) and cultured for 24 h without serum to avoid any effects on the growth factors in serum. FSH was used as a positive control [12]. The overexpression of Klf4 in the GCs was confirmed by real-time PCR (Supplementary Figure S1C). The CCK-8 assay was used to monitor the effect of Klf4 on GC viability. Klf4 overexpression decreased CCK-8 activity by 30% compared to the control (p = 0.035 vs. CT) (Figure 3A). Since the CCK-8 assay measures the metabolic activity of living cells and does not assess cell death, cell viability was measured by trypan blue exclusion. The percentage of viable cells fell by approximately 14% in Klf4-overexpressing cells (from 80.3 ± 6.4% to 66 ± 3.3%) (Figure 3B). We also analyzed apoptosis quantitatively by measuring caspase 3/7 activity. Transfected GCs were cultured with or without serum or follicle-stimulating hormone (FSH). The enzymatic activities that were measured in each group were compared with the activities in the control cells. As shown in Figure 3C, in the GCs cultured with 10% fetal bovine serum (FBS) and FSH, caspase 3/7 activity was 0.7-fold and 0.8-fold of the activity in the control, respectively (p < 0.05). The overexpression of Klf4 caused an approximately 1.3-fold increase in caspase-3/7 activity compared to the control cells (p = 0.05).
Figure 3.
Effects of Klf4 overexpression on cell viability and proliferation of preovulatory GCs. Cell viability was determined by (A) CCK-8 cell counting assay (ALX-850-039-0100, Enzo) and (B) trypan blue exclusion in cells transfected with Klf4 expression vector (300 ng/well) or empty vector (CT, control). * p < 0.05, ** p < 0.0001 vs. CT; (C) Analysis of caspase 3/7 activity. GCs transfected with Klf4 or empty vector were incubated without serum, and some control cells were treated with fetal bovine serum (FBS) or follicle-stimulating hormone (FSH) as positive controls for apoptosis suppressors. Twenty-four h later, caspase 3/7 activity was measured with the bioluminescence assay. FBS, 10% fetal bovine serum. * p < 0.05 vs. CT; (D) 5-bromo-2′deoxyuridine (BrdU)-incorporation assay. DNA labeling indices were determined by the scoring number of BrdU(+) nuclei per total nuclei, and are presented as means ± SDs. ** p < 0.001 vs. CT. Representative fluorescence microscopy images of BrdU-labeled GCs (100×) (lower panel). Cells were stained with BrdU antibody (B2531, Sigma, 1:200) to visualize BrdU-labeled cells (pink) and counterstained with 4′,6-diamidino-2-phenylindole (DAPI) to confirm nuclear status (blue). CT, control cells transfected with empty vector; FSH, follicle-stimulating hormone (50 ng/mL); KLF4, cells transfected with Klf4 (300 ng).
After FSH treatment, BrdU-labeled GCs increased twofold relative to non-treated control cells (p < 0.01), whereas they declined to half the control level of 10.1 ± 1.1% among the Klf4-overexpressing cells (4.8 ± 1.7%) (p < 0.01) (Figure 3D). These results indicate that Klf4 is implicated in the regulation of cell survival and proliferation in preovulatory GCs.
2.4. Klf4 Overexpression Induces Apoptosis and Promotes Cell Cycle Arrest in GCs
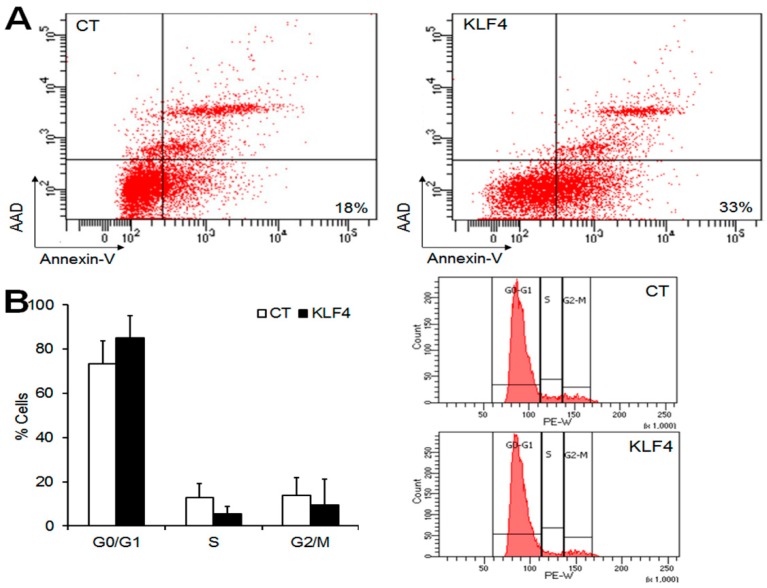
As Klf4 overexpression reduced the expression of anti-apoptotic factors and cell growth, we examined its impact on apoptosis and the cell cycle in GCs. To estimate the pro-apoptotic effect of Klf4 in GCs, apoptotic cells were measured by Annexin V and 7-aminoacinomycin D (AAD) staining and the flow cytometry of GCs transfected with Klf4 or an empty vector. The percentage of apoptotic cells (Annexin V+/AAD−) increased up to 33% in Klf4-transfected cells compared with empty vector-transfected controls (18%) (Figure 4A). As shown in Figure 4B, Klf4 overexpression also increased the proportion of G0/G1 phase cells (CT, 73.3 ± 10.4 vs. Klf4, 84.9 ± 10.1%), and reduced the proportion of S-phase cells compared to the control (CT, 12.7 ± 6.4 vs. Klf4, 5.5 ± 3.2%), although statistical significance was not attained (vs. CT, p = 0.05). The percentage of cells in the G2/M phase was comparable in the two groups. These results indicate that Klf4 is involved in the induction of apoptosis, and cell cycle arrest in G0/G1, in preovulatory GCs.
Figure 4.
Effect of Klf4 on apoptosis and the cell cycle of preovulatory GCs in vitro. Flow cytometry assays were performed to analyze apoptosis and the cell cycle in preovulatory GCs overexpressing Klf4. (A) The flow cytometric detection of apoptosis in GCs transfected with empty vector (CT, control) (left panel), or Klf4 expression vector (300 ng) (right panel). The cells were stained with Annexin-V and 7-aminoacinomycin D (AAD). The percentages indicate the proportions of late apoptotic cells (right lower quadrant) (Annexin-V+/AAD−). Representative images of three separate experiments are shown; (B) Analysis of the subpopulations of cells in cell cycle phases G0/G1, S, and G2/M. The cell cycle distributions were calculated and expressed as means ± SDs of three independent experiments. Representative Fluorescence-activated cell sorting (FACS) profiles are shown (right panel). CT, control cells transfected with empty vector; KLF4, cells transfected with Klf4 (300 ng).
3. Discussion
The transient induction of specific transcription factors in periovulatory follicles by the LH surge play an important role in cellular division, apoptosis, and differentiation [3]. In this study, we showed that LH significantly increased Klf4 expression in preovulatory GCs, and that Klf4 inhibited cell growth and induced apoptosis in these cells, indicating a role for Klf4 as a regulator of cell growth and apoptosis during the periovulatory period.
Using DNA chip technology, the Klf4 gene has been identified in mice ovarian follicles as one of the folliculogenesis-related genes [13], and its transcript level was markedly increased in Graafian follicles compared to preantral follicles [13]. In addition, LH stimulation rapidly increased Klf4 transcripts within 2 h in porcine granulosa-lutein cells in vitro [9]. Consistent with this, we observed that LH treatment led to a dose-dependent increase of the Klf4 transcript level in preovulatory GCs (Figure 1A), suggesting that the ovulatory LH stimulus could be an initial trigger of Klf4 expression.
Luteinizing GCs exit the cell cycle and stop dividing, and this is accompanied by the rapid down-regulation of the cell cycle gene, cyclin D2, as well as activation of the cell cycle inhibitors, p21 and p27 [14]. Our results confirmed that ovulatory doses of LH decreased the expression of cyclin D2 and increased p21 mRNA in GCs, with no significant change in p27 mRNA (Figure 1C). We also observed a decrease in the expression of cyclin D1, which is another cell cycle gene [15].
Since Klf4 has been known to cause G1/S arrest through the transcriptional activation of p21 or p27 and by the direct inhibition of cyclin D1 in various types of non-gonadal cells [16,17,18,19,20], we analyzed the expression of these cycle regulators. In addition, cyclin D2, a potent activator of the cell cycle in the G1/S phase, was also evaluated because it is selectively expressed in the GCs of growing follicles in the ovary [14]. As expected, Klf4 overexpression decreased cyclins D1 and D2 transcripts and increased those of p21 in preovulatory GCs (Figure 2B), and Klf4 knockdown increased cyclin D2 expression (Figure 2D). However, Klf4 knockdown had no effects on the p21 mRNA expression; considering that Klf4 expression was not completely blocked by siRNA treatment in the present study (Supplementary Figure S1-B,D), it is possible that only the complete absence of Klf4 would have an effect on p21 gene expression.
In agreement with the effects in cell cycle regulators, the overexpression of Klf4 inhibited GC proliferation (Figure 3D) and cell cycle progression at the G1/S transition (Figure 4B). p27 is also known as a potent inhibitor of cyclin-dependent kinases in the G1 phase of the cell cycle and its mRNA increases in response to Klf4 overexpression in non-gonadal cells [16,17,18,19,20,21]. However, changes in Klf4 expression had no effects on p27 mRNA levels in GCs (Figure 2B,D). Since p27 is known to be regulated primarily at the post-translational level [22,23], its mRNA level may not change after Klf4 induction in GCs.
We also characterized the LH-induced changes in the expression of the Bcl-2 proto-oncogene (an inhibitor of apoptosis) and Bax (an inducer of apoptosis), which are involved in GC apoptosis [24]. The observed LH-induced decrease in Bcl-2 expression coupled with constant Bax mRNA levels (Figure 1B) may be associated with the transient induction of GCs apoptosis before the luteal transition. Since periovulatory GCs are more prone to apoptosis than immature GCs [25,26,27], Klf4 may also be implicated in the increased susceptibility of preovulatory GCs to apoptosis during this transition period. In fact, evidence from various mammalian cell types indicates that Klf4 induces apoptosis by regulating the transcription of Bax and Bcl-2 [10], and we also observed that Klf4 overexpression decreased in Bcl-2 expression without changing Bax mRNA levels, thus lowering the Bcl-2/Bax ratio (Figure 2A). Another important regulator of the apoptotic process is the caspase family of proteases [28]. The increased permeability of mitochondria under the influence of Bax results in the activation of caspases 3 and 7 as the final common pathway of apoptosis [29,30]. In addition to Bcl-2 and Bax regulation by Klf4 in GCs, we demonstrated the pro-apoptotic effects of Klf4 as shown in Figure 3C, although these were not statistically significant. Along with this, Klf4 overexpression reduced cell viability and increased the proportion of apoptotic cells (Figure 3A and Figure 4A). Previous studies also found that Klf4 overexpression suppressed Bcl-2 expression in mice small follicles and in the GCs isolated from them [10]. However, they also reported the Klf4-induced activation of Bax expression. This discrepancy may be due to the differences in cell maturity or sensitivity to apoptosis as a function of follicle growth. Since Klf4 is highly expressed in Graafian follicles [13] and regulated by LH treatment in GCs, using preovulatory GCs might be more appropriate for analyzing the effect of Klf4 on GCs than whole ovaries or small follicles. In ovarian cancer cells, Klf4 overexpression increased Bcl-2 expression, but had no effect on cell viability [31]. Previous studies indicate that Klf4 may switch from anti-apoptotic to pro-apoptotic depending on the context [32,33,34]. Thus, Klf4 seems to regulate Bcl-2 or Bax differently in ovarian cancer cells than in normal ovarian cells due to unidentified cellular factors.
Considering that the massive apoptosis of preovulatory GCs in vitro is suppressed by an ovulatory dose of LH [35], redundant mechanisms to ensure their survival may exist. Follicle maturation and ovulation are critically dependent on gonadotropins that work in concert with intraovarian factors to suppress the cell death program [36]. The signaling pathway and potencies of various survival factors are different according to the follicle growth [36]. For instance, the activation of the Phosphoinositide 3-kinase/Protein kinase B (PI3K/Akt) pathway by ghrelin is involved in the regulation of cell survival and anti-apoptotic protection in bovine cumulus cells [37,38].
Collectively, our results suggest that Klf4 may influence the susceptibility of preovulatory GCs to apoptosis at a number of points, including the regulation of upstream regulator (down-regulating Bcl-2, thereby decreasing the Bcl-2/Bax ratio) and downstream effectors (caspase 3 and 7). In addition, Klf4 may contribute to the LH-induced cell cycle exit by altering cell cycle regulators at the G1/S checkpoint after the LH surge, and some time before the complete luteal transition. Thus, Klf4 is a transcriptional regulator influencing the fate of preovulatory GCs by reducing cell growth and increasing apoptosis during the luteal transition.
4. Materials and Methods
4.1. Animals and Reagents
Immature female Sprague–Dawley rats were obtained from Orient Biokorea (Kyunggi-do, South Korea) and housed under controlled conditions (22–24 °C, humidity 40–50%, 12-h light–dark cycle), with free access to food and water. Animal care was consistent with institutional guidelines, and the Hanyang University Animal Care and Use committee approved all of the procedures involving animals (HY-IACUC-16-0013, approval date: 24 February 2014). McCoy’s 5a medium (modified) and Leibovitz L-15 medium were obtained from GIBCO (Santa Clara, CA, USA). Penicillin and streptomycin were obtained from Sigma (St. Louis, MO, USA) and recombinant human FSH (Org 32489E) and ovine LH (NIH-LH-23) were from NV Organon (Oss, The Netherlands) and the National Hormone and Pituitary Distribution Program (Baltimore, MD, USA), respectively.
4.2. Preparation and Culture of Preovulatory Granulosa Cells
Animals aged 26 days (body weight 55–60 g) were injected intraperitoneally with 10 International Unit (IU) of pregnant mare serum gonadotropin (PMSG) (Sigma-Aldrich, St. Louis, MO, USA) to induce the growth of multiple preovulatory follicles. Forty-eight h later, ovaries were dissected, and preovulatory follicles were punctured to obtain granulosa cells. Ovarian debris and small follicles were removed, and GCs were collected by low-speed centrifugation at 500 × g for 10 min, dispersed by repeated washing and suspended in culture medium (McCoy’s 5a supplemented with two mM of L-glutamine, 100 U/mL of penicillin, and 100 μg/mL of streptomycin). To evaluate the effect of LH on expression of Klf4 on apoptosis-related and cell cycle-related genes, cells were plated in a 24-well culture dish (5 × 104 viable cells/well) and cultured in the absence or presence of LH (100 and 200 ng/mL) for 45 min. At the end of the experiment, they were frozen for RNA extraction.
4.3. Plasmid Construction and Transfection
The pCMV3×FLAG-Klf4 (FLAG-Klf4) construct was made by subcloning full-length rat Klf4 cDNA (kindly provided by H. Kook; Chonnam National University Medical School, Gwangju, Korea). Klf4-specific siRNA (Klf4-siRNA) was synthesized by Genepharma (Shanghai, China); the sequences of the 25-nucleotide sense and antisense RNAs were: 5′-CCAUUAUCAAGAGCUCAUGCCACCG-3′ (sense) and 5′-CGGUGGCAUGAGCUCUUGAUAAUGG-3′ (antisense) (accession no.: NM_053713). Non-targeting control siRNA (Universal Scrambled siRNA) was purchased from Genepharma (Shanghai, China). For transient transfections, GCs were resuspended in electroporation buffer (MPK1025; Thermo Fisher Scientific, Waltham, MA, USA), mixed with expression vectors or siRNA, and electroporated with a single pulse of 1000 V, 40 milliseconds, using a Neon™ Transfection System (MPK5000, Thermo Fisher Scientific, Waltham, MA, USA). This procedure was based on the results of preliminary optimization experiments that achieved a transfection efficiency of approximately 70%. The transfected cells (1 × 105 cells/well) were resuspended in culture medium, plated in 24-well culture plates, collected after 24–36 h, and snap-frozen for the isolation of RNA or proteins.
4.4. Total RNA Extraction and Real-Time Quantitative RT-PCR Analysis
Total RNA was isolated with an RNeasy extraction kit (Qiagen Inc., Valencia, CA, USA). One-µg aliquots of total RNA were annealed (five min at 70 °C) to oligo(dT)18 primers and reverse transcribed using cDNA synthesis platinum master mix (GenDEPOT, Katy, TX, USA). Primers were designed for the mRNA sequences of Klf4, Bax, Bcl-2, cyclin D1, cyclin D2, p21, and p27 using the Primers Express program (PE Applied Biosystems, Foster City, CA, USA) (Table 1). Real-time PCR reactions were carried out in total volumes of 20 μL, with Prime Q-Master Mix (with SYBR Green I) (GeNet Bio Inc., Daejeon, South Korea) using a LightCycler 480 II System (Roche Molecular Diagnostics, Indianapolis, IN, USA). The PCR cycle conditions were as follows: 10 min at 95 °C, 45 cycles of 95 °C for 10 s, 58~60 °C for 10 s, and 72 °C for 10 s. 18S rRNA was amplified and used to normalize each reaction. Samples were run in triplicate (Roche Molecular Diagnostics, Indianapolis, IN, USA), and mean values were compared with the control values to calculate relative amounts of transcript. Data are expressed as means ± standard deviations (SDs) of duplicate or triplicate measurements in four independent experiments.
Table 1.
Primer sequences used for real-time PCR amplification.
| Gene | Primer Sequence (5′-3′) | Product Size (bp) | Accession Number |
|---|---|---|---|
| Klf4 | F: GAGAGGAACTCTCTCACATGAAGC | 185 | NM_053713.1 |
| R: AAGGATAAAGTCTAGGTCCAGGAGA | |||
| Bax | F: TGTTTGCTGATGGCAACTTC | 104 | NM_017059.1 |
| R: GATCAGCTCGGGCACTTTAG | |||
| Bcl-2 | F: GGGATGCCTTTGTGGAACTA | 138 | NM_016993.1 |
| R: CTCACTTGTGGCCCAGGTAT | |||
| Cyclin D1 | F: TCAAGTGTGACCCGGACTG | 213 | NM_171992.4 |
| R: CACTACTTGGTGACTCCCGC | |||
| Cyclin D2 | F: CGATGATCGCAACTGGAAGC | 232 | NM_022267.1 |
| R: TGGTCCGGATCTTCCACAGA | |||
| p21Cip1 | F: TGAGGGACCAGTACATGAGAACT | 192 | NM_031515.1 |
| R: GAGCCTGTTTCGTGTCTACTGTT | |||
| p27Kip1 | F: TCGCGGCTCCGAGACTT | 204 | NM_031762.3 |
| R: TCTCCAAGTCCCGGGTTAGT | |||
| 18S rRNA | F: CGCGGTTCTATTTTGTTGGT | 218 | M11188.1 |
| R: AGTCGGCATCGTTTATGGTC |
F, forward; R, reverse; p21Cip1, cyclin-dependent kinase-interaction protein 1; p27Kip1, cyclin-dependent kinase inhibitor 1B.
4.5. Western Blot Analysis
Transfected GCs were lysed in Laemmli buffer containing β-mercaptoethanol (BioRad, Hercules, CA, USA). Samples were resolved by 8% Sodium dodecyl sulfate-polyacrylamide (SDS-PAGE) gel electrophoresis, transferred to nitrocellulose membranes (Amersham Pharmacia Biotech, Arlington Heights, IL, USA), and immunoblotted with anti-KLF4 antibody (4038, Cell Signaling Tech., Inc., Danvers, MA, USA) diluted 1:200. The membranes were washed and blotted with peroxidase-conjugated donkey anti-rabbit secondary antibody (1:2000) (Boehringer Mannheim, Indianapolis, IN, USA). Immunolabeled proteins were detected using an enhanced chemiluminescence kit (Pierce Chemical Co., Rockford, IL, USA). The 65-kDa KLF4 protein is indicated in Supplementary Figure S1D. To ensure that the lysates were loaded equally, the blots were stripped and incubated with an anti-β-actin antibody (1:4000) (Sigma, St. Louis, MO, USA).
4.6. Cell Counting Kit-8 Assays
Cell viability was determined using a CCK-8 (Enzo, Ann Arbor, MI, USA). Dehydrogenase enzymes present in metabolically active cells convert water-soluble tetrazolium-8 salt (WST-8) [2-(2-methoxy-4-nitrophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium, monosodium salt] to a water-soluble formazan dye. The amount of the formazan dye that is generated is proportional to the number of living cells. GCs transfected with empty vector (EV) or Flag-Klf4 (300 ng/well) were plated in 96-well plates (1 × 104 cells/well) and cultured in McCoy 5a medium containing 1% fetal bovine serum (FBS). Sixteen to 24 h after transfection, WST-8 solution was added and incubation continued in serum-free medium for 2 h. The quantity of formazan product was measured at 450 nm with an ELISA plate reader (BioRad, Hercules, CA, USA). The experiment was repeated three times, and the data are expressed as fold changes relative to the control values measured with empty vector-transfected cells. Cell viability was evaluated using trypan blue staining [39]. Equal volumes of 0.4% trypan blue were added to cell suspensions, and viable (unstained) and dead (stained) cells were counted using a hemocytometer. The data are expressed as percentages of viable cells. All of the experiments were replicated at least three times to assure reproducibility (Louis, MO, USA).
4.7. Caspase 3/7 Assays
Apoptosis was also assessed with the caspase 3/7 activity assay using a luminescence assay kit (Caspase-Glo 3/7 assay; Promega, Madison, WI, USA). GCs were cultured in McCoy 5a serum-free medium with or without FSH or 10% FBS for 24 h after transfection. Then, proluminescent Caspase-Glo 3/7 reagent was added to each well, and the cells were gently mixed and incubated at room temperature for one h. Luminescence was measured on a Monolight 2010 luminometer (Analytical Luminescence Laboratory, San Diego, CA, USA). The data were expressed as fold changes relative to the empty vector transfected cell (control). All of the experiments were replicated three times.
4.8. 5-Bromo-2′-Deoxyuridine (BrdU) Incorporation
Cells were plated onto 1.2 × 1.2 cm coverslips in 24-well plates (1 × 105 cells/well). After 16–24 h, 5-bromo-2′-deoxyuridine (BrdU, 3 µg/mL) (B5002, Sigma, St. Louis, MO, USA) was added and incubation continued for three h, followed by fixation with methanol for 10 min at −20 °C and three washes with phosphate-buffered saline (PBS). The slides were dehydrated, pretreated with 2N HCl at 37 °C for 15 min, washed twice with 0.2% Triton X in PBS (pH 7.4), and incubated with 1% normal goat serum/PBS for one h at room temperature, followed by anti-BrdU antibody (B2531, Sigma, St. Louis, MO, USA) diluted 1:250 in 1% normal goat serum at 4 °C overnight. The slides were then washed with PBS and incubated with 1:200 goat anti-mouse Alexa Fluor 488® IgG (Thermo Fisher Scientific, Waltham, MA, USA) for one h at 37 °C, and nuclear DNA was counterstained with 4′,6-diamidino-2-phenylindole (DAPI) (Vector Laboratories, Inc., Burlingame, CA, USA). Five magnified fields (100×) were randomly selected, and the number of BrdU-positive cells per field was counted under a fluorescence microscopy (Leica, Heidelberg, Germany). The proliferation rate (%) was defined as the percentage of total cells that were BrdU-positive.
4.9. Flow Cytometric Analysis
For assessing apoptosis, cells were washed twice with cold PBS, resuspended in 1X binding buffer (1 × 106 cells) (BD Biosciences, San Jose, CA, USA) and stained with APC-Annexin V and 7-AAD (BD Biosciences, San Jose, CA, USA) for 15 min in the dark at room temperature. Unstained cells were used to set negative gates, and each cell that was stained with either Allophycocyanin (APC)-Annexin V or 7-Aminoactinomycin D (7-AAD) alone was used to set the compensation parameters. Apoptosis rates were measured using a flow cytometer (FACSCalibur, BD Biosciences, San Jose, CA, USA) and analyzed with Cell Quest software (BD Biosciences, San Jose, CA, USA).
For cell cycle analysis, cells (1 × 106 cells) were harvested, fixed in 70% (v/v) ethanol for two h at −20 °C, washed twice with PBS, treated with RNase A (iNtRON Biotechnology, Seoul, Korea), and stained with propidium iodide (Sigma, St. Louis, MO, USA) at room temperature for 30 min in the dark. Using flow cytometry, the proportion of cells in each phases (G0-G1, S, and G-M) were analyzed.
4.10. Statistical Analysis
All of the results were expressed as means ± SDs of experiments and repeated at least three times. All of the data were analyzed using IBM SPSS statistics 21 for Windows (SPSS Inc., Chicago, IL, USA). Statistical significance was determined by the Mann–Whitney U test for two-group comparisons, and Kruskal–Wallis one-way analysis of variance for multiple comparisons. Significance was accepted at p < 0.05.
5. Conclusions
We conclude that Klf4 increases the susceptibility of preovulatory GCs to apoptosis by down-regulating Bcl-2 and promotes LH-induced cell cycle exit. It appears to be a key regulator induced by the LH surge that determines the fate of GCs in preovulatory follicles during the luteal transition.
Acknowledgments
We thank Hyun Kook (Medical Research Center for Gene Regulation, Chonnam National University Medical School, Gwangju, Korea) for a gift of the pCMV3×FLAG-KLF4 plasmid construct.
Supplementary Materials
Supplementary materials can be found at http://www.mdpi.com/1422-0067/20/1/87/s1.
Author Contributions
H.C. participated in the experiments, data collection and analysis; J.R. contributed to the design of the study, data analysis, supervision and development of the manuscript. J.R. takes responsibility for the integrity of the data analysis. All authors read and approved the final manuscript.
Funding
This study was supported by the Ministry of Education and Science (NRF-20090074679).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Hirshfield A.N. Development of follicles in the mammalian ovary. Int. Rev. Cytol. 1991;124:43–101. doi: 10.1016/s0074-7696(08)61524-7. [DOI] [PubMed] [Google Scholar]
- 2.Leo C.P., Pisarska M.D., Hsueh A.J. DNA array analysis of changes in preovulatory gene expression in the rat ovary. Biol. Reprod. 2001;65:269–276. doi: 10.1095/biolreprod65.1.269. [DOI] [PubMed] [Google Scholar]
- 3.Espey L.L., Richards J.S. Temporal and spatial patterns of ovarian gene transcription following an ovulatory dose of gonadotropin in the rat. Biol. Reprod. 2002;67:1662–1670. doi: 10.1095/biolreprod.102.005173. [DOI] [PubMed] [Google Scholar]
- 4.Black A.R., Black J.D., Azizkhan-Clifford J. Sp1 and krüppel-like factor family of transcription factors in cell growth regulation and cancer. J. Cell. Physiol. 2001;188:143–160. doi: 10.1002/jcp.1111. [DOI] [PubMed] [Google Scholar]
- 5.Ghaleb A.M., Nandan M.O., Chanchevalap S., Dalton W.B., Hisamuddin I.M., Vincent W.Y. Krüppel-like factors 4 and 5: The yin and yang regulators of cellular proliferation. Cell Res. 2005;15:92–96. doi: 10.1038/sj.cr.7290271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ohnishi S., Ohnami S., Laub F., Aoki K., Suzuki K., Kanai Y., Haga K., Asaka M., Ramirez F., Yoshida T. Downregulation and growth inhibitory effect of epithelial-type Krüppel-like transcription factor KLF4, but not KLF5, in bladder cancer. Biochem. Biophys. Res. Commun. 2003;308:251–256. doi: 10.1016/S0006-291X(03)01356-1. [DOI] [PubMed] [Google Scholar]
- 7.Wang J., Place R.F., Huang V., Wang X., Noonan E.J., Magyar C.E., Huang J., Li L.-C. Prognostic value and function of KLF4 in prostate cancer: RNAa and vector-mediated overexpression identify KLF4 as an inhibitor of tumor cell growth and migration. Cancer Res. 2010;70:10182–10191. doi: 10.1158/0008-5472.CAN-10-2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ji J., Wang H.-S., Gao Y.-Y., Sang L.-M., Zhang L. Synergistic anti-tumor effect of KLF4 and curcumin in human gastric carcinoma cell line. Asian Pac. J. Cancer Prev. 2014;15:7747–7752. doi: 10.7314/APJCP.2014.15.18.7747. [DOI] [PubMed] [Google Scholar]
- 9.Natesampillai S., Kerkvliet J., Leung P.C., Veldhuis J.D. Regulation of Kruppel-like factor 4, 9, and 13 genes and the steroidogenic genes LDLR, StAR, and CYP11A in ovarian granulosa cells. Am. J. Physiol. Endocrinol. Metab. 2008;294:E385–E391. doi: 10.1152/ajpendo.00480.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu L., Sun H., Zhang M., Jiang Y., Zhang C., Zhou J., Ding L., Hu Y., Yan G. MicroRNA-145 protects follicular granulosa cells against oxidative stress-induced apoptosis by targeting Krüppel-like factor 4. Mol. Cell. Endocrinol. 2017;452:138–147. doi: 10.1016/j.mce.2017.05.030. [DOI] [PubMed] [Google Scholar]
- 11.Williams G.T., Smith C.A. Molecular regulation of apoptosis: Genetic controls on cell death. Cell. 1993;74:777–779. doi: 10.1016/0092-8674(93)90457-2. [DOI] [PubMed] [Google Scholar]
- 12.Hirshfield A.N. Effect of a low dose of pregnant mare’s serum gonadotropin on follicular recruitment and atresia in cycling rats. Biol. Reprod. 1986;35:113–118. doi: 10.1095/biolreprod35.1.113. [DOI] [PubMed] [Google Scholar]
- 13.Liu H.-C., He Z., Rosenwaks Z. Application of complementary DNA microarray (DNA chip) technology in the study of gene expression profiles during folliculogenesis. Fertil. Steril. 2001;75:947–955. doi: 10.1016/S0015-0282(01)01706-X. [DOI] [PubMed] [Google Scholar]
- 14.Robker R.L., Richards J.S. Hormone-induced proliferation and differentiation of granulosa cells: A coordinated balance of the cell cycle regulators cyclin D2 and p27Kip1. Mol. Endocrinol. 1998;12:924–940. doi: 10.1210/mend.12.7.0138. [DOI] [PubMed] [Google Scholar]
- 15.Sherr C.J. D-type cyclins. Trends Biochem. Sci. 1995;20:187–190. doi: 10.1016/S0968-0004(00)89005-2. [DOI] [PubMed] [Google Scholar]
- 16.Zhang W., Geiman D.E., Shields J.M., Dang D.T., Mahatan C.S., Kaestner K.H., Biggs J.R., Kraft A.S., Yang V.W. The Gut-Enriched Kruppel-Like Factor (Kruppel-Like Factor 4) Mediates the Transactivating Effecto of p53 on the p21WAF1/Cip1 Promoter. J. Biol. Chem. 2000;275:18391–18398. doi: 10.1074/jbc.C000062200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shie J.-L., Chen Z.Y., Fu M., Pestell R.G., Tseng C.-C. Gut-enriched Krüppel-like factor represses cyclin D1 promoter activity through Sp1 motif. Nucleic Acids Res. 2000;28:2969–2976. doi: 10.1093/nar/28.15.2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen Z.Y., Shie J.-L., Tseng C.-C. Gut-enriched Krüppel-like factor represses ornithine decarboxylase gene expression and functions as checkpoint regulator in colonic cancer cells. J. Biol. Chem. 2002;277:46831–46839. doi: 10.1074/jbc.M204816200. [DOI] [PubMed] [Google Scholar]
- 19.Wei D., Kanai M., Jia Z., Le X., Xie K. Krüppel-like factor 4 induces p27Kip1 expression in and suppresses the growth and metastasis of human pancreatic cancer cells. Cancer Res. 2008;68:4631–4639. doi: 10.1158/0008-5472.CAN-07-5953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoon H.S., Chen X., Yang V.W. Krüppel-like factor 4 mediates p53-dependent G1/S cell cycle arrest in response to DNA damage. J. Biol. Chem. 2003;278:2101–2105. doi: 10.1074/jbc.M211027200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rowland B.D., Peeper D.S. KLF4, p21 and context-dependent opposing forces in cancer. Nat. Rev. Cancer. 2006;6:11–23. doi: 10.1038/nrc1780. [DOI] [PubMed] [Google Scholar]
- 22.Toyoshima H., Hunter T. p27, a novel inhibitor of G1 cyclin-Cdk protein kinase activity, is related to p21. Cell. 1994;78:67–74. doi: 10.1016/0092-8674(94)90573-8. [DOI] [PubMed] [Google Scholar]
- 23.Hengst L., Reed S.I. Translational control of p27Kip1 accumulation during the cell cycle. Science. 1996;271:1861–1864. doi: 10.1126/science.271.5257.1861. [DOI] [PubMed] [Google Scholar]
- 24.Hussein M.R. Apoptosis in the ovary: Molecular mechanisms. Hum. Reprod. Update. 2005;11:162–178. doi: 10.1093/humupd/dmi001. [DOI] [PubMed] [Google Scholar]
- 25.Robker R.L., Richards J.S. Hormonal control of the cell cycle in ovarian cells: Proliferation versus differentiation. Biol. Reprod. 1998;59:476–482. doi: 10.1095/biolreprod59.3.476. [DOI] [PubMed] [Google Scholar]
- 26.Chaffin C.L., Schwinof K.M., Stouffer R.L. Gonadotropin and steroid control of granulosa cell proliferation during the periovulatory interval in rhesus monkeys. Biol. Reprod. 2001;65:755–762. doi: 10.1095/biolreprod65.3.755. [DOI] [PubMed] [Google Scholar]
- 27.Quirk S., Cowan R., Harman R., Hu C.-L., Porter D. Ovarian follicular growth and atresia: The relationship between cell proliferation and survival. J. Anim. Sci. 2004;82:E40–E52. doi: 10.2527/2004.8213_supplE40x. [DOI] [PubMed] [Google Scholar]
- 28.Markström E., Svensson E.C., Shao R., Svanberg B., Billig H. Survival factors regulating ovarian apoptosis—Dependence on follicle differentiation. Reproduction. 2002;123:23–30. doi: 10.1530/rep.0.1230023. [DOI] [PubMed] [Google Scholar]
- 29.Oltvai Z.N., Milliman C.L., Korsmeyer S.J. Bcl-2 heterodimerizes in vivo with a conserved homolog, Bax, that accelerates programmed cell death. Cell. 1993;74:609–619. doi: 10.1016/0092-8674(93)90509-O. [DOI] [PubMed] [Google Scholar]
- 30.Lakhani S.A., Masud A., Kuida K., Porter G.A., Booth C.J., Mehal W.Z., Inayat I., Flavell R.A. Caspases 3 and 7, key mediators of mitochondrial events of apoptosis. Science. 2006;311:847–851. doi: 10.1126/science.1115035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoon O., Roh J. Downregulation of KLF4 and the Bcl-2/Bax ratio in advanced epithelial ovarian cancer. Oncol. Lett. 2012;4:1033–1036. doi: 10.3892/ol.2012.834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Z., Zhao J., Li Q., Yang W., Song Q., Li W., Liu J. KLF4 promotes hydrogen-peroxide-induced apoptosis of chronic myeloid leukemia cells involving the bcl-2/bax pathway. Cell Stress Chaperones. 2010;15:905–912. doi: 10.1007/s12192-010-0199-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whitlock N.C., Bahn J.H., Lee S.-H., Eling T.E., Baek S.J. Resveratrol-induced apoptosis is mediated by early growth response-1, Krüppel-like factor 4, and activating transcription factor 3. Cancer Prev. Res. 2011;4:116–127. doi: 10.1158/1940-6207.CAPR-10-0218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang B., Zhao M.-Z., Cui N.-P., Lin D.-D., Zhang A.-Y., Qin Y., Liu C.-Y., Yan W.-T., Shi J.-H., Chen B.-P. Krüppel-like factor 4 induces apoptosis and inhibits tumorigenic progression in SK-BR-3 breast cancer cells. FEBS Open Bio. 2015;5:147–154. doi: 10.1016/j.fob.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chun S.Y., Billig H., Tilly J.L., Furuta I., Tsafriri A., Hsueh A.J. Gonadotropin suppression of apoptosis in cultured preovulatory follicles: Mediatory role of endogenous insulin-like growth factor I. Endocrinology. 1994;135:1845–1853. doi: 10.1210/endo.135.5.7525255. [DOI] [PubMed] [Google Scholar]
- 36.Kaipia A., Hsueh A.J. Regulation of ovarian follicle atresia. Annu. Rev. Physiol. 1997;59:349–363. doi: 10.1146/annurev.physiol.59.1.349. [DOI] [PubMed] [Google Scholar]
- 37.Chouzouris T.M., Dovolou E., Krania F., Pappas I.S., Dafopoulos K., Messinis I.E., Anifandis G., Amiridis G.S. Effects of ghrelin on activation of Akt1 and ERK1/2 pathways during in vitro maturation of bovine oocytes. Zygote. 2017;25:183–189. doi: 10.1017/S096719941700003X. [DOI] [PubMed] [Google Scholar]
- 38.Dovolou E., Messinis I.E., Periquesta E., Dafopoulos K., Gutierrez-Adan A., Amiridis G.S. Ghrelin accelerates in vitro maturation of bovine oocytes. Reprod. Domest. Anim. 2014;49:665–672. doi: 10.1111/rda.12344. [DOI] [PubMed] [Google Scholar]
- 39.Strober W. Trypan blue test of cell viability. Curr. Protoc. Immunol. 2001;21:A.3B.1–A.3B.2. doi: 10.1002/0471142735.ima03bs21. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.